Abstract
Wastewater-based epidemiology has been used as a tool for surveillance of COVID-19 infections. This approach is dependent on the detection and quantification of SARS-CoV-2 RNA in untreated/raw wastewater. However, the quantification of the viral RNA could be influenced by the physico-chemical properties of the wastewater. This study presents the first use of Adaptive Neuro-Fuzzy Inference System (ANFIS) to determine the potential impact of physico-chemical characteristics of wastewater on the detection and concentration of SARS-CoV-2 RNA in wastewater. Raw wastewater samples from four wastewater treatment plants were investigated over four months. The physico-chemical characteristics of the raw wastewater was recorded, and the SARS-CoV-2 RNA concentration determined via amplification with droplet digital polymerase chain reaction. The wastewater characteristics considered were chemical oxygen demand, flow rate, ammonia, pH, permanganate value, and total solids. The mean SARS-CoV-2 RNA concentrations ranged from 648.1(±514.6) copies/mL to 1441.0(±1977.8) copies/mL. Among the parameters assessed using the ANFIS model, ammonia and pH showed significant association with the concentration of SARS-CoV-2 RNA measured. Increasing ammonia concentration was associated with increasing viral RNA concentration and pH between 7.1 and 7.4 were associated with the highest SARS-CoV-2 concentration. Other parameters, such as total solids, were also observed to influence the viral RNA concentration, however, this observation was not consistent across all the wastewater treatment plants. The results from this study indicate the importance of incorporating wastewater characteristic assessment into wastewater-based epidemiology for a robust and accurate COVID-19 surveillance.
Keywords: Wastewater-based epidemiology, SARS-CoV-2, Wastewater, Droplet digital PCR, Adaptive neuro-fuzzy inference system model
1. Introduction
SARS-CoV-2 is an enveloped, single-stranded, positive-sense, RNA virus responsible for the COVID-19 pandemic (Zafar, 2020; Munir et al., 2020; Mousavizadeh and Ghasemi, 2021). The SARS-CoV-2 virus particle (the virion) consists of the nucleic acid encapsulated in an outer protein coat called the capsid and a lipid envelope (Wang et al., 2020). This coronavirus virion has glycoprotein spikes on the outer surface, with which it neutralises antibody, attaches and enters a host (Ou et al., 2020). The SARS-CoV-2 is majorly a respiratory pathogen, and mainly spread from person-to-person via direct or indirect contact between people (Li et al., 2020; Sommerstein et al., 2020; Rabaan et al., 2021). Shedding of the viral particle via urine and feces of infected persons has been confirmed and raises concern for possible oral-faecal route of transmission (Collivignarelli et al., 2020; Sun et al., 2020; Xiao et al., 2020).
The viral particle shed from an infected individual, via feces and urine, end up mostly in sewage systems (Graham et al., 2021). Different polymerase chain reaction (PCR)-based methods including reverse-transcriptase PCR (RT-PCR), reverse-transcriptase loop mediated amplification (RT-LAMP) and reverse-transcriptase droplet digital PCR (RT–ddPCR) have been employed for detection or quantification of SARS-CoV-2 in wastewater (Ahmed et al., 2020a, 2020b; Graham et al., 2021; D'Aoust et al., 2021; Amoah et al., 2021). The detection of this virus in wastewater via the use of these molecular methods forms the basis for wastewater-based epidemiology surveillance of COVID-19.
Wastewater-based epidemiology is currently being proposed as a cost-effective complimentary approach for COVID-19 surveillance at the community-level (Olga and Halden, 2020; Gonzalez et al., 2020; Daughton, 2020; Aguiar-Oliveira et al., 2020; Spurbeck et al., 2021). The clinical-based surveillance and screening has been limited by cost, turnaround time and possible underestimation of the severity of the infection spread occasioned by asymptomatic COVID-19 cases (Olga and Halden 2020; Lodder and de Roda Husman. 2020); Randazzo et al., 2020). It has been shown by various researchers that the SARS-CoV-2 genetic markers in the untreated/raw wastewater can be used to track infection rate within the wastewater treatment plant (WWTP) catchment of the community in real time (Kumar et al., 2020; Ahmed et al., 2020a, 2020b; Graham et al., 2020; Peccia et al., 2020; D'Aoust et al., 2021). This method captures the totality of symptomatic, pre-symptomatic and asymptomatic carriers within specific community, which is usually not the case with clinical surveillance (Kumar et al., 2020; Bivins et al., 2020). The incorporation of population normalization to the wastewater-based epidemiology data provides the additional opportunity for determination of patterns, trends and possible comparison across catchments with different population sizes, ultimately helping in infection hotspot identification (Medema et al., 2020a, 2020b).
Different authors have employed the wastewater-based epidemiology approach for COVID-19 surveillance around the globe with some level of success achieved. However, there are still challenges to overcome for this approach to become more useful and widely acceptable. Viruses are generally susceptible to inactivation once released into the environment and factors including temperature, UV light, pH, inorganic cations and anions, antagonistic microbial interaction, among others can have virucidal effect on these viral particles (Joiner et al., 2020; Auffret et al., 2019). The wastewater environment is a complex matrix with different components that can affect the stability of virus. Enteric viruses are known to be more resistant to environmental factors and tend to survive longer than enveloped viruses (Hurst and Gerba, 1989). Gundy at al. (2009) noted that the Human coronavirus (HCoV-229 E), a causative agent of common cold could survive in wastewater up to 2–4 days under optimal conditions. They also noted that coronaviruses inactivation in the test water was highly dependent on temperature, level of organic matter, and presence of antagonistic bacteria. Olga and Halden (2020) also identified temperature, average in-sewer travel time and per-capita water use as key variables that affects SARS-CoV-2 estimation in wastewater. This indicates that the wastewater-based epidemiology approach for SARS-CoV-2 surveillance needs to factor in the impact of the wastewater environment on the stability of the virus. However, there is lack of information on the impact of wastewater characteristics on estimation of SARS-CoV-2 viral load in wastewater. Hence, this study investigated the effect of physico-chemical characteristics of wastewater on the estimation of SARS-CoV-2 for wastewater-based epidemiology.
An Adaptive Neuro-Fuzzy Inference System (ANFIS) model was used to identify the impact of selected physico-chemical parameters on the concentration of SARS-CoV-2 detected. The study therefore makes a valuable contribution towards the optimization of wastewater-based epidemiology for COVID-19 surveillance.
2. Methodology
2.1. Wastewater sampling
Four municipal wastewater treatment plants (WWTPs) from two of the most COVID-19 affected districts (eThekwini and Umgungundlovu)) in KwaZulu- Natal (KZN) province of South Africa were chosen for this study. In the eThekwini district, the Isipingo and Central WWTPs were selected. These WWTPs treat an average of 14 ML/d and 80 ML/d of wastewater, respectively. Similarly, two WWTPs; Darvill and Howick WWTPs, treating an average of 70 ML/d and 6 ML/d of wastewater respectively, were chosen from the Umgungundlovu district. Grab samples (2 L) of raw sewage were collected at the head of works (post-primary screening) for each of the WWTPs weekly between the hours of 7:00–11:00 a.m. Sampling was carried out for approximately four months, from July to October 2020. Full personal protective equipment (PPE) (Face shield, FFP2 face mask, waterproof coveralls, and safety boots) was worn during each sampling event.
