Abstract
Genetic testing is widely used in breast cancer and has identified a lot of susceptibility genes and single nucleotide polymorphisms (SNPs). However, for many SNPs, evidence of an association with breast cancer is weak, underlying risk estimates are imprecise, and reliable subtype-specific risk estimates are not in place. A recent genome-wide long non-coding RNA (lncRNA) association study in Chinese Han has verified a genetic association between rs12537 and breast cancer. This study is aimed at investigating the association between rs12537 and the phenotype. We collected the clinical information of 5,634 breast cancer patients and 6,308 healthy controls in the early study. And χ2 test was used for the comparison between different groups in genotype. The frequency of genotypic distribution among SNP rs12537 has no statistically significant correlation with family history (p = 0.8945), menopausal status (p = 0.3245) or HER-2 (p = 0.2987), but it is statistically and significantly correlated with ER (p = 0.004006) and PR (p = 0.01379). Most importantly, compared to the healthy control, rs12537 variant is significantly correlated with ER positive patients and the p-value has reached the level of the whole genome (p = 1.66E-08 <5.00E-08). Furthermore, we found rs12537 associated gene MTMR3 was lower expressed in breast cancer tissues but highly methylated. In conclusion, our findings indicate that rs12537 is a novel susceptibility gene in ER positive breast cancer in Chinese Han population and it may influence the methylation of MTMR3.
Keywords: breast cancer, rs12537, phenotype, estrogen receptor, MTMR3
Introduction
The burden of breast cancer is increasing worldwide. Among the 19.3 million new cases reported by the GLOBOCAN 2020, breast cancer patients account for 11.7% (1). China is undergoing cancer transition with an increasing burden of breast cancer, and the incidence of breast cancer arrives at 18.41%. In China, female breast cancer patients took up approximately 18% of breast cancer deaths across the world (2). Many sequencing methods such as genome-wide association studies (GWASs), exome and lncRNA sequencing are used to identify SNPs/loci/genes related to the occurrence, development, prognosis and drug resistance of breast cancer (3–7). Breast cancer is a heterogeneous and polygenic disease, and breast cancer susceptibility SNPs and genes are closely related to molecular subtype and clinical phenotypes (8–10). However, for many SNPs, evidence of an association with cancer is often weak, and accurate estimates of the cancer risks associated with variants are often not available (11). In our previous study, we performed a genome-wide lncRNA association study, and reported a suggestive SNP, rs12537 (p = 8.84E-07), which may be associated with breast cancer susceptibility (12). rs12537 variant was reported to be associated with IgA nephropathy in Han Chinese, and rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE) in Egyptian patients (13, 14). Moreover, rs12537 variant was also found associated with significantly increased gastric cancer risk (15, 16). However, the relationship between rs12537 and breast cancer remains unknown.
To investigate the association between rs12537 on 22q12.2 and breast cancer susceptibility as well as the clinical phenotype including familial history, menopausal status, estrogen receptor (ER), progestogen receptor (PR), human epidermal growth factor receptor 2 (HER-2) and molecular subtypes of breast cancer patients in Chinese Han population, we conducted a genotype-phenotype analysis to clarify the association of rs12537 with breast cancer phenotypes in Chinese Han population. Moreover, we tried to analyze rs12537 associated genes and breast cancer based on public databases.
Materials and Methods
Subjects
We collected the genotyping data from our previous data (including GWAS stage data and replication stage genotyping data) and clinical data (including age of onset, family history, menopausal status, ER, PR and HER-2) of a total of 5,634 patients (12). Immunohistochemical analysis was employed to evaluate the ER, PR and HER-2 status of breast tissue of biopsies. Each case was diagnosed and confirmed by at least two oncologists. And their clinical information was collected by investigators with a comprehensive clinical check-up. We also collected the genotyping data and age of 6,308 healthy controls, and they were clinically determined to be free of breast cancer, other neoplastic disease, systemic disorders, and to have no family history of cancer (including first-, second- and third-degree relatives). All participants provided written informed consent. This study was approved by the institutional ethics committee of each hospital and was conducted according to the Declaration of Helsinki principles.
