Abstract
Repetitive transcranial magnetic stimulation (rTMS) of the dorsolateral prefrontal cortex (DLPFC) is an established treatment for refractory depression, however, therapeutic outcomes vary. Mounting evidence suggests that clinical response relates to functional connectivity with the subgenual cingulate cortex (SGC) at the precise DLPFC stimulation site. Critically, SGC‐related network architecture shows considerable interindividual variation across the spatial extent of the DLPFC, indicating that connectivity‐based target personalization could potentially be necessary to improve treatment outcomes. However, to date accurate personalization has not appeared feasible, with recent work indicating that the intraindividual reproducibility of optimal targets is limited to 3.5 cm. Here we developed reliable and accurate methodologies to compute individualized connectivity‐guided stimulation targets. In resting‐state functional MRI scans acquired across 1,000 healthy adults, we demonstrate that, using this approach, personalized targets can be reliably and robustly pinpointed, with a median accuracy of ~2 mm between scans repeated across separate days. These targets remained highly stable, even after 1 year, with a median intraindividual distance between coordinates of only 2.7 mm. Interindividual spatial variation in personalized targets exceeded intraindividual variation by a factor of up to 6.85, suggesting that personalized targets did not trivially converge to a group‐average site. Moreover, personalized targets were heritable, suggesting that connectivity‐guided rTMS personalization is stable over time and under genetic control. This computational framework provides capacity for personalized connectivity‐guided TMS targets to be robustly computed with high precision and has the flexibly to advance research in other basic research and clinical applications.
Keywords: connectivity, depression, neuroimaging, personalization, precision psychiatry, transcranial magnetic stimulation
Transcranial magnetic stimulation (TMS) provides an important therapeutic option for treatment resistant depression. Prior research demonstrates that clinical outcomes to TMS could likely be enhanced by personalized treatment that is targeted to specific brain connections. Here we designed innovative methodology which enables these connections to be identified and targeted using TMS at a person‐specific level with unprecedented precision.

1. INTRODUCTION
Major depressive disorder (MDD) is the leading cause of years lived with disability and remains poorly understood. As few as 30% of patients achieve remission with initial treatment, even after 8–12 months (Rush, 2007). Repetitive transcranial magnetic stimulation (rTMS) at the dorsolateral prefrontal cortex (DLPFC) has emerged as a noninvasive antidepressant approach for individuals who do not respond to first‐line behavioral and pharmacological therapies. While rTMS is life‐changing for some individuals, approximately 50% with similar clinical profiles will not meet response criteria (Berlim, van den Eynde, Tovar‐Perdomo, & Daskalakis, 2014; Chen et al., 2013).
The heterogeneity of rTMS treatment outcomes is thought to be driven in part by suboptimal targeting of the DLPFC (Cash, Weigand, et al., 2020; Cash et al., 2019; Downar & Daskalakis, 2013; Fitzgerald et al., 2009; Fox, Buckner, White, Greicius, & Pascual‐Leone, 2012; Weigand et al., 2018). Determining the optimal site for therapeutic rTMS in the treatment of MDD remains a major goal in psychiatry, as it holds the promise of delivering improved and more consistent clinical outcomes. Current scalp‐based measures typically select a stimulation site that is 5–6 cm anterior to the motor cortex, but this approach is imprecise and yields a target outside the DLPFC in some individuals (Herwig, Padberg, Unger, Spitzer, & Schonfeldt‐Lecuona, 2001), unintentionally stimulating other brain networks (Opitz, Fox, Craddock, Colcombe, & Milham, 2016; Rastogi et al., 2017). While sophisticated targeting heuristics are available (e.g., “Beam F3”, Beam, Borckardt, Reeves, & George, 2009), the optimal stimulation site within the relatively large expanse of the DLPFC remains unknown.
Mounting evidence indicates that the clinical effects of brain stimulation are related, if not dictated, by the connectivity of the target site, and that this finding transcends different stimulation modalities (Cash, Cocchi, Lv, Fitzgerald, & Zalesky, 2020; Cash, Weigand, et al., 2020; Cocchi & Zalesky, 2018; Fox et al., 2014; Halko, Farzan, Eldaief, Schmahmann, & Pascual‐Leone, 2014). This represents a broader transition from a view that psychiatric disorders, and clinical outcomes, are dictated by focal abnormalities in neural activity, to recognition that these are related to broader disturbances, and normalization, of distributed network connectivity. Multiple retrospective studies, replicated internationally across different clinical cohorts and targeting methodologies have demonstrated that antidepressant outcomes to TMS were better when stimulation was by chance delivered at sites of the DLPFC that displayed stronger negative (“anticorrelated”) functional connectivity (FC) with the subgenual cingulate cortex (SGC) (Cash et al., 2019; Fox et al., 2012; Weigand et al., 2018). This presents a potential opportunity to enhance antidepressant outcomes by prospectively delivering TMS according to functional connectivity with the SGC. However, prefrontal regions including the DLPFC show the highest levels of interindividual variation in terms of cytoarchitecture, structural morphology, neural function, and connectivity (Doucet, Lee, & Frangou, 2019; Finn et al., 2015; Fischl et al., 2008; Fox, Liu, & Pascual‐Leone, 2013; Hill et al., 2010; Miranda‐Dominguez et al., 2014; Mueller et al., 2013; Rajkowska & Goldman‐Rakic, 1995). While targeting a single DLPFC coordinate for all individuals based on group‐average connectivity patterns has been proposed and tested (Blumberger et al., 2018; Fox et al., 2012; Li, Cheng, et al., 2019), this “one‐site‐fits‐all” approach does not account for interindividual differences in network architecture. Instead, individual‐specific connectivity‐guided targeting may be necessary to realize the full clinical potential of rTMS (Cash, Cocchi, et al., 2020).
The principal challenge for the implementation of connectivity‐guided treatment personalization is that individual functional connectivity measures are noisy (Dubois & Adolphs, 2016), particularly within subcortical regions such as the SGC (Ojemann et al., 1997). While individual differences in functional network organization have recently been illustrated, these were based on several hours of scan data (Gordon, Laumann, Adeyemo, et al., 2017; Gordon, Laumann, Gilmore, et al., 2017; Gratton et al., 2018). Moreover, demonstrating robust interindividual variation does not imply that it is actually feasible to pinpoint stable and reproducible personalized connectivity‐guided treatment coordinates within a clinically feasible acquisition time. Indeed, recent work indicates that the margin of error for defining connectivity‐based DLPFC targets is several centimeters, rendering treatment personalization effectively impossible with conventional methodology (Ning, Makris, Camprodon, & Rathi, 2019). Consequently, conventional scalp‐based targeting methodologies remain the most prevalent approach in clinical practice despite their limitations (Cash, Weigand, et al., 2020).
On this basis, we aimed to develop a computational framework to enable connectivity‐guided personalization of rTMS treatment. We specifically aimed to evaluate and enhance the feasibility, reliability, and precision of personalization methodology. We further aimed to characterize the extent of interindividual variation in treatment targets and whether this variation was mediated by familial factors. We demonstrate that using the proposed methodology, it is feasible to pinpoint personalized connectivity‐guided targets that remain highly reproducible across repeat scans with a median accuracy equivalent to a single voxel (~2 mm), given a sufficiently long acquisition. This represents a significant advance toward the feasible implementation of personalized and connectivity‐guided precision TMS.
2. METHODS AND MATERIALS
2.1. Participants
rfMRI images from 1,000 healthy adults participating in the Human Connectome Project (HCP, 532 Female, age 22–35) were analyzed (Glasser et al., 2013; Smith et al., 2013). HCP datasets are available for download to anyone agreeing to the open access data use terms (https://db.humanconnectome.org/).
2.2. Neuroimaging acquisition
Minimally preprocessed rfMRI images were sourced from the HCP. Acquisition and image preprocessing procedures were described in detail elsewhere (Glasser et al., 2013). Two fMRI scans per day were acquired on consecutive days for each individual (four scans per participant). In addition, 45 of these individuals underwent scanning 365 days after the initial scan. Data collection was performed in a customized Siemens Skyra 3T scanner according to the following parameters: gradient‐echo EPI sequence, 720 ms TR, 33.1 ms TE, 52° flip angle, 208 × 180 mm FOV, 104 × 90 matrix, 2.0 mm slice thickness, 72 slices, 2.0 mm isotropic voxels, 8 multiband factor and 0.58 ms echo spacing. Each of the two data acquisition sessions comprised two 14m33s runs (right‐to‐left and left‐to‐right phase encodings, 1,200 volumes each), with eyes open with relaxed fixation on a projected bright cross‐hair on a dark background (and presented in a darkened room). For each session, the two 14m33s runs per day were temporally concatenated to result in 29 min of data. To minimize temporal discontinuity, the mean was removed from each time series prior to concatenation.
2.3. Neuroimaging preprocessing
Acquired images were preprocessed by the HCP according to the HCP functional preprocessing pipeline, which involves: (1) spatial and gradient distortion corrections, (2) correction of head movement, (3) intensity normalization, (4) single spline resampling of EPI frames into 2 mm isotropic MNI space and (5) HCP's FIX+ICA pipeline for the removal of temporal artifacts. Refer to Glasser et al. (2013) and Smith et al., (2013) for further details on HCP resting‐state functional MRI acquisition and preprocessing.
In addition to the HCP minimal preprocessing, global signal regression (GSR) was implemented prior to band pass temporal filtering (BPTF; 0.01–0.1 Hz). GSR is a denoising strategy that remains controversial (Glasser et al., 2018; Li, Bolt, et al., 2019), and where the choice of implementation should be considered depending on study aims (Murphy & Fox, 2017). GSR was implemented here as the aim of the present work was to generate a pipeline which enables prospective fMRI personalization in a way that directly links to existing evidence. All prior work in relation to DLPFC‐SGC FC and rTMS antidepressant outcome has implemented GSR (Cash, Cocchi, et al., 2020; Cash et al., 2019; Fox et al., 2012; Weigand et al., 2018), and this step has been proposed for optimal evaluation of anti‐correlated connectivity patterns (Fox, Zhang, Snyder, & Raichle, 2009) and brain‐behavior relationships (Li, Kong, et al., 2019). Moreover, GSR has been proposed as the single most efficacious strategy for de‐noising (Ciric et al., 2017). The effect of head motion artifacts is mitigated via implementation of ICA‐FIX and GSR, but this point was not specifically investigated in the present work.
After initial exploration of the effect of smoothing (full width half maximum [FWHM] kernel sizes 4, 5, 6, 8, 10, 12, 16, 20 mm), all analyses, unless otherwise reported, were performed on minimally smoothed data (4 mm FWHM) to reduce loss of spatial information and spurious shifts in boundaries between gray and white matter (Coalson, Van Essen, & Glasser, 2018).
2.4. Computation of personalized DLPFC treatment coordinates
2.4.1. Overview
For each individual, we computed the optimal scan coordinate for two separate scan sessions, operationalized as the precise DLPFC coordinate at which SGC connectivity was most anti‐correlated (Fox et al., 2012). We developed new methodologies to improve the accuracy and robustness of this basic targeting strategy (Figure 1). We developed new assessment criteria to evaluate the reproducibility, precision and functional fidelity of each approach across successive scans (Table 1). We also assessed the extent to which individual heterogeneity was preserved by each method. Lastly, we determined the optimal computational parameters for each method.
FIGURE 1.
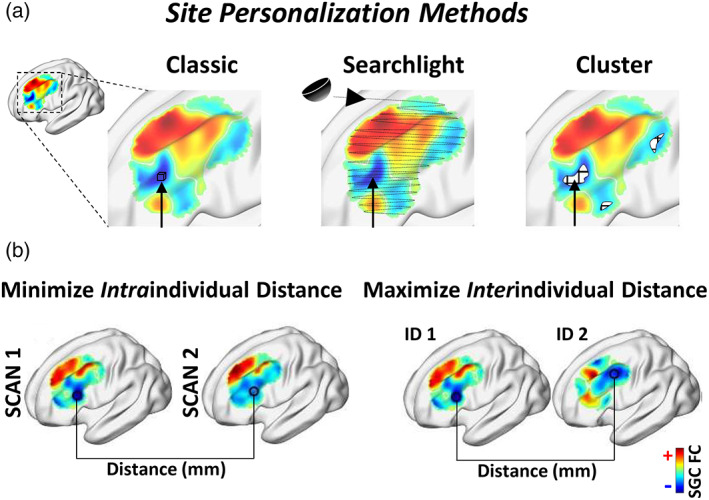
Schematic of experimental design, including personalization methodologies and evaluation metrics. (a) Illustration of the “classic”, “searchlight” and “cluster” based approaches for identifying the personalized stimulation target. The 'classic' method involves selecting the single most anticorrelated voxel within the DLPFC. The 'searchlight' method involves computing SGC FC within half‐spheres centerd at each voxel within the DLPFC. These half‐spheres are weighted by their proximity to the cortical surface and the most anticorrelated site is selected. The cluster approach involves retaining only a specified portion (between 0.1 and 50%) of the most negative voxels; these are then spatially clustered and the center‐of‐gravity of the largest cluster is defined as the target coordinate. (b) Measures quantifying the reliability of each personalization methodology. Intraindividual distance refers to the Euclidean distance between the target coordinate identified using two separate rfMRI scans from the same individual. Interindividual distance is defined as the distance between target coordinates from distinct individuals. Ideally, personalized targets should show high intraindividual precision while retaining a high degree of interindividual variation
TABLE 1.
