Abstract
Microbial forensics represents a promising tool to strengthen traditional forensic investigative methods and fill related knowledge gaps. Large-scale microbiome studies indicate that microbial fingerprinting can assist forensics in areas such as trace evidence, source tracking, geolocation, and circumstances of death. Nevertheless, the majority of forensic microbiome studies focus on soil and internal organ samples, whereas the microbiome of skin, mouth, and especially vaginal samples that are routinely collected in sexual assault and femicide cases remain underexplored. This review discusses the current and emerging insights into vaginal, skin, and salivary microbiome-modulating factors during life (e.g., lifestyle and health status) and after death (e.g., environmental influences and post-mortem interval) based on next-generation sequencing. We specifically highlight the key aspects of female reproductive tract, skin, and mouth microbiome samples relevant in forensics. To fill the current knowledge gaps, future research should focus on the degree to which the post-mortem succession rate and profiles of vaginal, skin, and saliva microbiota are sensitive to abiotic and biotic factors, presence or absence of oxygen and other gases, and the nutrient richness of the environment. Application of this microbiome-related knowledge could provide valuable complementary data to strengthen forensic cases, for example, to shed light on the circumstances surrounding death with (post-mortem) microbial fingerprinting. Overall, this review synthesizes the present knowledge and aims to provide a framework to adequately comprehend the hurdles and potential application of vaginal, skin, and salivary post-mortem microbiomes in forensic investigations.
Keywords: post-mortem microbiome, trace evidence, microbial forensics, sexual assault, femicide, next-generation sequencing, thanatomicrobiome, epinecrotic communities
Introduction
Violence against women is an urgent global problem, as more than one-third of women worldwide has been victim of physical and/or sexual violence in their lifetime (Devries et al., 2013; World Health Organization, 2013). Annually, 66,000 women and girls are victims of femicide, that is, intentional murder of women and girls because they are female (Geneva Declaration, 2011; World Health Organization and Pan American Health Organization, 2012). Remarkably, only 25% of reported rape cases in Europe lead to a conviction, often due to the difficulty of providing evidence (Lovett and Kelly, 2009). Moreover, forensic experts are convinced that a large number of undetected homicides are misclassified in annual death statistics as natural deaths, suicides, or accidents (Karger et al., 2004; Ferguson and McKinley, 2019). The major obstacle is the difficulty of elucidating the circumstances surrounding death, including cause and manner (Pechal et al., 2018; Kaszubinski et al., 2020b). Traditional forensic techniques, such as human DNA profiling, can provide critical evidence by linking biological traces to crime scenes and individuals or through victim identification (Franzosa et al., 2015). However, they occasionally fall short because of human DNA degradation (Sijen, 2015; Ranjan and Surajit, 2018) and need to be complemented with alternative techniques.
Recent advances in microbial profiling have uncovered that each individual is home to complex microbial communities (Oh et al., 2016; Gilbert and Stephens, 2018). These communities inhabit all surfaces of the human body (for example, orogastrointestinal tract, respiratory tract, urogenital tract, skin) and collectively represent the human microbiota, with their microbial DNA signatures forming the microbiome (Ursell et al., 2012; Berg et al., 2020). Recent research suggests that the microbiome could greatly aid forensic casework (Clarke et al., 2017; Hampton-Marcell et al., 2017; Metcalf et al., 2017; Oliveira and Amorim, 2018; Bishop, 2019). For example, the microbiome can serve as a personal microbial fingerprint that not only can associate individuals to objects (Lax et al., 2015) and geographical locations that they came in contact with (Knight et al., 2016), but also can provide identifiable characteristics (Franzosa et al., 2015). Moreover, many microbial cells contain robust cell walls that leave them better protected against degradation compared to human cells (Sijen, 2015). Nevertheless, the focus of microbial forensics to date has been predominantly on gut and soil samples (Metcalf et al., 2013, Metcalf et al., 2016; Pechal et al., 2014; Javan et al., 2016b; Burcham et al., 2019; DeBruyn et al., 2021), whereas research on forensic implementation of the (post-mortem) microbiome of the female reproductive tract, skin, and oral cavity is lagging behind. Therefore, in this review, we provide a critical assessment of current research on the vaginal, skin, and oral/salivary microbiome in relation to their potential application in forensics, especially sexual assault and femicide cases.
Relevance of Vaginal, Skin, and Salivary Microbiome During Life for Forensic Casework
Vaginal, skin, and saliva samples represent some of the most commonly collected samples in forensic casework, including sexual assault cases (World Health Organization, 2003; Mont and White, 2007) and cases involving touch evidence (Burrill et al., 2019; Oorschot et al., 2019). These mucocutaneous niches are shaped by several microbiome-influencing factors (e.g., pH and oxygen) (Rojo et al., 2017; Burcham et al., 2019). However, which of these factors have the largest effect is still unknown.
Vaginal samples are routinely collected in sexual assault cases (Quaak et al., 2018; Ghemrawi et al., 2021). While their microbiome is generally neglected in forensics, the less diverse composition of vaginal microbiota, its high microbial biomass, and protected anatomical location translate into its unique potential for microbial fingerprinting (Younes et al., 2018). Depending on the women’s ethnicity, the vaginal microbiome is generally dominated by Gram-positive Lactobacillus genera covered by a thick cell wall (i.e., Lactobacillus crispatus, Lactobacillus iners, Lactobacillus gasseri, and Lactobacillus jensenii) or a diverse microbiota dominated by non-lactobacilli such as Bifidobacterium, Gardnerella, Atopobium, and Prevotella (Ravel et al., 2010). Also fungal taxa mostly represented by Candida are detected, but generally in low abundances in healthy women (Chew et al., 2016). Ongoing research suggests that the vaginal microbiome composition can be correlated to individual characteristics valuable in forensics, such as health status (Ceccarani et al., 2019), ethnicity (Borgdorff et al., 2017; Gupta et al., 2017), sexual habits (Noyes et al., 2018), contraceptive use (Song et al., 2020), and pregnancy (Serrano et al., 2019), with various effect sizes that are not yet well mapped (Figure 1). For example, a longitudinal study found that sexual activity within 24 h of sampling has a significant negative impact on vaginal microbiome constancy as measured via the log Jensen–Shannon divergence rate (i.e., vaginal community deviation from constancy), independent of time in the menstrual cycle (Gajer et al., 2012). The vaginal microbiome could thus represent trace evidence in sexual assault cases indicating sexual intercourse in the last 24 h, in addition to providing links with other identifiable individual characteristics. However, whether this conclusion can be drawn from single, non-longitudinal samples after sexual intercourse needs to be investigated.
