Abstract
Rapid detection and quantification of bacterial foodborne pathogens are crucial in reducing the incidence of diseases associated with meat products contaminated with pathogens. For the identification, discrimination and quantification of Salmonella Typhimurium contamination in pork samples, a commercial electronic nose with ten (10) metal oxide semiconductor sensor array is applied. Principal component analysis was successfully applied for discrimination of inoculated samples and inoculated samples at different contaminant levels. Support vector machine regression (SVMR) together with a metaheuristic framework using genetic algorithm (GA), particle swarm optimization (PSO), and grid searching (GS) optimization algorithms were applied for S. Typhimurium quantification. Although SVMR results were satisfactory, SVMR hyperparameter tuning (c and g) by PSO, GA and GS showed superior performance of the models. The order of the prediction accuracy based on the prediction set was GA-SVMR (R2P = 0.989; RMSEP = 0.137; RPD = 14.93) > PSO-SVMR (R2P = 0.986; RMSEP = 0.145; RPD = 14.11) > GS-SVMR (R2P = 0.966; RMSEP = 0.148; RPD = 13.82) > SVMR (R2P = 0.949; RMSEP = 0.162; RPD = 12.63). GA-SVMR’s proposed approach was fairly more effective and retained an excellent prediction accuracy. A clear relationship was identified between odor analysis results, and reference traditional microbial test, indicating that the electronic nose is useful for accurate microbial volatile organic compound evaluation in the quantification of S. Typhimurium in a food matrix.
Electronic supplementary material
The online version of this article (10.1007/s13197-020-04847-y) contains supplementary material, which is available to authorized users.
Keywords: Salmonella, Foodborne pathogens, Electronic nose, Chemometric algorithms, Longissimus pork muscle, Metaheuristic algorithms
Introduction
Foodborne bacterial infections continue to be one of the world’s major causes of disease and death. Despite stringent inactivation control measures, such as pasteurization, and ultra-high temperature (UHT) treatment, numerous outbreaks of foodborne diseases have been reported due to the consumption of contaminated meat products (Nadi et al. 2020).
According to the 2015 report of the World Health Organization (WHO), as many as 600 million people worldwide annually become ill after eating contaminated food, of these, there are 420,000 recorded deaths, including 125,000 children under the age of 5 years (Havelaar et al. 2015). Most large-scale meat recalls have been caused by bacterial contamination of processed food with these major recalls leading to enormous financial costs as well as meat wastage (Pozo and Schroeder 2016). Pathogen identification for the meat industry, and for that matter, the entire food industry has emerged as the highest scientific and technological priority in the industry.
The global food microbiology test for pathogens amounted to 280 million tests in 2016, a market valued at $1.8 billion, according to a recent report (https://www.foodsafetymagazine. com/magazine-archive1/februarymarch-2017/a-look-at-the-microbiology-testing-market/). It reflects a 23.2% increase in the amount of testing over a span of 3 years, with Salmonella representing 43% of all tests performed followed by Listeria and Listeria monocytogenes (41%), pathogenic Escherichia coli (14%), and Campylobacter (2%). The trend observed for all four priority pathogens over the past two decades was a definite shift from traditional methods to rapid methods.
To sustain food safety and human health, the monitoring of food safety indicators, especially foodborne pathogens, is crucial. Rapid analytical methods that are sensitive, accurate, cost-effective, and easy-to-use for process control and foodborne pathogen detection are needed to ensure the quality and safety of meat products.
Systems for the identification of volatile and non-volatile characteristic compounds related to microbial growth in meat products have been studied previously. As markers of microbial contamination, microbial volatile organic compounds (MVOCs) have already played an essential role in clinical diagnosis, environment monitoring and in recent years and have been applied to determine microbial contamination of food (Wang et al. 2016).
An electronic nose (E-nose) is a non-invasive technique for detecting volatile analytes using a range of chemical sensors (Chang et al. 2020; Huang et al. 2019). E-nose can identify microbial genera via detailed MVOCs profiles in food samples (Zambotti et al. 2014). E-nose devices are currently used in the identification foodborne microbial pathogens (Balbin et al. 2017; Bonah et al. 2019a, b; Gobbi et al. 2015; Wang et al. 2016) together with multivariate statistical analysis tools. Many of these research concentrated on bacterial discrimination, prediction and classification. However, no study on foodborne bacteria quantification based on e-nose sensor responses have been reported in meat products. An earlier study utilized E-nose sensor responses to predict E. coli numbers in packaged alfalfa sprouts with a regression coefficient (R2) = 0.903 (Siripatrawan et al. 2006).
Multivariate statistical analysis is applied to obtain key information on the relationship between MVOCs and microbial genera in food. These computational intelligence techniques, such as support vector machines (SVMs) and artificial neural networks (ANN), have been widely applied in E-nose studies. SVMs are the newly developed machine learning methods that have achieved enormous prominence in identification, pattern recognition, and regression (Elbisy 2015). SVM provides a more robust model with a higher generalization error, which also shows that SVMs are not subject over-fitting relative to ANN (Patil et al. 2012).
