Abstract
Viruses are natural supramolecular nanostructures that form spontaneously by molecular self-assembly of complex biomolecules. Peptide self-assembly is a versatile tool that allows mimicking viruses by creating their simplified versions through the design of functional, supramolecular materials with modularity, tunability, and responsiveness to chemical and physical stimuli. The main challenge in the design and fabrication of peptide materials is related to the precise control between the peptide sequence and its resulting supramolecular morphology. We provide an overview of existing sequence patterns employed for the development of spherical and fibrillar peptide assemblies that can act as viral mimetics, offering the opportunity to tackle the challenges of viral infections.
Keywords: self-assembly, viral mimetics, peptides, minimalistic, co-assembly
Introduction
Designed bio-nanomaterials are often inspired by basic processes found in nature such as molecular recognition and self-assembly (Lehn, 2002; Whitesides and Grzybowski, 2002; Yang et al., 2020a). Viruses present a great source of inspiration for the design of life-like materials (Whitesides, 2015; Maslanka Figueroa et al., 2021) as they constitute simple, yet sophisticated supramolecular assemblies that contain genetic code and present well-defined rod-like or spherical morphologies. In addition, they show the ability to self-replicate, respond to physical and chemical stimuli, adapt to the environment, and evade the immune system which makes them ideal candidates to be manipulated and repurposed.
A variety of virus-mimetic materials have been developed for biological and chemical sensing (Mao et al., 2009), drug delivery (Li et al., 2016), cancer immunotherapy (Mohsen et al., 2020) and vaccine design (Abudula et al., 2020). Virus-like particles (VLPs), formed by the multimeric self-assembly of expressed viral structural proteins in absence of genetic material, are the most studied ones (Ludwig and Wagner, 2007; Ferreira and Martins, 2017; Roldão et al., 2019). The complexity of their fabrication, that requires fully folded proteins and efficient upstream and downstream strategies, impacts the production yields, and is associated to high costs. Other examples include polymer peptide nanogels (Lee et al., 2008), dendritic lipopeptides (Liang et al., 2019), iron oxide-lactoferrin magneto-responsive nanocapsules (Fang et al., 2015), peptide-DNA condensates (Cao et al., 2018), rabies-inspired gold nanorods (Lee et al., 2017) or metal–organic frameworks (Qiao et al., 2020). However, the potential of minimalistic, purely peptidic, supramolecular nanostructures to resemble the morphology and/or functionality of viruses has not been fully exploited yet.
Several peptide-based therapeutics have reached the market while others are in various phases of clinical development for the treatment of cancer and metabolic disorders (Vlieghe et al., 2010; Craik et al., 2013). Compared to their protein counterparts, peptides are easier to synthesize and more stable under harsh conditions (Fosgerau and Hoffmann, 2015). Furthermore, peptides can be exploited as building blocks for the fabrication of highly ordered nanostructures with varying morphologies and surface functionalities, developed for drug delivery, tissue engineering, and regenerative medicine due to their inherent biocompatibility and biodegradability (Zhang, 2003; Collier et al., 2010; Woolfson and Mahmoud, 2010; Frederix et al., 2015; Smith et al., 2015; Slocik and Naik, 2017; Lampel et al., 2018; Sharma et al., 2021).
Peptide-based nanomaterials offer simple and low costs alternatives to VLPs (Matsuura, 2012; Hendricks et al., 2017; Singh et al., 2017; Cai et al., 2020). When designing peptide-based virus mimetics, the main strategy is capsid reconstruction through the formation of supramolecular assemblies based on peptide segments with the goal of mimicking the viral architecture and functionality of efficient cell entry, immune evasion, and targeted cargo delivery. In here, we provide an overview of sequence patterns that drive peptide self-assembly, followed by the potential to achieve dimensional control through co-assembly. Finally, examples of peptide-based building blocks used in the design of supramolecular virus mimetics are discussed.
Morphological Control Through Sequence Design
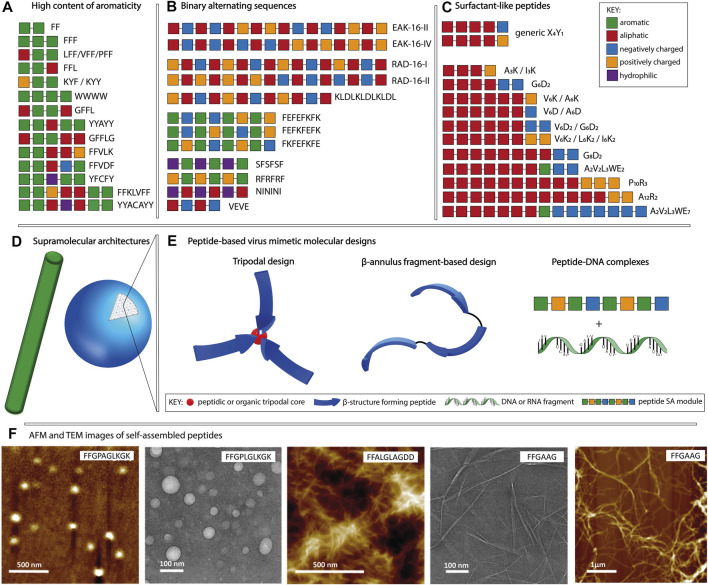
In the context of molecular self-assembly, the composition and the physico-chemical properties of amino acid side chains dictate their behavior in different environments. In a hydrophilic environment, aromatic amino acids tend to aggregate due to hydrophobic interactions and π-π stacking, whereas polar and charged amino acids promote nanostructure formation through hydrogen bonds and electrostatic interactions, respectively. In addition, the position of a particular amino acid within the sequence, as well as the type of neighboring residues, affect the formation of supramolecular assemblies and their morphologies. Although it is possible to identify distinct sequence patterns with the tendency to form a particular nanostructure, it is challenging to attribute a supramolecular morphology based solely on the amino acid composition. Peptide sequences can self-assemble into a variety of shapes, including spheres, fibers, vesicles and tubes, with diameters in the 10–100 nm range and in the case of nanofibers, reaching micrometers in length (Gazit, 2007a; Zhao et al., 2008). In this section, we will focus on three main patterns used in the design of purely peptidic materials (Figures 1A–C): (i) high content of aromaticity, (ii) binary alternating hydrophobic-hydrophilic and (iii) surfactant-like.
FIGURE 1.
Schematic representation of the main sequence patterns found in self-assembling peptides, namely (A) high content of aromaticity, (B) binary alternating patterns of hydrophobic-hydrophilic residues and (C) surfactant-like, followed by (D) the most common supramolecular morphologies. (E) Molecular designs used for peptide-based virus mimetics based on capsid-like materials (tripodal and β-annulus designs) and peptide-DNA/RNA complexes. (F) AFM and TEM example images of fibrillar and spherical peptide nanostructures, adapted with permission from (Son et al., 2019). Copyright 2019 American Chemical Society.
Peptides With High Content of Aromaticity
Peptides composed of aromatic amino acids preferentially self-assemble into nanofibers with high intermolecular β-sheet content. Short motifs such as FF, YY, and WW constitute fundamental building blocks for self-assembly, with diphenylalanine being the most widely studied one (Gazit, 2007b; Frederix et al., 2015; Tao et al., 2017). Depending on the combination of amino acids adjacent to the FF motif and its position within the sequence, various morphologies such as fibrous and plate-like assemblies for FFF, nanospheres for CFF, helical fibrils for PFF and heterogeneous nanostructures for FFV, VFF, and LFF, are observed (Reches and Gazit, 2004; Tamamis et al., 2009; Marchesan et al., 2012; Frederix et al., 2015; Bera et al., 2019). Examples of fiber forming peptides (Figure 1A) having longer sequences include FFKLVFF, GFFLG, GFFL, FFAGL, FFVLK, FFVDF, and WWWW (Kalafatovic et al., 2016; Diaferia et al., 2018; Son et al., 2019; Yang et al., 2020b). Moreover, tyrosine-rich sequences including YYAYY, YYACAYY, YFCFY, KYF, and KYY were found to assemble into nanosheets, nanocapsules or nanofibers (Frederix et al., 2015; Lee et al., 2019; Sloan-Dennison et al., 2021). Amyloid-like peptides find applications in plaque-associated neurodegenerative diseases research or as biosensors and nanocarriers (Gazit, 2007b; Al-Halifa et al., 2019).
