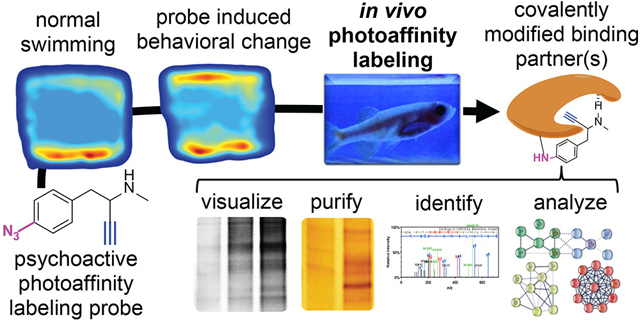
Abstract
Photoaffinity labeling (PAL) remains one of the most widely utilized methods of determining protein targets of drugs. Although useful, the scope of this technique has been limited to in vitro applications due to the inability of UV light to penetrate whole organisms. Herein, pigment-free Casper zebrafish were employed to allow in vivo PAL. A methamphetamine related phenethylamine PAL probe, designated here as 2, demonstrated dose-dependent effects on behavior similar to methamphetamine and permitted concentration dependent labeling of protein binding partners. Click chemistry was used to analyze binding partners via fluorimaging. Conjugation to a biotin permitted streptavidin pull-down and proteomic analysis to define direct binding partners of the methamphetamine probe. Bioinformatic analysis revealed the probe was chiefly bound to proteins involved in phagocytosis and mitochondrial function. Future applications of this experimental paradigm combining examination of drug-protein binding interactions alongside neurobehavioral readouts via in vivo PAL will significantly enhance our understanding of drug targets, mechanism(s) of action, and toxicity/lethality.
Keywords: PAL, target identification, methamphetamine, phenethylamine, novel tank test, Casper zebrafish, arylazide
Graphical Abstract
INTRODUCTION
Western civilization is engulfed in a substance abuse epidemic which has fueled a global synthetic drug revolution.1 Industrial scale chemistry facilities in developing nations are producing kilograms of novel synthetic psychoactive agents which infiltrate the mainstream marketplace and are accepted with little hesitation by much of America’s youth.2 Modern synthetic organic chemistry methodologies and internet ‘how-to’ websites have likely made it impossible to prevent the presence of synthetic psychoactive agents in our society.3 Gaining a more complete understanding of the drug targets, mechanism of action (MOA), and toxicity associated with these agents is an important step to effectively treat substance abuse disorders and episodic toxicity. To this end, this work defines a novel platform for the study of psychoactive drug targets by combining techniques in chemical biology with a model system capable of providing a neurobehavioral readout.
One of the most commonly abused psychoactive agents is methamphetamine (1, MA). MA use continues to increase among ages 12 and above, and emergency room admissions have also seen a spike from 17.5 to 28.4% from 2008 to 2017.4 This increase is attributed to the effects experienced by MA abusers, specifically euphoria, arousal, and increased energy.5–6 The drug remains in the bloodstream for a much longer period (half-life = 10–12 hr) than the short-lived psychoactive effects, necessitating repeated dosing to maintain a euphoric state. This leads to high levels of MA in the blood stream, which when coupled with long term use can induce cytotoxicity and negatively affect the central nervous system (CNS).5, 7 Studies in various animal models, mostly rats and mice, have also shown that prolonged use of MA reduces levels of dopamine (DA) and DA transporters (DAT) by approximately 50 % in the striatum, frontal cortex, hippocampus, and amygdala.8 Although these animal models have provided vital information about the mechanism of action underlying the rewarding and reinforcing effects of MA, the mechanisms underlying MA toxicity are still poorly understood. Based on these considerations of 1) prevalence of abuse, and 2) well-studied, but incomplete knowledge, MA was selected as the agent for development of our in vivo PAL target identification model.
Over the past decade, advances in chemical biology have led to a renaissance in small molecule target identification. The primary goal in target identification is to define drug-receptor binding interactions in dynamic living systems. To define binding interactions, activity-based probes can be designed to include three key components: 1) a reactive functionality for labeling interacting proteins, 2) a conjugation site for modification with a chemical biology reporter, and 3) a pharmacophore with validated efficacy. The use of terminal aliphatic alkynes as conjugation groups takes advantage of relatively selective Huisgen copper (Cu) assisted “click chemistry” and has demonstrated broad applicability as an appropriate component in probes due to its small size, efficient reaction rates in aqueous solutions, and negligible impact on the activity of many pharmacophore scaffolds.9–10 When appropriate, the ligand can be designed to include chemically stable probes which react with binding partners only after input from the experimenter, such as PAL (PAL) groups (Figure 1A). PAL groups allow for transient UV-light induced covalent (irreversible) attachment to binding partners in dynamic cellular environments. PAL groups allow probes sufficient time to travel to their site-of-action prior to UV activation and covalent attachment.11–12 Arylazides are particularly useful when the pharmacophore of interest contains an aryl system that tolerates substitution - as is present in MA. Furthermore, arylazides have relatively short photolytic half- lives compared to other common PAL groups (such as diazirines and benzophenones), making them ideal for applications which need rapid photoactivation. An essential step for PAL involves irradiation of the binding interaction with UV light (250–360 nm [depending on the PAL group being employed]).11–12 As such, PAL applications have, in the past, been limited to cell-based models due to the inability of UV light to penetrate the skin. To address this limitation, we have utilized a specific type of pigment-free zebrafish for in vivo PAL target identification studies.
Figure 1.
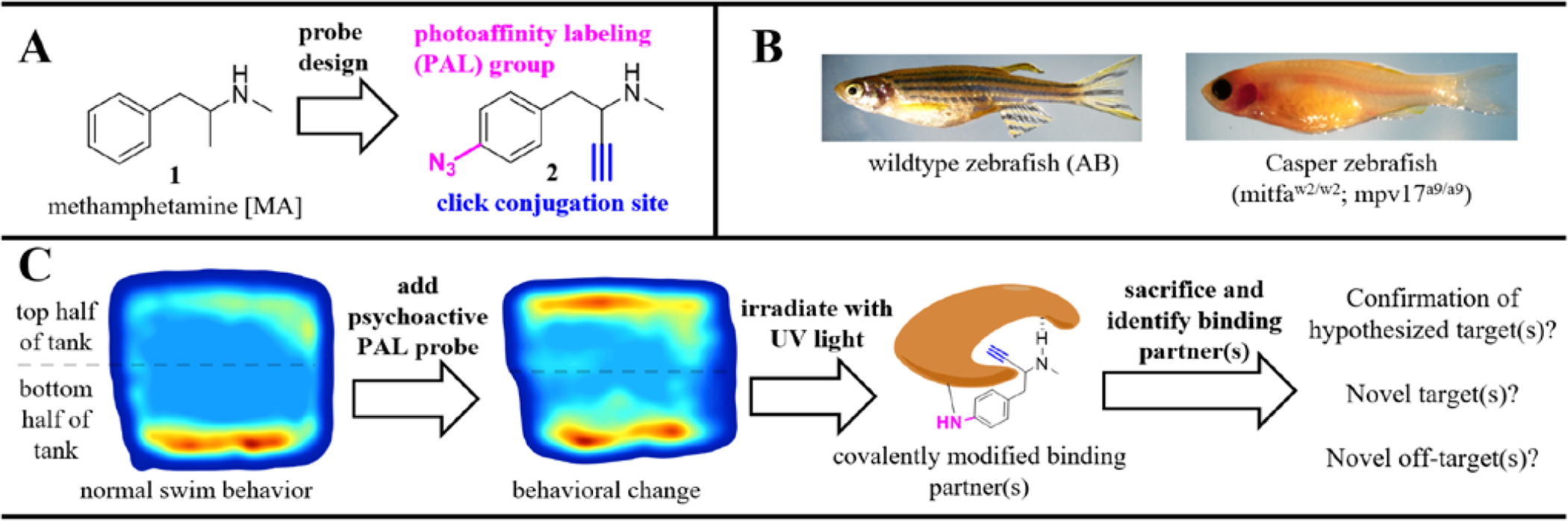
A) Rationale for modifications of methamphetamine (MA) to produce the photoaffinity labeling (PAL) probe, 2. B) Comparison of the pigmentation between adult wildtype zebrafish and Casper zebrafish. The lack of pigmentation makes Casper zebrafish an ideal species capable of providing a neurobehavioral readout whilst allowing the penetration of UV light for PAL. C) Rationale for in vivo PAL approach to covalently modify binding partners in concert with drug-induced behavioral changes. Density plots are used to demonstrate amount of time spent in different tank regions (e.g. top half versus bottom half). After UV irradiation and euthanasia, labeled proteins in the homogenized brain can be conjugated to chemical reporters for visualization of protein labeling and/or purification of labeled proteins followed by proteomic analysis to define binding partners.
Zebrafish present a model system for studying the psychological effects of psychoactive agents, such as MA, by analyzing movement patterns of fish in a 3D space.13 Zebrafish have been used to examine the phenotypic effects of various stimulants like cocaine, nicotine, amphetamine and MA by evaluating their time spent at the top or bottom of their tanks.14–18 Longer time spent at the bottom of a tank is associated with avoidance, anxiety, and stress.19 Zebrafish are an effective and validated model for the study of psychostimulant effects, mainly because of their high genetic and physiological homology to mammals.13, 20–21 While previous research has shown behavioral and physiological effects MA on zebrafish behavior that are similar to mammals, the target(s) are still poorly understood.18, 22 A mutant zebrafish strain known as Casper zebrafish (mitfaw2/w2; mpv17a9/a9) provides a viable model for in vivo fluorescent analysis.23 These mutations eliminate melanophores and iridophores, thus making this genetic strain transparent as adult zebrafish (Figure 1B).23 In contrast to the wildtype AB zebrafish, the translucent nature of Casper zebrafish permits UV light penetration, potentially allowing PAL for identification of drug targets. Herein we describe a new platform for identification of targets associated with psychoactive agents using Casper zebrafish in the first report of in vivo PAL in such a platform. A generalized overview of the potential utility of combining in vivo PAL with neurobehavioral analysis is provided in Figure 1C.
RESULTS AND DISCUSSION
Design, Synthesis, and Photolytic Characterization.
The structure activity relationship of MA (and related phenethylamines) is somewhat nuanced, with even small changes resulting in impact on pharmacological action. Nonetheless, substitution of the aryl system and/or homologation (i.e., lengthening) of the alkyl side chain are generally tolerated, albeit with diminished potency.24–26 A potential decrease in potency relative to MA was not a chief concern during design, so long as the probe could affect behavior without marked toxicity. MA itself is quite small (molecular weight = 149 g/mol). Hence, we used the smallest probe moieties possible to limit impact of the modifications on binding interactions and biodistribution. A para-azido group was placed on the aryl system as the photolytic group and an ethynyl group was used as the conjugation site.
Synthesis of the desired methamphetamine probe, 2, required the use of a novel synthetic route employing common synthetic organic transformations (Scheme 1). The synthesis began with protection of the aldehyde, 3, using ethylene glycol with catalytic p-toluenesulfonic acid (PTSA), followed by azidonation of the para-substituted bromine 4, yielding 5.27 The acetal was deprotected via PTSA to reveal the aldehyde, 6, which was then subjected to a Grignard reaction to install the alkyne moiety generating 7.24 The alcohol was mesylated followed by an ami-nation to yield the desired probe, 2.
