Abstract
Background
The ongoing search for viable treatment options to curtail Epstein Barr Virus (EBV) pathogenicity has necessitated a paradigmatic shift towards the design of peptide-based vaccines. Potential B-cell and T-cell epitopes were predicted for nine antigenic EBV proteins that mediate epithelial cell-attachment and spread, capsid self-assembly, DNA replication and processivity.
Methods
Predictive algorithms incorporated in the Immune Epitope Database (IEDB) resources were used to determine potential B-cell epitopes based on their physicochemical attributes. These were combined with a string-kernel method and an antigenicity predictive AlgPred tool to enhance accuracy in the end-point selection of highly potential antigenic EBV B-cell epitopes. NetCTL 1.2 algorithms enabled the prediction of probable T-cell epitopes which were structurally modeled and subjected to blind peptide-protein docking with HLA-A*02:01. All-atom molecular dynamics (MD) simulation and Molecular Mechanics Generalized-Born Surface Area methods were used to investigate interaction dynamics and affinities of predicted T-cell peptide-protein complexes.
Results
Computational predictions and sequence overlapping analysis yielded 18 linear (continuous) and discontinuous (conformational) subunit epitopes from the antigenic proteins with characteristic surface accessibility, flexibility and antigenicity, and predictive scores above the threshold value (1) set. A novel site was identified on HLA-A*02:01 with preferential affinity binding for modeled BMRF2, BXLF1 and BGLF4 T-cell epitopes. Interaction dynamics and energies were also computed in addition to crucial residues that mediated complex formation and stability.
Conclusion
This study implemented an integrative meta-analytical approach to model highly probable B-cell and T-cell epitopes as potential peptide-vaccine candidates for the treatment of EBV-related diseases.
Keywords: Immunoinformatics, Epstein-bar virus, Epitopes, Major histocompatibility complex 1, Antigenic proteins, Peptide-based vaccine
At a glance of commentary
Scientific background on the subject
The design of peptide-based vaccines could engender the treatment of Epstein Barr Virus (EBV) infection since no cure exists till date. Hence, predicting highly probable B- and T-cell epitopes from a cohort of antigenic EBV proteins could be vital for the design and development of highly effective anti-EBV vaccines.
What this study adds to the field
Our thorough and combinatorial investigative approaches yielded B-cell and T-cell epitopes with favorable physico-chemical attributes that make them potential anti-EBV vaccine candidates. Interestingly, we were able to define a novel site on HLA-A*02:01 that could be further exploited for high-affinity T-cell epitope binding.
Epstein Barr virus (EBV) is a ubiquitous lymphotropic gamma-1 herpesvirus that belongs to the Herpesviridae family. EBV is also known as human herpesvirus 4 (HHV-4) which is widespread in humans infecting over 90% of the adult population globally. More specifically, in developing countries, EBV mediates an asymptomatic infection that usually occurs at the early infancy stage while there is a delay in acute infection until the early adulthood stage in developed countries with about 25% cases resulting into infectious mononucleosis (IM) with clinical features such as fever, swollen lymph nodes, severe fatigue, sore throat and immunologic dysfunction [1].
EBV is the principal cause of IM while it has also been implicated in several human cancers. About 125,000 new cases of IM are reported annually in the United States while about 200,000 new cases of malignancies associated with EBV occur yearly worldwide [2]. EBV has been associated with distinct forms of cancer such as nasopharyngeal carcinoma, stomach cancer, Burkitt's lymphoma, Hodgkin's lymphoma and some entities of B-/T-cell lymphomas [[3], [4], [5], [6], [7]]. Also, the virus has been linked with autoimmune diseases such as multiple sclerosis [8].
EBV has a life-long persistence in host and it is usually transmitted via the oral route. Upon close contact, viral particles are transferred from the saliva of infected individuals to the new host, entering the oral cavity after which the virus is amplified by a lytic (replicative) infection in the permissive cells of the oropharynx. Subsequently, the virus is shed in vast amounts into the throat followed by a simultaneous infection of the B cells, initiating a latent, persistent infection of the pool of B cells [[9], [10], [11]].
Amplification of the viral load follows the viral transmission to a native host and occurs via replicative (lytic) infection in a permissive cell type. This then has a life-long persistence in the host as an asymptomatic latent infection in a secondary cell type which is occasionally reactivated into the lytic cycle to release infectious virions that can be further transferred into a new host [9]. Regardless, the virus persists whether the infection is accompanied by a disease or not.
EBV is classified based on two types; 1 and 2, based on genetic variations in sequences for the EBV nuclear antigens (EBNA)-2 and EBNA-3. Comparatively, type 1 EBV strains exhibit higher prevalence and cell-immortalizing ability than the type 2 forms [12]. Although both types are detected across the world, type 1 is the most dominant in many populations [13].
The life cycle of EBV constitutes to widely distinct stages which include the latency and lytic stages. In the former, only a subset of genes is expressed which include six nuclear proteins, three latent membrane proteins, two EBV-encoded small non-coding RNAs and the BamHI A rightward transcripts (BARTs). The lytic stage typifies the viral replication stage (lytic reactivation) and involves the production of infectious viral particles. This constitutes about 80 proteins which are categorized into immediate-early, early and late proteins [8,[14], [15], [16]]. Lytic proteins are also classified based on their functionalities; capsid, tegument, envelope (glycoproteins - gp) among many others. These antigenic proteins mediate a wide range of function which includes B-cell transformation, viral attachment, membrane fusion, entry, replication and spread [8,[15], [16], [17], [18], [19]]. EBV antigens such as EBNA-1, -2, -3(A-C), -LP-, LMP1, LMP-2A, -2B and non-coding RNAs (EBERs and BARTs) are crucial for the efficient transformation of infected B-cells and subsequent lymphomagenesis [[20], [21], [22], [23]]. These sums the oncogenic mechanisms (Latency III) of EBV in cancer development. Moreover, B- and T-cell epitopic predictions for most of these antigens have been previously carried out [9,14,24,25].
There have been extensive studies on immune protection against EBV invasion, which involves both protective arms of the immune systems; innate and adaptive. A major component of the early innate immune response is the Natural killer (NK) cells, which produce interferon-gamma (IFN-γ) to delay or prevent the transformation of B-cells by EBV [26,27]. B-cells and T-cells, as components of the adaptive system also mediate strong immune responses since they are activated in the presence of a foreign pathogen. The presence of EBV antigens triggers potent B-cell mediated humoral immune responses. The induction of IgM antibodies against virus capsid antigen (VCA) with a subsequent switch to an IgG isotype characterizes acute primary EBV infection. This results in the formation of non-neutralizing IgG anti-VCA antibodies which exist for a lifetime. The major EBV glycoprotein (gp350) also facilitates the induction of neutralizing IgG antibodies, a process that only occurs after the infection has been resolved [28]. In addition to the aforementioned, other antibodies that target antigens that are non-neutralizing also do not appear until the primary infection is resolved. These non-neutralizing antigens include intracellularly located viral proteins [28,29]. Cytotoxic CD8 T cells also mediate strong adaptive immune responses triggered by EBV antigens. This response is directed towards the elimination of cells infected by the virus, which are recognized due to EBV peptide antigens bound by MHC-1 molecules located at the cellular surface; descriptive of the antigenic presentation pathway [30,31]. Previous studies have reported a dramatic pattern in IM patients whereby EBV CD8 T-cell epitopes derived from immediate-early or early antigens are recognized by about 50% CD8 T-cells [9,24]. Similarly, EBV peptide antigens bound to surface-expressing MHC-II molecules are recognized by CD4 T-cells but in a less-dramatic and focused manner after which the complex is committed into various cytokine-producing T-helper cells (Th) phenotypes that regulate immune responses. In most cases, antiviral responses from most CD4 T-cells that are EBV-specific are characterized by the production of IFN-γ and tumor necrosis factor-alpha (TNF-α). In contrast, only a small proportion of these CD4 T-cells produces IL-2 even though it is the typical and most expected Th1-mediated antiviral response [32].
Currently, no medicinal cure for EBV exists neither are there any prophylactic or therapeutic vaccines available. Numerous vaccine-development efforts to curtail EBV infection have been directed towards these antigenic proteins, particularly gp350 since it is the primary target of naturally occurring neutralizing antibodies and mostly abundant gp present on the virus and in infected cells [2]. Several clinical trials have been carried out for vaccine development, including a phase 2 trial for an EBV vaccine from gp350 protein, which reduced the rate of IM but not viral infection [2,[33], [34], [35]]. Suggestively, this limitation could be due to variations in vaccine attributes such as efficacy and duration of protection towards EBV transmission and disease progression. From previous reports, most of the potential vaccines have a short duration efficiency in which they may delay EBV infection but possibly cannot prevent the increases in IM cases over a long period [36].
Dosing is also an important factor to be considered in EBV vaccination. Taking a cue from a closely-related Varicella-zoster virus (VZV), the efficacy of its approved vaccine is based on dosing as studies have reported that about 85% efficacy is recorded with a single dose while a double dose increased the vaccine efficacy to 94% [[37], [38], [39]]. Relative to EBV infection, a logical explanation could be that current vaccines with shorter duration are efficient until they lapse after which a loss of protection is recorded in congruence with increased susceptibility to EBV-disease such as IM at a later age in patients [33,34,40,41]. Presumably, given the short duration of protection for current EBV (gp350) vaccines, the need for increased single or double doses may be considered in future clinical evaluations [34,42].
The ability of EBV to evade immune response is another factor that may account for the reduced efficacy of potential EBV vaccines. While most vaccines are focused on gp350, certain antigens have been identified for their crucial roles in immune evasion. These include BGLF5, BZLF1, BNLF2a, BILF1, EBNA1, EBNA2 among others [10,43]. Possible inclusion of these antigens to consist of a prophylactic vaccine may further improve efficacy if evaluated in future clinical trials [44].
Vaccination has served as a viable therapeutic option for the prevention and treatment of several microbial infections with a common goal of inducing adaptive immune responses by presenting antigenic fragments to the immune system [[45], [46], [47]].
Over the years, the use of peptide-based vaccines has been promising due to their safety and easy preparation [[48], [49], [50], [51]] and are designed based on different B- and T-cell epitopes. This is possible due to the antigenic nature of proteins secreted by the virus [15,52,53], hence the prediction of antibody epitopes using in silico methods have evolved as a crucial step in vaccine design [[53], [54], [55], [56], [57]].
Epitopic prediction has been made possible via the implementation of immunoinformatics methods which contributes strongly towards the development of fast, reliable and effective peptide-based vaccines. These involve the identification of linear (continuous) or non-linear (non-continuous) epitopes for B-cells and T-cells for in silico vaccine design with a desirable reduction in cost and resources compared to immunological experimentations which are tedious and costly.
These predictions are usually centered on the physicochemical properties of these epitopes and their tendencies to provoke desired immunological responses mediated by B- and T-cells [58]. Therefore, immunoinformatics prediction of potential B and T-cell epitopes for viral antigens remains a prominent way-forward for peptide vaccine development [59,60]. Moreover, tools for predicting T-cell epitopes help identify allele-specific peptides, thereby increasing the possibility of selecting and characterizing peptides that are potential vaccine candidates.
These approaches have been successfully implemented for vaccine prediction against multiple HPV, Ebola, HIV, Nipah, and Zika viruses among others [31,53,56,[61], [62], [63], [64], [65], [66], [67]].
Moreover, immunoinformatics methods have also been employed to predict antibody epitopes for some major latent and lytic antigens that play important roles in EBV virulence [14,53].
Complementarily, in this study, we carried out exhaustive B- and T-cell epitopic predictions of antigenic EBV proteins involved in capsid self-assembly, viral DNA synthesis/replication, and oral epithelial cell attachment/cell-to-cell spread as efficient sub-unit vaccine candidates (Table 1) (Fig. 1). This study is highly essential and could serve as an avenue to achieve the design of highly effective and dynamic prophylactic vaccines having a characteristic mix of multiple EBV antigens with diverse functions over the lytic stage of viral infection [68].
Table 1.
Showing detailed information on selected EBV proteins investigated in this study.
| EBV proteins | Functions | Expression stage | Accession number | Residue count | References |
|---|---|---|---|---|---|
| BMRF2 | Viral entry, attachment to oral epithelial cells, manipulates cellular actin cell-to-cell spread | Lytic/structural | P03192 | 357 | [[132], [133], [134]] |
| BDLF2 | Interacts with BMRF2, rearranges cellular actin to promote viral cell-to-cell spread in epithelial cells | Lytic/structural | P03225 | 420 | [18,135] |
| BORF1 (Triplex Protein 1; TRX1) | Capsid assembly (minor capsid protein-binding protein; mCPBP). Required for efficient nuclear transport of Triplex protein 2 (TRX2) | Early lytic/structural | P03187 | 364 | [18] |
| BcLF1 | EBV capsid self-assembly (Major capsid protein; MCP) | Early lytic/structural | P03226 | 1381 | [18,136] |
| BDLF1 (Triplex Protein 2; TRX2) | minor capsid protein (mCP) required for capsid assembly in the nucleus | Early lytic/structural | P25214 | 301 | [136] |
| BFRF3 | Small capsomere-interacting protein (sCP) involved in capsid assembly | Early lytic/structural | P14348 | 176 | [18] |
| BXLF1 (EBV Thymidine kinase; TK) | Involved in the activation and re-activation of viral DNA replication | Early lytic | Q3KSQ2 | 607 | [123,137,138] |
| BGLF4 | Serine/threonine protein kinase. Mediates the phosphorylation of host nucleoporins. Enhances nuclear export of EBV lytic proteins, facilitates virion production | Early lytic | P13288 | 429 | [[138], [139], [140], [141], [142], [143], [144]] |
| BMRF1 | DNA polymerase processivity factor. Plays essential roles in lytic DNA activation and replication. Major phosphoryl target for BGLF4. Interacts with other EBV DNA-binding proteins | Early lytic |
P03191 PDB ID: 2Z0L |
404 | [83,[145], [146], [147], [148]] |
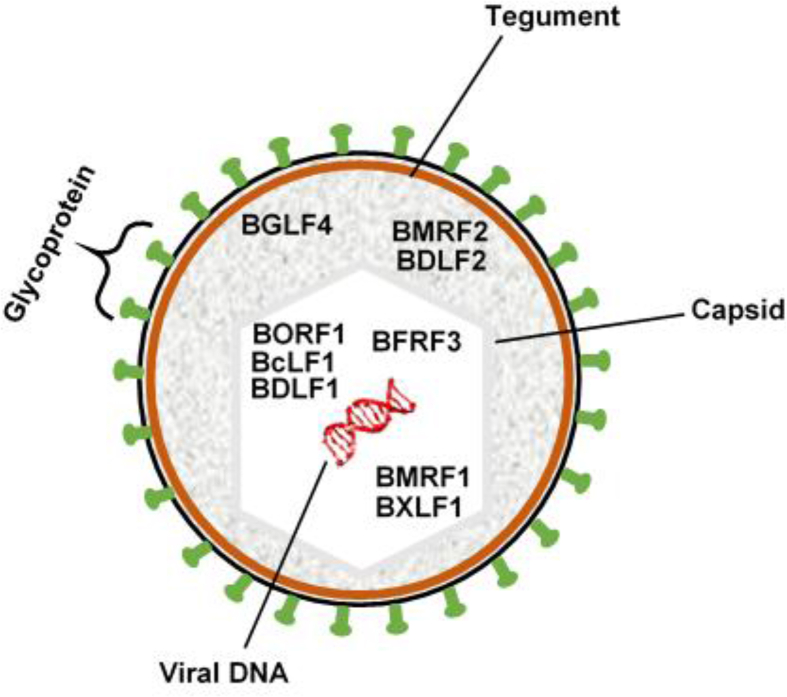
Fig. 1.
