Abstract
Coronavirus disease 2019 (COVID-19) is a viral pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that led to more than 800,00 deaths and continues to be a major threat worldwide. The scientific community has been studying the risk factors associated with SARS-CoV-2 infection and pathogenesis. Recent studies highlight the possible contribution of atmospheric air pollution, specifically particulate matter (PM) exposure as a co-factor in COVID-19 severity. Hence, meaningful translation of suitable omics datasets of SARS-CoV-2 infection and PM exposure is warranted to understand the possible involvement of airborne exposome on COVID-19 outcome. Publicly available transcriptomic data (microarray and RNA-Seq) related to COVID-19 lung biopsy, SARS-CoV-2 infection in epithelial cells and PM exposure (lung tissue, epithelial and endothelial cells) were obtained in addition with proteome and interactome datasets. System-wide pathway/network analysis was done through appropriate software tools and data resources. The primary findings are; 1. There is no robust difference in the expression of SARS-CoV-2 entry factors upon particulate exposure, 2. The upstream pathways associated with upregulated genes during SARS-CoV-2 infection considerably overlap with that of PM exposure, 3. Similar pathways were differentially expressed during SARS-CoV-2 infection and PM exposure, 4. SARS-CoV-2 interacting host factors were predicted to be associated with the molecular impact of PM exposure and 5. Differentially expressed pathways during PM exposure may increase COVID-19 severity. Based on the observed molecular mechanisms (direct and indirect effects) the current study suggests that airborne PM exposure has to be considered as an additional co-factor in the outcome of COVID-19.
Abbreviations: COVID19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; PM, particulate matter; ACE2, angiotensin-converting enzyme 2; TMPRSS2, transmembrane protease, serine 2; GEO, Gene Expression Omnibus; CTSB, cathepsin B; CTSL, cathepsin L; DEG, differentially expressed genes; PPI, protein-protein interaction; PTM, post-translational modification; X2K, eXpression2Kinases; GSEA, gene set enrichment analysis; TNF, tumor necrosis factor; PPAR, peroxisome proliferator-activated receptors; IL-17, interleukin-17; TLR, Toll-like receptor; VEGF, vascular endothelial growth factor
Keywords: COVID-19, Particulate matter, Omics, Microarray, RNA-seq, Proteome, Pathway analysis
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic caused by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to more than 500,000 deaths worldwide (“WHO COVID-19 Statistics,” n.d.). SARS-CoV-2 infection caused clusters of severe respiratory illness similar to severe acute respiratory syndrome coronavirus (SARS) and was associated with high mortality (Huang et al., 2020; Wang et al., 2020). In the throes of the COVID-19 crisis, Conticini et al. conclude that the high level of atmospheric pollution in northern Italy should be considered an additional co-factor of the high level of lethality recorded in that area (Conticini et al., 2020). In China, based on the daily confirmed cases, air pollution concentration and meteorological variables obtained from 120 cities indicate that there was a significant relationship between air pollution and COVID-19 infection (Zhu et al., 2020). Further, a study that investigated the geographical expansion of the infection and correlated it with the annual indexes of air quality observed from the Sentinel-5 satellite provide initial evidence of higher morbidity and mortality due to SARS-CoV-2 in regions with poor air quality (Pansini and Fornacca, 2020). Moreover, a nationwide cross-sectional study in the USA also found that a small increase in long-term exposure to PM2.5 (diameters of 2.5 μm or less) leads to a large increase in the COVID-19 death rate (X. Wu et al., 2020).
Although COVID-19 is well known to cause substantial respiratory pathology, the study of the association of particulate air pollution in the precise context of infection and pathogenesis is warranted. The mechanisms of SARS-CoV-2 and SARS-CoV infections closely resemble each other, with strong inflammatory responses being implicated resulting in the impairment of the respiratory system (Tay et al., 2020). Also, cumulative observation of several studies has now established that lung inflammation and hyperinflammatory response induced by SARS-CoV-2 are major contributors to disease severity and death in infected patients (Merad and Martin, 2020; C. Wu et al., 2020). SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells along with innate immune genes (Sungnak et al., 2020). Apart from epithelial cells, recent findings have shown the presence of viral elements within endothelial cells and an accumulation of inflammatory cells, with evidence of endothelial and inflammatory cell death (Varga et al., 2020). A recent study shows that SARS-CoV-2 utilizes the SARS-CoV receptor ACE2 for its entry and TMPRSS2, a serine protease for S protein priming (Hoffmann et al., 2020). Long-term exposure to ambient PM 2.5 is often connected to with faster deterioration of lung function along with a higher risk of the occurrence of lung diseases (Guo et al., 2018). Notably, previous in vivo and in vitro studies highlight the up-regulation of ACE2 (SARS-CoV-2 entry factor) expression during airborne particulate matter exposure (Lin et al., 2018; Miyashita et al., 2020).
The possible role of the airborne exposome in promoting resilience or susceptibility to SARS-CoV-2 infection has been recently highlighted (Naughton et al., 2020). Therefore, a systems approach that aims to target a list of disease molecular features, such as gene expression signatures, can be applied to understand the possible involvement of airborne particulate exposure in COVID19 pathogenesis and severity. Hence, the current study attempts an integrative analysis of omics big data (transcriptome, proteome and interactome) available for COVID19 pathogenesis and PM exposure in order to understand the possible molecular links between them and highlight the need to consider the physiological effects of air pollution during COVID19 clinical management.
2. Materials and methods
2.1. Gene expression – microarray datasets
Publicly available microarray datasets of various in vivo (mouse and rat) and human in vitro (epithelial and endothelial cells) studies related to particulate matter exposure were obtained from Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/), a public functional genomics data repository (Clough and Barrett, 2016). Table 1 demonstrates the details of microarray datasets with appropriate experimental comparison used in the current study.
Table 1.
Microarray experimental comparisons and logFC gene expression of SARS-CoV-2 entry factors.
| S. no | GEO ID | Comparison | SARS-CoV-2 viral entry receptors/factors |
|||
|---|---|---|---|---|---|---|
| ACE2 | TMPRSS2 | CTSB | CTSL | |||
| 1 | GSE7010 | Control vs coarse PM | 1.31 | 0.68 | −0.35 | 0.18 |
| Control vs fine PM | 1.03 | −0.43 | −0.68 | 0.55 | ||
| Control vs ultrafine PM | 1.38 | −0.81 | −0.33 | 0.71 | ||
| 2 | GSE17478 | PBS vs PM exposed lung | −1.20 | −0.23 | 1.18 | 0.30 |
| 3 | GSE38172 | Control vs day 1 PM exposure | −0.03 | −0.02 | −0.07 | 0.53 |
| Control vs day 4 PM exposure | −0.24 | 0.04 | 0.07 | 0.81 | ||
| 4 | GSE34607 | Control vs 4 h | −0.06 | 0.09 | −0.2 | −0.09 |
| Control vs 10 h | −0.29 | 0.02 | 0.24 | 0.12 | ||
| Control vs 24 h | −0.23 | 0.3 | −0.05 | 0.02 | ||
| 5 | GSE9465 | Control vs PM | 0.02 | −0.22 | 0.11 | −0.05 |
| Ova vs PM ova | −0.66 | 0.38 | 0.11 | 0.12 | ||
| 6 | GSE6584 | Control vs DEP 5 | 0.09 | −1.55 | −0.02 | −0.3 |
| Control vs DEP 25 | 0.23 | −2.07 | 0.09 | 0.57 | ||
| 7 | GSE4567 | Control vs UFP exposed | −0.75 | 0.22 | −0.3 | 0.03 |
| 8 | GSE93329 | Control vs PM 2.5 exposed | −0.002 | 0.03 | 0.44 | 0.23 |
Microarray experimental comparisons tabulated with corresponding log fold change (logFC) in expression of SARS-CoV-2 entry factors. For genes with multiple probes only probes with lowest FDR values were included. None of the expression values of entry factors passed the differential expression cut-off value (FDR < 0.05 and logFC>1).
2.2. Differential expression of SARS-CoV-2 entry factors
Differential gene expression analysis across experimental conditions was performed using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) by comparing two or more experimental conditions. GEO2R employs the GEOquery and limma R packages from the Bioconductor project (https://www.bioconductor.org/) and the results have been presented as a table of genes ordered by statistical significance. Expression of SARS-CoV-2 entry factors as mentioned in (www.covid19cellatlas.org) such as angiotensin-converting enzyme 2 (ACE2), transmembrane serine protease 2 (TMPRSS2), cathepsin B (CTSB) and cathepsin L (CTSL) were queried against the list of differentially expressed genes (DEGs) of the mouse, rat and human microarray datasets (control vs particulate exposure). The expression is considered as significant if genes met the stringent cut-off (adj. p < 0.05 and logFC>1) criterion.