It must be noted that SVM is constrained since the efficiency of SVM models relies heavily on setting the SVM hyper-parameters and SVM kernel parameters properly. Therefore, to determine the values of these parameters, which lead to the lowest generalization error, an automated, effective and relatively rapid approach is required. Most of the techniques used to solve this problem are based on metaheuristics such as genetic algorithm (GA) and particle swarm optimization (PSO).
In this work, E-nose measurements of the MVOCs sensor response profiles of Salmonella Typhimurium from inoculated pork samples at varying concentrations were used to estimate and predict pathogen concentration by support vector machine regression (SVMR). The optimization ability of different metaheuristic algorithms was investigated to optimize the models and improve the prediction accuracy of bacterial concentration in the inoculate pork sample. Therefore, our specific goal was to apply three-parameter optimization approaches (GA-SVMR, GS-SVMR and PSO-SVMR) and compare the performance to the ordinary SVMR models for the prediction of Salmonella Typhimurium.
Materials and methods
Sample preparation
The fresh longissimus pork meat was obtained from a Zhenjiang local supermarket. The samples were immediately packed without cleaning into commercial food-grade polymer wraps. The meat was minced and processed with a household mincing machine stored at 4 °C. To prevent contamination, mincer parts were cleaned and disinfected with detergent, chlorine (1000 ppm) and ethanol. All meat batches used were tested to ensure that they were clear of Salmonella Typhimurium according to the ISO 16654/2001, ISO 6579:2002 and National Standard of the People’s Republic of China, Standard of Pathogenic Limits for Food GB 29921-2013) standard for Salmonella detection in food and were determined to be negative in 25 g.
Bacterial preparation and inoculation
Salmonella Typhimurium CICC 22956 were acquired from the Center of Industrial Culture Collection. Beijing, China. Frozen culture on slant agar was activated in Tryptic Soy Broth (TSB, AOBOX, Beijing) at 37° C for 24 h. In other to achieve inoculation levels of ~ 102, ~ 104 and ~ 107 CFU/g, the appropriate serial dilutions of fresh S. Typhimurium culture was manually mixed with 100 g of thawed samples at 4° C.
Electronic nose system and data acquisition
This investigation was performed by a PEN3 electronic nose (Airsense Analytics GmbH, Schwerin/Germany), consisting of 10 metal–oxide–semiconductor (MOS) sensors with different selectivity to volatiles (Aheto et al. 2020; Giungato et al. 2019). Sampling, headspace generation parameters and system settings used in this study are described in detail by Bonah et al. (2019b). Furthermore, the ten (10) sensor response values were recorded for 120 s. Five (5) grams of the sample was transferred into a 20 mL vial and sealed with a magnetic screw cap and septum for analysis. Before sampling, 10 min was allowed to ensure the gas was saturated at the top of the sealed bottle. The PEN3 Win Muster v. 1.6.2 program was used for data acquisition, pattern recognition and interpretation.
Microbial enumeration
Evaluation of pork meat pathogen contamination was conducted using simultaneous E-nose measurements in parallel with microbiological analysis using the plate count method. Bacterial enumeration was performed according to Osaili et al. (2020) using Xylose Lysine Deoxycholate Agar (Sorbitol). Typical colonies were counted after 48 h of incubation at 37 °C.
HS-GC-IMS analysis
HS-GC-IMS (Headspace- gas chromatography-ion mobility spectrometry) was used as the reference method. GC-IMS analysis was performed with a FlavourSpec® (G.A.S. Gesellschaft für analytische Sensorsysteme mbH Dortmund, Germany) fitted with a nonpolar GC column (SE-54-CB from CS-Chromatographie Service GmbH (Düren, Germany) consisting of 0.25 μm film thickness (94% methyl-5% phenyl-1% vinyl silicone).
The sample was transferred into a 20 mL vial and sealed with a magnetic screw cap and septum for analysis. Automatic injection of a 500-μL aliquot of the headspace was carried out after the ampoules were heated (37 °C, 3 min) with an agitation speed of 500 rpm in the heated injector of the GC-IMS apparatus. The separation was achieved by means of a 30 m length nonpolar GC column composed of 94% methyl-5% phenyl-1% vinyl silicone after which the Volatile Organic Compounds trapped were thrust into the GC column (40 °C) through nitrogen (N2) carrier gas with a purity of ≥ 99.999%. The flow rate of the N2 carrier gas was set at a commencement rate of 2 ml.min−1 and programmed as follows: flow rate of 2 mL.min−1 (0–2 min), 2–5 mL.min−1 (2 –5 min), 5–50 mL.min−1 (5–10 min), and 50–150 mL.min−1 (10–15 min).
Afterwards, the VOCs are ionized in the ionization chamber, after been split up in the GC column (40 °C). With constant temperature and voltage (40 °C, 400 V cm−1) in the drift region, the ions the reach the IMS detector via a shutter grid with a grid pulse width of 100 μs. The drift tube length is 10-cm and a drift gas(N2) flow rate = 150 mL.min−1.
Data acquisition was performed with the IMS Control TFTP Server Software. To identify particular compounds signal drift and retentions times are compared at their coordinate locations. The data signals obtained are then compared to the GC × IMS Library version 1.01 (G.A.S. Gesellschaft für analytische Sensorsysteme mbH).
