Abstract
Increasing evidence has indicated that microRNAs (miRNAs/miRs) play an important role in the occurrence and development of various types of cancer. The aim of the present study was to investigate the role and underlying molecular mechanisms of miR-195-5p in laryngeal cancer cell proliferation, migration and invasion. Reverse transcription-quantitative PCR (RT-qPCR) was performed to measure the expression levels of miR-195-5p in laryngeal carcinoma cell lines. The expression levels of miR-195-5p and E2F transcription factor 3 (E2F3) were modified by transfection with miR-195-5p mimics and pcDNA3.1-E2F3. A luciferase reporter assay was used to verify the association between miR-195a-5p and E2F3. Cell Counting Kit-8, cell wound healing and Transwell invasion assays were used to detect the biological functions of laryngeal cancer cells. The expression of epithelial-mesenchymal transition (EMT)-associated genes was evaluated by western blotting and RT-qPCR. The results revealed that the expression of miR-195-5p was decreased in laryngeal cancer cell lines. The overexpression of miR-195-5p inhibited the proliferation, migration, invasion and EMT of laryngeal cancer cells. Dual-luciferase reporter assays revealed that miR-195-5p could directly target E2F3 and that there was a negative association between them. E2F3 overexpression significantly attenuated the inhibitory effects of the overexpression of miR-195-5p on the proliferation, migration, invasion and EMT of laryngeal cancer cells. Collectively, the findings of the present study demonstrated that the overexpression of miR-195-5p significantly inhibited the progression of laryngeal cancer cells, and these effects may be mediated via the downregulation of the expression of E2F3.
Keywords: microRNA-195-59, E2F3, laryngeal cancer, proliferation, migration, invasion
Introduction
Laryngeal cancer is a common tumour type with the highest degree of malignancy of head and neck tumours and is associated with high mortality and morbidity rates (1). The occurrence and development of laryngeal cancer is a multifactorial and multistep complex process, and may be associated with several complications, including dyspnoea, coughing and dysphagia, which severely affect the quality of life of the patients and render perioperative intervention difficult (2,3). At present, surgery and radiotherapy are the main treatment strategies for laryngeal cancer, despite major advances in therapeutic techniques. However, as the early symptoms of laryngeal cancer are not obvious, early diagnosis and treatment remain a challenge (4,5). Therefore, elucidating the molecular pathogenesis of laryngeal cancer is of utmost importance for the development of novel treatment strategies.
Studying the genome of laryngeal cancer is helpful for identifying novel therapeutic targets and understanding the molecular pathology of the disease. MicroRNAs (miRNAs/miRs) are a type of small single-stranded non-coding RNA molecules that regulate the proliferation, differentiation and apoptosis of multiple types of cells, and play an important role in the development of multiple tumours, such as esophageal squamous cell carcinoma, breast cancer, pancreatic cancer and laryngeal cancer (6-9). It has been reported that a variety of miRNAs are abnormally expressed in laryngeal cancer cells and serve as important biomarkers in laryngeal cancer and other head and neck tumours (10-12). For example, the expression of miR-21-5p is upregulated in laryngeal cancer cells, and promotes the proliferation, migration and invasion of laryngeal cancer cells by inhibiting the expression of laryngeal oncogene laryngeal carcinoma-related gene 1(13). Moreover, miR-34a and miR-34c have been shown to inhibit the proliferation of laryngeal cancer cells by negatively regulating polypeptide N-acetylgalactosaminyltransferase 7 in HEp-2 cells (14). In addition, the upregulation of miR-375 can suppress the proliferation and invasion of squamous cell laryngeal carcinoma (15). miR-195 is one of the most important members of the miR-15/16 family and is widely considered to exert antitumor effects. Studies have demonstrated that miR-195-5p is expressed at low levels in several types of cancer, such as melanoma, cervical cancer and colorectal cancer, which may be associated with tumour resistance, metabolism and proliferation (16-18). However, the role and mechanisms of miR-195-5p in laryngeal cancer remain unclear.
The present study confirmed the role of miR-195-5p in laryngeal cancer by detecting the expression of miR-195-5p in laryngeal cancer cell lines, as well as its effects on the proliferation, migration and invasion of laryngeal cancer cells. In addition, through database search, it was found that E2F3 may be a potential target gene of miR-195-5p and the effects of miR-195-5p on the biological function of laryngeal cancer cells via the regulation of E2F3 were analysed. The results may provide a theoretical basis for the important role of miR-195-5p/E2F3 in the diagnosis and treatment of laryngeal cancer.
Materials and methods
Cells and cell culture
A normal immortalized nasopharyngeal epithelial cell line (NP-69), laryngeal carcinoma cell lines (AMC-HN-8 and SNU-899) and oropharyngeal squamous cell line (M4E) were purchased from the The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. NP-69, AMC-HN-8 and M4E cells were cultured in DMEM (HyClone; Cytiva) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). SNU-899 cells were cultured in RPMI-1640 medium containing 10% FBS and 2% glutamine (Gibco; Thermo Fisher Scientific, Inc.). All cells were cultured in a 37˚C humidified incubator with 5% CO2. Cells in the logarithmic growth phase were used in subsequent experiments.
