Abstract
Background
Primary ciliary dyskinesia (PCD) is a ciliopathy with diverse clinical and genetic findings caused by abnormal motile cilia structure and function. In this study, we describe the clinical characteristics of confirmed PCD cases in our population and report the radiological, genetic, and laboratory findings.
Methods
This was a retrospective, observational, single-centre study. We enrolled 18 patients who were diagnosed with confirmed PCD between 2015 and 2019. We then analyzed their data, including clinical findings and workup.
Results
In our cohort, 56% of patients had molecularly confirmed PCD, and RSPH9 was the most common gene identified. Transmission electron microscopy (TEM) showed an ultrastructural defect in 64% of samples, all of which matched the genetic background of the patient. Situs inversus (SI) was observed in 50% of patients, and congenital heart disease was observed in 33%. The median body mass index (BMI) was 15.87 kg/m2, with a median z score of -1.48. The median FEV1 value was 67.6% (z score - 2.43). Radiologically, bronchiectasis was noted in 81% of patients at a variable degree of severity. Lung bases were involved in 91% of patients. We were unable to correlate the genotype-phenotype findings.
Conclusion
We describe the clinical and molecular characteristics of patients with confirmed PCD in a tertiary centre in Saudi Arabia and report 9 new pathogenic or likely pathogenic variants in one of the PCD-associated genes.
Keywords: Primary ciliary dyskinesia, Bronchiectasis, Situs inversus, Electron microscopy, Genetic testing
Abbreviations: PCD, Primary ciliary dyskinesia; TEM, Transmission electron microscopy; BMI, body mass index; HSVA, high-speed video microscopy analysis; nNO, nasal nitric oxide; WES, whole-exome sequencing; NGS, next-generation sequencing; ODA, outer dynein arm; IDA, inner dynein arm
1. Background
Primary ciliary dyskinesia (PCD) is a ciliopathy that affects the structure and function of motile cilia. The prevalence is estimated to be 1:15,000–30,000 [1], and as expected, it is high in areas with a high rate of consanguinity [2]. The clinical presentation of PCD includes neonatal respiratory distress, recurrent respiratory infections, sinusitis and bronchiectasis due to impaired mucociliary clearance in the upper and lower airways. Male and female subfertility also results from defective cilia in sperm flagella and fallopian tubes, respectively. Approximately 50% of affected children have situs inversus (SI). Other clinical manifestations include hydrocephalus and complex heart disease due to defective ependymal cilia and nodal cilia, respectively [3].
Transmission electron microscopy (TEM) is used as the mainstay to diagnose PCD. Approximately 30% of PCD cases have normal or nondiagnostic TEM findings [4]. Other diagnostic modalities include high-speed video microscopy analysis (HSVA), nasal nitric oxide (nNO) and abnormal ciliary protein patterns on immunofluorescent staining. With recent advances in molecular diagnostic techniques, including whole exome sequencing (WES), several novel PCD-associated genes are being increasingly reported. More than 40 different genes are already known to be associated with PCD. Nevertheless, 20%–30% of individuals with PCD do not have identifiable pathogenic variants in any of the associated genes [5,6].
PCD is underdiagnosed, and diagnosis is often delayed due to inadequate awareness, the complexity of diagnostic testing and lack of a consensus on a gold standard diagnostic test. This is unfortunate as a delayed diagnosis results in increased long-term pulmonary morbidity. The aim of this study was to identify the molecular and clinical characteristics of paediatric patients with PCD in Saudi Arabia.
2. Methods
We conducted a retrospective, observational, single-centre study. We enrolled all patients diagnosed with confirmed PCD between 2015 and 2019 who were followed up in our PCD programme at Children Specialized Hospital (CSH), King Fahad Medical City (KFMC), a tertiary centre in Saudi Arabia. Ethical approval was obtained from the institutional review board (IRB Log No. 19–524). Patient data were collected from our electronic health information medical system.
A total of 32 patients were followed up in our PCD programme. These patients were categorised as confirmed, likely or unlikely PCD in accordance with recent guidelines by the European Respiratory Society (ERS) and American Thoracic Society (ATS) [7]. Patients who had pathogenic/likely pathogenic biallelic variants in one of the PCD-associated genes were categorised as confirmed PCD (18 patients). Patients with clinical and radiological findings consistent with PCD but negative genetic tests were categorised as likely PCD after excluding other causes of chronic suppurative lung disease (14 patients). These 14 patients were excluded from our study. We calculated the PICADAR score, a recommended screening tool to predict the likelihood of PCD [8].
