Abstract
Remdesivir, a monophosphate prodrug of nucleoside analog GS-441524, is widely used for the treatment of moderate to severe COVID-19. It has been suggested to use GS-441524 instead of remdesivir in the clinic and in new inhalation formulations. Thus, we compared the anti-SARS-CoV-2 activity of remdesivir and GS-441524 in Vero E6, Vero CCL-81, Calu-3, Caco-2 cells, and anti-HCoV-OC43 activity in Huh-7 cells. We also compared the cellular pharmacology of these two compounds in Vero E6, Vero CCL-81, Calu-3, Caco-2, Huh-7, 293T, BHK-21, 3T3 and human airway epithelial (HAE) cells. Overall, remdesivir exhibited greater potency and superior intracellular metabolism than GS-441524 except in Vero E6 and Vero CCL-81 cells.
Keywords: COVID-19, Antiviral agents, Coronavirus, Anti-SARS-CoV-2, Remdesivir, GS-441524, HCoV-OC43, Pharmacology
Abbreviations: COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; HAE, human airway epithelial; WHO, World Health Organization; CES1, carboxylesterase 1; CatA, cathepsin A; HINT1, histidine triad nucleotide-binding protein 1; MP, monophosphate; DP, diphosphate; TP, triphosphate; NTP, nucleoside triphosphate; icSARS-CoV-2-mNG, SARS-CoV-2 infectious clone virus containing mNeonGreen reporter; ACE2, angiotensin-converting enzyme 2
Graphical abstract
Highlights
-
•
Anti-SARS-CoV-2 potency of remdesivir is similar to GS-441524 in Vero cells, but higher in Calu-3 and Caco-2 cells.
-
•
Anti-HCoV-OC43 activity of remdesivir is higher than GS-441524 in Huh-7 cells.
-
•
Cellular level of GS-443902 positively correlates with the anti-coronavirus activity in Calu-3, Caco-2 and Huh-7 cells.
-
•
Remdesivir exhibits superior cellular metabolism than GS-441524 in 293T, BHK-21, 3T3 and human airway epithelial cells.
1. Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was declared a pandemic by the World Health Organization (WHO) on March 11, 2020 (Cucinotta and Vanelli, 2020). Ever since the initial outbreak in December 2019 in Wuhan, China, the global scientific community has been actively developing and testing therapeutic agents and vaccines against SARS-CoV-2 (Ciotti et al., 2020; Nitulescu et al., 2020; Asselah et al., 2020). Remdesivir was initially developed by Gilead Sciences as an anti-Ebola agent (Warren et al., 2016). In record time, remdesivir was found to have anti-SARS-CoV-2 efficacy in vitro (Wang et al., 2020a), and clinical trials were initiated and conducted to evaluate its safety and efficacy in COVID-19 patients (Eastman et al., 2020). On May 1, 2020, remdesivir was issued FDA Emergency Use Authorization for COVID-19 patients hospitalized with severe disease for its potential to shorten the time to recovery (FDA, 2020a) and on October 22, 2020, FDA approved remdesivir for intravenous administration in adult and pediatric patients (>12 years old, > 40 kg) for treatment of COVID-19 requiring hospitalization (FDA, 2020b). The drug does not appear to reduce mortality in patients with severe symptoms (Dyer, 2020; Beigel et al., 2020) and there is some controversy regarding its value for the treatment of COVID-19 by the WHO (Dyer, 2020). It is worth noting that, unlike the Adaptive COVID-19 Treatment Trial-1 (ACTT-1) study which showed significant reduction in the duration of patient hospitalization, the WHO study was not performed under rigorous placebo controlled double-blind study conditions.
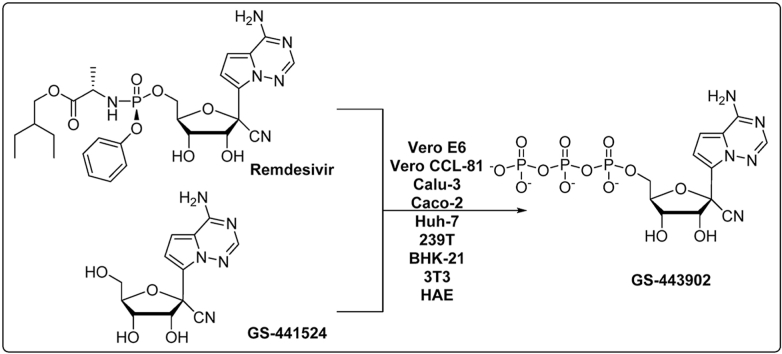
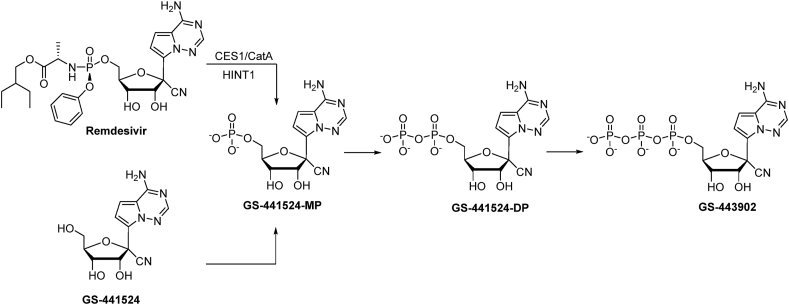
As a McGuigan-type adenosine monophosphate prodrug, remdesivir is hydrolyzed to the corresponding monophosphate (GS-441524-MP) and then intracellularly phosphorylated to its active metabolite GS-441524-triphosphate (GS-443902) (Warren et al., 2016; Li et al., 2021a, Li et al., 2021b) (Fig. 1). It has been reported that remdesivir exhibits a short plasma half-life of about 0.4 h in monkeys (Warren et al., 2016), 1 h in humans (Humeniuk et al., 2020), and that GS-441524 is the main metabolite. Interestingly, GS-441524 showed comparable efficacy to remdesivir against SARS-CoV-2 in Vero E6 African green monkey kidney cells and Calu3 2B4 human lung epithelial cells (Pruijssers et al., 2020). However, as GS-441524 has an easier synthetic route, potentially better safety profile, and may not require intravenous administration (Warren et al., 2016), some researchers in the field are advocating for the alternate use of GS-441524 for patients with COVID-19 (Yan and Muller, 2020). Thus, a side-by-side evaluation of these two compounds was warranted to determine advantages and differences between remdesivir and GS-441524.
