Abstract
Early detection and diagnosis of coronary artery disease could reduce the risk of developing a heart attack. The coronary arteries are optimally visualised using computed tomography coronary angiography (CTCA) imaging. These images are reviewed by specialist radiologists who evaluate the coronary arteries for potential narrowing. A lack of radiologists in the UK is a constraint to timely diagnosis of coronary artery disease, particularly in the acute accident and emergency department setting. The development of automated methods by which coronary artery narrowing can be identified rapidly and accurately are therefore timely. Such complex computer based tools also need to be sufficiently computationally efficient that they can run on servers typically found in hospital settings, where graphical processing units for example are unavailable. We propose a fully automatic two-dimensional Unet model to segment the aorta and coronary arteries on CTCA images. Two models are trained to segment two regions of interest, (1) the aorta and the coronary arteries or (2) the coronary arteries alone. Our method achieves 91.20% and 88.80% dice similarity coefficient accuracy on regions of interest 1 and 2 respectively. Compared with a semi-automatic segmentation method, our model performs better when segmenting the coronary arteries alone. The performance of the proposed method is comparable to existing published two-dimensional or three-dimensional deep learning models. Importantly, the algorithmic and graphical processing unit memory efficiencies are maintained such that the model can be deployed without requiring graphical processing units, and therefore can be used in a hospital setting.
Keywords: Aorta, computed tomography coronary angiography, coronary artery, deep learning, segmentation
I. Introduction
Coronary artery disease (CAD) is one of the leading causes of death in the UK [1] and worldwide. The lumen of the coronary arteries can narrow as a result of build-up of atheromatous plaque within the artery wall. Reductions in local blood flow as a result of vessel lumen narrowing can starve the heart muscle of oxygen. Vulnerable plaques can rupture, occlude the vessel lumen and result in cardiac muscle ischaemia/death, which manifests clinically as a heart attack. Early detection of the presence of atheromatous plaque and vessel stenosis [2] could allow early medical intervention and potentially reduce the risk of heart attack. Currently, several non-invasive imaging modalities are available to clinicians for visualising the anatomy of the coronary arteries as well as delineating the severity of vessel stenosis. These imaging modalities are Stress Echocardiography [3], Cardiac magnetic resonance imaging (MRI) [4] and Computed Tomography Coronary Angiography (CTCA) [5]. CTCA is the quickest of these methods and offers high sensitivity and specificity for detection and exclusion of significant coronary stenosis [6]. As a result CTCA is the preferred first-line option for the assessment of stable cardiac disease in the National Institute for Health and Care Excellence guidelines for the UK [7]. CTCA has the ability to identify calcified, non-calcified coronary plaque and mixed-attenuation plaques which can help clinicians characterise plaques and formulate management strategies.
To assess the severity of CAD, one approach involves visual estimation of stenosis severity on CTCA scans. This requires the geometrical information of the coronary arteries to be provided in order to accurately assess the severity of the stenosis. This approach however is subjective, time consuming and requires specialist radiologists, who are in short supply, to interpret the CTCA images [8]. Accurate interpretation and diagnosis of CAD is heavily reliant on the experience and expertise of individual clinicians [9], [10]. The diagnostic outcomes can differ between newly trained clinicians compared to experienced specialists.
An alternative approach for assessing CAD severity involves performing computational fluid dynamics (CFD) on the target arteries [11]. It first requires identification of accurate geometries of the aorta and coronary arteries. A set of partial differential equations of blood flow are then solved numerically given the boundary conditions and the geometries. Once blood flow is estimated, the useful clinical predictor, Fractional Flow Reserve (FFR) [12]–[14] can be derived. Provided accurate geometries of the aorta and coronary arteries are available, this approach provides an objective estimation of stenosis and does not require additional imaging, which makes it particularly attractive to clinicians.
Deriving accurate geometrical information of the aorta and coronary arteries is important for the above approaches. It is achieved by delineating the outline of the vessels, termed segmentation. Blood vessel segmentation can be performed manually, semi-automatically or automatically [15]–[20]. Manual segmentation is subjective and time consuming, requiring pixel-by-pixel labelling of individual vessels. Semi-automatic and automatic segmentation methods are objective and quicker, though they can require manual correction for under- or over- segmented vessels. There remains an unmet need to develop fast, objective and accurate automated computer-derived coronary artery segmentation algorithms that can be deployed in a hospital setting to assist clinicians diagnose CAD. This is especially relevant in Accident and Emergency (A+E) departments where CTCA reviews are often delayed due to a lack of available specialists to read the CTCAs [21]. This delay in turn slows patient management in A+E, increases resource utilisation and results in excess costs to the health service.
Deriving accurate geometries of the aorta and coronary arteries are crucial components of the clinical information required by radiologists and cardiologists to evaluate the severity of coronary artery disease. The current approaches used to segment the coronary arteries and the aorta have varying accuracy, can be time consuming and most importantly require extensive computational resources. Most hospital systems in the UK do not have access to the required computational resources such as GPUs required for machine learning analyses. We have proposed a deep learning approach to improve the accuracy and speed of cardiac vessel analysis. Our proposed approach is fast, accurate and fully automatic. But most importantly, our method requires only low computational resources, which are feasible to be implemented within a hospital network.
A fully automatic detection and classification system for CAD using computer-based deep learning algorithms is a way to achieve the above goal. The first stage of the process initially requires segmentation (identifying the outlines) of the aorta and coronary arteries on CTCA images. The second stage involves classification of disease severity performed on segmented CTCA images. The segmentation task has be considered in two ways in the literature. The first involves segmentation of both the aorta and the coronary arteries. For example, Gu et al. [22] proposed a 3D deep learning model to perform this task. The other strategy is to segment just the coronary arteries alone. For example, Huang et al. [23] suggested a 3D deep learning method with centreline to segment the coronary arteries. In general, the performance of aorta and coronary artery segmentation is better than segmentation of the coronary arteries alone. A more detailed summary regarding existing deep learning-based segmentations is discussed in the following paragraph.
The utility of deep learning applied to medical images [24] and relevant work in medical image segmentation have been extensively discussed in the literature [25]–[27]. The works related to artery segmentation on CTCA images have been discussed in two review papers [20], [28]. They provide a review of 3D vessel lumen segmentation techniques and deep learning methods for cardiac image segmentation. The focus of the current study relates to deep learning methods for coronary artery segmentation and we briefly summarise the work published in the following paragraph.
Several deep learning techniques have been proposed to segment the aorta and/or coronary arteries. Moeskops et al. [29] investigated a single convolutional neural network trained to segment coronary arteries in cardiac CTA images. The training dice similarity coefficient (DSC) accuracy was around 65%. A 3D-convolutional neural network was presented by Merkow et al. [30], which demonstrated that processing the volumetric data in 3D could improve the segmentation performance compared to 2D processing. However, the performance of the coronary artery segmentation model was not reported. Kjerland [31] adopted a 3D DeepMedic network to segment both the aorta and coronary arteries. The reported DSC accuracy was between 75%–78%. Huang et al. [23] examined a 3D Unet with/without a centerline to segment the coronary artery. The DSC accuracy was between 71%–78%.
Recently, a 3D multi-channel Unet has been proposed by Chen et al. [32], which had a DSC accuracy of 80% for coronary artery segmentation. Shen et al. [33] proposed a 3D fully convolutional network with attention gates to segment both the aorta and coronary artery. The boundary of the segmented artery was smoothed by a level set function. The average DSC accuracy was about 90%. Lee et al. [34] introduced a template transformer network where a shape template is deformed to match the underlying structure of interest through an end-to-end trained spatial transformer network for coronary artery segmentation. The DSC accuracy is between 76%–78%. Wolterink et al. [35] proposed using graph convolutional networks to predict the spatial location of vertices in a tubular surface mesh that segments the coronary artery lumen. The average DSC is 74%. Mirunalini et al. [36] proposed a two-stage approach to segment the coronary artery. The first stage adopted a 2D Recurrent Convolutional Neural Network to detect the artery in the slice, then a 2D residual Unet was used to segment the coronary artery. The intersection over union (IoU) was reported, which was 84%. Lei et al. [37] developed a 3D Attention Fully Convolutional Network model to automatically segment the aorta and coronary artery for CCTA. The mean DSC is 83%. Gu et al. [22] recently published a 3D global feature embedded network with active contour loss to segment the aorta and coronary artery. The reported average DSC is 91.43%.
Recently, a multi-objective clustering and toroidal model-guided tracking method has been used to segment the coronary artery tree automatically [38]. Gao et al. [39] have proposed a novel deep neural network solution (TreeVes-Net) that allows machines to perceive FFR values directly from static coronary CT angiography images.
There is a lack of efficient deep learning methods (which only require low computational resources) that can be used within typical hospital networks. For example, 3D deep learning models require a high computational resource. Though a 2D deep learning method has been described in the literature [36], the second additional component that was part of this model would increase the computational complexity of the model and hence decrease the algorithmic efficiency of the method. We propose a 2D Unet to perform aorta and/or coronary artery segmentation and demonstrate that this 2D Unet is practically feasible to be implemented given that the computational resources are limited to those available in a hospital network whilst maintaining a good segmentation accuracy. Additionally, we emphasise that we are the first to employ a 2D technique (2D Unet) for artery segmentation directly on the CTCA image. Most of the previous studies evaluating coronary artery segmentation have utilised 3D models. For the study that employed a 2D approach, where an additional sequence model was required for segmentation, the coronary artery segmentation was performed indirectly.
In this study, we propose a modified U-Net [40] model: (1) a batch normalization layer is added to the convolution block; (2) a dropout layer is added before each convolution block and evaluate its performance for the automated segmentation of the aorta and coronary arteries on CTCA images. We then retrain our model to segment the coronary arteries alone and demonstrate improved performance.
The main contributions of this study are:
-
•
The first study to propose a modified 2D Unet that directly segments the aorta and/or coronary arteries on CTCA scans
-
•
The method is practically feasible to be implemented within clinical systems where the available computational resources are limited
-
•
Importantly, our technique works well when the coronary arteries alone are segmented (accuracy ~89%).
II. Method
All the patients in the study had presented with chest pain and associated symptoms that indicated an intermediate risk of coronary artery disease. All patients underwent cardiac CT angiography for anatomical assessment of their coronary arteries and risk stratification for coronary artery disease. 71 cases were selected out of a total of 101 cases. 30 cases were not usable mainly due to major motion artefacts, artefacts on the image or the presence of coronary stents or bypass grafts distorting the coronary anatomy. The Simpleware ScanIP algorithm also failed in vessel segmentation on two of the selected cases. To segment the aorta and/or coronary artery we have proposed a modified 2D Unet model where: (1) a batch normalization layer has been added to the convolution block; (2) a dropout layer is added before each convolution block. The proposed method is compared to semi-automatic segmentation (Simpleware-ScanIP) and 2D/3D deep learning methods. The segmentation performance and time taken for vessel analysis are then evaluated.
