Summary
The organization of genomic DNA into defined nucleosomes has long been viewed as a hallmark of eukaryotes. This paradigm has been challenged by the identification of “minimalist” histones in archaea and more recently by the discovery of genes that encode fused remote homologs of the four eukaryotic histones in Marseilleviridae, a subfamily of giant viruses that infect amoebae. We demonstrate that viral doublet histones are essential for viral infectivity, localize to cytoplasmic viral factories after virus infection, and ultimately are found in the mature virions. Cryogenic electron microscopy (cryo-EM) structures of viral nucleosome-like particles show strong similarities to eukaryotic nucleosomes despite the limited sequence identify. The unique connectors that link the histone chains contribute to the observed instability of viral nucleosomes, and some histone tails assume structural roles. Our results further expand the range of “organisms” that require nucleosomes and suggest a specialized function of histones in the biology of these unusual viruses.
Keywords: NCLDV, giant virus, viral nucleosome, doublet histone, Analytical Ultracentrifugation, cryo-EM, viral factory, histone tail, acidic patch, non-eukaryotic nucleosome, nucleosome-like-particle, Melbournevirus genetics, KO fitness impact
Graphical abstract

Highlights
-
•
Marseilleviridae encode proteins that resemble fused histones H4-H3 and H2B-H2A
-
•
These histone doublets assemble into unstable nucleosome-like particles in vitro
-
•
Histone doublets localize to the viral factory and are highly abundant in the virus
-
•
They are essential for viral fitness and infectivity, a first for any virus
Some viruses can package DNA into particles that resemble eukaryotic nucleosomes, with noteworthy similarities and differences.
Introduction
The organization of genomic DNA with histones into distinct complexes known as nucleosomes is a universal and highly conserved feature of all eukaryotes. The eukaryotic nucleosome core invariably comprises two copies each of the four unique histones H2A, H2B, H3, and H4. Each protein has a histone fold (HF) region that is structurally conserved between the four histones, as well as additional HF extensions and highly charged cationic tails that are unique to each (Luger and Richmond, 1998b). H2B-H2A and H4-H3 form obligate heterodimers that assemble into an octamer that wraps 147 bp DNA to form nucleosomes (Luger et al., 1997).
The evolutionary origin of eukaryotic chromatin organization is widely thought to lie in the archaeal domain of life (Talbert et al., 2019). Archaeal histones are limited to the HF and are encoded by only one gene or a few closely related genes. They bind and bend DNA as homodimers or quasi-symmetric heterodimers using architectural principles also seen in eukaryotic histones but lack the ability to form defined particles and instead exist in “slinky-like” assemblies that organize between 90 and ∼600 bp DNA (Bowerman et al., 2021; Mattiroli et al., 2017).
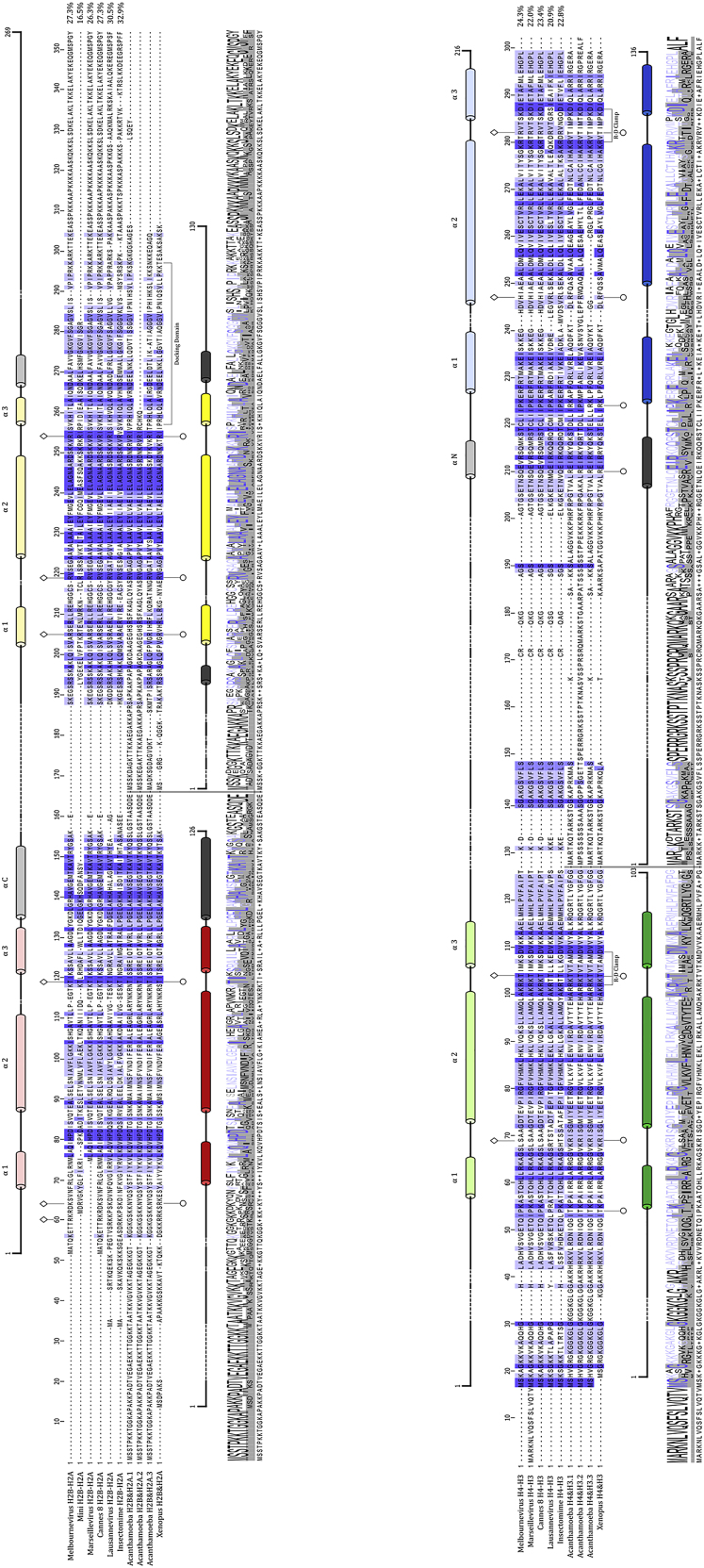
The genomes of some viruses (in particular nuclear DNA viruses and retroviruses that need to evade the host DNA damage recognition machinery) are organized into nucleosomes by appropriating eukaryotic histones and the host nucleosome assembly machinery during the latent and early lytic phase (e.g., Oh et al., 2015; reviewed in Lieberman, 2008). With the exception of SV40 (Polisky and McCarthy, 1975), viral genomes are not organized with nucleosomes in the capsid, and they do not encode viral histone homologs (Lieberman, 2008). In contrast, distinct histone-like proteins with homology to eukaryotic H2A, H2B, H3, and H4 have been identified in the genomes of some nucleocytoplasmic large DNA viruses (NCLDVs). Several members of the Marseilleviridae isolated from the Amoeba Acanthamoeba castellanii encode homologs of the four histone proteins in native doublet form, where H4 is fused to H3 and H2B is fused to H2A (Figure 1A) (Thomas et al., 2011). These proteins are present in Marseillevirus virions (Boyer et al., 2009; Fabre et al., 2017; Okamoto et al., 2018), where they might participate in the organization of the large (>300 kb) viral genomes. The arrangement of histones in doublets, the low level (<30%) of sequence similarity with eukaryotic histones, and their high degree of conservation within the family of Marseilleviridae (Figures 1A and S1A) suggest that they have evolved to fulfill viral functions other than (or in addition to) DNA compaction. Here, we show that viral histones are essential for viral infectivity, co-localize in the viral factory, and are in the same order of abundance as the major capsid protein in the viral capsids. Using cryogenic electron microscopy (cryo-EM), we demonstrate that viral histone doublets indeed interact with DNA to form nucleosome-like particles with distinct structural properties not seen in eukaryotes.
Figure 1.
MV histone doublets are re-localized to viral factories (VFs)
(A) Histone dimer pairs (H2B-H2A and H4-H3) within Eukarya were aligned against the doublet Marseilleviridae histones using HHpred’s multiple sequence alignment tool (ClustalΩ). Known α helices from the histone fold (HF) domain in Eukarya are dark-colored tubes (H2B, red; H2A, yellow; H4, green; H3, blue; and additional helices, gray). Predicted α helices in MV histones were generated using HHrped’s Quick 2D prediction web server (shown in lighter coloration) within the Marseilleviridae histone doublets. Known R-T pairs and DNA binding residues are shown in Eukarya histones along with their conservation within Marseilleviridae histones; additional predicted DNA-binding residues are shown (positions demonstrated by lollipops).
(B) Light microscopy fluorescence images (scale bar, 10 μm) of A. castellanii cells transfected with GFP-A. castellanii-H2A, MV-H2B-H2A-GFP, and MV-H3-H4-GFP, non-infected and infected with MV at 4 h PI. While GFP-A. castellanii-H2A concentrates only in the nucleus (N) of the non-infected cells, MV-H2B-H2A-GFP and MV-H3-H4-GFP are scattered in the entire cell (including the nucleus). Upon virus infection, GFP-A. castellanii-H2A remains in the nucleus (yellow arrows), while MV-H2B-H2A-GFP and MV-H3-H4-GFP re-localize to the VF (cyan arrows). DAPI staining remains in the nucleus all along the infection, but the intense fluorescence in the late VF hides the staining of the nucleus at 4 h PI.
See also Figure S1.
Figure S1.
Localization of fluorescently labeled MV histones in virus-infected Amoeba
(A) Complete sequence alignment of H2B-H2A (top) and H4-H3 (bottom) from Marseilleviridae, the host Acanthamoeba castellanii, and Xenopus laevis. Predicted α helices of H2B-H2A (light red and yellow) and H4-H3 (light green and blue) Melbournevirus histone doublets were generated using HHPRED’s Quick 2D prediction web server. Histone dimer pairs H2B-H2A and H4-H3 of Acanthamoeba castellanii and Xenopus laevis were each aligned against their respective Marseilleviridae histone doublets using HHPRED’s multiple sequence alignment tool, ClustalΩ. Conservation of each specific residue in each alignment is denoted by blue shading, with greater conservation being represented by darker blue. Known R-D clamp, R-T pairs, and DNA binding residues are indicated for Xenopus laevis histone pairs with their conservation within Melbournevirus histones. Marseilleviridae histones are 16%–33% conserved to Xenopus laevis histones (shown on far right of alignment). Known α helices from the histone fold domain in Xenopus laevis are shown as dark colored tubes; H2B are red, H2A are yellow, H4 are green, H3 in blue, and additional helices in gray. Logo plot demonstrating residue conservation among the alignments provided by ClustalΩ tool is shown below. Light microscopy fluorescence images (scale bar 10 μm) of A. castellanii cells transfected with (B) GFP-A. castellanii-H2A, C) MV-H2B-H2A-GFP, D) MV-H4-H3-GFP and E) MV-miniH2B-H2A-GFP, non-infected (NI) and infected with Melbournevirus at 30 min, 1h, 2h, 3h and 4h pi. B) GFP-A. castellanii-H2A concentrates only in the nucleus (N) of the non-infected cells and all along the infectious cycle. (C) MV-H2B-H2A-GFP, (D) MV-H4-H3-GFP and (E) MV-miniH2B-H2A-GFP are scattered in the entire cell (including the nucleus) in the non-infected cells. Between 1h and 2h pi, the viral histones start accumulating in the early viral factories (eVF). At 4h pi, the fluorescence is predominantly concentrated in the mature viral factory (VF). DAPI staining remains at the nucleus all along the infection but the intense fluorescence in the late VF hides the staining of the nucleus at 4h pi. (F). MV H2B-H2A-GFP and MV H4-H3-mRFP are co-localized in the viral factory, along with the viral DNA at 4h pi. (G). DAPI staining of Acanthamoeba cells infected with Melbournevirus at 4h pi. The host nucleus does not disappear upon the infectious cycle. Due to the large accumulation of viral DNA in the viral factories, exposure time for the DAPI fluorescence was optimized to visualize both the viral factory (VF) and the nucleus (N) of the cell (typically between 400ms and 1 s). (H) Schematic depicting the strategy for analysis of histone gene knockouts. Related to Figures 1 and 2.