Spirometry results (FEV1 and FEF25-75), body mass index (BMI), respiratory secretion cultures (sputum and nasopharyngeal aspirate) and radiological findings were taken into account to describe the phenotype. Spirometry values are expressed in litres as the predicted percentages of an age-matched healthy population (Global Lung Initiative references). ATS standards were adhered to when spirometry manoeuvres were performed. The z score for BMI was calculated, and GLI equations were used to calculate spirometry indices.
TEM samples were obtained by ENT surgeons who are part of the PCD programme. Nasal brush biopsy was the standard method used to obtain samples. If a sample was inadequate, a repeat sample was taken through nasal punch biopsy. Bronchial brush biopsy was obtained if bronchoscopy was performed for any other reason. Samples were maintained in glutaraldehyde solution until fixation. All samples were interpreted by a single histopathologist to ensure consistency of the results.
In most patients, next-generation sequencing panels for PCD were performed in commercial laboratories. WES was performed using Agilent SureSelect version 5 kits on an Illumina HiSeq 4000 system to an average depth of coverage of 150x with automated adapter trimming of the fast sequences (BGI Europe). DNA sequence quality metrics were assessed using FastQC version 0.11.7 at King Fahad Medical City. Alignment, quality filtering and variant identification were undertaken using commercially available algorithms (DNAStar and Qiagen Clinical Insight-Interpret software). Human reference assemblies were aligned against GRCh37.
3. Results
Eighteen patients (56%) were categorised as confirmed PCD based on positive genetic tests showing homozygous pathogenic or likely pathogenic variants in one of the PCD-associated genes (Table 1, Table 2). Additionally, two patients (patients 9 and 19) were homozygous for variants of unknown significance (VUSs) (DNAH5: c.972+5G > A (software prediction tool predicted Splicing impact) and DNAL1 c.529G > C (p.Asp177His) (In silico prediction tools predicted a damaging effect)). In the remainder of patients, test results were negative in three individuals, while others were heterozygous for one or more variants, but none were compound heterozygous for variants in the same gene. TEM samples were obtained from 14 patients. Samples were obtained nasally in 79% of patients and 21% through a bronchial brush sample. Nine patients (64%) had positive findings, while the remaining had either inadequate findings or technical challenges. All results matched the genetic background of the patients (Table 1).
Table 1.
Summary of the clinical and investigation findings in patients with confirmed PCD. IDA: inner dynein arm and ODA: outer dynein arm.
| Genetic defect | Mean Age of diagnosis (years) | PICADAR score | Situs inversus (n) | Cardiac defect (n) | BMI | Mean Spirometry |
CT chest |
TEM | ||
|---|---|---|---|---|---|---|---|---|---|---|
| FEV1 (%) | Z score | Timing (years) | Findings | |||||||
| RSPH9 (n = 3) | 9.5 | 6.3 | 0/3 | 1/3 (ASD) | 15.9 | 64.9 (3/3) | −3.35 | 10 | Bronchiectasis (cystic) (n = 3) | Absent central microtubules and supernumerary |
| CCNO (n = 2) | 5.2 | 5.5 | 0/2 | 0/2 | 12.1 | 56.3 (1/2) | −3.43 | 6 | Bronchiectasis (cylindrical) (n = 2) | No cilia (n = 2) |
| ZMYND10 (n = 3) | 7.6 | 9.7 | 2/3 | 1/3 (ASD) | 17.7 | 74 (3/3) | −1.49 | 9.7 | Early bronchiectatic changes (Signet rings) (n = 2) Bronchiectasis (cylindrical) (n = 1) |
IDA and ODA |
| DNAAF3 (n = 2) | 0.2 | 11 | 2/2 | 0/2 | 15.5 | 47 (1/2) | −4.54 | 13 | Bronchiectasis (cylindrical) (n = 1) | IDA and ODA |
| DNAH5 (n = 1) | 0.2 | 13 | 1/1 | 1/1 (ASD) | 16 | 71.6 (1/1) | −2.05 | 5 | Normal (n = 1) | NA |
| DNAAF4 (n = 2) | 0.2 | 11 | 2/2 | 1/2 (PDA) | 14.5 | 83.4 (1/2) | −1.11 | 7 | Normal (n = 1) | IDA and ODA |
| DNAI1 (n = 1) | 0.1 | 14 | 1/1 | 1/1 (ASD) | 16 | NA | NA | 4 | Normal (n = 1) | NA |
| CCDC151 (n = 2) | 7.1 | 8 | 1/2 | 0/2 | 18.4 | 79 (1/2) | −0.46 | 8 | Bronchiectasis (cylindrical) (n = 2) | ODA |
| DNAAF1 (n = 1) | 6.4 | 8 | 0/1 | 0/1 | 14.3 | 59.6 (1/1) | −3.05 | 10 | Bronchiectasis (cystic) (n = 1) | IDA |
| CCDC39 (n = 1) | 2.6 | 6 | 0/1 | 1/1 (PDA) | 12 | NA | NA | 5 | Bronchiectasis (cylindrical) (n = 1) | IDA/Abnormal ciliary orientation |
| Total (n = 18) | 4.74 | 8.89 | 9 | 6 | 15.47 | 67.6 | −2.4 | 8.19 | 9 | |
Table 2.