Statistical Analysis
To identify which phenotypes were associated with the specific SNP rs12537, we performed case-control and case-only analysis to examine the risk conferred by the suggestive SNP on different phenotypes of breast cancer. PLINK1.07 software (developed by Christopher Chang and others) and SPSS16.0 (IBM, https://www.ibm.com) were used to perform chi-square test and logistic regression analysis to explore the correlation between rs12537 and breast cancer susceptibility, as well as the different phenotypes of breast cancer. Allele frequency and genotype frequency were calculated by direct counting method, χ2 significance test was carried out, and the relative risk was evaluated by Odds ratio (OR) and 95% confidence interval (95% CI), with the difference being statistically significant (p < 0.05), a remarkable deviation from Hardy-Weinberg equilibrium in the controls (p > 0.05) during each stage.
Results
Sample Characteristics
All subjects involved in this study were from our early genome-wide lncRNA association study (12). The clinical features of 5,634 cases and 6,308 controls are intact in this study. The average age of onset of 5,634 female breast cancer patients was 50.7 ± 11.1. 4.70% (265) of these patients had familial history of cancer, 50.15% (2,118) were diagnosed with premenopausal breast cancer, 65.05% (2,773) were ER positive, 63.44% (2,704) were PR positive, and 26.99% (1,148) were HER-2 positive. For molecular subtypes, data of 1,538 of these patients were missing, 26.56% (1,088), 34.55% (1,415) and 15.38% (630) were lumina A, lumina B and HER-2 amplified breast cancer carriers, respectively, and 23.51% (963) were diagnosed with basal-like breast cancer. The average age of 6,308 female healthy controls was 47.4 ± 12.8 (Table 1).
Table 1.
Baseline characteristics of breast cancer patients and healthy controls.
| Characteristics | Total Samples | GWAS Samples | Replication Samples |
|---|---|---|---|
| Cases | |||
| Sample size | 5,634 | 1,496 | 4,138 |
| Mean age at onset (SD) | 50.7 ± 11.1 | 49.3 ± 10.9 | 51.2 ± 11.2 |
| Mean age (SD) | 51.0 ± 11.1 | 50.0 ± 10.9 | 51.3 ± 11.1 |
| Familial history of cancer | |||
| Familial (%) | 265 (4.70%) | 24 (1.60%) | 241 (5.82%) |
| Sporadic (%) | 5,369 (95.30%) | 1472 (98.40%) | 3,897 (94.18%) |
| Menopausal status | 4,223 | 849 | 3,374 |
| Premenopausal (%) | 2,118 (50.15%) | 471 (55.48%) | 1,647 (48.81%) |
| Postmenopausal (%) | 2,105 (49.85%) | 378 (44.52%) | 1,727 (51.19%) |
| ER | 4,263 | 703 | 3,560 |
| Positive (%) | 2,773 (65.05%) | 456 (64.86%) | 2,317 (65.08%) |
| Negative (%) | 1,490 (34.95%) | 247 (35.14%) | 1,243 (34.92%) |
| PR | 4,262 | 702 | 3,560 |
| Positive (%) | 2,704 (63.44%) | 413 (58.83%) | 2,291 (64.35%) |
| Negative (%) | 1,558 (36.56%) | 289 (41.17%) | 1,269 (35.65%) |
| HER-2 | 4,254 | 698 | 3,556 |
| Positive (%) | 1,148 (26.99%) | 136 (19.48%) | 1,012 (28.46%) |
| Negative (%) | 3,106 (73.01%) | 562 (80.52%) | 2,544 (71.54%) |
| Molecular subtypes | 4,096 | 687 | 3,409 |
| Luminal A breast cancer | 1,088 (26.56%) | 207 (30.13%) | 881 (25.84%) |
| Lumina B breast cancer | 1,415 (34.55%) | 244 (35.52%) | 1,171 (34.35%) |
| HER-2 amplified breast cancer | 630 (15.38%) | 93 (13.54%) | 537 (15.75%) |
| Basal-like breast cancer | 963 (23.51%) | 143 (20.82%) | 820 (24.05%) |
| Controls | |||
| Sample size | 6,308 | 1,257 | 5,051 |
| Mean age (SD) | 47.4 ± 12.8 | 37.2 ± 11.9 | 50.3 ± 10.6 |
Genotypic and Phenotype Analysis
To further explore the relationship between suggestive SNP rs12537 and breast cancer susceptibility, we combined our GWAS and replication data to perform a genotypic and phenotype analysis based on the clinical information we collected. The results show that the suggestive SNP rs12537 is not related to the familial history of cancer, menopausal status, HER-2 and the four molecular subtypes of breast cancer patients. And there is a statistical difference between PR positive patients and PR negative patients (p = 0.01379, OR = 0.8536, 95% CI: 0.7525–0.9638) in rs12537 variant (Table 2 and Supplementary Table 1). And we also performed genotypic and phenotype analysis on the other three SNPs, rs9397435, rs11066150 and rs62112521 in Chinese Han women (12), but we found no correlation between these three SNPs and the clinical characteristics of breast cancer (Supplementary Table 2).
