Abstract
Current evolutionary scenarios posit the emergence of Mycobacterium tuberculosis from an environmental saprophyte through a cumulative process of genome adaptation. Mycobacterium riyadhense, a related bacillus, is being increasingly isolated from human clinical cases with tuberculosis-like symptoms in various parts of the world. To elucidate the evolutionary relationship between M. riyadhense and other mycobacterial species, including members of the M. tuberculosis complex (MTBC), eight clinical isolates of M. riyadhense were sequenced and analyzed. We show, among other features, that M. riyadhense shares a large number of conserved orthologs with M. tuberculosis and shows the expansion of toxin/antitoxin pairs, PE/PPE family proteins compared with other non-tuberculous mycobacteria. We observed M. riyadhense lacks wecE gene which may result in the absence of lipooligosaccharides (LOS) IV. Comparative transcriptomic analysis of infected macrophages reveals genes encoding inducers of Type I IFN responses, such as cytosolic DNA sensors, were relatively less expressed by macrophages infected with M. riyadhense or M. kansasii, compared to BCG or M. tuberculosis. Overall, our work sheds new light on the evolution of M. riyadhense, its relationship to the MTBC, and its potential as a system for the study of mycobacterial virulence and pathogenesis.
INTRODUCTION
The Mycobacterium tuberculosis complex (MTBC) is a group of genetically related pathogens that cause tuberculosis (TB) in mammalian species. The hallmark member, Mycobacterium tuberculosis, is the single most deadly bacterial pathogen, causing over 1.4 million deaths globally in 2019. Current evolutionary scenarios posit the evolution of the MTBC from an environmental saprophyte through a cumulative process of genome adaptation. Such scenarios envisage intermediate mycobacterial species with increasing pathogenic potential for humans, the vestiges of which should be present in extant mycobacterial species. Comparative genomic analyses between the MTBC members and opportunistic mycobacterial pathogens may therefore reveal the key evolutionary steps involved in the emergence of the MTBC, as well as illuminating virulence mechanisms across mycobacterial pathogens as a whole.
Non-tuberculous mycobacteria (NTMs) are naturally occurring environmental bacteria commonly found in water and soil. A wide range of animal and environmental sources (aquaria, swimming pools) act as reservoirs for NTMs, and several human disease outbreaks caused by exposure to environmental NTMs have been described (1). Little is known about the natural reservoir of M. riyadhense except for two studies that describe M. riyadhense being isolated from water (2) and soil (3). Since its discovery in 2009, M. riyadhense has been revealed to be a clinically important pathogen, able to cause disease in both immunocompromised and immunocompetent individuals (4–6), The clinical and radiologic characteristics of pulmonary infection caused by M. riyadhense are indistinguishable from those caused by M. tuberculosis, the most important human pathogen of the MTBC (4,7).
Similar to M. tuberculosis, M. riyadhense grows at 37°C and requires 2–3 weeks (8) to form visible colonies on agar media. No evidence of human-to-human transmission yet reported. Infections with M. riyadhense have been reported in Asia and Europe in countries including Bahrain, South Korea, France, Italy and Germany (7,9,10), although most of the recent cases originated in patients from Saudi Arabia. Indeed, the very first case of acute M. riyadhense infection was initially misdiagnosed as a case of M. tuberculosis infection in a Saudi hospital using commercially available diagnostic tests (4).
It is postulated that M. tuberculosis evolved from a free-living environmental ancestor into an obligate pathogen (11). In a recent publication, Sapriel and Brosch (12) showed the close phylogenetic relationship of M. riyadhense to the MTBC, suggesting that it forms part of an MTB-associated phylotype (MTBAP) with Mycobacterium decipiens, Mycobacterium lacus and Mycobacterium shinjukuense. The MTBAP have only been isolated from human clinical samples, and share specific traits that differentiate them from environmental NTMs; they hence likely serve as evolutionary intermediates between the environmental mycobacteria and the pathogenic members of the MTBC.
Mycobacterium species have a complex cell wall that contains unusual lipids, such as lipo-oligosaccharides (LOSs), and functions as a permeability barrier. Studies have shown that the LOS-deficient rough variant of M. kansasii persists longer in infected mice compared with the LOS-producing smooth strain (13). Defects in higher order LOS biosynthesis have also been correlated with smooth-wrinkled phenotype (14). Modern M. tuberculosis complex strains are more hydrophobic compared to M. canettii that contains abundant LOSs. This change of polarity/hydrophobicity is thought to increase their capability for aerosol transmission, affecting their virulence and pathogenicity (15). It is thus speculated that the loss of LOSs was an important step in the emergence of pathogenic mycobacteria.
Mycobacteria contain specialized Type VII secretion (T7S) systems (ESX1-5) for transportation of proteins (harbouring WXG motifs) through the hydrophobic cell wall (16). ESX systems mediate a broad range of functions in mycobacteria. ESX-1 in M. tuberculosis that is linked with growth within macrophages (17), cytosolic translocation (18) and antigen presentation (19). It has also been shown that ESX-1 plays a role in DNA transfer in M. smegmatis (20). ESX-3 contributes to iron acquisition in mycobacteria, as well as cell survival (21) and virulence (22) in pathogenetic mycobacteria. ESX-5 plays a major role in the capsule integrity through its substrate PPE10 (23). The functions of ESX-2 and ESX-4 remain to be fully elucidated.
To study the distribution of the virulence factors in M. riyadhense and other mycobacteria from an evolutionary context, we investigated the presence and absence of several virulence factors using comparative genomics approaches. Pathogenic Mycobacterium species have developed strategies to avoid or modulate the host immune response through virulence factors that include lipids, secretion systems and their secreted effectors, and cell surface molecules. For example, various PE/PPE family proteins, named after the conserved proline-glutamate (PE) or proline-proline-glutamate (PPE) motifs at their N-terminus, have been shown to be expressed by M. tuberculosis upon infection of macrophages and play critical roles in virulence, antigenic diversity and modulation of the host immune response (24). Another group of virulence factors is the mammalian cell entry (mce) proteins, which play an important role in the host cell signalling modulation as well as their primary role as lipid/sterol transporters (25).
Toxin/antitoxin (T/A) systems were first found on plasmids or plasmid-derived chromosomal loci where they were implicated in plasmid maintenance in bacterial populations (26). When compared to other mycobacteria, the MTBC members are remarkable for the extensive repertoire of T/A systems. A hallmark of M. tuberculosis infection is the ability to survive long-term in host granulomas and develop a latent stage of infection. The molecular mechanisms and cellular components that are involved in the persistence of M. tuberculosis are still poorly understood, but several T/A systems have been implicated in the pathogenicity of M. tuberculosis (27).
To investigate the MTBC evolutionary history, we report the characterization of eight clinical isolates of M. riyadhense, compare their genomes to members of the MTBC, and provide insights into the speciation of the MTBC from MTBAP and environmental bacteria. Due to their suggested role in virulence, we examined the LOS profiles of both rough and smooth variants of M. riyadhense, comparing them to those of other related mycobacteria. We furthermore analyzed the comparative transcriptional response of immunity-related host genes in a murine macrophage infection model after infection with M. riyadhense, M. kansasii, M. bovis BCG or M. tuberculosis.
To facilitate ease of identification of M. riyadhense, we also built on our genomic analyses to develop a simple PCR-based diagnostic test for the rapid and accurate identification of M. riyadhense so as to minimize the risk of misdiagnosis in a clinical setting.
Our analyses provide a comprehensive description of the hallmarks of M. riyadhense that make it one of the closest known environmental relatives of the MTBC, and that can serve to illuminate studies into the evolution and pathogenesis of the MTBC.
MATERIALS AND METHODS
Ethics statement
The research protocol was approved by the Institutional Review Board of King Fahad Medical City (Riyadh, Saudi Arabia; #16–345) and the Institutional Biosafety and Bioethics Committee of King Abdullah University of Science and Technology (Jeddah, Saudi Arabia; #18IBEC23). All adult subjects provided informed and written consent. A parent or guardian of any child participant provided informed consent on their behalf.
Culturing, DNA isolation and sequencing of bacteria
Eight M. riyadhense strains were collected in Riyadh, Saudi Arabia, between June 2011 and March 2016 from patients with a positive culture for M. riyadhense isolated from the microbiology laboratory at the King Fahad Medical City (KFMC) in Riyadh, Saudi Arabia. The M. riyadhense strains were grown on Lowenstein Jensen (LJ) slants at 37°C for 2 weeks, DNA was extracted using a phenol-chloroform protocol, and the quality was measured by Qubit. Twenty micrograms of high-molecular-weight DNA from the eight M. riyadhense strains were sequenced using a PacBio RSII sequencer (Pacific Biosciences, Menlo Park, U.S.A.) with a 10 kb library. A NEBNext Ultra II DNA library preparation kit (New England BioLabs, Massachusetts, U.S.A.) was used to prepare libraries according to the manufacturer’s instructions, and sequences from each library were generated for all M. riyadhense strains using the Illumina HiSeq 4000 platform (Illumina, San Diego, U.S.A.).
Genome assembly and annotation
The Illumina short reads were trimmed, and low-quality reads were removed by Trimmomatic (28). We did de novo assemblies of the eight consensus genomes of each M. riyadhense strain with the PacBio long reads using the Canu assembler (29). After assembly, the draft genomes were subsequently corrected with short Illumina reads using the Pilon (30) software. The circularity of assemblies was checked by Gepard (31), and assemblies were annotated by Prokka (32). A circular map of the chromosome was compared with that of M. tuberculosis H37Rv and visualized with BRIG (33). For the comparative genomics analysis, the genome of the M. riyadhense MR226 strain was used as a high-quality representative reference in this study. The overall study design flowchart is illustrated in Supplementary Figure S1.
Phylogenetic, comparative genomics and M. tuberculosis T/A orthologs analysis
We obtained draft genomes from 146 Mycobacterium species from the National Center for Biotechnology Information (NCBI) database and independently annotated by Prokka (32) (except for the M. tuberculosis H37Rv, M. marinum M, M. kansasii ATCC12478 and the genomes of MTBC members) for phylogenetic analysis and to identify the members of the genes belonging to the mce and the pe/ppe families. Accession numbers for these genomes can be found in Supplementary Table S1.
