ABSTRACT
Postweaning diarrhea in pigs is mainly caused by pathogenic Escherichia coli and is a major source of revenue loss to the livestock industry. Bacteriophages dominate the gut virome and have the potential to regulate bacterial communities and thus influence the intestinal physiology. To determine the biological characterization of intestinal coliphages, we isolated and identified the fecal coliphages of healthy preweaned and postweaned piglets from the Nanjing and Chuzhou pig farms. First, ahead of coliphage isolation, 87 E. coli strains were isolated from healthy or diarrheal fecal samples from three pig farms, of which 8 were pathogenic strains, including enterotoxigenic E. coli (ETEC) and enteropathogenic E. coli (EPEC). Of the E. coli strains, 87.3% possessed drug resistance to three antibiotics. Using these 87 E. coli strains as indicator hosts, we isolated 45 coliphages and found a higher abundance in the postweaning stage than in the preweaning stage (24 versus 17 in the Nanjing and 13 versus 4 in the Chuzhou farm). Furthermore, each farm had a single most-prevalent coliphage strain. Pathogenic E. coli-specific bacteriophages were commonly detected (9/10 samples in the Nanjing farm and 7/10 in the Chuzhou farm) in guts of sampled piglets, and most had significant bacteriostatic effects (P < 0.05) on pathogenic E. coli strains. Three polyvalent bacteriophages (N24, N30, and C5) were identified. The N30 and C5 strains showed a genetic identity of 89.67%, with mild differences in infection characteristics. Our findings suggest that pathogenic E. coli-specific bacteriophages as well as polyvalent bacteriophages are commonly present in piglet guts and that weaning is an important event that affects coliphage numbers.
IMPORTANCE Previous studies based on metagenomic sequencing reported that gut bacteriophages profoundly influence gut physiology but did not provide information regarding the host range and biological significance. Here, we screened coliphages from the guts of preweaned and postweaned piglets against indicator hosts, which allowed us to identify the pathogenic E. coli-specific bacteriophages and polyvalent bacteriophages in pig farms and quantify their abundance. Our approach complements sequencing methods and provides new insights into the biological characterizations of bacteriophage in the gut along with the ecological effects of intestinal bacteriophages.
KEYWORDS: pre- and postweaned piglets, coliphage numbers, pathogenic E. coli-specific bacteriophages, polyvalent bacteriophages
INTRODUCTION
The mammalian gut is a complex ecosystem that supports a diverse microbial community including bacteria, archaea, viruses, fungi, and other eukaryotes (1–3). While the physiological relevance of intestinal bacteria is well established (4–6), little is known regarding the function of archaea, eukaryotes, and viruses. The gut viral community (virome) is dominated by bacteriophages. Recent sequencing-based approaches show that gut bacteriophages profoundly influence gut physiology (7, 8) and host health (9, 10). The healthy gut phageome that includes core and common bacteriophage communities in humans is conserved among healthy individuals worldwide and plays an important role in maintaining gut microbiome structure and function (11). In contrast, some phageome members such as lytic phages have undergone rapid evolution and alterations over time (12). Significant changes in bacteriophage abundance and diversity are associated with Crohn’s disease, ulcerative colitis (7, 8), and Clostridium difficile infection (CDI) (13).
The relationship between gut bacteriophages and bacteria is dynamic. During early intestinal development, it consists mostly of predator and prey interactions characterized by rapid fluctuations in bacterial and phage populations in terms of both abundance and diversity (14–16). Since the resident bacterial community is unstable in the early stages of intestinal development, bacteriophage infection at this stage rapidly decreases the prey population, which allows competitive bacteria to colonize the gut, and then leads to the increase of new bacteriophages with a new infection cycle (17). Furthermore, bacteriophages provide defense against bacterial pathogens through lytic infection (18) and can adhere to intestinal mucosal surfaces, where they are more likely to encounter and kill invading bacteria (19, 20). Therefore, bacteriophages play a pivotal role in establishing the early gut microbiota.
Porcine diarrhea coincides with weaning and causes considerable economic losses to the livestock industry (21). The sudden dietary transition and environmental changes during weaning lead to intestinal dysbiosis in piglets, which not only allows potential endogenous pathogens to proliferate but also increases the risk of exogenous infections that eventually lead to postweaning diarrhea (22). Coliform bacilli are one of the predominant diarrheal pathogens in mammals (23). For instance, the enterotoxigenic Escherichia coli (ETEC) is the major causative agent of colibacillosis diarrhea in piglets (22). Considering the importance of the bacteriophage’s lytic mode of infection in shaping the gut microbiota during early development, it is possible that coliphages strongly influence the pathogenicity of coliform bacilli in the piglet intestine and affect the incidence of postweaning diarrhea. However, the effect of weaning on the intestinal coliphages is currently unknown.
Previous studies on gut bacteriophages are mainly based on metagenomic sequencing, which provides information on phage communities but leaves important gaps in understanding the biological properties of phages such as host ranges, bacteriostatic effects, and efficiency of plating. In the present study, we investigated coliphages from preweaned and postweaned piglet gut by quantifying their abundance, isolating them, and identifying their biological characteristics. Our approach complements sequencing methods and provides new guidelines for shaping the gut microbiota and manipulating the gut health of piglets.
RESULTS
Antimicrobial resistance patterns and phylogenetic group categorization of isolated strains.
