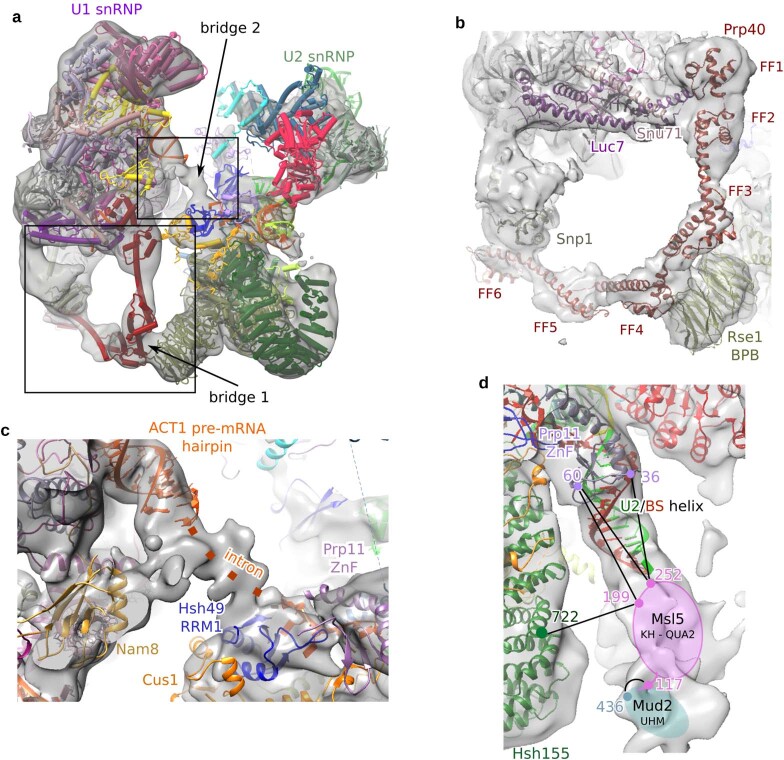
Extended Data Fig. 6. Molecular bridges connecting the U1 and U2 snRNPs in the pre-A complex.
a, Fit of the molecular model of the entire pre-A complex into the EM density (low-pass filtered). The two main bridges that connect the U1 and U2 snRNPs are indicated by arrows. The boxes indicate the regions expanded in b, c. b, Close-up of bridge 1 formed mainly by the interaction of Prp40 with Rse1. Bridge 1 is disrupted during the transition from the pre-A to the A complex (see Fig. 3 and Supplementary Video 1). Deletion analyses of Prp40 showed that although FF domains 3–6 are dispensable for yeast viability, they convey a considerable growth disadvantage when absent46. There is also evolutionary conservation of the presence of four or more of the FF domains in Prp40 from various organisms46, suggesting that FF3 and FF4 have important roles during spliceosome assembly and/or splicing. Our crosslinking data indicate that Snu71 also extensively contacts Rse1BPB and FF2 of Prp40 (see Extended Data Fig. 5d). It is conceivable that, in the absence of Prp40 FF3–FF4, Snu71 still interacts with Rse1BPB, the latter being a protein–protein interaction domain that interacts with different proteins in the subsequently formed B and Bact spliceosomal complexes. c, Close up of bridge 2 that is formed by intron nucleotides between the U2–BS helix and the 5′-ss. By analogy to the situation in later spliceosomal complexes12, these intron nucleotides are likely to be chaperoned by Hsh49RRM1 and Prp11ZnF. Intron nucleotides of the Act pre-mRNA (but not of other pre-mRNAs such as Ubc4) form a hairpin that can be localized adjacent to the U1 snRNP already in the E complex18. The resolution is not sufficient to determine the exact intron nucleotides comprising this stem. Note that the intron hairpin is not part of bridge 2. d, A lower threshold reveals EM density below the U2–BS helix, adjacent to the open Hsh155HEAT domain, that probably corresponds to the Mud2–Msl5 dimer. The EM map is low-pass filtered to 30 Å resolution. Protein crosslinks supporting the localization of Msl5–Mud2 adjacent to the U2–BS helix are shown. Numbers (colour coded to match protein colours) indicate the positions of crosslinked lysine residues, which are connected by black lines. Msl5–Mud2 could not be precisely modelled into the EM density, presumably because of their structural flexibility. However, on the basis of CXMS data, we tentatively position Msl5–Mud2 into weak density directly downstream of the branch site, close to the U2–BS helix, with Mud2 being bound to the 3′-end of the intron. Formation of the U2–BS helix requires that Msl5 hands the branch site over to the U2 snRNA, and thus Msl5 should already be displaced from the branch site in the pre-A complex. Therefore, retention of Msl5–Mud2 close to the U2–BS helix would be consistent with the binding of Mud2 to the intron downstream of the branch site.