Furthermore, PRTools 5.0 toolkit (Delft University of Technology, Netherlands) and MATLAB R2018a software (The Mathworks Inc., Natick, USA) was employed for ion mobility characterization, data visualization as well as feature extraction for chemometric analysis.
Data pre-processing and feature extraction
Electronic nose sampling showed different sensor response rates for different contaminant levels. The mean-differential coefficient value can be used to describe the sensor’s average response velocity and its main characteristics (Xu et al. 2018). Therefore, the mean differential coefficient value M(i)ave was taken as the characteristic value of the sensor response curve.
| 1 |
n = number of test points (n = 120)
i = sample number of each variety
xiz = zth response value of the ith sample
△t = time difference of adjacent test points (△t = 1 s).
Support vector machine regression
SVMR (Cortes and Vapnik 1995) was used to predict bacterial populations on pork meat samples based on sensor responses from E-nose measurement. The basic idea in SVM regression is to map the input data (nonlinear regression problem) into a higher-dimensional function space and solve a linear regression problem (Shokri et al. 2015). In the formulation of SVMR, several loss functions such as the Laplacian, Huber, Gaussian, and ε-insensitive can be applied. The stable ε-insensitive loss function (L ε) is more generally used among these loss functions (Cortes and Vapnik 1995). The SVR loss function is given as:
| 2 |
Where ε is a precision parameter that represents the tube radius around the regression function f(x)
The aim of using the ε—insensitive loss function is to find a function capable of fitting current training data with a deviation less than or equal to ε. The problem of optimization can be reformulated as
| 3 |
| 4 |
The distance between the actual values and the respective limit values of the ε—tube are represented by the positive slack variables. The constant is a parameter that determines the interaction and trade-off between the empirical risk and the flatness of the model.
Standard kernel functions that are implemented are stated below, where γ, r and d are kernel parameters.
Radial basis function (RBF) kernel:
| 5 |
Linear kernel:
| 6 |
Polynomial kernel:
| 7 |
Sigmoid kernel:
| 8 |
The RBF kernel is highly recommended for its complexity and performance (Bao et al. 2013). The kernel parameter specification implicitly specifies the layout of the high-dimensional feature space ϕ(x) which governs the nature of the final solution (Tay and Cao 2001).
Grid search and heuristic algorithms employed for SVMR optimization problem
The advantage of the SVM algorithm; is that it is not affected by local minima, nor does it suffer from the curse of high-dimensionality due to the use of support vectors. Unfortunately, the SVM performance depends heavily on the parameter setting and the selection of the kernel (Phan et al. 2017).
The quality of the selection of SVM parameters and kernel functions affects the performance of learning and generalization. The optimization of the model structure is crucial for developing high-quality SVM models with excellent generalization capabilities (Herceg et al. 2019). However, there is no exact procedure for achieving the maximal range of SVMR hyperparameters. Search algorithms must be applied to generate the optimum set of hyperparameters to guarantee the maximum possible accuracy of the final model. Search algorithms based on grid searches (Mohandes et al. 2004) where the parameter search space is split into groups of potential parameters to be evaluated, usually in a uniform manner and metaheuristics algorithms (Genetic algorithms, Particle swarm optimization, ant colony optimization etc.) is applied to conduct a robust investigation on the hyper-parameter search space.
While being successful in solving complex problems, metaheuristic approaches do not guarantee overall optimum results. In solving complex multi-modal problems, these methods may be stuck in local optima (Saddique et al. 2020). Furthermore, their convergence speed depends on the proper adjustment of the parameters associated with each metaheuristic approach (Moscato and Cotta 2019).
Model evaluation
Regression model accuracy and performance was measured by the root- mean -square error of cross-validation (RMSECV), the root-mean-squared error of calibration on the calibration set (RMSEC), root mean square error of prediction (RMSEP). The residual predictive deviation (RPD) was also calculated to examine the predictive ability of the model.
Data processing, classification and performance characterization
The application platform was implemented in Matlab 2018a in windows 10, which is a universal mathematical development tool. The Libsvm (version 3.23.) initially designed by (Chang and Chih-Jen 2011)was used for SVM classification and parameter optimization. The empirical evaluation was performed using Intel (R) Core (TM) i3-4010U CPU @ 1.70 GHz with 8.00 GB of RAM.
SPXY (sample set partitioning based on joint x–y distances) method(Galvão et al. 2005) was applied to split the acquired sample spectra from 120 samples per each bacteria into two parts that were subsequently used for model calibration (72) and prediction (48) respectively. Overall correct classification rate (OCCR), was employed to establish the performance of all models developed.
Results and discussion
Microbiological reference analysis
The reference testing for S. Typhimurium contamination was performed using the above methods, and the numeric results were reported in Table 1. The mean concentration of each bacterial inoculation levels was ~ 102(2.14 ± 0.42), ~ 104 (4.21 ± 0.32), and ~ 107(7.33 ± 0.36) Log CFU/g. The predictive models established in the study, as can be seen in Table 1, encompassed broad data ranges and can, therefore, produce relatively better results than the narrow data coverage for online applications.
Table 1.