2.2. Wastewater characteristics data collection and SARS-CoV-2 viral load determination
Wastewater characteristics such as chemical oxygen demand (COD), flow rate (FR), ammonia (NH3), pH, permanganate value (presented as PV4) and total solids (TS) were provided by the operators of the WWTPs from their routine wastewater analysis. Therefore, the choice of physico-chemical parameters to consider was based on the availability of data, during the study period. The viral recovery was performed using the Centricon® Plus-70 centrifugal ultrafilter according to the procedure published by Medema et al., 2020a, 2020b. Prior to the ultrafiltration the wastewater samples were heat-treated at 60 °C for 90 min immediately upon arrival in the laboratory (within 2 h of sampling). Samples were then centrifuged at 4750×g for 30 min and approximately, and 60 ml of supernatant was used for ultrafiltration.
RNA was extracted using the QiAmp Viral RNA MiniKit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The quality and quantity of the extracted RNA was determined using the Implen Nanophotometer® and extracted RNA stored at −80 °C until further analysis.
2.3. Droplet digital PCR
Detection and quantification of SARS-CoV-2 in the extracted RNA was performed using the droplet digital PCR (ddPCR) platform. The One-Step RT-ddPCR Advanced Kit for Probes from Biorad (USA) together with primers and probes targeting the N2 region of the viral (SARS-CoV-2) genome was used. The choice of N2 as the target gene was based on the preliminary results obtained, where the N2 had the lowest limit of detection and gave reproducible results. This finding has been supported by other researchers, where N2 was identified as a good target for amplification compared to either N1 or N3 (Randazzo et al., 2020, Shirato et al., 2020). The ddPCR was carried out in a 22 μl reaction mixture contained: 5 μl supermix, 2 μl reverse transcriptase, 1 μl dithiothreitol, 1.98 μl each of the forward and reverse primers (10 μM), 0.55 μl of 10 μM probe, 2.49 μl nuclease-free water, and 7 μl template RNA (1 ng). The forward primer sequence used was; 5′- TTACAAACATTGGCCGCAAA-3′, the reverse primer sequence was; 5′- GCGCGACATTCCGAAGAA-3′ and the probe was' 5′- ACAATTTGC(ZEN)CCCCAGCGCTTCAG-3′ with 5′ Modification with FAM and 3′ Modification with Iowa Black® FQ (Giri et al., 2020; Barra et al., 2020). The thermal cycling conditions were; Reverse transcription at 50 °C for 1 h, enzyme activation at 95 °C for 10 min, 40 cycles of denaturation at 94 °C for 30 s and annealing at 55 °C for 60 s. Deactivation of enzymes at 98 °C for 10 min and stabilization of the droplets at 4 °C for 30 min with a ramp rate of 2 °C/second. The results after thermal cycling were read in the QX200 Droplet Reader, using the QuantaSoft 1.7 software (Biorad, USA) while further analysis was carried out using the QuantaSoft Analysis Pro 1.0 software (Biorad, USA).
2.4. Determination of recovery efficiency
The efficiency of the methods used in recovering SARS-CoV-2 from wastewater has been reported previously (Pillay et al., 2021). Briefly, 400 ml of raw wastewater was spiked with a 200 μl suspension of inactivated SARS-CoV-2 strain USA/WA1/2020 (Microbiologics, USA). Wastewater samples were then processed using the methodology described above. Unspiked wastewater samples were also analyzed to determine the background concentrations of SARS-CoV-2 and to help in accurate estimation of recovery efficiency. Determination of possible impact of inhibitors in the wastewater was also assessed by spiking 60 μl of inactivated SARS-CoV-2 into 120 ml of sterile MilliQ water and processed using the same methodology. All spiking was done in triplicate. The percentage of SARS-CoV-2 recovered was then determined using the equation (Equation 1);
| (1) |
The concentration of SARS-CoV-2 in spiked wastewater or Milli-Q water is represented by C SW , C UW represents the concentration of SARS-CoV-2 in the un-spiked wastewater or Milli-Q water and the concentration of SARS-CoV-2 spiked into the wastewater or Milli-Q water is represented by C SC. The efficiency of the methods in recovering SARS-CoV-2 from wastewater was determined to be at 62.90 (±12.8) %.
2.5. Modeling the relationship between wastewater characteristics and detection of SARS-CoV-2 in wastewater
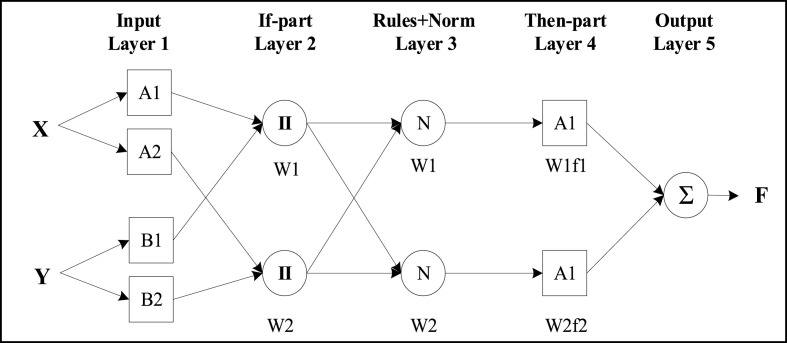
The Adaptive Neuro-Fuzzy Inference System (ANFIS) model was used to model the relationship between the wastewater characteristics and the detection of SARS-COV-2. ANFIS is a kind of artificial neural network (ANN), which is based on implementing the Takagi–Sugeno (TS) fuzzy approach, as shown in Fig. 1 . ANFIS implements fuzzy logic (FL) in the framework of ANN (Abunama et al., 2021). The development process of ANFIS modeling involves identifying the most relevant inputs that correlated to a targeted output (Shah et al., 2021). The optimum rules, types, and numbers of the associated membership functions (MFs), aiming at selecting the optimum ANFIS model structure with the lowest modeling errors was used (Abunama et al., 2018). As an example, two TS fuzzy sets of “if-then” rules in a typical ANFIS structure as following:
-
•
Rule 1: If x 1 is A 1 and x 2 is B 1; then f 1 = p 1 x 1 + q 1 x 2 + r 1
-
•
Rule 2: If x 1 is A 2 and x 2 is B 2; then f 2 = p 2 x 1 + q 2 x 2 + r 2
Where, p 1 ; q 1 ; p 2 and q 2 are ANFIS parameters, while Ai and Bj are the linguistic labels or grades (Ying, 1998). ANFIS architecture consists of five layers as shown in Fig. 1 (Abunama et al., 2018). A brief description of the role of these layers are descripted as follows:
-
•
Layer 1 (fuzzification layer): receives the input values and identifies the associated MFs.
-
•
Layer 2 (rule layer): generates the firing strengths for the rules.
-
•
Layer 3 (normalization layer): normalizes the computed strengths.
-
•
Layer 4: receives the normalized values and the consequence parameter sets.
-
•
Layer 5 (defuzzification layer): returns the values to the final output.
Fig. 1.
A typical ANFIS architecture, adopted from (Abunama et al., 2018).