A. Clinical Data
The final study data represented CTCA scans performed on 69 subjects with chest pain. The scans were acquired at University College Hospital London and Barts Health NHS Trust using different CT scanners and acquisition protocols. All the patients in the study had presented with chest pain and associated symptoms that indicated an intermediate risk of coronary artery disease. All patients underwent cardiac CT angiography for anatomical assessment of their coronary arteries and risk stratification for coronary artery disease. The study was carried out in accordance with the recommendations of the South East Research Ethics Committee, Aylesford, Kent, UK, with written informed consent from all subjects in accordance with the Declaration of Helsinki. An example of a CTCA scan is shown in Fig. 1.
FIGURE 1.
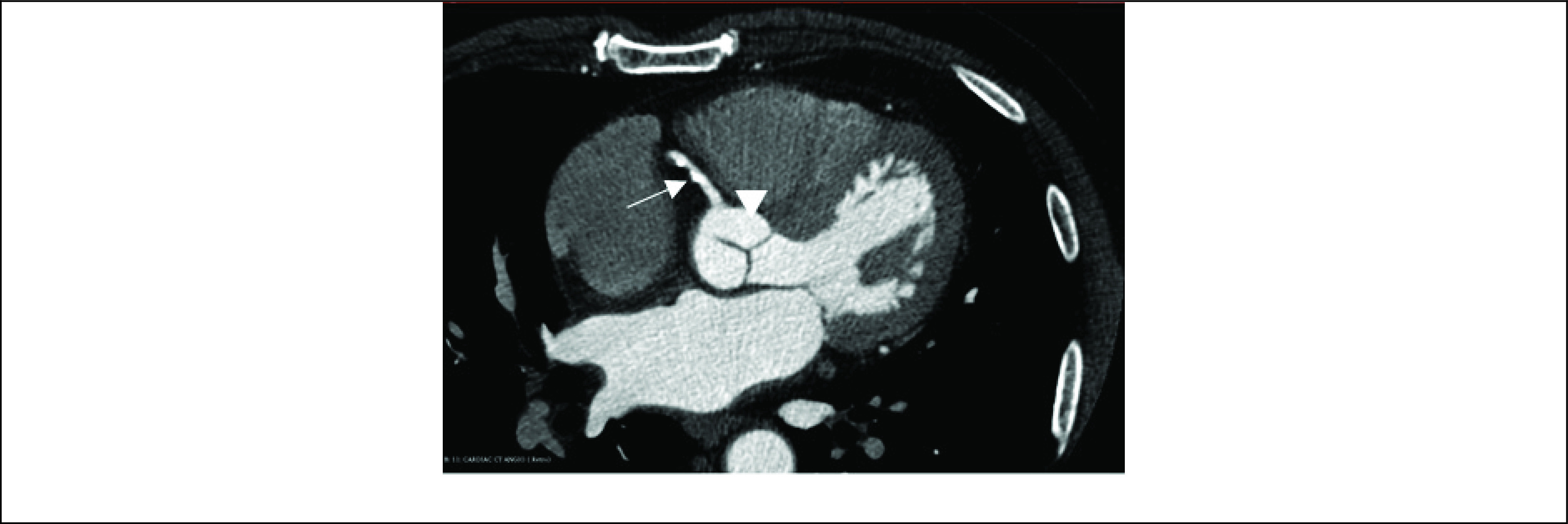
A CTCA scan of a patient. Arrow = right coronary artery; arrowhead = ascending aorta.
B. Data Pre-Processing
The original Digital Imaging and Communications in Medicine (DICOM) data were pre-processed using 3D Slicer. The brightness of the CTCA image was adjusted by windowing (window center = 40, window width = 400). The image was saved as a NIfTI Data Format. The image size was 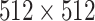 pixels. The image was then processed by ImageJ [41]. The pixel intensity was normalised by using linear histogram stretch and then rescaled to between 0 to 255. The final images were converted to 8-bit Portable Network Graphics (PNG) for training, validation and testing. The pre-processing technique is simple and quick to perform.
pixels. The image was then processed by ImageJ [41]. The pixel intensity was normalised by using linear histogram stretch and then rescaled to between 0 to 255. The final images were converted to 8-bit Portable Network Graphics (PNG) for training, validation and testing. The pre-processing technique is simple and quick to perform.
C. Semi-Automatic Segmentation
Initial annotation was performed using Simpleware-ScanIP (Version 2018.12; Synopsys, Inc., Mountain View, USA). The segmentation procedure consisted of thresholding, background flood-fill and split algorithms. Firstly, thresholding was applied such that only regions containing contrast were considered. Secondly, a seed point was placed within the aorta, and the background flood-fill algorithm was able to segment the coronary arteries and cardiac chambers which were connected to the aorta. Lastly, the split algorithm was performed such that the aorta and the coronary arteries were separated from the cardiac chambers. It should be noted that the split operation may be repeated such that all connected chambers are separated. The workflow of these procedures is displayed in Fig. 2.
FIGURE 2.
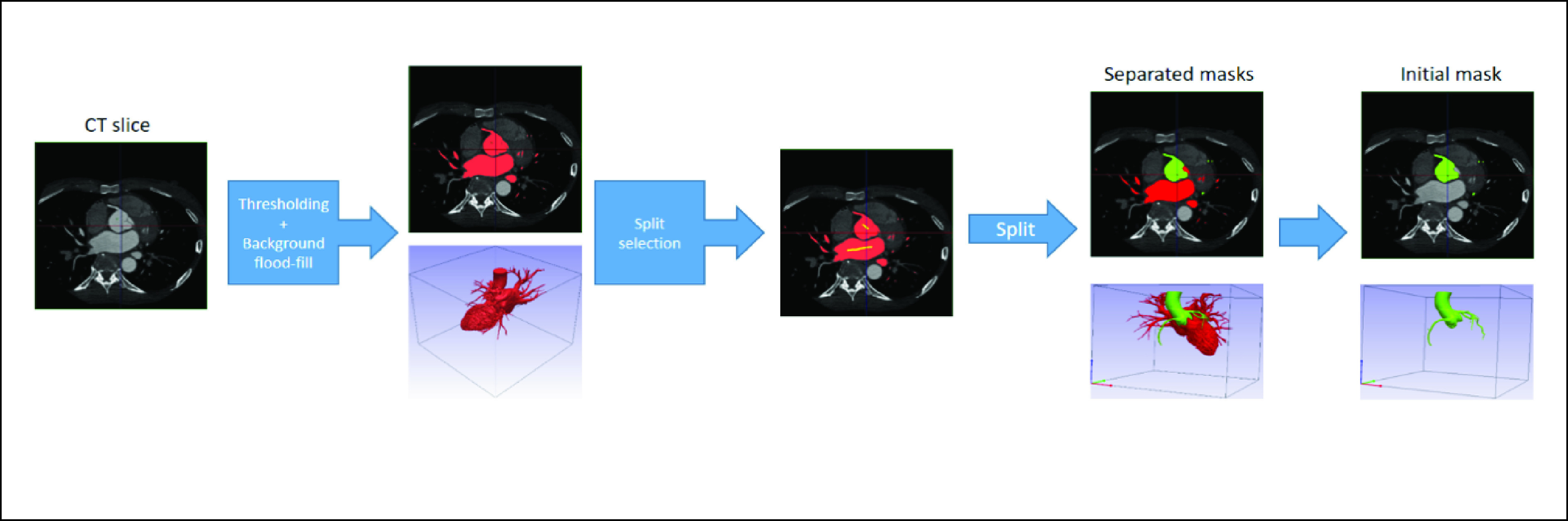
The workflow of initial annotation by using Simpleware-ScanIP.
The initial mask contained the ascending aorta (AA), right coronary artery (RCA), left circumflex artery (LCX) and left coronary artery (LCA). The mask was then fine-tuned manually using 3D Slicer [42]. To acquire the mask containing just the coronary arteries, the AA was removed to leave the RCA, LCX and LCA only. An example of initial and final masks is shown in Fig. 3.
FIGURE 3.
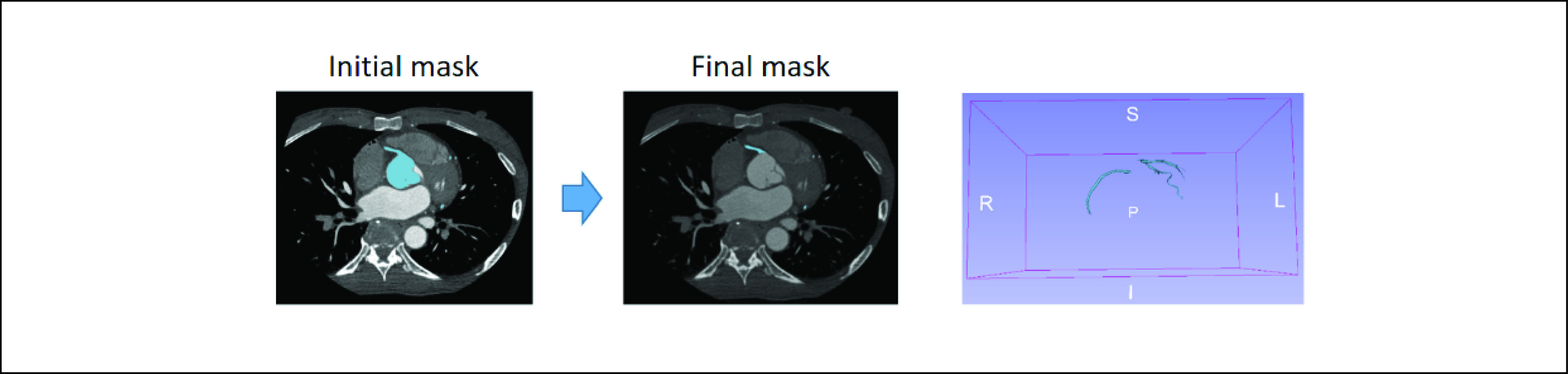
The final coronary artery mask fine-tuned using 3D slicer.
D. Manual Segmentation
The manual segmentation was implemented in Slicer 3D. The annotator highlighted the vessel by identifying the contrast within the CTCA image given an initial mask. The segmented masks were used as the optimum manual (ground-truth) labels.
E. Segmentation Methods
1). Aorta and Coronary Artery Segmentation
The regions of interest that the current work focusses on are the aorta and coronary arteries. There are two scenarios where these masks are used. (1) The combined mask of the aorta and coronary arteries is useful for blood flow estimation using computational fluid dynamics. Aorta segmentations can produce continuous 3D measurements of aortic size and shape which are objective and allow detailed longitudinal comparisons of subtle changes in aortic morphology for various disease states of the aorta. (2) The mask of coronary arteries alone is useful for cardiologists to assess the degree of stenosis in areas where CAD has developed. The proposed method was therefore evaluated on these two segmented masks.