Protein homology of M. tuberculosis H37Rv esx1-5 loci were detected by Proteinortho (34) across all the mycobacterial genomes. The presence of esx loci was defined such that at least three orthologs of genes present in M. tuberculosis esx loci were in close proximity to one another in the genome. The PhyoPhlAn2 (35) pipeline was used construct the phylogeny of the 147 (including the M. riyadhense strain MR226) genomes and Nocardia abscessus was used as the out-group. PhyloPhlAn2 was used to generate the phylogenetic tree by concatenating the informative amino-acid positions from 400 marker genes and aligned separately by MUSCLE (36) and reconstructed into trees using FastTree (37) and RaxML (38).
A focused OrthoMCL (39) comparison was performed between (i) M. riyadhense, M. marinum, M. kansasii, M. szulgai and M. tuberculosis and (ii) M. riyadhense and five species from the MTBC, namely, M. tuberculosis, M. bovis BCG, M. canettii, M. mungi and M. africanum.
To study the presence of M. tuberculosis orthologs in the genus Mycobacterium, 158 T/A proteins belonging to the VapBC, RelEF, HigBA, MazEF, ParDE and UCAT families that have been described previously (40) were retrieved from the M. tuberculosis H37Rv assembly. The M. tuberculosis T/A orthologs from all of the 147 species were calculated by Proteinortho (34) (Supplementary Material online).
SNP calling and phylogeny based on SNPs
The corrected Illumina reads were mapped using BWA (41) onto the M. riyadhense MR226 genome assembly. Picard tool (42) was used to clean SAM files, fix mate-pair information and mark duplicates. SNPs were called for two iterations and filtered according to the Genome Analysis Toolkit (GATK) (43) pipeline recommendations. Phylogeny was generated by RaxML (44) with the TVM model.
Infection of the RAW 246.7 cell line
The murine macrophage RAW264.7 cell line was obtained from the American Type Culture Collection (ATCC, Manassas, U.S.A.) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) (ThermoFisher Scientific, Waltham, U.S.A.) supplemented with 10% FCS, streptomycin and penicillin. Mycobacterium riyadhense MR226, M. kansasii (subtype I), M. bovis BCG Denmark and M. tuberculosis H37Rv strains were grown in Middlebrook 7H9 liquid medium after single-colony isolation from LJ slants or 7H10 agar. 7H9 was supplemented with 10% albumin, dextrose and catalase (ADC), while 7H10 was supplemented with oleic acid, albumin, dextrose and catalase (OADC) in addition to 0.2% glycerol. The infection experiment was carried out according to the experimental conditions described previously (45). The supernatant was removed after 3 h, and the infected cells were washed with PBS to remove extracellular bacteria. Subsequently, the cells were incubated in fresh DMEM with 10% FCS for 24 and 48 h. All of the experiments were carried out in three different independent batches and used as triplicates in the study. For harvesting, 400 μl of TRIzol (ThermoFisher Scientific, Waltham, U.S.A.) was added to the wells at each time point, and the adherent cells were scraped out and stored at -80°C for RNA extraction. Each bacterial infection was performed in triplicate, in addition to the non-infected controls.
RNA preparation and RNAseq analysis
RNA was isolated from the samples using the Direct-zol™ RNA Miniprep kit (Zymo Research, Irvine, U.S.A.) according to the manufacturer’s instructions. An Agilent RNA 6000 Nano kit was used to check the quality and quantity of the total RNA. The RNA libraries were prepared by following the Illumina TruSeq RNA Sample Preparation Kit v2 protocol.
The Illumina short reads were trimmed, and low-quality reads were removed by Trimmomatic (28). The RNAseq reads were mapped to GRCm38 reference genome with HISAT2 (46).The number of reads that mapped to each annotated gene was counted using the HTSeq (47) with ‘union’ overlap resolution mode. Data were normalized and the DE genes were identified using DESeq2 package (48). A fold-change in expression ≥2 with an adjusted P-value (padj) < 0.01 was considered as significant, and genes fulfilling these criteria were considered to be differentially expressed (DE) upregulated genes. The functional annotation of the DE genes were then analysed using the Metascape (49).
Thin-layer chromatography analysis of lipooligosaccharides in M. riyadhense, M. kansasii and M. marinum
For TLC analysis, mycobacterial strains were grown at 30°C (M. marinum M) or 37°C (M. smegmatis MC-1551, M. kansasii subtype I, M. riyadhense MR193 (R) and MR226 (S)) on LJ slants, and after sufficient incubation, grown cells were collected and washed once with PBS. Apolar and polar lipids were extracted from the cell pellets using methods described by Dobson et al. (50). Polar lipids were analyzed by 2D-TLC using solvent system E, which is designed to separate phospholipids and LOSs (50). Glycolipids were visualized by charring following staining with either molybdophophoric acid (MPA) or alpha-napthol (for glycolipids).
Diagnostic PCR markers for M. riyadhense
To develop diagnostic markers for M. riyadhense for distinguishing M. riyadhense from MTBC, M. kansasii, M, szulgai and M. angelicum, unique regions within the M. riyadhense reference genome compared to that of 152 other mycobacterial species were detected using Shustring (51). These regions were also examined by Proteinortho (34) and Blastn. MR_00036, MR_00263, MR_00606 and MR_01005 were selected as the amplification targets. Two primers for each gene were designed in this study:
MRDP-MR_00036-F (5′-TTCGTTGTCGGTTTCGTCGC-3′) and MRDP-MR_00036-R (5′-GCGTCAGCTCCACCGAAAAC-3′);
MRDP-MR_00263-F (5′-CCACCGCTGTTGGCGA-3′) and MRDP-MR_00263-R (5′-TTCGTCCCGTTGATCCCGTT-3′);
MRDP-MR_00606-F (5′-AACCTGCCCGATACGCACTT-3′) and MRDP-MR_00606-R (5′-ACTGTTCCTCCGTGGGGTTG-3′);
MRDP-MR_01005-F (5′-GACTGTGGGGTAACGGTGGA-3′) and MRDP-MR_01005-R (5′-CCGGTGATGTCGCCTACTCC-3′).
PCR was performed in a 25 μl reaction volume with 12.5 μl of GoTaq® Green Master Mix (Promega, U.S.A.), 1 μl of 100 ng/μl gDNA, 1 μl with 10 nmol of forward and reverse primers, 3 μl of dimethyl sulfoxide (DMSO) and 19 μl of nuclease-free water. The PCR mixture was denatured for 5 min at 94°C; followed by 35 cycles of amplification involving a denaturation step at 94°C for 30 s, a primer annealing step at 59°C for 45 s and a primer extension step at 72°C for 45 s, and a final extension step at 72°C for 7 min. The ITS-F/mycom-2 primer set, which is a Mycobacterium genus-specific primer set, was used as a control, with amplification conditions as described previously (52). The products were electrophoresed in a 2% agarose gel for 60 min and visualized.
RESULTS
Assembly and annotation of the M. riyadhense genomes
Between April 2011 and March 2017, eight clinical cases of infection with M. riyadhense were recorded in male patients aged from 8 to 82 years. The comparison of different assemblies and genomic variations of all sequenced M. riyadhense strains is listed in Table 1. We obtained chromosomes of all eight isolates in single contiguous sequences for genome comparison at a high resolution. The circular nature of the chromosome and the circular plasmids (pMR01, pMR02 and pMR03) were demonstrated through Gepard (31).
Table 1.
Comparison of M. riyadhense strains’ assemblies and genomic variations
| Type strain assembly GCA_002101845.1 | MR193 | MR206 | MR210 | MR222 | MR226 | MR244 | MR246 | MR1023 | |
|---|---|---|---|---|---|---|---|---|---|
| Assembly size (bp) | 6 269 850 | 6 695 517 | 6 835 855 | 6 528 955 | 6 533 138 | 6 888 178 | 6 744 506 | 6 916 580 | 6 906 827 |
| Chromosome size (bp) | 6 269 850 | 6 129 600 | 6 288 531 | 6 034 715 | 5 960 707 | 6 243 587 | 6 258 360 | 6 289 824 | 6 306 178 |
| Contigs | 263 | 7 | 6 | 2 | 7 | 3 | 9 | 4 | 9 |
| Gaps in chromosome | 262 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| pMRLP | A | P | P | P | P | P | P | P | P |
| pRAW-like plasmid | A | P | A | A | P | P | A | A | A |
| CDS | 5168 | 5901 | 6124 | 5800 | 5852 | 6077 | 6064 | 6060 | 6185 |
P: Presence; A: Absence.
In this study, we further identified the potential linear plasmids present in all 8 strains and circular plasmids (present in 3 strains as shown in Table 1) in M. riyadhense (Supplementary Figures S2 and S3). The pMRLP plasmids that are present in all 8 strains have no significant repetitive sequence that would support a circular topology and hence they likely represent linear plasmids. When compared with the circular plasmids of other species, such as pRAW in M. marinum (53), pMAH135 (54) and pMA100 (55) of M. avium, pMyong1 from Mycobacterium yongonense (56), pMK12478 (57) from M. kansasii and several plasmids from Mycobacterium chimaera (58), a high similarity was observed. The plasmid pMRLP01 in MR226 contains a pair of partitioning genes (parA/parB) that are involved in active segregation and thus stabilize the inheritance of the plasmid (59). The latter are known to contribute to genome evolution by active DNA transfer and exchange (60). The progressive alignments of the assembled chromosomes (Supplementary Figure S3A), pMRLP plasmids (Supplementary Figure S3B) and circular plasmids (Supplementary Figure S3C) of each M. riyadhense strain show that the chromosomes are relatively conserved; however, the likely linear plasmids present in all eight sequenced isolates are diverse from both structural and similarity perspectives, while the pRAW-like plasmids are present in only the MR226, MR193 and MR222 strains.
