Abstract
Malignant neoplasms are characterized by poor therapeutic efficacy, high recurrence rate, and extensive metastasis, leading to short survival. Previous methods for grouping prognostic risks are based on anatomic, clinical, and pathological features that exhibit lower distinguishing capability compared with genetic signatures. The update of sequencing techniques and machine learning promotes the genetic panels-based prognostic model development, especially the RNA-panel models. Gliomas harbor the most malignant features and the poorest survival among all tumors. Currently, numerous glioma prognostic models have been reported. We systematically reviewed all 138 machine-learning-based genetic models and proposed novel criteria in assessing their quality. Besides, the biological and clinical significance of some highly overlapped glioma markers in these models were discussed. This study screened out markers with strong prognostic potential and 27 models presenting high quality. Conclusively, we comprehensively reviewed 138 prognostic models combined with glioma genetic panels and presented novel criteria for the development and assessment of clinically important prognostic models. This will guide the genetic models in cancers from laboratory-based research studies to clinical applications and improve glioma patient prognostic management.
1. Introduction
Malignant tumors are characterized by therapeutic resistance, frequent recurrence, and distant metastasis, which cause difficulties in treating by either surgical resection or adjunctive therapies, leading to poor prognosis. For better clinical management, many prognostic models were proposed to analyze survival [1]. Previous models with unformulated predictors stratified patients into relative risk groups. However, they did not provide quantitative results or absolute risk stratification. Machine learning algorithms can identify critical patterns through big and complex data with high accuracy [2]. Common algorithms applied in cancer prediction include weighted gene coexpression network analysis (WGCNA), L1-penalized least absolute shrinkage selection operator (LASSO), Cox proportional hazards (PH) model [3], Neural Network [2], and Elastic regression [4]. Based on these machine learning algorithms, risk scores and further pictorial nomograms are constructed for addressing this issue [5, 6]. Risk score models are calculated with a spectrum of parameters in predicting clinical outcome risks. Samples are from local patient cases or online data. Establishing a model requires two main steps: development and validation [5]. First, predictors are selected and risks are calculated; thereafter, performance estimations are performed to assess the predictive quality. Second, the model is validated internally and externally (independent data set). Performance estimation is also performed [7].
Gliomas are common intracranial tumors causing the highest mortality rates in all cancer types [8]. Routine treatments for gliomas include surgical excision, radiotherapy, and chemotherapy. The world health organization (WHO) grouped gliomas into four grades based on their histological features: WHO grades I, II, III, and IV [9]. Low-grade gliomas (LGGs) comprise WHO grades I and II, showing a relatively good prognosis. High-grade gliomas (HGGs) consist of WHO grades III and IV, manifesting worse survival outcomes [9]. Histological classification provides an understanding of glioma behavior. However, molecular groups differentiate prognostic groups more accurately [10] (Figure 1). The 2016 Central Nervous System WHO classification, therefore, provided molecular features including IDH-mutant/wildtype astrocytoma and glioblastoma, IDH-mutant/wildtype, and 1p/19q-codeleted/nonco-deleted oligodendroglioma, H3K27M-mutant/wildtype diffuse midline glioma, and RELA fusion-positive/negative ependymoma to precisely classify gliomas [11]. These and other markers such as CDKN2A and EGFR were combined to develop prognostic models for glioma patients [12].
Figure 1.
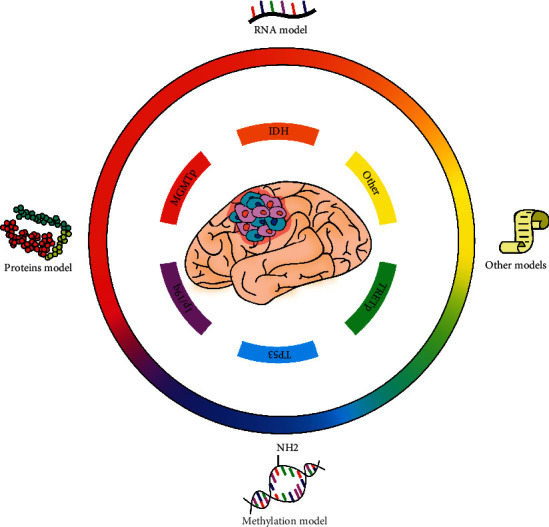
Glioma markers and genetic prediction models can stratify patient risks precisely and improve prognostic management. Glioma patient prognosis can be stratified and precisely predicted via prognostic markers and markers-based prediction models and, therefore, improved by adopting corresponding treatment strategies. Markers with prognostic values include IDH, MGMTp, 1p19q, TP53, TERTp, and many other molecules. Prediction models are based on RNA, protein, methylation, and other types of signatures. IDH, isocitrate dehydrogenase; MGMTp, O-6-methylguanine-DNA methyltransferase promoter; TP53, tumor protein p53; TERTp, telomerase reverse transcriptase promoter.
The poor prognosis of gliomas triggered the development of a clinically useful and effective model to assess survival risks for subsequent therapeutic strategies. This review systematically summarized and compared all 138 risk score models for gliomas with multivariable markers (Figure 2). It also presented the clinical significance of some frequently reported predictors, thus guiding the advanced prediction models for glioma. This will be conducive to translation from laboratory-based models to clinical available tools and clinical prognosis management improvement.
Figure 2.
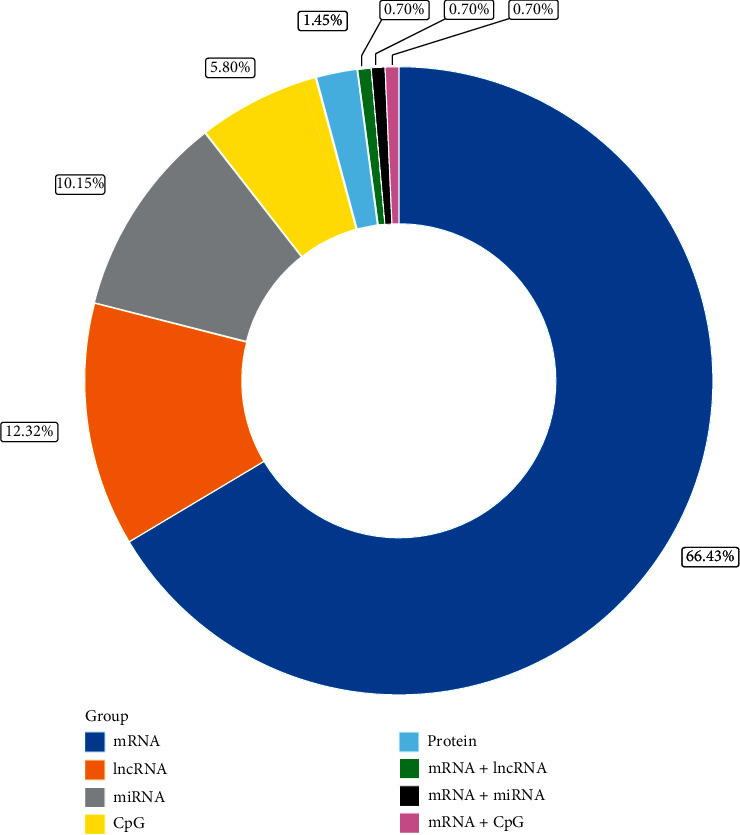
Proportion of published models based on signatures of different molecular types. The largest proportion of 94 mRNA models achieved 68% of all the 138 genetic prognostic models, followed by 17 lncRNA and 14 miRNA models, accounting for 12% and 10%, respectively. mRNA, message RNA; lncRNA, long noncoding RNA; miRNA, microRNA.
2. Rules for Evaluating Model Quality and Exclusion Criteria
The TRIPOD statement standardized reports on the prediction models. It proposes a checklist of 22 items required in the development and validation process [13]. However, it does not judge model quality. Thus, we screened through the 138 models and proposed novel criteria that classified them into different quality groups (Figure 3); the criteria are listed in Table 1. After quality division, the high- and medium-quality models were discussed, and the low-quality group models were not presented (Table S1).
Figure 3.
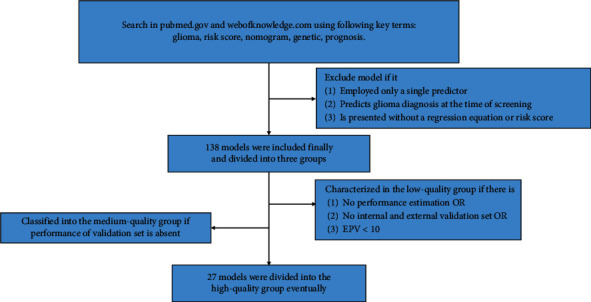
Procedure to screen and classify the genetic prediction models of glioma. The exclusion criteria and grouping rules are proposed firstly in this review, and details are described in Section 2. EPV, events per variable.
Table 1.
