Abstract
The omega-3 fatty acids eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) affect cell function and metabolism, but the differential effects of EPA and DHA are not known. In a randomized, controlled, double-blind, crossover study, we assessed the effects of 10-week supplementation with EPA-only and DHA-only (3 g/d), relative to a 4-week lead-in phase of high oleic acid sunflower oil (3 g/day, defined as baseline), on fasting serum metabolites in 21 subjects (9 men and 12 post-menopausal women) with chronic inflammation and some characteristics of metabolic syndrome. Relative to baseline, EPA significantly lowered the tricarboxylic acid (TCA) cycle intermediates fumarate and α-ketoglutarate and increased glucuronate, UDP-glucuronate, and non-esterified DHA. DHA significantly lowered the TCA cycle intermediates pyruvate, citrate, isocitrate, fumarate, α-ketoglutarate, and malate, and increased succinate and glucuronate. Pathway analysis showed that both EPA and DHA significantly affected the TCA cycle, the interconversion of pentose and glucuronate, and alanine, and aspartate and glutamate pathways (FDR < 0.05) and that DHA had a significantly greater effect on the TCA cycle than EPA. Our results indicate that EPA and DHA exhibit both common and differential effects on cell metabolism in subjects with chronic inflammation and some key aspects of metabolic syndrome.
Subject terms: Cell biology, Diseases, Biochemistry, Lipids, Metabolomics
Introduction
Chronic inflammation is a hallmark of several common cardiometabolic diseases such as metabolic syndrome, obesity, type 2 diabetes mellitus and cardiovascular disease (CVD)1. Recent advances in metabolomics tools and analysis have increased our ability to uncover metabolic pathways affected by these diseases and by treatments that ameliorate them2,3. Metabolic syndrome, which is highly prevalent in obesity and predisposes to both diabetes and CVD, has been associated with plasma levels of various metabolites: biogenic amines such as choline, L-carnitine and trimethylamine- N-oxide (TMAO), amino acids such as valine, alanine, glutamate and tyrosine, glucose, and the intermediates of energy metabolism such as pyruvate4,5. In animal models of diet-induced obesity and CVD, the long chain omega-3 fatty acids (n-3 FA) eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) have been shown to reduce inflammation and improve insulin sensitivity and glucose metabolism6. However, in human prospective and randomized intervention studies the evidence of these beneficial effects of n-3 FA supplementation on glucose homeostasis is mixed6,7. Nevertheless, reduction in CVD risk by n-3 FA, more by EPA than DHA, has been reported8,9, Within cells, n-3 FA are not only important components of the cell membrane, but also have important roles in a number of metabolic processes by regulating the activity of enzymes, acting as signaling molecules, and serving as ligands for transcription factors10. A direct comparison between EPA and DHA regarding their effects on metabolism in humans has not been performed.
The objective of this study was to gain a better understanding of the common and differential effects of EPA and DHA on the serum metabolome and associated metabolic pathways in individuals with chronic inflammation and multiple features of metabolic syndrome.
Results
Subjects’ characteristics
The mean age (± SD) of subjects who completed the study (n = 21) was 61 ± 6 years (Table 1). All subjects were overweight or obese, with a mean body mass index (BMI) of 32.2 kg/m2 and mean waist circumference above the sex-specific values for abdominal obesity. Subjects had borderline high fasting plasma blood glucose and triglyceride (TG) concentrations.
Table 1.
Characteristics of subjects at screening visit.
| All (n = 21) | Male (n = 9) | Female (n = 12) | |
|---|---|---|---|
| Age (y) | 61 ± 6 | 59 ± 5 | 64 ± 6 |
| Weight (kg) | 92.7 ± 20.4 | 102.5 ± 17.0 | 85.3 ± 20.1 |
| BMI (kg/m2) | 32.2 ± 6.6 | 32.2 ± 5.6 | 32.2 ± 7.5 |
| Waist circumference (cm) | 104 ± 15 | 111 ± 11 | 100 ± 15 |
| SBP (mmHg) | 130 ± 16 | 132 ± 10 | 128 ± 19 |
| DBP (mmHg) | 79 ± 12 | 87 ± 8 | 73 ± 11* |
| Fasting glucose (mg/dL) | 100 ± 10 | 101 ± 11 | 99 ± 10 |
| Fasting TG (mg/dL) | 144 ± 46 | 129 ± 31 | 155 ± 53 |
Values are reported as mean ± SD.
*P < 0.05, women versus men.
BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; TG, triglyceride.
Screening values in men and women were similar, except for diastolic blood pressure, which was significantly lower in women than men (P = 0.003).
Effects of EPA and DHA on biochemical parameters
We have previously reported the effect of EPA and DHA supplementation on the plasma phospholipid content of arachidonic acid (AA), EPA and DHA11. The median molar percent (mol%) of AA at baseline and at the end of the EPA and DHA phases was 11.5, 10.5 and 9.5, respectively. The median mol% of EPA was 0.7, 5.3 and 1.5, respectively, and of DHA was 2.8, 3.1 and 7.7, respectively.
Biochemical parameters at baseline and after EPA and DHA supplementation are shown in Table 2. Relative to baseline, EPA supplementation significantly increased serum concentrations of lactate dehydrogenase whereas DHA supplementation significantly increased serum albumin concentrations. There was no significant difference between the EPA- and DHA-induced changes.
Table 2.
Serum concentrations of biochemical parameters at baseline and after EPA and DHA supplementation1.
| Baseline | EPA | ∆EPA from baseline2 | DHA | ∆DHA from baseline2 | FDR-∆EPA vs. ∆DHA3 | |
|---|---|---|---|---|---|---|
| Albumin (g/dL) | 4.03 ± 0.20 | 4.11 ± 0.18 | 0.08 ± 0.20 | 4.16 ± 0.22 | 0.12 ± 0.17* | 0.36 |
| Total protein (g/dL) | 6.60 ± 0.30 | 6.71 ± 0.33 | 0.10 ± 0.31 | 6.78 ± 0.38 | 0.17 ± 0.31 | 0.36 |
| SGPT (mU/mL) | 6.48 ± 2.99 | 7.87 ± 3.20 | 1.33 ± 3.94 | 8.10 ± 3.95 | 1.62 ± 3.53 | 0.64 |
| SGOT (mU/mL) | 12.38 ± 3.34 | 13.24 ± 3.97 | 0.86 ± 3.18 | 13.57 ± 3.38 | 1.19 ± 2.56 | 0.82 |
| LDH (U/L) | 110.62 ± 21.37 | 120.38 ± 22.28 | 9.76 ± 17.18* | 118.33 ± 21.44 | 7.71 ± 15.14 | 0.35 |
| ALP (U/L) | 58.90 ± 16.62 | 61.71 ± 18.52 | 2.81 ± 10.91 | 57.76 ± 17.60 | − 1.14 ± 10.10 | 0.07 |
| BUN (mg/dL) | 16.43 ± 4.23 | 15.43 ± 3.68 | − 1.00 ± 2.57 | 16.38 ± 5.17 | − 0.05 ± 3.17 | 0.46 |
| Creatinine (mg/dL)4 | 0.87 ± 0.19 | 0.89 ± 0.19 | 0.01 ± 0.08 | 0.89 ± 0.19 | 0.02 ± 0.19 | 0.79 |
| Glucose (mg/dL) | 98.43 ± 8.07 | 100.29 ± 7.29 | 1.86 ± 5.75 | 98.95 ± 9.19 | 0.52 ± 6.10 | 0.35 |
| NEFA (mmol/L) | 0.18 ± 0.09 | 0.21 ± 0.13 | 0.03 ± 0.09 | 0.20 ± 0.13 | 0.02 ± 0.11 | 0.70 |
| Uric acid (mg/dL)4 | 5.86 ± 1.34 | 5.79 ± 1.41 | − 0.07 ± 0.76 | 5.79 ± 1.30 | − 0.07 ± 0.83 | 0.77 |
| Total bilirubin (mg/dL) | 0.56 ± 0.17 | 0.59 ± 0.19 | 0.03 ± 0.17 | 0.58 ± 0.16 | 0.02 ± 0.19 | 0.88 |
1Values are reported as unadjusted mean ± SD. To correct for skewed distribution, log-transformation was applied before analysis.