Table 2.
The genotypic and allelic frequency of rs12537.
| Groups | Allele frequency (%) | p-value | OR (95% CI) | p-hwe | ||
|---|---|---|---|---|---|---|
| T | C | |||||
| Family history | Positive vs. Control | 0.175 | 0.205 | 0.1095 | 0.8226 (0.6475-1.045) | 0.6475 |
| Negative vs. Control | 0.1774 | 0.205 | 1.44E-06 | 0.8362 (0.7775–0.8994) | 0.7775 | |
| Positive vs. Negative | 0.175 | 0.1774 | 0.8945 | 0.9837 (0.7719–1.254) | 0.7719 | |
| Menopausal status | Positive vs. Control | 0.1751 | 0.205 | 9.89E-05 | 0.8231 (0.7462–0.908) | 0.7462 |
| Negative vs. Control | 0.1843 | 0.205 | 0.008677 | 0.8764 (0.7941–0.9672) | 0.7941 | |
| Positive vs. Negative | 0.1751 | 0.1843 | 0.3245 | 0.9393 (0.8292–1.064) | 0.8292 | |
| ER | Positive vs. Control | 0.1664 | 0.205 | 1.66E-08 | 0.7744 (0.7085–0.8464) | 0.7085 |
| Negative vs. Control | 0.1938 | 0.205 | 0.2052 | 0.9321 (0.8360–1.0390) | 0.836 | |
| Positive vs. Negative | 0.1664 | 0.1938 | 0.004006 | 0.8309 (0.7323–0.9427) | 0.7323 | |
| PR | Positive vs. Control | 0.1678 | 0.205 | 6.13E-08 | 0.7822 (0.7155–0.8550) | 0.7155 |
| Negative vs. Control | 0.1911 | 0.205 | 0.114 | 0.9163 (0.8222–1.021) | 0.8222 | |
| Positive vs. Negative | 0.1678 | 0.1911 | 0.01379 | 0.8536 (0.7525–0.9683) | 0.7525 | |
| HER-2 | Positive vs. Control | 0.1832 | 0.205 | 0.02393 | 0.8699 (0.7708–0.9818) | 0.7708 |
| Negative vs. Control | 0.1728 | 0.205 | 1.14E-06 | 0.8101 (0.7441–0.8819) | 0.7441 | |
| Positive vs. Negative | 0.1832 | 0.1728 | 0.2987 | 1.0740 (0.9388–1.2280) | 0.9388 | |
Surprisingly, ER positive patients and healthy controls also differ statistically (p = 1.66E-8, OR = 0.7744, 95% CI: 0.7085–0.8464) in rs12537 variant, and this difference has reached the level of the whole genome for p <5.00E-8. Moreover, there is also a statistical difference between ER positive patients and ER negative patients (p = 0.004006, OR = 0.8309, 95% CI: 0.7323–0.9427) in rs12537 variant (Table 2). To exclude the influence of clinical features other than ER status on the results, we further analyzed and compared the clinical differences between ER positive, ER negative breast cancer patients and healthy controls (Supplementary Table 3), and we found that age, family history and HER-2 expression were not correlated with ER. So rs12537 is a novel ER positive breast cancer associated SNP variant in Chinese Han women.
