Abstract
Spices are often used in dried form, sometimes with significant microbial contamination including pathogenic and food spoilage bacteria. The antibiotic resistance represents an additional risk for food industry, and it is worthy of special attention as spices are important food additives. During our work, we examined the microbiological quality of 50 different spices with cultivation methods on diverse selective media. The identification of the most representative bacteria was carried out using 16S rDNA gene sequence analysis. Antibiotic resistance profiling of twelve identified Bacillus species (B. subtilis subsp. stercoris BCFK, B. licheniformis BCLS, B. siamensis SZBC, B. zhangzhouensis BCTA, B. altitudinis SALKÖ, B. velezensis CVBC, B. cereus SALÖB isolate, B. tequilensis KOPS, B. filamentosus BMBC, B. subtilis subsp. subtilis PRBC2, B. safensis BMPS, and B. mojavensis BCFK2 isolate) was performed using the standard disk‐diffusion method against 32 antibiotics. The study showed that the majority resistance was obtained against penicillin G (100%), oxacillin (91.67%), amoxyclav (91.67%), rifampicin (75%), and azithromycin (75%). Our findings suggest that spices harbor multidrug‐resistant bacteria.
Keywords: antibiotic resistance, Bacillus spp., food, pathogenic bacteria, spices, spoilage bacteria
During our work, we examined the microbiological quality of 50 different spices with cultivation methods on diverse selective media. The identification of the most representative bacteria was carried out using 16S rDNA gene sequence analysis. Antibiotic resistance profiling of twelve identified Bacillus species was performed using the standard disk‐diffusion method against 32 antibiotics. Our findings suggest that spices harbor multidrug‐resistant bacteria
1. INTRODUCTION
Spices are various plant parts that contain flavor and aroma compounds that in small amounts enhance the enjoyment value of foods. Nowadays, spices play a key role in modern food preparation as they contribute to salt reduction, reduce the utilization of artificial additives, impart natural color to foods, and act as natural antioxidants (Clemenson, 2019). Spices have been found to provide health benefits based on their bioactive ingredients. Diverse spices and herbs, namely anise, cinnamon, black cumin, curry, coriander, ginger, fenugreek, turmeric, garlic, mustard, pepper, and onion, are potentially involved in diabetes control (Sanlier & Gencerb, 2020).
Spices are additives used in small quantities, owing to a high potential for microbial contamination as they are used in many food commodities. This possibility may be attributed to the existing various critical points (vulnerabilities) of microbial contamination during plant cultivation and product supply chain (Alegbeleye et al., 2018; Székács et al., 2018). Even though the spices have low water activity, they could be harbored by diverse pathogenic and spoilage microorganisms (Costa et al., 2020; Melo González et al., 2017). As dried herb products, they may be contaminated by different molds too.
Due to the high tolerance to dehydration stress, Salmonella species can survive longer in dry products such as spices. The contamination can occur at several phases of the production including cultivation, harvesting, processing, storage, packaging, and distribution (Zweifel & Stephan, 2012). Different spices and spiced food caused Salmonella outbreaks (Sagoo et al., 2009).
Bacillus species are among the most usually detected bacteria species from diverse spices samples with different geographical origin (Antai, 1988; Banerjee & Sarkar, 2003; Chakraborty et al., 2020; Hariram & Labbé, 2015). Contamination of spices with molds and spore‐forming B. cereus is attributed to environmental conditions of manufacturing and distribution (Fogele et al., 2018). The survival of these microorganisms in spices and herbs is attributed to the high resistance of spores to different stress conditions including excessive pH values, heat, and low water activity (Frentzel et al., 2018). The result of improper food handling is that B. cereus, originating from herbs and spices, can reach the 105–106 CFU/g causing food poisoning (Sagoo et al., 2009). Some strains are capable of producing nonhemolytic enterotoxin, hemolysin BL, and cytotoxin K responsible for diarrhea and emesis causing heat stable cereulide (Frentzel et al., 2018).
Among the spore‐forming bacteria also, Bacillus subtilis and Clostridium perfringens are often present (Sagoo et al., 2009). Other bacteria strains like Staphylococcus aureus, Escherichia coli, and Shigella spp. were detected in different spices (Banerjee & Sarkar, 2003). In contrast to Bacillus spores, S. aureus has a short life cycle even though it supported by the low water activity conditions (Thanh et al., 2018).
In dried vegetables and spices, certain lactic acid bacteria were found, such as Enterococcus spp., Leuconostoc spp., Lactobacillus spp., Weissella confusa, W. cibaria, W. paramesenteroides, Pediococcus acidilactici, and P. pentosaceus (Säde et al., 2016).
The herbs in many cases are used as a fresh cut product without any or with improper treatment to combat microbial contamination. The contaminated plant material can take part in the transmission of pathogens to foods that provide suitable growth conditions and toxin production (Thanh et al., 2018). This increases the health hazard to consumers. The risk is increased in the case of multidrug‐resistant strains present on the plant surface.
The overuse of antibiotics selected the antimicrobial resistance of bacteria, and it represents a virulence factor resulting in a worldwide health threat to humans. Bacterial strains with drug resistance are listed as causes of human mortality (Bennani et al., 2020; Thapa et al., 2020). Bacterial strains with antibiotic resistance can enter the food supply chain from different environments (Dutta & Ramamurthy, 2020; Thapa et al., 2020). Diverse food commodities may act as a reservoir and transmission vector of antibiotic resistance due to contamination by bacteria (Mercimek Takci et al., 2020; Navaneethan & Effarizah, 2021). In food producing animals, bacterial strains like Campylobacter jejuni and Campylobacter coli resistant to quinolones and fluoroquinolones were determined. It was shown that E. coli, Salmonella spp., and C. jejuni harbor resistance to tetracyclines. Another association was found between macrolides and E. coli, Salmonella enterica serovar Heidelberg, and C. coli (Bennani et al., 2020).