Reverse transcription-quantitative PCR (RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract total RNA from the cells according to the manufacturer's instructions. RNA (2 µg) was transcribed into cDNA using the PrimeScript RT Reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd.). The RT conditions were as follows: 70˚C for 5 min, 37˚C for 5 min and 42˚C for 1 h. qPCR was conducted using an ABI 7300 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The miRNA expression levels were quantified using the miScript SYBR-Green PCR kit (Toyobo Life Science). The qPCR reaction conditions were as follows: Pre-denaturation at 95˚C for 5 min, followed by 36 cycles of denaturation at 95˚C for 15 sec, annealing at 60˚C for 30 sec and extension 72˚C for 30 sec. Relative expression was calculated using the 2-ΔΔCq method (19). U6 or GAPDH were used as the internal references. The primer sequences were: miR-195-5p forward, 5'-GAATTCGCCTCAAGAGAACAAAGTGGAG-3'; miR-195-5p reverse, 5'-AGATCTCCCATGGGGGCTCAGCCCCT-3'; E2F3 forward, 5'-CCCTAAACCCGCTTCC-3'; E2F3 reverse, 5'-GTTCACAAACGGTCCTTCTA-3'; E-cadherin forward 5'-CGAGAGCTACACGTTCACGG-3'; E-cadherin reverse 5'-GGGTGTCGAGGGAAAAATAGG-3'; vimentin forward, 5'-GACGCCATCAACACCGAGTT-3'; vimentin reverse, 5'-CTTTGTCGTTGGTTAGCTGGT-3'; N-cadherin forward, 5'-AGCTCCATTCCGACTTAGACA-3'; N-cadherin reverse, 5'-CAGCCTGAGCACGAAGAGTG-3'; matrix metalloproteinase (MMP)2 forward, 5'-GGCCAGGTGGTATCTTAGGC-3'; MMP2 reverse, 5'-AGCTGACCAGTGTTCATTCTTG-3'; MMP9 forward, 5'-GGGACGCAGACATCGTCATC-3'; MMP9 reverse, 5'-TCGTCATCGTCGAAATGGGC-3'; snail forward, 5'-TCGGAAGCCTAACTACAGCGA-3'; snail reverse, 5'-AGATGAGCATTGGCAGCGAG-3'; GAPDH forward, 5'-AGGTGGTCTCCTCTGACTTCAA-3'; GAPDH reverse, 5'-TTCGTTGTCATACCAGGAAATG-3'; U6 forward, 5'-GCTTCGGCAGCACATATACTAAAAT-3'; and U6 reverse, 5'-CGCTTCACGAATTTGCGTGTCAT-3'.
Cell transfection
miR-195-5p mimic and the negative control miRNA (miRNA-NC) were obtained from Guangzhou RiboBio Co., Ltd. The E2F3 overexpression vector pcDNA-E2F3 and empty control vector pcDNA-NC were constructed by Shanghai GenePharma Co., Ltd. The sequences were as follows: miR-195-5p mimic sense, 5'-UAGCAGCACAGAAAUAUUGGC-3'; miR-195-5p mimic antisense, 5'-CAAUAUUUCUGUGCUGCUAUU-3'; mimics negative control (miR-NC) sense, 5'-UUCUCCGAACGUGUCACGUTT-3'; miR-NC antisense, 5'-ACGUGACACGUUCGGAGAATT-3'. Cells were plated into 6-well plates (1x106 cells per well), and transfection was performed when cells at the logarithmic growth phase reached 80% confluence. According to the product instructions, AMC-HN-8 cells were transfected with miR-195-5p mimic (50 nM), miR-NC (50 nM), pcDNA-E2F3 (100 nM) and pcDNA-NC (100 nM) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The transfection efficiencies were assessed by RT-qPCR at 48 h after transfection.
Cell counting kit-8 (CCK-8) assay
Cell proliferation was detected using a CCK-8 assay (Dojindo Molecular Technologies, Inc.) according to the manufacturer's instructions. AMC-HN-8 cells in each group were inoculated into the 96-well plate at a density of 2x104 cells/ml and then cultured for 24, 48 and 72 h. Subsequently, 10 µl CCK-8 solution were added to each well followed by routine culture for 2 h at 37˚C. The optical density at a 480 nm wavelength was measured using a microplate reader (BioTek Instruments, Inc.).
Wound-healing assay
AMC-HN-8 cells were inoculated in 6-well plates at a density of 2x105 cells/well. A linear scratch was created on the surface of the cell monolayer using a sterile 20-µl pipette tip perpendicular to the 6-well plate after the cell growth density had reached ~100%. The surface of cell culture plate was washed twice with phosphate buffered solution (PBS). The cells were then routinely cultured in serum-free DMEM. After 24 h, the cell migration rate was calculated under an inverted light microscope (magnification, x100; Olympus Corporation). Quantitative analysis of the wound healing area was performed using ImageJ software (version 1.52r; National Institutes of Health). The relative cell migration rate of each group (distance at 24 h-initial distance) was normalized according to the average migrated distance of the control.
Transwell assay
A Transwell chamber (8 µm; BD Biosciences) was used to detect the invasive ability of the AMC-HN-8 cells. Cells (1x105 cells/ml) were added to the upper Transwell chamber coated with Matrigel (BD Biosciences) and cultured in serum-free DMEM. DMEM (800 µl) containing 10% FBS was added to the lower chamber. Following culture at room temperature for 24 h, the cells were fixed with 4% formaldehyde for 40 min at room temperature and then stained with 0.5% crystal violet for 1 h at room temperature. The number of invading cells were observed and counted in four representative fields using an inverted light microscope (magnification, x100; Olympus Corporation).