rs12537 Associated Gene MTMR3 and Breast Cancer
Expression quantitative trait locus (eQTL) has become a common tool to interpret the regulatory mechanisms of the variants associated with complex traits through genome-wide association studies (GWAS) (17, 18). To identify rs12537 associated genes, based on the eQTLGen database (https://www.eqtlgen.org/), we identified 8 cis-eQTL effects genes (ASCC2, NIPSNAP1, MTMR3, ZMAT5, SEC14L3, DUSP18, SF3A1 and THOC5) and 6 trans-eQTL effects genes (FHIT, CCR7, EPHX2, LEF1, PAQR8 and TRABD2A) (Table 3).
Table 3.
eQTL analysis to identify rs12537 associated genes.
| p-value | Gene ID | Gene Symbol | Chr | Pos (hg19) | Z-score | FDR |
|---|---|---|---|---|---|---|
| 4.91E-71 | ENSG00000100325 | ASCC2a | 22 | 30209434 | −17.8204 | 0 |
| 1.08E-34 | ENSG00000184117 | NIPSNAP1a | 22 | 29964061 | −12.2859 | 0 |
| 8.72E-30 | ENSG00000100330 | MTMR3a | 22 | 30352999 | −11.3358 | 0 |
| 4.64E-23 | ENSG00000100319 | ZMAT5a | 22 | 30144972 | 9.8893 | 0 |
| 1.15E-08 | ENSG00000100012 | SEC14L3a | 22 | 30855991 | −5.707 | 6.43E-05 |
| 6.63E-08 | ENSG00000167065 | DUSP18a | 22 | 31055957 | 5.401 | 0.000253 |
| 1.42E-06 | ENSG00000099995 | SF3A1a | 22 | 30740457 | −4.8224 | 0.003981 |
| 9.10E-06 | ENSG00000100296 | THOC5a | 22 | 29926536 | 4.4376 | 0.023729 |
| 3.85E-08 | ENSG00000189283 | FHITb | 3 | 60486084 | 5.4974 | 0.000499 |
| 6.03E-08 | ENSG00000126353 | CCR7b | 17 | 38715872 | 5.418 | 0.000806 |
| 4.52E-07 | ENSG00000120915 | EPHX2b | 8 | 27375688 | 5.0458 | 0.004067 |
| 7.11E-07 | ENSG00000138795 | LEF1b | 4 | 109029406 | 4.9584 | 0.00614 |
| 1.28E-06 | ENSG00000170915 | PAQR8b | 6 | 52249397 | 4.8428 | 0.010193 |
| 8.11E-06 | ENSG00000186854 | TRABD2Ab | 2 | 85091453 | 4.4624 | 0.04903 |
cis-eQTL effects genes.
trans-eQTL effects genes.
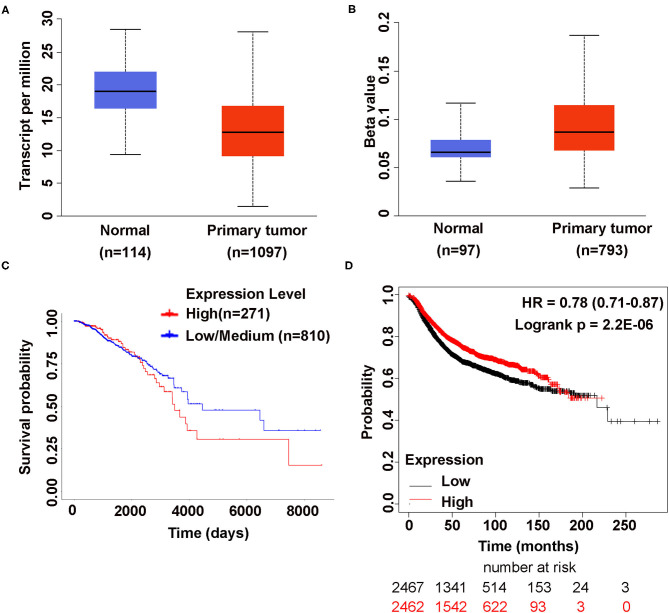
rs12537 associated gene, MTMR3, was reported to be associated with RA and SLE, gastric cancer and breast cancer (14, 15, 19). Therefore, we try to investigate the expression of MTMR3 in the cancer genome atlas (TCGA) database by UALCAN (20), discovering that compared to normal tissues MTMR3 was lower expressed in primary tumor tissues (p <1E-12), but the promoter methylation level was higher (p = 2.66E-02). MTMR3 expression was not associated with overall survival (OS) (p = 0.44) (Figures 1A–C). Moreover, based on Kaplan-Meier Plotter (www.kmplot.com) (21), we found highly expressed MTMR3 could improve patients relapse free survival (RFS) (p = 2.2E-06) (Figure 1D), but there was no correlation between MTMR3 expression and the OS, postoperative survival (PPS) as well as distant metastasis-free survival (DMFS) in breast cancer patients (Supplementary Figure 1).