Scheme 1. Synthesis of the photoaffinity labeling probe, 2.
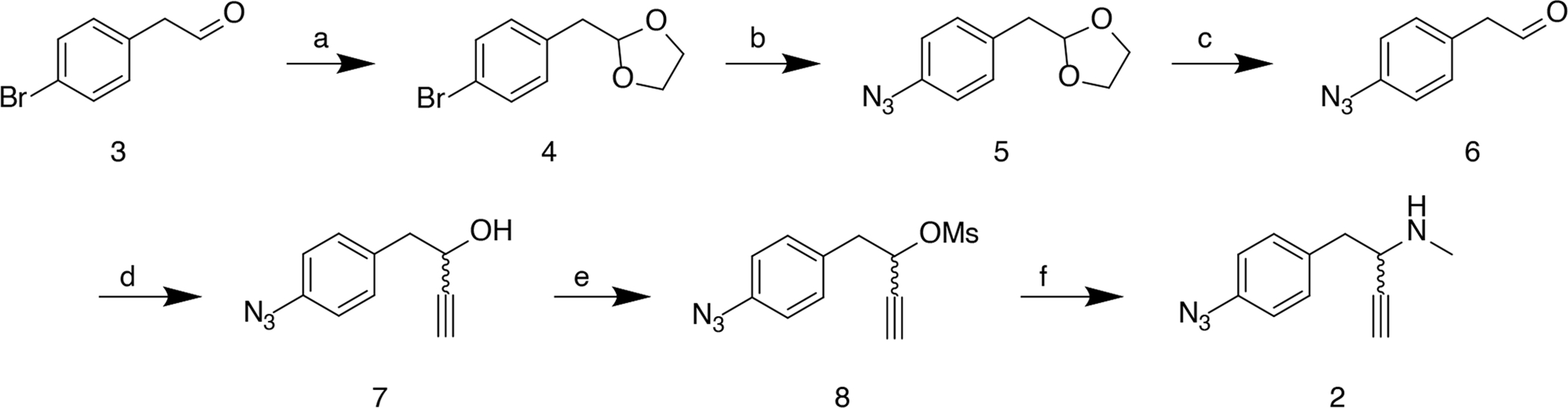
Reagents and conditions : (a) ethylene glycol, PTSA, reflux (50 %); (b) NaN3, CuI, sodium ascorbate, (1R,2R)-N1,N2-dimethylcyclohexane-1,2-diamine, reflux (77 %); (c) PTSA, reflux (44 %); (d) ethynylmagnesium bromide, −78 °C to r.t. (39 %); (e) methanesulfonyl chloride, TEA, 0 °C to r.t. (47%); (f) methylamine, reflux (21 %).
The photolytic half-life of 2 was examined to establish photo-activation parameters. A solution of 2 was irradiated with 312 nm UV light for 30 min, with aliquots taken every 5 min for analysis of starting material remaining by HPLC-PDA. HPLC analysis indicated a photolytic half-life ~ 3.2 min (Supplemental Figure S2). Short-term irradiation (< 15 min) with 312 nm UV light is generally considered to have minimal detrimental impact on biological systems. Hence, this probe was judged as having acceptable photolytic properties to advance to in vivo testing.
Zebrafish Behavioral Analysis.
The dose-dependent effect of MA and 2 on zebrafish behavior were assessed during exposure and via the novel tank test (NTT). The NTT has been extensively used to study anxiolytic behavior of adult zebrafish. In the NTT, zebrafish are introduced into a novel tank to gauge their level of stress or anxiety based on location (top or bottom of tank), locomotion, and freezing. Zebrafish begin to explore the novel environments after a few seconds of transfer, with anxiety-like behavior reducing over time, with initial reduced exploration, thigmotaxis, and geotaxis reflecting anxiety as they do in mammals.13, 28–29 Since elevated anxiety is known to be a symptom of high-dose MA use, the NTT is a reasonable behavioral test to understand the effects of MA compared to 2.18
Zebrafish behavioral studies followed the experimental workflow outlined in Figure 2A. Dosing choices were informed by a previous report demonstrating effects in adult zebrafish for a similar agent, 3,4-Methylenedioxymethamphetamine (MDMA).30 Adult Casper zebrafish are placed for 20 min in an exposure chamber containing either vehicle, MA (5 mg/L [26.9 μM]); 20 mg/L [107 μM]; 40 mg/L [215 μM]; and 65 mg/L [350 μM]) or equimolar doses of 2 (6 mg/L [26.9 μM]); 25 mg/L [107 μM]; 50 mg/L [215 μM]). Importantly, the exposure chamber contains a much smaller total volume of water than the novel tank (10 ml versus 1.7 L), making it possible to have high exposure concentrations in the tank water whilst using reasonably small quantities of each agent. Behavior during exposure was recorded and an experimenter blinded to the treatment groups characterized swim patterns based on previously established criteria.31 These categories included: 1) erratic movement, indicating a general anxiety/fear state, 2) freeze/drift, seen as a complete cessation of movement (except for gills and eyes) that is associated with increased anxiety, 3) controlled swim, seen as an exploratory state that is associated with decreased anxiety, and 4) freezing/recovery, seen as loss of balance during a freezing bout with a subsequent recovery maneuver, as part of the righting reflex. Vehicle-treated fish showed sustained erratic movement for a majority of the exposure time (~80% of the exposure period) with intermittent bouts of controlled swimming towards the end of the exposure period (summarized for entire exposure duration in Figure 2B, see Supplemental Figure S3 for time-course). These results suggest that when administering in tank water, high concentrations of MA are required to elicit a neurobehavioral response, in agreement with past reports using MDMA, for which an effect in the NTT was observed at 80 and 120 mg/L but not at 10 or 40 mg/L. Importantly, MA and 2 both demonstrated the ability to diminish the erratic movements observed in the vehicle cohort, with both compounds doing so in a concentration dependent manner (Figure 2C). However, whereas MA shifted behavior toward controlled swimming, 2 elicited freezing with drifting and recovery. These findings demonstrated that the probe has similar, but not identical, efficacy as MA. Nonetheless, the probe certainly showed psychoactive effects based on the results from the exposure chamber.
Figure 2.
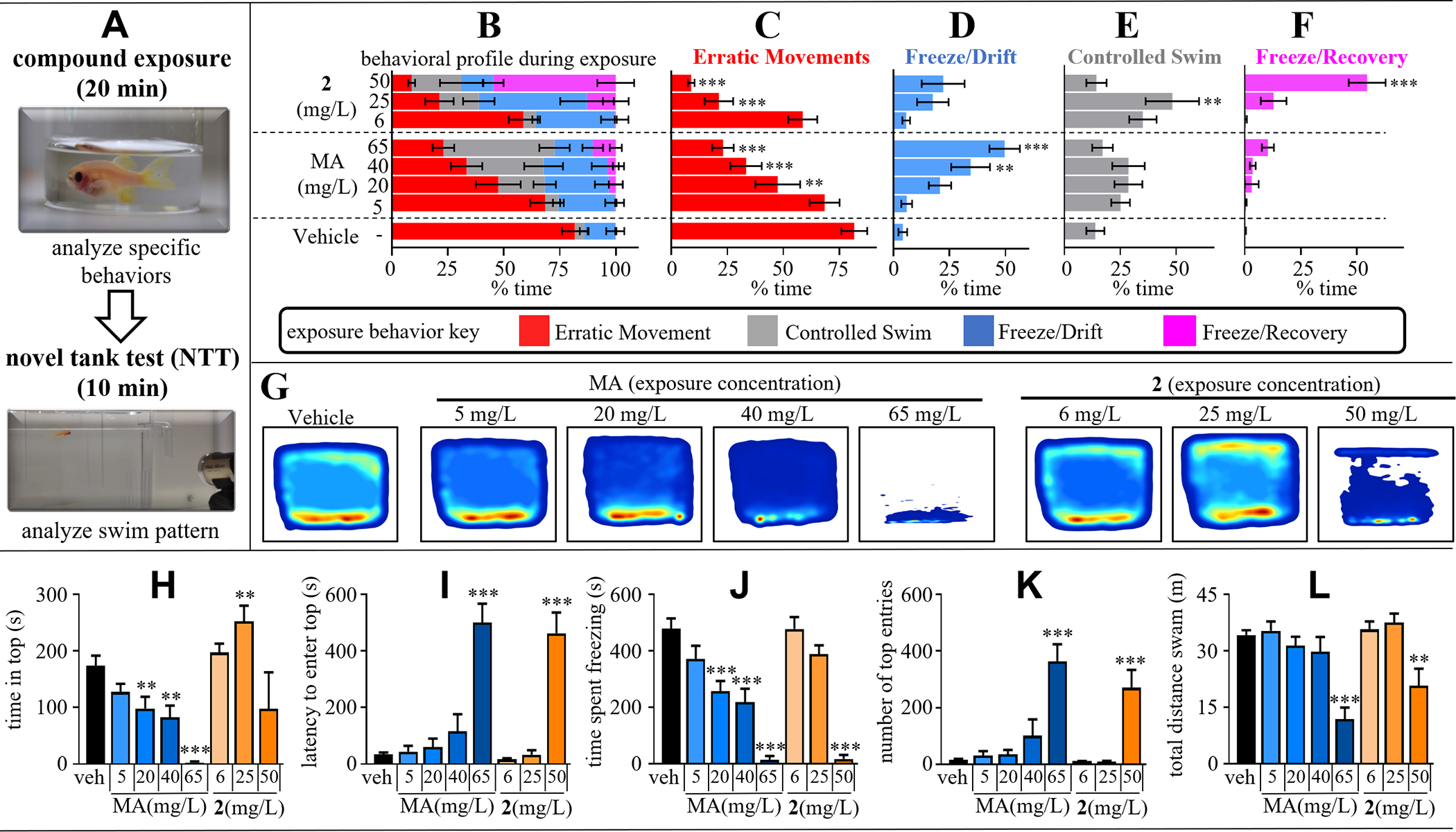
A) Experimental workflow for assessing the effects of varying concentrations of MA or 2 (5–65 mg/mL) on the behavior of adult Casper zebrafish. The exposure concentration/dose (mg/L) was adjusted to give equimolar exposure for MA and 2 respectively, 5 or 6 mg/L = 26.9 μM; 20 or 25 mg/L = 107 μM; 40 or 50 = 215 μM; and 65 mg/L = 350 μM (2 not tested at 350 μM). BF) Swimming in the exposure chamber was recorded for 20 min and behavior categorized as either erratic movement, freezing/drifting, controlled swimming, and freezing with recovery. Data are represented as percentage of time exhibiting a specific behavior. After drug exposure each fish was transferred to the novel tank test and swim patterns were recorded for 10 min. G) Bivariate kernel density estimations of NTT behavior. Density plots represent median swim pattern of each treatment cohort. H-L) Analysis of specific behavioral readouts related to swimming in the novel tank test. All data represented as mean ± s.e.m. (N = 8–10 per cohort [randomly selected / sex balanced]). *= p< 0.05, **= p<0.01, ***= p<0.001 compared to veh control using one-way ANOVA with Dunnett’s post hoc test.