Binary Alternating Sequences of Hydrophobic-Hydrophilic Residues
Peptides with the binary-alternating patters rely on hydrogen bonds and/or electrostatic interactions for the formation of supramolecular assemblies. The first reported self-assembling peptide, EAK16-II (AEAEAKAKAEAEAKAK) is a repetitive segment derived from a natural yeast protein consisting of alternating hydrophobic and hydrophilic, positively, and negatively charged amino acids (Zhang et al., 1993; Zhang, 2017). It was shown that the disposition of amino acids within the sequence and the pH of the environment influence the supramolecular morphology of EAK16 (Hong et al., 2003). At neutral pH, EAK16-II formed fibrils and its analogue EAK16-IV (AEAEAEAEAKAKAKAK) formed globular assemblies, whereas both peptides showed fibrillar assemblies at conditions above or below the neutral. Other examples (Figure 1B) including RAD16-I (RADARADARADARADA), RAD16-II (RARADADARARADADA), KLDLKLDLKLDL, FKFEFKFE, FEFKFEFK, FEFEFKFK, VEVE, SFSFSF, RFRFRF, and NININI have been reported to self-assemble into fibers (Kisiday et al., 2002; Marini et al., 2002; Yokoi et al., 2005; Cui et al., 2009; Guilbaud et al., 2010; Mandal et al., 2014; Do et al., 2016; Gao et al., 2017; Pelin et al., 2020). In addition to linear sequences, cyclic peptides have been used as building blocks allowing for manipulation of the supramolecular morphology through monomer design (Mandal et al., 2013). Cyclic peptides having the [WR]n structure, where n ∈ {3,4,5}, favor the formation of vesicle-type assemblies, unlike the linear designs with the binary alternating pattern, that preferentially assemble into fibrillar morphologies. The main applications of peptides classified in this category are related to their ability to form hydrogels. Such biomaterials can serve as scaffolds for tissue engineering, bioprinting, cell proliferation, regenerative medicine and drug delivery (Liu and Zhao, 2011; Levin et al., 2020; Gelain et al., 2021).
Surfactant-Like Peptides
Surfactant-like peptides formed by combining aliphatic and charged segments have been also reported as self-assembly units. Their design is based on a hydrophobic tail composed of V, I, L, G, A or P followed by a charged head group containing K, D, R or E (Figure 1C). Examples include V6K, V6K2, V6D, V6D2, I3K, I6K2, A3K, A6K, A6D, G4D2, G6D2, G8D2, A12R2, A2V2L3WE2/7, P10R3, etc. (Vauthey et al., 2002; van Hell et al., 2007; Yoon et al., 2008; Wang et al., 2009; Zhao, 2009; Xu et al., 2010; Hamley et al., 2013). These sequences can assemble into various morphologies comprising micelles, fibers, vesicles and tubes. The size and shape of the formed supramolecular assemblies depend on the type of amino acids used to constitute the amphiphile as well as the number of aliphatic and charged residues. In addition, factors such as temperature, solution pH and ionic strength affect the self-assembly process. The ability to form lipid bilayer-like assemblies makes them ideal for applications in immunotherapy, gene and drug delivery. Moreover, they can be used as protective envelopes for the delivery of enzymes and other biomolecules (Dasgupta and Das, 2019).
Dimensional Control Through Co-Assembly
Compared to the unimolecular assemblies described above, supramolecular co-assemblies lead to the formation of nanostructures with increased chemical diversity and structural complexity that can resemble natural systems. Peptides can co-assemble in cooperative, orthogonal, disruptive or random manner (Makam and Gazit, 2018). It is possible to fine-tune the morphology and dimension of nanostructures, and consequently their chemical, mechanical and physical properties, by controlling the mixing ratio of the individual building blocks. For example, the co-assembly of FF and FFF can be tuned to obtain nanorods, spherical nanovesicles, hollow nanotubes and toroid-like nanostructures (Guo et al., 2016). The disruptive co-assembly of the FF motif with its capped version Boc-FF allowed for the precise control of the nanotube length from 12 to 8 µm by changing the mixing ratio from 20:1 to 5:1, respectively (Adler-Abramovich et al., 2016). The cooperative co-assembly of dendrimeric poly (lysine) hydrophilic heads with linear poly (leucine) hydrophobic tails allowed the morphology control of the formed peptidosomes by alternating the ratio of dendrimeric to linear component. When the ratio changed from 10:1 to 1:5, the size of the nanoparticles increased from 300 to 800 nm and their morphology changed from spherical to fusiform (Xu et al., 2012). The ability to achieve dimensional control constitutes a promising tool for the design of peptide supramolecular materials where specific morphologies or dimensions are required. However, the co-assembly of peptide-based nanomaterials has not been researched extensively and constitutes an opportunity to improve the future design of peptide materials (Sasselli et al., 2017).
Viral Mimetics
Recently, the concept of mimicking viral capsids by creating their simplified versions through molecular self-assembly following the bottom-up strategy has emerged. Peptide self-assembly is a powerful tool to create biocompatible, tunable, low-cost supramolecular materials. It allows the conversion of chemically simple building blocks into a wide range of supramolecular architectures featuring modularity, functional diversity, adaptability and responsiveness to stimuli (Lampel, 2020). Peptides are versatile molecules for the design of virus mimetics as they can act as structural components as well as functional domains that favor selective binding, cell entry, endosomal escape or possess a specific activity (e.g., antimicrobial or catalytic). Short peptides offer the possibility to use minimal recognition modules for the design of functional materials and offer unique platforms for mimicking complex systems (Levin et al., 2020).
In this review, we distinguish the role of peptidic components used for the fabrication of virus mimetics into structural and functional modules (table 1). The structural modules are based on patterns that drive the formation of ordered supramolecular nanostructures having spherical or fibrillar morphologies (Figure 1D, F), dictated by the sequence, as described in section 2. In addition, trigonal cores (peptidic or organic), linkers (glycine or alkyl), cysteine residues serving as conjugation points, coiled coil, α-helical or β-annulus segments contribute to the design of structural modules. On the other hand, functional modules, related to the inherent biological signaling typical for peptides, are based on sequences with known activities such as cell penetration, integrin binding, DNA condensation and antimicrobial activity. Accordingly, peptide-based viral mimetic designs are divided into: i) capsid reconstruction strategies where structural modules contribute to the final supramolecular morphology and ii) simplified virus-like complexes where structural and functional peptidic modules are complexed with DNA or RNA fragments.
TABLE 1.
Examples of peptide-based viral mimetic design strategies.