2Comparison of changes from baseline to EPA or DHA was assessed using a liner mixed-effects model.FDR-adjusted P values; *FDR < 0.05.
3Pairwise comparison between EPA and DHA (changes form baseline) were conducted using the lsmeansLT function from the lmerTest package for the linear mixed effect model.
4Creatinine (p = 0.0231) and uric acid (p = 0.04) showed a significant sequence effect; therefore, only data from the first period was used to run ANCOVA analysis, with baseline value as a covariate.
SGPT, serum glutamic-pyruvic transaminase; SGOT, serum glutamic-oxaloacetic transaminase; LDH, lactate dehydrogenase; ALP, alkaline phosphatase; BUN, blood urea nitrogen; NEFA, non-esterified fatty acids.
Effects of EPA and DHA on serum primary metabolites
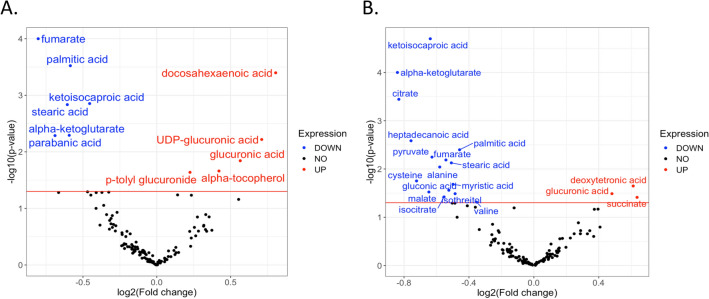
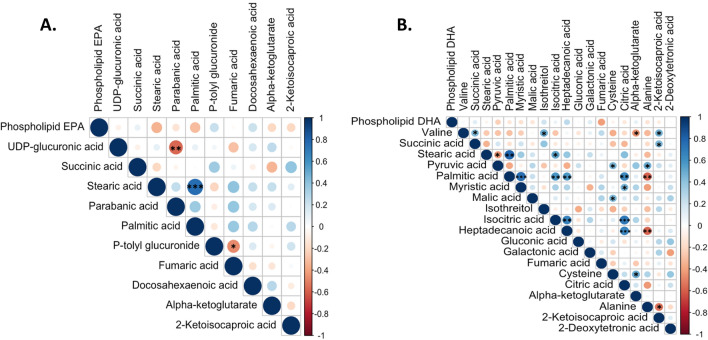
The PLS-DA plot of metabolites at baseline and after EPA and DHA supplementation is shown in Fig. 1. Relative to baseline, EPA supplementation significantly altered 11 known and 15 unknown serum primary metabolites (Table 3). EPA significantly increased non-esterified DHA and the glucose metabolites UDP-glucuronic acid and glucuronic acid. EPA lowered the tricarboxylic acid (TCA) cycle intermediate metabolites α-ketoglutarate and fumarate, the saturated fatty acids stearic acid and palmitic acid, and the leucine metabolite ketoisocaproic acid, in addition to parabanic acid (Table 3 and Fig. 2). The change in fumarate was inversely associated with the change in p-tolyl-glucuronide (Fig. 3).
Figure 1.
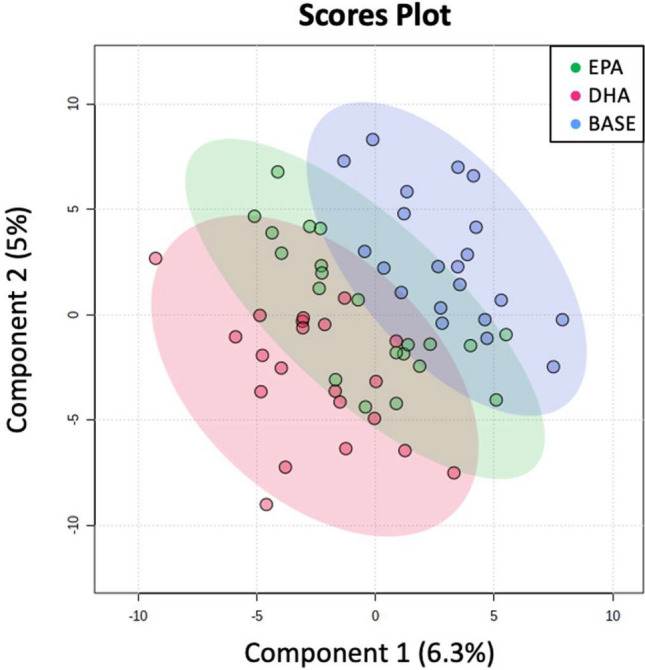
PLS-DA score plot. Primary metabolites are shown at baseline (blue) and at the end of the EPA (green) and DHA (red) supplementation phases.
Table 3.
Peak height values of serum primary metabolites at baseline and after EPA and DHA supplementation (alphabetic order)1.