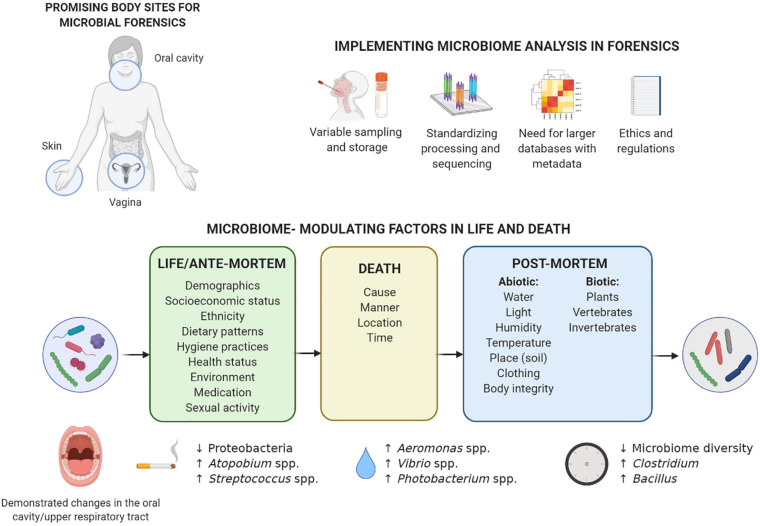
FIGURE 1.
Considerations for implementation of microbiome analysis in forensics and overview of factors in life and death that can influence the microbiome composition. The microbiome of the three body sites understudied in forensics (vagina, skin, and oral cavity) can be influenced by various individual factors during life, and these influences can sometimes also be detected after death. Death forms a turning point for the microbiome: the post-mortem microbiome is much more influenced by a range of environmental factors. Previously described microbiome-modulating factors in life and death are summarized, with some of the most prominent examples given of specific taxa in the oral cavity/upper respiratory tract associated with smoking during life (Wu et al., 2016), drowning as cause of death (Uchiyama et al., 2012; Benbow et al., 2015), and time that has passed since death (Adserias-Garriga et al., 2017b; Javan et al., 2017; García et al., 2020). More examples for the other body sites can be found in the text, although, to the best of our knowledge, no data are available on the post-mortem microbiome of the vagina. Implementation of microbiome analysis in forensics still requires extensive standardization of sampling and processing, as well as larger reference databases with metadata and an adjusted regulatory and ethical framework. Created with BioRender.com.
While vaginal samples are especially useful in sexual assault cases, the skin is probably the most commonly used source of forensic trace evidence, including skin under the victim’s fingernails (Metcalf et al., 2017; Burrill et al., 2019). However, the unique microbial trail left behind by skin shedding (Bishop, 2019; Hampton-Marcell et al., 2020) is often overlooked. The skin microbiome is dominated by Gram-positive Staphylococcus, Corynebacterium, Cutibacterium, Streptococcus, or Micrococcus, although Gram-negative Acinetobacter are also frequently isolated (Grice and Segre, 2011). The skin microbiome composition varies depending on the body location (Costello et al., 2009; Grice and Segre, 2011), host characteristics (e.g., age, lifestyle, and cohabitation) (Ross et al., 2017), and skin care (Bouslimani et al., 2019). Importantly, the skin microbiome is at an interface between the outside world and the body that undergoes most interactions with the environment. In fact, detectable amounts of skin microbiota can be transferred to objects such as a computer keyboard and mouse (Fierer et al., 2010), shoes and phones (Lax et al., 2015), door handle, toilet seat, etc. (Flores et al., 2011). Remarkably, this is not only limited to touched surfaces, but also extends to inhabited spaces through microbial clouds which are detectable within just a few hours (Lax et al., 2014; Meadow et al., 2015). However, whether built environment microbiota can be used as trace evidence remains to be substantiated (Hampton-Marcell et al., 2020). An important knowledge gap is to what extent the skin microbiome can be detected on touched objects after a certain amount of time, potentially even after death. Establishing this link is complicated, as it depends on environmental parameters (e.g., temperature, moisture, and UV radiation) (Fierer et al., 2010), individual shedder status (i.e., amount of epithelium deposited on a substrate) (Lowe et al., 2002; Kanokwongnuwut et al., 2018), and surface characteristics of the object (Meadow et al., 2014).
Saliva, as the primary oral cavity sample, is another widely used trace in forensic casework, especially for skin bite marks in sexual assault and child abuse cases (Chávez-Briones et al., 2015; Leake et al., 2016). The salivary microbiome is mostly dominated by the Gram-negative Neisseria, Prevotella, or Veillonella but also contains large proportions of Gram-positive Streptococcus taxa (e.g., Streptococcus salivarius and Streptococcus oralis) (Willis et al., 2018). Oral Gram-positive bacteria have recently been described as robust markers for highly degraded saliva samples, because of their higher resistance to degradation treatment (e.g., heat denaturation, microbial decomposition, and ultraviolet irradiation) compared to Gram-negative salivary bacteria, salivary α-amylase, and human DNA (Ohta and Sakurada, 2019). Individual characteristics that can influence the salivary microbiome composition include smoking (Belstrøm et al., 2014; Wu et al., 2016), dental hygiene (Mashima et al., 2017; Burcham et al., 2020), general and oral health (Zhou et al., 2016; Goodson et al., 2017), and socioeconomic status (Belstrøm et al., 2014; Figure 1). Importantly, shared environment at household level appears to more significantly determine the salivary microbiome than individual genetics (Shaw et al., 2017). Also intimate contact relevant in forensics, such as kissing (i.e., mixing of saliva), has been proposed to impact salivary microbial composition. Specifically, a transfer of approximately 80 million marker bacteria per intimate kiss of 10 s is observed, and partners have a more similar microbial community compared to unrelated individuals (Kort et al., 2014). Further research into factors influencing the vaginal, skin, and oral/salivary microbiomes during life will allow their more targeted implementation in forensic casework.
The Detrimental Effects of Death on the Human Microbiome
While the human microbiome during life is widely studied, we are just beginning to understand the post-mortem microbial community dynamics and how it can be influenced by ante-mortem microbial communities and modulating factors. During different stages after death, many anatomical and immunological barriers break, causing fluids, chemicals, and microorganisms that normally would not interact, to come into contact with each other (Gunn and Pitt, 2012). The post-mortem process facilitates the proliferation and relocation of microorganisms throughout the body and opens a gateway of cross-kingdom ecological interactions (Goff, 2009; García et al., 2020).
The decomposition of a human body is a continuous process caused by enzymatic reactions, (bio)chemical metabolic pathways and the activity of vertebrates and invertebrates (Pechal et al., 2018). This process is divided into a series of observable stages: fresh, active decay (including bloating and leakage of effusion), advanced decay, and dry remains/skeletonized (Goff, 2009; García et al., 2020). The course of these stages is partially determined by the diverse microbial communities occupying various internal and external body sites (Hauther et al., 2015; Javan et al., 2016a). Most studies on the post-mortem microbiome focus on estimating the minimum period of time since death [i.e., post-mortem interval (PMI)] (Goff, 2009). These studies aim to predict changes in the microbial composition of internal organs (e.g., gut, brain, liver, spleen, heart, etc.), also referred to as the post-mortem microbial clock (Metcalf et al., 2013; Finley et al., 2015).