In this study, ANFIS edit toolbox and coding in MATLAB 2019b environment was used to train and develop the proposed models. As mentioned earlier, sewage parameters were used as input variables, and related to the targeted SARS-CoV-2 RNA concentration determined with ddPCR. ANFIS learning process was repeated with many epochs aiming at minimizing the yielded errors between the observed actual values and the output of ANFIS model. Upon defining the best input combination, the development of the best ANFIS structure was conducted by applying various types and number of membership functions (MFs), and different rules and epochs' numbers. This was performed aiming at testing all possibilities of ANFIS parameters and compare their ability in modeling the SARS-CoV-2 RNA concentration. In the modeling, 70 % of the data was used in the training phase, while the rest was used to check or test the model's performances.
2.6. ANFIS models validation
The obtained results from the model were evaluated using numerous statistical checks. R squared (R2) was used to evaluate the relationship between observed values and predicted values (Ansari et al., 2018). The equation for calculating R2 is denoted as equation (2) as follows.
| (2) |
In equation (1), n, t and y are number of observed data, observed values and predicted values, respectively. Whereas and are the average of observed and predicted values. The range of R2 values lies between zero and 1, with 1 as highest accurate relationship possible. However, the values of R2 greater than 0.7 is considered highly reliable in engineering models (Abunama et al., 2019).
Furthermore, other validation criteria such as the mean absolute error (MAE) and the root mean square error (RMSE) were used to evaluate the accuracy of the developed models. Equations (3), (4) shows the estimation of both criteria. One of the main advantages of using RMSE is to assign higher weightage (as it contains square) to larger errors (Seyam et al., 2020). Equation (3) shows the mathematical expression for the calculation of RMSE.
| (3) |
| (4) |
Further assessment using the percent relative error (RE%) was used to evaluate the percentage errors in which the developed models underestimated or overestimated the SARS-CoV-2 concentration. Negative RE% values meant that the algorithm overestimated the actual SARS-CoV-2 concentration, while positive values indicated underestimation of the actual SARS-CoV-2 concentration. The RE% was determined as shown in Equation (5):
| (5) |
3. Results
3.1. Physico-chemical characteristics of the wastewater and SARS-CoV-2 concentration at the various wastewater treatment plants
Physico-chemical characteristics of the untreated wastewater at the different WWTPs varied considerably ranging from an average of 13.4 mg/L to 34.9 mg/L. For instance, the highest ammonia concentration of 34.9(±15.2) mg/L was recorded at the Isipingo WWTP (Table 1 ). However, the difference in ammonia concentration were not statistically significant between Central, Isipingo and Darvil WWTPs. While, Howick WWTPs had significantly lower concentration of ammonia (13.4(±7.3) mg/L) compared to the other three plants. A similar trend was observed for the pH of the wastewater, where Howick WWTP recorded the highest of 8.6 (±1.0) mg/L (Table 1). In addition, the flow rate data also showed that the volume of wastewater treated daily by the Howick WWTP was significantly lower than the other plants. For instance, the flow rate at Howick WWTP was 4.9(±1.1) m3/day, compared to 56.4(±16.4) m3/day and 63.7(±6.3) m3/day at Central and Darvil WWTPs, respectively.
Table 1.
Mean concentration (±standard deviation) of measured physico-chemical characteristics and SARS-CoV-2 in wastewater at the four wastewater treatment plants.
| WWTP | NH3(mg/L) | pH (pH@25 °C) | COD (mg/L) | FR (m3/day) | TS (mg/L) | PV4 | SARS-CoV-2 concentration (copies/mL) |
|---|---|---|---|---|---|---|---|
| Central | 32.5(±4.7) | 7.2(±0.3) | 554.5(±150.3) | 56.4(±16.4) | 953.9(±209.9) | 39(±8.8) | 1441.0(±1977.8) |
| Isipingo | 34.9(±15.2) | 7.4(±0.2) | 556.6(±125.1) | N/D* | 558.3(±181.9) | 42 (±17) | 1060.3(±1126.8) |
| Darvil | 30.3(±10.3) | 7.4(±0.2) | 702.7(±273.6) | 63.7(±6.3) | N/D | N/D | 791.8(±814.7) |
| Howick | 13.4(±7.3) | 8.6(±1.0) | 993.2(±596.3) | 4.9(±1.1) | N/D | N/D | 648.1(±514.6) |
*N/D = not measured. The physico-chemical data was provided by the operators of the wastewater treatment plants., therefore some parameters were considered important for their routine operations and were therefore not measured.
The concentration or copies of SARS-CoV-2 detected in the wastewater also varied between the WWTPs. The highest concentration of 1441.0(±1977.8) copies/mL was detected in wastewater from Central WWTP and the lowest of 648.1(±514.6) copies/mL from Howick WWTP. Despite the observed differences in the concentration of SARS-CoV-2 in the wastewater, there was no statistical difference based on the Kruskal-Wallis test, followed by the Dunn's Multiple comparison test.
3.2. Modeled association between physico-chemical characteristics and concentration of SARS-CoV-2 RNA detected
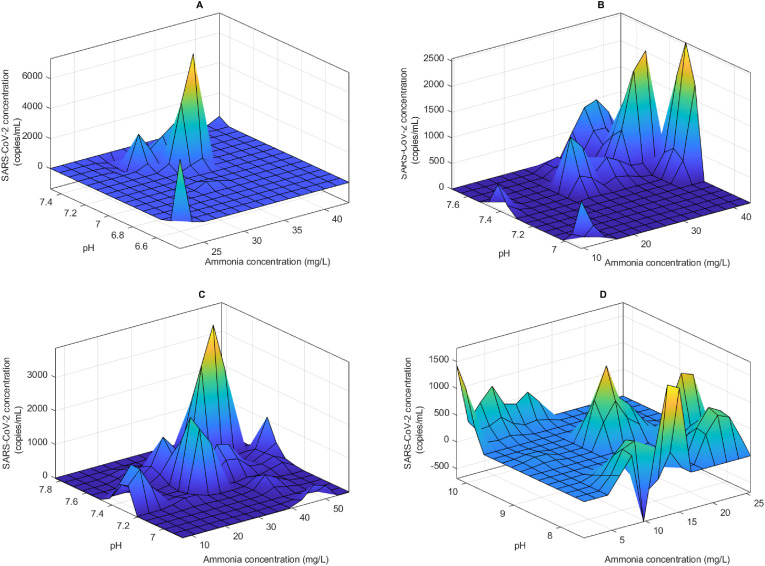
The ANFIS model outputs show that the impact of physico-chemical characteristics on the detection of SARS-CoV-2 varied between the WWTPs and the parameter measured. Ammonia and pH were observed to have a relationship with the concentration of the viral RNA measured. For instance, at the Central WWTP, ammonia concentration between 30 and 43 mg/L and pH between 7.1 and 7.4 was associated with high concentration of SARS-CoV-2 (Fig. 2 A). This shows the association of high viral RNA with increasing ammonia concentration (Table 1), because the range of ammonia concentration at this WWTP was 22–43 mg/L. Similar trend was observed with the other WWTPs except Howick WWTP. At this WWTP (Howick WWTP) the highest ammonia concentration was 25.6 mg/L, where no distinct association with SARS-CoV-2 concentration was observed (Fig. 2D).
Fig. 2.
Modeled impact of ammonia and pH on the detection of SARS-CoV-2 in wastewater from Central (A), Darvil (B), Isipingo(C) and Howick (D) wastewater treatment plants.
