Abstract
Invasive holoparasitic plants of the genus Cuscuta (dodder) threaten African ecosystems due to their rapid spread and attack on various host plant species. Most Cuscuta species cannot photosynthesize and hence rely on host plants for nourishment. After attachment through a peg-like organ called a haustorium, the parasites deprive hosts of water and nutrients, which negatively affects host growth and development. Despite their rapid spread in Africa, dodders have attracted limited research attention, although data on their taxonomy, host range, and epidemiology are critical for their management. Here, we combine taxonomy and phylogenetics to reveal the presence of field dodder (Cuscuta campestris) and C. kilimanjari (both either naturalized or endemic to East Africa), in addition to the introduction of the giant dodder (C. reflexa), a south Asian species, in continental Africa. These parasites have a wide host range, parasitizing species across 13 angiosperm orders. We evaluated the possibility of C. reflexa to expand this host range to tea (Camelia sinensis), coffee (Coffea arabica), and mango (Mangifera indica), crops of economic importance to Africa, for which haustorial formation and vascular-bundle connections in all three crops revealed successful parasitism. However, only mango mounted a successful postattachment resistance response. Furthermore, species distribution models predicted high habitat suitability for Cuscuta spp. across major tea- and coffee-growing regions of Eastern Africa, suggesting an imminent risk to these crops. Our findings provide relevant insights into a poorly understood threat to biodiversity and economic wellbeing in Eastern Africa, and provide critical information to guide development of management strategies to avert Cuscuta spp. spread.
Microscopy and habitat suitability modeling provide an early warning that dodder’s invasion in Eastern Africa poses a threat to important cash crops.
Introduction
In Africa, parasitic plants, such as Striga spp., are relatively well researched due to their direct negative impacts on major food cereal staples (reviewed by Parker, 2012). However, others, such as dodder (Cuscuta spp.), noxious vines of the Convolvulaceae family that are currently on the rise in the region, have received little attention. The genus Cuscuta comprises over 200 species of obligate parasites that infect a wide range of herbaceous and woody plants, including important crop species (Lanini and Kogan, 2005). Although some Cuscuta species, such as C. australis, C. campestris, and C. chinensis have been shown to arrest exotic weed invasions in their native ranges (Shen et al. 2006; Li and Dong 2009; Yu et al. 2009), most dodders are considered noxious, owing to their negative impacts on agriculture and biodiversity (reviewed by Press and Phoenix, 2005).
Cuscuta spp. are widely distributed worldwide and reportedly colonize a wide range of hosts across various habitats (Lanini and Kogan, 2005). Overall, members of this genus occur on all continents, except Antarctica, with most species reported in the Americas and Mexico, which are also considered their center of diversity (Yuncker, 1932; Stefanović et al., 2007). In Africa, only a handful of studies have reported dodder occurrence (Zerman and Saghir, 1995; García, 1999; García and Martin, 2007; García et al., 2014). However, their distribution patterns remain unknown. According to the Flora of Tropical East Africa (Verdcourt, 1963), several species, namely C. australis, C. campestris Yuncker, C. suaveolens Seringe, C. kilimanjari Oliv, C. hyalina Roth, C. Engelm, C. epilinum, and C. planiflora Tenore, are either endemic to or naturalized in Eastern Africa. However, some non-native species are currently on the rise in the region, although they have not been identified and it is not clear how and when they were introduced.
The widespread success of Cuscuta is attributed to its parasitic life history strategy and ability to steal most of the resources needed for growth and reproduction from its hosts. Particularly, most Cuscuta spp. do not photosynthesize due to reduced levels (or lack thereof) of chlorophylls, although some show localized photosynthesis (Hibberd et al., 1998; van der Kooij et al., 2000; Revill et al., 2005). Consequently, they entirely depend on their hosts for nourishment. Cuscuta life cycle follows a systematic pattern that begins with seed germination, attachment, and penetration of a suitable host (through a specialized organ called haustorium), development of vegetative tissues, flowering, and seed production. Due to a limited amount of food reserves in their seeds, seedlings must attach to an appropriate host within 3–5 d of germination (Lanini and Kogan, 2005), and establish vascular-bundle connections that act as a conduit for siphoning water, nutrients, and photo-assimilates. Thereafter, the parasite develops flowers and eventually produces viable seeds that shed back to the soil (Dawson et al., 1994; Albert et al., 2008).
Morphological identification of Cuscuta spp. is difficult, mainly because all members comprise slender vines, with scale-like leaves and no roots. Nevertheless, previous research works have used morphological descriptors, mainly floral and fruit characters, to distinguish species. The early monograph by Engelmann (1857) categorized Cuscuta into three groups based on stigma and style morphology. These were later adopted by Yuncker (1932) as subgenera. Specifically, subgenus Monogynella is characterized by fused styles, whereas subgenera Cuscuta and Grammica have two distinct styles, distinguished by respective elongate and globose stigmas. Yuncker later revised the monograph and subdivided these subgenera into 8 sections based on fruit dehiscence and 29 subsections based on a combination of characters, such as flower numbers, size, texture, and shape, as well as density of inflorescence among others.
We hypothesized that different Cuscuta spp. currently occur in Eastern Africa, and their distribution patterns are due to various biotic factors, such as presence of suitable hosts and interactions with the environment. This is because, in general, occurrence of a species in a particular locality is shaped by life history characteristics, environmental requirements, population genetics, and their associations with ecology over time. Therefore, we first used a combination of morphological descriptors and sequencing of the plastid locus (Ribulose bisphosphate carboxylase large—rbcL and trnL) and nuclear ITS region to identify Cuscuta spp. presently invading ecosystems in Kenya. We then determined their host range by compiling a comprehensive list of current Cuscuta hosts, and extrapolated the possibility of the parasite to expand this range to crop trees by infecting coffee (Coffea arabica), tea (Camelia sinensis), and mango (Mangifera indica) under greenhouse conditions. We selected coffee, tea, and mango because of their agricultural/economic importance; they contribute to the GDP of Kenya through export earnings and cover an estimated area of 114,700, 218,538, and 60,497 ha, respectively (FAO, 2017). Finally, we used geographical information system-based species distribution modeling (SDM) to estimate geographical distribution of the identified Cuscuta spp. across Eastern Africa, based on current climatic conditions and vegetation. Specifically, we adopted presence-only SDMs using the maximum entropy (MaxEnt) algorithm, which combines occurrence records with environmental variables to build correlative models for predicting habitat suitability for a species (Phillips et al., 2006). This algorithm has been previously used to predict distribution of parasitic plants, such as C. chinensis (Ren et al., 2020), Striga hermonthica (Cotter et al., 2012), and mistletoes (Viscum spp.; Zhang et al. 2016). We found that the current dodder invasion in Kenya (1) comprises C. campestris, C. kilimanjari, and C. reflexa; (2) has a wide host range that could potentially include tea and coffee; and (3) has a wide distribution with potential to invade new habitats. These findings will inform policies for management of the parasite in Eastern Africa.
