Abstract
Objectives
To evaluate changes in the characteristics of patients with coronavirus disease 2019 (COVID-19) after the emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant of concern (VOC) P.1 (Gamma), by comparing the clinical, demographic, and laboratory profiles of patients hospitalized during the first (May to July 2020) and second (December 2020 to February 2021) pandemic waves.
Methods
Data were collected from the records of COVID-19 patients (n = 4164) admitted to a single hospital in Salvador, Northeast Brazil. SARS-CoV-2 genome sequencing was performed on nasopharyngeal swab samples from 12 patients aged <60 years admitted to the intensive care unit (ICU) in February 2021.
Results
Between June 2020 and February 2021, the median age of patients admitted to the ICU decreased from 66 to 58 years (P < 0.05). This was accompanied by an increased proportion of patients without comorbidities (15.32% vs 32.20%, P < 0.0001). A significant reduction in the cycle threshold values of SARS-CoV-2 RT-PCR tests was observed in the second wave (P < 0.0001). Sequencing analysis detected lineage Gamma in all 12 ICU patients sampled in February 2021.
Conclusions
The results of this study demonstrated an increased proportion of younger adults without comorbidities with severe disease during the second COVID-19 wave, shortly after the confirmation of local Gamma circulation.
KEYWORDS: COVID-19, SARS-CoV-2, Variants of concern, P.1, Gamma
1. Introduction
There is concern regarding the increased infectivity and possible immune escape of new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants (Chakraborty et al., 2021). According to the World Health Organization (WHO), there are currently four variants of concern (VOC), all with key mutations in the receptor binding domain (RBD) of the spike protein (Korber et al., 2020). These are the Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), and Delta (B.1.617) lineages.
In Brazil, two SARS-CoV-2 lineages – B.1.1.28 and B.1.1.33 – were predominant during the first surge of infections in the first 6 months of 2020 (Resende et al., 2021, Candido et al., 2020). The emerging Brazilian Gamma variant, derived from the B.1.1.28 lineage, began spreading rapidly later in 2020. Initially detected in the state of Amazonas, Gamma was responsible for a public health calamity in that state (Faria et al., 2021, Naveca et al., 2021, Sabino et al., 2021). The Gamma variant started to be detected at increasing rates from January 2021 onwards throughout Brazil, and became the predominant lineage associated with the second wave of infections (http://www.genomahcov.fiocruz.br). Following an acceleration of the transmission rates in 2021, Brazil became the epicenter of the coronavirus disease 2019 (COVID-19) pandemic, with over 13 million confirmed cases and 350 000 deaths (https://www.covid.saude.gov.br).
Gamma has 21 lineage-defining mutations, including 10 in the spike protein, three of them in the RBD (K417T, E484K and N501Y), showing a surprising convergence with the B.1.351 RBD. These three mutations in the RBD combined have been shown to increase the receptor binding affinity (Nelson et al., 2021). The mutations found in Gamma have been associated with increased transmissibility (Naveca et al., 2021), a higher viral load (Faria et al., 2021) and propensity for immune evasion (Dejnirattisai et al., 2021), and SARS-CoV-2 reinfection (Naveca et al., 2021).
Along with the dissemination of the Gamma variant, there have also been reports of an increased percentage of young patients developing severe disease (Dejnirattisai et al., 2021). However, many other questions remain unanswered, including a possible increased fatality rate, increased disease severity in people without known SARS-CoV-2 risk comorbidities, reduced time from symptom onsent to hospitalization.
The public health authorities in the state of Bahia (Northeast Brazil) confirmed the circulation of Gamma in the state in February, 2021. The first reports of Gamma detection in Salvador date from late December 2020 and early January 2021, and these were linked to individuals with a history of travel to Manaus (Tosta et al., 2021). This was followed by a rampant increase in the number of hospitalizations and deaths due to COVID-19 in Bahia in February and March 2021 (https://bi.saude.ba.gov.br/transparencia/). However, the clinical and demographic features of this epidemic second wave, as well as the association with SARS-CoV-2 lineages, still need to be characterized. Herein, we report the changes in the profile of patients admitted to the intensive care unit (ICU) of a private hospital in Salvador, Bahia capital city, in February 2021, due to COVID-19 with possible involvement of the locally circulating Gamma VOC.