2.3. SARS-CoV-2 and human PPI
A list of 332 high confidence host proteins that interacted with SARS-CoV-2 proteins was obtained from a previous study by Gordon et al. In this study 26 of 29 SARS-CoV-2 proteins were cloned, tagged and expressed individually in HEK293T cells and the protein–protein interactions were measured by mass spectrometry (Gordon et al., 2020). It is reasonable to consider the effects of PM exposure on PPI network since therapeutic perspectives indicate that altering the expression of human genes encoding prey proteins for SARS-CoV-2 may interfere with the viral replication (Glinsky, 2020). In order to know the interaction among the host proteins, STRING resource (https://string-db.org/) was employed for creating the molecular action network with 0.4 as confidence level (Szklarczyk et al., 2019). The list of host proteins was compared against the list of DEGs (adj. p < 0.1 and logFC >1) associated with PM exposure. In this case, experiments related to PM exposure conducted on cells of human origin such as GSE4567 (endothelial cells) and GSE38172-4thday (pulmonary epithelial cells) were considered. Comparison of gene list used InteractiVenn (http://www.interactivenn.net/), a web-based tool for the analysis of sets through Venn diagrams (Heberle et al., 2015). Overall, the SARS-CoV-2 interacting host factors and possible effects of PM exposure highlighted in two aspects. 1. Predicting the possible post-translational modification (PTM) interactions (signaling events) among the host factors and impact of PM exposure in the pathophysiological context. 2. Highlighting the host factor genes that are differentially regulated during PM exposure obtained from transcriptomic studies.
2.4. SARS-CoV-2 RNA-Seq datasets and DEGs
Transcriptome profiles of lung samples derived from COVID19 deceased patient matched with uninfected lung biopsy (n = 2 for each group) and in vitro SARS-CoV-2 infection experiments conducted on Calu-3 cells with appropriate controls (n = 3 for each group) generated in a previous study (Blanco-Melo et al., 2020) were obtained from GEO database (GSE147507). Reanalysis of the datasets was performed for the exploration of DEGs between appropriate conditions through RNASeq2G (http://174.129.138.34:3838/rnaseq2g/) web server with default methods (Zhang et al., 2017). Differential analysis of count data was performed using DESeq2, which uses shrinkage estimation for dispersions and fold changes to improve stability and interpretability of estimates as described in earlier studies (Love et al., 2014). From this analysis, genes that passed the cut-off false-discovery rate (FDR) <0.05 and logFC >1in expression were considered as differentially expressed.
2.5. Proteomic data analysis and metabolic pathway analysis
Data from a previous proteomic study were obtained for this analysis (Bojkova et al., 2020). The authors used a TMT-labelling quantitative proteomics approach to understand SARS-CoV-2 infected Caco-2 cells. Proteins expressed with p < 0.05 and logFC > 0.5 were considered as significant as previously published (Bock and Ortea, 2020). Further, the pathway enrichment analysis of the proteomic dataset highlighted the elevation of various energy metabolism-related pathways during SARS-CoV-2 infection. Therefore, with the enriched terms, up-regulated proteins involved in carbon metabolism, citric acid (TCA) cycle and respiratory electron transport, citrate cycle, 2-oxocarboxylic acid metabolism, and glucose metabolism were collected. Comparison of differentially expressed proteins and metabolic enzymes against the DEGs (with FDR < 0.1 and logFC > 1) from PM exposed pulmonary epithelial (GSE38172-4thday) and endothelial cells (GSE4567) was done using InteractiVenn web tool.
2.6. Causal pathways prediction
To infer the mechanistic origins of transcriptomic dysregulation, upstream signaling pathway activities were predicted from the transcriptome data of COVID19 lung biopsy, SARS-CoV-2 infected lung-derived Calu-3 cells and PM exposed cells (epithelial-GSE38172-4thday and endothelial cells - GSE4567). In the current analysis, we submitted the significantly up-regulated genes derived from the above datasets to SPEED2 (https://speed2.sys-bio.net/). SPEED2 is a web server that utilizes pathway signatures derived from a large number of manually curated pathway perturbation experiments across many different cell types in human cells (Rydenfelt et al., 2020). It allows inferring signaling pathways that likely caused these genes to be deregulated. In this analysis, the Bates test was selected as test statistics of enrichment.
2.7. Upstream regulatory network – kinase signaling
In order to link the expression signature to upstream cell signaling kinase networks, the current study employed the eXpression2Kinases (https://amp.pharm.mssm.edu/X2K/) web server which computationally predicts involvement of upstream kinase signaling pathways. Current analysis utilized the list of significantly up-regulated genes derived from RNA-Seq datasets of COVID19 lung biopsy and SARS-CoV-2 infected Calu-3 cells. Additionally, significantly up-regulated genes (FDR < 0.1 and logFC > 1) obtained from PM exposed microarray datasets (GS2E4567 and GSE38172-4th day) were also included. This study did not consider the down regulated genes since analysis with less than 20 genes may produce inaccurate results in the X2K analysis. The X2K tool first computes enrichment of transcription factors that are likely to regulate the expression of the differentially expressed genes followed by connecting the transcription factors through known protein-protein interactions (PPIs) to construct a sub-network. Finally, it performs kinase enrichment analysis on the members of the sub-network and the complete upstream pathway is constructed via the eXpression2Kinases (X2K) algorithm (Clarke et al., 2018).
2.8. Pathway enrichment analysis
In this study, among the list of human microarray datasets, two of the real-world particulate matter exposed experimental designs were preferred to represent the pulmonary and vascular system (GSE38172-4th day and GSE4567). The list of DEGs was obtained between appropriate experimental conditions using moderate cut-off values of FDR <0.1 and logFC >0.8 for pathway enrichment analysis. In addition, the list of DEGs derived from RNA–Seq transcriptome profile of SARS-CoV-2 infected Calu-3 cells also utilized for pathway membership analysis. The up and down regulated genes were submitted to EnrichR (http://amp.pharm.mssm.edu/Enrichr), a comprehensive gene set enrichment analysis web server and the analysis was done through Kyoto Encyclopedia of Genes and Genomes (KEGG) human pathways module (Kuleshov et al., 2016). In this case, only the top 10 KEGG pathways based on the score in which terms with adjusted p value of <0.05 were prioritized.
2.9. Gene ontology and clustering
This analysis anticipated to explore the overlapping pathological mechanisms between particulate matter exposed bronchial epithelial cells and endothelial cells and COVID19 affected lungs. The differentially expressed genes (with FDR < 0.05 and logFC > 1) from RNA-Seq (GSE147507-COVID19) and microarray datasets (GSE4567, GSE38172-Day 1 and 4) were submitted to ToppCluster (http://toppcluster.cchmc.org) tool whose functionalities enables the identification of specialized biological functions and regulatory networks and systems biology-based dissection of biological states (Kaimal et al., 2010). Cytoscape (https://cytoscape.org/), a software environment for integrated models of biomolecular interaction networks (Shannon et al., 2003) was utilized for visualizing the network with the enriched terms (FDR < 0.05) between experimental conditions.
2.10. Gene set enrichment analysis
Gene set enrichment analysis (GSEA) designed to analyze ranked lists of all available genes and do not require a threshold and whole-genome ranked list is suitable for input into pathway enrichment analysis (Reimand et al., 2019; Subramanian et al., 2005). In this study, the genes from transcriptome experiments (GSE147507-COVID19 lung biopsy, GSE4567, GSE38172 - Day 1 and 4) were ranked by using sign (fold change) multiplied by (−log10(p-value)). The pre-ranked genes were subjected to GSEA with C2 pathways and C5 gene ontology biological processes from the MSigDb (v6.0) collection. Gene sets with a size between 15 and 300 were included and the network was constructed with gene sets having FDR < 0.05 using EnrichmentMap application (Merico et al., 2010) in Cytoscape.
3. Results
3.1. Expression of SARS-CoV-2 entry factors upon PM exposure
We first checked the expression of SARS-CoV-2 viral entry receptors upon particulate matter exposure. Among the 15 particulate exposure experimental conditions compared, none of the four SARS-CoV-2 viral entry receptors/factors (ACE2, TMPRSS2, CTSB and CTSL) passed the significant expression criteria (Table 1).