Luciferase reporter assay
E2F3 was predicted to be the target gene of miR-195-5p by TargetScan (www.targetscan.org) and ENCORI (http://starbase.sysu.edu.cn/), which was in accordance with the previous study (20). Firstly, wild-type (WT) and mutant (MUT) E2F3 3'untranslated region (UTR) luciferase reporter vectors were constructed by Shanghai GenePharma Co., Ltd.. The AMC-HN-8 cells were inoculated in 24-well plates at a density of 2x105 cells/well. miR-195-5p mimics or negative control and the WT and MUT 3'UTR of E2F3 were co-transfected into the AMC-HN-8 cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h following transfection, the luciferase activity was detected strictly according to the instructions provided with the Dual-Luciferase reporter assay kit (Promega Corporation). Firefly luciferase activity was normalized to Renilla luciferase activity.
Western blot analysis
Cells were lysed with RIPA lysis buffer (Beyotime Institute of Biotechnology) and the protein concentration was determined using a BCA protein assay kit (Thermo Fisher Scientific, Inc.). The proteins (40 µg/lane) were separated by 10% SDS-PAGE and then transferred to PVDF membranes (EMD Millipore). Skimmed milk (5%, dissolved in PBS) was used to block the PVDF membranes for 2 h at 37˚C. The membranes were then incubated with primary antibodies against E-cadherin (1:1,000; cat. no. 14472), vimentin (1:1,000; cat. no. 5741), N-cadherin (1:1,000; cat. no. 13116), snail (1:1,000; cat. no. 3879T), MMP-2 (1:1,000; cat. no. 40994), MMP-9 (1:1,000; cat. no. 13667) and GAPDH (1:1,000; cat. no. 5174) (all from Cell Signaling Technology, Inc.) at 4˚C overnight. After washing the membranes twice with PBS for 10 min each time, they were incubated with HRP-conjugated secondary antibody (1:5,000; cat. no. sc-2357; Santa Cruz Biotechnology, Inc.) for 1.5 h at room temperature. The protein bands were detected using ECL detection reagent (Cytiva). GAPDH was used as an internal reference. The intensity of the bands was semi-quantified using ImageJ software (version 1.52r; National Institutes of Health). Experiments were performed in triplicate followed by statistical analysis.
Statistical analysis
All experiments were repeated at least three times. SPSS software 20.0 (IMB Corp.) was used for the statistical analysis of data and all the experimental data are expressed as the mean ± standard deviation (SD). An unpaired Student's t-test was used for comparison between two groups, and comparisons among more than two groups were analyzed using the one-way analysis of variance followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-195-5p expression is downregulated in laryngeal cancer cell lines
Firstly, the expression of miR-195-5p in laryngeal carcinoma cell lines was detected by RT-qPCR. As shown in Fig. 1, the expression of miR-195-5p in the laryngeal cancer cell lines was significantly lower compared with that in the normal cell line. Of note, the expression of miR-195-5p was the lowest in the AMC-HN-8 cells. Therefore, the AMC-HN-8 cells were used in the subsequent experiments. These results indicated that miR-195-5p was expressed at low levels in laryngeal cancer cell lines.
Figure 1.
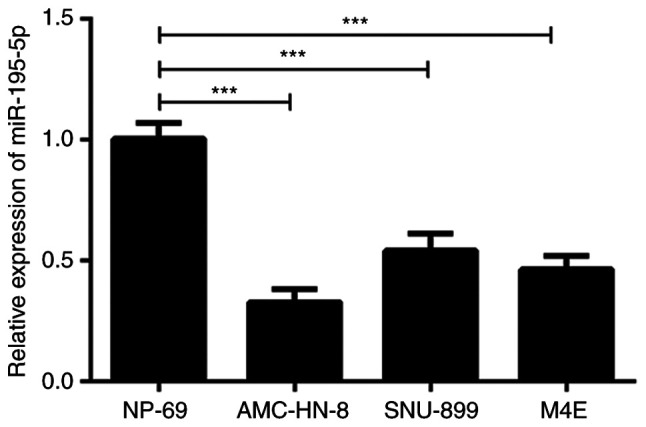
miR-195-5p expression is downregulated in laryngeal cancer cell lines. The expression of miR-195-5p in laryngeal carcinoma cell lines (AMC-HN-8 and SNU-899), oropharyngeal squamous cell line (M4E) and one normal immortalized nasopharyngeal epithelial cell line (NP-69) was determined with reverse transcription-quantitative PCR. Data are presented as the mean ± standard deviation. ***P<0.001. miR, microRNA.
