Abstract
The kidney is among the most complex organs in terms of the variety of cell types. The cellular complexity of human kidneys is not fully unraveled and this challenge is further complicated by the existence of multiple progenitor pools and differentiation pathways. Researchers disagree on the variety of renal cell types due to a lack of research providing a comprehensive picture and the challenge to translate findings between species. To find an answer to the number of human renal cell types, we discuss research that used single-cell RNA sequencing on developing and adult human kidney tissue and compares these findings to the literature of the pre-single-cell RNA sequencing era. We find that these publications show major steps towards the discovery of novel cell types and intermediate cell stages as well as complex molecular signatures and lineage pathways throughout development. The variety of cell types remains variable in the single-cell literature, which is due to the limitations of the technique. Nevertheless, our analysis approaches an accumulated number of 41 identified cell populations of renal lineage and 32 of non-renal lineage in the adult kidney, and there is certainly much more to discover. There is still a need for a consensus on a variety of definitions and standards in single-cell RNA sequencing research, such as the definition of what is a cell type. Nevertheless, this early-stage research already proves to be of significant impact for both clinical and regenerative medicine, and shows potential to enhance the generation of sophisticated in vitro kidney tissue.
Subject terms: Cell biology, Kidney, Regenerative medicine, Tissue engineering
The need for well-characterized renal cell types
A detailed understanding of the variety of cell types within the healthy kidney and their molecular composition will benefit scientists aiming to treat patients with kidney failure. The ideal treatment is kidney transplantation from a healthy immune-matching donor. However, donor kidney availability is far from meeting its demand, and waiting times are long so that many patients die while waiting for transplantation. There is great demand for new therapies, as current treatment methods, such as dialysis, do not provide all essential functionalities of a kidney and are not long-term solutions1. For example, toxins are insufficiently removed and the sodium and fluid homeostasis are distorted by intermittent treatment, while the metabolic and endocrine functions are completely neglected2,3. Dialysis can replace many functions of the kidney but is associated with high morbidity, mortality, reduced quality of life, and high costs. Currently, the prevalence of patients with chronic kidney disease is as high as 9.1% with an age-dependent mortality rate of 1.5–6.3%4. Recent projections indicate that by 2030, nearly 5.5 million people worldwide could depend on renal replacement therapy5
Due to the aforementioned shortcomings, clinicians and scientists aim to understand hurdles such as kidney development and regeneration, factors involved in kidney failure and disease, how to improve survival on dialysis and prevent donor tissue rejection. Furthermore, joint forces of (tissue) engineers, material scientists, and (developmental) biologists are working worldwide on artificial and bioengineered kidneys as a replacement for current dialyses. They have made major advances in the de- and re-cellularization of rodent and human kidneys3, renal tubule assist devices6, lightweight miniaturized kidneys7, and implantable bioartificial devices7 and tissues8, among others. All these approaches would benefit from a greater understanding of the cellular and molecular composition of developing an adult kidney.
Creating kidneys in vitro is, however, a challenging task that involves the coordination of complex cellular and molecular events9. Starting in 2014, major breakthroughs were published on the creation of miniaturized self-assembled kidney tissue (organoids) in vitro from induced pluripotent stem cells10–12. These organoids contain various renal cell types, patterned similarly to in vivo tissue. Nevertheless, they are incomplete and essential questions remain, such as: if the developmental lineage steps are being followed, why do some cell types emerge (e.g., podocytes and endothelial cells) while others do not (e.g., mesangial cells (MCs) and parietal epithelial cells (PECs), why does the tissue deteriorate after a few weeks in culture, and why does it not mature beyond the first trimester13,14, unless transplanted in vivo15. Finally, how can we achieve and maintain a complex 3D architecture of a nephron, tubulo-interstitium and vasculature? In order to answer such questions and to develop more mature tissues for transplantation, there is a greater understanding needed of the large variety of renal cell types, their plasticity, lineage, cell state, phenotype switching, and functionality.
Divergent numbers of renal cell types
Outstanding research has been conducted in the past decades to unravel mammalian kidney development and nephrogenesis and to characterize adult kidneys16–21. However, the extraordinary developmental complexity has made scientists across fields struggle to determine the number of renal cell types and to replicate these cell types in vitro. Heterogeneity is a regular term appearing in these publications, being a clear indication of one of the major difficulties in kidney research. So far, the field generally agrees on certain heterogeneity between individuals such as the timing of kidney development (e.g., cessation of nephrogenesis from 3222 to 37 weeks23 of gestation), numbers of nephrons (210,000–1,825,000 nephrons per kidney with an average of 600,000–800,00024,25), cell numbers per segment (e.g., 431–746 podocytes per adult glomerulus26) and anatomical differences (e.g., number of renal papillae27). However, there is still little consensus on the number or range of distinct renal cell types; neither in humans nor in rodents.
More than 18−26 cell types have been described in mammalian mature kidneys21,28–36, of which at least half are epithelial and/or located within nephrons37. While the source of these numbers is challenging to trace, possible reasons for the range can include the unclear definition of what is a distinct cell type, incomplete knowledge of cell-specific markers, technical limitations, a certain degree of variability in healthy subjects, and the difficulty of defining cell identities and distinguishing cell types. Additionally, inter-species differences could underlie these ranges, as often no species is mentioned apart from “mammalian kidneys”. Clearly, part of the challenge is a lack of consensus on the definition of what constitutes a distinct cell type and the resolution of cell-specific characteristics before we can elucidate the renal cellular complexity in humans in a comprehensive way.
Defining cell identity, plasticity, and maturity
It remains questionable if it is possible to state a specific number of renal cell types and which techniques would lend the most appropriate data to do so. A specific cell number would imply a clear-cut definition of what is considered a distinct cell type. While researchers try to define cellular plasticity38, this central, measurable definition of “cell type” has not yet been determined. Earlier, an evolutionary perspective was suggested, which states that a cell type can be defined by a core regulatory complex (CoRC), which is a set of transcription factors and their interacting factors39. In a recent publication, Morris40 proposes a more complex viewpoint made of three major components: (1) phenotype and function (physical, molecular and functional features), (2) lineage, and (3) state (changes in cell state in response to stimuli). The general idea is that cell types cannot be clearly identified by solely assessing only one of the three components.
Many existing techniques for cell type detection and characterization have technical limitations. Initially, histological stains were used, followed by techniques like immunostaining, (fluorescent) in situ hybridization (FISH), and flow cytometry. However, these techniques are biased by the existing knowledge of cellular markers, are by definition not designed to discover larger scales of novel markers, and are limited in the number of markers that can be simultaneously analyzed. This makes it technically impossible to assess a single cell on all three components of cellular identity. More recently, bulk RNA sequencing allowed the exploration of novel markers by providing average gene expression across a large population of cells41–43. However, the expression of low abundance genes is underrepresented in bulk sequencing and cellular lineage cannot be determined41,44. Now, in the era of single-cell RNA (scRNA) sequencing, lineages can be determined and rare gene detection is possible. The heterogeneity within cell populations can be detected in high throughput41,42, as can the profiling of cell states and transitions during differentiation41,45–47. Therefore, while not free of limitations, scRNA sequencing is a promising technique to answer the long-standing question of the number of renal cell types.
This review aims to investigate the variety of renal cell types in the developing and adult human kidney by discussing common and conflicting findings of scRNA sequencing studies. Given our focus on regenerative medicine and tissue engineering, we excluded publications on renal pathology and drug testing. Briefly, the review first addresses the major stages of nephrogenesis to discuss the cellular lineages within these stages, which are important for researchers aiming to replicate development in vitro. Subsequently, findings on the cellular variety of adult kidneys will be discussed. The discovery of novel cell types and markers, subpopulations, intermediate cell states, segment transitions, as well as phenotype-specific expression patterns, and developmental trajectories will be covered. Finally, shortcomings and opportunities of scRNA sequencing will be discussed in the context of kidney research.
Main analysis
Structural development of the human metanephric kidney
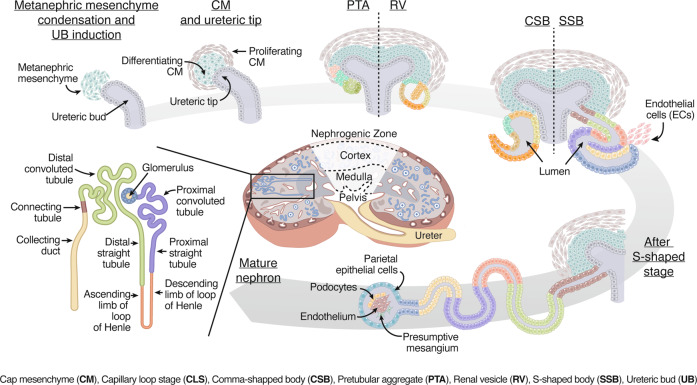
Here we highlight the major stages of development, knowledge needed for the remainder of this review. Development of the human metanephric kidney begins around four weeks of gestation17 from a close interaction of the metanephric mesenchyme (MM) and the ureteric bud (UB) (Fig. 1). While both MM and UB derive from the intermediate mesoderm, it is in the MM where nephron progenitor cells (NPCs) differentiate into nephron tubules, the glomerulus, and the renal stroma; whereas the UB gives rise to the collecting duct and ureter18. Simultaneous to the mesenchyme differentiation, UB differentiation and proliferation occur on the tip of the UB and lead to branching (bifurcation) into a tree-like pattern with later extensive elongation of the ureteric trunk48,49. The MM undergoes morphological changes during differentiation characterized as pretubular aggregate (PTA), renal vesicle (RV), comma-shaped bodies (CSB), and subsequently S-shaped bodies (SSB) before entering the capillary loop stage (CLS)16,50,51. The first SSB is detected around Carnegie stage (CS) 18–1917, from which further differentiation occurs.