| Baseline | EPA | ∆EPA from baseline2 | DHA | ∆DHA from baseline2 | FDR ∆EPA vs. ∆DHA3 | |
|---|---|---|---|---|---|---|
| -Ketoglutarate | 1572 (1374, 2098) | 1344 (820, 1699) | − 546 (− 848, 156)** | 1011 (814, 1303) | − 574 (− 1095, − 25)*** | 0.70 |
| -Tocopherol | 17,567 (15,450, 21,920) | 21,166 (17,377, 24,927) | 3599 (244, 4918)* | 21,490 (15,673, 23,604) | 3495 (− 430, 5599) | 0.042 |
| Citric acid | 67,091 (58,034, 76,758) | 63,059 (55,048, 74,021) | − 5585 (− 12,624, 8242) | 60,541 (46,556, 65,524) | − 10,005 (− 21,349, − 845)*** | 0.05 |
| Cysteine | 30,308 (28,004, 40,007) | 31,598 (25,512, 38,123) | − 1645 (− 6057, 10,161) | 28,160 (25,635, 30,844) | − 2545 (− 11,847, 3008)* | 0.47 |
| Docosahexaenoic acid | 1324 (1002, 1617) | 1833 (1384, 2472) | 493 (272, 1180)*** | 1611 (1214, 1840) | 278 (− 32, 572) | 0.51 |
| Deoxytetronic acid | 1323 (681, 1690) | 1235 (635, 1666) | − 88 (− 504, 273) | 1485 (1306, 1948) | 274 (− 66, 835)* | 0.0342 |
| Fumaric acid4 | 5539 (3793, 6117) | 4073 (3408, 5116) | − 871 (− 2260, − 14)*** | 4216 (3605, 5460) | − 104 (− 1516, 754)** | 0.0042 |
| Gluconic acid | 1288 (1105, 1608) | 1269 (1055, 1558) | 161 (− 164, 247) | 1217 (1042, 1649) | − 45 (− 119, 145)* | 0.23 |
| Glucuronic acid | 1856 (1481, 2717) | 2782 (2254, 3370) | 654 (101, 1153)* | 2697 (2145, 3088) | 580 (− 49, 1371)* | 0.56 |
| Heptadecanoic acid | 7790 (6494, 8727) | 7017 (6143, 8826) | − 560 (− 2055, 458) | 6604 (5490, 7788) | − 697 (− 2400, 344)** | 0.19 |
| Isocitric aicd | 1094 (1001, 1341) | 1146 (948, 1281) | − 94 (− 241, 138) | 979 (808, 1117) | − 152 (− 390, 24)* | 0.23 |
| Isothreitol | 2072 (1580, 2886) | 2418 (1580, 2972) | 85 (− 318, 735) | 1654 (1088, 2221) | − 330 (− 1013, 101)* | 0.41 |
| Ketoisocaproic acid | 50,177 (42,537, 57,751) | 48,120 (36,122, 50,704) | − 6050 (− 10,576, − 1164)** | 41,822 (33,509, 50,332) | − 5555 (− 16,107, 100)*** | 0.92 |
| Malic acid | 1350 (1233, 1731) | 1598 (1261, 1852) | − 94 (− 352, 427) | 1386 (1040, 1600) | − 253 (− 444, 111)* | 0.89 |
| Myristic acid | 7063 (5741, 7953) | 6388 (4678, 7191) | − 769 (− 1620, 583) | 6069 (5771, 8255) | − 798 (− 2198, 1192)* | 0.73 |
| P-tolyl glucuronide | 232 (164, 364) | 294 (197, 569) | 67 (− 64, 350)* | 248 (187, 408) | 19 (− 64, 120) | 0.0381 |
| Palmitic acid | 143,747 (130,251, 170,438) | 134,008 (103,508, 143,473) | − 20,098 (− 31,503, − 4319)*** | 135,581 (118,709, 161,108) | − 13,121 (− 29,561, 12,833)** | 0.13 |
| Parabanic acid | 4087 (2895, 4648) | 3369 (2586, 4401) | − 718 (− 1379, − 6)** | 3576 (2957, 4704) | 322 (− 849, 845) | 0.11 |
| Phenylacetic acid | 803 (612, 1223) | 874 (671, 996) | − 29 (− 205, 348) | 973 (762, 1241) | 190 (9, 464) | 0.0276 |
| Phenylethylamine | 374 (294, 513) | 586 (317, 724) | 130 (− 117, 379) | 358 (221, 549) | − 22 (− 152, 76) | 0.0011 |
| Pyruvic acid | 39,697 (20,995, 49,411) | 27,746 (17,769, 39,011) | − 9691 (− 21,665, − 363) | 14,831 (9284, 25,225) | − 15,509 (− 24,309, 1969)** | 0.83 |
| Serotonin | 1845 (1009, 2652) | 1864 (994, 2670) | − 96 (− 616, 678) | 2204 (1543, 3054) | − 158 (− 341, 739) | 0.99 |
| Stearic acid | 448,176 (414,026, 561,336) | 460,347 (352,958, 512,616) | − 48,364 (− 97,126, − 6545)** | 430,408 (409,245, 499,584) | 474 (− 124,502, 51,301)** | 0.86 |
| Succinic acid | 3303 (2833, 3744) | 3440 (3037, 3823) | 505 (− 233, 1029) | 3560 (3328, 4422) | 579 (3, 1716)* | 0.93 |
| UDP-glucuronic acid | 797 (611, 1103) | 1111 (935, 1578) | 389 (14, 587)** | 1107 (576, 1382) | 320 (− 286, 643) | 0.54 |
| Valine | 1,248,470 (1,134,926, 1,308,718) | 1,234,454 (1,088,759, 1,356,740) | 2072 (− 90,478, 102,381) | 1,134,770 (1,095,117, 1,297,538) | − 11,483 (− 151,936, 71,861)* | 0.68 |
1Values are unadjusted and reported as median (25th, 75th). To correct for skewed distribution, generalized log-transformation and auto-scaling were applied before analysis.
2Comparison of changes from baseline to EPA or DHA was assessed using a liner mixed-effects model. FDR-adjusted P value: * < 0.05, ** < 0.01, *** < 0.001.
3Pairwise comparison between EPA and DHA (changes form baseline) were conducted using the lsmeansLT function from the lmerTest package for the linear mixed effect model.
4Fumaric acid showed significant sequence effect (p = 0.0073). When there was a sequence effect, data from the first period is extracted to run ANCOVA analysis, with baseline value and age as covariates. EPA supplementation reduced level of fumaric acid (p = 0.0107) when sequence effect was taken into account.
Figure 2.
Volcano plots of changes in primary metabolites, relative to baseline, after EPA (a) and DHA (b) supplementation. Metabolites are shown in red if significantly increased following supplementation and in blue if significantly reduced following supplementation. Red line represents the cut-off point for FDR < 0.05.
Figure 3.
Correlation among metabolite changes after EPA and DHA supplementation. Metabolites shown to have significant changes after EPA and DHA supplementation were included for the Pearson’s correlation analysis. Positive and negative correlation are presented in blue and red, respectively. (a) Changes in metabolites and phospholipid EPA concentrations after EPA supplementation; (b) Changes in metabolites and phospholipid DHA concentrations after DHA supplementation. *P < 0.05, **P < 0.01, ***P < 0.001.
Relative to baseline, DHA significantly affected 19 known and 22 unknown metabolites. DHA increased glucuronic acid, the TCA cycle metabolite succinic acid, and deoxytetronic acid and reduced the amino acids alanine, valine and cysteine, the TCA intermediate metabolites pyruvate, α-ketoglutarate, citric acid, isocitrate, fumarate and malate, and the saturated fatty acids myristic acid, palmitic acid and stearic acid, in addition to phenylethylamine (Table 3 and Fig. 2). During DHA supplementation, the reduction in citrate was significantly associated with the reduction in myristic, palmitic and heptadecanoic acids and isocitric acid (Fig. 3). Non-esterified DHA concentrations did not change significantly after DHA supplementation (FDR = 0.07).
DHA supplementation, relative to EPA, led to a more pronounced reduction in citric acid (− 15% vs. − 8%, respectively; FDR = 0.05) and greater increases in phenylacetic acid (+ 21% vs. + 9%, FDR = 0.027) and deoxytetronic acid (+ 22% vs. − 6%, FDR = 0.034). On the other hand, EPA had more prominent effects than DHA in decreasing fumarate (− 16% vs. − 2%, respectively; FDR = 0.004) and increasing p-tolyl β-D-glucuronide (+ 27% vs. + 7%, FDR = 0.038) and phenylethylamine (+ 56% vs. -5%, FDR = 0.002).