2. Methods
2.1. Study design and procedures
A cross-sectional study was performed at São Rafael Hospital, a private general reference hospital in Salvador, Bahia, Northeast Brazil. Two time periods with increased numbers of hospital admissions were selected for data analysis, corresponding to the first wave (May to July 2020) and second wave (December 2020 to February 2021) of COVID-19 hospital admissions.
Data regarding the number of confirmed COVID-19 patients admitted to the hospital, along with their clinical and demographic characteristics, were obtained from the hospital health information system and electronic medical records (n = 4164). Data on confirmed COVID-19 cases in the city of Salvador and the state of Bahia were obtained from https://bi.saude.ba.gov.br.
The nasopharyngeal swabs were obtained and tested by multiplex real-time PCR Allplex SARS-CoV-2 assay (Seegene Inc., Seoul, Korea) in the laboratory of São Rafael Hospital, as part of routine diagnostic procedures. Cycle threshold (Ct) values were evaluated in positive samples from the peak months of each wave, June 2020 and February 2021 (n = 2360). Nucleic acid sequencing was performed of viral RNA extracted from 12 nasal swab samples selected from patients aged less than 60 years, who were admitted to the São Rafael Hospital ICU in February 2021. Written informed consent was obtained. The SARS-CoV-2 viral genome sequencing was performed in accordance with the manufacturer's instructions, using the Ion Torrent PGM system (Life Technologies, USA). cDNA was synthesized with the SuperScript VILO reverse transcriptase kit (Invitrogen, USA). The libraries were prepared using the Ion AmpliSeq SARS-CoV-2 assay panel, the Ion AmpliSeq Library Kit, and Ion Torrent PGM (Thermo Fisher Scientific, USA). Genomes were submitted to the Nextclade tool (https://clades.nextstrain.org) for quality control assessment and the Pangolin tool (https://pangolin.cog-uk.io/) for SARS-CoV-2 lineage assignment. Maximum likelihood (ML) phylogenetic analysis was performed in IQ-TREE for all detected Gamma variant genomes, and tree branch support was calculated with the Shimodaira–Hasegawa approximate likelihood ratio test (SH-aLRT), as described previously (Minh et al., 2020). Background high quality (>29 000 bp and <1% N) Brazilian Gamma variant sequences were obtained from the EpiCoV database in GISAID, as available on March 25, 2021 (Supplementary Material Table S1) and aligned with the genomes generated here in MAFFT (Katoh and Standley, 2013). The ML tree was visualized using FigTree v1.4 (http://tree.bio.ed.ac.uk/software/figtree/).
2.2. Statistical analysis
Categorical variables were compared using the Fisher test. Continuous data were presented as the median and 95% confidence interval (CI). The Mann–Whitney test and Kruskal–Wallis test were used for comparisons of non-parametric data (Ct values and median age). P-values <0.05 were considered significant. Data were analyzed with Prism software v.9.1 (GraphPad).
3. Results
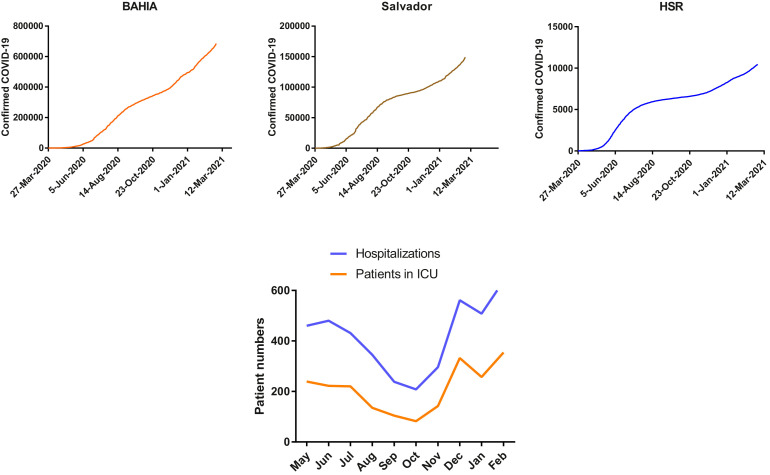
Accompanying the evolution of the pandemic in the city of Salvador, the data analysis demonstrated two waves of admission to the hospital due to COVID-19, the first during May to July 2020 and the second during December 2020 to February 2021. A similar pattern of cumulative confirmed COVID-19 cases reported by Bahia State, Salvador, and São Rafael Hospital in Salvador in the northeast of Brazil was observed (Figure 1 A). There was a rapid increase in patients requiring hospitalization and ICU treatment in February 2021 (Figure 1B).
