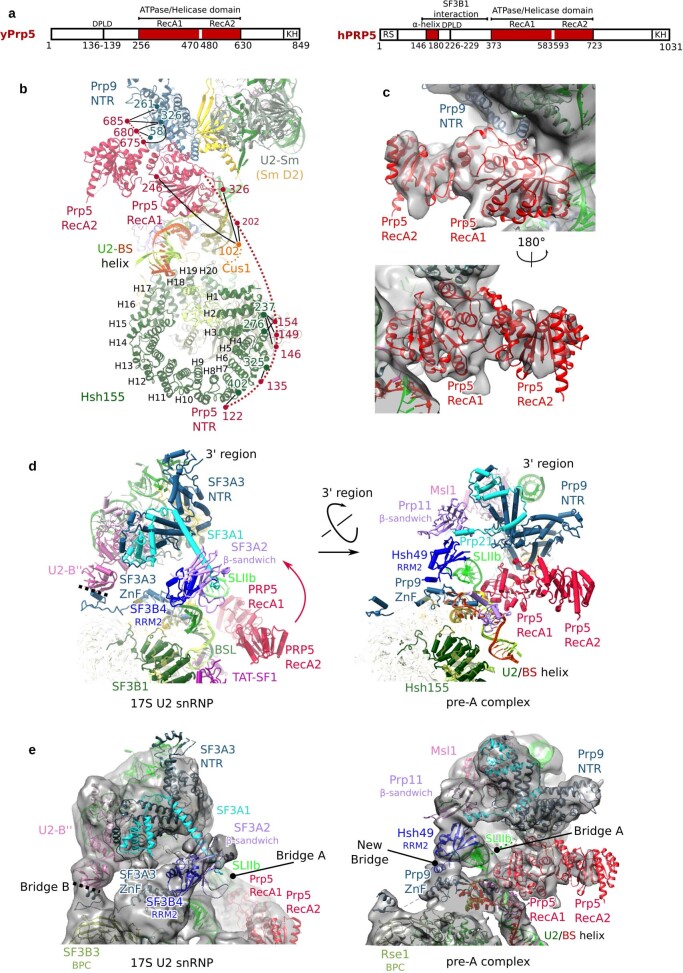
Extended Data Fig. 4. Repositioning of the Prp5 RecA domains and the U2 3′-region during formation of the pre-A complex.
a, Domain organization of the S. cerevisiae (y) and human (h) DEAD-box helicase Prp5, with the amino-acid boundaries of each domain indicated below. b, Protein crosslinks support the positions of the Prp5 NTR and RecA domains in the pre-A complex. Numbers (colour coded to match protein colours) indicate the positions of crosslinked lysine residues, which are connected by black lines. The proposed path of Prp5 amino acids located more N-terminally of the RecA domains is indicated by a dashed line. That the Prp5 NTR and RecA1 domains, but not RecA2 (and presumably also its C-terminal region), interact with other pre-A components is consistent with previous studies showing that, after destabilization of the U2 BSL, Prp5NTR and Prp5RecA1 are sufficient for the subsequent ATP-independent function of Prp5 during A-complex formation14. c, Two different views of the fit of the Prp5 RecA1 and RecA2 domains in an open conformation into the pre-A EM density (low-pass filtered to 10 Å). A closed conformation of the Prp5RecA domain does not fit well to the EM density (not shown). The open conformation of Prp5 found in the pre-A complex indicates that, after ATP hydrolysis, the RecA domains are able to transit spontaneously from the closed conformation back to the open conformation while probably remaining bound to U2. d, e, The positions of the Prp5RecA domains and the U2 3′-region plus SF3a proteins, relative to SF3b, are different in the human 17S U2 snRNP and the yeast pre-A complex. Aligned via U2 SLIIa and HR 19–20 of SF3B1/Hsh155. A cryo-EM structure of an isolated S. cerevisiae U2 snRNP is currently lacking. However, the high conservation of the sequence of yeast U2 proteins and their human homologues, and the similar structures of their conserved domains, suggests that the molecular architecture of the isolated U2 snRNP is similar in S. cerevisiae and humans. Thus, a comparison of the structures of the human 17S U2 snRNP and yeast pre-A complex reveals structural remodelling that the U2 snRNP most likely undergoes during formation of the pre-A complex. An alignment of the U2 5′-region in both complexes suggests that the U2 3′-region is repositioned after U2 stably interacts during formation of the pre-A complex. Specifically, the U2 3′-domain (that is, the 3′-region minus the SF3a core) and the Prp9NTR rotate towards the Prp5RecA domains, whereas the Prp11 β-sandwich and Hsh49RRM2 move towards Prp9ZnF. The shifted position of the U2 3′-region is stabilized by different molecular bridges formed between the U2 3′- and 5′-regions. In the pre-A complex, the bridge formed by U2-B′′ RRM2, Prp9 (human SF3A3) and Rse1BPC (human SF3B3) in the 17S U2 snRNP (denoted bridge B) is disrupted, which allows the 3′-region to move further away from the Rse1BPC. This then allows Hsh49RRM2 to dock on top of the Prp9ZnF, and by binding to Prp9 on one side and the Prp11 β-sandwich domain on the other, a new bridge involving Hsh49RRM2 is formed. Moreover, in the isolated human 17S U2 snRNP, U2 SLIIb forms a second bridge (denoted bridge A) between the U2 3′- and 5′-regions that is not stabilized and is only poorly resolved. By contrast, in the pre-A complex, Hsh49RRM2 now binds to the loop of SLIIb and thereby stabilizes the position of SLIIb.