The spread of antibiotic resistance genes is an increasing problem, and it is also indicated that clinically relevant resistance could be detected in nonpathogenic bacterial strains as in the case of different Bacillus spp. strains originating from natural ecosystems (Berić et al., 2018). The occurrence of the B. cereus group as opportunistic pathogen has been associated with vegetables. These bacterial groups display resistance to commonly used antibiotics. Multidrug‐resistant strains were also identified among them. These bacteria can contribute to the transfer of antibiotic resistance genes in the food chain (Fiedler et al., 2019; Navaneethan & Effarizah, 2021). According to Zarzecka et al., (2020), there is a risk of transmission of antibiotic resistance genes to pathogenic bacteria like S. aureus, B. cereus and others.
The utilization of antimicrobials exceeding the therapeutic use in food production and agriculture may promote the transmission of antibiotic genes (Wang et al., 2019). To prevent the dissemination of antibiotic resistance, the knowledge and estimation of allochthonous bacteria and their antibiotic pattern are necessary.
This study aimed to evaluate the microbiological quality of different spices and to determine the antibiotic resistance of the most representative bacterial isolates originating from the studied spice. With this study, we attempt to find an answer to the question of whether spices could represent a reservoir of multidrug‐resistant strains and whether their spread in the food supply chain is possible.
2. MATERIALS AND METHODS
2.1. Isolation of bacterial strains
The microbial contamination of 50 different commercially available spices (allspice, Cayenne pepper, marjoram, coriander, basil, cinnamon, granulated garlic, ground garlic, bay leaf, ground pepper, Provence spice mix, rosemary, chili peppers, ginger, star anise, savory, oregano, caraway seeds, ground cumin, tarragon, pepper mix, white mustard, parsley leaf, ground hot peppers, juniper berries, ground turmeric, sweet peppers, cloves, lovage, nutmeg, ground sage, crushed sage, ground vanilla, spice mixture, pepper+garlic mixture, curry, ground anise, green pepper, peppermint, cardamom, mustard seeds, ground cloves, dried celery, dill, parsley) was determined with cultivation methods on different selective media. During the microbiological assay, the total mesophilic aerobic bacteria on Nutrient agar (Himedia) medium were first determined. The incubation was performed at 37℃ for 48 hr. Thereafter, we determined the presence of aerobic spore‐forming B. cereus, E. coli, Salmonella spp., and Pseudomonas species on selective agar mediums. The detection of C. perfringens was carried out in Clostridial Differential Broth (Biolab). In the case of a positive result, a confirmation test (with regenerated milk) was conducted and the enumeration of C. perfringens was determined with the most probable number method. In 5 test tubes containing 10 ml of double concentrated Clostridial Differential Broth, it was added 10 ml of stock suspension from the samples. In 5 test tubes containing 10 ml of Clostridial Differential Broth, it was added 1 ml of stock suspension. 1–1 ml from 10–1 serial dilutions was transferred in 5 test tubes containing 10 ml of standard Clostridium selective broth. In the inoculated tubes, it was added paraffin and incubated for 10 min at 80℃ in water bath. After cooling, it was incubated for 48 hr at 44℃. The positive tubes were counted, and the number of Clostridium perfringens was determined from the MPN table (Drăgan‐Bularda, 2000). For the detection and enumeration of aerobic spore‐forming B. cereus, we used ChromoBio® Cereus Base Agar (Biolab). For the detection and enumeration of E. coli, TBX Chromo Agar (Oxoid) was used. Brilliance TM Salmonella Agar Base (Oxoid) was used for the detection of Salmonella species and Pseudomonas Isolation Agar Base (Himedia) for the detection and enumeration of Pseudomonas spp. The incubation was performed at 37℃ for 48 hr. Bacterial colonies with high number and characteristic colony morphology were isolated, and pure cultures were made.
2.2. Identification of the isolated bacterial strains
The identification of these bacteria was done using 16S rDNA gene sequence analysis. Genomic DNA isolation was carried out with AccuPrep® Genomic DNA Extraction Kit from Bioneer according to the manufacturer's protocol. An universal primer set 27f and 1492r (5’ AGAGTTTGATCMTGGCTCAG 3’, 5’ TACGGYTACCTTGTTACGACTT 3’) was used to amplify one part of the bacterial 16S rDNA gene. The amplification reaction was carried out with an initial denaturation at 94℃ for 5 min, followed by 30 cycles consisting of denaturation at 94℃ for 30 s, primer annealing at 55℃ for 30 s, and primer extension at 72℃ for 1 min, and a final extension at 72℃ for 7 min. Sequencing of the amplified PCR products of the isolated strains was performed by commercial service of Biomi KFT (Hungary) (György et al., 2020). The resulted sequences were edited and aligned with Chromas (Technelysium Pty. Ltd., South Brisbane, Australia); Molecular Evolutionary Genetics Analysis 4 system (www.megasoftware.net) was used forphylogenetic analyses. The isolated bacteria were identified through comparison of the sequences using the EzTaxon server based on the 16SrDNA sequence data (www.ezbiocloud.net/eztaxon).
2.3. Determination of antibiotic resistance
The next part of the research was the determination of antimicrobial resistance of isolated and identified 12 bacterial species belonging to the Bacillus genus (B. cereus SALÖB isolate, B. licheniformis BCLS, B. altitudinis SALKÖ, B. safensis BMPS, B. filamentosus BMBC, B. zhangzhouensis BCTA, B. velezensis CVBC, B. tequilensis KOPS, B. siamensis SZBC, B. mojavensis BCFK2 isolate, B. subtilis subsp. subtilis PRBC2, and B. subtilis subsp. stercoris BCFK) from various spices. The antibiotic susceptibility testing was performed by the disk diffusion method according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines.