Fig. 1. Schematic representation of nephrogenesis, starting with the interaction of the cap mesenchyme (CM) and ureteric bud (UB) in the nephrogenic zone.
Differentiation takes place along morphologically determined stages known as pretubular aggregate (PTA), comma-shaped body (CSB), renal vesicle (RV), S-shaped body (SSB), and capillary loop stage (CLS) nephron. The mature nephron is located throughout the cortex, whereas the Loop of Henle and collecting duct extend into the medulla.
Cells of the distal nephron segment start invading the collecting duct epithelium at the SSB stage to connect the nephron to the collecting duct52. Subsequently, endothelial cells start to invade the proximal segment of the SSB, initiating the CLS. This stage is further characterized by the development of the vascular system, including the glomerular capillaries, arteries, veins, and the appearance of the primitive loop of Henle (LoH)53. The first generation of glomeruli appears to be mature around week 954 and shortly after, glomerular filtration begins55. Eventually, 8–12 generations of glomeruli are formed, leading to nephrons in different developmental stages. Nephrogenesis ceases before birth, by week 38–4123.
Nephrogenesis is further supported by the surrounding interstitium and extracellular matrix56. The term interstitium describes a tissue with a variety of cell types such as renal fibroblasts, pericytes, and cells with endocrine functionalities (renin- and erythropoietin-producing cells), with possible distinct origins57. Although the discussion of the origin of renal interstitium is ongoing, one perspective is that it emerges early in nephrogenesis from a FOXD1+ subpopulation of the MM57,58.
Renal cell type discoveries by single-cell RNA sequencing
Hereinafter, we highlight discoveries from scRNA studies on fetal and adult human renal tissue and contrast the findings with earlier literature using techniques like histology, immunostaining, and FISH. We aim to elucidate the divergent numbers of clusters/cell types of the different scRNA and single nucleus RNA sequencing studies performed on human renal tissue to date, as summarized in Table 1 and Supplementary Table 2, and abbreviated as “scRNA sequencing papers” throughout this publication. Given the comprehensive topic, we decided to focus on kidney cells derived from the same intermediate mesoderm progenitor (excluding interstitial cells (ICs)) and summarize all findings in an updated lineage tree in Fig. 2. Consequently, the discussion of the variety of immune cells, vascular and blood cells present in developing and adult kidney goes beyond the scope of this review; however, all findings of these cell types are summarized in Fig. 3 and Supplementary Table 1.
Table 1.
Donor information and detected cell clusters from single-cell RNA sequencing studies of healthy human kidney tissue.
| Publication | Donor age (fetal in weeks; adult in years)a | Tissue source | Number of clusters | Number of clusters of metanephric mesenchyme lineage |
|---|---|---|---|---|
| Young, et al.67 | Fetal (8, 9) | Whole kidneys | 7 | 6 |
| Menon, et al.61 | Fetal (12.4, 15, 15.7, 16.4, 18.8) | Whole kidneys | 11 | 18 |
| Lindstrom, et al.64 | Fetal (16) | Nephrogenic zone | 12/15 | 10 |
| Wang, et al.68 | Embryonic/fetal (7–10, 13, 19, 22, 24, 25) | Whole kidneys | 13 | 10 |
| Combes, et al.89 | Fetal (16) | Nephrogenic zone | 16 | 13 |
| Lindström, et al.78 (preprint) | Fetal (14–17) | Nephrogenic zone | 18 | 18 |
| Stewart, et al.88 | Fetal (7 [F], 8 [M], 9 [F], 12 [M], 13 [F], 16 [F]) | Whole kidneys | 21 | 11 |
| Tran, et al.65 | Fetal (15, 17) | Inner and outer cortex sections | 21 | 18 |
| Hochane, et al.59 | Fetal (9 [M], 11 [M], 13 [F], 15 [F], 16 [M], 18 [F]) | Whole kidneys | 22 | 19 |
| Lindstrom, et al.66 | Fetal (17) | Nephrogenic zone | 22 | 15 |
| Liao, et al.100 | Adult (57–65 [2 × M + 1 × F]) | Whole kidney sections | 10 | 8 |
| Wu, et al.118 | Adult (70 [M]) | Biopsy | 16 | 6 |
| Wu, et al.104 | Adult (62 [M]) | Cortex | 17 | 7 |
| Young, et al.67 | Adult (72) | Interface or region-specific biopsies | 19 | 10 |
| Stewart, et al.88 | Adult (44–72 [M + F]) | Cortex, medulla, pelvis biopsy | 25 | 12 |
| Sivakamasundari, et al.101 (preprint) | Adult (62–66 [M + F]) | Resection samples | 27 | 11 |
| Kuppe, et al.86 | Adult (50–84) | Healthy tumor nephrectomy tissue | 27 | 23 |
| Lake, et al.103 | Adult (>50 [M + F]) | Cortex and medulla | 30 | 19 |
aBiological sex (M = male/F = female) is indicated when known.
A more detailed table can be found in the supplementary files (Supplementary Table 2).
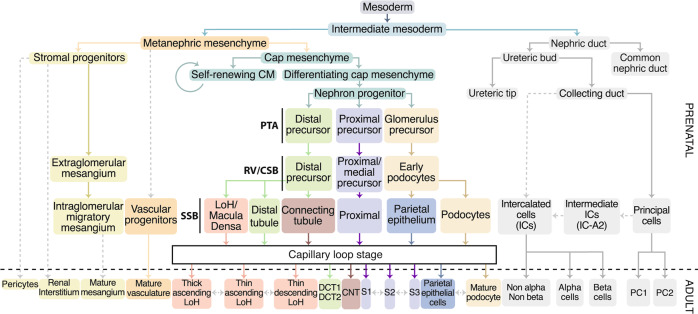
Fig. 2. Lineage tree of renal cell types in development based on non-scRNA sequencing and scRNA sequencing literature.
Dotted arrows indicate possible lineage relationships, which are not fully proven or not highlighted in the discussed scRNA sequencing publications. Pretubular aggregate (PTA), renal vesicle (RV), comma-shaped body (CSB), S-shaped body (SSB), Loop of Henle (LoH), connecting tubule (CNT), distal convoluted tubule 1 and 2 (DCT1/2), segment 1/2/3 (S1/S2/S3), intercalated cells (ICs), principal cells (PCs).
Fig. 3. The variety of different renal cell populations identified by single-cell RNA sequencing to date. All cell types detected (accumulated) by all discussed scRNA sequencing publications.
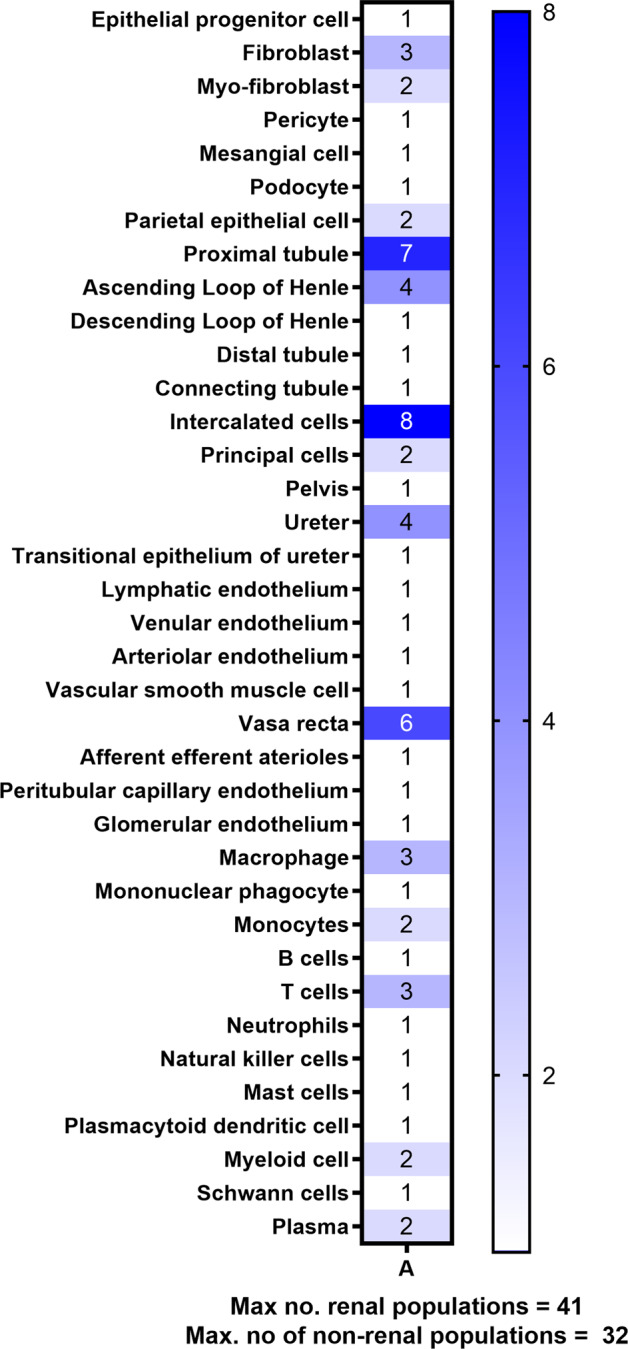
The number indicates the highest number of clusters distinguished by one or more of the discussed publications and with this, the total sum of populations was calculated. For the adult kidney, 41 renal and 32 non-renal populations were detected.