A previous study showed that M. tuberculosis genomes are highly conserved (61) in comparison to M. canettii (62). A much higher number of SNPs were detected in Smooth Tubercle Bacilli (STB) genomes compared to those of MTB strains (61,62). To study the genetic variations of the M. riyadhense strains, we called SNPs from all the strains and generated the SNP-based phylogeny using 43 136 polymorphic sites (Supplementary Figure S4). The SNP-based phylogeny revealed a group of five closely related M. riyadhense strains and three more distantly related strains, each of which is separated from the other strains by a long branch. The pair-wise comparison of SNPs between any 2 phylotypes is greater than the diversity between M. tuberculosis strains (61), while it is smaller than that seen within M. canettii strains (62), and the variation between the five closely related M. riyadhense strains is comparable to the SNP variation in M. tuberculosis strains. Currently, information on the genomic diversity of M. riyadhense strains is available only from a few isolates representing restricted geographic locations such as Saudi Arabia. Hence, we speculate that the current phylotype information may be subject to revision once genomic data become available for a larger cohort of M. riyadhense strains from wider geographic locations and ecological niches.
Regions of difference (RDs) in M. riyadhense
The RDs were originally described as genomic regions present in virulent M. bovis and M. tuberculosis but absent from the M. bovis BCG genome (63). RD loci were subsequently described across the MTBC (64) and contain functions believed to contribute to pathogenicity (65–67) and may link to the evolution of MTBC species (68). Mycobacterium riyadhense was found to harbor most of the RD loci (RD1, RD3-R11, R13-RD16) that are also intact in M. tuberculosis, while 2 of the RDs show unique deletions, RD2riyadh (Supplementary Figure S5A) and RD12riyadh (Supplementary Figure S5B).
RD2 was originally described as deleted in M. bovis BCG vaccine strains. Subsequently, it was shown that disruption of RD2 in M. tuberculosis led to decreased proliferation in vivo and impaired modulation of the innate immune response (66). The RD2 locus in M. riyadhense also harbours a deletion, but one that is distinct from that in M. bovis BCG. It is a larger deletion than the originally described RD2BCG as RD2riyadh contains 29 genes (M. tuberculosis coordinates were used, rv1971∼rv2000, location 2,216,498∼2,246,766); eight genes within this locus (M. tuberculosis coordinates were used, rv1978, rv1979c, rv1980c, rv1981c, rv1983, rv1984, rv1987, rv1988) have orthologs elsewhere in the M. riyadhense genome (MR_05764, MR_05852, MR_02310, MR_02993, MR_00486, MR_02995, MR_02325, MR_02349, MR_01747), suggesting possible functional redundancy.
The RD12 locus shows deletions across MTBC members, including M. tuberculosis (69), M. bovis (70), Mycobacterium caprae and M. orygis (71). Mycobacterium canettii isolates (except group B (72)) also show an independent deletion at the RD12 locus named RD12can (3,479,430∼3,491,866, rv3111∼rv3126), which is distinct from RD12bovis (3,484,740∼3,487,515, rv3117∼rv3121). We identified another unique deletion at the RD12 locus in M. riyadhense, designated RD12riyadh, which encompasses a larger region than RDcan and RD12bovis, encompassing rv3108-rv3127 (3,477,171∼3,492,150) (Supplementary Figure S5B). It is intriguing that multiple mycobacteria show independent deletion events at the RD2 and RD12 loci, suggesting selective forces may play a role in this variation.
Comparative phylogeny of M. riyadhense with other Mycobacteria
The phylogenetic tree shows that the slow-growers and rapid-growers are separated into two different clades and that the early branching species are fast-growers (Figure 1). The overall topology of our tree is similar to that of previously published phylogenetic trees (73–75); however, there are a number of features in our analysis that are worth highlighting. Hence, our current phylogeny comprises a large set of mycobacterial taxa (147 genomes) and was inferred with a robust phylogenetic approach (maximum likelihood-based phylogeny) using a substantially larger set of amino acid sequences to previous studies. By way of comparison, Tortoli et al. generated their phylogeny with a matrix of ANI-divergence scores (73), which is not based on specific evolutionary models and is less reliable than the maximum likelihood-based phylogeny used here. Furthermore, while Sapriel and Brosch used a similar methodology to the one we employed, they used a smaller data set (75). The phylogenetic analysis by Fedrizzi et al. (74) is similar in scale and methodology to that in the current manuscript, although our approach was broader. Given these improvements, our current study provides a strong confirmation of prior results and reveals several new perspectives.
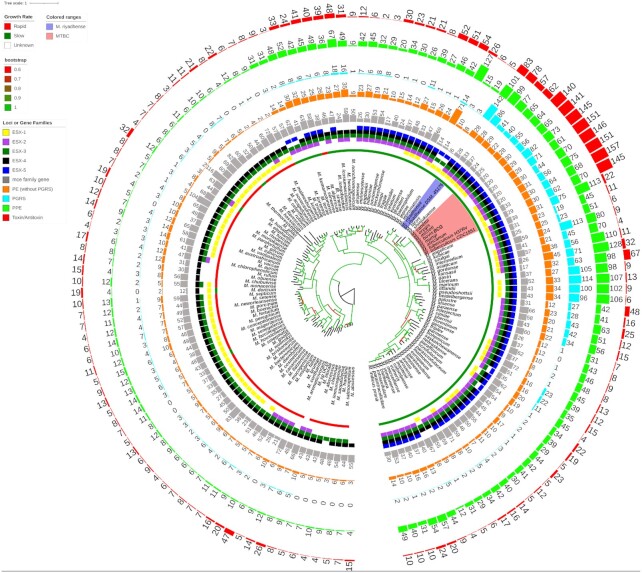
Figure 1.
A phylogenetic tree of 147 mycobacterial genomes showing close relationship of M. riyadhense and MTBC. The phylogeny was constructed using 147 available genomes by concatenating and aligning amino acid positions across 400 shared proteins automatically identified in the chosen genomes by PhyloPhlAn2 (35). Coloured shades highlight the M. riyadhense strains and MTBC. We annotated the growth rate, presence/absence of ESX systems (based on homology analysis), the number of annotated gene family members, the M. tuberculosis T/A orthologs for each strain external to the phylogeny. Branches are coloured based on the bootstrap values as shown in the legend in the left. The colour code of the gene families, ESX systems, growth rate and bootstrap range is listed on the left side figure legend.
Our phylogenetic analysis shows that M. riyadhense along with M. lacus, M. shinjukuense and MTBC form a monophyletic group that has descended from a common ancestor. Tortoli et al. found that M. riyadhense, M. lacus, M. shinjukuense, M. decipiens, M. szulgai, M. angelicum and MTBC form a monophyletic group (74) which differs from our findings that M. szulgai, M. angelicum are from the sister group. A wide evolutionary gap between the M. leprae clade and MTBAP in our phylogenetic analysis contradicts Fedrizzi et al. (74), where they showed the most recent common ancestor of the MTB and M. leprae clade was more recent than the MRCA of MTB and the MTBAP clade. Mycobacterium riyadhense is located within the same clade as obligate and opportunistic mycobacterial pathogens that include the MTBC, M. marinum, M. kansasii, M. leprae and related host-restricted mycobacteria with reduced genomes and decreased survivability in the environment.
The PE/PPE and mce family genes are known to be important for host adaptation (76) and pathogenicity (77). We observed an expansion of PE/PPE family genes in the MTBC members but also in M. riyadhense MR226 and other pathogenic species, such as M. kansasii and M. ulcerans. The number of mce family gene orthologs has not significantly changed across mycobacterial genomes (Figure 1). Our results agree with previous findings that during their evolution, the ESX systems were derived from the ancestor ESX-4, as shown in Figure 1 at the root node, and then ESX-3, ESX-1, ESX-2 and ESX-5 evolved by horizontal transfer (78).
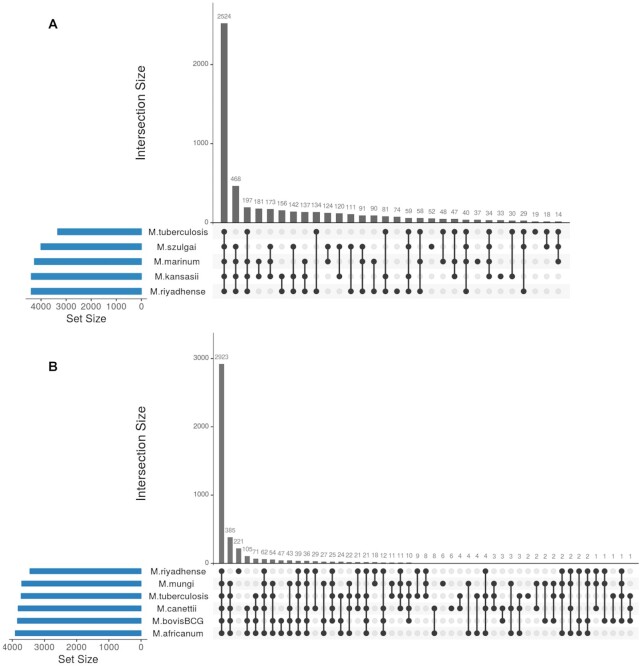
M. riyadhense shares a larger number of orthologs (3,122) with M. tuberculosis than with M. kansasii (2,978 orthologs), M. marinum (2,962 orthologs) and M. szulgai (2,724 orthologs) amongst the NTM that are closely related to the MTBC (Figure 2A). A total of 134 orthologs are exclusively shared between M. riyadhense and M. tuberculosis, while the number of orthologs exclusively shared between M. tuberculosis and M. kansasii (30 orthologs), M. marinum (48 orthologs) and M. szulgai (18 orthologs) is less (Figure 2A). It is notable that 34 T/A genes are shared exclusively between M. riyadhense and M. tuberculosis within this comparison.
Figure 2.
UpSet plot showing the comparison of orthologous groups. UpSet plot showing the orthologous groups total set size and overlaps between the (A) M. riyadhense, M. tuberculosis H37Rv, M. marinum M, M. kansasii 12478 and M. szulgai and (B) M. riyadhense MR226 and five species within the MTBC.
The comparative analysis of the orthologous groups of M. riyadhense and the MTBC is informative. Firstly, 385 orthologous groups present across all MTBC species are absent from M. riyadhense. Secondly, 221 protein groups uniquely present in M. riyadhense are not found amongst the MTBC (Figure 2B). This latter group of M. riyadhense unique protein groups are likely required for M. riyadhense to maintain a broad functional repertoire so as to secure its survival. In contrast, the MTBC species have lost genes involved in free-living survival due to their obligate pathogen lifestyle.