A total of 32 different antibiotic disks containing the antibiotics nalidixic acid 30 µg (NA), imipenem 10 µg (IPM), meropenem 10 µg (MRP), rifampicin 5 µg (RIF), tigecycline 15 µg (TGC), erythromycin 15 µg (E), colistin (methane sulfonate) 10 µg (CL), levofloxacin 5 µg (LE), amikacin 30 µg (AK), linezolid 30 µg (LZ), kanamycin 30 µg (K), cefotaxime 30 µg (CTX), ofloxacin 5 µg (OF), clindamycin 2 µg (CD), ceftazidime 30 µg (CAZ), ceftriaxone 30 µg (CTR), teicoplanin 30 µg (TEI), tobramycin 10 µg (TOB), cefoperazone 75 µg (CPZ), azithromycin 15 µg (AZM), oxocillin 1 µg (OX), amoxyclav (amoxicillin/clavulonic acid) 30 µg (AMC), ciprofloxacin 5 µg (CIP), trimethoprim 5 µg (TR), streptomycin 10 µg (S), cefoxitin 30 µg (CX), ampicillin 10 µg (AMP), vancomycin 30 µg (VA), tetracycline 30 µg (TE), gentamicin 10 µg (GEN), penicillin G 10 unite (P), and chloramphenicol 30 µg (C) were used. Onto the surface of Mueller‐Hinton agar medium (Himedia), 0.1ml suspension of bacteria (108 CFU/ml) taken in study was inoculated and the antibiotic disks were placed. This was followed by incubation for 24 hr at 37℃. The diameter (mm) of inhibition zone was measured, and the results of antimicrobial resistance test in accordance with interpretive guidelines and breakpoints were described as susceptible (S), intermediate (I), and resistant (R) (EUCAST, 2016).
2.4. Determination of multiple antibiotic resistance index
The multiple antibiotic resistance (MAR) index of the screened bacterial strains was calculated with the help of the following formula: MAR index =x/y, where (x) is the number of antibiotics, to which the bacteria species was resistant, and (y) refers to the number of tested antibiotics defined for multidrug resistance (Costa et al., 2020).
2.5. Statistical analysis
The principal component analysis was performed with Statistica 8.0 (StatSoft, Inc., Oklahoma, USA).
3. RESULTS
During the evaluation of the microbiological quality of spices, it was determined the aerobic mesophilic bacteria count. If this is high, it can be assumed that pathogenic bacteria may also be present in the samples. The highest total plate count was detected in the case of ground pepper 1⋅5∙107 CFU/g, but a high total colony count was characteristic for turmeric (4⋅105 CFU/g), tarragon (3.8⋅104 CFU/g), and savory1 (1⋅7.104 CFU/g) samples (Table 1). The number of the mesophilic aerobic bacteria was low in green pepper, vanilla, oregano1, spice mixture, and white mustard samples. The plate count of aerobic spore‐forming bacteria was highest in the allspice sample (3.5⋅104 CFU/g), and the lowest count was detected in vanilla and mustard seed samples.
TABLE 1.
Allochthonous bacteria count in studied spices (CFU/g)
| Spice sample and origin | Aerobic mesophilic bacteria | Aerobic spore‐forming bacteria | Bacillus cereus | Escherichia coli | Salmonella spp. | Pseudomonas spp. |
|---|---|---|---|---|---|---|
| Allspice (Hungary) | 2.8⋅104 | 3⋅5.104 | 1.3⋅102 | <10 | <10 | <10 |
| Basil (Egypt) | 9.9⋅103 | 8.7⋅103 | 2⋅10 | <10 | <10 | <10 |
| Bay leaf (Turkey) | 1.4⋅102 | 7.1⋅102 | <10 | <10 | <10 | <10 |
| Caraway seed (Poland) | 1⋅10 | 2⋅10 | <10 | <10 | <10 | <10 |
| Cardamom (Austria) | 1.6⋅10 | 6.6⋅102 | <10 | <10 | <10 | <10 |
| Cayenne pepper (non‐EU) | 3.8⋅103 | 3.1⋅102 | 1⋅10 | <10 | <10 | <10 |
| Chili peppers (Poland) | 2⋅10 | 4⋅10 | <10 | <10 | <10 | <10 |
| Cinnamon (Indonesia) | 5.8⋅10 | 1.1⋅102 | 1⋅10 | <10 | <10 | <10 |
| Cloves1 (Zanzibar) | 3⋅10 | 1⋅10 | <10 | <10 | <10 | <10 |
| Cloves2 (Zanzibar) | 1.1⋅10 | 8⋅10 | <10 | <10 | <10 | <10 |
| Coriander1 (Austria) | 5⋅102 | 4⋅103 | <10 | <10 | <10 | <10 |
| Coriander2 (Austria) | 5.6⋅10 | 2⋅10 | <10 | <10 | <10 | 3⋅2.102 |
| Curry (Poland) | 2.4⋅102 | 2.9⋅102 | <10 | <10 | <10 | <10 |
| Dill (Czech Republic) | 1⋅10 | 1.5⋅102 | <10 | <10 | <10 | <10 |
| Dried celery (Poland) | 2.4⋅10 | 4⋅10 | <10 | <10 | <10 | <10 |
| Ginger (Nigeria) | 3.5⋅10 | 9⋅10 | <10 | <10 | <10 | <10 |
| Granulated garlic (Austria) | 2.5⋅10 | 2.1⋅102 | <10 | <10 | <10 | <10 |
| Green pepper (Austria) | <10 | 1.3⋅102 | <10 | <10 | <10 | <10 |
| Ground anise (Hungary) | 3.3⋅104 | 2.4⋅103 | 6⋅10 | 3.10 | 7⋅102 | <10 |
| Ground cloves (Zanzibar) | 3⋅102 | 7.6⋅102 | 4⋅10 | <10 | <10 | <10 |