| Strategy | Peptidic component / sequence | Role (structural (s) / functional (f)) | Supramolecular Morphology | Development Stage | Ref. |
|---|---|---|---|---|---|
| Capsid mimicking nanomaterials with C3 symmetry (trigonal or based on α-helical or β-sheet forming peptides) | |||||
| Trigonal (trimesoyl) peptide conjugate | Ci-FKFEFKFEii | i) Conjugation to core molecule (s) | Nanospheres | Biophysical data | Matsuura et al. (2005) |
| Ci-KTWTWTEiii | ii) β-sheet self-assembly unit (s) | Matsuura et al. (2011) | |||
| (γE-Ci-G)iv | iii) Tryptophane zipper based β-sheet self-assembly (s) | Matsuura et al. (2009) | |||
| iv) self-assembly unit (s) | |||||
| Wheel (trimesoyl) peptide conjugate | (FKFE-Ci-KFE)ii | i) Conjugation to core molecule (s) | Nanofibers | Biophysical data | Murasato et al. (2008) |
| ii) β-sheet self-assembly unit (s) | |||||
| Trigonal (tertiary amine) dipeptide conjugate | WW | β-sheet self-assembly unit (s) | Nanospheres | Biophysical data | Ghosh et al., (2007) |
| FF | Nanotubes | ||||
| Trigonal (ethyl benzene)-peptide conjugate | (γE-Ci-G)ii | i) Conjugation to core molecule (s) | Nanospheres | Biophysical data | Matsuura et al., (2010a) |
| ii) Self-assembly unit (s) | |||||
| Peptide triskelion (trilateral honeycomb symmetry) | βAKKi-(RRWTWE)3ii,iii | i) Trigonal core (s) | Nanocapsules | Cell assays (RNA delivery, antimicrobial activity) | Castelletto et al. (2016) |
| ii) Tryptophane zipper based β-sheet self-assembly (s) | |||||
| iii) Antimicrobial activity (f) | |||||
| Trigonal peptidic coiled coil heterodimers | KβAKβAKi-(KIAKLKQKIQKLKAKIAKLKQ)3 ii | i) Trigonal core (s) | Nanospheres | Cell assays (RNA delivery, antimicrobial activity) | De Santis et al., (2017) |
| CβAEISALEQEIASLEQEISALEQiii | ii) Cationic, covalently bound antimicrobial component (s) | ||||
| iii) Anionic component for heterodimer formation (s) | |||||
| β-annulus fragment from TBSV capsid | INHVGGTGGAIMAPVAVTRQLVGSi | i) β-annulus segment (s) | Hollow nanocapsules | Biophysical data | Matsuura et al. (2010b); Fujita and Matsuura (2017) |
| INHVGGTGGAIMAPVAVTRQLVGGi-CGGGKIAALKKKNAALKQKIAALKQii | ii) Cationic component covalently bound to β-annulus (s) | Nanospheres | |||
| EIAALEKENAALEQEIAALEQiii | iii) Anionic component for heterodimer formation (s) | ||||
| β-annulus fragment from SMV | GISMAPSAQGAMi-FKFEii | i) β-annulus segment (s) | Nanospheres | Biophysical data | Matsuura et al., (2016) |
| ii) β-sheet self-assembly unit (s) | |||||
| Tecto-dendrimeric design | Ci-GGii-EIARLEQEIARLEQEIARLEYEIARLEiii | i) Disulfide crosslinking (s) | Spherical particles | Cell assays (gene transfection) | Noble et al., (2016) |
| ii) Glycine linker (s) | |||||
| iii) α-helical conformation promoting sequence (s) | |||||
| Multicomponent peptide-DNA complexes | |||||
| Surfactant-like sequences | I3V3A3G3 i-K3ii | i) β-sheet self-assembly unit (s) | Nanosheets for peptidic component | Cell assays (gene transfection) | Cao et al., (2018) |
| ii) DNA condensing (f) | Heterogeneous morphologies via condensation with DNA | ||||
| Multicomponent, glucose-peptide conjugate | GSGSGSi-K8 ii-GGSGGSiii-(WKWE)3WGiv | i) linker (s) | β-nanoribbons for peptidic component and for complexes with siRNA and dsDNA | Cells assays (siRNA transfection) | Lim et al., (2008) |
| ii) siRNA binding site (f) | |||||
| iii) linker (s) | |||||
| iv) β-sheet self-assembly unit (s) | |||||
| Cocoon-like viral mimics based on β-sheet forming sequences (C6 = alkyl linker) | K3 i-C6-WLVFFAQQii-Giii-SPDiv | i) cationic, DNA binding region (f) | Nanoribbons for peptidic component | Biophysical data | Ni and Chau, (2014) |
| ii) amyloid / β-sheet segment (s) | Nanococoons via condensation with DNA | ||||
| K3 i-C6-Xii-Giii-SPDiv where X ∈ {L8, L6, L4, A8, A6, (L2A2)2} | iii) glycine linker (s) | Nanofibers for peptidic component | Cell assays (gene transfection) | Ni and Chau, (2017) | |
| iv) hydrophilic segment (s) | Nanococoons via condensation with DNA for L8, L6, L4, (L2A2)2 | ||||
| Tat–LK15 conjugate | (RKKRRQRRRGGGi-KLLKLLLKLLLKLLKii)iii | i) Cell penetrating (f) | Peptide-DNA complex (morphology not determined) | Cells assays (gene transfection) | Saleh et al. (2010) |
| ii) Membrane lytic, amphipathic (f) | |||||
| iii) DNA binding (f) | |||||
| Multicomponent | K6 i-GGFLGii-FWRGENGRKTRSAYERMCNILKGKiii | i) DNA binding (f)ii) Enzyme cleavable (s/f)iii) Influenza-derived epitope (f) | Dimer formation through disulphide linkage for peptide component | Cells assays (gene transfection) | Haines et al. (2001) |
| Spherical aggregates in presence of DNA | |||||
| Multicomponent, bola-amphiphile | RGDi-GPLGLAGii-I3 iii-G-R8 iv | i) Integrin binding (f) | Nanospheres for peptidic component only | Cells assays (gene transfection) | Wang et al. (2020) |
| ii) enzyme cleavable (hydrophobic) (s/f) | Rod-like or spherical shapes in presence of DNA | ||||
| iii) structural (hydrophobic) (s) | |||||
| iv) Cell penetrating, DNA binding (f) | |||||
| Bi-functional bola-amphiphile with hydrocarbon (C12) core | RGDi-C12-R8 ii | i) Integrin binding (f) | Spherical nanoparticles via condensation with DNA | Cells assays (gene transfection) | Chen et al., (2013) |
| ii) Cell penetrating, DNA binding (f) | |||||
Capsid-Like Nanomaterials
Viral capsids with icosahedral symmetry formed through the assembly of multiple protein subunits have inspired the design of artificial, peptide-based nanostructures for applications in gene delivery and cancer immunotherapy (Matsuura, 2018; Cai et al., 2020). In capsid reconstruction, it is important to maintain the n-fold rotational symmetry with n ∈ {3,5}. The C3 assembly can be achieved at the molecular level by designing trigonal conjugates or through folding-assembly pathways of peptides with helical conformations or β-annulus segments found on capsid-forming proteins (Figure 1E).
Trigonal designs induce the symmetry through the manipulation of the tripodal core (organic or peptidic) conjugated to peptidic structural modules that favor β-sheet-like self-assembly including the WTW tryptophane zipper and the FKFE-based binary alternating pattern. Examples are trymesoyl conjugates bearing three β-sheet-forming sequences (CFKFEFKFE or CKTWTWTE) attached through the C-terminal cysteine, that assemble into spherical morphologies (Matsuura et al., 2005, 2011). Similarly, a wheel-like trigonal design where the same core is conjugated to FKFECKFE through the central cysteine residue, self-assembled into fibers (Murasato et al., 2008). A clathrin triskelion-inspired conjugate, having a tris(2-aminoethyl)amine core linked to three aromatic di-tryptophan modules, self-assembled into nanospheres. In contrast, the conjugate containing the FF motif linked to the same core resulted in the formation of nanotubes, indicating that the morphology of the assemblies could be tuned through the dipeptide sequence (Ghosh et al., 2007). Furthermore, the choice of the core molecule can influence the properties of the obtained assemblies. For glutathione (γECG) attached to two different cores, the 1,3,5-tris(aminomethyl)-2,4,6-triethyl benzene showed improved conformational rigidity compared to the trimesoyl, giving rise to nanospheres with narrow size distribution (Matsuura et al., 2009; Matsuura et al., 2010a).
A purely peptidic triskelion, designed by conjugating each amino acid of the core sequence βAKK to the antimicrobial RRWTWE peptide containing the virus-derived tryptophane zipper, self-assembled into nanocapsules with dual function consisting of siRNA delivery and intrinsic antimicrobial activity (Castelletto et al., 2016). In this case, the RRWTWE sequence contains both the structural (β-sheet-forming) and functional (antimicrobial) modules. In another example, the core KβAKβAK sequence was conjugated to a positively charged antimicrobial (KIAKLKQKIQKLKAKIAKLKQ) peptide to form a trigonal conjugate, that upon addition of a complementary anionic sequence (CβAEISALEQEIASLEQEISALEQ), assembled in a coiled-coil hetero dimer. The resulting C3 subunit gave rise to capsid-like nanomaterials with antimicrobial activity (De Santis et al., 2017).