Statistical characteristics of microbiological analysis reference values (Log CFU/g) of S. Typhimurium in pork samples
| Pathogen | Number of samples | Range | |
|---|---|---|---|
| S. Typhimurium | Calibration set | 72 | 2.33–7.53 |
| Prediction set | 48 | 2.12–7.36 |
Volatile organic compound analysis via E-nose and HS-GC-IMS
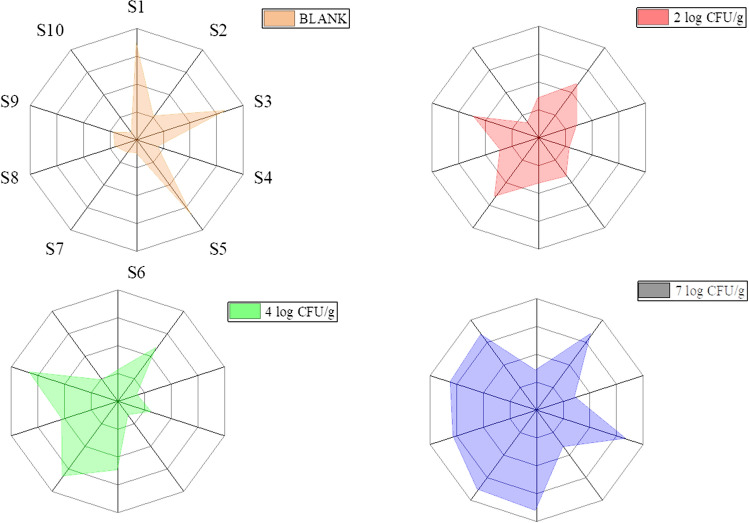
The resistance values (Go/G) of all the sensors rise sharply and decrease rapidly in the presence of pork samples inoculated with bacteria odors within 20–30 s as demonstrated (Fig. 1). Different sensors showed different responses in volatile substances in the pork samples inoculated with different concentrations of pathogens. In particular, significant variation in responses of S2, S4, S6, S7, S8 and S9 sensors varied considerably. Moreover, sensors S1, S3 and S5 had no response and remained unchanged, and the S10 sensors had few alterations (Fig. 1). The broad range of contaminated sample E-nose patterns is attributed to various concentrations of inoculum and increasing microorganism (S1)
Fig. 1.
Radar plot of sensor responses S. Typhimurium inoculated pork samples
The detection, discrimination and quantification of volatile compounds in pork samples with specific bacterial concentration by an electronic nose was therefore mostly dependent on the six sensors, including S2, S4, S6, S7, S8 and S9 sensors which were responsive to nitrogen oxides, hydrogen gas (H2), methane (wide-ranging organic compounds), inorganic sulfur compounds, alcohol and aromatic compounds respectively.
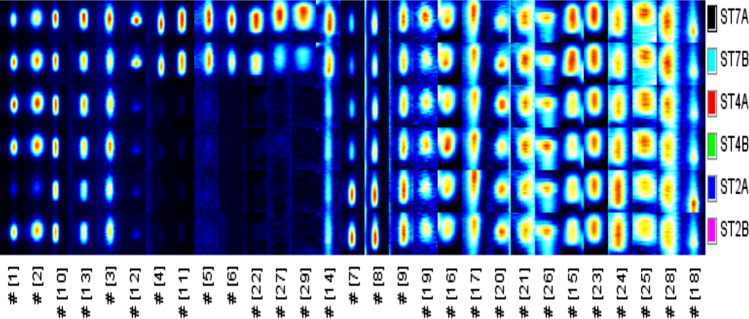
The E-nose results were further validated by the result from the HS-GC-IMS, with more than 29 compounds in the samples defined and measured based on HS-GC-IMS analysis. Figure 2 shows the 29 VOCs identified as distinctive ion transport peaks as well as the horizontal cross-section view corresponding to the inoculated bacterial meat samples at different concentrations. Only part (15 out of 29) of the distinctive ion transport peaks could be recognized in the NIST2014 spectral database, as shown in S2(Supplementary). From the table, more alcohols (7) were detected. Our study was consistent with the study by Timsorn et al. (2016) showed alcohols to be among the most prevalent volatiles in meat products when detecting bacteria population with an electronic nose.
Fig. 2.
Fingerprint comparison of volatile organic compounds (VOCs) by different concentrations of S. Typhimurium inoculated pork samples
Different intensities of VOCs identified in each bacterial sample are also shown in Fig. 2 to enable differentiation of bacteria according to their concentrations. As can be seen, the signal intensity of some VOCs was much higher or lower at different contaminant levels, while others exhibited minor changes in intensity. This could be due to growing or decreasing metabolic and catabolic activity such as proteolysis, glycolysis and lipolysis of the microbes (Audrain et al. 2015).
Qualitative discrimination based on E-nose response
VOCs are regarded as markers of microbial growth during primary and secondary metabolism as by-products and are primarily influenced by the form of microbial species and strains involved (Wang et al. 2016).
We conclude that the rise in sensor signal (E-nose) and color intensities (HS-GC-IMS) should also be closely related to bacterial concentration. The findings indicate that E-nose could qualitatively reflect bacterial concentration based on the levels of VOCs produced and should be successful in assessing pathogen presence in meat samples.