Outline of key terms for TMS personalization
| Term | Description | Research question | Answer | |
|---|---|---|---|---|
| Metrics | SGC FC reproducibility | Correlation of FC values across all voxels within the DLPFC for each individual across resting‐state fMRI sessions 1 and 2. For personalization to be viable, SGC FC needs to be consistent across sessions on different days. | Are SGC connectivity maps reproducible across sessions? | Highly reproducible (R = 0.94) given sufficient data quantity (acquisition time ~15–25 min, multiband sequence), quality and methodology. |
| Intraindividual distance | Distance between optimal coordinates from data acquired in the same individual on different days. Ideally this distance will be low, indicating high reproducibility. | How reproducible are connectivity‐guided targets within individuals over time? | Targets can be reproduced with a median variation of only 2.2 ± 0.4 mm between scans, subject to the factors noted above and using the cluster‐seedmap method. | |
| Interindividual distance | Distance between personalized coordinates across different individuals. Higher values indicate better preservation of individual differences or reduced accuracy. | How much variation is there across individuals? Is personalization justified on this basis? | Targets scatter broadly across the spatial extent of the DLPFC. The median interindividual distance was between 16 and 27 mm depending on methodology. | |
| Ratio of interindividual‐to‐intraindividual distance | Ratio between the variation across individuals to the variation within individuals. Ideally this ratio will be high, reflecting high interindividual distances (i.e., preservation of individual differences), and low intraindividual distances (i.e., high reliability). | Which method best preserves individual differences whilst also providing high intraindividual reproducibility. | This ratio was highest for the combined cluster and seedmap methodology. This ratio improves as acquisition time increases. | |
| Intrascan FC | The SGC FC value at the optimal DLPFC coordinate within a single neuroimaging session. This value should be negative as methods were designed to identify the site of maximal anticorrelated (i.e., negative) FC with DLPFC. Values should remain negative during the scan session. | Does a selected target maintain its functional fidelity (i.e., negative SGC‐FC) during the scan session? | All methods identified coordinates that remained negative throughout the scan. | |
| Interscan FC | This metric assesses whether the target from one scan retains its functional fidelity (negative SGC FC) in a scan performed on a separate day. | Will a selected target maintain its functional fidelity (negative SGC FC) over time? | Using appropriate methodology, the target identified in one session displayed negative SGC FC in 100% of individuals in a second scan. | |
| Other key terms | Seedmap method | A method to increase the signal‐to‐noise ratio of subcortical structures such as the SGC. | Are the SGC FC maps derived from seed and seedmap methodologies genuinely comparable and how does the seedmap approach impact the above metrics? | Seedmap derived SGC FC maps faithfully reflected seed‐based maps. The seedmap method improved several measures and did not result in homogenization of target sites. |
| Cluster threshold | DLPFC voxels are ranked in order of decreasing values of negative SGC FC. According to the cluster threshold, only the top 0.1 to 50% of most negative voxels are retained, before clustering. Spatial precision decreased as cluster threshold and the resultant cluster size increased (SI results). | What is the optimal cluster threshold? | The optimal threshold depends on data quality and methodology. The optimal threshold was 10% and 0.5% respectively for seed and seedmap based methods. | |
| Smoothing | Spatial smoothing involves running a small Gaussian kernel across the image to average the intensities of neighboring voxels. Spatial smoothing aims to reduce random noise from individual voxels, while retaining real signal, thereby improving the signal‐to‐noise ratio. | Does heavy smoothing facilitate the identification of optimal personalized target sites, as previously proposed? | No, excessive smoothing introduces a detrimental loss of spatial information and specificity. Minimal smoothing is preferable. |
2.4.2. DLPFC region of interest (ROI)
The DLPFC was defined as the combined extent of 20 mm radius spheres centered along the left hemisphere at BA9 (MNI −36, 39, 43), BA46 (MNI −44, 40, 29), the “5 cm” TMS site (MNI −41, 16, 54) (Fox et al., 2013), and the F3 Beam group‐average stimulation site (MNI −37, 26, 49) (Cash et al., 2019).
2.4.3. Computation of SGC time series
Conventional seed‐based approach: The SGC time series was determined separately for each individual by spatially averaging the rfMRI data across all voxels comprising a spherical mask of radius 10 mm positioned at the center of the SGC (MNI 6, 16, −10). The center of this ROI represents the average coordinates of treatment‐related decreases in SGC activity tied to antidepressant effect (Fox et al., 2012).
Seedmap approach: To increase the signal to noise ratio, the SGC time series was computed as a weighted spatial average of the fMRI data across all gray matter voxels (Fox et al., 2013), excluding the DLPFC. Gray matter voxels were weighted according to their group‐averaged connectivity with the SGC time series. The single group‐average connectivity map, which was used to weight each voxel's contribution to the SGC time series, was derived from 2000 scans (1,000 individuals × 2 scans). The signal to noise ratio improved because data from most of gray matter was used to estimate the SGC time series, as opposed to the approximately 20 voxels comprising the SGC.
2.4.4. Computation of personalized DLPFC treatment target
We investigated three methodologies to determine the individualized DLPFC stimulation target. An individualized site was separately computed for each of two rfMRI sessions to evaluate intersession reliability.
“Classic” method (Ning et al., 2019): The coordinate corresponding to the location of the single most anticorrelated voxel within the DLPFC ROI was selected (Figure 1a).
“Searchlight” method (Fox et al., 2012): Connectivity was computed for half‐spheres positioned at the surface of the DLPFC (Figure 1a). Half spheres were centered at each voxel of the DLPFC. The DLPFC time series for each individual was determined by spatially averaging the rfMRI data across all voxels comprising the half sphere. The target was selected as the center of the sphere at which connectivity was most anti‐correlated with the SGC. Each half sphere was weighted by proximity to the cortical surface, to mimic the linear decay of the TMS field (Fox et al., 2013). The influence of sphere dimension was characterized at diameters of 1, 3, 6, 12 and 20 mm.
New “cluster‐based” method: Contiguous clusters of DLPFC voxels at which connectivity was most anti‐correlated with the SGC were identified (Figure 1a). The center‐of‐gravity of the largest such cluster was defined as the target coordinate. Clusters were delineated among the top x% most anti‐correlated DLPFC voxels, where the x threshold was incrementally varied between 0.1 and 50%. Clusters were defined among the supra‐threshold voxels based on standard 26‐voxel neighborhoods.
2.5. Evaluation of each personalization methodology
We first determined the optimal level of smoothing, as recent work indicated that larger smoothing kernels (kernel diameter >12 mm) yield higher intersession reproducibility (Ning et al., 2019). We generated SGC FC maps using the seed and seedmap approaches at FWHM kernel sizes of 4, 5, 6, 8, 10, 12, 16 and 20 mm. All subsequent analyses focused on minimally smoothed data (4 mm FWHM).
We next evaluated the reproducibility, precision, interindividual heterogeneity and functional fidelity of each approach. These performance criteria are elucidated in Table 1.
The Pearson correlation coefficient was computed for SGC connectivity values across all DLPFC voxels between MRI sessions for each individual to examine the overall reproducibility of SGC connectivity maps over time. The reproducibility of SGC connectivity maps was compared between the two methods used to compute the SGC time series (i.e., conventional seed and seedmap methodologies).
To evaluate the precision of site personalization methodologies, we evaluated two criteria (Table 1): First, the optimal site should be reliably identifiable in rfMRI data acquired in the same individual on different days. Therefore, the distance between optimal coordinates identified using two separate rfMRI scans from the same individual should be minimal. We referred to this as the intraindividual distance. Second, if personalized targeting is justified and does not simply recapitulate a fixed group‐average target, the optimal site should exhibit spatial variation between individuals. Therefore, the distance between optimal coordinates from pairs of distinct individuals should be larger than the targeting specificity of TMS (Figure 1b). We refer to this as the interindividual distance. We thus evaluated the site computed from each personalization methodology based on the ratio of interindividual‐to‐intraindividual distance. At one extreme, a group‐level coordinate (i.e., no personalization) would yield an interindividual distance of zero, and thus a ratio of zero. At the other extreme, simple, but potentially unreliable, personalization methodologies based on selection of the single DLPFC voxel that is most anticorrelated with the SGC would yield a relatively high intraindividual distance, and thus also yield a low ratio. A robust personalization methodology should yield low intraindividual distances (i.e., high reliability) and comparatively high interindividual distances (i.e., preservation of individual differences), resulting in a high ratio.
Individualization is sensible only if identified targets reliably maintain their functional fidelity over time. To this end, we computed the percentage of individuals for whom the DLPFC target located in one scan was a site of negative SGC‐FC in a different scan. We also computed intrascan and interscan functional connectivity as follows. After determining the optimal stimulation target for Session 1 and 2, we computed the SGC connectivity values at these coordinates within the same scan (intrascan) and in a separate scan (interscan). Session 1 and 2 values were averaged.
2.6. Genetic determinants of personalized targets
The contribution of familial relationships to the localization of personalized targets was investigated. Specifically, the interindividual distance between personalized coordinates was computed between twin pairs, sibling pairs, and 1,000 pairs of unrelated individuals selected at random. The primary statistical comparison of interest was whether the proximity between monozygotic (MZ) twin pairs was less than for dizygotic (DZ) twin pairs, as this contrast indicates a genetic influence above and beyond environmental factors.
The dataset comprised MZ twins (133 pairs), DZ twins (70 pairs), non‐twin siblings (180 pairs) and unrelated individuals (n = 429). These individuals were defined as MZ and DZ twins, non‐twin siblings and unrelated individuals based primarily on genotype and familial information provided in the HCP dataset. For individuals where genotype information was unavailable or undefined, status was defined based on self‐report and family coding.
To derive an unbiased result, median interindividual distance data were computed after first concatenating distance values across classic, cluster and searchlight methods in combination with the seedmap approach. Statistical comparisons were also computed for the interindividual distance of MZ twin pairs compared to non‐twin siblings and unrelated individuals, using one‐tailed tests, on the a priori assumption that MZ twin pairs would have more similar target coordinates due to their shared genetic and environmental influences, if personalized targets are non‐random.
3. RESULTS
3.1. Development and evaluation of personalization methodologies
3.1.1. Overview
The topography of SGC functional connectivity across the DLPFC was consistent within individuals across separate days but highly variable between individuals (Figure 2). Computational methodologies varied widely in their capacity to pinpoint reproducible personalized connectivity‐guided targets: conventional methods did not perform better than selecting two points at random within the DLPFC; by comparison the new methodology described here enabled individualized connectivity‐guided stimulation targets to be computed with millimeter precision.
FIGURE 2.
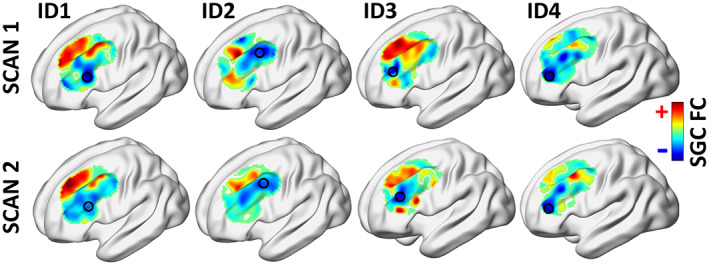
Functional connectivity between subgenual cingulate cortex and dorsolateral prefrontal cortex (DLPFC) displayed across the spatial extent of DLPFC for four representative individuals. Personalized target sites (circled) were computed based on the cluster and seedmap method. Target sites are highly variable between individuals but are consistent within individuals across separate days. Red and blue denote DLPFC regions of positive or negative SGC functional connectivity, respectively
3.1.2. Detrimental effect of smoothing
Initial exploration of the effect of smoothing (full width half maximum [FWHM] kernel sizes 4, 5, 6, 8, 10, 12, 16, 20 mm), demonstrated a loss of spatial information and specificity in the SGC connectivity map as FWHM kernel size was increased from 4 to 20 mm that would be detrimental to target‐site personalization (Figure 3). Therefore, subsequent analyses, unless otherwise specified, were performed on minimally smoothed data (4 mm FWHM).