The reconstruction of capsid morphology based on the assembly of β-annulus peptide segments from Tomato bushy stunt virus (INHVGGTGGAIMAPVAVTRQLVG) and Sesbania mosaic virus (GISMAPSAQGAM) is able to maintain the C3 symmetry while allowing for introduction of surface modifications (Matsuura et al., 2010b; Matsuura et al., 2016). Among others, these include coating with gold nanoparticles to enhance the imaging efficiency (Matsuura et al., 2015) or with albumin to confer greater serum stability without eliciting immune response or toxicity (Matsuura and Honjo, 2019).
At the sequence level, the β-annulus segments can be modified with β-sheet promoting sequences (FKFE) to improve their assembly propensity into spherical morphologies (Matsuura et al., 2016). Moreover, with the intention of mimicking spike-bearing viruses such as Influenza and SARS-CoV-2, the β-annulus segment covalently linked to a cationic, coiled-coil-forming sequence at the C-terminus (CGGGKIAALKKKNAALKQKIAALKQ) gives rise to nanospheres. In the presence of a complementary anionic peptide (EIAALEKENAALEQEIAALEQ) and depending on the ratio of the cationic to anionic component, spherical (4:1) or fibrillar (1:1) assemblies with surface-exposed dimeric coiled coils are obtained (Fujita and Matsuura, 2017).
Another strategy is the use of a tecto-dendrimeric architecture as template to achieve C3 assembly into spherical particles for gene delivery. The design is based on structural coiled-coil subunits (CGG-EIARLEQEIARLEQEIARLEYEIARLE) configured into helical wheels, containing a GG spacer motif adjacent to a cysteine residue allowing for disulfide crosslinking (Noble et al., 2016).
Multicomponent Peptide-DNA Complexes
Virus-mimicking nanostructures can be formed through the complexation of peptides with DNA or RNA (Figure 1E), simulating the co-assembly of capsid proteins with viral genomes. Predominantly positively charged peptides have the tendency to condense negatively charged gene fragments making the resulting virus mimicking nanostructures ideal candidates for gene delivery (Miyata et al., 2012). Compared to conventional, cytotoxic DNA condensation agents such as polyelectrolytes and lipidic surfactants, short peptides have higher biocompatibility and consequently lower toxicity. Moreover, their structure can easily be modified to obtain high affinity DNA binders (Wang et al., 2020). Furthermore, the condensation with the peptidic vector confers protection from DNases. Several peptide-DNA/RNA co-assemblies have been reported containing structural or functional modules or their combination resulting in multicomponent designs.
Peptide-DNA condensates composed of lysine modified surfactant-like, binary alternating or amyloid-like structural modules, have been reported. While the cationic region drives the binding to DNA or RNA through electrostatic attraction, peptide self-assembly and β-sheet formation takes place via hydrogen bonds and hydrophobic interactions. Surfactant-like sequences, obtained by varying the position of aliphatic amino acids (A, G, I, and L) as well as the position of the cationic (K3) region from N- to C- terminus, including cone-like (G3A3V3I3K3, K3I3V3A3G3), dumbbell-like (I3V3A3G3K3, K3G3A3V3I3) and irregular shaped sequences (V3G3I3A3K3, K3A3I3G3V3) gave rise to nanorods, nanosheets and nanofibrils, respectively. The I3V3A3G3K3 was the most efficient one in inducing DNA condensation showing high content of ordered domains (Cao et al., 2018). This example shows that the supramolecular morphology and content of ordered domains could be fine-tuned through sequence engineering. Furthermore, a glucose-peptide conjugate [Glucose-GSGSGS-K8-GGSGGS-(WKWE)3WG] containing a functional, cationic segment (K8) for siRNA binding positioned between two linkers (GSGSGS and GGSGGS) and a binary alternating structural motif (WKWE)3, assembled into bilayered β-nanoribbons. The carbohydrate ligand exhibited the dual function of maintaining the β-nanoribbons neutrally charged while enhancing the cell binding through glucose transporters (Lim et al., 2008). Therefore, this design offers the formation of a controllable filamentous morphology able to bind RNA while presenting surface functionalization that yields high transfection efficiency.
Another example is the design of the cocoon-like virus mimetics based on a sequence (K3-C6-WLVFFAQQGSPD) containing the cationic, DNA binding region (K3) at the N-terminus, followed by the alkyl linker (C6) and three structural components, namely, the amyloid-like motif (LVFFA), the glycine linker and the hydrophilic (SPD) region (Ni and Chau, 2014). The β-sheet forming segment can be modified from amyloid to aliphatic (L8, L6, L4, and L2A2L2A2) while maintaining the self-assembly propensity of the whole sequence. The peptides alone self-assemble into fibrillar aggregates, while their interaction with DNA in various ratios induces condensation into nanococoons (Ni and Chau, 2017).
Cell penetrating peptides including the arginine-rich, R8 and the HIV-1 derived, Tat (RKKRRQRRRGGG) constitute the main functional modules used for the design of DNA condensates (Kalafatovic and Giralt, 2017). The covalent conjugation of Tat to the amphipathic LK15 sequence (KLLKLLLKLLLKLLK) resulted in improved cellular uptake and transfection efficiency, compared to Tat or LK15 alone (Saleh et al., 2010). CL22 (K6-GGFLG-FWRGENGRKTRSAYERMCNILKGK) is an example of purely peptidic design containing an enzyme cleavable segment adjacent to the DNA binding region at the N-terminus and the Influenza nucleoprotein-derived sequence at the C-terminus. It assembles into spherical aggregates in the presence of DNA and attains maximum gene transfection efficiency upon spontaneous dimerization through the disulfide bond between cysteines at the C-terminus (Haines et al., 2001). Bola amphiphiles, composed of a central hydrophobic segment flanked by two hydrophilic ones, have the ability to self-assemble into fibrillar or spherical nanostructures depending on the sequence design (Chen et al., 2013). Examples are the purely peptidic RGD-GPLGLAG-I3-G-R8 (Wang et al. 2020) and the fatty acid containing RGD-C12-R8 (Chen et al., 2013) that accommodate both functional and structural motifs, where RGD is crucial for integrin-binding and R8 for cell penetration. Additionally, the PLGLA sequence serves as an enzyme-cleavable segment, while I3 confers hydrophobicity. The main drawback of peptide-DNA/RNA co-assemblies, mainly based on functional modules, is that oppositely charged polyions often form heterogeneous aggregates. The challenges resulting from the lack of control over their morphology, degree of order and size, often hamper the efficiency of gene transfection or delivery.
The DNA fragment length and composition can affect the formation of peptide-DNA complexes but also their morphology. The mechanism of formation depends on the peptides’ intrinsic ability to self-assemble. Self-assembling peptide sequences condense the DNA by reorganizing to a final morphology that is often different from the one formed by the peptide alone. On the other hand, predominantly cationic and/or cell-penetrating peptides, unable to self-assemble, tend to form irregular aggregates in the presence of DNA. Moreover, the size of the complex can be controlled by varying the length of the DNA fragment. For example, the I3V3A3G3K3–DNA complex size decreased from 122 to 85 nm by shortening the DNA fragment from 2000 to 300 bp (base pairs). Even though most examples use λ-DNA (∼4.8 kbp), shorter DNA fragments (2000—300 bp) were explored with the intention to improve the DNA delivery efficiency (Cao et al., 2018).
However, the key factor influencing the morphology of peptide-DNA complexes is the R + / − ratio of the positively charged peptide residues to the negatively charged DNA fragments. A stable peptide-DNA complex is formed when all the negative charges are successfully neutralized. For example, RGD-GPLGLAG-I3-G-R8 that self-assembles into spheres, upon the interaction with DNA and depending on the R + / - values forms thread-like (R + / - = 0.5) complexes or highly condensed rod-like or spherical (R + / - = 3) nanostructures (Wang et al., 2020). In another example, the R + / - of 10 is the minimum requirement for DNA condensation with K3C6SPD, where the peptide alone self-assembles into nanoribbons. However, upon DNA addition, the electrostatic interactions drive the self-assembly into amorphous aggregates (R + / - = 5), or agglomerations with small striped nanococoons (R + / - = 10). The R + / - = 20 presents the optimal ratio for nanococoon formation, while at R + / - of 25 and 50 both nanococoons and filamentous nanoribbons are formed (Ni and Chau, 2014).