A total of 87 E. coli isolates, one Proteus mirabilis isolate, one Citrobacter freundii isolate, and one Klebsiella pneumoniae isolate were recovered from the 80 examined samples collected from 3 different pig farms ahead of coliphage isolation. The sequenced 16S rRNA GenBank accession numbers are listed below under “Data availability.” We detected antibiotic resistance against six common antibiotics and found that 86.7% of the 90 isolates (87.3% in 87 E. coli strains) possessed drug resistance against at least three antibiotics (see Table S1 in the supplemental material). Most strains were resistant to ampicillin (AMP) (84/90), tetracycline (TE) (84/90), and/or chloramphenicol (CHL) (65/90). E. coli strains are categorized into eight phylogenetic groups: A, B1, B2, C, D, E, F, and clade I. Strains responsible for extraintestinal infection are more likely to be members of phylogroup B2, D, or F, which is a sister group of B2 (24). Our data showed that most isolates belonged to groups A (17/87), B1 (34/87), and C (26/87), whereas only three isolates belonged to groups D (2/87) and F (1/87). We did not observe any isolates belonging to group B2 (Table S1). These data indicated that antibiotic-resistant strains were prevalent and extraintestinal E. coli strains were rare in piglet gut.
Correlation between pathogenic E. coli and piglet diarrhea.
We evaluated the toxin genes of diarrhoeagenic E. coli in the 87 isolates and found a total of 8 pathogenic strains of which 6 were ETEC and 2 were enteropathogenic E. coli (EPEC) (Table 1). Notably, only the Maanshan farm had pigs with diarrhea and the ETEC strains were detected in both healthy and diarrhea samples from this farm. At the Nanjing and Nantong farms, no pigs with diarrhea were present at the time of sampling despite the detection of EPEC strains. Therefore, we suspected that ETEC was the predominant coliform that caused diarrhea in pigs, which was in line with previous reports (25, 26).
TABLE 1.
Toxin genes of 8 pathogenic E. coli strains
| Strain | Toxin gene(s) | Pathogroup |
|---|---|---|
| E. coli 101 | estla, hly | ETEC |
| E. coli 102 | estlb, elt | ETEC |
| E. coli 103 | estla, elt | ETEC |
| E. coli 104 | estlb | ETEC |
| E. coli 105 | estlb, elt | ETEC |
| E. coli 106 | estlb, elt | ETEC |
| E. coli 143 | eaeA, escV, hly | EPEC |
| E. coli 162 | eaeA, escV, ent | EPEC |
Changes in coliphage numbers between preweaned and postweaned piglet gut.
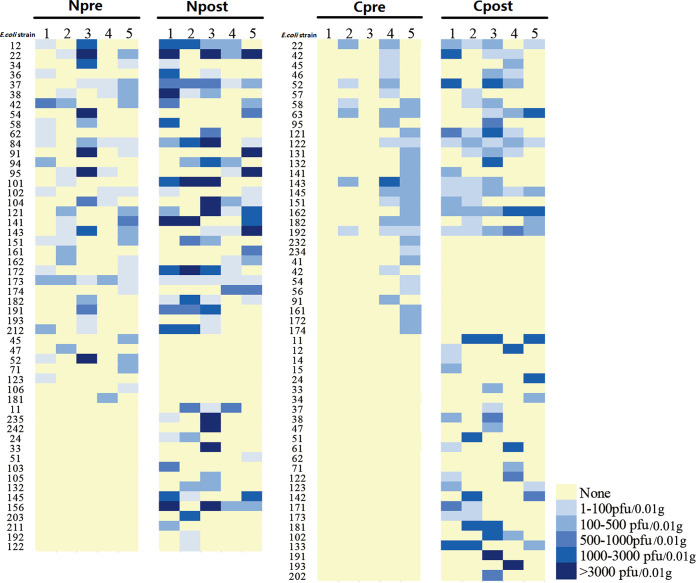
Using the 87 E. coli strains isolated above as indicator hosts, we isolated coliphages from another two farms. The number of plaques in each E. coli strain plate is shown in Fig. 1. At both farms, a significantly higher abundance of coliphages (31,868 ± 9,538 versus 6,163 ± 4,326 PFU/0.01 g, P = 0.04, in the Nanjing farm and 10,656 ± 2,251 versus 1,670 ± 879 PFU/0.01 g, P = 0.006, in the Chuzhou farm) and more plaque-positive E. coli strains (20 ± 2 versus 14 ± 2, P = 0.057, in the Nanjing farm and 18 ± 2 versus 9 ± 4, P = 0.067, in the Chuzhou farm) were found in postweaned fecal samples than in preweaned samples. Interestingly, two preweaned samples from the Chuzhou farm showed a complete absence of coliphages. Subsequently, we selected a clear plaque from each E. coli strain plate to isolate the coliphages and found that several coliphages from different E. coli strain plates were identified as the same strain. In total, 30 and 15 different coliphages were isolated from the Nanjing and Chuzhou farms, respectively. Consistent with the results described above, in both farms more coliphage strains were isolated from postweaned than preweaned piglet gut (24 versus 17 in the Nanjing farm and 13 versus 4 in the Chuzhou farm). Furthermore, 17 and 10 coliphage isolates from the Nanjing and Chuzhou farms, respectively, were present in multiple samples, whereas 13 and 5 coliphage isolates appeared only in a single sample (Tables 2 and 3). Among the widespread coliphage isolates, a few were detected only in either the preweaned or postweaned gut and some were common in both pre- and postweaning stages (Tables 2 and 3). The most frequently detected coliphages at the Nanjing and Chuzhou farms were N7 (9/10 samples) and C1 (7/10 samples), respectively. Transmission electron microscopy (TEM) examination indicated that the N7 and C1 strains were both members of the order of Caudovirales and belonged to the families of Podoviridae and Siphoviridae, respectively (Fig. 2). The sequences of the N7 and C1 strains were deposited in GenBank, which indicated that N7 was a P22-like temperate phage and C1 was a virulent phage. Taken together, our results suggest that weaning affects coliphage numbers in piglet gut.
FIG 1.
Coliphage abundance in plaque-positive E. coli strains in preweaned (pre) and postweaned (post) fecal samples. N, Nanjing; C, Chuzhou.
TABLE 2.