List of patients homozygous for variants in known PCD genes. Patients 4 and 32 are siblings and patients 6 and 15 are cousins. LP: likely pathogenic, P: pathogenic and VUS: Variants of unknown significance.
| Patient no. | Gene | DNA nucleotide change | Protein amino acid change | ACMG classification | Note |
|---|---|---|---|---|---|
| 1 | RSPH9 | c.804_806delGAA | p.Lys268del | P | – |
| 2 | DNAAF1 | Exon 5 deletion | NA | LP | Deletion including exon 5 was reported (Davis et al., 2019) |
| 3 | CCDC39 | c.1061del | p.Glu354Glysfs∗2 | LP | Previously unpublished |
| 4 | CCDC151 | c.925G > T | p.Glu309∗ | P | – |
| 5 | CCNO | c.425del | p.Pro142Argfs∗15 | LP | – |
| 6 | ZMYND10 | c.155_158del | p.Val52Alafs∗23 | P | Previously unpublished |
| 7 | RSPH9 | c.804_806delGAA | p.Lys268del | P | – |
| 9 | DNAH5 | c.9720+5G > A | NA | VUS | Previously unpublished, reported in ClinVar (Accession: VCV000454819) |
| 11 | DNAAF3 | c.1105C > T | p.Gln369∗ | LP | Previously unpublished |
| 14 | RSPH9 | c.804_806delGAA | p.Lys268del | P | – |
| 15 | ZMYND10 | c.155_158del | p.Val52Alafs∗23 | P | Previously unpublished |
| 16 | DNAAF4 | c.271+1G > T | NA | LP | Previously unpublished |
| 17 | DNAH5 | c.2278_2279del | p.Gln760Gulfs∗11 | LP | Previously unpublished |
| 19 | DNAL1 | c.529G > C | p.Asp177His | VUS | Previously unpublished |
| 22 | CCNO | c.833dup | p.Tyr278∗ | LP | Previously unpublished |
| 24 | DNAAF4 | c.1111C > T | p.Arg371∗ | LP | Previously unpublished |
| 27 | DNAL1 | c.1311+2T > A | NA | LP | Previously unpublished |
| 29 | ZMYND10 | c.1091C > G | p.Ser364∗ | LP | Previously unpublished |
| 30 | DNAAF3 | c.469C > T | p.Arg157∗ | P | – |
| 32 | CCDC151 | c.925G > T | p.Glu309∗ | P | – |
In patients with confirmed PCD, the mean age at diagnosis was 4.7 years (2 weeks-11.7 years), whereas the mean age at the last follow-up was 9.2 years (4 months-14 years). Unexplained neonatal respiratory distress was reported in 83% of patients (n = 15). All patients with confirmed PCD had chronic cough and chronic rhinitis. Moreover, 50% had SI and 33% had congenital heart disease in the form of septal defects. The average number of hospitalisations was 4.7 per patient. The average PICADAR score was 8.9, with 94% of patients scoring ≥5 and 39% scoring ≥10. The median BMI was 15.87 kg/m2, and the median z score was −1.48.