Figure 1.
Confirmed COVID-19 hospitalizations. (A) Cumulative confirmed COVID-19 cases in Bahia State, Salvador, and patients admitted to the São Rafael Hospital (HSR) since May 2020. (B) The total number of patients hospitalized (n = 4164, blue line) and admitted to the ICU (n = 2087, orange line) from May 2020 to February 2021.
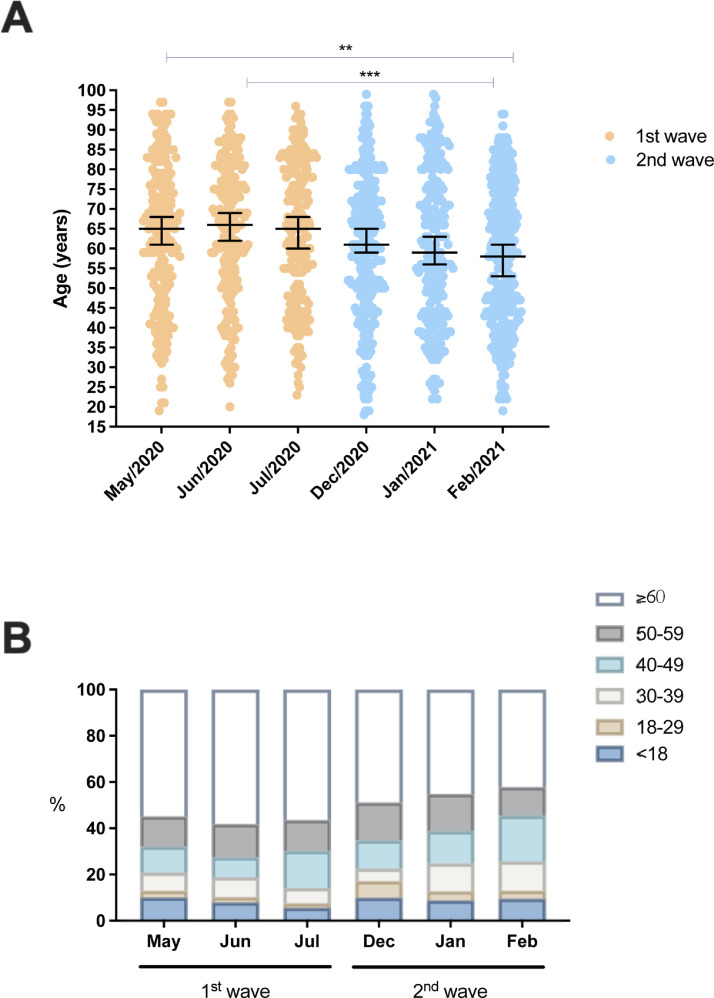
From May 2020 to February 2021, 2087 patients were admitted to the ICU of São Rafael Hospital; 672 were admitted during the first wave (May to July 2020) and 943 in the second wave (December 2020 to February 2021). The median age of the patients hospitalized and admitted to the ICU was lower in February 2021 (58 years) when compared to May and June 2020 (66 years), and this decrease in median age was found to be statistically significant (Figure 2 A). A progressive increase in the percentage of patients aged <60 years being admitted to the ICU was observed in the second wave, differing from the demographic pattern observed in the first wave (Figure 2B).
Figure 2.