Coliphages isolated from the Nanjing pig farm
| Sample | Coliphage(s) detected ina: |
|
|---|---|---|
| Multiple samples | Single samples | |
| Preweaned | ||
| Npre-1 | N1,N7, N8, N10, N15 | N22 |
| Npre-2 | N2,N7, N8, N12 | N25, N26, N28 |
| Npre-3 | N1, N3, N5, N6, N7, N12 | N27 |
| Npre-4 | N7, N8, N16 | |
| Npre-5 | N2, N3, N4, N6, N7 | |
| Postweaned | ||
| Npost-1 | N1, N3, N4, N5, N6, N7, N8, N9, N13, N30 | |
| Npost-2 | N3, N4, N8, N9, N10, N11, N13, N15, N30 | N19, N24 |
| Npost-3 | N3, N4, N5, N7, N8, N10, N11, N14, N30 | N17, N20, N21, N29 |
| Npost-4 | N6, N7, N12, N15 | N18 |
| Npost-5 | N4, N7, N12, N14, N16 | N23 |
Coliphages present in most samples are indicated by boldface. Coliphages specific for pathogenic E. coli are indicated by underlining.
TABLE 3.
Coliphages isolated from the Chuzhou pig farm
| Sample | Coliphage(s) detected ina: |
|
|---|---|---|
| Multiple samples | Single samples | |
| Preweaned | ||
| Cpre-1 | No phage isolated | |
| Cpre-2 | C6 | |
| Cpre-3 | No phage isolated | |
| Cpre-4 | C1, C6 | C12 |
| Cpre-5 | C1, C4 | |
| Postweaned | ||
| Cpost-1 | C1,C2, C5, C7, C8, C10 | |
| Cpost-2 | C1,C2, C4 | C15 |
| Cpost-3 | C1,C2, C3, C8, C10 | C14 |
| Cpost-4 | C1,C2, C3, C9, C10 | |
| Cpost-5 | C1,C2, C4, C5, C7, C9 | C11, C13 |
Coliphages present in most samples are indicated by boldface. Coliphages specific for pathogenic E. coli are indicated by underlining.
FIG 2.
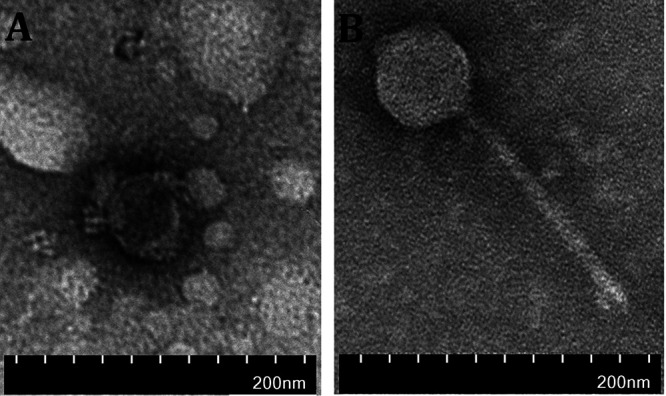
Transmission electron microscope images of coliphages N7 and C1 detected in farm samples. (A) Coliphage N7 was found in 9/10 samples from the Nanjing farm. (B) Coliphage C1 was found in 7/10 samples from the Chuzhou pig farm.
Identification of pathogenic E. coli-specific bacteriophages isolated from piglet gut.
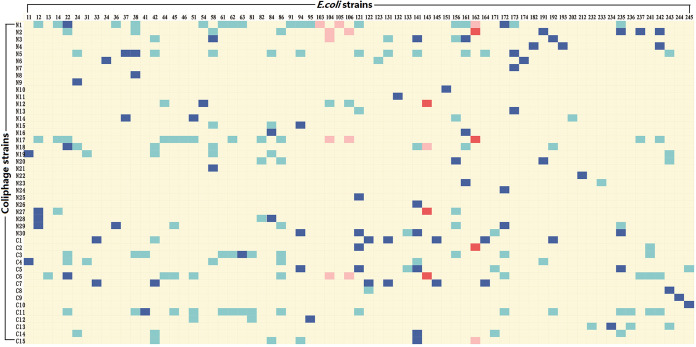
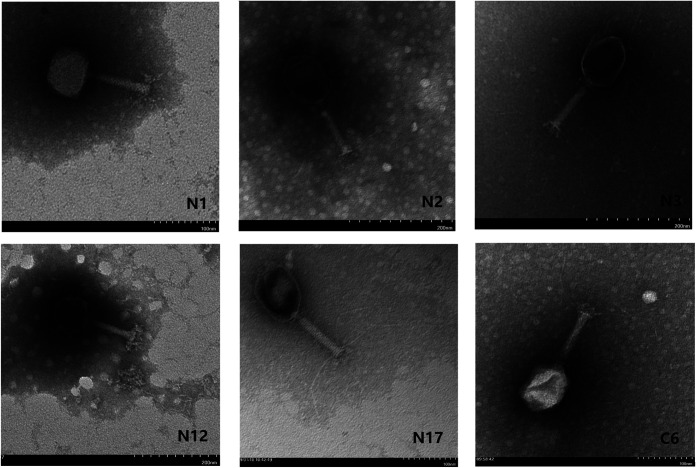
For most coliphage isolates, a broader host range was demonstrated by the spot assay than the efficiency of plating (EOP) assay (Fig. 3). To exclude the possibility that the lysis in the spot assay was caused by colicin from host bacteria, the culture supernatant of each host bacterium was tested on lysed bacteria, and none showed lytic activity. Using spot assay, 7 coliphages (N1, N5, N17, N18, C3, C6, and C11) were found to lyse more than 10 E. coli strains (Fig. 3). Moreover, we observed 10 coliphages capable of lysing pathogenic E. coli with some possessing the capability to lyse multiple pathogenic E. coli strains. The EOP assay indicated that only 6 coliphages (N2, N12, N17, N27, C2, and C6) infected pathogenic E. coli strains (Fig. 3). Five of the 6 coliphages had significant bacteriostatic effects (P < 0.05) on pathogenic E. coli strains at a multiplicity of infection (MOI) of 0.1 (Fig. 4) except coliphage C6, as the EOP of coliphage C6 on EPEC 143 was only 0.02. Although none of the isolated coliphages were capable of infecting ETEC, 6 coliphages (N1, N2, N3, N12, N17, and C6) showed the ability to lyse certain ETEC strains (Fig. 5). The phenomenon of lysis without infection can be attributed to “lysis from without” or “abortive infection” as described previously (27, 28). Unsurprisingly, these 6 coliphages needed high MOIs to induce a bacteriostatic effect on ETEC, and most coliphages showed better lytic activity at an MOI of 100 compared to 10 (Fig. 5). In contrast, the coliphages N17 and N3 poorly lysed ETEC strains 106k88 and 104, respectively, regardless of the MOI. TEM examination and PCR confirmation of the coliphages capable of lysing ETEC showed that all were T4-like bacteriophages except for N1 and N12 (Fig. 6 and 7), which belonged to the family Myoviridae. Tracing coliphages back to their host piglet (Tables 2 and 3), we found that coliphages specific for pathogenic E. coli. were common in piglet gut (9/10 samples in the Nanjing farm and 7/10 in the Chuzhou farm).