Following the 20-min exposure, zebrafish were removed from their respective exposure solutions and placed into the novel tank and swimming was recorded for 10 min prior to UV irradiation. It is difficult to summarize dynamic data within a static publication format. Kernel density plots (Figure 2G) help to illustrate the median swim paths of each treatment cohort (N = 8–10 per cohort). Vehicle treated zebrafish showed initial preference for the bottom of the tank and gradually began to explore the top region of the novel environment, interpreted as decreased anxiety. As seen in the density plots of Fig.2G and specific quantitative analysis criteria (Fig. 2H–L), MA significantly reduced activity in the top of the tank, even at the lowest concentration tested (5 mg/L) and provided a dose-dependent effect on the time spent, and number of entries into, the top of the tank, both of which decreased with stepwise increases in concentration. By contrast, 2 at the intermediate concentration (25 mg/L) displayed significantly increased time in the top region of the tank, indicating an anxiolytic-like effect that was not observed, for MA. Despite this difference in response at intermediate concentrations, at high concentrations, freezing behavior and corresponding increases in latency to enter the top was seen for both MA and 2. The time-course of response in the NTT behavioral endpoints was further assessed for habituation by comparing the first and last minute of each respective cohort (Supplemental Figure S4). The vehicle cohort showed significant differences in top half entries and time in top half when comparing the first minute to the last minute. Since 2 at 50 mg/L (215 μM) demonstrated a similar phenotype to the highest dose of MA (65 mg/L [350 μM), 2 was not tested at 350 μM. Both MA and 2 impaired the zebrafish ability to habituate at all tested concentrations for both top half entries and latency to enter the top.
Quantitation of MA in Zebrafish Brains using LC-MS/MS.
Immediately following the NTT, each fish was transferred to a small irradiation chamber (See Supplemental Figure S5) and UV crosslinking was initiated by irradiation with 312 nm UV light for 10 min (wavelength and duration parameters based on HPLC analysis of photolysis rate). After UV irradiation each fish was immediately euthanized, the brain removed and flash frozen in liquid N2 for subsequent analysis. Brain specimens were randomly selected from within each cohort for analysis for either LC-MS/MS quantitation of MA in the brain, or for chemical biology studies.
The thigmotaxis and geotaxis tracking data provided strong evidence that both agents are psychoactive. Hence, we pursued follow-up studies to directly measure each agent in the zebrafish brains. For MA, this involved direct measurement using LC-MS/MS via multiple reaction monitoring (MRM) analysis, with absolute quantitation enabled by an isotope labeled internal standard (methamphetamine-d5). Brains were extracted using the QuEChERS technique, dispersive solid phase extraction, prior to analysis by LC-MS/MS. To maintain consistency in our methodology, all fish were irradiated with UV light regardless of the presence of a PAL group, including the vehicle and MA-treated cohorts. Only MA-treated cohorts were analyzed by LC-MS/MS, whereas cohorts treated with 2 were reserved for chemical biology studies.
MA was observed in every brain that was analyzed by LC-MS, including the following median concentrations for each cohort: 0.12 μM ± 0.03 for the 5 mg/L cohort; 1.58 μM ± 0.41 for the 20 mg/L cohort; 13.9 μM ± 1.80 for the 40 mg/L cohort; and 42.7 μM ± 6.82 for the 65 mg/L (Figure 3A), represented as mean and standard error of mean (s.e.m.). Changes in MA concentration in the brain did not demonstrate a linear relationship relative to drug treatment. For instance, increasing from 5 to 20 mg/L represents a four times increase in drug treatment, but an approximately ten times increase of MA was observed in the brain. Similarly, increasing from 20 to 40 mg/L represents a doubling of drug exposure but an approximately ten times increase of MA was observed in the brain. The exact reason for this non-linear relationship is unclear but we hypothesize that the trend is related to saturation (or overwhelming) of systems involved in drug efflux, metabolism, and biodistribution. The concept of correlating drug exposure with behavioral responses has been for the most part lacking in all preclinical research, including studies involving zebrafish. Our findings provide an example of directly measuring dose-dependent drug distribution in target tissue alongside efficacy to inform future studies.
Figure 3.
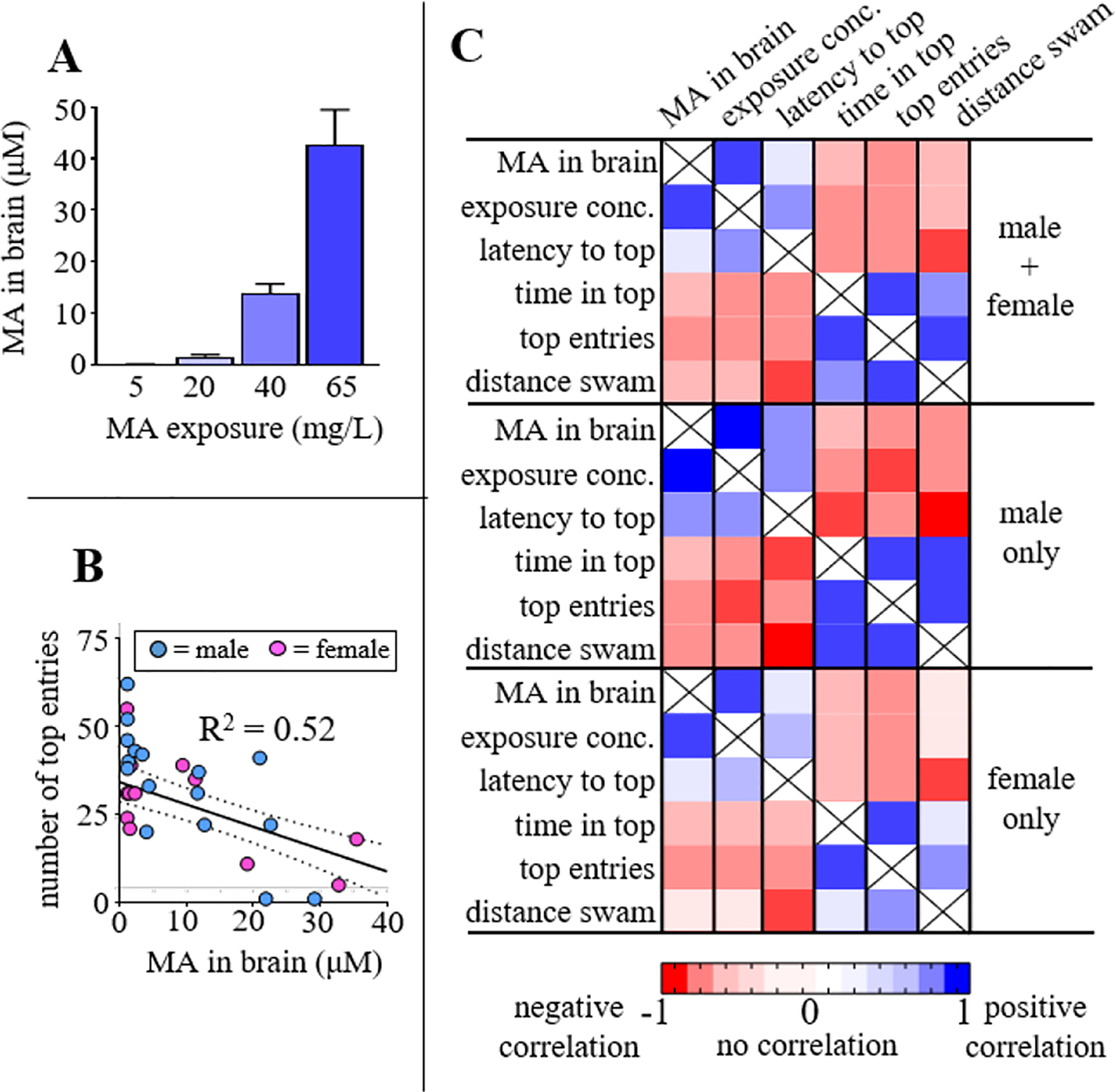
A) Subsequent to behavioral testing and UV irradiation, brains were dissected out, spiked with methamphetamine-d5 (internal standard), extracted, and MA quantified by LC-MS/MS. 8 fish from each MA treatment cohort were analyzed, represented as mean ± s.e.m., of analyte area under the curve (AUC) normalized to the internal standard AUC. B) Correlation between drug concentration measured in the brain and number of top entries in behavioral screen. Trendline represents correlation for all fish analyzed (N = 32 total [16 male + 16 female]). C) Correlation matrix for MA and behavioral readouts from NTT. In general, correlations are improved when only considering the male subjects. Correlations assumed Gaussian distribution and are displayed based on Pearson correlation coefficients (r).
We analyzed correlations between behavioral readouts, drug exposure, and the concentration of MA found in the brain. Fish were randomly selected for treatment and cohorts are for the most part sex-balanced, although there is some deviation due to the random selection process (Supplemental Table S1 catalogs sex of each cohort). In general, responses in female and male fish trended in the same direction, however, the differences were primarily quantitative. This is observed for the relationship between top entries and MA concentration in the brain (Figure 3B). This represents the strongest negative correlation between a behavioral readout and MA in the brain, where r = −0.60 for the entire cohort, compared to r = −0.77 [male only] and r = −0.55 [female only]. Figure 3C illustrates a correlation matrix with computed Pearson correlation coefficients, r. As anticipated, the strongest positive correlation was found between drug exposure and concentration of MA in the brain, with r = 0.84 when considering the entire cohort; r = 0.93 for males only; and r = 0.86 for females only. Latency to the top was the behavioral readout with the strongest positive correlation relative to MA concentration in the brain (r = 0.72 [entire cohort]; r = 0.75 [male only]; r = 0.76 [female only]). Interestingly, the strongest correlation from the entire matrix was between two behavioral readouts (total distance swam versus latency to the top), which had a negative correlation of r = −0.89 (entire cohort), r = −0.96 for male only, and r = −0.83 in females. This correlation reflects decreases in swimming at high concentrations of MA which cause increased time freezing (Fig. 2K) and corresponding increases in the latency to enter the top (Fig. 2I) and a decrease in locomotor activity (Fig. 2L).
In Vivo PAL to Irreversibly Attach the Probe to Binding Partners.