Results
MV histone doublets are essential for viral propagation and accumulate in viral factories and mature virions
Melbournevirus (MV) encodes three putative histone doublets (MEL_369, MEL_368, and MEL_149, here named MV-H2B-H2A, MV-H4-H3, and MV-miniH2B-H2A). Putative histone domains in each doublet are linked by a ∼20-amino-acid connector (Figure 1A). Although these doublet histone proteins share less than 30% amino acid sequence identity with eukaryotic histones, they are highly conserved within the group of Marseilleviridae (Figure S1A). Secondary structure prediction suggests that viral histones form HFs (α1-L1-α2-L2-α3) but have either longer (MV-H2B-H2A) or shorter (MV-miniH2B-H2A) H2A C-terminal tails than their eukaryotic counterparts (Figure 1A). The H3 αN helix (which organizes the terminal turn of eukaryotic nucleosomal DNA) is predicted to be present in the viral H4-H3 doublets, while the sequence of the H2A docking domain (which tethers the H2A-H2B dimers to the (H3-H4)2 tetramer in eukaryotic nucleosomes) diverges from eukaryotic H2A (Figure S1A). Many of the “signature” amino acids that are important for DNA binding in eukaryotic histones are conserved in viral histones. These include arginine side chains that reach into the DNA minor groove and, in MV-H4-H3, a threonine from the paired L1 loop (R-T pair), as well as many other basic amino acids that are also interacting with the DNA (Figures 1A and S1A).
To identify the localization of MV histone doublets during viral assembly, we imaged transfected cells expressing fluorescently (GFP) tagged MV-histone doublets as well as tagged amoeba histones at different time points post-infection (PI) and compared their localization with non-infected cells. Amoeba GFP-H2A concentrated only in nuclei, where it presumably associates with genomic DNA to form nucleosomes, regardless of MV infection (Figures 1B and S1B). In contrast, unique and distinct patterns were observed for the transfected MV-histone doublets upon virus infection, particularly in the viral factory (VF; foci in the amoeba cytoplasm, where viral transcription, replication, and assembly take place). MV-H2B-H2A-GFP and MV-H4-H3-GFP were initially dispersed throughout the whole cell, including the nucleus, but their localization started to change between 1 and 2 h PI (Figures S1C and S1D). Eventually, MV-H2B-H2A-GFP and MV-H4-H3-GFP accumulated and concentrated predominantly as foci in the viral factory (Figures 1B, S1C, and S1D, cyan arrows), although they were still present in nuclei at much lower concentrations (Figure 1B, yellow arrows). Similar localization patterns were obtained with MV-miniH2B-H2A-GFP (Figure S1E). In addition, co-localization of MV-H2B-H2A-GFP and MV-H4-H3-mRFP along with viral DNA was observed in the viral factory, indicating that MV histones are recruited for viral DNA packaging and virion production (Figure S1F). Thousand copies of the AT-rich 360-kb viral genome are produced in the viral factory to produce approximately thousands of mature virions, compared to the 24 copies of a 46.7-Mb GC-rich amoeba genome. As such, DAPI staining for the viral factory overpowers the weaker staining of the nucleus, which is not a sign of host genomic DNA disappearance during the infectious cycle (Figure S1G).
Mass spectrometry proteomic analysis of the purified MV virions was performed (Fabre et al., 2017) (Table 1). MV-histone doublets are among the most abundant proteins in the MV viral proteome, with the number of copies per capsid in the range of the major capsid protein. Importantly, the amount of histone doublets (even when using the most conservative estimate), is sufficient to package the entire viral genome with nucleosomal particles with a repeat length of ∼180 bp (∼4,000 histone doublets per 360 kilo base pairs (kbp) viral genome).
Table 1.
Mass spectrometry proteomics of the purified MV virions
| Protein ID | Annotation | Final rank | iBAQ R1 | Copy R1 | Rank R1 | iBAQ R2 | Copy R2 | Rank R2 |
|---|---|---|---|---|---|---|---|---|
| MEL_236 | hypothetical protein | 1 | 1,7E+09 | 14,087 | 1 | 3E+09 | 15,107 | 1 |
| MEL_342b | hypothetical protein | 2 | 1,2E+09 | 9,916 | 2 | 2E+09 | 12,017 | 2 |
| MEL_368 | histone H4-H3 doublet∗ | 3 | 9,8E+08 | 8,362∗ | 4 | 2E+09 | 10,344∗ | 3 |
| MEL_305 | major capsid protein | 4 | 1,1E+09 | 9,240 | 3 | 2E+09 | 9,240 | 4 |
| MEL_274 | hypothetical protein | 5 | 5,7E+08 | 4,835 | 5 | 1E+09 | 5,385 | 5 |
| MEL_369 | histone H2B-H2A doublet∗ | 6 | 5E+08 | 4,223∗ | 6 | 8E+08 | 3,895∗ | 6 |
| MEL_247 | papain-like cysteine protease | 7 | 3,5E+08 | 2,935 | 7 | 8E+08 | 3,812 | 7 |
| MEL_280 | hypothetical protein | 8 | 2,2E+08 | 1,862 | 8 | 5E+08 | 2,429 | 8 |
| MEL_234 | hypothetical protein | 9 | 2,1E+08 | 1,776 | 9 | 5E+08 | 2,354 | 9 |
| MEL_241 | hypothetical protein | 10 | 1,8E+08 | 1,569 | 10 | 4E+08 | 2,208 | 10 |
| MEL_358 | hypothetical protein | 11 | 1,4E+08 | 1,211 | 11 | 3E+08 | 1,454 | 11 |
| MEL_207 | thioredoxin | 12 | 1,3E+08 | 1,083 | 12 | 3E+08 | 1,424 | 12 |
| MEL_020 | hypothetical protein | 13 | 1,2E+08 | 1,053 | 13 | 2E+08 | 1,252 | 13 |
| MEL_202 | hypothetical protein | 14 | 9,7E+07 | 828 | 15 | 2E+08 | 1,072 | 14 |
| MEL_260 | hypothetical protein | 15 | 9,8E+07 | 835 | 14 | 2E+08 | 1,004 | 15 |
| MEL_200 | transmembrane-domain-containing protein | 16 | 7,9E+07 | 676 | 16 | 2E+08 | 851 | 16 |
| MEL_065 | hypothetical protein | 17 | 7E+07 | 596 | 17 | 2E+08 | 775 | 17 |
| MEL_025 | peptidase | 18 | 6,2E+07 | 531 | 19 | 1E+08 | 640 | 19 |
| MEL_097 | hypothetical protein | 19 | 6,3E+07 | 534 | 18 | 1E+08 | 613 | 21 |
| MEL_282b | hypothetical protein | 20 | 5,4E+07 | 463 | 24 | 1E+08 | 657 | 18 |
| MEL_181 | ATP-dependent helicase | 21 | 5,7E+07 | 482 | 21 | 1E+08 | 634 | 20 |
| MEL_354 | transmembrane-domain-containing protein | 22 | 5,8E+07 | 493 | 20 | 1E+08 | 604 | 22 |
| MEL_365b | hypothetical protein | 23 | 5,6E+07 | 477 | 22 | 1E+08 | 601 | 24 |
| MEL_196 | hypothetical protein | 24 | 5,5E+07 | 468 | 23 | 1E+08 | 601 | 23 |
| MEL_294 | thioredoxin | 25 | 5,4E+07 | 463 | 25 | 1E+08 | 583 | 25 |
| MEL_360 | AAA-family ATPase | 26 | 5,1E+07 | 435 | 27 | 1E+08 | 543 | 27 |
| MEL_244 | hypothetical protein | 27 | 5,1E+07 | 437 | 26 | 1E+08 | 532 | 28 |
| MEL_185 | hypothetical protein | 28 | 4,6E+07 | 389 | 29 | 1E+08 | 572 | 26 |
| MEL_275 | hypothetical protein | 29 | 4,9E+07 | 420 | 28 | 1E+08 | 505 | 29 |
| MEL_269 | transmembrane-domain-containing protein | 30 | 4,2E+07 | 354 | 30 | 9E+07 | 460 | 32 |
| MEL_228 | transmembrane-domain-containing protein | 31 | 3,9E+07 | 333 | 32 | 9E+07 | 476 | 30 |
| MEL_168 | lipase | 32 | 3,9E+07 | 328 | 33 | 9E+07 | 476 | 31 |
| MEL_041 | peptidase | 33 | 3,9E+07 | 334 | 31 | 8E+07 | 381 | 36 |
| MEL_331 | hypothetical protein | 34 | 3,7E+07 | 312 | 34 | 8E+07 | 396 | 34 |
| MEL_352 | lectin-domain-containing protein | 35 | 3,1E+07 | 263 | 38 | 9E+07 | 442 | 33 |
| MEL_211 | transmembrane-domain-containing protein | 36 | 3,5E+07 | 297 | 35 | 7E+07 | 379 | 37 |
| MEL_380 | serine/threonine protein kinase | 37 | 3,3E+07 | 279 | 37 | 8E+07 | 393 | 35 |
| MEL_149 | mini H2B-H2A histone∗ | 38∗ | 3,4E+07∗ | 289∗ | 36∗ | 7E+07∗ | 362∗ | 39 |
| MEL_089 | hypothetical protein | 39 | 2,9E+07 | 249 | 39 | 7E+07 | 368 | 38 |
| MEL_216 | helicase | 40 | 2,8E+07 | 241 | 40 | 6E+07 | 303 | 40 |
| MEL_235 | serine/threonine protein kinase | 41 | 2,7E+07 | 233 | 41 | 6E+07 | 282 | 44 |
| MEL_338 | hypothetical protein | 42 | 2,3E+07 | 199 | 44 | 6E+07 | 296 | 41 |
| MEL_301 | transmembrane-domain-containing protein | 43 | 2,5E+07 | 209 | 43 | 6E+07 | 282 | 43 |
| MEL_231 | hypothetical protein | 44 | 2,5E+07 | 216 | 42 | 5E+07 | 266 | 45 |
| MEL_223b | membrane protein | 45 | 2,1E+07 | 180 | 45 | 6E+07 | 288 | 42 |
| MEL_297 | hypothetical protein | 46 | 2E+07 | 167 | 49 | 5E+07 | 265 | 46 |
| MEL_213b | hypothetical protein | 47 | 2,1E+07 | 178 | 47 | 5E+07 | 247 | 47 |
| MEL_215 | hypothetical protein | 48 | 2,1E+07 | 177 | 48 | 5E+07 | 237 | 48 |
| MEL_255 | papain-like cysteine protease | 49 | 2,1E+07 | 179 | 46 | 5E+07 | 234 | 49 |
| MEL_387 | hypothetical protein | 50 | 1,9E+07 | 159 | 52 | 4E+07 | 212 | 50 |
| MEL_219 | ribonuclease III | 51 | 1,9E+07 | 163 | 50 | 4E+07 | 207 | 53 |
| MEL_278 | disulfideoxidoreductase | 52 | 1,9E+07 | 160 | 51 | 4E+07 | 208 | 51 |
Asterisks indicate histone doublets.