Future Perspectives
The idea of exploiting known principles of peptide self-assembly to obtain spherical or fibrillar nanostructures by including important features such as cell penetration, antimicrobial activity or viral transfection is conceptually attractive. Such systems are promising as they can be easily engineered and modified to include specific sequences found on the receptor binding domains of spike proteins. In addition, they can be designed as vehicles able to deliver cargo into cells. So far, morphology rather than functionality has been mimicked and it constitutes an advantage from the point of view of easy production compared to VLPs. A step towards functionality of peptide materials is their ability to enhance viral transfection by increasing the β-sheet content of supramolecular nanostructures (Sieste et al., 2021). However, efforts are needed to achieve controllable and complex functions such as self-replication and catalysis in the future. Although largely unexplored for clinical use, because of the multiscale and multiparameter optimization challenges of supramolecular nanostructures (Sieste et al., 2021), we envision that peptides have great potential in becoming future nanotechnological solutions in covid-19 therapy and diagnostics.
The intention of this review is to emphasize the increasing importance of peptide self-assembly in the design and fabrication of minimalistic, synthetic models applicable to a variety of viral infections. We expect that future research in this field will deliver simple and cost-effective viral mimetics composed of peptide modules found on the surface of specific viruses, rationally designed to assemble into multivalent and multifunctional nanostructures able to selectively bind receptors of interest, penetrate cells and carry cargos. In addition to mimicking the viral morphology, such systems would partly resemble basic functionality through the display of known functional modules and their combinations aiming for possible synergistic effects. Such an approach could lead to the development of efficient and safe platforms to study viral infections without the need of complicated genetic manipulations. Moreover, the developed models will provide screening platforms that can be rationally designed, allowing for rapid discovery of potential inhibitors or surface protein binders. Therefore, they could be used as safe alternatives for antiviral drug discovery or as vehicles for mRNA vaccines.
Acknowledgments
The authors would like to acknowledge the Centre for Artificial intelligence and cyber security (AIRI) and the Center for Advanced Computing and Modelling (CNRM) at the University of Rijeka.
Author Contribution
PJ and DK conceived and designed the review, PJ, IŠ, AP, and DK analysed the literature and wrote the manuscript. All authors have read and approved the final version of the manuscript.
Funding
This work was supported by the University of Rijeka (uniri-COV-1), the Foundation of the Croatian Academy of Sciences and Arts (HAZU) and by the Croatian Science Foundation/Hrvatska zaklada za znanost (UIP-2019-04-7999).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abudula T., Bhatt K., Eggermont L. J., O'Hare N., Memic A., Bencherif S. A. (2020). Supramolecular Self-Assembled Peptide-Based Vaccines: Current State and Future Perspectives. Front. Chem. 8, 1–11. 10.3389/fchem.2020.598160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler-Abramovich L., Marco P., Arnon Z. A., Creasey R. C. G., Michaels T. C. T., Levin A., et al. (2016). Controlling the Physical Dimensions of Peptide Nanotubes by Supramolecular Polymer Coassembly. ACS Nano. 10, 7436–7442. 10.1021/acsnano.6b01587 [DOI] [PubMed] [Google Scholar]
- Al-Halifa S., Babych M., Zottig X., Archambault D., Bourgault S. (2019). Amyloid Self-Assembling Peptides: Potential Applications in Nanovaccine Engineering and Biosensing. Pept. Sci. 111, e24095–12. 10.1002/pep2.24095 [DOI] [Google Scholar]
- Bera S., Mondal S., Xue B., Shimon L. J. W., Cao Y., Gazit E. (2019). Rigid Helical-like Assemblies from a Self-Aggregating Tripeptide. Nat. Mater. 18, 503–509. 10.1038/s41563-019-0343-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Ran W., Zhai Y., Wang J., Zheng C., Li Y., et al. (2020). Recent Progress in Supramolecular Peptide Assemblies as Virus Mimics for Cancer Immunotherapy. Biomater. Sci. 8, 1045–1057. 10.1039/c9bm01380f [DOI] [PubMed] [Google Scholar]
- Cao M., Wang Y., Zhao W., Qi R., Han Y., Wu R., et al. (2018). Peptide-Induced DNA Condensation into Virus-Mimicking Nanostructures. ACS Appl. Mater. Inter. 10, 24349–24360. 10.1021/acsami.8b00246 [DOI] [PubMed] [Google Scholar]
- Castelletto V., De Santis E., Alkassem H., Lamarre B., Noble J. E., Ray S., et al. (2016). Structurally Plastic Peptide Capsules for Synthetic Antimicrobial Viruses. Chem. Sci. 7, 1707–1711. 10.1039/c5sc03260a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-X., Xu X.-D., Yang S., Yang J., Zhuo R.-X., Zhang X.-Z. (2013). Self-Assembled BolA-like Amphiphilic Peptides as Viral-Mimetic Gene Vectors for Cancer Cell Targeted Gene Delivery. Macromol. Biosci. 13, 84–92. 10.1002/mabi.201200283 [DOI] [PubMed] [Google Scholar]
- Collier J. H., Rudra J. S., Gasiorowski J. Z., Jung J. P. (2010). Multi-component Extracellular Matrices Based on Peptide Self-Assembly. Chem. Soc. Rev. 39, 3413–3424. 10.1039/b914337h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik D. J., Fairlie D. P., Liras S., Price D. (2013). The Future of Peptide-Based Drugs. Chem. Biol. Drug Des. 81, 136–147. 10.1111/cbdd.12055 [DOI] [PubMed] [Google Scholar]
- Cui H., Muraoka T., Cheetham A. G., Stupp S. I. (2009). Self-assembly of Giant Peptide Nanobelts. Nano Lett. 9, 945–951. 10.1021/nl802813f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A., Das D. (2019). Designer Peptide Amphiphiles: Self-Assembly to Applications. Langmuir 35, 10704–10724. 10.1021/acs.langmuir.9b01837 [DOI] [PubMed] [Google Scholar]
- De Santis E., Alkassem H., Lamarre B., Faruqui N., Bella A., Noble J. E., et al. (2017). Antimicrobial Peptide Capsids of De Novo Design. Nat. Commun. 8, 1–11. 10.1038/s41467-017-02475-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaferia C., Balasco N., Sibillano T., Giannini C., Vitagliano L., Morelli G., et al. (2018). Structural Characterization of Self-Assembled Tetra-Tryptophan Based Nanostructures: Variations on a Common Theme. ChemPhysChem 19, 1635–1642. 10.1002/cphc.201800026 [DOI] [PubMed] [Google Scholar]
- Do T. D., De Almeida N. E. C., Lapointe N. E., Chamas A., Feinstein S. C., Bowers M. T. (2016). Amino Acid Metaclusters: Implications of Growth Trends on Peptide Self-Assembly and Structure. Anal. Chem. 88, 868–876. 10.1021/acs.analchem.5b03454 [DOI] [PubMed] [Google Scholar]