Temporal changes in the demographic profile of the patients admitted to the ICU due to COVID-19. (A) Individual age values (≥18 years old) are represented, along with median and 95% CI. The first wave includes the months of May, June, and July 2020; the second wave includes the months of December 2020, January 2021, and February 2021. (B) Total numbers of patients admitted to the ICU stratified by age group. **P < 0.001, ***P < 0.0001.
Along with the increase in percentage of young and middle-aged patients, there was also an increase in the percentage of patients entering the ICU without known comorbidities (cardiovascular diseases, hypertension, diabetes, obesity, liver diseases, asthma, kidney diseases, and immunosuppression). Patients without comorbidities represented 32.20% of hospitalized COVID-19 patients admitted to the ICU due to COVID-19 in February 2021, compared to 15.32% in June 2020 (P < 0.0001). Table 1 shows the frequency of patients with different comorbidities stratified by age group.
Table 1.
Comorbidity assessment in patients entering the ICU during the first wave (June 2020) and second wave (February 2021), stratified by age group.
| Comorbidities | 18–29 years |
30–39 years |
40–49 years |
50–59 years |
≥60 years |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Admission period | P-valuea | Admission period | P-valuea | Admission period | P-valuea | Admission period | P-valuea | Admission period | P-valuea | ||||||
| Jun2020(n = 3) | Feb2021(n = 10) | Jun2020(n = 19) | Feb2021(n = 40) | Jun2020(n = 17) | Feb2021(n = 66) | Jun2020(n = 29) | Feb2021(n = 41) | Jun2020(n = 118) | Feb2021(n = 123) | ||||||
| Obesity | - | 20.0% | 0.999 | 31.6% | 22.5% | 0.528 | 41.2% | 10.6% | 0.007* | 17.2% | 19.5% | 0.999 | 16.9% | 9.8% | 0.1 |
| Cardiovascular diseases | - | - | 26.3% | 12.5% | 0.266 | 47.0% | 24.2% | 0.078 | 48.3% | 51.2% | 0.999 | 85.6% | 69.1% | 0.002* | |
| Hematological | - | - | - | 2.5% | 0.999 | - | - | 3.4% | - | 0.414 | 3.4% | 1.6% | 0.439 | ||
| Liver disease | - | - | 5.3% | - | 0.322 | - | 4.5% | 0.999 | - | - | 2.5% | - | 0.116 | ||
| Asthma | - | - | 5.3% | - | 0.322 | 11.8% | 1.5% | 0.105 | - | - | 3.4% | 3.3% | 0.999 | ||
| Diabetes | - | - | - | 2.5% | 0.999 | 17.6% | 15.2% | 0.724 | 27.6% | 20.0% | 0.461 | 44.9% | 40.5% | 0.49 | |
| Neurological | - | 10.0% | 0.999 | 10.5% | - | 0.999 | - | - | - | 2.4% | 0.999 | 13.5% | 9.8% | 0.357 | |
| Lung disease | - | - | - | - | - | - | - | - | 6.8% | 4.1% | 0.351 | ||||
| Immunosuppression | - | - | - | - | 5.9% | - | 0.205 | 6.9% | 2.4% | 0.566 | 3.4% | 1.6% | 0.439 | ||
| Kidney disease | - | - | - | - | 17.6% | 3.0% | 0.056 | 13.8% | 4.9% | 0.224 | 17.8% | 5.7% | 0.003* | ||
ICU, intensive care unit.
P-value of the Chi-square test or Fisher's exact test; *P-value <0.05, significant.