Criteria for model quality estimation.
| Group | Items |
|---|---|
| High-quality group | EPV ≥10 AND |
| Variable in final model <10 AND AUC/c-index estimated for both training and external or internal validation sets | |
|
| |
| Medium-quality group | EPV ≥10 AND |
| Presence of external or internal validation set but only training set is estimated by receiver operating characteristic curve/c-index | |
|
| |
| Low-quality group | No performance estimation OR |
| No internal and external validation set OR EPV <10 | |
EPV, event per variable.
Performance estimation, validation, and EPV are vital factors to assess the model quality. Performance includes discrimination and calibration. Discrimination refers to the capability that differentiates patients from events that happen or not, which functions as the most essential quality results [14]. Calibration compares estimated event rates with observed rates [15]. Less efficacy arises when discrimination is confused with performance. Secondly, most studies lack calibration results [16]. For discrimination, we defined area under the curve (AUC), or c-index ≥ 0.80 as high accuracy, AUC ≥ 0.70 as acceptable, and AUC < 0.7 as low accuracies. The repeatability and transportability of the model should be verified through internal and external validation before clinical application [15]. Proper EPV requires 10 minimally determined by rule of thumb, but when many low-prevalence predictors are included, EPV should be up to 20 [17]; otherwise, it is considered an overfitting model.
Other aspects including variable number, missing value, outcome definition, reference genome, and annotation update also contribute to the model assessment. Excessive variables increase detective costs and decrease practicability. For most models contain less than 10 predictors and no certain conclusion have been presented, we defined less than 10 appropriate predictors. Missing data can lead to bias when not handled properly. Outcome definition is clearer and more informative with specific follow-up time than overall survival (OS). Reference genome and annotation updates can cause reversed individual risk prediction due to multiple gene expression diversity [18]. The inconsistent cutoff values between training and validation sets, screening method, and the threshold for predictors are complex questions and remain unresolved; they were not reviewed.
We excluded studies if (1) a single genetic predictor was employed with or without other types of predictors, (2) the model predicts glioma diagnosis at the time of screening, and (3) the model is inadequately presented without a regression equation or risk score.
3. RNA Models
3.1. mRNAs
Message RNA (mRNA) plays a critical role in central dogma that controls protein synthesis and decides cellular biology and behavior [19]. Many biological processes of mRNA in cancers, such as mRNA splicing, methylation, and interfering, can modulate the mRNA level and alter the cancer property; thereby, mRNA modulation has been a therapeutic target since early times. The levels of mRNAs in cancers, including gliomas, represent the expression of genes that are connected to the prognosis [11]. For glioma prognostic models, the largest proportion of models (78 models) are constructed based on mRNA sequencing data (Figure 2), of which 18 models are in the high-quality group.
3.1.1. LGGs
Low-grade gliomas (LGGs) refer to WHO I, II gliomas [9]. Twenty-one mRNA models for LGGs have been reported. Among them, ten are in the high-quality group and nine are in the medium-quality group.
In the high-quality group, nine [20–28] and one [29] confer high and acceptable accuracy, respectively. Common advantages and additional luminous points in the high-quality group are shown. The corresponding AUC for multiple outcome events of 1, 3, and 5 years' survival in the 4 models [22–25] functions more powerful than many other models that predict merely OS. Two models were validated and assessed highly accurately both internally and externally [21, 22]. Su's model [25] is highly accurate, except when it predicted a 5-year survival training set (AUC was 0.711). For Zeng's model [24], despite its lower AUC for the external validation set, the nomogram performed better via c-index and calibration curve.
For the medium-quality group, five models exhibit high accuracy [30–34], three are acceptable [35–37], and one is lowly accurate [38]. Song et al. constructed a 21- [33] and a 20-gene model [34] both internally and externally, and the former was validated via four independent datasets. But the many numbers of predictors should be further streamlined. Hsu et al. [31] and Cheng et al. [30] values for AUC are extremely high (above 0.98 and 0.93, respectively). For deficiencies in this group, the seven models [30–32, 35–38] lack performance estimation in external or internal validation sets, and Ni et al. did not validate their 25-gene model [37].
3.1.2. HGGs
High-grade gliomas (HGGs) comprise WHO III and Glioblastoma (GBM). A total of 44 HGGs models have been constructed but show a few satisfactory studies. The high-quality and medium-quality groups contain 7 and 11 models, respectively.
The high-quality models were designed for GBM [39–45]. 4-gene [44] and 3-gene [43] models associated with autophagy were both validated via two independent datasets. The two risk scores' predictive discrimination varies among survival rates in different years and datasets, while their nomograms stabilized the accuracy above 0.72, which integrated risk scores with other common factors. Nomograms' superior predictive accuracy to risk scores can also be observed in a study by Wang, the nomogram is highly accurate (from 0.77 to 0.85) compared with the AUC of risk score (from 0.67 to 0.79). Of note, nomogram performance decreased when estimated by c-index [39]. However, in a study by Zhu, the risk score outperformed the nomogram. AUC of risk score is 0.781 and 0.771 for 2 and 3 years of survival in the discovery cohort [41], while the c-indices of nomogram are also less than 0.70 [41]. Nomograms benefit from combining risk scores with other predictors, but whether their accuracy increases depends on the factors' quality. Moreover, the calibration curves were plotted and verified the reliability in the four studies [39, 41, 43, 44]. Additionally, 2329 samples from multiple cohorts in Zhu's study are the largest sample size in the currently existing models, improving its repeatability and transportability [41].
The medium-quality group is better since eight models are of high accuracy [46–53]. The AUC values in models developed by Zhang et al. [47], Hou et al. [52], and Chai et al. [48] are 0.93, 0.95, and 0.93, respectively. Two models achieved acceptable accuracy [54, 55], whereby Cheng's final model attained 0.81 when absorbing clinical features and 1p/19q status [55]. One model is lowly accurate [56]. This group is limited by a lack of performance estimation for the validation set except for Hou's model, but the weakness of Hou's model is the many predictors (14 genes) [52].
3.1.3. Other Classified Gliomas Clusters
Three models and ten models are grouped into the high-quality and medium-quality groups from the total 28 models reviewed, respectively. The three high-quality models are highly accurate [57–59]. Particularly, Wang's model for diffuse glioma exhibits excellent discrimination with AUC from 0.874 to 0.950 [58]. But it was only internally validated.
Nine medium-quality models show high accuracy [60–68], whereby three models for diffuse glioma estimated accuracy level in training and validation sets despite the large size of predictors [64, 66, 67]. Notably, Sun's model has high accuracy when compared with all the signatures randomly derived from the screen method and outperformed the other three signatures in predicting drug sensitivity [66]. Additionally, it achieved higher accuracy when combined with age, grade, and another signature in the validation set for 3 and 5 years' survival [66]. Other 3 studies (two for all diffuse glioma [60, 65], and one for 1p/19q codeletion diffuse glioma) [62] together with a low-accuracy model for all glioma [69] in the medium-quality group are characterized by missing accuracy estimation in the validation set and excessive predictors.
Conclusively, most studies were designed for diffuse gliomas, and our criteria characterized 3 high-quality models. Besides, many models show high predictive accuracy when subjected to training. However, the absence of accuracy of validation sets failed to affirm the obtained discrimination results.
3.2. LncRNAs
Long noncoding RNAs (lncRNAs) are transcripts more than 200 nt in length. LncRNAs lack significant protein-coding capacity, but their regulatory functions are widely engaged from gene expression to protein translation. In gliomas, the lncRNAs function in stemness, drug resistance, blood–tumor barrier permeability, angiogenesis, and motility cancer phenotypes [70]. lncRNA is the second major hotspot in model research, after mRNA (Figure 2).
Seventeen lncRNA-signature models on different gliomas and one model on diffuse intrinsic pontine glioma have been reported. Three are high-quality models, and five are medium-quality models.
All three high-quality models exhibit high accuracy [71–73]. Except for the slightly decreased AUC values (0.722) when submitted to external validation based on Chen's study [73], the other two models show similar high accuracy in training, internal validation, and the entire set with AUC from 0.84 to 0.91 [71, 72].
The highest AUC value in the medium-quality group was obtained in Wang's model (0.942) for anaplastic glioma [74]. This is followed by Kiran's UVA8 model (8-lncRNA signature) [75] acceptably test for 5-year survival. The AUC values for the other two models were 0.68 and 0.70, respectively [76, 77]. In the five medium-quality models, they were externally validated, but three lacked internal validation [74, 77, 78]. Moreover, Kiran's study reported the UVA8 model and compared it with other predictors and models [75]. The UVA8 accuracy [75] is higher than other clinical features or IDH status. It outperformed the 5 published signatures in the training dataset by c-indices [75]. While the 6 models were designed for a diverse class of gliomas and different prognostic events and to validate various datasets, this positive result in Kiran's study was inevitably questionable due to incomparability.
Internal validation is absent in the 9 low-quality models (Table S1), which is vital to address the stability in selecting predictors and the quality of predictions before clinical application [79]. Cross-validation or bootstrapping methods should be employed to achieve complete internal validation.
3.3. miRNAs
MicroRNAs (miRNAs) are a class of noncoding RNA that binds to complementary target mRNAs. This results in mRNA translational inhibition or degradation. In gliomas, miRNAs are involved in various tumor-associated activities, including immune response, hypoxia, tumor plasticity, and resistance to therapy through multigene targets [80], indicating miRNA-based models as a promising strategy for glioma prognosis.