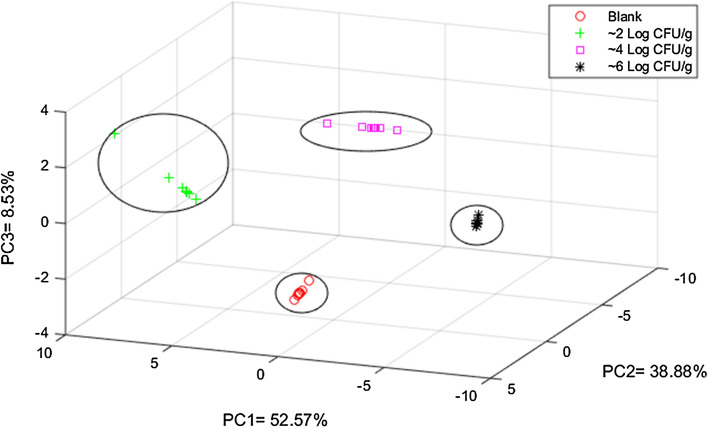
Principal component analysis (PCA) was applied to analyze the inoculated bacteria samples and the control group. E-nose PCA results showed that three main components (PC1-52.57%, PC2-38.88%, PC-8.53%) were maintained and constituted 99. 99% of the overall variation. The results from the PCA plot (Fig. 3) showed that the three levels of bacteria inoculated sample and control group were clustered at different positions, clearly showing that the influence of bacterial contamination on volatiles produce by the pork samples was sufficient to discriminate between the contaminated groups and control group.
Fig. 3.
Principal Component Analysis (PCA) of final classification model based on data fusion a 2logCFU/ml, b 4logCFU/ml c 7logCFU/ml
Furthermore, the output of the PCA may also be used to test the quality of the dataset before being applied as an input in a quantitative model. The above results show good sensor responses to bacterial contamination, since there is no significant overlap between different sample groups, with each group showing a unique pattern in PCA results.
Quantitative discrimination
Chemometric and enzyme-generated MVOCs are currently the two types of strategies employed to obtain vital information that can demonstrate the relationship between MVOCs and microbial genera in foods (Wang et al. 2016). In this study, statistical analysis was also applied to MVOC data analysis for quantitative analysis. The viability of using the E-nose signals extracted from the pork inoculated with the bacteria for colony count prediction was studied.
In other to identify the right SVM kernel parameter which improves the overall performance of SVMR model to quantify the BFP, radial basis function (RBF), linear, polynomial and sigmoid kernel functions were applied simultaneously (Andrew 2001). Each kernel function was performed five (5) times, and the results averaged. From the results (Table 2), the RBF function showed the lowest RMSE (0.217) and the highest accuracy for prediction (0.937) and was therefore used during the optimization stage.
Table 2.
Prediction accuracy of SVMR optimized by PSO, GA, and GS on S. Typhimurium contaminated pork e-nose dataset
| Model | Calibration | Cross validation | Prediction | RPD | |||
|---|---|---|---|---|---|---|---|
| RMSEC | R2C | RMSECV | R2Cv | RMSEP | R2P | ||
| PSO-SVMR | 0.065 | 0.991 | 0.183 | 0.962 | 0.145 | 0.986 | 14.11 |
| GA-SVMR | 0.036 | 0.991 | 0.163 | 0.964 | 0.137 | 0.989 | 14.93 |
| GS-SVMR | 0.058 | 0.986 | 0.177 | 0.959 | 0.148 | 0.966 | 13.82 |
| SVMR | 0.136 | 0.968 | 0.217 | 0.937 | 0.162 | 0.949 | 12.63 |
Although the SVMR model result obtained above could accurately predict bacterial foodborne pathogen concentrations in the pork samples, the results were not excellent due to poor hyperparameter tuning (the best combination of C and g parameters to create a good SVM regression model). The grid search (GS), genetic algorithm (GA) and particle swarm optimization (PSO) method were applied to optimize the C and g parameters for modifying the RBF kernel function for influencing the SVMR model (Hu et al. 2019).
Parameter optimization in multivariate regression is very valuable for enhancing the model’s predictive ability (Xu et al. 2017). The data from the E-nose signal is optimized and tested against the calculated microbial reference data using GA-SVMR, GS-SVMR and PSO-SVMR models. The results from the three optimized SVMR models offered varying but higher predictive accuracy relative to the SVMR model
SVM hyperparameter optimization
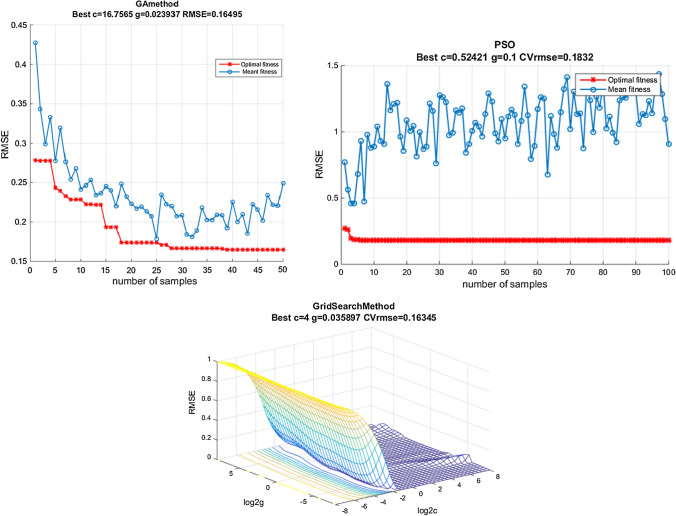
GS works on the basis of constructing a grid with all the c and g value points partitioned in a certain area, as shown in the three dimensional (3D) view in Fig. 4c. K‐fold cross-validation approach is then used to assess prediction accuracy on the basis of a given c and g category. The best results of optimization were obtained when c = 4 and g = 0.036 were used to obtain the best precision of SVM regression results with a lower mean square error (MSE). In this study the root mean square (RMSE) was applied for comparison, thus the MSE was recomputed where the RMSE = √MSE
Fig. 4.