Pseudo-time trajectory follows the developmental flow
Pseudo-time analysis is a computational method to establish a dynamic process experienced by cells and arranges these cells based on their progression through this process. Pseudo-time trajectory analysis suggested that the identified kidney cells generally seem to follow the known developmental stages from NPCs, CSB, and SSB to mature, differentiated cell types59. Interestingly, this analysis did not describe CLS as a separate stage during differentiation. The identified literature on fetal kidney (Table 1) agreed that podocytes emerge first from the SSB nephron progenitors, followed by proximal and distal tubules, and finally LoH. This confirms earlier knowledge on the timing of nephron patterning60, where proximal, distal segments, and podocytes followed distinct lineages, and the distal tubule further differentiated into LoH and the connecting tubule (CNT). To our knowledge, the early podocyte emergence has, however, not been described before the scRNA sequencing era. Additionally, Menon, et al.61 distinguished the UB, stroma, and nephron as three distinct developmental trajectories. Although podocytes in the same study had strong stress-related signaling, possibly a consequence of their dissociation, pseudo-time indicated an extraordinarily complex differentiation trajectory59. Needless to say, more studies are needed to confirm these findings to exclude interindividual differences and dissociation biases. Future research could consider reducing dissociation artifacts by, for instance, the addition of cold-active proteases62.
The balance of self-renewal and differentiation in early kidney development
The cap mesenchyme (CM) is considered the renal stem cell niche from which, in interaction with the ureteric epithelium, the mammalian kidney develops. Throughout development, the CM is maintained to repeatedly supply NPCs to undergo mesenchymal-to-epithelial transition (MET) and form the epithelial RV63. The number of different cell types at this stage has not yet been elucidated and scRNA sequencing research reveals that this will be a challenging task.
CM and NPCs populations were detected to various extents in the scRNA sequencing publications, with most papers describing one or the other, but not both populations (Supplementary Table 1). This could be explained by the different fetal ages. Studies of week 15–17 fetuses64–66 might be less likely to detect CM clusters, since the CM population decreased with further differentiation and the markers are less likely to be detected compared to analyses at week 7–1067,68. However, differences can also be attributed to the process of assigning markers differently to certain populations as well as the definition of CM and NPC. The clear differences in markers used to identify both CM and NPC in the various publications indicate the challenge of distinguishing cell populations in early development (Supplementary Table 3). Clearly, both a consensus on the main population markers as well as terminology is required in order to ensure comparability of this complex research and provide a common ground for future studies.
Irrespective of the definition of CM and NPC, both differentiating and pluripotent populations were detected. For instance, Wang, et al.68 distinguished two transcriptionally distinct populations, which are in line with earlier mouse18,69–72 and human studies73 investigating the role of SIX2 and MUC1 expression in the progenitor pool, respectively. One such population expressed markers SIX2, EYAI, and COL1A1, which are involved in the induction of CM and associated with the self-renewal capacity of stem cells indicated by high HMGA1 and HMGA2 expression68. Pseudo-time analysis suggested the self-renewing population sustained its proliferative capacity throughout nephrogenesis68. The second population was identified as gradually going through MET and expressing epithelial markers like CLDN11 as well as NPHS2, indicating the onset of podocyte differentiation. The fact that Hochane, et al.59 detected a self-renewing cluster clustered as NPCs could also indicate a gradual differentiation from CM to NPCs.
In terms of novel markers of the CM, Hochane, et al.59 described UNCX as a novel marker for the CITED1+ self-renewing population after validating their transcriptomic data with immunostainings. Although UNCX was found in early mouse studies74–76, where it was described as a transcription factor involved in distal RV differentiation that disappears around E17.5, this seems to be the first study showing UNCX gene and protein expression in human tissue. Future studies might consider investigating its role by analyzing spatiotemporal expression along with common CM markers in human tissue. In summary, various scRNA sequencing studies in renal development support the presence of a self-renewing and differentiating population within the CM, while distinct NPC populations seem yet to emerge, and the determination of specific cell types is challenging.
Segmentation and differentiation in early stages of developing nephrons
As development continues, the relative expression of CM genes decreases sharply after approximately week 10 with further cellular differentiation of NPCs and morphological organization into PTAs (Fig. 1). At this stage in mice, PTAs are already well known to be segmented into proximal and distal segments77. Three-dimensional imaging of the developing human kidney revealed that NPCs already assign to certain lineages at the onset of the PTA. SIX1+ NPCs segment into two layers upon recruiting, where the earliest recruited NPCs are of distal lineage and the latest are of proximal lineage. The last NPCs recruited are hypothesized to be of parietal lineage. The spatiotemporal location of NPCs within the early developing nephron could thus have an impact on their subsequent respective lineage. The latest research from the same group confirmed three distinct populations within the PTA-expressing proximal and distal markers78.
Simultaneously with NPC recruitment, the connection between NPC and PTA is gradually reduced until the late renal vesicle stage when it is broken and the SIX2+ CM remains on the tip of the UB66. In this process, the PTA undergoes MET, polarizes, forms a lumen and cadherin-mediated cell–cell contacts emerge to finally result in the RV. Earlier research shows that the RV is segmented into proximal and distal parts in mice and shows priming for podocytes, parietal epithelial cells, proximal tubules (PTs), and distal tubules (DTs)79. Few scRNA sequencing studies of human tissue detected the RV and only one detected more than one cluster, namely five in total, which were defined by patterns of CDH1, JAG1, and WT1 expression78. Since the latter study sequenced the nephrogenic zone only, it is clearly a challenge to detect the RV or even distinguish populations within whole kidneys.
In the final stage of nephrogenesis, the SSB stage, further segmentation of the nephron starts to be noticeable. Divided into proximal, medial, and distal segments, each segment differentiates further into distinct epithelia79. Few scRNA sequencing studies identified the SSB. Like in RVs, MAFB expression was associated with the podocyte progenitors within the proximal segment59. Correlation of scRNA sequencing and immunofluorescence confirmed the presence of precursors of distal, proximal, LoH populations and renal corpuscle within the SSB66. In a preprint, Lindström, et al.78 identified six distinct populations by three-dimensional spatial mapping of single-cell transcriptomes, which agreed with the previously identified populations and additionally detected parietal epithelium, CNT, and macula densa/LoH. Interestingly, a single tubular progenitor population initially exists next to podocytes and parietal epithelium. As development proceeds, this tubular population differentiates further into distal and medial domains. Congruence in gene expression with the distinct adult cell types supports the assumption that these early populations are precursors that start to express some transcription factors and genes associated with specific cell type functionality as known from mature cells.
To conclude, while the SSB is distinguished by some scRNA sequencing papers, it clearly remains challenging to distinguish PTAs, RVs, and CSBs. Identification by morphology and correlation to their respective transcriptome was done to overcome this challenge. This is an ideal example of the difficulty of distinguishing cells with subtle differences in the transcriptome following a gradual differentiation during development. However, the preprint of Lindström, et al.78 indicates that better molecular and temporal resolution could be achieved by spatially mapping transcriptomes. Alternatively, pseudo-depth analysis could be applied. Furthermore, in combination with markers for the G2/M phase of mitosis, a distinction might be made between RV and CSB based on an assumption of reduced proliferation after the RV stage59. By fine-tuning the in vitro replication of kidney development based on such data, more diverse cell types could be generated according to natural lineage trajectories.
Mesangial cells in development and adulthood
The stromal population comprises a diverse cellular composition. Functionally, it guides nephrogenesis and provides essential signaling around mature nephrons. However, the diversity and functionality of stromal cells are not fully unraveled. ICs, MCs, juxtaglomerular cells, fibroblasts, pericytes, and smooth muscle cells belong to the stromal populations, and all derive from a common FOXD1+ precursor80. MCs, nowadays considered a special type of pericyte, are located within the glomerulus in direct contact with glomerular endothelial cells, while extra-glomerular endothelial cells are located at the stalk81,82. Less than half of the discussed scRNA sequencing publications detected a mesangial cluster, of which just one discriminated intra- and extraglomerular mesangium68. This is because the molecular signature of e-MCs is not yet well defined and distinguishing them from intraglomerular MCs (i-MCs) is only possible by analyzing the microanatomy. This correlation of the two techniques has led to interesting results.
