ABSTRACT
Avian pathogenic Escherichia coli (APEC), an extraintestinal pathogenic E. coli (ExPEC), causes colibacillosis in chickens and is reportedly associated with urinary tract infections and meningitis in humans. Development of resistance is a major limitation of current ExPEC antibiotic therapy. New antibacterials that can circumvent resistance problem such as antimicrobial peptides (AMPs) are critically needed. Here, we evaluated the efficacy of Lactobacillus rhamnosus GG (LGG)-derived peptides against APEC and uncovered their potential antibacterial targets. Three peptides (NPSRQERR [P1], PDENK [P2], and VHTAPK [P3]) displayed inhibitory activity against APEC. These peptides were effective against APEC in biofilm and chicken macrophage HD11 cells. Treatment with these peptides reduced the cecum colonization (0.5 to 1.3 log) of APEC in chickens. Microbiota analysis revealed two peptides (P1 and P2) decreased Enterobacteriaceae abundance with minimal impact on overall cecal microbiota of chickens. Bacterial cytological profiling showed peptides disrupt APEC membranes either by causing membrane shedding, rupturing, or flaccidity. Furthermore, gene expression analysis revealed that peptides downregulated the expression of ompC (>13.0-fold), ompF (>11.3-fold), and mlaA (>4.9-fold), genes responsible for the maintenance of outer membrane (OM) lipid asymmetry. Consistently, immunoblot analysis also showed decreased levels of OmpC and MlaA proteins in APEC treated with peptides. Alanine scanning studies revealed residues crucial (P1, N, E, R and P; P2, D and E; P3, T, P, and K) for their activity. Overall, our study identified peptides with a new antibacterial target that can be developed to control APEC infections in chickens, thereby curtailing poultry-originated human ExPEC infections.
IMPORTANCE Avian pathogenic Escherichia coli (APEC) is a subgroup of extraintestinal pathogenic E. coli (ExPEC) and considered a foodborne zoonotic pathogen transmitted through consumption of contaminated poultry products. APEC shares genetic similarities with human ExPECs, including uropathogenic E. coli (UPEC) and neonatal meningitis E. coli (NMEC). Our study identified Lactobacillus rhamnosus GG (LGG)-derived peptides (P1 [NPSRQERR], P2 [PDENK], and P3 [VHTAPK]) effective in reducing APEC infection in chickens. Antimicrobial peptides (AMPs) are regarded as ideal candidates for antibacterial development because of their low propensity for resistance development and ability to kill resistant bacteria. Mechanistic studies showed peptides disrupt the APEC membrane by affecting the MlaA-OmpC/F system responsible for the maintenance of outer membrane (OM) lipid asymmetry, a promising new druggable target to overcome resistance problems in Gram-negative bacteria. Altogether, these peptides can provide a valuable approach for development of novel anti-ExPEC therapies, including APEC, human ExPECs, and other related Gram-negative pathogens. Furthermore, effective control of APEC infections in chickens can curb poultry-originated ExPEC infections in humans.
KEYWORDS: peptides, ExPEC, APEC, MlaA-OmpC/F, public health, resistance, chickens
INTRODUCTION
Avian pathogenic Escherichia coli (APEC), an extraintestinal pathogenic E. coli (ExPEC), is a severe and recalcitrant bacterial pathogen of poultry worldwide (1, 2). Despite improvements in the poultry production systems over the years, APEC continues to remain as a serious problem to the poultry industry worldwide (3). APEC causes multiple extraintestinal infections (yolk sac infection, omphalitis, respiratory tract infection, swollen head syndrome, septicemia, polyserositis, coligranuloma, enteritis, cellulitis, and salpingitis) in poultry, collectively referred to as avian colibacillosis (1, 4). Colibacillosis results in significant morbidity and mortality (up to 20%) and decreased meat (2% decline in live weight) and egg (up to 15%) production (4). Furthermore, in young chickens, APEC can be associated with up to 53.5% mortality (4) and can result in up to 36% to 43% carcass condemnation at slaughter (4). Thus, colibacillosis results in multimillion-dollar annual losses to the poultry industry and remains as a serious impediment to sustainable poultry production worldwide. Recently, APEC has also been reported as a foodborne human uropathogen which can be transmitted to humans through cross-contamination of food products while handling raw poultry meat or by consumption of undercooked poultry products (5). Colicin V (ColV) plasmids from poultry-associated APEC have been detected in E. coli isolates isolated from human patients with urinary tract infections, suggesting foodborne transmission of APEC from poultry to humans (5). Furthermore, APEC is also considered a source of antibiotic resistance genes (ARGs) to human pathogens, which can make the human infections difficult to treat; thus, APEC is a threat to both animal and human health (4, 6).
At present, antibiotics are commonly used to control APEC infections in poultry (2, 7, 8). However, APEC isolates are becoming more resistant to antibiotics, suggesting that the control of APEC infections will be challenging in the future (9). To date, APEC resistance to multiple antibiotics, including but not limited to tetracyclines, sulfonamides, aminoglycosides, quinolones, and β-lactams, has been reported worldwide (10). Moreover, the control of APEC infections is complicated by increased restrictions on antibiotic usage worldwide (particularly in the United States and European countries) in food-producing animals, including poultry, to reduce the emergence and transmission of antibiotic-resistant bacteria to humans (11). However, limiting on-farm use of antibiotics could significantly increase disease occurrence, leading to morbidity and mortality of food animals and thereby compromising production efficiency, food security, and food safety (12). Therefore, there is an urgent need for developing new antibacterials that can circumvent the resistance problem, such as antimicrobial peptides (AMPs), which will consequently promote sustainable poultry production as well as benefit public health.
Antimicrobial peptides (AMPs) are regarded as a new category of therapeutic agents as well as promising natural alternatives to conventional antibiotics (13, 14). A major strength of AMPs is their ability to kill antibiotic-resistant bacteria (15, 16). AMPs are relatively small (10 to 50 amino acid residues), easy to synthesize, and have fast and selective antimicrobial action with a low propensity for the development of resistance, which make them ideal candidates for antibacterial development (13, 17). The development of bacterial resistance to AMPs is less likely, due to AMPs’ mechanism of action involving multiple low-affinity targets rather than one defined high-affinity target, which is a characteristic of conventional antibiotics (14). The multiple low-affinity targets make it more difficult for bacteria to defend against AMPs by a single resistance mechanism (14). Furthermore, AMPs, unlike antibiotics, do not elicit bacterial stress pathways such as SOS and rpoS responsible for inducing bacterial mutations and resistance (17). AMPs therefore can be used to exploit weaknesses in antibiotic resistance mechanisms, considered the Achilles’ heel of antibiotic resistance (17). Most AMPs exhibit direct and rapid antimicrobial activity by disrupting the integrity of the bacterial membrane and/or by translocating into the cytoplasm of bacteria to act on intracellular targets (14). The differences between bacterial and mammalian membranes enable selective action of AMPs to bacterial membranes (14). Besides antimicrobial activity, AMPs also exhibit immunomodulatory activities, including suppression of proinflammatory responses, antiendotoxin activity, stimulation of chemotaxis, and differentiation of immune cells, thereby contributing to bacterial clearance by the host (13, 14, 17).
To date, few AMPs derived from soil bacteria such as colistin, gramicidin, vancomycin, and daptomycin have been successfully used as antibacterials to treat antibiotic-resistant bacteria (14, 17). Nisin, a polycyclic antibacterial peptide derived from probiotic Lactococcus lactis, has been used as food preservative and sanitizer (17). Similarly, multiple AMPs (pexiganan, omiganan, Lytixar [LTX-109], hlF1-11, Novexatin [NP-213], CZEN-002, LL-37, PXL01, iseganan [IB-367], and PAC-113) derived from natural (human, bovine, porcine, and frog) and synthetic sources are in clinical development, with indications against different bacterial pathogens (14, 17). The synthetic AMPs ZY4, SAAP-148, arenicin-3, AMPR-11, and CSP-4 are effective against multidrug-resistant (MDR) bacterial infections (15, 16, 18–20). Furthermore, AMPs (A3, P5, colicin E1, cecropin AD, cecropin A-D-Asn, cipB-lactoferricin [LFC]-lactoferrampin [LFA], sublancin, and cLF36) have shown efficacy in decreasing E. coli and Clostridium burden in the gut as well as enhancing the performance and immune status in pigs and chickens (21–23). Moreover, the efficacy of short or small AMPs (8 to 12 residues) has been also shown against Gram-negative sepsis (24), staphylococcal skin infections (25), and bone infections (26).
In this study, we measured the efficacy of probiotic Lactobacillus rhamnosus GG (LGG)-derived small peptides (NPSRQERR [P1], PDENK [P2], and VHTAPK [P3]) against APEC in vitro and in cultured chicken macrophage HD11 cells, wax moth (Galleria mellonella) larva, and chickens. These peptides were identified through liquid chromatography-mass spectrometry (LC-MS) analysis of LGG culture supernatant (27). We uncovered antibacterial targets of peptides using bacterial cytological profiling, gene expression, and immunoblot approaches and probed their structure-activity relationship by an alanine scanning mutagenesis method. We identified peptides effective in reducing APEC colonization in chickens and uncovered their likely mechanism of action, which involves maintenance of an outer membrane (OM) lipid asymmetry (MlaA-OmpC/F) system in APEC.
RESULTS
Three peptides (P1, P2, and P3) displayed bactericidal activity against APEC O78.
To assess the anti-APEC activity of peptides, peptides (NPSRQERR [P1], PDENK [P2], VHTAPK [P3], MLNERVK [P4], YTRGLPM [P5], and GKLSNK [P6]) were added to APEC suspension at different concentrations and incubated for 12 h at 37°C. The initial testing at 6 mM and 12 mM concentrations revealed 4 peptides (P2 > P1 > P4 > P3) inhibiting APEC growth (>10%) (see Fig. S1A in the supplemental material). P2 inhibited 26.6% of APEC growth at 6 mM, whereas, P1 inhibited 10% of APEC growth. Similarly, at 12 mM, P2 inhibited 100% of APEC growth, whereas, P1, P4, and P3 inhibited 36.8%, 24%, and 11.2% of APEC growth, respectively (Fig. S1B).
Furthermore, to determine the MIC, a dose-response study was conducted using different concentrations (6 mM, 9 mM, 12 mM, 15 mM, and 18 mM) of peptides. P1, P2, and P3 displayed 100% APEC growth inhibition (i.e., MIC) at 18 mM, 12 mM, and 18 mM, respectively (Fig. 1A to C). However, P4 did not display 100% APEC growth inhibition at concentrations up to 18 mM (Fig. 1D); therefore, only three peptides (P1, P2, and P3) were selected for further studies. The MIC50 (concentration that inhibits 50% of APEC growth) values of the peptides are displayed in Table S1. Additionally, the APEC cultures treated with peptides at their MICs were plated on LB agar plates to determine their bacteriostatic/bactericidal activity. No viable APEC colonies were observed (data not shown), indicating bactericidal activity of the peptides.
FIG 1.
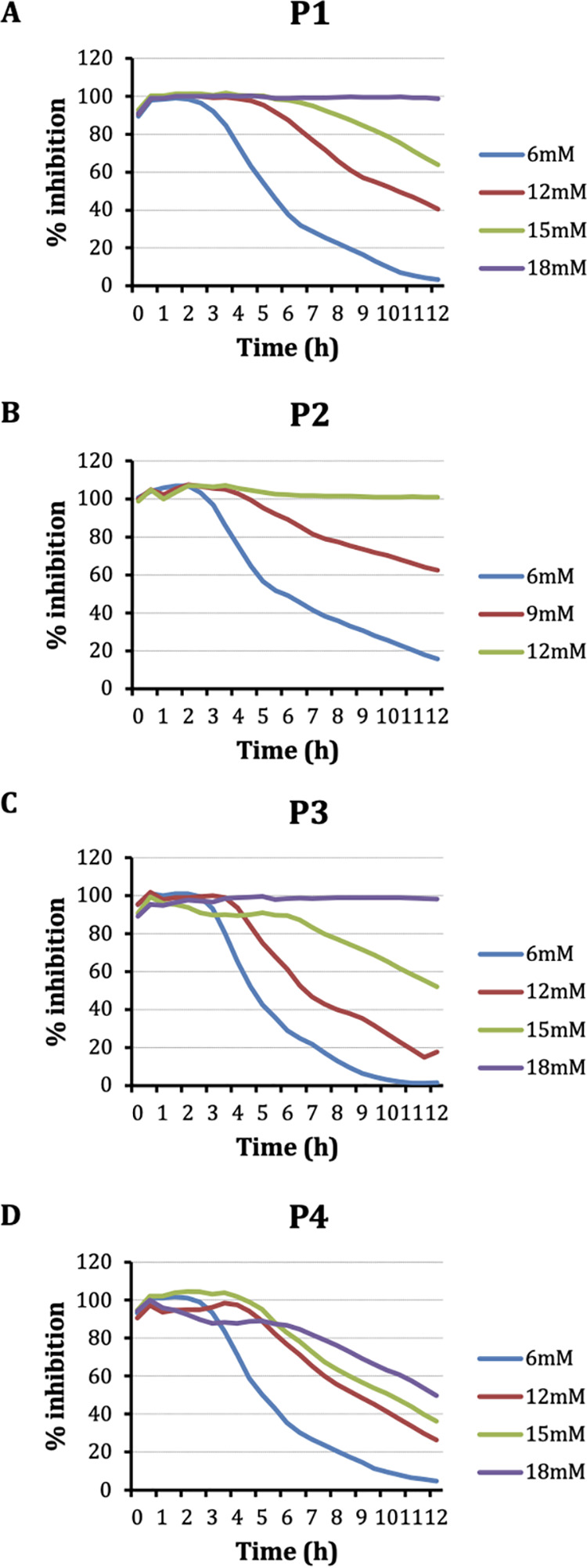
Inhibition of APEC growth by peptides P1 (A), P2 (B), P3 (C), and P4 (D) at different concentrations. Peptides were added to the wells of a 96-well plate containing APEC suspension and incubated at 37°C in a TECAN Sunrise absorbance microplate reader with kinetic absorbance measurement set at every 30 min for 12 h. The inhibition (%) was calculated using the formula (OD600 of the DMSO-treated well − OD600 of the peptide-treated well)/OD600 of the DMSO-treated well × 100. Two independent experiments were conducted.