Results
Floral morphological characters reveal three Cuscuta species
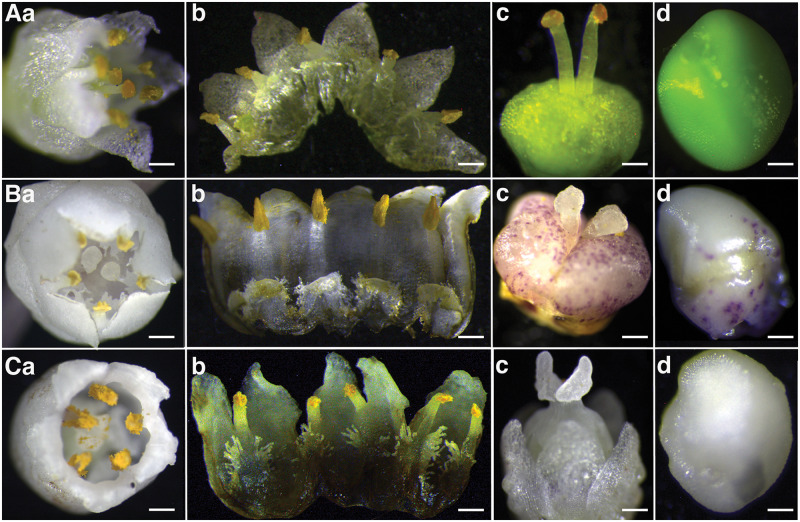
Floral characters revealed three distinct Cuscuta species in accessions collected from Kenya (Figure 1). Summarily, flowers across all specimens had clusters comprising five petals, five sepals, and five stamens, and were identified to the species level as follows;
Figure 1.
Profiles of floral morphology among Cuscuta accessions collected across Kenya, showing variations in gynoecia, ovule shape, size, and color across species. Aa–d, C. campestris: evidenced by small, white flowers with separate styles that bear globose stigmas; Ba–d, C. kilimanjari: confirmed by thick, separate styles with spherical stigmas; Ca–d, C. reflexa: evidenced by short, fused styles that bear ligulate stigmas. Bars Aa = 0.4 mm; Ab = 0.4 mm; Ac = 0.4 mm; Ad = 0.1 mm Ba = 0.8 mm; Bb = 0.8 mm; Bc = 1 mm; Bd = 1 mm; Ca = 1.2 mm; Cb = 1.2 mm; Cc = 1 mm, and Cd = 0.2 mm.
Cuscuta campestris Yuncker (subgenus Grammica); accessions here comprised slender, threadlike yellow to orange stems with a diameter of about 0.3 mm (Supplemental Figure S1A). Flowers were small, white, about 2 mm in diameter, with greenish-yellow capsules that appeared in compact cymose clusters. Calyx lobes were obtuse, or somewhat acute, whereas corolla lobes were triangular. Stamens were shorter than the lobes, with filaments of about 1 mm. They had two separate slender styles, about 1-mm long, with globose stigmas that did not split at the base. Four ovules, about 1-mm long, were present (Figure 1A a–d).
Cuscuta kilimanjari Oliv; accessions here had thick, coarse, purple vines, about 1 mm in diameter (Supplemental Figure S1B). Flowers were pale white, waxy, about 4-mm wide. Both sets of calyx and corolla were obtuse, whereas stamens were shorter than the lobes, with short, thick filaments. This category had two separate short, thick styles, less than 1-mm long and 0.3-mm wide. Styles bore white, spherical stigmas, with ovaries that had purple spots (Figure 1B a–d).
Cuscuta reflexa Roxb; vines were greenish-yellow, >2 mm in diameter (Supplemental Figure S1C), with large flowers about 6-mm wide. The flowers had a single thick, short style, with two elongated stigmas, and ovaries that contained four white ovules of different sizes (Figure 1C a–d).
Phylogenetic analysis
Parsimony consensus trees for accessions under this study, alongside other Cuscuta spp. from GenBank, revealed consistent topologies across all three (rbcL, trnL, and ITS) regions (Figure 2). In brief, three major clades, corresponding to the three Cuscuta subgenera, were resolved across the datasets with good bootstrap support. Specifically, C. campestris accessions from the current study were resolved in the first major clade with other reported species of subgenus Grammica, sister to C. campestris taxa from GenBank (Figure 2). Within the same clade, our C. kilimanjari accessions were resolved and nested with a South American clade of subgenus Grammica. The second major clade comprised members of subgenus Cuscuta, with emphasis on species previously reported to occur in Africa. The third major clade comprised subgenus Monogynella, with our C. reflexa taxa nested inside a Genbank-derived C. reflexa group and basal to both subgenera Cuscuta and Grammica. Unrooted Maximum Likelihood gene trees confirmed that subgenus Monogynella were basal to subgenus Grammica across all genes tested, as well as in the combined dataset (Supplemental Figure S2).
Figure 2.