Fourteen studies on miRNA signatures, one on LGG [81], and others on HGG were reported. Three models were characterized in the high-quality group [81–83], whereas the other models were classified in the low-quality group.
The three high-quality models have respective advantages. The 5-miRNA model by Cheng et al. [83] adopted complete internal and external validations, and the AUC values increased from 0.649 and 0.756 to 0.847 and 0.909 after integrating age and chemotherapy, respectively, although only 19 samples were submitted to external validation. Chen's model [82] was internally and externally validated and achieved similar AUC for disease-free survival and OS in three datasets. Besides, Qian's study [81] established a nomogram that predicted a 1-, 2-, 3-, and 5-year survival rate, which is informative for prognostic management. However, its drawbacks include the absence of internal validation and low accuracy in the training set (c-index = 0.68).
4. Methylation Models
Cytosine-phosphate-guanine (CpG) islands are a cluster of CpG sites located at or near the transcription start regions of genes and gene promoters. CpG island methylation is the most common epigenetic type of cancer. The CpG island methylator phenotype that comprises several CpG islands is an interesting topic in cancer epigenetics [84]. The glioma CpG island methylator phenotype is associated with gliomas tumorigenesis and is an independent biomarker stratifying gliomas into epigenetic subtypes [85]. Currently, 8 models have been reported and none of them is a high-quality model; 3 of the models are classified into the medium-quality group [86–88].
The two medium-quality models have acceptable discriminations from 0.71 to 0.77 but were not internally validated [86, 87]. Yin's 6-CpG risk score [86] achieved a higher AUC value (0.734) for patients receiving all treatment integrated with the CpG island methylator phenotype. Moreover, higher AUC (0.771) was achieved with the MGMT status combination for those receiving radiation therapy/ temozolomide. Besides, the prediction accuracy rate of the 6-CpG signature (87%) was validated via the support vector machines model.
5. Other Multimolecular Models
Two protein-signature models based on reverse phase protein array were constructed. Stetson's 13-protein risk model [89] applied c-index to estimate the model's accuracy in both training and validating sets for GBM (0.63 and 0.60, respectively) and IDH-wildtype LGG (0.82 and 0.70, respectively), but the shortage is the low EPV (less than 10). Patil and Mahalingam developed another 4-protein model, but without external validation and performance estimation [90]. Both two models are of low quality.
Three mixed models of different classes of molecular signatures were presented for GBMs prognosis. The mixed model for mRNA and lncRNA is of high quality [91], and the other two are medium-quality models [92, 93]. The three models were fully validated both internally and externally. They were estimated using receiver operating characteristic curve or c-index; however, the validation set lacked estimation, and there was a low c-index value (0.68) in the training set from Etcheverry's study [92].
6. Biofunction and Clinical Significance of Frequently Reported Molecules
Molecular signatures reviewed consist of mutated genes, noncoding RNAs, and proteins. Currently, the star markers including IDH, MGMT, 1p/19q, H3K27M, TERT, and ATRX are known to exhibit significant prognostic value. The IDH-mutant with 1p/19q codeletion and MGMT promoter methylation are favorable prognostic factors. The H3K27M-mutant and ATRX alteration are associated with higher risk whereas TERT has a dichotomous prognostic effect [94]. Besides, other molecular signatures collected from 138 published models were reported to contribute to prognostic risk estimation. The prognostic value for most molecular biomarkers has not been validated. Therefore, we analyzed 138 models to select the most overlapping biomarkers (Table 2) with known evidence from researches to determine their biofunctions and potential prognostic values in gliomas. The predictors that presented repeatedly more than twice in 138 model studies are listed in Table 2. Predictors that overlapped less than three times were not reviewed.
Table 2.
Highly overlapping genes and miRNAs.
| Overlapping times | Molecule numbers | Genes | miRNAs |
|---|---|---|---|
| 6 | 1 | IGFBP2 | — |
| 5 | 3 | HDAC, STAT1 | miR-222 |
| 4 | 6 | GPNMB, VEGFA, EFEMBP2, ISG20, FZD7, EGFR, | — |
| 3 | 23 | MDK, CHI3L1, LGALS3, IFI44, OSMR, MYC, TOP2A, CD44, LDHA, BMP2, KI67, BUB1, LAMB1, MAP2K3, KCNB1, KCNJ10, IRF1, ASF1A, SOCS3, KCNAB1 | miR-148a, miR-15b, miR-145, miR-20a |
6.1. IGFBP2
IGFBP2 regulates insulin-like growth factors' distribution and biofunction by interacting with the insulin-like growth factor system. High aberrant IGFBP2 expression was detected in HGG [95]. This played critical roles in glioma progression and was correlated with poor prognosis [95]. Besides, IGFBP2 downregulation was reported specifically in IDH-mutant gliomas [96]. Besides, EGFR (epidermal growth factor receptor) and integrinβ can integrate with IGFBP2 to promote tumor progression [95]. The IGFBP2 gene, therefore, presents prognostic value and functions as a potential immunotherapeutic target for GBM in the future clinical trials [95, 96].
6.2. HDAC Families and CD44
Histone deacetylase (HDAC) is a vast family of enzymes that mainly exert a repressive influence on transcription [97]. It blocks gene transcription by inhibiting histone acetylation and compacts the DNA/histone complex. In gliomas, HDAC functions to bridge the xCT-CD44 complex with malignant glioma cells and various tumor zones [98]. Currently, HDAC inhibitors exhibit unfavorable therapeutic efficacy in glioma patients. Researchers have reported a more beneficial strategy that adopted HDAC inhibitors in combined therapy [99]. Moreover, many clinical trials (Table 3) are ongoing to test their application prospects in treating diffuse intrinsic pontine glioma. and HGG. Currently, HDAC inhibitors are characterized by HDACs risk factors; however, their predictive ability has not been verified. In-depth explorations on their therapeutic efficacy and prognostic value are required.
Table 3.
Completed and active trials primarily completed in 5 years of HDAC, VEGF, and EGFR related therapy.
| Gene | Agent name | Glioma type | Clinical trials | Development phase | Status |
|---|---|---|---|---|---|
| HDAC | Vorinostat | DIPG | NCT01189266 | Phase I/II | Active |
| DIPG | NCT02420613 | Phase I | Active | ||
| HGG | NCT01236560 | Phase II/III | Completed | ||
| HGG, recurrent GBM | NCT01266031 | Phase I/II | Completed | ||
| Belinostat | GBM | NCT02137759 | Phase II | Active | |
|
| |||||
| VEGF | Bevacizumab | Anaplastic glioma, recurrent GBM | NCT01836536 | Unknown | Completed |
| Recurrent ependymoma, WHO II, III glioma | NCT00381797 | Phase II | Completed | ||
| GBM | NCT01091792 | Early phase I | Completed | ||
| HGG, recurrent GBM | NCT01266031 | Phase I/ II | Completed | ||
| HGG | NCT01236560 | Phase II/III | Completed | ||
| Recurrent GBM | NCT01648348 | Phase I/II | Completed | ||
| GBM | NCT01498328 | Phase II | Completed | ||
| GBM, oligodendroglioma | NCT01609790 | Phase II | Active | ||
| Recurrent GBM | NCT02142803 | Phase I | Active | ||
| Recurrent GBM | NCT02974621 | Phase II | Active | ||
|
| |||||
| EGFR | Erlotinib | HGG | NCT01257594 | Phase I | Completed |
| Lapatinib | Recurrent HGG | NCT02101905 | Phase I | Active | |
| Unknown† | GBM | NCT01454596 | Phase I/II | Completed | |
| AMG 595 | Recurrent GBM, AA | NCT01475006 | Phase I | Completed | |
| Sym004 | Recurrent GBM | NCT02540161 | Phase II | Completed | |
| Depatux-M | GBM | NCT02343406 | Phase II | Completed | |
| GBM | NCT02573324 | Phase II/III | Active | ||
| GBM | NCT03419403 | Phase III | Terminated | ||
| GBM | NCT01800695 | Phase I | Completed | ||
| Dacomitinib | Recurrent GBM | NCT01520870 | Phase II | Completed | |
| GBM | NCT01112527 | Phase II | Completed | ||
| Tesevatinib | GBM | NCT02844439 | Phase II | Completed | |
| Afatinib | GBM | NCT00977431 | Phase I | Completed | |
| Cetuximab | GBM, anaplastic astrocytoma | NCT01238237 | Phase I | Completed | |
| Rindopepimut | GBM | NCT01498328 | Phase II | Completed | |
| GBM | NCT01480479 | Phase III | Completed | ||
DIPG, diffuse intrinsic pontine glioma; HGG, high-grade glioma; GBM, glioblastoma; AA, anaplastic astrocytoma. †Epidermal growth factor receptor (EGFRv) III; chimeric antigen receptor (CAR); transduced peripheral blood lymphocytes (PBL).