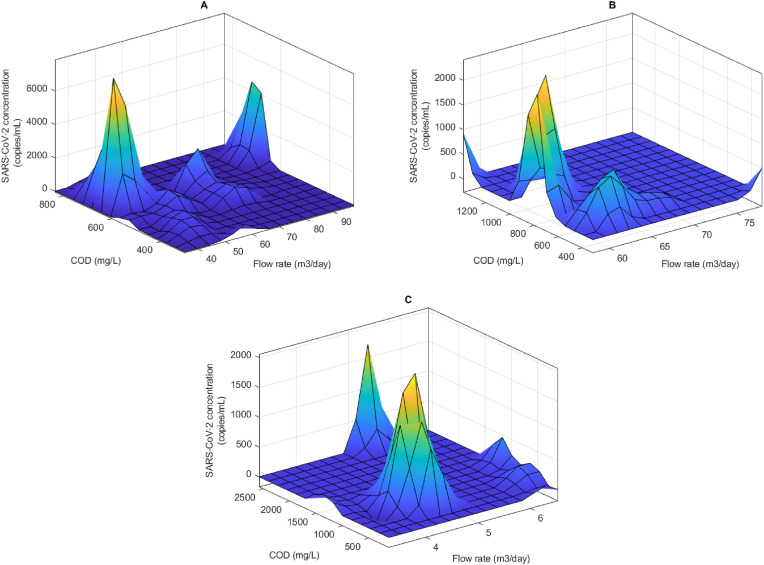
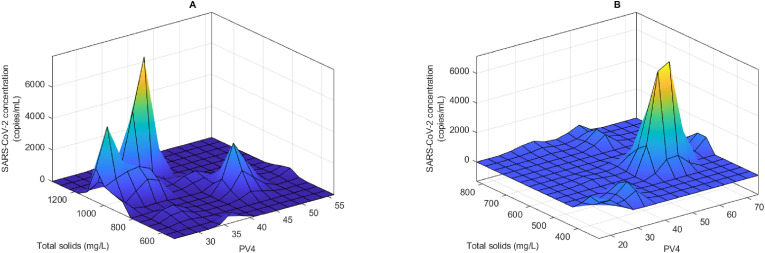
The detection of the viral RNA could also be attributed to the flow rate or COD of the wastewater. . In contrast to the impact of pH and ammonia a clear trend was not observed for the impact of the COD and flow rate on the detection of the viral RNA. In the three WWTPs with a complete set of these two parameters (Central, Darvil and Howick WWTPs), it was observed that the impact varies depending on the WWTP. For instance, in Central WWTP it was observed that the detection of high concentration of SARS-CoV-2 was within three flow rate ranges. These were 40–60 m3/day, 64–76 m3/day and 88–100 m3/day, with COD between 600 and 780 mg/L associated with high viral RNA concentration (Fig. 3 A). However, at Darvil WWTP there was a clear impact of flow rate on the concentration of SARS-CoV-2 RNA detected. The highest concentration of the viral RNA was associated with flow rates between 60 and 70 m3/day with a peak at 65 m3/day (Fig. 3B). However, in contrast to Central WWTP, the association of COD was not as clear, with high viral RNA detected at 400–700 m3/day and then again between 800 and 1200 m3/day (Fig. 3B). Similar unclear relationship was observed when considering the Howick WWTP as shown in Fig. 3C. Total solid concentration in the raw wastewater was also observed to have an influence on the concentration of SARS-CoV-2 measured (Fig. 4 ). At both the Central and Isipingo WWTPs increasing total solid concentrations were associated with high SARS-CoV-2 viral concentrations. For instance, at Central WWTP, the total solid concentrations ranged from 514 to 1354 mg/L, the ANFIS model determined that high concentration of SARS-CoV-2 viral RNA was associated with total solid range between 900 and 1200 mg/L (Fig. 4A). A similar trend was observed at the Isipingo WWTP, which had total solids ranging from 310 to 822 mg/L with high viral concentration associated with total solid concentrations from 500 to 700 mg/L (Fig. 4B).
Fig. 3.
Modeled impact of flow rate and COD on the detection of SARS-CoV-2 in wastewater from Central (A), Darvil (B) and Howick (C) wastewater treatment plants.
Fig. 4.
Modeled impact of permanganate value (PV4) and total solids on the detection of SARS-CoV-2 in wastewater from Central (A) and Isipingo (B) wastewater treatment plants.
3.3. Model validation
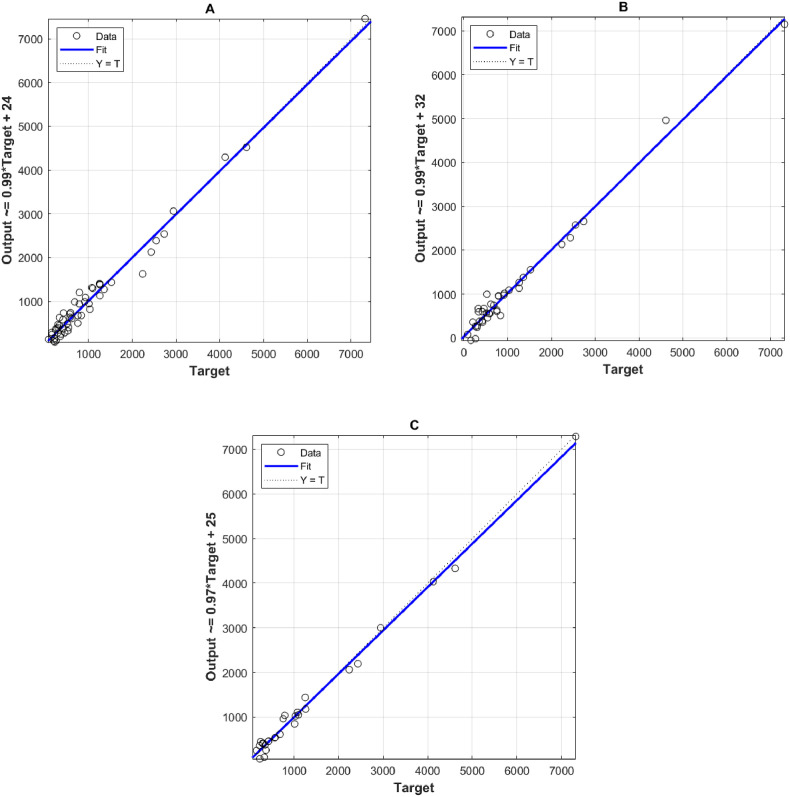
To evaluate the efficiency of the previously developed models, various evaluation criteria were tested as shown in Table 2 . Furthermore, the correlation diagrams for each inputs scenario are illustrated in Fig. 5 , ammonia and pH (A), flow rate and COD (B), and PV4 and total solids (C). Based on these results (Table 2), ANFIS modeling showed excellent accuracies in modeling SARS-CoV-2 concentrations for each input scenario. Coefficient of determination (R2) results were mostly greater than 0.9, with superiority to scenario (C) with inputs permanganate value (PV4) and Total solids. However, the modeling errors represented by RMSE and MAE, from lowest to highest, were using inputs scenario (C), (B) and (A), respectively. In general, ANFIS model was capable to simulate the variability in SARS-CoV-2 concentrations using wastewater physico-chemical parameters.
Table 2.
Results of models’ validation criteria.
| Models inputs | R | R2 | RMSE | MAE |
|---|---|---|---|---|
| (A) ammonia and pH | 0.989 | 0.979 | 184.52 | 154.37 |
| (B) Flow rate and COD | 0.992 | 0.984 | 168.05 | 126.95 |
| (C) PV4 and Total solids | 0.996 | 0.993 | 139.75 | 115.07 |
Fig. 5.