Interestingly, studies on internal organs of mice (Metcalf et al., 2013; Burcham et al., 2019), swine (Carter et al., 2015), and human bodies (Hyde et al., 2013; Can et al., 2014; Hauther et al., 2015) have observed a shift in microbial communities from predominant aerobic microorganisms such as Staphylococcus and Enterobacteriaceae to more facultative anaerobic bacteria such as Proteobacteria, Firmicutes, and Bacteroidetes to obligate anaerobic organisms such as Clostridium, and finally spore-forming microorganisms such as Clostridium and Bacillus (Figure 1; Hyde et al., 2015; Javan et al., 2016b, 2019; García et al., 2020). According to the “post-mortem Clostridium effect,” Clostridium species can be considered important drivers of this microbial shift due to their lipolytic enzymes (Janaway et al., 2009), proteolytic functions, and rapid generation time (Javan et al., 2017). However, whether the findings from studies focusing on internal organs can be extended to mucocutaneous niches entails a different narrative.
Surprisingly, to date, no post-mortem microbiome studies have examined the vaginal microbiome succession after death (Table 1). This can be explained by the limited population of decomposing human bodies (mostly white males; >65 years old) studied at anthropological research facilities (Pechal et al., 2018). Nevertheless, Lutz et al. (2020) found that reproductive organs (i.e., uterus and prostate) were the last internal organs to decay. Particularly, for the nulligravid uterus (i.e., never been pregnant), the post-mortem Clostridium effect was not observed in contrast to the prostate and other internal organs. Of note, during life, the uterine microbiome is distinct from the vagina with a significantly lower microbial biomass and colonization by Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria (Garcia-grau et al., 2019), and it is not clear to what degree bacterial transfer from the vagina to the uterus occurs after death. This highlights the underexplored potential of the female reproductive tract in post-mortem research.
TABLE 1.
List of human post-mortem microbiome studies which include female reproductive tract, skin and/or oral cavity samples in the last 5 years.
| Niches | Study aim | Sequencing | Population and sample size | Main outcome | Main pitfall | References |
| Brain, heart, liver, spleen, prostate, and uterus | Estimating minimum PMI and cause of death | 16S rRNA gene amplicon sequencing | 158 samples | Reproductive organs (uterus and prostate) were the last internal organs to decay during human decomposition | Larger population size is needed to further account for variation due to (a)biotic factors | Lutz et al., 2020 |
| 40 human bodies (14 female, 26 male) | ||||||
| 6 body sites | ||||||
|
| ||||||
| Skin: nose and ear | Estimating minimum PMI | 16S rRNA gene amplicon sequencing | 144 samples | Machine learning model predicted the PMI with an average error of 2 days | Model was based on only four human bodies that were sampled longitudinally | Johnson et al., 2016 |
| 21 human bodies | ||||||
| 2 body sites | ||||||
|
| ||||||
| Skin: left hip, right hip, left bicep, right bicep, left upper hip, right upper hip, left knee, groin, head | Understanding microbially mediated processes during decomposition on different soil substrates | 16S rRNA gene amplicon sequencing 18S rRNA gene amplicon sequencing ITS amplicon sequencing | 2 human bodies during winter | Soil type was not a dominant factor driving community development in the process of decomposition | Limited population size with no information on sex | Metcalf et al., 2016 |
| 3 skin sites | ||||||
| 143 days | ||||||
| 2 human bodies during spring | ||||||
| 8 skin sites | ||||||
| 82 days | ||||||
|
| ||||||
| Eyes, ears, mouth, nose, rectum, thigh skin | Estimating minimum PMI for buried bodies | 16S rRNA gene amplicon sequencing | 2 male bodies | Multidisciplinary methodology identified temporal changes in morphology, skeletal muscle protein decomposition, entomology, and microbiome for buried bodies | Model was based on only two human bodies of which multiple samples were taken | Pittner et al., 2020 |
| 10 timepoints | ||||||
|
| ||||||
| Skin: right hand palm | Linking objects at the death scene to deceased individuals | 16S rRNA gene amplicon sequencing | 11 male bodies | Objects could be traced to deceased individual 75% of the time | Ante-mortem population was not always a demographic representation of the deceased study population | Kodama et al., 2019 |
| 5 female bodies | ||||||
| 30 living individuals | ||||||
| 79 skin samples | ||||||
| 98 object samples | ||||||
|
| ||||||
| Eyes, nose,ears, mouth, umbilicus rectum | 1. Predicting the ante-mortem health condition of the deceased 2. Comparing three machine learning methods to predict PMI, location of death, and manner of death 3. Predicting cause and manner of death | 16S rRNA gene amplicon sequencing | 47 male bodies 141 female bodies 6 body sites 1 timepoint | 1. Microbial biodiversity from the mouth could predict ante-mortem host health condition (e.g., heart disease) 2. Analysis of post-mortem microbiota from more than thee anatomic areas had limited additional value 3. Beta-dispersion, and case demographic data reflected forensic death determination | Only one timepoint (majority of cases with estimated PMI of <72 h) which does not account for variability within a body | 1. Pechal et al., 2018 2. Zhang et al., 2019 3. Kaszubinski et al., 2020a |
|
| ||||||
| Mouth: palate, tongue, inner cheek mucosa and tooth surfaces | Estimating minimum PMI | 16S rRNA gene amplicon sequencing | 1 male body | Post-mortem microbial succession in the oral cavity changed in a temporal way according to oxygen availability | Limited population size with large variability | Adserias-Garriga et al., 2017b |
| 2 female bodies | ||||||
| 8 timepoints | ||||||
| 5 body sites | ||||||
|
| ||||||
| External auditory canal, eyes, nares, mouth, umbilicus, and rectum | Studying the impact of coexisting conditions such as frozen affect the human microbiome at the time of discovery | 16S rRNA gene amplicon sequencing | 1 male body 1 female body 3 timepoints |
The microbial diversity increased throughout the thawing process | Association with time since death or cause of death | Pechal et al., 2017 |
|
| ||||||
| Blood, brain, buccal cavity, heart, liver, and spleen | Estimating minimum PMI | 16S rRNA gene amplicon sequencing | 66 samples | Microbial communities demonstrated time-, organ-, and sex-dependent changes | Niche sampling was not equal for all deceased individuals | Javan et al., 2016b |
| 27 human bodies (12 female, 15 male) | ||||||
| 6 body sites | ||||||
|
| ||||||
| Mouth, external left/right cheeks external left/right bicep region, torso, and rectum | Studying outdoor decomposition under natural conditions | 16S rRNA gene amplicon sequencing and 454 pyro- sequencing | 1 male body | Shifts in community structure were recorded and associated with major decomposition and related events | Limited population size with large variability | Hyde et al., 2015 |
| 1 female body | ||||||
| 10 timepoints | ||||||
The bold terms refer to the most relevant niches.