FIGURE 3.
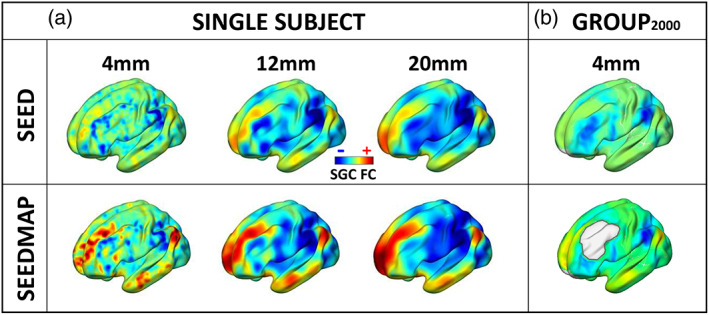
Influence of smoothing and comparison of SGC FC maps generated using seed and seedmap methodologies. (a) SGC FC maps are shown for a single representative participant, computed using either a conventional seed‐based approach (top row) or seedmap methodology (bottom row). The seedmap method generates a faithful representation of the conventional seed‐based SGC FC map. Finer details of the SGC FC map become evident when the seedmap method is utilized, likely because this enhances the signal‐to‐noise ratio of the SGC. Moving left to right, increasing the width of smoothing kernels (FWHM 4–20 mm) results in a pronounced loss of spatial information. (b) The group‐average SGC FC map (top row) derived from 2,000 brain scans (session 1 and 2 from 1,000 individuals). The seedmap method can be used to compute an individual's SGC time series as a weighted spatial average of the fMRI data across all gray matter voxels excluding the DLPFC
3.1.3. Intraindividual distance
Intraindividual (Session 1 vs. Session 2) distance between coordinates was lowest (i.e., interscan reproducibility was highest) when using the combination of cluster and seedmap methodologies (2.2 ± 0.4 mm) (Figure 4a–c, Table 2), and decreased (i.e., improved) with increasing acquisition time (SI Figure S1a). Intraindividual seed‐derived distances were halved (i.e., reproducibility doubled) when utilizing the cluster method, relative to classic and searchlight methods. Critically, the intraindividual distance for classic and searchlight seed‐based methods was in the range that would be obtained when selecting two coordinates at random within the DLPFC (>18 mm). The intraindividual distance of personalized targets between sessions was further reduced from 10 to 26 mm for the conventional seed approach, to the range of 2–6 mm using the seedmap method. The cluster method reduced the range of intraindividual distances between sessions across the 1,000 individuals, indicating substantially greater intersession consistency with this method (Figure 4c, SI Figure S3).
FIGURE 4.

Precision of rTMS personalization. Intraindividual distances between personalized targets (illustrated in the inset, a) are displayed for different methodologies and acquisition times of (a) 14 and (b) 28 min, that is, half and full scan duration (T14, T28). Overall, individual target site coordinates were most reproducible when using the combination of cluster and seedmap methodologies. Notably, when generating the SGC FC map using a conventional seed approach, the classic and searchlight methods did not perform better than selecting two points at random within the DLPFC, even at an acquisition time of 28 min. The intraindividual distance was approximately halved using the cluster approach. The horizontal line reflects the average distance obtained if two points are selected at random within the DLPFC (n = 1,000). The lower edge of gray shading represents the lower bound of the 95% confidence interval when two points are selected at random within the DLPFC (denoted as “Chance”). The red arrow on the y‐axis indicates the most recent benchmark for intraindividual accuracy (3.5 cm) for pinpointing individualized targets across two successive scans (Ning et al., 2019). Intraindividual distance was further reduced to the range of millimeters using the seedmap approach, and was again lowest for the cluster‐based method. (c) Beyond the quantitative reduction in median intraindividual distance (shown in a and b), the consistency of accurately identifying reproducible targets across the population was substantially enhanced when using the combined cluster and seedmap methodologies. This is illustrated here by the tighter distribution, lower median and much lower maximum intraindividual distance between scans. Other methods are shown to generate highly divergent targets between repeat scans for some individuals. (d, e) Ratio between interindividual and intraindividual distance. This ratio provides a summary measure of the capacity to identify unique individual targets (interindividual distance) while also reproducibly identifying each target with high intraindividual precision (intraindividual variation). This ratio reached a maximum of 6.85 for the cluster combined with seedmap approach
TABLE 2.
Quantitative assessment of personalization strategies
| Methodology | Reproducibility of SGC FC maps | Intraindividual distance [mm] | Interindividual distance [mm] | Ratio of interindividual‐to‐intraindividual distance | Intrasession FC | Intersession FC | Intersession FC (percent) | |
|---|---|---|---|---|---|---|---|---|
| Seed | T14 | 0.205 ± 0.008 | ||||||
| Classic | 24.454 ± 0.446 | 26.077 ± 0.438 | 1.07 ± 0.03 | −0.137 ± 0.001 | −0.03 ± 0.002 | 74.00 | ||
| Searchlight | 25.612 ± 0.513 | 26.907 ± 0.446 | 1.05 ± 0.04 | −0.085 ± 0.001 | −0.031 ± 0.001 | 80.16 | ||
| Cluster | 14.253 ± 0.382 | 17.251 ± 0.351 | 1.21 ± 0.05 | −0.08 ± 0.001 | −0.028 ± 0.001 | 84.40 | ||
| T28 | 0.379 ± 0.008 | |||||||
| Classic | 22.361 ± 0.476 | 23.58 ± 0.426 | 1.05 ± 0.04 | −0.11 ± 0.001 | −0.044 ± 0.002 | 85.20 | ||
| Searchlight | 20.785 ± 0.506 | 23.065 ± 0.444 | 1.11 ± 0.04 | −0.072 ± 0.001 | −0.039 ± 0.001 | 88.73 | ||
| Cluster | 10.61 ± 0.373 | 14.111 ± 0.359 | 1.33 ± 0.06 | −0.066 ± 0.001 | −0.034 ± 0.001 | 92.4 | ||
| Seedmap | T14 | 0.895 ± 0.002 | ||||||
| Classic | 6.325 ± 0.442 | 16.971 ± 0.418 | 2.68 ± 0.1 | −0.629 ± 0.002 | −0.565 ± 0.003 | 100.00 | ||
| Searchlight | 4.472 ± 0.408 | 16.125 ± 0.415 | 3.6 ± 0.14 | −0.528 ± 0.003 | −0.479 ± 0.004 | 99.85 | ||
| Cluster | 3.928 ± 0.387 | 14.908 ± 0.360 | 3.8 ± 0.12 | −0.578 ± 0.002 | −0.527 ± 0.003 | 100.00 | ||
| T28 | 0.941 ± 0.001 | |||||||
| Classic | 3.464 ± 0.408 | 15.748 ± 0.417 | 4.54 ± 0.17 | −0.610 ± 0.002 | −0.574 ± 0.003 | 100.00 | ||
| Searchlight | 2.828 ± 0.393 | 15.748 ± 0.396 | 5.56 ± 0.19 | −0.511 ± 0.003 | −0.485 ± 0.004 | 100.00 | ||
| Cluster | 2.158 ± 0.356 | 14.791 ± 0.380 | 6.85 ± 0.20 | −0.562 ± 0.002 | −0.534 ± 0.003 | 100.00 | ||
| 1 year (Seedmap) | T14 | 0.893 ± 0.009 | ||||||
| Classic | 6.000 ± 1.315 | 16.125 ± 0.813 | 2.69 ± 0.33 | −0.645 ± 0.010 | −0.556 ± 0.016 | 100.00 | ||
| Searchlight | 4.472 ± 1.090 | 16.125 ± 0.655 | 3.61 ± 0.35 | −0.533 ± 0.016 | −0.488 ± 0.018 | 100.00 | ||
| Cluster | 3.609 ± 1.178 | 14.073 ± 0.619 | 3.90 ± 0.89 | −0.592 ± 0.011 | −0.534 ± 0.015 | 100.00 | ||
| T28 | 0.935 ± 0.006 | |||||||
| Classic | 6.000 ± 1.385 | 15.231 ± 0.746 | 2.54 ± 0.35 | −0.619 ± 0.011 | −0.559 ± 0.013 | 100.00 | ||
| Searchlight | 3.464 ± 1.223 | 17.088 ± 0.843 | 4.93 ± 0.48 | −0.527 ± 0.015 | −0.501 ± 0.017 | 100.00 | ||
| Cluster | 2.655 ± 1.185 | 14.313 ± 0.794 | 5.39 ± 1.22 | −0.566 ± 0.011 | −0.534 ± 0.014 | 100.00 |
Note: “Intersession FC” refers to the percentage of individuals for whom the DLPFC target located in one scan was a site of negative SGC‐FC in a different scan. Other measures are detailed in the main text and Table 1.
3.1.4. Interindividual distance
Interindividual distance was greater for the seed‐based compared to seedmap approach (SI Figures S2a,b and S3, Table 2). This can likely be attributed to a lower signal to noise ratio with the seed‐based approach, which is consistent with the lower intersession reproducibility of the seed‐based approach. The broad distribution of individualized targets across the spatial extent of the DLPFC is illustrated for 100 individuals in Figure 5.
FIGURE 5.
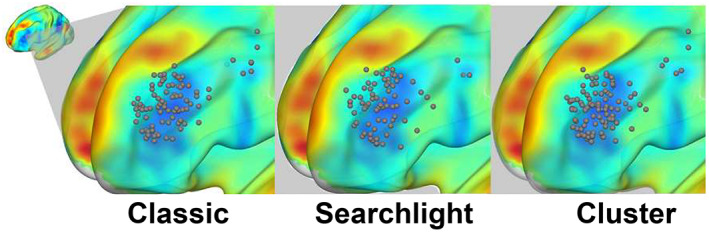
Distribution of personalized targets across the spatial extent of the DLPFC. These are shown for 100 individuals, as computed using the classic, searchlight, and cluster methodologies combined with the seedmap approach
3.1.5. Ratio of Interindividual to Intraindividual distance
The cluster method performed best at pinpointing individual‐specific sites while maintaining reproducibility across sessions, as indicated by higher interindividual‐to‐intraindividual distance ratios (Figure 4d,e, Table 2). This ratio was also substantially elevated using the seedmap approach (range 2.68–6.85). The ratio increased with acquisition time (SI Figure S1b) and reached a maximum at 28 min, at which interindividual variation was on average 6.85 times greater than intraindividual variation between scans, when utilizing cluster and seedmap methodologies (Figure 4e).
3.1.6. Interscan and intrascan functional connectivity at the personalized coordinate
Individualization is sensible only if identified targets maintain their functional fidelity over time. Notably, personalized DLPFC targets displayed negative SGC‐FC in 100% of individuals in a subsequent scan (interscan functional connectivity) when employing the seedmap approach (Table 2; SI Figures S1d; S2c,d, S3). Similarly, all methods identified coordinates that remained negative throughout the scan (intrascan functional connectivity; Table 2; Figure 1c, SI Figure S2e,f). The negative FC value for optimal coordinates was higher when employing the seedmap, compared to seed approach.
3.1.7. Relation to MRI acquisition time
The appropriate scan duration for accurate identification of personalized coordinates is of critical interest. The metrics outlined above were reported at acquisition times of 14 and 28 min (half and full duration) and are illustrated at 60 s intervals in SI Figures [Link], [Link] and S6. The intraindividual between‐scan voxel‐wise spatial correlation (reproducibility) of SGC FC across the spatial extent of the DLPFC continued to increase with increasing scan duration for the seed method, but reached an upper maximum around 20 min for the seedmap method (SI Figure S4). Intraindividual distance and the ratio of interindividual to intraindividual distance continued to improve with increasing scan duration, also reaching an asymptote at approximately 20–25 min (SI Figures S1a,b). Measures of functional fidelity (Interscan and intrascan negative functional connectivity) reached an asymptote by 14 min (SI Figure S1c,d).