Graph for optimizing SVM parameters by genetic algorithm, particle swarm algorithm and Grid search
In the PSO process, the number of iterations was set at 200 with the learning factor cl and c2 set at 1.5 and 1.7, respectively. From Fig. 4b PSO showed a faster convergence rate with the best optimization results and lower MSE achieved when c = 0.52 and g = 0.1.
In GA selection of the c and g, the number of iteration, population size, cross probability and mutation probability was set as 200,20, 04 and 0.02 respectively. From Fig, 4a, the convergence rate of GA was slower than PSO and became stable in the 17th generation and arrive at a lower MSE. GA requires a complete evaluation of all the items in one generation, and the best optimization results were achieved when c = 16.75 and g = 0.023
Table 2 highlights statistical measurements computed using GA-SVMR, GS-SVMR and PSO-SVMR train and test data. We improved again the RMSE of our support vector regression model with GA, GS and PSO optimization, respectively.
GA-SVMR shows a slightly higher RMSE 0.02896 and 0.03671 log CFU/g for the train, cross-validation and test data respectively. The correlation coefficients of GA-SVMR (Rc2, R2Cv, R2P) were slightly higher compared with the other models (GA-SVMR > PSO-SVMR > GS-SVMR > SVMR). From the results is observed that the efficiency and accuracy of the models rely on better SVM and kernel parameter selection. The overall performance of GA-SVMR model (Rc2 = 0.9910, R2Cv = 0.964, R2P = 0.989) performed better compared with the other models with a higher RPD of 14.11. Validation of model was subsequently performed using independent test data (Table 3). The aim of model validation is to compare model predictions with a real-world and unknown dataset for model accuracy and predictive ability assessment. Test data is used to assess the final chosen model and estimate the error of prediction. Test data will not be used until after the end of the model construction and selection process. Test results inform you how good your model is able to generalize; i.e. how great your model is performing for fresh data. The test data consisting of 45 samples inoculated at ~ 102(15 samples), ~ 104 (15 samples), and ~ 107 (15 samples) CFU/ g was used. The quantification of regression correlation values (R2 values) and root mean squares error of prediction (RMSEP) was used to measure the multivariate model. Evidently, the predictive performance of Prediction set (0.989) was higher than that of independent testing data (0.9825), presumably because the samples attributed to testing data did not belong to the same set and were not used in the calibration model development at all.
Table 3.
Prediction accuracy of SVMR optimized by GA on S. Typhimurium contaminated pork E-nose independent dataset
| Model | Prediction | RPD | |
|---|---|---|---|
| RMSEP | R2P | ||
| GA-SVMR | 0.1958 | 0.9825 | 13.24 |
E-nose nose has been widely used to compare in the past, volatile profiles to microbial counts, especially in the prediction of total viable counts. This study shows improved predictive ability of bacterial population in a food matrix and broth culture as compared to similar studies by Timsorn et al. (2016) (R2 = 0.94) and Siripatrawan (2008) (R2 = 0.96).E-nose has demonstrated to be an effective microbiological screening tool, while the test is limited by the production of volatile metabolites, which can occur after a few hours of growth.
Conclusion
The study presented was developed to identify the volatile compounds common to Salmonella Typhimurium inoculate in pork muscle. Results showed that the presence of Salmonella Typhimurium could be classified and quantified by a fast and simple analysis of the relative content of their volatile compounds using an electronic nose and GC-IMS
Previous research on the use of an electronic nose in meat products for the identification of bacterial foodborne pathogen had not been released. As a non-destructive technique, we show the electronic nose’s experimental conditions and parameter settings to obtain better response values. E-nose sensor response curve is first combined with PCA model for detection of bacterial presence/absence as well as discrimination of bacterial concentrations in meat.
The finding also shows that the e-nose, combined with machine learning algorithms and metaheuristic optimization techniques, is capable of quantifying bacterial concentrations in a food matrix, and is an efficient tool for rapid detection and quantification of bacteria pathogens in meat samples. Furthermore, E-nose can be incorporated as a process analytical technology (PAT) through the development of mini sensors for onsite process monitoring of bacterial pathogens in the meat industry. Potential technology improvements currently under study, the incorporation of sensors that are much more sensitive, can lead to a reduction in detection limits and thus the detection time.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (No. 2017YFD04001002).