Analysis of published transcriptomic data obtained for a member of the Marseilleviridae during its infectious cycle in A. castellanii (Rodrigues et al., 2020) sheds light on the possible contribution of the host to viral nucleosome assembly (Figure 2A). As expected, the viral DNA polymerase is expressed before the viral histones, between 1 and 2 h PI. Expression of viral histones begins 2–4 h PI and continues along the infectious cycle. Host histone expression, however, decreases after 2 h PI, as is the case for most of the A. castellanii genes (see, for reference, the actin gene in gray). The cellular histone chaperones are either never expressed (FUN_000176, FUN_002445, FUN_006481, FUN_014954, FUN_006327 lanes in gray at the bottom of the figure) or have their expression decreased after 2 h PI. No obvious homologs to the cellular histone chaperones were detected in the Marseilleviridae genome.
Figure 2.
MV histone doublets are essential for virus fitness
(A) Histone (MV and host) expression and host histone chaperones expression profile during the MV infectious cycle. Host actin genes expression were used for reference.
(B) Schematic representation of the strategy used to generate the recombinant mel_368 or mel_369 KO. Expected PCR product size are also shown. Primer a: HB146 or HB147 and primer b: HB145 or HB148 for mel_368 and mel_369 respectively; primer c, GFP reverse; primer d, GFP forward (Table S1). goi, gene of interest.
(C) PCR demonstrates correct integration of vectors in the locus of mel_368 or mel_369. Analysis of fitness changes associated with histone KO was analyzed by virus competition assay in wild-type or complemented amoebas. Mock cells were incubated with Superfect in the absence of plasmid. a+c, 5′ integration; d+b, 3′ integration; a+b, wild-type locus. HR, homologous recombination.
In order to assess the fitness cost associated with the impairment of potential nucleosome formation in MV capsids, we designed a strategy to knock out either mel_369 or mel_368 (Figures 2B and S1H). A. castellanii cells were transfected with linearized plasmid prior to infection with MV particles. Recombinant viruses were quickly detected by PCR upon 3–5 h PI (primers listed in Table S1). Expression of GFP could not be detected at 3h PI, likely due to the late expression profile of the histone promoters. The presence of mutant viral particles was followed during two generations by infecting wild-type amoeba compared to amoeba expressing an ectopic copy of the histones for trans-complementation (Figure 2C). Importantly, mutant viral particles were quickly outcompeted by wild-type MV in absence of trans-complementation, while growth of the mutant virus was sustained when the missing copy of the histone gene was stably expressed in the amoeba (Figure 2C). This demonstrates that both mel_369 and mel_368 are essential, and their knockout (KO) is associated with a strong fitness cost to the virus.
Altogether, the distinct localization and expression patterns reveal specific targeting of MV histone doublets to the viral factory and the viral particle.
MV doublet histones form defined, unstable nucleosome-like particles
To determine what types of complexes MV-H2B-H2A, MV-miniH2B-H2A, and MV-H4-H3 doublets form with DNA, we expressed, purified, and refolded the proteins from E. coli (Figure S2A). When combined with DNA (“601” nucleosome positioning sequence) of varying lengths, and using the classic salt-gradient nucleosome reconstitution protocol (Dyer et al., 2004), MV-H2B-H2A and MV-H4-H3 form defined nucleosome-like particles (MV-NLPs) that are stable at 37°C, whereas individual histone doublets fail to form defined bands on DNA (Figure 3A). The composition of the MV-NLP was confirmed by sucrose gradient fractionation (Figure S2B). MV histones also formed defined particles on a native 181 bp DNA fragment derived from the MV genome (GC content = 45%). Irrespective of DNA sequence, and unlike X. laevis nucleosomes (a representative of eNuc), MV-NLPs dissociate upon heat treatment at 55°C (Figure S2C). MV-miniH2B-H2A alone binds DNA poorly, and no homogeneous MV-NLPs were formed with MV-H4-H3 and DNA (Figure S2D). We therefore focused on MV-NLPs with full-length MV-H2B-H2A and MV-H4-H3.
Figure S2.
MV-histones form nucleosome-like particles
(A) SDS-PAGE of purified Melbournevirus (MV) histone doublets. (B) Sucrose gradient sedimentation of MV NLPs with 147 or 207 bp DNA. The compositions of MV NLPs were analyzed by native- and SDS-PAGE. (C) MV-NLP and eNuc were reconstituted on Widom ‘601’ DNA and Melbournevirus native DNA, respectively. The MV-NLP and eNuc were heat treated at 37 and 55°C. (D) Native PAGE of reconstituted MV mini-NLP (mini H2B-H2A instead of H2B-H2A) with 147 bp DNA. (E) GraFix of MV-NLPs with 207 bp DNA (top two panels) and 147 bp DNA (bottom two panels. Native- and SDS- PAGE of the crosslinked MV-NLP fractions representing successful crosslinking, compared to the native MV-NLP input. (F) Representative AFM images: Samples were diluted in TCS buffer and applied to APTES coated mica, rinsed with water and imaged on a NanoWizard Sa with a TAP300-GD cantilever. Samples include 147 bp DNA only, eNuc147, MV-H4-H3 with 147 bp DNA, MV-NLP147, MV-NLP207 and MV-NLP207 GraFix. Related to Figure 3.
Figure 3.
Histone doublets in Marseilleviridae form nucleosome-like particles (MV-NLPs)
(A) Native PAGE of reconstituted MV-NLPs with “601” DNA of various lengths. Left panel: individual and combinations of histone doublets reconstituted onto 147 bp DNA; right panel: MV-H2B-H2B and MV-H4-H3 reconstituted onto 601 DNA of varying lengths.
(B) Sedimentation velocity analytical ultracentrifugation (SV-AUC) of MV-NLPs. Left: van Holde-Weischet plot of eukaryotic nucleosomes (eNuc), histone-DNA complexes with individual MV histone doublets, and native MV-NLPs with 147 bp DNA (no GraFix); right: van Holde-Weischet plot of crosslinked (GraFix-ed) MV-NLPs with 123, 147, or 207 bp DNA.
(C) Height profile of MV-NLPs with 147 or 207 bp DNA, obtained by atomic force microscopy (AFM). The average height profiles with standard deviations of the particles from each sample are shown; representative, original images are shown in Figure S2, and statistics are shown in Table S2.
Sedimentation velocity analytical ultracentrifugation (SV-AUC) yields information on the size and shape distribution of macromolecular assemblies in solution. The diffusion-corrected sedimentation value of a particle is proportional to its mass, and inversely proportional to its viscous drag. The sedimentation behavior of MV-histone doublets assembled onto 147 bp DNA was compared to that of X. laevis histones reconstituted on the same DNA (eNuc147). The eNuc147 particle sediments at 11 S (Wang et al., 2018), while MV-NLP147 sediments at ∼8 S, with a “tail” toward lower S values, indicating dissociation of DNA at the relatively low (300 nM) concentrations used for AUC experiments (Figure 3B). Particles reconstituted with individual MV-histone doublets sediment at ∼5.5 S (Table 2). The AUC-derived molecular mass calculated for MV-NLP147 (Figure 3B, red arrow) suggests that at least one copy of the H2B-H2A doublet has been lost due to the sample dilution in AUC (Table 2). To counteract the apparent dissociation of MV-NLPs, we employed gradient fixation (GraFix) with glutaraldehyde (Kastner et al., 2008). Histone doublets were efficiently crosslinked, and the position of the main peak fraction in the gradient as well as migration on a native gel was unchanged upon crosslinking, suggesting little to no structural changes (Figure S2E). Particles reconstituted on 123, 147, and 207 bp DNA fragments sedimented between 8.8 and 9.9 S after crosslinking (Figure 3B, Table 2). The frictional ratio (f/f0) (a measure of particle “extension,” where larger values indicate increased drag, slowing sedimentation) of all MV-NLPs was higher than that of eNuc147, and f/f0 values increase in correlation with DNA length (Table 2). MV-NLP123 has the highest S-value among MV-NLP constructs, despite having the smallest mass, because of its significantly lower f/f0 value. This suggests that the additional DNA in MV-NLP147 and MV-NLP207 is extended.
Table 2.
S values (S(20,W)), frictional ratios (f/f0), and calculated molecular weights (including confidence intervals) of histone-DNA complexes derived from SV-AUC
| Sample | S(20,W) | f/f0 | Molecular weight (kDa) experimental/theoretical | |
|---|---|---|---|---|
| No GraFix | 147bp DNA | 5.10 (5.09, 5.10) | 3.09 (3.09, 3.09) | 117.13 (116.9, 117.3)/97 |
| MV-H2B-H2A 147 | 5.68 (4.61, 6.75) | 2.66 (2.51, 2.81) | 134.61 (89.83, 179.38)/150 | |
| MV-H4-H3 147 | 5.28 (4.59, 5.96) | 2.79 (2.70, 2.87) | 120.93 (104.14, 137.71)/146 | |
| MV-NLP147 | 7.78 (7.74, 7.82) | 2.0 (1.96, 2.04) | 179.17 (173.55, 185.80)/206 | |
| eNuc147 | 11.16 (11.15, 11.16) | 1.54 (1.52, 1.56) | 218.5 (214.4, 222.5)/210 | |
| GraFix | MV-NLP123 (nucleosome-like) | 9.86 (7.99, 11.72) | 1.77 (1.52, 2.02) | 221.46 (170.93, 271.99)/190 |
| MV-NLP147 (nucleosome-like) | 8.79 (7.30, 10.29) | 2.01 (1.86, 2.14) | 217.79 (140.57, 295.00)/206 | |
| MV-NLP207 (nucleosome-like) | 9.01 (7.48, 10.55) | 2.26 (2.00, 2.51) | 256.58 (194.54, 318.62)/245 | |
We visualized MV-NLPs by atomic force microscopy (AFM). Particles reconstituted with either MV-H4-H3 alone or the full complement of histone doublets were deposited onto mica surfaces, and their height profiles were compared to eNuc147 (Figures 3C and S2F; Table S2). The majority of eNuc147 survived the low concentration (2 nM) required for AFM imaging and presented the characteristic height profile of ∼2.1 nm (White et al., 2016). In contrast, over 30% of the observed particles heights indicative of free DNA, indicating complete MV-NLP207 disassembly, and the remaining particles had lower height profiles (∼1.3 nm) than the eNuc147 complex (Figures 3C and S2F; Table S2). This reduced height suggests that MV-NLP207 transiently disassemble at the low concentration required for AFM. Crosslinking MV-NLP prevented disassembly and resulted in particles comparable in height to eNuc147.