Schematic representation of the EBV virion showing structural components and their respective locations. Also, possible locations of antigenic proteins investigated in this study are respectively shown to depict their functionalities.
Interestingly, these antigenic EBV proteins have not been previously examined using immunoinformatics method despite their crucial roles in EBV virulence.
Immunogenicity of the potential T-cell epitopes was also predicted, which entailed their binding strength to human MHC-1 molecule; HLA-A*02:01. This allele (HLA-A*02) is usually reported as the most frequent across various populations and locations, hence our focus on the *02 haplotype in this study. Likewise, MHC-I HLA-B*35, MHC-II DRB1*13, and HLA-DRB1*15 molecules have been pin-pointed for their frequencies in human populations, according to previous reports [[69], [70], [71], [72]].
The response of the immune system to a pathogen in different populations is largely influenced by host genetic polymorphisms. This is because, in the human genome, the HLA genes are the most polymorphic. This has posed a serious challenge in vaccine design and population coverage since each allele interacts with a certain group of peptides [[73], [74], [75]]. Most HLA-I alleles have been grouped based on their ability to bind similar peptides, with this classification covering over 80% of the HLA-A and HLA-B alleles. This classification is referred to as supertypes which consist of HLA molecules that bind similar peptide sets [31].
Both classes I and II MHC molecules present T-cell epitopes that are recognized by the CD8 and CD4 subsets of T-cells respectively, indicating there are CD8 and CD4 T-cell epitopes [30]. Upon recognition of their corresponding epitopes, CD8 T-cells are translated into cytotoxic T lymphocytes (CTL) while primed CD4 T-cells are changed into helper (Th) or regulatory (T-reg) T-cells [76]. MHC-I presented peptides generated from the intracellular region are commonly recognized by the CD8 CTLs while those that originate from the extracellular region and presented by the MHC-II molecules are mostly recognized by the CD4 helper T lymphocytes (HTLs) as earlier stated. According to previous reports, epitopes of CD8 cytotoxic CTLs are highly immunogenic and their presence in vaccine formulation has been linked to the induction of a robust immune response. Epitopes of the CD4 T cell subclass also play important augmentative roles when added relative to the duration and strength of the vaccine [[77], [78], [79]].
Immunogenic prediction is important for surface antigenic presentation relative to T-cell induced cytotoxic cell death [30,80]. Interactions between the peptide-MHC complexes were also modeled and investigated using peptide docking and molecular dynamics simulation algorithms to identify potential epitopic candidates for peptide/multi-peptide vaccine development. Current findings will contribute to the development of possible prophylactic vaccine candidates against EBV infections.
Materials and methods
Retrieval of EBV-target sequences
The respective sequences of the selected proteins were obtained from the Universal Protein Knowledgebase (Uniprot) database [81] using corresponding accession numbers presented in Table 1. The FASTA formats were retrieved and used for antigenic epitopic predictions and structural modeling. Structural modeling was performed using the Local Meta-Threading Protein Fold Recognition Algorithm [82], which applies to non-resolved structures while the structurally resolved protein; BMRF1, was retrieved from the Protein Data Bank (PDB) with entry 2Z0L [83]. Validation of the modeled structures was done using PROCHECK [84] and RAMPAGE [85].
B-cell epitopic prediction and validation
Linear (continuous) B-cell epitopes were predicted from the retrieved antigenic protein sequences while the 3D structures of the respective proteins were used to predict non-linear (discontinuous) epitopes.
Integrative methods were used for linear B-cell epitopic predictions based on the physicochemical attributes of the innate antigenic sequences. These attributes include surface exposure (accessibility), flexibility, antigenicity, and hydrophilicity. These were estimated from the linear antigenic sequence using Emini Surface Accessibility [86], Karplus & Schulz flexibility [87], Kolaskar & Tongaonkar Antigenicity [88] and Parker hydrophilicity [89] predictive tools on Immune Epitope Database (IEDB) analysis resources [90]. High thresholds range of 0.9–1.0 were selected for epitopic prediction.
The B-cell epitope prediction method (BCPRED) was also employed for linear epitope prediction and scoring [91]. The epitope lengths were varied from 12 to 22 to obtain epitopes within the high threshold range of 0.9–1.0.
Using the 3D structures of the respective proteins, linear and discontinuous B-cell epitopes were predicted with ElliPro which functions based on structural protrusion measured by residual protrusion indices (PI), an approximation of protein shape together with the final neighboring residual clustering [92].
Multiple prediction methods were employed for cross-validation and end-point selection of the most probable antigenic EBV B-cell epitopes based on their physicochemical attributes.
Allergenicity predictions and mapping of probable IgE epitopes from the designed peptides were performed using support machine vector, motif-based and BLAST-search algorithms integrated with AlgPred web server, with a reported 85% accuracy [93].
Prediction of potential cytotoxic T-cell epitopes and MHC-1 processing
The NetCTL 1.2 method [94] was used to predict probable CTL epitopes from the selected antigenic EBV proteins. Moreover, this method integrates the prediction of the three main steps of the MHC-1 antigenic presentation pathways which include proteasomal processing, transporter-associated with antigen processing (TAP) and MHC-1 binding. While TAP transport efficiency is predicted using the weight matrix, MHC-peptide binding and proteasomal C-terminal cleavage were predicted using an artificial neural network [95,96]. Default values of 0.15 and 0.05 were used for weights on C terminal cleavage and TAP transport efficiency while the threshold for epitope identification was set at 1.00 to enhance predictive accuracy.
Prediction of MHC-1 immunogenicity
This was essential to further identify epitopic from non-epitopic peptides from the predicted sequences. This explains their ability to be recognized by T cells. According to previous studies, amino acid residues with large aromatic side chains have a higher propensity to induce T-cell immunogenicity. Moreover, highly critical positions in a peptide for provoking immunogenic responses are positions P4-6 [97]. Immunogenic determination of the predicted epitopes was carried out using an integrated IEDB method [97], which determines potential immunogenicity of a peptide-MHC complex (pMHC) based on amino acid properties and location in the queried peptide sequence.
Construction of selected peptide library and blind peptide-protein docking
The PEP-FOLD algorithm was employed for the de-novo structural prediction of selected epitopes. This stochastic procedure entails a Hidden Markov Model-derived Structural Alphabet (SA) [98] which relies on the sOPEP coarse-grained forcefield for 3D sampling and generation, [99] and the Monte-Carlo procedure for final refinement. This is the final step wherein clusters are identified and sorted while resulting conformations are scored [100].
The best-ranked peptide models were then docked to HLA-A*02:01 (A2 supertype) which is a highly frequent human MHC-I allele [31] with a well-resolved X-ray crystal structure; 4UQ3 as retrieved from PDB [101]. Since the predicted epitopes could possess varying binding site affinities for the selected MHC-1 molecule (HLA-A*02:01), a blind peptide-protein docking approach was employed using the pep-ATTRACT algorithm (http://bioserv.rpbs.univ-paris-diderot.fr/services/pepATTRACT/) [102,103]. This incorporates a three-step procedure involving ATTRACT, iATTRACT and AMBER modules. The procedure is initiated by ATTRACT which executes rigid-body docking using the coarse-grained ATTRACT forcefield after which resulting models were ranked by their respective ATTRACT scores [104]. Afterward, the best 1000 models were firstly refined using iATTRACT [103], which is a flexible interface refinement method and then secondly by molecular dynamics simulation using AMBER14 [105]. Then, the final models were clustered and ranked by the average energy of top-ranked four models [106,107].
Taken together, this method performs a rigid-body global search of the surfaces of receptor proteins provided as inputs and identifies the most appropriate sites for peptide binding.
Moreover, the docked complexes with the highest global energy scores were analyzed and further simulated in explicit solvent using Molecular Dynamics (MD) simulation methods according to protocols employed in our previous studies [108,109].
MD simulation of peptide-protein complexes and interaction analyses
To further explore the interaction dynamics between the modeled complexes, we employed the GPU-PMEMD engine of AMBER18 MD package together with its interaction modules [110]. The complexes were parameterized using the FF14SB forcefield while the tleap module was used to neutralize the systems by the addition of Na+/Cl− counter ions. More so, the systems were explicitly solvated in a TIP3P water box of sizes 10 Å. Resulting complexes were subjected to an initial 2500 energy minimization steps with a restraint potential of 500 kcal/mol. This was followed by a second full minimization of 5000 steps without restraints. The systems were then heated gradually from 0 → 310K for 50ps to maintain a fixed volume and atomic number. The solutes were restrained with a potential harmonic restraint of 1ps and maintained at a collision frequency of 1ps. Long-range interactions were included using the SHAKE algorithm and Particle-Mesh Ewald (PME) method while a nonbonded interaction cut-off radius of 12 Å was employed. The systems were then equilibrated for 500ps time at constant temperature (310K) and pressure with fixed atoms, simulating the isobaric-isothermal NPT ensemble. The Berendsen barostat was used to maintain the pressure at 1 bar [111]. MD production run of 100ns was then initiated for the respective epitopic-HLA complexes while resulting trajectories were saved at every 2ps. The integrated CPPTRAJ and PTRAJ modules were used to analyze these trajectories at the end of the simulation.
Post–MD analytical methods employed in this study include root mean square deviation (RMSD) and root mean square fluctuation (RMSF). Cα RMSD measures time-based deviations and motions that occur across the protein backbone atoms relative to the starting structures. This provides a clue into the degree of stability exhibited by the complex from the resulting frames, particularly when bound by the respective T-cell epitopes. The mathematical expression for RMSD is given below:
From the equation, N depicts the total number of atoms in the system while Ri represents the vector position of the Ca atom of particle i. Alignment of the initial (o) and final conformations with the least square fitting was used to predict Ri.
Moreover, the equation for calculating RMSF is shown below; and accordingly, to obtain the standardized RMSF (sRMSF), the average RMSF is deducted from RMSFi (RMSF of the ith residue) and further divided by its standard deviation. The RMSF parameter is used to monitor the motion or fluctuations of individual residues that constitute the protein molecule, giving overall insights into the characteristic flexibility or rigidity of such molecule [112,113].
3D visualization was done on the Graphical User Interfaces (GUIs) of UCSF Chimera, (BIOVIA) Discovery studio and Schrodinger Maestro while resulting data-plots were done using the Origin analytical tool [[114], [115], [116], [117]].
MM/GBSA peptide-protein interaction analysis
The interaction-free energies between the predicted T-cell epitopes and HLA-A*02:01 were estimated using the Molecular Mechanics Generalized-Born Surface Area (MM/GBSA) method which also enabled the estimation of other energy components such as van der Waals, electrostatic, polar and non-polar solvation energies [108,[118], [119], [120]]. The equations below mathematically describe total binding energy (ΔGbind) for complex formation:
| ΔGbind = Gcomplex − (Greceptor + Gpeptide) | (1) |
| ΔGbind = Egas + Gsol − TΔS | (2) |
| Egas = Eint + Evdw + Eele | (3) |
| Gsol = Gpol + GSA | (4) |
| GSA = γSASA | (5) |
From the equation, Gpeptide, Greceptor, and Gcomplex represent the absolute free energies of the peptides, unbound protein and peptide-protein complex respectively. Decomposition of the total energy (ΔGbind) as shown in equations (2), (3), (4), (5) results in the solvation energy term (Gsol) which sums up the polar (Gpol) and non-polar (GSA) solvation terms; the gas-phase (Egas) energy contribution which also sums up the non-bonded [van der Waals (Evdw) and electrostatics (Eele)] in addition to the bonded [internal energy (Eint)] energy terms, and the entropy term (TΔS), which was estimated using the quasi approximation method. The non-polar solvation term was determined by estimating the surface accessible surface area (SASA) using a water probe of 1.4 Å. Total binding energy (ΔGbind) for the peptide-protein complexes were computed in addition to other energy components. More so, to eliminate entropical effects, we selected frames at which the systems were structurally stable for MM/GBSA energy computations. 3D structures averaged over the 100ns simulation run were also used to investigate binding patterns and dynamics relative to complex formation.
Results
Sequence retrieval and 3D structural modeling
Accession numbers (AN) for the retrieved sequences are shown in Table 1, which also indicated the number of residues in the constituent proteins.
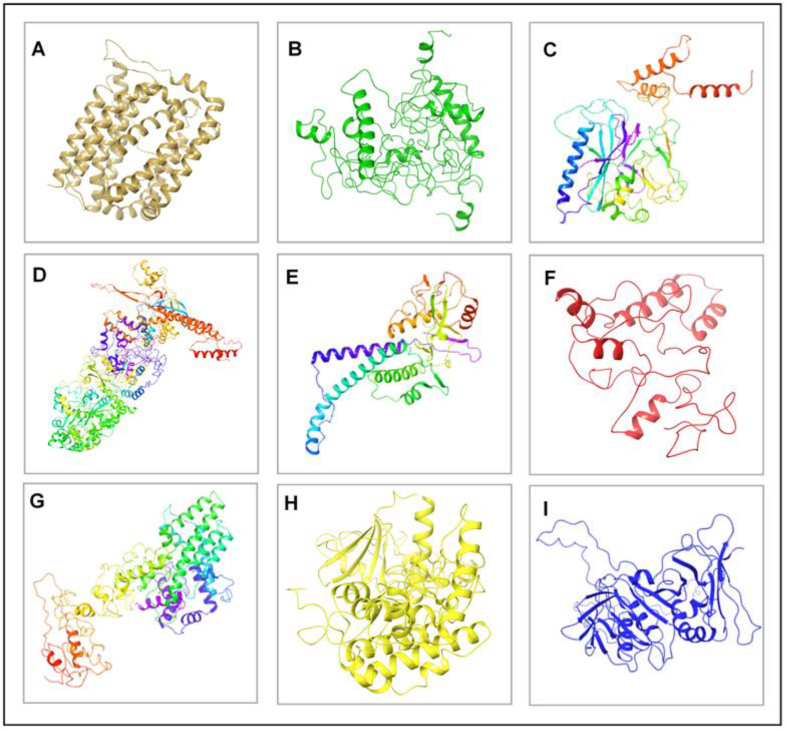
Also, the corresponding secondary structures of the proteins as modeled on LOMET webserver are shown in Fig. 2 including the crystal structure of BMRF1 obtained from PDB. Structural validation was carried out to verify the quality of the protein models used in this study. Ramachandran plots that estimate the percentage of residues in favored, allowed, expected and disallowed/outlier regions are shown in Supplementary Fig. S1.