Peptides displayed activity against multiple antibiotic-resistant APEC serotypes.
Peptides (P1, P2, and P3) were tested at their MICs against multiple APEC serotypes (O1, O2, O1-63, O2-211, O78-53, O8, O15, O18, O35, O109, O115, O78-X7122, O1-X7235, and O2-X7302) to determine their spectrum of activity. These tested APEC serotypes were previously found resistant to different antibiotics, including ampicillin, tetracycline, ciprofloxacin, and colistin (28). P1 and P2 displayed almost 100% (P1, 94.9% to 100%; P2, 94.8% to 100%) growth inhibition, whereas P3 displayed 43% to 90.4% growth inhibition against APEC serotypes (Fig. 2A, B, and C). These data suggest the broad spectral activity of peptides against multiple APEC serotypes irrespective of their resistant phenotype against antibiotics.
FIG 2.
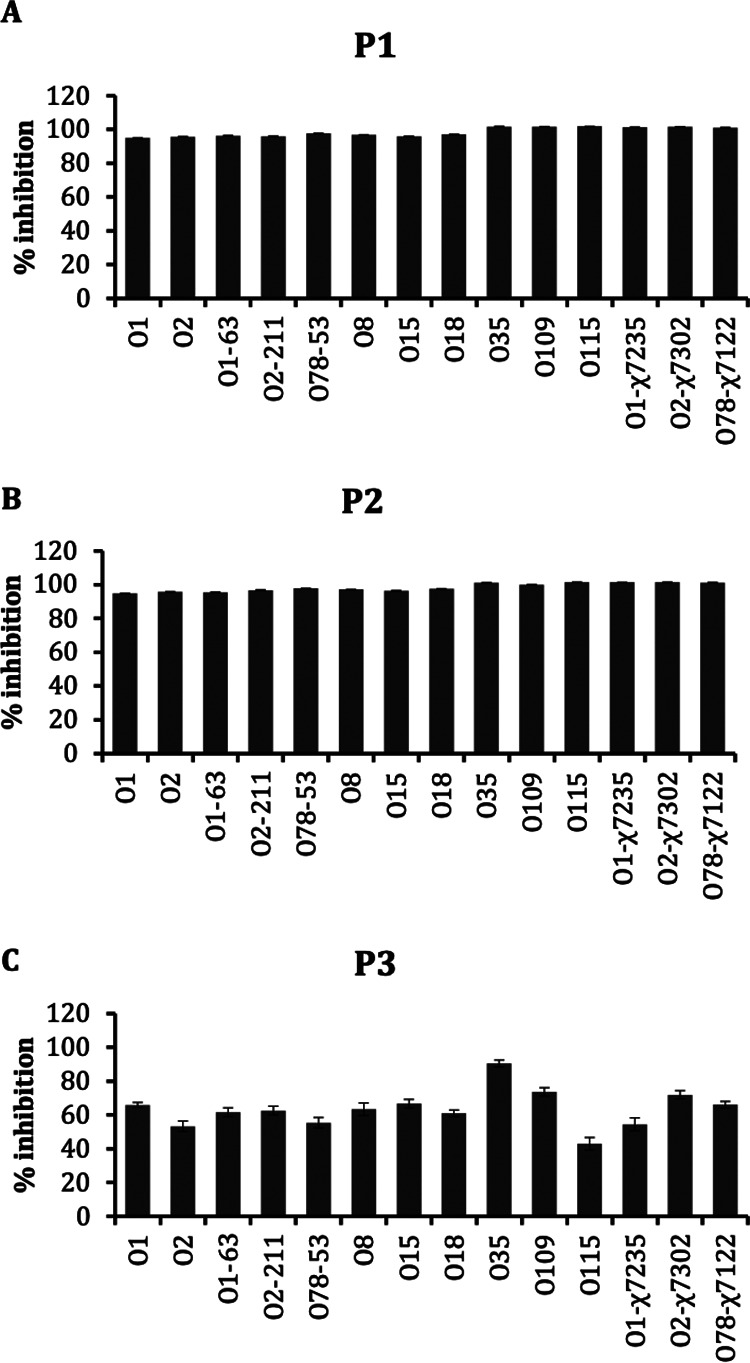
Inhibition of growth of different APEC serotypes/strains by peptides P1 (A), P2 (B), and P3 (C) at their MICs.
Peptides exhibited no effect against Gram-positive beneficial bacteria.
Peptides were tested at their MICs (P1 and P3, 18 mM; P2, 15 mM) against different commensal and probiotic bacteria to determine their specificity of activity. Interestingly, no effect on the growth of Gram-positive beneficial bacteria (Enterococcus faecalis, Streptococcus bovis, Lactobacillus rhamnosus GG (LGG), Lactobacillus acidophilus, Lactobacillus brevis, Bifidobacterium lactis Bb12, Bifidobacterium longum, and Bifidobacterium adolescentis) was observed when treated at the MICs of peptides. Growth of Gram-negative beneficial bacteria, particularly E. coli (Nissle 1917 and G58-1) but not the Bacteroides thetaiotaomicron, was inhibited at MICs of peptides, suggesting activity of peptides only against E. coli and related Gram-negative bacteria.
Peptides eradicated the biofilm-protected APEC in preformed biofilm and cleared intracellular APEC in macrophage cells.
The efficacy of peptides against biofilm-protected APEC was determined using the MBEC Assay. The preformed APEC biofilm in the pegs of the MBEC Assay device was treated with the peptides for 18 h at MICs followed by the enumeration of APEC. All peptide treatments (P1, P2, and P3) completely eradicated the biofilm-embedded APEC at their MICs (Table 1). The untreated pegs had 7.54 ± 0.07 log CFU/ml APEC in the biofilm.
TABLE 1.
Efficacy of peptides against biofilm protected and intracellular APEC O78 in macrophage cells
| Peptide | Efficacy (log CFU/ml)a |
Viability (%)b | |||
|---|---|---|---|---|---|
| Biofilm-embedded APEC | Intracellular APEC |
||||
| 12 mM | 15 mM | 18 mM | |||
| P1 | 0.00 ± 0.00c | 3.57 ± 0.07d | 0.00 ± 0.00c | 0.00 ± 0.00c | 96.5 |
| P2 | 0.00 ± 0.00c | 3.33 ± 0.03c | 0.00 ± 0.00c | 0.00 ± 0.00c | 98.5 |
| P3 | 0.00 ± 0.00c | 3.62 ± 0.06e | 2.75 ± 0.31d | 0.00 ± 0.00c | 99.1 |
| Untreated | 7.54 ± 0.07 | 3.75 ± 0.10 | 3.75 ± 0.10 | 3.75 ± 0.10 | 97.6 |
Values are means ± SDs.
Measured at 18 mM concentration.
P < 0.0001.
P < 0.01.
P < 0.05.
To measure the effect of peptides on the intracellular survival of APEC, a gentamicin protection assay was performed in chicken macrophage HD11 cells infected with APEC and treated with 12 mM, 15 mM, and 18 mM concentrations of peptides. The untreated HD11 cells had 3.75 ± 0.10 log CFU/ml APEC (Table 1). No intracellular APEC was recovered from cells treated with P1 and P2 at 15 mM and P3 at 18 mM. No effect on the viability of the HD11 cells was observed with treatment of up to 18 mM peptides when determined using the trypan blue exclusion test (Table 1).
Peptides disrupt the APEC membrane either by sloughing, rupturing, or inducing flaccidity.
To identify the mode of action (MOA) of peptides, a confocal fluorescence microscopy-mediated bacterial cytological profiling (BCP) method was utilized (29). The membrane stain FM4-64 (red) and nuclear stain SYTO-9 (green) were used. In the untreated APEC, a clearly demarcated APEC membrane encircling the chromosomes/nuclear material was visible (Fig. 3), whereas in P1-, P2-, and P3-treated APECs, no or minimally visible APEC membranes were observed, suggesting that peptides disrupted the APEC membrane. Consistent with the confocal images, in P1-, P2-, and P3-treated APECs, the membranes were either sloughed (or shed), ruptured, or flaccid, as observed by transmission electron microscopy (TEM), suggesting that the peptides affected the APEC membrane (Fig. 4). In untreated APEC, a clearly demarcated APEC membrane encircling the dense cytoplasmic contents was observed.
FIG 3.
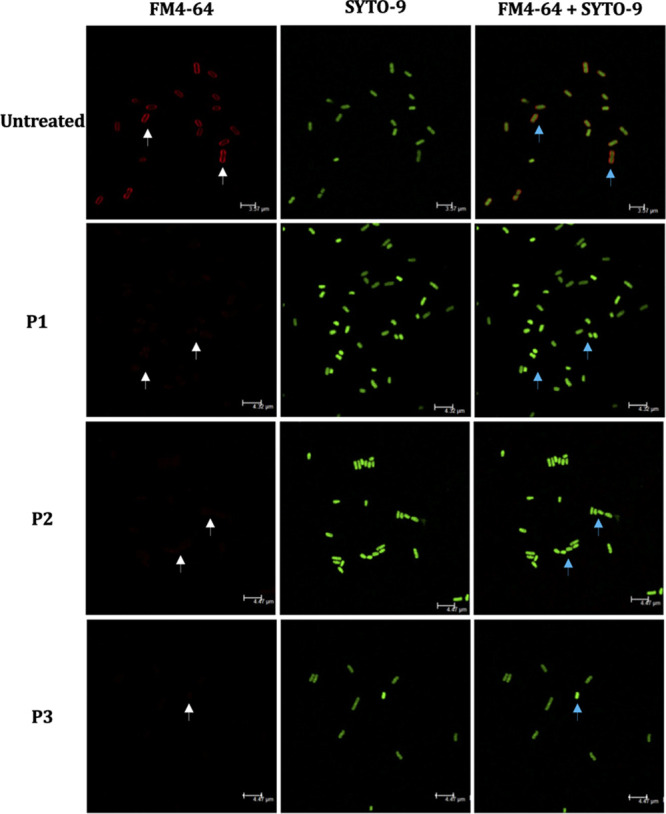
Confocal fluorescence images of APEC either untreated or treated with peptides. Red, FM4-64 (membrane stain); green, SYTO-9 (nuclear stain). APEC O78 cultures were treated with peptides (5× MIC), incubated (3 h), stained with FM4-64 and SYTO-9 (45 min), and imaged using a Leica TCS SP6 confocal scanning microscope. APEC membrane was clearly visible in untreated APEC (white arrows), whereas no or minimally visible membrane was observed in APEC treated with peptides. Superimposed images (FM4-64 plus SYTO-9) showed nuclear material of APEC enclosed by membrane in untreated APEC (blue arrows), whereas no membrane was visible covering the nuclear material in peptide-treated APEC.
FIG 4.
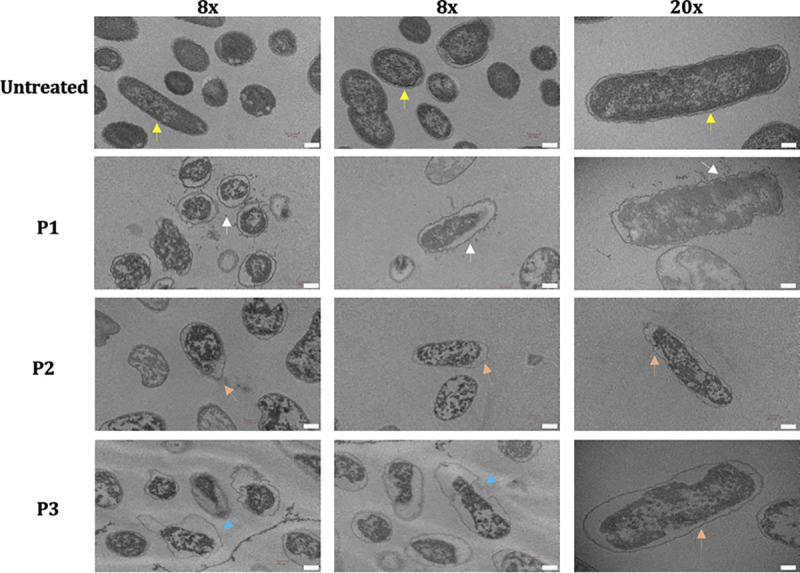
Transmission electron microscopy images (×8 and ×20 magnifications) of APEC either untreated or treated with peptides. APEC O78 cultures were treated with peptides (10× MIC), incubated (3 h), and imaged using a Hitachi H-7500 microscope. Clearly demarcated membrane encircling the dense cytoplasmic contents was observed in untreated APEC (yellow arrows), whereas the membrane was either sloughed/shed (white arrows), flaccid (orange arrows), or ruptured (blue arrows) in APEC treated with peptides. Bars, 0.2 μm.
Peptides protected wax moth larvae from APEC infection.
The in vivo efficacy and toxicity of peptides were measured in wax moth larvae. To measure the efficacy, larvae were pretreated with 25.5 mM peptides and infected with APEC, and larval survival was assessed for 72 h. APEC load in the larvae was quantified at 72 h postinfection. A 73.34% larval mortality was observed in the untreated group, whereas peptide treatments significantly (P < 0.001) increased the survival of larvae (Fig. 5A) with only 6.67%, 26.67%, and 6.67% mortality in P1-, P2-, and P3-treated groups, respectively. Peptides also significantly (P < 0.001) reduced the APEC load in the larvae (Fig. 5B). The untreated group had 7.4 ± 3.1 log CFU/larva of APEC, whereas no APEC was isolated in larvae treated with peptides, except for one larva in the P3-treated group.