Fig. 1.
Cellular metabolism of remdesivir and GS-441524 leading to the formation of bioactive 5′-triphosphate GS-443902. (CES1: carboxylesterase 1; CatA: cathepsin A; HINT1: histidine triad nucleotide-binding protein 1).
In this study, we compared the anti-coronavirus activity of remdesivir and GS-441524 in Vero E6, Vero CCL-81, Calu-3, Caco-2 and Huh-7 cells. Furthermore, we compared the cellular pharmacology of these two compounds in Vero E6, Vero CCL-81, Calu-3, Caco-2, Huh-7, 293T, BHK-21, 3T3 and human airway epithelial (HAE) cells. Our results in cellular models provide a reference set of data for a comparison between remdesivir and GS-441524.
2. Material and methods
2.1. Compounds and reagents
Remdesivir and GS-441524 were synthesized in house with a purity higher than 98%. GS-443902 was purchased from MedChem Express (Monmouth Junction, NJ). All other reagents were the highest quality available from Thermo Fisher Scientific (Waltham, MA).
2.2. Cells and virus
The African monkey kidney Vero E6 (ATCC® CRL-1586™) and Vero (ATCC® CCL-81™) cells, human lung epithelial cells Calu-3 (ATCC® HTB-55™), human epithelial colorectal adenocarcinoma cells Caco-2 (ATCC® HTB-37™), human kidney epithelial cells 293T/17 (ATCC® CRL-11268™), baby golden hamster kidney fibroblasts BHK-21 (ATCC® CCL-10™) and mouse embryonic fibroblasts 3T3 (ATCC® CRL-1658™) were purchased from ATCC (Manassas, VA). Human hepatocellular carcinoma cells Huh-7 (JCRB0403) was purchased from JCRB Cell Bank (Japan). Primary human airway epithelium cells (HAE) from healthy donors were obtained from Lonza Biosciences (CC-2540s) and cultured in air-liquid interface by Candela Manfredi in Dr. Eric Sorscher's group at Emory University. SARS-CoV-2 was provided by BEI Resources (NR-52281: USA-WA1/2020) and HCoV-OC43 was obtained from ATCC (Manassas, VA).
2.3. Antiviral activity and toxicity assays
To determine the antiviral activity of remdesivir and GS-441524 against SARS-CoV-2 in Vero E6, Vero CCL-81, Calu-3, and Caco-2 cells and against HCoV-OC43 in Huh-7 cells, confluent monolayers of cells in a 96-well cell culture microplate were treated with a range of concentrations of each compound in triplicate, followed by inoculation with virus MOI of 0.1 for Vero E6, Vero CCL-81, and Huh-7 or 0.01 for Calu-3 and Caco-2 cells, respectively. The production yield of progeny virus was assessed in cell supernatants collected optimally at 48 h (Vero) or 72 h (Calu-3 and Caco-2) post-infection using a specific q-RT PCR for SARS-CoV-2 and 72 h post-infection using a specific q-RT PCR for HCoV-OC43 (Zandi et al., 2020).
SARS-CoV-2 infectious clone virus containing mNeonGreen reporter (icSARS-CoV-2-mNG) inhibition assay (Xie et al., 2020) was further conducted in Caco-2 cells. Briefly, cells were treated with each compound in a range of concentrations in triplicate for 1 h prior to infection at 0.01 MOI. After 48 h, cells were fixed with 4% paraformaldehyde for 0.5–1 h, washed with PBS, permeabilized with 0.1% NP-40 in PBS and the nuclei stained with Hoechst 33342 (1:10,000 dilution in PBS) to facilitate autofocusing and imaging using a Cytation 5 cell imaging multi-mode reader (Biotek, Winooski, VT). Infection was quantified by counting mNeonGreen-expressing cells using Gen5 software (Biotek, Winooski, VT).
Cytotoxicity was evaluated using MTS method in Vero CCL-81, Calu-3, Caco-2, and Huh-7 cells as previously described. (Zandi et al., 2020).