CD44 was identified in GBM and from brain metastases [100]. It is a biomarker of the mesenchymal GBM subtype with the most aggressive growth patterns [101]. Besides, GBM progression was inhibited by inhibiting CD44 expression. This indicates the roles of CD44 in the tumor process [102].
6.3. MDK
MDK is a heparin-binding growth factor encoding gene extensively studied for its multiple functions in various tissues. MDK contributes to numerous tumor-related activities in glioma, and its overexpression is associated with poor prognosis [103]. However, no clinical trial has been performed to explain its therapeutic potential.
6.4. GPNMB
GPNMB encodes the type 1 transmembrane protein expressed mainly on the surface of cancer cells [104]. GPNMB promotes tumor progression through immune-microenvironment plasticity. It also enhances Wnt/β-catenin signaling pathway activation and interaction with Na+/K+-ATPase subunits [105–107]. Also, abnormally high GPNMB expression has been reported to be associated with unfavorable survival outcomes in GBM [108]. These mechanisms justify that GPNMB is a risk factor for glioma patients.
6.5. EGFR
It has been reported that EGFR signaling pathways are activated in the majority of GBM cells [109]. EGFR gene aberration contains rearrangement, amplification, and mutation. The EGFR variant III (EGFRvIII) is a common mutation product in GBM [110]. The EGFRvIII contributes to many tumor biological features [111], indicating a poor outcome. Besides, wild-type EGFR associated with tumor cell invasion and angiogenesis has been demonstrated in several in vivo and in vitro experiments [112]. Thus, EGFR and EGFRvIII have been identified as popular therapeutic targets for treating malignant glioma patients. However, current treatments that target EGFR have failed in clinical trials including the small molecule drugs and biologic antibodies [112]. While some trials are still ongoing (Table 3).
6.6. VEGFA
VEGFA, also known as VEGF, is a growth factor that promotes tumor angiogenesis and vascular permeability and regulates immune cell and fibroblastoma and microenvironment formation [113]. In glioma, VEGF acts as a regulatory growth factor secreted by glioma stem cells to promote the tumor vasculature [114]. Anti-VEGF-A antibody has been applied as a novel antiangiogenic therapy for malignant gliomas, as with bevacizumab [114]. However, it has not achieved considerable progression in treating gliomas. A report by Eskilsson et al. [112] indicates that causes of failure may be attributed to enhanced invasive properties of tumor cells by different mechanisms, for example, RTK signaling. Many trials using bevacizumab with other strategies, for example, EGFR inhibitors, showed improved efficacy (Table 3); some trials are underway. Since several VEGFA studies on gliomas exist, it is expected that it will work as an accurate and effective predictor for glioma prognosis. This is despite its current poor performance in antiangiogenesis therapy.
6.7. miR-221/miR-222
miR-221/miR-222 are two closely related miRNAs located in the genome region of the X chromosome. They have a similar sequence, structure, and biofunction and upregulate expression in various human cancers including glioma. Knocking down miR-221/miR-222 expression blocks cell cycle transition, suppresses tumor cell growth, and increases sensitivity to radiotherapy [115]. Also, miR-221/miR-222 is associated with tumor cell apoptosis by targeting the apoptosis-related gene [116]. Recent studies revealed its connection with glioma histology and patient prognosis [117]. This showed that either miR-221 or miR-222 expression was associated with a higher WHO grade thereby indicating a poor prognosis.
6.8. Other Significant Predictors
Apart from the above-discussed vital parameters, other factors play significant roles in glioma malignant properties and prognosis assessment. These predictors include MYC, KCNJ10, CHI3L1, STAT1, and FZD7. MYC amplification has been identified in many classes of cancers. It acted as a qualified prognostic prediction biomarker [118]. KCNJ10 encodes inwardly rectifying potassium channel Kir4.1 protein that is expressed exclusively in central nervous system glial cells. This establishes a hyperpolarized resting membrane potential and prevents glioma cell proliferation [119]. CHI3L1 overexpression is associated with poor survival [120] causing tumor invasion, migration, angiogenesis, and resistance to temozolomide therapy [121]. The aberrant STAT1 activation may cause oncogenesis [122]. Limited information on FZD7 activity in gliomas exists; however, the other four predictors have attracted extensive research.
7. Discussion
For model variables, previous studies focused on pathological, anatomic, and clinical predictors, but recently genetic data were widely recommended for enhancing predictive ability [123, 124]. Formulated genetic predictors harbor an advantage in calculating absolute risks based on marker expression levels via sequencing analysis and coefficients, thus providing stronger information compared with relative risk tools. However, they are still not recommended for clinical application, due to limitations, such as the lack of gold markers, difficulties of data collection, the complexity of analysis, and low adherence to complete and transparent reporting [125]. Our review confirmed a similar problem in gliomas. We found that only 27 models (20%) are classified into the high-quality group (Table 4), 31 models (22%) are in the medium-quality group, and 83 models (58%) are classified in the low-quality groups; none can be clinically applied according to our criteria, which was urgently required to address the issue that no method for assessing the model quality exists currently [125].
Table 4.
Characteristics of 27 high-quality models.
| Authors | Glioma type | Parameters | Sample sizea | Validationb | Performance estimationc | Accuracyd | Reference |
|---|---|---|---|---|---|---|---|
| Liu et al. | LGG | 2 mRNAs | 115/-/41 | E | T, E | Low (0.735) | [29] |
| Xiao et al. | LGG | 3 mRNAs | 456/-/159 | I, E | T, E | High (0.908, 0.878, 0.827)e | [23] |
| Chen et al. | LGG | 3 mRNAs | 164/-/599 | E | T, E | High (0.869) | [28] |
| Zeng et al. | LGG | 4 mRNAs | 172/-/451 | E | T, E | High (0.845, 0.890, 0.912)e | [24] |
| Zhang et al. | LGG | 4 mRNAs | 534/-/325 | E | T, E | High (0.858, 0.853, 0.837)e | [27] |
| Liu et al. | LGG | 5 mRNAs | 524/-/169 | I, E | T, E | High (0.887) | [26] |
| Li et al. | LGG | 4 mRNAs + 6mRNAs | 516/-/426 | E | T, E | High (0.84) | [20] |
| Zhang et al. | LGG | 6 mRNAs | 304/128/353 | I, E | T, I, E | High (0.914) | [21] |
| Zhang et al. | LGG | 7 mRNAs | 297/124/353 | I, E | T, I, E | High and acceptable (0.899, 0.875, 0.778)e | [22] |
| Su et al. | LGG | 8 mRNAs | 511/-/172 | E | T, E | High and acceptable (0.882, 0.831, 0.711)e | [25] |
| Qian et al. | LGG | 4 miRNAs | 100/-/420 | E | T, E | Low (0.680) | [81] |
| Wang et al. | GBM | 3 mRNAs | 155/-/216 | E | T, E | High (0.832)f | [43] |
| Wang et al. | GBM | 4 mRNAs | 241/160/- | I | T, I | High and acceptable (0.756, 0.821, 0.885)ef | [44] |
| Wang et al. | GBM | 5 mRNAs | 364/155/- | I | T, I | Acceptable (0.729)f | [39] |
| Zhao et al. | GBM | 6 mRNAs | 152/-/138 | E | T, E | High (0.908) | [40] |
| Zhu et al. | GBM | 5 mRNAs | 306/325/1957 | I, E | T, I, E | Acceptable (0.704)f | [41] |
| Zuo et al. | GBM | 6 mRNAs | 137/-/158 | E | T, E | Acceptable (1-year 0.699,2-year 0.779) | [42] |
| Gong et al. | GBM | 8 mRNAs | 151/-/138 | E | T, E | High (0.977) | [45] |
| Zhou et al. | GBM | 6 lncRNAs | 200/219/- | I | T, I | High (0.902) | [72] |
| Chen et al. | GBM | 4 lncRNAs | 240/-/80 | E | T, E | High (0.843) | [73] |
| Chen et al. | GBM | 7 miRNAs | 89/102/109 | I, E | T, I, E | Low (0.690) | [82] |
| Cheng et al. | GBM | 5 miRNAs | 75/75/19 | E | T, E | High (0.847)f | [83] |
| Gao et al. | GBM | 6 mRNAs + 5 lncRNAs | 76/77/80 | I, E | T, E | Acceptable (0.780) | [91] |
| Peng et al. | Diffuse glioma | 5 mRNAs | 641/-/319 | E | T, E | High (3-year 0.895, 5-year 0.864) | [59] |
| Wu et al. | Glioma | 9 mRNAs | 550/-/309 | E | T, E | High (0.860) | [57] |
| Wang et al. | Glioma | 5 mRNAs | 420/178/- | I | T, I | High (0.917, 0.950, 0.881)e | [58] |
| Hu et al. | Glioma | 5 lncRNAs | 70/70/- | I | T, I | High (0.910) | [71] |
aSample size of training set/internal validation set/external validation set. bI and E represent the presence of internal and external validation, respectively. cT, I, and E represent the presence of performance estimation in training, internal validation, and external validation sets, respectively. dModels were classified into high, acceptable, and low accuracy according to AUC or c-index as we present in rules for judging model quality and excision criteria part, and accuracy in only the training set is shown. eThree AUC values for prediction of 1-, 3-, and 5-year survival, respectively. fIf higher accuracy was achieved by nomogram or integrated model, the highest accuracy result was exhibited instead of the result of the original risk score.