3.1.8. Parameterization of searchlight and cluster‐based approaches
Cluster threshold: Increasing the cluster threshold from 0.1 to 50%, indicates that a greater percentage of voxels are retained prior to clustering, resulting in larger cluster sizes. Consequently, with higher thresholds the center of these clusters tends to gravitate toward the center of the DLPFC, resulting in a reduction in intraindividual and interindividual distance (SI Figure S5). Larger thresholds were, therefore, observed to increase reproducibility by increasing target homogeneity within and between individuals, at the cost of reducing true variation among individuals. As cluster size increases, precision is also reduced, such that the intrascan and interscan FC become less negative. The optimal threshold for voxel retention was effectively contingent on data quality: the best balance of intraindividual and interindividual distance was evident at a cluster threshold of around 10% for the seed approach and 0.5% for the seedmap approach. Based on these data, we employed a cluster threshold of 10% for the seed‐based data and 0.5% for the seedmap data.
Searchlight sphere dimensions: A similar loss of spatial precision is evident as the searchlight sphere size is increased (see also Fox et al., 2013). Intrascan and interscan negative FC become less negative with increasing sphere diameter. Intraindividual and interindividual distance were optimal around 3–6 mm. Based on these data (SI Figure S5), we utilized a searchlight sphere diameter of 6 mm.
3.1.9. Reproducibility at 1 year
Available data for 45 participants who underwent an additional scan 1 year after the first scan session indicate that connectivity‐based TMS target sites remain stable over extended periods of time. These data were computed only for the seedmap method based on its superior performance compared to the seed method, outlined above. Target sites remained stable, with a median intraindividual distance of 2.66 ± 1.19 mm for the cluster approach at an acquisition time of 28 min (Table 2, Figure 6, SI Figure S6). The intraindividual distance ranged up to 6.00 ± 1.31 mm for the classic method and 3.46 ± 1.22 for the searchlight method at this scan duration. The cluster method remained the most consistent at pinpointing individual‐specific sites while maintaining reproducibility over time, as indicated by higher inter‐to‐intraindividual distance ratios of 5.39 ± 1.22. This ratio was 4.93 ± 0.48 for the searchlight approach and 2.54 ± 0.35 for the classic method at T28. Importantly, in 100% of individuals personalized DLPFC targets displayed negative SGC‐FC in a subsequent scan 1 year later.
FIGURE 6.
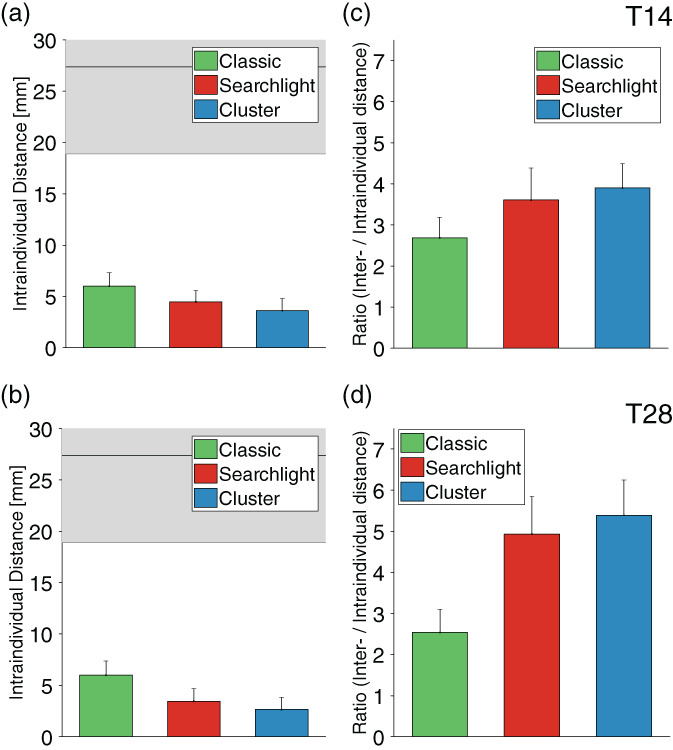
Reproducibility of targets after 1 year. These figures are derived from data for 45 individuals who underwent a repeat scan 365 days after their initial scan. All data were computed using seedmap methodology. (a, b) Target sites remained highly stable with acquisition times of 14 and 28 min, as indicated by a median intraindividual distance between scans that was as low as 2.7 mm for the cluster method at T28. (c, d) The ratio between interindividual and intraindividual distance remained high after 1 year reaching a maximum of 5.39 when the cluster method was applied to compute personalized targets
3.2. Optimized cortical treatment sites are genetically determined
The interindividual distance for stimulation coordinates was significantly shorter between monozygotic twins, compared to dizygotic twins (p = .008), non‐twin siblings (p = 3.2 × 10−5) and unrelated individuals (p = 1.3 × 10−19) (Figure 7).
FIGURE 7.

Personalized treatment sites are under genetic control. The genetic impact of personalized stimulation sites in the dorsolateral prefrontal cortex is indicated by increasing median interindividual distance with diverging familial status. The optimal stimulation target was most similar for monozygotic twins (MZ), and diverged increasingly for dizygotic twins (DZ), non‐twin siblings (NT) and unrelated individuals. Symbols represent significance values: *p = .008; # p = 3.2 × 10−5, Ψ p = 1.3 × 10−19
4. DISCUSSION
After 25 years of rTMS treatment for depression, clinical outcomes still have substantial margin for improvement, since as many as 50% of treated individuals do not achieve either response or remission (Brunoni et al., 2017; George, 2010). Mounting evidence now supports a robust relation between treatment response and network architecture, with greater benefits presumed to arise from personalized cortical TMS targets (Cocchi & Zalesky, 2018). Nonetheless, it remained unknown whether it was feasible to pinpoint stable optimal connectivity‐guided targets on an individual basis, and in the absence of reliable methodology, causal testing of the relation between treatment response and FC has not been possible.
Recent work indicated that the margin of error for defining connectivity‐based DLPFC targets was several centimeters, rendering treatment personalization effectively infeasible (Ning et al., 2019). We now demonstrate that it is possible to achieve reliable target selection with millimeter accuracy. While our novel cluster‐based approach provided substantial advances in intraindividual reproducibility (Figure 4c), doubling the precision of conventional seed‐based methodology (Figure 4b), the final precision achieved here is the strategic integration of this approach, together with new developments across preprocessing and noise‐extraction strategies (Ciric et al., 2017; Parkes, Fulcher, Yucel, & Fornito, 2018; Pruim et al., 2015), functional MRI acquisition (Smith et al., 2013) and signal‐to‐noise ratio elevation for subcortical structures (Fox et al., 2013). The capacity to robustly pinpoint optimal targets in the same individual across repeat scans with a median accuracy of just over 2 mm represents an important advance toward the feasible implementation of next‐generation precision therapy using TMS. This is an order of magnitude more accurate than the current benchmark of 3.5 cm and approximately equivalent to the dimensions of a single brain voxel, approaching the upper limit of 3T MRI spatial resolution. The fact that these targets remained highly stable, varying by an average of only 2.7 mm in a subsequent scan 1 year later, also represents a significant advance for this line of translational research.
Interestingly, interindividual variation across target sites was significantly mediated by familial genetic and environmental factors (Figure 6). While this might appear intuitive, prior research proposed that interindividual diversity at the PFC across microscopic, macroscopic and functional levels could be largely attributed to its greater complexity, relative to unimodal cortical regions, and that “successive propagation of errors” arising from its later stage of ontogenetic development might drive individual variation in this region (Fischl et al., 2008). Moreover, these data provide clear evidence that target site diversity is not driven by noise, but rather has a clear biological substrate, further underscoring the neurobiological rationale for personalization.
Personalization is only worthwhile if interindividual variation is greater than the variation within individuals. Our findings showed that personalized stimulation targets not only varied widely across individuals, but also that interindividual spatial variation was greater than intraindividual variation by a factor of up to 6.85. These data are consistent with recent evidence of unique and stable individual differences in network architecture (Finn et al., 2015; Fischl et al., 2008; Gordon, Laumann, Adeyemo, et al., 2017; Horien, Shen, Scheinost, & Constable, 2019). Critically, beyond spatial specificity, the optimal site identified by this computational methodology in one scan corresponded to a site of negative SGC FC site in a second scan in 100% of individuals when employing optimal methodology (Table 2). In other words, the functional fidelity of these targets remained stable over time.
Connectivity‐based target site personalization might be ineffective if the optimal target site were to shift spatially, either over time or during a course of rTMS treatment. A few lines of evidence suggest that FC‐based target sites are likely to be stable in both of these respects. First, personalized targets remained highly stable 1 year following the initial scan, varying by an average of less than 1.5 voxels (2.7 mm), and SGC FC maps across the spatial extent of the DLPFC remained highly reproducible (Table 2). Second, in related work, we found that there were no significant changes in DLPFC‐SGC FC in individuals with treatment resistant depression following a course of rTMS treatment (Cash, Cocchi, et al., 2020). Some studies have identified changes in the strength of DLPFC‐SGC FC over time, but not in its spatial topography (Siddiqi et al., 2019; Williams et al., 2018). Thirdly, the present data indicates that personalized targets are heritable, suggesting that these may be stable over time and under genetic control.
An important practical consideration is the acquisition time required to reliably pinpoint individualized TMS targets. All measures implemented here (i.e., reproducibility of FC, intraindividual interscan voxel‐wise spatial correlation of SGC FC across the spatial extent of the DLPFC), reproducibility of computed targets (intraindividual distance), capacity to pinpoint individual‐specific sites while maintaining reproducibility across sessions (ratio of interindividual to intraindividual distance), and functional fidelity of targets (interscan and intrascan functional connectivity at the personalized coordinate)) continued to improve with longer acquisition times, typically only reaching an asymptote at a scan duration of around 20 min. At this point the reproducibility of individualized targets also reached the upper limit of scanner resolution (i.e., a single 2 mm voxel). These methods therefore provide the capacity to systematically identify individual‐specific connectivity‐guided targets with excellent reproducibility within a time‐frame that is clinically feasible. While individual‐specific functional network features have recently been demonstrated (Braga & Buckner, 2017; Finn et al., 2015; Gratton et al., 2018; Horien et al., 2019), this has largely required many hours of scan data which is neither practical nor cost‐effective for clinical application. Beyond demonstrating robust intrinsic features of individual functional network architecture, the capacity to precisely pinpoint reproducible individual‐specific connectivity‐guided targets represents an important milestone with significant translational utility.
An additional technical consideration concerns the influence of spatial smoothing: recent work recommended larger smoothing kernels (kernel diameter >12 mm) as these yielded higher intraindividual reproducibility (Ning et al., 2019). Our findings demonstrated a detrimental loss of spatial information in the SGC connectivity map with increasing smoothing kernel size. The previously observed increase in intraindividual reproducibility with larger smoothing kernels is likely an incidental consequence of this reduction in spatial specificity. We recommend minimally smoothed data to reduce loss of spatial information as well as spurious shifts in boundaries between gray and white matter (Coalson et al., 2018).
A final consideration is whether rTMS should be delivered with greater or lesser spatial focality, and ultimately this can only be resolved by prospective clinical testing. Less focal stimulation could be achieved with certain coil designs and could likely be implemented without the costs and computations involved in target‐site personalization. However, the clinical efficacy of a less focal approach might be compromised by concurrent stimulation of regions of positive SGC‐FC, and confounded by a range of other networks impinging on the DLPFC (fig. 3 Gordon, Laumann, Adeyemo, et al., 2017; c.f. fig. 4 Opitz et al., 2016). A neurobiologically informed personalized precision medicine approach might convey additional advantages aside from the anticipated gains in treatment efficacy. RTMS that directly targets relevant neurobiological connections will likely permit the use of lower stimulation intensities, potentially reducing side‐effects (e.g., headache), increasing safety, and enhancing tolerability for uncomfortable targets (e.g., orbitofrontal cortex [Downar, 2019]) and the trend toward more intensive accelerated protocols. In addition, prospective individualized targeting enables direct and causal validation of whether clinical outcome is causally tied to functional network architecture or whether previous observations were epiphenomenal. Accurate network‐based targeting also provides capacity to accelerate the identification, refinement and robust validation of TMS targets for other psychiatric, cognitive and movement disorders, and to develop new knowledge around brain‐behavior relationships.
Prospective randomized trials are now warranted to test the clinical superiority of personalization (Cash, Weigand, et al., 2020). We anticipate that clinical benefits will mirror those recently observed with deep brain stimulation (DBS). Retrospective studies initially demonstrated a robust relation between network connectivity and DBS treatment response, whether utilizing connectivity or proximity‐based metrics (Horn, 2019; Horn, Neumann, Degen, Schneider, & Kuhn, 2017; Horn, Reich, et al., 2017; Horn et al., 2019). Thereafter, step‐wise gains in antidepressant efficacy were seen with a transition from conventional targeting (Morishita, Fayad, Higuchi, Nestor, & Foote, 2014), to a single anatomical target defined based on treatment response in a prior cohort (41% response rate at 6 months) (Holtzheimer et al., 2012) to individualized connectomic based targeting (73% response rate at 6 months) (Riva‐Posse et al., 2018). Preliminary studies in obsessive compulsive disorder report similar gains in DBS response rates with connectivity‐guided personalized targeting (Barcia et al., 2019).