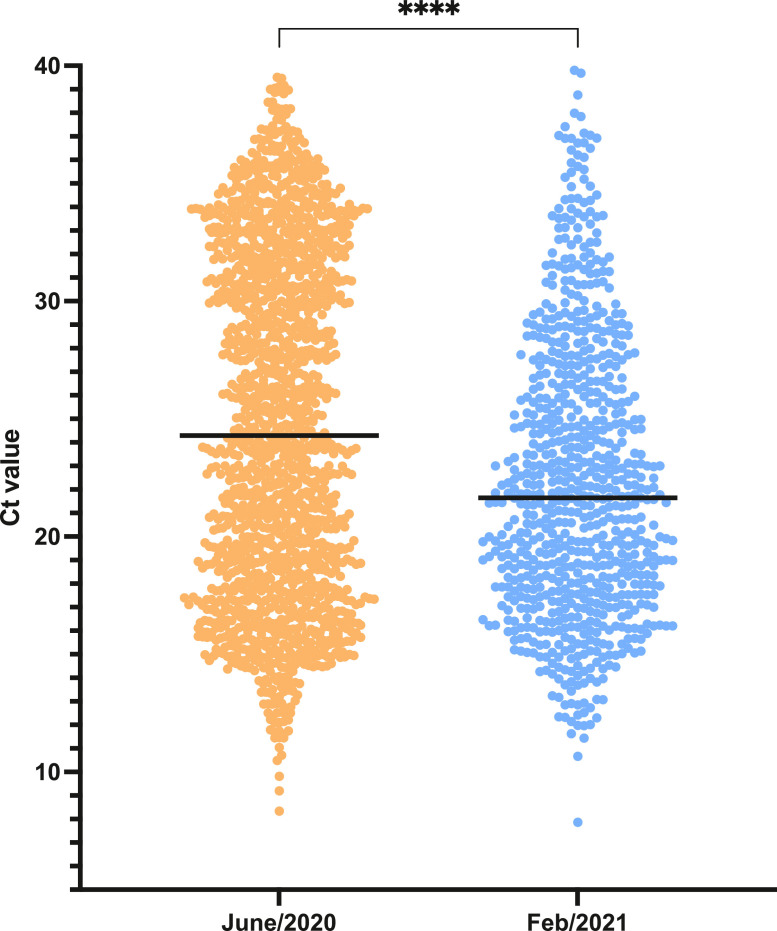
In addition to the changes in clinical and demographic profile found, possible alterations in the pattern of the results obtained in the RT-qPCR analysis were also evaluated. A significant reduction in the median Ct values from nasopharyngeal swab samples analyzed by RT-qPCR was observed when comparing June 2020 to February 2021 (P < 0.0001) (Figure 3 ). During the same periods, similar average values for the time between symptom onset and sample collection were observed: 4.4 days (95% CI 4.3–4.9 days) and 5.1 days (95% CI 4.8–5.4 days) for June 2020 and February 2021, respectively.
Figure 3.
Temporal change in the SARS-CoV-2 RT-PCR cycle threshold (Ct) values. N gene Ct values were evaluated and compared between samples from COVID-19 patients referred to the hospital in June 2020 (n = 1589) and in February 2021 (n = 771). Single Ct values and the median are plotted. ****P < 0.0001.
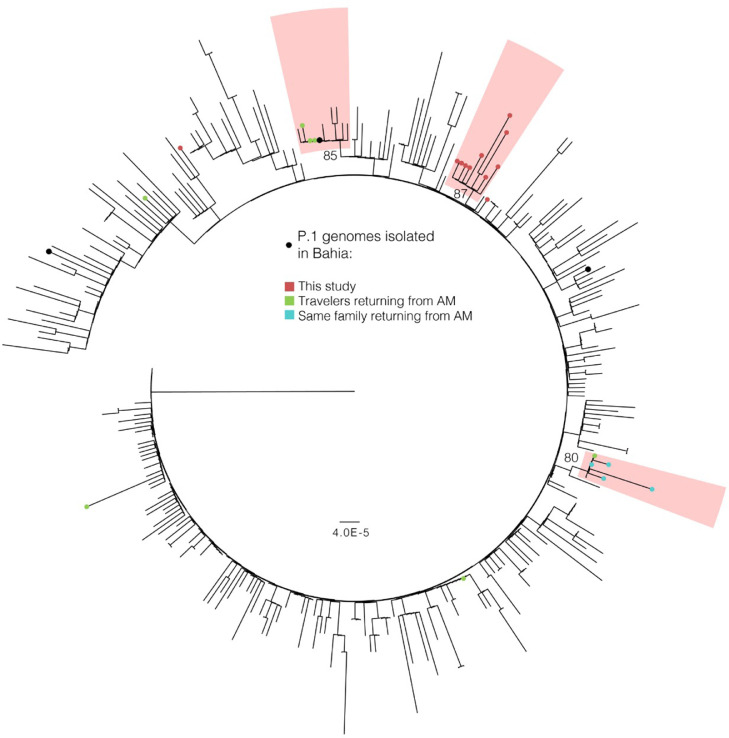
To investigate the viral diversity associated with the second COVID-19 wave in Salvador, 12 swab samples from patients admitted to the ICU in February 2021, who were within the age range of 18–59 years, were sequenced. The characteristics of these patients are shown in Table 2 . Five patients had no comorbidities. Half of the patients required invasive mechanical ventilation, while the other half were treated with non-invasive ventilation. Sequencing analysis identified the SARS-CoV-2 Gamma variant in all of the samples evaluated. Although all sequences were classified by the Pangolin tool as Gamma with high probability, one of the genomes presented low coverage and was not submitted to phylogenetic analysis. All of the genomes generated herein are available in the EpiCoV database, maintained by the GISAID initiative, with accession codes EPI_ISL_160861 to EPI_ISL_1608171. The ML phylogenetic tree revealed that nine of the 11 genomes clustered together with high support (SH-aLRT = 87), meaning that the Gamma lineage has already established local community transmission in Salvador (Figure 4 ). Two other highly supported clusters of Gamma genomes isolated in Bahia State were also observed in the tree, being composed of travelers returning from Amazonas (Tosta et al., 2021). These three clusters provide evidence of multiple introductions of the Gamma lineage into Bahia State.
Table 2.