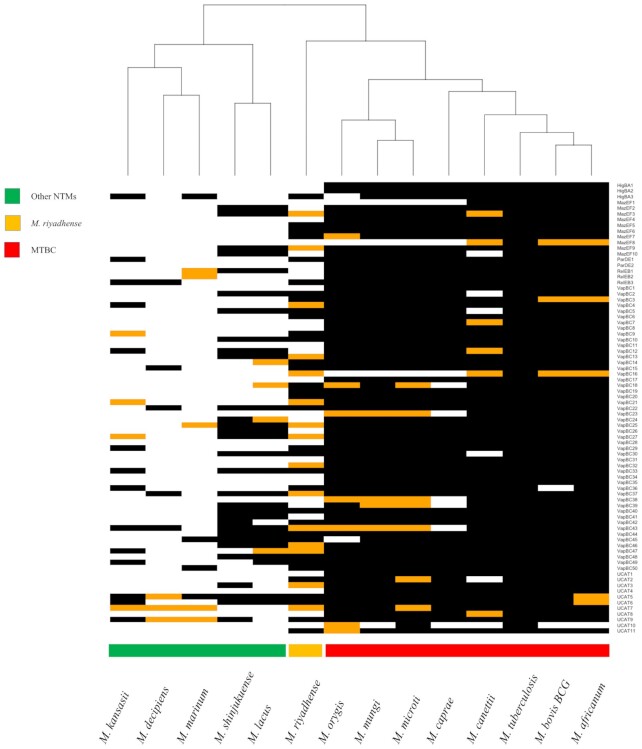
We compared the 79 pairs of T/A systems (belonging to the HigAB, MazEF, ParDE, RelEF, VapBC and UCAT families) in M. tuberculosis with the T/A pairs found in other members of the MTBC and NTMs. We observed an expansion (83 out of the 158 orthologs) of the T/A orthologs (Figures 1 and 3) in M. riyadhense MR226 strain compared to other NTMs including M. lacus, M. shinjukuense and M. decipiens. A detailed comparison of the T/A M. tuberculosis H37Rv orthologs across M. riyadhense strains is illustrated in Supplementary Figure S6.
Figure 3.
A hierarchical clustering of the presence (black) and absence (white) of M. tuberculosis H37Rv toxin/antitoxin orthologs in M. riyadhense, M. marinum, M. kansasii, M. shinjukuense, M. lacus, M. decipiens and MTBC species. The orange blocks denote the presence of either the toxin or antitoxin ortholog in a given pair of the T/A system. The black and white blocks represent presence and absence respectively. The name of the T/A system are shown for each row on the right.
M. riyadhense strains produce a distinct pattern of LOSs
It is well-known that the smooth morphotypes of mycobacteria, such as M. avium (79), M, abscessus (80) and M. kansasii (13), are less virulent than the rough morphotypes. It is noteworthy that we observed both smooth (MR210, MR222, MR226, MR244, MR246 and MR1023) (Figure 4A) and rough (MR193 and MR206) (Figure 4B) morphologies in M. riyadhense strains. The lipooligosaccharides are an important class of glycolipids that have been linked to diverse mycobacterial phenotypes including colony morphology. The LOS-biosynthesis gene locus has been characterised in M. marinum (14), M. kansasii and M. canettii (81). We therefore sought to first examine whether the genetic machinery for the production of LOS is present in the M. riyadhense genome, and then to follow up on the genome-level predictions with lipid analyses of rough and smooth variants using thin-layer chromatography (TLC).
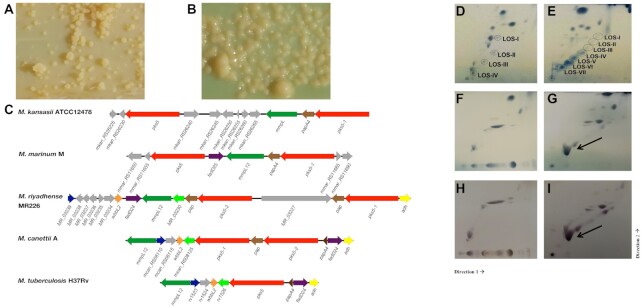
Figure 4.
pks5 loci in M. riyadhense and other related mycobacteria and 2D-TLC analysis of polar lipids extracted from selected M. riyadhense strains, M. kansasii (subtype I), M. marinum M, M. smegmatis MC-1551. (A) Rough-dry colony morphology (MR193) and (B) smooth morphology (MR226) of M. riyadhense. (C) Genetic locus map of the pks5 gene cluster from M. riyadhense MR226, M. marinum M (partial), and M. kansasii ATCC12478 (partial), M. canettii A and M. tuberculosis H37Rv (drawn to scale). The arrows show the direction of transcription and the genes are coloured according to the orthologous relationships. Mycobacterium tuberculosis H37Rv does not produce LOSs and the pks5 loci was drawn for genomic comparison purpose. Polar lipids from two known LOS producers, M. marinum (D) and M. kansasii (E), are included to illustrate the migration pattern of LOS species in System E. (F) 2D-TLC analysis of polar lipids extracted from select M. riyadhense rough strain or smooth (G) strain. A separate staining with alpha-napthol also confirmed that this was a glycolipid species from the same (H) rough and (I) smooth strain. (D–G) were charred after staining with MPA, while (H) and (I) were charred after staining with alpha napthol. LOS III from M. riyadhense is indicated by a solid arrow.
To investigate the genetic basis for the smooth and rough morphology in the M. riyadhense strains, we compared the LOS locus of the eight strains (Supplementary Figure S7A and Supplementary Table S3). We compared the locus map of the eight strains and have found several variations in the rough strains: (1) the LOS locus maps are the identical for strains MR226, MR246 and MR1023, which are all smooth strains; (ii) the fadD24 gene is intact except in strain MR206 (rough) (Supplementary Figure S7B) where a transposable element fragment was detected; fadD24 is a fatty-acid-AMP synthetase; a mutation in the fadD24 ortholog in M. marinum (fadD25, mmar_2341) generated a rough morphology, which matches our genotype prediction (14); (iii) absence of MR_03241 in both smooth (MR210) and rough (MR193) strains (Supplementary Figure S7A,S7C); (iv) a 23-gene insertion (several transposases, integrase and IS elements were detected in this region) was found in the rough strain MR193 (Supplementary Figure S7D), between MR193_03124 and MR193_03148. MR_03251, which is a glycotransferase without any orthologs in M. canettii, M. tuberculosis or M. marinum is missing in the LOS locus of MR193. Analysis of LOS profiles via mutagenesis/complementation of MR_03251 in M. riyadhense will help to confirm the function of this gene.
The pks5 locus is involved in polyketide metabolism and previous study has shown this locus regulates the S/R morphology of M. canettii. The pks5 and pap genes in the LOS locus are intact in M. riyadhense, as is the case in M. canettii, but not in M. tuberculosis, where the former is truncated and the latter deleted (81). Remarkably, the M. riyadhense pks5 gene locus layout is dissimilar to that in M. canettii, M. tuberculosis, M. kansasii and M. marinum (Figure 4C). A previous study speculated that wecE is involved in the biosynthesis of the unusual sugar residue of LOS-IV and a wecE mutant cannot generate LOS IV in M. marinum (14). We observed that the wecE and galE6 M. marinum orthologs are absent from the M. riyadhense genome (Supplementary Table S2). The same study also showed that a galE6 (UDP-glucose 4-epimerase) mutant was also not able to produce LOS II. Thus, absence of wecE and galE6 is likely to cause an accumulation of LOS II* and the lack of fully formed LOS IV, which have previously been shown to increase the pathogenicity of M. marinum (14).
To correlate rough versus smooth colony morphology with LOS production, we extracted polar lipids from the strains and analysed them by 2D-TLC using solvent system E (50), which is designed to separate phospholipids and LOSs. Charring of the TLC plates with alpha-napthol revealed glycolipids, including the accumulation of a species that migrated at a position similar to that of LOS III. This lipid was seen only in smooth strains; species with migration patterns similar to LOS I and LOS II were observed, while no spots were observed where LOS IV is expected to migrate. This result was not unexpected because all M. riyadhense strains lack wecE ortholog, which is required for the extension of LOS II to LOS IV (Figure 4). Additionally, the relative levels of the predominant LOS species in M. riyadhense seem to be quite high when compared to those seen in other LOS-producing mycobacteria (Figure 4D, E). Conversely, the rough strains did not produce any glycolipids that migrated in the positions corresponding to LOSs (Figure 4F, H).
PE-PGRS33 locus
The pe-pgrs33 (rv1818c) locus encodes the exported protein PE_PGRS33 that plays an important role in the pathogenesis of M. tuberculosis (82). A previous study (83) showed that pe-pgrs33 is present in all MTBC members but not in M. canettii, which implies a specific pe-pgrs33 insertion event in the ancestor of MTBC strains. In this study, we performed a comparison of the genomic context of the pe-pgrs33 locus using the Artemis Comparison Tool (ACT) (Supplementary Figure S8A), and precisely identified the breakpoint in the M. riyadhense MR226 genome (3,155,129bp-3,156,672bp, within the MR_02789 reading frame) and the MTB H37Rv genome (2,061,129bp-2,062,831bp). MR_02789 also contains orthologs in M. marinum M (mmar_2693) and M. kansasii ATCC12478 (mkan_RS00295). The overall synteny in the locus in MR226 also shares a high similarity with M. canettii PE-PGRS33 locus, and it is evident that the lactoylglutathione lyase gene (MR_02789) ortholog was lost before the MTBC speciation (Supplementary Figure S8B).