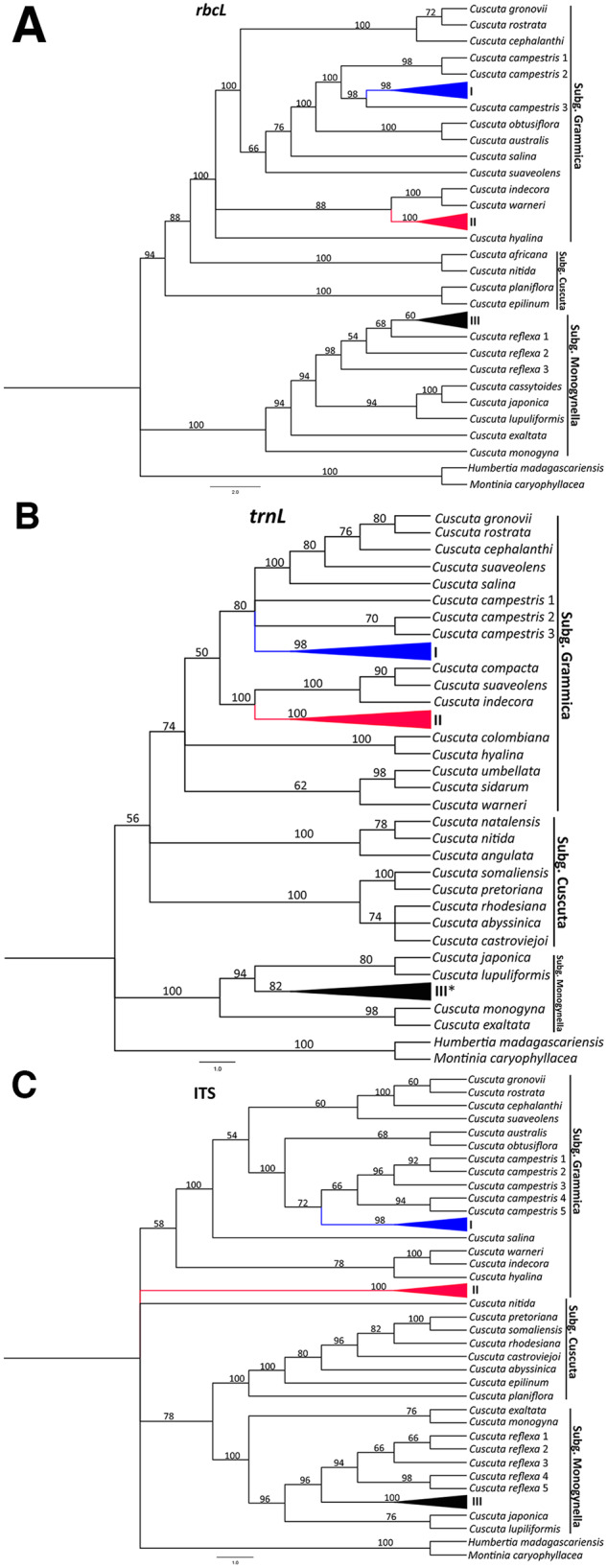
Phylogenetic reconstruction of Cuscuta species based on rbcL, trnL, and ITS regions. Maximum Parsimony bootstrap consensus trees (1,000 replicates) are shown, with bootstrap supports indicated above branches. I, II, and III represent Cuscuta taxa sequenced under this study, and denote C. campestris, C. kilimanjari, and C. reflexa, respectively. The asterisk (*) on the trnL tree implies that our C. reflexa taxa were collapsed with those from GenBank.
Dodder has a wide host range with potential to infect crops of great economic importance
In total, 26 host plant species across 13 angiosperm orders comprising shrubs (40%), trees (44%), and herbs (16%) were parasitized by the aforementioned Cuscuta spp. Fabales (20%) was the most parasitized order, followed by Lamiales (16%), Malpighiales, and Caryophyllales (the latter two with 12%), whereas the rest had a single species colonized by the parasite. With regards to host specificity, C. campestris and C. reflexa exhibited “generalist” behavior, indiscriminately parasitizing hosts across numerous orders. Among the parasitized species, Solanum incanum and Biancaea decapetala were the most suitable hosts for C. campestris, whereas Thevetia peruviana was predominantly colonized by C. reflexa. Conversely, C. kilimanjari exhibited “specialist” behavior, parasitizing host species across two orders only (Supplemental Table S1).
We further demonstrated the parasite’s potential threat to crops by infecting tea, coffee, and mango with C. reflexa, which was followed by histological analysis. We focused on C. reflexa for infections due to its invasiveness across the region, its wide host preference (perennial trees and shrubs), and because it was introduced to East Africa. In all instances (100%), C. reflexa successfully parasitized test plants and formed haustoria within 14 d of infection. Cross sections, performed 4 wk after infection, revealed successful penetration of the parasite into host tissues, which extended past sclerenchyma enabling successful formation of vascular-bundle connections (Figure 3). Interestingly, we observed a resistance response from an infected mango plant. Specifically, the infected point swelled, and exuded a sap-like substance that was deposited around the wounded area. This eventually led to death of the parasite (within 4 wk of attachment), with the infected area “healing” afterward (Supplemental Figure S3).
Figure 3.
Cuscuta parasitism and extent of ingression into host plants. The upper panel shows close-up photographs of infected test plants whereas the lower panel is toluidine blue-stained cross sections of the host-parasite interface. Red arrows indicate points of haustorium formation. Aa and Ab: coffee, Ba and Bb: tea, Ca and Cb: mango. P, parasite; H, host; HX, host xylem; PX, parasite xylem; SC, host sclerenchyma. Bar top panel = 10 mm, bottom panel = 5 mm.
To further evaluate the imminent danger posed by these parasites to tea, we sampled Kenyan locations where the ranges for tea and C. reflexa overlapped. We found that at a site in Kakamega, Western Kenya (https://earth.google.com/web/search/0+12+272+27+27N,+34+46+2721E/@0.202444,34.77314623,1583.10168457a,0d,15y,119.88850412h,90.54106391t,0r/data=CigiJgokCdGS14h74TRAEc6S14h74TTAGbGLz-dnrjzAIbWQBbGXbGDAIhoKFnVHbklCUWhkQ1BPdnMyZll4TTFqdFEQAg), Markhamia lutea, a host of C. reflexa, was infected and growing just next to a tea plantation, thus pointing to the definite possibility of tea infestation. A Google Earth image (using the Street View option) taken in June 2018 showed that the tree had not been infested, but by the time we visited the site in August 2019, the tree had heavy infestation that threatened to encroach the tea plantation (Figure 4). This indicates that C. reflexa is highly invasive with potential to rapidly infest new localities. In its native ranges of Asia, C. reflexa has been reported to parasitize a wide range of hosts, including coffee (Bhattarai et al., 1989). Additionally, dodder has been reported on coffee in Uganda (Jennipher Bisikwa, Personal Communication), and one of the records at the East African Herbarium (voucher number EA16731, collected in 1983) indicated that one of the C. kilimanjari specimens parasitized coffee.
Figure 4.
Cuscuta threat on tea. A, Google Earth™ (Street View) image of a tea plantation in western Kenya, taken in 2018. The circled bush represents M. lutea, a tree species commonly used as a windbreaker around plantations and later found to be a Cuscuta host. B, The windbreaker infested with C. reflexa, 1 year after the first image. Arrowheads indicate parasitic vines threatening to encroach into the tea plantation.
Predicted Cuscuta distribution and habitat suitability
Our SDMs had excellent predictive performances, with area under curve (AUC) values of 0.93 and 0.87 for C. reflexa built using occurrence records from Kenya and native ranges, respectively. On the other hand, models for C. campestris and C. kilimanjari had modest performances, with AUC values of 0.76 and 0.72, respectively (Supplemental Figures S4–S7). Precipitation of the warmest quarter (bio18 = 59.0+) and annual mean temperature (bio1 = 32.2%) were the most influential variables in the models for C. reflexa based on occurrences in Kenya and the native range, respectively. Conversely, precipitation of the driest quarter (bio17 = 46.2) and isothermality (bio3 = 47.6%) were the highest contributors to the models for C. campestris and C. kilimanjari, respectively (Table 1).
Table 1.
