ABSTRACT
Hexabromocyclododecanes (HBCDs) are widely used brominated flame retardants that cause antidiuretic hormone syndrome and even induce cancer. However, little information is available about the degradation mechanisms of HBCDs. In this study, genomic and proteomic analyses, reverse transcription-quantitative PCR, and gene knockout assays reveal that a cytochrome P450-encoding gene is responsible for HBCD catabolism in Pseudomonas aeruginosa HS9. The CO difference spectrum of the enzyme CYP168A1 was matched to P450 characteristics via UV visibility. We demonstrate that the reactions of debromination and hydrogenation are carried out one after another based on detection of the metabolites pentabromocyclododecanols (PBCDOHs), tetrabromocyclododecadiols (TBCDDOHs), and bromide ion. In the 18O isotope experiments, PBCD18OHs were only detected in the H218O group, proving that the added oxygen is derived from H2O, not from O2. This study elucidates the degradation mechanism of HBCDs by Pseudomonas.
IMPORTANCE Hexabromocyclododecanes (HBCDs) are environmental pollutants that are widely used in industry. In this study, we identified and characterized a novel key dehalogenase, CYP168A1, that is responsible for HBCD degradation from Pseudomonas aeruginosa strain HS9. This study provides new insights into understanding biodegradation of HBCDs.
KEYWORDS: biodegradation, cytochrome P450, hexabromocyclododecanes, mechanism
INTRODUCTION
Hexabromocyclododecanes (HBCDs) are the second most widely used brominated flame retardants (BFRs) and are utilized in building materials, electronics, textiles, and plastics (1). They are a threat to human health due to causing antidiuretic hormone syndrome and even inducing cancer. Microorganisms play important roles in degradation and detoxification of pollutants of HBCDs (2). However, little information is available about molecular and biochemical mechanisms, particularly how functional proteins relate to debromination. Only two dehalogenases, LinA and LinB from the hexachlorocyclohexane transformation strain Sphingobium indicum B90A, can convert HBCD to different debrominated products. LinA selectively catalyzes the transformation of β-HBCDs to 1E,5S,6S,9R,10S-pentabromocyclododecene (PBCDE), while LinB transforms all α-, β-, and γ-HBCD isomers to pentabromocyclododecanols (PBCDOHs) and even tetrabromocyclododecadiols (TBCDDOHs) (3, 4). The kinetics and stereochemistry of LinB-catalyzed γ-HBCD transformation have been described in detail, with Km, kcat, and kcat/Km values of 1.82 ± 0.60 μmol/liter, 0.25 ± 0.10 μmol/liter/h, and 13.0 ± 6.2 liter/mol/s. The results suggest that LinB has a strong ability to dehalogenate γ-HBCD (5).
Catalytic enzyme resources from bacteria are abundant, and cytochrome P450 enzymes (CYPs) are the key enzymes responsible for the degradation of numerous endogenous compounds. CYPs are involved in the degradation and detoxification of multiple toxicants, such as herbicides, xenobiotic polyaromatic hydrocarbons, halogenated aromatics, and polychlorinated biphenyls (6–10). Hydroxylation is the typical metabolic reaction of xenobiotics catalyzed by CYPs. Transformed CYP81As from Echinochloa phyllopogon decreased the susceptibility of Arabidopsis to clomazone (11, 12). Mammalian CYPs (CYP1 family) degrade dibenzo-p-dioxins (PCDDs) with efficient activity, and the rat CYP1A1 family also showed high activity toward 2,3,7-trichloro-dibenzo-p-dioxin, with the detection of hydroxylated products, i.e., 8-hydroxy-2,3,7-trichloro-dibenzo-p-dioxins (13). CYP101 dehalogenates hexachlorobenzene with a different metabolic method in which the halogen atoms are replaced by hydroxyl groups (14). CYP2E1 from Nicotiana tabacum, CYP3A4 from human liver, and CYPs (CYP71C3v2, CYP71C1, CYP81A1, and CYP97A16) from maize can metabolize HBCDs, and the hydroxylated metabolites OH-HBCDs, OH-PBCDs, and OH-TBCDs have been detected (15–20). However, the substitution reaction of HBCDs for CYPs is rarely reported.
Previous work by our research group on Pseudomonas aeruginosa HS9 indicated that HBCDs can be degraded to PBCDOHs. Strain HS9 was reported to be an HBCD-metabolizing bacterium based on its ability to convert HBCDs to PBCDOHs or tetrabromocyclododecene (TBCDe), dibromocyclododecadiene (DBCDi), and cyclododecatriene (CDT) (21). In this study, the whole-genome sequence of strain HS9 was sequenced and analyzed, and putative genes for HBCD degradation were elucidated. By combining metabolite analysis with real-time fluorescence quantification experiments (reverse transcription-quantitative PCR [RT-qPCR]), the cytochrome P450 enzyme CYP168A1 was considered the initial dehalogenase in HBCD metabolism. The gene cyp168A1 was cloned and expressed in Escherichia coli. The subsequent enzymatic properties were investigated in the purified CYP168A1.
RESULTS
Genomic and proteomic profiles of strain HS9.
Whole-genome sequencing was preformed, and the sequence was assembled into a single circular chromosome without gaps (see Fig. S1A in the supplemental material). The circular chromosome is 6,876,988 bp in size, with a G+C content of 66.2% and 6,421 coding sequences (CDSs). Further analysis indicated that there were 157 CDSs annotated as related to metabolism of aromatic compounds (Fig. S1B). To explore functional genes involved in HBCD degradation, a proteomic analysis was carried out to compare the expression of proteins from cells incubated in the presence or absence of 1 mg/liter HBCDs mineral salt medium (MSM). A total of 1,770 proteins were identified, accounting for 27.6% of the genomic putative CDSs in strain HS9. Normalization was performed to average the abundance of all peptides. Differentially expressed proteins were filtered if their fold changes were over 2.0-fold with significance of >20 (PEAKS significance B algorithm; P < 0.01) and if they had two unique peptides.
The expression of 277 proteins was significantly changed (≥2-fold change; P < 0.01), of which 190 proteins were upregulated and 87 were downregulated (Fig. S1C). The upregulated proteins were divided into 25 categories by Clusters of Orthologous Groups (COG) analysis (Fig. S1D). To narrow the search, the most significantly changed proteins (≥10-fold change; P < 0.01) are summarized in Fig. S2, and the HBCD-induced proteins are listed in Table S1. No annotated dehalogenases were identified among the 277 upregulated proteins, while the expression of the NADH reductase (HS5738), heme d1 biosynthesis protein (NirF) (HS1898), and iron(III) dicitrate transport protein (FecA) (HS1283) were upregulated with fold changes of 5.81, 217.80, and +∞, respectively. As many upregulated genes were related to electron donating, the functional genes that cooperated with electron donors were considered possible HBCD-degrading genes.