| Ground cumin (Hungary) | 4.5⋅103 | 6.4⋅103 | 2⋅10 | <10 | 1.1⋅102 | <10 |
| Ground garlic (China) | 2.7⋅102 | 2.3⋅103 | 6⋅10 | <10 | 3⋅10 | <10 |
| Ground hot peppers (Brazil) | 5⋅102 | 8.7⋅102 | 2⋅10 | <10 | <10 | 1.10 |
| Ground pepper (Brazil) | 1.5⋅107 | 1⋅104 | 5⋅102 | <10 | 3⋅104 | <10 |
| Juniper berries (Macedonia) | 3.5⋅102 | 3.8⋅103 | 1⋅102 | <10 | 3⋅10 | <10 |
| Lovage (Germany) | 1.2⋅10 | 3⋅10 | 1⋅10 | <10 | <10 | <10 |
| Marjoram (Germany) | 3.8⋅102 | 1.1⋅103 | <10 | <10 | <10 | <10 |
| Pepper mix (Poland) | 1⋅10 | 2⋅10 | <10 | <10 | <10 | <10 |
| Mustard seed (Moldova) | <10 | <10 | <10 | <10 | <10 | <10 |
| Nutmeg (Indonesia) | 4⋅102 | 2⋅102 | <10 | <10 | <10 | <10 |
| Oregano1 (Turkey) | <10 | 1⋅10 | <10 | <10 | <10 | <10 |
| Oregano2 (Turkey) | 4.3⋅104 | 1⋅102 | <10 | <10 | 4.4⋅103 | 1.1⋅102 |
| Parsley (Poland) | 6.2⋅103 | 3.1⋅103 | 5⋅102 | <10 | 7⋅10 | <10 |
| Parsley leaf (Poland) | 1.4⋅10 | 4⋅102 | <10 | <10 | <10 | <10 |
| Pepper +garlic (Poland) | 2.9⋅10 | 7⋅102 | <10 | <10 | <10 | <10 |
| Peppermint (Egypt) | 9.4⋅103 | 1.67⋅104 | 1.1⋅102 | <10 | 3.2⋅103 | 6⋅10 |
| Provence spice mix (Austria) | 3⋅102 | 5.6⋅103 | <10 | <10 | <10 | <10 |
| Rosemary (Morocco) | 3⋅10 | 3⋅10 | <10 | <10 | <10 | <10 |
| Sage1 (Turkey) | 1.1⋅102 | 1.4⋅102 | <10 | <10 | <10 | <10 |
| Sage2 (Austria) | 3.6⋅10 | 2.7⋅102 | <10 | <10 | <10 | <10 |
| Savory1 (Hungary) | 1.7⋅104 | 1.2⋅103 | <10 | 5⋅103 | 2.7⋅103 | 5⋅102 |
| Savory2 (Hungary) | 1.4⋅103 | 8⋅10 | <10 | <10 | <10 | <10 |
| Spice mixture (Poland) | <10 | 1.4⋅102 | <10 | <10 | <10 | <10 |
| Star anise (Vietnam) | 7.6⋅103 | 2.4⋅103 | <10 | 5⋅103 | <10 | <10 |
| Sweet pepper (China) | 6.2⋅102 | 5.3⋅102 | 7⋅10 | 5⋅10 | 9⋅10 | <10 |
| Tarragon1 (Poland) | 3.8⋅104 | 6⋅10 | <10 | 3⋅103 | 5.7⋅103 | 1.5⋅103 |
| Tarragon2 (Poland) | 3.3⋅102 | 1.8⋅102 | <10 | <10 | <10 | <10 |
| Turmeric (India) | 4⋅105 | 2.2⋅102 | <10 | 7⋅10 | 1.6⋅102 | 1.4⋅102 |
| Vanilla (Madagascar) | <10 | <10 | <10 | <10 | <10 | <10 |
| White mustard (Poland) | <10 | 1⋅10 | <10 | <10 | <10 | <10 |
Within the group of aerobic spore‐forming bacteria, typical B. cereus colonies were developed from 15 spice samples, and high count was detected in the case of ground pepper and parsley. Pseudomonas species, that play a role in food spoilage, were generally present in small numbers in the assayed spices, with the exception of tarragon1, coriander2, turmeric, savory1, and oregano2 samples. The hygienic indicator bacteria E. coli were detected in seven samples, whereas the colony count reached 103 CFU/g in samples star anise, savory1, and tarragon1. The presence of Salmonella spp. on the selective medium was found in 12 spice samples, and the highest count was detected in the case of ground pepper. Also, fecal origin C. perfringens, anaerobic spore‐forming bacterium, was detected in 7 spice samples: cinnamon, basil, tarragon1, ground pepper, ground cumin, peppermint and sage2. The MPNs of this bacteria were distributed from 5 (in cinnamon) to 1.6⋅103 (in ground cumin and peppermint), respectively, 9 in ground pepper and sage2. In tarragon1, the MPNs were 1.41⋅102 and 5.42⋅102 in basil. Among the examined spices, good microbiological quality with low or no microbiological load could be observed in the case of vanilla, rosemary, chili peppers, ginger, oregano1, cumin seeds, pepper mix, white mustard, cloves1, spice mixture, green pepper, and mustard seed samples.
In the case of some spices (star anise, peppermint, tarragon1, parsley, ground pepper, turmeric, allspice, ground anise), higher numbers of allochthonous bacteria were present, which may result from the poor conditions of cultivation, storage, and distribution.
A total number of 20 bacterial isolates originated from different spices on diverse selective mediums were identified by partial 16S rDNA sequencing. The bacterial isolates were selected based on colony morphology and provenience. The results show (Table 2) that the bacterial isolates belonged to 3 genera: 18 species from Bacillus genera, one species from Staphylococcus, and one species from Salinicoccus spp. Among the identified bacterial species, 15 different species were detected from the dried plant materials.
TABLE 2.