In our study, 12/18 patients with confirmed PCD could undergo spirometry reliably. The median values for FEV1 and FEF25-75 were 67.6% (z score −2.43) and 36.5% (z score −3.16), respectively. The z scores of FEV1 (r = 0.72) and FEF25-75 (r = 0.67) had a significant moderate positive correlation with the z score of BMI. The most common isolated organism was Haemophilus influenza (35%), followed by methicillin-resistant Staphylococcus aureus (26%) and Streptococcus pneumonia (22%). Other less frequently isolated organisms included methicillin-sensitive Staphylococcus aureus and Pseudomonas species.
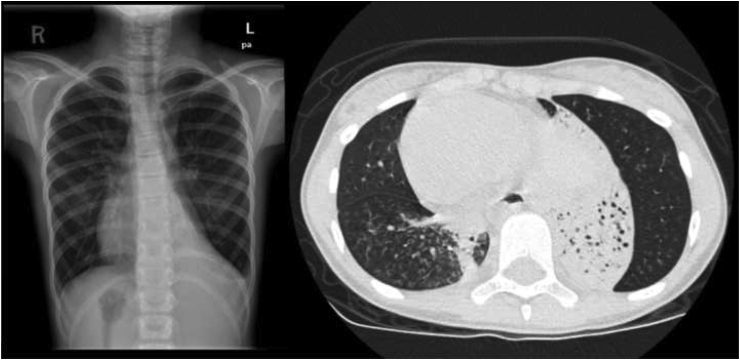
CT scans of the chest were performed in 16 patients with confirmed PCD at a mean age of 8.2 years. Bronchiectasis, including early bronchiectatic changes (peribronchial thickening and signet ring) (2/13), cylindrical bronchiectasis (7/13) and cystic bronchiectasis (4/13), was found in 13 (81%) patients at variable severity. The lung bases were likely to be affected (92% of patients), with the right middle and left lower lobes being the most commonly involved areas. A tree-in-bud appearance was observed in 77% of patients (Fig. 1).
Fig. 1.
Chest X-ray shows dextrocardia and hyperinflation with bronchial thickening. There is an atelectatic lung segment on the left lower lobe (arrow head). CT scan of the chest confirms these findings. There is an atelectatic bronchiectatic segment in the left lower lobe (arrow). A tree-in-bud appearance can be appreciated on the right lower lobe (star).
4. Discussion
PCD is a clinically and genetically heterogeneous disease that makes a confirmed diagnosis challenging [5]. An ultrastructural ciliary assessment through TEM was previously the gold standard for diagnosis. However, this changed recently as it has some drawbacks as a diagnostic test; some disease-causing mutations could result in a normal axonal structure, and there are difficulties to identify certain structural defects [4,9,10]. Extended gene panel testing is one of the best diagnostic modalities; however, it has limitations, including the interpretation of VUSs and the possibility of missing novel PCD genes that are not included in the panel [11]. Other diagnostic and screening tools are not without limitations. nNO requires a chemiluminescence analyser with a velum closure to yield accurate results and is technically a challenge. HSVM, which is used to measure ciliary beat frequency and pattern, requires a high degree of expertise and training [12,13].
In our practice, we utilised genetic tests as confirmatory diagnostic tests. TEM was used as a complementary test because it was recently introduced into our practice and because of the limited experience of our histopathologist. Unfortunately, we did not have access to HSVM or nNO, which is one of the limitations of our study. If confirmatory tests were positive, patients were categorised as confirmed PCD. Otherwise, they were categorised as likely PCD [7]. In the latter patients, we excluded other diagnoses by performing a CT chest scan, a sweat chloride test and an immunological work-up.
Eighteen (56%) patients had confirmed PCD. These patients had homozygous pathogenic or likely pathogenic variants in one of the PCD-associated genes (Table 2). Two patients in the likely PCD group (patients 9 and 19) were homozygous for previously unpublished VUSs. Both patients had recurrent sinopulmonary infection and bronchiectasis; one had SI and the other had two siblings with similar manifestations in addition to unexplained neonatal respiratory distress and SI. Further genetic testing and TEM are being conducted to help upgrade these variants.