After drug exposure and the NTT, each fish was irradiated for 10 min with 312 nm UV light. Fish brains were then processed for analysis according to Figure 4A. To identify the modified proteins, a copper-catalyzed alkyne-azide cycloaddi ton (i.e., a ‘click’ reaction) was utilized. Homogenized brain tissues were concentrated and normalized to have a consistent total protein concentration (~1 mg/ml). Homogenates were submitted to an in situ click with an Azido-Alexa647 fluorophore. Fluorescently labeled proteins were separated by SDS page and visualized by fluorimaging and Coomassie stain to compare relative amount of protein in each lane. PAL experiments were carried out several times during methodology development to confirm reproducibility, refine the technique, and to optimize signal-to-noise ratio. In general, techniques were adapted from previous procedures.32–33 Representative results are shown in Figure 4B. Treatment with increasing concentrations of 2 yielded increased fluorescent labeling of several proteins, especially in 25 and 50 mg/L cohorts. In addition to a few specific bands, substantial non-specific labeling was observed. Bands were determined to be background (e.g., non-specific) if they were present in brains of fish not treated with 2 and were found in gel regions associated with a large abundance of protein in the corresponding Coomasie stain of the gel, as seen for intense gel bands at 20, 25, 38, and 50 kDa (denoted in Figure 4B as ‘noise’). Much of this noise is associated with the non-specific binding of the fluorophore itself. Some non-specific PAL by 2 is also anticipated due to the presence of the unsubstituted arylazide in 2, which is known to produce PAL background based on ring expansion to a ketenimine intermediate.34–35 Despite this known drawback, the desire to maintain as similar a structure to MA as possible led to our choice not to block ring expansion with 3, 5 difluorination. Regardless, densitometry analysis suggests some specific bands at approximately 27, 45, and 55 kDa (denoted with yellow stars in Fig.4B), the authenticity of which was further informed by results from the biotin pull-down, vide infra.
Figure 4.
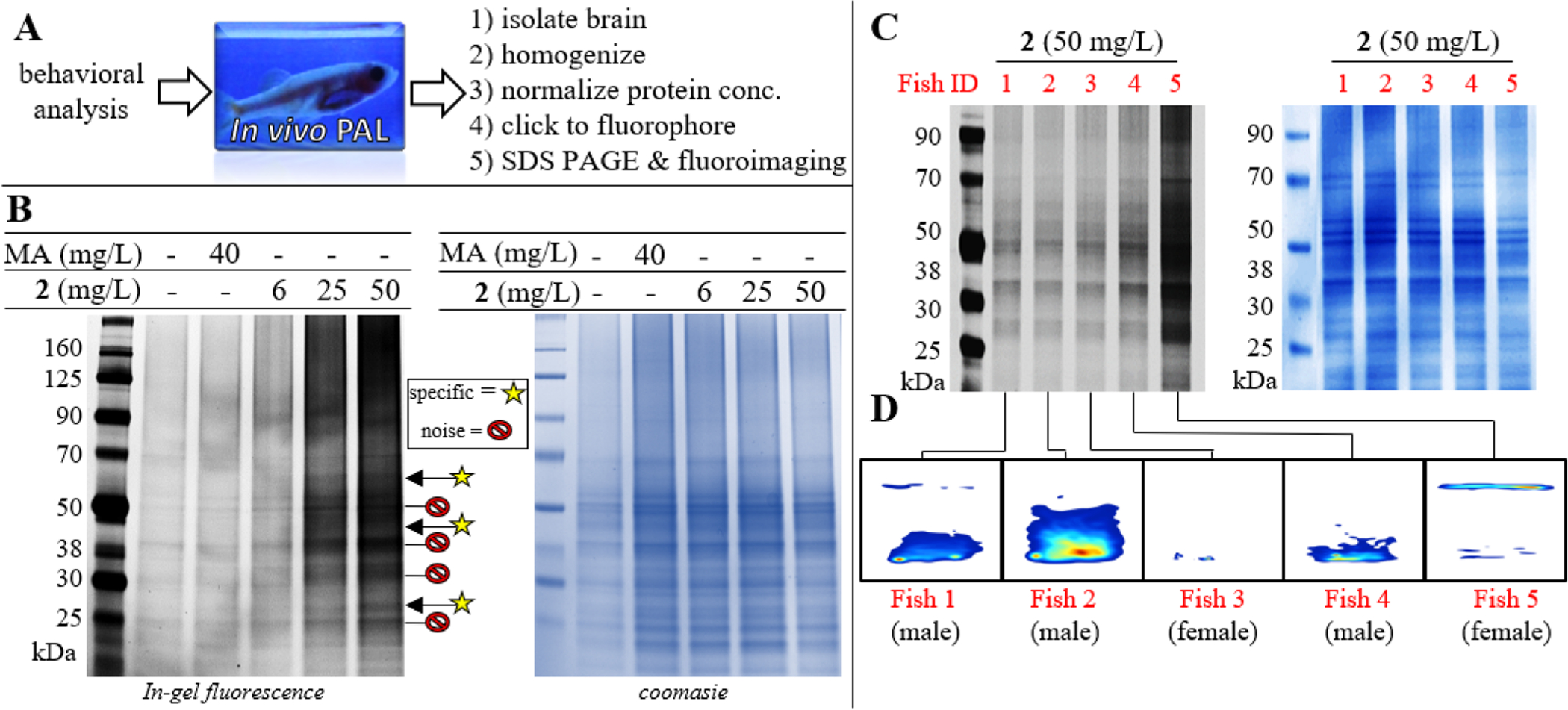
A) Overview of in vivo PAL and sample processing. B) In-gel fluorescence for labeled proteins in brain homogenates of fish treated with indicated concentration of 2, MA, or vehicle. Bands which are considered to be specific are indicated with a yellow star (at approximately 27, 45, and 55 kDa). Following fluoroimaging, each gel was stained by coomasie to confirm equivalent total protein concentrations loaded into each well (shown on right). C) Side-by-side comparison of labeling in five fish from the 50 mg/L cohort. D) Kernel density plot demonstrating behavior of each fish (designated as Fish 1–5) in the NTT prior to PAL.
As we developed our technique some inconsistencies were noticed in labeling which we have not previously encountered during cell-based PAL. Specifically, certain fish within a given treatment cohort presented significantly more PAL signal than other fish from the same cohort. For example, five fish from the 50 mg/ml cohort were analyzed side-by-side (Figure 4C). One fish, designated as Fish 5, showed significantly more labeling than any other fish in the group, despite all brain homogenates having approximately equivalent total protein concentrations after normalization prior to the click - as confirmed by Coomassie stain (Figure 4C). It is important to note that although there is much more PAL in Fish 5, the labeling profile (i.e., which bands are labeled) is approximately the same as the other fish. This was assessed based on inclusion of samples from Fish 1–4 on separate gels not containing Fish 5, since the signal from Fish 5 saturates the detector, limiting fluorimaging scan time/exposure to a point which is not sensitive enough to completely realize bands in Fish 1–4. For example, Fish 3 in Fig. 4D is the same fish analyzed in Fig. 4B (lane 5 [50 mg/L]). Interestingly, in addition to being an apparent outlier with regards to PAL, Fish 5 demonstrated significantly different behavior in the NTT compared to rest of the cohort, with most of its swimming taking place at the top of the tank (Figure 4D). Fish 5 was a female fish, which is in agreement with trends observed in the correlation matrix indicating more variability in females. Based on this PAL result, we anticipate the variability in females is likely related to differences in biodistribution or metabolism rather than differential receptor expression compared to males. The combined results in Figure 4C–D demonstrate the potential power of an in vivo PAL platform to study psychoactive agents, as it can potentially explain individual differences in behavioral observations based on patterns and/or abundance of PAL.
To validate that 2 was binding to the same proteins as MA, we carried out a competition experiment involving co-incubation of 2 (50 mg/L) with an equimolar dose of MA (40 mg/L). We hypothesized that since 2 and MA should have similar targets, the effects of the co-incubation should mirror the effect of fish if treated with 80 mg/L, although 80 mg/L was not tested due to observations of mild toxicity at the highest MA dose tested (65 mg/L). Interestingly, the co-incubation cohort displayed a similar exposure phenotype to 2 rather than MA (Figure 5A). Surprisingly, in the novel tank test the co-incubation did not perform similarly to either of the high dose effects seen for MA or 2. Instead, the fish most resembled behavior seen for the intermediate (25 mg/L) dose studied for 2, with a significant amount of activity in the top half of the tank. Qualitative differences between male and female were evident, with the males swimming mostly at the top whereas the females spent the majority of their time in the bottom of the tank (Figure 5B). Importantly, we observed a significant decrease in PAL in the competition experiment (Figure 5C). The competition experiment showed less PAL than the 6 mg/L, resulting in labeling closely matching the vehicle control. This result provides evidence that MA and 2 are competing for similar binding partners. No bands were competed off ‘specifically’, suggesting that MA itself may be quite promiscuous.
Figure 5.
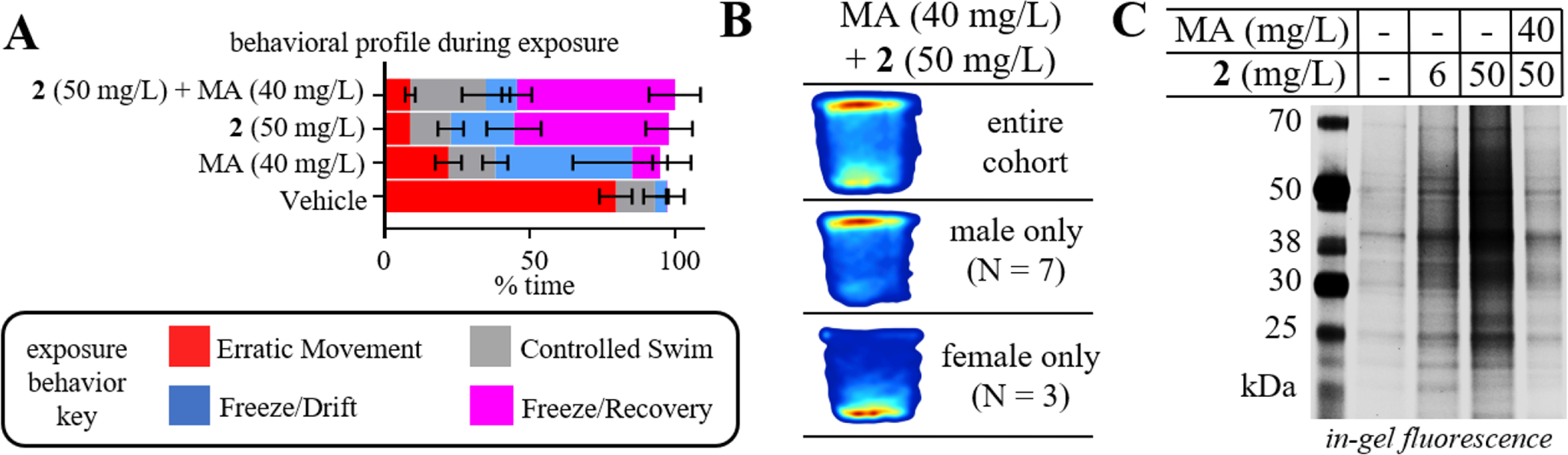
A) In-gel fluorescence for labeled proteins in brain homogenates of fish treated with indicated concentration of vehicle, 2 (6 or 50 mg/L), or co-incubation of 2 (50 mg/L [215 μM]) and MA (40 mg/L [215 μM]). B) Kernel density plot demonstrating behavior of zebrafish from co-incubation cohort. Data for 2, MA, and vehicle alone is shown here for comparison alongside co-incubation cohort and is duplicate from Figure 2B.
Identification of Protein Networks Covalently Modified During In Vivo PAL (PAL interactome).