Single-cell RNA sequencing revealed that e-MCs had higher proliferative potential, whereas i-MCs were more mature. Interestingly, until week 10, the expression of the mesenchymal cell marker PDGFRb and endothelial cell marker CD31 were restricted to the stromal compartment and later migrated into the glomeruli68. This migratory phenomenon has been described in rat development83 and aligns with the investigations of Menon, et al.61 who presented the expression of TAGL in migratory MCs. The molecular distinction of i-MCs and e-MCs was resolved via the co-expression of PDGFRb and LAM4A of i-MCs. Furthermore, PIEZO2, a gene encoding a stretch-gated ion channel, was identified in i-MCs of adult human kidneys. Stretch-gated ion channels in MCs were already described in 198984, but the research is very limited and PIEZO2 has not been described earlier in this context.
While it is beyond the scope of this review to discuss all stromal cell types in detail and few papers resolved a variety of stromal populations, we would like to highlight recent scRNA sequencing publications investigating the interstitial heterogeneity in both rodent85 and human kidneys86,87. A brief comparison with human datasets confirmed the conservation of this heterogeneity. Given the important role these cells play in extracellular matrix deposition and endocrine signaling, we hope that future research will continue in-depth characterization.
Development and maturation of glomerular cells
Pseudo-time trajectories suggest that podocytes are the first differentiated cell type to emerge and mature along a complex genetic trajectory61. Remarkably, 228 genes are involved only for podocytes to develop from SSB podocytes to mature podocytes59,61. Comparing different gestational weeks showed that podocytes emerge around week 9 of human development88. The podocyte-specific markers MAFB and TCF21 are first detected in renal vesicles overlapping with SIX2 and TMEM10065. Accordingly, nephron progenitor markers SIX2, EYA1, and MEOX1 are downregulated over time and diminish with the onset of early podocyte markers MAFB and TCF2165,66. The developmental trajectory becomes increasingly complex as it continues from this point.
Four studies independently provided evidence for a transitional, immature podocyte population59,61,65,89. A small subpopulation of cells, located in the visceral part of the proximal segment of the SSB, follows the podocyte trajectory while expressing a distinct set of markers in the SSB phase until the capillary loop phase59. While not all studies agree on all markers of either immature or mature podocytes (Table 2), there is a general consensus that the transitional, immature podocytes express OLFM3, and do not express well-known mature podocyte markers such as NPHS1, NPHS2, and PTPRO. Within each of these independent studies, the evidence confirms these findings as additional techniques such as single-molecule FISH and immunofluorescent labeling delivered supporting information59,65,66. The marker heterogeneity between the studies of this transient population could be due to age differences of the individuals, technical differences, and the spatial heterogeneity caused by the gradually increasing maturity towards the medulla. However, all except one study detected OLFM3 expression, confirming that the different studies found the same transitional podocyte cluster.
Table 2.
Overview of the number of clusters of podocyte progenitors and mature podocytes with their respective markers discovered using single-cell RNA sequencing of developing human kidney.
| Author | Tissue age | Podocyte progenitor clusters | Markers early podocytes/ transitional cells | Podocyte clusters | Markers mature podocytes |
|---|---|---|---|---|---|
| Hochane, et al. 59 | w9, w11, w13, w16, w18 | 1 | OLFM3+/MAFB+/FOXC2+/CLIC5-LOW/ PTPRO-LOW, NPHS1-LOW, NPHS2-LOW | 1 | MAFB-LOW, NPHS2+, PTPRO+, PODXL+ |
| Lindstrom, et al. 66 | w17 | 1 | CLDN5+, OLMF3+ | 1 | TCF21+, NPHS2+ |
| Lindstrom, et al. 64 | w16 | 1 | PODXL+, MAFB+, NPHS2+ | 0 | N/A |
| Menon, et al. 61 | w12.4, w15, w15.7, w16.4, w18.8 | 1 | OLFM3+/MAFB-LOW | 1 | PODXL+, NPHS1+, NPHS2+, CLIC5+ |
| Combes, et al. 89 | w16 | 1 | ON1 CTGF+, OLFM3+, MAFB+, NPHS1+, LHX1-LOW, PAX8-LOW | 2 | PTPRO+, SYNPO+, VEGFA+, WT1+ |
| Tran, et al. 65 | w15, w17 | 1 | LHX1+, PAX8+, FBLN2+, OLFM3+, PCDH9+, SLC16A1+, GFRA3+, LEFTY1+ | 2 | PLA2R1+, ARMH4+, F3+, SYNPO+, NPHS2+, MAFB+, TGFBR3+, COL4A3+, COL4A4+, TNNT2+, PLCE1+, ANXA1+ |
| Wang, et al. 68 | w7, w8, w9, w10, w13, w19, w22, w24, w25 | 0 | N/A | 1 | N/A |
| Stewart, et al. 88 | w7-16 | 0 | N/A | 1 | NPHS2+, PTPRO+, WT1+, TCF21+, PODXL+ |
| Young, et al. 67 | w8,9 | 0 | N/A | 0 | N/A |
| Lindström, et al. 78 | w14 | 1 | EFNB2+, BMP4+, OLFM3+, MAFB+ | 1 | N/A |
w weeks, N/A information not available. Gene symbols in italic.
Within the immature podocyte cluster, OLFM3 was most highly expressed compared to other genes, but diminished towards the CLS with the onset of mature podocyte markers. Proliferation markers had a low expression, which is expected for podocytes59,90. To date, no other publication has shown either OLFM3 expression or a transitional podocyte population in human kidneys. The only transitional cell type related to podocytes was described as CD133+/CD24+ progenitors, which are located at the urinary pole within the Bowman’s capsule and the PT and have been attributed to possess progenitor-like behavior by transdifferentiating into podocytes in case of injury91–93. However, this progenitor-like cell type is not to be confused with the OLFM3+ population as the latter is a transient population in development, while the former was found to be resident in adults.
Interestingly Brunskill, et al.94 extensively mapped the molecular signature of podocytes (144 podocyte-specific genes) in developing and adult mice and found a distinct set of podocyte-specific genes enriched in immature podocytes, with more mature markers emerging over time. These drastic gene expression changes can be associated with both the considerable morphological95 and functional changes in podocyte development, where the immature OLFM3+ population could represent pre-functional podocytes. Combes, et al.89 and Tran, et al.65 distinguished an additional maturing podocyte cluster that follows the transitional OLFM3+ subcluster. This might indicate even more complex developmental stages. Markers related to microtubule modulation, such as TUBA1A and STMN1, were upregulated in this additional cluster, indicating the extensive morphological transformations known for podocyte development96. The field would benefit from more in-depth studies elucidating the role of OLFM3 in the maturation of podocytes to support replication of differentiation in vitro.
Another interesting glomerular cell type is the PEC. Terminally differentiated PECs are located as a monolayer on top of the Bowman’s capsule and retain the capacity to proliferate. A variety of distinct PECs have been described according to the expression of different proteins, such as Pax-2 and claudin-1, but also their localization on the Bowman’s capsule97. Additionally, PECs have frequently been a discussion point in terms of their transdifferentiation into podocytes in glomerular diseases91,92,97–99. Single-cell RNA sequencing research on the nephrogenic zone showed the existence of a common progenitor of PECs and podocytes until the RV stage78. A few weeks later in development, sequencing of the whole kidney showed distinct gene expression profiles for PECs and podocytes in a different study61, indicating distinct maturation pathways. The potential to transdifferentiate has not been shown in the scRNA sequencing papers. We anticipate that the common progenitor is a starting point for more sophisticated research to investigate the role of PECs in injury and disease.
Surprisingly, only one of the published (single-cell) RNA sequencing papers could detect more than one PEC population; however, characterization of these clusters is still needed86. A few other papers found a single cluster of PECs in either fetal kidney61,65 or adult kidney100,101 with enhanced expression of CAV2, PTRF (CAVIN1), CLDN1, CLDN3, LIX1, CDH6 and KRT8, KRT18, CD24, VCAM1 respectively. However, there was no discrimination between subpopulations, while at least two transitional cell types (in between PECs and podocytes) have been described in earlier research97. Therefore, future research would benefit from defining the molecular signature of PECs to unravel their complete lineage trajectory and functionalities in adulthood.
Patterning of tubular epithelial cells
From around week 12 onwards, more cell types are detectable, such as PT, LoH, CD, and PE17,88. As previously described, PT and DT differentiate around the same stage and subsequently LoH and CNT emerge from the DT. While little information is provided on tubular maturation within the scRNA sequencing papers, interesting findings provide insight into segment transitions and molecular signatures related to cell functions.
Generally, little is known about the molecular signature of the distinct PT segments. The proximal convoluted tubule (PCT) includes the segments S1 (early PCT) and S2 (late PCT). The proximal straight tubule (PST) is composed of the S2 (cortical PST) and S3 (medullary PST). All scRNA sequencing studies of adult kidney detected at least one PT cluster in adult tissue, of which about half found distinct populations that correlated with the different segments S1–3. However, earlier research has indicated a lack of discrete transitions in morphology in between the distinct segments and the likelihood of intermediate cell types102. A continuum of gene expression, as shown in similarity weighted nonnegative embedding (SWNE) analysis, confirmed the existence of such continuous transitions from PT to DCT103. A recent publication distinguished seven PT populations of which three expressed markers of both S1 and S2, also confirming a gradual transition86. This phenomenon of gradual transition could also explain why some papers resolved only a single PT population101.