2). Fully Automatic Segmentation
a: Our Proposed Model
Our model is based on the 2D Unet [40]. A Unet is a deep convolutional neural network consisting of down-sample and up-sample paths. The first component of the network extracts spatial features and contexts, while the second component localizes the features by using transposed convolutions. A sigmoid function is used for the final background/foreground classification. We have modified the Unet model in two ways: (1) a batch normalization layer is added to the convolution block; (2) a dropout layer is added before each convolution block. The training process suffered from internal covariate shift. The batch normalisation was able to stabilise the training by normalising the inputs for each mini-batch which was performed by computing the mean and standard deviation of each input variable to a layer per mini-batch. The dropout layer was used to reduce overfitting by setting the weights to be zero randomly. This additional implementation improved the stability and performance of the proposed model. The details of our proposed model is shown in Fig. 4.
FIGURE 4.
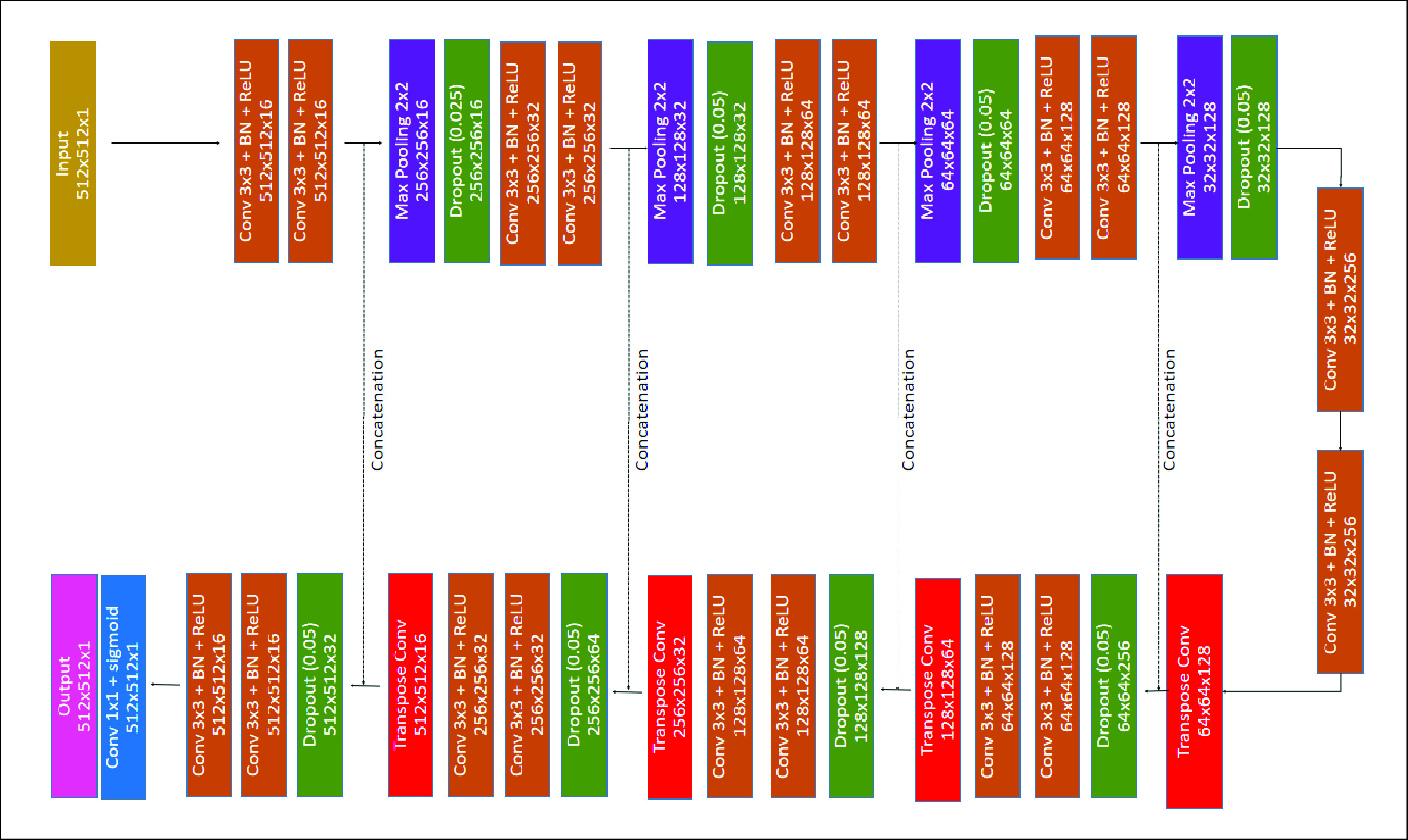
The network architecture of our proposed method.
b: Model Training
55 datasets (n = 13 with no coronary disease, n = 42 with coronary disease) were used for training (80%) and validation (20%) The test data set contained 14 datasets (n = 5 with no coronary disease, n = 9 with coronary disease). There were 11677 slices in the training dataset and 2920 slices in the validation dataset. Slice by slice training was adopted. Two models were trained by using the following optimum manual labels: (1) Aorta and coronary arteries (2) coronary arteries only.
c: Training Implementation
The proposed models were implemented in Tensorflow (v 2.1.0) and Keras (2.3.1) on Linux (Rocks 7). They were executed on a cluster (Intel Xeon Gold 5118, 2.3GHz) with a Tesla V100-PCIE-32GB GPU. The Adam algorithm was used to optimise the proposed models. The learning rate was initially set to 1e-5. 200 epochs were set for model training. The only hyperparameter optimised in this study was the learning rate. It was initially set as 1e-5. When the DSC had stopped improving after three epochs, the learning rate was reduced by a factor of 0.1 using Keras function – ReduceLROnPlateau. The setting was as follows: factor = 0.1, patience = 3, min_lr = 0.00001. Early stopping was executed when the loss was not reduced across 10 consecutive epochs.
d: Loss Function and Performance Evaluation
The combined binary cross entropy (BCE) and dice similarity coefficient (DSC) with equal weights were used as the loss function for deep learning. The segmentation performance was measured by using DSC and IoU metrics which are commonly used to measure the similarity between two segmentations.
e: The Segmentation Prediction Implementation
The prediction was performed by using the trained models above. It was implemented on Tensorflow (v 2.1.0) and Keras (v 2.3.1) on Windows 10 and executed on a machine (Intel i9-9960X, 3.1GHz) with a Nvidia Geforce RTX 2700 GPU. The time required for the prediction was also recorded on a per subject/patient basis.
3). Segmentation Performance and Time Evaluation
The accuracy of the segmentation performance of our proposed method was compared with published accuracies of existing 2D and 3D deep learning models. Our model was also compared with the standard Unet++ [43] and its variant that incorporating Xnet [44] with batch normalisation. It should be noted that the standard skip connections in Unet only combine the decoder feature maps with the same scale feature maps from the encoder, this could limit its ability to capture the intermediate features maps at multiscale levels. The redesigned skip connections mechanism in Unet++ could be used to overcome this limitation and hence a better artery segmentation might be obtained.
For the test dataset, the time required for segmentation and the segmentation performance for our method was compared to semi-automatic segmentation methods. The performance of the segmentation was evaluated by using the DSC and IoU metrics. The Mann-Whitney U Test was performed to evaluate whether there was any difference in segmentation time between automatic and semi-automatic approaches. The analyses were implemented on SPSS (IBM SPSS Statistics for Windows, version 25, IBM, Armonk, NY, USA).
III. Experiments and Results
A. Learning Curve
The learning curves of our model for two scenarios are shown in Fig. 5. No overfitting was found in the training for both scenarios. Some fluctuations of the loss function were found at an early stage of training, though the training became stable later on. This potentially reflects the fact that the training was performed in mini-batches. The training that used the aorta and coronary arteries as ground-truth labels took 125 epochs, while the training using coronary arteries alone as the ground-truth label took only 51 epochs. This indicates that the aorta and coronary arteries have distinct features that took longer to learn in the first scenario.
FIGURE 5.
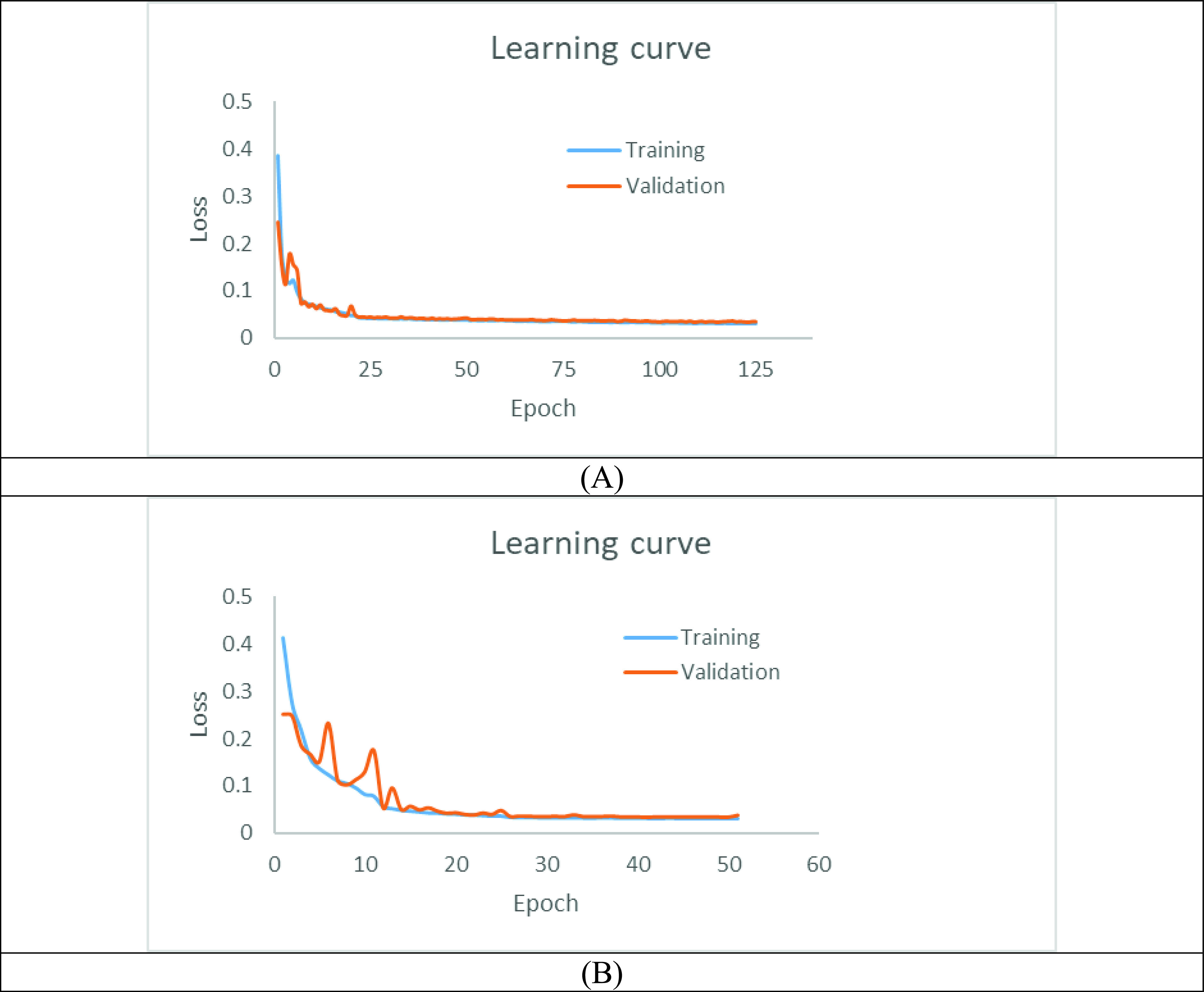
Learning curves: trained with (A) aorta and coronary arteries (B) coronary arteries only.