Affinity purification and proteomic analysis were employed to identify the proteins modified by the in vivo PAL, following the workflow described in Figure 6A. Normalized brain homogenates were first washed with a streptavidin resin to deplete endogenous biotin. The resulting mixture was clicked to an azidobiotin, followed by streptavidin pull-down of biotin conjugated proteins. Duplicate samples were run on two separate gels, and one gel was silver stained to yield semi-quantitative confirmation of protein labeling while the other gel was used for proteomic analysis. The silver stain indicated the presence of a few specific areas with protein enrichment compared to the control (Figure 6B). Specifically, selective pull-down was observed for molecular weights of approximately 22–24 kDa, 26 –28 kDa, and 51–53 kDa. These are the most abundant proteins from the pulldown, but it is possible that other proteins are present in lower abundance which are not observable by silver stain but may be detectable by LC-MS. Hence, two relatively large gel regions, 38–70 kDa (region 1) and 15–38 kDa (region 2) were selected for excision and proteomics analysis. These regions were excised from each lane and processed by in-gel tryptic digest, followed by proteomic analysis. LC-MS experimenters were blinded to the identity of samples. Proteins identified in both vehicle- and 2-treated fish were subtracted as background. In total, proteomics identified the selective pulldown of 30 proteins with 100 % certainty (Supplemental Table S2 shows full proteomics results). Within the 15–38 kDa region, 14 unique proteins were identified, whereas 16 proteins were found within the 38–70 kDa region. Protein clusters were assigned based on shared overlapping peptide evidence, resulting in the identification of clusters representing five high abundance proteins (summarized in Figure 6C).
Figure 6.
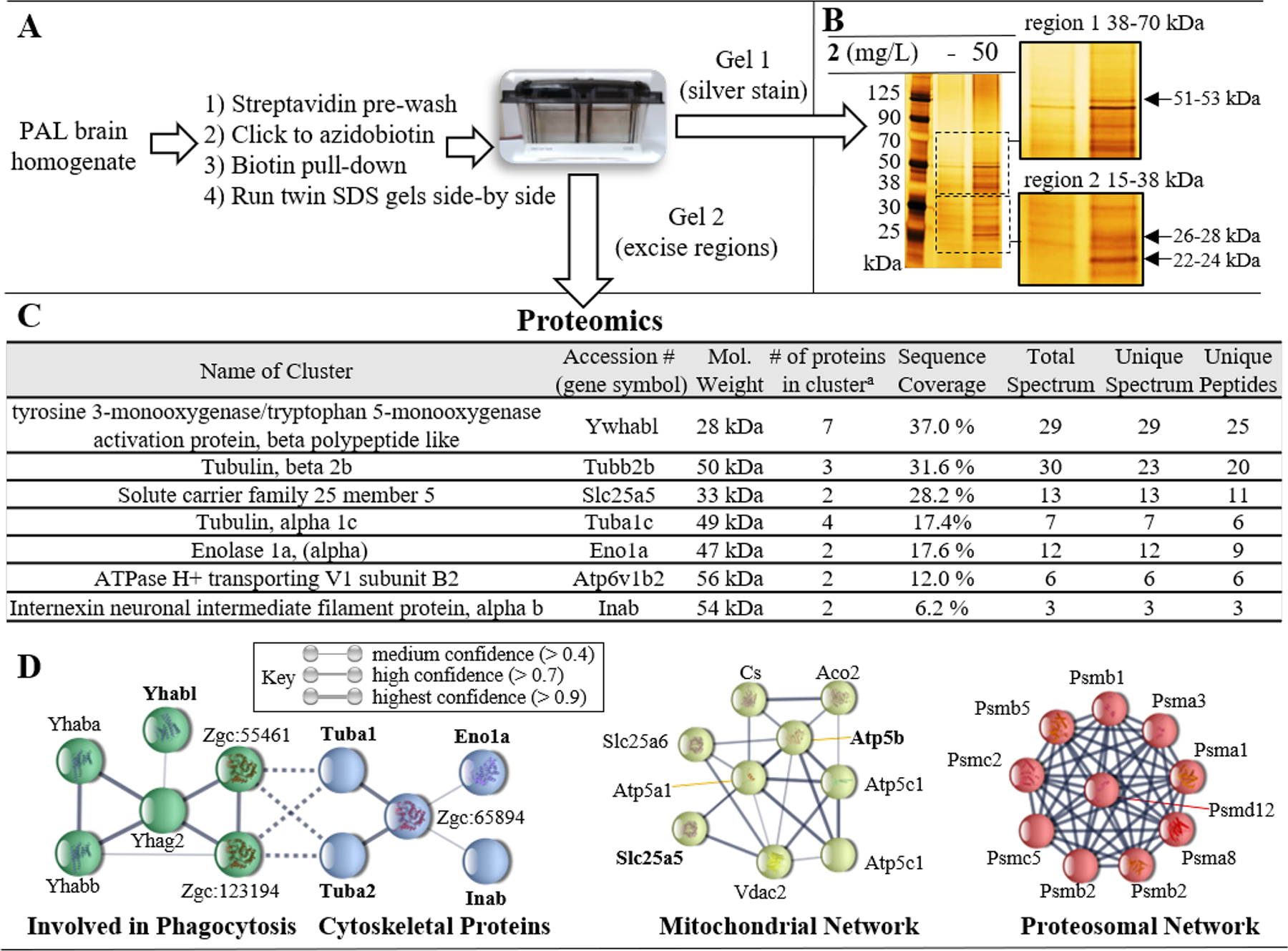
A) Experimental workflow for biotin pull-down. Two gels were run side-by-side. One gel was silver stained to visualize proteins and inform cutting of the unstained gel. B) Silver stain of the SDS PAGE using samples from the biotin pull-down for fish treated either with vehicle or 2 (50 mg/L). The gel regions (region 1 [38–70 kDa] and region 2 [15–38 kDa]) which were selected for excision from the unstained gel for submission to proteomics are magnified. C) Protein clusters identified during proteomic analysis which were not present in vehicle treated pulldown. D) Bioinformatic analysis of covalently modified binding partners identified with > 95 % certainty using STRING. Four interaction networks were identified, including: phagocytosis (green nodes); cytoskeletal proteins (blue nodes); mitochondrial proteins (yellow nodes); and proteasomal proteins (red nodes). Gene symbols shown in bold were those identified in proteomic cluster analysis. Unconnected nodes and low confidence interactions were omitted for clarity.
Tyrosine 3-monooxygenase/5-tyrptophan activating protein (Ywhabl) was detected as the strongest protein cluster, with 37.0 % sequence coverage. Accordingly, a band of approximately the same molecular weight as this protein (~27–28 kDa) was observed in the biotin pull-down (Figure 6B) and the fluorimaging (Figure 4B). This was the only instance of a protein for which the fluorimaging, silver stain, and proteomics were all in agreement. Of course, all techniques have limitations. The vehicle treated samples revealed non-specific labeling/pull-down in fluorimaging and biotin pull-down experiments. This background will mask labeling of some proteins. Furthermore, mechanistically important proteins detected by LC-MS may be present in low abundance such that they are not observable by silver stain and/or fluoroimaging. To account for these limitations, we carried out network analysis on the proteomics results to gain a more complete understanding of the phenethylamine interactome covalently labeled by 2.
We performed bioinformatic analysis of all binding partners identified with > 95 % confidence during proteomics analysis (See Supplemental Figure S7 for bioinformatic workflow). Uniprot accession numbers for each protein were translated into gene names which were analyzed to understand protein-protein interaction nodes, visualized using STRING (Figure 6D). Approximately half of the proteins analyzed (33 of 68) belonged to a few distinct networks. Two of these networks included protein associations related to phagocytosis and cytoskeletal proteins. Of course, there is evidence for cross-talk and interactions between these two networks based on the involvement of the cytoskeleton in physical processes involved in phagocytosis. The phagocytosis network included the protein which provided the strongest signal in proteomics (Ywhabl). The largest number of associations fell within the proteasomal network. Indeed, several proteasomal subunits (ATPase and non-ATPase) were identified during proteomics, even though no single proteasomal protein was identified during cluster analysis. A mitochondrial protein network was also identified, with an interactome containing four ATP synthase nodes, one voltage gated ion channel (Vdac2), and the cluster of solute carrier family 25, (members 5 and 6). It is noteworthy that several additional mitochondrial proteins were identified in proteomics but were omitted from Figure 6D because they failed to meet the bioinformatic threshold related to ‘experimental evidence’ of an association; including voltage dependent anion channel 1 (Vdac1).
The mechanisms underlying the low dose effects of amphetamine-like stimulants are generally well understood, but the mechanisms contributing to high dose effects are much less well known. This poor state of understanding of the mechanisms that might contribute to the toxic effects of drugs of abuse impacts upon our ability to mitigate the toxic and lethal effects of MA overdose. For this reason, in choosing doses for study in proteomic analyses, high doses of MA and 2 were intentionally chosen. One difficulty here is that drugs certainly become more promiscuous at higher doses. Although the proteomics analysis suggested that MA interacted with a number of novel targets, the cellular processes that this analysis suggested were affected have been implicated in many previous studies. Impairment of mitochondrial function has been implicated in the toxic effects of MA in the brain, heart and liver and have been reviewed in depth.36–38 The well-known neurotoxicity of MA has been linked to ubiquitin-proteasome dysfunction and autophagy39–40 and autophagy regulates phagocytosis.41–42 Perhaps the most surprising result is the observation of tubulin proteins in the proteomics analysis since these are usually only considered to be structural proteins, not signaling proteins. However, in the context of the forgoing links between autophagy, proteosomal inhibition and mitochondrial dysfunction, it is interesting to note that MA promotes α tubulin deacetylation.43 Although this might very well be associated with structural changes that occur in many cells in response to MA, it may also be related to cellular toxicity. Stabilization of microtubules can reduce axonal degeneration via increases in tubulin acetylation44 and inhibition of Vdac-tubulin interactions may promote cell death.45
The molecular target generally considered to be responsible for the psychostimulant and addictive effects of MA, the dopamine transporter (DAT), was not identified. Whether this is related to the techniques/model or DAT itself is unclear. There are several well-known obstacles associated with the isolation of transmembrane receptors, including the relatively low abundance of GPCR’s and the need for protein specific conditions optimized for the solubilization and purification of the transmembrane receptor of interest.46 Utilizing such specific conditions significantly biases the approach. An additional pitfall related to both the fluorimaging and biotin pull-down techniques is that less abundant proteins could be overlooked, especially when performing PAL at the high concentrations needed to study MA toxicity. Proteomics also carries drawbacks associated with the fact that ionization efficiency varies significantly different types of analytes. Additionally, the probe itself did not show strong affinity for human DAT when screened in the psychoactive drug screening program but did stimulate D1 and D5 (See Supplemental Figure S7). Of course, one possible explanation is that although MA certainly works thru DAT, the direct binding of MA to DAT in vivo could be less important, at least with regards to off-target toxicity at high concentrations. Our goal was to take an unbiased approach to identify potential novel targets associated with MA toxicity whilst employing a novel chemical biology target identification platform. To observe covalent modification of DAT it may be necessary to make a probe with high affinity for DAT and use a more selective/targeted detection method to directly observe DAT modification directly.