MV-NLPs resemble eukaryotic nucleosomes
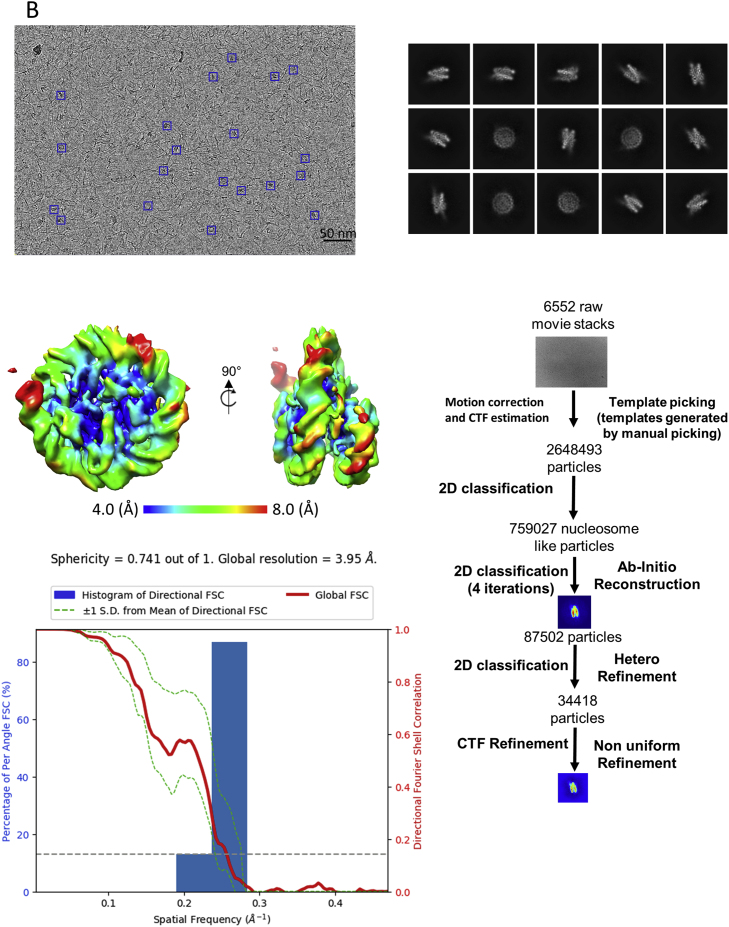
Single-particle cryogenic electron microscopy (cryo-EM) was used to visualize crosslinked MV-NLP207. GraFix was required as the majority of untreated MV-NLPs dissociate during plunge freezing. Raw images and 2D classes show that MV-NLPs share many characteristic features of eukaryotic nucleosomes (Figure S3A), with defined segments of extending linker DNA visible in many classes. After 3D classifications and refinement, we obtained a structure of the MV-NLP207 at 6.1 Å (Figure S3A; Table S3). A second dataset obtained with GraFix-treated particles reconstituted onto 147 bp DNA (MV-NLP147) yielded an improved overall resolution of ∼4.0 Å (Figure S3B; Table S3). In both structures, electron density for the DNA and histone helices are clearly distinguishable (Videos S1 and S2), which allowed the assignment of ∼120 bp of bound DNA and all histone chains (Figure 4A; Video S3).
Figure S3.
Cryo-EM data analysis of MV-NLP207 and MV-NLP147
(A) Raw micrograph of the Grafix-treated MV-NLP207 (Scale bar is 50 nm), 2D class averages generated from the dataset, 3D structure of the MV NLP, local resolution map and FSC curve, and data processing strategy flow chart. (B) Raw micrograph of the GraFix treated MV-NLP147 (Scale bar is 50 nm), 2D class averages generated from the dataset, 3D structure of the MV NLP, local resolution map and FSC curve, and data processing strategy flow chart. Related to Figure 4.
Figure 4.
Cryo-EM reveals that MV-histone doublets form nucleosome-like structures with asymmetrically extending DNA
(A) Overview of MV-NLP147 and electron density. The equivalent regions of MV H3, H4, H2A, and H2B are shown in blue, green, yellow, and red, respectively.
(B) Overlay of MV-NLP147 (blue) with eNuc (gray). Only 80 bp of DNA with associated histones are shown for clarity. Superhelix locations (SHLs) are numbered from 0 to 6 starting from the nucleosome dyad (φ).
(C) Comparison of the DNA path of eNuc147 (gray ribbon diagram), MV-NLP147 (blue electron density), and MV-NLP207 (green electron density).
(D) Charged surface representation of the histones for MV-NLP147 and eNuc147. Coordinates for eNuc147 were taken from 3LZ0.
See also Figures S3A and S3B and Table S3.
The equivalent regions of H3, H4, H2A, and H2B are shown in blue, green, yellow, and red, respectively.
The overall dimensions of the DNA superhelix, the path described by the DNA around the histone core, and the overall layout of HF helices are similar between MV-NLPs and eukaryotic nucleosomes (Figures 4A and 4B; Table S4). However, DNA wrapping is incomplete and asymmetric in both MV-NLPs, as only ∼120 bp are wrapped in most particles. This is more pronounced in MV-NLP207, where well-defined electron density describes a straight path for DNA extending away from the histone core at superhelical location (SHL) 4.5, whereas the DNA extending at the other side (from SHL 5.5) appears to be more disordered (Figure 4C; Video S1). In MV-NLP147 complex slightly more DNA is bound (up to SHL 5.5 on both sides), and only one arm of extending DNA has defined density (Figure 4C; Video S2). This asymmetry is likely a consequence of the asymmetric 601 DNA sequence (Chen et al., 2017; Chua et al., 2012).
Initial models of MV-histone doublets were generated though homology modeling (Waterhouse et al., 2018), where the ∼28- and ∼20-amino-acid connectors in H4-H3 and H2B-H2A were constructed through de novo methods (Webb and Sali, 2016). These were docked into the MV-NLP147 density, with good agreement with nucleosome-like configurations (Figures 4A and 4B; correlation coefficient between model and experimental density 0.766).
Overall, the MV-histone core is less positively charged than the eukaryotic histone octamer (pI of ∼9.5 versus ∼11.0; Figure 4D). While basic amino acids describe a distinct path for the DNA in both histone cores, and many amino acid side chains that engage DNA are conserved between viral and eukaryotic histones (Figures 1A and S1A), positive charges along the helical path are less pronounced in MV histones, in particular in the region formed by the H3-H3′ four-helix bundle (“dyad”). This contributes to the reduced stability of MV-NLPs compared to eNuc. Additionally, the acidic patch, the primary point of interactions between eukaryotic nucleosomes and nuclear proteins (Peng et al., 2020), differs in size and charge, due to fewer acidic residues in MV-H2A (Figure 4D).
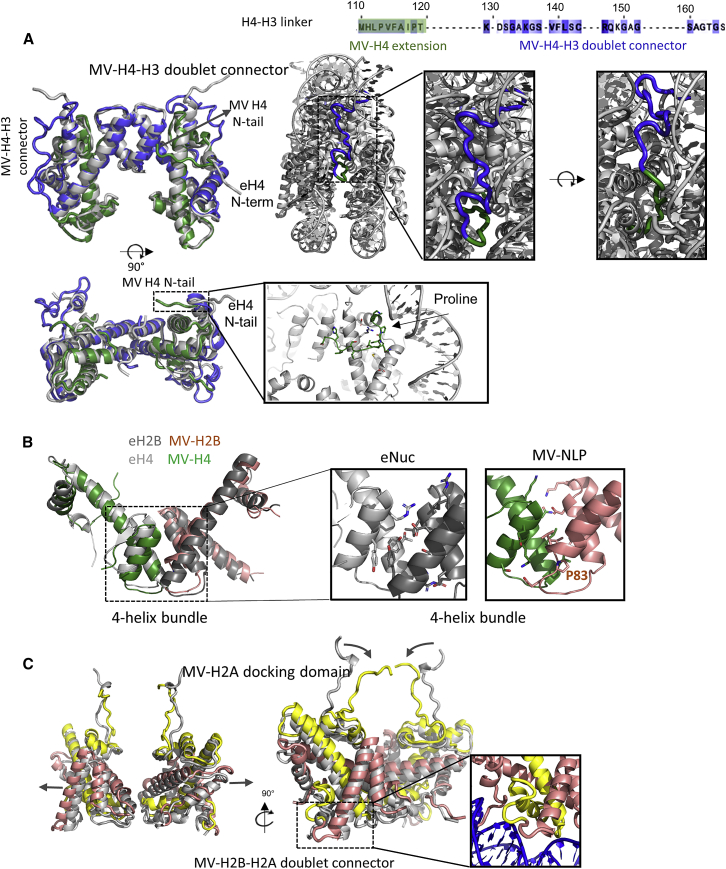
The two MV-H4-H3 doublets superimpose onto the eukaryotic (H4-H3)2 tetramer with a backbone root-mean-square deviation (RMSD) of ∼2 Å (Figure 5A; Table S4). The main-chain arrangements of the MV-H3-H3′ four-helix bundles at the nucleosomal dyad are similar as in eNuc147, but the interface lacks the hallmark histidine-cysteine configuration. Each MV-H2B-H2A doublet interfaces with the H4 portion of a MV-H4-H3 doublet through a four-helix bundle of similar architecture as the H4-H2B interface in eNuc147. However, the MV-H2B α2 helix is one turn shorter than the eukaryotic sequence (due to a conserved proline at position 83), and the tyrosines that form π-stacking interactions in eNuc systems are consistently absent in both MV-H4 and MV-H2B (Figure 5B). Additionally, while the main chains of the two H2A L1 loops (38NYAE41) are close to one another in eNuc147 and form direct interactions through their side chains, the distance between L1 loops in MV-NLP147 (153GGCS156; positions 214–217 in Figure S1A) is increased and shows no direct L1-L1 interactions. L1-L1 separation is accompanied by a similar increase in the distance between H2B-H2A centers (36.3 Å in eNuc147 versus 38.6 Å in MV-NLP147; Table S4). In addition to being further apart in the MV-NLP147 complex, the H2B-H2A HFs reorient away from the nucleosome core (Figures 5C and S4A; Table S4). Conversely, the H2A docking domains of MV-H2A are angled inward, toward the dyad axis, in an altogether different arrangement than what is seen in eNuc147 (Figure 5C; Table S4; 6–8 Å backbone RMSD for chains C and G, respectively). Together, these structural changes may contribute to the reduced stability and increased propensity to form sub-NLPs (hexasomes and tetrasomes) of MV-NLPs in comparison to the eNuc complex.
Figure 5.
Comparison of MV-NLP and eNuc histone structures
(A) Superposition of two MV-H4-H3 doublets (green and blue) with the eukaryotic (H3-H4)2 tetramer in gray (left), and a close-up of MV-H4-H3 doublet connector (right). Interactions of the MV-H4 N-terminal tail with the nucleosome are also shown.
(B) A comparison of the four-helix bundle structure formed by H4 and H2B.
(C) Superposition of two MV-H2B-H2A doublets (in red and yellow) with eukaryotic H2A-H2B dimers in gray (left), and a close-up of the MV-H2B-H2A doublet connector and docking domain.
See Table S4 for detailed information. Additional possible configurations for both connectors are shown in Figure S4.
Figure S4.
MV-nucleosome histone dimer arrangement and histone connectors
(A) Description of ϕ, the pseudo-dihedral angle measuring the relative orientations of the H2B-H2A subunits, between MV-NLP and eNuc. Larger magnitude values correspond to a dimer arrangement that is angled further away from the dyad axis. Fictitious particle locations are shown as black spheres, and histones (H3 in blue, H4 in green, H2A in yellow, and H2B in red) and DNA (silver and gold ribbons) are semi-transparent to allow for better visibility of each particle’s location. (B) Overlay of de novo loop models with feasible conformations connecting H2B to H2A and (C) H4 to H3 in the MV-histone doublets. Loop configurations were generated using Modeler (v9.20) and then placed in the nucleosome context. After manually removing physically non-relevant (knots) conformations and clashes identified with a cutoff distance of 0.8 Å using CPPTRAJ of the Amber MD package (v18), the three best loops are shown in different colors (cross correlation range against the simulated map is 0.614-0.623). Related to Figure 5.