The accuracy (DSC) during training is shown in Fig. 6. Within the same model, it can be seen that the segmentation performance improves as more features are learnt.
FIGURE 6.
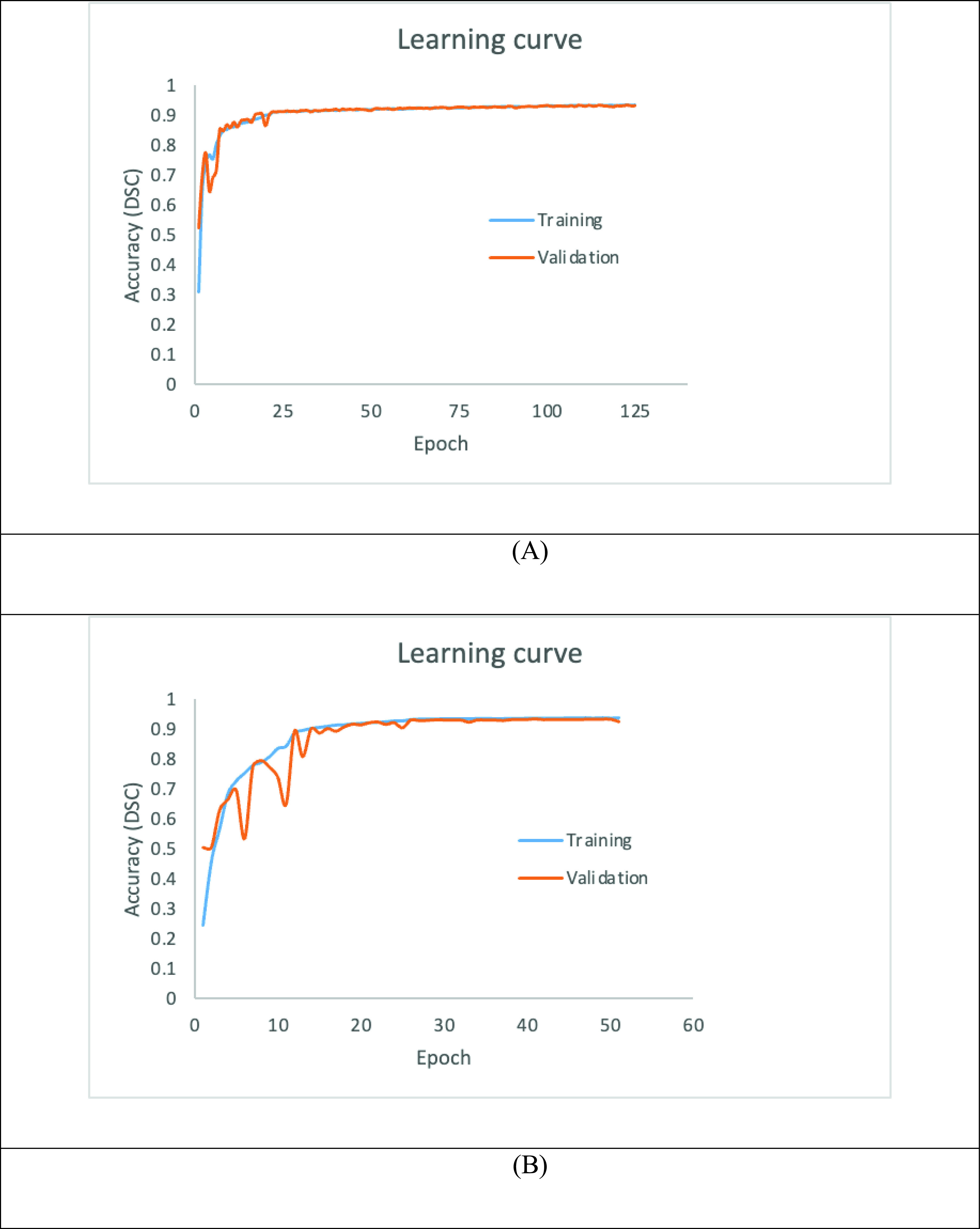
Learning curves – accuracy (DSC): trained with (A) aorta and coronary arteries (B) coronary arteries only.
B. Effect of Network Depth
In general, a deeper network would be able to capture the main/larger diameter artery on the CTCA image, while a shallow network would be able to capture the branch/narrow diameter artery on the CTCA image. For this work, an optimization study was performed (see Table 1). We found that the optimal results (performance on test set) were obtained when using a four-layer (number of layers over which skip connections were used) network.
TABLE 1. Effect of Network Depth to Segmentation Performance.
| Aorta and coronary artery | Coronary artery only | |||||
|---|---|---|---|---|---|---|
| DSC (average) | Training | Validation | Test | Training | Validation | Test |
| 1-layer | 27.35% | 26.95% | 24.14% | 51.48% | 50.62% | 48.14% |
| 2-layer | 66.80% | 67.13% | 67.60% | 66.46% | 68.65% | 73.34% |
| 3-layer | 86.24% | 86.79% | 85.36% | 87.74% | 87.51% | 82.63% |
| 4-layer | 93.62% | 93.33% | 91.20% | 93.82% | 93.41% | 88.80% |
| 5-layer | 95.06% | 94.98% | 88.78% | 94.63% | 94.49% | 88.16% |
| 6-layer | 95.37% | 95.13% | 87.81% | 94.94% | 94.55% | 88.22% |
| 7-layer | 94.83% | 94.99% | 88.97% | 94.56% | 94.39% | 87.58% |
C. Segmentation Performance
Table 2 shows the segmentation performance when the aorta and coronary arteries were segmented. The accuracy of our method and Simpleware-ScanIP are 91.20% and 99.40% respectively. The semi-automatic approach performed better than our method when both the aorta and coronary arteries were present in the mask.
TABLE 2. Segmentation Performance – Mask Contains Aorta and Coronary Arteries.
| With aorta | DSC (average) | IoU (average) |
|---|---|---|
| Simpleware-ScanIP | 99.40% | 98.81% |
| Our method | 91.20% | 83.82% |
The performance for segmentation of the coronary arteries alone is shown in Table 3. The accuracy of our method and Simpleware-ScanIP were 88.80% and 73.22% respectively. Our method performs better than the semi-automatic approach when just the coronary arteries are present in the mask.
TABLE 3. Segmentation Performance – Mask Contains Coronary Arteries Only.
| Without aorta | DSC (average) | IoU (average) |
|---|---|---|
| Simpleware-ScanIP | 73.22% | 57.75% |
| Our method | 88.80% | 79.85% |
The results demonstrate that a semi-automatic approach is good at segmenting the aorta. The semi-automatic approach was limited in its ability to segment the coronary arteries, but as the aorta occupied most of the volume of the mask, the overall segmentation accuracy remained high. Our method performed well when attempting to segment the coronary arteries alone. This suggests that our model has the ability to utilise other features (i.e. shape) to recognise the coronary arteries, while the semi-automatic approach relies solely on pixel density. If the contrast within the coronary artery is not bright enough, the semi-automatic approach will miss some segments of the coronary artery (See Fig. 7).
FIGURE 7.
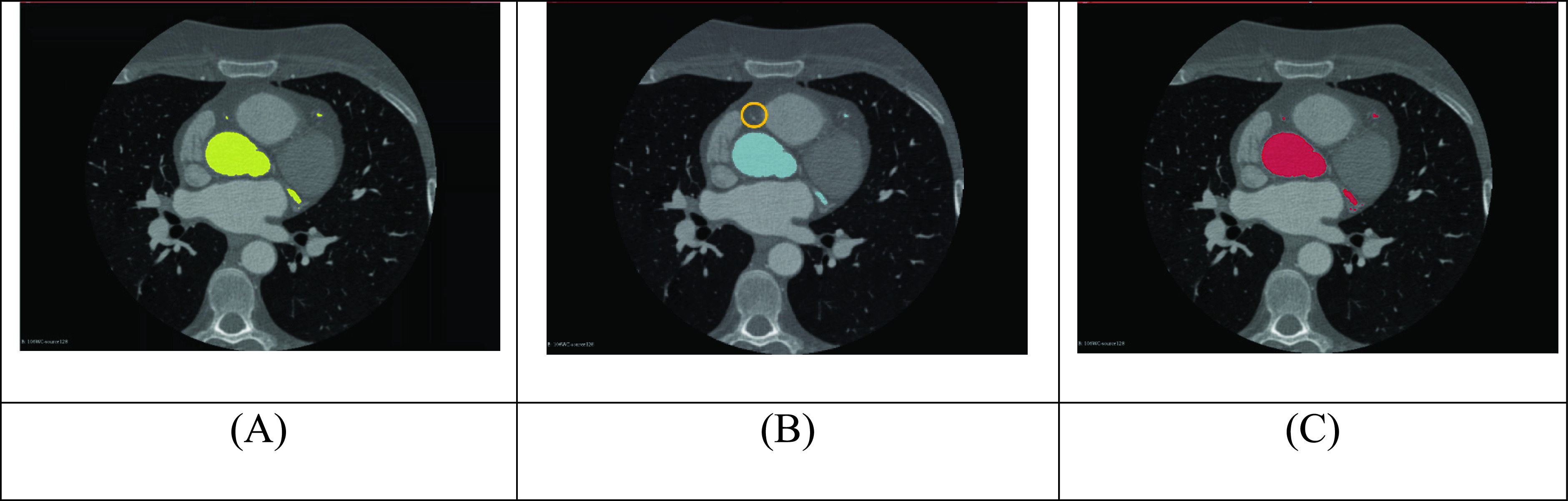
(A) Ground-truth mask (yellow) (B) Mask from Simpleware-ScanIP (blue) with a missing vessel (orange circle) (C) Mask from our model (red).