Figure 1.
The Association Between MTMR3 Gene Expression and Breast Cancer. (A) MTMR3 is highly expressed in normal tissues compared to primary tumor tissues (p <1E-12). (B) MTMR3 promoter is hypermethylated in primary tumor tissues compared to normal tissues (p = 2.66E-02). The Beta value indicates level of DNA methylation ranging from 0 (unmethylated) to 1 (fully methylated). (C) MTMR3 gene expression has no correlation with the overall survival of breast cancer patients (p =0.44). (D) Lower expressed MTMR3 could reduce patient's relapse free survival (RFS) (p =2.2E-06) (KIAA0371 202197_at).
Discussion
Breast cancer is a complex multifactorial disease, with high incidence, strong invasiveness, metastasis and heterogeneity (1, 22, 23). A large number of sequencing studies have identified more than 200 susceptibility SNPs/genes (24). By combining sequencing analysis with the clinical characteristics of breast cancer patients, more SNPs/genes that have a stronger correlation with clinical characteristics were identified, which provides important theoretical support for precision treatment of breast cancer (8, 11, 25).
It is reported that more than 60% of breast cancers, including Luminal A and Luminal B breast cancers, were ER positive (8, 10). And ER positive breast cancer is a highly heterogeneous disease comprising different histological and mutational patterns, with varied clinical courses and responses to systemic treatment. GWASs have identified a lot of ER positive breast cancer associated SNPs, such as rs112545418, rs17132398 in 4p16, rs116638271, rs77274510 and rs117564384 in 11q13 and rs10941679 in 5p12 (26, 27). In our previous study, we designed a lncRNA array independently, and then performed the first genome-wide lncRNA association study on Han Chinese women, identifying a novel breast cancer-associated susceptibility SNP, rs11066150, a previously reported SNP, rs9397435 and two suggestive SNPs rs12537 and rs62112521 (12), but our study revealed that rs11066150, rs9397435 and rs6211252 had no relationship with the clinical characteristics of breast cancer (Supplementary Table 2). In the present research, we identified rs12537 as a novel susceptibility SNP in ER positive breast cancer in Han Chinese women. And this is the first time that rs12537 has been reported to be associated with ER positive breast cancer. However, only 4,263 ER patients (65.05% ER positive, 34.56% ER negative) and 6,308 healthy controls were included in this study, and a larger and better-matched population (including age, familial history, menopausal status, ER, PR and HER-2) may be needed for further verification.
The SNP rs12537 present in the miR-181a-binding site in the 3' UTR of the MTMR3 gene (15) and T/C variant in MTMR3 were reported to be associated with IgA nephropathy, RA, SLE and gastric cancer (13–16). As an autophagy-related gene involved in the negative regulation of autophagy initiation (24), rs12537 T/T carriers were associated with lower serum MTMR3 expression and higher miR-181a expression than in other genotypes among SLE patients, and their interaction may lead to autophagy increasing (14). rs12537 CT genotype carriers in gastric cancer had low MTMR3 mRNA expression than CC genotype carriers (15). Ectopic expression of miR-181a mimics or introduction of MTMR3 small interfering RNA resulted in an increase in cell proliferation, colony formation, migration, invasion, as well as suppression of apoptosis in gastric cancer (28).