Many genes related to heavy metals were upregulated, including zinc and mercury transporting ATPase (EC 3.6.3.3), heavy-metal sensor histidine kinase, copper resistance protein (CopC), Na+/alanine symporter, iron(III) dicitrate transport protein (FecA), and zinc protease. Moreover, genes correlated with basic bioactivity, such as d-lactate dehydrogenase, l-lactate dehydrogenase (EC 1.1.2.3), l-lactate permease, and succinate dehydrogenase, were significantly upregulated.
Identification of HBCD-degrading genes.
To identify the possible genes involved in HBCD degradation in cooperation with ferredoxin-NADP(+) reductase, three cytochrome P450 (CYP) proteins were selected as candidates. RT-qPCR assays were carried out to further detect the mRNA expression levels of the potential genes (Fig. 1A). The cytochrome P450 coding gene cyp168A1 was upregulated in response to HBCDs with a fold change of 9.1. The HBCD consumption curve of strain HS9 was determined in a resting cell reaction system. The wild type of strain HS9 (WT) could degrade 1 mg/liter HBCDs within 8 h. In addition, we also deleted or complemented the gene cyp168A1, and the HBCD consumption capability of the mutant strain Mcyp168A1 was eliminated. When the cyp168A1 gene was complemented in Mcyp168A1, the HBCD degradation capability of strain Wcyp168A1 recovered to the same value as strain HS9 (Fig. 1B).
FIG 1.
Identification of functional proteins. (A) RT-qPCR verification of the proposed functional genes in degrading HBCDs. In RT-qPCR assays, the treatment group used HBCDs as the sole carbon source, and the control group used sodium citrate. HS651, putative cytochrome P450 hydroxylase; HS1037, cytochrome P450; HS6073 (cyp168A1), putative cytochrome P450 hydroxylase. (B) Comparison of the HBCD-degrading ability of the wild-type HS9 (WT), cyp168A1-deleted mutant strain (Mcyp168A1), and cyp168A1-complemented strain (Wcyp168A1).
Verification of electron donor capability of FdFNR cell-free system.
To confirm the donor-supplying ability of FdFNR [a 4Fe-4S ferredoxin (HS1040) and an NAD(P)H-dependent ferredoxin reductase], potassium ferricyanide (K3[Fe(CN)6]) was used as the receptor of the free donor. Compared to the control group, the absorbance of K3[Fe(CN)6] at 340 nm decreased to zero over 5 min, and the color feature (yellow) of K3[Fe(CN)6] disappeared (Fig. 2A). Results showed that the cell-free system was able to oxidize NADH to NAD+ with an electron accepter present.
FIG 2.
Characterization of CYP168A1. (A) Verification of the electron donor capability of the cell-free system FdFNR. The protein expression of FdFNR was measured and shown by SDS-PAGE, the color feature (yellow) of K3[Fe(CN)6] was captured, and the full wavelength scanning shows the concentration of K3[Fe(CN)6] in the cell-free system. (B) SDS-PAGE analysis of CYP168A1. M, protein marker; lane 1, supernatant of the sonicated Bl21-pET28a-cyp168A1; lane 2, column effluent; lane 3, 10 mM imidazole buffer washed effluent; lane 4, 45 mM imidazole washed effluent; lane 5, 70 mM imidazole washed effluent; lane 6, 100 mM imidazole washed effluent; lane 7, 150 mM imidazole washed effluent. (C) The CO difference spectrum of CYP168A1. (D) Kinetic analysis of CYP168A1 (fitted to the Michaelis-Menten kinetics).
CYP168A1 is an efficient debromination enzyme.
The gene cyp168A1 was amplified and expressed in pET28a in E. coli BL21(DE3). The heterologously expressed 6× His-CYP168A1 was successfully purified, with a molecular mass of 50 kDa (Fig. 2B), and the Western blot analysis demonstrated the purified protein (Fig. S3A). The CO difference spectrum showed that the purified protein has a strong absorbance at 450 nm (Fig. 2C). Results of enzyme activity detection showed that CYP168A1 degraded HBCDs in the presence of NADH in the FdFNR system. The optimal temperature for CYP168A1 activity was 30°C (Fig. S3B). The effect of temperature on CYP168A1 stability was monitored by circular dichroism spectroscopy (CDS) (Jasco, Japan), which showed that CYP168A1 began to degenerate at temperatures above 35°C (Fig. S3C). Kinetic analysis revealed that the Vmax and Km were 0.73 U/mg and 0.35 mM (Fig. 2D), respectively. Most metal ions, including Ca2+, Co2+, Cu2+, and MoO42+, enhanced enzyme activity, while Zn2+ and K+ did not (Fig. S3D).
Product analysis and 18O isotope experiments.
The products of the reaction catalyzed by CYP168A1 were identified using liquid chromatography-time-of-flight mass spectrometry (LC-TOF-MS), based on the mass spectra (m/z) of the target products. Products with molecular weights of [M-H]− 612.7000, 614.7000, and 616.7000 or 576.7240 and 578.7219 were detected. The results were matched to the previously reported standard compounds PBCDOH and two tetrabromocyclododecadiols (TBCDDOHs) (Fig. 3A and B). Products with molecular weights of [M-H]− 450.8949, 388.9792, 325.0679, and 263.1655 were also detected (Fig. 3C to F). The bromide was detected by ion chromatography analysis (Fig. S4). These products suggest that CYP168A1 can degrade HBCDs through a debromination and hydrogenation process, and one oxygen atom was added to the product in each step of the reaction. To determine the source of the oxygen that participates in the reaction, 18O isotope experiments were carried out as mentioned above. Products in the 18O2 group had molecular weights of [M-H]− 611.5232, 612.6262, and 613.5282, which match PBCD16OHs (Fig. 4A). In the non-16O-lyophilized and 16O-lyophilized groups, PBCD16OHs were detected (Fig. 4B and C). Thus, lyophilization would not inactivate the enzyme activity. In contrast, products containing 18O, with molecular weights of [M-H]− 612.7000, 614.7000, 616.7000, and 617.4908, were only detected in the H218O group (Fig. 4D). Comparing the results of Fig. 4A to C with Fig. 4D, 18O from H218O was added to PBCDOHs, and no 18O-labeled products formed in the 18O2 group. The results confirmed that the oxygen in PBCD18OHs was derived from H218O. All of the corresponding total ion chromatograph (TIC) spectra for product detection are shown in Fig. S5 and S6. The proposed pathway of HBCD degradation catalyzed by CYP168A1 is shown in Fig. 5.