The most common variant identified in our study was c.804_806delGAA (p.Lys268del) in the RSPH9 gene (observed in three different families). In a recent large cohort study conducted in Saudi Arabia, this variant was the most common, which accounted for 34% of families with molecularly confirmed PC, even though it depended on WES and our results were mainly through gene panels [14]. It is worth to mention that studies conducted in other Arab countries with different ancestries (Egypt and Tunisia) reported CCDC39 as the most commonly involved gene in their population [15,16]. This is in contrast to the American Thoracic Society report of DNAH5 and DNI1 being the most common genes associated with PCD [17]. All other pathogenic or likely pathogenic variants in our cohort were encountered in individual families. Nine of the 15 pathogenic or likely pathogenic variants that we report are novel (Table 2). Our cohort showed a high rate of parental consanguinity (83%), consistent with that reported in the cohort from Turkey but much higher than that reported in PCD cohorts from Belgium (19.6%) [18,19].
The identification of ciliary ultrastructure defects under TEM is one of the best diagnostic tools, with a detection rate of 74%, which is comparable to our rate of 69% [4,11]. We acquired samples through nasal brush biopsy. If a sample was inadequate, a repeat sample was taken through nasal punch biopsy. We performed bronchial brush biopsy only in patients who require bronchoscopy [20]. Ciliary ultrastructural defects are caused by alterations in different cytoplasmic proteins, and each is linked to a specific PCD genetic mutation [3,9]. In our study, combined outer dynein arm (ODA) and inner dynein arm (IDA) defects were the most common. These defects were identified in patients with mutations in the ZMYND10, DNAAF1, DNAAF3 and DNAAF4 genes [[21], [22], [23], [24]]. ODA defects alone were associated with CCDC151, IDA defects in combination with ciliary axonal disorientation were associated with CCDC39, and the absence of cilia was associated with CCNO [9,25,26]. Patients with RSPH9 mutations did not have the central microtubule complex; however, we could not appreciate the usual finding of radial spoke absence, which is difficult to distinguish because of the blurry, dense background [9,27].
Given the small number of patients, we could not ascertain the genotype-phenotype correlation. However, similar to reports in the literature, organ laterality was identified in 50% of patients, all of whom had SI [11,28,29]. Patients with situs solitus (SS) had a delayed mean age at diagnosis (6.8 vs 2.6 years) as compared to patients with SI (Table 3). The SS group had more severe lung disease even though the SI group was older at the final follow-up (9.8 vs 8.5 years). This finding was also evident by the average number of admissions (6.56 vs 2.89), mean BMI (14.3 vs 16.7), FEV1 (61.6 vs 74%) and advanced bronchiectasis on CT scans in the SS group. This observation is also supported by Keuhni et al., who advocated for more clinical awareness and not to delay the investigation of patients with a clinical suspicion of PCD because early detection and appropriate management may decrease recurrent infections, the loss of lung function and the progression of bronchiectasis [7,12,30].
Table 3.
Comparison between PCD patient with situs solitus and those with situs inversus.
| n | Age of diagnosis (years) | Average age of cases | PICADAR score | Recurrent admissions | Mean BMI | Spirometry |
Radiological evaluation | Average age ofstudy (years) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| FEV1 (%) | Z score | |||||||||
| Situs solitus | 9 | 6.9 | 9.8 | 6.6 | 6.56 | 14.3 | 61.6 | −3.1 | Cylindrical bronchiectasis (N = 5) | 6.4 |
| Cystic bronchiectasis (n = 4) | 10 | |||||||||
| Situs inversus | 9 | 2.6 | 8.5 | 11.2 | 2.89 | 16.7 | 74 | −1.8 | Normal (n = 3) | 5 |
| Early bronchiectatic changes (n = 2) | 10.5 | |||||||||
| Cylindrical bronchiectasis (N = 2) | 11 | |||||||||
The loss of lung function starts in paediatrics and accelerates in older patients [31]. This loss occurs at an annual decline of 0.56%–0.8% in the predicted FEV1 [32,33]. There are discrepancies in reports of the factors that determine the severity of lung function regression, with the most frequently described discrepancies being the genotype-phenotype association, delayed age at diagnosis and nutritional status [18,31,34]. Lung function is closely associated with the nutritional status and both go through a period of steady decline augmented by recurrent exacerbations. Our data validate the link between BMI and lung function, with the nutritionally affected group having the worst lung function and FEV1 showing the strongest correlation with BMI.