DNA methylation plays a crucial role in the formation and process of cancers and it could be potential candidate biomarkers for cancers (29). Based on TCGA database, we found that MTMR3 gene was lower expressed in breast cancer tissues than normal tissues and the promoter methylation level was higher. However, MTMR3 expression had no correlation with overall survival. Here, we hypothesize that rs12537 variant in ER positive breast cancer patients could regulate the methylation of MTMR3, and further studies are required to fully understand the mechanism.
In conclusion, the results of our study show that rs12537 is a novel susceptibility SNP in ER positive breast cancer in Chinese Han population. Moreover, rs12537 associated gene MTMR3 is lowly expressed but highly methylated in breast cancer. Considering that we have not found the correlation between MTMR3 expression and overall survival based on TCGA database, multicentric studies involving a larger number of cases and genotypic data are needed to verify this result.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by this study was approved by the Ethics Committee of Anhui Medical University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
JX, YC, and BZ conceived of the idea. JX, GL, and MC conducted statistical analyses. WL and YW collected clinical data. JX and GL wrote the manuscript with inputs from all authors. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank all the participating patients and healthy controls, as well as all the doctors and nurses who have contributed to this work.
Footnotes
Funding. This work was supported by Talent Funding of the First Affiliated Hospital of Anhui Medical University (1519) and Natural Science Foundation of Higher Education of Anhui Province of China (KJ2017A181).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.708644/full#supplementary-material
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J. (2021) 134:783–91. 10.1097/CM9.0000000000001474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. (2013) 154:26–46. 10.1016/j.cell.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milne RL, Kuchenbaecker KB, Michailidou K, Beesley J, Kar S, Lindstrom S, et al. Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat Genet. (2017) 49:1767–78. 10.1038/ng.3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart RL, Updike KL, Factor RE, Henry NL, Boucher KM, Bernard PS, et al. A multigene assay determines risk of recurrence in patients with triple-negative breast cancer. Cancer Res. (2019) 79:3466–78. 10.1158/0008-5472.CAN-18-3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang B, Chen MY, Shen YJ, Zhuo XB, Gao P, Zhou FS, et al. A large-scale, exome-wide association study of han chinese women identifies three novel loci predisposing to breast cancer. Cancer Res. (2018) 78:3087–97. 10.1158/0008-5472.CAN-17-1721 [DOI] [PubMed] [Google Scholar]
- 7.Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. (2018) 50:1705–15. 10.1038/s41588-018-0252-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y, Chen M, Liu C, Zhang X, Li W, Cheng H, et al. Association study confirmed three breast cancer-specific molecular subtype-associated susceptibility loci in chinese han women. Oncologist. (2017) 22:890–4. 10.1634/theoncologist.2016-0423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtit E, Pivot X, Henriques J, Paget-Bailly S, Fumoleau P, Rios M, et al. Assessment of the prognostic role of a 94-single nucleotide polymorphisms risk score in early breast cancer in the SIGNAL/PHARE prospective cohort: no correlation with clinico-pathological characteristics and outcomes. Breast Cancer Res. (2017) 19:98. 10.1093/annonc/mdw364.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Early Breast Cancer Trialists' Collaborative G. Davies C, Godwin J, Gray R, Clarke M, Cutter D, et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. (2011) 378:771–84. 10.1016/S0140-6736(11)60993-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorling L, Carvalho S, Allen J, González-Neira A, Luccarini C, Wahlström C, et al. Breast cancer risk genes - association analysis in More than 113,000 women. N Engl J Med. (2021) 384:428–39. 10.1056/NEJMoa1913948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu JK, Li GZ, Li Z, Li WJ, Chen RS, Zhang B, et al. Genome-wide long non-coding RNA association study on Han Chinese women identifies lncHSAT164 as a novel susceptibility gene for breast cancer. Chin Med J (Engl). (2021) 134:1138–45. 10.1097/CM9.0000000000001429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu XQ, Li M, Zhang H, Low HQ, Wei X, Wang JQ, et al. A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat Genet. (2011) 44:178–82. 10.1038/ng.1047 [DOI] [PubMed] [Google Scholar]