Single-cell RNA sequencing provides more insight into both molecular anatomy and (patho)physiology. With the PT being the most abundant cell type in the kidney, 54 transcription factors were discriminated, many of which are restricted to the PT104. Although markers and transcription factors are identified in various studies100,104, it is believed that differential gene expression along the PT trajectory indicates variation in metabolic processes and transport in the various segments103. Interestingly, next to metabolic markers, deep sampling revealed marker expressions with a role in immune defense against pathogens, as well as inflammation and regeneration following kidney injury103.
The DCT can be distinguished from the PT by a decrease in fatty acid, glucose, amino acid, and hormone metabolism68. Furthermore, the distal tubule, LoH, and CNT are thought to derive from the same distal precursor within the SSB, which is distinct from the PT precursor78. Only one scRNA sequencing publication distinguished more than one DCT population101, although previously two distinct populations have been confirmed. This might indicate a gradual transition between DCT cell types103. In contrast, there are indications for an abrupt transition of the thick ascending LoH into the DCT since there was no overlap of the segment-specific markers SLC12A1 and SLC12A3101. Indeed the same lack of double-positive cells could be seen in the data set of, for example, Lake, et al.103. Morphologically an abrupt transition was already shown in rodents several decades ago105.
The CD consists of the cortical collecting duct (CCD) and outer medullary collecting duct (OMCD) and comprises intercalated (ICs) and principal cells (PCs)102. Principal cells express the epithelial sodium channel (ENaC). Currently, the classification of intercalated cells takes into account the expression of the chloride–bicarbonate exchanger SLC4A1, the multi-subunit H+-ATPase, and pendrin (subtype of the chloride–bicarbonate exchanger). Consequently, IC-A cells apically express H+-ATPase, basolaterally express SLC4A1, and lack pendrin. IC-B express H+-ATPase at their basolateral pole and non-A, non-B cells express both H+-ATPase and pendrin at their apical membrane.
Non-A, non-B cells are, amongst others, located in the CNT106, which, against earlier belief, develops from the distal tubule and not the CD in both humans78 and mice79. The cellular complexity of the CNT could, however, only partly be resolved in the discussed papers. Only one CNT cluster has been reported throughout the studies, although the CNT is known to contain cell types with DCT2 and CD phenotypes103. This leads to the question of the identity of this single CNT cluster and why further clustering was or could not be shown. Future studies could include spatial information in their quest to distinguish subpopulations of the CNT and use spatial transcriptomics such as the Nanostring Whole Transcriptome Atlas (WTA) technology.
Nearly all scRNA sequencing papers of adult human kidney detected intercalated cells and most distinguished multiple populations and principal cells. Particularly, single nucleus RNA sequencing disclosed a remarkable resolution of the transitions from DCT to CD by distinguishing two PC and three IC populations. Within these transitions, IC-B, IC-A1, and PC-1 dominated within the cortex, while PC-3 and IC-A2 were highly abundant in the medulla. All IC and PC clusters expressed markers known for the collecting duct103. It remains to be determined if PCs and ICs located in the CNT can be distinguished on a transcriptional level. In the recent publication by Kuppe, et al.86 a surprising eight distinct IC populations were detected in adult kidneys. Future research could investigate if these clusters contain ICs located within the CNT. In sum, various scRNA sequencing papers succeeded in detecting more than the known cellular variety of populations within the CD, but limited populations within the CNT, which will be highly interesting to characterize further.
Interestingly, the PC marker AQP2 was also expressed in a subpopulation of IC-A, here named IC-A2, in two independent studies of human kidney68,103 as well as in mice107,108, leading to the hypothesis that IC cells derive from the first emerging PC cell population with a double-positive transitional/intermediate stage (Fig. 2). More characterization of the IC-A2 cell type is needed, particularly in terms of localization of SLC4A1 and H+-ATPase, to clearly identify this cluster since earlier research did not distinguish different IC-A populations106. No non-A, non-B cells were detected, which were previously defined as expressing both H+-ATPase and pendrin at their apical membrane and to be located in the CNT106. The detection of the additional IC subtype IC-A2 could inspire future research to investigate the lineage relationship between ICs and PCs.
All cell populations found in the discussed scRNA sequencing publications are summarized in Supplementary Table 1. Comparing the number of clusters for each cell type as described in each publication (excluding unassigned clusters such as “proliferating”, “differentiating” and too general clusters such as “epithelial cells”) provides insight into the cellular composition of developing and adult kidneys. Overall, 6–19 distinct cell populations of renal lineage were detected across the publications in the developing kidney and 6–33 distinct populations in the adult kidney, with a median of 14 populations in development and 13 in adult kidneys. Between 2–9 distinct non-renal populations were described, with a median of 4 in development and a range of 4–23 with a median of 7 in adult kidneys. Multiple factors could lead to these ranges including different tissue sources (biopsy, resection, whole kidney, etc.), tissue location (cortex vs medulla) or different digestion methods. Therefore, having the papers complement each other’s findings by taking the sum of all different populations of all publications, likely provides a more realistic number. Thus, we counted the largest number of clusters for specific cell types (i.e., seven PT clusters described by Kuppe, et al.86) and with this, the total sum was calculated. This approach helps us to provide an approximation for the variety of cell types within human kidneys of both renal and non-renal lineage; namely in total of 73 distinct populations in adult kidneys. Of this, 41 are populations of renal lineage and 32 are populations of non-renal lineage. (Fig. 3) The fetal kidney comprised approximately 32 cell populations of renal lineage; however, developing cells were likely counted repeatedly throughout the various stages of nephrogenesis. Clearly, these numbers are only an approximation of the true variety of cell types, due to the large variation in factors like tissue source, tissue age, dissociation method, terminology, and computational settings. Certain cell types, such as the large variety of stromal cells, are underrepresented in the current research and would need to be added. Furthermore, the definition of what defines a distinct cell type is yet unclear and heavily affects the number of identified cells. However, future studies with further developed protocols and novel technologies will certainly identify additional cell types, which cannot yet be resolved.
General discussion
In this review, we discuss new knowledge on the cellular composition of healthy developing and adult human kidneys based on scRNA sequencing studies. ScRNA sequencing has the unique potential to provide comprehensive information on the cellular and molecular complexity of various tissues. Therefore, we aimed to compare the current literature in the field to estimate the heterogeneity of kidney cell types. We conclude that scRNA sequencing is a valuable tool to detect a variety of cell types; for instance, transient cell populations such as developing podocytes and a new intercalated cell population.
However, there are also various issues with this technology such as the sparsity of the generated data. For instance, the biological sex of the studied patients was not considered in the analyses and has not been studied elsewhere on a single-cell transcriptomic level. Adding this could be of valuable impact. Differences in function, morphology, and responses to injury are already known to exist between females and males in human developing and adult kidneys23,109,110. Recent scRNA sequencing of rodent kidneys also reveals transcriptomic differences between sexes111. For instance, markers of PT functionality, including organic anion, amino acid transport, and drug and hormone metabolism, were differentially expressed in female and male mice. Future research could investigate conformity in humans and particularly elaborate on differences in the developmental trajectory in pseudo-time. Additionally, the variety of cell types detected with scRNA sequencing is strongly determined by factors such as computational settings, dissociation methods, tissue source, markers used to identify clusters, and patient age. Standardized reporting of factors such as gestational age would facilitate the integration of multiple datasets and allow further analysis of, for example, transcriptome development in relation to gestational age.
Certain differences of used terminologies, technical limitations, and the lack of the definition of a cell type impede the data comparability and integration. Several comprehensive reviews have been published on solving technical limitations of scRNA sequencing, such as distinguishing rare transcripts from noise, dropouts and computational challenges such as user-defined clustering42,112,113.
While many limitations are being addressed, additional techniques will be needed to confirm cellular identity besides the transcriptomic profile. These techniques could be applied to answer research questions in terms of the previously discussed three pillars of cellular identity. They could include anatomical localization of the scRNA sequencing markers by in situ hybridization, in situ sequencing, and studying ligand–cell interactions114. Determining cellular functionality and state in response to stimuli in human tissue will primarily require explantation since cell types outside their native environment, in 2D culture, will probably lose their phenotypic characteristics. Clearly, there is still no straightforward definition of a cell type, however, a combination of the aforementioned techniques and models could make a large step towards an atlas of functional human kidney cell types.