2.4. Cellular pharmacology studies
Vero E6, Vero CCL-81, Calu-3, Caco-2, Huh-7, 293T, BHK-21, 3T3 and HAE cells were seeded at 1 × 106 cells per well in 12-well plates or 0.15 × 106 cells per well in 24-well plates (HAE only) and incubated with 10 μM of each compound at 37 °C with 5% CO2. After 4 h, cells were washed twice by ice-cold PBS and resuspended in 70% ice-cold methanol overnight at −20 °C. The supernatants were then dried and reconstituted with HPLC mobile phase and then subjected to LC-MS analysis (Tao et al., 2019). Levels of the phosphorylated metabolites 5′-mono-, di-, and triphosphate (GS-441524-MP, GS-441524-DP, and GS-443902) were measured. GS-441524-MP and GS-441524-DP were semi-quantified using the calibration curve for GS-443902.
3. Results and discussion
3.1. Anti-coronavirus activity and cytotoxicity profile of remdesivir and GS-441524
Vero E6 and Vero CCL-81 cells are interferon deficient and do not secrete interferon alpha or beta when infected by viruses (Desmyter et al., 1968). Hence, these two cell lines are commonly used as in vitro models for virus research. As they are also highly susceptible to SARS-CoV-2 infection (Pruijssers et al., 2020), they have been used broadly to evaluate anti-SARS-CoV-2 compounds in vitro (Pruijssers et al., 2020; Zandi et al., 2020; Jeon et al., 2020; Fu et al., 2020). We compared the two compounds in both Vero E6 and Vero CCL-81 cells to see if there was any difference in these two cell lines in terms of activity. Calu-3 cells are used widely as an in vitro respiratory model for the SARS-CoV-2 infection (Pruijssers et al., 2020; Ko et al., 2021; Hoffmann et al., 2020a). In this study we also included Caco-2 cells since clinical reports indicate that individuals with COVID-19 can develop gastrointestinal symptoms (such as diarrhea) either alone or with respiratory symptoms (D'Amico et al., 2020). Indeed, recent studies indicated that SARS-CoV-2 can infect the gastrointestinal tract because of high expression of the angiotensin-converting enzyme 2 (ACE2) receptor and serine protease TMPRSS2 that activates the virus to a fusogenic state in enterocytes (Hoffmann et al., 2020b). Both of these host factors that are required for efficient SARS-CoV-2 infection are robustly expressed in Caco-2 cells (D'Amico et al., 2020). The median effective concentration (EC50) and the concentration with 90% inhibitory effect (EC90) against in vitro replication of SARS-CoV-2 in Vero E6, Vero CCL-81, Calu-3, Caco-3 cells are shown in Table 1. In both Vero E6 and Vero CCL-81 cells, remdesivir showed almost same potency as GS-441524, which is in agreement with previous reports (Pruijssers et al., 2020); and for each compound, similar potency were observed in Vero E6 and Vero CCL-81 cells. In Calu-3 cells, the EC50 value of GS-441524 was about 2 times higher than remdesivir, while interestingly, GS-441524 was about 80 times less potent than remdesivir in Caco-2 cells.
Table 1.
Anti-SARS-CoV-2 efficacy and cytotoxicity of remdesivir and GS-441524 in different cell linesa.
| Compound | EC50/90 (μM) |
Cytotoxicity: MTS CC50 (μM) |
||||||
|---|---|---|---|---|---|---|---|---|
| Vero E6 | Vero CCL-81 | Calu-3 | Caco-2 | Vero CCL-81 | Calu-3 | Caco-2 | Huh-7 | |
| Remdesivir | 1.0/3.1 | 0.7/1.7 | 0.11/0.49 | 0.001/0.022 | >100 | 72.8 | >100 | 2.1 |
| GS-441524 | 1.1/3.9 | 0.8/1.6 | 0.25/2.35 | 0.08/1.42 | >100 | >100 | >100 | >100 |
Antiviral assays conducted in triplicate at least three times.
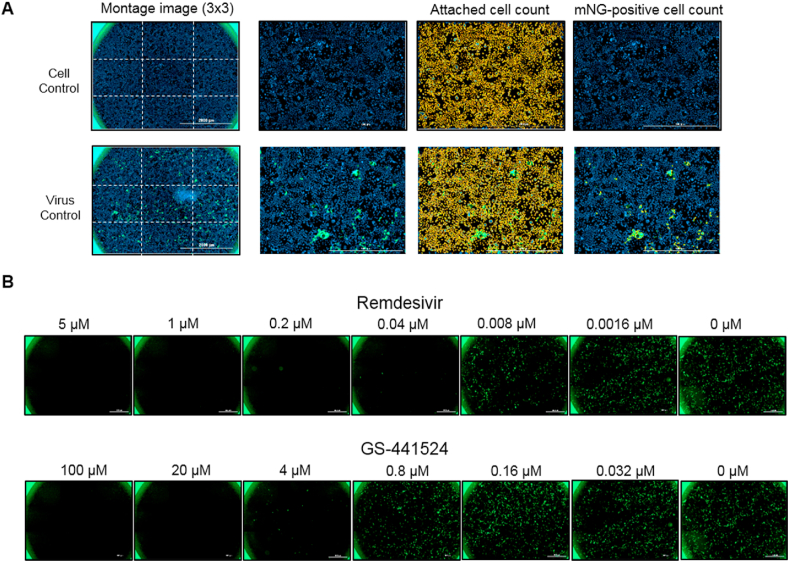
We further validated the higher potency of remdesivir versus GS-441524 in Caco-2 cells using icSARS-CoV-2-mNG inhibition assay (Fig. 2) (Xie et al., 2020). Resultant mean EC50 of remdesivir from three independent experiments was 0.018 μM while EC50 of GS-441524 was 1.3 μM, 70-fold lower compared to remdesivir. These data independently confirmed the antiviral results in Caco-2 cells determined by q-RT PCR assay (Table 1).