While current research has focused on the potential of the skin microbiome as trace evidence (Tozzo et al., 2020), to the best of our knowledge, only Kodama et al. (2019) have investigated whether actual objects from real death scenes (e.g., smoking pipes, medical devices, and phones) could be linked to the hand palm of the deceased through microbiome identification. The skin microbiome on the palm of the deceased remained stable up to 60 h after death, opening a window for individual microbiome identification even after death. It is noteworthy that this persistence of the skin microbiome into the early post-mortem period opens the possibility of also applying the post-mortem skin microbiome in PMI estimation. This is especially advantageous in cases where an autopsy is not requested, and a non-invasive microbiome sampling approach is best, because the most useful body sites for PMI estimation are external sites (e.g., skin).
Another body site easily accessible for microbiome and other sampling is the oral cavity. While its application for PMI estimation is yet to be studied in large populations, an increase of Firmicutes and Actinobacteria as the PMI increased was demonstrated (Hyde et al., 2013; Adserias-Garriga et al., 2017b). Interestingly, mouth samples pre-bloating resembled the oral microbiome during life, whereas the mouth samples post-bloating contained gut bacteria such as Tenericutes that possibly migrated from the large intestine (Adserias-Garriga et al., 2017b). Overall, studies that include more body sites, like Pechal et al. (2018) and others discussed in Table 1, could improve estimations.
The rate and pattern of decomposition are a mosaic system associated with biotic factors (e.g., individuality of the body, intrinsic and extrinsic bacteria, other microbes, and arthropods) and abiotic factors (e.g., weather, climate, humidity, and edaphic conditions) (Hyde et al., 2013; Carter et al., 2015; Newsome et al., 2021; Figure 1). It is yet to be elucidated how the contact of skin and natural body openings (mouth and vagina) with the outside environment (clothing, soil, aquatic ecosystems, etc.) can differentially influence the post-mortem body site-specific microbiome. For the latter, the application of epinecrotic communities such as aquatic microbes on the post-mortem submersion interval estimation could be highly relevant in aquatic death investigations (Benbow et al., 2015; Cartozzo et al., 2021; Randall et al., 2021). While the exact effect sizes are rarely reported (Meurs, 2016), abiotic factors, such as insects and soils beneath a decomposing body (Cobaugh et al., 2015; Metcalf et al., 2016; Adserias-Garriga et al., 2017a; Keenan et al., 2019), seasonal variation, and distinct climates (Carter et al., 2015), but also exposure and clothing (Goff, 2009), are some of the driving determinants of the microbial succession after death. Specifically, because of lack of thermoregulation, ambient temperatures (Goff, 2009) greatly affect the shift in nutrient availability and can thereby affect microbial community dynamics. Thus, while most studies have been performed in the United States (Metcalf et al., 2013; Pechal et al., 2018; DeBruyn et al., 2021) with a few in Australia, Japan, and the United Kingdom (García et al., 2020), post-mortem microbiome research in a wider range of climates should be encouraged.
Potential Hurdles and Considerations
Juries in the court of law have come to rely on physical evidence to corroborate a testimony (Shelton et al., 2007). However, before microbiome research can be reliably introduced into investigative and legislative casework, it has to be peer-reviewed, standardized, and accepted by the scientific community (Kiely, 2005).
Microbiome sequencing methods can be divided into those targeting specific parts of microbial DNA, such as the widely used 16S rRNA gene amplicon sequencing, or untargeted approaches, such as metagenomic shotgun sequencing. In-depth shotgun metagenomics is relatively new and currently more expensive than 16s rRNA gene amplicon sequencing, but it offers the advantage of sequencing the whole genetic content (microbial and human) of a sample with a higher taxonomic and functional resolution (Quince et al., 2017; Schmedes et al., 2017; Hillmann et al., 2018; Walker and Datta, 2019). However, currently, amplicon sequencing is most widely implemented and thus relies on larger available datasets with metadata on microbiome-modulating factors necessary for increasing method accuracy of machine learning–based tools (Clarke et al., 2017; Belk et al., 2018; Zhang et al., 2019), allowing for larger meta-analyses (Adams et al., 2015; Wang et al., 2018). Although both methods have specific limitations regarding taxonomic resolution, limit of specificity, and artificial bias are important when analyzing different types of samples. An integrative approach using both techniques could be implemented to rapidly advance the field, although this requires higher experimental costs (Metcalf et al., 2017; Hillmann et al., 2018).
Importantly, results of microbiome studies vary due to differences in sampling, storage, processing, and data analysis (e.g., machine learning classification models) (Clarke et al., 2017; Kaszubinski et al., 2020a). Thus, while the field expands, forensically oriented studies should contain standardized protocols with validated techniques. These methods should ensure reproducibility, sensitivity, and quantitative accuracy while defining and delineating the limitations (e.g., expected error rates, limit of detection, and limit of specificity) (Clarke et al., 2017; Metcalf et al., 2017). Minimizing the distinct impact of these variables on microbial profiling to reduce bias and skewing of the detected microbial composition is crucial in forensic evidence.
Future Perspectives and Concluding Remarks
Microbial forensics holds much potential; however, to integrate highly dimensional microbial data into routine investigative casework, several aspects need to be clarified. A key question is to what extent and for how long various individual factors shaping the vaginal, skin, and oral/salivary microbiome during life also play a role after death. These body sites are often inhabited by Gram-positive bacteria that are potentially more resistant to environmental and temporal degradation compared to Gram-negative bacteria and human DNA. In addition, vaginal, skin, and oral/saliva samples are routinely collected as critical components of sexual assault and femicide cases. Importantly, many sensitive individual characteristics can be associated with microbiome composition; however, the magnitude of these effects requires comprehensive investigation. A better understanding of the complex human body ecosystem during life and after death is necessary with the establishment of anthropological research facilities over different continents studying diverse populations and body sites. Hereby, we can facilitate discoveries especially related to female health and safety by comprehending how the post-mortem disturbance in the body homeostasis and its microbial communities make it more susceptible to the influences of the surrounding environment. While studies and regulations are complex specifically for the forensic field, the current and potential future possibilities of microbial forensics in phenotyping, identifying individuals, minimum PMI estimation, and the source of origin of a sample are highly important to consider and develop.
Author Contributions
SA, IS, and SL conceived and designed the manuscript. SA wrote the manuscript. IS made the figure. SA, IS, RD, EJ, and SL critically reviewed the manuscript and contributed with special attention towards their specific expertise. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We want to thank the entire research group ENdEMIC of the University of Antwerp, and the Forensic DNA Laboratories of the Antwerp University Hospital and University Hospitals of Leuven. We also want to thank the following funding agencies. SA was supported by the University Research Fund (BOF-DOCPRO 37054) of the University of Antwerp. IS was supported by the IOF-POC University of Antwerp funding ReLACT (FFI190115). SL was supported by the European Research Council (ERC) grant (Lacto-be 852600).