Fig. 2.
3D structures of EBV antigenic proteins. [A] BMRF2 [B] BDLF2 [C] BORF1 [D] BcLF1 [E] BDLF1 [F] BFRF3 [G] BXLF1 [H] BGLF4 [I] BMRF1.
Physicochemical characterization of linear B-cell epitopes
The selected epitopes were further analyzed for antigenic sequence characteristics using amino acid residue scale and Hidden Markov Models (HMMs) which incorporated integrative methods as earlier mentioned. The hydrophilicity of the predicted epitopes was accessed using the Parker hydrophilicity prediction method while graphical illustrations of corresponding residual hydrophilicity are presented in Supplementary Fig. S2. Regions with the highest score could correlate with high hydrophilicity. Threshold values of 1 were set to determine hydrophilic peptide sequences while those with the maximum and minimum scores were identified. Among the predicted epitopes, amino acid positions GEEDDDG54-60 for BXLF1 had the highest score while the minimum calculated score was −7.414 at positions FFIFLAL165-171 for BMRF2. Maximum and minimum hydrophilicity scores for other EBV antigenic proteins are presented in Table 2.
Table 2.
Characterization of predicted EBV B-cell epitopes based on their innate physicochemical properties.
| Physicochemical characterization | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Hydrophilicity |
Surface flexibility |
Surface accessibility |
Antigenicity |
Allergenicity |
|||||
| EBV proteins | Epitope | Score (Max/Min) | Epitope | Score (Max/Min) | Epitope | Score (Max/Min) | Epitope | Score (Max/Min) | AlgPred |
| BMRF2 | CARGDHS197-203 | 4.571 | LGSGSLA171-177 | 1.071 | RYKLKK320-325 | 10.315 | TVLVVCV225-231 | 1.3 | Non-allergenic |
| FFIFLAL165-171 | −7.414 | VVYMCCF336-342 | 0.876 | VLVVCV226-231 | 0.068 | AGARRNQ177-183 | 0.934 | ||
| BDLF2 | QDQNQNQ95-101 | 6.857 | DQNQNQT96-102 | 1.133 | QDQNQN95-100 | 4.805 | VFIYYCC190-196 | 1.253 | Non-allergenic |
| LAFLALL198-204 | −5.971 | RMAAMAP146-152 | 0.899 | CCYLAF195-200 | 0.070 | NRNRTFN298-304 | 0.868 | ||
| BORF1 | GGSGGGS73-79 | 5.929 | GGGSGGG72-78 | 1.142 | RRRLQR9-14 | 6.535 | VVHGPVV206-212 | 1.225 | Non-allergenic |
| LSLFILM129-135 | −6.071 | HCQIMNI286-292 | 0.892 | GGVCFV245-250 | 0.078 | GGGSGGG72-78 | 0.894 | ||
| BcLF1 | NGDHDHD785-791 | 6.700 | QKSNSPR1197-1203 | 1.115 | PRDRRE480-485 | 8.22 | VPLVSLC637-643 | 1.251 | Non-allergenic |
| IFYYVFL811-817 | −6.157 | IFYYVFL811-817 | 0.882 | ACVVSC1207-1212 | 0.052 | GNMDEMA767-773 | 0.869 | ||
| BDLF1 | STHGTQN33-39 | 5.386 | KLESNDV124-130 | 1.080 | KNKYQP94-99 | 8.702 | LKVVVSL3-9 | 1.227 | Non-allergenic |
| IVFPMVL132-138 | −5.129 | YFCIMVT208-214 | 0.883 | CIMVTL210-215 | 0.106 | RMRVMMT263-269 | 0.931 | ||
| BFRF3 | DTGSGGG155-161 | 6.357 | GSGGGGQ157-163 | 1.147 | DKRQRA75-80 | 4.673 | SYLVFLT46-52 | 1.151 | Non-allergenic |
| RSYLVFL45-51 | −3.214 | ASAAAAV148-154 | 0.912 | VAGAGA82-87 | 0.156 | DTGSGGG155-161 | 0.898 | ||
| BXLF1 | GEEDDDG54-60 | 8.143 | QKNGGKG204-210 | 1.126 | KKRGRK447-452 | 6.122 | VLLEVCL538-544 | 1.254 | Non-allergenic |
| VFPLMLL401-407 | −6.086 | LDCWILH385-391 | 0.880 | VLLEVC538-543 | 0.070 | GEEDDDG54-60 | 0.864 | ||
| BGLF4 | AEDGQDK127-133 | 6.757 | TNSSSSG12-18 | 1.155 | KKRFKE389-394 | 6.022 | VVLLEVL344-350 | 1.25 | Non-allergenic |
| VVLLEVL344-350 | −4.414 | YGIMHLA307-313 | 0.884 | AEVLAC349-354 | 0.117 | DTMWGMG207-213 | 0.867 | ||
| BMRF1 | SEPEDKS308-314 | 6.629 | RQQGSGG180-186 | 1.141 | RQKQKH387-392 | 6.086 | AHVVCSV237-243 | 1.249 | Non-allergenic |
| LSVILFN290-296 | −3.686 | CALMPYM143-149 | 0.899 | GIIAVV271-276 | 0.052 | TMEYDDK129-135 | 0.910 | ||
Surface flexibility prediction of the respective epitopes was predicted using the Karplus and Schulz flexibility method, which estimates atomic motions based on temperature or B-factor. While a high B-factor depicts a distorted structure, a low B-factor indicates a highly ordered or stable protein secondary structure [87]. Using threshold values set at 1, maximum predicted flexibility scores were 1.071 (LGSGSLA171-177), 1.133 (DQNQNQT96-102), 1.142 (GGGSGGG72-78), 1.115 (QKSNSPR1197-1203), 1.08 (KLESNDV124-130), 1.147 (GSGGGGQ157-163), 1.126 (QKNGGKG204-210), 1.155 (TNSSSSG12-18) and 1.141 (RQQGSGG180-186) respectively for BMRF2, BDLF2, BORF1, BcLF1, BDLF1, BFRF3, BGLF4 and BMRF1. Graphical illustrations coupled with the predicted minimum scores are shown in Supplementary Fig. S3. The Emini Surface Accessibility prediction method was used to determine the probable surface availability or exposure of the predicted epitopes on protein surfaces [86]. The threshold value was set at 1 which implies that epitopes with higher probability scores (>1) tend to localize on the surface of the respective antigenic proteins.
Among all predicted proteins, epitope RYKLKK320-325 from BMRF2 had the highest maximum score of 10.315. Moreover, predicted maximum scores for BDLF2, BORF1, BcLF1, BDLF1, BFRF3, BXLF1, BGLF4 and BMRF1 were 4.805 (QDQNQN95-100), 6.535 (RRRLQR9-14, RRLQRR10-15), 8.22 (PRDRRE480-485), 8.702 (KNKYQP94-99), 4.673 (DKRQRA75-80), 6.122 (KKRGRK447-452), 6.022 (KKRFKE389-394) and 6.086 (RQKQKH387-392) respectively. Predicted minimum scores are 0.068 (VLVVCV226-231) – BMRF2; 0.070 (CCYLAF195-200) – BDLF2; 0.078 (GGVCFV245-250) – BORF1; 0.052 (ACVVSC1207-1212) – BcLF1; 0.106 (CIMVTL210-215) – BDLF1; 0.156 (VAGAGA82-87) – BFRF3; 0.070 (VLLEVC538-543) – BXLF1; 0.117 (AEVLAC349-354) - BGLF4; and 0.052 (GIIAVV271-276) - BMRF1. Graphical illustrations showing the predicted surface accessibility of EBV are shown in Supplementary Fig. S4.
The ability of the epitopes to induce immune B-cell responses were also predicted using the Kolaskar & Tongaonkar antigenicity method. The epitopes with the maximum score were identified as those with greater probability to be immunogenic using a threshold of 1. As predicted, BMRF2 had the most antigenic epitope located at TVLVVCV225-231 with a score of 1.3 > 1 threshold set for predictions.
The allergenicity of the respective B-cell epitopes as predicted by AlgPred showed that all the proteins were non-allergenic as they do not match with any similar IgE epitopes. This is indicated in Table 2. Also, most antigenic epitopes for the other EBV proteins are presented in Table 2 together with their maximum and minimum scores. Graphical illustrations showing the degree of antigenicity per-residue are presented in Supplementary Fig. S5.
Prediction of linear B-cell epitopes
The most potentially antigenic B-cell epitopes selected were those with predictive values of 1 and are presented in Table 3. Using the BCPred server, a total of 16 epitopes were predicted for the nine antigenic EBV proteins, among which 14 had predictive scores of 1 and 13 were 20mer peptides. Two, with starting residue positions (62, 39) were predicted for BDLF2, three for BORF1 (positions 239, 71, 8); two for BcLF1 (positions 262, 475); two for BFRF3 (positions 157, 92); three for BXLF1 (positions 112, 2, 89); one for BGLF4 (position 10) and two for BMRF1 (positions 327, 349).
Table 3.
Predicted linear B-cell epitopes of antigenic EBV proteins.
| EBV proteins | Starting Position | Sequences | Epitope length | BCPred Score |
|---|---|---|---|---|
| BMRF2 | 90 | SLRVYSTSTWVSAPCL | 16 | 0.963 |
| 195 | IFCARGDHSVASLKET | 16 | 0.939 | |
| BDLF2 | 62 | PAAAAVPGPEPEPEPHPPMP | 20 | 1 |
| 39 | VFYKAPAPRPREGRASTFHD | 20 | 1 | |
| BORF1 | 239 | GGAGGGGGVCFVGGLGLQPC | 20 | 1 |
| 71 | AGGGSGGGSFSWSGWRPPVF | 20 | 1 | |
| 8 | DRRRLQRRIAGLLPPPARRL | 20 | 1 | |
| BcLF1 | 262 | KGVSTYTTAKGGEPVGGVFI | 20 | 1 |
| 475 | NAAPAPRDRRETYSLQHRRP | 20 | 0.998 | |
| BDLF1 | 189 | ALPEVPGPLGLA | 12 | 1 |
| BFRF3 | 157 | GSGGGGQPHDTAPRGARKKQ | 20 | 1 |
| 92 | GGSSATPVQQAQAAASAGTG | 20 | 1 | |
| BXLF1 | 112 | GAQPPAPAHQKPTAPTPKPR | 20 | 1 |
| 2 | AGFPGKEAGPPGGWRKCQED | 20 | 1 | |
| 89 | AVTSNTGNSPGSRHTSCPFT | 20 | 1 | |
| BGLF4 | 10 | SPTNSSSSGELSVSPEPPRE | 20 | 1 |
| BMRF1 | 327 | RPRHTVSPSPSPPPPPRTPT | 20 | 1 |
| 349 | SPARPETPSPAIPSHSSNTA | 20 | 1 |
BcLF1 KGVSTYTTAKGGEPVGGVFI475-494 was selected for its characteristic surface accessibility with a BCPred predictive score of 0.998. B-cell epitopes with predictive scores of 0.96 and 0.94 were selected for BMRF2 being the highest scores obtained for the 16mer peptides at positions (90,195). Moreover, a 12mer peptide was predicted for BDLF1 with a score of 1 at position (189). 75% specificity was used for scoring indicative of its degree of accuracy [91]. These were cross-referenced with inherent or overlapping residues that constitute linear B-cell epitopic peptides predicted by the Emini Surface Accessibility and Kolaskar & Tongaonkar Antigenicity methods. Relatively, we checked for surface accessibility and antigenic properties possessed by the 12-, 16- and 20-mer B-cell epitopes predicted by BCPred for the respective EBV proteins. Our results are presented in Fig. 3.
Fig. 3.
Sequential overlapping and analyses of predicted linear B-cell epitopes to identify surface accessible and antigenic attributes.
Prediction of B-cell conformational or discontinuous epitopes
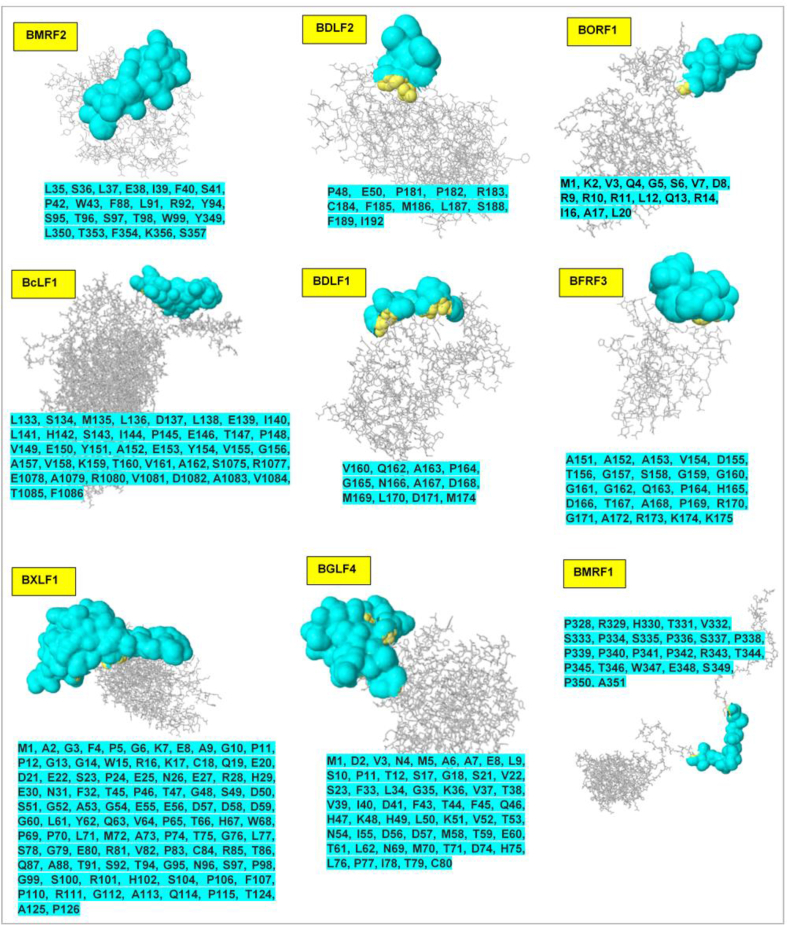
Discontinuous (non-linear) B-cell epitopes were predicted from the 3D structure of the respective antigenic proteins using the Ellipro method which functions based on structural protrusion measured by the protrusion index (PI). Minimum scores were varied from 0.5 to 1.0 to succinctly explore potential epitopes on the protein conformation. Predicted non-linear epitopes and 3D positional representation are shown in Fig. 4. Predicted discontinuous epitopes correlate with some of the linear epitopes earlier determined (surface accessibility and antigenicity) as presented earlier in Table 2.