Fig. 2.
Imaging and analysis of icSARS-CoV-2-mNG inhibition assay in Caco-2 cells.
It has been established that SARS-CoV-2 infects the liver and directly contributes to the liver injury in patients with COVID-19 (Wang et al., 2020b). Therefore, we also used the human hepatocarcinoma Huh-7 cells to evaluate compounds against closely related virus HCoV-1 strain OC43. Remdesivir was active against this virus with an EC50 of 0.01 μM while GS-441524 was markedly less active with an EC50 of 4.1 μM. Of note, Huh-7 cells have lower susceptibility to SARS-CoV-2 infection than Vero E6 cells, and therefore are not always selected as an in vitro model for anti-SARS-CoV-2 assays (Pruijssers et al., 2020).
Cytotoxicity was evaluated using MTS method in Vero CCL-81, Calu-3, and Caco-2 cells, and additionally Huh-7 cells, a cell line we use to routinely predict liver toxicity (Zandi et al., 2020). No relevant toxicity was detected for either compound in Vero CCL-81, Calu-3 and Caco-2 cells. Interestingly, remdesivir was quite toxic in Huh-7 cells (Table 1). Notably, liver injury has been reported in remdesivir-treated COVID-19 patients (Zampino et al., 2020). Fortunately, remdesivir is given intravenously bypassing first-pass metabolism in human liver and is only given for 10 days or less.
(A) Inhibition assay was performed in a 96-well format. Hoechst 33342-stained cells were imaged using a Cytation 5 cell imaging reader at 4× magnification with DAPI and GFP channels. Nine images were taken per well and stitched to form a 3 × 3 montage image for analysis of cell count per well. A representative image of cell control (top) or virus control (bottom) was shown next to the respective montage image. Hoechst 33342-stained cells (blue) were analyzed in Gen5 software to determine overall attached cell counts. Approximately 30,000 cells were counted per well. Next, mNG-positive cells (green) were analyzed to determine infected cell counts. Counted cells were encircled in yellow. (B) Representative montage images of Caco-2 cells treated with five-fold serial dilutions of remdesivir (top) or GS-441524 (bottom) and infected with icSARS-CoV-2-mNG for 48 h. Relative % infection was calculated by normalizing infected cell counts of inhibitor-treated wells to infected cell counts of virus control wells. Data were plotted in GraphPad Prism to determine EC50.
3.2. Cellular pharmacology of remdesivir and GS-441524 in different cell lines
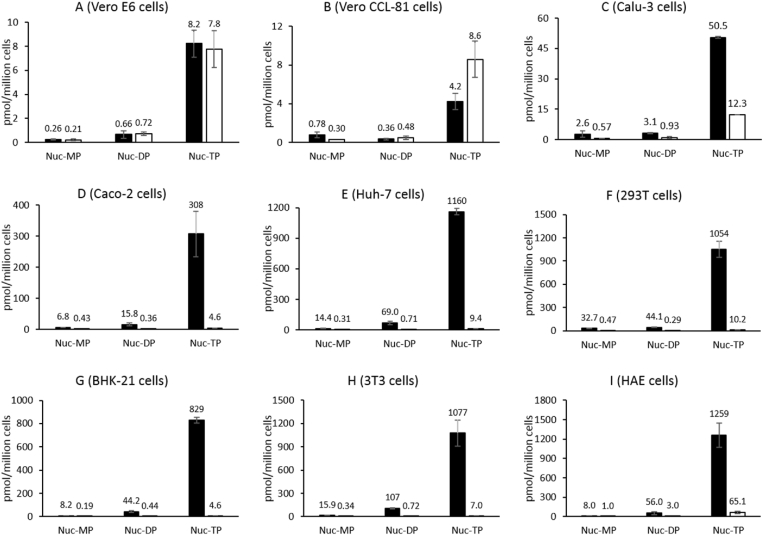
In addition to assessing the potency of the two compounds in Vero E6, Vero CCL-81, Calu-3, Caco-2 and Huh-7 cells, we compared their intracellular metabolism in these five cell lines, to determine if a correlation exists between antiviral activity and levels of the active nucleoside triphosphate (NTP) of GS-443902 produced and to further understand the cell-line dependent inhibition of SARS-CoV-2 by both compounds. Intracellular metabolites GS-441524-MP, GS-441524-DP and GS-443902 generated from remdesivir and GS-441524 after incubation in different cell lines at 10 μM for 4 h were quantified (Fig. 3). Among all the phosphorylated metabolites, the GS-443902 nucleotide was the most abundant intracellular metabolite following treatment with either compound. In Vero E6 cells, the level of GS-443902 formed from remdesivir was almost the same to that formed from GS-441524 at 4 h, and this correlated with similar antiviral activity of both compounds in Vero E6 cells (Table 1). Interestingly, in Vero CCL-81 cells, GS-441524 generated more GS-443902 than remdesivir (about 2 folds, p < 0.001, t-test) at 4 h while their anti-SARS-CoV-2 activity were similar. The antiviral assay was assessed at 48 h post infection while the cellular pharmacology was determined at 4 h. Thus, the level of active NTP at a specific time point could not be used as the only indicator for potency. In Calu-3 cells, the phosphorylated metabolites formed from remdesivir were about 4 folds higher (p < 0.001, t-test) than from GS-441524. With an EC50 of 0.11 and 0.25 μM in Calu-3 for remdesivir and GS-441524, respectively, our data are in accord with prior reports (Pruijssers et al., 2020; Li et al., 2021a), and positively correlates with the higher level of GS-443902 formed in these cells from remdesivir. In Caco-2 cells, the levels of GS-443902 formed from remdesivir were 67-fold higher (p < 0.001, t-test) than that from GS-441524 (Fig. 3). Anti-SARS-CoV-2 activity of remdesivir in Caco-2 cells is usually measured by analyzing cytopathic effect (EC50 ranging from 0.1 to 0.8 μM (Meyer et al., 2020; Ellinger et al., 2021; Bojkova et al., 2020)) or by assessing virus expression using RT-qPCR (EC50 = 46 nM) (Krüger et al., 2021). In our hands, we observed a correlation between the higher potency of remdesivir vs GS-441524 (EC50 of 1 nM vs 80 nM, respectively, or 0.018 μM vs 1.3 μM by icSARS-CoV-2-mNG inhibition assay) in Caco-2 cells and the higher levels of active NTP (308 vs 4.6 pmol/million cells, respectively) formed in this cell line. Interestingly, the levels of active NTP formed with GS-441524 only marginally fluctuate from cell line to cell line, which seemed to correlate with the relatively similar antiviral activity in Vero E6, Calu-3, and Caco-2 cells (Table 1). On the other hand, remdesivir produced significantly more GS-443902 in Calu-3 and Caco-2 cells than in Vero E6 cells (6- and 37-fold more, p < 0.001, t-test), a trend that follows the potency observed for that compound in the three different cell lines. We hypothesize that this difference is most likely due to a variation in the level of enzymes required to cleave remdesivir's prodrug moiety from one cell line to the other.
Fig. 3.
Intracellular levels of nucleoside analog GS-441524 (Nuc) -MP, -DP, and -TP produced from remdesivir (black bar) and GS-441524 (white bar) in Vero E6 (A), Vero CCL-81 (B), Calu-3 (C), Caco-2 (D), Huh-7 (E), 293T (F), BHK-21 (G), 3T3 (H), and HAE (I) cells. Ten μM of each compound was incubated with the nine cell lines/cells for 4 h at 37 °C. Values represent the means of three replicates.
This difference in anti-HCoV-OC43 potency between remdesivir and GS-441524 (0.01 μM vs 4.1 μM) correlated with the levels of active NTP formed, since incubation of Huh-7 cells with 10 μM remdesivir for 4 h generated more than 120-fold higher level of metabolite GS-443902 than incubation with 10 μM GS-441524. Notably, remdesivir was quite toxic in Huh-7 cells (Table 1), probably due to the very high intracellular levels (about 1160 pmol/million cells) of active NTP formed (Fig. 3).
In addition to Vero E6, Vero CCL-81, Calu-3, Caco-2 and Huh-7 cells, we also evaluated the intracellular metabolism of remdesivir and GS-441524 in 293T, BHK-21, 3T3 and HAE cells. 293T cell line is one of the most commonly used cell lines for in vitro SARS-CoV-2 studies because of its high expression of ACE2 and TMPRSS2 (Hoffmann et al., 2020b; Kumar et al., 2021). BHK-21 and 3T3 cells are also essential cell lines used for vaccine production (Kumar et al., 2021). Besides, BHK-21 cells support the replication of HCoV-OC43 (Shen et al., 2016). 3T3 cells had previously been modified for SARS-CoV-2 studies by transfection with a SARS-CoV-2 spike and ACE2 glycoprotein expression vector (Nunes-Santos et al., 2021). Recently, 293T and BHK-21 cell lines have been used by our group for anti-SARS-CoV-2 studies (unpublished data). The phosphorylated metabolites profiles of remdesivir and GS-441524 were very similar in 293T, BHK-21 and 3T3 cells, with the most abundant metabolite GS-443902 formed from remdesivir over 100-fold more (p < 0.001, t-test) than that from GS-441524. (Fig. 3). Finally, primary HAE cells have been used widely by researchers to study the SARS-CoV-2 (Sheahan et al., 2020; Pizzorno et al., 2020; Hattori et al., 2021; Mulay et al., 2021; Do et al., 2021). Researchers compared the anti-SARS-CoV-2 activity of remdesivir and GS-441524 in HAE models and showed that remdesivir was more potent than GS-441524 in human tracheal airway cultures (Do et al., 2021). In HAE cells at 4 h, remdesivir generated about 20-fold more (p < 0.001, t-test) GS-443902 than the parent nucleoside GS-441524 (Fig. 3), and these levels positively correlated with the higher potency. It has been reported that inhalation formulations are being developed for pulmonary administration (Sahakijpijarn et al., 2020; Sahakijpijarn et al., 2021) and clinical trials underway. With the higher potency against SARS-CoV-2 in HAE cells and superior cellular metabolism, remdesivir could be a better option than GS-441524 in the development of the inhalable formulation.
Overall, the large differences in active NTP formation after incubation with remdesivir or GS-441524 in different cell lines probably results from the fact that remdesivir is a nucleoside phosphoramidate prodrug that can be directly metabolized intracellularly to GS-441524 monophosphate, therefore bypassing the often-limiting monophosphorylation step (Varga et al., 2016). It is also worth noting that, if only passive permeation is considered, remdesivir, with an estimated LogP of 3.2 (ChemDraw Professional 16.0 - PerkinElmer), is more likely to effectively cross the hydrophobic phospholipid bilayer than the parent nucleoside GS-441524 (estimated LogP = −1.65) and therefore display higher potency.