Permutation importance of bioclimatic and vegetation variables
| Variable | Cuscuta campestris | Cuscuta kilimanjari | Cuscuta reflexa (invaded) | Cuscuta reflexa (native) |
|---|---|---|---|---|
| Annual mean temperature (bio1) | 4.4 | 0 | 0 | 32.2a |
| Isothermality (bio3) | 2.5 | 47.6a | 13.7 | 1.1 |
| Precipitation seasonality (bio15) | 0 | 12.7 | 0.5 | 15.6 |
| Precipitation of driest quarter (bio17) | 46.2a | 2.7 | 0.5 | 15.9 |
| Precipitation of warmest quarter (bio18) | 4.9 | 7.7 | 59.0a | 13.2 |
| Land cover fraction (grass; veg1) | 40.1 | 25.8 | 14.9 | 1.7 |
| Land cover fraction (shrub; veg2) | 0.3 | 0 | 0.8 | 11.2 |
| Land cover fraction (tree; veg3) | 1.5 | 3.5 | 10.6 | 9.1 |
Values represent percent (%) contributions of each variable to the model.
The highest contributing variable
Our models revealed different distribution patterns across Eastern Africa, with current estimates showing that all three species of this study can potentially colonize areas larger than the localities sampled herein (Figure 5). Overall, high habitat suitability for C. reflexa (log-transformed score >0.8) is predicted in Western and Central Kenya, Eastern Uganda, large parts of Rwanda and Burundi, and Central Ethiopia. Predicted habitat suitability for C. kilimanjari is higher than that observed for C. reflexa across the six countries. Particularly, Central and Western Kenya, Western Uganda, Rwanda and Burundi, and Central Ethiopia show high suitability. Conversely, moderate habitat suitability is predicted for C. campestris, with infestation likely to occur in Western and Central Kenya, Eastern Uganda, and in pockets of Ethiopia and Tanzania (Figure 5). Additionally, many major coffee- and tea-growing areas show high habitat suitability for all species. Our collected occurrence data indicate that C. reflexa seems to have already invaded many of these regions in Kenya (Figure 5). Projections of our models to other regions suggest that C. reflexa could become or already may be a concern in coffee- and tea-growing regions of Rwanda, eastern Uganda, and the Harar coffee zone of Ethiopia (Figure 5). Additionally, the predicted distributions are supported by various local press describing dodder infestations in the aforementioned countries including Uganda (https://www.newvision.co.ug/new_vision/news/1503872/dangerous-plant-invades-kampala-city) and Kenya (https://www.nation.co.ke/news/Dodder-plant-poses-threat-to-trees-and-crops/1056-5138904-aawhphz/index.html).
Figure 5.
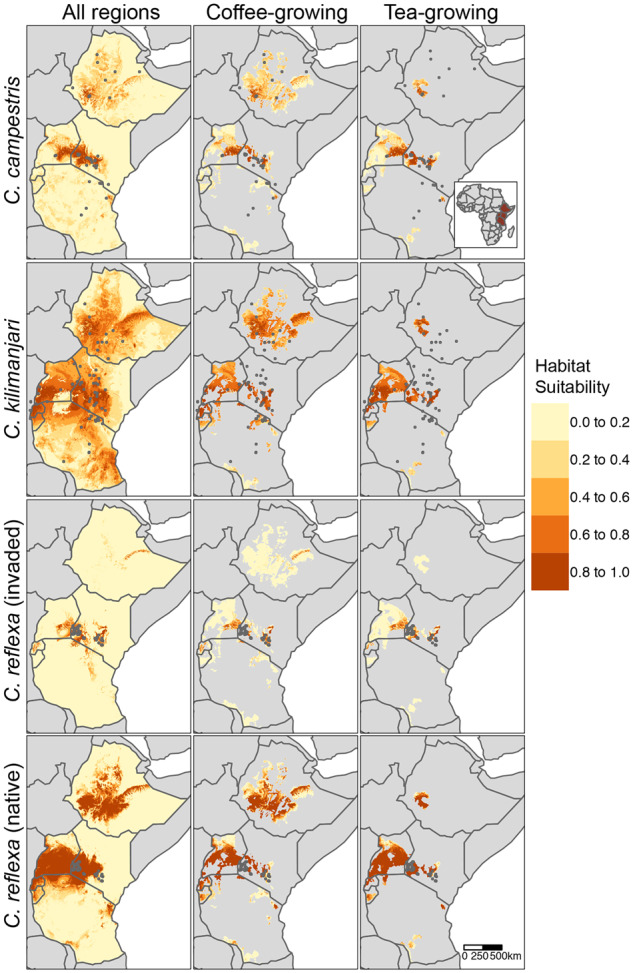
Habitat suitability for C. campestris, C. kilimanjari, and C. reflexa across Ethiopia, Kenya, Uganda, Rwanda, Burundi, and Tanzania. Dark-grey points indicate locations of occurrence records. For C. reflexa, species distribution models were trained using occurrences from the invaded range in Kenya (n = 66; dark-grey points) or native range from Afghanistan to Indo-China (n = 165; not shown), then projected to the six countries. Models for C. campestris and C. kilimanjari were constructed using a combination of occurrence records obtained from our sampling activities (in Kenya), as well as those obtained from GBIF and herbarium specimens at the East African Herbarium. Projections were masked to coffee- and tea-growing regions, estimated to produce >1 metric tons in 2017 (IFPR, 2020).
Discussion
Cuscuta identification
Interactions among global change components, such as land use, agricultural intensification, and invasions by non-native species, cause substantial changes to plant community composition worldwide. Consequently, plant communities that arguably play the biggest role in establishing and maintaining ecosystems are deteriorating. A key driver of such community change is invasion by alien plants that displace/arrest development of native species, subsequently reducing habitat quality, causing economic losses, and posing a serious threat to human wellbeing. Once established, the spread of invasive plants is extremely difficult to control or reverse. In African terrestrial ecosystems, a myriad of invasive plant species occurs, key among which is the current rapid spread of parasitic vines of the genus Cuscuta. We analyzed the potential threat of Cuscuta in Eastern Africa by describing their taxonomy, host range, and habitat suitability.
