Dear editor,
We read with interest the systematic review published by Walsh et al. in the Journal of Infection,1 focusing on the dynamics of SARS-CoV-2 RNA at the upper respiratory tract (URT). In this context, a novel SARS-CoV-2 variant lineage (B.1.1.7), first detected in the UK at the end of 2020 has transmission advantage over other lineages.2 Increased transmissibility of the B.1.1.7 variant has been linked to enhanced ACE2 affinity3 allegedly resulting in higher viral loads in URT, an observation that has been reported in some,3, 4, 5, 6 but not all7 large series published to date. In addition, longer duration of SARS-CoV-2 RNA shedding in URT has been reported in individuals infected by the B.1.1.7 variant as compared to controls;8 if proven, this may have important implications regarding isolation policies. The current retrospective, observational study was undertaken to gain further insight into the above issues. It included a convenience sample of 990 individuals (799 aged >18 years; 507 females) testing positive for SARS-CoV-2 RNA in nasopharyngeal specimens (NP) by the TaqPath COVID-19 Combo Kit (Thermo Fisher Scientific, MS, USA) between June 2020 and April 2021. The study was approved by INCLIVA Research Ethics Committee. A total of 338 subjects (median age, 38 years; range, 1–93 years) were infected by the SARS-CoV-2 lineage B.1.1.7 (179 had COVID-19-Supplementary Table 1). The study included a control group of 652 individuals, 339 presenting with COVID-19 (median age, 40.6 years; range, 0–95 years) infected by other variants, of which 390 were characterized by whole-genome sequencing (Supplementary Table 2). Individuals belonging to the control group were matched with the B.1.1.7 group for sex and age. Among patients with COVID-19, the time from symptoms onset to RT-PCR testing was 5 days (range, 1–10 days) in the B.1.1.7 group and 4 days (range, 1–10 days) in the control group, with no differences between children and adults. As for asymptomatic individuals (140 infected by the B.1.1.7 variant), RT-PCR testing was performed within the first 10 days since diagnosis (for household) or contact with (for non-household) the index case in individuals from both groups. A total of 1152 NP specimens (median 1 specimen/patient; range, 1–3) were included in the analyses described below.
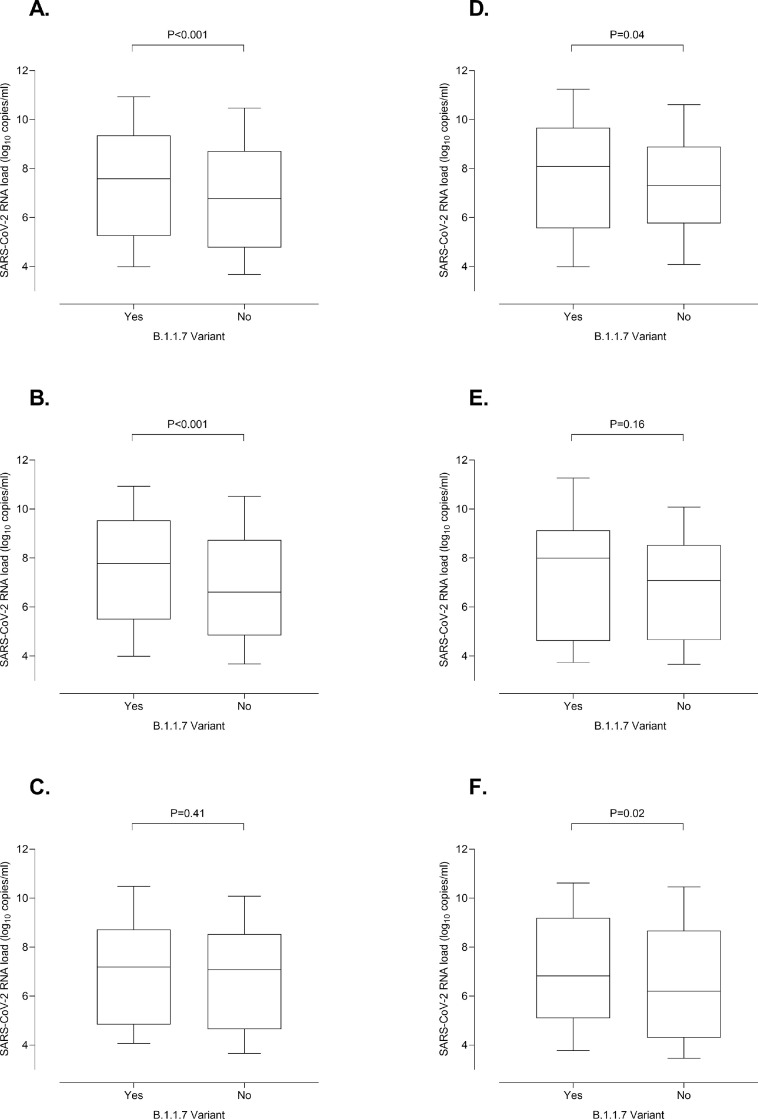
We found that SARS-CoV-2 B1.1.7-infected individuals displayed initial NP viral loads around 1 log10 higher than controls (median, 7.6 log10 copies/ml; range, 3.3–12.1 vs. 6.8 log10 copies/ml; range, 2.4–13.1; P < 0.001), a figure that concurs remarkably with that observed by Jones et al.,4 and overall support previous observations3, 4, 5, 6 reported in studies involving large cohorts, that were nevertheless poorly defined regarding subject age, individual clinical condition at diagnosis, timing of URT specimen collection or all of the above. A subanalysis including only SARS-CoV-2 B1.1.7-infected individuals as confirmed by whole-genome sequencing (n = 108) yielded comparable results (P < 0.001) (not shown). A novel observation was that the difference in viral load between B.1.1.7 and non-B.1.1.7 infected individuals