Discrimination of early nephrogenic stages and cell type precursors using scRNA sequencing appeared to be consistently challenging throughout various studies due to the subtle molecular and spatial changes. Comparing different studies is particularly difficult, since the transcriptome significantly changes during differentiation, and thus the time point of cell isolation is very important. Consequently, additional validation experiments are needed e.g., using staining of identified markers. Alternatively, differentiation paths in development could be regarded more as a continuum rather than distinct clusters115. For instance, podocytes would consequently not be distinguished as separate RV-, CSB-, SSB-podocytes, but as podocytes expressing gradients of transcriptomes throughout development. For this, trajectory interference could be a more suitable method. As such, pseudo-time trajectory analysis has shown that podocytes differentiate prior to the differentiation of tubular cell types and that there is a distinct differentiation pathway for proximal and distal tubular segments. Finding master regulators along these differentiation pathways can support both the understanding of disease as well as in vitro replication of development. Additionally, more insight could be generated into much earlier development on how the degeneration of temporary pro-and mesonephros could influence the pattering of the permanent metanephric kidney.
Sequencing cells from distinct anatomical locations, such as the nephrogenic zone as shown in various publications of Lindstrom, et al.66, can help to achieve a higher resolution of the early nephrogenesis. This approach might then also lead to the detection of the cells of the CLS. To conclude, the scRNA sequencing studies on human kidneys to date have shown a detailed transcriptomic complexity and future research will likely discover more.
Future clinical relevance and impact on regenerative medicine
ScRNA sequencing studies have already given us unprecedented insights into renal development and disease and the technology holds the promise to answer several longstanding questions in the field of nephrology. The higher resolution of molecular mechanisms could provide more insight into complex renal diseases86. Cell type-specific gene expression related to chronic kidney disease, diabetes and hypertension could for instance help to identify cell-specific and disease-specific targets and guide the development of urgently needed targeted therapeutics103. The diagnosis and treatment of various kidney diseases could be improved based on molecular data of research on pathogenic mechanisms and potential biomarker discovery43. ScRNA sequencing is already getting more patient-oriented by the possibility to sequence a variety of kidney cells from urine116.
Basic research to develop kidney grafts, such as kidney organoids from induced pluripotent stem cells for future transplantation rely on data on molecular signaling pathways of developing tissue. Currently, these models often lack certain cell types and segments such as the collecting duct, MCs and PECs. More information on the molecular pathways of these cell types is needed in order to reproduce these in vitro. ScRNA sequencing research on developing kidney can provide such data. Additionally, the transcriptome of developing kidneys and in vitro models can be compared as previously shown104 to confirm the formation of the appropriate cell types, cellular maturity, and timing. A more comprehensive discussion on this topic can be found in the recent review of Wu and Humphreys117.
Conclusion
In conclusion, we extensively discussed findings of all identified scRNA sequencing publications to date of healthy human developing and adult kidneys and compared their outcome in terms of differences in cellular variety. Together, the publications detected approximately 41 distinct cell populations of renal lineage in the adult human kidney. However, to determine a definite number of cell types, a clear definition of a cell type should be determined and various limitations of the technique need to be resolved. Due to the subtle and gradual changes in the transcriptome in development, we suggest that cells in developing kidneys may best be regarded as a continuum instead of distinct cell types. Detecting master regulators will then help to guide in vitro work along the same differentiation pathways. In the future, advanced technologies such as combined scRNA sequencing and measurement of DNA accessibility, increasing depth, as well as improved analysis methods will likely improve our understanding of the variety of cell types in the adult and developing nephron. Nevertheless, the studies to date provide essential insight into the molecular signature of a large variety of cell populations in adulthood and in development and therefore enabled us to present an updated lineage tree for cell types of renal lineage. On a final note, we trust that these studies will shape the future of regenerative medicine by unraveling lineage trees at an even higher resolution and thus help in organoid and tissue engineering approaches.
Supplementary information
Author contributions
A.S. analyzed and interpreted the data and wrote the manuscript. All authors provided critical feedback and helped shape the paper. All authors gave their final approval for the version to be published.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41536-021-00156-w.
References
- 1.van Gelder MK, et al. From portable dialysis to a bioengineered kidney. Expert Rev. Med Devices. 2018;15:323–336. doi: 10.1080/17434440.2018.1462697. [DOI] [PubMed] [Google Scholar]
- 2.Humes HD, Buffington D, Westover AJ, Roy S, Fissell WH. The bioartificial kidney: current status and future promise. Pediatr. Nephrol. 2014;29:343–351. doi: 10.1007/s00467-013-2467-y. [DOI] [PubMed] [Google Scholar]
- 3.Song JJ, et al. Regeneration and experimental orthotopic transplantation of a bioengineered kidney. Nat. Med. 2013;19:646–651. doi: 10.1038/nm.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bikbov B, et al. Global, regional, and national burden of chronic kidney disease, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2020;395:709–733. doi: 10.1016/S0140-6736(20)30045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liyanage T, et al. Worldwide access to treatment for end-stage kidney disease: a systematic review. Lancet. 2015;385:1975–1982. doi: 10.1016/S0140-6736(14)61601-9. [DOI] [PubMed] [Google Scholar]
- 6.Humes HD, et al. Initial clinical results of the bioartificial kidney containing human cells in ICU patients with acute renal failure. Kidney Int. 2004;66:1578–1588. doi: 10.1111/j.1523-1755.2004.00923.x. [DOI] [PubMed] [Google Scholar]
- 7.Huff C. How artificial kidneys and miniaturized dialysis could save millions of lives. Nature. 2020;579:186–188. doi: 10.1038/d41586-020-00671-8. [DOI] [PubMed] [Google Scholar]
- 8.Morizane R, Bonventre JV. Kidney Organoids: a Translational Journey. Trends Mol. Med. 2017;23:246–263. doi: 10.1016/j.molmed.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clapp, W. L. In Silva’s Diagnostic Renal Pathology (eds Tibor Nadasdy, Vivette D. D’Agati, Xin Jin Zhou & Zoltan G. Laszik) 1−56 (Cambridge University Press, 2017).
- 10.Takasato M, et al. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat. Cell Biol. 2014;16:118–126. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- 11.Lam AQ, et al. Rapid and efficient differentiation of human pluripotent stem cells into intermediate mesoderm that forms tubules expressing kidney proximal tubular markers. J. Am. Soc. Nephrol. 2014;25:1211–1225. doi: 10.1681/ASN.2013080831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taguchi A, et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014;14:53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Nishinakamura R. Human kidney organoids: progress and remaining challenges. Nat. Rev. Nephrol. 2019;15:613–624. doi: 10.1038/s41581-019-0176-x. [DOI] [PubMed] [Google Scholar]
- 14.Takasato M, Wymeersch FJ. Challenges to future regenerative applications using kidney organoids. Curr. Opin. Biomed. Eng. 2020;13:144–151. doi: 10.1016/j.cobme.2020.03.003. [DOI] [Google Scholar]
- 15.van den Berg CW, et al. Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation in vivo. Stem Cell Rep. 2018;10:751–765. doi: 10.1016/j.stemcr.2018.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindstrom NO, et al. Conserved and divergent molecular and anatomic features of human and mouse nephron patterning. J. Am. Soc. Nephrol. 2018;29:825–840. doi: 10.1681/ASN.2017091036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindstrom NO, et al. Conserved and divergent features of human and mouse kidney organogenesis. J. Am. Soc. Nephrol. 2018;29:785–805. doi: 10.1681/ASN.2017080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi A, et al. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costantini F, Kopan R. Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev. Cell. 2010;18:698–712. doi: 10.1016/j.devcel.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeshima A, Sakurai H, Nigam SK. Adult kidney tubular cell population showing phenotypic plasticity, tubulogenic capacity, and integration capability into developing kidney. J. Am. Soc. Nephrol. 2006;17:188–198. doi: 10.1681/ASN.2005040370. [DOI] [PubMed] [Google Scholar]
- 21.Challen GA, Bertoncello I, Deane JA, Ricardo SD, Little MH. Kidney side population reveals multilineage potential and renal functional capacity but also cellular heterogeneity. J. Am. Soc. Nephrol. 2006;17:1896–1912. doi: 10.1681/ASN.2005111228. [DOI] [PubMed] [Google Scholar]
- 22.Sutherland MR, et al. Accelerated maturation and abnormal morphology in the preterm neonatal kidney. J. Am. Soc. Nephrol. 2011;22:1365–1374. doi: 10.1681/ASN.2010121266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan D, et al. Development of the human fetal kidney from mid to late gestation in male and female infants. EBioMedicine. 2018;27:275–283. doi: 10.1016/j.ebiom.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoy WE, et al. A stereological study of glomerular number and volume: preliminary findings in a multiracial study of kidneys at autopsy. Kidney Int. Suppl. 2003;63:S31–S37. doi: 10.1046/j.1523-1755.63.s83.8.x. [DOI] [PubMed] [Google Scholar]
- 25.Hughson M, Farris AB, 3rd, Douglas-Denton R, Hoy WE, Bertram JF. Glomerular number and size in autopsy kidneys: the relationship to birth weight. Kidney Int. 2003;63:2113–2122. doi: 10.1046/j.1523-1755.2003.00018.x. [DOI] [PubMed] [Google Scholar]
- 26.Puelles VG, et al. Podocyte number in children and adults: associations with glomerular size and numbers of other glomerular resident cells. J. Am. Soc. Nephrol. 2015;26:2277–2288. doi: 10.1681/ASN.2014070641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cullen-McEwen, L., Sutherland, M. R. & Black, M. J. In Kidney Development, Disease, Repair and Regeneration (ed. Melissa H. Little) 27−40 (Academic Press, 2016).