FIG 3.
Identification of intermediates of HBCD degradation by LC-TOF-MS. (A) Mass spectra of PBCDOHs. (B) Mass spectra of TBCDDOHs. (C to F) Corresponding mass spectra of molecular weights m/z 450.8949, m/z 388.9792, m/z 325.0679, and m/z 263.1655.
FIG 4.
18O-labeled products of HBCD degradation. (A) Mass spectra extracted from the 18O group (PBCD16OHs). (B) Mass spectra extracted from the non-16O-lyophilized group (PBCD16OHs). (C) Mass spectra extracted from the 16O-lyophilized group (PBCD16OHs). (D) Mass spectra extracted from the 18O-lyophilized group (PBCD18OHs).
FIG 5.
Proposed pathway for HBCD degradation. HBCDs were dehalogenated by CYP168A1, with a sequential addition of hydroxide ions. The undetected oxohalonium metabolites have been drawn in frame.
Enhancement of degradation capacity of strain HS9.
To enhance the degradation capacity of strain HS9, the expression of gene cyp168A1 and the combined donor supplying system FdFNR were increased when promoter lacZ was added (Fig. 6A). The gene-engineering modes and comparison of the HBCD-degrading ability of wild-type HS9 (WT) and mutants PLAC-HS9, HS9-DW, and PLAC-DW are shown in Fig. 6B. The cell growth and degradation rates of the mutants were detected in the MSM-HBCD system, and the results showed that the degradation rates of strain HS9-DW were improved compared to those of the WT. However, increases in gene cyp168A1 expression cannot improve the degradation rate of HBCDs.
FIG 6.
Schematic for gene engineering (A) and comparison of the HBCD-degrading ability of the wild-type HS9 (WT) and genome-edited mutants PLAC-HS9, HS9-DW, and PLAC-DW (B).
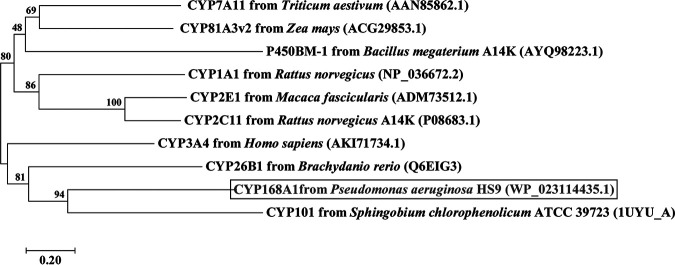
Phylogenetic analysis.
Several amino acid sequences of CYP dehalogenases, such as CYP7A11, CYP81A3v2 (17), CYP1A1, CYP2C11, CYP26B1 (13), CYP2E1 (15), CYP3A4 (17), CYP101 (14), and P450BM-1 (10), were compared, and the results showed that CYP168A1 was most closely related to CYP101 (Fig. 7).
FIG 7.
Phylogenetic tree analysis of CYP168A1 with reported cytochrome P450 enzymes that function in dehalogenation.
DISCUSSION
Hexabromocyclododecanes (HBCDs) have become a global research focus due to their widespread pollution and serious harm to human health, such as inducing cancer (22), disrupting liver and thyroid hormones (23, 24), and causing reproductive disorders (25). Several bacteria have been discovered from natural environments that can degrade HBCDs, such as Pseudomonas sp. strain HB01, Bacillus sp. strain HBCD-sjtu, Achromobacter sp. strain HBCD-1, Achromobacter sp. strain HBCD-2, and P. aeruginosa strain HS9 (26–29). Corresponding pathways have been proposed for these strains, but the specific molecular mechanisms of the degradation have not been revealed. In this study, the functional enzymes for HBCD degradation from P. aeruginosa strain HS9 were characterized.
Proteomic analysis comparing the expression of proteins of the cells incubated was carried out with MSM in the presence or absence of 1 mg/liter HBCDs, and the results showed that environmental stress response genes, like those encoding lactate dehydrogenases (LDH), were upregulated in the HBCD group. Enterococcus faecalis has general resistance to very different environmental stresses, depending on the ability to maintain redox balance via LDH (30). In addition, decreases and increases in salinity concentrations sharply increase the LDH activity of Neanthes arenaceodentata (31). Moreover, succinate dehydrogenase was upregulated, which could catalyze succinate to fumarate when an FADH was formed. Based on the above-described information, we propose that the resistance of strain HS9 to HBCD stress occurs due to maintaining the balance of reducing power in vivo, coupled with HBCD degradation. HBCD could induce the expression of cyp168A1 of strain HS9, while the CYP168A1 cooperation with electron donators and the electron transport and stress resistance reactions (related to lactate dehydrogenases or succinate dehydrogenase) were used to balance the electron supply in vivo.
In nature, cytochrome P450 (CYP) enzymes participate in degrading large amounts of environmental pollutants. Typically, CYP monooxygenases introduce a single oxygen atom into their substrates (32–34). However, there are few reports about CYP enzymes simultaneously catalyzing debromination and hydrogenation reactions (35). In this study, a novel CYP (CYP168A1) was shown to be the initial dehalogenase enzyme in HBCD biodegradation. The gene cyp168A1 was cloned and expressed in E. coli, and the enzymatic properties of the purified CYP168A1 were investigated. The Km of CYP168A1 for HBCDs was 0.35 mM, while the Km of γ-HBCD for LinB was 1.82 ± 0.60 μM. The affinity for HBCD to LinB is almost 1,000 times that of HBCD to CYP168A1. However, biochemistry information for the other two major HBCD isomers to LinB was still limited. The affinity of CYP168A1 to HBCDs was lower than that of LinA/B, and it matched the lower degradation rate of strain HS9, unlike other HBCD degraders.