Clinical and demographic characteristics of patients admitted to the ICU in February 2021 selected for sequencing.
| Sex | Age (years) | Symptom onset (days before ICU admission) | Ventilatory support | Comorbidities | Extent of ground glass opacitites (Thorax CT) |
|---|---|---|---|---|---|
| Male | 35 | 7 | Non-invasive ventilation | None | 25% |
| Male | 31 | 7 | Non-invasive ventilation | Obesity, hypertension, diabetes | 25–50% |
| Male | 41 | 9 | Non-invasive ventilation | None | 25–50% |
| Male | 59 | 11 | Non-invasive ventilation | Obesity, hypertension | 40% |
| Male | 37 | 10 | Invasive mechanical ventilation | Obesity | 25–50% |
| Male | 46 | 9 | Invasive mechanical ventilation | Obesity | >75% |
| Male | 44 | 5 | Invasive mechanical ventilation | None | 50% |
| Male | 41 | 10 | Non-invasive ventilation | None | 75% |
| Male | 56 | 8 | Invasive mechanical ventilation | Hypertension | >75% |
| Female | 36 | 5 | Non-invasive ventilation | Obesity | 50–75% |
| Male | 24 | 8 | Invasive mechanical ventilation | Obesity | 25–50% |
| Male | 35 | 7 | Invasive mechanical ventilation | None | 50% |
CT, computed tomography; ICU, intensive care unit.
Figure 4.
Circular maximum likelihood phylogenetic tree of the P.1 (Gamma) lineage diversity in Brazil. Samples isolated in Bahia are shown with circles, and the colors represent the origin of the sample, when available. Travel history data are those reported by Tosta et al., 2021 (Tosta et al., 2021). Three clusters with samples isolated in Bahia are highlighted and SH-aLRT support is shown. The tree was rooted in the oldest P.1 sampled genome.
4. Discussion
Here we report the identification of the P.1 or Gamma variant in all sequenced clinical samples from patients admitted to the ICU in February 2021, in Salvador, Brazil. These results suggest that the Gamma variant is responsible for a large share of the COVID-19 cases in Salvador, in line with different reports finding a high proportion of the Gamma variant across Brazil (Slavov et al., 2021).
Recently, researchers have reported a significant increase in case fatality rates among young and middle-aged adults in Parana, Brazil, which may be associated with the Gamma strain (de Oliveira et al., 2021). The data presented herein also give support to a role of the Gamma variant in the acceleration of the pandemic in Salvador and Bahia State seen in early 2021.