remained significant (P < 0.001) for adults (Fig. 1 B), but not for children (P = 0.41) (Fig. 1C). When symptomatic patients infected by either the B.1.1.7 or other variants were analyzed separately, initial viral loads were also higher (P = 0.04) for adults (Fig. 1D), but again not (P = 0.16) for children (Fig. 1E), although a trend towards higher viral loads was noticeable in B.1.1.7 infected subjects. We hypothesize that a more robust early innate immune response to SARS-CoV-2 in children as compared to adults may minimize the apparent replicative advantage of the B.1.1.7 variant over other less transmissible ones.9 Likewise, Asymptomatic adults infected by the B.1.1.7 variant had higher initial SARS-CoV-2 RNA loads (P = 0.02) than controls (Fig. 1F).
Fig. 1.
Whisker plots depicting initial SARS-CoV-2 RNA loads in nasopharyngeal specimens from individuals infected by either the B.1.1.7 variant or other variants. Participants were sampled at either primary care centers affiliated to the Clínico-Malvarrosa Health Department, which attends 350,000 inhabitants in Northwest Valencia (Spain), or at its tertiary reference hospital (Hospital Clínico Universitario de Valencia, Spain-HCU-). NP were collected by trained nurses, placed in 3 ml of Universal Transport Medium (UTM, Becton Dickinson, Sparks, MD, USA) and delivered to the Microbiology Service of HCU for testing. Specimens were analyzed by RT-PCR within 24 h of receipt. We used the TaqPath COVID-19 Combo Kit (Thermo Fisher Scientific, MS, USA), which targets SARS-CoV-2 ORF1ab, N and S genes, following RNA extraction carried out by using the Applied Biosystems™ MagMAX™ Viral/Pathogen II Nucleic Acid Isolation Kits coupled with Thermo Scientific™ KingFisher Flex automated instrument. The AMPLIRUN® TOTAL SARS-CoV-2 RNA Control (Vircell SA, Granada, Spain) was used as the reference material for estimating SARS-CoV-2 RNA load (in copies/mL, taking RT-PCR CT for the N gene). The B.1.1.7 lineage was confirmed by whole-genome sequencing in 108 cases, whereas in the remaining 230 it was inferred by S-gene target failure (SGTF) in the RT-PCR as within the timeframe of specimen collection (mid-February-April 2021) more than 95% of SGTFs detected in the Clínico-Malvarrosa Health Department belonged to that lineage (not shown). Control individuals were sampled before the SARS-CoV-2 B1.1.7 variant was first detected in our Health Department in early January 2021. Moreover, the SARS-CoV-2 RNA S-gene was detected in NP from all these participants. The data are depicted for all participants (A); adults (B), children (≤18 years old) (C); adults with COVID-19 (D); children with COVID-19 (E) and asymptomatic adults (F). The absence of non-B.1.1.7-infected asymptomatic children in the cohort precluded meaningful comparison of viral loads between these individuals and those infected by other variants. Differences between medians were compared using the Mann-Whitney U test. The Chi-squared test was used for frequency comparisons. Two-sided P-values < 0.05 were considered statistically significant. Statistical analyses were performed using the SPSS v.25 program. P values for comparisons across groups are shown.