Considering the toxicity of HBCDs to the environment and humans, comparative metabolism studies and in vitro activity tests indicated that the human liver CYP3A4, maize CYPs, and male rat CYPs can degrade different HBCD isomers. To trace the source of gene cyp168A1, phylogenetic analysis was carried out. Several mono- and dihydroxylated metabolites of HBCDs are formed through catalyzing with the human liver CYP3A4, maize CYPs, and male rat CYPs, with mono-OH-HBCDs detected as the major metabolites (6, 17, 34). However, the products of HBCDs catalyzed by CYP168A1 were PBCDOHs and TBCDDOHs, generated from the debromination and hydrogenation processes of HBCDs, of which PBCDOHs were also identical to that produced by strain HS9 in HBCD-MSM (21). This result revealed the difference in HBCD biodegradation between eukaryotic cells and prokaryotic microorganisms.
In the reactions of 1,2-halododecanoic acid oxidation catalyzed by both CYP4A (35) and CYP52A (36), oxygen in 1,2-hydroxydodecanoic acids derives from water, not from molecular oxygen, which was introduced by hydrolysis of an initially formed oxohalonium (R–X+–O−) metabolite. The results of 18O isotope labeling reactions showed that H2O serves as the source of the oxygen atom incorporated into PBCDOHs (Fig. 4). The mechanism of the oxygen addition was the same as oxidation of 1,2-halododecanoic acids by CYP4A and CYP52A. The present study revealed a novel mechanism of CYP to catalyze the brominated organic compounds.
In summary, this study reveals a new catalytic mechanism of CYP168A1 for the degradation of HBCDs in which the debromination and hydrogenation reactions are carried out one after another. The 18O isotope experiments show that the oxygen added to hydrated products was from H2O. Engineering mutants of strain HS9 not only supplies new insights into biochemical properties of protein CYP168A1 but also serves as a model for enhancing the abilities of this strain in bioremediation.
MATERIALS AND METHODS
Chemicals.
1,2,5,6,9,10-Hexabromocyclododecanes (HBCDs; ≥95% pure) were purchased from Anpel (NJ, USA). Hexachlorobenzene (HCB; ≥95% pure) was purchased from AccuStandard (CT, USA). Ethyl acetate, methanol, and all other reagents and solvents used in this study were of analytical grade.
Strains and culture media.
Pseudomonas aeruginosa HS9 was isolated by our research group in a previous work, and it can be obtained from the China Center for Type Culture Collection (CCTCC) under accession number M 2019094 (20). Escherichia coli DH5α and BL21(DE3) (Novagen, Inc. USA) were used for plasmid construction and protein expression, respectively. Lysogeny broth (LB), containing 5 g/liter yeast extraction, 10 g/liter tryptone, and 5 g/liter NaCl, or LB agar (1.5%, wt/vol) plates with appropriate antibiotics were used to culture E. coli (37). E. coli harboring each of the constructed plasmids was grown at 37°C, 200 rpm, with 50 mg/liter kanamycin or 100 mg/liter ampicillin for pET28a or pETduet-1 vectors. Strain HS9 was grown at 30°C in mineral salt medium (MSM) containing 5.0 g/liter K2HPO4, 3.7 g/liter KH2PO4, 1.0 g/liter Na2SO4, 0.2 g/liter MgSO4·7H2O, 2.0 g/liter NH4Cl, and 0.5 ml 2,000× trace element solution. The trace element solution consisted of 0.3 g/liter FeCl2·4H2O, 0.038 g/liter CaCl2·6H2O, 0.02 g/liter MnCl2·4H2O, 0.014 g/liter ZnCl2, 0.0124 g/liter H3BO3, 0.04 g/liter Na2MoO4·2H2O, and 0.0034 g/liter CuCl2·2H2O (21).
Genome sequencing and proteomic assay of strain HS9.
The genomic DNA of strain HS9 was extracted using a Wizard genomic purification kit (A1125; Promega, USA). Genome sequencing was performed on the Illumina HiSeq-2000 platform. Functional genes were predicted and annotated with the Rapid Annotations using Subsystems Technology (RAST) annotation server (38). Proteomic analysis comparing the protein expression of cells incubated in the presence or absence of 1 mg/liter HBCD-MSM was carried out as follows. Strain HS9 was cultured in 2-liter flasks containing 1 liter HBCD-MSM. As a control group, strain HS9 was grown in sodium citrate medium. A total of 10 liters of culture was collected during the exponential phase. Both groups were detected with three biological replicates (37).
RT-qPCR.
Total RNA was isolated from strain HS9 incubated in the presence or absence of 1 mg/liter HBCD MSM using a total RNA kit (Tiangen, China). Total cDNA was synthesized using a SuperScript III reverse transcriptase (Invitrogen, USA). The 20-μl reverse transcription reaction system contained 1.0 μg total RNA, 0.5 mM deoxynucleoside triphosphate mix, 200 U transcriptase, and 12.5 ng random primers. The reactions were performed according to the manufacturer’s protocols. RT-qPCR was then carried out using the CEX96 real-time PCR detection system (Bio-Rad) with a SYBR green I real master mix (TianGen, China). All data of candidate genes were normalized to the expression level of 16S rRNA and presented relative to the expression level in cells growing in the absence of HBCDs. All detections were performed with three replicates (38–40).
Expression and purification of heterologously expressed His-CYP168A1.
The DNA fragment of cyp168A1 was amplified by PFU DNA polymerase (New England Biolabs, Ipswich, MA) with primers Fcyp168A1 (CCGGAATTCCTACTCGCAGGTCTTCTGAG) and Rcyp168A1 (CCCAAGCTTATGGACGACGCATTCAGCGA), in which the enzyme digestion sites (EcoRI and HindIII) are underlined. The double enzyme-digested DNA fragments were ligated into expression vector pET28a, which incorporates 6× histidine tags. The constructed plasmid pET28a-cyp168A1 then was transferred into E. coli BL21 for heterologous expression. The culture was induced by adding 0.6 mM isopropyl β-d-thiogalactopyranoside (IPTG) after the optical density at 600 nm (OD600) reached 0.6 to 0.8. The culture then was incubated at 30°C for 10 h. E. coli was harvested by centrifuging at 4,000 rpm for 20 min, and the pellet was resuspended with nickel column balance buffer (20 mM NaH2PO4-Na2HPO4, 300 mM NaCl, 10 mM imidazole, 6 M urea, pH 8.0); urea was used to denature the proteins to enhance solubility (41). The cell suspension was broken by repetitive sonication at 4°C, and the cell debris was removed by centrifugation at 10,000 rpm for 40 min. The His-CYP168A1 was loaded into the nickel column and then washed by gradient imidazole buffers at 10, 40, 70, and 100 to 300 mM (42). The residual imidazole in the eluted buffer was removed by gradient dialysis from buffer I (20 mM KH2PO4-K2HPO4, 4 M urea, 5% glycerol, 1% glycine, 1‰ mercaptoethanol, pH 8.0) for 2 h, to buffer II (20 mM KH2PO4-K2HPO4, 2 M urea, 5% glycerol, 1% glycine, 1‰ mercaptoethanol, pH 8.0) for 2 h, and then to buffer III (20 mM KH2PO4-K2HPO4, 5% glycerol, 1% glycine, 1‰ mercaptoethanol, pH 8.0) for 3 h. CYP168A1 was successively refolded in situ through a gradient of decreased urea concentrations (41).