Conclusion
In vivo PAL was demonstrated in adult Casper zebrafish. The probe, 2, provided concentration dependent increases in labeling and a few relatively specific bands were observed in fluorimaging and after purification by biotin pull-down. Although the presence of no specific band could be correlated with behavioral changes, out-leir like behavior of individual fish was correlated with abundance of total global labeling relative to other fish within the same treatment cohort. This result highlighted the potential of the technique and the importance of considering responses in individuals relative to protein labeling when utilizing in vivo PAL. All readouts were more reliable in males than females. Competition experiments indicated that MA binds to the same proteins as 2 and is itself quite promiscuous. In the future, psychoactive probes with stronger binding affinity and better selectivity will take advantage of the ability of our approach to correlate neurobehavioral readouts with protein target identification.
A phenethylamine PAL probe analogous to MA was shown to be psychoactive in adult Casper zebrafish in a similar dose-de pendent manner to MA. Nuanced changes to the pharmacological profile compared to MA were observed during exposure and the novel tank test. Traditional drug target identification approaches often focus on labeling of a single (or a few) protein(s). However, using broader network analysis may be more appropriate to account for technique limitations and understand the impact of promiscuity on small molecule targets at high concentrations associated with adverse effects. Hence, we performed a network-based approach to gain a more comprehensive understanding of drug-target binding profile and the PAL interactome of 2. The proteomic analysis based on PAL labeling identified a network of associated proteins that is consistent with the high MA doses tested, connecting a network of proteins involved in vesicle trafficking and mitochondrial mediated toxicity. Deeper understanding of the interactions of MA with this network will provide novel targets that may be used to combat the effects of MA toxicity.
MATERIALS AND METHODS
All chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO), Fisher Scientific, or Click Chemistry Tools unless stated otherwise.
General Methods.
Reagents and solvents were purchased from common commercial suppliers (Fisher or Sigma) and used as received. All reactions were carried out under atmospheric conditions at rt unless otherwise indicated. Reactions were monitored by thin-layer chromatography (TLC, LuxPlate silica gel 60 F254 plates) and revealed by UV light (254 nm). Column chromatography was performed using Teledyne Combiflash Rf with RediSepRf Gold columns. HPLC analysis was performed using a Shimadzu Prominence HPLC (LCD-20AD) with temperature controlled autosampler (SIL-20AC), refractive index (RID-20A) and PDA (SPD-M20A) detectors. LC-MS/MS analysis was carried out with a Nexera XR UPLC coupled with a Shimadzu 8050 triple quadrupole mass spectrometer (ESI, positive mode). Separations utilized a Phenomenex Kinetix® core column (2.6 μm, C18, 100 Å, 100 × 4.6 mm column). HPLC conditions: Mobile phase A = H2O (0.1% formic acid [FA]) and mobile phase B = acetonitrile (0.1% FA); 1.0 mL/min at 30% B for 1 min followed by gradient increase to 95% B over 5 min followed by 1 min at 95% B and re-equilibration at 30% B for 4 min resulting in a total run time of 10 min. UPLC-MS/MS utilized same conditions substituting aq ammonium formate (25 mM, 0.1% FA) as mobile phase A. Purity of tested compounds was found to be > 95% pure at two wavelengths, 254 and 280 nm unless otherwise indicated. NMR (1H, 13C) was taken using a Bruker Avance 600 MHz spectrometer (cryoprobe). High resolution mass spectra (HRMS) was recorded using Waters Synapt high definition mass spectrometer (HDMS) equipped with nano-ESI source positive mode.
Synthetic chemistry 2-(4-bromobenzyl)-1,3-dioxolane (4).
2-(4-bromophenyl)acetaldehyde (3) (4.21 g, 21.2 mmol) and p-toluenesulfonic acid (PTSA) (728 mg, 4.2 mmol) were combined in an oven dried 2-necked flask, evacuated and flushed with argon. Ethylene glycol (6.56 g, 105.8 mmol) in toluene (100 mL, degassed and dried over molecular sieves) was introduced and the reaction was refluxed for 24 h connected to a Dean-Stark apparatus. The reaction mixture was then diluted in EA (100 mL), washed with sat. NaHCO3 (200 mL), dH2O (200 mL), and sat. brine (200 mL). The crude extract was concd in vacuo and purified via flash chromatography (Hex:EA, 4:1) to yield 4 as colorless crystal (2.55 g, 49.6%). 1H NMR (CDCl3, 400 MHz): 7.45–7.44 (2H, d, J = 5.56); 7.18–7.17 (2H, d, J = 5.60); 5.07–5.05 (1H, t); 3.95–3.3.93 (2H, m); 3.87–3.85 (2H, m); 2.95–2.94 (2H, d, J = 3.12). 13C NMR (CDCl3, 100 MHz): 135.0, 131.5, 131.4, 120.6, 104.2, 65.1, 40.1.
2-(4-azidobenzyl)-1,3-dioxolane (5).
4 (2.48 g, 10.2 mmol), NaN3 (1.33 g, 20.4 mmol), CuI (194 mg, 1.0 mmol), sodium ascorbate (101 mg, 0.5 mmol), and (1R,2R)-N1,N2-dimethylcyclohexane-1,2-diamine (218 mg, 1.5 mmol) were combined in an oven dried 2-necked flash, evacuated and flushed with argon. EtOH:dH2O (7:3, 20 mL, degassed) was introduced and immediately produced a blue solution. The reaction mixture was refluxed for 24 h, changing from blue to dark green. The reaction mixture was diluted in dH2O (200 mL) and extracted with DCM (200 mL × 2). The combined organic extracts were washed with brine (200 mL) and dried over Na2SO4. The crude extract was concd in vacuo and purified via flash chromatography (Hex:EA, 4:1) to yield 5 as yellow oil (1.62 g, 77.4%). 1H NMR (CDCl3, 400 MHz): 7.29–7.28 (2H, d, J = 5.64); 7.00–6.99 (2H, d, J = 5.68); 5.07–5.05 (1H, t); 3.96–3.94 (2H, m); 3.88–3.85 (2H, m); 2.97–2.96 (2H, d, J = 3.12). 13C NMR (CDCl3, 100 MHz): 138.4, 132.7, 131.1, 118.9, 104.4, 65.0, 40.1.
2-(4-azidophenyl)acetaldehyde (6).
5 (1.62 g, 7.9 mmol) and PTSA (1.5 g, 8.7 mmol) were combined in an oven dried 2-necked flash, evacuated and flushed with argon. Acetone:dH2O (15:1, 100 mL, degassed) was introduced and the reaction mixture was refluxed for 24 h, changing from pale yellow to dark yellow. The reaction mixture was diluted in EA (100 mL), washed with sat. NaHCO3 (150 mL), dH2O (150mL), and sat. brine (150 mL). The crude extract was concentrated in vacuo and purified via flash chromatography (Hex:EA, 4:1) to yield 6 as yellow oil (561 mg, 44.1%). 1H NMR (CDCl3, 400 MHz): 9.77–9.76 (1H, t); 7.23–7.22 (2H, d, J = 5.72); 7.06–7.05 (2H, d, J = 5.68); 3.71–3.71 (2H, d, J = 1.44). 13C NMR (CDCl3, 100 MHz): 199.0, 131.6, 131.0, 119.6, 49.9.
1-(4-azidophenyl)but-3-yn-2-ol (7).
6 (562 mg, 3.5 mmol) was dissolved in anhydrous THF (50 mL, degassed and dried over molecular sieves) in an oven dried, argon charged flask at −78 °C. Ethynyl magnesium bromide was added (541 mg, 4.2 mmol, 0.5 M in THF) dropwise at −78 °C and the reaction was allowed to warm to rt and stir for 4 h. The reaction was cooled to 0 °C and quenched with sat. NH4Cl (20 mL) and stirred for 5 min. The reaction mixture was diluted in dH2O and extracted with EA (100 mL × 3). The combined organic extract was washed with brine and dried over Na2SO4. The crude mixture was concentrated in vacuo and purified via flash chromatography (Hex:EA, 4:1) to yield 7 as yellow oil (254 mg, 38.9%). 1H NMR (CDCl3, 400 MHz): 7.31–7.29 (2H, d, J = 5.68); 7.02–7.00 (2H, d, J = 7.32); 4.60–4.58 (1H, td); 3.03–3.01 (2H, m); 2.52–2.52 (1H, d, J = 1.44). 13C NMR (CDCl3, 100 MHz): 138.8, 133.0, 131.0, 119.0, 83.9, 74.2, 62.9, 43.0.
1-(4-azidophenyl)but-3-yn-2-yl methanesulfonate (8).
Methanesulfonyl chloride (233 mg, 2.0 mmol) was added to a stirred solution of 7 (254 mg, 1.4 mmol) and trimethylamine (TEA) (411 mg, 4.1 mmol) in DCM at 0 °C under argon. The reaction was warmed to rt and allowed to stir for 12 h. The reaction mixture was diluted with DCM and washed with dH2O (50 mL × 3). The organic layer was then washed with brine and dried over Na2SO4. The crude mixture was concentrated in vacuo and purified via flash chromatography (Hex:EA, 3:1) to yield 8 as yellow oil (168 mg, 46.7%). 1H NMR (CDCl3, 400 MHz): 7.30–7.29 (2H, d, J = 5.64); 7.03–7.02 (2H, d, J = 5.64); 5.34–5.31 (1H, td); 3.20–3.19 (1H, dd); 3.04 (3H, s); 2.75–2.74 (1H, d, J = 1.44). 13C NMR (CDCl3, 100 MHz): 139.4, 131.3, 131.0, 119.2, 78.7, 77.9, 71.2, 41.1, 39.1.
1-(4-azidophenyl)-N-methylbut-3-yn-2-amine (2).
MeNH2 (596 mg, 6.3 mmol, 33% in absolute MeOH) was added to a stirred solution of the 8 in DCM at rt under argon. The reaction was refluxed for 12 h. MeNH2 (596 mg, 6.3 mmol, 33% in absolute MeOH) was added and the reaction continued to reflux for 12 h. The reaction mixture was diluted in dH2O and extracted in DCM (50 mL × 2). The combined organic extract was washed with brine. The crude mixture was concd in vacuo and purified via flash chromatography (Hex:EA, 70%) to yield 2 as yellow oil. 2 was acidified with 2 N HCl in diethyl ether to produce the salt form. The final product was isolated as a pale orange powder (32 mg, 21.4%). 1H NMR (CDCl3, 400 MHz): 7.30–7.29 (2H, d, J = 5.64); 7.00–6.99 (2H, d, J = 5.64); 3.56-F3.53 (1H, m); 2.99–2.90 (2H, qd); 2.51 (3H, s); 2.33–2.33 (1H, d, J = 1.40). 13C NMR (CDCl3, 100 MHz): 138.5, 134.2, 131.0, 118.9, 84.2, 72.7, 52.9, 41.1, 33.9. ESI-HRMS (m/z): [M+H]+ calculated for C11H12N4, 201.1140; observed, 201.1147.
Animals and Husbandry.