To gain insight into the unique pathways associated with the changes in primary metabolites induced by EPA and DHA supplementation, pathway analysis was performed. Compared with baseline, EPA supplementation significantly affected six pathways including: ascorbate and aldarate metabolism; pentose and glucuronate interconversions; TCA cycle; alanine, aspartate and glutamate metabolism; tyrosine metabolism; and amino sugar and nucleotide sugar metabolism (Table 4). DHA supplementation was significantly associated with four pathways, including: ascorbate and aldarate metabolism; TCA cycle; pentose and glucuronate interconversions; and alanine, aspartate and glutamate metabolism (Table 4).
Table 4.
Top pathways affected by EPA and DHA supplementation.
| Total cmpd | Hits N | Hits metabolites | Direction | FDR | Impact | |
|---|---|---|---|---|---|---|
| EPA, relative to baseline | ||||||
| Ascorbate and aldarate metabolism | 8 | 2 | Glucuronate | (+) | 0.01 | 1 |
| UDP-glucuronate | (+) | |||||
| Pentose and glucuronate interconversions | 18 | 2 | Glucuronate | (+) | 0.01 | 0.25 |
| UDP-glucuronate | (+) | |||||
| Citrate cycle (TCA cycle) | 20 | 2 | α-Ketoglutarate | (−) | 0.03 | 0.08 |
| Fumarate | (−) | |||||
| Alanine, aspartate and glutamate metabolism | 28 | 2 | α-Ketoglutarate | (−) | 0.03 | 0.05 |
| Fumarate | (−) | |||||
| Tyrosine metabolism | 42 | 1 | Fumarate | (−) | 0.023 | 0.02 |
| Amino sugar and nucleotide sugar metabolism | 37 | 1 | UDP-glucuronate | (+) | 0.023 | 0.02 |
| DHA, relative to baseline | ||||||
| Ascorbate and aldarate metabolism | 8 | 1 | Glucuronate | (+) | 0.042 | 0.50 |
| Citrate cycle (TCA cycle) | 20 | 7 | Pyruvate | (−) | 0.042 | 0.30 |
| Citrate | (−) | |||||
| Isocitrate | (−) | |||||
| α-Ketoglutarate | (−) | |||||
| Succinate | (+) | |||||
| Fumarate | (−) | |||||
| Malate | (−) | |||||
| Pentose and glucuronate interconversions | 18 | 1 | Glucuronate | (+) | 0.042 | 0.10 |
| Alanine, aspartate and glutamate metabolism | 28 | 6 | Alanine | (−) | 0.042 | 0.05 |
| Pyruvate | (−) | |||||
| Citrate | (−) | |||||
| α-Ketoglutarate | (−) | |||||
| Succinate | (+) | |||||
| Fumarate | (−) | |||||
| EPA vs. DHA | ||||||
| Citrate cycle (TCA cycle) | 20 | 2 | Citrate | (+) | 0.018 | 0.12 |
| Fumarate | (−) | |||||
| Glyoxylate and dicarboxylate metabolism | 32 | 1 | Citrate | (+) | 0.018 | 0.03 |
| Alanine, aspartate and glutamate metabolism | 28 | 2 | Citrate | (+) | 0.018 | 0.002 |
| Fumarate | (−) | |||||
Significant pathways (FDR < 0.05) with impact scores greater than 0 are presented.
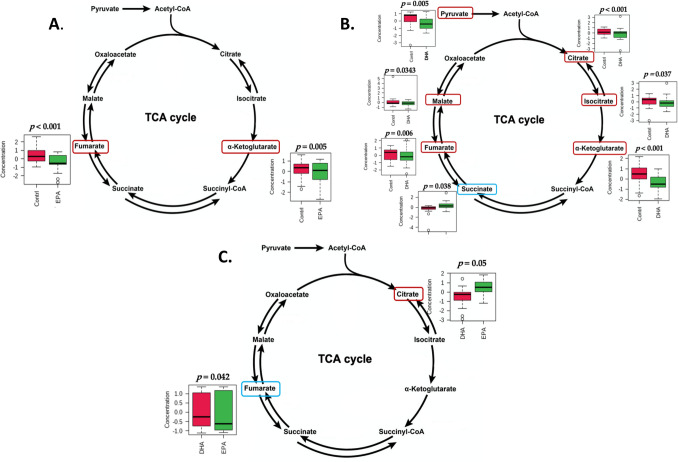
The TCA cycle was differentially affected by EPA and DHA supplementation, with a greater reduction of citrate by DHA and of fumarate by EPA (Table 4, Fig. 4).
Figure 4.
EPA and DHA modulation of TCA cycle metabolites. Significantly increased and decreased metabolites are circled in blue and red, respectively. (a) baseline versus EPA supplementation; (b) baseline versus DHA supplementation; (c) EPA versus DHA.
Effects of EPA and DHA on biogenic amines
Biogenic amine metabolites were measured in a subset of 10 subjects, whose characteristics are shown in Supplementary Tables 1 and 2. Compared to baseline, EPA supplementation significantly changed 255 metabolites (10 known and 245 unknown) (Supplementary Table 3). EPA significantly increased benzophenone and aminomethyl-cyclohexane-carboxylic acid and lowered alanine, creatinine, diamino-2-methylpropane, glutamate-glutamine, histamine, pyroglutamic acid, and stachydrine. DHA supplementation significantly affected a distinct set of metabolites (15 known and 343 unknown). DHA lowered acetyl-carnitine, L-propionylcarnitine, hydroxy-isovaleroylcarnitine, dihydrouracil, guanine, homoarginine, methylproline, methylumbelliferone, pyrazolyl-alanine, stachydrine, and triethanolamine, and increased serine, glycine and octanoylcarnitine.
DHA, relative to EPA, raised glycine (+ 29% vs. + 13%, respectively; FDR < 0.002) and serine (+ 12% vs. − 0.1%, FDR = 0.003), and reduced dihydrouracil (− 45% vs. − 20%, FDR < 0.0005) and stachydrine (− 31% vs. − 14%, FDR < 0.0001) to a greater extent than EPA.
There was no significant pathway associated with the changes in biogenic amines during EPA and DHA supplementation.
Discussion
At baseline, participating subjects had low-grade inflammation and some characteristics of metabolic syndrome. They presented with high AA and low EPA and DHA in plasma phospholipids, in agreement with their inflammatory status12. Supplementation with both EPA and DHA led to significant changes in the concentrations of several serum metabolites that are tightly associated with various biological and metabolic pathways, with DHA affecting a greater number of metabolites than EPA.
In the context of no significant change in total serum non-esterified FA (NEFA) by EPA and DHA, both n-3 FA lowered the non-esterified saturated myristic, palmitic and stearic acids but only EPA supplementation significantly increased non-esterified DHA. In the fasting state, serum NEFA are mostly derived from adipocyte TG lipolysis13. In TG, DHA is preferentially found at the sn-2 position, while saturated fatty acids can be found randomly at any position. Adipocyte TG lipase (ATGL), which catalyzes the first step of adipocyte lipolysis, preferentially cleaves fatty acids at the sn-2 position of TG14 and its activity has been shown to be regulated by n-3 FA15. The increase in serum non-esterified DHA observed after EPA supplementation, but not after DHA supplementation, may therefore signal a greater activation of ATGL by EPA. Alternatively, other enzymes such as phospholipase may also contribute to this effect.