Fig. 4.
Structural analysis of conformational or discontinuous B-cell epitopes (cyan). The locations of the respective epitopes (surface representation) are shown on the 3D structures of the selected EBV antigenic proteins. Corresponding amino acid sequences, as predicted, are also shown (cyan highlights).
Identification of potential T-cell epitopes
As earlier mentioned, the NetCTL method was used to predict epitopes for the selected antigenic proteins, a platform that incorporates the SNN, weight matrix and artificial neural network methods. Scoring of MHC-1 binding affinity was done against HLAs of the A2 supertype among which HLA-A*02:01 is reportedly the most frequent haplotype across various populations [[69], [70], [71], [72]].
Related antigenic processing stages were predicted using the proteasomal C-terminal cleavage and TAP scores, which altogether presents the possibility of identifying the most promising T-cell epitopes for the antigenic proteins. Peptides with nine residues (9mers) were selected since mostly presented peptides are of that length [121]. Possible MHC-1 ligands were identified (<- E) as indicated in Table 4 while the two highly ranked epitopes were selected and shown accordingly.
Table 4.
Prediction of antigenic processing and presentation for potential T-cell epitopes of EBV proteins.
| Antigenic proteins | Positions | Peptide sequences (T-cell epitopes) | MHC-1 binding affinity | Rescale binding affinity | Proteasomal C-terminal Cleavage | Transport affinity | Prediction score | MHC-1 binding |
|---|---|---|---|---|---|---|---|---|
| BMRF2 | 161–169 | FMSPFFIFL | 0.9130 | 1.361 | 0.9775 | 1.024 | 1.5588 | < - E |
| 342–350 | FVFTFCEYL | 0.7902 | 1.1779 | 0.7815 | 1.103 | 1.3503 | < - E | |
| BDLF2 | 246–254 | KVYTLIPAV | 0.8028 | 1.1967 | 0.4223 | 0.74 | 1.297 | < - E |
| 401–409 | RLTDRSFPA | 0.7341 | 1.0942 | 0.8579 | −0.447 | 1.2006 | < - E | |
| BORF1 | 199–207 | YLNKITTVV | 0.8500 | 1.2671 | 0.973 | 0.230 | 1.4245 | < - E |
| 157–165 | LLYDSPATL | 0.7634 | 1.1380 | 0.9742 | 1.306 | 1.3494 | < - E | |
| BcLF1 | 401–409 | MQYSYYFPV | 0.9188 | 1.3696 | 0.7904 | 0.756 | 1.526 | < - E |
| 1327–1335 | FLQEAFPAL | 0.8944 | 1.3332 | 0.9758 | 0.901 | 1.5247 | < - E | |
| BDLF1 | 266–274 | VMMTYLQSL | 0.8772 | 1.3076 | 0.9626 | 1.335 | 1.335 | < - E |
| 63–71 | YLSDCTLAV | 0.9317 | 1.3889 | 0.6197 | 0.451 | 1.5044 | < - E | |
| BFRF3 | 53–61 | SQFCYEEYV | 0.7511 | 1.1196 | 0.7174 | 0.5650 | 1.2554 | < - E |
| 122–131 | VAQSATPSV | 0.4544 | 0.6733 | 0.8997 | 0.5510 | 0.8398 | < - E | |
| BXLF1 | 552–560 | YLQFVLVDL | 0.702 | 1.0464 | 0.977 | 1.076 | 1.2468 | < - E |
| 548–556 | RTLAYLQFV | 0.788 | 1.1746 | 0.2347 | 0.565 | 1.2381 | < - E | |
| BGLF4 | 102–110 | KLYDSVTEL | 0.8824 | 1.3154 | 0.976 | 1.315 | 1.5276 | < - E |
| 336–344 | FLQFAAPKV | 0.8245 | 1.2291 | 0.4544 | 0.155 | 1.305 | < - E | |
| BMRF1 | 144–152 | ALMPYMPPA | 0.9089 | 1.3549 | 0.6638 | −0.3140 | 1.4388 | < - E |
| 32–40 | VLQVNLLSV | 0.6918 | 1.0312 | 0.8818 | 0.3340 | 1.1802 | < - E |
Structural modeling of selected epitopes and HLA-A*02:01 docking analysis
The selected T-cell epitopes (peptides) were structurally modeled using the PEPFOLD server before docking to HLA-A*02:01. pepATTRACT blind peptide-protein docking method was used to identify the most appropriate binding sites on HLA-A*02:01 for the respective epitopes into which they were docked simultaneously. 51 clusters were obtained for each peptide-protein complex and the best-docked complexes were selected based on global energy scoring and shown in Table 5. Moreover, 3D models of EBV T-cell epitopes with the highest energy scores are shown in Fig. 5.
Table 5.
Global energy estimations and MM/GBSA profiling of binding interactions between potential EBV T-cell epitopes and HLA-A*02:01. Estimations of associable energy components are also shown.
| Antigenic proteins | Peptide-MHC-1 complexes | pepATTRACT |
MM/GBSA estimations |
||||
|---|---|---|---|---|---|---|---|
| Global energy (kcal/mol) | ΔEvdw (kcal/mol) | ΔEele (kcal/mol) | ΔEGB (kcal/mol) | ΔESA (kcal/mol) | ΔGbind (kcal/mol) | ||
| BMRF2 | FMSPFFIFL161-169 | −18.44 | −11.24 | −58.83 | 61.37 | −7.44 | −10.18 |
| BDLF2 | KVYTLIPAV246-254 | −14.50 | −10.22 | −12.95 | 17.20 | −6.41 | −7.25 |
| BORF1 | YLNKITTVV199-207 | −15.08 | −10.42 | −11.55 | 15.85 | −6.51 | −7.43 |
| BcLF1 | FLQEAFPAL1327-1335 | −20.42 | −10.87 | −9.78 | 14.44 | −7.4 | −7.69 |
| BDLF1 | VMMTYLQSL266-274 | −14.37 | −8.4 | −15.03 | 17.02 | −5.9 | −7.59 |
| BFRF3 | SQFCYEEYV53-61 | −13.89 | −11.99 | −4.78 | 11.98 | −7.97 | −6.38 |
| BXLF1 | RTLAYLQFV548-556 | −14.79 | −16.69 | −58.64 | 64.04 | −10.85 | −13.47 |
| BGLF4 | FLQFAAPKV336-344 | −17.40 | −10.61 | −67.05 | 69.94 | −6.89 | −9.1 |
| BMRF1 | ALMPYMPPA144-152 | −18.54 | −10.36 | −14.07 | 16.84 | −6.6 | −8.88 |
ΔEele: electrostatic energy; ΔEvdW: van der Waals energy; ΔGbind: total binding free energy; ΔEGB: polar desolvation energy; ΔESA: non-polar solvation energy.
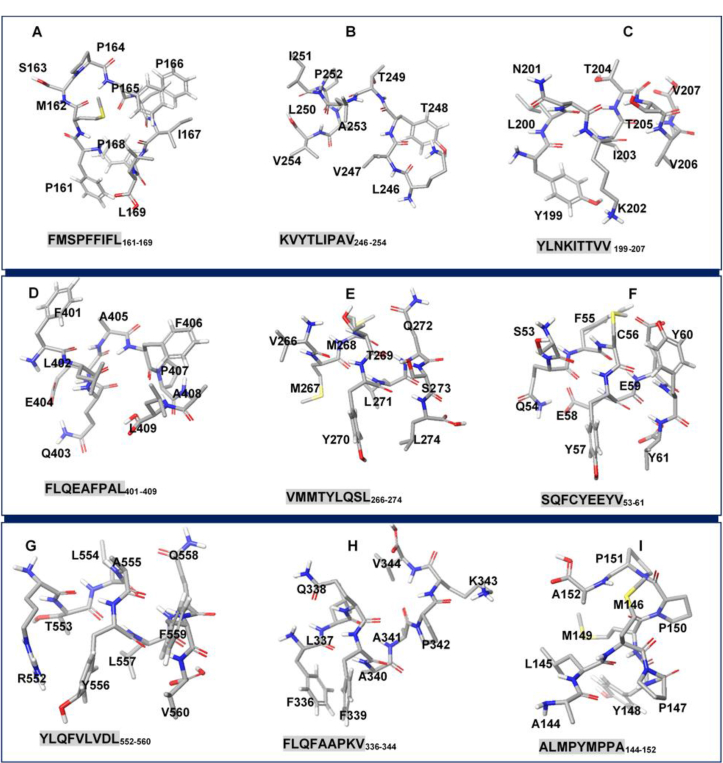
Fig. 5.
3D structures of potential EBV T-cell epitopes for [A] BMRF2 [B] BDLF2 [C] BORF1 [D] BcLF1 [E] BDLF1 [F] BFRF3 [G] BXLF1 [H] BGLF4 [I] BMRF1.
As estimated, BMRF2 T-cell epitope FMSPFFIFL161-169 had the most favorable docking energy of −18.4 kcal/mol compared to FVFTFCEYL342-350.
Likewise, the most favorable peptide-HLA-A*02:01 energy scores based on site binding affinities include −14.5 kcal/mol, −15.1 kcal/mol, −20.4 kcal/mol, −14.4 kcal/mol, −14.8 kcal/mol, −17.4 kcal/mol, −13.89 kcal/mol and −18.5 kcal/mol respectively for BDLF2 (KVYTLIPAV246-254), BORF1 (YLNKITTVV199-208), BcLF1 (FLQEAFPAL1327-1335), BDLF1 (VMMTYLQSL266-274), BXLF1 (RTLAYLQFV548-556), BGLF4 (FLQFAAPKV336-344), BFRF3 (SQFCYEEYV53-61), and BMRF1 (ALMPYMPPA144-152).
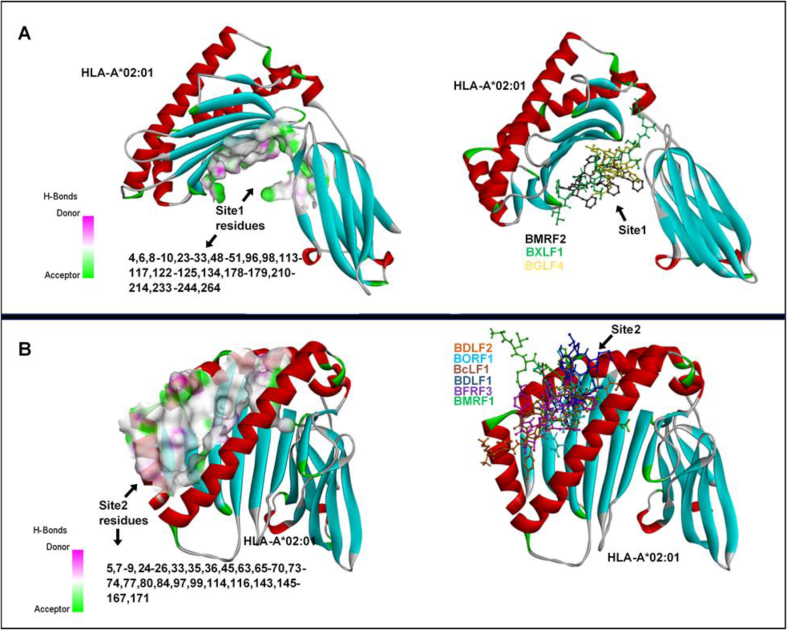
Amongst all predicted T-cell epitopes, BcLF1 epitope BcLF1 (FLQEAFPAL1327-1335) had the highest docking score. Also, we observed that the binding of all predicted T-cell epitopes occurred majorly across two sites on HLA-A*02:01 designated Site 1 and 2, which are depicted in Fig. 6. The predicted Site 2 into which some of the epitopes were docked correlates with the binding site determined in HLA-A*02:01 crystal structure (PDB ID: 4UQ3). As predicted, T-cell epitopes for EBV proteins; BMRF2, BXLF1, and BGLF4 exhibited a higher affinity for Site 1 where they were docked while epitopes for BDLF2, BFRF3, BORF1, BcLF1, BDLF1, BFRF3, and BMRF1 had a higher binding propensity for Site 2 as predicted by pepATTRACT.
Fig. 6.
Potential HLA-A*02:01 binding sites predicted for the respective EBV T-cell epitopes. [A] Site 1 and corresponding constituent residues [B] Site 2 and corresponding constituent residues. Also shown are the cohort of epitopes that bind with selective affinity to the respective sites as predicted by pepATTRACT blind-docking algorithm.
MD simulation and MM/GBSA-based interaction analyses
MD analyses were performed to obtain molecular insights into the binding and interaction dynamics of the predicted EBV T-cell epitopes and HLA-A*02:01. 100ns MD simulation runs were performed for the complexes and individual epitopes while resulting trajectories were analyzed.
This was performed using post-analytical metrics such as the root mean square deviation (RMSD) and root mean square fluctuation (RMSF) methods which were used to estimate trajectorial motions of constituent Cα atoms. Results showing estimated mean values for the epitopes and their corresponding MHC-1 complexes are presented in Table 6.
Table 6.