- 14.Senousy MA, Helmy HS, Fathy N, Shaker OG, Ayeldeen GM. Association of MTMR3 rs12537 at miR-181a binding site with rheumatoid arthritis and systemic lupus erythematosus risk in Egyptian patients. Sci Rep. (2019) 9:12299. 10.1038/s41598-019-48770-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y, Nie Y, Zhao J, Chen X, Ye M, Li Y, et al. Genetic polymorphism at miR-181a binding site contributes to gastric cancer susceptibility. Carcinogenesis. (2012) 33:2377–83. 10.1093/carcin/bgs292 [DOI] [PubMed] [Google Scholar]
- 16.Cipollini M, Landi S, Gemignani F. MicroRNA binding site polymorphisms as biomarkers in cancer management and research. Pharmgenomics Pers Med. (2014) 7:173–91. 10.2147/PGPM.S61693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. (2016) 48:481–7. 10.1038/ng.3538 [DOI] [PubMed] [Google Scholar]
- 18.Karlsson Linner R, Biroli P, Kong E, Meddens SFW, Wedow R, Fontana MA, et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. (2019) 51:245–57. 10.1038/s41588-018-0309-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B, Mao JH, Wang BY, Wang LX, Wen HY, Xu LJ, et al. Exosomal miR-1910-3p promotes proliferation, metastasis, and autophagy of breast cancer cells by targeting MTMR3 and activating the NF-kappaB signaling pathway. Cancer Lett. (2020) 489:87–99. 10.1016/j.canlet.2020.05.038 [DOI] [PubMed] [Google Scholar]
- 20.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. (2017) 19:649–58. 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. (2010) 123:725–31. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- 22.Long J, Cai Q, Sung H, Shi J, Zhang B, Choi JY, et al. Genome-wide association study in east Asians identifies novel susceptibility loci for breast cancer. PLoS Genet. (2012) 8:e1002532. 10.1371/journal.pgen.1002532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindstrom S, Thompson DJ, Paterson AD, Li J, Gierach GL, Scott C, et al. Genome-wide association study identifies multiple loci associated with both mammographic density and breast cancer risk. Nat Commun. (2014) 5:5303. 10.1038/ncomms6303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapoor PM, Lindstrom S, Behrens S, Wang X, Michailidou K, Bolla MK, et al. Assessment of interactions between 205 breast cancer susceptibility loci and 13 established risk factors in relation to breast cancer risk, in the breast cancer association consortium. Int J Epidemiol. (2020) 49:216–32. 10.1093/ije/dyz193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol. (2016) 18:213–24. 10.1038/ncb3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruiz-Narvaez EA, Sucheston-Campbell L, Bensen JT, Yao S, Haddad S, Haiman CA, et al. Admixture mapping of African-American women in the AMBER consortium identifies new loci for breast cancer and estrogen-receptor subtypes. Front Genet. (2016) 7:170. 10.3389/fgene.2016.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghoussaini M, French JD, Michailidou K, Nord S, Beesley J, Canisus S, et al. Evidence that the 5p12 variant rs10941679 confers susceptibility to estrogen-receptor-positive breast cancer through FGF10 and MRPS30 regulation. Am J Hum Genet. (2016) 99:903–11. 10.1016/j.ajhg.2016.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Y, Zhao J, Wang H, Cao J, Nie Y. miR-181a modulates proliferation, migration and autophagy in AGS gastric cancer cells and downregulates MTMR3. Mol Med Rep. (2017) 15:2451–6. 10.3892/mmr.2017.6289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Men C, Chai H, Song X, Li Y, Du H, Ren Q. Identification of DNA methylation associated gene signatures in endometrial cancer via integrated analysis of DNA methylation and gene expression systematically. J Gynecol Oncol. (2017) 28:e83. 10.3802/jgo.2017.28.e83 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.