The source and identification of the bacterial strains isolated from the different spices
| Bacterial isolate ID | Source of isolation | Isolation medium | Identified closely related species based on 16S rDNA | Sequence similarity% |
|---|---|---|---|---|
| BCFK | Spice mix | ChromoBio® Cereus Base | Bacillus subtilis subsp. stercoris | 98.39 |
| BCLS | Lovage | ChromoBio® Cereus Base | Bacillus licheniformis | 99.50 |
| ORPS2 | Oregano | Pseudomonas Isolation Agar Base | Bacillus tequilensis | 99.20 |
| KOPS | Coriander | Pseudomonas Isolation Agar Base | Bacillus tequilensis | 99.90 |
| CUBC | Curry | ChromoBio® Cereus Base | Bacillus velezensis | 99.30 |
| SALTARK | Tarragon | Brilliance TH Salmonella Agar Base | Bacillus zhangzhouensis | 99.30 |
| PRBC2 | Parsley leaf | ChromoBio® Cereus Base | Bacillus subtilis subsp. subtilis | 96.85 |
| SZBC | Ground cloves | ChromoBio® Cereus Base | Bacillus siamensis | 97.89 |
| PRBC1 | Parsley leaf | ChromoBio® Cereus Base | Bacillus zhangzhouensis | 98.91 |
| KNA | Dill | Nutrient Agar | Bacillus zhangzhouensis | 98.01 |
| BCFK2 | Spice mix | ChromoBio® Cereus Base | Bacillus mojavensis | 78.91 |
| BCTA | Tarragon | ChromoBio® Cereus Base | Bacillus zhangzhouensis | 99.50 |
| BCPR | Sweet peppers | ChromoBio® Cereus Base | Salinicoccus spp. | 84.62 |
| BMPS | Peppermint | Pseudomonas Isolation Agar Base | Bacillus safensis | 99.70 |
| BMBC | Peppermint | ChromoBio® Cereus Base | Bacillus filamentosus | 99.18 |
| ÁNTBX | Ground anise | TBX Chromo agar | Staphylococcus warneri | 96.00 |
| ÁNSAL | Ground anise | Brilliance TH Salmonella Agar Base | Bacillus altitudinis | 99.13 |
| BCKÖ | Ground cumin | ChromoBio® Cereus Base | Bacillus siamensis | 99.90 |
| SALÖB | Ground pepper | Brilliance TH Salmonella Agar Base | Bacillus cereus | 80.85 |
| SALKÖ | Ground cumin | Brilliance TH Salmonella Agar Base | Bacillus altitudinis | 100 |
Because the similarity percentage in the case of SALÖB isolate (B. cereus identity 80.85%) and BCFK2 isolate (B. mojavensis 78.91%) is very low, these bacteria are marked with isolate designation.
The principal component analysis (PCA) was carried out on the measured values (inhibition zone diameters). PCA was applied to classify the identified bacterial strains based on the inhibition of the different antibiotics. The results of multivariate data analysis give some indication of the complete data set, showing the variability among the bacterial strains regarding antibiotic pattern. The principal components, F1 accounts for 55.10% cumulative variability, F2 for 14.83% from total variability, explained 69.93% of total variability which was high enough to represent all the variables.
Half of the bacterial strains were clustered in the upper part of PC2 axis with high PC2 values in F1‐F2 biplot (Figure 1). B. tequilensis, B. subtilis subsp. subtilis, and B. mojavensis were situated in the upper left quadrant. 12 antibiotics did not affect the growth of these isolates. B. zhangzhouensis and B. altitudinis were clustered in the upper right zone, were characterized by almost the same mean of inhibition zone, except for penicillin G, imipenem, meropenem, streptomycin, erythromycin, amoxyclav, and oxacillin. B. filamentosus, B. cereus isolate, B. safensis, and B. velezensis were clustered on the left with low negative PC1 values. These bacteria were characterized by 0 mm zone size diameter or low values in the case of the majority of antibiotics. B. lichenformis, B. siamensis, and B. subtilis subsp. stercoris were in the lower right quadrant. All three displayed susceptibility to the same twenty antibiotics.
FIGURE 1.
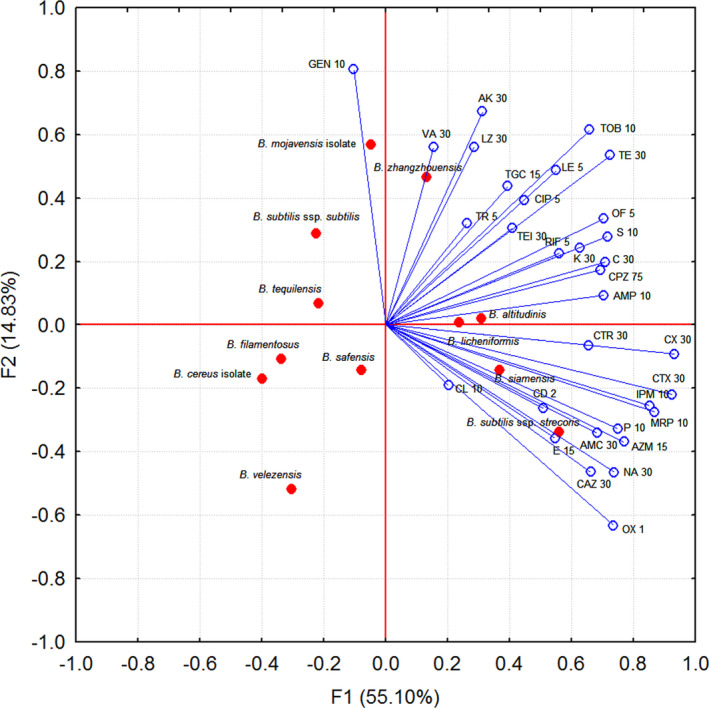
PCA analysis of antibiotic inhibition zones means of the bacterial strains isolated from spices
Because of the lack of EUCAST guidelines to determine resistance of Bacillus strains, the criteria for the Gram‐positive Staphylococcus group were used in our study. According to the results of antibiotic resistance, all the tested Bacillus species strains were generally resistant to penicillin G (100%), amoxyclav (91.67%), and oxacillin (91.7%). The majority of the strains (66.67%) showed resistance to clindamycin, imipenem, and meropenem. More than half of the tested bacterial strains displayed resistance to erythromycin, cephotaxime, cefoxitin, and nalidixic acid. Half of the strains exhibited resistance to ceftazidime, streptomycin, and cefoperazone. Kanamycin and chloramphenicol inhibited the growth of four bacterial strains, while ofloxacin inhibited the growth of one bacterial strain. All tested bacterial strains were highly sensitive (100%) to eight antibiotics: ciprofloxacin, linezolid, vancomycin, amikacin, trimethoprim, gentamicin, teicoplanin, tigecycline, and colistin (92.7%) (Table 3).