FIG 5.
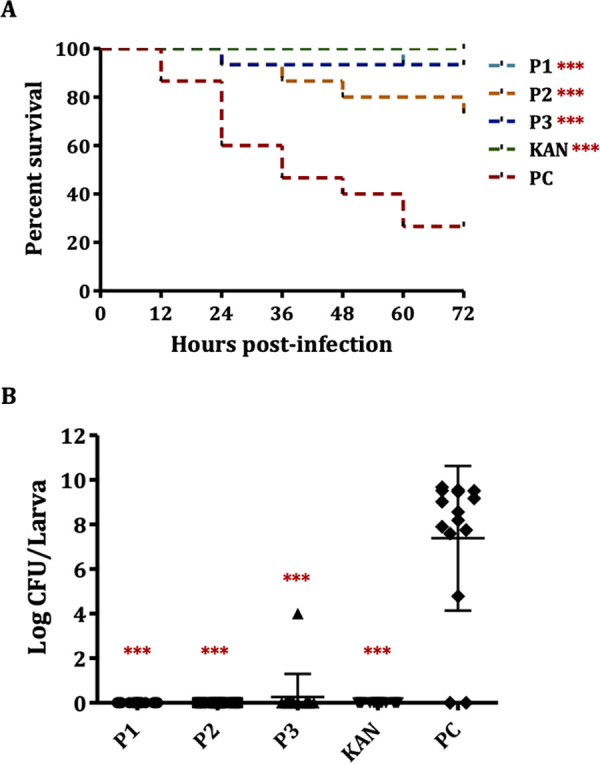
Efficacy of peptides in a wax moth (Galleria mellonella) larva model. (A) Survival curve of larvae either untreated or treated with peptides, ***, P < 0.001, log-rank test. (B) APEC load in larvae either untreated or treated with peptides. PC, infected and vehicle (sterile water containing DMSO)-treated larvae; KAN, infected and kanamycin (50 mg/kg body weight)-treated larvae; ***, P < 0.001, Tukey’s test.
To measure the toxicity, peptides were injected into larvae (noninfected) at 25.5 mM (>MIC), and larval survival was monitored for 72 h. No larval mortality was observed either in the control or in the peptide-treated groups in two independent experiments.
Alanine scanning revealed amino acid residues critical for anti-APEC activity of the peptides.
The alanine scanning libraries of peptides (see Table S2) were generated by substituting alanine at each amino acid residue to identify residues important for anti-APEC activity of peptides (30). In P1 (NPSRQERR), the relative importance of N (asparagine), P (proline), glutamate (E), and R (arginine) was >50%, whereas the relative importance of S (serine) and glutamine (Q) was <45% (Fig. 6A). Interestingly, relative importance of R (arginine) varied according to the position (16%, 59.1%, and 59.7% at positions 4, 7, and 8, respectively) in the peptide. In P2 (PDENK), the relative importance of D (aspartate) and E (glutamate) was very high (97.1%) compared to that of P (proline), N (asparagine), and K (lysine), whose relative importance was <40% (Fig. 6B). In P3 (VHTAPK), the relative importance of all amino acids, i.e., V (valine), H (histidine), T (tyrosine), P (proline), and K (lysine), was >50% (Fig. 6C).
FIG 6.
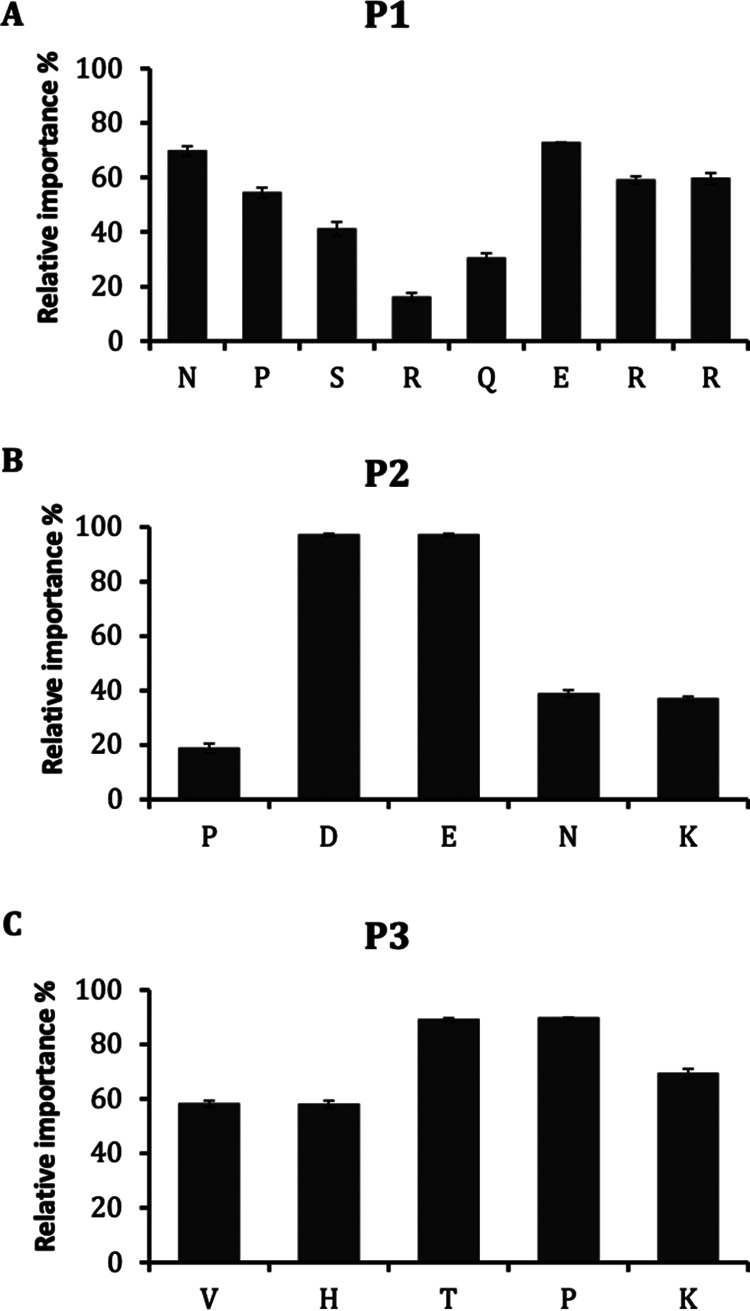
Relative importance (%) of each amino acid residue of peptides P1 (A), P2 (B), and P3 (C) determined through alanine scanning. Relative importance (%) was determined using the formula percent growth inhibition by peptide analogue/percent growth inhibition by original peptide × 100.
Arginine and lysine substitutions improved the anti-APEC activity of P1 and P2.
The amino acids not critical for antibacterial effect in each peptide were replaced with either arginine (R) or lysine (K) to enhance the anti-APEC activity (31). Compared to native peptide P1 (NPSRQERR), the MICs of NPRRQERR, NPSRRERR, and NPRRRERR were decreased by 3 mM to 6 mM (Table 2). Similarly, the MICs of KDENK and PDEKK were also decreased by 3 mM compared to that of the native peptide P2 (PDENK). However, the substitutions in peptide P3 (VHTAPK) increased the MICs of analogues.
TABLE 2.
MICs of the arginine/lysine-substituted peptide analoguesa
| Peptide | MIC (mM) |
|---|---|
| NPSRQERR | 18 |
| NPRRQERR | 12 |
| NPSRRERR | 15 |
| NPRRRERR | 12 |
| PDENK | 12 |
| KDENK | 9 |
| PDEKK | 9 |
| KDEKK | 12 |
| VHTAPK | 18 |
| KHTAPK | >18 |
| VKTAPK | >18 |
| VHTKPK | >18 |
| KKTKPK | >18 |
Amino acid residue in bold indicates the substituted amino acid in the peptide.
Peptides downregulated the expression of ompC, ompF, and mlaA genes responsible for the maintenance of OM lipid asymmetry.
The expression of genes essential for maintaining the OM integrity, including ompC, ompF, and mlaA, was quantified to determine the potential target(s) of the peptides (32–34). Interestingly, treatment with all peptides significantly downregulated the expression of ompC (13- to 31.4-fold), ompF (11.3- to 23.0-fold), and mlaA (4.9- to 6.3-fold), the genes responsible for the maintenance of OM lipid asymmetry in E. coli (33, 34) (Fig. 7A). No significant effect on the expression of other genes (lptD, bamA, lolB, and pbgA) was observed. Notably, the expression of the mlaC gene was only slightly affected (<1.5-fold), suggesting the peptides are likely affecting the OM.
FIG 7.
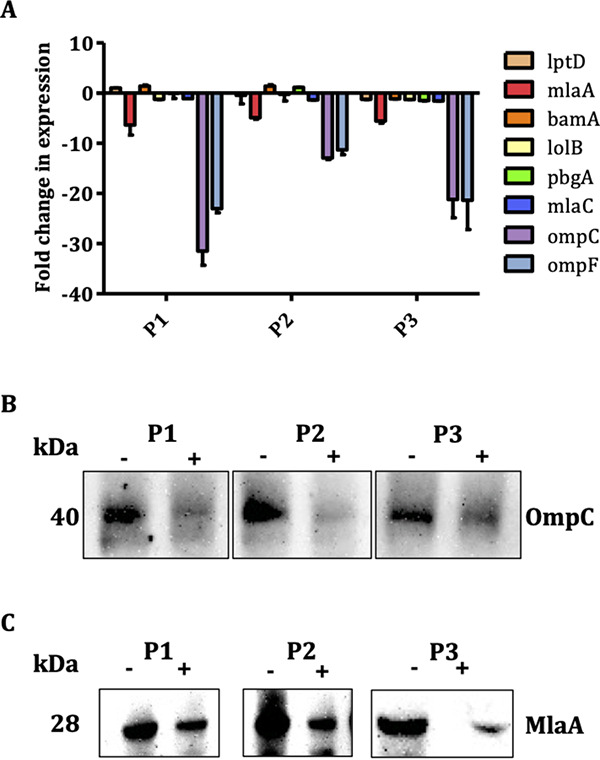
(A) Effect of peptides on the expression of genes essential for maintaining outer membrane integrity in APEC. APEC O78 cultures were treated with 50% lethal concentration of peptides, and RT-qPCR was performed. Fold change in expression was calculated using the ΔΔCT method with normalization to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (B and C) Effect of peptides on expression of OmpC and MlaA proteins. APEC O78 cultures were treated with 50% lethal concentration of peptides, membrane proteins were fractionated, and immunoblot was performed using anti-OmpC and anti-MlaA polyclonal antibodies. −, untreated, +, treated.
Peptides decreased the levels of OmpC and MlaA proteins.
The levels of OmpC and MlaA proteins in APEC treated with peptides were assessed using anti-OmpC and anti-MlaA polyclonal antibodies. Consistent with downregulation of the ompC gene, peptide treatments decreased the OmpC level in the OM of APEC compared to that in untreated APEC (Fig. 7B). Based on the densitometry analysis, the levels of OmpC were 4.76- to 9.27-fold (P1, 8.63-fold; P2, 9.27-fold; P3, 4.76-fold) lower in the peptide-treated APECs than in the untreated APEC. Similarly, peptide treatments also decreased the MlaA level (P1, 1.50-fold; P2, 1.40-fold; P3, 1.98-fold) in the OM of APEC compared to that in untreated APEC (Fig. 7C).
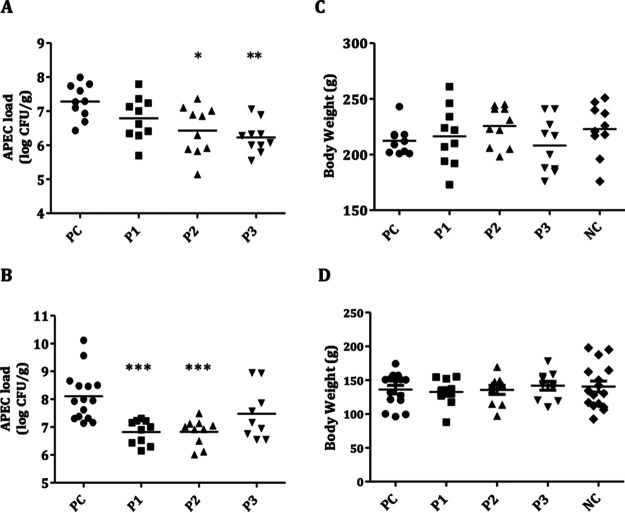
Peptides reduced APEC colonization in ceca of chickens.
Peptides were administered at 50-mg/kg body weight and 100-mg/kg body weight doses in two successive experiments in order to determine their efficacy. All peptides reduced the colonization of APEC in the ceca of chickens at both doses (Fig. 8). At the 50-mg/kg dose, P1, P2, and P3 reduced the colonization by 0.5, 0.9 (P < 0.05), and 1.1 (P < 0.01) log, respectively (Fig. 8A). At the 100-mg/kg dose, P1 (1.3 log, P < 0.001) and P2 (1.3 log, P < 0.001) showed better effects in reducing APEC colonization (Fig. 8B). Surprisingly, the efficacy of VHTAPK (0.6 log) was not enhanced with the increased dose. Notably, at the 50-mg/kg dose, peptides also reduced the APEC load in internal organs (lung, kidney, liver, and heart) of chickens, particularly in the lung (P < 0.05) (Table 3), whereas at the 100-mg/kg dose, with exception of P1, P2 and P3 also reduced the number of chickens positive for APEC in internal organs. Furthermore, at both doses, no significant effect on the body weights of chickens was observed (Fig. 8C and D).