FIG 3.
Host range of isolated coliphages on E. coli strains. Pathogenic strains susceptible to coliphage lysis and infection are indicated in light red and dark red, respectively. Other strains susceptible to coliphage lysis and infection are indicated in light blue and dark blue, respectively.
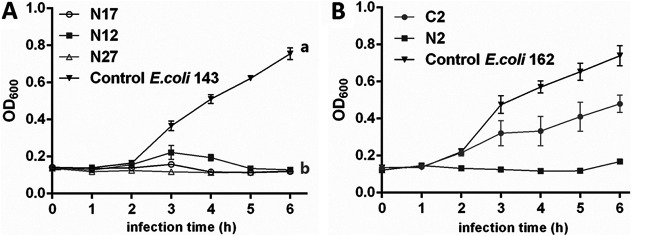
FIG 4.
Bacteriostatic curves of 5 coliphages that infected pathogenic E. coli. (A) Bacteriostatic curves of N12, N17, and N27 on EPEC 143 at an MOI of 0.1. (B) Bacteriostatic curves of N2 and C2 on EPEC 162 at an MOI of 0.1. Different letters denote statistically significant differences between groups at P < 0.05.
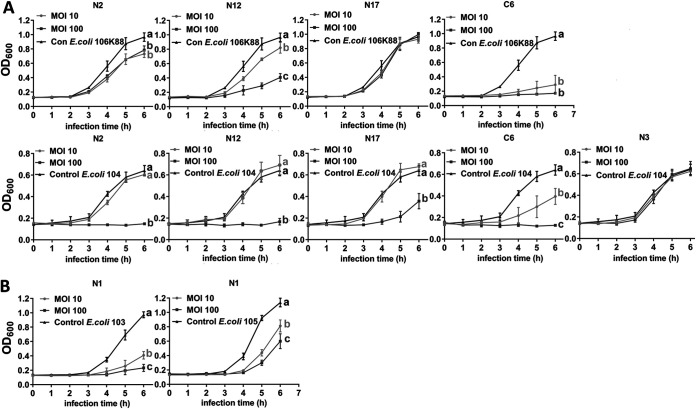
FIG 5.
Bacteriostatic curves of 6 coliphages that lysed but did not infect ETEC. (A) Bacteriostatic curves of N2, N12, N17, C6, and N3 on E. coli 106K88 and E. coli 104 at MOIs of 10 and 100. (B) Bacteriostatic curves of N1 on E. coli 103 and E. coli 105 at MOIs of 10 and 100. Different letters denote statistically significant differences between groups at P < 0.05.
FIG 6.
Transmission electron microscope images of 6 coliphages lysing but not infecting ETEC. N2, N3, N17, and C6 were T4-like bacteriophages.
FIG 7.
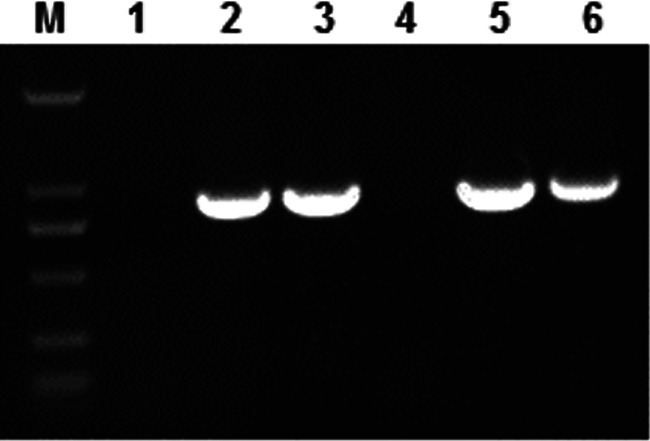
Identification of gene 23 in T4-like bacteriophages from 6 coliphages lysing but not infecting ETEC. Lanes: M, 2000 marker; 1, N1; 2, N2; 3, N3; 4, N12; 5, N17; 6, C6.
Identification of polyvalent coliphages isolated from piglet gut.
To detect whether there were polyvalent coliphages that infected other genera, the isolated coliphages were also tested against other common enteric pathogens like Salmonella enterica serovar Choleraesuis, P. mirabilis, Klebsiella pneumoniae, and C. freundii. Coliphages C5 and N30 were found to infect C. freundii, and coliphage N24 infected S. enterica serovar Choleraesuis. These results indicated that polyvalent bacteriophages existed in the piglet gut. Interestingly, we found that bacteriophages N30 and C5 had the same infection host range and also showed very similar lysis host ranges with the exception of E. coli 245 (Fig. 3). We sequenced the two bacteriophages, which were deposited in GenBank (see “Data availability” below). Both were T7-like phages and showed a genetic identity of 89.67%. The EOPs of N30 in all infected E. coli strains relative to C. freundii 113ju hosts were significantly lower than those of C5, especially in E. coli 94 (Table 4).