It is worth noting that during the preparation of this manuscript, Li et al. reported that GS-441524 effectively inhibits SARS-CoV-2 infection in a mouse model (Li et al., 2021a). Their reported in vitro data seems consistent with ours except that we observed a far greater difference in EC50 between GS-441524 and remdesivir in Caco-2 cells (80-fold vs 6-fold). It is not surprising to see some in vivo efficacy in a mouse model (transduced BALB/c mice with adenovirus associated virus vector expressing hACE2) with dosing at 25 mg/kg per day (ip for 8 days) started one day before infection. Based on that study, and without a side-by-side mouse experiment with remdesivir, no definitive conclusion can be made regarding a potential advantage of GS-441524 over remdesivir in mice.
4. Conclusion
In summary, we systematically evaluated the anti-coronavirus activity of remdesivir and its parent nucleoside GS-441524 in Vero E6, Vero CCL-81, Calu-3, Caco-2, and Huh-7 cells, and intracellular pharmacology of these two compounds in Vero E6, Vero CCL-81, Calu-3, Caco-2, Huh-7, 293T, BHK-21, 3T3 and HAE cells. Except for Vero E6 and Vero CCL-81 cells, remdesivir demonstrated greater antiviral potency and markedly higher phosphorylation efficiency than GS-441524 in all cells. These results show that the relative potency of the nucleoside prodrug and the parent nucleoside is cell-type dependent, emphasizing the fact that cell model selection is highly important for evaluation of antiviral activity, especially with nucleoside analogs (Mumtaz et al., 2017). Ideally, the best nucleoside analogs are those that are active in most susceptible relevant cell culture systems. Because of the broader phosphorylation profile and higher potency of remdesivir compared to GS-441524 in the several cell systems we studied, we anticipate that remdesivir will likely continue to be a better clinical option than GS-441524.
CRediT authorship contribution statement
Sijia Tao: Methodology, Investigation, Formal analysis, Data curation, Writing – original draft. Keivan Zandi: Methodology, Investigation, Data curation, Writing – review & editing. Leda Bassit: Methodology, Investigation, Data curation, Writing – review & editing. Yee Tsuey Ong: Methodology, Investigation, Data curation, Writing – original draft. Kiran Verma: Investigation. Peng Liu: Investigation. Jessica A. Downs-Bowen: Investigation. Tamara McBrayer: Investigation. Julia C. LeCher: Investigation, Writing – review & editing. James J. Kohler: Writing – review & editing. Philip R. Tedbury: Writing – review & editing. Baek Kim: Funding acquisition. Franck Amblard: Writing – review & editing. Stefan G. Sarafianos: Writing – review & editing, Funding acquisition. Raymond F. Schinazi: Conceptualization, Supervision, Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by NIH grants R01-AI-141327 (BK and RFS), and in part by the Emory Center for AIDS Research P30-AI-050409 (RFS) and R01-AI-121315 (SGS) grants. The Nahmias-Schinazi Chair fund is also acknowledged (SGS). We thank World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) and Dr. Pei-Yong Shi from the University of Texas Medical Branch for providing icSARS-CoV-2-mNG. This paper is dedicated to the memory of our friend and colleague Dr. John C. Martin whose leadership in the field of antiviral agents led to the development of remdesivir for Ebola and more recently for COVID-19.
References
- Asselah T., Durantel D., Pasmant E., Lau G., Schinazi R.F. COVID-19: discovery, diagnostics and drug development. J. Hepatol. 2020;74(1):168–184. doi: 10.1016/j.jhep.2020.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C., ACTT-1 Study Group Members Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., McGreig J.E., McLaughlin K.M., Masterson S.G., Widera M., Krähling V., Ciesek S., Wass M.N., Michaelis M., Cinatl J. SARS-CoV-2 and SARS-CoV differ in their cell tropism and drug sensitivity profiles. bioRxiv. 2020 doi: 10.1101/2020.04.03.024257. 2020.04.03.024257. [DOI] [Google Scholar]
- Ciotti M., Ciccozzi M., Terrinoni A., Jiang W.C., Wang C.B., Bernardini S. The COVID-19 pandemic. Crit. Rev. Clin. Lab Sci. 2020;57(6):365–388. doi: 10.1080/10408363.2020.1783198. [DOI] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91(1):157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amico F., Baumgart D.C., Danese S., Peyrin-Biroulet L. Diarrhea during COVID-19 infection: pathogenesis, epidemiology, prevention, and management. Clin. Gastroenterol. Hepatol. 2020;18(8):1663–1672. doi: 10.1016/j.cgh.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmyter J., Melnick J.L., Rawls W.E. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero) J. Virol. 1968;2(10):955–961. doi: 10.1128/JVI.2.10.955-961.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do T.N.D., Donckers K., Vangeel L., Chatterjee A.K., Gallay P.A., Bobardt M.D., Bilello J.P., Cihlar T., Jonghe S.D., Neyts J., Jochmans D. A robust SARS-CoV-2 replication model in primary human epithelial cells at the air liquid interface to assess antiviral agents. Antiviral Res. 2021;192:105122. doi: 10.1016/j.antiviral.2021.105122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer O. Covid-19: remdesivir has little or no impact on survival, WHO trial shows. BMJ. 2020;371 doi: 10.1136/bmj.m4057. [DOI] [PubMed] [Google Scholar]
- Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S., Hall M.D. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent. Sci. 2020;6(5):672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger B., Bojkova D., Zaliani A., Cinatl J., Claussen C., Westhaus S., Keminer O., Reinshagen J., Kuzikov M., Wolf M., Geisslinger G., Gribbon P., Ciesek S. A SARS-CoV-2 cytopathicity dataset generated by high-content screening of a large drug repurposing collection. Sci. Data. 2021;8(1):70. doi: 10.1038/s41597-021-00848-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA News Release Remdesivir EUA letter of authorization. 2020. https://doh.sd.gov/documents/COVID19/Remdesivir_EUA_LetterOfAuthorization.pdf May 1. accessed on June 15, 2021.