N-3 FA are activators of peroxisome proliferator-activated receptor γ (PPARγ), a transcriptional factor up-regulating key genes involved in adipocyte lipogenesis and TG storage16. The reduction in non-esterified saturated FA after EPA and DHA supplementation, even in the context of no significant change in total NEFA, may in part be explained by adipocyte PPARy activation17,18. In addition, since after DHA supplementation the reduction in myristic, palmitic and heptadecanoic acids was associated with the reduction in the TCA metabolite citrate, improved metabolic health due to greater mitochondrial efficiency and adipose tissue insulin sensitivity may explain our findings.
The TCA cycle, operating in the mitochondrial matrix, plays a critical role in the integration of several anabolic and catabolic pathways19. It serves as the primary source for the production of ATP, mostly through its linkage to oxidative phosphorylation20,21. In mice, excessive caloric intake or feeding a high-fat diet was associated with elevated TCA cycle activity, paralleled by mitochondrial dysfunction and increased oxidative stress and inflammation22. Our data show that 10-week supplementation with both EPA and DHA resulted in the reduction of two key TCA cycle intermediates, -ketoglutarate and fumarate, and that DHA supplementation led to a further significant reduction of four TCA cycle intermediates, including pyruvate, citrate, isocitrate and malate. Considering that our subjects were overweight or obese and displayed several characteristics of metabolic syndrome, the reduction in TCA metabolites during n-3 FA supplementation may indicate a greater efficiency of the TCA cycle, possibly due to improvements in mitochondrial function or insulin sensitivity. DHA is incorporated into the inner and outer mitochondrial membranes and thus may affect mitochondrial membrane organization and functions, including membrane potential, respiration and reactive oxygen species (ROS) production23. DHA also decreases membrane viscosity and potentially alter lipid-lipid and lipid-protein interactions23,24. The effect of DHA on membrane viscosity is greater than that of EPA25. Furthermore, as PPAR activators, EPA and DHA may regulate mitochondrial function through regulation of mitochondrial gene expression: PPARγ activation increases mitochondrial DNA copy number, mitochondrial biogenesis and the expression of genes in the fatty acid β-oxidation pathway26. The greater reduction in citrate by DHA than EPA is in accordance with a previous animal study which showed that DHA supplementation, rather than EPA, significantly lowered hepatic citrate compared to a control Western diet27.
In contrast to the other TCA cycle metabolites, succinate was increased after DHA, but not EPA, supplementation. Recent evidence indicates that succinate is not only a metabolite in the TCA cycle but also a gut microbiota metabolite28. Succinate is involved in multiple pathways and has been linked to both health and a range of pathologic conditions28. Through binding to a plasma membrane G-protein coupled receptor on macrophages29, succinate has been shown to initiate the process of active resolution of acute inflammation in the context of obesity30. Whether the upregulation of succinate by DHA is caused by succinate-producing bacteria or changes in the host metabolism is not known.
New roles of TCA cycle intermediates in cellular signaling and activation of immune cells have been documented31,32. Acetyl-CoA, isocitrate and citrate can alter both innate and adaptive immune responses and stem cell function, and α-ketoglutarate was shown to repress pro-inflammatory responses while improving the anti-inflammatory profile in macrophages32. In contrast, succinate and fumarate may work as oncometabolites that promote tumorigenesis through the suppression of histone and DNA demethylation32. Given that our data show significant changes in these metabolites, more studies are needed to further elucidate their systemic effects on non-metabolic signaling and their association with diseases.
Glucuronate was significantly affected by EPA and DHA supplementation in relation to both the ascorbate and aldarate metabolism and the pentose and glucuronate interconversion pathway. EPA demonstrated a higher impact by increasing both glucuronate and UDP-glucuronate, while DHA only raised glucuronate. These metabolites are generated from glucose through a series of enzymatic reactions33. Conjugation with glucuronic acid is the most common phase-II reaction, with UDP-glucuronosyltransferases catalyzing the formation of glucuronides from a large variety of xenobiotics and endogenous substrates like bilirubin, making them more hydrophilic and easily excreted by the kidneys34. At low doses, DHA has been shown to reduce the activity of the UDP-glucuronosyltransferase A1 in newborn mice, leading to increased blood bilirubin levels35. At higher doses, however, DHA rather activated the enzyme, possibly through PPAR induction35. In our study, bilirubin levels were not affected by either DHA or EPA supplementation. Therefore, the increase in UDP-glucuronate and glucuronate levels by n-3 FA may be due to increased synthesis from glucose.
In the subset of 10 participants who had serum biogenic amines assessed, several metabolites were differently affected by EPA and DHA supplementation. EPA significantly reduced alanine levels, possibly due to reduced muscle protein breakdown. Creatinine levels, a marker of kidney function, was also reduced, but reductions in serum creatinine may also indicate reduced protein breakdown.
DHA supplementation changed the concentration of a greater number of metabolites than EPA. Specifically, DHA reduced plasma levels of short-chain acylcarnitines, including acetylcarnitine, propionylcarnitine and hydroxyisovalerylcarnitine. Patients with insulin resistance and type 2 diabetes tend to present with elevated levels of these acylcarnitines, which are considered a marker of inefficient β-oxidation and of mitochondrial dysfunction36. Reduction in acetylcarnitine following n-3 FA supplementation has been previously reported37. Studies have demonstrated that acetyl-CoA accumulates in conditions of TCA cycle overwork, leading to increased formation of acetylcarnitine38. In addition, propionylcarnitine and hydroxyisovalerylcarnitine are metabolites of amino acid metabolism and their reduction may indicate reduced protein breakdown. Taken together, our findings suggest improved β-oxidation, and mitochondrial function, and reduced protein breakdown by DHA. On the other hand, the finding of increased octanoylcreatinine by DHA remains unexplained in the context of the reduction of the other acylcarnitines.
DHA supplementation was also associated with increased levels of glycine and serine. In the serine synthesis pathway, glucose is converted to 3P-pyruvate, and, with glutamate, 3P-pyruvate is converted to serine and then glycine39. In endothelial cells, this pathway leads to the synthesis of heme in the mitochondria and results in improved mitochondrial function and reduced oxidative stress39.
In conclusion, we observed significant and shared effects of EPA and DHA on metabolites related to lipid and adipocyte metabolism, TCA cycle efficiency, mitochondrial function and detoxification processes in subjects with chronic inflammation and some key aspects of metabolic syndrome. However, EPA and DHA also exhibited differential effects on the serum metabolome as evidenced by a greater number of metabolites affected by DHA than EPA and a significantly greater effect on selected metabolites by DHA or EPA.
Strengths of our study include the randomized, controlled, crossover study design, the high compliance of participants, and the assessment of the metabolic effects of EPA and DHA effects in subjects with chronic inflammation, a population at higher risk of cardiometabolic diseases. Limitations are the small sample size and the lack of a healthy control group.
Methods
Study design
The details of the study design and related primary outcomes have been published previously11. The study had a randomized, double-blind, controlled, crossover design and consisted of an initial 4-week lead-in phase during which the subjects were given 3 g/day of high-oleic acid sunflower oil as a control supplement, followed by randomization to a 10-week supplementation with either EPA or DHA (3 g/day) and crossover after a 10-week washout phase. The random allocation sequence was generated by REDCap. Subjects, study investigators and laboratory personnel were blind to treatment. The end of the 4-week lead-in phase was defined as baseline. Ethyl ester forms of EPA and DHA were provided in capsules containing 750 mg/capsule with a purity > 97% (Prevention Pharmaceuticals Inc, CT). Each participant was instructed to take a total of four capsules daily, two with the morning meal, and two with the evening meal. Subjects’ compliance was assessed by the number of returned capsules at the end of each phase and was greater than 80% in all participants. All research was performed in accordance with the Declaration of Helsinki.