Our work aims primarily to develop a method for connectivity‐based target site personalization that is reliable, precise, easy to implement, and can be flexibly adapted to other targets and respects interindividual variation. However, it is important to acknowledge that other forms of personalization have been proposed, both within and beyond the context of MDD (for review see Cash, Weigand, et al., 2020). These include targeting hypometabolic regions (Garcia‐Toro et al., 2006; Herwig et al., 2003; Paillere Martinot et al., 2010), sites identified as functionally relevant or aberrant according to task‐based fMRI (Bystritsky et al., 2008; Luber et al., 2008; Taylor et al., 2018), and sites other than the DLPFC showing strong FC to the SGC (Cash et al., 2019; Fox et al., 2012). Several network‐based approaches also deserve attention and could be implemented in a seedmap‐style manner to pinpoint individualized cortical target sites—these include the DLPFC‐rTMS responsive network (Tik et al., 2017), efficacy‐based networks (Fox et al., 2013), symptom‐specific networks (Siddiqi et al., 2020) and the “depression network” characterized by lesion‐network mapping (Padmanabhan et al., 2019). Another option is to parcellate the SGC and DLPFC into functional subunits prior to defining the optimal stimulation target (Cole et al., 2020; Williams et al., 2018). We considered a similar option here using Principal Component Analysis while Cole et al. (2020) implemented a hierarchical agglomerative clustering algorithm, but ultimately this step did not appear necessary to achieve high levels of accuracy in the present pipeline and introduced unnecessary complexity. Nonetheless, different methods will have different advantages and this area will continue to evolve. Lastly, we note that several proof‐of‐concept studies have now demonstrated unprecedented rTMS response and remission rates using connectivity‐based targets related to SGC‐FC, even in individuals who previously failed conventional nonpersonalized rTMS and electroconvulsive therapy (Barbour, Lee, Ellard, & Camprodon, 2019; Cole et al., 2020; Siddiqi et al., 2019; Williams et al., 2018).
In this rapidly accelerating area of research, we propose that the reproducibility, functional fidelity, precision, and retainment of individual differences for personalization strategies be openly documented prior to clinical implementation (Figure 8). This ensures that the potential benefits of connectivity‐based personalization are meaningfully translated into clinical practice. More specifically, it would be important to demonstrate that a given targeting methodology firstly generates reproducible targets within a single session and across repeat sessions; second, that these targets retain their functional fidelity over time (in this case negative SGC FC), third, that interindividual heterogeneity is retained (for example with the cluster method, this was contingent on cluster threshold, whereby if the threshold was too high, all coordinates gravitated toward the center of the DLPFC [Figure 8, 3rd panel]); and fourth, it is worth retrospectively examining whether closer proximity between actual clinically implemented and proposed individualized targets relates to better clinical outcome in an existing clinical dataset (e.g., see Cash, Cocchi, et al., 2020). Finally, it is critical to demonstrate that personalization is warranted based on an analysis of personalized versus group consensus targets (Cash, Cocchi, et al., 2020).
FIGURE 8.
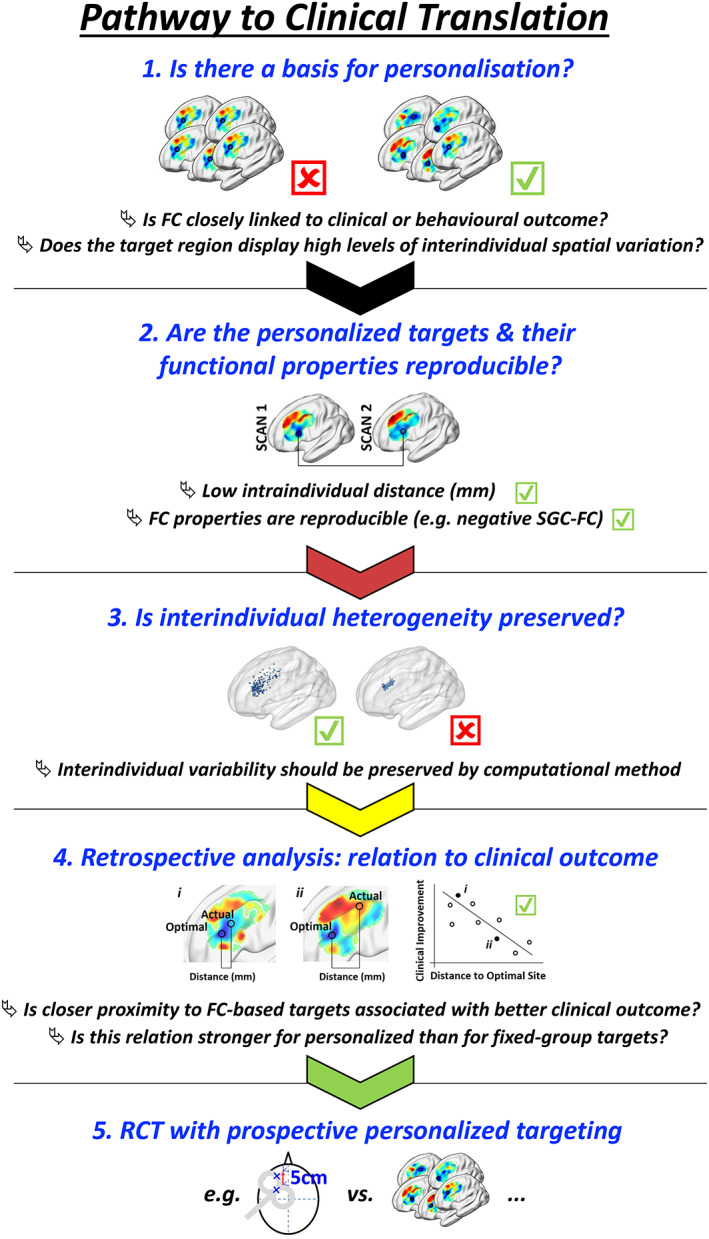
Ideal pathway to clinical translation. Important aspects for adoption of target site personalization are indicated. First, functional connectivity of the proposed target should be closely linked to relevant clinical or behavioral outcomes. Personalized targeting is most readily justified when there is substantial interindividual variation in relevant FC spatial topography. Second, personalized targets should be stable over time in terms of position and functional fidelity. Third, it is critical that any personalization methodology has the capacity to preserve underlying differences in interindividual FC topography—personalized targets should show a relatively broad spatial distribution. The result of ineffective personalization parameters is illustrated on the right: if the cluster threshold is too high, larger clusters were formed resulting in individualized coordinates gravitating toward the center of the DLPFC. Fourth, if possible retrospective analysis should be undertaken in an existing dataset to determine whether closer proximity between actual clinically implemented and proposed individualized targets relates to better treatment or behavioral outcomes. At this stage, it is also critical to demonstrate that personalization is warranted based on an analysis of personalized versus group consensus targets. Finally, the expense of target site personalization should be confirmed in a prospective randomized clinical trial. Note that each of these aspects will fail if MRI scan duration is too short
One limitation of imaging‐based targeting strategies is logistics, time and cost. It would be useful to propose a “best‐guess” approach that could be implemented in the absence of neuroimaging (Beam et al., 2009; Mir‐Moghtadaei et al., 2015). However, in our related retrospective work, we found that clinical outcome in a cohort of individuals with treatment resistant depression was not significantly related to the proximity between the actual clinical stimulation site and any previously proposed fixed group target, including those based on a group‐average SGC FC (Cash, Cocchi, et al., 2020). Instead, clinical outcome was significantly better when individuals were treated closer in proximity to their personalized connectivity‐based targets, computed using the cluster method described above. Moreover, personalized connectivity‐based targets were on average 30 mm from the actual clinically implemented TMS target. This indicates that personalization may be necessary and that a best‐guess approach may not suffice to improve treatment response, but this remains to be confirmed prospectively in larger cohorts. It is also worth noting that at least two studies have found antidepressant outcomes to be better at the group‐level with more anterolateral stimulation (Cash et al., 2019; Herbsman et al., 2009), although antidepressant outcomes reported in preliminary studies using personalized strategies appears to be higher still (for review see Cash, Weigand, et al., 2020). If MRI‐based personalization is proven to be warranted in large prospective clinical trials, then the logistics of clinical translation are likely to be facilitated by the fact that many rTMS clinics are already situated within health‐centers that have MRI access. The cost of fMRI also appears reasonable when considering for the total cost of TMS treatment (McClintock et al., 2018). Cost reductions could be found elsewhere, for example the duration of rTMS can be reduced to less than 4 min by reducing rTMS intertrain interval from 32 to 4 s (Cash et al., 2017) with equivalent clinical efficacy (Ke et al., 2020; Miron et al., 2019), and the requisite MRI acquisition time could be reduced by implementing multi‐echo T2* imaging sequences to enhance data quality and signal‐to‐noise ratio of subcortical regions (Puckett et al., 2018).
4.1. Conclusion
It appears increasingly likely that a fixed group‐level structural target may not appropriately account for the interindividual functional diversity of the prefrontal cortex. We demonstrate the capacity to robustly pinpoint unique and stable individualized optimal targets with high accuracy and reliability. Critically, these targets maintain their functional fidelity over time. These findings provide a strong foundation and direct capacity for personalized targeting of the DLPFC in rTMS treatment of depression, with principles likely generalizing and equally pertinent for other psychiatric disorders in which rTMS is applied. Future randomized clinical trials will be designed to directly quantify the advantages of this approach in terms of clinical improvement, but are beyond the scope of the present investigation. We anticipate that our work will accelerate research and clinical implementation of connectivity‐based personalized rTMS treatments in depression and other psychiatric disorders.
CONFLICT OF INTEREST
The authors report no competing interests.
Supporting information
Figure S1: Full characterization of the change in metrics with increasing acquisition time for seed (empty symbols) and seedmap methodologies (filled symbols). SEM is shaded in all figures, but very small. Dotted vertical lines indicate half and full scan duration (14 and 28 min respectively). (a) Intraindividual distance decreased throughout the acquisition for all approaches, but approached an asymptote at approximately 20–25 min when using the seedmap method. The black horizontal line reflects the average distance obtained if two points are selected at random within the DLPFC (average across 1,000 random permutations). The lower edge of gray shading represents the lower bound of the 95% confidence interval when two points are selected at random within the DLPFC. Although the searchlight and classic methodologies typically select meaningful targets (as indicated by the negative FC at these sites), the distance between these targets across successive scans was similar to selecting two sites at random within the DLPFC. (b) The ratio between intraindividual and interindividual distance reached a maximum at around 14 min for the seed and cluster combination. This ratio was substantially greater for the seedmap method and continued to climb throughout the acquisition for the cluster method, but reached an asymptote at approximately 20–25 min for classic and searchlight methods. (c, d) Functional connectivity (FC) at the optimal coordinate within a single scan (c) or in a separate scan (d) was substantially more negative for the seedmap approach and differed across the classic, cluster and searchlight methods. The classic method performed slightly better than the other approaches. However, it is important to note that our analysis approach here (c, d) simple computes FC at the optimal single voxel coordinate—this is likely to favor the classic approach because the classic method is directly designed to operate at this spatial scale (i.e., it is designed to identify the single most negative FC voxel). Other methods (searchlight and cluster) identify more spatially distributed areas of negative FC. To an extent, the analysis approach is circular for the classic approach, therefore apparently favoring this method, but in fact the single voxel scale of the classic method is least relatable to the spatially distributed TMS e‐field.
Figure S2: Characterization of the different personalization methodologies employed, quantified at acquisition times of 14 and 28 min. (a, b) Interindividual distance quantifies the average distance between personalized targets for 250 of the 1,000 participants. Interindividual distance was generally lower for the cluster approach. However, this is likely to relate to the superior accuracy in the identification of optimal rTMS targets as only the cluster approach identified targets at better than chance levels for the seed data (Figure 4, main text). (c, d) Interscan functional connectivity (FC) indicates how negative the FC value was at the optimal coordinate in a scan performed on a separate day. More negative FC values indicate better selection of a target region, although this approach considers only FC at the voxel represented by the optimal coordinate, rather than within the TMS e‐field. While the classic method potentially performed better than the other approaches, the searchlight and cluster approaches better mimic a spatially distributed TMS field. (e, f) Intrascan FC indicates how negative the FC value was at the optimal target within a single scan at an acquisition time of 14 and 28 min. A similar pattern was evident and the same limitations apply as for as for Interscan FC.