miR-195-5p overexpression inhibits the proliferation, migration and invasion of AMC-HN-8 cells
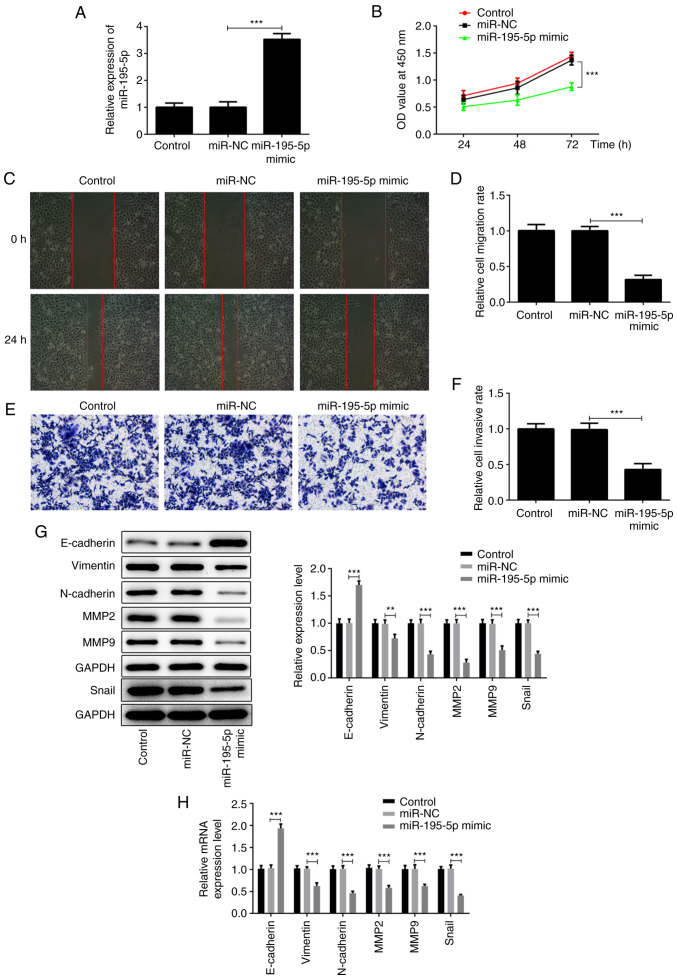
Due to the low expression of miR-195-5p in the AMC-HN-8 laryngeal cancer cell line, the expression of miR-195-5p was modified by transfection with miR-195-5p overexpression plasmid. As shown in Fig. 2A, the expression of miR-195-5p in the miR-195-5p mimic group was significantly higher compared with that of the miR-NC group. The aforementioned results indicated that the miR-195-5p overexpression plasmid was successfully constructed. Subsequently, CCK-8, wound healing and Transwell assays, and western blot analysis, were used to examine the effects of miR-195-5p overexpression on the proliferation, migration and invasion of laryngeal cancer cells. Pang et al (21) reported that miR-195 is associated with the cell viability of the AMC-HN-8 cell line. In the present study, the results of CCK-8 assay revealed that miR-195-5p overexpression significantly inhibited the proliferation of AMC-HN-8 cells compared with the control group (Fig. 2B). The wound-healing assay revealed that miR-195-5p overexpression significantly inhibited the migration of the AMC-HN-8 cells compared with the control group (Fig. 2C and D). Similar results were obtained by Transwell assay; miR-195-5p overexpression significantly lowered the invasive ability of the AMC-HN-8 cells (Fig. 2E and F). In addition, the results of western blot analysis and RT-qPCR assay demonstrated that, compared with the control group, miR-195-5p overexpression significantly increased the expression of E-cadherin, while it inhibited the expression levels of vimentin, N-cadherin, snail, MMP-2 and MMP-9, indicating that miR-195-5p overexpression can significantly suppress the epithelial-mesenchymal transition (EMT) of AMC-HN-8 cells (Fig. 2G and H). Taken together, these results suggest that miR-195-5p overexpression inhibits the proliferation, migration and invasion of laryngeal cancer cells.
Figure 2.
miR-195-5p overexpression inhibits the proliferation, migration and invasion of AMC-HN-8 cells. (A) miR-195-5p overexpression efficiency was detected by RT-qPCR. (B) Cell proliferation in each group following transfection was detected by the Cell Counting Kit-8 assay. (C and D) Cell migration in each group following transfection was detected by wound-healing assays. Magnification, x100. (E and F) Cell invasion in each group following transfection was detected by Transwell assays. Magnification, x100. The levels of the epithelial-mesenchymal transition-associated proteins (E-cadherin, vimentin, N-cadherin and snail) and MMP-2 and -9 were respectively examined by (G) western blot analysis and (H) RT-qPCR assay. **P<0.01; and ***P<0.001. miR, microRNA; RT-qPCR, reverse transcription-quantitative; NC, negative control; MMP, matrix metalloprotease.
E2F3 is the direct target gene of miR-195-5p
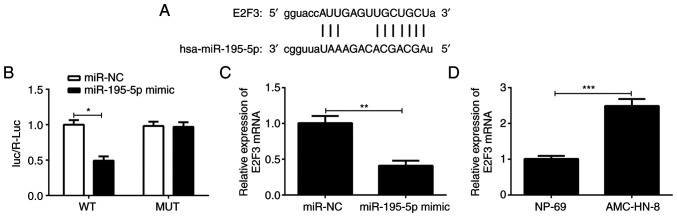
Subsequently, the target gene of miR-195-5p was predicted through the TargetScan and ENCORI website. As shown in Fig. 3A, E2F3 has potential sites for binding to miR-195-5p. Therefore, the association between miR-195-5p and E2F3 was determined using the dual-luciferase reporter assay. As shown in Fig. 3B, compared with the miR-NC group, the luciferase activity of the cells co-transfected with MUT E2F3 3'UTR reporter plasmid and miR-195-5p mimic was not altered; however, that of the cells transfected with the WT E2F3 3'UTR reporter plasmid and miR-195-5p mimic was significantly decreased. In addition, as exhibited in Fig. 3C, E2F3 expression was notably downregulated in AMC-HN-8 cells after transfection with miR-195-5p mimic compared with the miR-NC group. The results of Fig. 3D indicated that E2F3 level was markedly enhanced in AMC-HN-8 cells compared with that in the NP-69 cells. Collectively, the results indicate that E2F3 is a direct target of miR-195-5p.
Figure 3.