The observed increase in the percentage of patients being admitted to the ICU with no risk factors for severe COVID-19, such as increased age, hypertension, and diabetes, is notable (de Oliveira et al., 2021). However, these are preliminary data and more studies are urgently required to clarify whether changes in the virulence may be attributable to the Gamma variant.
It is also necessary to investigate the influence of the vaccine rollout, which started in mid-January, first recruiting healthcare workers and then progressing to elderly individuals older than 80 years of age by the end of February, utilizing either CoronaVac (a two-dose, 28-day interval vaccine scheme) or AstraZeneca ChAdOx nCoV-19 (a two-dose, 3-month interval vaccine scheme). Therefore, during the period evaluated in this study, a very low percentage of elderly patients had received two doses of the vaccine.
Previous studies have suggested an increased Gamma viral load in nasopharyngeal swab samples compared to previous SARS-CoV-2 lineages (Resende et al., 2021). The present study demonstrated that positive swab samples in February 2021 presented lower Ct values than samples evaluated in June 2020. Although Ct values are subject to intrinsic sample collection variability and do not utilize reference measurements, the comparison of the results found in the peaks of the first wave and the second wave suggest that patients presented increased viral loads in February 2021, which was also reported when Gamma samples were compared to non-Gamma samples (Nelson et al., 2021, Faria et al., 2021).
Confirming that Gamma is a highly transmissible variant, associated with increased viral replication and disease severity, would require restrictive control measures to be revisited in populations with confirmed Gamma variant circulation, to adequately prevent viral spread and pressure on the healthcare system, as has been observed in Brazil. The data presented herein do not give support to an increased mortality rate among hospitalized COVID-19 patients after the emergence of the Gamma variant, since mortality rates and invasive mechanical ventilation rates decreased during the period evaluated. Since these data may be influenced by other confounders, such as improved disease management and evolving therapeutic protocols, future properly designed studies will be required to determine whether the Gamma variant may be associated with any changes in disease severity.
In summary, the findings of this study contribute to describe the characteristics of the patients with severe COVID-19 during the second pandemic wave, which appears to be associated with an increased proportion of young and middle-aged adults. Additionally, the results suggest that the Gamma variant may have spread rapidly in Salvador, Brazil, leading to increased numbers of cases, hospitalizations, and ICU admissions shortly after its first detection in Bahia State. These preliminary findings reinforce the immediate need to adopt measures to reduce its spread and accelerate the vaccine rollout.
CRediT authorship contribution statement
Carolina Kymie Vasques Nonaka: Writing – original draft, Writing – review & editing, Investigation, Formal analysis, Methodology. Tiago Gräf: Writing – original draft, Writing – review & editing, Investigation, Formal analysis, Methodology. Camila Araújo de Lorenzo Barcia: Investigation, Methodology. Vanessa Ferreira Costa: Investigation, Methodology. Janderson Lopes de Oliveira: Investigation, Methodology. Rogério da Hora Passos: Investigation, Methodology. Iasmin Nogueira Bastos: Investigation, Methodology. Maria Clara Brito de Santana: Investigation, Methodology. Ian Marinho Santos: Investigation, Methodology. Karoline Almeida Felix de Sousa: Investigation, Methodology. Thamires Gomes Lopes Weber: Methodology. Isadora Cristina de Siqueira: Writing – review & editing. Clarissa Araújo Gurgel Rocha: Writing – original draft, Writing – review & editing, Investigation, Methodology. Ana Verena Almeida Mendes: Writing – review & editing, Conceptualization. Bruno Solano de Freitas Souza: Writing – original draft, Writing – review & editing, Conceptualization, Formal analysis, Funding acquisition.
Acknowledgments
Acknowledgements
We thank Ms Roquelina Assis for technical support, the team of the Molecular Biology Laboratory of São Rafael Hospital for sample handling, and Jessica Pronestino for assistance with the statistical analysis.
Funding
This study was funded by CNPq, Inova Fiocruz, the Serrapilheira Institute, and the D'Or Institute for Research and Education.
Ethics statement
This study was reviewed by local IRBs and received ethical approval by the National Committee for Ethics on Research (CONEP; CAAE: 29496920.8.0000.5262, 46821621.5.0000.0048 and 3.980.128 /2020). All sampled patients have provided written informed consent.