References
- Adams R. I., Bateman A. C., Bik H. M., Meadow J. F. (2015). Microbiota of the indoor environment: a meta-analysis. Microbiome 3:49. 10.1186/s40168-015-0108-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adserias-Garriga J., Hernández M., Quijada N. M., Rodríguez Lázaro D., Steadman D., Garcia-Gil J. (2017a). Daily thanatomicrobiome changes in soil as an approach of postmortem interval estimation: an ecological perspective. Forensic Sci. Int. 278 388–395. 10.1016/j.forsciint.2017.07.017 [DOI] [PubMed] [Google Scholar]
- Adserias-Garriga J., Quijada M., Hernandez N. M., Rodríguez Lázaro D., Steadman D., Garcia-Gil L. J. (2017b). Dynamics of the oral microbiota as a tool to estimate time since death. Mol. Oral Microbiol. 32 511–516. 10.1111/omi.12191 [DOI] [PubMed] [Google Scholar]
- Belk A., Xu Z. Z., Carter D. O., Lynne A., Bucheli S., Knight R., et al. (2018). Microbiome data accurately predicts the postmortem interval using random forest regression models. Genes (Basel) 9:104. 10.3390/genes9020104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belstrøm D., Holmstrup P., Nielsen C. H., Kirkby N., Heitmann B. L., Klepac-ceraj V., et al. (2014). Bacterial profiles of saliva in relation to diet, lifestyle factors, and socioeconomic status. J. Oral Microbiol. 6:10.3402/jom.v6.23609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbow M. E., Pechal J. L., Lang J. M., Erb R., Wallace J. R. (2015). The potential of high-throughput metagenomic sequencing of aquatic bacterial communities to estimate the postmortem submersion interval. J. Forensic Sci. 60 1500–1510. 10.1111/1556-4029.12859 [DOI] [PubMed] [Google Scholar]
- Berg G., Rybakova D., Fischer D., Cernava T., Vergès M. C., Charles T., et al. (2020). Microbiome definition re-visited: old concepts and new challenges. Microbiome 8:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A. H. (2019). The signatures of microorganisms and of human and environmental biomes can now be used to provide evidence in legal cases. FEMS Microbiol. Lett. 366:fnz021. 10.1093/femsle/fnz021 [DOI] [PubMed] [Google Scholar]
- Borgdorff H., van der Veer C., van Houdt R., Alberts C. J., de Vries H. J., Bruisten S. M., et al. (2017). The association between ethnicity and vaginal microbiota composition in Amsterdam, the Netherlands. PLoS One 12:e0181135. 10.1371/journal.pone.0181135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouslimani A., Silva R., Kosciolek T., Janssen S., Callewaert C., Amir A., et al. (2019). The impact of skin care products on skin chemistry and microbiome dynamics. BMC Biol. 17:47. 10.1186/s12915-019-0660-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcham Z. M., Garneau N. L., Comstock S. S., Tucker R. M., Knight R., Metcalf J. L., et al. (2020). Patterns of oral microbiota diversity in adults and children: a crowdsourced population study. Sci. Rep. 10:2133. 10.1038/s41598-020-59016-59010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcham Z. M., Pechal J. L., Schmidt C. J., Bose J. L., Rosch J. W., Benbow M. E., et al. (2019). Bacterial community succession, transmigration, and differential gene transcription in a controlled vertebrate decomposition model. Front. Microbiol. 10:745. 10.3389/fmicb.2019.00745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrill J., Daniel B., Frascione N. (2019). A review of trace “Touch DNA” deposits: variability factors and an exploration of cellular composition. Forensic Sci. Int. Genet. 39 8–18. 10.1016/j.fsigen.2018.11.019 [DOI] [PubMed] [Google Scholar]
- Can I., Javan G. T., Pozhitkov A. E., Noble P. A. (2014). Distinctive thanatomicrobiome signatures found in the blood and internal organs of humans. J. Microbiol. Methods 106 1–7. 10.1016/j.mimet.2014.07.026 [DOI] [PubMed] [Google Scholar]
- Carter D. O., Metcalf J. L., Bibat A., Knight R. (2015). Seasonal variation of postmortem microbial communities. Forensic Sci. Med. Pathol. 11 202–207. 10.1007/s12024-015-9667-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartozzo C., Simmons T., Swall J., Singh B. (2021). Postmortem submersion interval (PMSI) estimation from the microbiome of Sus scrofa bone in a freshwater river. Forensic Sci. Int. 318:110480. 10.1016/j.forsciint.2020.110480 [DOI] [PubMed] [Google Scholar]
- Ceccarani C., Foschi C., Parolin C., Antonietta D. A., Gaspari V., Consolandi C., et al. (2019). Diversity of vaginal microbiome and metabolome during genital infections. Sci. Rep. 9:14095. 10.1038/s41598-019-50410-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez-Briones M. L., Hernández-Cortés R., Jaramillo-Rangel G., Ortega-Martínez M. (2015). Relevance of sampling and DNA extraction techniques for the analysis of salivary evidence from bite marks: a case report. Genet. Mol. Res. 14 10165–10171. [DOI] [PubMed] [Google Scholar]
- Chew S. Y., Thian L., Than L. (2016). Vulvovaginal candidosis: contemporary challenges and the future of prophylactic and therapeutic approaches. Mycoses 56 262–273. 10.1111/myc.12455 [DOI] [PubMed] [Google Scholar]
- Clarke T. H., Gomez A., Singh H., Nelson K. E., Brinkac L. M. (2017). Integrating the microbiome as a resource in the forensics toolkit. Forensic Sci. Int. Genet. 30 141–147. 10.1016/j.fsigen.2017.06.008 [DOI] [PubMed] [Google Scholar]
- Cobaugh K. L., Schaeffer S. M., Debruyn J. M. (2015). Functional and structural succession of soil microbial communities below decomposing human cadavers. PLoS One 10:e0130201. 10.1371/journal.pone.0130201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello E., Lauber C., Hamady M., Fierer N., Gordon J., Knight R. (2009). Bacterial community variation in human body habitats across space and time. Science 326 1694–1697. 10.1126/science.1177486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruyn J. M., Hoeland K. M., Taylor L. S., Stevens J. D., Moats M. A., Bandopadhyay S., et al. (2021). Comparative decomposition of humans and pigs: soil biogeochemistry, microbial activity and metabolomic profiles. Front. Microbiol. 11:608856. 10.3389/fmicb.2020.608856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devries K. M., Mak J. Y. T., García-Moreno C., Petzold M., Child J. C., Falder G., et al. (2013). The global prevalence of intimate partner violence against women. Science 340 1527–1528. 10.1126/science.1240937 [DOI] [PubMed] [Google Scholar]
- Ferguson C., McKinley A. (2019). Detection avoidance and mis/unclassified, unsolved homicides in Australia. J. Crim. Psychol. 10 113–122. 10.1108/JCP-09-2019-2030 [DOI] [Google Scholar]
- Fierer N., Lauber C. L., Zhou N., Mcdonald D., Costello E. K., Knight R. (2010). Forensic identi fi cation using skin bacterial communities. Proc. Natl. Acad. Sci. U S A. 107 6477–6481. 10.1073/pnas.1000162107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley S. J., Benbow M. E., Javan G. T. (2015). Microbial communities associated with human decomposition and their potential use as postmortem clocks. Int. J. Legal Med. 129 623–632. 10.1007/s00414-014-1059-0 [DOI] [PubMed] [Google Scholar]