Problems in models consist of methodological deficiencies and clinical confirmation absence. The former was mainly attributed to the low adherence to guidelines like the TRIPOD statement [13], leading to series of deficiencies like the lack of performance assessment, validation that was also observed in other cancers [123, 126, 127]. A good example of a prostate cancer model was constructed in transparent and clear details [128], and its constructing details were listed in a table, facilitating the check of model reliability for users and avoiding methodological negligence during the establishment process. And for validating, age-related risk score [129] was analyzed through 2953 cases from 10 datasets; the large sample size and high AUCs verified its robustness. For improving the model methodology, complete and transparent reporting should be strictly ensured. Besides, Zhang et al. highlighted that reference genome and annotation updates cause inconsistent gene expression levels, leading to discordant individual risk grouping. Using up-to-date reference genome, stable gene in each annotation release (with consistent length and overlap), and gene pairs is helpful [18]. For clinical application, while decision curve analysis [130] compares the net benefit of the models with traditional approaches, the best methods for testing clinical significance are prospect clinical trials [123, 125, 126]. Health economic impact evaluations should also be considered [131], but we found that no study has reported the cost of predictor detection. Economic cost and effectiveness of models were critical for the medical decision-maker, like cost-effectiveness ratio; they also contributed to the optimization of risk thresholds. However, since few studies have conducted these methods, we emphasized increasing awareness of clinical considerations when a high-quality model was established.
Conclusively, we comprehensively reviewed all 138 machine learning genetic models in gliomas and proposed novel criteria that will foster the development or assessment of clinically important models for not only neurosurgeons but also researchers in other cancers. Given the current situation, a lot of effort should be put to standardize the model quality through adherence to complete and transparent reporting and promote model generalization by conducting prospective clinical trials, and economic effectiveness should be the following issue. Despite the various difficulties, future genetic models will lead the prognostic management, and novel gene-pair-based models deserve development.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NO. 82073893 to Q. C., NO. 81873635 to Z. L., and NO. 81703622 to Q. C.), the China Postdoctoral Science Foundation (NO. 2018M633002 to Q. C.), the Natural Science Foundation of Hunan Province (NO. 2018JJ3838 to Q. C. and NO. 2018SK2101 to Z. L.), the Hunan Provincial Health and Health Committee Foundation of China (NO. C2019186 to Q. C.), and Xiangya Hospital Central South University Postdoctoral Foundation (to Q. C.).
Abbreviations
- AUC:
Area under the curve
- EGFR:
Epidermal growth factor receptor
- EPV:
Events per variable
- GBM:
Glioblastoma multiforme
- HGG:
High-grade glioma
- LGG:
Low-grade glioma
- lncRNA:
Long noncoding RNA
- miRNA:
Micro RNA
- OS:
Overall survival
- WHO:
World Health Organization.
Contributor Information
Quan Cheng, Email: chengquan@csu.edu.cn.
Zhixiong Liu, Email: zhixiongliu@csu.edu.cn.
Data Availability
No data were used to support this study.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this article.
Authors' Contributions
Conception and design were done by Quan Cheng and Zhixiong Liu; administrative support was given by Quan Cheng and Zhixiong Liu; provision of study materials or patients was done by Xisong Liang and Zeyu Wang; collection and assembly of data were carried out by Ziyu Dai and Hao Zhang; data analysis and interpretation were performed by Xisong Liang and Zeyu Wang; manuscript writing was done by all authors (VII). All authors approved the final version of the manuscript.
Supplementary Materials
Table S1: characteristics of 73 models in the low-quality group.
References
- 1.Simmons C. P. L., McMillan D. C., McWilliams K., et al. Prognostic tools in patients with advanced cancer: a systematic review. Journal of Pain and Symptom Management. 2017;53(5):962–970. doi: 10.1016/j.jpainsymman.2016.12.330. [DOI] [PubMed] [Google Scholar]
- 2.Ngiam K. Y., Khor I. W. Big data and machine learning algorithms for health-care delivery. The Lancet Oncology. 2019;20(5):e262–e273. doi: 10.1016/s1470-2045(19)30149-4. [DOI] [PubMed] [Google Scholar]
- 3.Liang R., Zhi Y., Zheng G., Zhang B., Zhu H., Wang M. Analysis of long non-coding RNAs in glioblastoma for prognosis prediction using weighted gene co-expression network analysis, Cox regression, and L1-LASSO penalization. OncoTargets and Therapy. 2019;12:157–168. doi: 10.2147/OTT.S171957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang N., Dai Z., Wu W., et al. The predictive value of monocytes in immune microenvironment and prognosis of glioma patients based on machine learning. Frontiers in Immunology. 2021;12 doi: 10.3389/fimmu.2021.656541.656541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moons K. G. M., Royston P., Vergouwe Y., Grobbee D. E., Altman D. G. Prognosis and prognostic research: what, why, and how? BMJ. 2009;338:p. b375. doi: 10.1136/bmj.b375. [DOI] [PubMed] [Google Scholar]
- 6.Grimes D. A. The nomogram epidemic: resurgence of a medical relic. Annals of Internal Medicine. 2008;149(4):273–275. doi: 10.7326/0003-4819-149-4-200808190-00010. [DOI] [PubMed] [Google Scholar]
- 7.Steyerberg E. W., Harrell F. E., Jr. Prediction models need appropriate internal, internal-external, and external validation. Journal of Clinical Epidemiology. 2016;69:245–247. doi: 10.1016/j.jclinepi.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller J. J., Loebel F., Juratli T. A., et al. Accelerated progression of IDH mutant glioma after first recurrence. Neuro-Oncology. 2019;21(5):669–677. doi: 10.1093/neuonc/noz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louis D. N., Ohgaki H., Wiestler O. D., et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathologica. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brat D. J., Verhaak R. G., Aldape K. D., et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. New England Journal of Medicine. 2015;372(26):2481–2498. doi: 10.1056/nejmoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louis D. N., Perry A., Reifenberger G., et al. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathologica. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 12.Molinaro A. M., Wrensch M. R., Jenkins R. B., Eckel-Passow J. E. Statistical considerations on prognostic models for glioma. Neuro-Oncology. 2016;18(5):609–623. doi: 10.1093/neuonc/nov255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins G. S., Reitsma J. B., Altman D. G., Moons K. G. M. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. European Urology. 2015;67(6):1142–1151. doi: 10.1016/j.eururo.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 14.Alba A. C., Agoritsas T., Walsh M., et al. Discrimination and calibration of clinical prediction models. JAMA. 2017;318(14):1377–1384. doi: 10.1001/jama.2017.12126. [DOI] [PubMed] [Google Scholar]
- 15.Altman D. G., Vergouwe Y., Royston P., Moons K. G. M. Prognosis and prognostic research: validating a prognostic model. BMJ. 2009;338:p. b605. doi: 10.1136/bmj.b605. [DOI] [PubMed] [Google Scholar]
- 16.Wessler B. S., Lai Yh L., Kramer W., et al. Clinical prediction models for cardiovascular disease. Circulation: Cardiovascular Quality and Outcomes. 2015;8(4):368–375. doi: 10.1161/circoutcomes.115.001693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogundimu E. O., Altman D. G., Collins G. S. Adequate sample size for developing prediction models is not simply related to events per variable. Journal of Clinical Epidemiology. 2016;76:175–182. doi: 10.1016/j.jclinepi.2016.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z., Zhang S., Li X., et al. Reference genome and annotation updates lead to contradictory prognostic predictions in gene expression signatures: a case study of resected stage I lung adenocarcinoma. Briefings in Bioinformatics. 2020;22(3):p. bbaa081. doi: 10.1093/bib/bbaa081. [DOI] [PubMed] [Google Scholar]
- 19.Crick F. Central dogma of molecular biology. Nature. 1970;227(5258):561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 20.Li G., Jiang Y., Lyu X., et al. Gene signatures based on therapy responsiveness provide guidance for combined radiotherapy and chemotherapy for lower grade glioma. Journal of Cellular and Molecular Medicine. 2020;24(8):4726–4735. doi: 10.1111/jcmm.15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M., Wang X., Chen X., Zhang Q., Hong J. Novel immune-related gene signature for risk stratification and prognosis of survival in lower-grade glioma. Frontiers in Genetics. 2020;11:p. 363. doi: 10.3389/fgene.2020.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang M., Wang X., Chen X., Guo F., Hong J. Prognostic value of a stemness index-associated signature in primary lower-grade glioma. Frontiers in Genetics. 2020;11:p. 441. doi: 10.3389/fgene.2020.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao K., Liu Q., Peng G., Su J., Qin C.-Y., Wang X.-Y. Identification and validation of a three-gene signature as a candidate prognostic biomarker for lower grade glioma. PeerJ. 2020;8 doi: 10.7717/peerj.8312.e8312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng F., Liu X., Wang K., Zhao Z., Li G. Transcriptomic profiling identifies a DNA repair-related signature as a novel prognostic marker in lower grade gliomas. Cancer Epidemiology, Biomarkers & Prevention. 2019;28(12) doi: 10.1158/1055-9965.epi-19-0740. [DOI] [PubMed] [Google Scholar]