TABLE 4.
EOPs of 2 polyvalent bacteriophages on susceptible hosts
| Bacteriophage | EOP for host straina: |
||||
|---|---|---|---|---|---|
| C. freundii 113ju | E. coli 94 | E. coli 121 | E. coli 141 | E. coli 235 | |
| N30 | 1 | 0.0015 ± 0.00048 a | 0.53 ± 0.029 a | 0.66 ± 0.11 a | 0.62 ± 0.17 a |
| C5 | 1 | 0.93 ± 0.12 b | 1.47 ± 0.28 b | 1.45 ± 0.14 b | 1.33 ± 0.30 b |
| P value | <0.0001 | 0.004 | 0.017 | 0.021 | |
Data are expressed as arithmetic means ± standard deviations. Different lowercase letters denote statistically significant differences between groups at P < 0.05.
DISCUSSION
Our study on coliforms from pig gut showed that multidrug resistance to antibiotics caused by indiscriminate use was a serious problem in the swine industry. Since China has banned antibiotic additives to feed in 2020, the animal husbandry industry needs to find natural alternatives to antibiotics. As substitutes for antibiotics, bacteriophages have gained extensive attention in recent years. Before the use of bacteriophages as substitutes for antibiotics to control pathogenic microorganisms, it is essential to map the endogenous bacteriophages present in the animal gut. The weaning of mammals is characterized by a transition in the diet and environment of piglets, which can significantly alter the gut microbiota (22). Several studies (29–31) have reported weaning-related transitions in the gut bacterial community, but little is known regarding the effect of weaning on the gut virome of piglets. In this study, we isolated coliphages from the feces of pre- and postweaned piglets to determine their role in postweaning colibacillosis diarrhea.
Weaning was an important factor that affected coliphage numbers.
We initially isolated 45 coliphages from two farms using the double agar plate method. More coliphages were isolated from the postweaned piglet gut, and the novel coliphages were likely introduced by the environment and new diet or changes in host communities as well as via induction of the lytic cycle in prophages (32–34) due to changes in the gut environment after weaning. Longer surveillance periods would be required to determine whether the new coliphages fluctuate or remain stable over time. In each farm, a large number of coliphage isolates were present in most samples, indicating that pigs from the same farm may share a similar gut phageome. Interestingly, the coliphages N2 and C6 were isolated only from preweaned piglets. It is less certain whether both are introduced from the milk of the sow or the initial environment which cannot colonize the piglet’s intestinal tract or whether they just fluctuated and diminished without disappearing due to weaning. In addition, the weaning-induced dysbiosis may have eliminated the host bacteria of N2 and C6 and consequently resulted in the extermination of the phages.
Core and common coliphages may exist in pig gut with mild genetic divergence in different pig farms.
Coliphage N7 was detected in 9/10 samples from the Nanjing farm and coliphage C1 was present in 7/10 samples from the Chuzhou farm. Manrique and colleagues hypothesized that a healthy gut phageome is essential for a functional gut microbiome and consists of both core and common bacteriophages (11). Based on our findings, we presumed that the coliphages N7 and C1 may be the core bacteriophages in the piglet gut. However, investigations in more pig farms are required to confirm the core coliphages of piglet gut. It is worth noting that coliphage N7 is a temperate bacteriophage but we isolated it when it was in lytic cycle. Lysogenic cycle is reversible, and the circuit controlling lysogen-lysis is subtle. Longer surveillance would be needed to detect how coliphage N7 changed as age and diet changed. The bacteriophages N30 and C5 showed high genetic identity (89.67%) and were prevalent in postweaned piglet gut (3/5 samples from the Nanjing farm and 2/5 samples from the Chuzhou farm). Therefore, they may be considered part of the common phageome in the piglet gut. Furthermore, N30 and C5 were both polyvalent bacteriophages and their impact might be even more significant. Polyvalent bacteriophages that infect genetically diverse bacteria are often reported in the Enterobacteriaceae family (35–37). N30 and C5 showed mild differences in their genome and infection characteristics, which may be due to the discrepancies in the bacterial flora of the host gut. The interaction between bacteriophages and host bacteria in the normal gut is explained by the arms race dynamics (ARD) and fluctuating selection dynamics (FSD) models (17, 38). Both models associate the evolution of bacteriophages with mutations that occur in response to the selective pressure of predation.
Endogenous intestinal coliphages may provide a line of defense against pathogenic E. coli by infection or “lysis from without.”
Our findings also suggested that coliphages specific for pathogenic E. coli were a common occurrence in piglet gut. In addition, coliphages capable of infecting bacteria provided better protection than those lysing bacteria without infection, which may explain the differences in susceptibility of weaned piglet littermates to diarrhea. Studies show that plaque formation is a better indicator of productive phage infection than the spot inhibition on a bacterial lawn (39–41). Accordingly, we detected 10 coliphages capable of lysing pathogenic E. coli by spot assay but only 6 that could infect the bacteria. Early bacterial lysis without phage production may be caused by lysis from without (LO) or abortive infection (27, 28). LO in turn is induced by either high-multiplicity virion adsorption (LOV) or exogenously supplied phage lysin (LOL). LOV is frequently seen with T2 and T4 phages (28, 42), whereas LOL is common in Gram-positive bacteria due to the presence of lysin in the cell wall. As the coliphages N2 and C6 that lysed ETEC were T4-like phages and showed greater bacteriostasis at higher MOIs, they likely lysed the bacteria via LOV. Therefore, bacteriophages residing in the gut in abundant numbers profoundly influenced the bacterial communities not only by lysis via infection but also through other lytic methods. The endogenous coliphages may play a potential role in the defense against invading pathogens.
In conclusion, we found that weaning was an important factor that affected coliphage numbers and identified pathogenic E. coli-specific bacteriophages and polyvalent bacteriophages in piglet gut. Our study complements sequencing approaches to gain deeper insights into gut coliphages and their biological impact.
MATERIALS AND METHODS
Isolation and identification of Enterobacteriaceae strains.
We collected 80 fecal samples from 3 pig farms in Nanjing, Nantong, and Maanshan in 2015. In Nanjing, samples were collected from healthy pre- and postweaned piglets. In Nantong, samples were collected from healthy and growing (80 to 100 days) piglets. In Maanshan, postweaned samples were collected from healthy pigs and pigs with diarrhea. Enterobacteriaceae strains were isolated by plate streaking in eosin-methylene blue (EMB) medium. Colonies were then collected and identified on a genus level by PCR amplification of the 16S rRNA gene (43), and sequences were submitted to GenBank.
Characterization of Escherichia coli strains.
Phylogenetic grouping of E. coli was conducted by PCR as described previously (24). Antimicrobial susceptibilities to ampicillin (AMP), chloramphenicol (CHL), gentamicin (CN), enrofloxacin (ENR), ceftiofur (EFT), and tetracycline (TE) were tested using the disk diffusion susceptibility method described by Mahmoud and colleagues (35). Detection of virulence genes in diarrheagenic E. coli was performed as described previously (44).
Isolation, identification, and morphological characterization of coliphages.
A total of 20 fecal samples were collected from a small pig farm (100 sows) in Nanjing and a medium-sized pig farm (5,000 sows) in Chuzhou in July and September 2016, respectively. From each farm, samples were randomly selected from 5 preweaned (5 to 10 days before weaning) and 5 postweaned (5 to 10 days after weaning) piglets (Nanjing, Landrace × Yorkshire; Chuzhou, Duroc × Landrace) from different pens. Fecal samples were collected by massaging the rectum or immediately after defecation and stored at 4°C for no more than 3 days before coliphage isolation.
The coliphages in the fecal samples were isolated by plaque-forming assay from all E. coli isolates. Briefly, each sample was resuspended in saline magnesium (SM) buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris [pH 7.5], and 0.002% [wt/vol] gelatin) at a ratio of 1:20 (g/ml) and mixed followed by centrifugation at 10,000 × g for 30 min. The supernatant was passed through a 0.22-μm filter (Millipore Sterivex; Merck, Germany), and 100 μl filtrate was mixed with 100 μl logarithmic-phase E. coli suspension and 5 ml 55°C 0.7% LB agar. The mixture was poured on 1.5% solid agar, and the double-layered agar plates were incubated at 37°C for 5 h. First, the number of the plaques in each plate was recorded, and then the clear plaques were picked from each plate and individually replated on double-layered agar for phage enrichment. Purified phage particles were spotted on a copper grid and stained with phosphotungstic acid (PTA, 2% [wt/vol]) and observed using an HT7700 (Hitachi, Japan) transmission electron microscope (TEM).
Determination of host range.
The host range of the purified coliphages was determined by the spot test followed by the efficiency of plating (EOP) assay. Each coliphage was tested against all E. coli isolates. In addition, a Salmonella enterica serovar Choleraesuis strain (ATCC 13312) and other Enterobacteriaceae strains isolated from fecal samples in this study were also employed to detect the presence of polyvalent coliphages that infect other genera. Briefly, 100 μl target bacterial suspension was spread on LB plates and incubated for 30 min at 37°C and 10 μl purified coliphage (108 to 109 PFU/ml) was then spotted on each plate. After incubation for 6 h at 37°C, the plates were observed for clear zones, which were indicative of lysis. EOP was determined by calculating the ratio of the phage plaque titer obtained with the target bacterial strain to that obtained with host bacteria as previously described (27) with slight modifications. The lysed bacteria determined by the spot assay were defined as the target bacteria whereas the host bacteria were those used for coliphage amplification. Briefly, the coliphages were serially diluted (109 to 103 PFU/ml) and 100 μl of each dilution was tested against the target bacteria and host bacteria, respectively, and then the plaque numbers were counted for the calculation of the ratio. If the EOP of a strain was higher than 10−8, it was regarded as an infected strain. The experiments were performed in triplicate.
The coliphages with the same host range in one fecal sample were considered one strain. For those coliphages with the same host range but isolated from different samples, the DNA was extracted and digested with restriction endonucleases (see below). If the restriction endonuclease maps were the same, the coliphages were confirmed as one strain. The identified coliphages were then used to infect the host using the plaque assay described above.
Extraction of phage genomic DNA, restriction analysis, and sequencing.
The phage DNA was extracted using the SDS-proteinase K protocol as described previously (45). The DNA was digested with EcoRI, BamHI, XhoI, XbaI, HindIII, and AvaI according to the manufacturer’s instructions (TaKaRa Bio Inc., Japan) and separated by 0.8% agarose gel electrophoresis. Whole-genome sequencing was performed by Shanghai Biozeron Biotechnology Co. Ltd. using the Illumina HiSeq sequencing platform. The genome sequences were assembled using SOAPdenovo v2.04 software. Coding sequences were analyzed by Glimmer 3.02.
Pathogenic E. coli strain challenge test.
To determine the bacteriostatic effects of the coliphages on the 8 isolated pathogenic E. coli strains, the bacteria were cultured in LB medium up to the logarithmic phase and reinoculated in fresh medium containing the coliphage at multiplicities of infection (MOIs) of 0.1, 10, or 100. Bacterial growth was monitored every hour for 6 h by measuring the optical density (OD) at 600 nm. Experiments were performed in triplicate with different coliphages.
Detection of T4-like bacteriophage by PCR.
PCR was performed as described previously (46). The central region of gene 23 of various T4-type phages was amplified using the following primers: Mzia1 (5′-TGTTATIGGTATGGTICGICGTGCTAT-3′) and CAP8 (5′-TGAAGTTACCTTCACCACGACCGG-3′).
Data availability.
The sequences of the N7, C1, N30, and C5 bacteriophage strains were deposited in GenBank (accession numbers MH717096, MH717097, MH717098, and MH717099, respectively). For bacterial strains that were sequenced, the GenBank accession numbers are as follows: E. coli, MH671408 to MH671417, MH671419 to MH671423, MH671425 to MH671463, MH671465 to MH671479, MH671481 to MH671484, MH671486 to MH671497, MK156384, and MK615932; P. mirabilis, MH643694; C. freundii, MH643693; K. pneumoniae, MH643695.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31802102) and the Natural Science Foundation of Jiangsu Province (BK20180540).
We declare that we have no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Weiyun Zhu, Email: zhuweiyun@njau.edu.cn.
Danilo Ercolini, University of Naples Federico II.
REFERENCES
- 1.Laforest-Lapointe I, Arrieta MC. 2018. Microbial eukaryotes: a missing link in gut microbiome studies. mSystems 3:e00201-17. 10.1128/mSystems.00201-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vemuri R, Shankar EM, Chieppa M, Eri R, Kavanagh K. 2020. Beyond just bacteria: functional biomes in the gut ecosystem including virome, mycobiome, archaeome and helminths. Microorganisms 8:483. 10.3390/microorganisms8040483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. 2018. Current understanding of the human microbiome. Nat Med 24:392–400. 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heintz-Buschart A, Wilmes P. 2018. Human gut microbiome: function matters. Trends Microbiol 26:563–574. 10.1016/j.tim.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Ananthakrishnan AN. 2020. Microbiome-based biomarkers for IBD. Inflamm Bowel Dis 26:1463–1469. 10.1093/ibd/izaa071. [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Guryn K, Leone V, Chang EB. 2019. Regional diversity of the gastrointestinal microbiome. Cell Host Microbe 26:314–324. 10.1016/j.chom.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duerkop BA, Kleiner M, Paez-Espino D, Zhu W, Bushnell B, Hassell B, Winter SE, Kyrpides NC, Hooper LV. 2018. Murine colitis reveals a disease-associated bacteriophage community. Nat Microbiol 3:1023–1031. 10.1038/s41564-018-0210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, Stappenbeck TS, McGovern DP, Keshavarzian A, Mutlu EA, Sauk J, Gevers D, Xavier RJ, Wang D, Parkes M, Virgin HW. 2015. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160:447–460. 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen TS, Mentzel CMJ, Kot W, Castro-Mejia JL, Zuffa S, Swann JR, Hansen LH, Vogensen FK, Hansen AK, Nielsen DS. 2020. Faecal virome transplantation decreases symptoms of type 2 diabetes and obesity in a murine model. Gut 69:2122–2130. 10.1136/gutjnl-2019-320005. [DOI] [PubMed] [Google Scholar]
- 10.Ma Y, You X, Mai G, Tokuyasu T, Liu C. 2018. A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome 6:24. 10.1186/s40168-018-0410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ. 2016. Healthy human gut phageome. Proc Natl Acad Sci U S A 113:10400–10405. 10.1073/pnas.1601060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. 2013. Rapid evolution of the human gut virome. Proc Natl Acad Sci U S A 110:12450–12455. 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuo T, Wong SH, Lam K, Lui R, Cheung K, Tang W, Ching JYL, Chan PKS, Chan MCW, Wu JCY, Chan FKL, Yu J, Sung JJY, Ng SC. 2018. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut 67:634–643. 10.1136/gutjnl-2017-313952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan Mirzaei M, Khan MAA, Ghosh P, Taranu ZE, Taguer M, Ru J, Chowdhury R, Kabir MM, Deng L, Mondal D, Maurice CF. 2020. Bacteriophages isolated from stunted children can regulate gut bacterial communities in an age-specific manner. Cell Host Microbe 27:199–212.e195. 10.1016/j.chom.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, Warner BB, Tarr PI, Wang D, Holtz LR. 2015. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med 21:1228–1234. 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharon I, Morowitz MJ, Thomas BC, Costello EK, Relman DA, Banfield JF. 2013. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res 23:111–120. 10.1101/gr.142315.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan Mirzaei M, Maurice CF. 2017. Menage a trois in the human gut: interactions between host, bacteria and phages. Nat Rev Microbiol 15:397–408. 10.1038/nrmicro.2017.30. [DOI] [PubMed] [Google Scholar]
- 18.Gorski A, Weber-Dabrowska B. 2005. The potential role of endogenous bacteriophages in controlling invading pathogens. Cell Mol Life Sci 62:511–519. 10.1007/s00018-004-4403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almeida GMF, Laanto E, Ashrafi R, Sundberg LR. 2019. Bacteriophage adherence to mucus mediates preventive protection against pathogenic bacteria. mBio 10:e01984-19. 10.1128/mBio.01984-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, Stotland A, Wolkowicz R, Cutting AS, Doran KS, Salamon P, Youle M, Rohwer F. 2013. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci U S A 110:10771–10776. 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell JM, Crenshaw JD, Polo J. 2013. The biological stress of early weaned piglets. J Anim Sci Biotechnol 4:19. 10.1186/2049-1891-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gresse R, Chaucheyras-Durand F, Fleury MA, Van de Wiele T, Forano E, Blanquet-Diot S. 2017. Gut microbiota dysbiosis in postweaning piglets: understanding the keys to health. Trends Microbiol 25:851–873. 10.1016/j.tim.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Schokker D, Zhang J, Vastenhouw SA, Heilig HG, Smidt H, Rebel JM, Smits MA. 2015. Long-lasting effects of early-life antibiotic treatment and routine animal handling on gut microbiota composition and immune system in pigs. PLoS One 10:e0116523. 10.1371/journal.pone.0116523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 25.Messori S, Trevisi P, Simongiovanni A, Priori D, Bosi P. 2013. Effect of susceptibility to enterotoxigenic Escherichia coli F4 and of dietary tryptophan on gut microbiota diversity observed in healthy young pigs. Vet Microbiol 162:173–179. 10.1016/j.vetmic.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen UV, Coddens A, Melkebeek V, Devriendt B, Goetstouwers T, Poucke MV, Peelman L, Cox E. 2017. High susceptibility prevalence for F4(+) and F18(+) Escherichia coli in Flemish pigs. Vet Microbiol 202:52–57. 10.1016/j.vetmic.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 27.Kutter E. 2009. Phage host range and efficiency of plating. Methods Mol Biol 501:141–149. 10.1007/978-1-60327-164-6_14. [DOI] [PubMed] [Google Scholar]
- 28.Abedon ST. 2011. Lysis from without. Bacteriophage 1:46–49. 10.4161/bact.1.1.13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dou S, Gadonna-Widehem P, Rome V, Hamoudi D, Rhazi L, Lakhal L, Larcher T, Bahi-Jaber N, Pinon-Quintana A, Guyonvarch A, Huerou-Luron IL, Abdennebi-Najar L. 2017. Characterisation of early-life fecal microbiota in susceptible and healthy pigs to post-weaning diarrhoea. PLoS One 12:e0169851. 10.1371/journal.pone.0169851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levesque CL, Hooda S, Swanson KS, de Lange K. 2014. Alterations in ileal mucosa bacteria related to diet complexity and growth performance in young pigs. PLoS One 9:e108472. 10.1371/journal.pone.0108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konstantinov SR, Awati AA, Williams BA, Miller BG, Jones P, Stokes CR, Akkermans AD, Smidt H, de Vos WM. 2006. Post-natal development of the porcine microbiota composition and activities. Environ Microbiol 8:1191–1199. 10.1111/j.1462-2920.2006.01009.x. [DOI] [PubMed] [Google Scholar]
- 32.Duerkop BA, Clements CV, Rollins D, Rodrigues JL, Hooper LV. 2012. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc Natl Acad Sci U S A 109:17621–17626. 10.1073/pnas.1206136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh JH, Alexander LM, Pan M, Schueler KL, Keller MP, Attie AD, Walter J, van Pijkeren JP. 2019. Dietary fructose and microbiota-derived short-chain fatty acids promote bacteriophage production in the gut symbiont Lactobacillus reuteri. Cell Host Microbe 25:273–284.e276. 10.1016/j.chom.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 34.Mills S, Shanahan F, Stanton C, Hill C, Coffey A, Ross RP. 2013. Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes 4:4–16. 10.4161/gmic.22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahmoud M, Askora A, Barakat AB, Rabie OE, Hassan SE. 2018. Isolation and characterization of polyvalent bacteriophages infecting multi drug resistant Salmonella serovars isolated from broilers in Egypt. Int J Food Microbiol 266:8–13. 10.1016/j.ijfoodmicro.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 36.Park M, Lee JH, Shin H, Kim M, Choi J, Kang DH, Heu S, Ryu S. 2012. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157:H7. Appl Environ Microbiol 78:58–69. 10.1128/AEM.06231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamdi S, Rousseau GM, Labrie SJ, Tremblay DM, Kourda RS, Ben Slama K, Moineau S. 2017. Characterization of two polyvalent phages infecting Enterobacteriaceae. Sci Rep 7:40349. 10.1038/srep40349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scanlan PD. 2017. Bacteria-bacteriophage coevolution in the human gut: implications for microbial diversity and functionality. Trends Microbiol 25:614–623. 10.1016/j.tim.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 39.Letarov AV, Kulikov EE. 2018. Determination of the bacteriophage host range: culture-based approach. Methods Mol Biol 1693:75–84. 10.1007/978-1-4939-7395-8_7. [DOI] [PubMed] [Google Scholar]
- 40.Khan Mirzaei M, Nilsson AS. 2015. Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS One 10:e0118557. 10.1371/journal.pone.0118557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hyman P, Abedon ST. 2010. Bacteriophage host range and bacterial resistance. Adv Appl Microbiol 70:217–248. 10.1016/S0065-2164(10)70007-1. [DOI] [PubMed] [Google Scholar]
- 42.Tarahovsky YS, Ivanitsky GR, Khusainov AA. 1994. Lysis of Escherichia coli cells induced by bacteriophage T4. FEMS Microbiol Lett 122:195–199. 10.1111/j.1574-6968.1994.tb07164.x. [DOI] [PubMed] [Google Scholar]
- 43.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonkoungou IJ, Lienemann T, Martikainen O, Dembele R, Sanou I, Traore AS, Siitonen A, Barro N, Haukka K. 2012. Diarrhoeagenic Escherichia coli detected by 16-plex PCR in children with and without diarrhoea in Burkina Faso. Clin Microbiol Infect 18:901–906. 10.1111/j.1469-0691.2011.03675.x. [DOI] [PubMed] [Google Scholar]
- 45.Amarillas L, Rubi-Rangel L, Chaidez C, Gonzalez-Robles A, Lightbourn-Rojas L, Leon-Felix J. 2017. Isolation and characterization of phiLLS, a novel phage with potential biocontrol agent against multidrug-resistant Escherichia coli. Front Microbiol 8:1355. 10.3389/fmicb.2017.01355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tetart F, Desplats C, Kutateladze M, Monod C, Ackermann HW, Krisch HM. 2001. Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. J Bacteriol 183:358–366. 10.1128/JB.183.1.358-366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download AEM.00966-21-s0001.pdf, PDF file, 0.2 MB (216.1KB, pdf)
Data Availability Statement
The sequences of the N7, C1, N30, and C5 bacteriophage strains were deposited in GenBank (accession numbers MH717096, MH717097, MH717098, and MH717099, respectively). For bacterial strains that were sequenced, the GenBank accession numbers are as follows: E. coli, MH671408 to MH671417, MH671419 to MH671423, MH671425 to MH671463, MH671465 to MH671479, MH671481 to MH671484, MH671486 to MH671497, MK156384, and MK615932; P. mirabilis, MH643694; C. freundii, MH643693; K. pneumoniae, MH643695.