- Fang J.-H., Lee Y.-T., Chiang W.-H., Hu S.-H. (2015). Magnetoresponsive Virus-Mimetic Nanocapsules with Dual Heat-Triggered Sequential-Infected Multiple Drug-Delivery Approach for Combinatorial Tumor Therapy. Small 11, 2417–2428. 10.1002/smll.201402969 [DOI] [PubMed] [Google Scholar]
- Ferreira D., Martins I. M. (2017). “Artificial Virus Particles,” in Bioinspired Materials for Medical Applications (Elsevier; ), 427–450. 10.1016/B978-0-08-100741-9.00015-2 [DOI] [Google Scholar]
- Fosgerau K., Hoffmann T. (2015). Peptide Therapeutics: Current Status and Future Directions. Drug Discov. Today. 20, 122–128. 10.1016/j.drudis.2014.10.003 [DOI] [PubMed] [Google Scholar]
- Frederix P. W. J. M., Scott G. G., Abul-Haija Y. M., Kalafatovic D., Pappas C. G., Javid N., et al. (2015). Exploring the Sequence Space for (Tri-)peptide Self-Assembly to Design and Discover New Hydrogels. Nat. Chem. 7, 30–37. 10.1038/nchem.2122 [DOI] [PubMed] [Google Scholar]
- Fujita S., Matsuura K. (2017). Self-Assembled Artificial Viral Capsids Bearing Coiled-Coils at the Surface. Org. Biomol. Chem. 15, 5070–5077. 10.1039/c7ob00998d [DOI] [PubMed] [Google Scholar]
- Gao J., Tang C., Elsawy M. A., Smith A. M., Miller A. F., Saiani A. (2017). Controlling Self-Assembling Peptide Hydrogel Properties through Network Topology. Biomacromolecules 18, 826–834. 10.1021/acs.biomac.6b01693 [DOI] [PubMed] [Google Scholar]
- Gazit E. (2007b). Self Assembly of Short Aromatic Peptides into Amyloid Fibrils and Related Nanostructures. Prion 1, 32–35. 10.4161/pri.1.1.4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit E. (2007a). Self-assembled Peptide Nanostructures: The Design of Molecular Building Blocks and Their Technological Utilization. Chem. Soc. Rev. 36, 1263–1269. 10.1039/b605536m [DOI] [PubMed] [Google Scholar]
- Gelain F., Luo Z., Rioult M., Zhang S. (2021). Self-assembling Peptide Scaffolds in the Clinic. Npj Regen. Medmed. 6, 1–8. 10.1038/s41536-020-00116-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Reches M., Gazit E., Verma S. (2007). Bioinspired Design of Nanocages by Self-Assembling Triskelion Peptide Elements. Angew. Chem. Int. Ed. 46, 2002–2004. 10.1002/anie.200604383 [DOI] [PubMed] [Google Scholar]
- Guilbaud J.-B., Vey E., Boothroyd S., Smith A. M., Ulijn R. V., Saiani A., et al. (2010). Enzymatic Catalyzed Synthesis and Triggered Gelation of Ionic Peptides. Langmuir 26, 11297–11303. 10.1021/la100623y [DOI] [PubMed] [Google Scholar]
- Guo C., Arnon Z. A., Qi R., Zhang Q., Adler-Abramovich L., Gazit E., et al. (2016). Expanding the Nanoarchitectural Diversity through Aromatic Di- and Tri-peptide Coassembly: Nanostructures and Molecular Mechanisms. ACS Nano. 10, 8316–8324. 10.1021/acsnano.6b02739 [DOI] [PubMed] [Google Scholar]
- Haines A., Irvine A., Mountain A., Charlesworth J., Farrow N., Husain R., et al. (2001). CL22 - A Novel Cationic Peptide for Efficient Transfection of Mammalian Cells. Gene Ther. 8, 99–110. 10.1038/sj.gt.3301314 [DOI] [PubMed] [Google Scholar]
- Hamley I. W., Dehsorkhi A., Castelletto V., Seitsonen J., Ruokolainen J., Iatrou H. (2013). Self-assembly of a Model Amphiphilic Oligopeptide Incorporating an Arginine Headgroup. Soft Matter 9, 4794–4801. 10.1039/c3sm50303h [DOI] [Google Scholar]
- Hendricks M. P., Sato K., Palmer L. C., Stupp S. I. (2017). Supramolecular Assembly of Peptide Amphiphiles. Acc. Chem. Res. 50, 2440–2448. 10.1021/acs.accounts.7b00297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Legge R. L., Zhang S., Chen P. (2003). Effect of Amino Acid Sequence and pH on Nanofiber Formation of Self-Assembling Peptides EAK16-II and EAK16-IV. Biomacromolecules 4, 1433–1442. 10.1021/bm0341374 [DOI] [PubMed] [Google Scholar]
- Kalafatovic D., Giralt E. (2017). Cell-penetrating Peptides: Design Strategies beyond Primary Structure and Amphipathicity. Molecules 22, 1929–1938. 10.3390/molecules22111929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalafatovic D., Nobis M., Son J., Anderson K. I., Ulijn R. V. (2016). MMP-9 Triggered Self-Assembly of Doxorubicin Nanofiber Depots Halts Tumor Growth. Biomaterials 98, 192–202. 10.1016/j.biomaterials.2016.04.039 [DOI] [PubMed] [Google Scholar]
- Kisiday J., Jin M., Kurz B., Hung H., Semino C., Zhang S., et al. (2002). Self-assembling Peptide Hydrogel Fosters Chondrocyte Extracellular Matrix Production and Cell Division: Implications for Cartilage Tissue Repair. Proc. Natl. Acad. Sci. 99, 9996–10001. 10.1073/pnas.142309999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampel A. (2020). Biology-Inspired Supramolecular Peptide Systems. Chem 6, 1222–1236. 10.1016/j.chempr.2020.03.005 [DOI] [Google Scholar]
- Lampel A., Ulijn R. V., Tuttle T. (2018). Guiding Principles for Peptide Nanotechnology through Directed Discovery. Chem. Soc. Rev. 47, 3737–3758. 10.1039/c8cs00177d [DOI] [PubMed] [Google Scholar]
- Lee C., Hwang H. S., Lee S., Kim B., Kim J. O., Oh K. T., et al. (2017). Rabies Virus-Inspired Silica-Coated Gold Nanorods as a Photothermal Therapeutic Platform for Treating Brain Tumors. Adv. Mater. 29, 1605563–1605568. 10.1002/adma.201605563 [DOI] [PubMed] [Google Scholar]
- Lee E. S., Kim D., Youn Y. S., Oh K. T., Bae Y. H. (2008). A Virus-Mimetic Nanogel Vehicle. Angew. Chem. Int. Ed. 47, 2418–2421. 10.1002/anie.200704121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Ju M., Cho O. H., Kim Y., Nam K. T. (2019). Tyrosine-Rich Peptides as a Platform for Assembly and Material Synthesis. Adv. Sci. 6, 1801255. 10.1002/advs.201801255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehn J.-M. (2002). Toward Self-Organization and Complex Matter. Science 295, 2400–2403. 10.1126/science.1071063 [DOI] [PubMed] [Google Scholar]
- Levin A., Hakala T. A., Schnaider L., Bernardes G. J. L., Gazit E., Knowles T. P. J. (2020). Biomimetic Peptide Self-Assembly for Functional Materials. Nat. Rev. Chem. 4, 615–634. 10.1038/s41570-020-0215-y [DOI] [Google Scholar]
- Li Y., Lai Y., Xu X., Zhang X., Wu Y., Hu C., et al. (2016). Capsid-like Supramolecular Dendritic Systems as pH-Responsive Nanocarriers for Drug Penetration and Site-specific Delivery. Nanomedicine: Nanotechnology, Biol. Med. 12, 355–364. 10.1016/j.nano.2015.09.015 [DOI] [PubMed] [Google Scholar]
- Liang H., Hu A., Chen X., Jin R., Wang K., Ke B., et al. (2019). Structure Optimization of Dendritic Lipopeptide Based Gene Vectors with the Assistance from Molecular Dynamic Simulation. J. Mater. Chem. B. 7, 915–926. 10.1039/c8tb02650e [DOI] [PubMed] [Google Scholar]