Type VII secretion system of M. riyadhense
All five ESX systems (ESX1-ESX5), which are Type IV Secretion (T7S) systems, were found in the M. riyadhense strains (Supplementary Figure S9) with the same level of synteny except for espK which was truncated in MR206 (Supplementary Table S4). The overall gene arrangement of the ESX1-ESX5 loci is similar in both M. riyadhense and M. tuberculosis (Supplementary Figure S9) (84). The espACD operon, which is essential for secretion of virulence factors via ESX1, and eccB and eccC genes in ESX-2, are absent from the ESX-2 system in M. riyadhense compared to M. tuberculosis H37Rv. This conserved synteny reinforces the previous results of phylogenetically relatedness of M. riyadhense to MTBC. As noted before, the pMR01-03 plasmids also contain an extra ESX-P5 locus, which could indicate a role for this plasmid in mediating pathogenicity (Supplementary Figure S2).
Comparative transcriptional response of murine macrophages upon infection by M. riyadhense, M. tuberculosis, M. kansasii and M. bovis BCG
Our genomic analysis of M. riyadhense revealed a range of genes and potential gene networks that could play a role in host–pathogen interactions. We therefore sought to assess the initial interaction of M. riyadhense with macrophages, using the murine RAW264.7 cell line as our experimental model. As comparator strains in our analysis, we performed parallel infections with M. kansasii, an opportunistic pathogen that also contains an orthologous ESX-1 system (Figure 1), M. bovis BCG and M. tuberculosis. These comparisons allowed us to explore the M. riyadhense-triggered innate immune responses.
To compare the innate immune responses of macrophages infected with these mycobacterial isolates, their transcriptional profiles were analysed at 3, 24 and 48 h post infection (hpi) compared to uninfected controls. In total, 1496 genes, or 3.79% of the genes in the genome (39 504 genes in total), were upregulated when comparing infected cells to non-infected cells at each time point (Tables S5-S16; Log2Fold Change> = 2, padj<0.01).
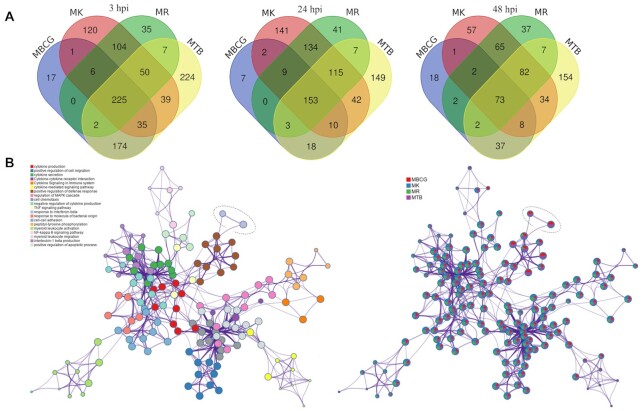
A large number of DE genes overlapped at each timepoint: 225, 153 and 73 (Figure 5A). These genes are responsible for general immune responses, such as proinflammatory genes associated with TLR signalling (e.g., upregulation of Il-1β, Tnf-α, Ccl4, Ptgs2 and Cxcl2, albeit to different absolute levels; Tables S5, S8, S11, S14) and cytokine activity-related genes (upregulation of Ccl7, Ccl2, Cd86, Ccl22, Il-1β and Ccl3). Genes uniquely triggered by M. riyadhense infection compared with other mycobacteria included Cd22, Gpr65, Adh7 and TrnT (Supplementary Table S11); however, no functional categories were statistically enriched from this category of genes.
Figure 5.
The transcriptional response of RAW264.7 macrophages to mycobacterial infections. (A) Venn diagram showing the overlap of differentially expressed genes upon M. riyadhense (MR), M. kansasii (MK), M. bovis BCG (MBCG) and M. tuberculosis (MTB) infection over 3, 24 and 48 hpi, and (B) Network of enriched terms for each bacterial infection upon 3 hpi. Each node represents an enriched term and the size of the circle represents the number of genes within that term as applied by the Metascape (49). Terms with a similarity > 0.3 are connected by edges. The left panel is coloured by cluster, while the right panel is colour-coded based on different bacterial infections. The nodes that represent the ‘response to interferon-beta’ are highlighted by a dotted circle.
The largest number of DE genes were identified from M. tuberculosis infected RAW264.7 cells 3 hpi, with 756 genes being upregulated. As expected, the transcriptional responses due to M. riyadhense and M. kansasii infections clustered more closely while the BCG and M. tuberculosis responses clustered together (Supplementary Figure S10). One of the key differences in the macrophage transcriptional responses between M. riyadhense or M. kansasii and M. bovis BCG or M. tuberculosis pairs at 3hpi was the differential induction of Type I IFN signaling-related genes (Figure 5B and Supplementary Figure S11A). The expression of Ifnb1 was relatively lower in macrophages infected with M. riyadhense or M. kansasii, and that of IFN-β-controlled genes was only significantly enriched with, or more strongly upregulated by, M. bovis BCG or M. tuberculosis (Figure 5B).
In addition, activation of the TLR4/Myd88/ NF-κB signalling axis triggers expression of Tnf-α, Il-1β and Cox-2. These genes were potently induced by all four species, but less by M. riyadhense (Supplementary Figure S11B), suggesting that M. riyadhense triggers reduced signalling through TLR4. We also observed that M. riyadhense or M. kansasii were stronger activators of Liver X Receptors (LXR), compared to M. bovis BCG or M. tuberculosis (Supplementary Figure S11B). This is shown by the relatively higher expression of Abca1 and Abcg1 in M. kansasii or M. riyadhense which could be a secondary effect of a less potent induction of NF-κB pathway (85).
Developing a rapid PCR-based diagnostic marker for M. riyadhense
Due to the issues previously encountered in diagnosing M. riyadhense infections (4,7), correct and prompt identification of cases upon presentation at healthcare units is of paramount importance. We therefore sought to translate our knowledge on the genome sequences into a PCR diagnostic test that could be used in a clinical microbiology setting to distinguish M. riyadhense from other mycobacteria, including the members of the MTBC.
By identifying unique K-mers ranging in size from 11 bp to 4209 bp (Supplementary Figure S12) in the assembled genome compared to the genomes of 152 other mycobacterial species, four primer sets were developed targeting the MR_00036, MR_00263, MR_00606 and MR_01005 genes. The MRDP primer pair MRDP-F/MRDP-R amplified a single product from each of the eight isolates of M. riyadhense (Supplementary Figure S13) but not from other mycobacterial species, including M. tuberculosis, M. bovis, M. kansasii, M. marinum, M. szulgai, M. avium and Mycobacterium angelicum. This result shows that the MRDP-F/MRDP-R primers are highly specific to M. riyadhense and form the basis for a simple diagnostic PCR that can inform appropriate treatment protocols.
DISCUSSION
Mycobacterium riyadhense has become a clinically relevant NTM species globally (4–7,9,10). Contrary to prior publications on M. riyadhense that have been based primarily on clinical case reports, here we present the largest and most comprehensive genomic study undertaken to date on clinical M. riyadhense isolates. The eight new M. riyadhense strains sequenced in this study originated from pulmonary infections, with some having additional extra-pulmonary involvement, fulfilling the American Thoracic Society/Infectious Diseases Society of America (ATS/IDSA) criteria for NTM infection (86).
It is well known that plasmids are important ‘vehicles’ for the exchange of genetic material between bacteria or between chromosomes and extra-chromosomal plasmids. In this study, we identified likely linear plasmids pMRLP (present in all 8 strains) and circular plasmids (present in 3 strains as shown in Table 1) in M. riyadhense (Supplementary Figures S3C and S4). Linear plasmids were first described in 1989 in maize (87) and have also been found in Actinomycetales and Mycobacterium species, such as Mycobacterium xenopi, Mycobacterium branderi and Mycobacterium celatum. They are often accompanied by a circular plasmid in the same host (88).
The pRAW-like circular plasmids (pMR01-03) identified in this study all harbour both a T4S (Type IV secretion system) and T7S systems, which are necessary for conjugation (53), and facilitate the exchange of genetic material between different species of slow-growing mycobacteria. We therefore speculate that pMR01-03 are likely novel conjugative plasmids.
Our comparative analysis of M. riyadhense genomes with the MTBC and a large collection of NTMs provide unequivocal evidence that M. riyadhense is one of the close known NTM species to the MTBC and forms a phylotype with M. lacus and M. shinjukuense. Indeed, while our manuscript was in preparation, independent work by Sapriel and Brosch (12) also showed the close phylogenetic relationship of M. riyadhense to the MTBC, suggesting that it forms part of an MTB-associated phylotype. Our analyses of multiple M. riyadhense isolates complements and extends the findings of Sapriel and Brosch by revealing that expansion of T/A pairs, modification of secretion systems, alterations in cell wall lipids all play key roles in the evolution of the MTBC.
Our study shows that M. riyadhense shares a larger number of orthologs with M. tuberculosis than M. kansasii and M. marinum, notably in the T/A gene family (Figures 2 and 3). Eighty-three T/A orthologs were found in M. riyadhense MR226, far greater than the number of orthologs observed in any of the other NTMs (Figures 1 and 3). The expansion of T/A genes among the MTBC offers additional evidence that suggests that the original acquisition of T/A modules into mycobacteria through lateral gene transfer played a key role in the development of pathogenicity (89,90). The other virulence-associated gene family compared in this study is the mce family. The number of mce gene orthologs show the plasticity of this gene family. A recent study has shown that the mce homology between rapid and slow-growing species was low (<50%), which may indicate a possible adaptation of mce homologs to an intracellular lifestyle in slow-growing mycobacteria (91).
Mycobacterium riyadhense strains appeared as both smooth and rough colony forms when grown on solid LJ media. The observation of smooth and rough colony variants is seen in other mycobacterial pathogens where it is linked to presence or absence of LOS. For example, the presence or absence of LOS from M. canettii causes a transition from smooth to rough colony variants, respectively, with rough variants showing increased virulence. MTBC strains lack LOS, and it has been suggested that the removal of LOS was a key event in the evolution of the MTBC species towards their current obligate pathogen status (81). In M. riyadhense smooth colony variants, we observed the presence of LOS I and LOS II but the absence of LOS IV. These biochemical observations agree with the genomic prediction that M. riyadhense strains lack a wecE ortholog, which is required for the extension of LOS II to LOS IV in M. marinum (14). Overall, our results show that M. riyadhense exhibits a LOS production phenotype distinct from that of other LOS-producing mycobacteria. The truncated fadD24 gene in MR206 and absence of glycotransferase MR_03251 is likely responsible for the rough morphology of MR193 and MR206 strains, but further complementation experiments are required to provide a definitive confirmation.