Cuscuta identification using morphological characters is challenging because members of this genus lack roots and their leaves are reduced to minute scales (Kuijt, 1969). However, diversity among floral components presents a unique opportunity for distinguishing species. In the present study, we adopted monographs by Engelmann (1857) and Yuncker (1932) to interpret stigma and style morphology, and consequently identified three Cuscuta species from two related subgenera (Grammica and Monogynella) in accessions collected from Kenya. Individuals in subgenus Grammica had two separate styles, with either globose or spherical stigmas, and were classified as C. campestris and C. kilimanjari, respectively, whereas those in subgenus Monogynella had elongate stigmas born on fused styles, typical of C. reflexa. We validated this identification by sequencing rbcL, trnL, and ITS regions from representative individuals, and performed phylogenetic reconstruction alongside other species from GenBank. These markers have been extensively used to infer phylogenetic relationships, character evolution, and biogeography across the genus (García and Martin, 2007; McNeal et al., 2007; García et al., 2014). In our case, all our sequences were resolved alongside respective C. campestris, C. kilimanjari, and C. reflexa taxa from GenBank, indicating that they indeed belonged to these species. Additionally, these phylogenetic reconstructions confirmed the monophyly of subgenus Monogynella, with all members basal to subgenera Cuscuta and Grammica, consistent with earlier reports (García and Martin, 2007; McNeal et al., 2007; Stefanović et al., 2007; García et al., 2014). Apart from these three, other Cuscuta species have also been reported in Africa, although most of them belong to subgenus Cuscuta (Zerman and Saghi, 1995; García, 1999; García and Martin, 2007).
SDM-based distribution patterns
Cuscuta’s damaging success and worldwide distribution are attributed to its ability to colonize a wide range of hosts and survive in areas with an array of environmental conditions (Parker and Riches, 1993). Additionally, most members are nonspecific, colonizing multiple host plants across various angiosperm families (Dawson et al., 1994; Lanini and Kogan, 2005; Kim and Westwood, 2015). Our SDMs, coupled with the observed host range and artificial infection assays, provided insights into the parasite’s potential distribution patterns in Eastern Africa. These models showed that the species are likely to be present in additional areas not covered by this study, as evidenced by areas of high habitat suitability. Three climatic variables, namely precipitation of the warmest quarter, annual mean temperature, and isothermality, had the highest contribution to our models, suggesting that they could play a significant role in the predicted distribution. These were also found to significantly contribute to habitat suitability for C. chinensis (Ren et al., 2020). With regards to land cover contribution, grass cover fraction was an important predictor for SDMs in all three species, with lower habitat suitability corresponding with areas of high grass cover. This finding is consistent with lower frequency of preferred Cuscuta hosts in grasslands and parasitism on diverse herbaceous plants as well as perennial shrubs and trees. Additional identification of lower risk areas for Cuscuta invasion is imperative to guiding preparedness, owing to scarcity of resources for managing invasive species. For example, our models predicted relatively low habitat suitability in central and western Uganda for C. reflexa (according to models based on occurrences from a restricted sampling area in the invaded range). However, SDMs based on occurrences throughout a broader set of environments in the native range suggested high habitat suitability in these same regions. Although there are many limits to spatial transferability of SDMs from native and introduced ranges (Liu et al., 2020), these findings suggest that current environments may not be an effective barrier to the spread and establishment of C. reflexa in many East Africa regions. Thus, active management of C. reflexa will be needed to prevent its spread.
The potential threat of Cuscuta to cash crops
Results from artificial infection of C. reflexa on coffee, tea, and mango revealed its potential to parasitize these economically important crops. Specifically, we observed attachment, haustoria, and vascular bundle formation, which are indicative of successful parasitism. Cuscuta parasitism on coffee and tea could have devastating impacts on the income generated by these crops in East African countries. In addition, the presence of a C. reflexa-infected M. lutea adjacent to a tea plantation is indicative that such an infestation may be imminent. Governments in East Africa will therefore be required to develop urgent interventions and appropriate policies to stop such eventualities, which would have devastating effects to farmers in affected areas.
Interestingly, we observed a resistance response in the mango genotype artificially infected with C. reflexa. Cross sections indicated ingression of parasitic haustoria into the host and successful establishment of vascular-bundle connections. However, this success was short-lived with the host initiating wound response and chemical deposition that resulted in death of the parasite and subsequent healing of the infected area. It is possible that this resistance response goes beyond the observed wounding, although this remains to be investigated. Such a phenomenon could also be key in determining dodder’s host preference, since plants that display resistance are avoided during parasitism (Kaiser et al., 2015). Previous studies have described this type of resistance (Albert et al., 2006) among other mechanisms, including incompatibility due to anatomical attributes (Dawson et al., 1994), induction of defense-related stress hormones (salicylic and jasmonic acid), and the use of mechanical barriers that block parasitic ingression into host vasculature (Kaiser et al., 2015). Consequently, species such as Gossypium hirsutum (Capderon et al., 1985), Solanum lycopersicum (Albert et al., 2006; Runyon et al., 2010), and some varieties of chickpea (Cicer arietinum; Goldwasser et al., 2012) have been reported to resist dodder infection.
Conclusions and future prospects
In summary, our findings reveal the presence of C. campestris, C. kilimanjari, and C. reflexa across various ecosystems in Kenya. C. campestris and C. kilimanjari, endemic to or naturalized in Eastern Africa, have been documented in the Flora of Tropical East Africa, alongside others such as C. australis, C. suaveolens Seringe, C. hyalina Roth, C. cassytoides Engelm, C. epilinum, and C. planiflora Tenore (Verdcourt, 1963). However, here we describe the occurrence of C. reflexa, a south Asian species, in continental Africa. These parasites have a broad host range, and infestation on crops of economic importance may be inevitable if urgent actions are not taken to stop their spread. In fact, our predictions show that many regions across Eastern Africa are characterized by highly suitable habitats for Cuscuta infestation and may already be infested. Therefore, this work will be critical in developing informed strategies for managing the parasite and averting the looming risk. This may potentially involve identifying resistant plant species and genotypes to aid development of cultural control and adaptation measures in agriculture and forestry within the region. Additionally, unraveling the physical, biochemical, and genetic factors controlling the observed resistance response in mango will provide insights into regulation of these resistance phenomena and guide future control strategies.
Materials and methods
Sample collection
We collected a total of 96 Cuscuta accessions across Kenya. In our case, an accession is defined as material from an individual plant from the same species found within a similar geographical area. Sampling was done between July and November 2018, with at least five individual accessions collected per location. Dodder flowers and vines were collected and immediately dried in silica gel to await morphological analysis and DNA isolation. Cuscuta-parasitized plants were also collected and identified to the species level, according to the keys of plant identification described in the Flora of Tropical East Africa (Verdcourt, 1963) and Pennsylvania State University (https://extension.psu.edu/plant-identification-preparing-samples-and-using-keys).
Morphological Cuscuta identification
Morphological identification was performed according to the keys of Cuscuta monograph constructed by Engelmann (1857) and Yuncker (1932). Briefly, flowers were either used immediately after collection or rehydrated before microscopy (for those kept in silica gel). To observe different parts, we examined a single full flower (sepals, petals, gynoecium, and androecium) under a Leica MZ10F stereomicroscope (Leica Microsystems, UK) and photographed it. Thereafter, we carefully dissected and photographed it, with focus given to the gynoecia, number of parts, fusion (or lack thereof) of the styles, and the size and shape of stigmas. Ovaries were also dissected, and then the number, size, and color of ovules were observed and photographed.