- FDA News Release FDA approves first treatment for COVID-19. 2020. https://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-covid-19 October 22. accessed on June 15, 2021.
- Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., Song H., Shi Y., Li X., Tan W., Qi J., Gao G.F. Both boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11(1):4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori S.I., Higashi-Kuwata N., Hayashi H., Allu S.R., Raghavaiah J., Bulut H., Das D., Anson B.J., Lendy E.K., Takamatsu Y., Takamune N., Kishimoto N., Murayama K., Hasegawa K., Li M., Davis D.A., Kodama E.N., Yarchoan R., Wlodawer A., Misumi S., Mesecar A.D., Ghosh A.K., Mitsuya H. A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2021;12(1):668. doi: 10.1038/s41467-021-20900-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Mösbauer K., Hofmann-Winkler H., Kaul A., Kleine-Weber H., Krüger N., Gassen N.C., Müller M.A., Drosten C., Pöhlmann S. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature. 2020;585(7826):588–590. doi: 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humeniuk R., Mathias A., Cao H., Osinusi A., Shen G., Chng E., Ling J., Vu A., German P. Safety, tolerability, and pharmacokinetics of remdesivir, an antiviral for treatment of COVID-19, in healthy subjects. Clin. Transl. Sci. 2020;13(5):896–906. doi: 10.1111/cts.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon S., Ko M., Lee J., Choi I., Byun S.Y., Park S., Shum D., Kim S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020;64(7):e00819–e00820. doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M., Jeon S., Ryu W., Kim S. Comparative analysis of antiviral efficacy of FDA-approved drugs against SARS-CoV-2 in human lung cells. J. Med. Virol. 2021;93(3):1403–1408. doi: 10.1002/jmv.26397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger J., Groß R., Conzelmann C., Müller J.A., Koepke L., Sparrer K.M.J., Schütz D., Seufferlein T., Barth T.F.E., Stenger S., Heller S., Münch J., Kleger A. Drug inhibition of SARS-CoV-2 replication in human pluripotent stem cell-derived intestinal organoids. Cell Mol. Gastroenterol. Hepatol. 2021;11(4):935–948. doi: 10.1016/j.jcmgh.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Sarma P., Kaur H., Prajapat M., Bhattacharyya A., Avti P., Sehkhar N., Kaur H., Bansal S., Mahendiratta S., Mahalmani V.M., Singh H., Prakash A., Kuhad A., Medhi B. Clinically relevant cell culture models and their significance in isolation, pathogenesis, vaccine development, repurposing and screening of new drugs for SARS-CoV-2: a systematic review. Tissue Cell. 2021;70:101497. doi: 10.1016/j.tice.2021.101497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Cao L., Li G., Cong F., Li Y., Sun J., Luo Y., Chen G., Li G., Wang P., Xing F., Ji Y., Zhao J., Zhang Y., Guo D., Zhang X. Remdesivir metabolite GS-441524 effectively inhibits SARS-CoV-2 infection in mice models. J. Med. Chem. 2021 doi: 10.1021/acs.jmedchem.0c01929. acs.jmedchem.0c01929. [DOI] [PubMed] [Google Scholar]
- Li R., Liclican A., Xu Y., Pitts J., Niu C., Zhang J., Kim C., Zhao X., Soohoo D., Babusis D., Yue Q., Ma B., Murray B.P., Subramanian R., Xie X., Zou J., Bilello J.P., Li L., Schultz B.E., Sakowicz R., Smith B.J., Shi P.Y., Murakami E., Feng J.Y. Key metabolic enzymes involved in remdesivir activation in human lung cells. Antimicrob. Agents Chemother. 2021 doi: 10.1128/AAC.00602-21. AAC0060221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S.D., Bojkova D., Cinatl J., Damme E.V., Buyck C., Loock M.V., Woodfall B., Ciesek S. Lack of antiviral activity of darunavir against SARS-CoV-2. Int. J. Infect. Dis. 2020;97:7–10. doi: 10.1016/j.ijid.2020.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulay A., Konda B., Garcia G., Jr., Yao C., Beil S., Villalba J.M., Koziol C., Sen C., Purkayastha A., Kolls J.K., Pociask D.A., Pessina P., de Aja J.S., Garcia-de-Alba C., Kim C.F., Gomperts B., Arumugaswami V., Stripp B.R. SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep. 2021;35(5):109055. doi: 10.1016/j.celrep.2021.109055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumtaz N., Jimmerson L.C., Bushman L.R., Kiser J.J., Aron G., Reusken C., Koopmans M.P.G., van Kampen J.J.A. Cell-line dependent antiviral activity of sofosbuvir against Zika virus. Antivir. Res. 2017;146:161–163. doi: 10.1016/j.antiviral.2017.09.004. [DOI] [PubMed] [Google Scholar]