Post–MD simulation analyses of structural motions and stability for the T-cell epitopes and peptide-protein complexes.
| Molecular dynamic simulation analyses | |||
|---|---|---|---|
| EBV proteins | T-cell Epitope RMSD (Å) | T-cell Epitope-MHC complex |
|
| Cα- RMSD (Å) | Cα- RMSF (Å) | ||
| BMRF2 [FMSPFFIFL161-169] | 2.4 ± 0.4 | 4.2 ± 1.0 | 1.9 ± 0.8 |
| BDLF2 [KVYTLIPAV246-254] | 4.2 ± 1.0 | 6.8 ± 1.9 | 3.0 ± 1.2 |
| BORF1 [YLNKITTVV199-207] | 4.1 ± 1.0 | 4.1 ± 1.7 | 3.0 ± 1.3 |
| BcLF1 [FLQEAFPAL1327-1335] | 2.7 ± 0.5 | 6.4 ± 3.5 | 4.2 ± 1.8 |
| BDLF1 [VMMTYLQSL266-274] | 3.9 ± 0.6 | 5.5 ± 2.3 | 3.3 ± 2.0 |
| BFRF3 [SQFCYEEYV53-61] | 3.2 ± 0.3 | 4.3 ± 2.4 | 3.1 ± 1.1 |
| BXLF1 [RTLAYLQFV548-556] | 4.9 ± 1.0 | 2.8 ± 0.4 | 1.9 ± 0.6 |
| BGLF4 [FLQFAAPKV336-344] | 3.6 ± 1.5 | 9.0 ± 3.0 | 3.9 ± 1.7 |
| BMRF1 [ALMPYMPPA144-152] | 3.5 ± 0.5 | 5.6 ± 2.9 | 3.7 ± 1.4 |
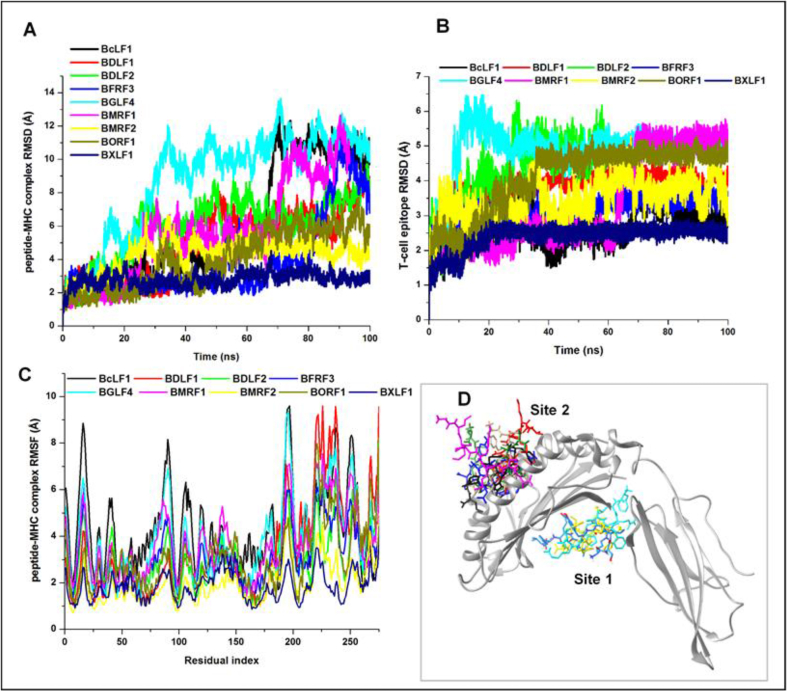
Comparatively Site 1–bound BXLF1 T-cell epitope [RTLAYLQFV548-556] exhibited the highest RMSD with a mean value of 4.9 Å while the lowest RMSD of 2.4 Å was exhibited by BMRF2 [FMSPFFIFL161-169], indicative of its highly stable motion at the target Site 1 region of HLA-A*02:01. The trajectorial motions of the respective epitopes at the target Sites (1 and 2) are shown in Fig. 7. Moreover, it can be deduced from the plot that while their motions were highly unstable at the initial periods of the MD simulation, they seemingly attained stability towards the end of the simulation (∼50–80ns) which could mediate optimal binding and interactions at their target sites. Dynamical motions of the peptide-MHC complexes were also measured using the RMSD and RMSF metrics. Mean values are also presented in Table 6 while corresponding plots are shown in Fig. 7. BXLF1 [RTLAYLQFV548-556]-MHC complex was more stable while the highest structural instability was exhibited by the BGLF4 [FLQFAAPKV336-344]-MHC complex.
Fig. 7.
Graphical plots depicting structural motions and residual fluctuations of potential EBV T-cell epitopes and protein complexes over 100ns MD simulation run [A] peptide-protein complex RMSD [B] peptide RMSD [C] peptide-protein complex RMSF [D] 3D representation of T-cell epitope binding on HLA-A*02:01 Sites 1 and 2.
Presumably, high RMSDs exhibited by the epitopes at the respective binding sites could correlate with systematic motions necessary to achieve optimal and high-affinity binding with target residues. Consequently, this could account for the complementary increase in structural motions and deviations observed for the peptide-MHC complexes until relative stability was attained.
Complementary interaction dynamics of the EBV T-cell epitopes were also investigated while binding affinities were measured using the MM/GBSA method. Findings revealed that predicted Site 1 HLA-A*02:01 binders; BMRF2 [FMSPFFIFL161-169], BXLF1 [RTLAYLQFV548-556] and BGLF4 [FLQFAAPKV336-344] had total binding energies (ΔGbind) of −10.18, −13.85 and −9.1 kcal/mol, which are higher when compared to other EBV T-cell epitopes that are Site 2 binders as shown in Table 5.
Estimations of other energy components revealed that both van der Waals (ΔEvdW) and electrostatic energies (ΔEele) contributed to the ΔGbind while positive polar desolvation energy values (ΔEGB) indicated that binding interactions were favorable in the hydrophobic regions. This is supported by the estimated negative non-polar energies (ΔESA) as shown.
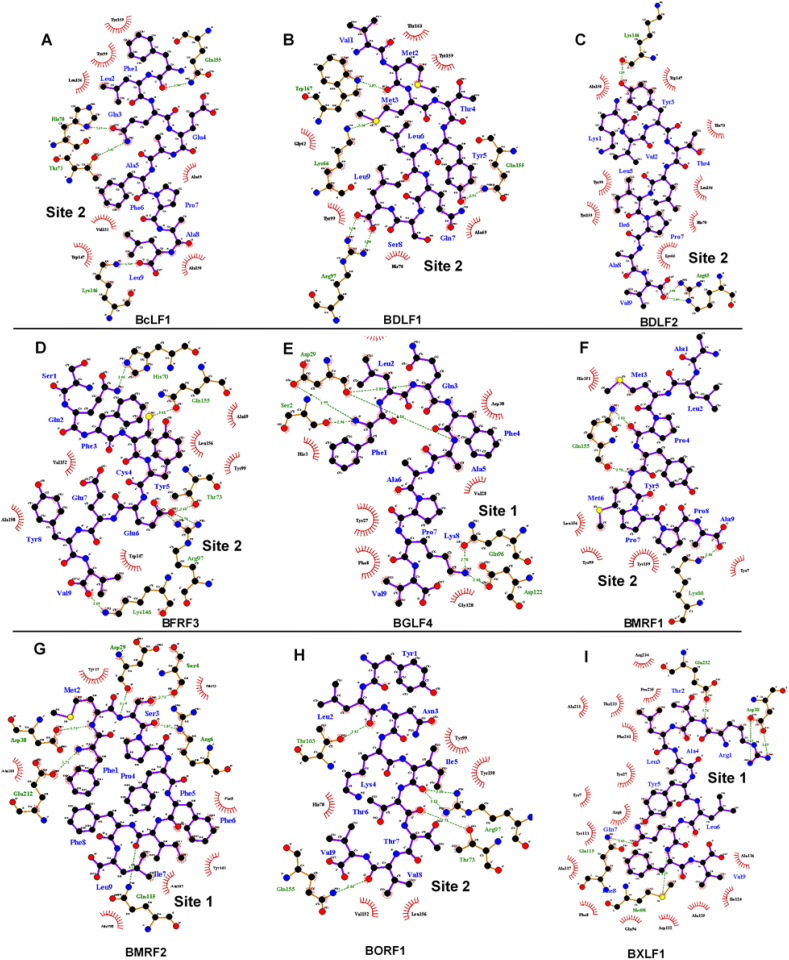
Analysis of the intermolecular interactions between the T-cell epitopes and target MHC-1 molecule revealed a similar binding pattern across both sites as shown in Fig. 8.
Fig. 8.
Analysis of intermolecular interactions mediated by potential EBV T-cell epitopes at their respective HLA-A*02:01 binding sites.
For Site 1 binders; amino acid residues Asp29, Asp30 and Gln115 (amongst other interacting residues) were crucial to the binding and stability of the epitopes; BMRF2 [FMSPFFIFL161-169], BXLF1 [RTLAYLQFV548-556] and BGLF4 [FLQFAAPKV336-344]. These include the formation of high-affinity hydrogen bonds with proximal residues of the T-cell epitopes. As shown, Site 1 residues; Asp29, Asp30 and Gln115 formed H bonds with Met2, Ser3 and Ile7 of BMRF2 [FMSPFFIFL161-169] respectively. Likewise, Asp30 and Gln115 recurrently interacted with Arg1 and Gln7 of BXLF1 [RTLAYLQFV548-556] respectively.
Similarly in BGLF4 [FLQFAAPKV336-344] complex, Asp29 formed two H bonds with Gln3 and Phe4 whilst also involved in H bond formation with Phe1. On the other hand, Asp30 existed in hydrophobic interactions with the hydrophobic side chain of Phe4.
Also, we identified important interactions that account for the stability of Site 2 binders at the target region (Fig. 8). A highly important residue that was uniformly involved in complementary H bonding with the respective EBV epitopes is Gln155. At the target Site 2, His70, Thr73, Lys246 and, Gln155 contributed to the binding of BcLF1 [FLQEAFPAL1327-1335] via H bond interactions while Gln155, Arg97, Lys66, Trp167 were involved in the binding of BDLF1 [VMMTYLQSL266-274]. Moreover, we observed that Lys146 and Arg65 played key roles in the binding of BDLF2 [KVYTLIPAV246-254] while BFRF3 [SQFCYEEYV53-61] binding was stabilized by interactions with His70, Gln155, Thr73, Arg97 and, Lys146. Gln155 and Lys66 were crucial for the binding of BMRF1 [ALMPYMPPA144-152] while constituent BORF1 [YLNKITTVV199-208] residues; Leu2, Ile5, Thr7 and, Val8 interacted with Site 2 residues; Thr163, Arg97, Thr73, and Gln155 respectively as shown in Fig. 8.
Discussion
Efforts to curtail the virulent activities of EBV have been ongoing requiring several therapeutic options ranging from the development of small molecule inhibitors to vaccination. While antagonists are directed towards crucial viral proteins [122,123], vaccines are developed to stimulate immunogenic B-cell or T-cell responses towards lytic and latent EBV proteins [2,15]. Although efforts to develop effective cures for EBV-mediated infections have been unsuccessful, a gp350 vaccine has made significant progress through clinical trials even though it does not curtail viral infections [2,[33], [34], [35]].
Among the various vaccine types, peptide or subunit vaccines have emerged as attractive candidates for treatment since there is a huge possibility of designing them based on essential information from viral pathogenic components. This is achievable via the implementation of immunoinformatics approaches which has been widely employed for B- and T-cells epitopic predictions from viral proteins [31,53,56,[61], [62], [63], [64], [65]]. The antigenic nature of viral proteins corresponds with their ability to elicit adaptive immunogenic responses by host B- and T-cells which could, in turn, lead to the death of infected cells. Molecular components (antigens) and not the whole pathogens are recognized by receptors on B- and T-cells; B-cell receptors (BCR) and T-cell receptors (TCR), a process that differs greatly for both cells. While BCRs recognize and bind surface-exposed antigens (epitopes) on viral proteins to secrete antibodies and mediate humoral adaptive immunity, TCRs recognize antigens presented on the surfaces of antigen-presenting cells (APCs) that are bound to major histocompatibility complex (MHC) molecules categorized into MHC-I and II. These are respectively recognized by CD8+ (cytotoxic) and CD4+ (helper) T-cells. Altogether, the pathway for antigenic presentation (at the extracellular surface) incorporates proteasomal degradation, TAP processing and MHC-1 [human leukocytes antigen – (HLA) in humans] binding [31,[124], [125], [126]]. Several crucial functional proteins (lytic and latent) constitute the EBV proteome and contribute to its virulence in host cells. Similar to EBV glycoproteins [8,53], proteins that regulate epithelial cell attachment, DNA synthesis/replication and, capsid assembly are highly important in the viral latent and lytic cycles. These proteins include BMRF2, BDLF2, BORF1, BcLF1, BFRF3, BXLF1, BGLF4, and BMRF1 (see Table 1). Therefore, immunoinformatics prediction of potential B-cell and T-cell (CD8+) epitopes for these proteins represent the highlight of this study.
Potential linear B-cell epitopes reported in this study were selected based on their characteristic surface-exposure/accessibility and antigenicity. These attributes are essential to facilitate their recognition and binding to paratopes on BCRs provoking humoral immunogenic responses. Therefore, linear (continuous) B-cell epitopes; BMRF2 [SLRVYSTSTWVSAPCL90-105], BDLF2 [PAAAAVPGPEPEPEPHPPMP62-81], BORF1 [DRRRLQRRIAGLLPPPARRL8-27], BcLF1 [KGVSTYTTAKGGEPVGGVFI262-281], BGLF1 [SPTNSSSGELSVSPEPPRE10-30] and [RPRHTVSPSPSPPPPPRTPT327-347] all possess high propensities for eliciting B-cell mediated immune responses. Since most B-cell epitopes are conformational or discontinuous, we also employed prediction algorithms for 3D conformational epitopes. As shown in Fig. 4, conformational epitopes for the respective EBV antigens are highly surface exposed. Antigenic and allergenic profiling was also carried out thereby presenting our predicted epitopes as potential candidates for the development of EBV B-cell vaccines. The presence of residues with hydrophilic, large and aromatic side chains could provide a clue into the characteristic surface-exposure and immunogenicity exhibited by the predicted epitopes [97].
T-cell epitope prediction was also carried out to identify the shortest peptides that constitute these antigens and can stimulate CD8 T-cells activation [56,127]. Herein, binding to a highly frequent human MHC I molecule (HLA-A*02:01), TAP processing and C-terminal cleavage were considered which resulted in the selection of T-cell epitopes; BMRF2 [FMSPFFIFL161-169], BDLF2 [KVYTLIPAV246-254], BORF1 [YLNKITTVV199-207], BcLF1 [FLQEAFPAL1327-1335], BDLF1 [VMMTYLQSL266-274], BFRF3 [SQFCYEEYV53-61], BXLF1 [RTLAYLQFV548-556], BGLF4 [FLQFAAPKV336-344] and BMRF1 [ALMPYMPPA144-152]. With a 75% threshold, higher predictive scores obtained could indicate the immunogenic propensities of these T-cell epitopes. Information on the peptide-blinding clefts of MHC molecules is also essential since they could be vital for the structure-based design of high-affinity and selective epitopes for peptide-vaccine design [56,128]. Without restraining epitopic binding to a particular site on HLA-A*02:01, two sites were identified using a blind peptide-protein docking algorithm, one of which corresponds to an experimentally defined site (PDB entry 4UQ3). These sites were identified based on a global search of the protein surface after which the peptides (conformation) were docked to the corresponding sites [102,103]. Investigating the binding dynamics, affinity, and stability of the T-cell epitopes at their target HLA-A*02:01 sites using MD and MM/GBSA methods further yielded interesting insights, particularly for the identification of crucial site residues that mediate high-affinity peptide-protein interactions.