3.2. SARS-CoV-2 and PM modulated pathways
In order to obtain insights into the pathways modulated upon SARS-CoV-2 infection and PM exposure, we performed enrichment analysis of the DEG from different datasets. There were 1179 upregulated and 581 down-regulated genes in Calu-3 cells upon COVID-19 exposure. Various KEGG pathways were significantly (FDR <0.05) enriched among the DEGs of SARS-CoV-2 infected Calu-3 cells and PM exposed cells. Table. 2 represents the top 10 pathways deregulated in SARS-CoV-2 infected Calu-3 cells. Among that, notably TNF signaling pathway, IL-17 signaling pathway and NF-kappa B signaling pathways were enriched. Table 3 illustrates the top 10 pathways enriched from differentially expressed endothelial genes upon PM exposure which including TNF signaling pathway, IL-17 signaling pathway and NF-kappa B signaling pathway. Moreover, pathways enriched with DEGs of PM exposed bronchial epithelial cells indicate that PPAR signaling, ferroptosis and ovarian steroidogenesis as significantly enriched (FDR < 0.05) among other pathways as presented in Table 4 .
Table 2.
KEGG pathways enriched for SARS-CoV-2 infected Calu3 cells.
| Index | Name | p-Value | Adjusted p-value |
|---|---|---|---|
| Upregulated pathways | |||
| 1 | TNF signaling pathway | 3.858e−26 | 1.188e−23 |
| 2 | Influenza A | 1.238e−15 | 9.531e−14 |
| 3 | Cytokine-cytokine receptor interaction | 4.351e−18 | 6.700e−16 |
| 4 | C-type lectin receptor signaling pathway | 1.730e−12 | 1.066e−10 |
| 5 | AGE-RAGE signaling pathway in diabetic complications | 2.091e−11 | 7.155e−10 |
| 6 | NF-kappa B signaling pathway | 3.253e−11 | 1.002e−9 |
| 7 | NOD-like receptor signaling pathway | 2.821e−12 | 1.241e−10 |
| 8 | Herpes simplex virus 1 infection | 6.111e−16 | 6.274e−14 |
| 9 | JAK-STAT signaling pathway | 1.108e−11 | 4.264e−10 |
| 10 | IL-17 signaling pathway | 6.385e−10 | 1.639e−8 |
| Downregulated pathways | |||
| 1 | Steroid biosynthesis | 7.363e−11 | 2.268e−8 |
| 2 | DNA replication | 6.333e−10 | 9.752e−8 |
| 3 | Glyoxylate and dicarboxylate metabolism | 2.572e−7 | 0.00002640 |
| 4 | Glycine, serine and threonine metabolism | 0.000003694 | 0.0002275 |
| 5 | Terpenoid backbone biosynthesis | 0.00005004 | 0.002569 |
| 6 | Cell cycle | 4.381e−7 | 0.00003373 |
| 7 | Tyrosine metabolism | 0.0001224 | 0.005387 |
| 8 | Phenylalanine metabolism | 0.001763 | 0.04524 |
| 9 | Base excision repair | 0.0005509 | 0.01885 |
| 10 | Pyrimidine metabolism | 0.0004314 | 0.01661 |
The table demonstrates the top 10 KEGG pathways enriched by up-regulated and down-regulated genes. Pathway terms with adjusted p-value < 0.05 were considered as significant.
Table 3.
KEGG pathways enriched for PM exposed endothelial cells.
| Index | Name | p-Value | Adjusted p-value |
|---|---|---|---|
| Upregulated pathways | |||
| 1 | Malaria | 0.000001499 | 0.0001539 |
| 2 | TNF signaling pathway | 1.350e−8 | 0.000004159 |
| 3 | IL-17 signaling pathway | 0.000001956 | 0.0001506 |
| 4 | NF-kappa B signaling pathway | 0.000002217 | 0.0001365 |
| 5 | Legionellosis | 0.00006931 | 0.002372 |
| 6 | Cytokine-cytokine receptor interaction | 2.448e−8 | 0.000003770 |
| 7 | AGE-RAGE signaling pathway in diabetic complications | 0.00005047 | 0.002221 |
| 8 | HIF-1 signaling pathway | 0.00005047 | 0.001943 |
| 9 | Kaposi sarcoma-associated herpesvirus infection | 0.000009629 | 0.0004943 |
| 10 | Ovarian steroidogenesis | 0.0009945 | 0.01914 |
| Downregulated pathways | |||
| 1 | Hematopoietic cell lineage | 0.008680 | 1.000 |
| 2 | Nicotine addiction | 0.05644 | 1.000 |
| 3 | Sphingolipid metabolism | 0.06600 | 1.000 |
| 4 | Malaria | 0.06871 | 1.000 |
| 5 | B cell receptor signaling pathway | 0.09806 | 1.000 |
| 6 | Synaptic vesicle cycle | 0.1072 | 1.000 |
| 7 | Insulin secretion | 0.1176 | 1.000 |
| 8 | GABAergic synapse | 0.1214 | 1.000 |
| 9 | Morphine addiction | 0.1240 | 1.000 |
| 10 | Aldosterone synthesis and secretion | 0.1329 | 1.000 |
The table demonstrates the top 10 KEGG pathways enriched by up-regulated and down-regulated genes. Pathway terms with adjusted p-value < 0.05 were considered as significant.
Table 4.
KEGG pathways enriched for PM exposed bronchial epithelial cells.
| S. no | Name | p-Value | Adjusted p-value |
|---|---|---|---|
| Upregulated pathways | |||
| 1 | PPAR signaling pathway | 0.000001372 | 0.0004226 |
| 2 | Ferroptosis | 0.0001530 | 0.02356 |
| 3 | Ovarian steroidogenesis | 0.0002807 | 0.02161 |
| 4 | Biosynthesis of unsaturated fatty acids | 0.002232 | 0.1146 |
| 5 | Synthesis and degradation of ketone bodies | 0.02570 | 0.4948 |
| 6 | Ubiquinone and other terpenoid-quinone biosynthesis | 0.02824 | 0.5116 |
| 7 | Toxoplasmosis | 0.0002122 | 0.02179 |
| 8 | Antigen processing and presentation | 0.001059 | 0.06522 |
| 9 | Mineral absorption | 0.007793 | 0.2400 |
| 10 | Nitrogen metabolism | 0.04331 | 0.6063 |
| Downregulated pathways | |||
| 1 | ECM-receptor interaction | 0.005453 | 1.000 |
| 2 | Vascular smooth muscle contraction | 0.01362 | 1.000 |
| 3 | Central carbon metabolism in cancer | 0.08419 | 1.000 |
| 4 | PI3K-Akt signaling pathway | 0.01174 | 1.000 |
| 5 | Acute myeloid leukemia | 0.08543 | 1.000 |
| 6 | Retinol metabolism | 0.08667 | 1.000 |
| 7 | Focal adhesion | 0.02937 | 1.000 |
| 8 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 0.09284 | 1.000 |
| 9 | p53 signaling pathway | 0.09284 | 1.000 |
| 10 | Bacterial invasion of epithelial cells | 0.09530 | 1.000 |
The top 10 KEGG pathways enriched by up-regulated and down-regulated genes. Pathway terms with adjusted p-value < 0.05 were considered as significant.
3.3. Causal pathway analysis
We predicted the possible upstream signaling pathways using SPEED. Upstream causal signaling pathway and kinase network associated with upregulated genes in SARS-CoV-2 infected Calu-3 cells and PM exposed bronchial epithelial cells have been illustrated in Fig. 1, Fig. 2 respectively. In this observation, the top 3 causal signaling pathways in COVID-19 affected lung (JAK-STAT, IL-1 and TLR signaling) and SARS-CoV-2 infected Calu-3 cells (IL-1, TLR and TNF-α signaling) were considered. In the case of PM exposed models as well, top 3 causal pathways in PM exposed endothelial cells (IL-1, VEGF and TNF-α signaling) and bronchial epithelial cells (Insulin, TNF-α and VEGF signaling) were prioritized. Based on the X2K derived kinase network, upstream kinases in COVID-19 lung (MAPK3, MAPK14, JNK1, CDK1, HIPK2 and ERK2) and Calu-3 cells (MAPK1, MAPK3, MAPK14, JNK1, ERK1, CK2ALPHA, CSNK2A1, DNAPK, IKKAPLHA and HIPK2) were prioritized. Additionally, various kinases associated with up-regulated genes in PM exposed bronchial epithelial cells (ERK1, ERK2, MAPK3, MAPK14, CDK1, CDK2, CSNK2A1 and HIPK2) and endothelial cells (CDK1, CDK2, CDK4, MAPK1, MAPK3, MAPK14 and GSK-3β) were also identified. The result also indicates the overlapping of kinases between multiple conditions (ex. MAPK14).
Fig. 1.
Upstream pathways. The graphic shows the top 3 upstream signaling pathways for target genes upregulated during SARS-CoV-2 infection and PM exposure in appropriate models. The pathways ranked by significance and the average score are shown, and the bars are colored according to the significance (FDR adjusted p-value using the Bates distribution).
Fig. 2.