Conflict of interest
The authors claim no conflict of interest.
Access to data
C.K.V.N. and A.V.A.M. have full access to the data and are the guarantor for the data. Data are available upon reasonable request.
References
- 4 Candido D.S, Claro IM, de Jesus JG, Souza W.M., Moreira F.R., Dellicour S. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369(6508):1255–1260. doi: 10.1126/science.abd2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai, W., Zhou, D., Supasa, P., Liu, C, Mentzer, A.J., Ginn, H. M. et al. Antibody evasion by the Brazilian P.1 strain of SARS-CoV-2. bioRxiv preprint. 2021. 10.1101/2021.03.12.435194
- 1 Chakraborty D., Agrawal A., Maiti S. Rapid identification and tracking of SARS-CoV-2 variants of concern. The Lancet. 2021;397(10282):1346–1347. doi: 10.1016/S0140-6736(21)00470-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5 Faria N.R, Mellan T.A, Whittaker C, Claro I.M., Candido D.D.S., Mishra S. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372(6544):815–821. doi: 10.1126/science.abh2644. http://www.ncbi.nlm.nih.gov/pubmed/33688664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17 Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. medRxiv. 2021 doi: 10.1101/2021.02.26.21252554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12 Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usa-bility. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2 Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11 Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., Von Haeseler A. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the Genomic Era. Molecular Biology and Evolution. 2020;37(5):1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveca, F., Nascimento, V., Souza, Corado, A., Nascimento, F., Silva, G. et al. Phylogenetic relationship of SARS-CoV-2 sequences from Amazonas with emerging Brazilian variants harboring mutations E484K and N501Y in the Spike protein. Virological.org 2021;
- 9 Naveca F., da Costa C., Nascimento V., Souza V., Corado A., Nascimento F. Three SARS-CoV-2 reinfection cases by the new Variant of Concern (VOC) P.1/501Y.V3. Res Sq. 2021 doi: 10.21203/rs.3.rs-318392/v1. [DOI] [Google Scholar]
- 7 Nelson G, Buzko O, Spilman P.R., Niazi K., Rabizadeh S., Soon-Shiong P.R. Molecular dynamic simulation reveals E484K mutation enhances spike RBD-ACE2 affinity and the 1 combination of E484K, K417N and N501Y mutations (501Y.V2 variant) induces conformational 2 change greater than N501Y mutant alone, potentially resulting in an escape mutant. bioRxiv. 2021 doi: 10.1101/2021.01.13.426558. [DOI] [Google Scholar]
- 16 de Oliveira M.H.S., Lippi G., Henry B.M. Sudden rise in COVID-19 case fatality among young and middle-aged adults in the south of Brazil after identification of the novel B. 1.1. 28.1 (P. 1) SARS-CoV-2 strain: analysis of data from the state of Parana. medRxiv. 2021 doi: 10.1101/2021.03.24.21254046. [DOI] [Google Scholar]
- 3 Resende P.C, Delatorre E, Gräf T, Mir D., Motta F.C., Appolinario L.R. Evolutionary dynamics and dissemination pattern of the SARS-CoV-2 lineage B.1.1.33 during the early pandemic phase in Brazil. Front Microbiol. 2021;11:1–14. doi: 10.3389/fmicb.2020.615280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15 Sabino E.C., Buss L.F., Carvalho M.P., Prete C.A., Crispim M.A., Fraiji N.A. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. The Lancet. 2021;397(10273):452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13 Slavov S.N., Patane J., Bezerra R.S., Giovanetti M., Fonseca V., Martins A.J. Genomic monitoring unveil the early detection of the SARS-CoV-2 B.1.351 lineage (20H/501Y.V2) in Brazil. medRxiv. 2021 doi: 10.1101/2021.03.30.21254591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10 Tosta S., Giovanetti M., Nardy V.B., da Silva L.R.D.O., Gomez M.K.A., Lima J.G. Early genomic detection of SARS-CoV-2 P. 1 variant in Northeast Brazil. medRxiv. 2021 doi: 10.1101/2021.02.25.21252490. [DOI] [Google Scholar]