MD simulation analyses revealed the dynamical motions of HLA-A*02:01 when bound by the predicted T-cell epitopes from the respective EBV antigens. According to previous reports, high structural motions are necessary for MHC-1 molecules and correlates with their ability to translocate the processed peptide (proteasomal C-terminal cleavage and TAP) from the endoplasmic reticulum (ER) lumen to the Golgi and ultimately to the extracellular space [30,128,129]. As shown in Fig. 7, the complexes demonstrated high RMSDs indicative of high structural motions which became more stable towards the end of the simulation period. These motion patterns were also reflected by the RMSF analysis which estimates residual fluctuations. While notably high motions were observed in HLA-A*02:01 bound by predicted T-cell epitopes from BcLF1 [FLQEAFPAL1327-1335], BGLF4 [FLQFAAPKV336-344], BMRF1 [ALMPYMPPA144-152] and BFRF3 [SQFCYEEYV53-61], low structural motions were observed in the BXLF1 complex. However, the T-cell epitope from BXLF1 [RTLAYLQFV548-556] exhibited the highest RMSD which could indicate its degree of motion at its target site (Site 1). Recurring inter-molecular interactions between specific HLA-A*02:01 site residues and the predicted T-cell epitopes further explained binding patterns relative to high-affinity binding and stabilization. Identification of a novel site (Site 1) for epitope binding on HLA-A*02:01 could also aid in the structure-based design of high-affinity immunogenic binders for the target MHC-I molecule. This is because the most favorable ΔGs estimated were for T-cell epitopes bound to Site 1; BMRF2 [SLRVYSTSTWVSAPCL90-105] (−10.18 kcal/mol), BGLF4 [SPTNSSSGELSVSPEPPRE10-30] (−9.1 kcal/mol) and BXLF1 [RTLAYLQFV548-556] (−13.471 kcal/mol) compared to those bound to Site 2.
As proposed in this study, while BMRF2 [SLRVYSTSTWVSAPCL90-105], BDLF2 [PAAAAVPGPEPEPEPHPPMP62-81], BORF1 [DRRRLQRRIAGLLPPPARRL8-27], BcLF1 [KGVSTYTTAKGGEPVGGVFI262-281], BGLF4 [SPTNSSSGELSVSPEPPRE10-30] and [RPRHTVSPSPSPPPPPRTPT327-347] depict potential epitopes that could elicit immunogenic B-cell responses based on physicochemical attributes, BMRF2 [FMSPFFIFL161-169], BDLF2 [KVYTLIPAV246-254], BORF1 [YLNKITTVV199-207], BcLF1 [FLQEAFPAL1327-1335], BDLF1 [VMMTYLQSL266-274], BFRF3 [SQFCYEEYV53-61], BXLF1 [RTLAYLQFV548-556], BGLF4 [FLQFAAPKV336-344] and BMRF1 [ALMPYMPPA144-152] have high propensities of provoking T-cell mediated cytotoxic immune responses towards infected cells.
An interesting futuristic approach is the possible derivation of highly selective antagonists from the predicted epitopes, as reported in a study by Abdul et al. [59]. This could present avenues to reverse-map highly interactive regions of the potential EBV epitopes and generate pharmacophore models that can be used to screen virtually for possible lead candidates. These derived compounds would presumably be able to achieve selective targeting and inhibition of crucial pathogenic machinery of EBV.
Also, more precise and accurate methods can be incorporated to distinguish epitopes from non-epitopes from the large pool of predicted epitopes. This is an effective approach as demonstrated in a previous study by Khanna and Rana where they implemented a machine learning model for determining epitopes of Mycobacterium tuberculosis [130].
Immunoinformatics predictions performed in this study covered multiple EBV antigens thereby presenting a possibility of developing highly effective anti-EBV vaccines consisting of a mixture of antigens from these pools of B- and T-cell epitopes [68].
Furthermore, while we propose the ability of these predicted epitopes to provoke efficacious immune responses, it is still necessary to validate their propensity using in vitro and in vivo techniques in the future. These could include binding assays for in vitro determination of peptide binding and/or ELISPOT or CFSE dilution assays for estimating T-cell recognition [131].
Conclusion
The recent paradigm shift towards peptide or subunit vaccine development has been highly resourceful, particularly with regards to provoking adaptive or cell-mediated immune responses. These are designed based on B-cell and T-cell epitopes derived as components of antigenic proteins secreted by viruses during their infectious life cycles. Although no significant progress has been made to date, vaccination remains a promising treatment option to curtail the virulence of Epstein Barr virus (EBV) implicated in infectious mononucleosis (IM) and several human cancers. In this study, we implement a computational meta-analysis paradigm that integrates immunoinformatics, blind peptide-protein docking, and molecular dynamics to predict potential B-cell and T-cell epitopes for antigenic EBV proteins that mediate epithelial cell–cell attachment/spread, capsid self-assembly, DNA synthesis, replication and, processivity. Predictive BCPred and Ellipro algorithms were used to rank linear and conformational B-cell epitopes based on their physicochemical properties such as surface-exposure or accessibility, flexibility, hydrophilicity and, antigenicity. More so, based on the Major Histocompatibility Complex (MHC) class 1 antigenic presentation pathway, potential T-cell epitopes were predicted using the integrative weight matrix and Artificial Neural Network (ANN) algorithm of the NetCTL 1.2 webserver. Peptide modeling and blind docking were used to identify probable binding sites on a highly frequent human MHC-1 molecule; HLA-A*02:01 while varying binding affinities for the peptide-MHC complexes were obtained. A novel high-affinity binding site (Site 1) was identified in addition to a structurally resolved site (Site 2). Dynamical studies revealed that the binding of the T-cell epitopes induced a high degree of structural motions in HLA-A*02:01 with favorable binding energy (ΔG) that correlates with their stabilities at their targeted sites. Our results suggest potential B- and T-cell epitopes that could serve as effective candidates for peptide-vaccine development towards the treatment of EBV-related diseases.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors thank the School of Health Sciences, University of KwaZulu-Natal for infrastructural support and the Center for High-Performance Computing (CHPC – www.chpc.ac.za), Cape Town, South Africa for computational resources.
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bj.2020.01.002.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Rist M.J., Neller M.A., Burrows J.M., Burrows S.R. T cell epitope clustering in the highly immunogenic BZLF1 antigen of epstein-barr virus. J Virol. 2015;89:703–712. doi: 10.1128/JVI.02642-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen J.I., Fauci A.S., Varmus H., Nabel G.J. Epstein-Barr virus: an important vaccine target for cancer prevention. Sci Transl Med. 2011;3:107fs7. doi: 10.1126/scitranslmed.3002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maeda E., Akahane M., Kiryu S., Kato N., Yoshikawa T., Hayashi N. Spectrum of Epstein-Barr virus-related diseases: a pictorial review. Jpn J Radiol. 2009;27:4–19. doi: 10.1007/s11604-008-0291-2. [DOI] [PubMed] [Google Scholar]
- 4.Cho W.C.S. Nasopharyngeal carcinoma: molecular biomarker discovery and progress. Mol Cancer. 2007;6:1. doi: 10.1186/1476-4598-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuckwiller L.S., Glaser R. Epstein-barr virus and nasopharyngeal carcinoma. Nasal Tumors Anim Man Vol III Exp Nasal Carcinog. 2017;372:20160270. [Google Scholar]
- 6.Taylor G.S., Long H.M., Brooks J.M., Rickinson A.B., Hislop A.D. The immunology of epstein-barr virus–induced disease. Annu Rev Immunol. 2015;33:787–821. doi: 10.1146/annurev-immunol-032414-112326. [DOI] [PubMed] [Google Scholar]
- 7.Rickinson A.B. Co-infections, inflammation and oncogenesis: future directions for EBV research. Semin Cancer Biol. 2014;26:99–115. doi: 10.1016/j.semcancer.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Hutt-Fletcher L.M. EBV glycoproteins: where are we now? Future Virol. 2015;10:1155–1162. doi: 10.2217/fvl.15.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hislop A.D., Taylor G.S., Sauce D., Rickinson A.B. Cellular responses to viral infection in humans: lessons from epstein-barr virus. Annu Rev Immunol. 2007;25:587–617. doi: 10.1146/annurev.immunol.25.022106.141553. [DOI] [PubMed] [Google Scholar]
- 10.Tangye S.G., Palendira U., Edwards E.S.J. Human immunity against EBV—lessons from the clinic. J Exp Med. 2017;214:269–283. doi: 10.1084/jem.20161846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long H.M., Taylor G.S., Rickinson A.B. Immune defence against EBV and EBV-associated disease. Curr Opin Immunol. 2011;23:258–264. doi: 10.1016/j.coi.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 12.Young L.S., Murray P.G. Epstein-Barr virus and oncogenesis: from latent genes to tumours. Oncogene. 2003;22:5108–5121. doi: 10.1038/sj.onc.1206556. [DOI] [PubMed] [Google Scholar]
- 13.Neves M., Marinho-Dias J., Ribeiro J., Sousa H. Epstein–Barr virus strains and variations: geographic or disease-specific variants? J Med Virol. 2017;89:373–387. doi: 10.1002/jmv.24633. [DOI] [PubMed] [Google Scholar]
- 14.Alonso-Padilla J., Lafuente E.M., Reche P.A. Computer-aided design of an epitope-based vaccine against epstein-barr virus. J Immunol Res. 2017;2017:9363750. doi: 10.1155/2017/9363750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Zyl D.G., Mautner J., Delecluse H.J. Progress in EBV vaccines. Front Oncol. 2019;9:104. doi: 10.3389/fonc.2019.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou L., Chen J.N., Qiu X.M., Pan Y.H., Zhang Z.G., Shao C.K. Comparative analysis of 22 epstein-barr virus genomes from diseased and healthy individuals. J Gen Virol. 2017;98:96–107. doi: 10.1099/jgv.0.000699. [DOI] [PubMed] [Google Scholar]
- 17.Kang M.S., Kieff E. Epstein-Barr virus latent genes. Exp Mol Med. 2015;47:e131. doi: 10.1038/emm.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johannsen E., Luftig M., Chase M.R., Weicksel S., Cahir-McFarland E., Illanes D. Proteins of purified Epstein-Barr virus. Proc Natl Acad Sci. 2004;101:16286–16291. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanfield B.A., Luftig M.A. Recent advances in understanding Epstein-Barr virus. F1000Res. 2017;6:386. doi: 10.12688/f1000research.10591.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farrell P.J. Epstein–Barr virus and cancer. Annu Rev Pathol Mech Dis. 2019;14:29–53. doi: 10.1146/annurev-pathmechdis-012418-013023. [DOI] [PubMed] [Google Scholar]
- 21.Hui-Yuen J., McAllister S., Koganti S., Hill E., Bhaduri-Mcintosh S. Establishment of Epstein-Barr virus growth-transformed lymphoblastoid cell lines. J Vis Exp. 2011 Nov:3321. doi: 10.3791/3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mrozek-Gorska P., Buschle A., Pich D., Schwarzmayr T., Fechtner R., Scialdone A. Epstein–Barr virus reprograms human B lymphocytes immediately in the prelatent phase of infection. Proc Natl Acad Sci. 2019;116:16046–16055. doi: 10.1073/pnas.1901314116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saha A., Robertson E.S. Mechanisms of B-cell oncogenesis induced by epstein-barr virus. J Virol. 2019;93 doi: 10.1128/JVI.00238-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brooks J.M., Long H.M., Tierney R.J., Shannon-Lowe C., Leese A.M., Fitzpatrick M. Early T cell recognition of B cells following epstein-barr virus infection: identifying potential targets for prophylactic vaccination. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tu J., Wang X., Geng G., Xue X., Lin X., Zhu X. The possible effect of B-cell epitopes of Epstein-Barr virus early antigen, membrane antigen, latent membrane protein-1, and -2A on systemic lupus erythematosus. Front Immunol. 2018;9:187. doi: 10.3389/fimmu.2018.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strowig T., Brilot F., Arrey F., Bougras G., Thomas D., Muller W.A. Tonsilar NK cells restrict B cell transformation by the epstein-barr virus via IFN-γ. PLoS Pathog. 2008;4:e27. doi: 10.1371/journal.ppat.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jud A., Kotur M., Berger C., Gysin C., Nadal D., Lünemann A. Tonsillar CD56brightNKG2A+ NK cells restrict primary Epstein-Barr virus infection in B cells via IFN-γ. Oncotarget. 2017;8:6130–6141. doi: 10.18632/oncotarget.14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bu W., Hayes G.M., Liu H., Gemmell L., Schmeling D.O., Radecki P. Kinetics of Epstein-Barr Virus (EBV) neutralizing and virus-specific antibodies after primary infection with EBV. Clin Vaccine Immunol. 2016;23:363–369. doi: 10.1128/CVI.00674-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 30.Lundegaard C., Lund O., Buus S., Nielsen M. Major histocompatibility complex class i binding predictions as a tool in epitope discovery. Immunology. 2010;130:309–318. doi: 10.1111/j.1365-2567.2010.03300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panahi H.A., Bolhassani A., Javadi G., Noormohammadi Z. A comprehensive in silico analysis for identification of therapeutic epitopes in HPV16, 18, 31 and 45 oncoproteins. PloS One. 2018;13 doi: 10.1371/journal.pone.0205933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amyes E., Hatton C., Montamat-Sicotte D., Gudgeon N., Rickinson A.B., McMichael A.J. Characterization of the CD4+ T cell response to epstein-barr virus during primary and persistent infection. J Exp Med. 2003;198:903–911. doi: 10.1084/jem.20022058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokal E.M., Hoppenbrouwers K., Vandermeulen C., Moutschen M., Léonard P., Moreels A. Recombinant gp350 vaccine for infectious mononucleosis: a phase 2, randomized, double-blind, placebo-controlled trial to evaluate the safety, immunogenicity, and efficacy of an epstein-barr virus vaccine in healthy young adults. J Infect Dis. 2007;196:1749–1753. doi: 10.1086/523813. [DOI] [PubMed] [Google Scholar]
- 34.Rees L., Tizard E.J., Morgan A.J., Cubitt W.D., Finerty S., Oyewole-Eletu T.A. A phase i trial of epstein-barr virus Gp350 vaccine for children with chronic kidney disease awaiting transplantation. Transplantation. 2009;88:1025–1029. doi: 10.1097/TP.0b013e3181b9d918. [DOI] [PubMed] [Google Scholar]
- 35.Elliott S.L., Suhrbier A., Miles J.J., Lawrence G., Pye S.J., Le T.T. Phase I trial of a CD8+ T-cell peptide epitope-based vaccine for infectious mononucleosis. J Virol. 2008;82:1448–1457. doi: 10.1128/JVI.01409-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goscé L., Winter J.R., Taylor G.S., Lewis J.E.A., Stagg H.R. Modelling the dynamics of EBV transmission to inform a vaccine target product profile and future vaccination strategy. Sci Rep. 2019;9:9290. doi: 10.1038/s41598-019-45381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.2019. Centers for Disease Control and Prevention Varicella vaccine effectiveness and duration of protection. https://www.cdc.gov/vaccines/vpd-vac/varicella/hcp-effective-duration.htm/;2019 [accessed 7 November 2019] [Google Scholar]
- 38.Vázquez M., LaRussa P.S., Gershon A.A., Niccolai L.M., Muehlenbein C.E., Steinberg S.P. Effectiveness over time of varicella vaccine. J Am Med Assoc. 2004;291:851–855. doi: 10.1001/jama.291.7.851. [DOI] [PubMed] [Google Scholar]
- 39.Kuter B., Matthews H., Shinefield H., Black S., Dennehy P., Watson B. Ten year follow-up of healthy children who received one or two injections of varicella vaccine. Pediatr Res. 2004;23:132–137. doi: 10.1097/01.inf.0000109287.97518.67. [DOI] [PubMed] [Google Scholar]