- Flores G. E., Bates S. T., Knights D., Lauber C. L., Stombaugh J., Knight R., et al. (2011). Microbial biogeography of public restroom surfaces. PLoS One 6:e28132. 10.1371/journal.pone.0028132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa E. A., Huang K., Meadow J. F., Gevers D., Lemon K. P. (2015). Identifying personal microbiomes using metagenomic codes. Proc. Natl. Acad. Sci. U S A. 112 E2930–E2938. 10.1073/pnas.1423854112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajer P., Brotman R. M., Bai G., Sakamoto J., Schütte U. M. E., Zhong X., et al. (2012). Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med. 4:132ra52. 10.1126/scitranslmed.3003605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García M. G., Pérez-Cárceles M. D., Osuna E., Legaz I. (2020). Impact of the human microbiome in forensic sciences: a systematic review. Appl. Environ. Microbiol. 86:e01451-20. 10.1128/AEM.01451-1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-grau I., Simon C., Moreno I. (2019). Uterine microbiome — low biomass and high expectations. Biol. Reprod. 101 1102–1114. 10.1093/biolre/ioy257 [DOI] [PubMed] [Google Scholar]
- Geneva Declaration (2011). When the Victim Is a Woman in Global Burden of Armed Violence 2011: Lethal Encounters. Cambridge: Cambridge University Press. [Google Scholar]
- Ghemrawi M., Ramírez A., Duncan G., Colwell R., Dadlani M., Mccord B. (2021). The genital microbiome and its potential for detecting sexual assault. Forensic Sci. Int. Genet. 51:102432. 10.1016/j.fsigen.2020.102432 [DOI] [PubMed] [Google Scholar]
- Gilbert J. A., Stephens B. (2018). Microbiology of the built environment. Nat. Rev. Microbiol. 16 661–670. 10.1038/s41579-018-0065-65 [DOI] [PubMed] [Google Scholar]
- Goff M. L. (2009). Early post-mortem changes and stages of decomposition in exposed cadavers. Exp. Appl. Acarol. 49 21–36. 10.1007/s10493-009-9284-9289 [DOI] [PubMed] [Google Scholar]
- Goodson J. M., Hartman M., Shi P., Hasturk H., Yaskell T., Vargas J., et al. (2017). The salivary microbiome is altered in the presence of a high salivary glucose concentration. PLoS One 12:e0170437. 10.1371/journal.pone.0170437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice E. A., Segre J. A. (2011). The skin microbiome. Nat. Rev. Microbiol. 9 244–253. 10.1038/nrmicro2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn A., Pitt S. J. (2012). Microbes as forensic indicators. Trop. Biomed. 29 311–330. [PubMed] [Google Scholar]
- Gupta V. K., Paul S., Dutta C. (2017). Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front. Microbiol. 8:1162. 10.3389/fmicb.2017.01162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton-Marcell J. T., Larsen P., Anton T., Cralle L., Sangwan N., Lax S., et al. (2020). Detecting personal microbiota signatures at artificial crime scenes. Forensic Sci. Int. 313:110351. 10.1016/j.forsciint.2020.110351 [DOI] [PubMed] [Google Scholar]
- Hampton-Marcell J. T., Lopez J. V., Gilbert J. A. (2017). The human microbiome: an emerging tool in forensics. Microb. Biotechnol. 10 228–230. 10.1111/1751-7915.12699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauther K. A., Cobaugh K. L., Jantz L. M., Ph D., Sparer T. E., Ph D., et al. (2015). Estimating time since death from postmortem human gut microbial communities. J. Forensic Sci. 60 1234–1240. 10.1111/1556-4029.12828 [DOI] [PubMed] [Google Scholar]
- Hillmann B., Al-ghalith G. A., Shields-cutler R. R., Zhu Q., Gohl D. M., Beckman K. B., et al. (2018). Evaluating the information content of shallow shotgun metagenomics. mSystems 3:e00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde E. R., Haarmann D. P., Bucheli S. R. (2015). Initial insights into bacterial succession during human decomposition. Int. J. Leg. Med. 129 661–671. 10.1007/s00414-014-1128-1124 [DOI] [PubMed] [Google Scholar]
- Hyde E. R., Haarmann D. P., Lynne A. M., Bucheli S. R., Petrosino J. F. (2013). The living dead: bacterial community structure of a cadaver at the onset and end of the bloat stage of decomposition. PLoS One 8:e77733. 10.1371/journal.pone.0077733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janaway R. C., Percival S. L., Wilson A. S. (2009). “Decomposition of human remains,” in Microbiology and Aging: Clinical Manifestations, ed. Percival S. L. (New York, NY: Springer; ), 313–334. 10.1007/978-1-59745-327-1_14 [DOI] [Google Scholar]
- Javan G. T., Finley S. J., Abidin Z., Mulle J. G. (2016a). The thanatomicrobiome: a missing piece of the microbial puzzle of death. Front. Microbiol. 7:225. 10.3389/fmicb.2016.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javan G. T., Finley S. J., Can I., Wilkinson J. E., Hanson J. D., Tarone A. M. (2016b). Human thanatomicrobiome succession and time since death. Sci. Rep. 6:29598. 10.1038/srep29598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javan G. T., Finley S. J., Smith T., Miller J., Wilkinson J. E., Cevallos M. A. (2017). Cadaver thanatomicrobiome signatures: the ubiquitous nature of clostridium species in human decomposition. Front. Microbiol. 8:2096. 10.3389/fmicb.2017.02096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javan G. T., Finley S. J., Tuomisto S., Hall A., Bendow M. E., Mills D. (2019). An interdisciplinary review of the thanatomicrobiome in human decomposition. Forensic Sci. Med. Pathol. 15 75–83. 10.1007/s12024-018-0061-0 [DOI] [PubMed] [Google Scholar]
- Johnson H. R., Trinidad D. D., Guzman S., Khan Z., Parziale V., Debruyn J. M., et al. (2016). A machine learning approach for using the postmortem skin microbiome to estimate the postmortem interval. PLoS One 11:e0167370. 10.1371/journal.pone.0167370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanokwongnuwut P., Martin B., Kirkbride K. P., Linacre A. (2018). Shedding light on shedders. Forensic Sci. Int. Genet. 36 20–25. 10.1016/j.fsigen.2018.06.004 [DOI] [PubMed] [Google Scholar]
- Karger B., Lorin de la Grandmaison G., Bajanowski T., Brinkmann B. (2004). Analysis of 155 consecutive forensic exhumations with emphasis on undetected homicides. Int. J. Legal Med. 118 90–94. 10.1007/s00414-003-0426-z [DOI] [PubMed] [Google Scholar]
- Kaszubinski S. F., Pechal J. L., Schmidt C. J., Jordan H. R., Benbow M. E., Meek M. H. (2020a). Evaluating bioinformatic pipeline performance for forensic microbiome analysis∗,†,‡. J. Forensic Sci. 65 513–525. 10.1111/1556-4029.14213 [DOI] [PubMed] [Google Scholar]