Study subjects
Subjects were studied at the Jean Mayer USDA Human Nutrition Research Center on Aging at Tufts University. Between April 2016 and November 2017, 24 participants were enrolled and 21subjects (9 men and 12 postmenopausal women) completed the study. Three subjects dropped out of the study during the placebo lead-in phase. Subjects, aged 50–75 years and meeting the following inclusion criteria, completed the study: (1) serum C-reactive protein (CRP) concentrations 2 g/mL; (2) fasting plasma TG concentrations between 90 and 500 mg/dL; and (3) at least one of the criteria for the modified definition of metabolic syndrome including abdominal obesity (waist circumference 102 cm for men and 89 cm for women), hypertension (blood pressure 130/80 mmHg or use of antihypertensive medications), and fasting blood glucose concentrations 100–125 mg/dL11. Subjects were excluded if they had consumed either fish more than 2 times/week or supplements containing fish oil or EPA/DHA during the 6 months prior to the trial; were allergic to sardines and/or sunflower oil; had regularly used anti-inflammatory medications such as NSAID or were under anticoagulant therapy; had other known diseases that might alter their metabolic status such as kidney or liver diseases; were alcohol users (> 7 drinks/week) or smokers. Subjects were instructed to follow a low-saturated fat diet (25–35% of calories from total fat, < 7% from saturated fat, and cholesterol < 200 mg/day) and keep their physical activity levels unchanged throughout the study. Subjects were also asked not to consume more than 2 fish meals/week and to abstain from taking fish oil supplements during the study. To enhance adherence to dietary and lifestyle instructions, consultations with a registered dietitian were carried out at baseline and at each study visit. The study protocol was approved by the Tufts Health Sciences Institutional Review Board and is registered at clinicialtrials.gov (NCT02670382, 01/02/2016). All study participants provided informed consent.
Blood sample collection
Twelve-hour fasting venous blood samples were collected into silica-coated tubes (Becton Dickinson, NJ) at baseline and the end of each supplementation phase. After being kept at room temperature for 20 min to allow coagulation to occur, blood was centrifuged (1,000 g for 15 min at 4 °C) and serum was aliquoted and immediately stored at − 80 °C for further analysis.
All methods used in the current study for the assessment of biochemical parameters and metabolites have been previously described and have been carried out in accordance with relevant guidelines and regulations. The effects of EPA and DHA on the serum metabolome was intended as an exploratory analysis.
Measurement of biochemical parameters
Serum concentrations of TG, glucose, albumin, and other blood chemistry analytes were assessed at the screening visit and at the end of each phase using commercial reagents on an AU480 Chemistry Analyzer (Beckman Coulter Inc, CA)11.
Metabolic profiling
Serum primary metabolites
Primary metabolites, including a panel of carbohydrates and sugar phosphates, amino acids, hydroxyl acids, free fatty acids, purines, pyrimidines, aromatics, and exposome-derived chemicals, were measured in fasting serum samples, obtained at baseline and at the end of each supplementation phase from all 21 subjects, by gas chromatography-time of flight mass spectrometry (GC-TOF MS) by West Coast Metabolomics40,41. Briefly, serum samples were extracted and derivatized using acetonitrile:isopropanol:water (3∶3∶2; v/v/v), centrifuged and dried. A set of 13 C8–C30 fatty acid methyl ester internal standards were added, and samples were derivatized by 10 µL methoxyamine hydrochloride in pyridine followed by 90 µL MSTFA. Analytes were separated using an Agilent 6890 gas chromatograph (Santa Clara, CA) equipped with a 30 m long, 0.25 mm i.d. Rtx5Sil-MS column with 0.25 µm 5% diphenyl film and additional 10 m integrated guard column (Restek, Bellefonte PA). Mass spectrometry was performed by a Leco Pegasus IV time of flight mass spectrometer (St. Joseph, MI). Mass spectra were acquired from m/z 85–500 at 17 spectra s−1 and 1850 V detector voltage. Raw data files were exported and validated using the metabolomics BinBase database (BinBase.com, Ltd)41, combined with the validity of chromatogram (< 10 peaks with intensity > 10^7 counts s-1), detection of unbiased retention index marker (MS similarity > 800 with high m/z marker ions) and retention index calculation by 5th order polynomial regression, as previously described40. This method identified 130 known and 276 unknown primary metabolites in all serum samples from all participants.
Serum biogenic amines
Serum biogenic amines were measured only in a subset of 10 randomly selected participants, due to budget restrictions. Biogenic amines, comprising acylcarnitines, TMAO, cholines, betaines, SAM, SAH, nucleotides and nucleosides and methylated and acetylated amines, were assessed by West Coast Metabolomics by HILIC-QTOF MS/MS, as previously described42. Hydrophilic interaction liquid chromatography (HILIC) method was used. Briefly, serum samples were extracted and derivatized. Five µL of re‐suspended samples were injected onto a Waters Acquity UPLC BEH Amide column (150 mm length × 2.1 mm id; 1.7 µm particle size) with an additional Waters Acquity VanGuard BEH Amide pre‐column (5 mm × 2.1 mm id; 1.7 µm particle size) coupled to an Agilent 1290 Infinity UHPLC. The mobile phases were prepared with 10 mm ammonium formate and 0.125% formic acid (Sigma‐Aldrich) in either 100% LC‐MS grade water for mobile phase (A) or acetonitrile: water (95:5, v/v) for mobile phase (B). Spectra were collected using Q Exactive HF mass spectrometer in positive ion mode. Quality control samples (Standard Reference Material NIST plasma samples) were included. For metabolite identification, HILIC spectral library of authentic standards was used in addition to m/z and retention time database. A total of 178 known and 2,946 unknown biogenic metabolites were measured.
Data analysis
Results are presented as mean and standard deviation (SD) or median (25th and 75th percentile). Metabolite data were subjected to generalized log transformation and auto-scaling before statistical analyses due to skewness. Statistical analyses, including partial least square discriminant analysis (PLS-DA), were conducted in R version 4.0.0 (R core team, 2020). Comparisons of the change in serum metabolites from baseline to EPA versus DHA supplementation were evaluated using a linear mixed-effects model, by the lmer function from the lme4 package. The model accounted for the random effect of subjects and the fixed effects of treatment, period, and sequence. Baseline values and age were included in the model as covariates. The effect of EPA or DHA supplementation versus baseline was assessed by using the lsmeansLT function from the lmerTest package, which allows for comparisons of least square means between baseline and each supplemental phase in the linear mixed model. When there was a significant sequence effect, data from only the first supplementation period was selected to perform ANCOVA for the comparisons between EPA and DHA and paired t-tests for the comparisons between baseline versus each treatment, with the inclusion of baseline concentration as a covariate. No sex differences were observed in the changes of metabolites following EPA and DHA supplementation, therefore men and women were combined in our analyses. To correct for multiple testing, false discovery rate (FDR) was used to determine adjusted p-values. Statistical significance was defined as FDR < 0.05.