Western blot analysis and UV-visible characterization of purified CYP168A1.
The purified CYP168A1 was determined by Western blotting using an anti-6× His tag antibody (Abcam, China). The purified CYP168A1 was diluted 10, 100, and 1,000 times, and 10 μl was transformed to polyvinylidene difluoride film. The carbon monoxide (CO) difference spectrum was performed in buffer (50 mM KH2PO4-K2HPO4, 1‰ mercaptoethanol, 5% glycerol, pH 8.0) at 20°C in a 0.5-ml quartz cuvette with a 1-mm path length. Protein CYP168A1 was reduced by adding 10 mM dithionite, and the CO complex was created by slow bubbling with CO gas for 90 s (42).
Construction of electron-supplying system and enzyme activity.
To test the in vitro activity of CYP168A1, sufficient electrons must be supplied to the reaction system. Therefore, an electron-supplying system (named FdFNR) was constructed by combining a 4Fe-4S ferredoxin (HS1040) (Fd) and an NAD(P)H-dependent ferredoxin reductase (HS6332) (FNR) with a glycine linker (GGGGG). The combined DNA fragment was ligated to expression vector pETduet-1 at the second multiple-cloning site (MCS). The protein was induced by adding 0.2 mM IPTG after the OD600 reached 0.6 to 0.8; the culture then was incubated at 16°C for 10 h. Ultimately, the cells were broken in PBS buffer with the same method as that for CYP168A1 purification, and it was used as the electron-supplying cell-free system. The electron-supplying ability was determined with potassium ferricyanide (K3[Fe(CN)6]) as the receptor of the free donor and NADPH as the source of donor. Absorbance of K3[Fe(CN)6] at 340 nm was measured, and the color feature (yellow) of K3[Fe(CN)6] was captured. To test the enzyme activity of CYP168A1 with HBCDs, 1 mg/liter HBCDs, 0.4 mM NADH, 5 μg purified CYP168A1, and 1 ml cell-free system were mixed, and the reaction system was incubated under different reaction conditions. The decrease in HBCD concentration was used to calculate enzyme activity. To test the effect of metal ions on enzyme activity, 10 mM chloride salts (NiCl2, CoCl2, CaCl2, CuCl2, MnCl2, ZnCl2, MgCl2, KCl, FeCl2 and NaMoO4) were separately added to the reaction system.
18O isotope experiments and analysis.
To confirm the source of the oxygen atom incorporated into the HBCD degradation products, 18O2 and H218O were used to supply oxygen atoms for CYP168A1 reactions. The 18O2 labeling reaction and anaerobic assay were performed in an anaerobic workstation AW200SG (Electrotek Ltd., UK). After excluding air for 1 h by N2 atmosphere, all the liquid (1 ml FdFNR buffer, 5 μg purified CYP168A1) was exposed to an N2 atmosphere for 30 min to remove O2. An activity assay system (equal to the system for enzyme activity detection) that was cell free and contained enzyme and NADH was dried and dissolved in H218O. All reactions were carried out at 30°C for 6 h. After terminating the reaction by adding 10 μl HCl (11.64 M) to the 1-ml reaction system, HBCDs were extracted by using an equal volume of ethyl acetate. The samples then were mixed using vortex oscillation for 30 s. Before detection, the upper organic phase was concentrated 30 times. Samples were analyzed using ultra-high-performance liquid chromatography/time-of-flight mass spectrometry (UPLC-TOF/MS).
HBCDs or products in activity assay experiments and the 18O isotope experiments were quantified by UPLC-TOF/MS, equipped with an Eclipse XDB C18 analytical column (5 μm; 4.6 by 150 μm; Keystone Scientific, Agilent). HBCDs in samples used for product detection were extracted as described above, and then the organic phase was concentrated about 1,000 times. A mobile phase of water and methanol at a flow rate of 0.25 ml/min was applied for the target compounds. The proportional gradient of the mobile phase was started at 95% methanol, increased linearly to 100% over 25 min, and then decreased directly to 95% for 10 min. For mass spectrometric analysis, the ionization source was run in negative mode, and MS detection was set from 0 to 1,700 m/z. All target compounds were extracted based on their hydrogen adduct ion [M+H]− at m/z and characterization of bromine isotope (43).
Bromide detection.
The detection of bromide was conducted on an ion chromatograph coupled with an AS11-HC negative ion column (ICS-5000+; Thermo Fisher, Germany). The samples were prepared by terminating the reaction by adding 10 μl HCl to the 1-ml reaction system, followed by centrifugation at 12,000 rpm for 5 min to remove the proteins.
Gene deletion and complementation.
The suicide vector pK18mobsacB-Gm, used for gene deletion, was derived from pK18mobsacB by replacing the kanamycin resistance gene with a gentamicin resistance gene (from plasmid pUCTn7T). Upstream (600 bp) and downstream (600 bp) fragments close to the target gene were amplified and aligned using fusion PCR. The primers and corresponding PCR functions used in this study are listed in Table 1. The constructed plasmid pK18mobsacB-Gm-cyp168A1AB was transferred from the E. coli donor strain S17-1 to P. aeruginosa HS9 by conjugal transfer. Donor strain S17-1 and recipient strain HS9 were mixed with a volume rate at 5:1 to 10:1 and cultured on LB solid medium without antibiotics at 37°C for 4 h and 30°C for 20 h, and then the mixture was spread on M9 solid plates, incubated at 30°C for 48 h with 50 mg/liter gentamicin, and resuspended and washed using saline solution. Finally, the correct transconjugants were washed and plated on LB-sucrose agar medium for plasmid elimination (44). The gene-engineered groups were obtained by inserting a lacZ promoter with the FdFNR genes before cyp168A1 (PLAC-HS9), inserting a single lacZ promoter with the FdFNR genes followed by cyp168A1 (HS9-DW), or combining the operations of PLAC-HS9 and HS9-DW to get PLAC-DW. The lacZ promoter sequence was amplified from clone vector pMD18T.