- 28.Takasato M, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564–568. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- 29.Little, M., Georgas, K., Pennisi, D. & Wilkinson, L. In Organogenesis in Development 90 Current Topics in Developmental Biology (ed Peter Koopman) 193−229 (Academic Press, 2010). [DOI] [PubMed]
- 30.Kretzler M, Menon R. Single-cell sequencing the glomerulus, unraveling the molecular programs of glomerular filtration, one cell at a time. J. Am. Soc. Nephrol. 2018;29:2036–2038. doi: 10.1681/ASN.2018060626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Wingert RA. Regenerative medicine for the kidney: stem cell prospects & challenges. Clin. Transl. Med. 2013;2:11. doi: 10.1186/2001-1326-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schutgens F, Verhaar MC, Rookmaaker MB. Pluripotent stem cell-derived kidney organoids: an in vivo-like in vitro technology. Eur. J. Pharmacol. 2016;790:12–20. doi: 10.1016/j.ejphar.2016.06.059. [DOI] [PubMed] [Google Scholar]
- 34.Thiagarajan RD, et al. Identification of anchor genes during kidney development defines ontological relationships, molecular subcompartments, and regulatory pathways. PLoS One. 2011;6:e17286. doi: 10.1371/journal.pone.0017286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Awqati Q, Oliver JA. Stem cells in the kidney. Kidney Int. 2002;61:387–395. doi: 10.1046/j.1523-1755.2002.00164.x. [DOI] [PubMed] [Google Scholar]
- 36.McMahon AP. Development of the mammalian kidney. Curr. Top. Dev. Biol. 2016;117:31–64. doi: 10.1016/bs.ctdb.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Humphreys BD. Mapping kidney cellular complexity. Science. 2018;360:709–710. doi: 10.1126/science.aat7271. [DOI] [PubMed] [Google Scholar]
- 38.Assmus AM, Mullins JJ, Brown CM, Mullins LJ. Cellular plasticity: a mechanism for homeostasis in the kidney. Acta Physiol. 2020;229:e13447. doi: 10.1111/apha.13447. [DOI] [PubMed] [Google Scholar]
- 39.Arendt D, et al. The origin and evolution of cell types. Nat. Rev. Genet. 2016;17:744–757. doi: 10.1038/nrg.2016.127. [DOI] [PubMed] [Google Scholar]
- 40.Morris SA. The evolving concept of cell identity in the single cell era. Development. 2019;146:dev169748. doi: 10.1242/dev.169748. [DOI] [PubMed] [Google Scholar]
- 41.Vallejos CA, Risso D, Scialdone A, Dudoit S, Marioni JC. Normalizing single-cell RNA sequencing data: challenges and opportunities. Nat. Methods. 2017;14:565–571. doi: 10.1038/nmeth.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Potter SS. Single-cell RNA sequencing for the study of development, physiology and disease. Nat. Rev. Nephrol. 2018;14:479–492. doi: 10.1038/s41581-018-0021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malone AF, Wu H, Humphreys BD. Bringing renal biopsy interpretation Into the molecular age with single-cell RNA sequencing. Semin Nephrol. 2018;38:31–39. doi: 10.1016/j.semnephrol.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lun AT, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hicks SC, Townes FW, Teng M, Irizarry RA. Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics. 2018;19:562–578. doi: 10.1093/biostatistics/kxx053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia B, Yanai I. A periodic table of cell types. Development. 2019;146:dev169854. doi: 10.1242/dev.169854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sweeney D, Lindstrom N, Davies JA. Developmental plasticity and regenerative capacity in the renal ureteric bud/collecting duct system. Development. 2008;135:2505–2510. doi: 10.1242/dev.022145. [DOI] [PubMed] [Google Scholar]
- 49.Michael L, Davies JA. Pattern and regulation of cell proliferation during murine ureteric bud development. J. Anat. 2004;204:241–255. doi: 10.1111/j.0021-8782.2004.00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dressler GR. Advances in early kidney specification, development, and patterning. Development. 2009;136:3863–3874. doi: 10.1242/dev.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Brien LL, McMahon AP. Induction and patterning of the metanephric nephron. Semin. Cell Dev. Biol. 2014;36:31–38. doi: 10.1016/j.semcdb.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kao RM, Vasilyev A, Miyawaki A, Drummond IA, McMahon AP. Invasion of distal nephron precursors associates with tubular interconnection during nephrogenesis. J. Am. Soc. Nephrol. 2012;23:1682–1690. doi: 10.1681/ASN.2012030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faa G, et al. Morphogenesis and molecular mechanisms involved in human kidney development. J. Cell. Physiol. 2012;227:1257–1268. doi: 10.1002/jcp.22985. [DOI] [PubMed] [Google Scholar]
- 54.Black MJ, et al. When birth comes early: effects on nephrogenesis. Nephrol. 2013;18:180–182. doi: 10.1111/nep.12028. [DOI] [PubMed] [Google Scholar]
- 55.Rosenblum S, Pal A, Reidy K. Renal development in the fetus and premature infant. Semin. Fetal Neonatal. Med. 2017;22:58–66. doi: 10.1016/j.siny.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Das A, et al. Stromal-epithelial crosstalk regulates kidney progenitor cell differentiation. Nat. Cell Biol. 2013;15:1035–1044. doi: 10.1038/ncb2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeisberg M, Kalluri R. Physiology of the renal interstitium. Clin. J. Am. Soc. Nephrol. 2015;10:1831–1840. doi: 10.2215/CJN.00640114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Little MH, McMahon AP. Mammalian kidney development: principles, progress, and projections. Cold Spring Harb. Perspect. Biol. 2012;4:a008300. doi: 10.1101/cshperspect.a008300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hochane M, et al. Single-cell transcriptomics reveals gene expression dynamics of human fetal kidney development. PLoS Biol. 2019;17:e3000152. doi: 10.1371/journal.pbio.3000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dressler GR. The cellular basis of kidney development. Annu. Rev. Cell Dev. Biol. 2006;22:509–529. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- 61.Menon R, et al. Single-cell analysis of progenitor cell dynamics and lineage specification in the human fetal kidney. Development. 2018;145:dev164038. doi: 10.1242/dev.164038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adam M, Potter AS, Potter SS. Psychrophilic proteases dramatically reduce single-cell RNA-seq artifacts: a molecular atlas of kidney. Devlopment. 2017;144:3625–3632. doi: 10.1242/dev.151142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park JS, et al. Six2 and Wnt regulate self-renewal and commitment of nephron progenitors through shared gene regulatory networks. Dev. Cell. 2012;23:637–651. doi: 10.1016/j.devcel.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lindstrom NO, et al. Conserved and divergent features of mesenchymal progenitor cell types within the cortical nephrogenic niche of the human and mouse kidney. J. Am. Soc. Nephrol. 2018;29:806–824. doi: 10.1681/ASN.2017080890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tran T, et al. In vivo developmental trajectories of human podocyte inform in vitro differentiation of pluripotent stem cell-derived podocytes. Dev. Cell. 2019;50:102–116. doi: 10.1016/j.devcel.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lindstrom NO, et al. Progressive recruitment of mesenchymal progenitors reveals a time-dependent process of cell fate acquisition in mouse and human nephrogenesis. Dev. Cell. 2018;45:651–660. doi: 10.1016/j.devcel.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Young MD, et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science. 2018;361:594–599. doi: 10.1126/science.aat1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang P, et al. Dissecting the global dynamic molecular profiles of human fetal kidney development by single-cell RNA sequencing. Cell Rep. 2018;24:3554–3567. doi: 10.1016/j.celrep.2018.08.056. [DOI] [PubMed] [Google Scholar]
- 69.Hendry C, Rumballe B, Moritz K, Little MH. Defining and redefining the nephron progenitor population. Pediatr. Nephrol. 2011;26:1395–1406. doi: 10.1007/s00467-010-1750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boyle SC, Kim M, Valerius MT, McMahon AP, Kopan R. Notch pathway activation can replace the requirement for Wnt4 and Wnt9b in mesenchymal-to-epithelial transition of nephron stem cells. Development. 2011;138:4245–4254. doi: 10.1242/dev.070433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Self M, et al. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 2006;25:5214–5228. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kobayashi A, et al. Identification of a multipotent self-renewing stromal progenitor population during mammalian kidney organogenesis. Stem Cell Rep. 2014;3:650–662. doi: 10.1016/j.stemcr.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fanni D, et al. MUC1 in mesenchymal-to-epithelial transition during human nephrogenesis: changing the fate of renal progenitor/stem cells. J. Matern Fetal Neonatal Med. 2011;24(Suppl. 2):63–66. doi: 10.3109/14767058.2011.613159. [DOI] [PubMed] [Google Scholar]