The segmentation results of patients 1 and 2 are displayed in Figs. 8 and 9. From Fig. 8, it is clear that our method can segment the aorta and coronary arteries, with a result very close to the optimal manual label. The Simpleware–ScanIP segments the aorta with good accuracy while some segments of the coronary arteries are missing.
FIGURE 8.
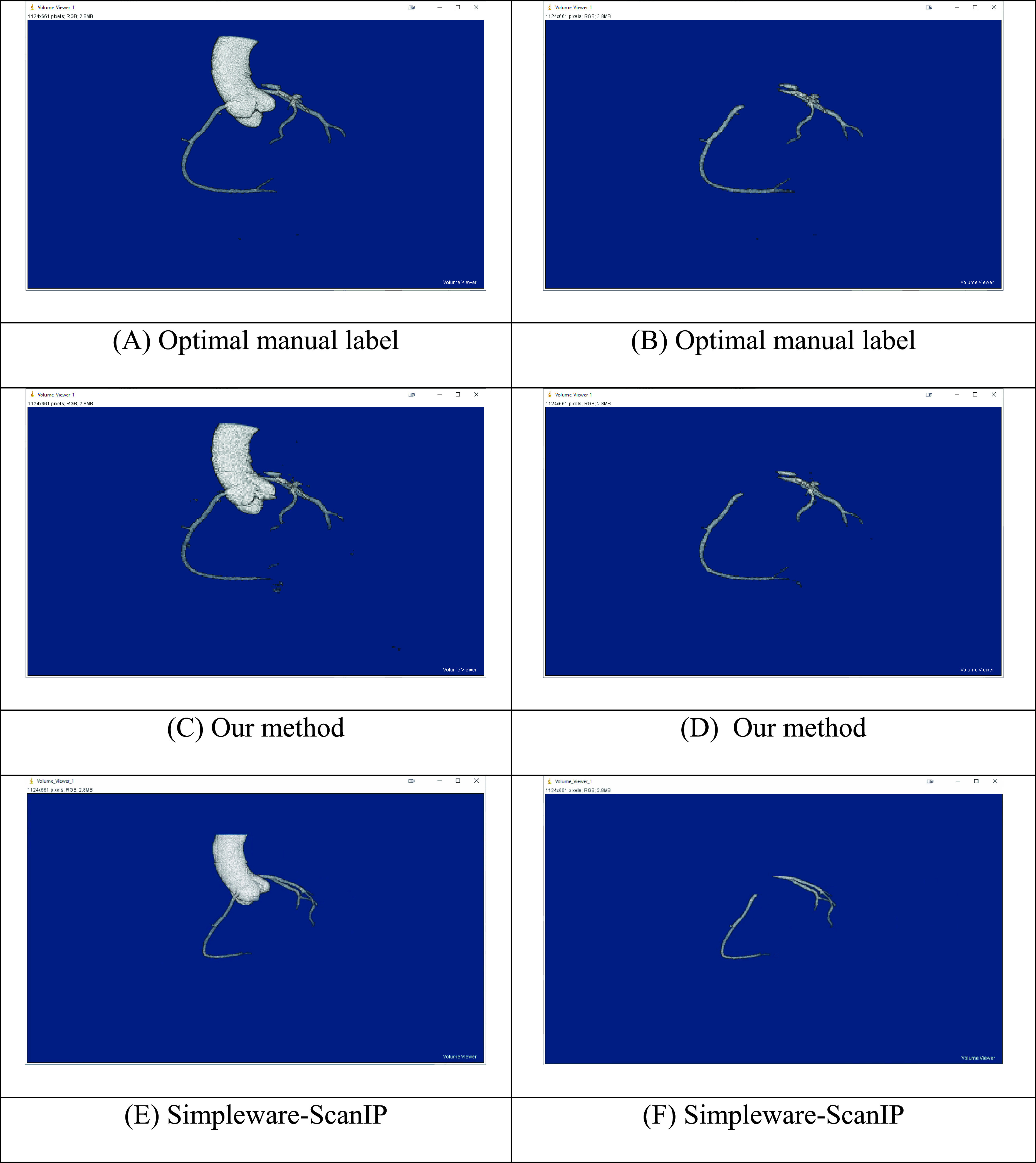
Segmentation results of patient 1. Segmentation of aorta and coronary arteries: (A) Optimal manual label, (C) Our method, (E) Simpleware–ScanIP. Segmentation of coronary arteries only: (B) Optimal manual label, (D) Our method, (F) Simpleware–ScanIP.
FIGURE 9.
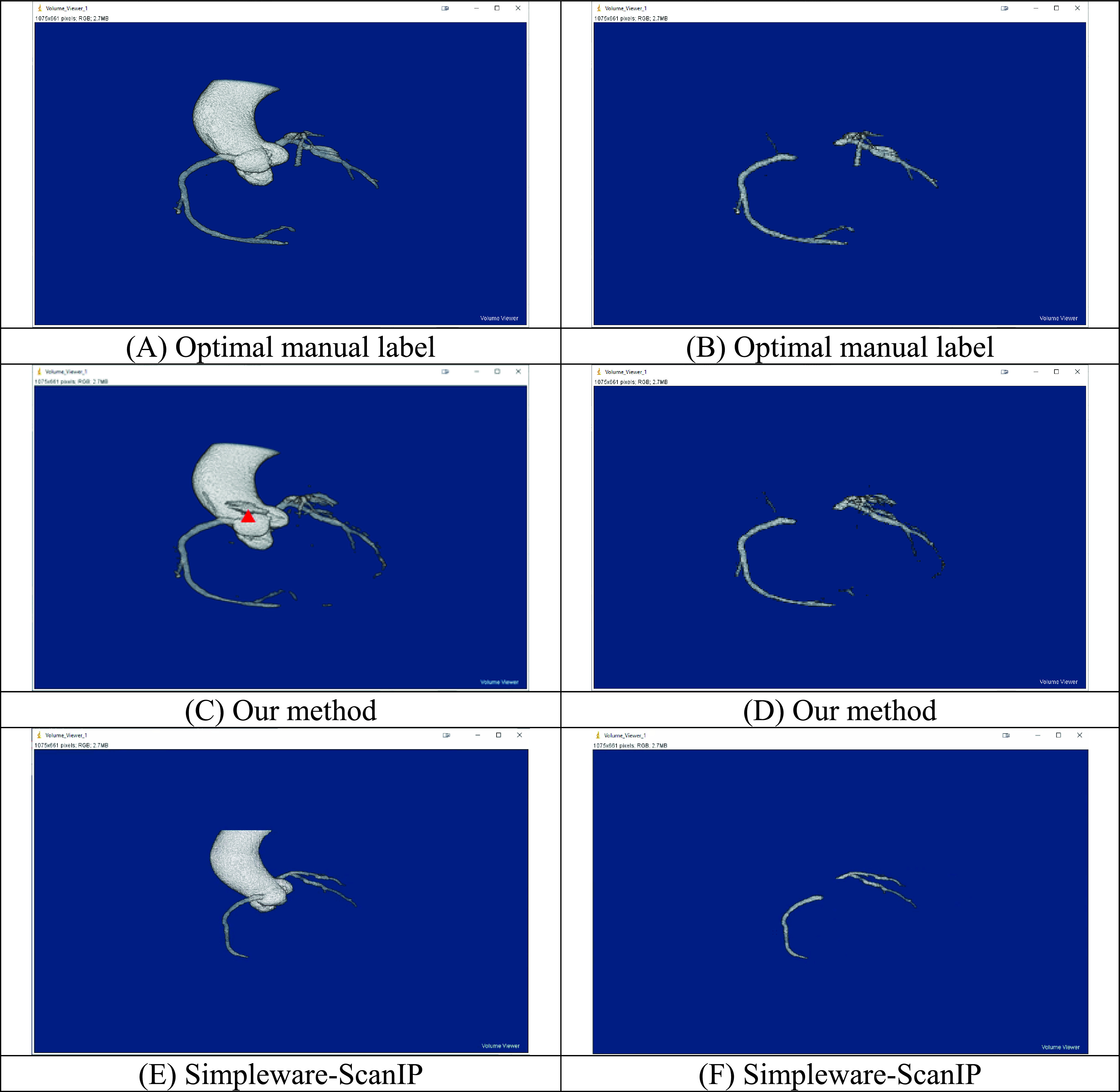
Segmentation results of Patient 2: with aorta and coronary arteries (A) Optimal manual label (C) Our method (E) Simpleware–ScanIP, with coronary arteries only (B) Optimal manual label (D) Our method (F) Simpleware–ScanIP.
For patient 2 (Fig. 9), our method can segment the aorta and coronary arteries well, but the segmentation incorporates an artefact (red arrowhead). When segmenting the coronary arteries alone, some segments of the coronary arteries are missing in the segmentation. As expected, the aorta segmentation is good when using the semi-automatic method, while the segmentation of the coronary arteries is relatively poor. It should be noted that the artefact present in the segmentation of the aorta and coronary arteries using our method can be easily removed by excluding the non-connected components of the mask.
D. Segmentation Performance Comparison With Unet++
Table 4 shows the segmentation performance of the standard Unet++ and its variant [Xnet with batch normalisation]. From Table 4, the performance of the standard Unet++ is the weakest. This may be due to the fact that batch normalisation has not been adopted to reduce the internal covariate shift which has affected the training significantly. While the Unet++ variant performed much better than standard Unet++, its performance remains slightly worse than our model. This suggests that learning through semantically similar feature maps may not be useful for artery segmentation. The segmentation of the aorta and/or coronary artery using the Unet++ [Xnet/BN] is shown in Fig. 10.
TABLE 4. Segmentation Performance of Standard Unet++ and its Variant [Xnet With Batch Normalisation].
| Aorta and coronary artery | Coronary artery only | ||||||
|---|---|---|---|---|---|---|---|
| DSC (average) | Training | Validation | Test | Training | Validation | Test | |
| Standard Unet++ (L1) | 27.47% | 26.95% | 24.14% | 51.48% | 50.62% | 48.14% | |
| Standard Unet++ (L4) | 27.47% | 26.95% | 24.14% | 51.48% | 50.62% | 48.14% | |
| Unet++ [Xnet/BN] (L4) | 85.81% | 79.61% | 86.27% | 86.67% | 76.27% | 78.11% | |
FIGURE 10.