It is of interest that SARS-CoV-2 RNA loads measured in the comparison groups were unlikely to be biased by differences in cellularity across NP specimens, as determined by amplification of the β‐glucuronidase housekeeping gene by RT-PCR10 in 50 randomly selected participants (24 infected by the B.1.1.7 variant and 26 from controls). In fact, median CT was similar (P = 0.43) between NP samples in both groups (not shown).
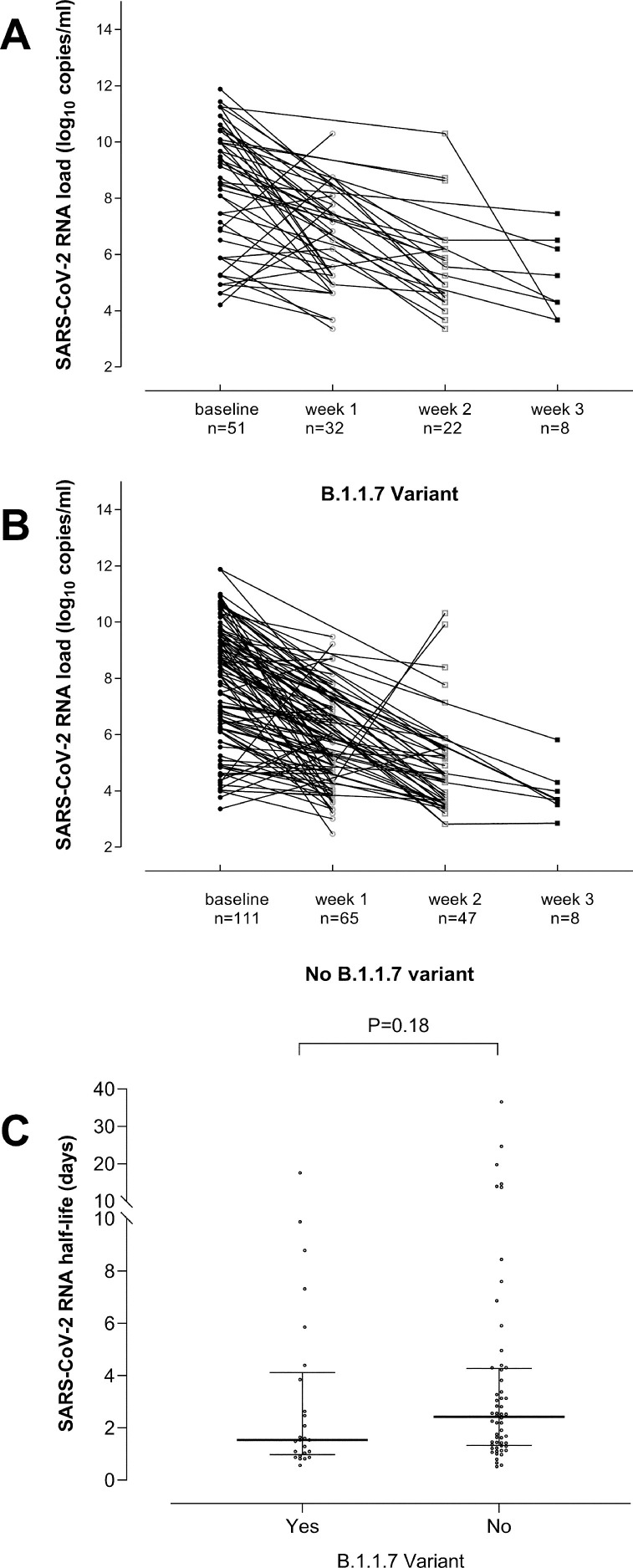
A total of 162 participants (51 infected by the B.1.1.7 variant and 111 controls), had 2 or more follow-up NP specimens collected within 3 weeks after diagnosis of SARS-CoV-2 infection. As shown in Fig. 2 , decreasing viral loads were observed in most participants, regardless of whether they were infected with the B.1.1.7 variant (2A) or not (2B). SARS-CoV-2 RNA half-life in URT could be calculated for 25 individuals infected by the B.1.1.7 variant and 56 controls. Most individuals in both comparison groups were adults and were matched (P > 0.5) by sex, the presence or absence of symptoms, hospitalization (Supplementary Table 3) and age (median, 38 years in both groups). As shown in Fig. 2C, SARS-CoV-2 RNA load half-life was similar (P = 0.18) among groups (B.1.1.7 infected: median 1.54 days; range, 0.5–17 days; controls: 2.42 days; range, 0.27–36 days). In contrast to our findings, Calistri et al.8 reported longer persistence of SARS-CoV-2 RNA in NP specimens in individuals infected with the B.1.1.7 variant (median 16 days) compared to controls (median 14 days). Nevertheless, the major drawbacks of that study were the lack of information on clinical status of participants and the absence of sequential specimens collected on an individual basis.
Fig. 2.