The connector linking the C terminus of MV-H2B with the N terminus of MV-H2A is easily accommodated, as these are near each other. A variety of favorable conformations are predicted through de novo modeling, with several different arrangements in agreement with observed weak density in MV-NLP147 (Figure 5C, inset, and Figure S4B). A conserved arginine is pointing into the minor groove, and main-chain phosphate interactions further hold the loop in place, echoing the contributions of the H2A N-terminal domain in eNuc. Connecting the MV-H4 C terminus with the N-terminal tail of MV-H3 is conceptually more difficult. In eukaryotic H4, the eight most C-terminal amino acids are engaged in contacts with H2A and H2B in the interior of the histone core. Our models show that the equivalent region in MV-H4 maintains docking domain interactions with MV-H2A. Physical constraints dictate that the missing linker segment must continue toward the DNA superhelix and project upward in the direction of MV-H3 αN, with which it connects (Figure 5A). Electron density attributed to the connector is visible between the DNA gyres on both sides and in both structures (MV-NLP207 and MV-NLP147). Although not sufficiently defined to allow modeling through real-space refinement, de novo generation of linker structures provided multiple feasible conformations (Figure S4C), and molecular dynamics flexible-fitting simulations confirmed that these orientations are physically relevant (Figure 5A). A stretch of 10 predominantly hydrophobic amino acids packs against the region connecting MV-H3αN with α3 to form a hydrophobic core (Figures 5A and S4C), and the connector is also positioned to engage in main-chain contacts with the DNA backbone at SHL ± 1.5. Mostly flexible and small amino acids (GAGSAGTGS) form the turn toward MV-H3 αN (Figure 5A). As such, the MV-H4-H3 connector (which is highly conserved among Marseilleviridae doublets) can be accommodated within the structural framework of a canonical nucleosome, although its presence likely contributes to the unwrapping of the terminal 10 bp of DNA observed in both MV-NLP densities.
The other MV-histone tails that are not engaged in doublet formation also differ from the histone tails in eNuc. The C-terminal tail of MV-H2A is significantly longer than that of eukaryotic H2A (Figure S1A). The MV-H4 N-terminal domain (equivalent to the first 26 amino acids in eH4) does not extend over the DNA but takes the opposite direction to interact with MV-H4 α2 and α3 (Figure 5A). This density is as well defined as that of the main HF helices, suggesting that this domain may consistently interact with its own histone core rather than with DNA or the acidic patch in neighboring particles, as observed for the eukaryotic H4 tail (Dorigo et al., 2003; Morrison et al., 2018). The amino acid residues in this region are highly conserved among Marseilleviridae and differ from the eH4 tail (Figure S1A). A conserved proline (P34) at the tip of MV-H4 α1 might be responsible for the redirection (Figure 5A).
As is the case with the eH4 tail, the first 20 amino acids of the MV-H4 tail are too disordered to be observed in the electron density map.
Discussion
The discovery that the genomes of some giant viruses encode histone-like proteins in which the dimerization partners are fused into a single chain was surprising (Thomas et al., 2011). Our results that these MV-histone doublets associate with the viral factory and indeed with the mature virus and that they form nucleosome-like particles with unique properties support the idea that they are involved in viral genome organization into the capsids. Instead of organizing DNA into nucleoprotein “core complexes” inside the viral capsid, Marseilleviridae utilize their own specialized histone doublets to form nucleosomes and possibly more compact chromatin within the viral particle. In the context of the ongoing debate regarding the role of viruses in the emergence of the eukaryotic nucleus, our findings also provide a new dimension to the diverse role of histones in genome organization of non-eukaryotic entities (Bell, 2001; Erives, 2017).
Marseilleviridae exhibit intermediate dependency on their host, as they develop their viral factory in the cytoplasm but transiently recruit host nuclear proteins to the viral factory to transcribe early genes until the virally encoded RNA polymerase has been synthesized (Fabre et al., 2017). This is achieved through a reorganized leaky nucleus during the early phase. Amoeba histones are exclusively localized in the host nucleus whether the cells are infected or not, whereas transfected viral histones are present in the nucleus and cytoplasm in uninfected cells but move into viral factories upon infection to integrate into mature virions. This suggests that amoeba histones remain bound to cellular DNA, while the viral histones that made it into the nucleus apparently do not interact with amoeba genomic DNA and can leave the nucleus to associate with the viral factory. As such, amoeba histones are likely targeted to the nucleus and assembled into chromatin by histone chaperones and transporters that mostly avoid viral histones. As is the case for the other nuclear proteins transiently leaving the cell nucleus upon Marseilleviridae infection, it remains to be seen whether transfected viral histones are actively or passively recruited to the viral factory. Many “nucleosome signature features” are maintained among archaeal, eukaryotic, and now viral nucleosomes, such as the overall geometry and arrangement of the DNA superhelix, achieved by interactions between the DNA minor groove backbone and the main chain of antiparallel L1L2′ loops (from H4-H3 and H2B-H2A pairs) and the utilization of positive α helix dipole moments to bind DNA (Luger and Richmond, 1998a). Several of the “sprocket arginines” (Hodges et al., 2015) and their pairing with a threonine from the dimerization partner, as well as the highly conserved R-D clamps stabilizing the HF, are conserved among eukaryotic, archaeal, and viral histones. However, MV-doublet histones stably bind only ∼120 bp DNA as opposed to the 147 bp in eukaryotic nucleosomes. In our cryo-EM structures, the unbound DNA is characterized by distinct density indicating that it is in a defined orientation, and in this, it differs from the structures of eukaryotic variant nucleosomes that are also characterized by organizing only ∼120 bp DNA (Pentakota et al., 2017; Zhou et al., 2021). Unique to MV-NLPs is also the packing of the H4 N-terminal tail onto the surface of the histone core, the overall less positive charge of the histone core surface including the acidic patch, and, most notably, the linking of MV-H4-H3 and MV-H2B-H2A to form doublet histones. Fused histone genes are also observed in some archaea (Talbert et al., 2019) but to date not in any eukaryotic genome. The amino acid linkers connecting the histone moieties to form the doublets, which are conserved in sequence across Marseilleviridae, can be accommodated in the structure. However, the MV-H4-H3 connector (which is conserved in length and amino acid sequence between members of the Marseilleviridae) likely contributes to destabilizing the last turn of DNA. Overall, MV-NLPs are significantly destabilized compared to eNuc, and this seems to be an intrinsic property related to their virus-specific function. While this article was in revision, the structure of MV nucleosomes was published, with identical results and conclusions regarding the structural composition of the particles (Valencia-Sánchez et al., 2021).
A common trait of DNA viruses is that they must actively package their DNA into capsids to protect it outside the host cell environment. On the other hand, they also need to make their genomes accessible to the transcription machinery immediately upon infection. Previous analyses of the infectious cycle of MVs revealed that the capsids appear to dissolve ∼2 h PI, leaving a spherical electron dense core in the cytoplasm (Fabre et al., 2017). Our study suggests that Marseilleviridae assemble virally encoded histones into nucleosomes to condense and protect their genome, to allow it to fit into the ∼200-nm-diameter icosahedron, also explaining the higher than expected electron density of the spherical core (Fabre et al., 2017). This process is essential for MV propagation, since mutant viral particles are rapidly lost in competition with wild-type viruses, strongly suggesting that they are noninfectious. Arguably, viral nucleosomes must be metastable to make the genome (once transferred into the cytoplasm) accessible to transcription by the host RNA polymerase recruited to the viral factory. This might explain the requirement for virus-encoded histones, as ATP-dependent remodelers utilized by eukaryotes to facilitate transcription through chromatin are localized mostly in the nucleus. The regions in MV histones that convey this metastability (such as the H4-H3 connector and the distinct makeup of the four-helix bundle regions) are highly conserved among Marseilleviridae. Why these histones exist as doublets, what determines their specificity for the viral genome, and which (if any) assembly factors they rely on for their association with the viral DNA are intriguing open questions that warrant further research.
Genetic manipulation of giant viruses has been a neglected topic of research since their discovery despite the immense richness in genes with unknown functions encoded by their genome. Homology recombination has been used as a powerful tool to modify the genome of other viruses (Gowripalan et al., 2020), and we show here that it can be efficiently utilized for genetic manipulation of MV. Moreover, the essentiality of genes that encode MV histones, like mel_369 and mel_368, can be overcome by trans-complementation, introducing ectopic copies of the gene in the amoeba genome. Future efforts will be directed to identify selection systems to isolate mutant viruses for phenotypic analysis.
Limitations of study
Our study demonstrates that virally encoded histone doublets assemble into nucleosome-like particles in vitro and that they are present in the virus at amounts sufficient to package the entire viral genome into nucleosomes. However, we have no direct proof that the histones are indeed bound to viral DNA in the capsid. This is because any attempt to open the extremely stable virus capsid also destroys chromatin structure. However, the presence of strong electron density in the capsid core suggests that the genome is highly compacted. Methods such as cryo-electron tomography or correlative light electron microscopy (that do not require opening of the capsid) might be needed to visualize the organization of chromatin in the capsid. Although co-localization of the two histone doublets suggests that they are forming nucleosomes in the viral factory to which they are recruited, we have no direct proof of this.
The timing of the association of viral histones with DNA in the VF (in particular when nucleosomes are formed and which chaperones, if any, assist in the process) has not yet been investigated, as it requires an infectious virus whose genome encodes tagged histones. These exciting experiments have only now become possible due to our breakthrough in being able to genetically manipulate the virus.
Finally, visualizing nucleosome-like particles on the cryo-EM grid required crosslinking (as did a related study describing the structure of the MV nucleosome that was published while this article was under review; Valencia-Sánchez et al., 2021). It is possible that some aspects of the structure are more dynamic than suggested in these two structures. The high mobility of the connectors, together with the overall resolution reported here (∼4 Å), precluded a detailed description of the main chain in the connecting regions. It is therefore gratifying that the two independently determined structures are very similar, even in the ill-defined regions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Rosetta 2(DE3)pLysS | EMD Millipore | 71401-4 |
| Melbournevirus | IGS lab | N/A, available upon request |
| Chemicals, peptides, and recombinant proteins | ||
| SuperFect transfection reagent | QIAGEN | Cat# 301305 |
| Glutaraldehyde | EMS | CAS #111-30-8 |
| APTES for AFM slide | Fischer Scientific | # AC151081000 |
| Formaldehyde | Sigma-Aldrich | 252549 |
| Vectashield mounting medium with DAPI | Vector Laboratories | H-1200 |
| Deposited data | ||
| MV NLP147 structure | This paper | PDB: 7N8N |
| EM map of MV NLP147 | This paper | EMD-24238 |
| MS proteomics data | This paper | ProteomeXchange Consortium |
| PRIDE accession code: PXD003910 | ||
| Transcriptome data |
Rodrigues et al., 2020 | NCBI Short Read Archive https://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi |
| Front Microbiol | SRA accession code | |
| https://doi.org/10.3389/fmicb.2020.00651 | ERP117375 | |
| Experimental models: Organisms/strains | ||
| Acanthamoba castellanii | ATCC | 30010 |
| Oligonucleotides | ||
| All listed in Table S1 | This paper | This paper Table S1 |
| Recombinant DNA | ||
| pET-MV-H2B-H2A | This paper | N/A, available upon request |
| pET-MV-H4-H3 | This paper | N/A, available upon request |
| pET-MV-miniH2B-H2A | This paper | N/A, available upon request |
| pEF1-Mel368-GFP (GeneticinR) | This paper | N/A, available upon request |
| pEF1-Mel369-GFP (GeneticinR) | This paper | N/A, available upon request |
| pEF1-Mel149-GFP (GeneticinR) | This paper | N/A, available upon request |
| pEF1-H2A(A.castellanii)-GFP (GeneticinR) | This paper | N/A, available upon request |
| pEF1-Mel368-mRFP (NourseothricinR) | This paper | N/A, available upon request |
| pEF1- Mel368 (NourseothricinR) | This paper | N/A, available upon request |
| pEF1-Mel369 (NourseothricinR) | This paper | N/A, available upon request |
| 5′ Mel368- GFP-3′ Mel368 | This paper | N/A, available upon request |
| 5′ Mel369- GFP-3′ Mel369 | This paper | N/A, available upon request |
| Software and algorithms | ||
| AxioVision rel. 4.8.2 Software | Carl Zeiss | https://www.zeiss.com/corporate/us/home.html |
| JPK SPM software for AFM image processing | V 6.1.158 | https://customers.jpk.com/ |
| GraphPad Prism | version 9.0.0 | https://www.graphpad.com:443/ |
| CryoSPARC | V2.1 | https://cryosparc.com/ |
| UCSF Chimera | V1.14 | https://www.cgl.ucsf.edu/chimera/ |
| winCOOT | V0.9.4.1 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| UltraScan III (ver 4.0) | Demeler et al., 2014 | https://ultrascan3.aucsolutions.com/ |
| UltraScan LIMS Cluster | Demeler et al., 2014 | https://uslims4.aucsolutions.com/ |
| SV-AUC Visualization Scripts (Python3) | Edwards et al., 2020 | https://github.com/Luger-Lab/AUC-analysis |
| SWISS-MODEL webserver | Waterhouse et al., 2018 | https://swissmodel.expasy.org/interactive |
| MODELER (ver 9.20) | Sali and Blundell, 1993 | https://salilab.org/modeller/ |
| NAMD (CUDA-accelerated, ver 2.13) | Chan et al., 2012 | https://www.ks.uiuc.edu/Research/namd/ |
| CHARMM36 Forcefield Parameters | Huang and MacKerell, 2013 | http://mackerell.umaryland.edu/charmm_ff.shtml#charmm |
| VMD (ver 1.9.3) | Humphrey et al., 1996. | https://www.ks.uiuc.edu/Research/vmd/ |
| CPPTRAJ (ver 18) | Roe and Cheatham, 2013 | https://ambermd.org/AmberTools.php |
Resource availability
Lead contact
The lead contact is Karolin Luger (karolin.luger@colorado.edu).