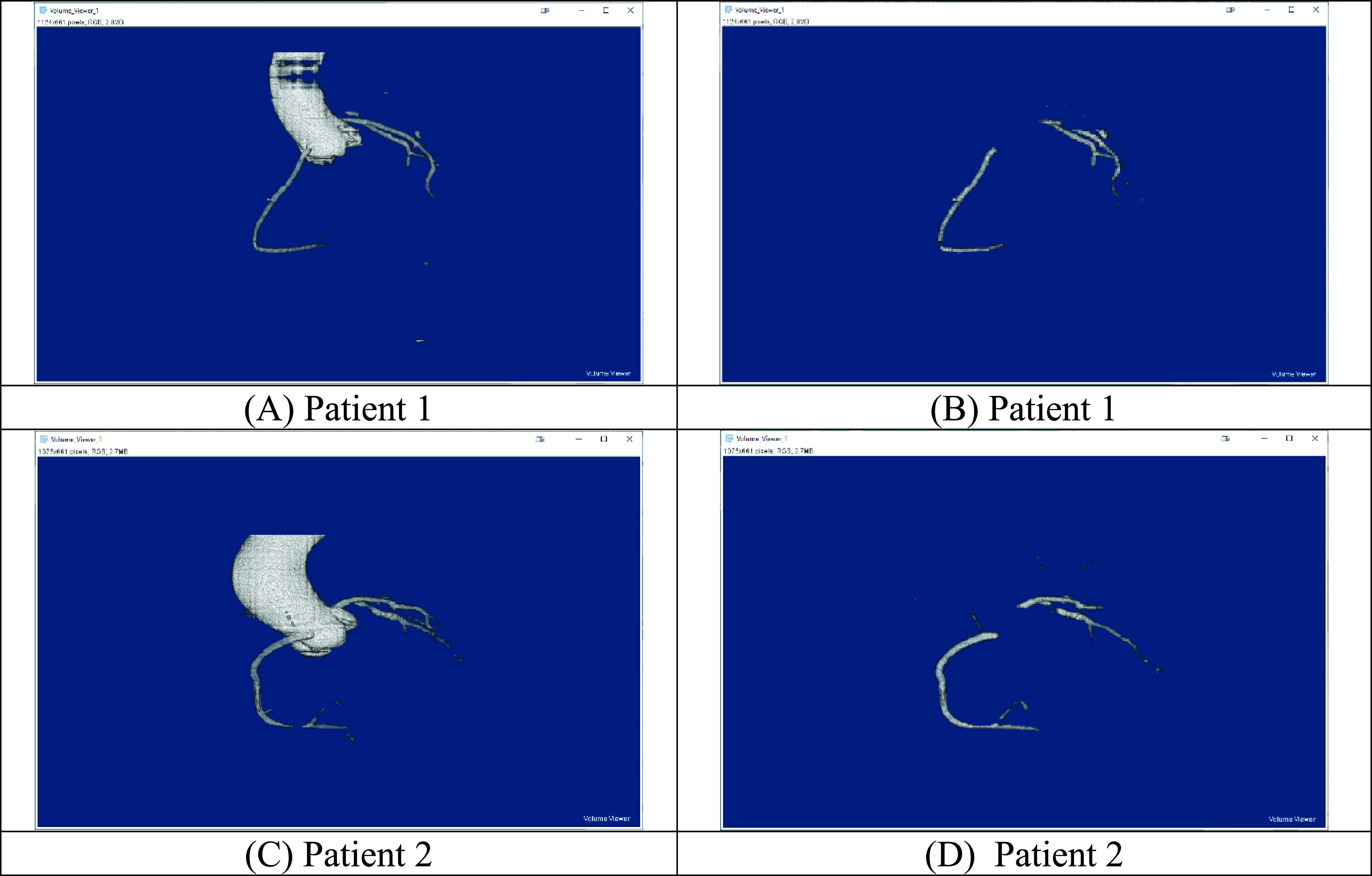
Segmentation results by Unet++ [Xnet/BN]. Segmentation of aorta and coronary arteries: (A) Patient 1, (C) Patient 2. Segmentation of coronary arteries only: (B) Patient 1, (D) Patient 2.
E. Segmentation Time
The segmentation time of our proposed method compared to semi-automatic segmentation is shown in Table 5 (aorta and coronary arteries) and Table 6 (coronary arteries only) respectively. The mask prediction using our method was significantly faster than the Simpleware-ScanIP for both segmentation scenarios (p-value < 0.001), taking less than 4 seconds on average to predict the aorta and/or coronary arteries masks. Additionally, the segmentation time was between 40s and 141s when run on a CPU with multi-cores.
TABLE 5. Segmentation Time – Mask Contains Aorta and Coronary Arteries.
| With aorta | Time (average) | Time (SD) |
|---|---|---|
| Simpleware-ScanIP | 203.07s | 101.69s |
| Our method (GPU) | 3.29s | 0.47s |
| Our method (CPU only – 2 cores, 4 threads) | 138.57s | 15.74s |
| Our method (CPU only – 16 cores, 32 threads) | 40.93s | 4.71s |
TABLE 6. Segmentation Time – Mask Contains Coronary Arteries Only.
| Without aorta | Time (average) | Time (SD) |
|---|---|---|
| Simpleware-ScanIP | 345.00s | 113.45s |
| Our method (GPU) | 3.36s | 0.50s |
| Our method (CPU only – 2 cores, 4 threads) | 140.07s | 16.90s |
| Our method (CPU only – 16 cores, 32 threads) | 40.93s | 4.75s |
IV. Discussions
A deep learning model based on a 2D Unet has been developed to segment the aorta and/or coronary arteries on cardiac CTCA images. Two models were trained to segment ROIs in two scenarios - (1) the aorta and coronary arteries (2) coronary arteries only. Our method demonstrates 91.20% and 88.80% DSC accuracy on scenarios 1 and 2 respectively. This suggests that our method can segment the aorta and/or coronary arteries with high accuracy.
Current deep learning approaches require high computational resources, which are difficult to implement in standard hospital networks. Therefore, we developed an alternative deep learning approach (2D Unet) that is feasible for implementation in a hospital network and which importantly maintains good segmentation accuracy.
Compared with a published 3D deep learning model [22] developed to segment the aorta and coronary arteries, which uses a 3D global feature embedded network with active contour loss, the performance of our method was similar (DSC: 91.20% (our method) vs 91.43% ([22])). Our proposed method utilised a smaller number of network parameters resulting in more efficient training and a faster prediction time compared to the published 3D model. Our method requires less GPU memory, which is a common limitation when training and implementing a 3D model. It should also be noted that our method does not require GPUs for deployment, which favours its application in hospital networks where typically only CPUs are available.
The performance of our method was also compared with the 2D RCNN + 2D Unet technique [36] for scenario 2. Our method showed comparable performance (IoU: 79.85% (our method) vs 84.36% ([36])), but does not require implementation of a sequence model to detect the coronary arteries. Implementing an additional sequence model would increase the computational complexity and hence decrease the algorithmic efficiency. It should be noted that the difference in reported accuracy between these methods are likely to relate to the different test sets that were evaluated.
Compared with semi-automatic methods, our model performance is degraded when segmenting the aorta and coronary arteries. However, our model gives improved accuracy when segmenting the coronary arteries alone. The findings highlight the importance of evaluating segmentation performance of large vessels and small vessels separately to reduce the potential bias of segmentation performance metrics. In terms of the prediction time, our proposed model provided the fastest prediction when compared with the semi-automatic method. Though the time difference is statistically significant, it may be negligible from a clinical perspective.
Our study utilised a larger sample size for training and prediction when compared to the published 2D approach. This allows for a more generalizable model and therefore a more reliable prediction. It should be noted that two cases were excluded in our analysis as the Simpleware ScanIP failed to segment the vessels. Our deep learning model however was still able to segment the aorta and coronary arteries adequately.
There are several limitations to this study. The design of the study was retrospective, and accordingly it may have suffered from patient selection bias. The ground-truth labels of the study were obtained by manual annotation and it is possible that the accuracy of the labels were potentially biased due to the annotator’s experience. The performance of our model was compared with existing models using different datasets. The lack of openly available code for other models precluded direct comparisons of models on the same dataset. Although our method can predict the segmented mask with good accuracy, visual inspection of the imaging by experts is still required. Currently, small regions of the proximal coronary artery are occasionally missed when using our models. Further improvements could be made by incorporating an attention gate to our model, which could allow the network to focus more closely on the coronary arteries during training.
V. Conclusion
Our study demonstrates that a 2D UNET model is able to segment the coronary arteries efficiently and with good accuracy. It has the advantage that it can be deployed within hospital computer networks where GPUs are not available. Our study is a first essential stage of work to develop fully automatic detection and classification systems for coronary artery disease by using computer-based deep learning algorithms. The aims of our future work would be to extend our model for coronary artery disease severity classification which could speed up the patient pathway for referrals with chest pain in A&E departments.
Acknowledgment
The authors would like to thank Mr Xiaoxia Yang for his assistance with data processing. For the purpose of open access, the author has applied a CC-BY public copyright licence to any author accepted manuscript version arising from this submission.
Biographies

Wing Keung Cheung received the B.Sc. and M.Phil. degrees in physics from the Hong Kong University of Science and Technology, in 2003 and 2005, respectively, and the Ph.D. degree in bioengineering from Imperial College London, in 2016. He was a Postdoctoral Research Assistant with the School of Engineering and Materials Science, Queen Mary University of London. He is currently a Research Fellow with the Centre of Medical Image Computing, University College London. His research interests include ultrasound imaging, ultrasound elastography, biomechanics, and medical image computing.

Robert Bell received the MRCP and Ph.D. degrees. He is the Principal Research Fellow with the Hatter Cardiovascular Institute and an Honorary Cardiology Consultant with University College London Hospitals, including the National Hospital for Neurology and Neurosurgery.
His research interests include cytoprotection and ischemia-reperfusion injury. His current projects are centered on the identification of novel cellular protection pathways and their clinical translation, as well as determining the impact of hyperglycemia upon myocardial infarction and the cardioprotective properties of sodium/glucose linked transporter (SGLT) inhibitors. From this work, he has developed a clinical interest in the translation of these protective strategies in both the heart and brain. In addition, his research interest includes red-cell cardiology, including Thalassemia, in which he works closely with Dr. Malcolm Walker.
Dr. Bell serves as the Chairman for the Barts Heart Centre Peer Review and BioResearch Boards and as an Active Member of the European Society of Cardiology Stroke Working Group, British Cardiology Society, British Society for Cardiovascular Research, and International Society for Heart Research.

Arjun Nair is currently a Consultant Radiologist with a sub-specialty interest in cardiothoracic imaging with University College London Hospital, following his initial appointment as a Consultant Radiologist at Guy’s and St. Thomas’ NHS Foundation Trust, in 2013. He qualified from the University of Edinburgh Medical School, in 2001, and was awarded the MRCP, in 2005. He undertook radiology training at St. George’s Hospital, passing the FRCR in 2008. He was awarded the Thoracic Imaging Fellowship at Royal Brompton Hospital, from 2011 to 2013, and during this time completed his thesis on “Computed Tomography Reading Strategies in Lung Cancer Screening,” for which he was awarded an M.D. degree by the University of Edinburgh in 2014. In the course of his research, He has been actively involved in the UK Lung Screening pilot trial. He has presented at various national and international meetings and has been awarded prizes for presentations at the Radiological Society of North America, the European Society of Thoracic Imaging, and the World Congress of Thoracic Imaging. He is the author/coauthor of several peer-reviewed scientific papers and book chapters on cardiothoracic imaging. He is currently an Advisory Editor to three scientific journals, namely, European Radiology, Clinical Radiology, and British Journal of Radiology.

Leon J. Menezes is currently a Lead in nuclear cardiology with the Bart’s Heart Centre, and a Consultant Radiologist and a Nuclear Medicine Physician with University College London Hospitals. He read physiology and medicine at Oxford. He is trained in clinical radiology at Guy’s & St. Thomas’ and in nuclear medicine with University College London Hospital. He was previously a European Society of Radiology Fellow in cardiac imaging with La Sapienza, Rome. He is currently the President of the British Nuclear Cardiology Society.
His clinical interests include multimodality and hybrid cardiac imaging, using SPECT and PET, with cardiac CT and cardiac MRI. His research interests include the application of these hybrid modalities to both rare and common cardiovascular diseases, such as sarcoidosis, amyloidosis, atherosclerosis, and infection, with funding from the Royal College of Radiologists and the British Heart Foundation.

Riyaz Patel received the degree from King’s College London, in 2001. He is a consultant cardiologist and clinical academic working with UCLH and Barts Health NHS trusts and University College London (UCL). He was subsequently elected to the Fellowship of the Royal College of Physicians and the European Society of Cardiology, in 2018. Having trained and worked across major teaching hospitals in London and the USA, he is highly experienced in managing all general cardiology conditions, with a focus on advanced cardiac imaging and premature coronary artery disease.
Aside from his role as a cardiologist, He is also a fully funded BHF Clinician Scientist and a Professor of cardiology with UCL, conducting research into the causes of heart disease, with a focus on cardiovascular risk and the genetics of coronary heart disease. He leads global initiatives and consortia in this field with a track record of research funding and high-profile research publications.
Prof. Patel clinically sees and manages patients with all general cardiology problems, including chest pain breathlessness, palpitations, and high blood pressure, in both outpatient and acute settings. He is a recognized specialist in advanced cholesterol management, including familial hypercholesterolemia (FH), being one of only a handful of cardiologists in the U.K. with this expertise. In addition, in line with his research expertise, he investigates and manages patients with possible genetically driven coronary artery disease, including early-onset heart disease or those who are well but have a strong family history of early heart disease. He founded and leads a new cardiovascular prevention service at the Barts Heart Centre, a service modeled on those available in the USA at centers such as the Mayo Clinic, to serve the local population of East London. Aside from general cardiology, he is also a fully accredited and recognized specialist in advanced echocardiography, which includes stress (exercise and pharmacological) and transoesophageal echocardiography (TOE), and cardiac and coronary CT angiography, and provides these tests with the Barts Heart Centre.

Simon Wan qualified in radiology from Kings College Hospital, London. Following a Fellowship in nuclear medicine, he joined the Institute of Nuclear Medicine as a Consultant Radionuclide Radiologist.

Kacy Chou received the B.Sc. degree in management science and the M.Sc. degree in business analytics with specialization in computer science from the University College London, in 2018 and 2019, respectively. She is currently working as a Software Engineer at J.P. Morgan, Hong Kong. Her research interests include computer vision and machine learning.

Jiahang Chen received the M.Sc. degree in mechanical engineering from University College London, in 2019. His research interests include the application of machine learning in medical image analysis and computational modeling, especially in the cardiovascular area.

Ryo Torii received the Ph.D. degree in mechanical engineering from the University of Tokyo, in 2003. He is currently an Associate Professor with the Department of Mechanical Engineering, University College London (UCL). Following the period that he spent as a Researcher with the University of Tokyo and Imperial College London, he joined UCL, in 2012. His expertise is the development and application of computational methods in cardiovascular biomedicine, with a special focus on aortic and coronary artery diseases.

Rhodri H. Davies received the B.Sc. degree in computer science, the M.Res. degree in informatics, and the Ph.D. degree in medical image analysis from The University of Manchester, in 1998, 1999, and 2002, respectively. After a brief Postdoctoral Fellowship with the Howard Florey Institute, he received the MBBCh degree (Hons.) in clinical medicine from Cardiff University, in 2009. He is currently a Consultant Cardiologist with University Hospital of Wales, specializing in cardiac MRI and echocardiography, and an Associate Professor with the Institute of Cardiovascular Science at University College London. His research centers on using machine learning techniques to improve the way we interpret medical images and find new ways of understanding normal physiology and disease.

James C. Moon is currently a Professor of cardiology, UCL, London, the Clinical Director of imaging with Barts Heart Centre (BHC), and the Medical Director of Chenies Mews Imaging Centre. Formerly, he set up and led the Heart Hospital Imaging Centre. Trained at Cambridge and Oxford, his M.D. degree at Imperial was “myocardial tissue characterization by CMR.” His current research interests include measuring diffuse myocardial fibrosis (health and disease), iron, fat infiltration, and amyloid using “T1 mapping.” He has published more than 300 articles and works in partnership with Pharma to develop surrogate endpoints for clinical trials including first in man studies. Another interest is in making scans faster/cheaper and in improving/maintaining quality as CMR expands. He sets up and leads the international T1 mapping development group aiming to “coordinate activity to the goal of being able to change therapy using CMR endpoints.” He is a former Board Member of SCMR and the immediate past President of the British equivalent, BSCMR. He is also a former member of NICE.

Daniel C. Alexander received the B.Sc. degree in mathematics from the University of Oxford, in 1993, and the M.Sc. and Ph.D. degrees in computer science from UCL, in 1994 and 1998, respectively. He is currently the Director of the UCL Centre for Medical Image Computing (CMIC), University College London (UCL), and a Professor of imaging science with the Department of Computer Science, UCL. His expertise is in computational modeling, machine learning, imaging, and image analysis. He worked in a postdoctoral position with the University of Pennsylvania, until 2000 when he returned to London to take up an academic position. He became a Full Professor, in 2010, and the Director of CMIC, in 2015. He is currently an Associate Editor for Magnetic Resonance in Medicine and a Senior Fellow of the International Society for Magnetic Resonance in Medicine.

Joseph Jacob received the M.D. degree (research) with Imperial College under Prof. David Hansell with Royal Brompton Hospital, in 2016. He qualified in medicine from Imperial College, worked for Medecins sans Frontieres for two years, and completed radiology training with Kings College Hospital, London. His research with the Centre for Medical Image Computing, University College London centers on computational image analysis of the lungs and heart on CT imaging. He has coauthored over 80 articles, won national and international awards for his work, and was awarded the 2017 Best Thesis Prize by the National Heart and Lung Institute. He was awarded a five-year Wellcome Trust Clinical Research Career Development Fellowship, in 2018, for his M.D. degree.
Funding Statement
This work was supported in part by the Wellcome Trust under Grant 209553/Z/17/Z, and in part by the National Institute for Health Research (NIHR) University College London Hospital (UCLH) Biomedical Research Centre, U.K. The work of Leon J. Menezes, Joseph Jacob, Wing Keung Cheung, Ryo Torii, Rhodri H. Davies, Riyaz Patel and Daniel C. Alexander was supported by the NIHR BRC at University College London.
Contributor Information
Wing Keung Cheung, Email: tony.cheung@ucl.ac.uk.
Joseph Jacob, Email: j.jacob@ucl.ac.uk.
References
- [1].World Health Organization. (2016). The Top 10 Causes of Death. [Online]. Available: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death
- [2].Bom M. J., van der Heijden D. J., Kedhi E., van der Heyden J., Meuwissen M., Knaapen P., Timmer S. A. J., and van Royen N., “Early detection and treatment of the vulnerable coronary plaque: Can we prevent acute coronary syndromes?” Circulation, Cardiovascular Imag., vol. 10, no. 5, May 2017, Art. no. e005973. [DOI] [PubMed] [Google Scholar]
- [3].Senior R., Monaghan M., Becher H., Mayet J., and Nihoyannopoulos P., “Stress echocardiography for the diagnosis and risk stratification of patients with suspected or known coronary artery disease: A critical appraisal. Supported by the British society of echocardiography,” Heart, vol. 91, no. 4, pp. 427–436, Apr. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Saeed M., Van T. A., Krug R., Hetts S. W., and Wilson M. W., “Cardiac MR imaging: Current status and future direction,” Cardiovascular Diagnosis Therapy, vol. 5, pp. 290–310, Aug. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Meyersohn N., Janjua S., Staziaki P., Bittner D., Takx R., Weiner R., Wasfy J., Hoffmann U., and Ghoshhajra B., “Medical management of non-obstructive coronary artery disease on coronary CT angiography: Insight from a modern emergency department clinical registry,” J. Amer. College Cardiol., vol. 67, no. 13, p. 1763, Apr. 2016. [Google Scholar]
- [6].Wallis A., Manghat N., and Hamilton M., “The role of coronary CT in the assessment and diagnosis of patients with chest pain,” Clin. Med., vol. 12, no. 3, pp. 222–229, Jun. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].National Institute for Health and Care Excellence (NICE). (2016). Recent-Onset Chest Pain of Suspected Cardiac Origin: Assessment and Diagnosis Clinical Guideline [CG95]. [Online]. Available: https://www.nice.org.uk/guidance/cg95 [PubMed]
- [8].(2020). Radiology Workforce Census. [Online]. Available: https://www.rcr.ac.uk/press-and-policy/policy-priorities/workforce/radiology-workforce-census
- [9].Pugliese F., Hunink M. G. M., Gruszczynska K., Alberghina F., Malagó R., van Pelt N., Mollet N. R., Cademartiri F., Weustink A. C., Meijboom W. B., Witteman C. L. M., de Feyter P. J., and Krestin G. P., “Learning curve for coronary CT angiography: What constitutes sufficient training?” Radiology, vol. 251, pp. 359–368, May 2009. [DOI] [PubMed] [Google Scholar]
- [10].Saur S. C., Alkadhi H., Stolzmann P., Baumüller S., Leschka S., Scheffel H., Desbiolles L., Fuchs T. J., Székely G., and Cattin P. C., “Effect of reader experience on variability, evaluation time and accuracy of coronary plaque detection with computed tomography coronary angiography,” Eur. Radiol., vol. 20, no. 7, pp. 1599–1606, Jul. 2010. [DOI] [PubMed] [Google Scholar]
- [11].Morris P. D., Narracott A., von Tengg-Kobligk H., Soto D. A. S., Hsiao S., Lungu A., Evans P., Bressloff N. W., Lawford P. V., Hose D. R., and Gunn J. P., “Computational fluid dynamics modelling in cardiovascular medicine,” Heart, vol. 102, pp. 18–28, Jan. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fearon W. F., Achenbach S., Engstrom T., Assali A., Shlofmitz R., Jeremias A., Fournier S., Kirtane A. J., Kornowski R., Greenberg G., and Jubeh R., “Accuracy of fractional flow reserve derived from coronary angiography,” Circulation, vol. 139, pp. 477–484, Jan. 2019. [DOI] [PubMed] [Google Scholar]
- [13].Li Z., Zhang J., Xu L., Yang W., Li G., Ding D., Chang Y., Yu M., Kitslaar P., Zhang S., Reiber J. H. C., Arbab-Zadeh A., Yan F., and Tu S., “Diagnostic accuracy of a fast computational approach to derive fractional flow reserve from coronary CT angiography,” JACC, Cardiovascular Imag., vol. 13, no. 1, pp. 172–175, Jan. 2020. [DOI] [PubMed] [Google Scholar]
- [14].Itu L., Rapaka S., Passerini T., Georgescu B., Schwemmer C., Schoebinger M., Flohr T., Sharma P., and Comaniciu D., “A machine-learning approach for computation of fractional flow reserve from coronary computed tomography,” J. Appl. Physiol., vol. 121, no. 1, pp. 42–52, Jul. 2016. [DOI] [PubMed] [Google Scholar]
- [15].Zhou C., Chan H.-P., Chughtai A., Patel S., Hadjiiski L. M., Wei J., and Kazerooni E. A., “Automated coronary artery tree extraction in coronary CT angiography using a multiscale enhancement and dynamic balloon tracking (MSCAR-DBT) method,” Comput. Med. Imag. Graph., vol. 36, no. 1, pp. 1–10, Jan. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shahzad R., Kirişli H., Metz C., Tang H., Schaap M., van Vliet L., Niessen W., and van Walsum T., “Automatic segmentation, detection and quantification of coronary artery stenoses on CTA,” Int. J. Cardiovascular Imag., vol. 29, no. 8, pp. 1847–1859, Dec. 2013. [DOI] [PubMed] [Google Scholar]
- [17].Öksüz İ., Ünay D., and Kadipaşaoğlu K., “A hybrid method for coronary artery stenoses detection and quantification in CTA images,” in Proc. MICCAI Workshop 3d Cardiovascular Imag., MICCAI Segmentation, 2012, pp. 1–8. [Google Scholar]
- [18].Kitamura Y., Li Y. Z., Ito W., and Ishikawa H., “Coronary lumen and plaque segmentation from CTA using higher-order shape prior,” in Medical Image Computing and Computer-Assisted Intervention—MICCAI 2014 (Lecture Notes in Computer Science), vol. 8673. Cham, Switzerland: Springer; 2014, pp. 339–347. [DOI] [PubMed] [Google Scholar]
- [19].Tian Y., Pan Y., Duan F., Zhao S., Wang Q., and Wang W., “Automated segmentation of coronary arteries based on statistical region growing and heuristic decision method,” BioMed Res. Int., vol. 2016, pp. 1–7, Jan. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lesage D., Angelini E. D., Bloch I., and Funka-Lea G., “A review of 3D vessel lumen segmentation techniques: Models, features and extraction schemes,” Med. Image Anal., vol. 13, no. 6, pp. 819–845, Dec. 2009. [DOI] [PubMed] [Google Scholar]
- [21].Clinical Radiology UK Workforce Census Report 2018. Accessed: Mar. 12, 2020. https://www.rcr.ac.uk/publication/clinical-radiology-uk-workforce-census-report-2018
- [22].Gu J., Fang Z., Gao Y., and Tian F., “Segmentation of coronary arteries images using global feature embedded network with active contour loss,” Comput. Med. Imag. Graph., vol. 86, Dec. 2020, Art. no. 101799. [DOI] [PubMed] [Google Scholar]
- [23].Huang W., Huang L., Lin Z., Huang S., Chi Y., Zhou J., Zhang J., Tan R.-S., and Zhong L., “Coronary artery segmentation by deep learning neural networks on computed tomographic coronary angiographic images,” in Proc. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc., Jul. 2018, pp. 608–611. [DOI] [PubMed] [Google Scholar]
- [24].Mahmud M., Kaiser M. S., McGinnity T. M., and Hussain A., “Deep learning in mining biological data,” Cogn Comput, vol. 13, pp. 1–33, Jan. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Litjens G., Kooi T., Bejnordi B. E., Setio A. A. A., Ciompi F., Ghafoorian M., van der Laak J. A. W. M., van Ginneken B., and Sánchez C. I., “A survey on deep learning in medical image analysis,” Med. Image Anal., vol. 42, pp. 60–88, Dec. 2017. [DOI] [PubMed] [Google Scholar]
- [26].Ali H. M., Kaiser M. S., and Mahmud M., “Application of convolutional neural network in segmenting brain regions from MRI data,” in Brain Informatics (Lecture Notes in Computer Science), vol. 11976. Cham, Switzerland: Springer, 2019, pp. 136–146. [Google Scholar]
- [27].Mahmud M., Kaiser M. S., Hussain A., and Vassanelli S., “Applications of deep learning and reinforcement learning to biological data,” IEEE Trans. Neural Netw. Learn. Syst., vol. 29, no. 6, pp. 2063–2079, Jun. 2018. [DOI] [PubMed] [Google Scholar]
- [28].Chen C., Qin C., Qiu H., Tarroni G., Duan J., Bai W., and Rueckert D., “Deep learning for cardiac image segmentation: A review,” Frontiers Cardiovascular Med., vol. 7, p. 25, Mar. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Moeskops P., Wolterink J. M., van der Velden B. H. M., Gilhuijs K. G. A., Leiner T., Viergever M. A., and Išgum I., “Deep learning for multi-task medical image segmentation in multiple modalities,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent., Cham, Switzerland, 2016, pp. 478–486. [Google Scholar]
- [30].Merkow J., Marsden A., Kriegman D., and Tu Z., “Dense volume-to-volume vascular boundary detection,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent., Cham, Switzerland, 2016, pp. 371–379. [Google Scholar]
- [31].Kjerland Ø., “Segmentation of coronary arteries from CT-scans of the heart using deep learning,” Norwegian Univ. Sci. Technol., Trondheim, Norway, Tech. Rep. ntnudaim:16918, 2017. [Google Scholar]
- [32].Chen Y.-C., Lin Y.-C., Wang C.-P., Lee C.-Y., Lee W.-J., Wang T.-D., and Chen C.-M., “Coronary artery segmentation in cardiac CT angiography using 3D multi-channel U-Net,” in Proc. MIDL, 2019, pp. 1–5. [Google Scholar]
- [33].Shen Y., Fang Z., Gao Y., Xiong N., Zhong C., and Tang X., “Coronary arteries segmentation based on 3D FCN with attention gate and level set function,” IEEE Access, vol. 7, pp. 42826–42835, 2019. [Google Scholar]
- [34].Lee M. C. H., Petersen K., Pawlowski N., Glocker B., and Schaap M., “TeTrIS: Template transformer networks for image segmentation with shape priors,” IEEE Trans. Med. Imag., vol. 38, no. 11, pp. 2596–2606, Nov. 2019. [DOI] [PubMed] [Google Scholar]
- [35].Wolterink J. M., Leiner T., and Išgum I., “Graph convolutional networks for coronary artery segmentation in cardiac CT angiography,” in Proc. Int. Workshop Graph Learn. Med. Imag., Cham, Switzerland, 2019, pp. 62–69. [Google Scholar]
- [36].Mirunalini P., Aravindan C., Nambi A. T., Poorvaja S., and Priya V. P., “Segmentation of coronary arteries from CTA axial slices using deep learning techniques,” in Proc. IEEE Region 10 Conf. (TENCON), Oct. 2019, pp. 2074–2080. [Google Scholar]
- [37].Lei Y., Guo B., Fu Y., Wang T., Liu T., Curran W., Zhang L., and Yang X., “Automated coronary artery segmentation in coronary computed tomography angiography (CCTA) using deep learning neural networks,” Proc. SPIE, vol. 11318, Mar. 2020, Art. no. 1131812. [Google Scholar]
- [38].Du H., Shao K., Bao F., Zhang Y., Gao C., Wu W., and Zhang C., “Automated coronary artery tree segmentation in coronary CTA using a multiobjective clustering and toroidal model-guided tracking method,” Comput. Methods Programs Biomed., vol. 199, Feb. 2021, Art. no. 105908. [DOI] [PubMed] [Google Scholar]
- [39].Gao Z., Wang X., Sun S., Wu D., Bai J., Yin Y., Liu X., Zhang H., and de Albuquerque V. H. C., “Learning physical properties in complex visual scenes: An intelligent machine for perceiving blood flow dynamics from static CT angiography imaging,” Neural Netw., vol. 123, pp. 82–93, Mar. 2020. [DOI] [PubMed] [Google Scholar]
- [40].Ronneberger O., Fischer P., and Brox T., “U-Net: Convolutional networks for biomedical image segmentation,” in Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015 (Lecture Notes in Computer Science), vol. 9351. Cham, Switzerland: Springer, 2015, pp. 234–241. [Google Scholar]
- [41].Schneider C. A., Rasband W. S., and Eliceiri K. W., “NIH Image to ImageJ: 25 years of image analysis,” Nature Methods, vol. 9, pp. 671–675, Jul. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fedorov A., Beichel R., Kalpathy-Cramer J., Fillion-Robin J.-C., Pujol S., Bauer C., Jennings D., Fennessy F., Sonka M., Buatti J., Aylward S., Miller J. V., Pieper S., and Kikinis R., “3D slicer as an image computing platform for the quantitative imaging network,” Magn. Reson. Imag., vol. 30, pp. 1323–1341, Nov. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhou Z., Siddiquee M. M. R., Tajbakhsh N., and Liang J., “UNet: Redesigning skip connections to exploit multiscale features in image segmentation,” IEEE Trans. Med. Imag., vol. 39, no. 6, pp. 1856–1867, Jun. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bullock J., Cuesta-Lázaro C., and Quera-Bofarull A., “XNet: A convolutional neural network (CNN) implementation for medical X-ray image segmentation suitable for small datasets,” Proc. SPIE, vol. 10953, Mar. 2019, Art. no. 109531Z. [Google Scholar]