Upstream signaling network. The figure illustrates the complete subnetwork with the kinases at the top, the intermediate proteins in the middle and the transcription factors at the bottom associated with the up-regulated genes in SARS-CoV-2 infected Calu-3 cells and PM exposed bronchial epithelial cells.
3.4. Proteome and metabolic pathways
The list of differentially expressed proteins during SARS-CoV-2 infection and the results of reactome pathway analysis (list of genes increased in protein level corresponding to major metabolic pathways) obtained from a previous study (Bojkova et al., 2020) were adapted for proteomic and metabolic pathway analysis. Among the differentially expressed proteins none of them overlapped with PM induced differentially expressed genes except for F3 (upregulated). Altogether 22 genes namely, ENO1, IDH2, GOT2, CS, HK2, IDH3B, PKM, HKDC1, GAPDH, PDHB, MDH2, IDH3G, NDUFA10, PDK1, SCO1, VDAC1, COX11, NDUFAF2, NDUFB9, NDUFA7, NUP93, POM121C were identified from enriched terminologies such as carbon metabolism, citric acid cycle and respiratory electron transport, citrate cycle (TCA cycle), 2-oxocarboxylic acid metabolism, and glucose metabolism. Among the up-regulated metabolic enzymes none of them were differentially (both up and down) regulated in PM exposed datasets of bronchial epithelial cells and endothelial cells considered.
3.5. SARS-CoV-2-human PPI network and PM exposure genes
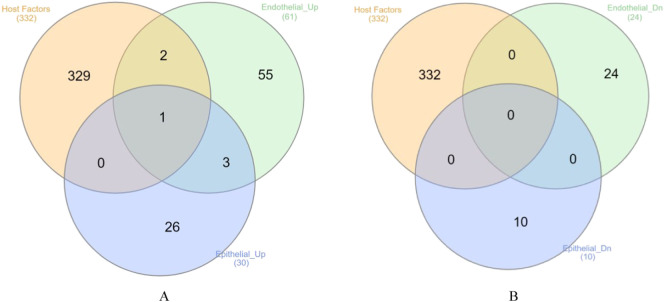
We studied PTM using the protein-protein interaction network. The protein-protein interaction network suggests that the possibility of interconnections among the various host proteins (Fig. 3 ). Among the SARS-CoV-2 hijacked host factors, the current study focuses on proteins involved in PTM events, especially if their possible interaction partner also exists in the predicted network. In the context of molecular action, 7 post-translational modification partners namely MARK3-PKP2, MARK2-RNF41, TBK1-MIB1, PRKACA-GNB1, PRKACA-GNG5, RHOA-PRKACA and PRKACA-ATP1B1 were predicted apart from the general interactions. While comparing the host genes against the PM exposed DEGs, totally 3 genes were found to be up-regulated in both the PM exposed conditions (Fig. 4 ). Bronchial epithelial cells showed upregulation of HMOX1 while in endothelial cells, HMOX1, NPTX1 and RDX were found to be upregulated among the host protein interactors. No overlap between host factors and downregulated genes was observed.
Fig. 3.
Protein-protein interaction (PPI) network. Molecular action network of SARS-CoV-2 interacting host factors. The colored nodes symbolize proteins included in the query. The color of the edges stands for different kinds of known protein-protein associations. Green: activation, red: inhibition, dark blue: binding, light blue: phenotype, dark purple: catalysis, light purple: post-translational modification. The nodes of light purple color interactions were selected and prioritized as possible PTM events.
Fig. 4.
PM induced expression of host factors. The Venn diagram shows the number of genes/proteins overlapping between SARS-CoV-2 interacting host factors (332 high confidence factors) with genes up-regulated (A) and down-regulated (B) upon PM exposure in bronchial epithelial cells and endothelial cells.
3.6. Clustering of COVID-19 and PM associated pathways
We used Toppcluster tool to perform clustering of COVID-19 and PM associated pathways. An overview of the Gene Ontology (GO) functional classification network constructed with the DEGs is illustrated in Fig. 5 . Specifically, functional categories were enriched under biological process (BP), molecular function (MF) and pathways. The network illustrates that various biological processes and molecular pathways enriched in COVID19 infected lungs overlap with the enrichment observed in PM exposed endothelial cells and bronchial epithelial cells. Remarkably, many of them were associated with immune and inflammatory response associated molecular signaling pathways.
Fig. 5.
Functional association. The figure indicates functional association analysis performed by ToppCluster based on pathway networks showing enriched terms from Gene Ontology and pathways. The association between terms enriched by differentially expressed genes in COVID-19 lung tissue, PM exposed bronchial epithelial cells - day 1 and 4 (GSE38172) and endothelial cells (GSE4567) has been illustrated.
3.7. Gene set enrichment analysis
In an attempt to connect the DEGs with a priori defined set of genes through GSEA for generating an added/improved result towards the system level understanding, we performed GSEA. GSEA identified several pathways that were highly enriched between genes of COVID-19 lung and PM exposed bronchial epithelial cells and endothelial cells (Fig. 6 ). Notably, it included innate immune response, cytokine signaling and stress response.
Fig. 6.
GSEA enrichment maps. Enrichment map from GSEA showing the interaction network with nodes of gene sets. The node size corresponds to the number of genes in each GO term and the proportion of clusters in the network is determined by the number of gene sets. Clustered subnetworks of nodes (depicted as circles) reflect generic functional networks. Association of enriched biological processes, pathways and molecular function among expressed genes from COVID-19 lung tissue, PM exposed bronchial epithelial cells - day 1 and 4 (GSE38172) and endothelial cells (GSE4567) has been illustrated.
4. Discussion
Currently, the effect of COVID-19 in people with other comorbidities remains inconclusive which necessitates suitable prospective studies. At the molecular level, genes/pathways closely linked with SARS-CoV-2 infection and/or associated pathological changes in lung and vascular cells need to be investigated. In this study, the plausible impact of particulate air pollution exposure on the progression of COVID-19 and associated pathophysiology of the pulmonary system has been discussed. While focusing on the expression of SARS-CoV-2 entry factors (ACE2, TMPRSS2, CTSB and CTSL), the current study did not observe any changes (especially up-regulation) in the existing microarray datasets related to particulate exposure. Whereas, a recent observation showed that traffic-related particulate matter upregulated the ACE2 expression in A549 cells (Miyashita et al., 2020) and another recent study suggests PM2.5 exposure may up-regulate ACE-2 a protein level (Borro et al., 2020). In large-scale gene expression experiments, such as transcriptomic studies, the false discover rate (FDR) correction and stringent fold change cut-off allow us to select only the genes showing a high confident expression change. Hence, a range of expression studies performed on various models clearly indicates that exposure to airborne PM pollution may not robustly deregulate the SARS-CoV-2 entry factors.
Studying the upstream signaling (infer the mechanistic origin of transcriptomic dysregulation of genes) and downstream (consequent effects) pathways of SARS-CoV-2 infected cells is key to understanding the molecular mechanism of SARS-CoV-2 viral infection. This is in turn essential for correlating the existing knowledge of particulate exposure associated pathogenic signaling with COVID-19. The prediction of upstream pathways in SARS-CoV-2 infected conditions prioritized the association of TLR, TNF-α, IL-1 and JAK-STAT signaling pathways with the up-regulated genes where some of them are shared with PM exposed conditions. While considering the pathway membership of deregulated genes, many of the up-regulated genes fall under pathways associated with viral pathogenesis and inflammatory signaling including TNF, IL-17 and NF-κB signaling pathways where overlapping of pathway terms between SARS-CoV-2 infected and PM exposure conditions are also observed. The above-mentioned pathways are known to be associated with chronic inflammation of the airways (Athari, 2019) and also involved in the activation of proinflammatory responses in cells of the airway mucosa by particulate matter (Øvrevik et al., 2015). Precisely, a previous study suggests that urban PM2.5 may aggravate allergic inflammation in the lungs via TLR2/TLR4/MyD88-signaling pathway in a mouse model (He et al., 2017). Exposure of human bronchial epithelial cells to airborne PM2.5 consistently up-regulated a number of genes including TNF-α implicated in the inflammatory response (Zhou et al., 2015). While focusing on IL-17 signaling, an earlier study indicates that PM2.5 treatment changed the gene expression associated with IL-17 signaling pathway in the lung (Jeong et al., 2019). In addition, cytokine expression profiling in bronchoalveolar lavage fluid (BALF) of animals exposed to traffic related PM10 also indicates increased expression of IL-17A and TNF-α (Huang et al., 2017). IL-17 is a pro-inflammatory cytokine which can induce A549 alveolar epithelial cells to undergo EMT via the TGF-β1 mediated Smad2/3 and ERK1/2 activation which is in turn associated with pulmonary fibrosis (T. Wang et al., 2017), while the role of IL-17 in COVID-19-related pathogenesis has been already highlighted (Pacha et al., 2020).