TABLE 3.
Antibiotic resistance pattern of bacterial strains isolated from spices
| Bacterial strain | Antibiotic resistance |
|---|---|
| Bacillus subtilis subsp. stercoris BCFK | P, CD |
| Bacillus licheniformis BCLS | P, CPZ, CD, C, AMC,OX,CL |
| Bacillus siamensis SZBC | P, RIF, AMC, OX |
| Bacillus zhangzhouensis BCTA | P, RIF,AZM, IPM, MRP,CTX, AMC, OX, NA |
| Bacillus altitudinis SALKÖ | P, RIF, AZM, S, E, AMC, OX |
| Bacillus velezensis CVBC | CX, P, CPZ, RIF, AZM, LE, IPM, MRP, S, C, TE, CTX, AMC, OF, OX, CTR, TOB, AMP |
| Bacillus cereus SALÖB | CX, P, CPZ, RIF, AZM, LE,CAZ, CD, IPM, MRP, S, C, E, CTX, AMC, K, OF, OX, TOB, AMP, NA |
| Bacillus tequilensis KOPS | CX, P, RIF, AZM, CAZ, CD, IPM, MRP, E, CTX, AMC, OF, OX, CTR, AMP, NA |
| Bacillus filamentosus BMBC | CX, P, CPZ, RIF, AZM, CAZ, CD, IPM, MRP, S, C, E, CTX, AMC, K, OX, TOB, NA |
| Bacillus subtilis subsp. subtilis PRBC2 | CX, P, RIF, AZM, CAZ, CD, IPM, MRP, S, E, CTX, AMC,K, OX, AMP, NA |
| Bacillus safensis BMPS | CX, P, RIF, AZM, CAZ, CD, IPM, MRP, E, AMC,K, OX, TOB, NA |
| Bacillus mojavensis BCFK2 | CX, P, AZM, CAZ, CD, IPM, MRP, S, E, CTX, AMC, OX, NA |
CX cefoxitin, P penicillin G, CPZ cefoperazone, RIF rifampicin, AZM azithromycin, LE levofloxacin, LZ linezolid, CAZ ceftazidime, CD clindamycin, IPM imipenem, MRP meropenem, S streptomycin, C chloramphenicol, TE tetracycline, E erythromycin, CTX cephotaxime, AMC amoxyclav, OF ofloxacin, OX oxacillin, CTR ceftriaxone, TOB tobramycin, AMP ampicillin, NA nalidixic acid, TEI teicoplanin, CL colistin.
Four bacterial strains were generally susceptible (33.3%) to seven antibiotics: cefoxitin, cefoperazone, imipenem, meropenem, erythromycin, cephotaxime, and nalidixic acid. Susceptible strains were also determined for levofloxacin (58.3%), ceftazidime (41.7%), chloramphenicol (66.7%), kanamycin (66.7%), ceftriaxone (41.7%), tobramycin (66.7%), and ampicillin (66.7%).
Strains with intermediate susceptibility were also determined for cefoxitin (8.33%), cefoperazone (16.7%), levofloxacin (25%), ceftazidime (8.33%), clindamycin (8.33%), streptomycin (25%), tetracycline (16.7%), erythromycin (8.33%), cephotaxime (8.33%), ceftriaxone (41.7%), and nalidixic acid (8.33%).
The multiple antibiotic resistance (MAR) index was taken into account (the antimicrobial categories and agents) used to define multidrug resistance (due to the lack of data for Bacillus strains regarding the antibiotics used to define MAR we applied the antibiotics proposed for S. aureus) (Magiorakos et al., 2012).
When a bacteria strain displays resistance to more than two antibiotics or to at least one antibiotic in three or more categories (Table 3), it can be noted as multiple antibiotic resistance (MAR) bacteria. The multiple antibiotic resistance (MAR) index of the twelve isolated and identified Bacillus strains is presented in Figure 2. MAR index of seven out of twelve bacterial strains (B. velezensis CVBC, B. cereus SALÖB isolate, B. tequilensis KOPS, B. filamentosus BMBC, B. subtilis subsp. subtilis PRBC2, B. safensis BMPS, and B. mojavensis BCFK2 isolate) is equal to or higher than 0.23. MAR index values in the case of five bacteria strains are below 0.2. B. zhangzhouensis BCTA, B. altitudinis SALKÖ, B. licheniformis BCLS, Bacillus siamensis SZBC, and Bacillus subtilis subsp. stercoris BCFK possess MAR index in the interval 0.08–0.15. Based on these criteria, seven bacteria strains can be defined as multi antibiotic resistant strains. The values of MAR index higher than 0.2 indicate high‐risk sources of contamination.
FIGURE 2.