- 25.Su J., Long W., Ma Q., et al. Identification of a tumor microenvironment-related eight-gene signature for predicting prognosis in lower-grade gliomas. Frontiers in Genetics. 2019;10:p. 1143. doi: 10.3389/fgene.2019.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu B., Liu J., Liu K., et al. A prognostic signature of five pseudogenes for predicting lower-grade gliomas. Biomedicine & Pharmacotherapy. 2019;117 doi: 10.1016/j.biopha.2019.109116.109116 [DOI] [PubMed] [Google Scholar]
- 27.Zhang C., Yu R., Li Z., et al. Comprehensive analysis of genes based on chr1p/19q co-deletion reveals a robust 4-gene prognostic signature for lower grade glioma. Cancer Management and Research. 2019;11:4971–4984. doi: 10.2147/cmar.s199396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen B., Liang T., Yang P., et al. Classifying lower grade glioma cases according to whole genome gene expression. Oncotarget. 2016;7(45):74031–74042. doi: 10.18632/oncotarget.12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Q., Wang K., Huang R., et al. A novel DNA damage response signature of IDH-mutant grade II and grade III astrocytoma at transcriptional level. Journal of Cancer Research and Clinical Oncology. 2020;146(3):579–591. doi: 10.1007/s00432-020-03132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng W., Ren X., Zhang C., Cai J., Han S., Wu A. Gene expression profiling stratifies IDH1-mutant glioma with distinct prognoses. Molecular Neurobiology. 2017;54(8):5996–6005. doi: 10.1007/s12035-016-0150-6. [DOI] [PubMed] [Google Scholar]
- 31.Hsu J. B.-K., Chang T.-H., Lee G. A., Lee T.-Y., Chen C.-Y. Identification of potential biomarkers related to glioma survival by gene expression profile analysis. BMC Medical Genomics. 2019;11(S7):p. 34. doi: 10.1186/s12920-019-0479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li G., Jiang Y., Lyu X., et al. Deconvolution and network analysis of IDH-mutant lower grade glioma predict recurrence and indicate therapeutic targets. Epigenomics. 2019;11(11):1323–1333. doi: 10.2217/epi-2019-0137. [DOI] [PubMed] [Google Scholar]
- 33.Song L.-R., Weng J.-C., Huo X.-L., et al. Identification and validation of a 21-mRNA prognostic signature in diffuse lower-grade gliomas. Journal of Neuro-Oncology. 2020;146(1):207–217. doi: 10.1007/s11060-019-03372-z. [DOI] [PubMed] [Google Scholar]
- 34.Song L. R., Weng J. C., Li C. B., et al. Prognostic and predictive value of an immune infiltration signature in diffuse lower-grade gliomas. JCI Insight. 2020;5(8) doi: 10.1172/jci.insight.133811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng W.-J., Yang Y.-L., Liu Z.-Z., et al. Integrative analysis of DNA methylation and gene expression identify a three-gene signature for predicting prognosis in lower-grade gliomas. Cellular Physiology and Biochemistry. 2018;47(1):428–439. doi: 10.1159/000489954. [DOI] [PubMed] [Google Scholar]
- 36.Chai R. C., Wang N., Chang Y. Z., et al. Systematically profiling the expression of eIF3 subunits in glioma reveals the expression of eIF3i has prognostic value in IDH-mutant lower grade glioma. Cancer Cell International. 2019;19:p. 155. doi: 10.1186/s12935-019-0867-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ni J., Liu S., Qi F., et al. Screening TCGA database for prognostic genes in lower grade glioma microenvironment. Annals of Translational Medicine. 2020;8(5):p. 209. doi: 10.21037/atm.2020.01.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W., Li J., Lin F., Guo J., Zhao J. Expression and prognostic value of mRNAs in lower grade glioma with MGMT promoter methylated. Journal of Clinical Neuroscience. 2020;75:45–51. doi: 10.1016/j.jocn.2020.03.037. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y., Liu X., Guan G., Zhao W., Zhuang M. A risk classification system with five-gene for survival prediction of glioblastoma patients. Frontiers in Neurology. 2019;10:p. 745. doi: 10.3389/fneur.2019.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J., Wang L., Hu G., Wei B. A 6-gene risk signature predicts survival of glioblastoma multiforme. Biomed Research International. 2019;2019 doi: 10.1155/2019/1649423.1649423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu C., Zou C., Guan G., et al. Development and validation of an interferon signature predicting prognosis and treatment response for glioblastoma. Oncoimmunology. 2019;8(9) doi: 10.1080/2162402x.2019.1621677.e1621677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuo S., Zhang X., Wang L. A RNA sequencing-based six-gene signature for survival prediction in patients with glioblastoma. Scientific Reports. 2019;9(1):p. 2615. doi: 10.1038/s41598-019-39273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z., Gao L., Guo X., et al. Development and validation of a nomogram with an autophagy-related gene signature for predicting survival in patients with glioblastoma. Aging. 2019;11(24):12246–12269. doi: 10.18632/aging.102566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., Zhao W., Xiao Z., Guan G., Liu X., Zhuang M. A risk signature with four autophagy-related genes for predicting survival of glioblastoma multiforme. Journal of Cellular and Molecular Medicine. 2020;24(7):3807–3821. doi: 10.1111/jcmm.14938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gong Z., Hong F., Wang H., Zhang X., Chen J. An eight-mRNA signature outperforms the lncRNA-based signature in predicting prognosis of patients with glioblastoma. BMC Medical Genetics. 2020;21(1):p. 56. doi: 10.1186/s12881-020-0992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Tayrac M., Aubry M., Saïkali S., et al. A 4-gene signature associated with clinical outcome in high-grade gliomas. Clinical Cancer Research. 2011;17(2):317–327. doi: 10.1158/1078-0432.ccr-10-1126. [DOI] [PubMed] [Google Scholar]
- 47.Zhang C.-B., Zhu P., Yang P., et al. Identification of high risk anaplastic gliomas by a diagnostic and prognostic signature derived from mRNA expression profiling. Oncotarget. 2015;6(34):36643–36651. doi: 10.18632/oncotarget.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chai R., Zhang K., Wang K., et al. A novel gene signature based on five glioblastoma stem-like cell relevant genes predicts the survival of primary glioblastoma. Journal of Cancer Research and Clinical Oncology. 2018;144(3):439–447. doi: 10.1007/s00432-017-2572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q., Cai J., Fang C., et al. Mesenchymal glioblastoma constitutes a major ceRNA signature in the TGF-β pathway. Theranostics. 2018;8(17):4733–4749. doi: 10.7150/thno.26550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin S., Qian Z., Liang T., et al. Identification of a DNA repair-related multigene signature as a novel prognostic predictor of glioblastoma. World Neurosurgery. 2018;117:E34–E41. doi: 10.1016/j.wneu.2018.05.122. [DOI] [PubMed] [Google Scholar]
- 51.Ye N., Jiang N., Feng C., et al. Combined therapy sensitivity index based on a 13-gene signature predicts prognosis for IDH wild-type and MGMT promoter unmethylated glioblastoma patients. Journal of Cancer. 2019;10(22):5536–5548. doi: 10.7150/jca.30614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hou Z., Yang J., Wang H., Liu D., Zhang H. A potential prognostic gene signature for predicting survival for glioblastoma patients. BioMed Research International. 2019;2019 doi: 10.1155/2019/9506461.9506461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang P., Chai Y., Zhao H., Wang G. Predictive analyses of prognostic-related immune genes and immune infiltrates for glioblastoma. Diagnostics (Basel, Switzerland) 2020;10(3):p. 177. doi: 10.3390/diagnostics10030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bao Z.-S., Zhang C.-B., Wang H.-J., et al. Whole-genome mRNA expression profiling identifies functional and prognostic signatures in patients with mesenchymal glioblastoma multiforme. CNS Neuroscience & Therapeutics. 2013;19(9):714–720. doi: 10.1111/cns.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng Q., Huang C., Cao H., et al. A novel prognostic signature of transcription factors for the prediction in patients with GBM. Frontiers in Genetics. 2019;10:p. 906. doi: 10.3389/fgene.2019.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang W., Zhao Z., Wu F., et al. Bioinformatic analysis of gene expression and methylation regulation in glioblastoma. Journal of Neuro-Oncology. 2018;136(3):495–503. doi: 10.1007/s11060-017-2688-1. [DOI] [PubMed] [Google Scholar]