In a precise perspective, the upstream kinase signaling network plausibly associated with the up-regulated genes during SARS-CoV-2 infection and PM exposure was found to be overlap. A recent differential proteome data obtained from COVID-19 patients reveal that activation of viral infection pathways (MAPK, ERK1/ERK2, JAK-STAT and PI3K) occurs in acute COVID-19 infection (Hou et al., 2020). An earlier study explored that PM exposure enhanced the airway inflammatory response significantly through ROS-mediated activation of MAPK (ERK, JNK and p38 MAPK) and downstream NF-κB signaling pathways (J. Wang et al., 2017). In addition, a study on human bronchial epithelial cells (HBECs) exposed to particulate matter observed an increased phosphorylation of ERK, JNK and PI3K which activating the associated signaling pathways in a time-dependent manner (C. Song et al., 2019). Further, PM exposure increase ICAM-1 expression in pulmonary epithelial cells in vitro and in vivo through the IL-6/AKT/STAT3/NF-κB signaling pathway (Liu et al., 2018). Studies on A549 lung cells showed that NRF2 regulates the steady-state levels of HIPK2 and their crosstalk shapes the cytoprotective responses (Torrente et al., 2017), where the inhaled particulate matter toxicity is associated with redox imbalance and NRF2 activation (Deng et al., 2013; Pardo et al., 2020). Further, PM2.5 rapidly induces severe cell cycle alterations including elevation of cyclin B1 level, which associates with CDK1 and drives the progression of cells through mitosis (Longhin et al., 2013). From the above findings, we suggest that host cell signaling pathways that might be modulated by SARS-CoV-2 to facilitate their own replication cycle and signaling may be synergistically impacted during particulate exposure in the pulmonary system.
The study also attempts to find clues from the microarray experiments to explore the possible effects of particulate pollution in the context of COVID-19 pathogenesis such as pulmonary dysfunction and endotheliitis. Pathway analysis showed that genes involved in PPAR signaling pathway, ferroptosis and steroidogenesis were upregulated where the source study indicates that genes involved in inflammation, cytokine production and cell metabolism were found to be deregulated in addition with the up-regulation of genes related to cholesterol and lipid biosynthesis in cells exposed to PM10 for 4 days (Sun et al., 2012). Apart from the enrichment of phenomena such as immune response and inflammatory signaling observed in various PM exposure models, the upregulation of ferroptosis related genes (HMOX1, SLC7A11 and GCLM) highlights the possible association of iron-dependent cell death (Hirschhorn and Stockwell, 2019). This is experimentally supported in endothelial cells where PM2.5 enhances ferroptosis sensitivity which is dependent on iron overload and oxidative stress (Wang and Tang, 2019). Moreover, a study on cigarette smoke treated lung epithelial cells also highlights the involvement of epithelial cell ferroptosis in chronic obstructive pulmonary disease pathogenesis (Yoshida et al., 2019). In this regard, a recent study also described the potential of iron chelation as an alternative beneficial adjuvant in treating SARS-CoV-2 infection (Liu et al., 2020).
In the setting of COVID-19, pulmonary and peripheral endothelial injury due to direct viral attack might be an important inducer of hypercoagulation (Cao and Li, 2020). Previous histopathological studies from COVID-19 patients have shown direct viral infection of endothelial cells, endotheliitis with diffuse endothelial inflammation and thrombosis in blood vessels (Pons et al., 2020; Varga et al., 2020). In vitro experiments highlights that SARS-CoV-2 has the ability to directly infect human blood vessel organoids (Monteil et al., 2020). An earlier study (source study of microarray data GSE4567) on human pulmonary artery endothelial cells (HPAEC) exposed to ultrafine particles (UFPs) indicates that PM may cause adverse cardiovascular health effects by activating the coagulation-inflammation circuitry (Karoly et al., 2007). In this regard, the current observation also highlights the elevation of TNF-α, IL-17 and NF-κB pathway components during PM exposure in endothelial cells. Notably, the blood coagulation pathway component F3 which is up-regulated in endothelial cells during UFPs exposure was also found to be elevated during SARS-CoV-2 infection in Caco-2 cells as evident from the proteome analysis (Bojkova et al., 2020).
In the host PPI network, few kinases associated with PTM events were prioritized among possibly hijacked signaling pathways associated with SARS-CoV-2 infection. A recent study showed that PRKACA is one among proteins possessing the highest degree centrality in SARS-CoV-2- host PPI network where GNB1 and GNG5 possess high centrality within the PPI network related to the immune system (Andrés et al., 2020). A recent observation identified that ARHGEF2 phosphorylation by MARK3 induced RHOA activation and a regulatory switch controlled by MARK3 couples the microtubule and actin cytoskeletons to establish polarity in epithelial cells (Sandi et al., 2017). Further, RNF41, an E3 ubiquitin ligase has been indicated as a novel MARK2 effector in the cell–ECM pathway (Lewandowski and Piwnica-Worms, 2014). In alveolar epithelial cells, TBK1–AKT–ERK signaling pathway modulates radiation-induced EMT, which suggests that TBK1 is a potential target in the prevention of pulmonary fibrosis (Qu et al., 2019) and increased levels of active TBK1 are implicated in inflammatory diseases (Yu et al., 2012). In the case of PKA signaling, protein kinase A-mediated CREB phosphorylation is a survival pathway in alveolar type II cells (Barlow et al., 2008). In the vasculature, PRKACA was identified to directly regulate 137 neighbouring genes being located at the center of the gene network in angiotensin II-induced arterial injury (Zhang et al., 2018). PM2.5 induced activation of the RhoA/ROCK pathway where ROCK inhibitor attenuated the PM2.5 induced inflammatory response, NF-кB activation and epithelial barrier dysfunction in human lung epithelial BEAS-2B cells (Yan et al., 2017). In cultured lung epithelial cells, chronic oxidative stress damages mitochondrial DNA (mtDNA) which in turn triggering inflammation via the TBK1/interferon regulatory factor 3 signaling pathway (Szczesny et al., 2018) wherein oxidative stress and mitochondrial DNA damage are considered as a significant consequence of PM exposure (Grevendonk et al., 2016; Hou et al., 2010).
Moreover, among the 332 high-confidence SARS-CoV-2 human protein interactors, HMOX1, NPTX1 and RDX were found to be up-regulated during PM exposure. The heme oxygenase 1 (HMOX1) acts as a critical mediator in ferroptosis induction and plays a causative factor in the progression of several diseases (Chiang et al., 2018). It is a major intracellular source of iron and an essential enzyme for iron-dependent lipid peroxidation during ferroptotic cell death (Kwon et al., 2015). Studies demonstrated that NPTX1 inhibited growth and promoted apoptosis in hepatocellular carcinoma (Zhao et al., 2019) and enhanced NPTX1 secretion and reactive oxygen species generation in human endometrial stromal cells impair the survival of endothelial cells resulting in the loss in vascular integrity (Guzeloglu-Kayisli et al., 2014). Phosphorylation of radixin (RDX) likely plays important roles in endothelial cell responses to TNF-α and modulates permeability increases in human pulmonary microvascular endothelial cells (Koss et al., 2006). Thus, the study hints that SARS-CoV-2 hijacked human factors involved in cell death and vascular integrity may modulated by PM exposure.
Evaluating disease similarity merely on the basis of shared genes can be misleading as variable combinations of genes may be associated with similar conditions. This holds true for complex diseases as well, it is ideal to look for common biological processes rather than comparing gene matches (Mathur and Dinakarpandian, 2012). Therefore, grouping of similar pathways and major biological themes (between disease or conditions) is necessary for the understanding of overlapping mechanisms of pathogenesis. A great deal of practical importance exists for computing mechanism based disease-disease similarities based on the shared underlying molecular processes (Hamaneh and Yu, 2016). In this study, a cluster of pathways and biological mechanisms indicate that both COVID-19 and PM exposure conditions share many similar terms including innate immunity associated processes, inflammatory signaling, viral replication and stress associated pathways. Previously, Wu et al. reported the inflammatory health effects of PM exposure and their substantial association with cardiopulmonary disease burden (Wu et al., 2018). In this regard, it was previously highlighted that, controlling the inflammatory response may be as important as targeting the SARS-CoV-2 virus where modulation of deregulated immune responses along with inhibiting viral infection may synergize to prevent COVID-19 pathologies at multiple steps (Tay et al., 2020). In addition, a recent study using RNA virus (influenza virus) indicates that PM exposure promotes viral infection through modulation of innate immune responses causing overall pathogenic burden in the lung cells (Mishra et al., 2020). Further, the interleukin-mediated inflammatory mechanisms stimulated by the inhalation of PM2.5 are sufficient to activate coagulation and inhibit fibrinolysis in the lung and systemically (Budinger et al., 2011). Meanwhile, the progressive increase of inflammation and an unusual trend of hypercoagulation have been noticed in severe and critical patients with COVID-19 (Cao and Li, 2020). In the aspect of stress response, the possible role of endoplasmic reticulum (ER) stress (Banerjee et al., 2020) and oxidative stress (Delgado-Roche and Mesta, 2020) in COVID-19 pathogenesis and the need of its mitigation is previously suggested (Csukasi et al., 2020) while the role of ER stress and oxidative stress in the lung during PM exposure is well demonstrated (Valavanidis et al., 2013; Watterson et al., 2009). Hence, along with the possible direct impact on the viral pathogenesis, the progression of indirect pathophysiological consequences due to PM exposure has to be considered for understanding the etiology of COVID-19.