FIG 8.
APEC load in ceca of chickens (at 7 dpi [days postinfection]) treated with peptides at 50-mg/kg body weight (A) and 100-mg/kg body weight (B) doses and body weights of chickens treated with peptides at 50-mg/kg body weight (C) and 100-mg/kg body weight (D) doses. *, P < 0.05; **, P < 0.01; ***, P < 0.001, Tukey’s test; PC, infected but not treated chickens; NC, noninfected and nontreated chickens.
TABLE 3.
Percentage of chickens positive for APEC in internal organs in different treatment groups
| Organ | % chickens positive at: |
|||||||
|---|---|---|---|---|---|---|---|---|
| 50 mg/kg peptide |
100 mg/kg peptide |
|||||||
| PCa | P1 | P2 | P3 | PC | P1 | P2 | P3 | |
| Lung | 90 | 20b | 40c | 30b | 46.67 | 60 | 30 | 40 |
| Kidney | 70 | 50 | 40 | 20c | 53.34 | 60 | 50 | 40 |
| Heart | 20 | 0 | 10 | 20 | 6.67 | 30 | 10 | 0 |
| Liver | 10 | 0 | 0 | 10 | 20 | 50 | 10 | 0 |
PC, untreated chickens.
P < 0.01.
P < 0.05.
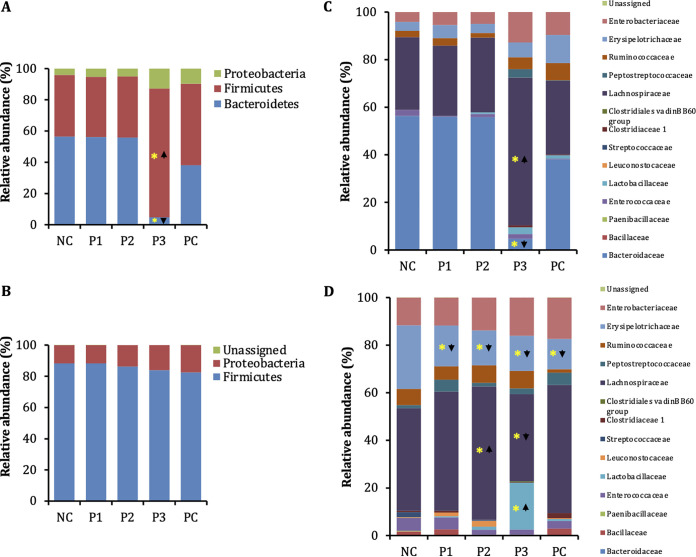
P1 and P2 decreased Enterobacteriaceae abundance with minimal impact on the cecal microbiota of chickens.
A metagenomic analysis of the cecal microbiota of chickens infected with APEC and treated with peptides was performed to investigate the effect of peptides on gut microbiota and identify the potential microbial markers associated with APEC infection in poultry (35). At the phylum level, no significant alteration was observed with the peptide treatment compared to the negative-control (NC; noninfected and nontreated) chickens, except in chickens treated with P3 at the 50-mg/kg dose (Fig. 9A and B). P3 (50 mg/kg) decreased Bacteroidetes abundance (56.39% to 4.78%) but increased Firmicutes (39.54% to 82.47%) abundance. At the class level, all peptide treatments (P1, P2, and P3) at the 100-mg/kg dose reduced Erysipelotrichia abundance (26.67% to 14.70%) compared to that in NC chickens, whereas P2 increased Clostridia (51.76% to 64.92%) abundance. P3 increased Bacilli abundance (7.28% to 22.17%) and decreased Clostridia (62.63% to 46.99%) abundance compared to that in the positive-control (PC; infected but not treated) chickens. On the other hand, peptide treatments (P1 and P2) at the 50-mg/kg dose reduced Erysipelotrichia abundance (11.82% to 3.83%) compared to that in PC chickens, whereas P3 increased Clostridia abundance (33.28% to 71.46%) and decreased Bacteroidia (38.09% to 4.78%) abundance compared to that in NC and PC chickens. At the family level, all peptide treatments (P1, P2, and P3) at the 100-mg/kg dose reduced Erysipelotrichaceae (26.67% to 14.70%) and Streptococcaceae (2.17% to 0%) abundance compared to that in NC chickens, whereas P2 increased Lachnospiraceae (43.05% to 56.10%) and Lactobacillaceae (0.01% to 1.38%) abundance and decreased Clostridiaceae 1 (0.53% to 0%) abundance (Fig. 9C and D). P3 increased Lactobacillaceae abundance (0.74% to 19.61%) and decreased Lachnospiraceae (53.78% to 36.58%) abundance compared to that in PC chickens, whereas Ruminococcaceae (1.40% to 7.35%) abundance was increased with all peptide treatments (P1, P2, and P3), and Leuconostocaceae abundance (0.31% to 0%) was decreased with P2 treatment. On the other hand, peptide treatments (P1 and P2) at the 50-mg/kg dose reduced Erysipelotrichaceae abundance (11.82% to 3.83%) compared to that in PC chickens, whereas P3 increased Lachnospiraceae abundance (30.95% to 62.09%) and decreased Bacteroidaceae (56.39% to 4.78%) abundance compared to that in NC and PC chickens (Fig. 9C and D). Interestingly, all peptide treatments (P1, P2, and P3) at 100-mg/kg and 50-mg/kg doses decreased Enterobacteriaceae abundance compared to that in PC chickens, except P3 treated at the 50-mg/kg dose (Fig. 9C and D, Table 4).
FIG 9.
Microbial relative abundance (at the phylum level) in ceca of chickens treated with peptides at 50-mg/kg (A) and 100-mg/kg (B) doses. NC, noninfected and nontreated chickens; PC, infected but not treated chickens. Microbial relative abundance (at the family level) in ceca of chickens treated with peptides at 50-mg/kg (C) and 100 mg/kg (D) doses. *, P < 0.05, Mann-Whitney U test.
TABLE 4.
Relative abundance of cecal bacteria at the genus level in different groups treated with peptides
| Organism and dose | Relative abundance (%) |
||||
|---|---|---|---|---|---|
| NC | P1 | P2 | P3 | PC | |
| 50-mg/kg dose | |||||
| Escherichia-Shigella | 4.07 | 4.95 | 4.95 | 9.17 | 8.14 |
| Erysipelatoclostridium | 3.79 | 5.53a | 3.84a | 6.22 | 11.82 |
| Ruminiclostridium 9 | 2.25 | 0.00 | 1.25 | 2.14 | 2.80 |
| Ruminiclostridium 5 | 0.00 | 1.69 | 0.23 | 0.00 | 0.00 |
| Oscillibacter | 0.08 | 0.18 | 0.00 | 0.71 | 1.08 |
| Flavonifractor | 0.35 | 0.63 | 0.68 | 1.39 | 0.65 |
| Anaerotruncus | 0.00 | 0.73 | 0.00 | 0.00 | 0.00 |
| Clostridioides | 0.00 | 0.00 | 0.00 | 0.00 | 0.19 |
| Lachnospiraceae (uncultured) | 2.16 | 0.00 | 2.30 | 7.40 | 1.65 |
| Ruminococcus torques group | 8.45 | 7.08 | 9.83 | 13.82 | 6.10 |
| Ruminococcus gnavus group | 0.52 | 0.00 | 0.00 | 0.00 | 0.00 |
| Clostridium sensu stricto 1 | 0.00 | 0.00 | 0.00 | 0.77 | 0.34 |
| Pediococcus | 0.00 | 0.00 | 0.74 | 2.88 | 1.35 |
| Enterococcus | 2.47 | 0.24 | 1.24 | 1.92 | 0.34 |
| Bacteroides | 56.40 | 56.08 | 55.87 | 4.79a,b | 38.09 |
| 100-mg/kg dose | |||||
| Escherichia-Shigella | 11.12 | 11.73 | 13.77 | 14.69 | 16.24 |
| Clostridium innocuum group | 2.03 | 0.00b | 0.00b | 0.00b | 0.00c |
| Erysipelatoclostridium | 24.65 | 17.11 | 14.71b | 14.78b | 12.68c |
| Eubacterium coprostanoligenes group | 0.00 | 0.00 | 0.00 | 0.21 | 0.00 |
| UBA1819 | 0.00 | 0.00 | 0.68 | 0.00 | 0.00 |
| Ruminococcaceae UCG-005 | 0.57 | 0.56 | 2.19 | 0.00 | 0.00 |
| Ruminiclostridium 9 | 0.00 | 0.00 | 0.78 | 1.52a,b | 0.00 |
| Ruminiclostridium 5 | 0.00 | 0.00 | 0.28 | 0.00 | 0.00 |
| Oscillibacter | 0.00 | 0.00 | 0.13 | 0.76 | 0.00 |
| Flavonifractor | 1.92 | 2.40 | 0.68b | 1.25 | 0.52c |
| Caproiciproducens | 0.43 | 0.00 | 0.00 | 0.34 | 0.00 |
| Butyricicoccus | 2.04 | 0.44b | 1.53 | 0.19b | 0.60 |
| Anaerotruncus | 1.47 | 0.00 | 0.00 | 1.65 | 0.12 |
| Clostridioides | 0.00 | 0.21 | 0.00 | 0.73 | 0.00 |
| Lachnospiraceae (uncultured) | 2.38 | 1.85 | 0.73 | 10.38b | 5.85 |
| Ruminococcus torques group | 9.31 | 7.62 | 10.37 | 8.44 | 10.93 |
| Sellimonas | 2.95 | 0.00b | 0.00b | 1.60a | 0.13c |
| Blautia | 0.00 | 0.00 | 0.00 | 1.07 | 0.00 |
| Clostridiales vadin BB60 group | 0.00 | 0.00 | 0.00 | 0.41a | 0.00 |
| Clostridium sensu stricto 1 | 0.53 | 0.68 | 0.00 | 0.21 | 2.16 |
| Lactococcus | 2.18 | 0.00b | 0.40 | 0.00 | 0.00c |
| Weissella | 0.32 | 1.60 | 2.43a | 0.00 | 0.40 |
| Pediococcus | 0.01 | 0.49 | 1.38b | 0.00 | 0.68 |
| Lactobacillus | 0.00 | 0.00 | 0.00 | 19.62a,b | 0.06 |
| Enterococcus | 5.37 | 4.99 | 2.01 | 2.14 | 3.16 |
| Paenibacillus | 0.27 | 0.00 | 0.00 | 0.00 | 0.00 |
| Bacillus | 1.74 | 2.64 | 0.37 | 0.42 | 2.98 |
Bacteria significantly (P < 0.05) altered in peptide-treated group compared to PC (infected but not treated) group.
Bacteria significantly (P < 0.05) altered in peptide-treated group compared to NC (noninfected and nontreated) group.
Bacteria significantly (P < 0.05) altered in PC group compared to NC group.
At the genus level, the abundance of Escherichia-Shigella was decreased at both 100-mg/kg (16.24% to 11.73%) and 50-mg/kg (8.14% to 4.95%) doses, except in chickens treated with P3 at the 50-mg/kg dose (Table 4). At the 50-mg/kg dose, P1 and P2 decreased Erysipelatoclostridium abundance (11.82% to 3.84%) compared to that in PC chickens, whereas P3 decreased Bacteroides abundance (56.04% to 38.09%) compared to that in PC and NC chickens (Table 4). At the 100-mg/kg dose, all peptides decreased the abundance of the Clostridium innocuum group (2.03% to 0.00%) compared to that in NC chickens (Table 4). Furthermore, P1 decreased the abundance of Butyricicoccus (2.04% to 0.44%), Sellimonas (2.95% to 0.00%), and Lactococcus (2.18 to 0.00%), whereas P2 decreased the abundance of Erysipelatoclostridium (24.65% to 14.71%), Flavonifractor (1.92% to 0.68%), and Sellimonas (2.95% to 0.00%) and increased the abundance of Pediococcus (0.01% to 1.38%) compared to that in NC chickens and increased the abundance of Weissella (0.40% to 2.43%) compared to that in PC chickens. P3 decreased the abundance of Erysipelatoclostridium (24.65% to 14.78%) and Butyricicoccus (2.04% to 0.19%) and increased the abundance of Lachnospiraceae (uncultured) (2.38% to 10.38%) compared to that in NC chickens but increased the abundance of Lactobacillus (0.00% to 19.62%) and Ruminiclostridium 9 (0.00% to 1.52%) compared to that in both PC and NC chickens and increased the abundance of Clostridiales vadin BB60 group (gut metagenome) (0.00% to 0.41%) and decreased the abundance of Sellimonas (2.95% to 1.60%) compared to that in PC chickens. Interestingly, compared to NC chickens, PC chickens showed decreased abundance of Clostridium innocuum group (2.03% to 0.00%), Erysipelatoclostridium (24.65% to 12.68%), Flavonifractor (1.92% to 0.52%), Sellimonas (2.95% to 0.13%), and Lactococcus (2.18% to 0.00%).
The analysis of alpha-diversity (Shannon index) revealed a significant difference (P < 0.05) in the microbial richness between the P1- and P2-treated chickens and the NC chickens, at both 100-mg/kg and 50-mg/kg doses (see Fig. S2). However, a significant difference (P < 0.01) was only observed between P3-treated chickens and NC chickens at the 50-mg/kg dose. Furthermore, the beta-diversity (weighted UniFrac) analysis showed that the microbial communities of P1- and P2-treated chickens were similar (P > 0.05) to that of NC chickens treated at the 50-mg/kg dose. In contrast, microbial communities of P3-treated and PC chickens were significantly different (P < 0.01) from that of NC chickens (see Fig. S3). At the 100-mg/kg dose, the microbial communities of P2- and P3-treated chickens and PC chickens were significantly different (P < 0.01) from that of NC chickens (Fig. S3). Overall, less impact on cecal microbiota was observed with P1 and P2 treatments than with the P3 treatment.
No resistance was observed in APEC against peptides in vitro or in vivo.
The acquisition of resistance by APEC to peptides was evaluated in sublethal and lethal resistance assays. No change in the MICs of peptides was observed against APEC cultures grown after 14 repeated passages in the presence of sublethal concentrations of peptides (0.75× MIC). Consistently, no resistant colonies were isolated when APEC cultures were plated on LB agar containing lethal concentrations (5× MIC) of peptides. Similarly, APEC colonies isolated from ceca and internal organs of chickens treated with peptides (described below) possessed the same MICs as described above.
Peptides bind with higher affinities to OmpC than OmpF and MlaA.
HPEPDOCK and PEP-SiteFinder tools were used to predict the affinity and binding sites of peptides with OmpC, OmpF, and MlaA proteins (36, 37). P1 bound with highest affinity (−207.969 kcal/mol) to OmpC followed by P3 (−168.294 kcal/mol) and P2 (−135.46 kcal/mol) (Fig. 10). Furthermore, the binding affinity of peptides to OmpF (P1, −197.146; P2, −129.838; P3, −146.798 kcal/mol) was lower than that to OmpC. Similarly, the binding propensities of peptides to OmpF (P1, 38; P2, 22.5; P3, 22) and MlaA (P1, 8; P2, 12.5; P3, 22) are lower than those to OmpC (P1, 50; P2, 30; P3, 34). P1 binds with a high propensity index (46 to 50) at W72, R174, Q61, and Q63 residues of the OmpC C chain and N46 residue of the B chain. P2 binds with a high propensity index (30 to 33) at E66, Q61, Q59, and W72 residues of the OmpC B chain. P3 binds with a high propensity index (34) at L20, Y22, Q33, Y35, R37, Q59, Q61, W72, R74, E109, F110, G112, G113, N113, S117, Q124, and Q123 residues of the OmpC B chain and E66 residues of the OmpC C chain.
FIG 10.
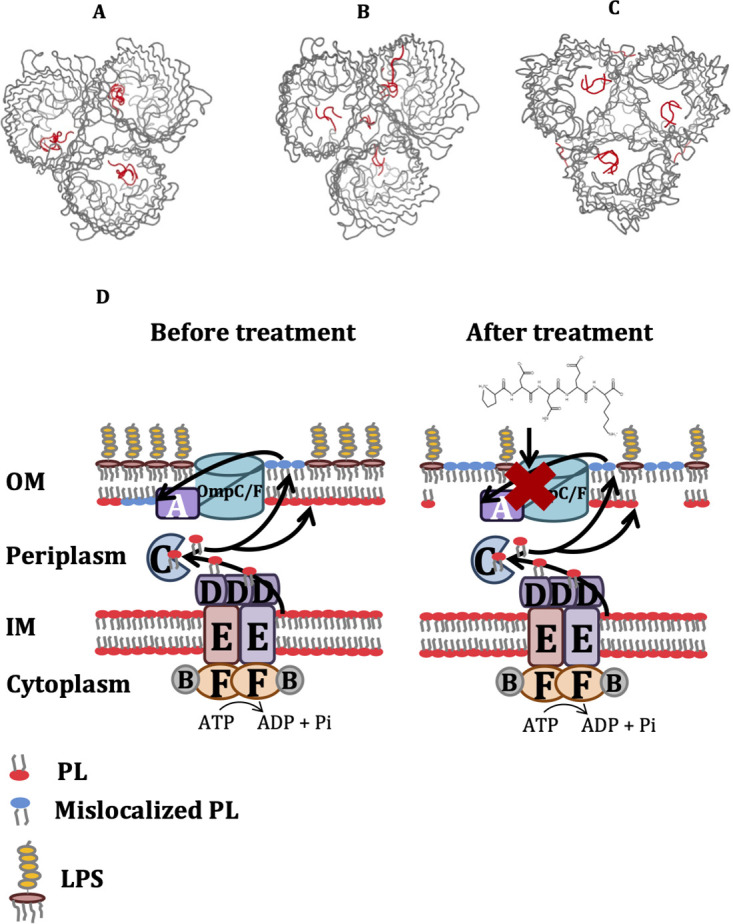
In silico prediction of binding sites (based on 10 best poses) of peptides P1 (A), P2 (B), and P3 (C) to trimeric OmpC (PDB 2J1N) using PEP-SiteFinder tool. (D) Hypothetical mechanism of action of peptides. Peptides impair APEC OM lipid asymmetry by inhibiting the function of the MlaA-OmpC/F system. MlaA-OmpC/F system retrogradely transports mislocalized phospholipid (PL) from the outer leaflet of the OM to inner leaflet of the OM in order to maintain the lipid asymmetry. PL, phospholipid, LPS, lipopolysaccharide.
DISCUSSION
Our study uncovered the potential of LGG-derived small peptides (P1, NPSRQERR; P2, PDENK; P3, VHTAPK) as new antibacterials against APEC infection in chickens (Fig. 8). To date, no small peptide-based (fewer than 10 amino acid residues) antibacterials have been evaluated specifically to treat bacterial infections in chickens. Several larger peptides (A3, P5, PABP, cecropin A-D-Asn, microcin J25, cLF36, cLFchimera, and piscidin) have been evaluated and shown efficacy to promote growth performance, improve nutrient digestibility and gut morphology, enhance intestinal mucosal immune responses, and modulate gut microbiota in chickens (22, 23, 46–48). Although not targeted to any specific bacterial pathogen, it is reported that treatment with these peptides favors the proliferation of beneficial microorganisms such as Lactobacillus and Bifidobacterium and suppression of harmful microorganisms such as coliforms and Clostridium spp. (22, 23, 46, 48). Therefore, our data suggest a new avenue for the development of small peptide-based antibacterials against bacterial infections in chickens.
Even though there are not many previous reports on treating bacterial infections in chickens using peptides, different peptides have shown efficacy against MDR bacterial infections in other animal models (15, 18–20). A cyclic peptide ZY4 resists MDR Pseudomonas aeruginosa and Acinetobacter baumannii infections by decreasing susceptibility to lung infection by P. aeruginosa and suppressing dissemination of P. aeruginosa and A. baumannii to target organs in a mouse septicemia model (18). Similarly, arenicin-3, an amphipathic peptide, is effective in reducing bacterial (MDR E. coli and P. aeruginosa) burden in mouse models of peritonitis, pneumonia, and urinary tract infection (19). AMPR-11, a peptide derived from Romo1 (reactive oxygen species modulator 1), also increased the survival of mice and decreased the bacterial load in murine model of sepsis with MDR bacteria, including methicillin-resistant Staphylococcus aureus (MRSA) and carbapenem-resistant P. aeruginosa, Klebsiella pneumoniae, and A. baumannii (20). More interestingly, SAAP-148, a synthetic peptide derived from LL-37, eradicated biofilm-associated infections with MRSA and MDR A. baumannii from wounded ex vivo human skin and murine skin in vivo (15). These studies indicate that peptides can be valuable antibacterial agents to control bacterial infections, including antibiotic-resistant infections.
Our study showed that all three peptides affected the MlaA-OmpC/F system in APEC, which impairs OM lipid asymmetry. The hypothetical mechanism of action of peptides is shown in Fig. 10. The MlaA-OmpC/F system maintains OM lipid asymmetry by retrogradely transporting mislocalized phospholipid (PL) from the outer leaflet of the OM to the inner leaflet or inner membrane (IM) and is regarded as a novel target for antibacterial drug development (32, 49, 50). It has been reported that MlaA interacts specifically with OmpC and OmpF and functions as a complex to maintain lipid asymmetry in the OM (50), which might be the reason behind peptides concurrently affecting the expression of mlaA, ompC, and ompF genes in our study. A recent study identified a peptide, arenicin-3, targeting the mla operon (mlaABCDEF) in uropathogenic E. coli (UPEC; NCTC 13441, ST131) (19) which thereby dysregulates PL transport and compromises the membrane integrity. Arenicin-3 also disrupts the bacterial membrane, with membrane debris surrounding the cells (19), which is similar to what we observed in our study and thus supporting the MlaA-OmpC/F system as the likely target of these peptides.
No resistance was observed in APEC against the peptides, which might be due to their MOA of targeting/disrupting the bacterial OM. The antibacterial agents that target/disrupt the OM of Gram-negative bacteria can counter the resistance problem by overcoming antibiotic inactivation determinants, decreasing the development of spontaneous resistance, and impairing biofilm formation (51, 52). Furthermore, resistance is less likely to occur against OM-targeting antibacterial agents, as they avoid the OM barrier and the action of efflux pumps that eliminate antibiotics from inside the bacterial cells, which are the most common mechanisms of antibiotic resistance (32, 51). Therefore, antibacterial agents targeting OM are suitable for development as novel antibacterials in Gram-negative bacteria such as APEC.
All three peptides downregulated the expression of ompC and ompF genes as well as decreased the OmpC level in the OM of APEC (Fig. 7B). This suggest that peptides not only inhibit APEC growth but also reduce APEC virulence (53). The role of ompC and ompF porins in APEC pathogenesis, including adhesion, invasion, colonization, and proliferation, has been reported in mouse brain microvascular endothelial (BMEC) bEnd.3 cells, ducks, and mouse models (53). Furthermore, the antibacterial agents that target the OM can act as antibiotic adjuvants (54); therefore, these peptides could also be used in combination therapy with conventional antibiotics to reduce the use of antibiotics and subsequent resistance.
Peptide treatments (P1 and P2) had minimal impact on cecal microbiota (Fig. 9; see also Fig. S2 and S3 in the supplemental material), which is very significant because the conventional antibiotics cause profound changes in the intestinal microbiota; particularly, they diminish the abundance of beneficial commensals and increase the abundance of potentially detrimental microorganisms (55). Thus, these peptides can be developed as safe antibacterials for use in chickens. A decreased abundance of Erysipelatoclostridium was observed in APEC-infected chickens compared to that in noninfected chickens in our study. Bacteria belonging to Erysipelotrichaceae are reported as performance enhancers in broiler chickens (56); therefore, the exposure of chickens to pathogens such as APEC can reduce the productivity by altering the Erysipelotrichaceae population in the guts of chickens. Furthermore, similar to the decreased Sellimonas abundance observed in APEC-infected chickens in our study, the abundance of Sellimonas was also decreased in Salmonella enterica serovar Typhimurium-infected hens (57), suggesting a potential role for Sellimonas in resisting APEC infection. The abundance of Flavonifractor, which is reported to provide colonization resistance against Salmonella enterica serovar Enteritidis (58), was also decreased in APEC-infected chickens in our study. These findings hint that genera belonging to Erysipelatoclostridium, Sellimonas, and Flavonifractor could be used as potential gut microbial markers to monitor APEC infection in chickens; however, further investigation is needed to identify specific species within these genera. Interestingly, peptide P2 increased the abundance of lactic acid bacteria Pediococcus and Weissella, which might be the reasons behind better anti-APEC activity than that of other peptides. Bacteria belonging to Pediococcus are regarded as good probiotic candidates for chickens (59, 60). Similarly, Weissella has been also used as a probiotic in chickens (60, 61). A greater reduction in APEC load in ceca of chickens was observed when chickens were treated with increased doses of P1 and P2; however, no such effect was observed with P3 treatment. This might be due to the large variability in the gut microbiota profile of P3-treated chickens at 50-mg/kg and 100-mg/kg doses (62). Gut microbiota can act as an invisible organ to modulate the functions of drugs by affecting drug absorption, toxicity, metabolism, and bioavailability (62).
In conclusion, our study identified peptides (P1, NPSRQERR; P2, PDENK; P3, VHTAPK) having effects on APEC in vitro, in vivo in a wax moth larva model, and in commercial broiler chickens. These peptides, therefore, can be developed as novel antibacterial agents against APEC as an alternative to antibiotics. Furthermore, our study revealed a promising new druggable target, the MlaA-OmpC/F system in APEC, which can facilitate further drug development against APEC and related pathogens such as human ExPECs and other pathogenic E. coli, including antibiotic-resistant strains. In our future studies, we will evaluate these peptides for oral delivery in water or feed to reduce APEC infection and assess their impact on performance parameters of chickens such as body weight gain and feed conversion ratio. Moreover, effective control of APEC infections in chickens can help in curbing poultry-originated ExPEC infections in humans.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and media.
APEC serotypes (O78, O1, O2, O8, O15, O18, O35, O109, and O115) provided by Tim Johnson (UMN, Saint Paul, MN), Lisa K. Nolan (UGA, Athens, GA), Catherine M. Logue (UGA, Athens, GA), and Roy Curtiss III (UF, Gainesville, FL) were used in this study (Table 5) (28). These serotypes are predominantly associated with colibacillosis cases in the field (1, 2, 7). Luria-Bertani (LB; BD Difco) broth was used for routine propagation of APEC serotypes. Briefly, APEC serotypes stored at −80°C in glycerol were inoculated in LB broth and grown overnight at 37°C with shaking at 180 rpm. LB agar plates were used for quantification of APEC, unless otherwise indicated.
TABLE 5.
List of APEC serotypes/strains and beneficial microbes used in this study
| Bacterial serotype or strain | Medium | Culture conditions | Source |
|---|---|---|---|
| APEC serotypes | |||
| APEC O78 | LB broth | 37°C, aerobic, 12 h, 180 rpm | Tim Johnson, UMN, MN |
| APEC O1 | Tim Johnson, UMN, MN | ||
| APEC O2 | Tim Johnson, UMN, MN | ||
| APEC O1-63 | Lisa K. Nolan and Catherine M. Logue, UGA, GA | ||
| APEC O2-211 | Lisa K. Nolan, Catherine M. Logue, UGA, GA | ||
| APEC O78-53 | Lisa K. Nolan, Catherine M. Logue, UGA, GA | ||
| APEC O8 | Lisa K. Nolan, Catherine M. Logue, UGA, GA | ||
| APEC O15 | Lisa K. Nolan, Catherine M. Logue, UGA, GA | ||
| APEC O18 | Lisa K. Nolan, Catherine M. Logue, UGA, GA | ||
| APEC O35 | Lisa K. Nolan, Catherine M. Logue, UGA, GA | ||
| APEC O109 | Lisa K. Nolan, Catherine M. Logue, UGA, GA | ||
| APEC O115 | Lisa K. Nolan, Catherine M. Logue, UGA, GA | ||
| APEC O1-X7235 | Roy Curtiss III, UF, FL | ||
| APEC O2-X7302 | Roy Curtiss III, UF, FL | ||
| APEC O78-X7122 | Roy Curtiss III, UF, FL | ||
| Beneficial microbes | |||
| Enterococcus faecalis | MRS broth | 37°C, anaerobic, 16–18 h | David Francis, SDSU |
| Streptococcus bovis | MRS broth | 37°C, anaerobic, 16–18 h | David Francis, SDSU |
| Lactobacillus brevis | MRS broth | 37°C, anaerobic, 1–2 days | David Francis, SDSU |
| Lactobacillus acidophilus | MRS broth | 37°C, anaerobic, 1–2 days | David Francis, SDSU |
| Lactobacillus rhamnosus GG | MRS broth | 37°C, anaerobic, 1–2 days | ATCC, Manassas, VA, USA |
| Bifidobacterium longum | MRS broth plus 0.05% cysteine | 37°C, anaerobic, 24 h | David Francis, SDSU |
| Bifidobacterium adolescentis | MRS broth plus 0.05% cysteine | 37°C, anaerobic, 24 h | David Francis, SDSU |
| Bifidobacterium lactis Bb12 | MRS broth plus 0.05% cysteine | 37°C, anaerobic, 24 h | Christian Hansen Ltd., Hørsholm, Denmark |
| Escherichia coli Nissle 1917 | LB broth | 37°C, aerobic, 12 h, 180 rpm | Ulrich Sonnenborn, Ardeypharm GmbH, Herdecke, Germany |
| Escherichia coli G58-1 | LB broth | 37°C, aerobic, 12 h, 180 rpm | David Francis, SDSU |
| Bacteroides thetaiotaomicron | MRS broth | 37°C, anaerobic, 4–5 days | David Francis, SDSU |
Peptide synthesis.
All peptides (NPSRQERR, P1; PDENK, P2; VHTAPK, P3; MLNERVK, P4; YTRGLPM, P5; GKLSNK, P6) used in this study were synthesized (>95% purity) by GenScript (NJ, USA). These peptides were selected because LGG showed activity against APEC in our study (data not published). Peptides were dissolved in 100% dimethyl sulfoxide (DMSO) at a concentration of 300 mM and stored at −80°C until further use.
Anti-APEC activity determination.
The anti-APEC activity of peptides was determined in LB broth according to CLSI guidelines (63). Briefly, overnight grown APEC O78 was adjusted to 5 × 105 CFU/ml in fresh LB medium. One-hundred microliters of APEC suspension was aliquoted into the wells of a 96-well plate. Peptides were added to the wells at 6 mM and 12 mM concentrations (27) and incubated at 37°C in a TECAN Sunrise absorbance microplate reader with kinetic absorbance measurement set at every 30 min for 12 h (28). Sterile medium and DMSO (solvent) were used as controls. The growth inhibitory activity of the peptides against APEC was calculated using the formula (optical density at 600 nm [OD600] of the DMSO-treated well − OD600 of the peptide-treated well)/OD600 DMSO-treated well × 100.
The MIC was determined for selected peptides (P1, P2, P3, and P4) using a similar assay as described above (28). Peptides were added to the wells of a 96-well plate at 6 mM, 9 mM, 12 mM, 15 mM, and 18 mM. These peptide concentrations were selected based on a previous study (27). The peptide concentration at which there was no visible APEC growth (no OD increase) was determined as the MIC. Two independent experiments were conducted.
Anti-APEC spectrum determination.
Peptides (P1, P2, and P3) were tested against multiple APEC serotypes/strains as listed in Table 5 to determine their spectrum of activity. Peptides were added at their MICs, and the growth inhibitory activity of the peptides was determined as described above.
Effect against beneficial microbes.
Peptides (P1, P2, and P3) were tested against different commensal and probiotic bacteria to determine their specificity of activity as described previously (28). The beneficial microbes used in this study along with their culture requirements are listed in Table 5. Peptides were added at their MICs to 100 μl of a known concentration of bacterial cultures, and cultures were incubated under required conditions. Following incubation, OD600 of cultures was measured to determine the effect of peptides on bacterial growth.
MBEC-HTP assay.
To test the efficacy of the peptides (P1, P2, and P3) against biofilm protected APEC, Innovotech’s MBEC Assay was conducted as described previously (28). Briefly, 150 μl of 0.05 OD600 (5 × 107 CFU/ml)-adjusted APEC suspension in LB medium was aliquoted into the wells of the MBEC Assay device containing polystyrene pegs and incubated for 36 h on a rocker platform at 37°C to allow biofilm formation. After biofilm formation, the pegs were washed to remove loosely adherent planktonic bacteria and transferred to a new 96-well plate (challenge plate) containing peptides at MICs in 200 μl diluted (25%) LB medium. The diluted LB medium mimics the minimal medium which stimulates biofilm formation and maintenance (28). The plate was incubated for 18 h at 37°C with rotation at 150 rpm. Sterile medium and DMSO (solvent) were used as controls. Following incubation, the MICs of peptides in the challenge plate were recorded. The peptide-exposed pegs were transferred to a new 96-well plate containing phosphate-buffered saline (PBS) and sonicated for 30 min to disrupt the biofilm. The sonicated suspensions were then 10-fold serially diluted and plated on LB agar plates to enumerate the biofilm-protected bacteria, and the minimum biofilm eradication concentrations (MBECs) of peptides were determined. The statistical significance (P < 0.05) of treatments on the reduction of biofilm-protected APEC was calculated using a Student’s t test. Three independent experiments were conducted.
Effect of peptides on intracellular survival of APEC in HD11 cells.
To determine the efficacy of peptides (P1, P2, and P3) against intracellular APEC, a gentamicin protection assay was conducted in HD11 (chicken macrophage) cells (42) as described previously (28, 43). The cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, and 2% penicillin-streptomycin (P/S) solution and maintained in a 37°C incubator with 5% CO2. Cells (approximately 104 cells/well) were seeded in a 96-well cell culture plate in 100 μl culture medium and incubated for 36 h prior to use for the assay. Following incubation, the cells were replenished with 100 μl of fresh cell culture medium with no FBS and antibiotic and incubated until infection (2 h). For the infection, the overnight-grown APEC O78 was subcultured to mid-logarithmic phase, centrifuged, washed twice with PBS, and adjusted to 1 × 107 CFU/ml. One hundred microliters of the APEC suspension (multiplicity of infection [MOI] of 100) was added to each well of the 96-well plate and incubated for 1 h to allow for invasion. The cells were washed twice and treated with gentamicin (150 μg/ml) to kill the extracellular APEC. The cells were treated with peptides at 12 mM, 15 mM, and 18 mM concentrations. Infected but not treated, infected and DMSO treated, and noninfected and not treated cells were used as controls. The statistical significance (P < 0.05) of treatments on intracellular survival of APEC was calculated using a Student’s t test. Three independent experiments were conducted.
Bacterial cytological profiling.
To identify the mode of action (MOA) of peptides (P1, P2, and P3), a BCP approach was used as described previously (28, 29). Briefly, 100 μl of APEC O78 cultures adjusted to an OD600 (5 × 108 CFU/ml) of 0.5 were treated with 5× MIC of peptides and incubated at 37°C for 3 h with shaking at 180 rpm. Following incubation, cultures were centrifuged, and bacteria were resuspended in 100 μl sterile water. FM4-64 (1 μg/ml; Invitrogen Molecular Probes) and Syto-9 (5 μM; Invitrogen Molecular Probes) stains were added to the bacterial cultures and incubated at 4°C for 45 min with shaking at 150 rpm. The stained bacteria were centrifuged, pelleted, and resuspended in 10 μl sterile water. The bacterial suspension (3 μl) was transferred onto glass slides containing a thin layer of 1.2% agar and 20% LB medium for confocal microscopy. Microscopy was performed using Leica TCS SP6 confocal scanning microscope (FM4-64 excitation at 515 nm and emission at 640 nm; Syto-9 excitation at 485 nm and emission at 498 nm) at the Molecular and Cellular Imaging Center (MCIC), The Ohio State University (https://mcic.osu.edu/microscopy). Untreated bacterial culture was used as control.
Transmission electron microscopy.
To visualize the effect of peptides (P1, P2, and P3) on APEC membranes, transmission electron microscopy (TEM) was performed as described previously (44). The peptide treatment was performed as described above. Briefly, 500 μl of APEC O78 cultures adjusted to an OD600 (1 × 109 CFU/ml) of 1.0 were treated with 10× MIC of peptides and incubated at 37°C for 3 h with shaking at 180 rpm. After incubation, cells were fixed overnight at room temperature with 3% glutaraldehyde and 2% paraformaldehyde, washed, and postfixed with 1% osmium tetroxide (OsO4). The cells were then dehydrated with a graded ethanol series, and embedding was performed using an Embed 812 kit (Electron Microscopy Sciences, PA). Ultrathin (70 nm) sections were prepared on Formvar carbon-coated grids and stained with uranyl acetate and lead citrate. Microscopy was performed using a Hitachi H-7500 microscope at MCIC. Untreated bacterial culture was used as control.
Efficacy and toxicity of peptides in a wax moth (Galleria mellonella) larva model.
To measure the efficacy and toxicity of peptides (P1, P2, and P3) in vivo, a wax moth larva model was used as described previously (28, 45, 64). Wax moth larvae were obtained from Vanderhorst Wholesale, Inc. (Saint Marys, OH) and were kept at 37°C overnight before use. Healthy larvae (creamy color with no blackening) were selected and kept in sterile petri dishes before inoculation. For toxicity evaluation, larvae (n = 10/group) were injected with 25.5 mM peptides (dissolved in sterile water containing DMSO) through the last left proleg using a PB600-1 repeating dispenser (Hamilton) attached to an insulin syringe (ReliOn; 31 gauge, 8 mm needle length). The injected larvae were incubated for 72 h at 37°C, and mortality was recorded every 12 h.
For efficacy evaluation, larvae (n = 15/group) were pretreated (as described above) and infected with rifampicin-resistant (Rifr) APEC O78 (2.3 × 104 CFU/larva) through last right proleg within 30 min of treatment. The larvae were incubated at 37°C for 72 h, and mortality was recorded as described above. At 72 h postinfection, all dead and live larvae were surface sterilized (70% ethanol followed by sterile water) and macerated in PBS. The macerated larval suspensions were 10-fold serially diluted and plated on MacConkey agar plates containing 50 μg/ml rifampicin. Noninjected larvae and larvae injected with vehicle (sterile water containing DMSO) were included as controls in both the experiments. Two independent experiments were conducted. The statistical significance (P < 0.05) of treatments on survival of larvae and APEC load inside larvae was calculated using log rank test and one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test, respectively.
Structure-activity relationship study.
To identify the crucial amino acid residues required for the peptide activity, alanine scanning libraries (30) of peptides (P1, P2, and P3) (https://www.genscript.com/alanine_scanning.html) were synthesized (see Table S2 in the supplemental material) and tested for anti-APEC activity as described above. The anti-APEC activity of peptide analogues was then compared with that of the original peptide, and the relative importance (percent growth inhibition by peptide analogue/percent growth inhibition by original peptide × 100) of each amino acid residue of the peptide was determined.
Arginine- and lysine-substituted peptide analogues.
The nonessential amino acid residues identified in each peptide were replaced with arginine/lysine to test whether these substitutions enhance the anti-APEC activity of peptides (31). The MICs of arginine and lysine substituted peptide analogues (NPRRQERR, NPSRRERR, NPRRRERR, KDENK, PDEKK, KDEKK, KHTAPK, VKTAPK, VHTKPK, and KKTKPK) were determined as described above and compared with original peptides.
Resistance studies.
To evaluate ability of APEC to develop resistance against peptides, sublethal and lethal resistance assays were performed as described previously (28, 65, 66). For the sublethal resistance assay, peptides (P1, P2, and P3) were added at subinhibitory (0.75× MIC) concentrations to 100 μl of APEC suspension (5 × 105 CFU/ml) in LB medium in 1.5-ml microcentrifuge tubes. The tubes were incubated at 37°C with shaking at 125 rpm for 24 h. After the first incubation, 20 μl of grown APEC culture was mixed with 80 μl of fresh LB medium, and peptides were added again at 0.75× MICs. This procedure was repeated 13 times. Following the 14th passage, the MIC of peptides was determined against APEC cultures grown from the 14th passage as described above. The tubes containing sterile LB medium and DMSO-treated APEC suspension were used as controls. For the lethal resistance assay, overnight-grown APEC cultures (108 CFU) were plated on LB agar plates mixed with lethal (5× MIC) concentrations of peptides (P1, P2, and P3) and incubated for 5 days at 37°C. Any colonies grown on the agar plate were assessed for resistance by determining the MIC as described above. The agar plates containing sterile LB medium and DMSO-mixed APEC cultures were used as controls. Experiments were performed in duplicates.
Efficacy of peptides in chickens.
The animal study was approved by The Ohio State University Institutional Animal Care and Use Committee (IACUC, protocol number 2010A00000149). Chickens were euthanized using CO2 according to the American Veterinary Medical Association (AVMA) guidelines. Standard animal husbandry practices were followed throughout the experiment. Feed and water were provided ad libitum. The efficacy of peptides (P1, P2, and P3) were assessed at 50-mg/kg and 100-mg/kg doses in two successive experiments using commercial broiler chickens (n = 10/group) (Cobb & Ross; Case Farms Ohio Hatchery, Strasburg, OH). The schematic diagram of the experimental design is displayed in Fig. S4. Briefly, peptides dissolved in water (5 mg [50 mg/kg] or 10 mg [100 mg/kg] in 100 μl) were administered orally twice a day from day 1 (1 day before APEC infection) to day 7 (5 days postinfection [dpi]). On day 2, chickens were infected orally with rifampicin-resistant (Rifr) APEC O78 (28) (1 × 109 to 2 × 109 CFU/chicken). At 7 dpi (day 9), chickens were euthanized and necropsied, and cecum and internal organs (lung, liver, heart, and kidney) were assessed for APEC load by plating on MacConkey agar plates containing 50 μg/ml rifampicin (28). The body weights of chickens were measured on the day of necropsy (day 9). The positive- (PC; infected but not treated) and negative-control (NC; noninfected and nontreated) chickens were included in both experiments. The statistical significance (P < 0.05) of treatment on the reduction of APEC load and effect on body weight was calculated using one-way ANOVA followed by Tukey’s post hoc test.
To test if APEC acquired resistance against peptides (P1, P2, and P3) after treatment in chickens, APEC colonies (n = 25/peptide/experiment) were randomly selected from agar plates that were plated with cecum and different internal organs at the end of the experiments. Bacteria were grown in LB medium, and MICs were determined as described above against respective peptides.
Cecal microbiome analysis.
To investigate the impact of peptide treatments (P1, P2, and P3) (50 mg/kg and 100 mg/kg) on the cecal microbiome of chickens, a 16S rRNA-based metagenomic study was conducted as previously described (67, 68). DNA was extracted from 0.2 g of cecal contents using a PureLink microbiome DNA purification kit (Thermo Fisher Scientific) and treated with RNase A (2 μl of 100 mg/ml solution per sample; Qiagen) to remove the RNA. DNA quantity and quality were measured using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific). The extracted DNA samples were subjected to 16S rRNA V4-V5 sequencing at the Molecular and Cellular imaging center (MCIC), Ohio Agricultural Research and Development Center (OARDC) (https://mcic.osu.edu/genomics/illumina-sequencing). Amplicon libraries were prepared using an IFU KAPA HiFi HotStart ReadyMix PCR kit (Roche), and PCR cleanup was performed using Agencourt AMPure XP beads (Beckman Coulter Life Sciences). A Nextera XT DNA library preparation kit (Illumina) was used to generate an Illumina library, and sequencing was performed using an Illumina MiSeq platform generating paired-end 300-bp reads.
For the metagenomic analysis, the QIIME (Quantitative Insights Into Microbial Ecology) 2 bioinformatics platform (69) (https://qiime2.org/) was used. Quality control of the raw reads was performed using FastQC 0.11.8 (Babraham Bioinformatics). Trimmomatic-0.33 was used to trim the adaptor and other Illumina-specific sequences (http://www.usadellab.org/cms/?page=trimmomatic). The trimmed sequences (fastq.gz) were then imported into the QIIME 2 as a manifest file format (PairedEndManifestPhred33V2). The feature table construction and additional filtering of the sequences was performed using DADA2 (70). The taxonomic analysis was performed using naive Bayes classifiers trained on the Silva 132 99% operational taxonomic units (OTUs) (silva-132-99-nb-classifier.qza) database. The phylogenetic diversity was analyzed using the align-to-tree-mafft-fasttree pipeline, and alpha (Shannon’s diversity index) and beta (Bray-Curtis distance) diversities were analyzed using the core-metrics-phylogenetic pipeline (https://docs.qiime2.org/2019.7/tutorials/moving-pictures/). The statistical difference (P < 0.05) in the taxonomic composition between the peptide-treated, PC, and NC groups was determined using the Mann-Whitney U test. The alpha and beta diversities were analyzed using Kruskal-Wallis and permutational multivariate analysis of variance (PERMANOVA) tests (P < 0.05), respectively.
Gene expression analysis.
To identify the potential target(s) of the peptides (P1, P2, and P3), the expression of genes essential for maintaining OM integrity (lptD, mlaA, bamA, lolB, pbgA, and mlaC) was quantitated as described previously (32, 43). Furthermore, the expression of ompC and ompF genes was also quantitated to confirm the peptides’ effect on the MlaA-OmpC/F system (33). APEC O78 cultures grown overnight were adjusted to an OD600 of 0.1 (1 × 108 CFU/ml) in LB medium, and 200 μl of the APEC suspension was treated with peptides (8 replicates) for 8 h (50% lethal concentration) at 37°C with shaking at 200 rpm, as previously described (38, 39). An APEC O78 culture treated with DMSO was used as the control. Posttreatment, total RNA was extracted using an RNeasy minikit (Qiagen). RNA quantity and quality were measured using a NanoDrop 2000c spectrophotometer. DNA traces were removed using a genomic DNA elimination mix (Qiagen). Five micrograms of purified RNA was used to synthesize cDNA using an RT2 first strand kit (Qiagen). Reverse transcription-quantitative PCR (RT-qPCR) was performed using Maxima SYBR green/ROX qPCR master mix (Thermo Fisher) in a RealPlex2 Mastercycler (Eppendorf), with a 55°C annealing temperature. The primers (Table 6) were designed using PrimerQuest Tool and obtained from Integrated DNA Technologies (IDT). The data were normalized to the housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and relative fold change was calculated using the comparative threshold cycle (ΔΔCT) method (40). Two independent experiments were conducted.
TABLE 6.
Primers used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| LptD_F | GCGATCAGAGCGACATCTATAA |
| LptD_R | TGGTTAGCGGAGGCAATAC |
| BamA_F | TCGGTGGTCGTCTCTTCTAT |
| BamA_R | CGTCACGTCTGTACCATAACTC |
| MlaA_F | TGATATGGCGGATGGTCTTTAC |
| MlaA_R | ACTGCGCACGAGTTTCTATC |
| MlaC_F | CTGCCATACGTACAGGTGAAA |
| MlaC_R | ACGGAAAGCGGCAAAGTA |
| LolB_F | AAGTGTACGCCCGTTTCTTC |
| LolB_R | GTTACCCGGTTGAGCATTCA |
| PbgA_F | TCAGCGTGCCGGTTATTT |
| PbgA_R | CGAGGCGATAAAGGCGATAA |
| OmpC_F | CTACACCGGTGGTCTGAAATA |
| OmpC_R | CCTACGCGAGTTGCGTTATAG |
| OmpF_F | ACACGACCAACGAAGAAGTC |
| OmpF_R | CCGTAACTACGGTGTGGTTTAT |
| GAPDH_F | CGGTACCGTTGAAGTGAAAGA |
| GAPDH_R | ACTTCGTCCCATTTCAGGTTAG |
Immunoblot analysis.
To corroborate the findings of the gene expression study, the levels of OmpC and MlaA proteins upon peptide treatments (P1, P2, and P3) were investigated as described previously (41). Peptide-treated cultures of APEC O78 were prepared as described above, and a DMSO-treated culture was used as the control. Posttreatment, the fractions of cytoplasmic and membrane proteins was prepared using ultracentrifugation as previously described (71). The concentrations of fractionated membrane proteins were then normalized based on 280-nm readings and separated on a 12% SDS-PAGE gel. For immunoblot analysis, membrane proteins (25 μg) resolved by SDS-PAGE described above were electrotransferred onto an Immun-Blot polyvinylidene difluoride (PVDF) membrane (Bio-Rad) and probed for OmpC and MlaA using anti-OmpC (1:2,000) (Thermo Fisher Scientific) and anti-MlaA (1:15,000; Thomas J. Silhavy) polyclonal antibodies and goat anti-rabbit IgG horseradish peroxidase (HRP)-conjugated secondary antibody (Sigma-Aldrich). The membrane was developed with Clarity Western ECL substrate (Bio-Rad) and visualized in a FluorChem Q (ProteinSimple) imager. The density of OmpC and MlaA proteins was quantified using ImageJ software.
Docking studies.
HPEPDOCK and PEP-SiteFinder tools were used to predict the affinity and binding sites of peptides to OmpC (PDB 2J1N), OmpF (PDB 3K1B) and MlaA (PDB 5NUO) proteins (36, 37).
Data availability.
Microbiome sequence data have been deposited in the BioProject database under accession number PRJNA735872.
ACKNOWLEDGMENTS
We thank Dhwani Parsana for technical assistance, Thomas J. Silhavy for kindly providing the anti-MlaA antibody, and the MCIC facility for microscopy and analysis of chicken samples for the microbiome study. We also thank Juliette Hanson, Megan Strother, Sara Tallmadge, and Ronna Wood for animal care assistance.
This research was supported by the U.S. Department of Agriculture (USDA) National Institute for Food and Agriculture (NIFA) (grant no. 2015-68004-23131, 2020-6701-31401) and The Ohio State University internal grants.
Footnotes
Supplemental material is available online only.
Contributor Information
Gireesh Rajashekara, Email: rajashekara.2@osu.edu.
Danilo Ercolini, University of Naples Federico II.
REFERENCES
- 1.Dziva F, Stevens MP. 2008. Colibacillosis in poultry: unravelling the molecular basis of virulence of avian pathogenic Escherichia coli in their natural hosts. Avian Pathol 37:355–366. 10.1080/03079450802216652. [DOI] [PubMed] [Google Scholar]
- 2.Kathayat D, Lokesh D, Ranjit S, Rajashekara G. 2021. Avian pathogenic Escherichia coli (APEC): an overview of virulence and pathogenesis factors, zoonotic potential, and control strategies. Pathogens 10:467. 10.3390/pathogens10040467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadeyen J-R, Kaiser P, Stevens MP, Dziva F. 2014. Analysis of immune responses induced by avian pathogenic Escherichia coli infection in turkeys and their association with resistance to homologous re-challenge. Vet Res 45:19. 10.1186/1297-9716-45-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mellata M. 2013. Human and avian extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog Dis 10:916–932. 10.1089/fpd.2013.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu CM, Stegger M, Aziz M, Johnson TJ, Waits K, Nordstrom L, Gauld L, Weaver B, Rolland D, Statham S, Horwinski J, Sariya S, Davis GS, Sokurenko E, Keim P, Johnson JR, Price LB. 2018. Escherichia coli ST131-H22 as a foodborne uropathogen. mBio 9:e00470-18. 10.1128/mBio.00470-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osman KM, Kappell AD, Elhadidy M, ElMougy F, El-Ghany WAA, Orabi A, Mubarak AS, Dawoud TM, Hemeg HA, Moussa IMI, Hessain AM, Yousef HMY. 2018. Poultry hatcheries as potential reservoirs for antimicrobial-resistant Escherichia coli: a risk to public health and food safety. Sci Rep 8:5859. 10.1038/s41598-018-23962-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dho-Moulin M, Fairbrother JM. 1999. Avian pathogenic Escherichia coli (APEC). Vet Res 30:299–316. [PubMed] [Google Scholar]
- 8.Agunos A, Léger D, Carson C. 2012. Review of antimicrobial therapy of selected bacterial diseases in broiler chickens in Canada. Can Vet J 53:1289–1300. [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson TJ, Wannemuehler Y, Doetkott C, Johnson SJ, Rosenberger SC, Nolan LK. 2008. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J Clin Microbiol 46:3987–3996. 10.1128/JCM.00816-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nhung NT, Chansiripornchai N, Carrique-Mas JJ. 2017. Antimicrobial resistance in bacterial poultry pathogens: a review. Front Vet Sci 4:126–126. 10.3389/fvets.2017.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R. 2015. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci U S A 112:5649–5654. 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao H, Cheng G, Iqbal Z, Ai X, Hussain HI, Huang L, Dai M, Wang Y, Liu Z, Yuan Z. 2014. Benefits and risks of antimicrobial use in food-producing animals. Front Microbiol 5:288–288. 10.3389/fmicb.2014.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baltzer SA, Brown MH. 2011. Antimicrobial peptides: promising alternatives to conventional antibiotics. J Mol Microbiol Biotechnol 20:228–235. 10.1159/000331009. [DOI] [PubMed] [Google Scholar]
- 14.Mahlapuu M, Håkansson J, Ringstad L, Björn C. 2016. Antimicrobial peptides: an emerging category of therapeutic agents. Front Cell Infect Microbiol 6:194–194. 10.3389/fcimb.2016.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Breij A, Riool M, Cordfunke RA, Malanovic N, de Boer L, Koning RI, Ravensbergen E, Franken M, van der Heijde T, Boekema BK, Kwakman PHS, Kamp N, El Ghalbzouri A, Lohner K, Zaat SAJ, Drijfhout JW, Nibbering PH. 2018. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci Transl Med 10:eaan4044. 10.1126/scitranslmed.aan4044. [DOI] [PubMed] [Google Scholar]
- 16.Lee J-K, Luchian T, Park Y. 2018. New antimicrobial peptide kills drug-resistant pathogens without detectable resistance. Oncotarget 9:15616–15634. 10.18632/oncotarget.24582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewies A, Du Plessis LH, Wentzel JF. 2019. Antimicrobial peptides: the Achilles' heel of antibiotic resistance? Probiotics Antimicrob Proteins 11:370–381. 10.1007/s12602-018-9465-0. [DOI] [PubMed] [Google Scholar]
- 18.Mwangi J, Yin Y, Wang G, Yang M, Li Y, Zhang Z, Lai R. 2019. The antimicrobial peptide ZY4 combats multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii infection. Proc Natl Acad Sci U S A 116:26516–26522. 10.1073/pnas.1909585117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott AG, Huang JX, Neve S, Zuegg J, Edwards IA, Cain AK, Boinett CJ, Barquist L, Lundberg CV, Steen J, Butler MS, Mobli M, Porter KM, Blaskovich MAT, Lociuro S, Strandh M, Cooper MA. 2020. An amphipathic peptide with antibiotic activity against multidrug-resistant Gram-negative bacteria. Nat Commun 11:3184. 10.1038/s41467-020-16950-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H-R, You D-g, Kim HK, Sohn JW, Kim MJ, Park JK, Lee GY, Yoo YD. 2020. Romo1-derived antimicrobial peptide is a new antimicrobial agent against multidrug-resistant bacteria in a murine model of sepsis. mBio 11:e03258-19. 10.1128/mBio.03258-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao H, Shao F, Wu M, Ren W, Xiong X, Tan B, Yin Y. 2015. The application of antimicrobial peptides as growth and health promoters for swine. J Anim Sci Biotechnol 6:19. 10.1186/s40104-015-0018-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang S, Zeng X, Yang Q, Qiao S. 2016. Antimicrobial peptides as potential alternatives to antibiotics in food animal industry. Int J Mol Sci 17:603. 10.3390/ijms17050603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daneshmand A, Kermanshahi H, Sekhavati MH, Javadmanesh A, Ahmadian M. 2019. Antimicrobial peptide, cLF36, affects performance and intestinal morphology, microflora, junctional proteins, and immune cells in broilers challenged with E. coli. Sci Rep 9:14176. 10.1038/s41598-019-50511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Jacob B, Jang M, Kwak C, Lee Y, Son K, Lee S, Jung ID, Jeong MS, Kwon S-H, Kim Y. 2019. Development of a novel short 12-meric papiliocin-derived peptide that is effective against Gram-negative sepsis. Sci Rep 9:3817. 10.1038/s41598-019-40577-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohamed MF, Abdelkhalek A, Seleem MN. 2016. Evaluation of short synthetic antimicrobial peptides for treatment of drug-resistant and intracellular Staphylococcus aureus. Sci Rep 6:29707. 10.1038/srep29707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bormann N, Koliszak A, Kasper S, Schoen L, Hilpert K, Volkmer R, Kikhney J, Wildemann B. 2017. A short artificial antimicrobial peptide shows potential to prevent or treat bone infections. Sci Rep 7:1506. 10.1038/s41598-017-01698-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu R, Fasano S, Madayiputhiya N, Morin NP, Nataro J, Fasano A. 2009. Isolation, identification, and characterization of small bioactive peptides from Lactobacillus GG conditional media that exert both anti-Gram-negative and Gram-positive bactericidal activity. J Pediatr Gastroenterol Nutr 49:23–30. 10.1097/MPG.0b013e3181924d1e. [DOI] [PubMed] [Google Scholar]
- 28.Kathayat D, Helmy YA, Deblais L, Rajashekara G. 2018. Novel small molecules affecting cell membrane as potential therapeutics for avian pathogenic Escherichia coli. Sci Rep 8:15329. 10.1038/s41598-018-33587-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nonejuie P, Burkart M, Pogliano K, Pogliano J. 2013. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc Natl Acad Sci U S A 110:16169–16174. 10.1073/pnas.1311066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Migoń D, Jaśkiewicz M, Neubauer D, Bauer M, Sikorska E, Kamysz E, Kamysz W. 2019. Alanine scanning studies of the antimicrobial peptide Aurein 1.2. Probiotics Antimicrob Proteins 11:1042–1054. 10.1007/s12602-018-9501-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Koh J-J, Liu S, Lakshminarayanan R, Verma CS, Beuerman RW. 2017. Membrane active antimicrobial peptides: translating mechanistic insights to design. Front Neurosci 11:73. 10.3389/fnins.2017.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi U, Lee CR. 2019. Antimicrobial agents that inhibit the outer membrane assembly machines of Gram-negative bacteria. J Microbiol Biotechnol 29:1–10. 10.4014/jmb.1804.03051. [DOI] [PubMed] [Google Scholar]
- 33.Hughes GW, Hall SCL, Laxton CS, Sridhar P, Mahadi AH, Hatton C, Piggot TJ, Wotherspoon PJ, Leney AC, Ward DG, Jamshad M, Spana V, Cadby IT, Harding C, Isom GL, Bryant JA, Parr RJ, Yakub Y, Jeeves M, Huber D, Henderson IR, Clifton LA, Lovering AL, Knowles TJ. 2019. Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat Microbiol 4:1692–1705. 10.1038/s41564-019-0481-y. [DOI] [PubMed] [Google Scholar]
- 34.Malinverni JC, Silhavy TJ. 2009. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc Natl Acad Sci U S A 106:8009–8014. 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maier L, Typas A. 2017. Systematically investigating the impact of medication on the gut microbiome. Curr Opin Microbiol 39:128–135. 10.1016/j.mib.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Zhou P, Jin B, Li H, Huang SY. 2018. HPEPDOCK: a web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res 46:W443–W450. 10.1093/nar/gky357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saladin A, Rey J, Thévenet P, Zacharias M, Moroy G, Tufféry P. 2014. PEP-SiteFinder: a tool for the blind identification of peptide binding sites on protein surfaces. Nucleic Acids Res 42:W221–W226. 10.1093/nar/gku404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaw KJ, Miller N, Liu X, Lerner D, Wan J, Bittner A, Morrow BJ. 2003. Comparison of the changes in global gene expression of Escherichia coli induced by four bactericidal agents. J Mol Microbiol Biotechnol 5:105–122. 10.1159/000069981. [DOI] [PubMed] [Google Scholar]
- 39.Kaldalu N, Mei R, Lewis K. 2004. Killing by ampicillin and ofloxacin induces overlapping changes in Escherichia coli transcription profile. Antimicrob Agents Chemother 48:890–896. 10.1128/AAC.48.3.890-896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Urfer M, Bogdanovic J, Lo Monte F, Moehle K, Zerbe K, Omasits U, Ahrens CH, Pessi G, Eberl L, Robinson JA. 2016. A peptidomimetic antibiotic targets outer membrane proteins and disrupts selectively the outer membrane in Escherichia coli. J Biol Chem 291:1921–1932. 10.1074/jbc.M115.691725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beug H, von Kirchbach A, Döderlein G, Conscience J-F, Graf T. 1979. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 18:375–390. 10.1016/0092-8674(79)90057-6. [DOI] [PubMed] [Google Scholar]
- 43.Helmy YA, Deblais L, Kassem II, Kathayat D, Rajashekara G. 2018. Novel small molecule modulators of quorum sensing in avian pathogenic Escherichia coli (APEC). Virulence 9:1640–1657. 10.1080/21505594.2018.1528844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartmann M, Berditsch M, Hawecker J, Ardakani MF, Gerthsen D, Ulrich AS. 2010. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob Agents Chemother 54:3132–3142. 10.1128/AAC.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai CJ-Y, Loh JMS, Proft T. 2016. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7:214–229. 10.1080/21505594.2015.1135289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang G, Song Q, Huang S, Wang Y, Cai S, Yu H, Ding X, Zeng X, Zhang J. 2020. Effect of antimicrobial peptide microcin J25 on growth performance, immune regulation, and intestinal microbiota in broiler chickens challenged with Escherichia coli and Salmonella. Animals (Basel) 10:345. 10.3390/ani10020345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tai H-M, Huang H-N, Tsai T-Y, You M-F, Wu H-Y, Rajanbabu V, Chang H-Y, Pan C-Y, Chen J-Y. 2020. Dietary supplementation of recombinant antimicrobial peptide Epinephelus lanceolatus piscidin improves growth performance and immune response in Gallus gallus domesticus. PLoS One 15:e0230021. 10.1371/journal.pone.0230021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daneshmand A, Kermanshahi H, Sekhavati MH, Javadmanesh A, Ahmadian M, Alizadeh M, Aldawoodi A. 2020. Effects of cLFchimera peptide on intestinal morphology, integrity, microbiota, and immune cells in broiler chickens challenged with necrotic enteritis. Sci Rep 10:17704. 10.1038/s41598-020-74754-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeow J, Tan KW, Holdbrook DA, Chong Z-S, Marzinek JK, Bond PJ, Chng S-S. 2018. The architecture of the OmpC-MlaA complex sheds light on the maintenance of outer membrane lipid asymmetry in Escherichia coli. J Biol Chem 293:11325–11340. 10.1074/jbc.RA118.002441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chong Z-S, Woo W-F, Chng S-S. 2015. Osmoporin OmpC forms a complex with MlaA to maintain outer membrane lipid asymmetry in Escherichia coli. Mol Microbiol 98:1133–1146. 10.1111/mmi.13202. [DOI] [PubMed] [Google Scholar]
- 51.Lehman KM, Grabowicz M. 2019. Countering Gram-negative antibiotic resistance: recent progress in disrupting the outer membrane with novel therapeutics. Antibiotics (Basel) 8:163. 10.3390/antibiotics8040163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacNair CR, Brown ED. 2020. Outer membrane disruption overcomes intrinsic, acquired, and spontaneous antibiotic resistance. mBio 11:e01615-20. 10.1128/mBio.01615-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hejair HMA, Zhu Y, Ma J, Zhang Y, Pan Z, Zhang W, Yao H. 2017. Functional role of ompF and ompC porins in pathogenesis of avian pathogenic Escherichia coli. Microb Pathog 107:29–37. 10.1016/j.micpath.2017.02.033. [DOI] [PubMed] [Google Scholar]
- 54.Kathayat D, Antony L, Deblais L, Helmy YA, Scaria J, Rajashekara G. 2020. Small molecule adjuvants potentiate colistin activity and attenuate resistance development in Escherichia coli by affecting pmrAB system. Infect Drug Resist 13:2205–2222. 10.2147/IDR.S260766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zimmermann P, Curtis N. 2019. The effect of antibiotics on the composition of the intestinal microbiota - a systematic review. J Infect 79:471–489. 10.1016/j.jinf.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 56.Stanley D, Hughes RJ, Geier MS, Moore RJ. 2016. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: challenges presented for the identification of performance enhancing probiotic bacteria. Front Microbiol 7:187–187. 10.3389/fmicb.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khan S, Chousalkar KK. 2020. Salmonella Typhimurium infection disrupts but continuous feeding of Bacillus based probiotic restores gut microbiota in infected hens. J Animal Sci Biotechnol 11:29. 10.1186/s40104-020-0433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu L, Lin L, Zheng L, Tang H, Fan X, Xue N, Li M, Liu M, Li X. 2018. Cecal microbiome profile altered by Salmonella enterica, serovar Enteritidis inoculation in chicken. Gut Pathog 10:34–34. 10.1186/s13099-018-0261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noohi N, Ebrahimipour G, Rohani M, Talebi M, Pourshafie MR. 2016. Evaluation of potential probiotic characteristics and antibacterial effects of strains of Pediococcus species isolated from broiler chickens. Br Poult Sci 57:317–323. 10.1080/00071668.2016.1169247. [DOI] [PubMed] [Google Scholar]
- 60.Reuben RC, Roy PC, Sarkar SL, Alam R-U, Jahid IK. 2019. Isolation, characterization, and assessment of lactic acid bacteria toward their selection as poultry probiotics. BMC Microbiol 19:253–253. 10.1186/s12866-019-1626-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baldwin S, Hughes RJ, Hao Van TT, Moore RJ, Stanley D. 2018. At-hatch administration of probiotic to chickens can introduce beneficial changes in gut microbiota. PLoS One 13:e0194825. 10.1371/journal.pone.0194825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li X, Liu L, Cao Z, Li W, Li H, Lu C, Yang X, Liu Y. 2020. Gut microbiota as an “invisible organ” that modulates the function of drugs. Biomed Pharmacother 121:109653. 10.1016/j.biopha.2019.109653. [DOI] [PubMed] [Google Scholar]
- 63.CLSI. 2018. Performance standards for antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 64.Ignasiak K, Maxwell A. 2017. Galleria mellonella (greater wax moth) larvae as a model for antibiotic susceptibility testing and acute toxicity trials. BMC Res Notes 10:428. 10.1186/s13104-017-2757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. 2015. A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459. 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deblais L, Helmy YA, Kumar A, Antwi J, Kathayat D, Acuna UM, Huang HC, de Blanco EC, Fuchs JR, Rajashekara G. 2019. Novel narrow spectrum benzyl thiophene sulfonamide derivatives to control Campylobacter. J Antibiot (Tokyo) 72:555–565. 10.1038/s41429-019-0168-x. [DOI] [PubMed] [Google Scholar]
- 67.Deblais L, Helmy YA, Kathayat D, Huang H-c, Miller SA, Rajashekara G. 2018. Novel imidazole and methoxybenzylamine growth inhibitors affecting Salmonella cell envelope integrity and its persistence in chickens. Sci Rep 8:13381. 10.1038/s41598-018-31249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Helmy YA, Kathayat D, Ghanem M, Jung K, ClossG, Jr, Deblais L, Srivastava V, El-Gazzar M, Rajashekara G. 2020. Identification and characterization of novel small molecule inhibitors to control Mycoplasma gallisepticum infection in chickens. Vet Microbiol 247:108799. 10.1016/j.vetmic.2020.108799. [DOI] [PubMed] [Google Scholar]
- 69.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sandrini SM, Haigh R, Freestone PPE. 2014. Fractionation by ultracentrifugation of Gram negative cytoplasmic and membrane proteins. Bio Protoc 4:e1287. 10.21769/BioProtoc.1287. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S4, Tables S1 and S2. Download AEM.00567-21-s0001.pdf, PDF file, 0.3 MB (279.8KB, pdf)
Data Availability Statement
Microbiome sequence data have been deposited in the BioProject database under accession number PRJNA735872.