MetaboAnalyst 4.0 (https://www.metaboanalyst.ca), a web-based platform for comprehensive analysis of quantitative metabolomic data, was used to perform pathway analysis and visualize the biological pathways associated with significantly affected metabolites. The pathway analysis algorithms are based on the hypergeometric test for over-representation analysis, and relative-betweenness centrality for pathway topology analysis. Hypergeometric test compares the number of significant metabolites within a specific pathway with the expected value43. Results included the impact value of the metabolic pathway based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) for Homo sapiens database that were most relevant to the selected metabolites44. Pathways with an impact score > 0 and FDR 0.05 were considered statistically significant.
To identify metabolites with similar or opposite changes after EPA and DHA supplementation, Pearson’s correlation coefficient analysis was conducted. Significantly changed metabolites from the linear-mixed model were chosen and the coefficients were calculated across metabolite pairs. In addition, the change in concentration of each metabolite was correlated with the change in plasma phospholipid EPA or DHA mol percentage. The correlation coefficients (r) of metabolites within each feature, which reflect the strength of their relationships, were ranked from high to low using the pattern searching function. Heatmaps were created to provide an overview and visualization of the correlation matrix. An absolute value of r that is greater than 0.6 indicated a moderate to strong linear relationship, and P < 0.05 was considered statically significant.
Supplementary Information
Author contributions
W.C.C.: Data Curation; Formal analysis; Methodology; Visualization; Roles/Writing—original draft. J.S.: Data curation; Investigation; Resources; Supervision; Writing—review & editing. S.L.-F.: Conceptualization; Funding acquisition; Investigation; Methodology; Project administration; Resources; Visualization; Supervision; Roles/Writing—original draft.
Funding
This work was supported by Agriculture and Food Research Initiative Grant 2015-67017-23142 from National Institute of Food and Agriculture to Dr. Lamon-Fava; the U.S. Department of Agriculture under agreement no. 58-1950-4-401; and the NIH CTSA award UL1TR002544. The funding sources were not involved in the study design, conduct of the study, or collection, management, analysis, or interpretation of the data or in the preparation or review of the manuscript and had no right to approve or disapprove of the submitted manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-95590-7.
References
- 1.Saltiel AR, Olefsky JM. Inflammatory mechanisms linking obesity and metabolic disease. J. Clin. Invest. 2017;127:1–4. doi: 10.1172/JCI92035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: A global biochemical approach to drug response and disease. Annu. Rev. Pharmacol. Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 3.Newgard CB. Metabolomics and metabolic diseases: Where do we stand? Cell Metab. 2017;25:43–56. doi: 10.1016/j.cmet.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lent-Schochet D, McLaughlin M, Ramakrishnan N, Jialal I. Exploratory metabolomics of metabolic syndrome: A status report. World J. Diabetes. 2019;10:23–36. doi: 10.4239/wjd.v10.i1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Surowiec I, Noordam R, Bennett K, Beekman M, Slagboom PE, Lundstedt T, van Heemst D. Metabolomic and lipidomic assessment of the metabolic syndrome in Dutch middle-aged individuals reveals novel biological signatures separating health and disease. Metabolomics. 2019;15:23. doi: 10.1007/s11306-019-1484-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lombardo YB, Chicco AG. Effects of dietary polyunsaturated n-3 fatty acids on dyslipidemia and insulin resistance in rodents and humans. A review. J. Nutr. Biochem. 2006;17:1–13. doi: 10.1016/j.jnutbio.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Flachs P, Rossmeisl M, Kopecky J. The effect of n-3 fatty acids on glucose homeostasis and insulin sensitivity. Physiol Res/Academia Scientiarum Bohemoslovaca. 2014;63:S93–118. doi: 10.33549/physiolres.932715. [DOI] [PubMed] [Google Scholar]
- 8.Yokoyama M, Origasa H, Matsuzaki M, Matsuzawa Y, Saito Y, Ishikawa Y, Oikawa S, Sasaki J, Hishida H, Itakura H, Kita T, Kitabatake A, Nakaya N, Sakata T, Shimada K, Shirato K, Japan EPAlisI Effects of eicosapentaenoic acid on major coronary events in hypercholesterolaemic patients (JELIS): A randomised open-label, blinded endpoint analysis. Lancet. 2007;369:1090–1098. doi: 10.1016/S0140-6736(07)60527-3. [DOI] [PubMed] [Google Scholar]
- 9.Bhatt DL, Steg PG, Miller M, Brinton EA, Jacobson TA, Ketchum SB, Doyle RT, Jr, Juliano RA, Jiao L, Granowitz C, Tardif JC, Ballantyne CM, Investigators R-I. Cardiovascular risk reduction with icosapent ethyl for hypertriglyceridemia. N Engl. J. Med. 2019;380:11–22. doi: 10.1056/NEJMoa1812792. [DOI] [PubMed] [Google Scholar]
- 10.Calder PC. Marine omega-3 fatty acids and inflammatory processes: Effects, mechanisms and clinical relevance. Biochim. Biophys. Acta. 2015;1851:469–484. doi: 10.1016/j.bbalip.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 11.So J, Wu D, Lichtenstein AH, Tai AK, Matthan NR, Maddipati KR, Lamon-Fava S. EPA and DHA differentially modulate monocyte inflammatory response in subjects with chronic inflammation in part via plasma specialized pro-resolving lipid mediators: A randomized, double-blind, crossover study. Atherosclerosis. 2021;316:90–98. doi: 10.1016/j.atherosclerosis.2020.11.018. [DOI] [PubMed] [Google Scholar]
- 12.Ferrucci L, Cherubini A, Bandinelli S, Bartali B, Corsi A, Lauretani F, Martin A, Andres-Lacueva C, Senin U, Guralnik JM. Relationship of plasma polyunsaturated fatty acids to circulating inflammatory markers. J. Clin. Endocrinol. Metab. 2006;91:439–446. doi: 10.1210/jc.2005-1303. [DOI] [PubMed] [Google Scholar]
- 13.Schweiger M, Eichmann TO, Taschler U, Zimmermann R, Zechner R, Lass A. Measurement of lipolysis. Methods Enzymol. 2014;538:171–193. doi: 10.1016/B978-0-12-800280-3.00010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichmann TO, Kumari M, Haas JT, Farese RV, Jr, Zimmermann R, Lass A, Zechner R. Studies on the substrate and stereo/regioselectivity of adipose triglyceride lipase, hormone-sensitive lipase, and diacylglycerol-O-acyltransferases. J. Biol. Chem. 2012;287:41446–41457. doi: 10.1074/jbc.M112.400416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, Zechner R. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 16.Sharma AM, Staels B. Review: Peroxisome proliferator-activated receptor gamma and adipose tissue–understanding obesity-related changes in regulation of lipid and glucose metabolism. J. Clin. Endocrinol. Metab. 2007;92:386–395. doi: 10.1210/jc.2006-1268. [DOI] [PubMed] [Google Scholar]
- 17.Rosen E, Eguchi J, Xu Z. Transcriptional targets in adipocyte biology. Exp. Opin Ther Targets. 2009;13:975–986. doi: 10.1517/14728220903039706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moseti D, Regassa A, and Kim WK. Molecular regulation of adipogenesis and potential anti-adipogenic bioactive molecules. Int. J. Mol. Sci. 2016;17. [DOI] [PMC free article] [PubMed]