Finally, the SARS-CoV-2 modulated metabolic pathways as mentioned in the proteomic study have been prioritized with the context of PM exposure. Although the genes of metabolic pathways did not overlap with DEG from bronchial epithelial and endothelial datasets, the possible links were obtained through a literature survey. In human lung epithelial cells (A549), the citrate cycle, amino acid biosynthesis and glutathione metabolism were the 3 major metabolic pathways perturbed by PM2.5 exposure (Huang et al., 2015). Further, seasonal PM2.5 modulated the metabolic rewiring and caused cardiac dysfunction in which a low dose of PM2.5 exposure induced the elevation of tricarboxylic acid cycle (TCA cycle), whereas a high dose of PM2.5 exposure mitigated TCA cycle, while increasing the glycolysis levels (Zhang et al., 2020). Specifically, metabolic remodeling from oxidative phosphorylation to glycolysis was also found in human lung bronchial epithelial (BEAS-2B) cells treated with PM2.5 (Y. Song et al., 2019). On the other hand, Bojkova et al. found that blocking glycolysis with hexokinase inhibitor, 2-deoxy-d-glucose (2-DG), prevented SARS-CoV-2 replication in Caco-2 cells suggesting glycolysis inhibitors as potential therapeutic agents for COVID-19 (Bojkova et al., 2020). Therefore, the modulation of glycolysis during air particulate exposure has to be taken into account due to its supportive role in SARS-CoV-2 replication.
In the exposome context, current perspectives focus on the possible role of exposome (including air pollution) in promoting resilience or susceptibility after SARS-CoV-2 infection (Naughton et al., 2020). Apart from this, variations in pathogenic potential of airborne PM may be originated from the seasonal and compositional variability (ions and organic compounds) and also by means of its potential to cause oxidative stress and inflammation (Al Hanai et al., 2019; Park et al., 2018).
5. Conclusion
The current study spotlights the possible role of PM exposure as an encouraging factor in SARS-CoV-2 pathogenesis through the evidence of predicted overlapping system-wide molecular mechanisms. Moreover, along with the possible direct impact on viral pathogenesis, the progression of indirect pathophysiological pulmonary and vascular consequences of PM exposure which may enhance the susceptibility leading to severe outcomes after SARS-CoV-2 infection need to be emphasized. Overall, the current study suggests that airborne PM exposure in the population need to be taken into account as a co-factor in determining resilience or susceptibility during COVID-19 management.
CRediT authorship contribution statement
Jeganathan Manivannan: Study design, Data analysis, Writing.
Lakshmikirupa Sundaresan: Data analysis, Reviewing and Editing.
Funding
None.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Al Hanai A.H., Antkiewicz D.S., Hemming J.D.C., Shafer M.M., Lai A.M., Arhami M., Hosseini V., Schauer J.J. Seasonal variations in the oxidative stress and inflammatory potential of PM2.5 in Tehran using an alveolar macrophage model; the role of chemical composition and sources. Environ. Int. 2019;123:417–427. doi: 10.1016/j.envint.2018.12.023. [DOI] [PubMed] [Google Scholar]
- Andrés, L.-C., Patricia, G.-R., Nikolaos C, K., Carlos, B.-O., Ángela León, C., Santiago, G., Cristian Robert, M., Eduardo, T., Esteban, O.-P., Doménica, C.-R., Ana María, G.J., Katherine, S.-R., Adriana, G.-M., Gabriela, P.-M., Jennyfer M., G.-C., Ana Karina, Z., Silvana, M., Yunierkis, P.-C., Alejandro, C.-A., Lourdes Puig San, A., Carolina, P.-C., Jhomayra, B., Nelson, V., Luis Abel, Q., Cesar, P.-M., 2020. In silico analyses of immune system protein interactome network, single-cell RNA Sequencing of human tissues, and artificial neural networks reveal potential therapeutic targets for drug repurposing against COVID-19. chemRxiv. doi:10.26434/chemrxiv.12408074.v1.
- Athari S.S. Targeting cell signaling in allergic asthma. Signal Transduct. Target. Ther. 2019;4:45. doi: 10.1038/s41392-019-0079-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Czinn S.J., Reiter R.J., Blanchard T.G. Crosstalk between endoplasmic reticulum stress and anti-viral activities: a novel therapeutic target for COVID-19. Life Sci. 2020;255 doi: 10.1016/j.lfs.2020.117842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow C.A., Kitiphongspattana K., Siddiqui N., Roe M.W., Mossman B.T., Lounsbury K.M. Protein kinase A-mediated CREB phosphorylation is an oxidant-induced survival pathway in alveolar type II cells. Apoptosis. 2008;13:681–692. doi: 10.1007/s10495-008-0203-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020 doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock J.-O., Ortea I. Re-analysis of SARS-CoV-2-infected host cell proteomics time-course data by impact pathway analysis and network analysis: a potential link with inflammatory response. Aging (Albany NY) 2020;12:11277–11286. doi: 10.18632/aging.103524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Münch C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borro M., Di Girolamo P., Gentile G., De Luca O., Preissner R., Marcolongo A., Ferracuti S., Simmaco M. Evidence-based considerations exploring relations between sars-cov-2 pandemic and air pollution: involvement of PM2.5-mediated up-regulation of the viral receptor ace-2. Int. J. Environ. Res. Public Health. 2020 doi: 10.3390/ijerph17155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budinger G.R.S., McKell J.L., Urich D., Foiles N., Weiss I., Chiarella S.E., Gonzalez A., Soberanes S., Ghio A.J., Nigdelioglu R., Mutlu E.A., Radigan K.A., Green D., Kwaan H.C., Mutlu G.M. Particulate matter-induced lung inflammation increases systemic levels of PAI-1 and activates coagulation through distinct mechanisms. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W., Li T. COVID-19: towards understanding of pathogenesis. Cell Res. 2020;30:367–369. doi: 10.1038/s41422-020-0327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang S.-K., Chen S.-E., Chang L.-C. A dual role of heme oxygenase-1 in cancer cells. Int. J. Mol. Sci. 2018;20:39. doi: 10.3390/ijms20010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke D.J.B., Kuleshov M.V., Schilder B.M., Torre D., Duffy M.E., Keenan A.B., Lachmann A., Feldmann A.S., Gundersen G.W., Silverstein M.C., Wang Z., Ma’ayan A. eXpression2Kinases (X2K) Web: linking expression signatures to upstream cell signaling networks. Nucleic Acids Res. 2018;46:W171–W179. doi: 10.1093/nar/gky458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E., Barrett T. Humana Press Inc; 2016. The Gene Expression Omnibus Database, in: Methods in Molecular Biology; pp. 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticini E., Frediani B., Caro D. Can atmospheric pollution be considered a co-factor in extremely high level of SARS-CoV-2 lethality in Northern Italy? Environ. Pollut. 2020 doi: 10.1016/j.envpol.2020.114465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csukasi F., Rico G., Becerra J., Duran I. Should we unstress SARS-CoV-2 infected cells? Cytokine Growth Factor Rev. 2020 doi: 10.1016/j.cytogfr.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Roche L., Mesta F. Oxidative stress as key player in severe acute respiratory syndrome coronavirus (SARS-CoV) infection. Arch. Med. Res. 2020 doi: 10.1016/j.arcmed.2020.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Zhang F., Rui W., Long F., Wang L., Feng Z., Chen D., Ding W. PM2.5-induced oxidative stress triggers autophagy in human lung epithelial A549 cells. Toxicol. Vitr. 2013;27:1762–1770. doi: 10.1016/j.tiv.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Glinsky G.V. Tripartite combination of candidate pandemic mitigation agents: vitamin D, quercetin, and estradiol manifest properties of medicinal agents for targeted mitigation of the COVID-19 pandemic defined by genomics-guided tracing of SARS-CoV-2 targets in human cells. Biomedicines. 2020;8 doi: 10.3390/BIOMEDICINES8050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y.F., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevendonk L., Janssen B.G., Vanpoucke C., Lefebvre W., Hoxha M., Bollati V., Nawrot T.S. Mitochondrial oxidative DNA damage and exposure to particulate air pollution in mother-newborn pairs. Environ. Heal. A Glob. Access Sci. Source. 2016;15 doi: 10.1186/s12940-016-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Zhang Z., Lau A.K.H., Lin C.Q., Chuang Y.C., Chan J., Jiang W.K., Tam T., Yeoh E.K., Chan T.C., Chang L.Y., Lao X.Q. Effect of long-term exposure to fine particulate matter on lung function decline and risk of chronic obstructive pulmonary disease in Taiwan: a longitudinal, cohort study. Lancet Planet. Heal. 2018;2:e114–e125. doi: 10.1016/S2542-5196(18)30028-7. [DOI] [PubMed] [Google Scholar]