Compliance with ethical standards
Conflict of interest
Authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aheto JH, Huang X, Tian X, Ren Y, Ernest B, Alenyorege EA, Dai C, Hongyang T, Xiaorui Z, Wang P. Multi-sensor integration approach based on hyperspectral imaging and electronic nose for quantitation of fat and peroxide value of pork meat. Anal Bioanal Chem. 2020;412:1169–1179. doi: 10.1007/s00216-019-02345-5. [DOI] [PubMed] [Google Scholar]
- Andrew AM (2001) An introduction to support vector machines and other kernel-based learning methods by Nello Christianini and John Shawe-Taylor, Cambridge University Press, Cambridge, 2000, xiii + 189 pp., ISBN 0-521-78019-5.Robotica 18:687-689. 10.1017/s0263574700232827
- Audrain B, Farag MA, Ryu C-M, Ghigo J-M. Role of bacterial volatile compounds in bacterial biology. FEMS Microbiol Rev. 2015;39:222–233. doi: 10.1093/femsre/fuu013. [DOI] [PubMed] [Google Scholar]
- Balbin JR, Sese JT, Babaan CVR, Poblete DMM, Panganiban RP, Poblete JG (2017) Detection and classification of bacteria in common street foods using electronic nose and support vector machine. In: 2017 7th IEEE international conference on control system, computing and engineering (ICCSCE), pp 247–252. 10.1109/iccsce.2017.8284413
- Bao Y, Hu Z, Xiong T. A PSO and pattern search based memetic algorithm for SVMs parameters optimization. Neurocomputing. 2013;117:98–106. doi: 10.1016/j.neucom.2013.01.027. [DOI] [Google Scholar]
- Bonah E, Huang X, Aheto JH, Osae R. Application of electronic nose as a non-invasive technique for odor fingerprinting and detection of bacterial foodborne pathogens: a review. J Food Sci Technol. 2019 doi: 10.1007/s13197-019-04143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonah E, Huang X, Yi R, Aheto JH, Osae R, Golly M. Electronic nose classification and differentiation of bacterial foodborne pathogens based on support vector machine optimized with particle swarm optimization algorithm. J Food Process Eng. 2019 doi: 10.1111/jfpe.13236. [DOI] [Google Scholar]
- Chang C, Chih-Jen L. LIBSVM : a library for support vector machines. ACM Trans Intell Syst Technol. 2011;2:27:21–27:27. doi: 10.1145/1961189.1961199. [DOI] [Google Scholar]
- Chang X, Huang X, Tian X, Wang C, Aheto JH, Ernest B, Yi R. Dynamic characteristics of dough during the fermentation process of Chinese steamed bread. Food Chem. 2020;312:126050. doi: 10.1016/j.foodchem.2019.126050. [DOI] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–297. doi: 10.1007/BF00994018. [DOI] [Google Scholar]
- Elbisy MS. Support Vector Machine and regression analysis to predict the field hydraulic conductivity of sandy soil. KSCE J Civ Eng. 2015;19:2307–2316. doi: 10.1007/s12205-015-0210-x. [DOI] [Google Scholar]
- Galvão RKH, Araujo MCU, José GE, Pontes MJC, Silva EC, Saldanha TCB. A method for calibration and validation subset partitioning. Talanta. 2005;67:736–740. doi: 10.1016/j.talanta.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Giungato P, Renna M, Rana R, Licen S, Barbieri P. Characterization of dried and freeze-dried sea fennel (Crithmum maritimum L.) samples with headspace gas-chromatography/mass spectrometry and evaluation of an electronic nose discrimination potential. Food Res Int. 2019;115:65–72. doi: 10.1016/j.foodres.2018.07.067. [DOI] [PubMed] [Google Scholar]
- Gobbi E, Falasconi M, Zambotti G, Sberveglieri V, Pulvirenti A, Sberveglieri G. Rapid diagnosis of Enterobacteriaceae in vegetable soups by a metal oxide sensor based electronic nose. Sens Actuators B Chem. 2015;207:1104–1113. doi: 10.1016/j.snb.2014.10.051. [DOI] [Google Scholar]
- Havelaar AH, Kirk MD, Torgerson PR, Gibb HJ, Hald T, Lake RJ, Praet N, Bellinger DC, de Silva NR, Gargouri N, Speybroeck N, Cawthorne A, Mathers C, Stein C, Angulo FJ, Devleesschauwer B, on behalf of World Health Organization Foodborne Disease Burden Epidemiology Reference G World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLOS Med. 2015;12:e1001923. doi: 10.1371/journal.pmed.1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herceg S, Ujević Andrijić Ž, Bolf N. Development of soft sensors for isomerization process based on support vector machine regression and dynamic polynomial models. Chem Eng Res Des. 2019;149:95–103. doi: 10.1016/j.cherd.2019.06.034. [DOI] [Google Scholar]
- Hu L, Yin C, Ma S, Liu Z. Vis-NIR spectroscopy combined with wavelengths selection by PSO optimization algorithm for simultaneous determination of four quality parameters and classification of Soy Sauce. Food Anal Methods. 2019;12:633–643. doi: 10.1007/s12161-018-01407-1. [DOI] [Google Scholar]