The T7S systems are key elements of mycobacterial virulence. All the 5 ESX T7S systems present in M. tuberculosis, and known to be involved in virulence and pathogenicity, were found in M. riyadhense with very similar gene arrangement. An additional ESX-P5 system was also found on the circular plasmid pMR01-03. pRAW-like plasmids have been shown in many different NTMs, including M. kansasii, M. marinum, M. chimaera and M. avium. The extensive presence of these plasmids may also explain the distribution of the T7S system during the evolution of the mycobacterial genus (53).
Differences in macrophage gene expression after infection with the mycobacterial strains used were most evident at 3hpi which is likely reflective of the initial macrophage response to infection. In support of this, the comparative transcriptomic response of macrophages after infection with M. riyadhense or M. kansasii versus M. bovis BCG or M. tuberculosis pairs showed very similar signatures, suggesting that the bacterial cell wall determinants, key determinants of the initial interactions these bacterial pairs with host cell receptors are well conserved.
We observed an increased abundance of Ifnb1 mRNA levels in all of the four Mycobacterial species (log2FC, M. kansasii: 7.1; M. riyadhense:5.3; M. tuberclosis:8.2; M. bovis BCG:7.4) at 3 hours post infection. However, Ifnb1 expression levels were comparatively lower in the M. riyadhense/M. kansasii pair in comparison to the M. bovis BCG/M. tuberculosis pair. It has been shown that M. kansasii and M. fortuitum induce stronger IFN-β responses in comparison with M. tuberculosis after infection of bone marrow-derived dendritic cells (BMDCs) at MOI of 10: 1 (92), while this was not observed in infection of the RAW264.7 cells in our study. This could be due to differences in the experimental systems used, or a post-transcriptional regulatory mechanism of Ifnb1 transcripts that modulates protein levels. Type I IFNs can have either beneficial or detrimental consequences in bacterial infections (93). While generally promoting the growth of M. tuberculosis they may have an opposite effect on NTMs (94), whether they have a protective or detrimental role in M. riyadhense infection remains to be addressed.
The clinical presentation of our cases was by and large indistinguishable from disease caused by M. tuberculosis, as reported earlier (7), but with a negative M. tuberculosis PCR. Due to the relatively recent emergence of M. riyadhense as an important clinical pathogen coupled with its misdiagnosis as M. tuberculosis by commercially available kits, we developed an accurate set of diagnostic markers based on the genomic datasets generated in this study. The primer sets accurately detect M. riyadhense in a mixed cocktail of closely related mycobacteria and can hence serve as part of an accurate and fast diagnostic protocol in clinical settings thus reducing the need for strict isolation, laborious contact tracing and inappropriate use of TB antimicrobials. The diagnostic primers are of immediate utility in distinguishing between MTBC and M. riyadhense and provide a basis towards developing a set of primers that can be used in broader epidemiological surveys. Our work, and that of previous studies, was based on M. riyadhense human clinical isolates. As only two studies have found the presence of M. riyadhense in water and soil without confirmation by bacterial culture, its environmental reservoir remains largely unknown. We believe that systematic screening of relevant environmental samples with the MRDP established in this study may help to identify the natural habitat of this bacterium and hence the source of human infections.
In conclusion, our study provides new insights into the evolution of the MTBC relative to extant NTMs, unveiling the genomic shifts from NTMs to MTBC before the speciation of the common ancestor of MTBC. Our work provides the underpinning data to support the use of M. riyadhense as a novel mycobacterium for the study of evolution, virulence and pathogenesis in the MTBC.
DATA AVAILIBILITY
The M. riyadhense dataset is available at European Nucleotide Archive (ENA) under the study accession no. PRJEB32162. The derived datasets, including phylogenetic trees, M. tuberculosis T/A orthologs, protein orthologs comparison between 8 M. riyadhense strains etc. have been deposited in Zenodo (10.5281/zenodo.4434791).
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank members of the Bioscience Core Lab (BCL) of King Abdullah University of Science and Technology for sequencing the bacterial genomic DNAs on the Illumina HiSeq 4000 and PacBio RSII platforms.
Author Contributions: A.P. conceived the comparative genomics part of the study, obtained the funding and supervised the work; M.G. and A.H. initiated the clinical part of the study; F.A., M.G., T.A., S.F., A.H., M.A.R., A.R. and T.S. collected the microbiological and clinical information; and A.P., Q.G., C.D., S.G., A.B. and F.A. designed the experiments. Q.G. performed the data analysis and prepared the initial draft of the manuscript, followed by edits from A.P., S.G., M.G., S.F., A.B. and C.M. Q.G., S.M., A.Sm and J.B. performed the transcriptome experiment. A.Si and A.B. performed the TLC analysis. Q.G., S.F. and T.L. performed the infections experiment and extracted the RNA. C.N. and Y.S. provided materials for the diagnostic markers and intellectual advice. All authors have commented on various sections of the manuscript, which were finally curated and incorporated in the final version by Q.G. and A.P.
Notes
Present address: Musa Garbati, Infectious Diseases Unit, Department of Medicine, University of Maiduguri, Nigeria.
Present address: Anwar Hoosen, Pathcare/Vermaak & Partners Pathologists, Pretoria, South Africa.
Contributor Information
Qingtian Guan, Pathogen Genomics Laboratory, BESE Division, King Abdullah University of Science and Technology (KAUST), Thuwal-Jeddah, 23955, Saudi Arabia.
Musa Garbati, King Fahad Medical City (KFMC), Riyadh, 11525, Saudi Arabia.
Sara Mfarrej, Pathogen Genomics Laboratory, BESE Division, King Abdullah University of Science and Technology (KAUST), Thuwal-Jeddah, 23955, Saudi Arabia.
Talal AlMutairi, King Fahad Medical City (KFMC), Riyadh, 11525, Saudi Arabia.
Thomas Laval, Immunobiology of Infection Unit, Institut Pasteur, INSERM U1221, Paris, France; Université Paris Diderot, Sorbonne Paris Cité, 75205 CEDEX 13, Paris, France.
Albel Singh, Institute of Microbiology and Infection, School of Biosciences, University of Birmingham, B15 2TT, Edgbaston, Birmingham, UK.
Shamsudeen Fagbo, One Health Unit, Executive Directorate for Surveillance and Response, Saudi Center for Disease Prevention and Control, 13352, Riyadh, Saudi Arabia; Department of Public Health, Nigerian Institute of Medical Research, P.M.B. 2013, Yaba, Lagos, Nigeria.
Alicia Smyth, UCD School of Veterinary Medicine, University College Dublin, Dublin, D04 W6F6, Ireland.
John A Browne, UCD School of Agriculture and Food Science, University College Dublin, Dublin, D04 W6F6, Ireland.
Muhammad Amin urRahman, King Fahad Medical City (KFMC), Riyadh, 11525, Saudi Arabia.
Alya Alruwaili, King Fahad Medical City (KFMC), Riyadh, 11525, Saudi Arabia.
Anwar Hoosen, King Fahad Medical City (KFMC), Riyadh, 11525, Saudi Arabia.
Conor J Meehan, School of Chemistry and Biosciences, University of Bradford, Bradford, BD7 1AZ, UK.
Chie Nakajima, Global Institution for Collaborative Research and Education, Hokkaido University, Kita 20 Nishi 10, Kita-ku, Sapporo, Japan; Research Center for Zoonosis Control, Hokkaido University, Kita 20 Nishi 10, Kita-ku, Sapporo, Japan.
Yasuhiko Suzuki, Global Institution for Collaborative Research and Education, Hokkaido University, Kita 20 Nishi 10, Kita-ku, Sapporo, Japan; Research Center for Zoonosis Control, Hokkaido University, Kita 20 Nishi 10, Kita-ku, Sapporo, Japan.
Caroline Demangel, Immunobiology of Infection Unit, Institut Pasteur, INSERM U1221, Paris, France.
Apoorva Bhatt, Institute of Microbiology and Infection, School of Biosciences, University of Birmingham, B15 2TT, Edgbaston, Birmingham, UK.
Stephen V Gordon, UCD School of Veterinary Medicine, University College Dublin, Dublin, D04 W6F6, Ireland; Global Institution for Collaborative Research and Education, Hokkaido University, Kita 20 Nishi 10, Kita-ku, Sapporo, Japan.
Faisal AlAsmari, King Fahad Medical City (KFMC), Riyadh, 11525, Saudi Arabia.