- Guzeloglu-Kayisli O., Basar M., Shapiro J.P., Semerci N., Huang J.S., Schatz F., Lockwood C.J., Kayisli U.A. Long-acting progestin-only contraceptives enhance human endometrial stromal cell expressed neuronal pentraxin-1 and reactive oxygen species to promote endothelial cell apoptosis. J. Clin. Endocrinol. Metab. 2014;99:E1957–E1966. doi: 10.1210/jc.2014-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaneh M., Yu Y. Mechanism-based disease similarity. J. Rare Dis. Res. Treat. 2016;1 doi: 10.29245/2572-9411/2016/3.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Ichinose T., Yoshida Y., Arashidani K., Yoshida S., Takano H., Sun G., Shibamoto T. Urban PM2.5 exacerbates allergic inflammation in the murine lung via a TLR2. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-11471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle H., Meirelles V.G., da Silva F.R., Telles G.P., Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16 doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn T., Stockwell B.R. The development of the concept of ferroptosis. Free Radic. Biol. Med. 2019 doi: 10.1016/j.freeradbiomed.2018.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L., Zhu Z.Z., Zhang X., Nordio F., Bonzini M., Schwartz J., Hoxha M., Dioni L., Marinelli B., Pegoraro V., Apostoli P., Bertazzi P.A., Baccarelli A. Airborne particulate matter and mitochondrial damage: a cross-sectional study. Environ. Heal. A Glob. Access Sci. Source. 2010;9 doi: 10.1186/1476-069X-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Nethery R.C., Sabath M.B., Braun D., Dominici F. Air pollution and COVID-19 mortality in the United States: Strengths and limitations of an ecological regression analysis. Sci. Adv. 2020;6(45) doi: 10.1126/sciadv.abd4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Zhang X., Wu X., Lu M., Wang D., Xu M., Wang H., Dai J., Duan H., Xu Y., Yu X., Li Y. Serum protein profiling reveals a landscape of inflammation and immune signaling in early-stage COVID-19 infection. medRxiv. 2020 doi: 10.1101/2020.05.08.20095836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO COVID-19 statistics. https://covid19.who.int/ URL. (WWW Document, n.d.)
- Huang Q., Zhang J., Luo L., Wang Xiaofei, Wang Xiaoxue, Alamdar A., Peng S., Liu L., Tian M., Shen H. Metabolomics reveals disturbed metabolic pathways in human lung epithelial cells exposed to airborne fine particulate matter. Toxicol. Res. (Camb). 2015;4:939–947. doi: 10.1039/c5tx00003c. [DOI] [Google Scholar]
- Huang K.L., Liu S.Y., Chou C.C.K., Lee Y.H., Cheng T.J. The effect of size-segregated ambient particulate matter on Th1/Th2-like immune responses in mice. PLoS One. 2017;12 doi: 10.1371/journal.pone.0173158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S., Park S.A., Park I., Kim P., Cho N.H., Hyun J.W., Hyun Y.-M. PM2.5 exposure in the respiratory system induces distinct inflammatory signaling in the lung and the liver of mice. J. Immunol. Res. 2019;2019:1–11. doi: 10.1155/2019/3486841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaimal V., Bardes E.E., Tabar S.C., Jegga A.G., Aronow B.J. ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res. 2010;38:W96–W102. doi: 10.1093/nar/gkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoly E.D., Li Z., Hyseni X., Huang Y.C.T. Up-regulation of tissue factor in human pulmonary artery endothelial cells after ultrafine particle exposure. Environ. Health Perspect. 2007;115:535–540. doi: 10.1289/ehp.9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koss M., Pfeiffer G.R., Wang Y., Thomas S.T., Yerukhimovich M., Gaarde W.A., Doerschuk C.M., Wang Q. Ezrin/radixin/moesin proteins are phosphorylated by TNF-α and modulate permeability increases in human pulmonary microvascular endothelial cells. J. Immunol. 2006;176:1218–1227. doi: 10.4049/jimmunol.176.2.1218. [DOI] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma’ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M.Y., Park E., Lee S.J., Chung S.W. Heme oxygenase-1 accelerates erastin-induced ferroptotic cell death. Oncotarget. 2015;6:24393–24403. doi: 10.18632/oncotarget.5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski K.T., Piwnica-Worms H. Phosphorylation of the E3 ubiquitin ligase RNF41 by the kinase Par-1b is required for epithelial cell polarity. J. Cell Sci. 2014;127:315–327. doi: 10.1242/jcs.129148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.I., Tsai C.H., Sun Y.L., Hsieh W.Y., Lin Y.C., Chen C.Y., Lin C.S. Instillation of particulate matter 2.5 induced acute lung injury and attenuated the injury recovery in ACE2 knockout mice. Int. J. Biol. Sci. 2018;14:253–265. doi: 10.7150/ijbs.23489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.W., Lee T.L., Chen Y.C., Liang C.J., Wang S.H., Lue J.H., Tsai J.S., Lee S.W., Chen S.H., Yang Y.F., Chuang T.Y., Chen Y.L. PM2.5-induced oxidative stress increases intercellular adhesion molecule-1 expression in lung epithelial cells through the IL-6/AKT/STAT3/NF-ΚB-dependent pathway. Part. Fibre Toxicol. 2018;15 doi: 10.1186/s12989-018-0240-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Zhang S., Nekhai S., Liu S. Depriving iron supply to the virus represents a promising adjuvant therapeutic against viral survival. Curr. Clin. Microbiol. Reports. 2020 doi: 10.1007/s40588-020-00140-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhin E., Holme J.A., Gutzkow K.B., Arlt V.M., Kucab J.E., Camatini M., Gualtieri M. Cell cycle alterations induced by urban PM2.5 in bronchial epithelial cells: characterization of the process and possible mechanisms involved. Part. Fibre Toxicol. 2013;10 doi: 10.1186/1743-8977-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur S., Dinakarpandian D. Finding disease similarity based on implicit semantic similarity. J. Biomed. Inform. 2012;45:363–371. doi: 10.1016/j.jbi.2011.11.017. [DOI] [PubMed] [Google Scholar]
- Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D., Isserlin R., Stueker O., Emili A., Bader G.D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R., Pandikannan K., Gangamma S., Raut A.A., Kumar H. Particulate matter (PM10) enhances RNA virus infection through modulation of innate immune responses. Environ. Pollut. 2020 doi: 10.1016/j.envpol.2020.115148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita L., Foley G., Semple S., Grigg J. Traffic-derived particulate matter and angiotensin-converting enzyme 2 expression in human airway epithelial cells. bioRxiv. 2020 doi: 10.1101/2020.05.15.097501. [DOI] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado del Pozo C., Prosper F., Romero J.P., Wirnsberger G., Zhang H., Slutsky A.S., Conder R., Montserrat N., Mirazimi A., Penninger J.M. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton S.X., Raval U., Harary J.M., Pasinetti G.M. The role of the exposome in promoting resilience or susceptibility after SARS-CoV-2 infection. J. Expo. Sci. Environ. Epidemiol. 2020 doi: 10.1038/s41370-020-0232-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Øvrevik J., Refsnes M., Låg M., Holme J.A., Schwarze P.E. Activation of proinflammatory responses in cells of the airway mucosa by particulate matter: oxidant- and non-oxidant-mediated triggering mechanisms. Biomolecules. 2015 doi: 10.3390/biom5031399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacha O., Sallman M.A., Evans S.E. COVID-19: a case for inhibiting IL-17? Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-020-0328-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansini R., Fornacca D. COVID-19 higher induced mortality in Chinese regions with lower air quality. medRxiv. 2020:1–16. doi: 10.1101/2020.04.04.20053595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo M., Qiu X., Zimmermann R., Rudich Y. Particulate matter toxicity is nrf2 and mitochondria dependent: the roles of metals and polycyclic aromatic hydrocarbons. Chem. Res. Toxicol. 