Kinetics of SARS-CoV-2 RNA load in nasopharyngeal specimens from individuals either infected by the B.1.1.7 variant (A) or by other variants (B) through week 3 after diagnosis of SARS-CoV-2 infection. Trajectories of SARS-CoV-2 RNA load in URT were classified as rising or decreasing, as the variation in the viral load between consecutive specimens was > or <0.5 log10 copies/ml (upper limit of intra-assay log10 variation), respectively. SARS-CoV-2 RNA half-life in the upper respiratory tract from individuals either infected by the B.1.1.7 variant or by other variants (C). The kinetics of SARS-CoV-2 RNA clearance followed a logarithmic decay curve in most individuals, expressed by the equation yt=y0e−kt, where y0 is the initial SARS-CoV-2 RNA load, t is time of follow-up specimen sampling since diagnosis (initial RT-PCR result) and k is the decay constant. SARS-CoV-2 RNA load half-life was then calculated using the equation ln2/k. SARS-CoV-2 RNA half-life in URT was calculated only for individuals (25 infected by the B.1.1.7 variant and 56 controls) that met two criteria: (i) displaying a descending trajectory throughout the study period (three weeks); (ii) the follow-up specimen used for calculations was collected within 4–10 days after the initial one. P value for comparisons across groups is shown.
The current study has several limitations, most notably its retrospective design, the lack of follow-up specimens from many participants, the low numbers of children with B.1.1.7 symptomatic infection and with non-B.1.1.7 asymptomatic infection and the lack of precise information on the timing of specimen collection in asymptomatic subjects following contact with the index case. Furthermore, the number of individuals from whom viral load half-life could be calculated was limited.