- 57.Wu F., Zhao Z., Chai R. C., et al. Prognostic power of a lipid metabolism gene panel for diffuse gliomas. Journal of Cellular and Molecular Medicine. 2019;23(11):7741–7748. doi: 10.1111/jcmm.14647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Liu X., Guan G., Xiao Z., Zhao W., Zhuang M. Identification of a five-pseudogene signature for predicting survival and its ceRNA network in glioma. Frontiers in Oncology. 2019;9:p. 1059. doi: 10.3389/fonc.2019.01059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng Z., Chen Y., Cao H., et al. Protein disulfide isomerases are promising targets for predicting the survival and tumor progression in glioma patients. Aging. 2020;12(3):2347–2372. doi: 10.18632/aging.102748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shahid M., Cho K. M., Nguyen M. N., et al. Prognostic value and their clinical implication of 89-gene signature in glioma. Oncotarget. 2016;7(32):51237–51250. doi: 10.18632/oncotarget.9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao K.-M., Chen X.-C., Zhang J.-X., Wang Y., Yan W., You Y.-P. A pseudogene-signature in glioma predicts survival. Journal of Experimental & Clinical Cancer Research. 2015;34(1):p. 23. doi: 10.1186/s13046-015-0137-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu X., Martinez-Ledesma E., Zheng S., et al. Multigene signature for predicting prognosis of patients with 1p19q co-deletion diffuse glioma. Neuro-Oncology. 2017;19(6):786–795. doi: 10.1093/neuonc/now285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chai R.-C., Wu F., Wang Q.-X., et al. m6A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging. 2019;11(4):1204–1225. doi: 10.18632/aging.101829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou Z., Huang R., Chai R., et al. Identification of an energy metabolism-related signature associated with clinical prognosis in diffuse glioma. Aging. 2018;10(11):3185–3209. doi: 10.18632/aging.101625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y.-Q., Chai R.-C., Wang Y.-Z., et al. Amino acid metabolism-related gene expression-based risk signature can better predict overall survival for glioma. Cancer Science. 2019;110(1):321–333. doi: 10.1111/cas.13878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun X., Liu X., Xia M., Shao Y., Zhang X. D. Multicellular gene network analysis identifies a macrophage-related gene signature predictive of therapeutic response and prognosis of gliomas. Journal of Translational Medicine. 2019;17(1):p. 159. doi: 10.1186/s12967-019-1908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y., Liu Y., Liu H., Zhao Z., Wu F., Zeng F. Clinical and biological significances of a methyltransferase-related signature in diffuse glioma. Frontiers in Oncology. 2020;10:p. 508. doi: 10.3389/fonc.2020.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeng F., Wang K., Liu X., Zhao Z. Comprehensive profiling identifies a novel signature with robust predictive value and reveals the potential drug resistance mechanism in glioma. Cell Communication and Signaling. 2020;18(1):p. 2. doi: 10.1186/s12964-019-0492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu W., Liu Z., Ren H., et al. Twenty metabolic genes based signature predicts survival of glioma patients. Journal of Cancer. 2020;11(2):441–449. doi: 10.7150/jca.30923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peng Z., Liu C., Wu M. New insights into long noncoding RNAs and their roles in glioma. Molecular Cancer. 2018;17(1):p. 61. doi: 10.1186/s12943-018-0812-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu C., Zhou Y., Liu C., Kang Y. Risk assessment model constructed by differentially expressed lncRNAs for the prognosis of glioma. Oncology Reports. 2018;40(5):2467–2476. doi: 10.3892/or.2018.6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou M., Zhang Z., Zhao H., Bao S., Cheng L., Sun J. An immune-related six-lncRNA signature to improve prognosis prediction of glioblastoma multiforme. Molecular Neurobiology. 2018;55(7):3684–3697. doi: 10.1007/s12035-017-0572-9. [DOI] [PubMed] [Google Scholar]
- 73.Chen G., Cao Y., Zhang L., Ma H., Shen C., Zhao J. Analysis of long non-coding RNA expression profiles identifies novel lncRNA biomarkers in the tumorigenesis and malignant progression of gliomas. Oncotarget. 2017;8(40):67744–67753. doi: 10.18632/oncotarget.18832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang W., Yang F., Zhang L., et al. LncRNA profile study reveals four-lncRNA signature associated with the prognosis of patients with anaplastic gliomas. Oncotarget. 2016;7(47):77225–77236. doi: 10.18632/oncotarget.12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kiran M., Chatrath A., Tang X., Keenan D. M., Dutta A. A prognostic signature for lower grade gliomas based on expression of long non-coding RNAs. Molecular Neurobiology. 2019;56(7):4786–4798. doi: 10.1007/s12035-018-1416-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang X.-Q., Sun S., Lam K.-F., et al. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiology of Disease. 2013;58:123–131. doi: 10.1016/j.nbd.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 77.Li D., Lu J., Li H., Qi S., Yu L. Identification of a long noncoding RNA signature to predict outcomes of glioblastoma. Molecular Medicine Reports. 2019;19(6):5406–5416. doi: 10.3892/mmr.2019.10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xia P., Li Q., Wu G., Huang Y. An immune-related lncRNA signature to predict survival in glioma patients. Cellular and Molecular Neurobiology. 2021;41(2):365–375. doi: 10.1007/s10571-020-00857-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Steyerberg E. W., Vergouwe Y. Towards better clinical prediction models: seven steps for development and an ABCD for validation. European Heart Journal. 2014;35(29):1925–1931. doi: 10.1093/eurheartj/ehu207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anthiya S., Griveau A., Loussouarn C., et al. MicroRNA-based drugs for brain tumors. Trends in Cancer. 2018;4(3):222–238. doi: 10.1016/j.trecan.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 81.Qian Z., Li Y., Fan X., et al. Prognostic value of a microRNA signature as a novel biomarker in patients with lower-grade gliomas. Journal of Neuro-Oncology. 2018;137(1):127–137. doi: 10.1007/s11060-017-2704-5. [DOI] [PubMed] [Google Scholar]
- 82.Chen W., Yu Q., Chen B., Lu X., Li Q. The prognostic value of a seven-microRNA classifier as a novel biomarker for the prediction and detection of recurrence in glioma patients. Oncotarget. 2016;7(33):53392–53413. doi: 10.18632/oncotarget.10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cheng W., Ren X., Cai J., et al. A five-miRNA signature with prognostic and predictive value for MGMT promoter-methylated glioblastoma patients. Oncotarget. 2015;6(30):29285–29295. doi: 10.18632/oncotarget.4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ushijima T., Suzuki H. The origin of CIMP, at last. Cancer Cell. 2019;35(2):165–167. doi: 10.1016/j.ccell.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 85.Malta T. M., De Souza C. F., Sabedot T. S., et al. Glioma CpG island methylator phenotype (G-CIMP): biological and clinical implications. Neuro-Oncology. 2018;20(5):608–620. doi: 10.1093/neuonc/nox183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yin A.-A., Lu N., Etcheverry A., et al. A novel prognostic six-CpG signature in glioblastomas. CNS Neuroscience & Therapeutics. 2018;24(3):167–177. doi: 10.1111/cns.12786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yin A. A., He Y. L., Etcheverry A., et al. Novel predictive epigenetic signature for temozolomide in non-G-CIMP glioblastomas. Clinical Epigenetics. 2019;11(1):p. 76. doi: 10.1186/s13148-019-0670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shukla S., Pia Patric I. R., Thinagararjan S., et al. A DNA methylation prognostic signature of glioblastoma: identification of NPTX2-PTEN-NF-κB nexus. Cancer Research. 2013;73(22):6563–6573. doi: 10.1158/0008-5472.can-13-0298. [DOI] [PubMed] [Google Scholar]
- 89.Stetson L. C., Dazard J.-E., Barnholtz-Sloan J. S. Protein markers predict survival in glioma patients. Molecular & Cellular Proteomics. 2016;15(7):2356–2365. doi: 10.1074/mcp.m116.060657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Patil V., Mahalingam K. A four-protein expression prognostic signature predicts clinical outcome of lower-grade glioma. Gene. 2018;679:57–64. doi: 10.1016/j.gene.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 91.Gao W.-Z., Guo L.-M., Xu T.-Q., Yin Y.-H., Jia F. Identification of a multidimensional transcriptome signature for survival prediction of postoperative glioblastoma multiforme patients. Journal of Translational Medicine. 2018;16(1):p. 368. doi: 10.1186/s12967-018-1744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Etcheverry A., Aubry M., Idbaih A., et al. DGKI methylation status modulates the prognostic value of MGMT in glioblastoma patients treated with combined radio-chemotherapy with temozolomide. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0104455.e104455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xiong J., Bing Z., Su Y., Deng D., Peng X. An integrated mRNA and microRNA expression signature for glioblastoma multiforme prognosis. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0098419.e98419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eckel-Passow J. E., Lachance D. H., Molinaro A. M., et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. New England Journal of Medicine. 2015;372(26):2499–2508. doi: 10.1056/nejmoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li T., Forbes M. E., Fuller G. N., Li J., Yang X., Zhang W. IGFBP2: integrative hub of developmental and oncogenic signaling network. Oncogene. 2020;39(11):2243–2257. doi: 10.1038/s41388-020-1154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cai J., Chen Q., Cui Y., et al. Immune heterogeneity and clinicopathologic characterization of IGFBP2 in 2447 glioma samples. Oncoimmunology. 2018;7(5) doi: 10.1080/2162402x.2018.1426516.e1426516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Haberland M., Montgomery R. L., Olson E. N. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature Reviews Genetics. 2009;10(1):32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Eyüpoglu I. Y., Savaskan N. E. Epigenetics in brain tumors: HDACs take center stage. Current Neuropharmacology. 2016;14(1):48–54. doi: 10.2174/1570159x13666151030162457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peters K. B., Lipp E. S., Miller E., et al. Phase I/II trial of vorinostat, bevacizumab, and daily temozolomide for recurrent malignant gliomas. Journal of Neuro-Oncology. 2018;137(2):349–356. doi: 10.1007/s11060-017-2724-1. [DOI] [PubMed] [Google Scholar]