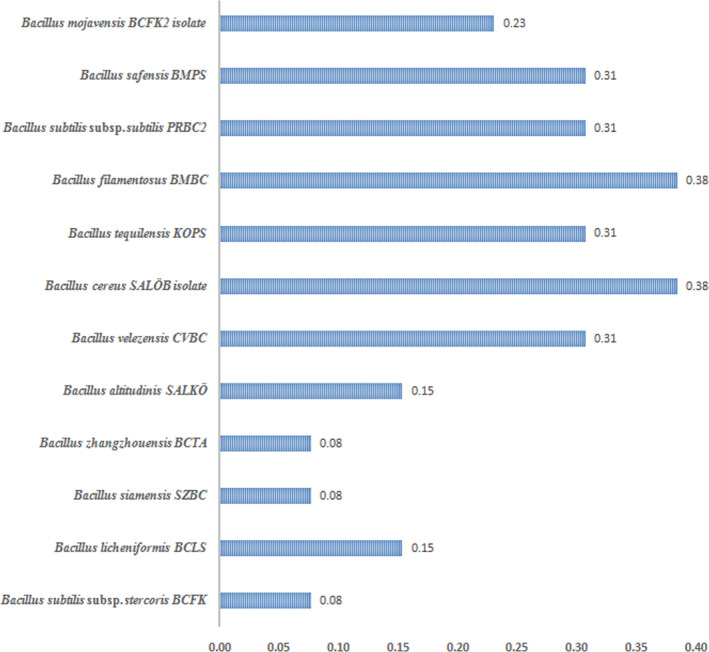
Multiple antibiotic resistance (MAR) index values of Bacillus species isolated from spices
4. DISCUSSION
Isolation and identification of antibiotic resistant bacteria from different ecosystems including food commodities are carried out by direct plating methods and 16S rRNA gene sequence analysis (Leboffe & Pierce, 2011; Poretsky et al., 2014; Sentausa & Fournier, 2013; Wang, Zhou, et al., 2019).
Microbial quality determinations have shown that spices and herbs can reach high microbial counts of up to 108 colony forming units (CFU/g). Among the isolated microorganisms, there are also pathogenic species that can be responsible for diverse foodborne diseases. The most relevant bacteria strains are B. cereus, C. perfringens, and Salmonella spp. Among the foodborne pathogens were detected S. aureus too (Thanh et al., 2018).
The lack of or the low microbial load of different spices could be attributed to the bioactive components with antimicrobial properties (Costa et al., 2020; Thanh et al., 2018). Bata‐Vidács et al., (2018) determining the microbiological quality of seventy‐one spice paprika samples from 10 countries (Hungary, China, Serbia, Spain, India, Bulgaria, Brazil, Peru, Kenya, Thailand, and unknown place) concluded that the dominant microorganisms in spice paprika samples are influenced by different factors such as the climate of cultivation. It was revealed that bacteria species are related to geographical origin. It was shown that B. mycoides and B. licheniformis take part in the characteristic microflora of spice paprika from Central Europe. The presence of B. safensis was determined in samples grown in the tropical monsoon climate. In samples from Spain was detected B. amyloliquefaciens subsp. plantarum, B. amyloliquefaciens subsp. amyloliquefaciens, and B. mojavensis. There were reported outbreaks caused by spices contaminated with B. cereus and B. subtilis (Moore et al., 2019). Some of these strains listed above were detected in our samples too. Bacillus strains detected in our samples possess different attributes described as appearing in different environmental conditions. B. licheniformis is often involved in food spoilage because of the extremely resistant endospore forming ability, like in milk powder. It was shown that the endospore is highly resistant to different factors like desiccation, heat, and irradiation. This resistance was attributed to the fact that spores germinate at a reduced rate compared to other bacteria belonging to the B. subtilis group (Aspholm et al., 2019; Wang et al., 2019). B. mojavensis was reported as endophyte bacteria, isolated from maize showing plant growth promoting benefits and antifungal activity.
The spreading of the antibiotic resistant bacteria and genes represents major public health concerns in the different environments of food commodities like fruits, vegetables, and foods from animals. Preventive measure has been proposed, including awareness of antibiotic resistance and monitoring (Mancilla‐Becerra, Lías‐Macías, Ramírez‐Jiménez, & León, 2019; Thapa et al., 2020). Consumption of foods with spices without any heat treatment that can harbor multidrug‐resistant bacteria represent a danger for immune depressed individuals. These bacteria strains may survive gastrointestinal tract complicating the treatment in persons with lowered immune function (Fiedler et al., 2019).
Antibiotic resistance develops when the bacteria with different mechanisms (target changes as modifications of the penicillin binding protein (PBP), enzymatic inhibition, porin mutations, efflux pumps) survive the effect of antibiotics. The antibiotic mechanisms are named: intrinsic resistance, acquired resistance, genetic changes in DNA, and DNA transfer (Breijyeh et al., 2020). It was confirmed that some classes of antibiotic resistance gene can be acquired. Acquired resistance has confirmed to the most of the standard antibiotics and evaluated in different bacteria (Gold & Moellering, 1996; Kapoor et al., 2017; Mancilla‐Becerra et al., 2019). According to WHO priority list of antibiotic resistant bacteria, S. aureus possess high priority, but other bacteria strains are also classified as serious (Breijyeh et al., 2020; Kumar et al., 2020).
Limited scientific information is available regarding the antibiotic resistance of Bacillus strains and the transfer of antibiotic resistance genes. According to Alanber et al., (2020), Bacillus spp. are able to transfer the antibiotic resistance genes. Agersø et al., (2019) suggested that in B. licheniformis, putative genes correlated with the resistance of streptomycin, erythromycin, and chloramphenicol are present in the chromosome and are intrinsic, which means that they are not transferable to other bacteria and consequently, will not take part in the pool of resistance genes. According to Muriuki et al., (2020), vended fast food carries antibiotic resistant bacteria as B. velezensis isolated from fish, with tetracycline, chloramphenicol, cefotaxime, and nalidixic acid resistance. B. subtilis isolated from vegetable salads exhibited resistance to streptomycin, gentamycin, amoxicillin, tetracycline, chloramphenicol, cefotaxime, nalidixic acid, and trimethoprim +sulfamethoxazole. B. siamensis B44, originating from pickled vegetables, displayed susceptibility to 14 antibiotics, whereas the other isolates as Bacillus spp. strain B51f were resistant to clindamycin, chloramphenicol, penicillin G, and cephalothin (Meidong et al., 2017).