Correlation plots for the inputs: (A) ammonia and pH (B) Flow rate and COD, and (C) PV4 and Total solids.
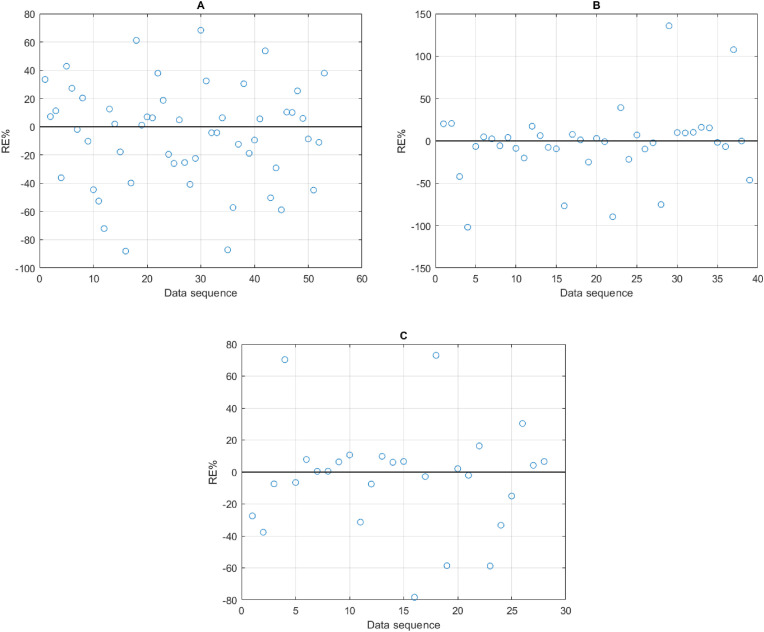
Fig. 6 illustrates the RE% results in modeling SARS-CoV-2 concentrations for each inputs scenario as follows: (A) ammonia and pH, (B) Flow rate and COD, and (C) PV4 and Total solids. The range of errors using Flow rate and COD were the widest for some points comparing to other inputs, however, most points located within ±50 % as presented in Fig. 6 (B). In contrast, using ammonia and pH (Fig. 6 (A)) had high RE% amplitudes comparedto using PV4 and Total solid as inputs (Fig. 6C).
Fig. 6.
Relative errors (%) plots for the inputs: (A) ammonia and pH (B) Flow rate and COD, and (C) PV4 and Total solids.
4. Discussion
The characteristics of wastewater is largely dependent on the source and type of the wastewater. Domestic wastewater generally contain approximately 99.9 % water (Sperling, 2007), with the remaining 0.1 % comprising of different substances, that are either biological, chemical or physical. These substances have the potential to influence the characteristics of the wastewater. However, in this study there was no observed significant impact of the source of wastewater on the characteristics measured. For instance, the concentration of ammonia, pH and COD in Isipingo WWTP did not differ significantly from Central and Darvil WWTPs. Although these WWTPs have a high component of industrial wastewater (Refer to Table 1). Howick and Isipingo WWTPs exclusively treat domestic wastewater, whilst Darvil and Central WWTPs have a mix of 80 % domestic wastewater. The variation in concentration of these characteristics could however be attributed to the size of the WWTPs. Howick WWTP is the smallest WWTP studied, with a flow rate of 4.9 (±1.1) m3/day compared to Central and Darvil WWTP with flow rates of 56.4(±16.4) m3/day and 63.7(±6.3) m3/day respectively. It also worth noting that sampling was done at time economic activities were either minimal or non-existent due to the COVID-19 lockdown regulations. This could potentially had contributed to low industrial activity, resulting in less wastewater input from the industries. The variations observed in the concentration of SARS-CoV-2 copies between and within the WWTPs could also be attributed to the size of the population served and shedding of the viral particle by infected individuals. For instance, the highest concentration of the viral RNA of 1441.0(±1977.8) copies/mL was detected in Central WWTP (Table 1), this also happens to be the biggest WWTP, with a designed capacity to treat 80 ML/d. During this study the clinically confirmed active cases of COVID-19 in South Africa was 600 000, with the province of KwaZulu-Natal, were this study was undertaken, reporting over 100 000 cases. The World Health Organization (WHO) estimates that between 2 and 27 % of COVID-19 patients have diarrhoea (WHO, 2020), resulting in shedding of the virus in feces. With estimated viral loads ranging from 6.3 × 106 to 1.26 × 108 gc/g of feces (Lescure et al., 2020). Therefore, more infected people commected to a WWTP could potentially lead to higher viral loads in the wastewater. This could be the reason for the high SARS-CoV-2 loads in wastewater from the Central WWTP, which serves a larger population.
The ANFIS model showed that the observed concentration of SARS-CoV-2 RNA in the wastewater described above could be potentially influenced by the physico-chemical characteristics such as pH, ammonia and to a lesser extent flow rate. The influence of pH on the concentration of the viral RNA was observed in all WWTPs except Howick WWTP. For instance, it was observed that the highest SARS-CoV-2 viral RNA were associated with pH between 7.1 and 7.4. This could be attributed to impact of pH on the viral particles. pH range between 5 and 7.4 is a stable range for coronaviruses (Casanova et al., 2009; Daniel and Talbot, 1987). Therefore, the higher SARS-CoV-2 RNA detected in the wastewater, with pH between 7.1 and 7.4, could be attributed to the stability of the viral particle at this range. Furthermore, a recent study by Varbanov et al. (2021) observed that a 10 min exposure of SARS-CoV-2 to pH of 9 or 10 could result in viral load decrease by more than 5 log10 units. These findings show that at alkaline pH SARS-CoV-2 viral particles could be inactivated, with low pH supporting increased stability. This could be the reason for the observed association of low pH with high SARS-CoV-2 concentrations in the wastewater.
Increasing ammonia concentration was also observed to be associated with increasing concentration of SARS-CoV-2 RNA. Ammonia in wastewater could be from several sources, most importantly, human urine. A large proportion of ammonia produced by humans end up in the WWTPs (Lee et al., 2021). Therefore, the association between increasing ammonia and increasing SARS-CoV-2 concentration could be attributed to increasing shedding of the viral particles by infected individuals in the connected populations. For instance, at Central WWTP, the mean ammonia was 32.5 (±4.7) mg/L, with a range of 22–43 mg/L. Therefore, the ANFIS model was able to show that increasing ammonia could be an indicator of increasing SARS-CoV-2 concentration. Similar trend was observed for all the other WWTPs except Howick WWTP, which could be attributed to the small nature of this WWTP as discussed above. As mentioned previously the size of the population connected to a WWTP could be a major contributory factor to the concentration of SARS-CoV-2 detected in the wastewater. The association of high ammonia with high SARS-CoV-2 concentration therefore shows that when there is an increase in the loading rate at the WWTP, it is associated with increasing viral particles. A larger population within the catchment of the WWTP indicates a potential high load of SAR-CoV-2 in the wastewater.