Host plant infection and histology
We evaluated whether dodder could expand its host range to tree crops of agricultural value, by artificially infecting tea (Camelia sinensis), coffee (Coffea arabica), and mango (Mangifera indica) with C. reflexa under controlled conditions in the greenhouse. Summarily, 3-month-old test seedlings were maintained in potted soil with regular watering then infected by winding a 30-cm piece of parasitic vine (that had at least one node) around their stems. Parasitism was determined by histological analysis of the host-parasite interface, 4 wk after infection as previously described (Gurney et al., 2003). Briefly, tissues at the interface were collected and fixed using Carnoy’s fixative (4:1 ethanol: acetic acid [v/v]), dehydrated with absolute ethanol then pre-infiltrated in Technovit solution (Haraeus Kulzer, GmbH). The tissues were embedded in 1.5-mL microcentrifuge tube lids containing Technovit/Hardener and left to set according to the manufacturer’s instructions, and then mounted onto histoblocks using the Technovit 3040 kit (Haraeus Kulzer GmbH). Microscopic sections (5-µm thick) of the tissues were cut using a microtome (Leica RM 2145), transferred onto glass slides and stained using 0.1% (w/v) Toluidine Blue O dye in phosphate buffer. After washing off excess dye and drying, slides were covered with slips containing a drop of DPX (BDH, Poole, UK), observed, and photographed using a Leica microscope mounted with a Leica MC190 HD camera (Leica, UK).
DNA extraction, PCR, and sequencing
We sequenced representative accessions from each of the aforementioned Cuscuta spp. following morphological characterization. We could not acquire material for DNA extraction from voucher specimens held at the East African Herbarium in Nairobi, Kenya, owing to the destructive nature of sampling involved. DNA was extracted from flowers and hanging vines, collected at least 10 cm away from the point of attachment to avoid host-DNA contamination. PCR amplification of the ITS region was done using ITS4 and ITS5 primers (Baldwin, 1992), whereas rbcL was amplified using rbcL-512F and rbcL-1392R primers (McNeal et al., 2007). Partial amplification of trnL was using trnLF- 5′-CGAAATCGGTAGACGCTACG-3′ and trnLR-5′-ATTTGAACTGGTGACACGAG-3′ primers, designed specifically for Cuscuta. PCRs were performed in 25-µL volumes using MyTaq DNA polymerase kit (Bioline, Meridian Biosciences) under the following conditions; 95°C for 1 min, followed by 35 cycles comprising 95°C for 15 s, each primer’s respective annealing temperature for 30 s, and a 72°C extension for 1 min. A final 10-min extension, at 72°C, was also included. PCR products were confirmed on a 1% (w/v) agarose gel, cleaned using the Qiaquick PCR purification kit (Qiagen), and sequenced on the ABI platform at Macrogen (Macrogen Inc).
Phylogenetic analysis
Sequences were edited in SeqMan Pro17 in Lasergene package (DNASTAR Inc., Madison, WI) to remove low-quality reads, then aligned using ClustaX version 2.0 (Larkin et al., 2007). Sequences were submitted to NCBI (Supplemental Table S2), then used as queries to identify similar taxa using the nucleotide BLAST algorithm at NCBI. Highly similar sequences across the three Cuscuta subgenera were retrieved for phylogenetic reconstruction and ancestry inferencing, with sequences for Montinia caryophyllacea and Humbertia madagascariensis included as outgroups. We first constructed unrooted Maximum Likelihood gene trees for the accessions under this study, and then generated Parsimony consensus trees (with 1,000 replications) for phylogenetic reconstruction of taxa from our species alongside those from NCBI. All phylogenetic analyses were performed in MEGAX (Kumar et al., 2018), and the trees visualized in FigTree version 1.4.4 (Rambaut, 2009).
Species distribution modeling
Occurrence records
We combined respective occurrence records for C. kilimanjari and C. campestris from our sampling efforts in Kenya with records from the Global Biodiversity Information Facility (GBIF); https://doi.org/10.15468/dl.muwe3t and https://doi.org/10.15468/dl.y8rtg4 for C. kilimanjari and C. campestris, respectively, as well as records for specimens held in the collection at the East African Herbarium (EA). This resulted in a total of 74 and 51 unique locations in East Africa for C. kilimanjari and C. campestris, respectively. We found no records of C. reflexa in neither GBIF nor in the EA collection, hence all occurrence records from East Africa were from localities sampled as part of this study (n = 66 unique locations across Kenya). We also built SDMs based on 165 unique occurrences of C. reflexa from its native range (Afghanistan to Indo-China) that were available from GBIF (https://doi.org/10.15468/dl.kaqby6) and then projected the models to East Africa. To characterize the background of the study, we randomly sampled 1,000 points from a radius of 300 km from known occurrences.
Environmental variables
Species distribution models were based on five bioclimatic variables, namely annual mean temperature (bio1), isothermality (bio3), precipitation seasonality (bio15), precipitation of the driest quarter (bio17), and precipitation of the warmest quarter (bio18), as well as three variables related to vegetation structure, namely land cover fraction of grass (veg1), shrub (veg2), and tree (veg3). Four of the bioclimatic variables (bio1, bio3, bio15, and bio18) were previously reported as important for species distribution models for C. chinensis (Ren et al., 2020), whereas the fifth (bio17) exhibited high feature importance in our preliminary analyses. Bioclimatic data were obtained from the CHELSA dataset (https://zenodo.org/record/3939050#.X3N49y2ZM8Z;Karger et al., 2017), whereas vegetation layers were from the Copernicus Global Land Service (Buchhorn et al., 2020). These layers were based on epoch 2019 from Collection 3 of the annual, global 100-m land cover maps, and were resampled to match resolution of the bioclimatic layers (1 km) using the bilinear interpolation method of the “resample” function from the R package “raster” (Hijmans 2019). The vegetation layers captured aspects of vegetation cover, which may be important for Cuscuta spp. parasitism on various herbaceous and woody host species. All variables had Pearson’s correlation coefficients less than 0.8 across background points of the study.