A bacterial isolate originating from a river was identified as B. altitudinis with resistance to three groups of antibiotics beta lactams, lincomycin, and polymyxin B (Lobova et al., 2015). A similar pattern of results was also obtained by Alanber et al., (2020). Generally, the isolated Bacillus strains from powdered infant milk and from spices (in our study) were susceptible including amikacin, gentamicin, and imipenem. The B. cereus displayed resistance to ceftazidime and B. licheniformis showed resistance to cefoperazone and B. subtilis was susceptible to ceftazidime. The results regarding the enterotoxin producing foodborne pathogen B. cereus are in accordance with findings confirmed by Fiedler et al., (2019). This bacteria strain was detected also in dried herbs and spices showing resistance to standard antibiotics including chloramphenicol, cefotaxime, erythromycin, penicillin, and imipenem. Fei et al., (2020) revealed a high B. cereus contamination level in powdered foods, where all the isolates displayed ampicillin, oxacillin, and rifampicin resistance, as in this study.
The multidrug resistance of 12 Bacillus strains against 32 antibiotics is impossible to visualize graphically without using dimension reduction statistical methods. For this purpose, PCA is a widely used and important multivariate statistical approach (Jolliffe & Cadima, 2016; Kassambara, 2017) and could be applied in classifying the antibiotic resistance bacteria. The PCA results showed different groupings of the tested bacteria. In some previous studies also, it applied this method for reduction of multivariate data to two or three principal components. Zhang et al., (2013) carried on PCA and cluster analysis in explaining antibiotic resistance indicators in E. coli isolated from aquaculture, indicating that MAR index defined detailed the antibiotic resistance profile. Mandal et al., (2020) performed PCA for assessing the resistance profiles and association of heavy metal tolerance and antibiotic resistance of clinically relevant bacteria, grouped into three factors with high total variance.
5. CONCLUSION
The study has revealed that the assayed 50 spices are moderately contaminated. It was shown that among the identified bacteria occur pathogenic and food spoilage bacteria, as B. cereus.
The isolated and identified Bacillus species exhibited different resistance to the tested antibiotics, showing resistance at least against two antimicrobials. All tested strains showed susceptibility to eight antibiotics. The presence of bacteria with multiple antibiotic resistance in the tested products should be considered a health concern. From the results of the present study, it can be concluded that improperly processed or unwashed raw herb spices could contribute to the transfer of multidrug resistance bacteria through the supply chain. Further studies are needed to confirm with accuracy the minimal inhibitory concentration and detection of genetic elements associated with antibiotic resistance in case of bacteria with human health concern.
CONFLICT OF INTEREST
The authors declare that they do not have any conflict of interest.
AUTHOR CONTRIBUTIONS
Éva György: Conceptualization (equal); Investigation (equal); Methodology (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Éva Laslo: Conceptualization (equal); Investigation (equal); Methodology (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Márta Antal: Investigation (equal). András Csaba Dezső: Data curation (equal); Formal analysis (equal); Visualization (equal).
György, É., Laslo, É., Antal, M., & András, C. D.. Antibiotic resistance pattern of the allochthonous bacteria isolated from commercially available spices. Food Science & Nutrition. 2021;9, 4550–4560. 10.1002/fsn3.2433
REFERENCES
- Agersø, Y., Bjerre, K., Brockmann, E., Johansen, E., Nielsen, B., Siezen, R., Stuer‐Lauridsen, B., Wels, M., & Zeidan, A. A. (2019). Putative antibiotic resistance genes present in extant Bacillus licheniformis and Bacillus paralicheniformis strains are probably intrinsic and part of the ancient resistome. PLoS One, 14(1), e0210363. 10.1371/journal.pone.0210363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alanber, M. N., Alharbi, N. S., & Khaled, J. M. (2020). Evaluation of multidrug‐resistant Bacillus strains causing public health risks in powdered infant milk formulas. Journal of Infection and Public Health, 13, 1462–1468. 10.1016/j.jiph.2019.11.013 [DOI] [PubMed] [Google Scholar]
- Alegbeleye, O. O., Singleton, I., & Sant'Ana, A. S. (2018). Sources and contamination routes of microbial pathogens to fresh produce during field cultivation: A review. Food Microbiology., 73, 177–208. 10.1016/j.fm.2018.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antai, S. P. (1988). Study of the Bacillus flora of Nigerian spices. International Journal of Food Microbiology, 6, 259–261. 10.1016/0168-1605(88)90018-9 [DOI] [PubMed] [Google Scholar]
- Aspholm, M. E., Kollerud, K. K., Hansen, H. C. H., Granum, P. E., Christie, G., & Lindback, T. (2019). Biochemical and mutational analysis of spore cortex‐lytic enzymes in the food spoiler Bacillus licheniformis . Food Microbiology, 84, 103259. 10.1016/j.fm.2019.103259 [DOI] [PubMed] [Google Scholar]
- Banerjee, M., & Sarkar, P. K. (2003). Microbiological quality of some retail spices in India. Food Research International, 36, 469–474. 10.1016/S0963-9969(02)00194-1 [DOI] [Google Scholar]
- Bata‐Vidács, I., Baka, E., Tóth, Á., Csernus, O., Sz, L., Adányi, N., Székács, A., & Kukolya, J. (2018). Investigation of regional differences of the dominant microflora of spice paprika by molecular methods. Food Control, 83, 109–117. 10.1016/j.foodcont.2017.04.030 [DOI] [Google Scholar]
- Bennani, H., Mateus, A., Mays, N., Eastmure, E., Stärk, K. D. C., & Häsler, B. (2020). Overview of evidence of antimicrobial use and antimicrobial resistance in the food chain. Antibiotics, 9, 49. 10.3390/antibiotics9020049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berić, T., Biočanin, M., Stanković, S., Dimkić, I., Janakiev, T., Fira, D., & Lozo, J. (2018). Identification and antibiotic resistance of Bacillus spp. isolates from natural samples. Archives of Biological Sciences, 70, 581–588. 10.2298/ABS180302019B [DOI] [Google Scholar]
- Breijyeh, Z., Jubeh, B., & Karaman, R. (2020). Resistance of gram‐negative bacteria to current antibacterial agents and approaches to resolve it. Molecules, 25(6), 1340. 10.3390/molecules25061340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, M., Afrin, T., & