ABSTRACT
The heat shock response (HSR) is a universal cellular response that promotes survival following temperature increase. In filamentous Streptomyces, which accounts for ∼70% of commercial antibiotic production, HSR is regulated by transcriptional repressors; in particular, the widespread MerR-family regulator HspR has been identified as a key repressor. However, functions of HspR in other biological processes are unknown. The present study demonstrates that HspR pleiotropically controls avermectin production, morphological development, and heat shock and H2O2 stress responses in the industrially important species Streptomyces avermitilis. HspR directly activated ave structural genes (aveA1 and aveA2) and H2O2 stress-related genes (katA1, catR, katA3, oxyR, ahpC, and ahpD), whereas it directly repressed heat shock genes (HSGs) (the dnaK1-grpE1-dnaJ1-hspR operon, clpB1p, clpB2p, and lonAp) and developmental genes (wblB, ssgY, and ftsH). HspR interacted with PhoP (response regulator of the widespread PhoPR two-component system) at dnaK1p to corepress the important dnaK1-grpE1-dnaJ1-hspR operon. PhoP exclusively repressed target HSGs (htpG, hsp18_1, and hsp18_2) different from those of HspR (clpB1p, clpB2p, and lonAp). A consensus HspR-binding site, 5′-TTGANBBNNHNNNDSTSHN-3′, was identified within HspR target promoter regions, allowing prediction of the HspR regulon involved in broad cellular functions. Taken together, our findings demonstrate a key role of HspR in the coordination of a variety of important biological processes in Streptomyces species.
IMPORTANCE Our findings are significant to clarify the molecular mechanisms underlying HspR function in Streptomyces antibiotic production, development, and H2O2 stress responses through direct control of its target genes associated with these biological processes. HspR homologs described to date function as transcriptional repressors but not as activators. The results of the present study demonstrate that HspR acts as a dual repressor/activator. PhoP cross talks with HspR at dnaK1p to coregulate the heat shock response (HSR), but it also has its own specific target heat shock genes (HSGs). The novel role of PhoP in the HSR further demonstrates the importance of this regulator in Streptomyces. Overexpression of hspR strongly enhanced avermectin production in Streptomyces avermitilis wild-type and industrial strains. These findings provide new insights into the regulatory roles and mechanisms of HspR and PhoP and facilitate methods for antibiotic overproduction in Streptomyces species.
KEYWORDS: HspR, Streptomyces avermitilis, avermectin, morphological development, heat shock response
INTRODUCTION
Streptomyces species are Gram-positive, filamentous soil bacteria that undergo complex morphological development involving the formation of substrate hyphae, aerial hyphae, and spore-bearing hyphae (1). They are an economically important group that produce a wide variety of secondary metabolites, notably, antibiotics having antibacterial, antifungal, antiviral, anthelmintic, anticancer, or immunosuppressive activity (2, 3). Antibiotic biosynthesis is generally associated with development and controlled by multiple levels of transcriptional regulators (TRs), including cluster-situated regulators (CSRs) and higher-level pleiotropic/global regulators, in response to environmental and physiological cues such as low nutrient availability, temperature changes, and pH changes (4–6).
All cellular organisms respond to sudden temperature increases by substantially raising levels of heat shock proteins (HSPs). HSPs are divided into two major classes: (i) molecular chaperones that promote correct folding of denatured and newly synthesized proteins and prevent formation of insoluble protein aggregates (e.g., DnaK, DnaJ, GrpE, GroES, and GroEL), and (ii) ATP-dependent proteases involved in the degradation of denatured proteins (e.g., ClpAP, ClpXP, ClpB, and Lon) (7–9). These HSPs are essential for normal cell growth and are synthesized at a basal level under physiological conditions. Induction of HSPs under stress conditions (particularly heat shock) is a universal response, but synthesis of HSPs is controlled by a variety of mechanisms depending on the bacterial species. In Escherichia coli, two alternative sigma factors, Sig32 and SigE, activate heat shock genes (HSGs) in response to denatured proteins generated in cytoplasm (Sig32) or periplasm (SigE) following stimulation (10). In Streptomyces, HSGs are mainly negatively regulated by three repressors: HrcA, RheA, and HspR (10). HrcA in Streptomyces albus represses hrcA-dnaJ2, groES-groEL1, and groEL2 operons (11). The RheA regulon of S. albus consists of its own gene rheA and nearby divergently transcribed gene hsp18 (12). The HspR repressor is a member of the MerR-family TRs that contain conserved helix-turn-helix (HTH) DNA-binding motif at the N terminus and function as homodimers. HspR represses the dnaK-grpE-dnaJ-hspR operon and clpB and lon genes by interacting with HspR-binding site HAIR (HspR-associated inverted repeat [5′-TTGAGY-N7-ACTCAA-3′]) at target promoter regions in Streptomyces coelicolor, S. albus, and Streptomyces lividans (13–17). The S. coelicolor DnaK chaperone functions as a corepressor of the HspR regulon by binding to HspR at HAIR sites (14, 18). HspR itself can bind DNA targets, but formation of a stable complex with the DNA targets requires HspR-DnaK interaction. During heat shock, DnaK is titrated by denatured proteins and unable to bind to HspR, leading to the induction of target HSGs (14, 18). The reported studies have only addressed the role of Streptomyces HspR in the heat shock response (HSR), and nothing is known regarding its role in Streptomyces development and antibiotic production.
HspR is present in other actinomycetes besides Streptomyces, including Mycobacterium tuberculosis (19). HSPs contribute to M. tuberculosis virulence and are induced following entry into host cells to protect M. tuberculosis against stress imposed by host macrophages. Among M. tuberculosis HSPs, Acr2 is a member of the α-crystallin family of molecular chaperones and plays a key role during the M. tuberculosis stress response. HspR functions as a repressor of acr2 expression (19). Another acr2 repressor, PhoP, the response regulator of the widespread PhoPR two-component system, interacts with HspR at the acr2 promoter region to coregulate acr2 expression (20). PhoP is not necessary for the DNA-binding activity of HspR; however, HspR-PhoP interaction stabilizes the higher-order DNA-protein complex to prevent access of RNA polymerase to the acr2 promoter, thus repressing the initiation of transcription. M. tuberculosis dnaK is repressed by HspR (19) but is not regulated by HspR-PhoP interactions (20).
The PhoPR two-component system, which senses and responds to phosphate limitation stress, is well conserved in Streptomyces. PhoP acts as a master regulator for the coordination of a variety of physiological processes, including phosphate metabolism, nitrogen metabolism, respiration, development, and antibiotic biosynthesis (21–24). PhoP cross talks with GlnR (main nitrogen metabolism regulator) and AfsR (response regulator of AfsRK two-component system that promotes antibiotic biosynthesis in S. coelicolor) by competing for binding to the same target promoter regions in control of nitrogen metabolism and antibiotic biosynthesis (25, 26). A role of PhoP in HSR and its cross talk with other regulators in Streptomyces has not been reported.
The industrial species Streptomyces avermitilis produces avermectins, a series of 16-membered macrocyclic anthelmintic antibiotics that are widely used in agricultural and medical fields and have great commercial importance (27, 28). The heat shock repressor HspR has not yet been studied in S. avermitilis. The role of HspR in antibiotic production and development in this species is of interest in view of previous findings that it is involved in stress response and that antibiotic biosynthesis in the genus is associated with development and occurs under stress conditions.
Here, we characterize HspR (SAV_4487) in S. avermitilis as a dual repressor/activator of avermectin production, development, and heat shock and H2O2 stress responses and identify HspR target genes associated with these biological processes. HspR interacted with PhoP at the dnaK1 regulatory region to corepress the dnaK1-grpE1-dnaJ1-hspR operon. PhoP had exclusive target HSGs (htpG, hsp18_1, and hsp18_2) different from those of HspR (clpB1p, clpB2p, and lonAp). We propose a novel strategy for enhancing antibiotic production through overexpression of the hspR gene.
RESULTS
HspR represses S. avermitilis development but activates avermectin production.
The gene hspR (sav_4487) in S. avermitilis contains 450 nucleotides (nt) and encodes a 16.8-kDa protein, HspR. The S. avermitilis chromosome has two copies of HSGs dnaK, grpE, and dnaJ. hspR is clustered with dnaK1 (sav_4484), grpE1 (sav_4485), and dnaJ1 (sav_4486) to form the operon dnaK1-grpE1-dnaJ1-hspR (Fig. 1A). Protein alignment study showed that HspR is a highly conserved protein in Streptomyces; S. avermitilis HspR has 90.8%, 90.3%, 90.3%, 92%, and 90.8% amino acid identity with its homologs in S. coelicolor, S. venezuelae, S. griseus, S. scabies, and S. lividans, respectively, reflecting the biological importance of this protein in the genus.
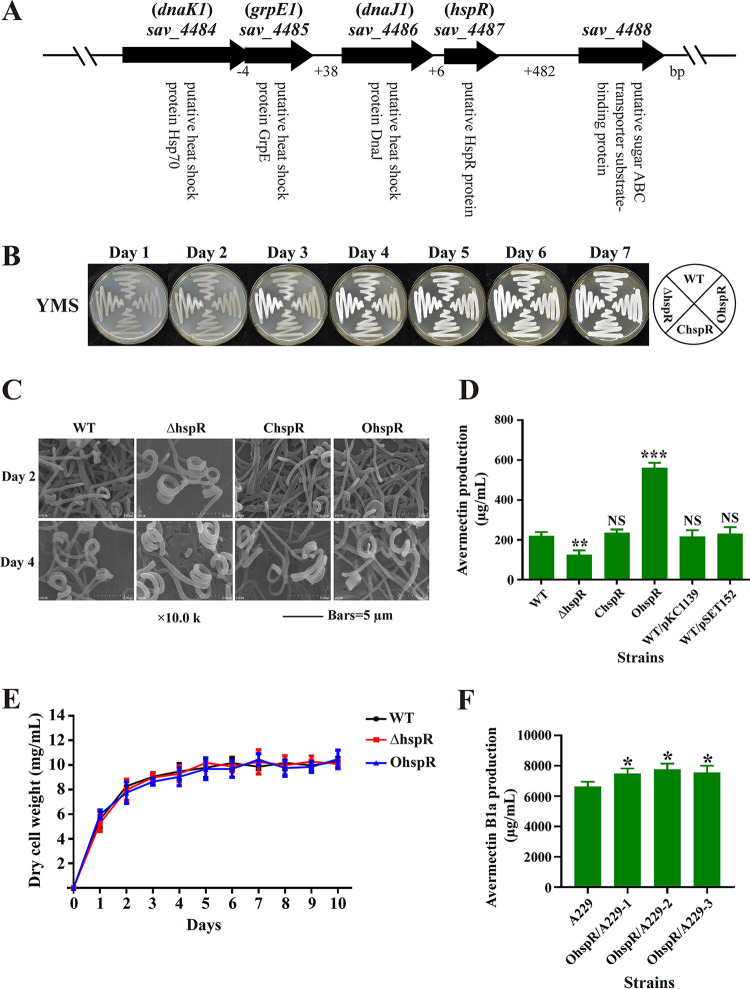
FIG 1.
Effects of HspR on morphological development, avermectin production, and cell growth in S. avermitilis. (A) Genetic organization of the dnaK1-grpE1-dnaJ1-hspR operon. (B) Phenotypes of WT strain, the hspR deletion mutant (ΔhspR), complemented strain (ChspR), and overexpression strain (OhspR) grown on YMS agar at 28°C. (C) SEM images of WT, ΔhspR, ChspR, and OhspR strains grown on YMS agar for 2 or 4 days. (D) Avermectin yield in WT, ΔhspR, ChspR, OhspR, and control strains (WT/pKC1139 and WT/pSET152) after 10-day culture in FM-I. (E) Growth curves of WT, ΔhspR, and OhspR strains cultured in FM-II. (F) Avermectin yield in industrial strain A229 and its derivatives OhspR/A229-1, -2, and -3 (hspR overexpression strains) after 10-day culture in FM-I. Error bars (panels D, E, and F) indicate standard deviations (SDs) from three replicates. NS, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001 for comparison with WT (D) or A229 (F) (Student’s t test).
To elucidate the functions of HspR in S. avermitilis, we constructed hspR deletion mutant ΔhspR (see Fig. S1 in the supplemental material), complemented strain ChspR, and overexpression strain OhspR. hspR transcription was undetectable in the ΔhspR strain as shown by reverse transcription and real-time quantitative PCR (RT-qPCR) analysis, and its levels were ∼1.8-fold higher on day 2 (exponential phase) and ∼3.1-fold higher on day 6 (stationary phase) in OhspR than in the wild-type (WT) strain (see Fig. S2), confirming the successful deletion or overexpression of hspR in the above-described strains.
The effect of HspR on morphological development was investigated by growing WT, ΔhspR, OhspR, and ChspR strains on solid sporulation yeast extract-malt-starch (YMS) plates. OhspR and ChspR strains were phenotypically similar to the WT, whereas the ΔhspR strain showed earlier and enhanced formation of spores (Fig. 1B). Spore numbers on days 2 and 4 were higher for the ΔhspR strain than for WT, as shown by scanning electron microscopy (SEM) (Fig. 1C). These findings indicate that HspR functions as a repressor during S. avermitilis development.
The effect of HspR on avermectin biosynthesis was investigated by high-performance liquid chromatography (HPLC) analysis of 10-day insoluble FM-I fermentation broth. In comparison to the WT value, the avermectin yield of the ΔhspR strain was ∼43% lower, that of the OhspR strain was ∼154% higher, and those of the ChspR and control strains (WT/pKC1139, WT/pSET152) were not significantly different (Fig. 1D and see Fig. S3). Time course measurements of growth in soluble FM-II showed that biomass (dry cell weight) values of the ΔhspR and OhspR strains were similar to that of the WT (Fig. 1E), ruling out the possibility that altered avermectin yields in the ΔhspR strain and the OhspR strain were due to changes in growth. These findings demonstrate that HspR functions as an activator in avermectin production.
In view of the finding that hspR overexpression increased avermectin yield in the WT, we transformed hspR overexpression vector pKC-erm-hspR into industrial strain A229 to construct OhspR/A229. Yield of avermectin B1a, the most effective component, was ∼14% to 17% higher for OhspR/A229 than in A229 (Fig. 1F). This finding indicates that avermectin yield could be substantially enhanced in high-producing industrial strains by hspR overexpression.
HspR directly activates ave structural genes.
The transcription profile of hspR in FM-I culture of the WT was monitored by RT-qPCR to further clarify the regulatory role of HspR in avermectin production. The hspR transcription level increased to maximum on day 5, followed by a gradual decrease, and the level on day 7 was similar to that on day 8 (Fig. 2A). The HspR protein expression level was examined concurrently by Western blotting. Avermectin production in strain ΔhspR/hspR-3FLAG (fusion protein HspR-3FLAG expressed in the ΔhspR strain) was similar to that in the WT (see Fig. S4), indicating that HspR-3FLAG complemented HspR function and that the HspR expression profile could be examined using anti-FLAG antibody against HspR-3FLAG in the ΔhspR/hspR-3FLAG strain. Consistent with the transcription profile, the HspR protein level was maximal on day 5 and then declined gradually (Fig. 2B). These findings suggest that HspR plays its regulatory role particularly during the middle fermentation stage or that there is something about this stage in growth that is particularly relevant to one of the stresses modulated by this protein.
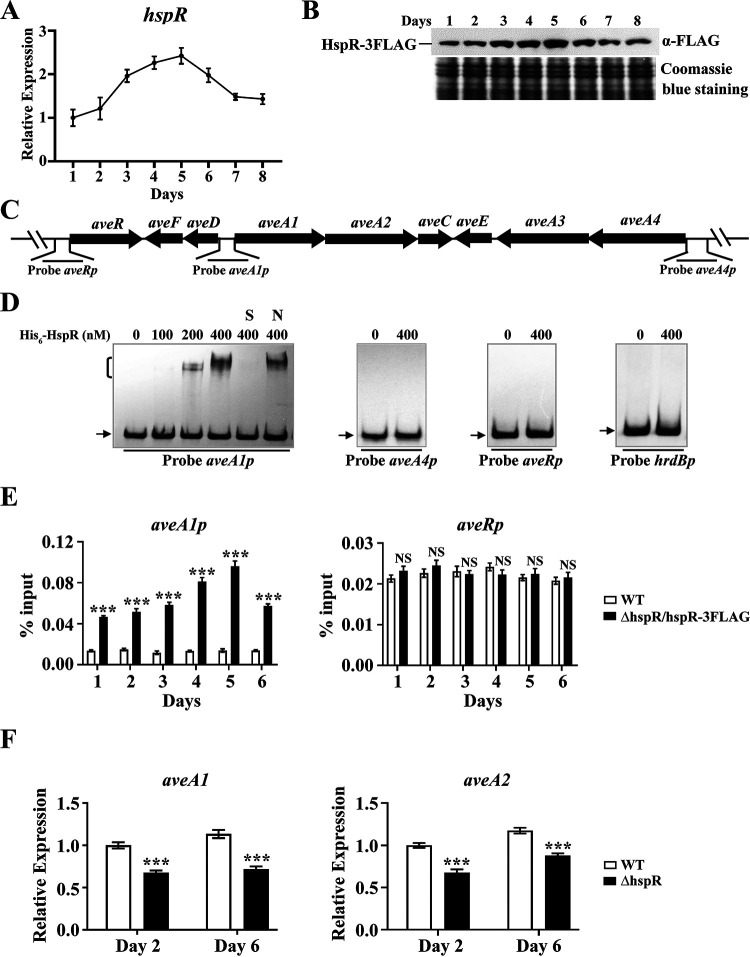
FIG 2.
Direct activation of aveA1-aveA2 by HspR. (A) RT-qPCR analysis of hspR transcriptional pattern in the WT grown in FM-I. The hspR transcription level on day 1 was set to 1. (B) Western blotting analysis of the HspR protein level during the fermentation process. The HspR expression level in strain ΔhspR/hspR-3FLAG grown in FM-I was determined using anti-FLAG MAb. (C) Schematic diagram of promoter probes used in EMSAs. (D) In vitro EMSAs of interactions of His6-HspR with indicated probes (hrdBp as negative-control probe). A 0.15 nM concentration of labeled probe and various amounts of His6-HspR were used for each binding reaction. In competition experiments, ∼300-fold unlabeled nonspecific probe hrdBp (lane N) or specific probe aveA1p (lane S) was used. Arrows indicate free probes. Bracket indicates the HspR-DNA complex. (E) In vivo ChIP-qPCR assays of HspR binding to aveA1p and aveRp using ΔhspR/hspR-3FLAG and WT (negative control) strains grown in FM-II for the indicated times. The y axis indicates the relative binding level of HspR-FLAG on each site, determined by recovery of target sequence with anti-FLAG MAb. (F) RT-qPCR analysis of aveA1 and aveA2 in WT and ΔhspR strains grown in FM-I. WT value on day 2 for each gene was set to 1. Error bars (panels A, E, and F) indicate SDs from three replicates. NS, not significant; ***, P < 0.001 (t test).
To determine whether HspR regulates avermectin production through CSR gene aveR (29, 30) or structural genes, we performed a series of in vitro electrophoretic mobility shift assays (EMSAs) using soluble His6-HspR expressed in and purified from E. coli. The four genes in the ave gene cluster (aveA1, aveA2, aveA3, and aveA4) encode polyketide synthases (PKSs) responsible for the synthesis of the avermectin polyketide backbone. aveA1 is cotranscribed with aveA2, and aveA4 is cotranscribed with aveA3 (31). We therefore designed promoter probes aveRp, aveA1p (for aveA1-aveA2), and aveA4p (for aveA4-aveA3) for EMSAs (Fig. 2C) and used nonspecific probe hrdBp as the control. His6-HspR clearly retarded aveA1p but did not bind to aveA4p, aveRp, or hrdBp (Fig. 2D). Binding specificity was confirmed by adding ∼300-fold unlabeled specific probe aveA1p (lane S), which abolished the retarded band, or unlabeled nonspecific probe hrdBp (lane N), which had no effect on the delayed signal. Because of the cotranscription of aveA1 and aveA2, aveA2 is also targeted by HspR. These findings indicate that HspR directly regulates ave structural genes (aveA1 and aveA2) but not CSR gene aveR.
In vivo binding of HspR to aveA1p was confirmed by chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) assays. Samples were taken from WT and ΔhspR/hspR-3FLAG strains grown in FM-II for various durations. HspR bound to aveA1p at various time points. Binding was strongest on day 5 (consistent with HspR expression profile), whereas HspR was not enriched on aveRp (Fig. 2E), indicating dynamic binding of HspR to the target promoter aveA1p in vivo.
The effect of HspR on expression of targeted aveA1 and aveA2 genes was assessed by RT-qPCR, using RNAs isolated from 2-day and 6-day FM-I cultures of WT and ΔhspR srains. aveA1 and aveA2 transcription levels were lower in the ΔhspR strain than in the WT on both days (Fig. 2F), consistent with avermectin yields for the strains, indicating that HspR activates transcription of these two genes.
Determination of precise HspR-binding site on the aveA1 promoter region.
To identify the precise HspR-binding site on aveA1p and clarify the mechanism whereby HspR regulates the aveA1 gene, we performed DNase I footprinting assays. HspR protected a 31-nt region containing a 19-nt sequence (TTGAACGTCTTCAACTCTT) (Fig. 3A) similar to the conserved HspR-binding site HAIR in S. coelicolor (14), suggesting that the DNA-binding property of HspR is conserved.
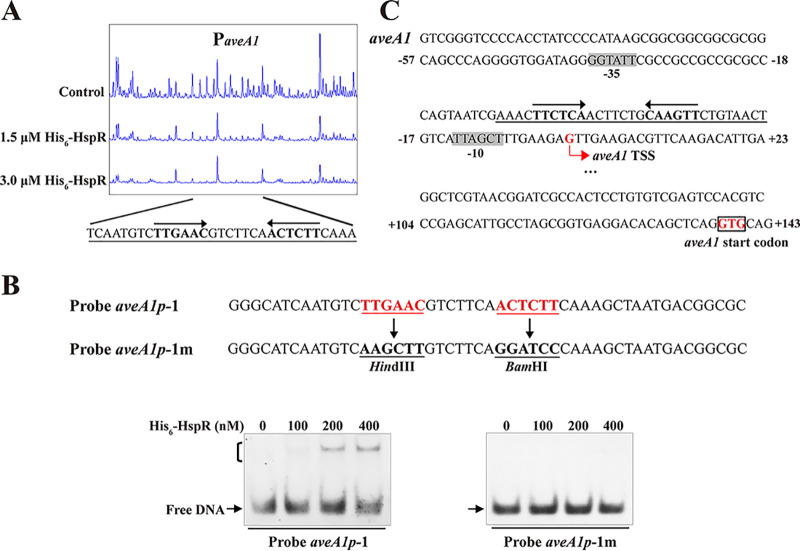
FIG 3.
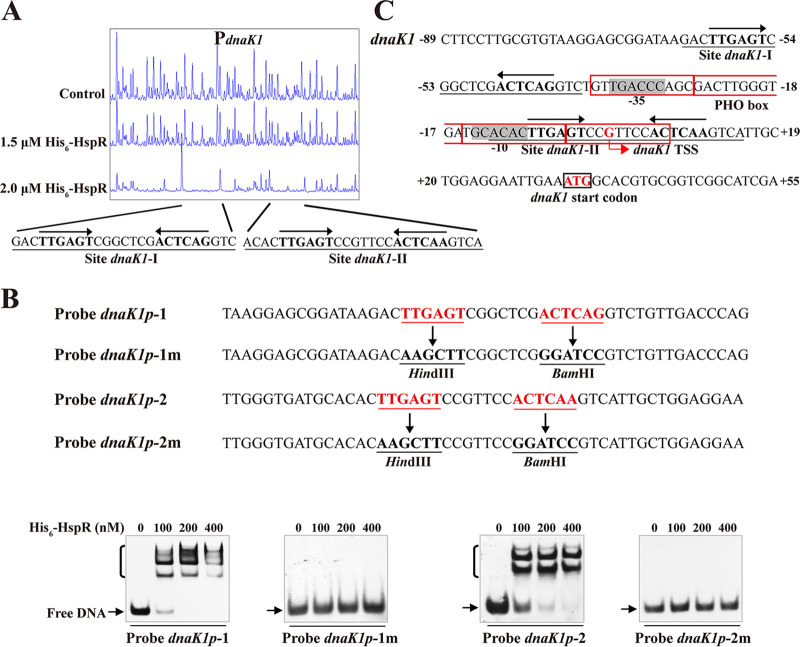
HspR-binding site on aveA1 promoter region. (A) DNase I footprinting assay of HspR on aveA1p. Protection patterns were acquired with increasing His6-HspR concentrations (reaction without His6-HspR used as a control). (B) EMSAs using 50-nt WT probe aveA1p-1 and its mutated probe aveA1p-1m. Imperfect inverted repeats in probe aveA1p-1 were replaced with HindIII and BamHI sites to produce mutated probe aveA1p-1m. Each lane contained 0.15 nM labeled probe. (C) Nucleotide sequences of aveA1 promoter region and HspR-binding site. Numbers indicate the distance (nucleotides) from aveA1 TSS. Box, aveA1 translational start codon (TSC); bent arrow, aveA1 TSS; shading, probable −10 and −35 regions; solid line, HspR-binding site; straight arrows, inverted repeats.
The importance of the 19-nt HAIR-like sequence in HspR binding was evaluated by performing EMSAs using 50-nt probes that contained either the intact sequence (termed probe aveA1p-1) or the mutated sequence (termed probe aveA1p-1m; lacking inverted repeats) (Fig. 3B). His6-HspR bound to WT probe aveA1p-1 but not to mutated probe aveA1p-1m (Fig. 3B), indicating that the 6-nt inverted repeats within the HAIR-like sequence are essential for HspR binding.
The transcriptional start site (TSS) of the aveA1 gene was determined by our group previously (32), and −35 and −10 promoter sequences were predicted on this basis (Fig. 3C). The 19-nt HspR-binding site on aveA1p is very close to the −10 region (3 nt downstream) and overlaps the aveA1 TSS (Fig. 3C). The HspR-binding site on aveA1p is unusual in regard to transcriptional activation; however, it is analogous to the binding sites of Streptomyces antibiotic regulatory protein (SARP)-family regulators AfsR (33) and OtcR (34), which are close to the −10 regions. These regulators presumably activate target transcription by recruiting RNA polymerase to the promoters. The mechanism of such activation of aveA1 by HspR remains to be elucidated.
Identification of S. avermitilis HspR target HSGs.
To examine response of HspR to heat shock stress in S. avermitilis, we measured the sensitivity of WT and ΔhspR strains to heat treatment. Growth of the ΔhspR strain was more resistant to heat shock stress than that of the WT (Fig. 4A), indicating that HspR represses the HSR, consistent with reported functions of its homologs in S. coelicolor and S. albus (13–16).
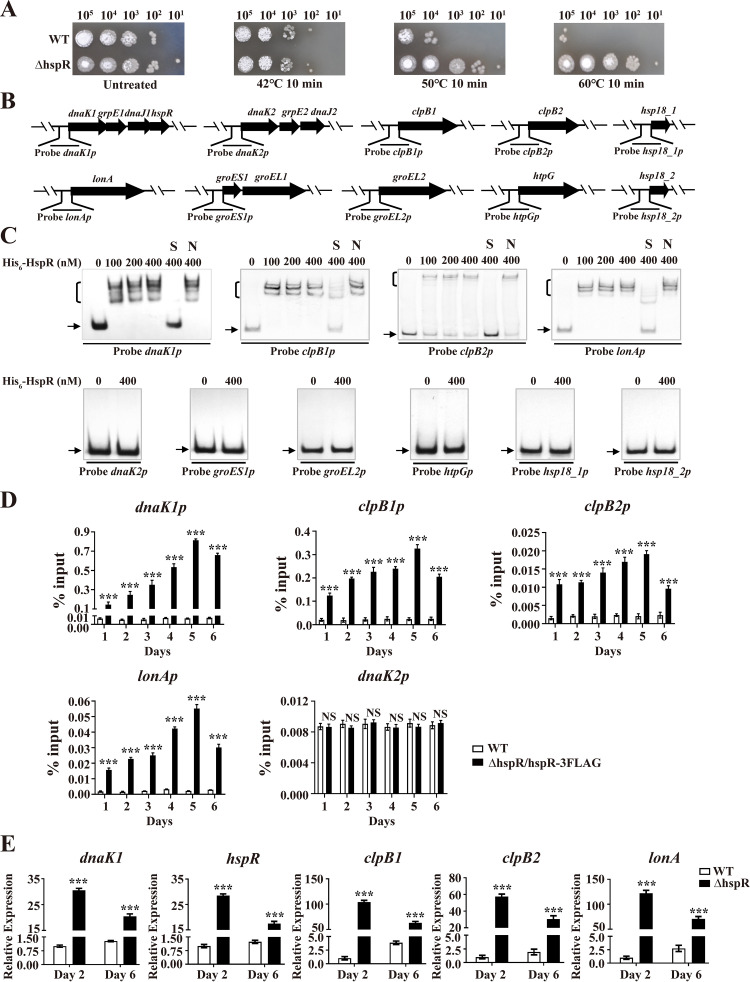
FIG 4.
Identification of HspR target HSGs. (A) Sensitivity of WT and ΔhspR strains to heat stress. Spore suspensions were treated at 42°C, 50°C, or 60°C for 10 min, spotted on YMS agar, and photographed after 3-day growth at 28°C. (B) Promoter probes of HSGs used in EMSAs. (C) EMSAs of His6-HspR with indicated probes. Notations as in Fig. 2D. (D) ChIP-qPCR assays of HspR binding to dnaK1p, clpB1p, clpB2p, lonAp, and dnaK2p. Notations as in Fig. 2E. (E) RT-qPCR analysis of five HspR target HSGs in WT and ΔhspR strains grown in FM-I. Error bars (panels D and E) indicate SDs from three replicates. ***, P < 0.001 (t test).
To identify HspR target HSGs, we performed EMSAs using His6-HspR and promoter probes of potential HSGs based on the S. avermitilis genome database (http://avermitilis.ls.kitasato-u.ac.jp): dnaK1p (for the dnaK1-grpE1-dnaJ1-hspR operon), dnaK2p (for the dnaK2-grpE2-dnaJ2 operon), clpB1p, clpB2p, lonAp, and groES1p (for the groES1-groEL1 operon), groEL2p, htpGp, hsp18_1p, and hsp18_2p (Fig. 4B). His6-HspR bound specifically to dnaK1p, clpB1p, clpB2p, and lonAp but not to other probes (Fig. 4C). hspR belongs to the dnaK1-grpE1-dnaJ1-hspR operon; therefore, HspR is autoregulated. Direct binding of HspR to dnaK1p, clpB1p, clpB2p, and lonAp in vivo was confirmed by ChIP-qPCR assays (Fig. 4D). Transcription levels of dnaK1, hspR, clpB1, clpB2, and lonA were shown by RT-qPCR analysis to be strongly upregulated in the ΔhspR strain on days 2 and 6 (Fig. 4E), indicating that HspR functions as a repressor of these target HSGs, consistent with the ΔhspR phenotype in heat stress assays.
To determine whether dnaK1, hspR, clpB1, clpB2, and lonA were induced in an HspR-dependent manner under heat stress, WT and ΔhspR strains were cultured in FM-II for 2 days, followed by heat (50°C) treatment for various durations. For the WT, heat treatment resulted in maximal induction of dnaK1 (∼49-fold), hspR (∼43-fold), clpB1 (∼84-fold), clpB2 (∼1.4-fold), and lonA (∼52-fold) within 10 min (see Fig. S5), indicating that HSPs DnaK1, ClpB1, and LonA play key roles in the HSR. For the ΔhspR strain, transcription levels of these five genes were all higher than the levels for the WT and also reached maximal values within 10 min of heat treatment. These findings confirm the negative role of HspR in controlling these target HSGs and suggest that these genes are also controlled by other regulator(s).
DNase I footprinting assays revealed that HspR protects two sites (dnaK1-I, dnaK1-II) on dnaK1p and that both sites contain a 19-nt HAIR-like sequence (Fig. 5A). The role of HAIR-like sequences in HspR binding was investigated by site-directed mutagenesis of 50-bp WT probes dnaK1p-1 and dnaK1p-2 on the inverted repeats to generate mutated probes dnaK1p-1m and dnaK1p-2m, respectively (Fig. 5B). EMSAs revealed that His6-HspR affinity for the mutated probes was abolished in comparison to that for the corresponding WT probes (Fig. 5B), reflecting an essential role of HAIR-like sequences in HspR binding. Site dnaK1-I extends from positions −63 to −39, and site dnaK1-II extends from positions −12 to +15 relative to the dnaK1 TSS (35) (Fig. 5C). Site dnaK1-I is close to the putative −35 region, and site dnaK1-II overlaps the putative −10 region of the dnaK1 promoter and dnaK1 TSS, indicating that HspR represses dnaK1 by blocking RNA polymerase access.
FIG 5.
HspR-binding site on dnaK1 promoter region. (A) DNase I footprinting assay of HspR on dnaK1p. (B) EMSAs using 50-nt WT probes dnaK1p-1 and dnaK1p-2 and their mutated probes dnaK1p-1m and dnaK1p-2m. Each lane contained 0.15 nM labeled probe. (C) Nucleotide sequences of dnaK1 promoter region and HspR-binding sites. Numbers indicate distance (nucleotides) from dnaK1 TSS. Black box, dnaK1 TSC; bent arrow, dnaK1 TSS; red boxes, PHO box. Other notations as in Fig. 3C.
The clpB1, clpB2, and lonA promoter regions also contain a 19-nt sequence similar to the HAIR motif, indicating that S. avermitilis HspR binds to these target DNAs at HAIR sites.
HspR responds to H2O2 stress.
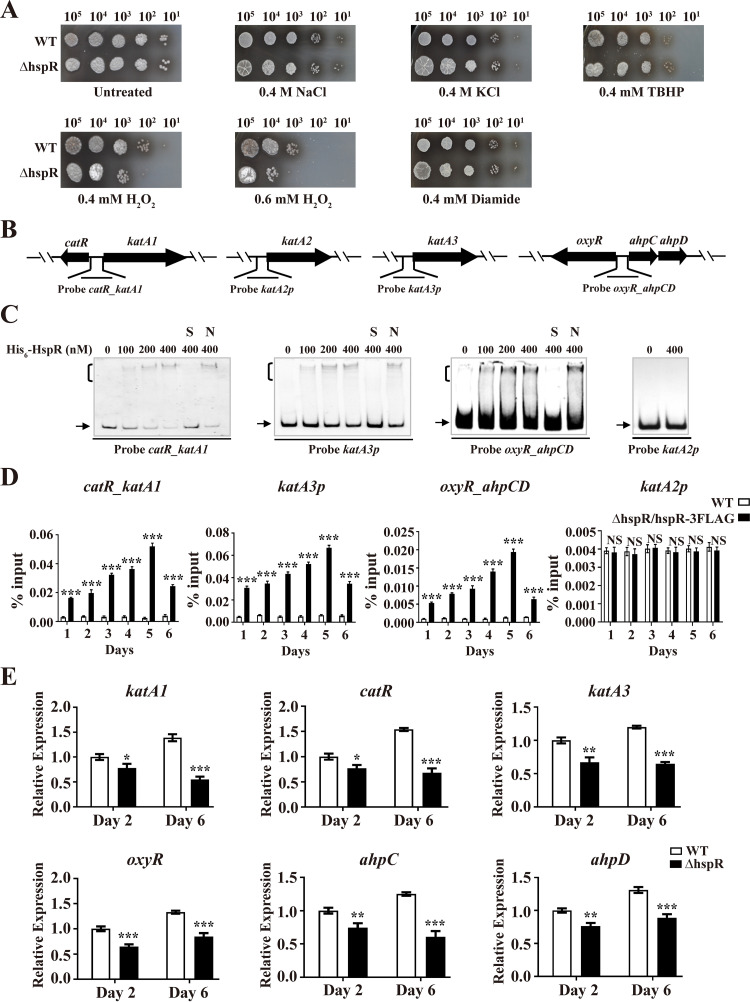
Possible regulation of other types of stress response by S. avermitilis HspR was evaluated on YMS plates. Relative to that of the WT, the ΔhspR strain showed greater sensitivity to H2O2 (which causes peroxidative stress) but similar sensitivity to tert-butyl hydroperoxide (TBHP; causes organic peroxidative stress), diamide (causes thiol-oxidative stress), and NaCl and KCl (cause osmotic stress) (Fig. 6A). S. avermitilis HspR evidently plays a role in resistance to H2O2 stress.
FIG 6.
Identification of HspR target genes involved in H2O2 stress response. (A) Sensitivity of WT and ΔhspR strains to various stress conditions. Spore suspensions were diluted serially and spotted on YMS agar containing NaCl, KCl, TBHP, H2O2, or diamide at the indicated concentrations. (B) Promoter probes of H2O2 stress-related genes used in EMSAs. (C) EMSAs of His6-HspR with indicated probes. Notations as in Fig. 2D. (D) ChIP-qPCR assays of HspR binding to katA2p, katA3p, catR_katA1, and oxyR_ahpCD. Notations as in Fig. 2E. (E) RT-qPCR analysis of six HspR target H2O2 stress-related genes in WT and ΔhspR strains grown in FM-I. Error bars (panels D and E) indicate SDs from three replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (t test).
Bacteria typically respond to H2O2 stress by producing peroxidases and catalases that degrade H2O2. In S. avermitilis, one ahpCD operon (for alkyl hydroperoxide reductase and alkylhydroperoxidase), three catalase genes (katA1, katA2, and katA3), and two TR genes (oxyR and catR) are involved in H2O2 stress response (36). Possible interactions of His6-HspR with promoter probes of these H2O2 stress-related genes (Fig. 6B) were investigated by EMSAs. His6-HspR bound specifically to probe katA3p and bidirectional promoter probes catR_katA1 and oxyR_ahpCD but not to katA2p (Fig. 6C). HspR-binding promoter regions katA3p, catR_katA1, and oxyR_ahpCD all contained HAIR-like sequences, as expected. Direct binding of HspR to katA3p, catR_katA1, and oxyR_ahpCD was confirmed in vivo by ChIP-qPCR assays (Fig. 6D). Consistent with the H2O2 stress phenotype for the ΔhspR strain, transcription levels of katA1, catR, katA3, oxyR, ahpC, and ahpD were all reduced in the ΔhspR strain relative to that in the WT grown in FM-I (Fig. 6E), indicating that HspR acts as an activator of these genes.
For analysis of H2O2 stress responses, WT and ΔhspR strains were treated with 0.6 mM H2O2 for various durations. In the WT, H2O2 treatment caused notable induction of katA1 (∼47-fold) and catR (∼8.7-fold) within 10 min and slight induction of katA3 (∼2.7-fold), ahpC (∼1.39-fold), and ahpD (∼1.36-fold) within 30 min and of oxyR (∼2.2-fold) within 40 min, whereas H2O2 treatment of the ΔhspR strain had a much lower inducing effect on katA1 (∼5-fold), catR (∼4.6-fold), and oxyR (∼1.7-fold) and no effect on katA3, ahpC, or ahpD expression (see Fig. S6). These findings indicate that HspR promotes H2O2 stress resistance in S. avermitilis by activating transcription of target genes (katA1, catR, katA3, oxyR, ahpC, and ahpD).
The effect of H2O2 on avermectin production was also investigated by HPLC analysis of 10-day cultures of the WT in FM-I containing various concentrations of H2O2, and the results showed that H2O2 addition did not promote avermectin production (see Fig. S7).
HspR interacts with PhoP at the dnaK1 promoter region.
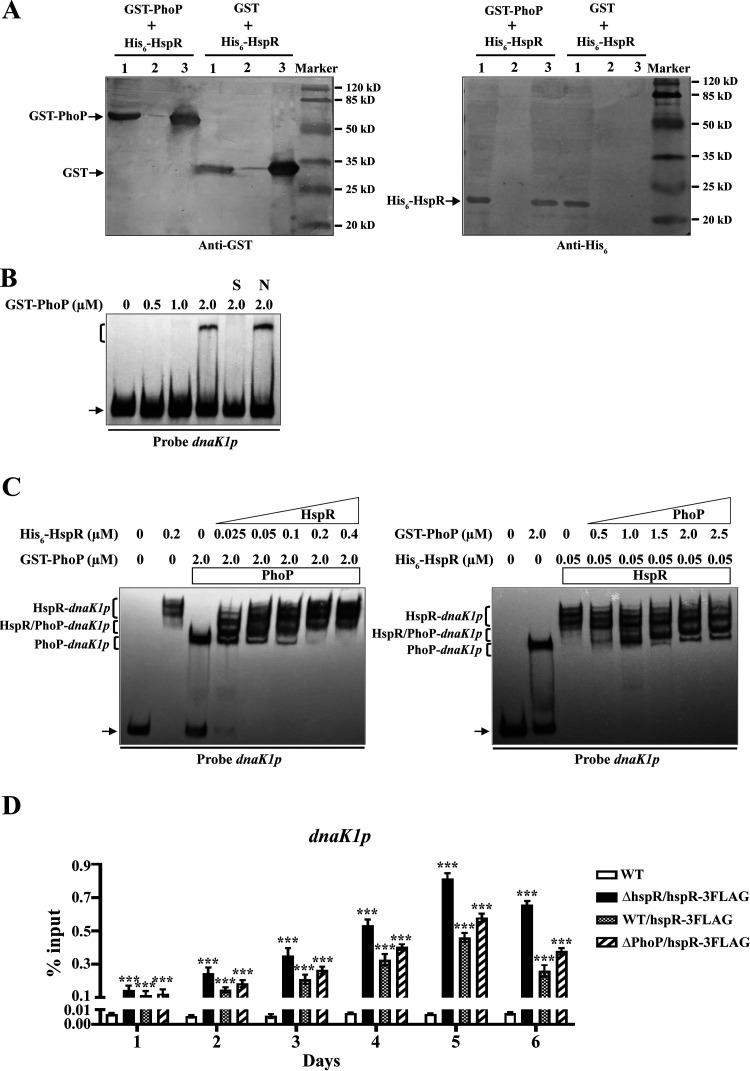
Cross talk between different regulatory systems commonly occurs in bacteria for the coordination of cell growth and metabolism. In M. tuberculosis, HspR interacts with PhoP to coregulate expression of acr2, which encodes an essential pathogenic determinant (20). We examined the possibility that HspR also interacts with PhoP in Streptomyces. The Streptomyces PhoP-binding sequence (termed PHO box) consists of 11-nt consecutive direct repeat units (DRus; GG/TTCAYYYRG/CG) (37). We found that dnaK1p contains a PHO box sequence formed by four DRus and that the PHO box overlaps site dnaK1-II (Fig. 5C), suggesting that HspR-PhoP interaction may occur at dnaK1p. To test this possibility, we coexpressed His6-HspR with glutathione transferase (GST) or GST-PhoP in E. coli and performed GST pulldown assays. His6-HspR was pulled down by GST-PhoP but not by GST tag (negative control) (Fig. 7A), indicating that HspR interacts with PhoP. Specific binding of purified GST-PhoP (38) to dnaK1p was confirmed by EMSAs (Fig. 7B).
FIG 7.
Interaction of HspR with PhoP at dnaK1p. (A) GST pulldown assays of HspR and PhoP from E. coli whole-cell lysate containing both His6- and GST-tagged proteins. Lane 1, flowthrough of cell lysate after incubation with glutathione-Sepharose beads; lane 2, washing buffer following fourth wash of beads; lanes 3, GST pulldown. (B) EMSAs of GST-PhoP with probe dnaK1p. A 0.15 nM concentration of labeled probe and various amounts of GST-PhoP were used for each binding reaction. For nonspecific (lane N) or specific (lane S) competition assays, ∼500-fold unlabeled competitor DNA was used. (C) Competitive EMSAs of probe dnaK1p with His6-HspR and GST-PhoP. A 0.15 nM concentration of labeled probe dnaK1p was incubated with indicated concentrations of His6-HspR and GST-PhoP. (D) ChIP-qPCR assays of HspR-3FLAG binding to dnaK1p in WT (negative control), ΔhspR/hspR-3FLAG, WT/hspR-3FLAG, and ΔphoP/hspR-3FLAG strains grown in FM-II for the indicated times. Error bars indicate SDs from three replicates. ***, P < 0.001 (t test).
The PhoP-binding site on dnaK1p (PHO box) overlaps the HspR-binding site dnaK1-II; these two regulators may therefore affect each other’s binding to dnaK1p. We examined this possibility by applying His6-HspR and GST-PhoP, both separately and together, with probe dnaK1p in EMSAs. When applied separately, both proteins retarded dnaK1p migration (Fig. 7C). In the presence of 2 μM GST-PhoP, increasing amounts of His6-HspR resulted in a reduction of the PhoP-dnaK1p complex, an increase of the HspR-dnaK1p complex, and formation of a new retarded band located between those for PhoP-dnaK1p and HspR-dnaK1p (Fig. 7C, left), which was presumably formed by interaction of the HspR-PhoP complex with dnaK1p. In the presence of 0.05 μM His6-HspR, increasing amounts of GST-PhoP resulted in reduction of the HspR-dnaK1p complex, appearance of PhoP-dnaK1p, and appearance of a new band, presumably HspR-PhoP-dnaK1p (Fig. 7C, right). These findings suggest that HspR and PhoP cooperate as a complex for DNA binding besides competing for binding to dnaK1p.
We used ChIP-qPCR assays to examine binding of HspR-3FLAG to dnaK1p in WT, ΔhspR/hspR-3FLAG, WT/hspR-3FLAG, and ΔphoP/hspR-3FLAG strains. HspR-3FLAG on dnaK1p was enriched (relative to that for negative-control WT) in ΔhspR/hspR-3FLAG, WT/hspR-3FLAG, and ΔphoP/hspR-3FLAG strains grown in FM-II for various durations (Fig. 7D), indicating that DNA-binding activity of HspR does not depend on PhoP. Enrichment levels of HspR-3FLAG on dnaK1p were highest in the ΔhspR/hspR-3FLAG strain and higher in the ΔphoP/hspR-3FLAG strain than in the WT/hspR-3FLAG strain at various time points (Fig. 7D), indicating competitive binding of HspR and PhoP to dnaK1p. HspR evidently plays a dominant role relative to PhoP in the regulation of dnaK1p.
PhoP represses target HSGs.
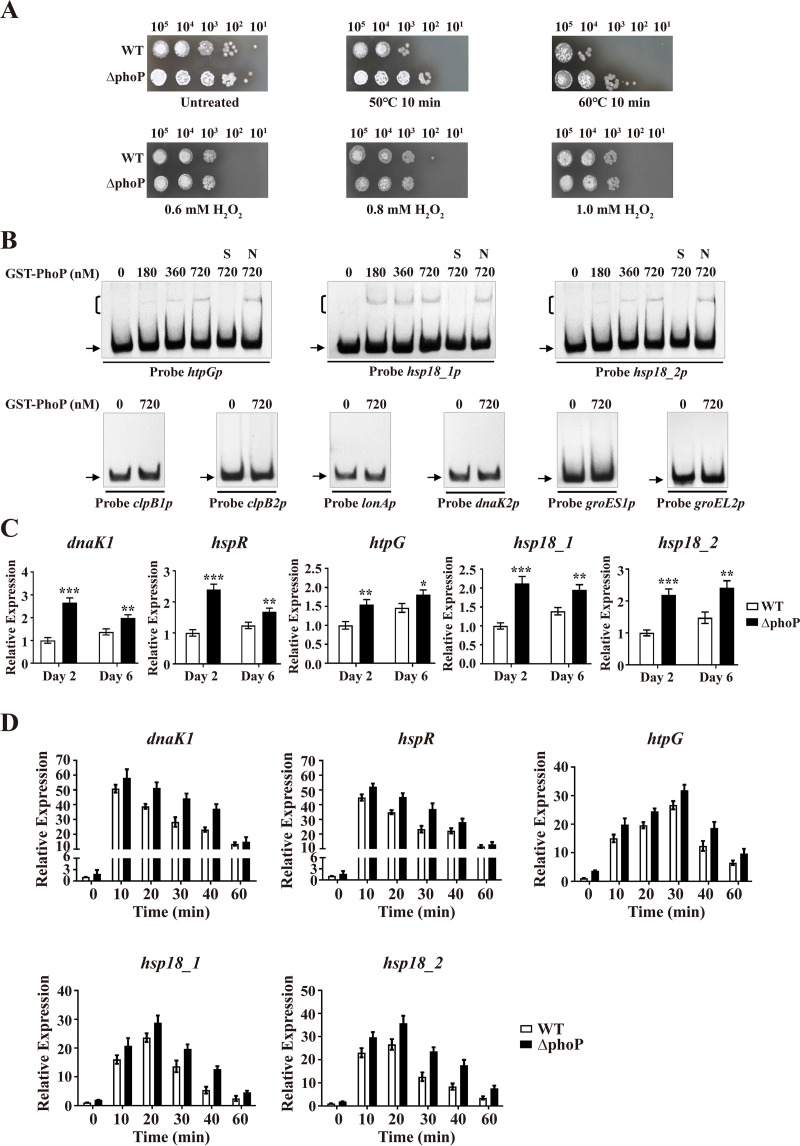
The observed binding of PhoP to dnaK1p suggests that PhoP may also be involved in the HSR. We evaluated this possibility by comparing the growth of WT and ΔphoP strains (38) on YMS plates following heat treatment. Resistance to heat shock stress was greater for the ΔphoP strain than for the WT (Fig. 8A), indicating that PhoP (like HspR) has a negative effect on HSR. In contrast to HspR, PhoP had no effect on the H2O2 stress response (Fig. 8A).
FIG 8.
Identification of PhoP target HSGs. (A) Sensitivity of WT and ΔphoP strains to heat and H2O2 stresses. (B) EMSAs of GST-PhoP with promoter probes of HSGs. Notations as in Fig. 7B. (C) RT-qPCR analysis of five PhoP target HSGs in WT and ΔphoP strains grown in FM-I. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (t test). (D) Induction of five HSGs by heat treatment (50°C) in WT and ΔphoP strains grown in FM-II. For each gene, the transcription level in the WT before temperature rise (0 min) was set to 1. Error bars (panels C and D) indicate SDs from three replicates.
PhoP bound to probe dnaK1p (Fig. 7B). EMSAs with GST-PhoP and other promoter probes of HSGs (Fig. 4B) revealed specific binding of PhoP to htpGp, hsp18_1p, and hsp18_2p but not to other tested probes (Fig. 8B). HspR and PhoP thus have exclusive target HSGs in addition to their common target HSGs (genes in the dnaK1-grpE1-dnaJ1-hspR operon); i.e., clpB1, clpB2, and lonA are targets of HspR but not of PhoP, whereas htpG, hsp18_1, and hsp18_2 are targets of PhoP but not of HspR.
Consistent with the ΔphoP phenotype observed in heat stress tests, transcription levels of PhoP target HSGs dnaK1, hspR, htpG, hsp18_1, and hsp18_2 were much higher in the ΔphoP strain than in the WT grown in FM-I (Fig. 8C), indicating negative regulation of these targets by PhoP. Transcription levels of these five genes in WT and ΔphoP strains were recorded under heat shock stress. In the WT, levels increased to maximum for dnaK1 (∼49-fold) and hspR (∼43-fold) within 10 min, for hsp18_1 (∼22-fold) and hsp18_2 (∼25-fold) within 20 min, and for htpG (∼25-fold) within 30 min (Fig. 8D), indicating important roles of DnaK1, Hsp18_1, Hsp18_2, and HtpG in HSR. In the ΔhspR strain, all five genes showed increased transcription levels (Fig. 8D), confirming repression of these genes by PhoP.
Prediction of HspR regulon and identification of HspR targets involved in development.
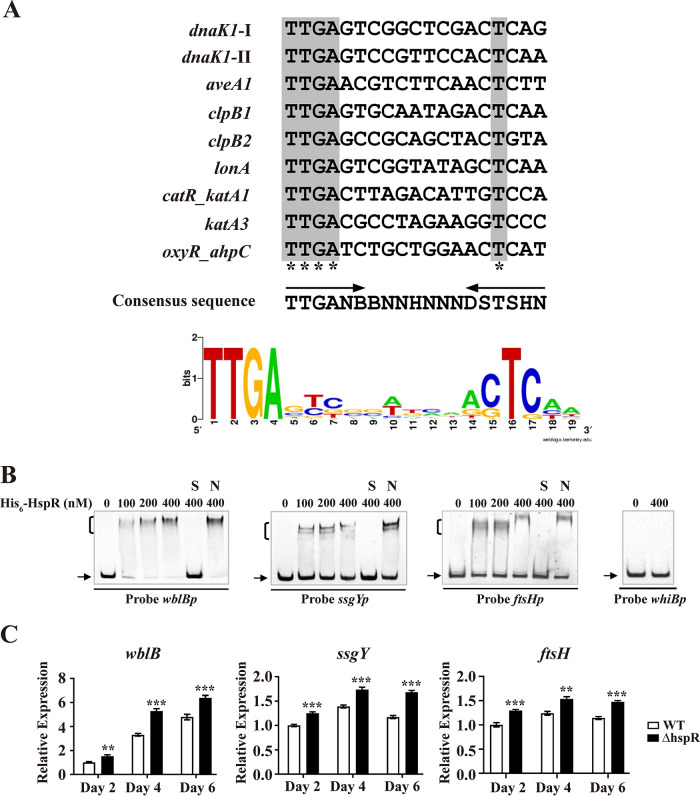
To further clarify the roles of HspR in S. avermitilis, we predicted its regulon. WebLogo (http://weblogo.berkeley.edu) analysis of the 19-nt HAIR sequences in the above-mentioned HspR-binding promoter regions (aveA1p, dnaK1p, clpB1p, clpB2p, lonAp, catR-katA1, katA3p, and oxyR-ahpCD) revealed a consensus sequence, 5′-TTGANBBNNHNNNDSTSHN-3′ (N is A/T/C/G, B is T/C/G, H is A/C/T, D is A/G/T, and S is C/G) (Fig. 9A). Scanning of the S. avermitilis genome with the 19-nt consensus HspR-binding sequence by PREDetector (39) identified 155 putative HspR target genes (cutoff; score ≥ 8) (see Table S1). Of these, 60 were unclassified or unknown, and the remaining 95 were assigned to 15 functional groups on the basis of the KEGG pathway database for S. avermitilis.
FIG 9.
Identification of HspR target genes associated with development. (A) Analysis of consensus HspR-binding sequence by WebLogo. Arrows, conserved 6-nt inverted repeats; asterisks, consensus bases. (B) EMSAs of His6-HspR with promoter probes of four developmental genes. Notations as in Fig. 2D. (C) RT-qPCR analysis of wblB, ssgY, and ftsH in WT and ΔhspR strains grown on YMS agar. Error bars show SDs from three replicates. **, P < 0.01; ***, P < 0.001 (t test).
Our phenotypic observations revealed the negative role of HspR in S. avermitilis development. We therefore performed EMSAs on several predicted HspR target developmental genes listed in Table S1: whiB (sav_5042) for sporulation regulator WhiB (40), wblB (sav_4997) for putative WhiB-family TR (putative control of cell cycle), ssgY (sav_4267) for putative sporulation-specific cell division protein SsgY, and ftsH (sav_4666) for putative cell division protein FtsH. His6-HspR bound to promoter probes wblBp, ssgYp, and ftsHp but not to probe whiBp (Fig. 9B). Transcription levels of wblB, ssgY, and ftsH were determined by RT-qPCR using RNAs prepared from WT and ΔhspR strains grown on YMS plates for 2 (aerial hypha growth stage), 4 (middle sporulation stage), or 6 (spore maturation stage) days. Levels of these three genes were higher in the ΔhspR strain than in the WT at three time points (Fig. 9C), consistent with the earlier differentiation phenotype of the ΔhspR strain. These findings indicate that HspR negatively regulates development by directly repressing wblB, ssgY, and ftsH.
DISCUSSION
HspR has been reported to function as an HSR repressor in S. coelicolor, S. albus, and S. lividans (13–17); however, its roles in antibiotic production, development, and other stress responses of Streptomyces had not been studied until now. Results of the present study of molecular mechanisms underlying HspR function in avermectin production, development, and heat shock and H2O2 stress responses in S. avermitilis demonstrate that HspR acts as a dual repressor/activator in these important physiological processes through control of corresponding target genes. HspR was shown to interact with another important regulator, PhoP, to coregulate the HSR. These findings bring to light previously unrecognized roles and regulatory mechanisms of HspR in this genus and provide a basis for the construction of antibiotic-overproducing strains in Streptomyces species.
S. avermitilis HspR directly activated avermectin production by interacting with the promoter region of structural genes aveA1 and aveA2 and strongly promoted avermectin production in both WT and industrial strains. HspR homologs are widely distributed among Streptomyces. The HspR-mediated activation of antibiotic production may occur in other Streptomyces species, and this possibility requires further investigations.
HspR was shown to play a negative regulatory role in S. avermitilis development, and three developmental genes (wblB, ssgY, and ftsH) were identified as HspR targets. wblB encodes a putative WhiB-family TR homologous to S. coelicolor WhiD, which is required for late-stage sporulation (41). ssgY encodes a putative cell division protein homologous to S. coelicolor SsgA, which is required for synthesis of sporulation septa (42). ftsH encodes a putative cell division protein homologous to E. coli FtsH. Enhanced expression of these three HspR target genes in the ΔhspR strain may account for the phenotype of this mutant. However, we cannot rule out possible contributions to the ΔhspR phenotype by other HspR target developmental genes; further studies are needed to resolve this point.
In regard to control of the HSR, S. avermitilis HspR acts as a direct repressor of several HSGs (the dnaK1-grpE1-dnaJ1-hspR operon, clpB1p, clpB2p, and lonAp), consistent with its reported role in HSRs in other Streptomyces species (13–17). The ability of HspR and chaperone DnaK to form a complex to corepress HspR target HSGs has been well established in S. coelicolor (14, 18). HspR and DnaK are both highly conserved in the genus, and it is therefore likely that HspR-DnaK interaction occurs in other species. The mechanism of such interaction was not investigated in the present study. Our findings reveal an additional layer of complex regulation of HSR, i.e., HspR cross talks with PhoP at dnaK1p to corepress the dnaK1-grpE1-dnaJ1-hspR operon. GST pulldown and competitive EMSAs revealed the existence of the HspR-PhoP complex and the reduction or disappearance of the HspR-PhoP-dnaK1p complex with increasing concentrations of one protein while concentration of the other protein remained constant, indicating that formation of the HspR-PhoP complex is a highly dynamic process. HspR and PhoP do not depend on each other for DNA binding, and they both have exclusive target HSGs (clpB1, clpB2, and lonA for HspR; htpG, hsp18_1, and hsp18_2 for PhoP). The HspR-PhoP complex may therefore play a regulatory role which neither protein by itself is capable; i.e., stabilization of DNA-protein structure to precisely regulate essential genes. Consistent with this possibility, the essential pathogenic determinant gene acr2 in M. tuberculosis is regulated by the HspR-PhoP complex (20). Notwithstanding their cooperative activity, HspR and PhoP compete for binding to dnaK1p, i.e., an increasing concentration of one protein suppresses formation of the complex with dnaK1p by the other protein, reflecting the delicate interplay between these two regulators in the control of the dnaK1-grpE1-dnaJ1-hspR operon. Our findings reveal a novel role of PhoP in the HSR and further demonstrate the importance of PhoP in Streptomyces. Coregulation of the dnaK1-grpE1-dnaJ1-hspR operon by HspR and PhoP suggests that HSGs within this operon play a dominant role over other HSGs in response to heat shock stress.
HspR has a negative regulatory role in the HSR, but we also observed a positive role in the H2O2 stress response, i.e., it directly activates the transcription of related genes. HspR target genes (katA1, catR, katA3, oxyR, ahpC, and ahpD) are well conserved in Streptomyces; such an HspR-based mechanism involved in the control of the H2O2 stress response is therefore likely to be present in other species. HspR also functions as an activator (it activates transcription of ave structural genes and H2O2 stress-related genes), but the mechanism underlying this function remains to be elucidated.
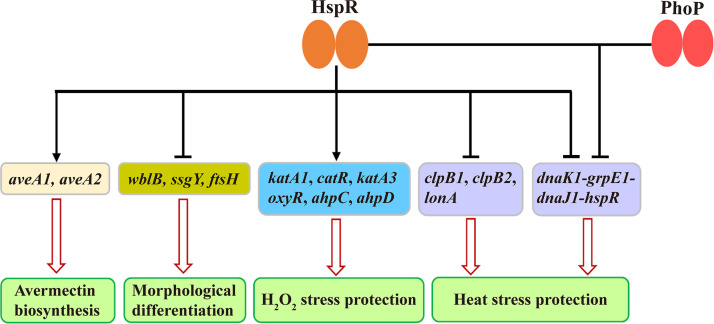
Our findings, taken together, provide a basis for a proposed model of the HspR-mediated regulatory network involved in development, avermectin production, and heat shock and H2O2 stress responses in S. avermitilis (Fig. 10). These important physiological processes are coordinated by HspR through its positive or negative effect on target genes. HspR also interacts with PhoP to corepress the dnaK1-grpE1-dnaJ1-hspR operon. Predicted HspR targets are involved in other essential functions such as primary metabolism (see Table S1 in the supplemental material). It is clear that the roles of HspR in Streptomyces are much broader than previously recognized. Identification of additional targets and related molecular processes will require extensive further study.
FIG 10.
Proposed model of HspR-mediated regulatory network in S. avermitilis. Solid arrows, direct activation; bars, direct repression; hollow arrows, avermectin biosynthesis, development, or response to stress.
MATERIALS AND METHODS
Strains, plasmids, primers, and growth conditions.
Strains and plasmids used in this study are listed in Table 1, and primers are listed in Table 2. Culture conditions for S. avermitilis and E. coli were described previously (43). YMS (44) agar was used for observation of the S. avermitilis phenotype. Insoluble FM-I fermentation medium and soluble FM-II fermentation medium (45) were used for routine avermectin production and growth analysis, respectively. FM-II was also used to grow mycelia for ChIP-qPCR assays and for RNA isolation following stress treatment.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| S. avermitilis | ||
| ATCC 31267 | Wild-type (WT) strain | Laboratory stock |
| A229 | Industrial strain | Qilu Pharmaceutical |
| ΔhspR | hspR deletion mutant | This study |
| ChspR | hspR complemented strain | This study |
| OhspR | hspR overexpression strain | This study |
| ΔphoP | phoP deletion mutant | 38 |
| ΔhspR/hspR-3FLAG | hspR complemented strain with HspR-3FLAG | This study |
| ΔphoP/hspR-3FLAG | phoP deletion mutant with HspR-3FLAG | This study |
| WT/hspR-3FLAG | WT strain with HspR-3FLAG | This study |
| WT/pKC1139 | WT strain carrying empty vector pKC1139 | This study |
| WT/pSET152 | WT strain carrying empty vector pSET152 | This study |
| OhspR/A229 | hspR overexpression strain based on A229 | This study |
| E. coli | ||
| JM109 | General cloning host for plasmid manipulation | Laboratory stock |
| BL21(DE3) | Host for protein expression | Novagen |
| Plasmids | ||
| pKC1139 | Multiple-copy temperature-sensitive E. coli-Streptomyces shuttle vector | 46 |
| pSET152 | Integrative E. coli-Streptomyces shuttle vector | 46 |
| pET-28a(+) | Vector for His6-tagged protein expression in E. coli | Novagen |
| pGEX-4T-1 | Vector for GST-tagged protein expression in E. coli | GE Healthcare |
| pCIMt005 | Multiple-copy temperature-sensitive E. coli-Streptomyces shuttle vector | 47 |
| pJL117 | pIJ2925 derivative carrying the Streptomyces strong constitutive promoter ermE*p | 49 |
| pKCΔhspR | hspR deletion vector based on pKC1139 | This study |
| pΔhspR | hspR deletion vector based on pCIMt005 | This study |
| pSET152-hspR | hspR complemented vector based on pSET152 | This study |
| pKC-erm-hspR | hspR overexpression vector based on pKC1139 | This study |
| pSET152-hspR- | hspR complemented vector with 3×FLAG-tagged | This study |
| 3FLAG | HspR on pSET152 | |
| pIJ10500 | Vector carrying 3×FLAG fragment | 50 |
| pET28-hspR | His6-HspR expression vector based on pET-28a(+) | This study |
| pGEX-phoP | GST-PhoP expression vector based on pGEX-4T-1 | 38 |
TABLE 2.
Primers used in this study
| Primer | DNA sequencea (5′→3′) | Use |
|---|---|---|
| Gene disruption, complementation, and overexpression | ||
| WQ11 | CGGGATCCGATCGCCGAGAACCCCTG (BamHI) | Deletion of hspR gene |
| WQ12 | GCTCTAGAGTACTGACGCAGGGTCTG (XbaI) | |
| WQ13 | GCTCTAGAACCAGGAGGTCCAGCAGA (XbaI) | |
| WQ14 | CCCAAGCTTAACGAGATGGCAACAGCA (HindIII) | |
| LXR101A | GTCTCGTACGAAGAGCTTTTATAAAAGCTTGATCGCCGAGAACCCCTG | |
| LXR101B | AAAATCCCTTAACGTGAGCCTAGGGCGTGCAACGAGATGGCAACAGCA | |
| WQ15 | CCTGCAAGGCCTGTTCGG | Confirmation of hspR deletion in ΔhspR strain |
| WQ16 | AGGGCTATGACGGTACGA | |
| WQ17 | ACCGTCTCGGCCTGGTCT | |
| WQ18 | CTCGCGCTGCTGCATCAT | |
| LXR133A | CCGGAATTCGATGGCGTCCTCGTGCTC (EcoRI) | Complementation of hspR in ΔhspR strain |
| LXR133B | CGGGATCCTTCAATTCCTCCAGCAA (BamHI) | |
| LXR134A | CGGGATCCGAGGAGATGGACGGCCGT (BamHI) | |
| LXR134B | CCCAAGCTTCGAAGCCGTCAGTCCGAG (HindIII) | |
| WQ25 | CGGGATCCCCGCGAAGGGAGCATGAGG (BamHI) | Overexpression of hspR in Streptomyces |
| WQ26 | CCCAAGCTTCTGGACAGTCTCGGTGCCG (HindIII) | |
| WQ27 | CCGGAATTCCATGAGGAGATGGACGGC (EcoRI) | Expression of His6-HspR protein in E. coli |
| WQ28 | CCCAAGCTTACATGCTGGACAGTCTCG (HindIII) | |
| LXR60A | CCCAAGCTTGGAGGTGGCATGGACTAC (HindIII) | Amplification of 3×FLAG fragment |
| LXR60B | CGGGATCCTCCGGTTGACCCTTATTT (BamHI) | |
| LXR136A | GCTCTAGAGATGGCGTCCTCGTGCTC (XbaI) | Complementation of hspR in ΔhspR strain with 3×FLAG-tagged HspR |
| LXR136B | TTCAATTCCTCCAGCAA | |
| LXR135A | TCATTGCTGGAGGAATTGAAGAGGAGATGGACGGCCGT | |
| LXR135B | CCCAAGCTTGTCCGAGGACTGGCGCTT (HindIII) | |
| EMSA | ||
| aveA1p-Fw | ATGGTCGGGAACCTCCGCAA | Probe aveA1p |
| aveA1p-Rev | CTGTGTCCTCACCGCTAGGC | |
| LXR59A | ATCTAGTCGTTTTTGAGT | Probe hrdBp |
| LXR59B | ACGAACAACCTCTCGGAA | |
| aveA4p-Fw | CGACAAGAGAAATCGGAAATT | Probe aveA4p |
| aveA4p-Rev | GCCTGCACCTGTGACAAG | |
| aveRp-Fw | CCGCACCGCCATACATAC | Probe aveRp |
| aveRp-Rev | GAAACTCCCTGCATGATGTTC | |
| LXR102A | GGGCATCAATGTCTTGAACGTCTTCAACTCTTCAAAGCTAATGACGGCGC | Probe aveA1p-1 |
| LXR102B | GCGCCGTCATTAGCTTTGAAGAGTTGAAGACGTTCAAGACATTGATGCCC | |
| LXR103A | GGGCATCAATGTCAAGCTTGTCTTCAGGATCCCAAAGCTAATGACGGCGC | Probe aveA1p-1m |
| LXR103B | GCGCCGTCATTAGCTTTGGGATCCTGAAGACAAGCTTGACATTGATGCCC | |
| WQ39 | AGGAGCGGATAAGACTTGAGT | Probe dnaK1p |
| WQ40 | TTAGTCGTGCCCAGGTCG | |
| LXR104A | TAAGGAGCGGATAAGACTTGAGTCGGCTCGACTCAGGTCTGTTGACCCAG | Probe dnaK1p-1 |
| LXR104B | CTGGGTCAACAGACCTGAGTCGAGCCGACTCAAGTCTTATCCGCTCCTTA | |
| LXR105A | TAAGGAGCGGATAAGACAAGCTTCGGCTCGGGATCCGTCTGTTGACCCAG | Probe dnaK1p-1m |
| LXR105B | CTGGGTCAACAGACGGATCCCGAGCCGAAGCTTGTCTTATCCGCTCCTTA | |
| LXR106A | TTGGGTGATGCACACTTGAGTCCGTTCCACTCAAGTCATTGCTGGAGGAA | Probe dnaK1p-2 |
| LXR106B | TTCCTCCAGCAATGACTTGAGTGGAACGGACTCAAGTGTGCATCACCCAA | |
| LXR107A | TTGGGTGATGCACACAAGCTTCCGTTCCGGATCCGTCATTGCTGGAGGAA | Probe dnaK1p-2m |
| LXR107B | TTCCTCCAGCAATGACGGATCCGGAACGGAAGCTTGTGTGCATCACCCAA | |
| LXR108A | CATGGGGCCGAACCTAGC | Probe clpB1p |
| LXR108B | TCGCTGTCTCCTCCTGTG | |
| LXR109A | AAGGAAGAGGCGGGCGTG | Probe clpB2p |
| LXR109B | CGACCGGATCCGACGCG | |
| LXR110A | GAAGCACCTGGTTCGCCT | Probe lonAp |
| LXR110B | AAGCCATGATCTCCCCTT | |
| SD335A | CGTTCTCCTCGAGCCTACAGG | Probe dnaK2p |
| SD335B | CTCCCACACGGCGATCAC | |
| SD339A | ATTGGCACTCCGCTTGAC | Probe groES1p |
| SD339B | GGCTTGATGGCAACCTTG | |
| SD337A | TTCAGATGGACGGCTACC | Probe groEL2p |
| SD337B | AGAGTGCTAACGCCAATGA | |
| SD340A | GGAAAGCCGCTGGTCAGAC | Probe htpGp |
| SD340B | GGGAGTCCATCGTCGCAG | |
| SD341A | ATCGTCCCTGGTCATCGG | Probe hsp18_1p |
| SD341B | ATCAACATCGTGAAACACCTCC | |
| SD342A | AGTGCTGTCGTCGCTCAT | Probe hsp18_2p |
| SD342B | CATCAACACGGTAAACACCTC | |
| SD123A | GCGCAGTCGTTCCAGTAG | Probe catR_katA1 |
| SD123B | GGACTCGCTGTTCTGATTGT | |
| SD124A | GGAAAGGTGGTCTGGTTC | Probe katA2p |
| SD124B | GGTGTACGGAGCCTTCTG | |
| SD122A | AGGTCCGTGGAGCGCCTC | Probe katA3p |
| SD122B | CGGTGGCTGAGTTCTGGTTGTC | |
| SD236A | GAGAAGCCGAGCACCTGG | Probe oxyR_ahpCD |
| SD236B | CGCTGGAGAAGGGCAATG | |
| SD210A | CCGATGGCTGTAGGCGTTTAT | Probe whiBp |
| SD210B | CCCTCGTCTGTCTTCGCG | |
| LXR111A | GAGCGCGTTTTCTCCGAG | Probe ssgYp |
| LXR111B | ATGCAGCCCCTGAATTTC | |
| LXR112A | ATCTTCGGCACTCTACGT | Probe wblBp |
| LXR112B | AAATCTGCCATTACGTGT | |
| SD115A | GCCCGGTTCTCGTAAGAC | Probe ftsHp |
| SD115B | CAGCACGATCCACATGAC | |
| DNase I footprinting | ||
| FAM-WQ45 | CCCAGCGCACGATTCATGA | aveA1 promoter region |
| WQ46 | CCGTCCATCCTCTGCACCTG | |
| FAM-WQ43 | GTCTCGGTCCTCGGGCTG | dnaK1 promoter region |
| WQ44 | TGCTGCGGGTTGAAGTCC | |
| RT-qPCR | ||
| 16S-QP-Fw | AGCGGAGCATGTGGCTTAAT | 16S rRNA ORF |
| 16S-QP-Rev | ACGTATTCACCGCAGCAATG | |
| GJ99 | CGGACAGGACTACGCACTTC | aveA1 ORF |
| GJ100 | ACGAGATACGACCGGAGATC | |
| LXR32A | ATCTCGTCAAGTCCCAGA | aveA2 ORF |
| LXR32B | GTGTAACTGGTTCCTGAG | |
| SD346A | CCAAGAACGGTGAGGTGC | dnaK1 ORF |
| SD346B | CTGCTGCGGGTTGAAGTC | |
| LXR113A | TCGGAACCCCTATGAACT | hspR ORF |
| LXR113B | GTACTGACGCAGGGTCTG | |
| LXR114A | CCTCAAGAACAAGCGGCT | clpB1 ORF |
| LXR114B | CGGCGAGCACGGTCTTCA | |
| LXR115A | ATGAACCGTCTCACCCAG | clpB2 ORF |
| LXR115B | TCCTCCTGATCGAGAAGT | |
| LXR116A | AAGGGGAGATCATGGCTT | lonA ORF |
| LXR116B | GGCGTCGTTCAGGTCCAG | |
| katA1-QP-Fw | ACCTCGTCGGCAACAACAC | katA1 ORF |
| katA1-QP-Rev | TGTACGGGTCGCGCTTCTG | |
| catR-QP-Fw | TGAAGCTGCCCGAGATCTCC | catR ORF |
| catR-QP-Rev | CTTGTCCGTGGCGACCTCC | |
| katA3-QP-Fw | CATCCACTCCCAGAAGCGC | katA3 ORF |
| katA3-QP-Rev | TCGCCCATCAGCCAGGTC | |
| oxyR-QP-Fw | GATGGAGGAGGCCGAGGC | oxyR ORF |
| oxyR-QP-Rev | GCAGGAGGTTGAGCTCACG | |
| ahpC-QP-Fw | CCAGGTGCTCGGCTTCTCC | ahpC ORF |
| ahpC-QP-Rev | GAAGCACGAGCTGATGCGC | |
| ahpD-QP-Fw | CTCTCGATGAACTGAAGG | ahpD ORF |
| ahpD-QP-Rev | TGCGGCAGGTCGGAGTTG | |
| LXR117A | TCACAGTCATCCCAGGCAT | htpG ORF |
| LXR117B | CTCGCGCAGATAGACCTT | |
| LXR118A | CGATGCCAATGGACGCCTA | hsp18_1 ORF |
| LXR118B | TCAGCATGTTCCGTTCGA | |
| LXR119A | ATCTGATGGGTCCGGGAA | hsp18_2 ORF |
| LXR119B | AAGGCGATGACGTACTGG | |
| LXR120A | CAGCTCGCTCTTCTTCCA | wblB ORF |
| LXR120B | TCATGCAGACCTCTTTGG | |
| LXR121A | AATGGATCATCGGACGAGA | ssgY ORF |
| LXR121B | TGTACGAGTCGGAGCGGTCAG | |
| SD247A | CGGCTACAAGACAGTGGACA | ftsH ORF |
| SD247B | GACCTTGATGACCTGCTCGT | |
| LXR122A | GCACCGTCCAGCAGTTGT | hspR deletion region |
| LXR122B | ACTCCAGTTCCACCACGC | |
| ChIP-qPCR | ||
| LXR123A | AGGGGTGGATAGGGGTATT | aveA1 promoter DNA |
| LXR123B | AAGAATGAAAGGAGCGCG | |
| LXR69A | CACTCGTGAATTGTGGAC | aveR promoter DNA |
| LXR69B | TGACTTTGATGATTAACT | |
| LXR124A | CGTGTAAGGAGCGGATAA | dnaK1 promoter DNA |
| LXR124B | TTCAATTCCTCCAGCAAT | |
| LXR125A | TTCCCGGCACCCCCGAA | clpB1 promoter DNA |
| LXR125B | TCTGACATTGGAAGCGTA | |
| LXR126A | ACGAGCGGCGGTGGCGGT | clpB2 promoter DNA |
| LXR126B | CGACCGGATCCGACGCG | |
| LXR127A | ATTTCCGATGATCGCTGA | lonA promoter DNA |
| LXR127B | AAGCCATGATCTCCCCTT | |
| LXR128A | TGTGACTTTGGAGCTTTA | catR_katA1 promoter DNA |
| LXR128B | TCCTGCGTCATTCCTGCAA | |
| LXR129A | GGAACCAACTTCAACAAA | katA3 promoter DNA |
| LXR129B | TTCGACATCGTGCACCTT | |
| LXR130A | AAGATGACCTTCCACTGA | oxyR_ahpC promoter DNA |
| LXR130B | TGACAAGTTCCCCGAGTT | |
| LXR131A | TTCGTCACCCTGGGGGTG | dnaK2 promoter DNA |
| LXR131B | ACTCTCCGGATCACGTTC | |
| LXR132A | CCCAGAACCGAACCGAAG | katA2 promoter DNA |
| LXR132B | AAAGAAGAGGGTCGTTAG |
Underlining indicates sequences for the restriction enzymes shown in parentheses.
Construction of S. avermitilis mutant strains.
To construct the hspR deletion mutant, a 594-bp 5′ flanking region (positions −527 to +67 relative to the hspR translational start codon [TSC]) and a 454-bp 3′ flanking region (positions +385 to +838) were generated by PCR from S. avermitilis WT genome with primers WQ11/WQ12 and WQ13/WQ14. The two fragments were digested with BamHI/XbaI and XbaI/HindIII, respectively, and then ligated into BamHI/HindIII-digested pKC1139 (46) to generate pKCΔhspR. Temperature-sensitive plasmid pCIMt005, containing the idgS gene that encodes indigoidine synthetase to make colonies blue, was used for hspR deletion by simple blue-white screening (47). A 1,048-bp DNA fragment containing 5′- and 3′-flanking regions of hspR was amplified from pKCΔhspR with primers LXR101A and LXR101B and ligated into NcoI-digested pCIMt005 to generate hspR deletion vector pΔhspR, which was transformed into WT protoplasts. The hspR deletion mutant was screened as described previously (48) and confirmed by colony PCR and DNA sequencing. Use of primers WQ15 and WQ16 (flanking the exchange regions) (see Fig. S1 in the supplemental material) resulted in the appearance of a 1,337-bp band, whereas a 1,655-bp band was observed in the WT. When primers WQ17 and WQ18 (located within the deletion region of hspR) were used, only the WT produced a 385-bp band. We thus generated the hspR deletion mutant (ΔhspR strain), in which a 275-bp fragment within hspR open reading frame (ORF) (positions +112 to +386 relative to the TSC) was deleted (see Fig. S1).
For complementation of ΔhspR, a 465-bp fragment containing the hspR ORF and a 446-bp fragment containing the dnaK1 promoter were amplified with primers LXR134A/LXR134B and LXR133A/LXR133B, respectively. The two PCR fragment were digested with BamHI/HindIII and EcoRI/BamHI, respectively, and ligated simultaneously into pSET152 (46) to generate hspR-complemented vector pSET152-hspR, which was then introduced into the ΔhspR strain to generate complemented strain ChspR.
For overexpression of hspR, a 586-bp fragment carrying the hspR ORF was amplified with primers WQ25 and WQ26 and ligated simultaneously with a 188-bp ermE*p (Streptomyces strong constitutive promoter) fragment from pJL117 (49) into pKC1139 to generate hspR-overexpressing vector pKC-erm-hspR, which was introduced into S. avermitilis WT and industrial strain A229 to generate hspR overexpression strains OhspR and OhspR/A229, respectively.
To express 3×FLAG-tagged HspR in S. avermitilis, the hspR ORF and dnaK1 promoter were amplified with primers LXR135A/LXR135B and LXR136A/LXR136B from WT genomic DNA, and the 3×FLAG fragment was amplified with primers LXR60A and LXR60B from plasmid pIJ10500 (50). The resulting 453-bp hspR, 446-bp dnaK1 promoter, and 100-bp FLAG fragments were cloned into pSET152 to generate pSET152-hspR-3FLAG, which was then transformed into WT, ΔhspR, and ΔphoP strains to generate recombinant strains WT/hspR-3FLAG, ΔhspR/hspR-3FLAG, and ΔphoP/hspR-3FLAG strains, respectively, for expression of C-terminally 3×FLAG-tagged HspR.
Scanning electron microscopy.
Mycelia and spores of S. avermitilis WT and ΔhspR grown on YMS plates for 2 or 4 days were observed by SEM. Samples were prepared and examined as described in our previous study (51).
Production and analysis of avermectins.
Avermectin yield from S. avermitilis fermentation culture was analyzed by HPLC as described previously (52).
Heterologous expression and purification of His6-HspR.
For expression of S. avermitilis HspR in E. coli, the 579-bp hspR ORF was generated by PCR using primers WQ27 and WQ28. The PCR fragment was digested with EcoRI/XhoI and ligated into pET-28a(+), generating pET28-hspR for overexpression of N-terminal His6-tagged HspR recombinant protein. Expression vector pET28-hspR was confirmed by sequencing and transformed into E. coli BL21(DE3). His6-HspR expression was induced by 3-h treatment with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C. Cell lysates were prepared by sonication on ice in lysis buffer (53) and centrifugation. Soluble His6-HspR was purified on an Ni-nitrilotriacetic acid (NTA) column (Bio-Works; Sweden) and eluted by lysis buffer with 250 mM imidazole. The purified protein was dialyzed in binding buffer for EMSAs (54) to eliminate imidazole and stored at −80°C until use.
Electrophoretic mobility shift assays.
Promoter probes of tested genes were amplified by PCR with corresponding primers listed in Table 2, and labeled at the 3′ end with nonradioactive digoxigenin-1-ddUTP. EMSAs were performed as described previously (52). Specificity of the HspR-probe interaction was tested by adding ∼300-fold excess of unlabeled nonspecific probe (hrdBp) or respective specific probe to the binding reaction system.
Reverse transcription and real-time quantitative PCR analysis.
Total RNAs were extracted from S. avermitilis cultures grown in FM-I or FM-II or on YMS plates for various durations. RNA isolation, removal of genomic DNA, and subsequent RT-qPCR analysis (using primers listed in Table 2) were performed as described previously (52). Relative expression levels of tested genes were normalized with 16S rRNA (housekeeping gene) as the internal control. Experiments were performed in triplicates.
Western blotting.
Total protein was prepared at various time points from ΔhspR/hspR-3FLAG mycelia grown in FM-I. Western blotting was performed as described in our previous study (54), using 1:3,300-diluted mouse anti-FLAG monoclonal antibody (MAb) (Sigma; USA).
ChIP-qPCR.
To prepare samples for ChIP-qPCR, S. avermitilis WT, WT/hspR-3FLAG, ΔhspR/hspR-3FLAG, and ΔphoP/hspR-3FLAG strains were cultured in FM-II, and mycelia were harvested at various time points and treated as described previously (29). Immunoprecipitation reactions and ChIP-qPCR data analysis were performed as described previously (32, 48). Relative protein enrichment of each site was normalized based on input chromosomal DNA, and binding level was shown as a percentage of input DNA. Experiments were performed in triplicates.
DNase I footprinting.
To determine HspR binding sites, 5′ 6-carboxyfluorescein (FAM)-labeled DNA fragments corresponding to aveA1 and dnaK1 promoter regions were PCR-synthesized using primers listed in Table 2 and then purified. DNase I footprinting assays were performed as described previously (51, 55), and data were analyzed using GeneMarker software program v2.2.0.
GST pulldown.
pET28-hspR (for His6-HspR expression) and pGEX-PhoP (for GST-PhoP expression) (38) were cotransformed into E. coli BL21(DE3). Bacteria containing pET28-hspR and pGEX-4T-1 (for GST tag expression) were used as negative controls. Following 0.4 mM IPTG induction for 3 h at 37°C, cells containing both His6- and GST-tagged proteins were sonicated in lysis buffer (53) on ice and centrifuged. One milliliter cell lysate and 50 μl 50% glutathione-Sepharose beads (equilibrated with lysis buffer) were incubated overnight at 4°C. Beads were washed four times with phosphate-buffered saline (PBS) (56), and the resulting pellet was boiled with 50 μl protein loading buffer. Eluted bound proteins were separated by electrophoresis on SDS-PAGE gels and identified by Western blotting using anti-His6 or anti-GST antibody (Tiangen; China).
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (grant 31872629) and the Project for Extramural Scientists of the State Key Laboratory of Agrobiotechnology (grant 2020SKLAB6-4).
We thank S. Anderson for English editing of the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Ying Wen, Email: wen@cau.edu.cn.
Maia Kivisaar, University of Tartu.
REFERENCES
- 1.Flardh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol 7:36–49. 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 2.Demain AL. 2002. Prescription for an ailing pharmaceutical industry. Nat Biotechnol 20:331. 10.1038/nbt0402-331. [DOI] [PubMed] [Google Scholar]
- 3.Challis GL, Hopwood DA. 2003. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc Natl Acad Sci U S A 100(Suppl 2):14555–14561. 10.1073/pnas.1934677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibb MJ. 2005. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 8:208–215. 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 5.Liu G, Chater KF, Chandra G, Niu G, Tan H. 2013. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev 77:112–143. 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon V, Nodwell JR. 2014. Activating secondary metabolism with stress and chemicals. J Ind Microbiol Biotechnol 41:415–424. 10.1007/s10295-013-1387-y. [DOI] [PubMed] [Google Scholar]
- 7.Georgopoulos C, Welch WJ. 1993. Role of the major heat shock proteins as molecular chaperones. Annu Rev Cell Biol 9:601–634. 10.1146/annurev.cb.09.110193.003125. [DOI] [PubMed] [Google Scholar]
- 8.Gottesman S, Wickner S, Maurizi MR. 1997. Protein quality control: triage by chaperones and proteases. Genes Dev 11:815–823. 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 9.Hendrick JP, Hartl FU. 1993. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem 62:349–384. 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- 10.Schumann W. 2016. Regulation of bacterial heat shock stimulons. Cell Stress Chaperones 21:959–968. 10.1007/s12192-016-0727-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grandvalet C, Rapoport G, Mazodier P. 1998. hrcA, encoding the repressor of the groEL genes in Streptomyces albus G, is associated with a second dnaJ gene. J Bacteriol 180:5129–5134. 10.1128/JB.180.19.5129-5134.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Servant P, Grandvalet C, Mazodier P. 2000. The RheA repressor is the thermosensor of the HSP18 heat shock response in Streptomyces albus. Proc Natl Acad Sci U S A 97:3538–3543. 10.1073/pnas.070426197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bucca G, Hindle Z, Smith CP. 1997. Regulation of the dnaK operon of Streptomyces coelicolor A3(2) is governed by HspR, an autoregulatory repressor protein. J Bacteriol 179:5999–6004. 10.1128/jb.179.19.5999-6004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bucca G, Brassington AM, Hotchkiss G, Mersinias V, Smith CP. 2003. Negative feedback regulation of dnaK, clpB and lon expression by the DnaK chaperone machine in Streptomyces coelicolor, identified by transcriptome and in vivo DnaK-depletion analysis. Mol Microbiol 50:153–166. 10.1046/j.1365-2958.2003.03696.x. [DOI] [PubMed] [Google Scholar]
- 15.Grandvalet C, Servant P, Mazodier P. 1997. Disruption of hspR, the repressor gene of the dnaK operon in Streptomyces albus G. Mol Microbiol 23:77–84. 10.1046/j.1365-2958.1997.1811563.x. [DOI] [PubMed] [Google Scholar]
- 16.Grandvalet C, de Crecy-Lagard V, Mazodier P. 1999. The ClpB ATPase of Streptomyces albus G belongs to the HspR heat shock regulon. Mol Microbiol 31:521–532. 10.1046/j.1365-2958.1999.01193.x. [DOI] [PubMed] [Google Scholar]
- 17.Sobczyk A, Bellier A, Viala J, Mazodier P. 2002. The lon gene, encoding an ATP-dependent protease, is a novel member of the HAIR/HspR stress-response regulon in actinomycetes. Microbiology (Reading) 148:1931–1937. 10.1099/00221287-148-6-1931. [DOI] [PubMed] [Google Scholar]
- 18.Bucca G, Brassington AM, Schonfeld HJ, Smith CP. 2000. The HspR regulon of Streptomyces coelicolor: a role for the DnaK chaperone as a transcriptional co-repressor. Mol Microbiol 38:1093–1103. 10.1046/j.1365-2958.2000.02194.x. [DOI] [PubMed] [Google Scholar]
- 19.Stewart GR, Wernisch L, Stabler R, Mangan JA, Hinds J, Laing KG, Young DB, Butcher PD. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology (Reading) 148:3129–3138. 10.1099/00221287-148-10-3129. [DOI] [PubMed] [Google Scholar]
- 20.Singh R, Anil Kumar V, Das AK, Bansal R, Sarkar D. 2014. A transcriptional co-repressor regulatory circuit controlling the heat-shock response of Mycobacterium tuberculosis. Mol Microbiol 94:450–465. 10.1111/mmi.12778. [DOI] [PubMed] [Google Scholar]
- 21.Martin JF. 2004. Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: an unfinished story. J Bacteriol 186:5197–5201. 10.1128/JB.186.16.5197-5201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin JF, Santos-Beneit F, Rodriguez-Garcia A, Sola-Landa A, Smith MCM, Ellingsen TE, Nieselt K, Burroughs NJ, Wellington EMH. 2012. Transcriptomic studies of phosphate control of primary and secondary metabolism in Streptomyces coelicolor. Appl Microbiol Biotechnol 95:61–75. 10.1007/s00253-012-4129-6. [DOI] [PubMed] [Google Scholar]
- 23.Martin JF, Rodriguez-Garcia A, Liras P. 2017. The master regulator PhoP coordinates phosphate and nitrogen metabolism, respiration, cell differentiation and antibiotic biosynthesis: comparison in Streptomyces coelicolor and Streptomyces avermitilis. J Antibiot 70:534–541. 10.1038/ja.2017.19. [DOI] [PubMed] [Google Scholar]
- 24.Allenby NEE, Laing E, Bucca G, Kierzek AM, Smith CP. 2012. Diverse control of metabolism and other cellular processes in Streptomyces coelicolor by the PhoP transcription factor: genome-wide identification of in vivo targets. Nucleic Acids Res 40:9543–9556. 10.1093/nar/gks766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sola-Landa A, Rodriguez-Garcia A, Amin R, Wohlleben W, Martin JF. 2013. Competition between the GlnR and PhoP regulators for the glnA and amtB promoters in Streptomyces coelicolor. Nucleic Acids Res 41:1767–1782. 10.1093/nar/gks1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santos-Beneit F, Rodriguez-Garcia A, Sola-Landa A, Martin JF. 2009. Cross-talk between two global regulators in Streptomyces: PhoP and AfsR interact in the control of afsS, pstS and phoRP transcription. Mol Microbiol 72:53–68. 10.1111/j.1365-2958.2009.06624.x. [DOI] [PubMed] [Google Scholar]
- 27.Burg RW, Miller BM, Baker EE, Birnbaum J, Currie SA, Hartman R, Kong YL, Monaghan RL, Olson G, Putter I, Tunac JB, Wallick H, Stapley EO, Oiwa R, Omura S. 1979. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob Agents Chemother 15:361–367. 10.1128/AAC.15.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egerton JR, Ostlind DA, Blair LS, Eary CH, Suhayda D, Cifelli S, Riek RF, Campbell WC. 1979. Avermectins, new family of potent anthelmintic agents: efficacy of the B1a component. Antimicrob Agents Chemother 15:372–378. 10.1128/AAC.15.3.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo J, Zhao J, Li L, Chen Z, Wen Y, Li J. 2010. The pathway-specific regulator AveR from Streptomyces avermitilis positively regulates avermectin production while it negatively affects oligomycin biosynthesis. Mol Genet Genomics 283:123–133. 10.1007/s00438-009-0502-2. [DOI] [PubMed] [Google Scholar]
- 30.Kitani S, Ikeda H, Sakamoto T, Noguchi S, Nihira T. 2009. Characterization of a regulatory gene, aveR, for the biosynthesis of avermectin in Streptomyces avermitilis. Appl Microbiol Biotechnol 82:1089–1096. 10.1007/s00253-008-1850-2. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda H, Nonomiya T, Usami M, Ohta T, Omura S. 1999. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc Natl Acad Sci U S A 96:9509–9514. 10.1073/pnas.96.17.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu X, Liu X, Chen Z, Li J, van Wezel GP, Chen W, Wen Y. 2020. The ROK-family regulator Rok7B7 directly controls carbon catabolite repression, antibiotic biosynthesis, and morphological development in Streptomyces avermitilis. Environ Microbiol 22:5090–5108. 10.1111/1462-2920.15094. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka A, Takano Y, Ohnishi Y, Horinouchi S. 2007. AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J Mol Biol 369:322–333. 10.1016/j.jmb.2007.02.096. [DOI] [PubMed] [Google Scholar]
- 34.Yin S, Wang W, Wang X, Zhu Y, Jia X, Li S, Yuan F, Zhang Y, Yang K. 2015. Identification of a cluster-situated activator of oxytetracycline biosynthesis and manipulation of its expression for improved oxytetracycline production in Streptomyces rimosus. Microb Cell Fact 14:46. 10.1186/s12934-015-0231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun D, Wang Q, Chen Z, Li J, Wen Y. 2017. An alternative sigma factor, σ8, controls avermectin production and multiple stress responses in Streptomyces avermitilis. Front Microbiol 8:736. 10.3389/fmicb.2017.00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Sun M, Cheng Y, Yang R, Wen Y, Chen Z, Li J. 2016. OxyR is a key regulator in response to oxidative stress in Streptomyces avermitilis. Microbiology (Reading) 162:707–716. 10.1099/mic.0.000251. [DOI] [PubMed] [Google Scholar]
- 37.Sola-Landa A, Rodriguez-Garcia A, Franco-Dominguez E, Martin JF. 2005. Binding of PhoP to promoters of phosphate-regulated genes in Streptomyces coelicolor: identification of PHO boxes. Mol Microbiol 56:1373–1385. 10.1111/j.1365-2958.2005.04631.x. [DOI] [PubMed] [Google Scholar]
- 38.Yang R, Liu X, Wen Y, Song Y, Chen Z, Li J. 2015. The PhoP transcription factor negatively regulates avermectin biosynthesis in Streptomyces avermitilis. Appl Microbiol Biotechnol 99:10547–10557. 10.1007/s00253-015-6921-6. [DOI] [PubMed] [Google Scholar]
- 39.Hiard S, Maree R, Colson S, Hoskisson PA, Titgemeyer F, van Wezel GP, Joris B, Wehenkel L, Rigali S. 2007. PREDetector: a new tool to identify regulatory elements in bacterial genomes. Biochem Biophys Res Commun 357:861–864. 10.1016/j.bbrc.2007.03.180. [DOI] [PubMed] [Google Scholar]
- 40.Bush MJ, Chandra G, Bibb MJ, Findlay KC, Buttner MJ. 2016. Genome-wide chromatin immunoprecipitation sequencing analysis shows that WhiB is a transcription factor that cocontrols its regulon with WhiA to initiate developmental cell division in Streptomyces. mBio 7:e00523-16. 10.1128/mBio.00523-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molle V, Palframan WJ, Findlay KC, Buttner MJ. 2000. WhiD and WhiB, homologous proteins required for different stages of sporulation in Streptomyces coelicolor A3(2). J Bacteriol 182:1286–1295. 10.1128/JB.182.5.1286-1295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. 2011. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev 25:89–99. 10.1101/gad.600211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu W, Zhang Q, Guo J, Chen Z, Li J, Wen Y. 2015. Increasing avermectin production in Streptomyces avermitilis by manipulating the expression of a novel TetR-family regulator and its target gene product. Appl Environ Microbiol 81:5157–5173. 10.1128/AEM.00868-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikeda H, Kotaki H, Tanaka H, Omura S. 1988. Involvement of glucose catabolism in avermectin production by Streptomyces avermitilis. Antimicrob Agents Chemother 32:282–284. 10.1128/AAC.32.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang L, Liu Y, Wang P, Wen Y, Song Y, Chen Z, Li J. 2011. Inactivation of the extracytoplasmic function sigma factor Sig6 stimulates avermectin production in Streptomyces avermitilis. Biotechnol Lett 33:1955–1961. 10.1007/s10529-011-0673-x. [DOI] [PubMed] [Google Scholar]
- 46.Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49. 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 47.Li P, Li J, Guo Z, Tang W, Han J, Meng X, Hao T, Zhu Y, Zhang L, Chen Y. 2015. An efficient blue-white screening based gene inactivation system for Streptomyces. Appl Microbiol Biotechnol 99:1923–1933. 10.1007/s00253-014-6369-0. [DOI] [PubMed] [Google Scholar]
- 48.Yan H, Lu X, Sun D, Zhuang S, Chen Q, Chen Z, Li J, Wen Y. 2020. BldD, a master developmental repressor, activates antibiotic production in two Streptomyces species. Mol Microbiol 113:123–142. 10.1111/mmi.14405. [DOI] [PubMed] [Google Scholar]
- 49.Li L, Guo J, Wen Y, Chen Z, Song Y, Li J. 2010. Overexpression of ribosome recycling factor causes increased production of avermectin in Streptomyces avermitilis strains. J Ind Microbiol Biotechnol 37:673–679. 10.1007/s10295-010-0710-0. [DOI] [PubMed] [Google Scholar]
- 50.Pullan ST, Chandra G, Bibb MJ, Merrick M. 2011. Genome-wide analysis of the role of GlnR in Streptomyces venezuelae provides new insights into global nitrogen regulation in actinomycetes. BMC Genomics 12:175. 10.1186/1471-2164-12-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun D, Zhu J, Chen Z, Li J, Wen Y. 2016. SAV742, a novel AraC-family regulator from Streptomyces avermitilis, controls avermectin biosynthesis, cell growth and development. Sci Rep 6:36915. 10.1038/srep36915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo S, Sun D, Zhu J, Chen Z, Wen Y, Li J. 2014. An extracytoplasmic function sigma factor, σ25, differentially regulates avermectin and oligomycin biosynthesis in Streptomyces avermitilis. Appl Microbiol Biotechnol 98:7097–7112. 10.1007/s00253-014-5759-7. [DOI] [PubMed] [Google Scholar]
- 53.Guo J, Zhang X, Lu X, Liu W, Chen Z, Li J, Deng L, Wen Y. 2018. SAV4189, a MarR-family regulator in Streptomyces avermitilis, activates avermectin biosynthesis. Front Microbiol 9:1358. 10.3389/fmicb.2018.01358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu J, Sun D, Liu W, Chen Z, Li J, Wen Y. 2016. AvaR2, a pseudo gamma-butyrolactone receptor homologue from Streptomyces avermitilis, is a pleiotropic repressor of avermectin and avenolide biosynthesis and cell growth. Mol Microbiol 102:562–578. 10.1111/mmi.13479. [DOI] [PubMed] [Google Scholar]
- 55.Zianni M, Tessanne K, Merighi M, Laguna R, Tabita FR. 2006. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J Biomol Tech 17:103–113. [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu J, Chen Z, Li J, Wen Y. 2017. AvaR1, a butenolide-type autoregulator receptor in Streptomyces avermitilis, directly represses avenolide and avermectin biosynthesis and multiple physiological responses. Front Microbiol 8:2577. 10.3389/fmicb.2017.02577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S7, Table S1. Download AEM.00473-21-s0001.pdf, PDF file, 0.7 MB (707.1KB, pdf)