- Lim Y.-b., Lee E., Yoon Y.-R., Lee M. S., Lee M. (2008). Filamentous Artificial Virus from a Self-Assembled Discrete Nanoribbon. Angew. Chem. 120, 4601–4604. 10.1002/ange.200800266 [DOI] [PubMed] [Google Scholar]
- Liu J., Zhao X. (2011). Design of Self-Assembling Peptides and Their Biomedical Applications. Nanomedicine 6, 1621–1643. 10.2217/nnm.11.142 [DOI] [PubMed] [Google Scholar]
- Ludwig C., Wagner R. (2007). Virus-like Particles-Universal Molecular Toolboxes. Curr. Opin. Biotechnol. 18, 537–545. 10.1016/j.copbio.2007.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makam P., Gazit E. (2018). Minimalistic Peptide Supramolecular Co-assembly: Expanding the Conformational Space for Nanotechnology. Chem. Soc. Rev. 47, 3406–3420. 10.1039/c7cs00827a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal D., Nasrolahi Shirazi A., Parang K. (2014). Self-assembly of Peptides to Nanostructures. Org. Biomol. Chem. 12, 3544–3561. 10.1039/c4ob00447g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal D., Tiwari R. K., Nasrolahi Shirazi A., Oh D., Ye G., Banerjee A., et al. (2013). Self-assembled Surfactant Cyclic Peptide Nanostructures as Stabilizing Agents. Soft Matter. 9, 9465–9475. 10.1039/c3sm50764e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C., Liu A., Cao B. (2009). Virus-based Chemical and Biological Sensing. Angew. Chem. Int. Ed. 48, 6790–6810. 10.1002/anie.200900231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesan S., Easton C. D., Kushkaki F., Waddington L., Hartley P. G. (2012). Tripeptide Self-Assembled Hydrogels: Unexpected Twists of Chirality. Chem. Commun. 48, 2195–2197. 10.1039/c2cc16609g [DOI] [PubMed] [Google Scholar]
- Marini D. M., Hwang W., Lauffenburger D. A., Zhang S., Kamm R. D. (2002). Left-Handed Helical Ribbon Intermediates in the Self-Assembly of a β-Sheet Peptide. Nano Lett. 2, 295–299. 10.1021/nl015697g [DOI] [Google Scholar]
- Maslanka Figueroa S., Fleischmann D., Goepferich A. (2021). Biomedical Nanoparticle Design: What We Can Learn from Viruses. J. Controlled Release. 329, 552–569. 10.1016/j.jconrel.2020.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura K. (2012). Construction of Spherical Virus-Inspired Peptide Nanoassemblies. Polym. J. 44, 469–474. 10.1038/pj.2012.16 [DOI] [Google Scholar]
- Matsuura K., Fujino K., Teramoto T., Murasato K., Kimizuka N. (2010a). Glutathione Nanosphere: Self-Assembly of Conformation-Regulated Trigonal-Glutathiones in Water. Bcsj 83, 880–886. 10.1246/bcsj.20100048 [DOI] [Google Scholar]
- Matsuura K., Hayashi H., Murasato K., Kimizuka N. (2011). Trigonal Tryptophane Zipper as a Novel Building Block for pH-Responsive Peptide Nano-Assemblies. Chem. Commun. 47, 265–267. 10.1039/c0cc01324b [DOI] [PubMed] [Google Scholar]
- Matsuura K., Honjo T. (2019). Artificial Viral Capsid Dressed up with Human Serum Albumin. Bioconjug. Chem. 30, 1636–1641. 10.1021/acs.bioconjchem.9b00327 [DOI] [PubMed] [Google Scholar]
- Matsuura K., Matsuyama H., Fukuda T., Teramoto T., Watanabe K., Murasato K., et al. (2009). Spontaneous Self-Assembly of Nanospheres from Trigonal Conjugate of Glutathione in Water. Soft Matter. 5, 2463–2470. 10.1039/b819472f [DOI] [Google Scholar]
- Matsuura K., Mizuguchi Y., Kimizuka N. (2016). Peptide Nanospheres Self-Assembled from a Modified β -annulus Peptide of Sesbania Mosaic Virus. Biopolymers 106, 470–475. 10.1002/bip.22774 [DOI] [PubMed] [Google Scholar]
- Matsuura K., Murasato K., Kimizuka N. (2005). Artificial Peptide-Nanospheres Self-Assembled from Three-Way Junctions of β-Sheet-Forming Peptides. J. Am. Chem. Soc. 127, 10148–10149. 10.1021/ja052644i [DOI] [PubMed] [Google Scholar]
- Matsuura K. (2018). Synthetic Approaches to Construct Viral Capsid-like Spherical Nanomaterials. Chem. Commun. 54, 8944–8959. 10.1039/C8CC03844A [DOI] [PubMed] [Google Scholar]
- Matsuura K., Ueno G., Fujita S. (2015). Self-assembled Artificial Viral Capsid Decorated with Gold Nanoparticles. Polym. J. 47, 146–151. 10.1038/pj.2014.99 [DOI] [Google Scholar]
- Matsuura K., Watanabe K., Matsuzaki T., Sakurai K., Kimizuka N. (2010b). Self-Assembled Synthetic Viral Capsids from a 24-mer Viral Peptide Fragment. Angew. Chem. Int. Ed. 49, 9662–9665. 10.1002/anie.201004606 [DOI] [PubMed] [Google Scholar]
- Miyata K., Nishiyama N., Kataoka K. (2012). Rational Design of Smart Supramolecular Assemblies for Gene Delivery: Chemical Challenges in the Creation of Artificial Viruses. Chem. Soc. Rev. 41, 2562–2574. 10.1039/c1cs15258k [DOI] [PubMed] [Google Scholar]
- Mohsen M. O., Speiser D. E., Knuth A., Bachmann M. F. (2020). Virus‐like Particles for Vaccination against Cancer. WIREs Nanomed Nanobiotechnol. 12, 1–17. 10.1002/wnan.1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murasato K., Matsuura K., Kimizuka N. (2008). Self-Assembly of Nanofiber with Uniform Width from Wheel-type Trigonal-β-Sheet-Forming Peptide. Biomacromolecules 9, 913–918. 10.1021/bm701302p [DOI] [PubMed] [Google Scholar]
- Ni R., Chau Y. (2014). Structural Mimics of Viruses through Peptide/DNA Co-assembly. J. Am. Chem. Soc. 136, 17902–17905. 10.1021/ja507833x [DOI] [PubMed] [Google Scholar]
- Ni R., Chau Y. (2017). Tuning the Inter-nanofibril Interaction to Regulate the Morphology and Function of Peptide/DNA Co-assembled Viral Mimics. Angew. Chem. 129, 9484–9488. 10.1002/ange.201703596 [DOI] [PubMed] [Google Scholar]
- Noble J. E., De Santis E., Ravi J., Lamarre B., Castelletto V., Mantell J., et al. (2016). A De Novo Virus-like Topology for Synthetic Virions. J. Am. Chem. Soc. 138, 12202–12210. 10.1021/jacs.6b05751 [DOI] [PubMed] [Google Scholar]
- Pelin J. N. B. D., Gerbelli B. B., Edwards-Gayle C. J. C., Aguilar A. M., Castelletto V., Hamley I. W., et al. (2020). Amyloid Peptide Mixtures: Self-Assembly, Hydrogelation, Nematic Ordering, and Catalysts in Aldol Reactions. Langmuir 36, 2767–2774. 10.1021/acs.langmuir.0c00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao C., Zhang R., Wang Y., Jia Q., Wang X., Yang Z., et al. (2020). Rabies Virus‐Inspired Metal-Organic Frameworks (MOFs) for Targeted Imaging and Chemotherapy of Glioma. Angew. Chem. Int. Ed. 59, 16982–16988. 10.1002/anie.202007474 [DOI] [PubMed] [Google Scholar]
- Reches M., Gazit E. (2004). Formation of Closed-Cage Nanostructures by Self-Assembly of Aromatic Dipeptides. Nano Lett. 4, 581–585. 10.1021/nl035159z [DOI] [Google Scholar]
- Roldão A., Silva A. C., Mellado M. C. M., Alves P. M., Carrondo M. J. T. (2017). Viruses and Virus-like Particles in Biotechnology: Fundamentals and Applications. Compr. Biotechnol., 633–656. 10.1016/B978-0-12-809633-8.09046-4 [DOI] [Google Scholar]
- Saleh A. F., Aojula H., Arthanari Y., Offerman S., Alkotaji M., Pluen A. (2010). Improved Tat-Mediated Plasmid DNA Transfer by Fusion to LK15 Peptide. J. Controlled Release. 143, 233–242. 10.1016/j.jconrel.2009.12.025 [DOI] [PubMed] [Google Scholar]