- 74.Neidhardt LM, Kispert A, Herrmann BG. A mouse gene of the paired-related homeobox class expressed in the caudal somite compartment and in the developing vertebral column, kidney and nervous system. Dev. Genes Evol. 1997;207:330–339. doi: 10.1007/s004270050120. [DOI] [PubMed] [Google Scholar]
- 75.Kispert A. T-Box genes in the kidney and urinary tract. Curr. Top. Dev. Biol. 2017;122:245–278. doi: 10.1016/bs.ctdb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 76.Brunskill EW, et al. Single cell dissection of early kidney development: multilineage priming. Development. 2014;141:3093–3101. doi: 10.1242/dev.110601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mugford JW, Yu J, Kobayashi A, McMahon AP. High-resolution gene expression analysis of the developing mouse kidney defines novel cellular compartments within the nephron progenitor population. Dev. Biol. 2009;333:312–323. doi: 10.1016/j.ydbio.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lindström, N. O. et al. Spatial transcriptional mapping of the human nephrogenic program. Preprint at bioRxiv10.1101/2020.04.27.060749 (2020). [DOI] [PMC free article] [PubMed]
- 79.Georgas K, et al. Analysis of early nephron patterning reveals a role for distal RV proliferation in fusion to the ureteric tip via a cap mesenchyme-derived connecting segment. Dev. Biol. 2009;332:273–286. doi: 10.1016/j.ydbio.2009.05.578. [DOI] [PubMed] [Google Scholar]
- 80.Sequeira-Lopez ML, et al. The earliest metanephric arteriolar progenitors and their role in kidney vascular development. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015;308:R138–R149. doi: 10.1152/ajpregu.00428.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schlondorff D, Banas B. The mesangial cell revisited: no cell is an island. J. Am. Soc. Nephrol. 2009;20:1179–1187. doi: 10.1681/ASN.2008050549. [DOI] [PubMed] [Google Scholar]
- 82.Goligorsky MS, et al. Role of mesangial cells in macula densa to afferent arteriole information transfer. Clin. Exp. Pharm. Physiol. 1997;24:527–531. doi: 10.1111/j.1440-1681.1997.tb01240.x. [DOI] [PubMed] [Google Scholar]
- 83.Daniel C, et al. Transgelin is a marker of repopulating mesangial cells after injury and promotes their proliferation and migration. Lab Invest. 2012;92:812–826. doi: 10.1038/labinvest.2012.63. [DOI] [PubMed] [Google Scholar]
- 84.Craelius W, el-Sherif N, Palant CE. Stretch-activated ion channels in cultured mesangial cells. Biochem. Biophys. Res. Commun. 1989;159:516–521. doi: 10.1016/0006-291X(89)90023-5. [DOI] [PubMed] [Google Scholar]
- 85.England AR, et al. Identification and characterization of cellular heterogeneity within the developing renal interstitium. Development. 2020;147:dev.190108. doi: 10.1242/dev.190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuppe C, et al. Decoding myofibroblast origins in human kidney fibrosis. Nature. 2021;589:281–286. doi: 10.1038/s41586-020-2941-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barwinska D, et al. Molecular characterization of the human kidney interstitium in health and disease. Sci. Adv. 2021;7:eabd3359. doi: 10.1126/sciadv.abd3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stewart BJ, et al. Spatiotemporal immune zonation of the human kidney. Science. 2019;365:1461–1466. doi: 10.1126/science.aat5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Combes AN, Zappia L, Er PX, Oshlack A, Little MH. Single-cell analysis reveals congruence between kidney organoids and human fetal kidney. Genome Med. 2019;11:3. doi: 10.1186/s13073-019-0615-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kriz W. The Inability of Podocytes to Proliferate: Cause, Consequences, and Origin. Anat. Rec. 2020;303:2588–2596. doi: 10.1002/ar.24291. [DOI] [PubMed] [Google Scholar]
- 91.Ronconi E, et al. Regeneration of glomerular podocytes by human renal progenitors. J. Am. Soc. Nephrol. 2009;20:322–332. doi: 10.1681/ASN.2008070709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Benigni A, Morigi M, Remuzzi G. Kidney regeneration. Lancet. 2010;375:1310–1317. doi: 10.1016/S0140-6736(10)60237-1. [DOI] [PubMed] [Google Scholar]
- 93.Romagnani P, Lasagni L, Remuzzi G. Renal progenitors: an evolutionary conserved strategy for kidney regeneration. Nat. Rev. Nephrol. 2013;9:137–146. doi: 10.1038/nrneph.2012.290. [DOI] [PubMed] [Google Scholar]
- 94.Brunskill EW, Georgas K, Rumballe B, Little MH, Potter SS. Defining the molecular character of the developing and adult kidney podocyte. PLoS One. 2011;6:e24640. doi: 10.1371/journal.pone.0024640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ichimura K, et al. Morphological process of podocyte development revealed by block-face scanning electron microscopy. J. Cell Sci. 2017;130:132–142. doi: 10.1242/jcs.187815. [DOI] [PubMed] [Google Scholar]
- 96.Ichimura K, et al. Three-dimensional architecture of podocytes revealed by block-face scanning electron microscopy. Sci. Rep. 2015;5:8993. doi: 10.1038/srep08993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shankland SJ, Smeets B, Pippin JW, Moeller MJ. The emergence of the glomerular parietal epithelial cell. Nat. Rev. Nephrol. 2014;10:158–173. doi: 10.1038/nrneph.2014.1. [DOI] [PubMed] [Google Scholar]
- 98.Kopp JB. Replenishment of the podocyte compartment by parietal epithelial cells. Kidney Int. 2015;88:934–935. doi: 10.1038/ki.2015.256. [DOI] [PubMed] [Google Scholar]
- 99.Romagnani P. Parietal epithelial cells: their role in health and disease. Contributions Nephrol. 2011;169:23–36. doi: 10.1159/000313943. [DOI] [PubMed] [Google Scholar]
- 100.Liao J, et al. Single-cell RNA sequencing of human kidney. Sci. Data. 2020;7:4. doi: 10.1038/s41597-019-0351-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sivakamasundari, V. et al. Comprehensive cell type specific transcriptomics of the human kidney. Preprint at bioRxiv10.1101/238063 (2017).
- 102.Chen L, et al. Renal-tubule epithelial cell nomenclature for single-cell RNA-sequencing studies. J. Am. Soc. Nephrol. 2019;30:1358–1364. doi: 10.1681/ASN.2019040415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lake BB, et al. A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat. Commun. 2019;10:2832. doi: 10.1038/s41467-019-10861-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu H, et al. Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics. Cell Stem Cell. 2018;23:869–881. doi: 10.1016/j.stem.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Madsen KM, Tisher CC. Structural-functional relationships along the distal nephron. Am. J. Physiol. 1986;250:F1–F15. doi: 10.1152/ajpcell.1986.250.1.C1. [DOI] [PubMed] [Google Scholar]
- 106.Roy A, Al-bataineh MM, Pastor-Soler NM. Collecting duct intercalated cell function and regulation. Clin. J. Am. Soc. Nephrol. 2015;10:305–324. doi: 10.2215/CJN.08880914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen L, et al. Transcriptomes of major renal collecting duct cell types in mouse identified by single-cell RNA-seq. Proc. Natl Acad. Sci. USA. 2017;114:E9989–E9998. doi: 10.1073/pnas.1710964114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Park J, et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360:758–763. doi: 10.1126/science.aar2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Si H, et al. Human and murine kidneys show gender- and species-specific gene expression differences in response to injury. PLoS One. 2009;4:e4802. doi: 10.1371/journal.pone.0004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sabolic I, et al. Gender differences in kidney function. Pflug. Arch. 2007;455:397–429. doi: 10.1007/s00424-007-0308-1. [DOI] [PubMed] [Google Scholar]
- 111.Ransick A, et al. Single-cell profiling reveals sex, lineage, and regional diversity in the mouse kidney. Dev. Cell. 2019;51:399–413. doi: 10.1016/j.devcel.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen G, Ning B, Shi T. Single-cell RNA-seq technologies and related computational data analysis. Front Genet. 2019;10:317. doi: 10.3389/fgene.2019.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kiselev VY, Andrews TS, Hemberg M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat. Rev. Genet. 2019;20:273–282. doi: 10.1038/s41576-018-0088-9. [DOI] [PubMed] [Google Scholar]
- 114.Ren X, et al. Reconstruction of cell spatial organization from single-cell RNA sequencing data based on ligand-receptor mediated self-assembly. Cell Res. 2020;30:763–778. doi: 10.1038/s41422-020-0353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Han X, et al. Construction of a human cell landscape at single-cell level. Nature. 2020;581:303–309. doi: 10.1038/s41586-020-2157-4. [DOI] [PubMed] [Google Scholar]
- 116.Abedini A, et al. Urinary single-cell profiling captures the cellular diversity of the kidney. J. Am. Soc. Nephrol. 2021;32:614–627. doi: 10.1681/ASN.2020050757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wu H, Humphreys BD. Single cell sequencing and kidney organoids generated from pluripotent stem cells. Clin. J. Am. Soc. Nephrol. 2020;15:550–556. doi: 10.2215/CJN.07470619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wu H, et al. Single-cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J. Am. Soc. Nephrol. 2018;29:2069–2080. doi: 10.1681/ASN.2018020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.