E2F3 is a direct target gene of miR-195-5p. (A) TargetScan and ENCORI databases were used to predict the binding sequence between miR-195-5p and E2F3. (B) Dual-luciferase reporter assay was used to analyze the association between miR-195-5p and E2F3. (C) RT-qPCR was used to evaluate the effect of miR-195-5p overexpression on E2F3 expression. (D) RT-qPCR was used to evaluate the expression of E2F3 in AMC-HC-8 cells. *P<0.05; **P<0.01; and ***P<0.001. miR, microRNA; E2F3, E2F transcription factor 3; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; WT, wild type; MUT mutant.
miR-195-5p suppresses the proliferation, migration and invasion of AMC-HN-8 cells by regulating E2F3 expression
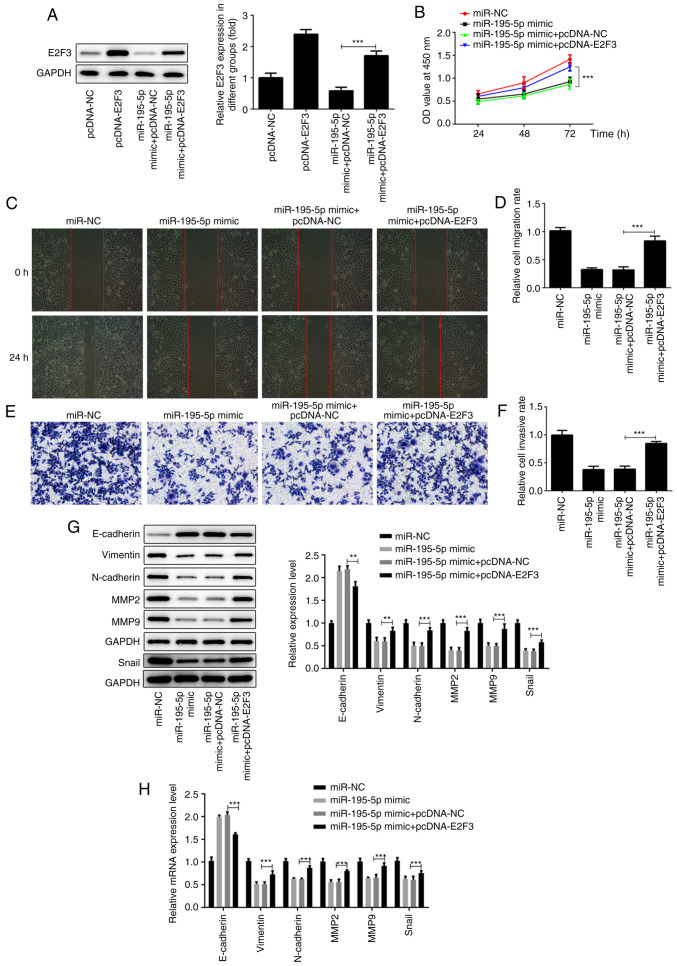
Based on the aforementioned results, it was suggested that miR-195-5p can affect the biological function of AMC-HN-8 cells by regulating E2F3. As shown in Fig. 4A, compared with the pcDNA-NC group, the expression of E2F3 in the cells transfected with pcDNA-E2F3 was significantly increased. Remarkably, cells co-transfected with miR-195-5p mimic and pcDNA-E2F3 displayed upregulated E2F3 expression compared with the miR-195-5p mimic + pcDNA-NC group. The results of the CCK-8 assay revealed that the overexpression of E2F3 in AMC-HN-8 cells transfected with miR-195-5p mimic reversed the inhibitory effect of miR-195-5p overexpression on the proliferation of AMC-HN-8 cells (Fig. 4B). The results of wound healing and Transwell assays demonstrated that the co-transfection with pcDNA-E2F3 and miR-195-5p mimic reversed the blocking effects of miR-195-5p overexpression alone on the migration and invasion of AMC-HN-8 cells (Fig. 4C-F). Similarly, the E2F3 upregulation attenuated the inhibitory effects of miR-195-5p overexpression on EMT of AMC-HN-8 cells transfected with miR-195-5p mimic, obtained by downregulated expression of E-cadherin, upregulated expression of vimentin, N-cadherin, snail, MMP-2 and MMP-9 (Fig. 4G and H). In general, the results suggest that miR-195-5p may suppress the progression of laryngeal cancer by downregulating E2F3.
Figure 4.
miR-195-5p suppresses the proliferation, migration and invasion of AMC-HN-8 cells by regulating E2F3 expression. (A) E2F3 overexpression efficiency was detected by western blot analysis. (B) miR-195-p mimic and pcDNA-E2F3 were co-transfected into AMC-HN-8 cells and cell proliferation was detected by Cell Counting Kit-8 assays. (C and D) Cell migration in each group following transfection with miR-195-5p and pcDNA-E2F3 was detected by wound-healing assays. Magnification, x100. (E and F) Cell invasion in each group following transfection with miR-195-5p and pcDNA-E2F3 was detected by Transwell assays. Magnification, x100. (G) Western blot analysis and (H) RT-qPCR assay were respectively used to measure the protein and mRNA levels of the epithelial-mesenchymal transition-associated proteins (E-cadherin, vimentin, N-cadherin and snail) and MMP-2 and -9. **P<0.01; ***P<0.001. miR, microRNA; E2F3, E2F transcription factor 3; miR, microRNA; NC, negative control; MMP, matrix metalloprotease.
Discussion
It is well known that the occurrence and development of laryngeal cancer is closely associated with a number of factors, and its pathogenesis is complex, affected and restricted by a variety of regulatory networks (22-25). miRNAs are a type of gene expression regulators in laryngeal cancer cells and play an important role in the occurrence and development of laryngeal cancer (26). Therefore, the study of miRNAs may provide new insights into the research and development of drugs for the targeted treatment of laryngeal cancer. The results of the present study revealed that the expression of miR-195-5p in laryngeal cancer cells was lower compared with that in normal cells. In addition, miR-195-5p overexpression significantly inhibited the proliferation, migration and invasion of AMC-HN-8 cells, suggesting that miR-195-5p may be a potential target for the diagnosis and treatment of laryngeal cancer.
miR-195, located on chromosome 17, is unanimously considered as an inhibitor of tumour proliferation and has broad prospects as a tumour suppressor (27). It has been demonstrated that miR-195-5p is downregulated in breast, ovarian, colorectal and liver cancer, indicating that miR-195-5p is associated with tumour cell invasion and metastasis (28). In non-small cell lung cancer, the low expression of miR-195-5p inhibited cell migration and invasion (29). Moreover, miR-195-5p overexpression has been shown to suppress the proliferation of lung cancer cells, and to induce apoptosis and G0/G1 phase arrest (30). Importantly, miR-195 is reported to be associated with regulating the pathophysiologic process of human laryngeal squamous cell carcinoma (21). In the present study, the expression of miR-195-5p in laryngeal cancer cells was found to be significantly lower compared with that in normal cells, which was consistent with the expression of miR-195-5p in other types of tumours. The proliferation, migration and invasion of AMC-HN-8 cells were significantly suppressed upon miR-195-5p mimic transfection. It is thus suggested that miR-195-5p mainly functions as a tumour suppressor, and can inhibit the proliferation, migration and invasion of laryngeal cancer cells.
EMT is the process through which the cell phenotype changes from an epithelial to a stromal phenotype, accompanied by the decreased expression of epithelial cell markers (E-cadherin) and the increased expression of stromal cell markers (N-cadherin, vimentin and snail) (31). EMT decreases cell-cell and cell-matrix adhesion, so as to enhance the ability of cell migration, invasion and metastasis (32). MMP-2 and MMP-9 are the most extensively investigated members of the MMP family, which can degrade the basement membrane and the extracellular matrix, and promote tumour cell migration and invasion (33-35). In the present study, it was demonstrated that miR-195-5p overexpression significantly promoted the expression of E-cadherin, and inhibited the expression of N-cadherin, vimentin and snail, while MMP-2 and MMP-9 were also suppressed, suggesting that miR-195-5p overexpression inhibited the EMT process in laryngeal cancer cells.
E2F3 was predicted to be the target gene of miR-195-5p by the bioinformatics algorithms miRTarBase, DIANA-tools and miRDB in the previous study (20). The transcription factor E2F3 plays an important role in cell cycle progression, tumorigenesis and cancer development. E2F3 is involved in the regulation of the cell cycle by forming dimers with cyclin D1, and is associated with a variety of carcinogenic and tumour suppressor genes (36,37). Previous studies have found that miR-449a and miR-125b decrease cell proliferation and induce apoptosis by inhibiting E2F3 (38,39). Although E2F3 showed no differential expression in colorectal cancer cells after transfection with miR-195-5p mimic (20), E2F3 was the downstream target gene of miR-195-5p and the functional mediator of miR-195-5p in laryngeal cancer cells in the present study. Further analysis indicated that miR-195-5p overexpression inhibited the proliferation, migration and invasion of laryngeal cancer cells by downregulating E2F3. The mechanism of miR-195-5p downregulation in laryngeal cancer might be associated with the regulatory effects of competing endogenous RNAs.
miR-195-5p is a type of inhibitory miRNA of laryngeal cancer. The overexpression of miR-195-5p can inhibit the proliferation, migration and invasion of laryngeal cancer cells. Further analyses revealed that miR-195-5p suppressed the activity of laryngeal cancer cells by negatively regulating E2F3. However, the present study had certain limitations. miRNAs can regulate multiple genes and one gene can also be regulated by multiple miRNAs. Therefore, there may be other genes regulated by miR-195-5p that are involved in the proliferation, migration and invasion of laryngeal cancer cells. Thus, these issues need to be resolved progressively in future research, in order to provide further in-depth knowledge of the regulatory pathway(s) of miR-195-5p. Moreover, the effect of miR-195-5p on apoptosis and cell cycle of laryngeal cancer cells will be investigated in the following studies. Furthermore, whether miR-195-5p can regulate the progression of laryngeal cancer in vivo remains to be elucidated in the next investigation.
In conclusion, the findings of the present study demonstrated that the upregulation of miR-195-5p decreases the expression of E2F3, and inhibits the proliferation, migration and invasion of laryngeal cancer cells, suggesting that the miR-195-5p/E2F3 axis may be used as a potential target for the treatment of laryngeal cancer.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
MZ and XX made substantial contributions to the conception and design of the present study. MZ and YW designed the study; MZ, YW, CZ, MQ, MY and LS performed the experiments; MZ and YW analyzed the data and wrote the manuscript. MZ and XW confirm the authenticity of all the raw dat. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Thompson LD. Laryngeal dysplasia, squamous cell carcinoma, and variants. Surg Pathol Clin. 2017;10:15–33. doi: 10.1016/j.path.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Echanique KA, Desai SV, Marchiano E, Spinazzi EF, Strojan P, Baredes S, Eloy JA. Laryngeal verrucous carcinoma: A systematic review. Otolaryngol Head Neck Surg. 2017;156:38–45. doi: 10.1177/0194599816662631. [DOI] [PubMed] [Google Scholar]
- 3.Kolator M, Kolator P, Zatoński T. Assessment of quality of life in patients with laryngeal cancer: A review of articles. Adv Clin Exp Med. 2018;27:711–715. doi: 10.17219/acem/69693. [DOI] [PubMed] [Google Scholar]
- 4.Feng W, Yang Y, Xin ZH, Ni XG. Misdiagnosis experience of laryngeal carcinoma with acute laryngitis as the first symptom. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2018;53(640) doi: 10.3760/cma.j.issn.1673-0860.2018.08.018. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 5.Obid R, Redlich M, Tomeh C. The treatment of laryngeal cancer. Oral Maxillofac Surg Clin North Am. 2019;31:1–11. doi: 10.1016/j.coms.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Chang HY, Lee CH, Li YS, Huang JT, Lan SH, Wang YF, Lai WW, Wang YC, Lin YJ, Liu HS, Cheng HC. MicroRNA-146a suppresses tumor malignancy via targeting vimentin in esophageal squamous cell carcinoma cells with lower fibronectin membrane assembly. J Biomed Sci. 2020;27(102) doi: 10.1186/s12929-020-00693-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escuin D, López-Vilaró L, Bell O, Mora J, Moral A, Pérez JI, Arqueros C, Ramón Y, Cajal T, Lerma E, Barnadas A. MicroRNA-1291 is associated with locoregional metastases in patients with early-stage breast cancer. Front Genet. 2020;11(562114) doi: 10.3389/fgene.2020.562114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu WG, Kuang YM, Wang D, Li Z, Xia RP. Effect of miR-210 on proliferation and migration of pancreatic cancer cells through regulating Runx3 level. J Biomater Tissue Eng. 2020;10:1827–1831. [Google Scholar]
- 9.Chen L, Sun DZ, Fu YG, Yang PZ, Lv HQ, Gao Y, Zhang XY. Upregulation of microRNA-141 suppresses epithelial-mesenchymal transition and lymph node metastasis in laryngeal cancer through HOXC6-dependent TGF-β signaling pathway. Cell Signal. 2020;66(109444) doi: 10.1016/j.cellsig.2019.109444. [DOI] [PubMed] [Google Scholar]
- 10.Yang B, Zang J, Yuan W, Jiang X, Zhang F. The miR-136-5p/ROCK1 axis suppresses invasion and migration, and enhances cisplatin sensitivity in head and neck cancer cells. Exp Ther Med. 2021;21(317) doi: 10.3892/etm.2021.9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu YB, Wang Y, Zhang MD, Yue W, Sun CN. MicroRNA-29a functions as a tumor suppressor through targeting STAT3 in laryngeal squamous cell carcinoma. Exp Mol Pathol. 2020;116(104521) doi: 10.1016/j.yexmp.2020.104521. [DOI] [PubMed] [Google Scholar]
- 12.Cossu AM, Mosca L, Zappavigna S, Misso G, Bocchetti M, De Micco F, Quagliuolo L, Porcelli M, Caraglia M, Boccellino M. Long non-coding RNAs as important biomarkers in laryngeal cancer and other head and neck tumours. Int J Mol Sci. 2019;20(3444) doi: 10.3390/ijms20143444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erkul E, Yilmaz I, Gungor A, Kurt O, Babayigit MA. MicroRNA-21 in laryngeal squamous cell carcinoma: Diagnostic and prognostic features. Laryngoscope. 2017;127:E62–E66. doi: 10.1002/lary.26226. [DOI] [PubMed] [Google Scholar]
- 14.Li W, Ma HP, Sun J. MicroRNA-34a/c function as tumor suppressors in Hep-2 laryngeal carcinoma cells and may reduce GALNT7 expression. Mol Med Rep. 2014;9:1293–1298. doi: 10.3892/mmr.2014.1929. [DOI] [PubMed] [Google Scholar]
- 15.Guo Y, An R, Zhao R, Sun Y, Liu M, Tian L. miR-375 exhibits a more effective tumor-suppressor function in laryngeal squamous carcinoma cells by regulating KLF4 expression compared with simple co-transfection of miR-375 and miR-206. Oncol Rep. 2016;36:952–960. doi: 10.3892/or.2016.4852. [DOI] [PubMed] [Google Scholar]
- 16.Chai L, Kang XJ, Sun ZZ, Zeng MF, Yu SR, Ding Y, Liang JQ, Li TT, Zhao J. MiR-497-5p, miR-195-5p and miR-455-3p function as tumor suppressors by targeting hTERT in melanoma A375 cells. Cancer Manag Res. 2018;10:989–1003. doi: 10.2147/CMAR.S163335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng C, Zhang L, Sun Y, Li X, Zhan L, Lou Y, Wang Y, Liu L, Zhang Y. GDPD5, a target of miR-195-5p, is associated with metastasis and chemoresistance in colorectal cancer. Biomed Pharmacother. 2018;101:945–952. doi: 10.1016/j.biopha.2018.03.028. [DOI] [PubMed] [Google Scholar]