TABLE 1.
Primers used in this study
| Name | Sequencea (3′–5′) | Function |
|---|---|---|
| GmF | CCCAAGCTTATGTTACGCAGCAGCAACGA | Replacement of resistance gene |
| GmR | CTAGCTAGCTTAGGTGGCGGTACTTGGGT | Replacement of resistance gene |
| Fcyp168A1 | CCGGAATTCATGGACGACGCATTCAGCGA | Construction of pET28a-cyp168A1 |
| Rcyp168A1 | CCCAAGCTTCTCGCAGGTCTTCTGAGCGT | Construction of pET28a-cyp168A1 |
| AFcyp168A1 | TATGACATGATTACGAATTCATGGACGACGCATTCAGCGA | Gene knockout |
| ARcyp168A1 | GTTATAAATTTGGAGTGTGAGCACGGCGTCGGGGCCGAAG | Gene knockout |
| BFcyp168A1 | TCACACTCCAAATTTATAACGCGGCGAACGCGGTGGAGGA | Gene knockout |
| BRcyp168A1 | AGGTCGACTCTAGAGGATCCCTCGCAGGTCTTCTGAGCGT | Gene knockout |
| Uplac-A1F | TGACATGATTACGAATTCCGAATACCAGAACCAGGGCA | Construction of PLAC-HS9/PLAC-DW |
| Uplac-A1R | TGAGTGAGCTAACTCACATTGGCCCTTGCTCCGCTGGGTT | Construction of PLAC-HS9/PLAC-DW |
| UplacF | AATGTGAGTTAGCTCACTCA | Construction of PLAC-HS9/PLAC-DW |
| UplacR | TCGCTGAATGCGTCGTCCATGGCGTAATCATGGTCATAGC | Construction of PLAC-HS9/PLAC-DW |
| Uplac-B1F | ATGGACGACGCATTCAGCGA | Construction of PLAC-HS9/PLAC-DW |
| Uplac-B1R | GCAGGTCGACTCTAGAGGATCCCCCGGCATCGCCGTGGCTGG | Construction of PLAC-HS9/PLAC-DW |
| Dplac-A2F | TGACATGATTACGAATTCCCAGCCACGGCGATGCCGGG | Construction of HS9-DW/PLAC-DW |
| Dplac-A2R | TGAGTGAGCTAACTCACATTCTACTCGCAGGTCTTCTGAG | Construction of HS9-DW/PLAC-DW |
| Dplac-F | AATGTGAGTTAGCTCACTCA | Construction of HS9-DW/PLAC-DW |
| Dplac-R | TCCAGCACGACGAAGGTCATGGCGTAATCATGGTCATAGC | Construction of HS9-DW/PLAC-DW |
| DFERF | ATGACCTTCGTCGTGCTGGA | Construction of HS9-DW/PLAC-DW |
| DFERR | GGCAGCCGGCTGATCCTGCGTCACTTCTCGACGAAGGCGC | Construction of HS9-DW/PLAC-DW |
| Dplac-B2F | CGCAGGATCAGCCGGCTGCC | Construction of HS9-DW/PLAC-DW |
| Dplac-B2R | GCAGGTCGACTCTAGAGGATCCGAGGCCGACGACTTCATGGA | Construction of HS9-DW/PLAC-DW |
| cyp168A1cF | GGTCGACTCTAGAGGATCCCATGGACGACGCATTCAGCGA | Gene complementation |
| cyp168A1cR | AGGTCGACTCTAGAGGATCCCTCGCAGGTCTTCTGAGCGT | Gene complementation |
The underlining represents homologous sequences to the constructed vector.
The expression vector pUC18k was used for gene complementation in P. aeruginosa. The new expression plasmid pUC18k-cyp168A1 was electrotransferred to strain HS9. Electrocompetent cells were prepared as follows. First, the strain was cultured in LB medium at 30°C after the OD600 reached 0.6, and then the cells were pelleted at 4,000 rpm for 10 min after incubation on ice for 20 min; finally, the cell pellets were washed twice using electroporation buffer (10% glycerol) (45). Correct transformants were verified by PCR, and the cells were further incubated in LB medium with corresponding antibiotics.
Data availability.
This whole-genome sequence project was submitted to the NCBI database under accession number GCA_003319235.1.
ACKNOWLEDGMENTS
This study was supported by grants from the National Key Research and Development Project (2018YFA0901200), from the Shanghai Excellent Academic Leaders Program (20XD1421900), from the Shuguang Program (17SG09) supported by Shanghai Education Development Foundation and Shanghai Municipal Education Commission, from the National Natural Science Foundation of China (31770114), and from the Science and Technology Commission of Shanghai Municipality (17JC1403300).
We have no competing interests to declare.
L.H. and H.T. conceived and designed experiments. L.H. and W.W. performed experiments. H.T. and P.X. contributed reagents and materials. L.H., H.T., G.Z., and P.X. wrote the paper. All authors discussed and revised the manuscript. All authors commented on the manuscript before submission. All authors read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Hongzhi Tang, Email: tanghongzhi@sjtu.edu.cn.