Materials availability
Histone expression constructs, cell lines, viruses, as well as vectors are available from the lead contact upon request.
Data and code availability
Coordinates and density maps have been deposited in the PDB and EMDB databases (access codes PDB ID 7N8N, EMD-24238). All other data is available in the main text or supplemental data. Mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with accession code PXD003910.
Method details
Histone sequence alignment and secondary structure prediction
Predicted Marseilleviridae histone-like proteins were aligned with eukaryotic histone proteins with HHpred’s Multiple Alignment using Fast Fourier Transform (MAFFT) with a 1.53 gap open penalty. Lineages A, B, and C of the Marseilleviridae family were included, lineage D was excluded due to sequence similarity with lineage C (∼90%) and lineage E was excluded due to the lack of annotated histone-like proteins. Using the MAFFT alignment, protein secondary structures were predicted using HHpred’s Quick 2D structural prediction webserver to demonstrate structural conservation of the histone fold domain between Eukarya and Marseilleviridae (Zimmermann et al., 2018).
Fluorescence localization of Melbournevirus histones in infected A. castellanii cells
The three Melbournevirus (AIT54904) histones encoding genes Mel-368 (MV-H4-H3), Mel-369 (MV-H2B-H2A) and Mel-149 (MV-miniH2B-H2A) were amplified by PCR from genomic DNA and cloned into an in-house modified plasmid derived from the pGAPDH-GFP amoebal expression plasmid (Bateman, 2010). Briefly, the GAPDH promoter was replaced by the stronger EF1 gene promoter and the genes were inserted using the NdeI restriction site to yield C-terminally GFP-tagged proteins. The A. castellanii histone H2A (2) gene (ACA1_364730A) was amplified from Acanthamoeba castellanii (Douglas) Neff (ATCC 30010TM) genomic DNA and cloned into the same plasmid engineered to yield a N-terminally GFP-tagged protein.
Acanthamoeba castellanii cells were transfected with 6 μg of each plasmid using Superfect (QIAGEN). Selection of transformed cells was initially performed at 30 μg/mL Neomycin and increased up to 100 μg/mL within a couple of weeks. Transfected A. castellanii cells were grown on poly-L-lysine coating coverslips in a 12-well plate and infected with Melbournevirus at a MOI of 50 except for the negative control. At 30 min, 1h, 2h, 3h and 4h post-infection (pi), cells were fixed with PBS containing 3.7% formaldehyde for 20 min at room temperature. After one wash with PBS buffer, coverslips were mounted on a glass slide with 4 μl of VECTASHIELD mounting medium with DAPI and the fluorescence was observed using a Zeiss Axio Observer Z1 inverted microscope using a 63x objective lens associated with a 1.6x Optovar for DIC, DAPI or GFP fluorescence recording.
To perform the co-localization experiment, the previous plasmid was modified to replace GFP with mRFP. We screened several selection markers to allow the selection of the co-transfected cells and Nourseothricin was the only antibiotics highly efficient on Acanthamoeba cells. The Neomycin-resistance gene (neo) was thus replaced by the Nourseothricin-resistance gene (nat1) in the previous plasmid. Briefly, the plasmid was digested using NdeI and XbaI to remove the GFP and the linearized plasmid was gel purified. The mRFP gene was amplified and inserted by homologous recombination using the In-Fusion kit (Takara Bio) according to the manufacturer protocol. The subsequent plasmid was amplified by PCR to get rid of the Neomycin-resistance gene and Nourseothricin-resistance gene was inserted by homologous recombination. The Melbournevirus gene encoding the H3-H4 histone was then cloned using the NdeI site to yield a C-terminally mRFP-tagged protein. The structure of the construct was confirmed by sequencing. Acanthamoeba castellanii cells expressing the MV-H2A-H2B-GFP proteins were transfected with 6μg of the MV-H3-H4-mRFP plasmid and selection of the co-transfected cells was performed using 100μg/mL Neomycin and 30 to 50 μg/mL Nourseothricin. Cells were grown in a 96-well glass bottom plate and infected at a MOI of 50. At 4h pi, the cells were washed with PBS containing 10 μg/mL of DAPI and observed for DIC, DAPI, GFP or mRFP fluorescence recording.
Mass spectroscopy proteomic analyses
The mass spectrometry proteomic of the purified virions was performed as previously described (Fabre et al., 2017). Data are available at the ProteomeXchange Consortium (proteomecentral.proteomexchange.org) via the PRIDE partner repository with the accession code PXD003910. Based on the structure of Melbournevirus (Okamoto et al., 2018) exhibiting a triangulation number T = 309 corresponding to 9240 Major Capsid Proteins, the copy numbers for each protein in the viral proteome was estimated using the computed intensity-based absolute quantification (iBAQ) values.
Marseillevirus-Acanthamoeba castellanii transcriptome
The time-course transcriptome analysis of A. castellanii cells infected by Marseillevirus was performed using the RNA-seq data from (Rodrigues et al., 2020). We first annotated the Marseillevirus genome (accession GU071086) as described (Blanca et al., 2020), as well as the A. castellanii Neff host genome (assembly GCA_000193105.1) using the Funannotate pipeline (version 1.7; https://github.com/nextgenusfs/funannotate). We then mapped the RNA-seq reads corresponding to 30 min pi (ERR3528397), 1 h pi (ERR3528398), 2 h pi (ERR3528399), 4 h pi (ERR3528400), 5 h pi (ERR3528401), 6 h pi (ERR3528402), 8 h pi (ERR3528403), 10 h pi (ERR3528404) and 12 h pi (ERR3528405) to the host and viral transcripts using Bowtie (Langmead and Salzberg, 2012) (version 2.3.4.1). Transcript quantification was done using RSEM (Li and Dewey, 2011) (version 1.3.0) and data normalization using the edgeR R package (Robinson et al., 2010). Host and viral mapped reads were normalized separately with the “TMMwsp” method and then combined to a single matrix. Expression values were then expressed in log transformed counts per million of mapped reads (CPM). Finally, we scaled each transcript expression values to a Z-score and produced a heatmap for the genes of interest. A hierarchical clustering of the heatmap was obtained using a Pearson correlation distance and the “Wards.D2” method.
Knockout generation cloning strategies and analysis
Vectors for knockout of MEL_369 or MEL_368 were generated by inserting 500 base pairs homology arms into the pGAPDH-EGFP vector (Bateman, 2010). Homology arms were amplified from Melbournevirus genomic DNA produced by Wizard genomic DNA purification kit (PROMEGA) according to manufacturer’s specifications. Primers used are presented in Table S1 (HB112-115 and HB120-123 for amplification of homology arms of MEL_368 or MEL_369, respectively), and inserted into the NdeI and XbaI sites by infusion (Takara). The vectors were digested with NdeI and XbaI (NEB) prior to transfection into A. castellanii. A. castellanii cells were transfected with 8 μg of each linearized plasmid using Superfect (QIAGEN) and infection by Melbournevirus was performed 1-hour post transfection. One well was used to analyze the genomic integration using primers presented in Table S1 after 5-hours post-infection. Another well was kept to analyze the infectiousness of neo-synthesized viral particles. To remove the excess of viral particle used for infection, the growth medium was removed 1-hour post-infection and fresh growth medium was added to the culture. Washing of the cells was repeated 5 times. After lysis, the viral progeny was recovered and kept for additional round of infection.
Vector plasmids for trans-complementation were generated by directed mutagenesis using infusion (Takara) utilizing the primers HB155-HB160 to insert untagged genes into the pEF1-mRFP-NAT target vector. 6 μg of circular vectors were transfected into A. castellanii using Superfect (QIAGEN). Selection of transformed cells was initially performed at 30 μg/mL Nourseothricin and increased up to 100 μg/mL within a week post transfection. Competition assay were performed by passaging the recovered transfected viruses subsequently. Again, the presence of recombinant DNA was performed by PCR using infected cells 5-hours post infection. Extracellular virions were removed by washing 5 times. Genomic DNA was obtained by Wizard genomic DNA purification kit (PROMEGA) according to manufacturer’s specifications.
Melbournevirus histone doublet purification
Melbournevirus ORF 369 (H2B-H2A), ORF 368 (H4-H3) and ORF 149 (miniH2B-H2A) were cloned into pET-28 plasmid for expression and purification from E.coli, using protocols adapted from eukaryotic histones (Dyer et al., 2004). 6L of E. coli cells were re-suspended and lysed with 80 mL of guanidinium lysis buffer (6 M Guanidinium HCl, 20 mM sodium phosphate pH 6.8 and 2 M NaCl) and sonicated for 30 s at 50% strength for 4 times. Lysate was spun at 16,000 rpm for 25 minutes, and the supernatant was incubated with Nickel NTA Agarose Beads (GoldBio) for approx. 45 minutes at RT. The nickel beads were pelleted, and the supernatant was discarded. The beads were resuspended in 8 M Urea lysis buffer (8 M Urea, 20 mM sodium phosphate pH 6.8, and 2 M NaCl) and incubated for approximately 1 hour at room temperature.
Proteins were eluted using histone elution buffer (8 M Urea, 20 mM sodium phosphate pH 6.8, 2 M NaCl, and 500 mM Imidazole). For protein refolding, histones were dialyzed against 1 M Urea Buffer (1 M urea, 1 mM EDTA, 20 mM Tris-Cl pH 7.5 and 2 M NaCl) for 6 hr, followed by overnight dialysis against the same buffer without urea (1 mM EDTA, 20 mM Tris-Cl pH 7.5 and 2 M NaCl). Precipitated protein was removed by centrifugation, and the supernatant was concentrated and run over a size-exclusion column S200 equilibrated in 1 mM EDTA, 20 mM Tris-Cl pH 7.5 and 2 M NaCl. Samples were stored in 20% glycerol at −80°C.