A total of 90 adult (7 to 9-month-old) male and female Casper zebrafish were produced and raised in the University of Toledo zebrafish core facility. All fish were housed in groups of 10–20 fish per 9.6 L tank on a recirculated H2O system (Aquaneering, USA) and feed live brine shrimp (Artemia sp.) b.i.d. The housing room was maintained on a 14:10 h cycle at 26–28 °C. Fish were acclimated to the testing room conditions the evening prior to testing. All testing took place during the 14 hr light cycle after the lights had been on for at least 2 hr. All experiments and husbandry practices were approved by the Institutional Animal Care and Use Committee (IACUC #108667).
Zebrafish PAL Novel Tank Test (NTT).
Adult Casper zebrafish were transferred from the home tank into an exposure chamber for 20 min with varying equimolar conc of either MA (5–65 mg/L) or 2 (6.3–50.9 mg/L). Fish were then transferred to a novel tank and their behavior tracked for 10 min via a collection of freeware programming functions (available at http://iEthology.com/). Following the NTT behavioral recording, fish were transferred to a custom-built acrylic container and UV irradiated for 10 min (λ-312 nm). Immediately following irradiation, fish were euthanized by cold shock immersion, brain tissue collected, and frozen in liquid nitrogen for subsequent analysis.
Zebrafish Behavior Statistical Analysis.
All data for exposure and the NTT behavioral endpoints were analyzed by one-way ANOVA (factor: drug conc) followed by a Dunnett’s multiple comparisons test for post-hoc significance between drug conc and control group. NTT habituation data was analyzed by the paired t-test (1 min vs. 10 min of the respective drug conc) Data is expressed as mean ± SEM with significance set at P<0.05.
Lysis.
Dissected brain was lysed in buffer (500 μL, 1% Tritonx-100 and 100 mM Na3PO4) was added to the pellet. This was incubated on ice for 1 h and vortexed 6 × within that time frame. The precipitate was transferred into a 1.5 mL centrifuge tube. The tube was centrifuged at 14,000 × g for 30 min at 4 °C. Carefully, the supernatant was transferred to a new centrifuge tube and stored at −80 °C till use. The pellet was discarded. A Bradford assay was performed after centrifugation. Lysate conc lower than 1 mg/mL after Bradford was concentrated using 3K Amicon Ultra 0.5 mL centrifugal filter until desire conc was achieved.
Click Chemistry.
A modified form of Head and Liu’s method was used.32 Cell lysate (15 μL, 1 mg/mL) was transferred into a 1.5 mL centrifuge tube. AFDye647 Azide (0.1 μL, 1 mM stock), TCEP and NaOH premix (0.29 μL, 100 mM TCEP and 400 mM NaOH), THPTA (1.69 μL, 1.7 mM stock), and CuSO4. 5H2O (0.57 μL, 50 mM stock) were added respectively. The solution was briefly vortexed after the addition of each reagent and incubated at rt for 30 min, while covered with aluminum foil. After incubation, the reaction was quenched with 4x SDS loading buffer (12.5 μL). Sample was boiled for 10 min, span done in a tabletop centrifuge and loaded (5 μL) on a 4–20% Tris-glycine gel. The gel was resolved at 120 V for 54 min. Image was obtained using a GE Typhoon Trio Imager.
Biotin/Streptavidin pull-down.
Bradford assay was performed on cell lysate to ensure conc was at 1 mg/mL. High capacity streptavidin beads (100 μL) were washed with DPBS (1 mL), pH 7.4 for 1 h at 4 °C. Cell lysate (200 μL) was added to the beads and incubated at 4 °C for 1 h. After 1 h, Bradford assay was performed again. Conc below 1 mg/mL was further concentrated to obtain the desired value. Pre-cleared lysate (30 μL) was clicked to biotin using the same conditions stated above. After the 30 min incubation at rt, the solution was added to prewashed streptavidin beads (25 μL) and the vol was adjusted by adding DPBS (200 μL). Then incubated at 37 °C with rotation for 1 h. After 1 h the supernatant was aspirated, and pellet washed with DPBS (200 μL). Supernatant was aspirated 30 min after incubation. 4x SDS loading buffer (30 μL) was added to the beads, vortexed briefly and boiled by 10 min. Supernatant was transferred into a new tube and stored at −80 °C for later use. Streptavidin beads were disposed, resolving the supernatant on a 4–20% Tris-glycine gel.
Silver Staining.
The silver stain (SilverXpress) kit used in this process was purchased from Thermofisher. The gel was removed from cassette and placed into fixative (MeOH [135 mL], acetic acid [30 mL], and ultrapure H2O [135 mL]) for 10 min. After fixation, the gel was incubated in sensitizing solution (315 mL; ultrapure H2O [157.5 mL], MeOH [150 mL] and sensitizer [7.5 mL]) for 10 min. The sensitizing step was performed twice. The gel was then washed with ultrapure H2O (300 mL × 2) for 5 min, followed by incubation in a staining solution (150 mL; stainer A [7.5 mL], stainer B [7.5 mL], and ultrapure H2O [135 mL]) for 15 min. The solution was then decanted and washed in ultrapure H2O (300 mL × 2). The H2O was disposed, and gel immersed in a developing solution (developer [7.5 mL] and ultrapure H2O [142.5 mL]) for 6.5 min. When the desired intensity was achieved, stopper solution (7.5 mL) was added to the developing mixture to stop the reaction. Gel was then washed with ultrapure H2O (300 mL × 3) for 10 min, and image obtained using G: Box Western Blot Imager.
Proteomics.
Unstained gel sections of biotin pull-down samples were excised, blinded, and sent to the University of Michigan’s Proteomics and Peptide Synthesis Core for analysis. Briefly, in-gel digestion with trypsin was performed on small section of each submitted sample with a ProGest robot (Digi-Lab) with the following protocol: washed with 25mM ammonium bicarbonate followed by acetonitrile; reduced with 10mM dithiothreitol at 60 °C followed by alkylation with 50mM iodoacetamide at room temperature; digested with sequencing grade trypsin (Promega) at 37 °C for 4 h; and quenched with formic acid and the supernatant was analyzed directly without further processing. Half of each digested sample was analyzed by nano LC-MS/MS with a Waters M-Class HPLC system interfaced to a ThermoFisher Fusion Lumos. Peptides were loaded on a trapping column and eluted over a 75 μm analytical column at 350 nL/min; both columns were packed with Luna C18 resin (Phenomenex). The mass spectrometer was operated in data-dependent mode, with the Orbitrap operating at 60,000 FWHM and 15,000 FWHM for MS and MS/MS respectively. The instrument was run with a 3s cycle for MS and MS/MS and APD was enabled. Data were searched using Mascot (Matrix Science) with the following parameters: Enzyme: Trypsin/P; Database: UniProt Zebra fish or SwissProt Human (concatenated forward and reverse plus common contaminants); Fixed modification: Carbamidomethyl (C); Variable modifications: Oxidation (M), Acetyl (N-term), Pyro-Glu (N-term Q), Deamidation (N/Q); Mass values: Monoisotopic; Peptide Mass Tolerance: 10 ppm; Fragment Mass Tolerance: 0.02 Da; Max. Missed cleavages: 2. Resulting Mascot DAT files were parsed into Scaffold (Proteome Software) for validation, filtering and to create a non-redundant list per sample. Data were filtered using at 1% protein and peptide FDR and requiring at least two unique peptides per protein. See Supplemental Table S2 for full proteomics results.
Bioinformatics.
Background and non-specific interactions were removed to subtract baseline (control) peptide abundance from studied group. Proteins identified with > 95 % certainly in Scaffold were selected for analysis. Zebrafish Uniprot numbers were converted to gene symbols for downstream analysis. String was used for protein-protein interaction analysis47–48. Our cleaned protein set was used as input for String clustering and pathway analysis. Active interaction sources used were experiments, databases, co-expression, neighborhood, and gene fusion evidence with a cutoff interaction score of at least 0.4 (medium confidence) to generate our clustering and pathways.
Supplementary Material
ACKNOWLEDGMENT
We thank the Shimadzu Laboratory for Pharmaceutical Research Excellence at The University of Toledo for mass spec-trometry instrumentation used in this study. We thank the Center for Drug Design and Development at The University of Toledo for use of the zebrafish core facilities. Receptor binding profile was generously provided by the National Institute of Mental Health’s Psychoactive Drug Screening Program, Contract # HHSN-271-2018-00023-C (NIMH PDSP). The NIMH PDSP is Directed by Bryan L. Roth MD, PhD at the University of North Carolina at Chapel Hill and Project Officer Jamie Driscoll at NIMH, Bethesda MD, USA. We thank Dr. William S. Messer Jr. for his helpful insight regarding the data and revision of the manuscript. We thank the Henriette Renner for proteomics services at the University of Michigan Biomedical Research Core Facilities Proteomics and Peptide Synthesis Core.
Funding Sources
This work was supported by the National Institute on Drug Abuse (R03DA045833). Student stipend support was provided by the Office of Undergraduate Research at the University of Toledo.
ABBREVIATIONS
- 5-HTT
Serotonin
- CNS
central nervous system
- CuAAC
copper-catalyzed alkyne-azide cycloadditon
- DA
Dopamine
- DAT
Dopamine transporter
- HPLC
high performance liquid chromatography
- MA
Methamphetamine
- NE
Norepinephrine
- NTT
novel tank test
- PAL
photoaffnity labeling
- PTSA
p-toluenesulfonic acid
- TCEP
tris(2-carboxyethyl)phosphine
- THPTA
tris-hydroxypropyltriazolylmethylamine
Footnotes
Supporting Information. Supplemental Figures S1–S7; Supplemental Table S1; and Supplemental Table S2. This material is available free of charge via the Internet at http://pubs.acs.org.
Any additional relevant notes should be placed here.