Model building and prediction of suitable habitats
Species distribution models were built using the Maxent algorithm (Phillips et al., 2006). The Models were tuned and evaluated with R version 3.6.1 with ENMeval (Muscarella et al., 2014) using the checkerboard2 method for partitioning occurrence data into training and test sets. To determine overlap between Cuscuta spp. distributions with major coffee- and tea-growing areas, we used crop production maps from IFPRI (2020; https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/FSSKBW) to mask Cuscuta spp. distribution models to just those areas estimated to produce at least one metric ton of coffee or tea in 2017.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers listed in Supplemental Table S2 (ITS [EF194641.1; EF194642; AY554399; EF194652.1; MT363349.1; KT383247.1; KT383097.1MG669320.1MN718805.1KT383096.1KJ025065.1; MT947605MT947606; MT947607; EF194707.1; EF194541; MT952140MT952141MT952142; EF194549; KX683007; DQ924573.1; DQ924627.1; DQ924630.1; DQ924640.1; DQ924638.1; DQ924631.1; DQ924610.1; KF805106.1; EU330323.1; DQ924569.1; EU330322.1AY558823.1HQ728506.1HQ728505.1HQ728504.1; MW080817MW080818MW080819; DQ924571.1; DQ924570.1; KJ400198.1; KJ400199] trnL [EF194417.1; EF194428.1; AY558829.1; EF194497.1; MH795068.1; KT371734.1; AJ428057.1; MH392436.1; KU761501.1; KT371719.1; MH392404.1; MW086603; MW086604; MW086605; EF194497.1; EF194291.1; MW086607; MW086608; MW086609; AY558837.1; EF194318.1; AJ457118.1; EF152073.1; EF152075.1; EF152074.1; EF152069.1; EF152067.1; AJ430075.1; AJ457096.1; EU189132.1; MH392459.1; AM711640.1; AY558843.1; AY558859.1; MW115588; MW115589; MW115590; EF152064.1; AY558838.1; AY101171.1; AY101173.1] rbcL [AM711639.1; EU330263.1; EU330264.1; KJ436718.1; KJ436607.1; KJ436693.1; MN708214.1; EU330268.1; KJ436614.1; MF135355.1; KX430890.1; MW078922; MW078923; MW078924; EU883466.1; KJ436735.1; MW078930; MW078931; MW078932; KJ436664.1; KJ436661.1; EU330275.1; KJ436602.1; KJ436641.1; KJ436702.1; KJ436645.1; AY558871.1; KJ436708.1; AM711640.1; X61698.1; MW078927; MW078928; MW078929; MH780080.1; KJ436617.1; KJ436680.1; AY101062.1; L11194.2]).
Supplemental data
The following supplemental materials are available in the online version of this article.
Supplemental Figure S1. Categories of Cuscuta species observed parasitizing various susceptible host plants in Kenya.
Supplemental Figure S2. Unrooted Maximum Likelihood trees based on rbcL, trnL, ITS, and a combination of the three regions.
Supplemental Figure S3. Resistance response exhibited by a mango (Mangifera indica) genotype under C. reflexa infection.
Supplemental Figure S4. AUC values for C. campestris.
Supplemental Figure S5. AUC values for C. kilimanjari.
Supplemental Figure S6. AUC values for C. reflexa.
Supplemental Figure S7. AUC values for C. reflexa.
Supplemental Table S1. Occurrence records of Cuscuta species collected from Kenya.
Supplemental Table S2. List of accession numbers.
Supplementary Material
Acknowledgments
We thank the National Museums of Kenya, through the East Africa Herbarium, for providing occurrence records. We acknowledge Prof. Claude dePamphilis (Pennsylvania State University-USA) for fruitful scientific discussions and Prof. Alistair Jump (University of Stirling-UK) for thoughtful reviews.
Funding
We acknowledge financial support from Kenyatta University through the Vice Chancellors Research grant number KU/DVCR/VRG/VOL.11/216. J.M.’s Ph.D. is funded by the National Research Fund (NRF) grant number NRF/PhD/02/76.
Conflict of interest statement. The authors have no conflicts of interest to declare.
S.R. conceived the study, guided fieldwork, and oversaw experimental work. J.M. carried out fieldwork, sequence analysis, infection assays, and wrote the draft manuscript. B.N.M carried out morphological characterization. W.K performed histological analysis. P.S. curated environmental data. E.S.B. and M.W carried out species distribution modelling. R.O, M.P, A.A, and P.O assisted with conceptual support and manuscript editing. All authors read and approved the final manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Steven Runo (runo.steve@ku.ac.ke).
References
- Albert M, Belastegui-Macadam XM, Bleischwitz M, Kaldenhoff R (2008) Cuscuta spp: parasitic plants in the spotlight of plant physiology, economy and ecology. In, Lüttge U, Beyschlag W, Murata J, eds, BT - Progress in Botany, Springer, Berlin, Germany, pp 267–277 [Google Scholar]
- Albert M, Belastegui-Macadam X, Kaldenhoff R, (2006) An attack of the plant parasite Cuscuta reflexa induces the expression of attAGP, an attachment protein of the host tomato. Plant J 48: 548–556 [DOI] [PubMed] [Google Scholar]
- Baldwin BG (1992) Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal dna in plants; an example from the compositae. Mol Phylogenet Evol 1: 3–16 [DOI] [PubMed] [Google Scholar]
- Bhattarai T, Bhandary H, Shrestha P (1989) Host range of Cuscuta reflexa Roxb. in the Kathmandu Valley, Nepal. Plant Protect Q 4: 78–80 [Google Scholar]
- Buchhorn M, Lesiv M, Tsendbazar NE, Herold M, Bertels L, Smets B (2020) Copernicus global land cover layers—collection 2. Remote Sens 12: 1044 [Google Scholar]
- Capderon M, Fer A, Ozenda P (1985) About an unreported system leading to the expulsion of a parasite—Cuscuta on cotton-plant (Cuscuta lupuliformis Krock on Gossypium hirsutum-L). Comptes rendus de l’Académie des Sciences 3: 227–232 [Google Scholar]
- Cotter M, Renzoandre P-L, Sauerborn J (2012) Understanding the present distribution of the parasitic weed Striga hermonthica and predicting its potential future geographic distribution in the light of climate change. In Julius-Kühn-Archiv 434:630–636 [Google Scholar]
- Dawson JH, Musselman LJ, Wolswinkel P, Dörr I (1994) Biology and control of Cuscuta. Rev Weed Sci 6: 265–317 [Google Scholar]
- Engelmann G (1857) Botanical notebook 23: Cuscuta. https://www.biodiversitylibrary.org/item/234938 (August 10, 2020)
- FAO (2017) Food and Agriculture Organization. http://www.fao.org/faostat/en/#data/QC. (September 2019)
- García MA (1999) Cuscuta subgenus Cuscuta (Convolvulaceae) in Ethiopia, with the description of a new species. Ann Bot Fenn 36: 165–170 [Google Scholar]