The association of ammonia and pH with high SARS-CoV-2 concentration was observed in all WWTPs, with the exception of Howick WWTP. This could be attributed to the small nature of the WWTP. For instance, the mean pH at Howick WWTP was 8.6(±1.0), the highest among all the WWTPs. This could be attributed to the impact of alkaline conditions on SARS-CoV-2 stability. For instance, as reported by Varbanov et al. (2021), pH of 9 could potentially result in a 5 log10 unit decrease in viral load. Furthermore, the standard deviation of 1.0, shows the high variability in pH at this WWTP. This observation could be due to small nature of the plant, which makes it easily influenced by small changes in wastewater characteristics related to water use.
Total solids concentration was also observed to influence SARS-CoV-2 concentration, however to a much lesser extent compared to pH and ammonia. It was observed that increase in total solids correspond to increasing SARS-CoV-2 RNA. This could be attributed to the attachment of the viral particles to solids in the wastewater, implying that more solids may result in higher attachment of the viral particle to the soilds resulting in higher recovery from the wastewater. For instance, Forés et al. (2021) reported that about 23 % of SARS-CoV-2 in wastewater were attached to the soild fraction. Therefore, an increasing total solid concentration could be associated with increasing recovery of SARS-CoV-2 leading to higher viral concentration.
The results from the ANFIS model therefore shows that the most fundamental parameters that potentially have an association with concentration of SARS-CoV-2 in wastewater are the shedding of the virus by infected individuals, demonstrated by the association with ammonia, the stability of the virus, demonstrated by the association with pH between 7.1 and 7.3 and the total solids in the wastewater. It is worth noting that other properties of the wastewater could potentially affect the concentration of SARS-CoV-2, however in this study the main factors were pH, ammonia and total solids.
5. Conclusion
The detection of SARS-CoV-2 in untreated wastewater could be affected by the physico-chemical characteristics of the wastewater. This has the potential to significantly impact the utility of wastewater-based epidemiology. This study presented the determination of the impact of these wastewater characteristics on SARS-CoV-2 viral concentration in wastewater. The Adaptive Neuro-Fuzzy Inference System (ANFIS) model showed that ammonia, pH and to a lesser extent total solids are the major parameters that could affect the concentration of SARS-CoV-2 RNA in wastewater. The highest concentration of SARS-CoV-2 was determined to be associated with low pH between 7.1 and 7.4 and associated with increasing ammonia concentration. In addition to these parameters with an observed association in almost all the WWTPs, we also observed that there are unique characteristics within each WWTP that has an impact on the concentration of the viral RNA detected. This study therefore shows that SARS-CoV-2 concentration in wastewater could be due to the shedding rate in the connected population, as reported in literature and corroborated in this study via the observed association with increasing ammonia concentration. Additionally, we observed that the stability of the viral particle in wastewater could be impacted by pH and solids in the wastewater. This calls for the inclusion of wastewater physico-chemical characteristic assessment in each WWTP, especially during the method optimization stages, to ensure accurate estimation of viral concentration and contribute to the utility of wastewater-based epidemiology for decision-making. Additionally, future studies could also explore more characteristics and the cumulative impact of these different wastewater characteristics on SARS-CoV-2 detection and quantification.
6. Study limitations
The findings presented in this study are useful for the further development of wastewater-based epidemiology. However, some limitations of the study have been identified and elaborated below;
-
1.
Impact of heat treatment on viral load: Wastewater samples were heat treated prior to concentration and RNA extraction to provide protection to laboratory personnel. However, there are concerns that this heat treatment could reduce the viral load. Therefore, a limitation of the current approach is the possible reduction in SARS-CoV-2 concentration measured due to the impact of the heat treatment process.
-
2.
Grab sampling of wastewater samples: Samples were collected using the grab sampling approach, this could result in different viral load compared to composite sampling approach. The data used in this study was not from a one-time sampling event, therefore by using samples taken over several months from four wastewater treatment plants, the impact of grab sampling is minimized. However, it is worth noting this could potentially impact the results obtained.
-
3.
Limited physico-chemical parameters: The selection of physico-chemical characteristics for this study was limited by the available data. The study relied on data collected by the various WWTPs as part of their routine analysis, therefore there is the potential that other parameters may have an impact on SARS-CoV-2 concentration. This is therefore a limitation that needs to be highlighted and considered for future studies.
Funding sources
We acknowledge the financial support from the South African Research Chair Initiative (SARChI) of the Department of Science and Technology, and the National Research Foundation of South Africa (NRF). This study was also funded partially by Umgeni Water and TA has additional funding from NRF (Grant Numbers: 84166).
Authors contributions
Isaac Dennis Amoah: conceptualization, methodology and writing original draft; Taher Abunama: conceptualization, analysis and modeling, and writing original draft; Oluyemi Olatunji Awolusi: data collection, writing-review and edit; Leanne Pillay: data collection, writing-review and edit; Kriveshin Pillay: data collection, writing-review and edit; Sheena Kumari: writing-review and edit; Faizal Bux: supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We are grateful to the eThekwini Water and Sanitation (EWS) department, most especially Dr Xolani Nocanda, Umgeni water for the support and supply of the relevant wastewater characteristics data. We also acknowledge the assistance of the superintendents, operators and workers of the various wastewater treatment plants during the sample collection.
Appendix A.
RAW DATA.
| CENTRAL WWTP |
|||||||
|---|---|---|---|---|---|---|---|
| Date | Flow rate | COD | Ammonia | pH | Total suspended solids | Permanganate value (PV4) | SARS-CoV-2 copies/mL |
| 7-Jul | 72.46 | 518 | 33 | 7.32 | 1034 | 34 | 753 |
| 14-Jul | 76.03 | 528 | 38 | 7.405 | 986.8 | 40 | 300 |
| 21-Jul | 34.92 | 538 | 43 | 7.49 | 939.6 | 45 | 420 |