- Huang X, Yu S, Xu H, Aheto JH, Bonah E, Ma M, Wu M, Zhang X. Rapid and non-destructive detection of freshness quality of postharvest spinaches based on machine vision and electronic nose. J Food Saf. 2019;39:e12708. doi: 10.1111/jfs.12708. [DOI] [Google Scholar]
- Mohandes MA, Halawani TO, Rehman S, Hussain AA. Support vector machines for wind speed prediction. Renew Energy. 2004;29:939–947. doi: 10.1016/j.renene.2003.11.009. [DOI] [Google Scholar]
- Moscato P, Cotta C. An accelerated introduction to memetic algorithms. In: Gendreau M, Potvin J-Y, editors. Handbook of Metaheuristics. Cham: Springer; 2019. pp. 275–309. [Google Scholar]
- Nadi ZR, Salehi TZ, Tamai IA, Foroushani AR, Sillanpaa M, Dallal MMS. Evaluation of antibiotic resistance and prevalence of common Salmonella enterica serovars isolated from foodborne outbreaks. Microchem J. 2020;155:104660. doi: 10.1016/j.microc.2020.104660. [DOI] [Google Scholar]
- Osaili TM, Hasan F, Dhanasekaran DK, Obaid RS, Al-Nabulsi AA, Rao S, Fatima H, Ayyash M, Savvaidis I, Holley R. Thermal inactivation of Escherichia coli O157:H7 strains and Salmonella spp. in camel meat burgers. LWT. 2020;120:108914. doi: 10.1016/j.lwt.2019.108914. [DOI] [PubMed] [Google Scholar]
- Patil SG, Mandal S, Hegde AV. Genetic algorithm based support vector machine regression in predicting wave transmission of horizontally interlaced multi-layer moored floating pipe breakwater. Adv Eng Softw. 2012;45:203–212. doi: 10.1016/j.advengsoft.2011.09.026. [DOI] [Google Scholar]
- Phan AV, Nguyen ML, Bui LT. Feature weighting and SVM parameters optimization based on genetic algorithms for classification problems. Appl Intell. 2017;46:455–469. doi: 10.1007/s10489-016-0843-6. [DOI] [Google Scholar]
- Pozo VF, Schroeder TC. Evaluating the costs of meat and poultry recalls to food firms using stock returns. Food Policy. 2016;59:66–77. doi: 10.1016/j.foodpol.2015.12.007. [DOI] [Google Scholar]
- Saddique MS, Bhatti AR, Haroon SS, Sattar MK, Amin S, Sajjad IA, ul Haq SS, Awan AB, Rasheed N. Solution to optimal reactive power dispatch in transmission system using meta-heuristic techniques-Status and technological review. Electr Power Syst Res. 2020;178:106031. doi: 10.1016/j.epsr.2019.106031. [DOI] [Google Scholar]
- Shokri S, Sadeghi MT, Marvast MA, Narasimhan S. Improvement of the prediction performance of a soft sensor model based on support vector regression for production of ultra-low sulfur diesel. Pet Sci. 2015;12:177–188. doi: 10.1007/s12182-014-0010-9. [DOI] [Google Scholar]
- Siripatrawan U. Rapid differentiation between E. coli and Salmonella Typhimurium using metal oxide sensors integrated with pattern recognition. Sensors Actuators B Chem. 2008;133:414–419. doi: 10.1016/j.snb.2008.02.046. [DOI] [Google Scholar]
- Siripatrawan U, Linz JE, Harte BR. Detection of escherichia coli in packaged alfalfa sprouts with an electronic nose and an artificial neural network. J Food Prot. 2006;69:1844–1850. doi: 10.4315/0362-028X-69.8.1844. [DOI] [PubMed] [Google Scholar]
- Tay FEH, Cao L. Application of support vector machines in financial time series forecasting. Omega. 2001;29:309–317. doi: 10.1016/S0305-0483(01)00026-3. [DOI] [Google Scholar]
- Timsorn K, Thoopboochagorn T, Lertwattanasakul N, Wongchoosuk C. Evaluation of bacterial population on chicken meats using a briefcase electronic nose. Biosys Eng. 2016;151:116–125. doi: 10.1016/j.biosystemseng.2016.09.005. [DOI] [Google Scholar]
- Wang Y, Li Y, Yang J, Ruan J, Sun C. Microbial volatile organic compounds and their application in microorganism identification in foodstuff. TrAC Trends Anal Chem. 2016;78:1–16. doi: 10.1016/j.trac.2015.08.010. [DOI] [Google Scholar]
- Xu S, Zhao Y, Wang M, Shi X. Determination of rice root density from Vis–NIR spectroscopy by support vector machine regression and spectral variable selection techniques. CATENA. 2017;157:12–23. doi: 10.1016/j.catena.2017.05.008. [DOI] [Google Scholar]
- Xu S, Zhou Z, Tian L, Lu H, Luo X, Lan Y. Study of the similarity and recognition between volatiles of brown rice plant hoppers and rice stem based on the electronic nose. Comput Electron Agric. 2018;152:19–25. doi: 10.1016/j.compag.2018.06.047. [DOI] [Google Scholar]
- Zambotti G, Sberveglieri V, Gobbi E, Falasconi M, Nunez E, Pulvirenti A. Fast identification of microbiological contamination in vegetable soup by electronic nose. Procedia Eng. 2014;87:1302–1305. doi: 10.1016/j.proeng.2014.11.686. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.