Figure S3. Distribution of outcomes across the sample population. These figures illustrate the distribution of values for each metric across the population (n = 1,000) acquisition times of 14 and 28 min (left and right columns respectively). (a, b) Intraindividual distances between personalized targets. For the cluster and seedmap methodologies a tighter distribution, lower median and much lower maximum intraindividual distance between scans is evident compared to other methodologies. Other methods generate highly divergent targets between repeat scans for some individuals. (c, d) Interindividual distance was lower for the seedmap compared to seed method and was reduced slightly further using the cluster method. This likely reflects superior accuracy and reduced random variability of this approach. Interscan (e, f) and intrascan (g, h) functional connectivity was much more negative for the seedmap compared to seed approach suggesting greater success in identifying a DLPFC site that consistently displays negative SGC FC between and within scans respectively. The classic approach appeared to identify slightly more negative targets, but these two metrics are also biased toward the classic method—the classic method is designed to find the single most negative coordinate whereas the cluster and seedmap methods identify spatially broader sites of negative SGC FC which may better mimic a spatially distributed TMS field. Note that the distribution of the ratio metric cannot be faithfully displayed because for individuals where the intraindividual distance (denominator) between scans is zero, the division cannot be computed.
Figure S4: Relation between the reproducibility of SGC FC topography and scan duration. Intraindividual reproducibility of SGC FC maps across separate scans increases with scan acquisition time. Reproducibility reaches an upper limit at approximately 20 min when SGC FC is computed using the seedmap method, but continues to rise with the seed‐based method which achieves much lower levels of intraindividual reproducibility. To generate this figure, the voxel‐wise correlation of SGC FC across spatial extent of the DLPFC was computed for each individual (n = 1,000) between scan sessions. The group average correlation is indicated (median ± SEM, shaded; N.B. SEM is displayed but very small). Smoothing was applied at FWHM 4 mm.
Figure S5. Influence of Cluster threshold and Searchlight sphere diameter on the precision and functional fidelity of connectivity‐based targets. (a) Within subject intraindividual distance generally decreased with increasing cluster threshold. Higher thresholds resulted in larger clusters, which while more reproducible, lost their functional fidelity (i.e., less negative FC value, targets trended toward a central DLPFC coordinate with decreasing interindividual distance). For the seedmap approach, there was an optimal “sweetspot” at 0.5%, at which intraindividual distance was low but functional fidelity was high. Dotted lines at 0.5 and 10% represent the thresholds chosen for the seedmap and seed methodologies respectively. For A and B, the black horizontal line reflects the average distance obtained if two points are selected at random within the DLPFC (n = 1,000 permutations). The lower edge of gray shading represents the lower bound of the confidence interval when two points are selected at random within the DLPFC. (b) Intraindividual distance decreased with increasing searchlight sphere diameter for the seed approach, but reached a minimum point at 3–6 mm for the seedmap with no further decrease as diameter was increased. Dotted line corresponds to a sphere diameter of 6 mm. (c, d) The SGC FC value at the optimal target within scan and (e, f) in a separate scan became less negative as the cluster threshold and searchlight sphere diameter were increased. This represents a decrease in functional fidelity of targets, that is, they are less likely to represent a desired site of negative FC. (g) Interindividual distance decreased with increasing cluster threshold, representing a trend toward a more central DLPFC coordinate. (h) Interindividual distance varied only marginally for searchlight approaches but was generally greatest in the range of 1–6 mm. (i) The ratio between intraindividual and interindividual distance was much greater for the seedmap approach, reaching a maximum at a cluster threshold of 0.5% (25 voxels). For the seed approach, the ratio was highest at thresholds between 10 and 20% (495–990 voxels). (j) For the seedmap approach, the ratio was greatest at searchlight diameters between 3 and 6 mm. For the seed approach, this ratio increased marginally between 1 and 20 mm.
Figure S6: Change in metrics with increasing acquisition time (1–28 min) at an interscan interval of 365 days. These figures indicate the same relation between increasing acquisition time and improvements in (a) intraindividual distance, (b) the ratio between interindividual to intraindividual distance and (c) interscan FC. Note that intrascan FC is not shown as this metric would not add any information regarding reproducibility over the 1‐year period.
ACKNOWLEDGMENTS
We thank Aaron Kucyi, Caio Seguin, Ye Tian and Sina Mansour for helpful discussions. We thank participants and all those involved in the Human Connectome Project, WU–Minn Consortium (1U54MH091657; Principal Investigators David Van Essen and Kamil Ugurbil) funded by the 16 National Institutes of Health (NIH) institutes and centers that support the NIH Blueprint for Neuroscience Research, and by the McDonnell Center for Systems Neuro‐science at Washington University. RFHC is funded by the Australian Research Council (DE200101708). L.C. is supported by the Australian National Health and Medical Research Council (APP1099082 and APP1138711). A.Z. was supported by the Australian National Health and Medical Research Council (NHMRC) Senior Research Fellowship B (ID: 1136649). PBF has received equipment for research from Cervel Neurotech, Medtronic Ltd., MagVenture A/S and Brainsway Ltd. Part of these data were presented in poster format at the Biological Psychiatry Australia conference, Melbourne, 2019 and Organization for Human Brain Mapping Conference, Montreal, 2020, and 7th International Conference on Non‐invasive Brain Stimulation, virtual, 2020.
Cash RFH, Cocchi L, Lv J, Wu Y, Fitzgerald PB, Zalesky A. Personalized connectivity‐guided DLPFC‐TMS for depression: Advancing computational feasibility, precision and reproducibility. Hum Brain Mapp. 2021;42:4155–4172. 10.1002/hbm.25330
Funding information Australian Research Council, Grant/Award Number: DE200101708; Australian National Health and Medical Research Council (NHMRC), Grant/Award Numbers: APP1099082, APP1138711, 1136649
DATA AVAILABILITY
HCP datasets are available for download to anyone agreeing to the open access data use terms (https://db.humanconnectome.org/).
REFERENCES
- Barbour, T., Lee, E., Ellard, K., & Camprodon, J. (2019). Individualized TMS target selection for MDD: Clinical outcomes, mechanisms of action and predictors of response. Brain Stimulation: Basic, Translational, and Clinical Research in Neuromodulation, 12(2), 516. [Google Scholar]
- Barcia, J. A., Avecillas‐Chasin, J. M., Nombela, C., Arza, R., Garcia‐Albea, J., Pineda‐Pardo, J. A., … Strange, B. A. (2019). Personalized striatal targets for deep brain stimulation in obsessive‐compulsive disorder. Brain Stimulation, 12(3), 724–734. 10.1016/j.brs.2018.12.226 [DOI] [PubMed] [Google Scholar]
- Beam, W., Borckardt, J. J., Reeves, S. T., & George, M. S. (2009). An efficient and accurate new method for locating the F3 position for prefrontal TMS applications. Brain Stimulation, 2(1), 50–54. 10.1016/j.brs.2008.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlim, M. T., van den Eynde, F., Tovar‐Perdomo, S., & Daskalakis, Z. J. (2014). Response, remission and drop‐out rates following high‐frequency repetitive transcranial magnetic stimulation (rTMS) for treating major depression: A systematic review and meta‐analysis of randomized, double‐blind and sham‐controlled trials. Psychological Medicine, 44(2), 225–239.S0033291713000512. 10.1017/S0033291713000512 [DOI] [PubMed] [Google Scholar]
- Blumberger, D. M., Vila‐Rodriguez, F., Thorpe, K. E., Feffer, K., Noda, Y., Giacobbe, P., … Downar, J. (2018). Effectiveness of theta burst versus high‐frequency repetitive transcranial magnetic stimulation in patients with depression (THREE‐D): A randomised non‐inferiority trial. Lancet, 391(10131), 1683–1692. 10.1016/S0140-6736(18)30295-2 [DOI] [PubMed] [Google Scholar]
- Braga, R. M., & Buckner, R. L. (2017). Parallel Interdigitated distributed networks within the individual estimated by intrinsic functional connectivity. Neuron, 95(2), 457–471 e455. 10.1016/j.neuron.2017.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoni, A. R., Chaimani, A., Moffa, A. H., Razza, L. B., Gattaz, W. F., Daskalakis, Z. J., & Carvalho, A. F. (2017). Repetitive Transcranial magnetic stimulation for the acute treatment of major depressive episodes: A systematic review with network meta‐analysis. JAMA Psychiatry, 74(2), 143–152. 10.1001/jamapsychiatry.2016.3644 [DOI] [PubMed] [Google Scholar]
- Bystritsky, A., Kaplan, J. T., Feusner, J. D., Kerwin, L. E., Wadekar, M., Burock, M., … Lacoboni, M. (2008). A preliminary study of fMRI‐guided rTMS in the treatment of generalized anxiety disorder. Journal of Clinical Psychiatry, 69(7), 1092–1098. [DOI] [PubMed] [Google Scholar]
- Cash, R. F. H., Cocchi, L., Lv, J., Fitzgerald, P., & Zalesky, A. (2020). Functional magnetic resonance imaging‐guided personalization of Transcranial magnetic stimulation treatment for depression. JAMA Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash, R. F. H., Dar, A., Hui, J., De Ruiter, L., Baarbé, J., Fettes, P., … Chen, R. (2017). Influence of inter‐train interval on the plastic effects of rTMS. Brain Stimulation: Basic, Translational, and Clinical Research in Neuromodulation, 10(3), 630–636. 10.1016/j.brs.2017.02.012 [DOI] [PubMed] [Google Scholar]
- Cash, R. F. H., Weigand, A., Zalesky, A., Siddiqi, S. H., Downar, J., Fitzgerald, P. B., & Fox, M. D. (2020). Using brain imaging to improve spatial targeting of TMS for depression. Biological Psychiatry In Press. [DOI] [PubMed] [Google Scholar]
- Cash, R. F. H., Zalesky, A., Thomson, R. H., Tian, Y., Cocchi, L., & Fitzgerald, P. B. (2019). Subgenual functional connectivity predicts antidepressant treatment response to transcranial magnetic stimulation: independent validation and evaluation of personalization. Biological Psychiatry, 86(2), e5–e7. [DOI] [PubMed] [Google Scholar]
- Chen, J., Zhou, C., Wu, B., Wang, Y., Li, Q., Wei, Y., … Zou, D. (2013). Left versus right repetitive transcranial magnetic stimulation in treating major depression: A meta‐analysis of randomised controlled trials. Psychiatry Research, 210(3), 1260–1264. [DOI] [PubMed] [Google Scholar]
- Ciric, R., Wolf, D. H., Power, J. D., Roalf, D. R., Baum, G. L., Ruparel, K., … Satterthwaite, T. D. (2017). Benchmarking of participant‐level confound regression strategies for the control of motion artifact in studies of functional connectivity. NeuroImage, 154, 174–187. 10.1016/j.neuroimage.2017.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coalson, T. S., Van Essen, D. C., & Glasser, M. F. (2018). The impact of traditional neuroimaging methods on the spatial localization of cortical areas. Proceedings of the National Academy of Sciences of the United States of America, 115(27), E6356–E6365. 10.1073/pnas.1801582115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchi, L., & Zalesky, A. (2018). Personalized Transcranial magnetic stimulation in psychiatry. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, 3(9), 731–741. 10.1016/j.bpsc.2018.01.008 [DOI] [PubMed] [Google Scholar]
- Cole, E. J., Stimpson, K. H., Bentzley, B. S., Gulser, M., Cherian, K., Tischler, C., … Aaron, H. (2020). Stanford accelerated intelligent neuromodulation therapy for treatment‐resistant depression. American Journal of Psychiatry appi. ajp. 2019.19070720. [DOI] [PubMed] [Google Scholar]
- Doucet, G. E., Lee, W. H., & Frangou, S. (2019). Evaluation of the spatial variability in the major resting‐state networks across human brain functional atlases. Human Brain Mapping, 40(15), 4577–4587. 10.1002/hbm.24722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downar, J. (2019). Orbitofrontal cortex: A 'Non‐rewarding' new treatment target in depression? Current Biology, 29(2), R59–R62. 10.1016/j.cub.2018.11.057 [DOI] [PubMed] [Google Scholar]
- Downar, J., & Daskalakis, Z. J. (2013). New targets for rTMS in depression: A review of convergent evidence. Brain Stimulation, 6(3), 231–240.S1935‐861X(12)00154‐4. 10.1016/j.brs.2012.08.006 [DOI] [PubMed] [Google Scholar]