- 100.Bradshaw A., Wickremsekera A., Tan S. T., Peng L., Davis P. F., Itinteang T. Cancer stem cell hierarchy in glioblastoma multiforme. Frontiers in Surgery. 2016;3:p. 21. doi: 10.3389/fsurg.2016.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Verhaak R. G. W., Hoadley K. A., Purdom E., et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mooney K. L., Choy W., Sidhu S., et al. The role of CD44 in glioblastoma multiforme. Journal of Clinical Neuroscience. 2016;34:1–5. doi: 10.1016/j.jocn.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 103.Filippou P. S., Karagiannis G. S., Constantinidou A. Midkine (MDK) growth factor: a key player in cancer progression and a promising therapeutic target. Oncogene. 2020;39:2040–2054. doi: 10.1038/s41388-019-1124-8. [DOI] [PubMed] [Google Scholar]
- 104.Weterman M. A. J., Ajubi N., Van Dinter I. M. R., et al. nmb, a novel gene, is expressed in low-metastatic human melanoma cell lines and xenografts. International Journal of Cancer. 1995;60(1):73–81. doi: 10.1002/ijc.2910600111. [DOI] [PubMed] [Google Scholar]
- 105.Bao G., Wang N., Li R., Xu G., Liu P., He B. Glycoprotein non-metastaticmelanoma protein B promotes glioma motility and angiogenesis through the Wnt/β-catenin signaling pathway. Experimental Biology and Medicine. 2016;241(17):1968–1976. doi: 10.1177/1535370216654224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ono Y., Chiba S., Yano H., et al. Glycoprotein nonmetastatic melanoma protein B (GPNMB) promotes the progression of brain glioblastoma via Na+/K+-ATPase. Biochemical and Biophysical Research Communications. 2016;481(1-2):7–12. doi: 10.1016/j.bbrc.2016.11.034. [DOI] [PubMed] [Google Scholar]
- 107.Szulzewsky F., Pelz A., Feng X., et al. Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1. PLoS One. 2015;10(2) doi: 10.1371/journal.pone.0116644.e0116644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Taya M., Hammes S. R. Glycoprotein non-metastatic melanoma protein B (GPNMB) and cancer: a novel potential therapeutic target. Steroids. 2018;133:102–107. doi: 10.1016/j.steroids.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brennan C. W., Verhaak R. G., McKenna A., et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Weller M., Wick W., Aldape K., et al. Glioma. Nature Reviews Disease Primers. 2015;1(1):p. 15017. doi: 10.1038/nrdp.2015.17. [DOI] [PubMed] [Google Scholar]
- 111.Keller S., Schmidt M. H. H. EGFR and EGFRvIII promote angiogenesis and cell invasion in glioblastoma: combination therapies for an effective treatment. International Journal of Molecular Sciences. 2017;18(6):p. 1295. doi: 10.3390/ijms18061295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Eskilsson E., Røsland G. V., Solecki G., et al. EGFR heterogeneity and implications for therapeutic intervention in glioblastoma. Neuro-Oncology. 2018;20(6):743–752. doi: 10.1093/neuonc/nox191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Goel H. L., Mercurio A. M. VEGF targets the tumour cell. Nature Reviews Cancer. 2013;13(12):871–882. doi: 10.1038/nrc3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jhaveri N., Chen T. C., Hofman F. M. Tumor vasculature and glioma stem cells: contributions to glioma progression. Cancer Letters. 2016;380(2):545–551. doi: 10.1016/j.canlet.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 115.Zhang Z., Cui B. Z., Wu L. H., Xu Q. L., Wang Z., Yang B. The inhibition effect of expressions of miR-221 and miR-222 on glioma and corresponding mechanism. Bratislava Medical Journal. 2014;115(11):685–691. doi: 10.4149/bll_2014_133. [DOI] [PubMed] [Google Scholar]
- 116.Brognara E., Fabbri E., Montagner G., et al. High levels of apoptosis are induced in human glioma cell lines by co-administration of peptide nucleic acids targeting miR-221 and miR-222. International Journal of Oncology. 2016;48(3):1029–1038. doi: 10.3892/ijo.2015.3308. [DOI] [PubMed] [Google Scholar]
- 117.Xue L., Wang Y., Yue S., Zhang J. The expression of miRNA-221 and miRNA-222 in gliomas patients and their prognosis. Neurological Sciences. 2017;38(1):67–73. doi: 10.1007/s10072-016-2710-y. [DOI] [PubMed] [Google Scholar]
- 118.Ramaswamy V., Remke M., Bouffet E., et al. Risk stratification of childhood medulloblastoma in the molecular era: the current consensus. Acta Neuropathologica. 2016;131(6):821–831. doi: 10.1007/s00401-016-1569-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nwaobi S. E., Cuddapah V. A., Patterson K. C., Randolph A. C., Olsen M. L. The role of glial-specific Kir4.1 in normal and pathological states of the CNS. Acta Neuropathologica. 2016;132(1):1–21. doi: 10.1007/s00401-016-1553-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Qin G., Li X., Chen Z., et al. Prognostic value of YKL-40 in patients with glioblastoma: a systematic review and meta-analysis. Molecular Neurobiology. 2017;54(5):3264–3270. doi: 10.1007/s12035-016-9878-2. [DOI] [PubMed] [Google Scholar]
- 121.Batista K. M. P., Eulate-Beramendi S. A. D., Pińa K. Y. Á. R. D., et al. Mesenchymal/proangiogenic factor YKL-40 related to glioblastomas and its relationship with the subventricular zone. Folia Neuropathologica. 2017;55(1):14–22. doi: 10.5114/fn.2017.66709. [DOI] [PubMed] [Google Scholar]
- 122.Swiatek-Machado K., Kaminska B. STAT signaling in glioma cells. Advances in Experimental Medicine and Biology. 2013;986:189–208. doi: 10.1007/978-94-007-4719-7_10. [DOI] [PubMed] [Google Scholar]
- 123.Kluth L. A., Black P. C., Bochner B. H., et al. Prognostic and prediction tools in bladder cancer: a comprehensive review of the literature. European Urology. 2015;68(2):238–253. doi: 10.1016/j.eururo.2015.01.032. [DOI] [PubMed] [Google Scholar]
- 124.Bradley A., Van Der Meer R., McKay C. J. A systematic review of methodological quality of model development studies predicting prognostic outcome for resectable pancreatic cancer. BMJ Open. 2019;9(8) doi: 10.1136/bmjopen-2018-027192.e027192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Klatte T., Rossi S. H., Stewart G. D. Prognostic factors and prognostic models for renal cell carcinoma: a literature review. World Journal of Urology. 2018;36(12):1943–1952. doi: 10.1007/s00345-018-2309-4. [DOI] [PubMed] [Google Scholar]
- 126.Mahar A. L., Compton C., Halabi S., Hess K. R., Weiser M. R., Groome P. A. Personalizing prognosis in colorectal cancer: a systematic review of the quality and nature of clinical prognostic tools for survival outcomes. Journal of Surgical Oncology. 2017;116(8):969–982. doi: 10.1002/jso.24774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Phung M. T., Tin Tin S., Elwood J. M. Prognostic models for breast cancer: a systematic review. BMC Cancer. 2019;19(1):p. 230. doi: 10.1186/s12885-019-5442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Veen K. M., De Angst I. B., Mokhles M. M., et al. A clinician’s guide for developing a prediction model: a case study using real-world data of patients with castration-resistant prostate cancer. Journal of Cancer Research and Clinical Oncology. 2020;146(8):2067–2075. doi: 10.1007/s00432-020-03286-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xiao G., Zhang X., Zhang X., et al. Aging-related genes are potential prognostic biomarkers for patients with gliomas. Aging. 2021;13(9):13239–13263. doi: 10.18632/aging.203008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vickers A. J., Elkin E. B. Decision curve analysis: a novel method for evaluating prediction models. Medical Decision Making. 2006;26(6):565–574. doi: 10.1177/0272989x06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Van Giessen A., Peters J., Wilcher B., et al. Systematic review of health economic impact evaluations of risk prediction models: stop developing, start evaluating. Value in Health. 2017;20(4):718–726. doi: 10.1016/j.jval.2017.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: characteristics of 73 models in the low-quality group.
Data Availability Statement
No data were used to support this study.