Arnab Pain, Pathogen Genomics Laboratory, BESE Division, King Abdullah University of Science and Technology (KAUST), Thuwal-Jeddah, 23955, Saudi Arabia; Global Institution for Collaborative Research and Education, Hokkaido University, Kita 20 Nishi 10, Kita-ku, Sapporo, Japan.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
KAUST [BAS/1/1020-01- 01]; BBSRC [BB/N004574/1 to S.G., A.B.]; SFI [SFI/15/IA/3154 to S.G.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Falkinham J.O.Ecology of nontuberculous mycobacteria-where do human infections come from. Semin. Respir. Crit. Care Med. 2013; 34:95–102. [DOI] [PubMed] [Google Scholar]
- 2.King H.C., Khera-Butler T., James P., Oakley B.B., Erenso G., Aseffa A., Knight R., Wellington E.M., Courtenay O.. Environmental reservoirs of pathogenic mycobacteria across the Ethiopian biogeographical landscape. PLoS One. 2017; 12:e0173811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Narendrula-Kotha R., Nkongolo K.K.. Microbial response to soil liming of damaged ecosystems revealed by pyrosequencing and phospholipid fatty acid analyses. PLoS One. 2017; 12:e0168497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Ingen J., Al-Haijoj S.A.M., Boeree M., Al-Rabiah F., Enaimi M., de Zwaan R., Tortoli E., Dekhuijzen R., van Soolingen D. Mycobacterium riyadhense sp. nov., a non-tuberculous species identified as Mycobacterium tuberculosis complex by a commercial line-probe assay. Int. J. Syst. Evol. Microbiol. 2009; 59:1049–1053. [DOI] [PubMed] [Google Scholar]
- 5.Saad M.M., Alshukairi A.N., Qutub M.O., Elkhizzi N.A., Hilluru H.M., Omrani A.S.. Mycobacterium riyadhense infections. Saudi Med. J. 2015; 36:620–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alenazi T.H., Alanazi B.S., Alsaedy A., Khair A., Solomon R., Al Johani S.M.. Mycobacterium riyadhense as the opportunistic infection that lead to HIV diagnosis: A report of 2 cases and literature review. J. Infect. Public Health. 2019; 12:285–288. [DOI] [PubMed] [Google Scholar]
- 7.Choi J.I., Lim J.H., Kim S.R., Lee S.H., Park J.S., Seo K.W., Jeon J.B., Jeong J.. Lung infection caused by Mycobacterium riyadhense confused with Mycobacterium tuberculosis: The first case in Korea. Ann. Lab. Med. 2012; 32:298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varghese B., Enani M.A., Althawadi S., Johani S., Fernandez G.M., Al-Ghafli H., Al-Hajoj S.. Mycobacterium riyadhense in Saudi Arabia. Emerg Infect Dis. 2017; 23:1732–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godreuil S., Marchandin H., Michon A.L., Ponsada M., Chyderiotis G., Brisou P., Bhat A., Panteix G.. Mycobacterium riyadhense pulmonary infection, France and Bahrain. Emerg. Infect. Dis. 2012; 18:176–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van der Werf M.J., Ködmön C., Katalinić-Janković V., Kummik T., Soini H., Richter E., Papaventsis D., Tortoli E., Perrin M., van Soolingen D.et al.. Inventory study of non-tuberculous mycobacteria in the European Union. BMC Infect. Dis. 2014; 14:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murty D.S., Prasanthi K.. A brief review on ecology and evolution of mycobacteria. Mycobact. Dis. 2014; 4:4–6. [Google Scholar]
- 12.Sapriel G., Brosch R.. Shared pathogenomic patterns characterize a new phylotype, revealing transition towards host-adaptation long before speciation of Mycobacterium tuberculosis. Genome Biol. Evol. 2019; 11:2420–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belisle J.T., Brennan P.J.. Chemical basis of rough and smooth variation in mycobacteria. J. Bacteriol. 1989; 171:3465–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Der Woude A.D., Sarkar D., Bhatt A., Sparrius M., Raadsen S.A., Boon L., Geurtsen J., Van Der Sar A.M., Luirink J., Houben E.N.G.et al.. Unexpected link between lipooligosaccharide biosynthesis and surface protein release in Mycobacterium marinum. J. Biol. Chem. 2012; 287:20417–20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Vilanova A., Chan J., Torrelles J.B.. Underestimated manipulative roles of mycobacterium tuberculosis cell envelope glycolipids during infection. Front. Immunol. 2019; 10:2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdallah A.M., Gey van Pittius N.C., DiGiuseppe Champion P.A., Cox J., Luirink J., Vandenbroucke-Grauls C.M.J.E., Appelmelk B.J., Bitter W.. Type VII secretion - Mycobacteria show the way. Nat. Rev. Microbiol. 2007; 5:883–891. [DOI] [PubMed] [Google Scholar]
- 17.Stanley S.A., Raghavan S., Hwang W.W., Cox J.S.. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc. Natl. Acad. Sci. USA. 2003; 100:13001–13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houben E.N.G., Bestebroer J., Ummels R., Wilson L., Piersma S.R., Jiménez C.R., Ottenhoff T.H.M., Luirink J., Bitter W.. Composition of the type VII secretion system membrane complex. Mol. Microbiol. 2012; 86:472–484. [DOI] [PubMed] [Google Scholar]
- 19.Sreejit G., Ahmed A., Parveen N., Jha V., Valluri V.L., Ghosh S., Mukhopadhyay S.. The ESAT-6 Protein of Mycobacterium tuberculosis Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage. PLoS Pathog. 2014; 10:e1004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coros A., Callahan B., Battaglioli E., Derbyshire K.M.. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol. Microbiol. 2008; 69:794–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tinaztepe E., Wei J.R., Raynowska J., Portal-Celhay C., Thompson V., Philipsa J.A.. Role of metal-dependent regulation of ESX-3 secretion in intracellular survival of Mycobacterium tuberculosis. Infect. Immun. 2016; 84:2255–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tufariello J.A.M., Chapman J.R., Kerantzas C.A., Wong K.W., Vilchèze C., Jones C.M., Cole L.E., Tinaztepe E., Thompson V., Fenyö D.et al.. Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence. Proc. Natl. Acad. Sci. USA. 2016; 113:E348–E357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ates L.S., van der Woude A.D., Bestebroer J., van Stempvoort G., Musters R.J.P., Garcia-Vallejo J.J., Picavet D.I., van de Weerd R., Maletta M., Kuijl C.P.et al.. The ESX-5 system of pathogenic mycobacteria is involved in capsule integrity and virulence through its substrate PPE10. PLoS Pathog. 2016; 12:e1005696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akhter Y., Ehebauer M.T., Mukhopadhyay S., Hasnain S.E.. The PE/PPE multigene family codes for virulence factors and is a possible source of mycobacterial antigenic variation: Perhaps more?. Biochimie. 2012; 94:110–116. [DOI] [PubMed] [Google Scholar]
- 25.Fenn K., Wong C.T., Darbari V.C.. Mycobacterium tuberculosis uses mce proteins to interfere with host cell signaling. Front. Mol. Biosci. 2020; 6:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerdes K., Rasmussen P.B., Molin S.. Unique type of plasmid maintenance function: Postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. U. S. A. 1986; 83:3116–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slayden R.A., Dawson C.C., Cummings J.E.. Toxin-antitoxin systems and regulatory mechanisms in Mycobacterium tuberculosis. Pathog. Dis. 2018; 76: 10.1093/femspd/fty039. [DOI] [PubMed] [Google Scholar]
- 28.Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M.. Canu: Scalable and accurate long-read assembly via adaptive κ-mer weighting and repeat separation. Genome Res. 2017; 27:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S.K.et al.. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014; 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krumsiek J., Arnold R., Rattei T.. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007; 23:1026–1028. [DOI] [PubMed] [Google Scholar]
- 32.Seemann T.Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014; 30:2068–2069. [DOI] [PubMed] [Google Scholar]
- 33.Alikhan N.F., Petty N.K., Ben Zakour N.L., Beatson S.A.. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011; 12:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lechner M., Findeiß S., Steiner L., Marz M., Stadler P.F., Prohaska S.J.. Proteinortho: Detection of (Co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011; 12:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segata N., Börnigen D., Morgan X.C., Huttenhower C.. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 2013; 4:2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar R.C.MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004; 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price M.N., Dehal P.S., Arkin A.P.. FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS One. 2010; 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamatakis A.RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006; 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- 39.Li L., Stoeckert C.J., Roos D.S.. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003; 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gupta A., Venkataraman B., Vasudevan M., Gopinath Bankar K.. Co-expression network analysis of toxin-antitoxin loci in Mycobacterium tuberculosis reveals key modulators of cellular stress. Sci. Rep. 2017; 7:5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Broad Institute Picard tools. 2017; 27 March 2017, date last accessedhttps://broadinstitute.github.io/picard/.