- 18.Liu X, Zhou Y, Ning YE, Gu H, Tong Y, Wang N. MiR-195-5p inhibits malignant progression of cervical cancer by targeting YAP1. OncoTargets Ther. 2020;13:931–944. doi: 10.2147/OTT.S227826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Poel D, Boyd LNC, Beekhof R, Schelfhorst T, Pham TV, Piersma SR, Knol JC, Jimenez CR, Verheul HMW, Buffart TE. Proteomic analysis of miR-195 and miR-497 replacement reveals potential candidates that increase sensitivity to oxaliplatin in MSI/P53wt colorectal cancer cells. Cells. 2019;8(1111) doi: 10.3390/cells8091111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pang H, Xu X, Dai L, Wang K, Yao X. MicroRNA-195 is associated with regulating the pathophysiologic process of human laryngeal squamous cell carcinoma. Mol Med Rep. 2018;17:5283–5291. doi: 10.3892/mmr.2018.8523. [DOI] [PubMed] [Google Scholar]
- 22.Cheng JZ, Chen JJ, Wang ZG, Yu D, Zu YZ. The functional role of microRNAs in laryngeal carcinoma. Open Life Sci. 2017;12:460–464. [Google Scholar]
- 23.Wang PP, Ding SY, Sun YY, Li YH, Fu WN. MYCT1 inhibits the adhesion and migration of laryngeal cancer cells potentially through repressing collagen VI. Front Oncol. 2021;10(564733) doi: 10.3389/fonc.2020.564733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song Y, Yang M, Zhang H, Sun Y, Tao Y, Li H, Zhang J, Li Y, Yang J. IL-17 affects the progression, metastasis, and recurrence of laryngeal cancer via the inhibition of apoptosis through activation of the PI3K/AKT/FAS/F pathways. J Immunol Res. 2020;2020(2953191) doi: 10.1155/2020/2953191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Li D, Wang P, Zhu W, Yin W. Tetrandrine partially reverses multidrug resistance of human laryngeal cancer cells. J Int Med Res. 2020;48(300060520944706) doi: 10.1177/0300060520944706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shang Y, Wang LQ, Guo QY, Shi TL. MicroRNA-196a overexpression promotes cell proliferation and inhibits cell apoptosis through PTEN/Akt/FOXO1 pathway. Int J Clin Exp Pathol. 2015;8:2461–2472. [PMC free article] [PubMed] [Google Scholar]
- 27.Jin Y, Wang M, Hu H, Huang Q, Chen Y, Wang G. Overcoming stemness and chemoresistance in colorectal cancer through miR-195-5p-modulated inhibition of notch signaling. Int J Biol Macromol. 2018;117:445–453. doi: 10.1016/j.ijbiomac.2018.05.151. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Di C, Li W, Cai W, Tan X, Xu L, Yang L, Lou G, Yan Y. Oncomirs miRNA-221/222 and tumor suppressors miRNA-199a/195 are crucial miRNAs in liver cancer: A systematic analysis. Dig Dis Sci. 2016;61:2315–2327. doi: 10.1007/s10620-016-4156-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu X, Zhang Y, Ma X, Pertsemlidis A. miR-195 potentiates the efficacy of microtubule-targeting agents in non-small cell lung cancer. Cancer Lett. 2018;427:85–93. doi: 10.1016/j.canlet.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng J, Xu T, Chen F, Zhang Y. MiRNA-195-5p functions as a tumor suppressor and a predictive of poor prognosis in non-small cell lung cancer by directly targeting CIAPIN1. Pathol Oncol Res. 2019;25:1181–1190. doi: 10.1007/s12253-018-0552-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest. 2003;112:1776–1784. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Wang F, Zhong W, Ling H, Wang J, Cui J, Xie T, Wen S, Chen J. RNA-binding protein RBM6 as a tumor suppressor gene represses the growth and progression in laryngocarcinoma. Gene. 2019;697:26–34. doi: 10.1016/j.gene.2019.02.025. [DOI] [PubMed] [Google Scholar]
- 34.Hwang KE, Kim HJ, Song IS, Park C, Jung JW, Park DS, Oh SH, Kim YS, Kim HR. Salinomycin suppresses TGF-β1-induced EMT by down-regulating MMP-2 and MMP-9 via the AMPK/SIRT1 pathway in non-small cell lung cancer. Int J Med Sci. 2021;18:715–726. doi: 10.7150/ijms.50080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karmakar D, Maity J, Mondal P, Shyam Chowdhury P, Sikdar N, Karmakar P, Das C, Sengupta S. E2F5 promotes prostate cancer cell migration and invasion through regulation of TFPI2, MMP-2 and MMP-9. Carcinogenesis. 2020;41:1767–1780. doi: 10.1093/carcin/bgaa043. [DOI] [PubMed] [Google Scholar]
- 36.Miles WO, Tschöp K, Herr A, Ji JY, Dyson NJ. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev. 2012;26:356–368. doi: 10.1101/gad.182568.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang W, Tang J, Qin J, Geng Q, Zhou Z, Li B, Zhang J, Chen H, Xia Y, Wang X. Involvement of down-regulated E2F3 in Hirschsprung's disease. J Pediatr Surg. 2013;48:813–817. doi: 10.1016/j.jpedsurg.2012.10.048. [DOI] [PubMed] [Google Scholar]
- 38.Huang L, Luo J, Cai Q, Pan Q, Zeng H, Guo Z, Dong W, Huang J, Lin T. MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3. Int J Cancer. 2011;128:1758–1769. doi: 10.1002/ijc.25509. [DOI] [PubMed] [Google Scholar]
- 39.Ren XS, Yin MH, Zhang X, Wang Z, Feng SP, Wang GX, Luo YJ, Liang PZ, Yang XQ, He JX, Zhang BL. Tumor-suppressive microRNA-449a induces growth arrest and senescence by targeting E2F3 in human lung cancer cells. Cancer Lett. 2014;344:195–203. doi: 10.1016/j.canlet.2013.10.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.