A high-resolution chest CT scan might be helpful in making a PCD diagnosis by allowing for the detection of SI with bronchiectasis, and CT scan findings are useful for determining disease severity, which is correlated with FEV1 [35]. Santamaria et al. reported the presence of bronchiectasis in 71% of paediatric patients [36]. However, in our study, 81% of patients had bronchiectasis, with an average age of 8.2 years. Bronchiectasis is almost always central or diffuse and involves the middle and lower lobes. Other common features include a tree-in-bud appearance, atelectasis and mucus plugging [35,37,38]. These findings are similar to those found in our population.
5. Conclusion
In summary, we describe the clinical and molecular characteristics of patients with confirmed PCD in a tertiary centre in Saudi Arabia. We report nine new pathogenic or likely pathogenic variants. We found that RSPH9 is the most common PCD-causing gene in our region. We recognise that there are some limitations to our study, i.e. the lack of other diagnostic tests and the small sample size. However, we believe that it is essential to report new PCD genetic mutations to contribute to the PCD genetic pool.
Declaration of competing interest
The authors declare that they have no competing interests.
Footnotes
Peer review under responsibility of King Faisal Specialist Hospital & Research Centre (General Organization), Saudi Arabia.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpam.2021.03.002.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpam.2021.03.002.
Contributor Information
Mohammed Alzaid, Email: Mazaid123@gmail.com.
Khalid Al-Mobaireek, Email: khalidfm1@yahoo.com.
Mohammed Almannai, Email: MAlMannai@kfmc.med.sa.
Gawahir Mukhtar, Email: jmukhtar@kfmc.med.sa.
Safa Eltahir, Email: seltahir@kfmc.med.sa.
Adnan Zafar, Email: azafar@kfmc.med.sa.
Abdulali P. Zada, Email: azada@kfmc.med.sa.
Wadha Alotaibi, Email: walotaibi@kfmc.med.sa.
Funding
Not applicable.
Ethics approval and consent to participate
The Institutional Review Board (IRB) in King Fahad Medical City approved this study (IRB Log No. 19–524). The need to receive an informed consent from all patients was waived due to the retrospective nature of the study.
Availability of data and materials
The datasets generated and/or analysed during the current study are not publicly available due to confidentiality agreements but are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Visual abstract
The following are the Supplementary data to this article:
Fig S1.
References
- 1.Bush A., Chodhari R., Collins N. Primary ciliary dyskinesia: current state of the art. Arch Dis Child. 2007;92(12):1136–1140. doi: 10.1136/adc.2006.096958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Callaghan C., Chetcuti P., Moya E. High prevalence of primary ciliary dyskinesia in a British Asian population. Arch Dis Child. 2010;95(1):51–52. doi: 10.1136/adc.2009.158493. [DOI] [PubMed] [Google Scholar]
- 3.Horani A., Ferkol T.W., Dutcher S.K., Brody S.L. Genetics and biology of primary ciliary dyskinesia. Paediatr Respir Rev. 2016;18:18–24. doi: 10.1016/j.prrv.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kouis P., Yiallouros P.K., Middleton N., Evans J.S., Kyriacou K., Papatheodorou S.I. Prevalence of primary ciliary dyskinesia in consecutive referrals of suspect cases and the transmission electron microscopy detection rate: a systematic review and meta-analysis. Pediatr Res. 2017;81(3):398–405. doi: 10.1038/pr.2016.263. [DOI] [PubMed] [Google Scholar]
- 5.Storm van’s Gravesande K., Omran H. Primary ciliary dyskinesia: clinical presentation, diagnosis and genetics. Ann Med. 2005;37(6):439–449. doi: 10.1080/07853890510011985. [DOI] [PubMed] [Google Scholar]
- 6.Noone P.G., Leigh M.W., Sannuti A. Primary ciliary dyskinesia: diagnostic and phenotypic features. Am J Respir Crit Care Med. 2004;169(4):459–467. doi: 10.1164/rccm.200303-365OC. [DOI] [PubMed] [Google Scholar]