- 19.Akram M. Citric acid cycle and role of its intermediates in metabolism. Cell Biochem. Biophys. 2014;68:475–478. doi: 10.1007/s12013-013-9750-1. [DOI] [PubMed] [Google Scholar]
- 20.Wilson DF. Oxidative phosphorylation: regulation and role in cellular and tissue metabolism. J. Physiol. 2017;595:7023–7038. doi: 10.1113/JP273839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osellame LD, Blacker TS, Duchen MR. Cellular and molecular mechanisms of mitochondrial function. Best Pract. Res. Clin. Endocrinol. Metab. 2012;26:711–723. doi: 10.1016/j.beem.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satapati S, Sunny NE, Kucejova B, Fu X, He TT, Mendez-Lucas A, Shelton JM, Perales JC, Browning JD, Burgess SC. Elevated TCA cycle function in the pathology of diet-induced hepatic insulin resistance and fatty liver. J. Lipid Res. 2012;53:1080–1092. doi: 10.1194/jlr.M023382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan EM, Pennington ER, Green WD, Beck MA, Brown DA, Shaikh SR. Mechanisms by which dietary fatty acids regulate mitochondrial structure-function in health and disease. Adv. Nutr. 2018;9:247–262. doi: 10.1093/advances/nmy007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbst EA, Paglialunga S, Gerling C, Whitfield J, Mukai K, Chabowski A, Heigenhauser GJ, Spriet LL, Holloway GP. Omega-3 supplementation alters mitochondrial membrane composition and respiration kinetics in human skeletal muscle. J. Physiol. 2014;592:1341–1352. doi: 10.1113/jphysiol.2013.267336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherratt SCR, Mason RP. Eicosapentaenoic acid and docosahexaenoic acid have distinct membrane locations and lipid interactions as determined by x-ray diffraction. Chem. Phys. Lipids. 2018;212:73–79. doi: 10.1016/j.chemphyslip.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Corona JC, Duchen MR. PPARgamma as a therapeutic target to rescue mitochondrial function in neurological disease. Free Rad. Biol. Med. 2016;100:153–163. doi: 10.1016/j.freeradbiomed.2016.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Depner CM, Traber MG, Bobe G, Kensicki E, Bohren KM, Milne G, Jump DB. A metabolomic analysis of omega-3 fatty acid-mediated attenuation of western diet-induced nonalcoholic steatohepatitis in LDLR-/- mice. PLoS ONE. 2013;8:e83756. doi: 10.1371/journal.pone.0083756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez-Veledo S, Vendrell J. Gut microbiota-derived succinate: Friend or foe in human metabolic diseases? Rev. Endocr. Metab. Disord. 2019;20:439–447. doi: 10.1007/s11154-019-09513-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Castro FM, Aguiar CJ, da Rocha Franco JA, Gingold RN, Leite MF. GPR91: expanding the frontiers of Krebs cycle intermediates. Cell Commun. Signal. 2016;14:3. doi: 10.1186/s12964-016-0126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keiran N, Ceperuelo-Mallafre V, Calvo E, Hernandez-Alvarez MI, Ejarque M, Nunez-Roa C, Horrillo D, Maymo-Masip E, Rodriguez MM, Fradera R, de la Rosa JV, Jorba R, Megia A, Zorzano A, Medina-Gomez G, Serena C, Castrillo A, Vendrell J, Fernandez-Veledo S. SUCNR1 controls an anti-inflammatory program in macrophages to regulate the metabolic response to obesity. Nat. Immunol. 2019;20:581–592. doi: 10.1038/s41590-019-0372-7. [DOI] [PubMed] [Google Scholar]
- 31.Ryan DG, Murphy MP, Frezza C, Prag HA, Chouchani ET, O'Neill LA, Mills EL. Coupling Krebs cycle metabolites to signalling in immunity and cancer. Nat. Metab. 2019;1:16–33. doi: 10.1038/s42255-018-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Reyes I, Chandel NS. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020;11:102. doi: 10.1038/s41467-019-13668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eisenberg F, Jr, Dayton PG, Burns JJ. Studies on the glucuronic acid pathway of glucose metabolism. J. Biol. Chem. 1959;234:250–253. doi: 10.1016/S0021-9258(18)70282-2. [DOI] [PubMed] [Google Scholar]
- 34.Chowdhury JR, Jansen PL, Fischberg EB, Daniller A, Arias IM. Hepatic conversion of bilirubin monoglucuronide to diglucuronide in uridine diphosphate-glucuronyl transferase-deficient man and rat by bilirubin glucuronoside glucuronosyltransferase. J. Clin. Invest. 1978;62:191–196. doi: 10.1172/JCI109105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shibuya A, Itoh T, Tukey RH, Fujiwara R. Impact of fatty acids on human UDP-glucuronosyltransferase 1A1 activity and its expression in neonatal hyperbilirubinemia. Sci. Rep. 2013;3:2903. doi: 10.1038/srep02903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bene J, Hadzsiev K, Melegh B. Role of carnitine and its derivatives in the development and management of type 2 diabetes. Nutr. Diabetes. 2018;8:8. doi: 10.1038/s41387-018-0017-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rudkowska I, Paradis AM, Thifault E, Julien P, Tchernof A, Couture P, Lemieux S, Barbier O, Vohl MC. Transcriptomic and metabolomic signatures of an n-3 polyunsaturated fatty acids supplementation in a normolipidemic/normocholesterolemic Caucasian population. J. Nutr. Biochem. 2013;24:54–61. doi: 10.1016/j.jnutbio.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 38.Meienberg F, Loher H, Bucher J, Jenni S, Krusi M, Kreis R, Boesch C, Betz MJ, Christ E. The effect of exercise on intramyocellular acetylcarnitine (AcCtn) concentration in adult growth hormone deficiency (GHD) Sci. Rep. 2019;9:19431. doi: 10.1038/s41598-019-55942-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandekeere S, Dubois C, Kalucka J, Sullivan MR, Garcia-Caballero M, Goveia J, Chen R, Diehl FF, Bar-Lev L, Souffreau J, Pircher A, Kumar S, Vinckier S, Hirabayashi Y, Furuya S, Schoonjans L, Eelen G, Ghesquiere B, Keshet E, Li X, Vander Heiden MG, Dewerchin M, Carmeliet P. Serine synthesis via PHGDH is essential for heme production in endothelial cells. Cell Metab. 2018;28:573–587. doi: 10.1016/j.cmet.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 40.Fiehn O. Metabolomics by gas chromatography-mass spectrometry: Combined targeted and untargeted profiling. Curr. Protoc. Mol. Biol. 2016;114:3041–30432. doi: 10.1002/0471142727.mb3004s114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiehn O, Garvey WT, Newman JW, Lok KH, Hoppel CL, Adams SH. Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese African–American women. PLoS ONE. 2010;5:e15234. doi: 10.1371/journal.pone.0015234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blazenovic I, Oh YT, Li F, Ji J, Nguyen AK, Wancewicz B, Bender JM, Fiehn O, Youn JH. Effects of gut bacteria depletion and high-Na(+) and low-K(+) Intake on circulating levels of biogenic amines. Mol. Nutr. Food Res. 2019;63:1801184. doi: 10.1002/mnfr.201801184. [DOI] [PubMed] [Google Scholar]
- 43.Poisson LM, Sreekumar A, Chinnaiyan AM, Ghosh D. Pathway-directed weighted testing procedures for the integrative analysis of gene expression and metabolomic data. Genomics. 2012;99:265–274. doi: 10.1016/j.ygeno.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.