Marseillevirus histone-DNA complex (nucleosome) reconstitution
Melbournevirus H2B-H2A, H4-H3 and DNA of the specified lengths were mixed at a ratio of DNA to MV-H4-H3 to MV-H2B-H2A = 1.0: 2.4: 2.4. Widom 601 DNA at 147 bp and 207bp, and 181 bp of a conserved Melbournevirus native DNA fragment selected from its genome (GC cotent = 45%) were used. Viral NLPs and X.laevis nucleosomes were both reconstituted by gradient dialysis (Dyer et al., 2004). Reconstituted products were analyzed by 5% native-PAGE.
Sucrose gradient sedimentation and gradient fixiation (GraFix) crosslinking
To form the sucrose gradient, 6 ml top solution (50 mM NaCl, 20 mM HEPES, 1 mM EDTA, pH 7.5 and 5% sucrose) was added to a tube (Beckman, 331372). Then 6 mL bottom solution (50 mM NaCl, 20 mM HEPES, 1 mM EDTA, pH 7.5 and 40% sucrose) was slowly added to the bottom until it reached the halfway mark. The tubes were placed on a gradient maker (BioComp Gradient Master) to form a continuous gradient. 200 μl of sample at 4 μM was loaded on top and spun at 4 °C for 18 h at 30,000 rpm (Beckman, Rotor SW-41Ti). Fractioned samples were dialyzed against 50 mM NaCl, 20 mM Tris-Cl, 1 mM EDTA, pH 7.5 and 1 mM DTT to remove the sucrose.
For GraFix, the same procedure was applied, except that 10% to 30% of glycerol was used instead of sucrose. In addition, 0.15% of glutaraldehyde was added to the bottom solution to form the continuous density and crosslinker gradient. After GRAFIX, the samples were eluted and dialyzed against 50 mM NaCl, 20 mM Tris-Cl, 1 mM EDTA, pH 7.5 and 1 mM DTT to quench the crosslinking reaction and remove glycerol.
Sedimentation velocity analytical ultracentrifugation (SV-AUC)
SV-AUC with absorbance optics (λ = 260 nm) was used to evaluate the homogeneity of the complexes in solution and to infer their molecular size and shape. Samples (composition as indicated) at 300 nM were spun at 30–35,000 rpm at 20°C in a Beckman XL-A ultracentrifuge in 50 mM NaCl, 20 mM Tris-Cl, 1 mM EDTA, pH 7.5 and 1 mM DTT, using an An60Ti rotor. Partial specific volumes of samples were estimated using UltraScan III version 4.0; this program was also used for all data analysis (Demeler et al., 2014; Gorbet et al., 2014).
Time-invariant and radial-invariant noise contributions were subtracted from the experimental sedimentation velocity data by 2-dimensional Spectrum Analysis (2DSA), followed by refinement using the Genetic Algorithm-Monte Carlo (GA-MC) approach (Brookes et al., 2010). Sedimentation coefficients and molecular weights in Table 1 were extracted from resulting GA-MC models. Modeling calculations were performed on the UltraScan LIMS clusters at the Bioinformatics Core Facility at University of Texas Health Science Center at San Antonio and Lonestar cluster at Texas Advanced Computing Center. Integral sedimentation coefficient distributions (G(s)) were obtained from noise-corrected experimental data with the Enhanced van Holde-Weischet Analysis method. G(s) plot figures were created using python (Edwards et al., 2020) (https://github.com/Luger-Lab/AUC-analysis).
Atomic force microscopy (AFM)
All samples were imaged in air on JPK/Bruker NanoWizard Sa with TAP300-Gold (Ted Pella) cantilevers using fast scan glass block. Images were usually scanned at 1.5 × 1.5 μm at 1- 3 Hz. All buffers were filtered through 0.02 μM filters (Anotop) before use. Mica was freshly cleaved, treated with APTES for 30 min, rinsed with water and then dried with nitrogen gas utilizing a 0.22 μm PES filter. Samples were serially diluted in TE (20 mM Tris-HCl pH 7.5, 1 mM EDTA) to a final concentration of 2 nM. Within 2 minutes of final dilution, 30 μl sample was placed on the ATPES mica slide for 2 minutes, rinsed with 1.0 mL of water and dried with filtered N2 gas. Images were analyzed using JPK SPM software V 6.1.158. Each 1.5 × 1.5 μm image was divided into 4 quadrants and digitally zoomed to obtain clearer visuals of separated particles. Lines were drawn and dragged through each particle with the greatest height of each recorded. Analysis of heights were completed in either MS Excel and/or GraphPad Prism.
Single particle cryo electron microscopy (cryo EM) and data processing
Complexes of MV-NLPs from GraFix were concentrated to 2-3 μM using Amicon Ultra-4 centrifugal filters (Ultracel 50K, Millipore). C-Flat 1.2/1.3 (Au) grids were glow discharged (EMItec, Lohmar, DE) at 40 mA for 30 s. 4 μl sample was applied onto the grid before manual plunge freezing into ethane. Images for MV-NLP with 147 bp “601” DNA were acquired at nominal magnification of 64000x on a FEI Titan Krios (300 kV), equipped with a Gatan K3 Summit direct detector. Pixel size was 1.065 Å. The movies were captured in super resolution mode with electron dose rate at 10.5 electrons per pixel per second for 5.4 s and 0.108 s per frame. Defocus range was −0.8 to −2.0 μm. Images for MV-NLP with 207 bp “601” DNA were acquired at nominal magnification of 29000x on a FEI Tecani F20 (200 kV), equipped with a Gatan K3 Summit direct detector. Pixel size was 1.219 Å. The movies were captured in super resolution mode with electron dose rate at 10 electrons per pixel per second for 8 s and 0.2 s per frame. Defocus range was −1.0 to −2.5 μm.
Both datasets were processed (Motion correction and CTF estimation) by cryoSPARC (v2.12.4) (Zhang, 2016; Zheng et al., 2017). Images were evaluated by inspecting the CTF fitting resolution. Roughly 1000 particles were then manually picked for generating autopicking templates through 2D classification in CryoSPARC. For MV-NLP with 147bp DNA, this yielded 2,648,493 particles across 6,552 micrographs, and these particles were subjected to four iterative rounds of 2D classification in order to discard bad particles. Two ab initio models were then generated, followed by heterogeneous refinement of the nucleosome-like class (34,418 particles). Non-uniform refinement, as well as global and local CTF refinements, were performed to improve the density to the final resolution, as estimated by GFSC. For MV-NLP with 207bp DNA, template-based particle picking yielded ∼2,131,563 particles across 3,495 micrographs, and these particles were subjected to four iterative rounds of 2D classifications to get best nucleosome particles. Two ab initio models were then generated, followed by heterogeneous refinement of the nucleosome like class. Non-uniform refinement with 377,702 particles, were performed to improve the density to the final resolution, as estimated by GFSC.
Initial models were built by fitting MV histone homology models (see below) into the final 3D electron maps in UCSF Chimera, yielding an initial correlation coefficient of 0.7086 (Pettersen et al., 2004). These models were then iteratively modified and locally refined in COOT (Emsley and Cowtan, 2004).Molecular dynamics flexible-fitting (MDFF, details below) was then used utilized to simultaneously balance correlation with the EM density and atomic-level energy evaluation (electrostatic configurations and removal of steric overlaps).
Homology modeling
Initial homology models of Melbournevirus ORF 369 (H2B-H2A) and ORF 368 (H4-H3) doublets were constructed using SWISSMODEL, and the histone structures from the Xenopus laevis nucleosome (PDB 1AOI) were used as a reference (Bienert et al., 2017; Guex et al., 2009; Waterhouse et al., 2018). Using Modeler (v9.20), 10,000 loops of each doublet (H2B-H2A Y152 to S192 and H4-H3 F122 to T203, with sequence numbers identified as in Figure S1A) were generated (Sali and Blundell, 1993). To eliminate improbable loops within each doublet, clash identification was performed using CPPTRAJ of the Amber MD package (v18), where a cutoff distance of 0.8 Å between potentially over-lapping atoms was used (Roe and Cheatham, 2013). Any loops identified by CPPTRAJ without a clash were manually parsed through to eliminate loops that were not sterically overlapping but also not physically relevant, such as those forming “knots” in the histone folds. While several configurations with reasonable qualitative agreement to the EM density were identified (Figure S4), the loop with the overall best correlation with the density was selected as the initial conformation for further refinement via COOT (Emsley et al., 2010), PHENIX (Adams et al., 2010), and MDFF (see below) (McGreevy et al., 2016). Figures were rendered using Chimera and VMD (Humphrey et al., 1996).
Molecular dynamics flexible fitting (MDFF) protocol
MDFF simulations were initialized from the PHENIX-refined structure based upon our initial homology modeling. This method improves fitting to EM data by introducing external forces, which bias the structure from its initial conformation into the empirical density. Simulations were conducted using the GPU-enhanced NAMD engine (v2.13) with the CHARMM36 forcefield in an implicit solvent environment (Chan et al., 2012; Huang and MacKerell, 2013), biasing forces were applied to the protein backbone and heavy atoms of the DNA, in accordance with the level of resolution provided by the density. The simulation timestep was 2 fs, and a total simulation time of 5 ns was sufficient for convergence into the empirical density. Distance- and angle-based restraints of 20 kcal/mol/Å2 and 20 kcal/mol/degree2 were applied to structured regions to prevent over-fitting of the density by breaking of domains with known secondary structure, and dihedral restraints of 50 kcal/mol/degree2 were applied to maintain proper chirality during the biasing process. Over the course of the simulation, correlation between the model and experimental density only showed a modest improvement from 0.761 to 0.766, but significant benefits to residue conformations were observed, such as the removal of drastic Φ and Ψ orientations or the repositioning of sidechains that were originally used to explain main chain densities due to overfitting by real-space refinements in PHENIX.
Structural characterization of the MV-NLP147 model
Comparisons between MV-NLP147 and eNuc147 (PDB 3LZ0; (Vasudevan et al., 2010)) were conducted using VMD, and the protein backbone (N, C, and Cα) and DNA sugar ring (C1’, C2′, C3′, C4’, and C5′) atoms were used for RMSD and center-of-geometry calculations (Table S3). To minimize discrepancies between each system, the eNuc147 molecule was least-squares fit to the MV-NLP147 using the core α helices and central 121 bp of DNA as a reference. In this way, RMSD values represent the changes in the global positioning of each constituent, rather than measuring the conformational rearrangement within each individual piece, which was altogether quite low (typically ∼1-2 Å). MV-H2B-H2A dimer reorientation, relative to the eukaryotic H2A-H2B dimer, was determined by placing fictitious particles at the geometric centers of the first and last turns of the H2B α2 helix and calculating the dihedral angle describing this imaginary atom arrangement (Figure S4A). In the eNuc147 system, the fictitious atoms describing the C-terminal end of each helix were not located at the final turn of the helix, but rather at the turn within the helix that would yield an analogous length of the H2B α2 helix in MV-NLP147, to provide the best direct comparison of the two systems. Larger magnitude values in this measurement correspond to dimers that are angled further away from the dyad-axis, as well as from one another.
Quantification and statistical analysis
Statistical analyses and software used, as well as statistical details are found in the figure and table legends, as well as in the STAR Methods.