- Kaszubinski S. F., Pechal J. L., Smiles K., Schmidt C. J., Allen M. S. (2020b). Dysbiosis in the dead: human postmortem microbiome beta-dispersion as an indicator of manner and cause of death. Front. Microbiol. 11:555347. 10.3389/fmicb.2020.555347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan S. W., Schaeffer S. M., Debruyn J. M. (2019). Spatial changes in soil stable isotopic composition in response to carrion decomposition. Biogeosciences 16 3929–3939. 10.5194/bg-16-3929-2019 [DOI] [Google Scholar]
- Kiely T. F. (2005). Forensic Evidence: Science and the Criminal Law, 2nd Edition. Boca Raton, FL: CRC Press. [Google Scholar]
- Knight R., Kelley S. T., Siegel J., Gregory J. (2016). Geography and location are the primary drivers of office microbiome composition. mSystems 1:e00022-16. 10.1128/mSystems.00022-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama W. A., Xu Z., Metcalf J. L., Song S. J., Harrison N., Knight R., et al. (2019). Trace evidence potential in postmortem skin microbiomes: from death scene to morgue. J. Forensic Sci. 64 791–798. 10.1111/1556-4029.13949 [DOI] [PubMed] [Google Scholar]
- Kort R., Caspers M., Graaf A., Van De, Egmond W., Van, et al. (2014). Shaping the oral microbiota through intimate kissing. Microbiome. 2:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax S., Hampton-Marcell J. T., Gibbons S. M., Colares G. B., Smith D., Eisen J. A., et al. (2015). Forensic analysis of the microbiome of phones and shoes. Microbiome 3:21. 10.1186/s40168-015-0082-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax S., Smith D. P., Hampton-Marcell J., Owens S. M., Handley K. M., Scott N. M., et al. (2014). Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345 1048–1052. 10.1126/science.1254529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leake S. L., Pagni M., Falquet L., Taroni F., Greub G. (2016). The salivary microbiome for differentiating individuals: proof of principle. Microbes Infect. 18 399–405. 10.1016/j.micinf.2016.03.011 [DOI] [PubMed] [Google Scholar]
- Lovett J., Kelly L. (2009). Different Systems, Similar Outcomes? Tracking Attrition in Reported Rape Cases Across Europe. London: Child and Women Abuse Studies Unit. [Google Scholar]
- Lowe A., Murray C., Whitaker J., Tully G., Gill P. (2002). The propensity of individuals to deposit DNA and secondary transfer of low level DNA from individuals to inert surfaces. Forensic Sci Int. 129 25–34. 10.1016/s0379-0738(02)00207-4 [DOI] [PubMed] [Google Scholar]
- Lutz H., Vangelatos A., Gottel N., Osculati A., Visona S., Finley S. J., et al. (2020). Effects of extended postmortem interval on microbial communities in organs of the human cadaver. Front. Microbiol. 11:569630. 10.3389/fmicb.2020.569630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashima I., Theodorea C. F., Thaweboon B., Thaweboon S., Scannapieco F. A., Nakazawa F. (2017). Exploring the salivary microbiome of children stratified by the oral hygiene index. PLoS One 12:e0185274. 10.1371/journal.pone.0185274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadow J. F., Altrichter A. E., Bateman A. C., Stenson J., Brown G. Z., Green J. L. (2015). Humans differ in their personal microbial cloud. Peer J 3:e1258. 10.7717/peerj.1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadow J. F., Altrichter A. E., Kembel S. W., Moriyama M., Connor T. K. O., Womack A. M., et al. (2014). Bacterial communities on classroom surfaces vary with human contact. Microbiome 2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf J. L., Parfrey L. W., Gonzalez A., Lauber C. L., Knights D., Ackermann G., et al. (2013). A microbial clock provides an accurate estimate of the postmortem interval in a mouse model system. eLife 2:e01104. 10.7554/eLife.01104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf J. L., Xu Z. Z., Bouslimani A., Dorrestein P., Carter D. O., Knight R. (2017). Microbiome tools for forensic science. Trends Biotechnol. 35 814–823. 10.1016/j.tibtech.2017.03.006 [DOI] [PubMed] [Google Scholar]
- Metcalf J. L., Xu Z. Z., Weiss S., Lax S., Treuren W., Van, et al. (2016). Microbial community assembly and metabolic function during mammalian corpse decomposition. Science 351 158–162. 10.1126/science.aad2646 [DOI] [PubMed] [Google Scholar]
- Meurs J. (2016). The experimental design of postmortem studies: the effect size and statistical power. Forensic Sci. Med. Pathol. 12 343–349. 10.1007/s12024-016-9793-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mont J. D., White D. (2007). The Uses and Impacts of Medico-legal Evidence in Sexual Assault Cases: A Global Review. Geneva: World Health Organization. [Google Scholar]
- Newsome T. M., Barton B., Buck J. C., DeBruyn J., Spencer E., Ripple W. J., et al. (2021). Monitoring the dead as an ecosystem indicator. Ecol. Evol. 11 5844–5856. 10.1002/ece3.7542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes N., Cho K.-C., Ravel J., Forney L. J., Abdo Z. (2018). Associations between sexual habits, menstrual hygiene practices, demographics and the vaginal microbiome as revealed by Bayesian network analysis. PLoS One 13:e0191625. 10.1371/journal.pone.0191625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., Byrd A. L., Park M., Kong H. H., Segre J. A., Oh J., et al. (2016). Temporal stability of the human skin microbiome article temporal stability of the human skin microbiome. Cell 165 854–866. 10.1016/j.cell.2016.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta J., Sakurada K. (2019). Oral gram-positive bacterial DNA-based identification of saliva from highly degraded samples. Forensic Sci. Int. Genet. 42 103–112. 10.1016/j.fsigen.2019.06.016 [DOI] [PubMed] [Google Scholar]
- Oliveira M., Amorim A. (2018). Microbial forensics: new breakthroughs and future prospects. Appl. Microbiol. Biotechnol. 102 10377–10391. 10.1007/s00253-018-9414-9416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oorschot R. A. H., Van, Goray M., Szkuta B., Meakin G. E., Kokshoorn B. (2019). DNA transfer in forensic science: a review. Forensic Sci. Int. Genet. 38 140–166. 10.1016/j.fsigen.2018.10.014 [DOI] [PubMed] [Google Scholar]
- Pechal J. L., Crippen T. L., Benbow M. E., Tarone A. M., Dowd S., Tomberlin J. K. (2014). The potential use of bacterial community succession in forensics as described by high throughput metagenomic sequencing. Int. J. Legal Med. 128 193–205. 10.1007/s00414-013-0872-1 [DOI] [PubMed] [Google Scholar]
- Pechal J. L., Schmidt C. J., Jordan H. R., Benbow M. E. (2017). Frozen: thawing and its effect on the postmortem microbiome in two pediatric. J. Forensic Sci. 62 1399–1405. 10.1111/1556-4029.13419 [DOI] [PubMed] [Google Scholar]