2020 doi: 10.1021/acs.chemrestox.0c00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M., Joo H.S., Lee K., Jang M., Kim S.D., Kim I., Borlaza L.J.S., Lim H., Shin H., Chung K.H., Choi Y.H., Park S.G., Bae M.S., Lee J., Song H., Park K. Differential toxicities of fine particulate matters from various sources. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-35398-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons S., Fodil S., Azoulay E., Zafrani L. The vascular endothelium: the cornerstone of organ dysfunction in severe SARS-CoV-2 infection. Crit. Care. 2020 doi: 10.1186/s13054-020-03062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu H., Liu L., Liu Z., Qin H., Liao Z., Xia P., Yang Y., Li B., Gao F., Cai J. Blocking TBK1 alleviated radiation-induced pulmonary fibrosis and epithelial-mesenchymal transition through Akt-Erk inactivation. Exp. Mol. Med. 2019;51 doi: 10.1038/s12276-019-0240-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J., Isserlin R., Voisin V., Kucera M., Tannus-Lopes C., Rostamianfar A., Wadi L., Meyer M., Wong J., Xu C., Merico D., Bader G.D. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019;14:482–517. doi: 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydenfelt M., Klinger B., Klünemann M., Blüthgen N. SPEED2: inferring upstream pathway activity from differential gene expression. Nucleic Acids Res. 2020;48:W307–W312. doi: 10.1093/nar/gkaa236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandi M.J., Marshall C.B., Balan M., Coyaud E., Zhou M., Monson D.M., Ishiyama N., Chandrakumar A.A., La Rose J., Couzens A.L., Gingras A.C., Raught B., Xu W., Ikura M., Morrison D.K., Rottapel R. MARK3-mediated phosphorylation of ARHGEF2 couples microtubules to the actin cytoskeleton to establish cell polarity. Sci. Signal. 2017;10 doi: 10.1126/scisignal.aan3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Liu L., Chen J., Hu Y., Li J., Wang B., Bellusci S., Chen C., Dong N. Evidence for the critical role of the PI3K signaling pathway in particulate matter-induced dysregulation of the inflammatory mediators COX-2/PGE2 and the associated epithelial barrier protein Filaggrin in the bronchial epithelium. Cell Biol. Toxicol. 2019 doi: 10.1007/s10565-019-09508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Li R., Zhang Y., Wei J., Chen W., Chung C.K.A., Cai Z. Mass spectrometry-based metabolomics reveals the mechanism of ambient fine particulate matter and its components on energy metabolic reprogramming in BEAS-2B cells. Sci. Total Environ. 2019;651:3139–3150. doi: 10.1016/j.scitotenv.2018.10.171. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Shamy M., Kluz T., Muñoz A.B., Zhong M., Laulicht F., Alghamdi M.A., Khoder M.I., Chen L.C., Costa M. Gene expression profiling and pathway analysis of human bronchial epithelial cells exposed to airborne particulate matter collected from Saudi Arabia. Toxicol. Appl. Pharmacol. 2012;265:147–157. doi: 10.1016/j.taap.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny B., Marcatti M., Ahmad A., Montalbano M., Brunyánszki A., Bibli S.I., Papapetropoulos A., Szabo C. Mitochondrial DNA damage and subsequent activation of Z-DNA binding protein 1 links oxidative stress to inflammation in epithelial cells. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-19216-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J., Mering C. von. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020:1–12. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrente L., Sanchez C., Moreno R., Chowdhry S., Cabello P., Isono K., Koseki H., Honda T., Hayes J.D., Dinkova-Kostova A.T., De La Vega L. Crosstalk between NRF2 and HIPK2 shapes cytoprotective responses. Oncogene. 2017;36:6204–6212. doi: 10.1038/onc.2017.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valavanidis A., Vlachogianni T., Fiotakis K., Loridas S. Pulmonary oxidative stress, inflammation and cancer: respirable particulate matter, fibrous dusts and ozone as major causes of lung carcinogenesis through reactive oxygen species mechanisms. Int. J. Environ. Res. Public Health. 2013 doi: 10.3390/ijerph10093886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., Mehra M.R., Schuepbach R.A., Ruschitzka F., Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020 doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang M. PM2.5 induces ferroptosis in human endothelial cells through iron overload and redox imbalance. Environ. Pollut. 2019;254 doi: 10.1016/j.envpol.2019.07.105. [DOI] [PubMed] [Google Scholar]
- Wang J., Huang J., Wang L., Chen C., Yang D., Jin M., Bai C., Song Y. Urban particulate matter triggers lung inflammation via the ROS-MAPK- NF-κB signaling pathway. J. Thorac. Dis. 2017;9:4398–4412. doi: 10.21037/jtd.2017.09.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Liu Y., Zou J.F., Cheng Z.S. Interleukin-17 induces human alveolar epithelial to mesenchymal cell transition via the TGF-β1 mediated Smad2/3 and ERK1/2 activation. PLoS One. 2017;12 doi: 10.1371/journal.pone.0183972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA - J. Am. Med. Assoc. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson T.L., Hamilton B., Martin R., Coulombe R.A. Urban particulate matter causes ER stress and the unfolded protein response in human lung cells. Toxicol. Sci. 2009;112:111–122. doi: 10.1093/toxsci/kfp186. [DOI] [PubMed] [Google Scholar]
- Wu W., Jin Y., Carlsten C. Inflammatory health effects of indoor and outdoor particulate matter. J. Allergy Clin. Immunol. 2018 doi: 10.1016/j.jaci.2017.12.981. [DOI] [PubMed] [Google Scholar]
- Wu C., Chen X., Cai Y., Xia J., Zhou Xing, Xu S., Huang H., Zhang L., Zhou Xia, Du C., Zhang Y., Song J., Wang S., Chao Y., Yang Z., Xu J., Zhou Xin, Chen D., Xiong W., Xu L., Zhou F., Jiang J., Bai C., Zheng J., Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020 doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Lai C.H., Lung S.C.C., Chen C., Wang W.C., Huang P.I., Lin C.H. Industrial PM2.5 cause pulmonary adverse effect through RhoA/ROCK pathway. Sci. Total Environ. 2017;599–600:1658–1666. doi: 10.1016/j.scitotenv.2017.05.107. [DOI] [PubMed] [Google Scholar]
- Yoshida M., Minagawa S., Araya J., Sakamoto T., Hara H., Tsubouchi K., Hosaka Y., Ichikawa A., Saito N., Kadota T., Sato N., Kurita Y., Kobayashi K., Ito S., Utsumi H., Wakui H., Numata T., Kaneko Y., Mori S., Asano H., Yamashita M., Odaka M., Morikawa T., Nakayama K., Iwamoto T., Imai H., Kuwano K. Involvement of cigarette smoke-induced epithelial cell ferroptosis in COPD pathogenesis. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-10991-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T., Yi Y.-S., Yang Y., Oh J., Jeong D., Cho J.Y. The pivotal role of TBK1 in inflammatory responses mediated by macrophages. Mediat. Inflamm. 2012;2012:1–8. doi: 10.1155/2012/979105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang Y., Evans P., Chinwalla A., Taylor D. RNA-Seq 2G: online analysis of differential gene expression with comprehensive options of statistical methods. bioRxiv 122747. 2017 doi: 10.1101/122747. [DOI] [Google Scholar]
- Zhang Y.L., Zhi L.Y., Zou L.X., Chen C., Bai J., Lin Q.Y., Lai S., Wang L., Liu Y., Li H.H. Analysis of genes related to angiotensin II-induced arterial injury using a time series microarray. Cell. Physiol. Biochem. 2018;48:983–992. doi: 10.1159/000491966. [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhou Q., Su R., Sun Z., Zhang W., Jin X., Zheng Y. Cardiac dysfunction and metabolic remodeling due to seasonally ambient fine particles exposure. Sci. Total Environ. 2020;721 doi: 10.1016/j.scitotenv.2020.137792. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Yu Y., Zhao W., You S., Feng M., Xie C., Chi X., Zhang Y., Wang X. As a downstream target of the AKT pathway, NPTX1 inhibits proliferation and promotes apoptosis in hepatocellular carcinoma. Biosci. Rep. 2019;39 doi: 10.1042/BSR20181662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Liu Y., Duan F., Qin M., Wu F., Sheng W., Yang L., Liu J., He K. Transcriptomic analyses of the biological effects of airborne PM2.5 exposure on human bronchial epithelial cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0138267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Xie J., Huang F., Cao L. Association between short-term exposure to air pollution and COVID-19 infection: evidence from China. Sci. Total Environ. 2020;727 doi: 10.1016/j.scitotenv.2020.138704. [DOI] [PMC free article] [PubMed] [Google Scholar]