In summary, our data support that initial SARS-CoV-2 load is higher in B.1.1.7-infected adult individuals than in those infected by other variants, regardless of presence or absence of symptoms, but not in symptomatic children. Nevertheless, the data did not suggest an extended duration of SARS-CoV-2 RNA shedding in the URT in individuals infected with the B.1.1.7 variant.
CRediT authorship contribution statement
Rosa Costa: Methodology, Validation, Conceptualization, Writing – original draft. Felipe Bueno: Methodology, Validation, Writing – original draft. Estela Giménez: Methodology, Validation, Writing – original draft. Alma Bracho: Methodology, Validation, Writing – original draft. Eliseo Albert: Methodology, Validation, Writing – original draft. Diego Carretero: . Paula de Michelena: Methodology, Validation, Writing – original draft. Cecilia Martínez-Costa: Methodology, Validation, Writing – original draft. Fernando González-Candelas: Methodology, Validation, Conceptualization, Writing – original draft. David Navarro: Conceptualization, Supervision, Writing – original draft.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgments
Acknowledgments
We are grateful to all personnel working in the Clínico-Malvarrosa Health Department and at Clinic University Hospital, in particular those at the Microbiology laboratory, for their commitment in the fight against COVID-19. Eliseo Albert holds a Juan Rodés Contract (JR20/00011) from the Health Institute Carlos III. Estela Giménez holds a Juan Rodés Contract (JR18/00053) from the Health Institute Carlos III.
Financial support
This work received no public or private funds.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2021.08.015.
Appendix. Supplementary materials
References
- 1.Walsh K.A., Jordan K., Clyne B., Rohde D., Drummond L., Byrne P. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect. 2020;81:357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramanathan M., Ferguson I.D., Miao W., Khavari P.A. SARS-CoV-2 B1.1.7 and B.1.351 spike variants bind human ACE2 with increased affinity. Lancet Infect Dis. 2021;S1473-3099(21):00262–00263. doi: 10.1016/S1473-3099(21)00262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones T.C., Biele G., Mühlemann B., Veith T., Schneider J., Beheim-Schwarzbach J. Estimating infectiousness throughout SARS-CoV-2 infection course. Science. 2021:eabi5273. doi: 10.1126/science.abi5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kidd M., Richter A., Best A., Cumley N., Mirza J., Percival B. S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral load in samples tested by TaqPath polymerase chain reaction. J Infect Dis. 2021;223:1666–1670. doi: 10.1093/infdis/jiab082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ratcliff J., Nguyen D., Fish M., Rynne J., Jennings A., Williams S. Virological and serological characterization of critically ill patients with COVID-19 in the UK: interactions of viral load, antibody status and B.1.1.7 variant infection. J Infect Dis. 2021:jiab283. doi: 10.1093/infdis/jiab283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.S.A. Walker, K.D. Vihta, O. Gethings, E. Pritchard, J. Jones, T. House, et al. Increased infections, but not viral burden, with a new SARS-CoV-2 variant. medRxiv 2021.01.13.21249721; 10.1101/2021.01.13.2124972.
- 8.Calistri P., Amato L., Puglia I., Cito F., Di Giuseppe A., Danzetta M.L. Infection sustained by lineage B.1.1.7 of SARS-CoV-2 is characterised by longer persistence and higher viral RNA loads in nasopharyngeal swabs. Int J Infect Dis. 2021;105:753–755. doi: 10.1016/j.ijid.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tosif S., Neeland M.R., Sutton P., Licciardi P.V., Sarkar S., Selva K.J. Immune responses to SARS-CoV-2 in three children of parents with symptomatic COVID-19. Nat Commun. 2020;11:5703. doi: 10.1038/s41467-020-19545-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albert E., Ferrer B., Torres I., Serrano A., Alcaraz M.J., Buesa J. Amplification of human β-glucuronidase gene for appraising the accuracy of negative SARS-CoV-2 RT-PCR results in upper respiratory tract specimens. J Med Virol. 2021;93:48–50. doi: 10.1002/jmv.26112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.