- García MA, Costea M, Kuzmina M, Stefanovic S (2014) Phylogeny, character evolution, and biogeography of Cuscuta (Dodders; Convolvulaceae) inferred from coding plastid and nuclear sequences. Am J Bot 101: 670–690 [DOI] [PubMed] [Google Scholar]
- García MA, Martin MP (2007) Phylogeny of Cuscuta Subgenus Cuscuta (Convolvulaceae) Based on nrDNA ITS and Chloroplast trnL Intron Sequences. Syst Bot 32: 899–916 [Google Scholar]
- GBIF.org (October 07, 2020) 10.15468/dl.muwe3t [DOI]
- GBIF.org (October 07, 2020) 10.15468/dl.y8rtg4 [DOI]
- GBIF.org (October 07, 2020) 10.15468/dl.kaqby6 [DOI]
- Goldwasser Y, Miryamchik H, Sibony M, Rubin B (2012) Detection of resistant chickpea (Cicer arietinum) genotypes to Cuscuta campestris (field dodder). Weed Res 52: 122–130 [Google Scholar]
- Gurney AL, Grimanelli D, Kanampiu F, Hoisington D, Scholes JD, Press MC (2003) Novel sources of resistance to Striga hermonthica in Tripsacum dactyloides, a wild relative of maize. New Phytol 160: 557–568 [DOI] [PubMed] [Google Scholar]
- Hibberd JM, Bungard RA, Press MC, Jeschke WD, Scholes JD, Quick WP (1998) Localization of photosynthetic metabolism in the parasitic angiosperm Cuscuta reflexa. Planta 205: 506–513 [Google Scholar]
- Hijmans RJ (2019) raster: Geographic Data Analysis and Modelling. R package version 3.0-7. https://CRAN.R-project.org/package=raster (October 2, 2020)
- IFPR (2020) International Food Policy Research Institute; "Spatially-Disaggregated Crop Production Statistics Data in Africa South of the Saharan for 2017" 10.7910/DVN/FSSKBW, Harvard Dataverse, V1 (September 12, 2020)
- Kaiser B, Vogg G, Fürst UB, Albert M (2015) Parasitic plants of the genus Cuscuta and their interaction with susceptible and resistant host plants. Front Plant Sci 6: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karger D, Nikol OC, Jürgen B, Tobias K, Holger K, Rodrigo WS, Niklaus EZ, Linde HP, Michael K (2017) Climatologies at High Resolution for the Earth’s Land Surface Areas. Sci Data 4: 170122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G, Westwood JH (2015) Macromolecule exchange in Cuscuta— host plant interactions. Curr Opin Plant Biol 26:20–25 [DOI] [PubMed] [Google Scholar]
- Kuijt J (1969) The biology of parasitic flowering plants. University of California Press, Berkeley, CA [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol 35: 1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanini WT, Kogan M (2005) Biology and Management of Cuscuta in Crops. Ciencia e Investigación Agraria 32: 165–179 [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X Version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li JM, Dong M (2009) Fine-scale clonal structure and diversity of invasive plant Mikania micrantha H.B.K. and its plant parasite Cuscuta campestris Yunker. Biol Invasions 11: 687–695 [Google Scholar]
- Liu C, Wolter C, Xian W, Jeschke JM (2020) Species distribution models have limited spatial transferability for invasive species. Ecol Lett 23:1682–1692 [DOI] [PubMed] [Google Scholar]
- McNeal JR, Arumugunathan K, Kuehl JV, Boore JL, dePamphilis CW (2007) Systematics and plastid genome evolution of the cryptically photosynthetic parasitic plant genus Cuscuta (Convolvulaceae). BMC Biol 5: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscarella R, Galante PJ, Soley-Guardia M, Boria RA, Kass JM, Uriarte M, Anderson RP (2014) ENMeval: an R Package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent Ecological Niche Models. Methods Ecol Evol 5: 1198–1205 [Google Scholar]
- Parker C (2012) Parasitic weeds: a world challenge. Weed Sci 60: 269–276 [Google Scholar]
- Parker C, Riches CR (1993). Parasitic weeds of the world: Biology and control. CAB International, Wallingford, UK, p 332 [Google Scholar]
- Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modelling of species geographic distributions. Ecol Model 190: 231–59 [Google Scholar]
- Press MC, Phoenix GK (2005) Impacts of parasitic plants on natural communities. New Phytol 166: 737–751 [DOI] [PubMed] [Google Scholar]
- Rambaut A (2009) FigTree v1.2.2. http://tree.bio.ed.ac.uk/software/FigTree/
- Ren Z, Zagortchev L, Ma J, Yan M, Li J (2020) Predicting the potential distribution of the parasitic Cuscuta chinensis under global warming. BMC Ecol 20: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revill MJW, Stanley S, Hibberd JM (2005) Plastid genome structure and loss of photosynthetic ability in the parasitic genus Cuscuta. J Exp Bot 56: 2477–2486 [DOI] [PubMed] [Google Scholar]
- Runyon JB, Mescher MC, Felton GW, De Moraes CM (2010) Parasitism by Cuscuta pentagona sequentially induces JA and SA defence pathways in tomato. Plant Cell Environ 33:290–303 [DOI] [PubMed] [Google Scholar]
- Shen H, Ye W, Hong L, Huang H, Wang Z, Deng X, Yang Q, Xu Z (2006) Progress in parasitic plant biology: host selection and nutrient transfer. Plant Biol 8:175–185 [DOI] [PubMed] [Google Scholar]
- Stefanović S, Kuzmina M, Costea M (2007) Delimitation of major lineages within Cuscuta subgenus Grammica (Convolvulaceae) using plastid and nuclear DNA sequences. Am J Bot 94: 568–589 [DOI] [PubMed] [Google Scholar]
- van der Kooij T, Krause K, Dörr I (2000) Molecular, functional and ultrastructural characterisation of plastids from six species of the parasitic flowering plant genus Cuscuta. Planta 210: 701–707 [DOI] [PubMed] [Google Scholar]
- Verdcourt B (1963) Convolvulaceae. InHubbard CE, Milne-Redhead E, eds, Flora of tropical East Africa, Crown Agents for Overseas Governments and Administrations, London, UK, pp 1–16 [Google Scholar]
- Yu H, He WM, Liu J, Miao SL, Dong M (2009) Native Cuscuta campestris restrains exotic Mikania micrantha and enhances soil resources beneficial to natives in the invaded communities. Biol Invasions 11: 835–844 [Google Scholar]
- Yuncker TG (1932) The Genus Cuscuta. Torrey Botanical Soc 18: 113–331 [Google Scholar]
- Zerman N, Saghir AR (1995) The genus Cuscuta in Algeria. Arab J Plant Prot 13: 69–75 [Google Scholar]
- Zhang C, Chen L, Tian CM, Li T, Wang R, Yang QQ (2016). Predicting the distribution of dwarf mistletoe (Arceuthobium sichuanense) with GARP and Maxent models. J Beijing For Univ 38: 23–32 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.