REFERENCES
- 1.Palamar JJ; Le A, Use of new and uncommon synthetic psychoactive drugs among a nationally representative sample in the United States, 2005–2017. Hum. Psychopharmacol 2019, 34 (2), e2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sexton JD; Nichols CD; Hendricks PS, Population Survey Data Informing the Therapeutic Potential of Classic and Novel Phenethylamine, Tryptamine, and Lysergamide Psychedelics. Front. Psychiatry 2019, 10, 896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolesnikova TO; Khatsko SL; Demin KA; Shevyrin VA; Kalueff AV, DARK Classics in Chemical Neuroscience: α-Pyrrolidinovalerophenone (“Flakka”). ACS Chem. Neurosci 2019, 10 (1), 168–174. [DOI] [PubMed] [Google Scholar]
- 4.Jones CM; Olsen EO; O’Donnell J; Mustaquim D, Resurgent Methamphetamine Use at Treatment Admission in the United States, 2008–2017. Am. J. Public Health 2020, 110 (4), 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panenka WJ; Procyshyn RM; Lecomte T; MacEwan GW; Flynn SW; Honer WG; Barr AM, Methamphetamine use: A comprehensive review of molecular, preclinical and clinical findings. Drug Alcohol Depend. 2013, 129 (3), 167–179. [DOI] [PubMed] [Google Scholar]
- 6.Krasnova IN; Cadet JL, Methamphetamine toxicity and messengers of death. Brain Res. Rev 2009, 60 (2), 379–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Logan BK, Methamphetamine - Effects on Human Performance and Behavior. Forensic Sci. Rev 2002, 14 (1–2), 133–51. [PubMed] [Google Scholar]
- 8.Itzhak Y; Achat-Mendes C, Methamphetamine and MDMA (ecstasy) neurotoxicity: ‘of mice and men’. IUBMB Life 2004, 56 (5), 249–55. [DOI] [PubMed] [Google Scholar]
- 9.Huisgen R, Kinetics and Mechanism of 1,3-Dipolar Cycloadditions. Angew. Chem., Int. Ed. Engl 1963, 2 (11), 633–645. [Google Scholar]
- 10.Kolb HC; Finn MG; Sharpless KB, Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem., Int. Ed 2001, 40 (11), 2004–2021. [DOI] [PubMed] [Google Scholar]
- 11.Murale DP; Hong SC; Haque MM; Lee JS, Photoaffinity labeling (PAL) in chemical proteomics: a handy tool to investigate protein-protein interactions (PPIs). Proteome Sci 2016, 15, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brunner J, New photolabeling and crosslinking methods. Annu. Rev. Biochem 1993, 62, 483–514. [DOI] [PubMed] [Google Scholar]
- 13.Stewart AM; Braubach O; Spitsbergen J; Gerlai R; Kalueff AV, Zebrafish models for translational neuroscience research: from tank to bedside. Trends Neurosci. 2014, 37 (5), 264–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darland T; Dowling JE, Behavioral screening for cocaine sensitivity in mutagenized zebrafish. Proc. Natl. Acad. Sci. U. S. A 2001, 98 (20), 11691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez-Patino MA; Yu L; Cabral H; Zhdanova IV, Anxiogenic effects of cocaine withdrawal in zebrafish. Physiol. Behav 2008, 93 (1–2), 160–71. [DOI] [PubMed] [Google Scholar]
- 16.Wronikowska O; Michalak A; Skalicka-Wozniak K; Crawford AD; Budzynska B, Fishing for a deeper understanding of nicotine effects using zebrafish behavioural models. Prog. Neuropsychopharmacol. Biol. Psychiatry 2020, 98, 109826. [DOI] [PubMed] [Google Scholar]
- 17.Duarte T; Fontana BD; Muller TE; Bertoncello KT; Canzian J; Rosemberg DB, Nicotine prevents anxiety-like behavioral responses in zebrafish. Prog. Neuropsychopharmacol. Biol. Psychiatry 2019, 94, 109655. [DOI] [PubMed] [Google Scholar]
- 18.Mi G; Gao Y; Yan H; Jin X; Ye E; Liu S; Gong Z; Yang H; Yang Z, l-Scoulerine attenuates behavioural changes induced by methamphetamine in zebrafish and mice. Behav. Brain. Res 2016, 298 (Pt A), 97–104. [DOI] [PubMed] [Google Scholar]
- 19.Stewart A; Gaikwad S; Kyzar E; Green J; Roth A; Kalueff AV, Modeling anxiety using adult zebrafish: a conceptual review. Neuropharmacology 2012, 62 (1), 135–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalueff AV; Stewart AM; Gerlai R, Zebrafish as an emerging model for studying complex brain disorders. Trends Pharmacol. Sci 2014, 35 (2), 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacRae CA; Peterson RT, Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov 2015, 14 (10), 721–31. [DOI] [PubMed] [Google Scholar]
- 22.Jiang M; Chen Y; Li C; Peng Q; Fang M; Liu W; Kang Q; Lin Y; Yung KK; Mo Z, Inhibiting effects of rhynchophylline on zebrafish methamphetamine dependence are associated with amelioration of neurotransmitters content and down-regulation of TH and NR2B expression. Prog. Neuropsychopharmacol. Biol. Psychiatry 2016, 68, 31–43. [DOI] [PubMed] [Google Scholar]
- 23.White RM; Sessa A; Burke C; Bowman T; LeBlanc J; Ceol C; Bourque C; Dovey M; Goessling W; Burns CE; Zon LI, Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2008, 2 (2), 183–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battisti UM; Sitta R; Harris A; Sakloth F; Walther D; Ruchala I; Negus SS; Baumann MH; Glennon RA; Eltit JM, Effects of N-Alkyl-4-Methylamphetamine Optical Isomers on Plasma Membrane Monoamine Transporters and Abuse-Related Behavior. ACS Chem. Neurosci 2018, 9 (7), 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang QY; Kolanos R; De Felice LJ; Glennon RA, Structural analysis of dopamine- and amphetamine-induced depolarization currents in the human dopamine transporter. ACS Chem. Neurosci 2015, 6 (4), 551–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shulgin AT; Shulgin A, Pihkal : a chemical love story. Transform Press: Berkeley, CA, 1991. [Google Scholar]
- 27.Andersen J; Madsen U; Bjoerkling F; Liang X, Rapid Synthesis of Aryl Azides from Aryl Halides under Mild Conditions. Synlett 2005, 14, 2209–2213. [Google Scholar]
- 28.Egan RJ; Bergner CL; Hart PC; Cachat JM; Canavello PR; Elegante MF; Elkhayat SI; Bartels BK; Tien AK; Tien DH; Mohnot S; Beeson E; Glasgow E; Amri H; Zukowska Z; Kalueff AV, Understanding behavioral and physiological phenotypes of stress and anxiety in zebrafish. Behav. Brain. Res 2009, 205 (1), 38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blaser RE; Rosemberg DB, Measures of anxiety in zebrafish (Danio rerio): dissociation of black/white preference and novel tank test. PLoS One 2012, 7 (5), e36931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart A; Riehl R; Wong K; Green J; Cosgrove J; Vollmer K; Kyzar E; Hart P; Allain A; Cachat J; Gaikwad S; Hook M; Rhymes K; Newman A; Utterback E; Chang K; Kalueff AV, Behavioral effects of MDMA (‘ecstasy’) on adult zebrafish. Behav. Pharmacol 2011, 22 (3), 275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalueff AV; Gebhardt M; Stewart AM; Cachat JM; Brimmer M; Chawla JS; Craddock C; Kyzar EJ; Roth A; Landsman S; Gaikwad S; Robinson K; Baatrup E; Tierney K; Shamchuk A; Norton W; Miller N; Nicolson T; Braubach O; Gilman CP; Pittman J; Rosemberg DB; Gerlai R; Echevarria D; Lamb E; Neuhauss SC; Weng W; Bally-Cuif L; Schneider H, Towards a comprehensive catalog of zebrafish behavior 1.0 and beyond. Zebrafish 2013, 10 (1), 70–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Au - Head SA; Au - Liu JO, Identification of Small Molecule-binding Proteins in a Native Cellular Environment by Live-cell Photoaffinity Labeling. J. Visualized Exp 2016, (115), e54529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aboukhatwa SM; Hanigan TW; Taha TY; Neerasa J; Ranjan R; El-Bastawissy EE; Elkersh MA; El-Moselhy TF; Frasor J; Mahmud N; McLachlan A; Petukhov PA, Structurally Diverse Histone Deacetylase Photore-active Probes: Design, Synthesis, and Photolabeling Studies in Live Cells and Tissue. ChemMedChem 2019, 14 (11), 1096–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kym PR; Carlson KE; Katzenellenbogen JA, Evaluation of a highly efficient aryl azide photoaffinity labeling reagent for the progesterone receptor. Bioconjugate Chem. 1995, 6 (1), 115–22. [DOI] [PubMed] [Google Scholar]
- 35.Pinney KG; Carlson KE; Katzenellenbogen BS; Katzenellenbogen JA, Efficient and selective photoaffinity labeling of the estrogen receptor using two nonsteroidal ligands that embody aryl azide or tetrafluoroaryl azide photoreactive functions. Biochemistry 1991, 30 (9), 2421–31. [DOI] [PubMed] [Google Scholar]
- 36.Varga ZV; Ferdinandy P; Liaudet L; Pacher P, Drug-induced mitochondrial dysfunction and cardiotoxicity. Am. J. Physiol.: Heart Circ. Physiol 2015, 309 (9), H1453–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbosa DJ; Capela JP; Feio-Azevedo R; Teixeira-Gomes A; Bastos Mde L; Carvalho F, Mitochondria: key players in the neurotoxic effects of amphetamines. Arch. Toxicol 2015, 89 (10), 1695–725. [DOI] [PubMed] [Google Scholar]
- 38.Eskandari MR; Rahmati M; Khajeamiri AR; Kobarfard F; Noubarani M; Heidari H, A new approach on methamphetamine-induced hepatotoxicity: involvement of mitochondrial dysfunction. Xenobiotica 2014, 44 (1), 70–6. [DOI] [PubMed] [Google Scholar]
- 39.Lin M; Chandramani-Shivalingappa P; Jin H; Ghosh A; Anantharam V; Ali S; Kanthasamy AG; Kanthasamy A, Methamphetamine-induced neurotoxicity linked to ubiquitin-proteasome system dysfunction and autophagy-related changes that can be modulated by protein kinase C delta in dopaminergic neuronal cells. Neuroscience 2012, 210, 308–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moszczynska A; Yamamoto BK, Methamphetamine oxidatively damages parkin and decreases the activity of 26S proteasome in vivo. J. Neurochem 2011, 116 (6), 1005–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonilla DL; Bhattacharya A; Sha Y; Xu Y; Xiang Q; Kan A; Jagannath C; Komatsu M; Eissa NT, Autophagy regulates phagocytosis by modulating the expression of scavenger receptors. Immunity 2013, 39 (3), 537–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang CS; Rodgers M; Min CK; Lee JS; Kingeter L; Lee JY; Jong A; Kramnik I; Lin X; Jung JU, The autophagy regulator Rubicon is a feedback inhibitor of CARD9-mediated host innate immunity. Cell Host Microbe 2012, 11 (3), 277–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandes S; Salta S; Summavielle T, Methamphetamine promotes alpha-tubulin deacetylation in endothelial cells: the protective role of acetyl-l-carnitine. Toxicol. Lett 2015, 234 (2), 131–8. [DOI] [PubMed] [Google Scholar]
- 44.Tian N; Hanson KA; Canty AJ; Vickers JC; King AE, Microtubule-dependent processes precede pathological calcium influx in excitotoxin-induced axon degeneration. J. Neurochem 2020, 152 (5), 542–555. [DOI] [PubMed] [Google Scholar]
- 45.Fang D; Maldonado EN, VDAC Regulation: A Mitochondrial Target to Stop Cell Proliferation. Adv. Cancer Res 2018, 138, 41–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grisshammer R, Purification of recombinant G-protein-coupled receptors. Methods Enzymol. 2009, 463, 631–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szklarczyk D; Gable AL; Lyon D; Junge A; Wyder S; Huerta-Cepas J; Simonovic M; Doncheva NT; Morris JH; Bork P; Jensen LJ; Mering CV, STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47 (D1), D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szklarczyk D; Morris JH; Cook H; Kuhn M; Wyder S; Simonovic M; Santos A; Doncheva NT; Roth A; Bork P; Jensen LJ; von Mering C, The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45 (D1), D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