- Dubois, J., & Adolphs, R. (2016). Building a science of individual differences from fMRI. Trends in Cognitive Sciences, 20(6), 425–443. 10.1016/j.tics.2016.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, E. S., Shen, X., Scheinost, D., Rosenberg, M. D., Huang, J., Chun, M. M., … Constable, R. T. (2015). Functional connectome fingerprinting: Identifying individuals using patterns of brain connectivity. Nature Neuroscience, 18(11), 1664–1671. 10.1038/nn.4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl, B., Rajendran, N., Busa, E., Augustinack, J., Hinds, O., Yeo, B. T., … Zilles, K. (2008). Cortical folding patterns and predicting cytoarchitecture. Cerebral Cortex, 18(8), 1973–1980. 10.1093/cercor/bhm225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, P. B., Hoy, K., McQueen, S., Maller, J. J., Herring, S., Segrave, R., … Daskalakis, Z. J. (2009). A randomized trial of rTMS targeted with MRI based neuro‐navigation in treatment‐resistant depression. Neuropsychopharmacology, 34(5), 1255–1262. 10.1038/npp.2008.233 [DOI] [PubMed] [Google Scholar]
- Fox, M. D., Buckner, R. L., Liu, H., Chakravarty, M. M., Lozano, A. M., & Pascual‐Leone, A. (2014). Resting‐state networks link invasive and noninvasive brain stimulation across diverse psychiatric and neurological diseases. Proceedings of the National Academy of Sciences of the United States of America, 111(41), E4367–E4375. 10.1073/pnas.1405003111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, M. D., Buckner, R. L., White, M. P., Greicius, M. D., & Pascual‐Leone, A. (2012). Efficacy of transcranial magnetic stimulation targets for depression is related to intrinsic functional connectivity with the subgenual cingulate. Biological Psychiatry, 72(7), 595–603. 10.1016/j.biopsych.2012.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, M. D., Liu, H., & Pascual‐Leone, A. (2013). Identification of reproducible individualized targets for treatment of depression with TMS based on intrinsic connectivity. NeuroImage, 66, 151–160. 10.1016/j.neuroimage.2012.10.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, M. D., Zhang, D., Snyder, A. Z., & Raichle, M. E. (2009). The global signal and observed anticorrelated resting state brain networks. Journal of Neurophysiology, 101(6), 3270–3283. 10.1152/jn.90777.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Toro, M., Salva, J., Daumal, J., Andres, J., Romera, M., Lafau, O., … Aguirre, I. (2006). High (20‐Hz) and low (1‐Hz) frequency transcranial magnetic stimulation as adjuvant treatment in medication‐resistant depression. Psychiatry Research, 146(1), 53–57. 10.1016/j.pscychresns.2004.08.005 [DOI] [PubMed] [Google Scholar]
- George, M. S. (2010). Transcranial magnetic stimulation for the treatment of depression. Expert Review of Neurotherapeutics, 10(11), 1761–1772. 10.1586/ern.10.95 [DOI] [PubMed] [Google Scholar]
- Glasser, M. F., Coalson, T. S., Bijsterbosch, J. D., Harrison, S. J., Harms, M. P., Anticevic, A., … Smith, S. M. (2018). Using temporal ICA to selectively remove global noise while preserving global signal in functional MRI data. NeuroImage, 181, 692–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser, M. F., Sotiropoulos, S. N., Wilson, J. A., Coalson, T. S., Fischl, B., Andersson, J. L., … Consortium, W. U.‐M. H. (2013). The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage, 80, 105–124. 10.1016/j.neuroimage.2013.04.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, E. M., Laumann, T. O., Adeyemo, B., Gilmore, A. W., Nelson, S. M., Dosenbach, N. U. F., & Petersen, S. E. (2017). Individual‐specific features of brain systems identified with resting state functional correlations. NeuroImage, 146, 918–939. 10.1016/j.neuroimage.2016.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, E. M., Laumann, T. O., Gilmore, A. W., Newbold, D. J., Greene, D. J., Berg, J. J., … Dosenbach, N. U. F. (2017). Precision functional mapping of individual human brains. Neuron, 95(4), 791–807 e797. 10.1016/j.neuron.2017.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratton, C., Laumann, T. O., Nielsen, A. N., Greene, D. J., Gordon, E. M., Gilmore, A. W., … Petersen, S. E. (2018). Functional brain networks are dominated by stable group and individual factors, not cognitive or daily variation. Neuron, 98(2), 439–452 e435. 10.1016/j.neuron.2018.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halko, M. A., Farzan, F., Eldaief, M. C., Schmahmann, J. D., & Pascual‐Leone, A. (2014). Intermittent theta‐burst stimulation of the lateral cerebellum increases functional connectivity of the default network. The Journal of Neuroscience, 34(36), 12049–12056. 10.1523/JNEUROSCI.1776-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbsman, T., Avery, D., Ramsey, D., Holtzheimer, P., Wadjik, C., Hardaway, F., … Nahas, Z. (2009). More lateral and anterior prefrontal coil location is associated with better repetitive transcranial magnetic stimulation antidepressant response. Biological Psychiatry, 66(5), 509–515. 10.1016/j.biopsych.2009.04.034 [DOI] [PubMed] [Google Scholar]
- Herwig, U., Lampe, Y., Juengling, F. D., Wunderlich, A., Walter, H., Spitzer, M., & Schonfeldt‐Lecuona, C. (2003). Add‐on rTMS for treatment of depression: A pilot study using stereotaxic coil‐navigation according to PET data. Journal of Psychiatric Research, 37(4), 267–275. [DOI] [PubMed] [Google Scholar]
- Herwig, U., Padberg, F., Unger, J., Spitzer, M., & Schonfeldt‐Lecuona, C. (2001). Transcranial magnetic stimulation in therapy studies: Examination of the reliability of "standard" coil positioning by neuronavigation. Biological Psychiatry, 50(1), 58–61. [DOI] [PubMed] [Google Scholar]
- Hill, J., Dierker, D., Neil, J., Inder, T., Knutsen, A., Harwell, J., … Van Essen, D. (2010). A surface‐based analysis of hemispheric asymmetries and folding of cerebral cortex in term‐born human infants. The Journal of Neuroscience, 30(6), 2268–2276. 10.1523/JNEUROSCI.4682-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzheimer, P. E., Kelley, M. E., Gross, R. E., Filkowski, M. M., Garlow, S. J., Barrocas, A., … Mayberg, H. S. (2012). Subcallosal cingulate deep brain stimulation for treatment‐resistant unipolar and bipolar depression. Archives of General Psychiatry, 69(2), 150–158. 10.1001/archgenpsychiatry.2011.1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horien, C., Shen, X., Scheinost, D., & Constable, R. T. (2019). The individual functional connectome is unique and stable over months to years. NeuroImage, 189, 676–687. 10.1016/j.neuroimage.2019.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, A. (2019). The impact of modern‐day neuroimaging on the field of deep brain stimulation. Current Opinion in Neurology, 32(4), 511–520. 10.1097/WCO.0000000000000679 [DOI] [PubMed] [Google Scholar]
- Horn, A., Neumann, W. J., Degen, K., Schneider, G. H., & Kuhn, A. A. (2017). Toward an electrophysiological "sweet spot" for deep brain stimulation in the subthalamic nucleus. Human Brain Mapping, 38(7), 3377–3390. 10.1002/hbm.23594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, A., Reich, M., Vorwerk, J., Li, N., Wenzel, G., Fang, Q., … Fox, M. D. (2017). Connectivity predicts deep brain stimulation outcome in Parkinson disease. Annals of Neurology, 82(1), 67–78. 10.1002/ana.24974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, A., Wenzel, G., Irmen, F., Huebl, J., Li, N., Neumann, W. J., … Kuhn, A. A. (2019). Deep brain stimulation induced normalization of the human functional connectome in Parkinson's disease. Brain, 142(10), 3129–3143. 10.1093/brain/awz239 [DOI] [PubMed] [Google Scholar]
- Ke, J., Zou, X., Huang, M., Huang, Q., Li, H., & Zhou, X. (2020). High‐frequency rTMS with two different inter‐train intervals improves upper limb motor function at the early stage of stroke. Journal of International Medical Research, 48(6), 0300060520928737. [Google Scholar]
- Li, C.‐T., Cheng, C.‐M., Chen, M.‐H., Juan, C.‐H., Tu, P.‐C., Bai, Y.‐M., … Su, T.‐P. (2019). Antidepressant efficacy of prolonged intermittent theta burst stimulation Monotherapy for recurrent depression and comparison of methods for coil positioning: A randomized, double‐blind, Sham‐Controlled Study. Biological Psychiatry. [DOI] [PubMed]
- Li, J., Bolt, T., Bzdok, D., Nomi, J. S., Yeo, B. T., Spreng, R. N., & Uddin, L. Q. (2019). Topography and behavioral relevance of the global signal in the human brain. Scientific Reports, 9(1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Kong, R., Liegeois, R., Orban, C., Tan, Y., Sun, N., … Yeo, B. T. T. (2019). Global signal regression strengthens association between resting‐state functional connectivity and behavior. NeuroImage, 196, 126–141. 10.1016/j.neuroimage.2019.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luber, B., Stanford, A., Bulow, P., Nguyen, T., Rakitin, B., Habeck, C., … Lisanby, S. (2008). Remediation of sleep‐deprivation–induced working memory impairment with fMRI‐guided transcranial magnetic stimulation. Cerebral Cortex, 18(9), 2077–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, S. M., Reti, I. M., Carpenter, L. L., McDonald, W. M., Dubin, M., Taylor, S. F., … Wall, C. (2018). Consensus recommendations for the clinical application of repetitive Transcranial magnetic stimulation (rTMS) in the treatment of depression. The Journal of Clinical Psychiatry, 79(1). 10.4088/JCP.16cs10905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda‐Dominguez, O., Mills, B. D., Carpenter, S. D., Grant, K. A., Kroenke, C. D., Nigg, J. T., & Fair, D. A. (2014). Connectotyping: Model based fingerprinting of the functional connectome. PLoS One, 9(11), e111048. 10.1371/journal.pone.0111048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir‐Moghtadaei, A., Caballero, R., Fried, P., Fox, M. D., Lee, K., Giacobbe, P., … Downar, J. (2015). Concordance between BeamF3 and MRI‐neuronavigated target sites for repetitive Transcranial magnetic stimulation of the left dorsolateral prefrontal cortex. Brain Stimulation, 8(5), 965–973. 10.1016/j.brs.2015.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron, J. P., Feffer, K., Cash, R. F. H., Derakhshan, D., Kim, J. M. S., Fettes, P., … Downar, J. (2019). Safety, tolerability and effectiveness of a novel 20 Hz rTMS protocol targeting dorsomedial prefrontal cortex in major depression: An open‐label case series. Brain Stimulation, 12(5), 1319–1321. 10.1016/j.brs.2019.06.020 [DOI] [PubMed] [Google Scholar]
- Morishita, T., Fayad, S. M., Higuchi, M.‐a., Nestor, K. A., & Foote, K. D. (2014). Deep brain stimulation for treatment‐resistant depression: Systematic review of clinical outcomes. Neurotherapeutics, 11(3), 475–484. 10.1007/s13311-014-0282-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, S., Wang, D., Fox, M. D., Yeo, B. T., Sepulcre, J., Sabuncu, M. R., … Liu, H. (2013). Individual variability in functional connectivity architecture of the human brain. Neuron, 77(3), 586–595. 10.1016/j.neuron.2012.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, K., & Fox, M. D. (2017). Towards a consensus regarding global signal regression for resting state functional connectivity MRI. NeuroImage, 154, 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning, L., Makris, N., Camprodon, J. A., & Rathi, Y. (2019). Limits and reproducibility of resting‐state functional MRI definition of DLPFC targets for neuromodulation. Brain Stimulation, 12(1), 129–138. 10.1016/j.brs.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojemann, J. G., Akbudak, E., Snyder, A. Z., McKinstry, R. C., Raichle, M. E., & Conturo, T. E. (1997). Anatomic localization and quantitative analysis of gradient refocused echo‐planar fMRI susceptibility artifacts. NeuroImage, 6(3), 156–167. 10.1006/nimg.1997.0289 [DOI] [PubMed] [Google Scholar]
- Opitz, A., Fox, M. D., Craddock, R. C., Colcombe, S., & Milham, M. P. (2016). An integrated framework for targeting functional networks via transcranial magnetic stimulation. NeuroImage, 127, 86–96. 10.1016/j.neuroimage.2015.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan, J. L., Cooke, D., Joutsa, J., Siddiqi, S. H., Ferguson, M., Darby, R. R., … Fox, M. D. (2019). A human depression circuit derived from focal brain lesions. Biological Psychiatry, 86, 749–758. 10.1016/j.biopsych.2019.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillere Martinot, M. L., Galinowski, A., Ringuenet, D., Gallarda, T., Lefaucheur, J. P., Bellivier, F., … Martinot, J. L. (2010). Influence of prefrontal target region on the efficacy of repetitive transcranial magnetic stimulation in patients with medication‐resistant depression: A [(18)F]‐fluorodeoxyglucose PET and MRI study. The International Journal of Neuropsychopharmacology, 13(1), 45–59. 10.1017/S146114570900008X [DOI] [PubMed] [Google Scholar]
- Parkes, L., Fulcher, B., Yucel, M., & Fornito, A. (2018). An evaluation of the efficacy, reliability, and sensitivity of motion correction strategies for resting‐state functional MRI. NeuroImage, 171, 415–436. 10.1016/j.neuroimage.2017.12.073 [DOI] [PubMed] [Google Scholar]