Emma R. Master, University of Toronto
REFERENCES
- 1.Fonseca VM, Fernandes VJ, Araujo AS, Carvalho LH, Souza AG. 2005. Effect of halogenated flame-retardant additives in the pyrolysis and thermal degradation of polyester sisal composites. J Therm Anal Calorim 79:429–433. 10.1007/s10973-005-0079-x. [DOI] [Google Scholar]
- 2.Tang H, Wang L, Wang W, Yu H, Zhang K, Yao Y, Xu P. 2013. Systematic unraveling of the unsolved pathway of nicotine degradation in Pseudomonas. PLoS Genet 9:e1003923. 10.1371/journal.pgen.1003923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heeb NV, Wyss SA, Geueke B, Fleischmann T, Kohler HPE, Lienemann P. 2014. LinA2, a HCH-converting bacterial enzyme that dehydrohalogenates HBCDs. Chemosphere 107:194–202. 10.1016/j.chemosphere.2013.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Heeb NV, Zindel D, Geueke B, Kohler HPE, Lienemann P. 2012. Biotransformation of hexabromocyclododecanes (HBCDs) with linB—an HCH-converting bacterial enzyme. Environ Sci Technol 46:6566–6574. 10.1021/es2046487. [DOI] [PubMed] [Google Scholar]
- 5.Heeb NV, Manuel M, Simon W, Birgit G, Hans-Peter EK, Peter L. 2018. Kinetics and stereochemistry of LinB-catalyzed δ-HBCD transformation: comparison of in vitro and in silico results. Chemosphere 207:118–129. 10.1016/j.chemosphere.2018.05.057. [DOI] [PubMed] [Google Scholar]
- 6.Dimaano NG, Yamaguchi T, Fukunishi K, Tominaga T, Iwakami S. 2020. Functional characterization of cytochrome P450 CYP81A subfamily to disclose the pattern of cross-resistance in Echinochloa phyllopogon. Plant Mol Biol 102:403–416. 10.1007/s11103-019-00954-3. [DOI] [PubMed] [Google Scholar]
- 7.Ding J, Guotao LG, Huang Z. 2018. Research progress in microbial cytochrome P450 and xenobiotic metabolism. Chinese J Appl Environ Biol 3:657–662. [Google Scholar]
- 8.Karl F, Roger B. 2000. Cytochrome P4501A induction potencies of polycyclic aromatic hydrocarbons in a fish hepatoma cell line: demonstration of additive interactions. Environ Toxicol Chem 19:2047–2058. [Google Scholar]
- 9.Brack W, Schirmer K, Kind T, Schrader S, Schüürmann G. 2002. Effect-directed fractionation and identification of cytochrome P4501 A-inducing halogenated aromatic hydrocarbons in a contaminated sediment. Environ Toxicol Chem 21:2654–2662. 10.1002/etc.5620211218. [DOI] [PubMed] [Google Scholar]
- 10.Sakaki T, Yamamoto K, Ikushiro S. 2013. Possibility of application of cytochrome P450 to bioremediation of dioxins. Biotechnol Appl Biochem 60:65–70. 10.1002/bab.1067. [DOI] [PubMed] [Google Scholar]
- 11.Guo F, Iwakami S, Yamaguchi T, Uchino A, Sunohara Y, Matsumoto H. 2019. Role of CYP81A cytochrome P450s in clomazone metabolism in Echinochloa phyllopogon. Plant Sci 283:321–328. 10.1016/j.plantsci.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Iwakami S, Kamidate Y, Yamaguchi T, Ishizaka M, Endo M, Suda H, Nagai K, Sunohara Y, Toki S, Uchino A, Tominaga T, Matsumoto H. 2019. CYP81A P450s are involved in concomitant cross-resistance to ALS and ACCase herbicides in Echinochloa phyllopogon. New Phytol 221:2112–2122. 10.1111/nph.15552. [DOI] [PubMed] [Google Scholar]
- 13.Sakaki T, Shinkyo R, Takita T, Ohta M, Inouye K. 2002. Biodegradation of polychlorinated dibenzo-p-dioxins by recombinant yeast expressing rat CYP1A subfamily. Arch Biochem Biophys 401:91–98. 10.1016/S0003-9861(02)00036-X. [DOI] [PubMed] [Google Scholar]
- 14.Yan D, Liu H, Zhou NY. 2006. Conversion of Sphingobium chlorophenolicum ATCC 39723 to a hexachlorobenzene degrader by metabolic engineering. Appl Environ Microbiol 72:2283–2286. 10.1128/AEM.72.3.2283-2286.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh S, Sherkhane PD, Kale SP, Eapen S. 2011. Expression of a human cytochrome P450 2E1 in Nicotiana tabacum enhances tolerance and remediation of γ-hexachlorocyclohexane. Nat Biotechnol 28:423–429. 10.1016/j.nbt.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 16.Huang H, Wang D, Wen B, Lv J, Zhang S. 2019. Roles of maize cytochrome P450 (CYP) enzymes in stereo-selective metabolism of hexabromocyclododecanes (HBCDs) as evidenced by in vitro degradation, biological response and in silico studies. Sci Total Environ 656:364–372. 10.1016/j.scitotenv.2018.11.351. [DOI] [PubMed] [Google Scholar]
- 17.Erratico C, Zheng X, Nele VDE, Tomy GT, Covaci A. 2016. Stereo-selective metabolism of α-, β- and γ-hexabromocyclododecanes (HBCDs) by human liver microsomes and CYP3A4. Environ Sci Technol 50:8263–8273. 10.1021/acs.est.6b01059. [DOI] [PubMed] [Google Scholar]
- 18.Esslinger S, Becker R, Maul R, Nehls I. 2011. Hexabromocyclododecane enantiomers: microsomal degradation and patterns of hydroxylated metabolites. Environ Sci Technol 45:3938–3944. 10.1021/es1039584. [DOI] [PubMed] [Google Scholar]
- 19.Zheng X, Erratico C, Abdallah MA, Negreira N, Luo X, Mai B, Covaci A. 2015. In vitro metabolism of BDE-47, BDE-99, and α-, β-, γ-HBCD isomers by chicken liver microsomes. Environ Res 143:221–228. 10.1016/j.envres.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 20.Zheng X, Erratico C, Luo X, Mai B, Covaci A. 2016. Oxidative metabolism of BDE-47, BDE-99, and HBCDs by cat liver microsomes: implications of cats as sentinel species to monitor human exposure to environmental pollutants. Chemosphere 151:30–36. 10.1016/j.chemosphere.2016.02.054. [DOI] [PubMed] [Google Scholar]
- 21.Huang L, Wang W, Shah SB, Hu H, Xu P, Tang H. 2019. The HBCDs biodegradation using a Pseudomonas strain and its application in soil phytoremediation. J Hazard Mater 380:e120833. 10.1016/j.jhazmat.2019.120833. [DOI] [PubMed] [Google Scholar]
- 22.Yvonne F, Inga B. 2009. Technical pentabromodipheny ether and hexabromocyclododecane as activators of the pregnane-X-receptor (PXR). Toxicol 29:656–661. 10.1016/j.tox.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Palace VP, Pleskach K, Halldorson T, Danell R, Wautier K, Evans B, Alaee M, Marvin C, Tomy GT. 2008. Biotransformation enzymes and thyroid axis disruption in juvenile rainbow trout (Oncorhynchus mykiss) exposed to hexabromocyclododecane diastereoisomers. Environ Sci Technol 42:1967–1972. 10.1021/es702565h. [DOI] [PubMed] [Google Scholar]