- 40.Gu S.Y., Huang T.M., Ruan L., Miao Y.H., Lu H., Chu C.M. First EBV vaccine trial in humans using recombinant vaccinia virus expressing the major membrane antigen. Dev Biol Stand. 1995;84:171–177. [PubMed] [Google Scholar]
- 41.Moutschen M., Léonard P., Sokal E.M., Smets F., Haumont M., Mazzu P. Phase I/II studies to evaluate safety and immunogenicity of a recombinant gp350 Epstein-Barr virus vaccine in healthy adults. Vaccine. 2007;25:4697–4705. doi: 10.1016/j.vaccine.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 42.Haigh T.A., Lin X., Jia H., Hui E.P., Chan A.T.C., Rickinson A.B. EBV latent membrane proteins (LMPs) 1 and 2 as immunotherapeutic targets: LMP-specific CD4 + cytotoxic T cell recognition of EBV-transformed B cell lines. J Immunol. 2008;180:1643–1654. doi: 10.4049/jimmunol.180.3.1643. [DOI] [PubMed] [Google Scholar]
- 43.Ressing M.E., Gram A.M., Gram A.M., Hooykaas M.J.G., Piersma S.J., Wiertz E.J.H.J. Immune evasion by Epstein-Barr virus. Curr Top Microbiol Immunol. 2015;391:355–381. doi: 10.1007/978-3-319-22834-1_12. [DOI] [PubMed] [Google Scholar]
- 44.Leskowitz R.M., Zhou X.Y., Villinger F., Fogg M.H., Kaur A., Lieberman P.M. CD4+ and CD8+ T-cell responses to latent antigen EBNA-1 and lytic antigen BZLF-1 during persistent lymphocryptovirus infection of rhesus macaques. J Virol. 2013;87:8351–8362. doi: 10.1128/JVI.00852-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cunha-Neto E., Rosa D.S., Harris P.E., Olson T., Morrow A., Ciotlos S. An approach for a synthetic CTL vaccine design against Zika flavivirus using class I and class II epitopes identified by computer modeling. Front Immunol. 2017;8:640. doi: 10.3389/fimmu.2017.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bello M., Campos-Rodriguez R., Rojas-Hernandez S., Contis-Montes de Oca A., Correa-Basurto J. Predicting peptide vaccine candidates against H1N1 influenza virus through theoretical approaches. Immunol Res. 2015;62:3–15. doi: 10.1007/s12026-015-8629-1. [DOI] [PubMed] [Google Scholar]
- 47.Cohen J.I. Epstein–barr virus vaccines. Clin Transl Immunol. 2015;4:e32. doi: 10.1038/cti.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hemmati M, Raoufi E, Fallahi H. Predicting candidate epitopes on Ebola virus for possible vaccine development [Internet]. London, UK: InTech; 2018 [cited 2019 Oct 14]. Available from: https://www.intechopen.com/books/advances-in-ebola-control/predicting-candidate-epitopes-on-ebola-virus-for-possible-vaccine-development.
- 49.Chakraborty S. Ebola vaccine: multiple peptide-epitope loaded vaccine formulation from proteome using reverse vaccinology approach. Int J Pharm Pharm Sci. 2014;6:407–412. [Google Scholar]
- 50.Patronov A., Doytchinova I. T-cell epitope vaccine design by immunoinformatics. Open Biol. 2013;3:120139. doi: 10.1098/rsob.120139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vici P., Pizzuti L., Mariani L., Zampa G., Santini D., Di Lauro L. Targeting immune response with therapeutic vaccines in premalignant lesions and cervical cancer: hope or reality from clinical studies. Expert Rev Vaccines. 2016;15:1327–1336. doi: 10.1080/14760584.2016.1176533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khatoon N., Pandey R.K., Prajapati V.K. Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci Rep. 2017;7:8285. doi: 10.1038/s41598-017-08842-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ali A., Khan A., Kaushik A.C., Wang Y., Ali S.S., Junaid M. Immunoinformatic and systems biology approaches to predict and validate peptide vaccines against Epstein–Barr virus (EBV) Sci Rep. 2019;9:720. doi: 10.1038/s41598-018-37070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de R.K., Tomar N. 2nd ed. Human Press; New York: 2014. Immunoinformatics. [Google Scholar]
- 55.Dubey K.K., Luke G.A., Knox C., Kumar P., Pletschke B.I., Singh P.K. Vaccine and antibody production in plants: developments and computational tools. Brief Funct Genomics. 2018;17:295–307. doi: 10.1093/bfgp/ely020. [DOI] [PubMed] [Google Scholar]
- 56.Sanchez-Trincado J.L., Gomez-Perosanz M., Reche P.A. Fundamentals and methods for T- and B-cell epitope prediction. J Immunol Res. 2017;2017:2680160. doi: 10.1155/2017/2680160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sirskyj D., Diaz-Mitoma F., Golshani A., Kumar A., Azizi A. Innovative bioinformatic approaches for developing peptide-based vaccines against hypervariable viruses. Immunol Cell Biol. 2011;89:81–89. doi: 10.1038/icb.2010.65. [DOI] [PubMed] [Google Scholar]
- 58.Khan A., Junaid M., Kaushik A.C., Ali A., Ali S.S., Mehmood A. Computational identification, characterization and validation of potential antigenic peptide vaccines from hrHPVs E6 proteins using immunoinformatics and computational systems biology approaches. PloS One. 2018;13 doi: 10.1371/journal.pone.0196484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wadood A., Mehmood A., Khan H., Ilyas M., Ahmad A., Alarjah M. Epitopes based drug design for dengue virus envelope protein: a computational approach. Comput Biol Chem. 2017;71:152–160. doi: 10.1016/j.compbiolchem.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 60.Mehmood A., Kaushik A.C., Wei D. Prediction and validation of potent peptides against herpes simplex virus type 1 via immunoinformatic and systems biology approach. Chem Biol Drug Des. 2019;94:1868–1883. doi: 10.1111/cbdd.13602. [DOI] [PubMed] [Google Scholar]
- 61.Ip P., Nijman H., Daemen T. Epitope prediction assays combined with validation assays strongly narrows down putative cytotoxic T lymphocyte epitopes. Vaccines. 2015;3:203–220. doi: 10.3390/vaccines3020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tarek M.M., Shafei A.E., Ali M.A., Mansour M.M. Computational prediction of vaccine potential epitopes and 3-dimensional structure of XAGE-1b for non-small cell lung cancer immunotherapy. Biomed J. 2018;41:118–128. doi: 10.1016/j.bj.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kozlova E.E.G., Cerf L., Schneider F.S., Viart B.T., NGuyen C., Steiner B.T. Computational B-cell epitope identification and production of neutralizing murine antibodies against Atroxlysin-I. Sci Rep. 2018;8:14904. doi: 10.1038/s41598-018-33298-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Damilano G.D., Sued O., Ruiz M.J., Ghiglione Y., Canitano F., Pando M. Computational comparison of availability in CTL/gag epitopes among patients with acute and chronic HIV-1 infection. Vaccine. 2018;36:4142–4151. doi: 10.1016/j.vaccine.2018.04.086. [DOI] [PubMed] [Google Scholar]
- 65.Khan M.A., Hossain M.U., Rakib-Uz-Zaman S.M., Morshed M.N. Epitope-based peptide vaccine design and target site depiction against Ebola viruses: an immunoinformatics study. Scand J Immunol. 2015;82:25–34. doi: 10.1111/sji.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ali M.T., Morshed M.M., Hassan F. A computational approach for designing a universal epitope-based peptide vaccine against Nipah virus. Interdiscip Sci Comput Life Sci. 2015;7:177–185. doi: 10.1007/s12539-015-0023-0. [DOI] [PubMed] [Google Scholar]
- 67.Pradhan D., Yadav M., Verma R., Khan N.S., Jena L., Jain A.K. Discovery of T-cell driven subunit vaccines from Zika virus genome: an immunoinformatics approach. Interdiscip Sci Comput Life Sci. 2017;9:468–477. doi: 10.1007/s12539-017-0238-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang M., Jiang S. The development of prophylactic EBV vaccines. Immunol Disord Immunother. 2015;1:e101. [Google Scholar]
- 69.de C Salvadori L., de S Santana F.C., Marcos E.V.C. Frequency of alleles and haplotypes of the human leukocyte antigen system in Bauru, São Paulo, Brazil. Rev Bras Hematol Hemoter. 2014;36:108–114. doi: 10.5581/1516-8484.20140026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.del Pilar Fortes M., Gill G., Paredes M.E., Gamez L.E., Palacios M., Blanca I. Allele and haplotype frequencies at Human leukocyte antigen class i and II genes in Venezuela's population. Ann Biol Clin. 2012;702:175–181. doi: 10.1684/abc.2012.0663. [DOI] [PubMed] [Google Scholar]
- 71.Costantino P.R., Zeck S.C., da Silva W.A., da G Bicalho M. Human leukocyte antigen allele linkage disequilibrium and haplotype structure in volunteer bone marrow donors of Paraná state. Rev Bras Hematol Hemoter. 2017;39:229–236. doi: 10.1016/j.bjhh.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Esmaeili A.R., Zamani Taghizadeh Rabe S., Mahmoudi M., Rastin M. Frequencies of HLA-A, B and DRB1 alleles in a large normal population living in the city of mashhad, northeastern Iran. Iran J Basic Med Sci. 2017;20:940–943. doi: 10.22038/IJBMS.2017.9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paris R., Bejrachandra S., Thongcharoen P., Nitayaphan S., Pitisuttithum P., Sambor A. HLA class II restriction of HIV-1 clade-specific neutralizing antibody responses in ethnic Thai recipients of the RV144 prime-boost vaccine combination of ALVAC-HIV and AIDSVAX ® B/E. Vaccine. 2012;30:832–836. doi: 10.1016/j.vaccine.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 74.Singh S.P., Mishra B.N. Major histocompatibility complex linked databases and prediction tools for designing vaccines. Hum Immunol. 2016;77:295–306. doi: 10.1016/j.humimm.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 75.Bui H.H., Sidney J., Dinh K., Southwood S., Newman M.J., Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinf. 2006;7:153. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun B., Zhang Y. Overview of orchestration of CD4+ T cell subsets in immune responses. Adv Exp Med Biol. 2014;840:1–13. doi: 10.1007/978-94-017-9487-9_1. [DOI] [PubMed] [Google Scholar]
- 77.Abul K., Abbas A.H.L., Pillai Shiv. 9th ed. Philadelphia: Saunders Elsevier; 2018. Cellular and molecular immunology. [Google Scholar]
- 78.Rosa D.S., Ribeiro S.P., Cunha-Neto E. CD4+ T cell epitope discovery and rational vaccine design. Arch Immunol Ther Exp. 2010;58:121–130. doi: 10.1007/s00005-010-0067-0. [DOI] [PubMed] [Google Scholar]
- 79.Ribeiro S.P., Rosa D.S., Fonseca S.G., Mairena E.C., Postól E., Oliveira S.C. A vaccine encoding conserved promiscuous HIV CD4 epitopes induces broad T cell responses in mice transgenic to multiple common HLA class II molecules. PloS One. 2010;5 doi: 10.1371/journal.pone.0011072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lazarski C.A., Chaves F.A., Jenks S.A., Wu S., Richards K.A., Weaver J.M. The kinetic stability of MHC class II:Peptide complexes is a key parameter that dictates immunodominance. Immunity. 2005;23:29–40. doi: 10.1016/j.immuni.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 81.Bateman A., Martin M.J., O'Donovan C., Magrane M., Alpi E., Antunes R. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu S., Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35:3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murayama K., Nakayama S., Kato-Murayama M., Akasaka R., Ohbayashi N., Kamewari-Hayami Y. Crystal structure of Epstein-Barr virus DNA polymerase processivity factor BMRF1. J Biol Chem. 2009;284:35896–35905. doi: 10.1074/jbc.M109.051581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M., IUCr PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 85.Lovell S.C., Davis I.W., Arendall W.B., de Bakker P.I.W., Word J.M., Prisant M.G. Structure validation by Cα geometry: ϕ,ψ and Cβ deviation. Proteins Struct Funct Bioinforma. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 86.Emini E.A., Hughes J.V., Perlow D.S., Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 1985;55:836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karplus P.A., Schulz G.E. Prediction of chain flexibility in proteins - a tool for the selection of peptide antigens. Naturwissenschaften. 1985;72:212–213. [Google Scholar]
- 88.Kolaskar A.S., Tongaonkar P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172–174. doi: 10.1016/0014-5793(90)80535-q. [DOI] [PubMed] [Google Scholar]
- 89.Parker J.M.R., Guo D., Hodges R.S. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry. 1986;25:5425–5432. doi: 10.1021/bi00367a013. [DOI] [PubMed] [Google Scholar]
- 90.Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R. The immune epitope database (IEDB): 2018 update. Nucleic Acid Res. 2019;47:D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.El-Manzalawy Y., Dobbs D., Honavar V. Predicting linear B-cell epitopes using string kernels. J Mol Recogn. 2008;21:243–255. doi: 10.1002/jmr.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ponomarenko J., Bui H.H., Li W., Fusseder N., Bourne P.E., Sette A. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saha S., Raghava G.P.S. AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acid Res. 2006;34:W202–W209. doi: 10.1093/nar/gkl343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Larsen M.V., Lundegaard C., Lamberth K., Buus S., Lund O., Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinf. 2007;8:424. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Peters B., Bulik S., Tampe R., van Endert P.M., Holzhutter H.-G. Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J Immunol. 2003;171:1741–1749. doi: 10.4049/jimmunol.171.4.1741. [DOI] [PubMed] [Google Scholar]
- 96.Nielsen M., Lundegaard C., Lund O., Keşmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57:33–41. doi: 10.1007/s00251-005-0781-7. [DOI] [PubMed] [Google Scholar]
- 97.Jja Calis, Maybeno M., Greenbaum J.A., Weiskopf D., De Silva A.D., Sette A. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Camproux A.C., Gautier R., Tufféry P. A hidden Markov model derived structural alphabet for proteins. J Mol Biol. 2004;339:591–605. doi: 10.1016/j.jmb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 99.Maupetit J., Tuffery P., Derreumaux P. A coarse-grained protein force field for folding and structure prediction. Proteins Struct Funct Genet. 2007;69:394–408. doi: 10.1002/prot.21505. [DOI] [PubMed] [Google Scholar]
- 100.Thévenet P., Shen Y., Maupetit J., Guyon F., Derreumaux P., Tufféry P. PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acid Res. 2012;40:W288–W293. doi: 10.1093/nar/gks419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Choo J.A.L., Thong S.Y., Yap J., Van Esch W.J.E., Raida M., Meijers R. Bioorthogonal cleavage and exchange of major histocompatibility complex ligands by employing azobenzene-containing peptides. Angew Chem Int Ed. 2014;53:13390–13394. doi: 10.1002/anie.201406295. [DOI] [PubMed] [Google Scholar]
- 102.De Vries S.J., Rey J., Schindler C.E.M., Zacharias M., Tuffery P. The pepATTRACT web server for blind, large-scale peptide-protein docking. Nucleic Acid Res. 2017;45:W361–W364. doi: 10.1093/nar/gkx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schindler C.E.M., De Vries S.J., Zacharias M. Fully blind peptide-protein docking with pepATTRACT. Structure. 2015;23:1507–1515. doi: 10.1016/j.str.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 104.Zacharias M. Protein-protein docking with a reduced protein model accounting for side-chain flexibility. Protein Sci. 2003;12:1271–1282. doi: 10.1110/ps.0239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Case DA, Jtb VB, Betz RM, Cai Q, Cerutti DS, Cheatham TE, et al. Amber 14. San Fr Univ California 2014[cited 2019 Oct 14 ]. Available from https://www.researchgate.net/publication/313508727_AMBER_14_University_of_California_San_Francisco.