- Pruim, R. H., Mennes, M., van Rooij, D., Llera, A., Buitelaar, J. K., & Beckmann, C. F. (2015). ICA‐AROMA: A robust ICA‐based strategy for removing motion artifacts from fMRI data. NeuroImage, 112, 267–277. 10.1016/j.neuroimage.2015.02.064 [DOI] [PubMed] [Google Scholar]
- Puckett, A. M., Bollmann, S., Poser, B. A., Palmer, J., Barth, M., & Cunnington, R. (2018). Using multi‐echo simultaneous multi‐slice (SMS) EPI to improve functional MRI of the subcortical nuclei of the basal ganglia at ultra‐high field (7T). NeuroImage, 172, 886–895. [DOI] [PubMed] [Google Scholar]
- Rajkowska, G., & Goldman‐Rakic, P. S. (1995). Cytoarchitectonic definition of prefrontal areas in the normal human cortex: II. Variability in locations of areas 9 and 46 and relationship to the Talairach coordinate system. Cerebral Cortex, 5(4), 323–337. 10.1093/cercor/5.4.323 [DOI] [PubMed] [Google Scholar]
- Rastogi, A., Cash, R., Dunlop, K., Vesia, M., Kucyi, A., Ghahremani, A., … Chen, R. (2017). Modulation of cognitive cerebello‐cerebral functional connectivity by lateral cerebellar continuous theta burst stimulation. NeuroImage, 158, 48–57. 10.1016/j.neuroimage.2017.06.048 [DOI] [PubMed] [Google Scholar]
- Riva‐Posse, P., Choi, K. S., Holtzheimer, P. E., Crowell, A. L., Garlow, S. J., Rajendra, J. K., … Mayberg, H. S. (2018). A connectomic approach for subcallosal cingulate deep brain stimulation surgery: Prospective targeting in treatment‐resistant depression. Molecular Psychiatry, 23(4), 843–849. 10.1038/mp.2017.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush, A. J. (2007). Limitations in efficacy of antidepressant monotherapy. The Journal of Clinical Psychiatry, 68(Suppl 10), 8–10. [PubMed] [Google Scholar]
- Siddiqi, S. H., Taylor, S., Cooke, D., Pascual‐Leone, A., George, M. S., & Fox, M. D. (2020). Distinct symptom‐specific treatment targets for circuit‐based neuromodulation. The American Journal of Psychiatry, In Press, 177, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi, S. H., Trapp, N. T., Hacker, C. D., Laumann, T. O., Kandala, S., Hong, X., … Brody, D. L. (2019). Repetitive transcranial magnetic stimulation with resting‐state network targeting for treatment‐resistant depression in traumatic brain injury: A randomized, controlled, double‐blinded pilot study. Journal of Neurotrauma, 36(8), 1361–1374. 10.1089/neu.2018.5889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. M., Beckmann, C. F., Andersson, J., Auerbach, E. J., Bijsterbosch, J., Douaud, G., … Consortium, W. U.‐M. H. (2013). Resting‐state fMRI in the human Connectome project. NeuroImage, 80, 144–168. 10.1016/j.neuroimage.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S. F., Ho, S. S., Abagis, T., Angstadt, M., Maixner, D. F., Welsh, R. C., & Hernandez‐Garcia, L. (2018). Changes in brain connectivity during a sham‐controlled, transcranial magnetic stimulation trial for depression. Journal of Affective Disorders, 232, 143–151. 10.1016/j.jad.2018.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tik, M., Hoffmann, A., Sladky, R., Tomova, L., Hummer, A., Navarro de Lara, L., … Windischberger, C. (2017). Towards understanding rTMS mechanism of action: Stimulation of the DLPFC causes network‐specific increase in functional connectivity. NeuroImage, 162, 289–296. 10.1016/j.neuroimage.2017.09.022 [DOI] [PubMed] [Google Scholar]
- Weigand, A., Horn, A., Caballero, R., Cooke, D., Stern, A. P., Taylor, S. F., … Fox, M. D. (2018). Prospective validation that Subgenual connectivity predicts antidepressant efficacy of Transcranial magnetic stimulation sites. Biological Psychiatry, 84(1), 28–37. 10.1016/j.biopsych.2017.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, N. R., Sudheimer, K. D., Bentzley, B. S., Pannu, J., Stimpson, K. H., Duvio, D., … Schatzberg, A. F. (2018). High‐dose spaced theta‐burst TMS as a rapid‐acting antidepressant in highly refractory depression. Brain, 141(3), e18. 10.1093/brain/awx379 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Full characterization of the change in metrics with increasing acquisition time for seed (empty symbols) and seedmap methodologies (filled symbols). SEM is shaded in all figures, but very small. Dotted vertical lines indicate half and full scan duration (14 and 28 min respectively). (a) Intraindividual distance decreased throughout the acquisition for all approaches, but approached an asymptote at approximately 20–25 min when using the seedmap method. The black horizontal line reflects the average distance obtained if two points are selected at random within the DLPFC (average across 1,000 random permutations). The lower edge of gray shading represents the lower bound of the 95% confidence interval when two points are selected at random within the DLPFC. Although the searchlight and classic methodologies typically select meaningful targets (as indicated by the negative FC at these sites), the distance between these targets across successive scans was similar to selecting two sites at random within the DLPFC. (b) The ratio between intraindividual and interindividual distance reached a maximum at around 14 min for the seed and cluster combination. This ratio was substantially greater for the seedmap method and continued to climb throughout the acquisition for the cluster method, but reached an asymptote at approximately 20–25 min for classic and searchlight methods. (c, d) Functional connectivity (FC) at the optimal coordinate within a single scan (c) or in a separate scan (d) was substantially more negative for the seedmap approach and differed across the classic, cluster and searchlight methods. The classic method performed slightly better than the other approaches. However, it is important to note that our analysis approach here (c, d) simple computes FC at the optimal single voxel coordinate—this is likely to favor the classic approach because the classic method is directly designed to operate at this spatial scale (i.e., it is designed to identify the single most negative FC voxel). Other methods (searchlight and cluster) identify more spatially distributed areas of negative FC. To an extent, the analysis approach is circular for the classic approach, therefore apparently favoring this method, but in fact the single voxel scale of the classic method is least relatable to the spatially distributed TMS e‐field.
Figure S2: Characterization of the different personalization methodologies employed, quantified at acquisition times of 14 and 28 min. (a, b) Interindividual distance quantifies the average distance between personalized targets for 250 of the 1,000 participants. Interindividual distance was generally lower for the cluster approach. However, this is likely to relate to the superior accuracy in the identification of optimal rTMS targets as only the cluster approach identified targets at better than chance levels for the seed data (Figure 4, main text). (c, d) Interscan functional connectivity (FC) indicates how negative the FC value was at the optimal coordinate in a scan performed on a separate day. More negative FC values indicate better selection of a target region, although this approach considers only FC at the voxel represented by the optimal coordinate, rather than within the TMS e‐field. While the classic method potentially performed better than the other approaches, the searchlight and cluster approaches better mimic a spatially distributed TMS field. (e, f) Intrascan FC indicates how negative the FC value was at the optimal target within a single scan at an acquisition time of 14 and 28 min. A similar pattern was evident and the same limitations apply as for as for Interscan FC.
Figure S3. Distribution of outcomes across the sample population. These figures illustrate the distribution of values for each metric across the population (n = 1,000) acquisition times of 14 and 28 min (left and right columns respectively). (a, b) Intraindividual distances between personalized targets. For the cluster and seedmap methodologies a tighter distribution, lower median and much lower maximum intraindividual distance between scans is evident compared to other methodologies. Other methods generate highly divergent targets between repeat scans for some individuals. (c, d) Interindividual distance was lower for the seedmap compared to seed method and was reduced slightly further using the cluster method. This likely reflects superior accuracy and reduced random variability of this approach. Interscan (e, f) and intrascan (g, h) functional connectivity was much more negative for the seedmap compared to seed approach suggesting greater success in identifying a DLPFC site that consistently displays negative SGC FC between and within scans respectively. The classic approach appeared to identify slightly more negative targets, but these two metrics are also biased toward the classic method—the classic method is designed to find the single most negative coordinate whereas the cluster and seedmap methods identify spatially broader sites of negative SGC FC which may better mimic a spatially distributed TMS field. Note that the distribution of the ratio metric cannot be faithfully displayed because for individuals where the intraindividual distance (denominator) between scans is zero, the division cannot be computed.
Figure S4: Relation between the reproducibility of SGC FC topography and scan duration. Intraindividual reproducibility of SGC FC maps across separate scans increases with scan acquisition time. Reproducibility reaches an upper limit at approximately 20 min when SGC FC is computed using the seedmap method, but continues to rise with the seed‐based method which achieves much lower levels of intraindividual reproducibility. To generate this figure, the voxel‐wise correlation of SGC FC across spatial extent of the DLPFC was computed for each individual (n = 1,000) between scan sessions. The group average correlation is indicated (median ± SEM, shaded; N.B. SEM is displayed but very small). Smoothing was applied at FWHM 4 mm.
Figure S5. Influence of Cluster threshold and Searchlight sphere diameter on the precision and functional fidelity of connectivity‐based targets. (a) Within subject intraindividual distance generally decreased with increasing cluster threshold. Higher thresholds resulted in larger clusters, which while more reproducible, lost their functional fidelity (i.e., less negative FC value, targets trended toward a central DLPFC coordinate with decreasing interindividual distance). For the seedmap approach, there was an optimal “sweetspot” at 0.5%, at which intraindividual distance was low but functional fidelity was high. Dotted lines at 0.5 and 10% represent the thresholds chosen for the seedmap and seed methodologies respectively. For A and B, the black horizontal line reflects the average distance obtained if two points are selected at random within the DLPFC (n = 1,000 permutations). The lower edge of gray shading represents the lower bound of the confidence interval when two points are selected at random within the DLPFC. (b) Intraindividual distance decreased with increasing searchlight sphere diameter for the seed approach, but reached a minimum point at 3–6 mm for the seedmap with no further decrease as diameter was increased. Dotted line corresponds to a sphere diameter of 6 mm. (c, d) The SGC FC value at the optimal target within scan and (e, f) in a separate scan became less negative as the cluster threshold and searchlight sphere diameter were increased. This represents a decrease in functional fidelity of targets, that is, they are less likely to represent a desired site of negative FC. (g) Interindividual distance decreased with increasing cluster threshold, representing a trend toward a more central DLPFC coordinate. (h) Interindividual distance varied only marginally for searchlight approaches but was generally greatest in the range of 1–6 mm. (i) The ratio between intraindividual and interindividual distance was much greater for the seedmap approach, reaching a maximum at a cluster threshold of 0.5% (25 voxels). For the seed approach, the ratio was highest at thresholds between 10 and 20% (495–990 voxels). (j) For the seedmap approach, the ratio was greatest at searchlight diameters between 3 and 6 mm. For the seed approach, this ratio increased marginally between 1 and 20 mm.
Figure S6: Change in metrics with increasing acquisition time (1–28 min) at an interscan interval of 365 days. These figures indicate the same relation between increasing acquisition time and improvements in (a) intraindividual distance, (b) the ratio between interindividual to intraindividual distance and (c) interscan FC. Note that intrascan FC is not shown as this metric would not add any information regarding reproducibility over the 1‐year period.
Data Availability Statement
HCP datasets are available for download to anyone agreeing to the open access data use terms (https://db.humanconnectome.org/).