- 7.Shoemark A., Dell S., Shapiro A., Lucas J.S. ERS and ATS diagnostic guidelines for primary ciliary dyskinesia: similarities and differences in approach to diagnosis. Eur Respir J. 2019;54(3) doi: 10.1183/13993003.01066-2019. [DOI] [PubMed] [Google Scholar]
- 8.Behan L., Dimitrov B.D., Kuehni C.E. PICADAR: a diagnostic predictive tool for primary ciliary dyskinesia. Eur Respir J. 2016 doi: 10.1183/13993003.01551-2015. Published online February 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shapiro A.J., Leigh M.W. Value of transmission electron microscopy for primary ciliary dyskinesia diagnosis in the era of molecular medicine: genetic defects with normal and non-diagnostic ciliary ultrastructure. Ultrastruct Pathol. 2017;41(6):373–385. doi: 10.1080/01913123.2017.1362088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucas J.S., Barbato A., Collins S.A. European Respiratory Society guidelines for the diagnosis of primary ciliary dyskinesia. Eur Respir J. 2016 doi: 10.1183/13993003.01090-2016. Published online November 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shapiro A.J., Davis S.D., Polineni D. Diagnosis of primary ciliary dyskinesia. An official American thoracic society clinical practice guideline. Am J Respir Crit Care Med. 2018;197(12):e24–e39. doi: 10.1164/rccm.201805-0819ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rumman N., Jackson C., Collins S., Goggin P., Coles J., Lucas J.S. Diagnosis of primary ciliary dyskinesia: potential options for resource-limited countries. Eur Respir Rev. 2017;26(143) doi: 10.1183/16000617.0058-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leigh M.W., Hazucha M.J., Chawla K.K. Standardizing nasal nitric oxide measurement as a test for primary ciliary dyskinesia. Ann Am Thorac Soc. 2013;10(6):574–581. doi: 10.1513/AnnalsATS.201305-110OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shamseldin H.E., Al Mogarri I., Alqwaiee M.M. An exome-first approach to aid in the diagnosis of primary ciliary dyskinesia. Hum Genet. 2020 doi: 10.1007/s00439-020-02170-2. Published online May 4. [DOI] [PubMed] [Google Scholar]
- 15.Fassad M.R., Shoman W.I., Morsy H. Clinical and genetic spectrum in 33 Egyptian families with suspected primary ciliary dyskinesia. Clin Genet. 2020;97(3):509–515. doi: 10.1111/cge.13661. [DOI] [PubMed] [Google Scholar]
- 16.Mani R., Belkacem S., Soua Z. Primary ciliary dyskinesia gene contribution in Tunisia: identification of a major Mediterranean allele. Hum Mutat. 2020;41(1):115–121. doi: 10.1002/humu.23905. [DOI] [PubMed] [Google Scholar]
- 17.Zariwala M.A., Omran H., Ferkol T.W. The emerging genetics of primary ciliary dyskinesia. Proc Am Thorac Soc. 2011;8(5):430–433. doi: 10.1513/pats.201103-023SD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emiralioğlu N., Taşkıran E.Z., Koşukcu C. Genotype and phenotype evaluation of patients with primary ciliary dyskinesia: first results from Turkey. Pediatr Pulmonol. 2020;55(2):383–393. doi: 10.1002/ppul.24583. [DOI] [PubMed] [Google Scholar]
- 19.Boon M., Smits A., Cuppens H. Primary ciliary dyskinesia: critical evaluation of clinical symptoms and diagnosis in patients with normal and abnormal ultrastructure. Orphanet J Rare Dis. 2014;9:11. doi: 10.1186/1750-1172-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adil E.A., Kawai K., Dombrowski N., Irace A.L., Cunningham M.J. Nasal versus tracheobronchial biopsies to diagnose primary ciliary dyskinesia: a meta-analysis. Laryngoscope. 2017;127(1):6–13. doi: 10.1002/lary.26070. [DOI] [PubMed] [Google Scholar]
- 21.Zariwala M.A., Gee H.Y., Kurkowiak M. ZMYND10 is mutated in primary ciliary dyskinesia and interacts with LRRC6. Am J Hum Genet. 2013;93(2):336–345. doi: 10.1016/j.ajhg.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartill V.L., van de Hoek G., Patel M.P. DNAAF1 links heart laterality with the AAA+ ATPase RUVBL1 and ciliary intraflagellar transport. Hum Mol Genet. 2018;27(3):529–545. doi: 10.1093/hmg/ddx422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchison H.M., Schmidts M., Loges N.T. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat Genet. 2012;44(4):381. doi: 10.1038/ng.1106. S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarkar A., Loges N.T., Slagle C.E. DYX1C1 is required for axonemal dynein assembly and ciliary motility. Nat Genet. 2013;45(9):995–1003. doi: 10.1038/ng.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hjeij R., Onoufriadis A., Watson C.M. CCDC151 mutations cause primary ciliary dyskinesia by disruption of the outer dynein arm docking complex formation. Am J Hum Genet. 2014;95(3):257–274. doi: 10.1016/j.ajhg.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antony D., Becker-Heck A., Zariwala M.A. Mutations in CCDC39 and CCDC40 are the major cause of primary ciliary dyskinesia with axonemal disorganisation and absent inner dynein arms. Hum Mutat. 2013;34(3):462–472. doi: 10.1002/humu.22261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kott E., Legendre M., Copin B. Loss-of-Function mutations in RSPH1 cause primary ciliary dyskinesia with central-complex and radial-spoke defects. Am J Hum Genet. 2013;93(3):561–570. doi: 10.1016/j.ajhg.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shapiro A.J., Davis S.D., Ferkol T. Laterality defects other than situs inversus totalis in primary ciliary dyskinesia: insights into situs ambiguus and heterotaxy. Chest. 2014;146(5):1176–1186. doi: 10.1378/chest.13-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lucas J.S., Burgess A., Mitchison H.M., Moya E., Williamson M., Hogg C. Diagnosis and management of primary ciliary dyskinesia. Arch Dis Child. 2014;99(9):850–856. doi: 10.1136/archdischild-2013-304831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuehni C.E., Frischer T., Strippoli M.-P.F. Factors influencing age at diagnosis of primary ciliary dyskinesia in European children. Eur Respir J. 2010;36(6):1248–1258. doi: 10.1183/09031936.00001010. [DOI] [PubMed] [Google Scholar]
- 31.Halbeisen F.S., Jose A., Jong C de. Spirometric indices in primary ciliary dyskinesia: systematic review and meta-analysis. ERJ Open Res. 2019;5(2) doi: 10.1183/23120541.00231-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Werner C., Lablans M., Ataian M. An international registry for primary ciliary dyskinesia. Eur Respir J. 2016;47(3):849–859. doi: 10.1183/13993003.00776-2015. [DOI] [PubMed] [Google Scholar]
- 33.Marthin J.K., Petersen N., Skovgaard L.T., Nielsen K.G. Lung function in patients with primary ciliary dyskinesia: a cross-sectional and 3-decade longitudinal study. Am J Respir Crit Care Med. 2010;181(11):1262–1268. doi: 10.1164/rccm.200811-1731OC. [DOI] [PubMed] [Google Scholar]
- 34.Cohen-Cymberknoh M., Simanovsky N., Hiller N., Hillel A.G., Shoseyov D., Kerem E. Differences in disease expression between primary ciliary dyskinesia and cystic fibrosis with and without pancreatic insufficiency. Chest. 2014;145(4):738–744. doi: 10.1378/chest.13-1162. [DOI] [PubMed] [Google Scholar]
- 35.Kennedy M.P., Noone P.G., Leigh M.W. High-resolution CT of patients with primary ciliary dyskinesia. Am J Roentgenol. 2007;188(5):1232–1238. doi: 10.2214/AJR.06.0965. [DOI] [PubMed] [Google Scholar]
- 36.Santamaria F., Montella S., Tiddens H.A.W.M. Structural and functional lung disease in primary ciliary dyskinesia. Chest. 2008;134(2):351–357. doi: 10.1378/chest.07-2812. [DOI] [PubMed] [Google Scholar]
- 37.Dettmer S., Ringshausen F., Vogel-Claussen J. Computed tomography in adult patients with primary ciliary dyskinesia: typical imaging findings. PloS One. 2018;13(2) doi: 10.1371/journal.pone.0191457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jain K., Padley S.P.G., Goldstraw E.J. Primary ciliary dyskinesia in the paediatric population: range and severity of radiological findings in a cohort of patients receiving tertiary care. Clin Radiol. 2007;62(10):986–993. doi: 10.1016/j.crad.2007.04.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are not publicly available due to confidentiality agreements but are available from the corresponding author on reasonable request.