- 24.Ven L, Verhoef A, Kuil TVD, Slob W, Leonards PEG, Visser TJ, Hamers T, Herlin M, Hakansson H, Olausson H, Piersma A, Vos J. 2006. A 28-day oral dose toxicity study enhanced to detect endocrine effects of hexabromocyclododecane in Wistar rats. Toxicol Sci 94:281–292. 10.1093/toxsci/kfl113. [DOI] [PubMed] [Google Scholar]
- 25.Makoto E, Sakiko F, Mutsuko HK, Mariko M. 2008. Two-generation reproductive toxicity study of the flame retardant hexabromocyclododecane in rats. Reprod Toxicol 25:335–351. https://www.sciencedirect.com/science/article/abs/pii/S0890623807003383. [DOI] [PubMed] [Google Scholar]
- 26.Gao Y, Zhang X, Yang C. 2011. Photodegadation of hexabromocyclododecane in water. Environ Chem 30:598–603. [Google Scholar]
- 27.Zhao YY, Zhang XH, Sojinu OS. 2010. Thermodynamics and photochemical properties of alpha, beta, and gamma-hexabromocyclododecanes: a theoretical study. Chemosphere 80:150–156. 10.1016/j.chemosphere.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Zhou D, Wu Y, Feng X, Chen Y, Wang Z, Tao T, Wei D. 2014. Photodegradation of hexabromocyclododecane (HBCD) by Fe(III) complexes/H2O2 under simulated sunlight. Environ Sci Pollut Res Int 21:6228–6233. 10.1007/s11356-014-2553-0. [DOI] [PubMed] [Google Scholar]
- 29.Nyholm J, Lundberg C, Andersson PL. 2010. Biodegradation kinetics of selected brominated flame retardants in aerobic and anaerobic soil. Environ Pollut 158:2235–2240. 10.1016/j.envpol.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 30.Rana NF, Sauvageot N, Laplace JM, Bao Y, Nes I, Rince A, Posteraro B, Sanguinetti M, Hartke A. 2013. Redox balance via lactate dehydrogenase is important for multiple stress resistance and virulence in Enterococcus faecalis. Infect Immun 81:2662–2668. 10.1128/IAI.01299-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cripps RA, Reish DJ. 1973. The effect of environmental stress on the activity of malate dehydrogenase and lactate dehydrogenase and lactate dehydrogenase in Neanthes arenacedentata (Annelida: Polychaeta). Comp Biochem Phys B 46:123–133. 10.1016/0305-0491(73)90052-7. [DOI] [PubMed] [Google Scholar]
- 32.Durairaj P, Hur JS, Yun H. 2016. Versatile biocatalysis of fungal cytochrome P450 monooxygenases. Microb Cell Fact 15:125. 10.1186/s12934-016-0523-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urlacher VB, Girhard M. 2019. Cytochrome P450 monooxygenases in biotechnology and synthetic biology. Trends Biotechnol 37:882–897. 10.1016/j.tibtech.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Hakk H. 2016. Comparative metabolism studies of hexabromocyclododecane (HBCD) diastereomers in male rats following a single oral dose. Environ Sci Technol 50:89–96. 10.1021/acs.est.5b04510. [DOI] [PubMed] [Google Scholar]
- 35.He X, Cryle MJ, De Voss JJ, Ortiz MPR. 2005. Calibration of the channel that determines the ω-hydroxylation regiospecificity of cytochrome P4504A1. J Biol Chem 280:22697–22705. 10.1074/jbc.M502632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D, Cryle MJ, De Voss JJ, Ortiz MPR. 2007. Functional expression and characterization of cytochrome P450 52A21 from Candida albicans. Arch Biochem Biophys 464:213–220. 10.1016/j.abb.2007.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang H, Yao Y, Zhang D, Meng X, Wang L, Yu H, Ma L, Xu P. 2011. A novel NADH-dependent and FAD-containing hydroxylase is crucial for nicotine degradation by Pseudomonas putida. J Biol Chem 286:39179–39187. 10.1074/jbc.M111.283929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu X, Wang W, Zhang L, Hu H, Xu P, Wei T, Tang H. 2019. Molecular mechanism of N,N-dimethylformamide degradation in Methylobacterium sp. strain DM1. Appl Environ Microbiol 85:e00275-19. 10.1128/AEM.00275-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao X, Tao F, Zhang K, Tang H, Xu P. 2017. Multiple roles for two efflux pumps in the polycyclic aromatic hydrocarbon-degrading Pseudomonas putida strain B6-2 (DSM 28064). Appl Environ Microbiol 83:e01882-17. 10.1128/AEM.01882-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu H, Tang H, Zhu X, Li Y, Xu P. 2015. Molecular mechanism of nicotine degradation by a newly isolated strain, Ochrobactrum sp. strain SJY1. Appl Environ Microbiol 81:272–281. 10.1128/AEM.02265-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu J, Zhang Y, Wang Z. 2018. Chicken (Gallus gallus) HNF1α expression in Escherichia coli and its purification. J Agric Biotechnol 3:e1. [Google Scholar]
- 42.Funhoff EG, Bauer U, García-Rubio I, Witholt B, van Beilen JB. 2006. CYP153A6, a soluble p450 oxygenase catalyzing terminal-alkane hydroxylation. J Bacteriol 188:5220–5227. 10.1128/JB.00286-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu H, Hausinger RP, Tang H, Xu P. 2014. Mechanism of the 6-hydroxy-3-succinoyl-pyridine 3-monooxygenase flavoprotein from Pseudomonas putida S16. J Biol Chem 289:29158–29170. 10.1074/jbc.M114.558049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu Y, Ma Q, Liu Z, Wang W, Tang H, Zhou J, Xu P. 2017. Unveiling the biotransformation mechanism of indole in a Cupriavidus sp. strain. Mol Microbiol 106:905–918. 10.1111/mmi.13852. [DOI] [PubMed] [Google Scholar]
- 45.Jiang Y, Tang H, Wu G, Xu P. 2015. Functional identification of a novel gene, moaE, for 3-succinoylpyridine degradation in Pseudomonas putida S16. Sci Rep 5:13464. 10.1038/srep13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S6, Table S1. Download AEM.00826-21-s0001.pdf, PDF file, 2.4 MB (2.4MB, pdf)
Data Availability Statement
This whole-genome sequence project was submitted to the NCBI database under accession number GCA_003319235.1.