- 106.Rodrigues J.P.G.L.M., Trellet M., Schmitz C., Kastritis P., Karaca E., Melquiond A.S.J. Clustering biomolecular complexes by residue contacts similarity. Proteins Struct Funct Bioinforma. 2012;80:1810–1817. doi: 10.1002/prot.24078. [DOI] [PubMed] [Google Scholar]
- 107.Trellet M., Melquiond A.S.J., Bonvin A.M.J.J. A unified conformational selection and induced fit approach to protein-peptide docking. PloS One. 2013;8 doi: 10.1371/journal.pone.0058769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Olotu F.A., Munsamy G., Soliman M.E.S. Does size really matter? Probing the efficacy of structural reduction in the optimization of bioderived compounds – a computational “proof-of-concept”. Comput Struct Biotechnol J. 2018;16:573–586. doi: 10.1016/j.csbj.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Olotu F.A., Agoni C., Adeniji E., Abdullahi M., Soliman M.E. Probing gallate-mediated selectivity and high-affinity binding of epigallocatechin gallate: a way-forward in the design of selective inhibitors for anti-apoptotic bcl-2 proteins. Appl Biochem Biotechnol. 2019;187:1061–1080. doi: 10.1007/s12010-018-2863-7. [DOI] [PubMed] [Google Scholar]
- 110.Case D., Ben-Shalom I., Brozell S., Cerutti D., Cheatham I.I.I. T., Cruzeiro V. Vol. 18. 2018. (AMBER). University of California, San Francisco. [Google Scholar]
- 111.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., Dinola A., Haak J.R., Berendsen H.J.C. Molecular dynamics with coupling to an external bath Molecular dynamics with coupling to an external bath. J Chem Phys. 2012;3684:926–935. [Google Scholar]
- 112.Weber F., Brune S., Börgel F., Lange C., Korpis K., Bednarski P.J. Rigidity versus flexibility: is this an issue in σ 1 receptor ligand affinity and activity? J Med Chem. 2016;59:5505–5519. doi: 10.1021/acs.jmedchem.6b00585. [DOI] [PubMed] [Google Scholar]
- 113.Karshikoff A., Nilsson L., Ladenstein R. Rigidity versus flexibility: the dilemma of understanding protein thermal stability. FEBS J. 2015;282:3899–3917. doi: 10.1111/febs.13343. [DOI] [PubMed] [Google Scholar]
- 114.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C. UCSF Chimera - a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 115.Biovia DS. San Diego: Dassault Systemes. Discovery Studio Visualizer v16.1.0.15350 [software]. 2016 [cited 2019 Oct 14]. Available from: https://www.3ds.com/products-services/biovia/products/#_ga=2.32243323.451562452.1625461054-f3f89660-dd4c-11eb-a73f-7b3fc7418fa5/.
- 116.Seifert E. OriginPro 9.1: scientific data analysis and graphing software—software review. J Chem Inf Model. 2014;54:1552. doi: 10.1021/ci500161d. [DOI] [PubMed] [Google Scholar]
- 117.Schrödinger. New York: Maestro schrödinger, schrödinger release. Version 2019.1 [Software]. 2018 [cited 2019 Oct 4]. Available from: https://www.schrodinger.com/products/maestro/.
- 118.Weng G., Wang E., Wang Z., Liu H., Zhu F., Li D. HawkDock: a web server to predict and analyze the protein–protein complex based on computational docking and MM/GBSA. Nucleic Acid Res. 2019;47:W322–W330. doi: 10.1093/nar/gkz397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ylilauri M., Pentikäinen O.T. MMGBSA as a tool to understand the binding affinities of filamin-peptide interactions. J Chem Inf Model. 2013;53:2626–2633. doi: 10.1021/ci4002475. [DOI] [PubMed] [Google Scholar]
- 120.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expet Opin Drug Discov. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reche P.A., Reinherz E.L. 2007. Prediction of peptide-MHC binding using profiles Methods Mol Biol. 409:185–200. [DOI] [PubMed] [Google Scholar]
- 122.Procko E., Berguig G.Y., Shen B.W., Song Y., Frayo S., Convertine A.J. A computationally designed inhibitor of an Epstein-Barr viral Bcl-2 protein induces apoptosis in infected cells. Cell. 2014;157:1644–1656. doi: 10.1016/j.cell.2014.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gustafson E.A., Chillemi A.C., Sage D.R., Fingeroth J.D. The Epstein-Barr virus thymidine kinase does not phosphorylate ganciclovir or acyclovir and demonstrates a narrow substrate specificity compared to the herpes simplex virus type 1 thymidine kinase. Antimicrob Agents Chemother. 1998;42:2923–2931. doi: 10.1128/aac.42.11.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jensen P.E. Recent advances in antigen processing and presentation. Nat Immunol. 2007;8:1041–1048. doi: 10.1038/ni1516. [DOI] [PubMed] [Google Scholar]
- 125.Vartak A., Sucheck S. Recent advances in subunit vaccine carriers. Vaccines. 2016;4:12. doi: 10.3390/vaccines4020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kloetzel P.M. The proteasome and MHC class I antigen processing. Biochim Biophys Acta Mol Cell Res. 2004;1695:225–233. doi: 10.1016/j.bbamcr.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 127.Ahmad T.A., Eweida A.E., El-Sayed L.H. T-cell epitope mapping for the design of powerful vaccines. Vaccine Rep. 2016;6:13–22. [Google Scholar]
- 128.Wieczorek M., Abualrous E.T., Sticht J., Álvaro-Benito M., Stolzenberg S., Noé F. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front Immunol. 2017;8:292. doi: 10.3389/fimmu.2017.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yaneva R., Schneeweiss C., Zacharias M., Springer S. Peptide binding to MHC class I and II proteins: new avenues from new methods. Mol Immunol. 2010;47:649–657. doi: 10.1016/j.molimm.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 130.Khanna D., Rana P.S. Ensemble technique for prediction of T-cell Mycobacterium tuberculosis epitopes. Interdiscip Sci Comput Life Sci. 2018;11:611–627. doi: 10.1007/s12539-018-0309-0. [DOI] [PubMed] [Google Scholar]
- 131.Sabah S.N., Gazi M.A., Sthity R.A., Husain A.B., Quyyum S.A., Rahman M. Designing of epitope-focused vaccine by targeting E6 and E7 conserved protein sequences: an immuno-informatics approach in human papillomavirus 58 isolates. Interdiscip Sci Comput Life Sci. 2018;10:251–260. doi: 10.1007/s12539-016-0184-5. [DOI] [PubMed] [Google Scholar]
- 132.Gill M.B., Edgar R., May J.S., Stevenson P.G. A gamma-herpesvirus glycoprotein complex manipulates actin to promote viral spread. PloS One. 2008;3:e1808. doi: 10.1371/journal.pone.0001808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xiao J., Palefsky J.M., Herrera R., Berline J., Tugizov S.M. The Epstein-Barr virus BMRF-2 protein facilitates virus attachment to oral epithelial cells. Virology. 2008;370:430–442. doi: 10.1016/j.virol.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xiao J., Palefsky J.M., Herrera R., Berline J., Tugizov S.M. EBV BMRF-2 facilitates cell-to-cell spread of virus within polarized oral epithelial cells. Virology. 2009;388:335–343. doi: 10.1016/j.virol.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gore M., Hutt-Fletcher L.M. The BDLF2 protein of Epstein-Barr virus is a type II glycosylated envelope protein whose processing is dependent on coexpression with the BMRF2 protein. Virology. 2009;383:162–167. doi: 10.1016/j.virol.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Henson B.W., Perkins E.M., Cothran J.E., Desai P. Self-assembly of epstein-barr virus capsids. J Virol. 2009;83:3877–3890. doi: 10.1128/JVI.01733-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gill M.B., Kutok J.L., Fingeroth J.D. Epstein-Barr virus thymidine kinase is a centrosomal resident precisely localized to the periphery of centrioles. J Virol. 2007;81:6523–6535. doi: 10.1128/JVI.00147-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Meng Q., Hagemeier S.R., Fingeroth J.D., Gershburg E., Pagano J.S., Kenney S.C. The epstein-barr virus (EBV)-Encoded protein kinase, EBV-PK, but not the thymidine kinase (EBV-TK), is required for ganciclovir and acyclovir inhibition of lytic viral production. J Virol. 2010;84:4534–4542. doi: 10.1128/JVI.02487-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Asai R., Kato A., Kato K., Kanamori-Koyama M., Sugimoto K., Sairenji T. Epstein-Barr virus protein kinase BGLF4 is a virion tegument protein that dissociates from virions in a phosphorylation-dependent process and phosphorylates the viral immediate-early protein BZLF1. J Virol. 2006;80:5125–5134. doi: 10.1128/JVI.02674-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee C.P., Chen J.Y., Wang J.T., Kimura K., Takemoto A., Lu C.C. Epstein-Barr virus BGLF4 kinase induces premature chromosome condensation through activation of condensin and topoisomerase II. J Virol. 2008;82:3162. doi: 10.1128/JVI.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gershburg E., Raffa S., Torrisi M.R., Pagano J.S. Epstein-Barr virus-encoded protein kinase (BGLF4) is involved in production of infectious virus. J Virol. 2007;81:5407–5412. doi: 10.1128/JVI.02398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lee C.P., Huang Y.H., Lin S.F., Chang Y., Chang Y.H., Takada K. Epstein-Barr virus BGLF4 kinase induces disassembly of the nuclear lamina to facilitate virion production. J Virol. 2008;82:11913–11926. doi: 10.1128/JVI.01100-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chang C.W., Lee C.P., Huang Y.H., Yang P.W., Wang J.T., Chen M.R. Epstein-Barr virus protein kinase BGLF4 targets the nucleus through interaction with nucleoporins. J Virol. 2012;86:8072–8085. doi: 10.1128/JVI.01058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chang C.W., Lee C.P., Su M.T., Tsai C.H., Chen M.R. BGLF4 kinase modulates the structure and transport preference of the nuclear pore complex to facilitate nuclear import of epstein-barr virus lytic proteins. J Virol. 2015;89:1703–1718. doi: 10.1128/JVI.02880-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yang P.W., Chang S.S., Tsai C.H., Chao Y.H., Chen M.R. Effect of phosphorylation on the transactivation activity of Epstein-Barr virus BMRF1, a major target of the viral BGLF4 kinase. J Gen Virol. 2008;89:884–895. doi: 10.1099/vir.0.83546-0. [DOI] [PubMed] [Google Scholar]
- 146.Zeng Y., Middeldorp J., Madjar J.J., Ooka T. A major DNA binding protein encoded by BALF2 open reading frame of Epstein-Barr Virus (EBV) forms a complex with other EBV DNA-binding proteins: DNAase, EA-D, and DNA polymerase. Virology. 1997;239:285–295. doi: 10.1006/viro.1997.8891. [DOI] [PubMed] [Google Scholar]
- 147.Zhang Q., Hong Y., Dorsky D., Holley-Guthrie E., Zalani S., Elshiekh N.A. Functional and physical interactions between the Epstein-Barr virus (EBV) proteins BZLF1 and BMRF1: effects on EBV transcription and lytic replication. J Virol. 1996;70:5131–5142. doi: 10.1128/jvi.70.8.5131-5142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Neuhierl B., Delecluse H.J. The epstein-barr virus BMRF1 gene is essential for lytic virus replication. J Virol. 2006;80:5078–5081. doi: 10.1128/JVI.80.10.5078-5081.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.