- Nitulescu G.M., Paunescu H., Moschos S.A., Petrakis D., Nitulescu G., Ion G.N.D., Spandidos D.A., Nikolouzakis T.K., Drakoulis N., Tsatsakis A. Comprehensive analysis of drugs to treat SARS-CoV-2 infection: mechanistic insights into current COVID19 therapies. Int. J. Mol. Med. 2020;46(2):467–488. doi: 10.3892/ijmm.2020.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Santos C.J., Kuehn H.S., Rosenzweig S.D. N-Glycan Modification in Covid-19 Pathophysiology: in vitro structural changes with limited functional effects. J. Clin. Immunol. 2021;41(2):335–344. doi: 10.1007/s10875-020-00905-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzorno A., Padey B., Julien T., Trouillet-Assant S., Traversier A., Errazuriz-Cerda E., Fouret J., Dubois J., Gaymard A., Lescure F.X., Dulière V., Brun P., Constant S., Poissy J., Lina B., Yazdanpanah Y., Terrier O., Rosa-Calatrava M. Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. Cell Rep. Med. 2020;1(4):100059. doi: 10.1016/j.xcrm.2020.100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijssers A.J., George A.S., Schafer A., Leist S.R., Gralinksi L.E., Dinnon K.H., Yount B.L., Agostini M.L., Stevens L.J., Chappell J.D., Lu X., Hughes T.M., Gully K., Martinez D.R., Brown A.J., Graham R.L., Perry J.K., Du Pont V., Pitts J., Ma B., Babusis D., Murakami E., Feng J.Y., Bilello J.P., Porter D.P., Cihlar T., Baric R.S., Denison M.R., Sheahan T.P. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep. 2020;32(3):107940. doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakijpijarn S., Moon C., Koleng J.J., Christensen D.J., Williams R.O., III Development of remdesivir as a dry powder for inhalation by thin film freezing. Pharmaceutics. 2020;12(11):1002. doi: 10.3390/pharmaceutics12111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakijpijarn S., Moon C., Warnken Z.N., Maier E.Y., DeVore J.E., Christensen D.J., Koleng J.J., Williams R.O., III In vivo pharmacokinetic study of remdesivir dry powder for inhalation in hamsters. Int. J. Pharm. X. 2021;3:100073. doi: 10.1016/j.ijpx.2021.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12(541) doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Yang Y., Ye F., Liu G., Desforges M., Talbot P.J., Tan W. Safe and sensitive antiviral screening platform based on recombinant human coronavirus OC43 expressing the luciferase reporter gene. Antimicrob. Agents Chemother. 2016;60(9):5492–5503. doi: 10.1128/AAC.00814-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao S., Zhou L., Zhang H., Zhou S., Amiralaei S., Shelton J., Ehteshami M., Jiang Y., Amblard F., Coats S.J., Schinazi R.F. Intracellular metabolism and potential cardiotoxicity of a β-D-2'-C-methyl-2,6-diaminopurine ribonucleoside phosphoramidate that inhibits hepatitis C virus replication. Nucleos Nucleot. Nucleic Acids. 2019;39(1–3):204–224. doi: 10.1080/15257770.2019.1671594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga A., Lionne C., Roy B. Intracellular metabolism of nucleoside/nucleotide analogues: a bottleneck to reach active drugs on HIV reverse transcriptase. Curr. Drug Metabol. 2016;17(3):237–252. doi: 10.2174/1389200217666151210141903. [DOI] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu S., Liu H., Li W., Lin F., Jiang L., Li X., Xu P., Zhang L., Zhao L., Cao Y., Kang J., Yang J., Li L., Liu X., Li Y., Nie R., Mu J., Lu F., Zhao S., Lu J., Zhao J. SARS-CoV-2 infection of the liver directly contributes to hepatic impairment in patients with COVID-19. J. Hepatol. 2020;73(4):807–816. doi: 10.1016/j.jhep.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Van Tongeren S.A., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Gomba L., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.S., Fearns R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531(7594):381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V., Plante K.S., Weaver S.C., Makino S., LeDuc J.W., Menachery V.D., Shi P. An infectious cNDA clone of SARS-CoV-2. Cell Host Microbe. 2020;27(5):841–848. doi: 10.1016/j.chom.2020.04.004. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan V.C., Muller F.L. Advantages of the parent nucleoside GS-441524 over remdesivir for Covid-19 treatment. ACS Med. Chem. Lett. 2020;11(7):1361–1366. doi: 10.1021/acsmedchemlett.0c00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi K., Amblard F., Musall K., Downs-Bowen J., Kleinbard R., Oo A., Cao D., Liang B., Ollinger-Russell O., McBrayer T., Bassit L., Kim B., Schinazi R.F. Repurposing nucleoside analogs for human coronaviruses. Antimicrob. Agents Chemother. 2020;65(1) doi: 10.1128/AAC.01652-20. e01652-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampino R., Mele F., Florio L.L., Bertolino L., Andini R., Galdo M., Rosa R.D., Corcione A., Durante-Mangoni E. Liver injury in remdesivir-treated COVID-19 patients. Hepatol. Int. 2020;14(5):881–883. doi: 10.1007/s12072-020-10077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]