- 43.Alkan C., Coe B.P., Eichler E.E.. GATK toolkit. Nat. Rev. Genet. 2011; 12:363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stamatakis A., Ludwig T., Meier H.. RAxML-III: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005; 21:456–463. [DOI] [PubMed] [Google Scholar]
- 45.Magee D.A., Conlon K.M., Nalpas N.C., Browne J.A., Pirson C., Healy C., McLoughlin K.E., Chen J., Vordermeier H.M., Gormley E.et al.. Innate cytokine profiling of bovine alveolar macrophages reveals commonalities and divergence in the response to Mycobacterium bovis and Mycobacterium tuberculosis infection. Tuberculosis. 2014; 94:441–450. [DOI] [PubMed] [Google Scholar]
- 46.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L.. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016; 11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anders S., Pyl P.T., Huber W.. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love M.I., Anders S., Huber W.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K.. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019; 10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobson G., Minnikin D.E., Minnikin S.M., Parlett M., Goodfellow M., Ridell M., Magnusson M.. Systematic analysis of complex mycobacterial lipids. Chemical Methods in Bacterial Systematics. 1985; London: Academic Press; 237–265. [Google Scholar]
- 51.Haubold B., Pierstorff N., Möller F., Wiehe T.. Genome comparison without alignment using shortest unique substrings. BMC Bioinformatics. 2005; 6:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park H., Jang H., Kim C., Chung B., Chang C.L., Park Soon Kew, Song S. Detection and identification of mycobacteria by amplification of the internal transcribed spacer regions with genus- and species-specific PCR primers. J. Clin. Microbiol. 2000; 38:4080–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ummels R., Abdallah A.M., Kuiper V., Aâjoud A., Sparrius M., Naeem R., Spaink H.P., Bitter W., Pain A., Bitter W.. Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. MBio. 2014; 5:e01744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uchiya K.I., Takahashi H., Nakagawa T., Yagi T., Moriyama M., Inagaki T., Ichikawa K., Nikai T., Ogawa K.. Characterization of a novel plasmid, pMAH135, from Mycobacterium avium subsp. hominissuis. PLoS One. 2015; 10:e0117797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.da Silva Rabello M.C., Matsumoto C.K., de Almeida L.G.P., Menendez M.C., de Oliveira R.S., Silva R.M., Garcia M.J., Leão S.C.. First description of natural and experimental conjugation between mycobacteria mediated by a linear plasmid. PLoS One. 2012; 7:e29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim B.-J., Kim B.-R., Lee S.-Y., Seok S.-H., Kook Y.-H., Kim B.-J.. Whole-Genome Sequence of a Novel Species, Mycobacterium yongonense DSM 45126 T. Genome Announc. 2013; 1:604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J., McIntosh F., Radomski N., Dewar K.. Insights on the emergence of Mycobacterium tuberculosis from the analysis of Mycobacterium kansasii. Genome Biol. Evol. 2015; 7:857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Ingen J., Kohl T.A., Kranzer K., Hasse B., Keller P.M., Katarzyna Szafrańska A., Hillemann D., Chand M., Schreiber P.W., Sommerstein R.et al.. Global outbreak of severe Mycobacterium chimaera disease after cardiac surgery: a molecular epidemiological study. Lancet Infect. Dis. 2017; 17:1033–1041. [DOI] [PubMed] [Google Scholar]
- 59.Surtees J.A., Funnell B.E.. Plasmid and chromosome traffic control: How ParA and ParB drive partition. Curr. Top. Dev. Biol. 2003; 56:145–180. [DOI] [PubMed] [Google Scholar]
- 60.Zrimec J., Lapanje A.. DNA structure at the plasmid origin-of-Transfer indicates its potential transfer range. Sci. Rep. 2018; 8:1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jia X., Yang L., Dong M., Chen S., Lv L., Cao D., Fu J., Yang T., Zhang J., Zhang X.et al.. The bioinformatics analysis of comparative genomics of mycobacterium tuberculosis complex (MTBC) provides insight into dissimilarities between intraspecific groups differing in host association, virulence, and epitope diversity. Front. Cell. Infect. Microbiol. 2017; 7:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Supply P., Marceau M., Mangenot S., Roche D., Rouanet C., Khanna V., Majlessi L., Criscuolo A., Tap J., Pawlik A.et al.. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat. Genet. 2013; 45:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahairas G.G., Sabo P.J., Hickey M.J., Singh D.C., Stover C.K.. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 1996; 178:1274–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gordon S. V., Brosch R., Billault A., Garnier T., Eiglmeier K., Cole S.T.. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 1999; 32:643–655. [DOI] [PubMed] [Google Scholar]
- 65.Van Ingen J., De Zwaan R., Dekhuijzen R., Boeree M., Van Soolingen D. Region of difference 1 in nontuberculous Mycobacterium species adds a phylogenetic and taxonomical character. J. Bacteriol. 2009; 191:5865–5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kozak R.A., Alexander D.C., Liao R., Sherman D.R., Behr M.A.. Region of difference 2 contributes to virulence of mycobacterium tuberculosis. Infect. Immun. 2011; 79:59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ru H., Liu X., Lin C., Yang J., Chen F., Sun R., Zhang L., Liu J.. The impact of genome region of difference 4 (RD4) on mycobacterial virulence and BCG efficacy. Front. Cell. Infect. Microbiol. 2017; 7:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brosch R., Gordon S. V., Marmiesse M., Brodin P., Buchrieser C., Eiglmeier K., Garnier T., Gutierrez C., Hewinson G., Kremer K.et al.. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. 2002; 99:3684–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duffy S.C., Srinivasan S., Schilling M.A., Stuber T., Danchuk S.N., Michael J.S., Venkatesan M., Bansal N.. Articles Reconsidering Mycobacterium bovis as a proxy for zoonotic tuberculosis : a molecular epidemiological surveillance study. The Lancet Microbe. 2019; 1:e66–e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marmiesse M., Brodin P., Buchrieser C., Gutierrez C., Simoes N., Vincent V., Glaser P., Cole S.T., Brosch R.. Macro-array and bioinformatic analyses reveal mycobacterial ‘core’ genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology. 2004; 150:483–496. [DOI] [PubMed] [Google Scholar]
- 71.Alexander K.A., Laver P.N., Michel A.L., Williams M., van Helden P.D., Warren R.M., van Pittius N.C.G.. Novel mycobacterium tuberculosis complex pathogen, M. Mungi. Emerg. Infect. Dis. 2010; 16:1296–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cristina G., Brisse S., Brosch R., Fabre M., Omaïs B., Marmiesse M., Supply P., Vincent V.. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 2005; 1:0055–0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tortoli E., Fedrizzi T., Meehan C.J., Trovato A., Grottola A., Giacobazzi E., Serpini G.F., Tagliazucchi S., Fabio A., Bettua C.et al.. The new phylogeny of the genus Mycobacterium: The old and the news. Infect. Genet. Evol. 2017; 56:19–25. [DOI] [PubMed] [Google Scholar]
- 74.Fedrizzi T., Meehan C.J., Grottola A., Giacobazzi E., Fregni Serpini G., Tagliazucchi S., Fabio A., Bettua C., Bertorelli R., De Sanctis V.et al.. Genomic characterization of Nontuberculous Mycobacteria. Sci. Rep. 2017; 7:45258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sapriel G., Brosch R.. Shared pathogenomic patterns characterize a new phylotype, revealing transition toward Host-Adaptation long before speciation of mycobacterium tuberculosis. Genome Biol. Evol. 2019; 7:45258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Delogu G., Brennan M.J.. Comparative immune response to PE and PE _ PGRS antigens of mycobacterium tuberculosis comparative immune response to PE and PE _ PGRS antigens of mycobacterium tuberculosis. Infect. Immun. 2001; 69:5606–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Isom G.L., Davies N.J., Chong Z.S., Bryant J.A., Jamshad M., Sharif M., Cunningham A.F., Knowles T.J., Chng S.S., Cole J.A.et al.. MCE domain proteins: Conserved inner membrane lipid-binding proteins required for outer membrane homeostasis. Sci. Rep. 2017; 7:8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dumas E., Boritsch E.C., Vandenbogaert M., De La Vega R.C.R., Thiberge J.M., Caro V., Gaillard J.L., Heym B., Girard-Misguich F., Brosch R.et al.. Mycobacterial pan-genome analysis suggests important role of plasmids in the radiation of type VII secretion systems. Genome Biol. Evol. 2016; 8:387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kansal R.G., Gomez-Flores R., Mehta R.T.. Change in colony morphology influences the virulence as well as the biochemical properties of the Mycobacterium avium complex. Microb. Pathog. 1998; 25:203–214. [DOI] [PubMed] [Google Scholar]
- 80.Catherinot E., Clarissou J., Etienne G., Ripoll F., Emile J.F., Daffé M., Perronne C., Soudais C., Gaillard J.L., Rottman M.. Hypervirulence of a rough variant of the Mycobacterium abscessus type strain. Infect. Immun. 2007; 75:1055–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boritsch E.C., Frigui W., Cascioferro A., Malaga W., Etienne G., Laval F., Pawlik A., Le Chevalier F., Orgeur M., Ma L.et al.. Pks5-recombination-mediated surface remodelling in Mycobacterium tuberculosis emergence. Nat. Microbiol. 2016; 1:15019. [DOI] [PubMed] [Google Scholar]
- 82.Cohen I., Parada C., Acosta-Gío E., Espitia C.. The PGRS domain from PE_PGRS33 of Mycobacterium tuberculosis is target of humoral immune response in mice and humans. Front. Immunol. 2014; 5:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boritsch E.C., Supply P., Honoré N., Seeman T., Stinear T.P., Brosch R.. A glimpse into the past and predictions for the future: The molecular evolution of the tuberculosis agent. Mol. Microbiol. 2014; 93:835–852. [DOI] [PubMed] [Google Scholar]
- 84.Houben E.N.G., Korotkov K. V., Bitter W.. Take five - Type VII secretion systems of Mycobacteria. Biochim. Biophys. Acta - Mol. Cell Res. 2014; 1843:1707–1716. [DOI] [PubMed] [Google Scholar]
- 85.Castrillo A., Joseph S.B., Vaidya S.A., Haberland M., Fogelman A.M., Cheng G., Tontonoz P.. Crosstalk between LXR and Toll-like receptor signaling mediates bacterial and viral antagonism of cholesterol metabolism. Mol. Cell. 2003; 12:805–816. [DOI] [PubMed] [Google Scholar]
- 86.Griffith D.E., Aksamit T., Brown-Elliott B. a., Catanzaro A., Daley C., Gordin F., Holland S.M., Horsburgh R., Huitt G., Iademarco M.F.et al.. An Official ATS/IDSA Statement: Diagnosis, Treatment, and Prevention of Nontuberculous Mycobacterial Diseases. Am. J. Respir. Crit. Care Med. 2007; 175:367–416. [DOI] [PubMed] [Google Scholar]
- 87.Leon P., Virginia W., Patricia B.. Molecular analysis of the linear 2.3 kb plasmid of maize mitochondria: Apparent capture of tRNA genes. Nucleic Acids Res. 1989; 17:4089–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Picardeau M., Vincent V.. Characterization of large linear plasmids in mycobacteria. J. Bacteriol. 1997; 179:2753–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stinear T.P., Seemann T., Harrison P.F., Jenkin G., Davies J.K., Johnson P.D.R., Abdellah Z., Arrowsmith C., Chillingworth T., Churcher C.et al.. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008; 18:729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Becq J., Gutierrez M.C., Rosas-Magallanes V., Rauzier J., Gicquel B., Neyrolles O., Deschavanne P.. Contribution of horizontally acquired genomic islands to the evolution of the tubercle bacilli. Mol. Biol. Evol. 2007; 24:1861–1871. [DOI] [PubMed] [Google Scholar]
- 91.Bachmann N.L., Salamzade R., Manson A.L., Whittington R., Sintchenko V., Earl A.M., Marais B.J.. Key transitions in the evolution of rapid and slow growing mycobacteria identified by comparative genomics. Front. Microbiol. 2020; 10:3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shah S., Bohsali A., Ahlbrand S.E., Srinivasan L., Rathinam V.A.K., Vogel S.N., Fitzgerald K.A., Sutterwala F.S., Briken V.. Cutting Edge: Mycobacterium tuberculosis but not nonvirulent mycobacteria inhibits IFN-β and AIM2 Inflammasome–Dependent IL-1β production via its ESX-1 secretion system. J. Immunol. 2013; 191:3514–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McNab F., Mayer-Barber K., Sher A., Wack A., O’Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015; 15:87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ruangkiattikul N., Nerlich A., Abdissa K., Lienenklaus S., Suwandi A., Janze N., Laarmann K., Spanier J., Kalinke U., Weiss S.et al.. cGAS-STING-TBK1-IRF3/7 induced interferon-β contributes to the clearing of non tuberculous mycobacterial infection in mice. Virulence. 2017; 8:1303–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.