| 27-Jul | 79.27 | 548 | 36 | 7.34 | 892.4 | 51 | 330 |
| 4-Aug | 49.23 | 832 | 32 | 6.83 | 845.2 | 56 | 350 |
| 11-Aug | 44.09 | 742 | 30 | 7.27 | 798 | 46 | 2236 |
| 18-Aug | 41.934 | 651 | 33 | 7.12 | 1132 | 36 | 7320 |
| 25-Aug | 54.02 | 561 | 29 | 7.31 | 926 | 25 | 1029 |
| 1-Sep | 56.28 | 300 | 35 | 7.16 | 720 | 32 | 577 |
| 7-Sep | 44.42 | 382 | 29 | 7.23 | 514 | 37 | 682 |
| 15-Sep | 48.05 | 464 | 31 | 7.3 | 988 | 42 | 789 |
| 22-Sep | 42.04 | 546 | 34 | 7.19 | 1354 | 37 | 427 |
| 29-Sep | 69.79 | 628 | 33 | 7.28 | 968 | 32 | 2431 |
| 6-Oct | 97.55 | 710 | 22 | 6.4 | 1050 | 27 | 4610 |
| ISIPINGO WWTP |
||||||
|---|---|---|---|---|---|---|
| Date | COD | Ammonia | pH | Total suspended solids | Permanganate value (PV4) | SARS-CoV-2 copies/mL |
| 7-Jul | 710 | 47 | 6.85 | 824 | 52 | 568 |
| 14-Jul | 534 | 48 | 7.84 | 572 | 47 | 250 |
| 21-Jul | 525.5 | 57 | 7.47 | 558 | 74 | 1075 |
| 27-Jul | 517 | 7.2 | 7.39 | 544 | 33 | 230 |
| 4-Aug | 731 | 23 | 7.62 | 530 | 58 | 225 |
| 11-Aug | 702 | 46 | 7.64 | 516 | 56 | 4120 |
| 18-Aug | 673 | 30 | 7.34 | 530 | 55 | 2940 |
| 25-Aug | 644 | 41 | 7.4 | 742 | 53 | 1009 |
| 1-Sep | 602 | 23 | 7.22 | 782 | 37 | 155 |
| 7-Sep | 532 | 53 | 7.34 | 822 | 35 | 299 |
| 15-Sep | 477 | 31 | 7.42 | 382 | 32 | 1250 |
| 22-Sep | 422 | 44 | 7.15 | 346 | 22 | 368 |
| 29-Sep | 377 | 29 | 7.53 | 310 | 19 | 1095 |
| 6-Oct | 332 | 10 | 7.33 | 342 | 16 | 1260 |
| DARVIL WWTP |
|||||
|---|---|---|---|---|---|
| Date | Flow rate | COD | Ammonia | pH | SARS-CoV-2 copies/mL |
| 7-Jul | 58 | 1388 | 39.3 | 7.7 | 927 |
| 14-Jul | 68 | 505 | 27.4 | 7.4 | 335 |
| 21-Jul | 61 | 549 | 32.3 | 7.4 | 415 |
| 27-Jul | 61 | 559 | 28.3 | 7.6 | 265 |
| 4-Aug | 62 | 414 | 26.8 | 7.4 | 615 |
| 11-Aug | 58 | 816 | 39.4 | 7.5 | 2730 |
| 18-Aug | 62 | 975 | 43.4 | 7.3 | 2547 |
| 25-Aug | 60 | 860 | 37.4 | 7.6 | 525 |
| 1-Sep | 63 | 832 | 41.3 | 7 | 0 |
| 7-Sep | 61 | 876 | 35.8 | 7.7 | 834 |
| 15-Sep | 64 | 543 | 10.1 | 6.9 | 788 |
| 22-Sep | 59 | 696 | 28.3 | 7.5 | 589 |
| 29-Sep | 77 | 517 | 24.9 | 7.4 | 0 |
| 6-Oct | 78 | 308 | 9.46 | 7.4 | 515 |
| HOWICK WWTP |
|||||
|---|---|---|---|---|---|
| Date | Flow rate | COD | Ammonia | pH | SARS-CoV-2 copies/mL |
| 7-Jul | 5.41 | 2580 | 12.7 | 7.5 | 1520 |
| 14-Jul | 3.29 | 1346.5 | 11.02 | 7.55 | 205 |
| 21-Jul | 6.26 | 113 | 9.34 | 7.6 | 160 |
| 27-Jul | 4.13 | 1119 | 8.77 | 8.01 | 305 |
| 4-Aug | 6.28 | 889 | 4.78 | 9.055 | 85 |
| 11-Aug | 4.5 | 659 | 0.79 | 10.1 | 1259 |
| 18-Aug | 4.49 | 742 | 5.99 | 10.15 | 753 |
| 25-Aug | 4.02 | 825 | 11.2 | 10.2 | 540 |
| 1-Sep | 4.75 | 924 | 15.05 | 9.05 | 1356 |
| 7-Sep | 3.7 | 1023 | 18.9 | 7.9 | 0 |
| 15-Sep | 4.44 | 825.5 | 22.25 | 7.85 | 911 |
| 22-Sep | 6.37 | 628 | 25.6 | 7.8 | 263 |
| 29-Sep | 4.51 | 897 | 22.4 | 8.45 | 1260 |
| 6-Oct | 6.52 | 1166 | 19.2 | 9.1 | 456 |
References
- Abunama T., Ansari M., Awolusi O.O., Gani K.M., Kumari S., Bux F. Fuzzy inference optimization algorithms for enhancing the modelling accuracy of wastewater quality parameters. J. Environ. Manag. 2021;293:112862. doi: 10.1016/j.jenvman.2021.112862. [DOI] [PubMed] [Google Scholar]
- Abunama T., Othman F., Ansari M., El-Shafie A. Leachate generation rate modeling using artificial intelligence algorithms aided by input optimization method for an MSW landfill. Environ. Sci. Pollut. Res. 2019;26(4):3368–3381. doi: 10.1007/s11356-018-3749-5. [DOI] [PubMed] [Google Scholar]
- Abunama T., Othman F., Younes M.K. Predicting sanitary landfill leachate generation in humid regions using ANFIS modeling. Environ. Monit. Assess. 2018;190(10):597. doi: 10.1007/s10661-018-6966-y. [DOI] [PubMed] [Google Scholar]
- Aguiar-Oliveira M.D.L., Campos A., R Matos A., Rigotto C., Sotero-Martins A., Teixeira P.F., Siqueira M.M. Wastewater-based epidemiology (wbe) and viral detection in polluted surface water: a valuable tool for COVID-19 surveillance—a brief review. Int. J. Environ. Res. Publ. Health. 2020;17(24):9251. doi: 10.3390/ijerph17249251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Angel N., Bibby K., Bivins A., Dierens L., Edson J., Ehret J., Gyawali P., Hamilton K.A., Hosegood I., Hugenholtz P., Jiang G., Kitajima M., Sichani H.T., Shi J., Shimko K.M., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travellers. J. Trav. Med. 2020;27:1–11. doi: 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoah I.D., Mthethwa N.P., Pillay L., Deepnarain N., Pillay K., Awolusi O.O., Kumari S., Bux F. RT-LAMP: a cheaper, simpler and faster alternative for the detection of SARS-CoV-2 in wastewater. Food Environ Virol. 2021;1–10 doi: 10.1007/s12560-021-09489-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari M., Othman F., Abunama T., El-Shafie A. Analysing the accuracy of machine learning techniques to develop an integrated influent time series model: case study of a sewage treatment plant, Malaysia. Environ. Sci. Pollut. Res. 2018;25(12):12139–12149. doi: 10.1007/s11356-018-1438-z. [DOI] [PubMed] [Google Scholar]
- Auffret M.D., Brassard J., Jones T.H., Gagnon N., Gagné M.J., Muehlhauser V., Masse L., Topp E., Talbot G. Impact of seasonal temperature transition, alkalinity and other abiotic factors on the persistence of viruses in swine and dairy manures. Sci. Total Environ. 2019;659:640–648. doi: 10.1016/j.scitotenv.2018.12.306. [DOI] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A., Castiglioni S., Cetecioglu Gurol Z., Chakraborty S., Costa F., Curcio S., De Los Reyes F.L., Delgado Vela J., Farkas K., Fernandez-Casi X., Gerba C., Gerrity D., Girones R., Gonzalez R., Haramoto E., Harris A., Holden P.A., Islam M.T., Jones D.L., Kasprzyk-Hordern B., Kitajima M., Kotlarz N., Kumar M., Kuroda K., La Rosa G., Malpei F., Mautus M., McLellan S.L., Medema G., Meschke J.S., Mueller J., Newton R.J., Nilsson D., Noble R.T., Van Nuijs A., Peccia J., Perkins T.A., Pickering A.J., Rose J., Sanchez G., Smith A., Stadler L., Stauber C., Thomas K., Van Der Voorn T., Wigginton K., Zhu K., Bibby K. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020:7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43(7):1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collivignarelli M.C., Collivignarelli C., Carnevale Miino M., Abbà A., Pedrazzani R., Bertanza G. SARS-CoV-2 in sewer systems and connected facilities. Process Saf. Environ. Protect. 2020;143:196–203. doi: 10.1016/j.psep.2020.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Mercier E., Montpetit D., Jia J.J., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Langlois M.A., Servos M.R., MacKenzie M., Figeys D., MacKenzie A.E., Graber T.E., Delatolla R. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188:116560. doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel C., Talbot P.J. Physico-chemical properties of murine hepatitis virus, strain A 59. Brief report. Arch. Virol. 