Acknowledgments
We thank Garry Morgan (CU Boulder), Rui Yan, and Zhiheng Yu (Janelia Research Campus cryo-EM facility) for help with data collection. We also thank Matthieu Legendre for performing the expression profile analyses. This study was funded by the Howard Hughes Medical Institute (Y.L., C.M.T., K.Z., S.B., A.W., G.E., and K.L.), an NIGMS NSRA fellowship (F32GM137496) to S.B., and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No 832601; N.P., S.J., and C.A.). H.B. is the recipient of an EMBO Long-Term Fellowship (ALTF 979-2019). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
Y.L., conceptualization, methodology, validation, formal analysis, investigation, data curation, writing (original draft, review, and editing), visualization, and supervision; C.M.T., methodology, validation, formal analysis, investigation, data curation, writing (review & editing), and visualization; H.B., investigation (in vivo data) and data analysis; N.P., investigation (in vivo data) and data analysis; S.J., investigation (in vivo data) and data analysis; K.Z., methodology, validation, formal analysis, data curation, writing, visualization; S.B., methodology, software, formal analysis, investigation, data curation, writing - review & editing, supervision, and funding acquisition; A.W., investigation, formal analysis, and data curation; G.E., investigation, formal analysis, and data curation; C.A., conceptualization, formal analysis, writing (discussion and editing), supervision, project administration, and funding acquisition; K.L., conceptualization, writing (original draft, review, and editing), supervision, project administration, and funding acquisition. The two corresponding authors collaborated closely on the scientific content of this manuscript. The work is clearly split between structural “in vitro” (K.L.) and in vivo experiments (C.A.), and each corresponding author fulfilled responsibilities 1–9 listed in information to authors.
Declaration of interests
The authors declare no competing interests.
Published: July 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.06.032.
Contributor Information
Chantal Abergel, Email: chantal.abergel@igs.cnrs-mrs.fr.
Karolin Luger, Email: karolin.luger@colorado.edu.
Supplemental information
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman E. Expression plasmids and production of EGFP in stably transfected Acanthamoeba. Protein Expr. Purif. 2010;70:95–100. doi: 10.1016/j.pep.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell P.J. Viral eukaryogenesis: was the ancestor of the nucleus a complex DNA virus? J. Mol. Evol. 2001;53:251–256. doi: 10.1007/s002390010215. [DOI] [PubMed] [Google Scholar]
- Bienert S., Waterhouse A., de Beer T.A., Tauriello G., Studer G., Bordoli L., Schwede T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017;45(D1):D313–D319. doi: 10.1093/nar/gkw1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanca L., Christo-Foroux E., Rigou S., Legendre M. Comparative Analysis of the Circular and Highly Asymmetrical Marseilleviridae Genomes. Viruses. 2020;12:1270. doi: 10.3390/v12111270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman S., Wereszczynski J., Luger K. Archaeal chromatin ‘slinkies’ are inherently dynamic complexes with deflected DNA wrapping pathways. eLife. 2021;10:e65587. doi: 10.7554/eLife.65587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer M., Yutin N., Pagnier I., Barrassi L., Fournous G., Espinosa L., Robert C., Azza S., Sun S., Rossmann M.G. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. USA. 2009;106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes E., Demeler B., Rocco M. Developments in the US-SOMO bead modeling suite: new features in the direct residue-to-bead method, improved grid routines, and influence of accessible surface area screening. Macromol. Biosci. 2010;10:746–753. doi: 10.1002/mabi.200900474. [DOI] [PubMed] [Google Scholar]
- Chan K.Y., Trabuco L.G., Schreiner E., Schulten K. Cryo-electron microscopy modeling by the molecular dynamics flexible fitting method. Biopolymers. 2012;97:678–686. doi: 10.1002/bip.22042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Tokuda J.M., Topping T., Meisburger S.P., Pabit S.A., Gloss L.M., Pollack L. Asymmetric unwrapping of nucleosomal DNA propagates asymmetric opening and dissociation of the histone core. Proc. Natl. Acad. Sci. USA. 2017;114:334–339. doi: 10.1073/pnas.1611118114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua E.Y., Vasudevan D., Davey G.E., Wu B., Davey C.A. The mechanics behind DNA sequence-dependent properties of the nucleosome. Nucleic Acids Res. 2012;40:6338–6352. doi: 10.1093/nar/gks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeler B., Nguyen T.L., Gorbet G.E., Schirf V., Brookes E.H., Mulvaney P., El-Ballouli A.O., Pan J., Bakr O.M., Demeler A.K. Characterization of size, anisotropy, and density heterogeneity of nanoparticles by sedimentation velocity. Anal. Chem. 2014;86:7688–7695. doi: 10.1021/ac501722r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorigo B., Schalch T., Bystricky K., Richmond T.J. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- Dyer P.N., Edayathumangalam R.S., White C.L., Bao Y., Chakravarthy S., Muthurajan U.M., Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- Edwards G.B., Muthurajan U.M., Bowerman S., Luger K. Analytical Ultracentrifugation (AUC): An Overview of the Application of Fluorescence and Absorbance AUC to the Study of Biological Macromolecules. Curr. Protoc. Mol. Biol. 2020;133:e131. doi: 10.1002/cpmb.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erives A.J. Phylogenetic analysis of the core histone doublet and DNA topo II genes of Marseilleviridae: evidence of proto-eukaryotic provenance. Epigenetics Chromatin. 2017;10:55. doi: 10.1186/s13072-017-0162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E., Jeudy S., Santini S., Legendre M., Trauchessec M., Couté Y., Claverie J.M., Abergel C. Noumeavirus replication relies on a transient remote control of the host nucleus. Nat. Commun. 2017;8:15087. doi: 10.1038/ncomms15087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbet G., Devlin T., Hernandez Uribe B.I., Demeler A.K., Lindsey Z.L., Ganji S., Breton S., Weise-Cross L., Lafer E.M., Brookes E.H., Demeler B. A parametrically constrained optimization method for fitting sedimentation velocity experiments. Biophys. J. 2014;106:1741–1750. doi: 10.1016/j.bpj.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowripalan A., Smith S., Stefanovic T., Tscharke D.C. Rapid poxvirus engineering using CRISPR/Cas9 as a selection tool. Commun. Biol. 2020;3:643. doi: 10.1038/s42003-020-01374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N., Peitsch M.C., Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009;30(Suppl 1):S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- Hodges A.J., Gallegos I.J., Laughery M.F., Meas R., Tran L., Wyrick J.J. Histone Sprocket Arginine Residues Are Important for Gene Expression, DNA Repair, and Cell Viability in Saccharomyces cerevisiae. Genetics. 2015;200:795–806. doi: 10.1534/genetics.115.175885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Kastner B., Fischer N., Golas M.M., Sander B., Dube P., Boehringer D., Hartmuth K., Deckert J., Hauer F., Wolf E. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods. 2008;5:53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman P.M. Chromatin organization and virus gene expression. J. Cell. Physiol. 2008;216:295–302. doi: 10.1002/jcp.21421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Richmond T.J. DNA binding within the nucleosome core. Curr. Opin. Struct. Biol. 1998;8:33–40. doi: 10.1016/s0959-440x(98)80007-9. [DOI] [PubMed] [Google Scholar]
- Luger K., Richmond T.J. The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Mattiroli F., Bhattacharyya S., Dyer P.N., White A.E., Sandman K., Burkhart B.W., Byrne K.R., Lee T., Ahn N.G., Santangelo T.J. Structure of histone-based chromatin in Archaea. Science. 2017;357:609–612. doi: 10.1126/science.aaj1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGreevy R., Teo I., Singharoy A., Schulten K. Advances in the molecular dynamics flexible fitting method for cryo-EM modeling. Methods. 2016;100:50–60. doi: 10.1016/j.ymeth.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison E.A., Bowerman S., Sylvers K.L., Wereszczynski J., Musselman C.A. The conformation of the histone H3 tail inhibits association of the BPTF PHD finger with the nucleosome. eLife. 2018;7:e31481. doi: 10.7554/eLife.31481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., Sanders I.F., Chen E.Z., Li H., Tobias J.W., Isett R.B., Penubarthi S., Sun H., Baldwin D.A., Fraser N.W. Genome wide nucleosome mapping for HSV-1 shows nucleosomes are deposited at preferred positions during lytic infection. PLoS ONE. 2015;10:e0117471. doi: 10.1371/journal.pone.0117471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Miyazaki N., Reddy H.K.N., Hantke M.F., Maia F.R.N.C., Larsson D.S.D., Abergel C., Claverie J.M., Hajdu J., Murata K., Svenda M. Cryo-EM structure of a Marseilleviridae virus particle reveals a large internal microassembly. Virology. 2018;516:239–245. doi: 10.1016/j.virol.2018.01.021. [DOI] [PubMed] [Google Scholar]
- Peng Y., Markov Y., Goncearenco A., Landsman D., Panchenko A.R. Human Histone Interaction Networks: An Old Concept, New Trends. J. Mol. Biol. 2020;433:16684. doi: 10.1016/j.jmb.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentakota S., Zhou K., Smith C., Maffini S., Petrovic A., Morgan G.P., Weir J.R., Vetter I.R., Musacchio A., Luger K. Decoding the centromeric nucleosome through CENP-N. Elife. 2017;6:e33442. doi: 10.7554/eLife.33442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Polisky B., McCarthy B. Location of histones on simian virus 40 DNA. Proc. Natl. Acad. Sci. USA. 1975;72:2895–2899. doi: 10.1073/pnas.72.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues R.A.L., Louazani A.C., Picorelli A., Oliveira G.P., Lobo F.P., Colson P., La Scola B., Abrahão J.S. Analysis of a Marseillevirus Transcriptome Reveals Temporal Gene Expression Profile and Host Transcriptional Shift. Front. Microbiol. 2020;11:651. doi: 10.3389/fmicb.2020.00651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe D.R., Cheatham T.E., 3rd PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Talbert P.B., Meers M.P., Henikoff S. Old cogs, new tricks: the evolution of gene expression in a chromatin context. Nat. Rev. Genet. 2019;20:283–297. doi: 10.1038/s41576-019-0105-7. [DOI] [PubMed] [Google Scholar]
- Thomas V., Bertelli C., Collyn F., Casson N., Telenti A., Goesmann A., Croxatto A., Greub G. Lausannevirus, a giant amoebal virus encoding histone doublets. Environ. Microbiol. 2011;13:1454–1466. doi: 10.1111/j.1462-2920.2011.02446.x. [DOI] [PubMed] [Google Scholar]
- Valencia-Sánchez M.I., Abini-Agbomson S., Wang M., Lee R., Vasilyev N., Zhang J., De Ioannes P., La Scola B., Talbert P., Henikoff S. The structure of a virus-encoded nucleosome. Nat. Struct. Mol. Biol. 2021;28:413–417. doi: 10.1038/s41594-021-00585-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan D., Chua E.Y.D., Davey C.A. Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J. Mol. Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- Wang T., Liu Y., Edwards G., Krzizike D., Scherman H., Luger K. The histone chaperone FACT modulates nucleosome structure by tethering its components. Life Sci. Alliance. 2018;1:e201800107. doi: 10.26508/lsa.201800107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B., Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinformatics. 2016;54 doi: 10.1002/cpbi.3. 5.6.1–5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A.E., Hieb A.R., Luger K. A quantitative investigation of linker histone interactions with nucleosomes and chromatin. Sci. Rep. 2016;6:19122. doi: 10.1038/srep19122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Dai L., Li C., Shi L., Huang Y., Guo Z., Wu F., Zhu P., Zhou Z. Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2. EMBO J. 2021;40:e105907. doi: 10.15252/embj.2020105907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann L., Stephens A., Nam S.Z., Rau D., Kübler J., Lozajic M., Gabler F., Söding J., Lupas A.N., Alva V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018;430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The equivalent regions of H3, H4, H2A, and H2B are shown in blue, green, yellow, and red, respectively.
Data Availability Statement
Coordinates and density maps have been deposited in the PDB and EMDB databases (access codes PDB ID 7N8N, EMD-24238). All other data is available in the main text or supplemental data. Mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with accession code PXD003910.