- Pechal J. L., Schmidt C. J., Jordan H. R., Benbow M. E. (2018). A large-scale survey of the postmortem human microbiome, and its potential to provide insight into the living health condition. Sci. Rep. 8:5724. 10.1038/s41598-018-23989-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittner S., Bugelli V., Benbow M. E., Ehrenfellner B., Zissler A., Campobasso C. P., et al. (2020). The applicability of forensic time since death estimation methods for buried bodies in advanced decomposition stages. PLoS One 15:e0243395. 10.1371/journal.pone.0243395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaak F. C. A., van Duijn T., Hoogenboom J., Kloosterman A. D., Kuiper I. (2018). Human-associated microbial populations as evidence in forensic casework. Forensic Sci. Int. Genet. 36 176–185. 10.1016/j.fsigen.2018.06.020 [DOI] [PubMed] [Google Scholar]
- Quince C., Walker A. W., Simpson J. T., Loman N. J., Segata N. (2017). Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 35 833–844. 10.1038/nbt.3935 [DOI] [PubMed] [Google Scholar]
- Randall S., Cartozzo C., Simmons T., Swall J. L., Singh B. (2021). Prediction of minimum postmortem submersion interval (PMSImin) based on eukaryotic community succession on skeletal remains recovered from a lentic environment. Forensic Sci. Int. 323:110784. 10.1016/j.forsciint.2021.110784 [DOI] [PubMed] [Google Scholar]
- Ranjan H., Surajit D. (2018). Microbial degradation of forensic samples of biological origin: potential threat to human DNA typing. Mol. Biotechnol. 60 141–153. 10.1007/s12033-017-0052-55 [DOI] [PubMed] [Google Scholar]
- Ravel J., Gajer P., Abdo Z., Schneider G. M., Koenig S. S. K., Mcculle S. L., et al. (2010). Vaginal microbiome of reproductive-age women. PNAS 108 4680–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo D., Celia M., Raczkowska B. A., Ferrer M., Barbas C., Bargiela R., et al. (2017). Exploring the human microbiome from multiple perspectives: factors altering its composition. FEMS Microbiol. Rev. 41 453–478. 10.1093/femsre/fuw046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A. A., Doxey A. C., Neufeld J. D. (2017). The skin microbiome of cohabiting couples. mSystems 2:e00043-17. 10.1128/mSystems.00043-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmedes S. E., Woerner A. E., Budowle B. (2017). Forensic human identification using skin microbiomes. Appl. Environ. Microbiol. 83:e01672-17. 10.1128/AEM.01672-1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M. G., Parikh H. I., Brooks J. P., Edwards D. J., Arodz T. J., Edupuganti L., et al. (2019). Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy. Nat. Med. 25 1001–1011. 10.1038/s41591-019-0465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L., Ribeiro A. L. R., Levine A. P., Pontikos N. (2017). The human salivary microbiome is shaped by shared environment rather than genetics: evidence from a large family of closely related individuals. mBio 8:e01237-17. 10.1128/mBio.01237-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton H. D. E., Kim Y. S., Barak G. (2007). A study of juror expectations and demands concerning scientific evidence: does the “csi effect” exist? vanderbilt. J. Entertain. Technol. Law 9 331–368. [Google Scholar]
- Sijen T. (2015). Molecular approaches for forensic cell type identification: on mRNA, miRNA, DNA methylation and microbial markers. Forensic Sci. Int. Genet. 18 21–32. 10.1016/j.fsigen.2014.11.015 [DOI] [PubMed] [Google Scholar]
- Song S. D., Acharya K. D., Zhu J. E., Deveney, Christen M., Walther-Antonio M. R. S., et al. (2020). Daily vaginal microbiota fluctuations associated with natural hormonal cycle, contraceptives, diet, and exercise. mSphere 5:e00593-20. 10.1128/mSphere.00593-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozzo P., Angiolella G. D., Brun P., Castagliuolo I., Gino S., Caenazzo L. (2020). Skin microbiome analysis for forensic human identification: what do we know so far? Microorganisms 8:873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama T., Kakizaki E., Kozawa S., Nishida S., Imamura N., Yukawa N. (2012). A new molecular approach to help conclude drowning as a cause of death: simultaneous detection of eight bacterioplankton species using real-time PCR assays with TaqMan probes. Forensic Sci. Int. 222 11–26. 10.1016/j.forsciint.2012.04.029 [DOI] [PubMed] [Google Scholar]
- Ursell L. K., Metcalf J. L., Parfrey L. W., Knight R. (2012). Defining the human microbiome. Nutr. Rev. 70 38–44. 10.1111/j.1753-4887.2012.00493.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A. R., Datta S. (2019). Identification of city specific important bacterial signature for the MetaSUB CAMDA challenge microbiome data. Biol. Direct. 14:11. 10.1186/s13062-019-0243-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Kurilshikov A., Radjabzadeh D., Turpin W., Croitoru K., Bonder M. J., et al. (2018). Meta-analysis of human genome-microbiome association studies: the MiBioGen consortium initiative. Microbiome 6:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis J. R., González-Torres P., Pittis A. A., Bejarano L. A., Cozzuto L., Andreu-Somavilla N., et al. (2018). Citizen science charts two major “stomatotypes” in the oral microbiome of adolescents and reveals links with habits and drinking water composition. Microbiome 6:218. 10.1186/s40168-018-0592-593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2003). Guidelines for Medico-legal Care for Victims of Sexual Violence. Geneva: World Health Organization. [Google Scholar]
- World Health Organization (2013). Global and Regional Estimates of Violence Against Women: Prevalence and Health Effects of Intimate Partner Violence and Non-partner Sexual Violence. Geneva: World Health Organization. [Google Scholar]
- World Health Organization, and Pan American Health Organization (2012). Understanding and Addressing Violence Against Women: Femicide. Geneva: World Health Organization. [Google Scholar]
- Wu J., Peters B. A., Dominianni C., Zhang Y., Pei Z., Yang L., et al. (2016). Cigarette smoking and the oral microbiome in a large study of American adults. ISME J. 10 2435–2446. 10.1038/ismej.2016.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes J. A., Lievens E., Hummelen R., van der Westen R., Reid G., Petrova M. I. (2018). Women and their microbes: the unexpected friendship. Trends Microbiol. 26 16–32. 10.1016/j.tim.2017.07.008 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Id J. L. P., Schmidt C. J., Jordan H. R., Wang W., Id M. E. B., et al. (2019). Machine learning performance in a microbial molecular autopsy context: a cross-sectional postmortem human population study. PLoS One 14:e0213829. 10.1371/journal.pone.0213829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Jiang N., Wang S., Hu X., Jiao K., He X. (2016). Exploration of human salivary microbiomes — insights into the novel characteristics of microbial community structure in caries and caries-free subjects. PLoS One 11:e0147039. 10.1371/journal.pone.0147039 [DOI] [PMC free article] [PubMed] [Google Scholar]