1987;96:241–248. doi: 10.1007/BF01320963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Graber T.E., Mercier E., Montpetit D., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Servos M.R. Catching a resurgence: increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci. Total Environ. 2021;770:145319. doi: 10.1016/j.scitotenv.2021.145319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C.G. Wastewater surveillance for population-wide COVID-19: the present and future. Sci. Total Environ. 2020;736:139631. doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forés E., Bofill-Mas S., Itarte M., Martínez-Puchol S., Hundesa A., Calvo M., Borrego C.M., Corominas L.L., Girones R., Rusiñol M. Evaluation of two rapid ultrafiltration-based methods for SARS-CoV-2 concentration from wastewater. Sci. Total Environ. 2021;768:144786. doi: 10.1016/j.scitotenv.2020.144786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186:116296. doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., Mendoza Grijalva L.M., Roldan-Hernandez L., Langenfeld K., Wigginton K.R., Boehm A.B. SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ. Sci. Technol. 2020;55(1):488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Hurst C.J., Gerba C.P. Fate of viruses during wastewater sludge treatment processes. Crit. Rev. Environ. Sci. Technol. 1989;18(4):317–343. doi: 10.1080/10643388909388352. [DOI] [Google Scholar]
- Joiner C.L., Oidtmann B.C., Rimmer G.S., McPherson N.J., Dixon P.F., Paley R.K. Survival of viral haemorrhagic septicaemia virus and infectious haematopoietic necrosis virus in the environment and dried on stainless steel. Transbound Emerg Dis. 2020:1–13. doi: 10.1111/tbed.13888. 00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. The first proof of the capability of wastewater surveillance for COVID-19 in India through the detection of the genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746:141326. doi: 10.1101/2020.06.16.20133215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., An S., Choi Y. Ammonia harvesting via membrane gas extraction at moderately alkaline pH: a step toward net-profitable nitrogen recovery from domestic wastewater. Chem. Eng. Technol. 2021;405:126662. [Google Scholar]
- Lescure F.X., Bouadma L., Nguyen D., Parisey M., Wicky P.H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q., Enouf V. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020;20(6):697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wang Y., Ji M., Pei F., Zhao Q., Zhou Y., Hong Y., Han S., Wang J., Wang Q., Li Q. Transmission routes analysis of SARS-CoV-2: a systematic review and case report. Front. Cell Dev. Biol. 2020;8:618. doi: 10.3389/fcell.2020.00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;1253(20):30087. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Been F., Heijnen L., Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr. Opin. Environ. Sci. Health. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Mousavizadeh L., Ghasemi S. Genotype and phenotype of COVID-19: their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2021;54(2):159–163. doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir K., Ashraf S., Munir I., Khalid H., Muneer M.A., Mukhtar N., Amin S., Ashraf S., Imran M.A., Chaudhry U., Zaheer M.U. Zoonotic and reverse zoonotic events of SARS-CoV-2 and their impact on global health. Emerg. Microb. Infect. 2020;9(1):2222–2235. doi: 10.1080/22221751.2020.1827984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38(10):1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay L., Amoah I.D., Deepnarain N., Pillay K., Awolusi O.O., Kumari S., Bux F. Monitoring changes in COVID-19 infection using wastewater-based epidemiology: a South African perspective. Sci. Total Environ. 2021;786:147273. doi: 10.1016/j.scitotenv.2021.147273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaan A.A., Al-Ahmed S.H., Al-Malkey M.K., Alsubki R.A., Ezzikouri S., Al-Hababi F.H., Sah R., Al Mutair A., Alhumaid S., Al-Tawfiq J.A., Al-Omari A. Airborne transmission of SARS-CoV-2 is the dominant route of transmission: droplets and aerosols. Infez Med. 2021;29:10–19. [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyam M., S Alagha J., Abunama T., Mogheir Y., Affam A.C., Heydari M., Ramlawi K. Investigation of the influence of excess pumping on groundwater salinity in the Gaza Coastal Aquifer (Palestine) using three predicted future scenarios. Water. 2020;12(8):2218. [Google Scholar]
- Shah M.I., Abunama T., Javed M.F., Bux F., Aldrees A., Tariq M.A.U.R., Mosavi A. Modeling surface water quality using the adaptive neuro-fuzzy inference system Aided by input optimization. Sustainability. 2021;13(8):4576. [Google Scholar]
- Sommerstein R., Fux C.A., Vuichard-Gysin D., Abbas M., Marschall J., Balmelli C., Troillet N., Harbarth S., Schlegel M., Widmer A. Risk of SARS-CoV-2 transmission by aerosols, the rational use of masks, and protection of healthcare workers from COVID-19. Antimicrob. Resist. Infect. Contr. 2020;9(1):1–8. doi: 10.1186/s13756-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling M.V. vol. 6. Water Intelligence Online; 2007. (Wastewater Characteristics, Treatment and Disposal). [Google Scholar]
- Spurbeck R.R., Minard-Smith A., Catlin L. Feasibility of neighborhood and building scale wastewater-based genomic epidemiology for pathogen surveillance. Sci. Total Environ. 2021;789:147829. doi: 10.1016/j.scitotenv.2021.147829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhu A., Li H., Zheng K., Zhuang Z., Chen Z., Shi Y., Zhang Z., Chen S., Liu X., Dai J., Li X., Huang S., Huang X., Luo L., Wen L., Zhuo J., Li Yuming, Wang Y., Zhang L., Zhang Y., Li F., Feng L., Chen X., Zhong N., Yang Z., Huang J., Zhao J., Li Yi min. Isolation of infectious SARS-CoV-2 from urine of a COVID-19 patient. Emerg. Microb. Infect. 2020;9:991–993. doi: 10.1080/22221751.2020.1760144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varbanov M., Bertrand I., Philippot S., Retourney C., Gardette M., Hartard C., Jeulin H., Duval R.E., Loret J.F., Schvoerer E., Gantzer C. Somatic coliphages are conservative indicators of SARS-CoV-2 inactivation during heat and alkaline pH treatments. Sci. Total Environ. 2021 doi: 10.1016/j.scitotenv.2021.149112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.Y., Zhao R., Gao L.J., Gao X.F., Wang D.P., Cao J.M. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO-World Health Organization. Water, Sanitation, Hygiene and Waste Management for COVID-19: Technical Brief, 03 March 2020 (No. WHO/2019-NcOV/IPC_WASH/2020.1). World Health Organization.
- Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., Zhao Jingxian, Huang J., Zhao Jincun. Infectious SARS-CoV-2 in feces of patient with severe COVID-19 - volume 26, number 8—august 2020 - emerging infectious diseases journal - CDC. Emerg. Infect. Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafar T. Novel COVID-19 outbreak: the pandemic of the decade. Bull. World Health Organ. 2020;98(3):150. [Google Scholar]