- Sasselli I. R., Moreira I. P., Ulijn R. V., Tuttle T. (2017). Molecular Dynamics Simulations Reveal Disruptive Self-Assembly in Dynamic Peptide Libraries. Org. Biomol. Chem. 15, 6541–6547. 10.1039/c7ob01268c [DOI] [PubMed] [Google Scholar]
- Sharma P., Pal V. K., Roy S. (2021). An Overview of Latest Advances in Exploring Bioactive Peptide Hydrogels for Neural Tissue Engineering. Biomater. Sci. 9, 3911–3938. 10.1039/D0BM02049D [DOI] [PubMed] [Google Scholar]
- Sieste S., Mack T., Lump E., Hayn M., Schütz D., Röcker A., et al. (2021). Supramolecular Peptide Nanofibrils with Optimized Sequences and Molecular Structures for Efficient Retroviral Transduction. Adv. Funct. Mater. 31, 2009382. 10.1002/adfm.202009382 [DOI] [Google Scholar]
- Singh N., Kumar M., Miravet J. F., Ulijn R. V., Escuder B. (2017). Peptide-Based Molecular Hydrogels as Supramolecular Protein Mimics. Chem. Eur. J. 23, 981–993. 10.1002/chem.201602624 [DOI] [PubMed] [Google Scholar]
- Sloan‐Dennison S., Lampel A., Raßlenberg E., Ulijn R. V., Smith E., Faulds K., et al. (2021). Elucidation of the Structure of Supramolecular Polymorphs in Peptide Nanofibres Using Raman Spectroscopy. J. Raman Spectrosc., 1–7. 10.1002/jrs.6121 [DOI] [Google Scholar]
- Slocik J. M., Naik R. R. (2017). Sequenced Defined Biomolecules for Nanomaterial Synthesis, Functionalization, and Assembly. Curr. Opin. Biotechnol. 46, 7–13. 10.1016/j.copbio.2016.11.025 [DOI] [PubMed] [Google Scholar]
- Smith D. J., Brat G. A., Medina S. H., Tong D., Huang Y., Grahammer J., et al. (2015). A Multiphase Transitioning Peptide Hydrogel for Suturing Ultrasmall Vessels. Nat. Nanotech. 11, 95–102. 10.1038/nnano.2015.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J., Kalafatovic D., Kumar M., Yoo B., Cornejo M. A., Contel M., et al. (2019). Customizing Morphology, Size, and Response Kinetics of Matrix Metalloproteinase-Responsive Nanostructures by Systematic Peptide Design. ACS Nano. 13, 1555–1562. 10.1021/acsnano.8b07401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamis P., Adler-Abramovich L., Reches M., Marshall K., Sikorski P., Serpell L., et al. (2009). Self-assembly of Phenylalanine Oligopeptides: Insights from Experiments and Simulations. Biophysical J. 96, 5020–5029. 10.1016/j.bpj.2009.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K., Makam P., Aizen R., Gazit E. (2017). Self-assembling Peptide Semiconductors. Science 358, eaam9756. 10.1126/science.aam9756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hell A. J., Costa C. I. C. A., Flesch F. M., Sutter M., Jiskoot W., Crommelin D. J. A., et al. (2007). Self-Assembly of Recombinant Amphiphilic Oligopeptides into Vesicles. Biomacromolecules 8, 2753–2761. 10.1021/bm0704267 [DOI] [PubMed] [Google Scholar]
- Vauthey S., Santoso S., Gong H., Watson N., Zhang S. (2002). Molecular Self-Assembly of Surfactant-like Peptides to Form Nanotubes and Nanovesicles. Proc. Natl. Acad. Sci. 99, 5355–5360. 10.1073/pnas.072089599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe P., Lisowski V., Martinez J., Khrestchatisky M. (2010). Synthetic Therapeutic Peptides: Science and Market. Drug Discov. Today. 15, 40–56. 10.1016/j.drudis.2009.10.009 [DOI] [PubMed] [Google Scholar]
- Wang J., Han S., Meng G., Xu H., Xia D., Zhao X., et al. (2009). Dynamic Self-Assembly of Surfactant-like Peptides A6K and A9K. Soft Matter. 5, 3870–3878. 10.1039/b901653h [DOI] [Google Scholar]
- Wang Y., Nie Y., Ding Z., Yao M., Du R., Zhang L., et al. (2020). An Amphiphilic Peptide with Cell Penetrating Sequence for Highly Efficient Gene Transfection. Colloids Surf. A: Physicochemical Eng. Aspects. 590, 124529. 10.1016/j.colsurfa.2020.124529 [DOI] [Google Scholar]
- Whitesides G. M. (2015). Bioinspiration: Something for Everyone. Interf. Focus. 5, 20150031. 10.1098/rsfs.2015.0031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesides G. M., Grzybowski B. (2002). Self-assembly at All Scales. Science 295, 2418–2421. 10.1126/science.1070821 [DOI] [PubMed] [Google Scholar]
- Woolfson D. N., Mahmoud Z. N. (2010). More Than Just Bare Scaffolds: Towards Multi-Component and Decorated Fibrous Biomaterials. Chem. Soc. Rev. 39, 3464–3479. 10.1039/c0cs00032a [DOI] [PubMed] [Google Scholar]
- Xu H., Wang Y., Ge X., Han S., Wang S., Zhou P., et al. (2010). Twisted Nanotubes Formed from Ultrashort Amphiphilic Peptide I3K and Their Templating for the Fabrication of Silica Nanotubes. Chem. Mater. 22, 5165–5173. 10.1021/cm101019p [DOI] [Google Scholar]
- Xu X., Yuan H., Chang J., He B., Gu Z. (2012). Cooperative Hierarchical Self-Assembly of Peptide Dendrimers and Linear Polypeptides into Nanoarchitectures Mimicking Viral Capsids. Angew. Chem. Int. Ed. 51, 3130–3133. 10.1002/anie.201106080 [DOI] [PubMed] [Google Scholar]
- Yang J., Zhang X., Liu C., Wang Z., Deng L., Feng C., et al. (2021a). Biologically Modified Nanoparticles as Theranostic Bionanomaterials. Prog. Mater. Sci. 118, 100768. 10.1016/j.pmatsci.2020.100768 [DOI] [Google Scholar]
- Yang P., Li Y.-J., Cao Y., Zhang L., Wang J.-Q., Lai Z.-W., et al. (2020b). Rapid Discovery of Self-Assembling Peptides with One-Bead One-Compound Peptide Libraries. Res. Sq., 1–15. 10.21203/rs.3.rs-109949/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi H., Kinoshita T., Zhang S. (2005). Dynamic Reassembly of Peptide RADA16 Nanofiber Scaffold. Proc. Natl. Acad. Sci. 102, 8414–8419. 10.1073/pnas.0407843102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y.-R., Lim Y.-b., Lee E., Lee M. (2008). Self-assembly of a Peptide Rod-Coil: A Polyproline Rod and a Cell-Penetrating Peptide Tat Coil. Chem. Commun., 1892–1894. 10.1039/b719868j [DOI] [PubMed] [Google Scholar]
- Zhang S. (2017). Discovery and Design of Self-Assembling Peptides. Interf. Focus. 7, 20170028. 10.1098/rsfs.2017.0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. (2003). Fabrication of Novel Biomaterials through Molecular Self-Assembly. Nat. Biotechnol. 21, 1171–1178. 10.1038/nbt874 [DOI] [PubMed] [Google Scholar]
- Zhang S., Holmes T., Lockshin C., Rich A. (1993). Spontaneous Assembly of a Self-Complementary Oligopeptide to Form a Stable Macroscopic Membrane. Proc. Natl. Acad. Sci. 90, 3334–3338. 10.1073/pnas.90.8.3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. (2009). Design of Self-Assembling Surfactant-like Peptides and Their Applications. Curr. Opin. Colloid Interf. Sci. 14, 340–348. 10.1016/j.cocis.2009.07.002 [DOI] [Google Scholar]
- Zhao X., Pan F., Lu J. R. (2008). Recent Development of Peptide Self-Assembly. Prog. Nat. Sci. 18, 653–660. 10.1016/j.pnsc.2008.01.012 [DOI] [Google Scholar]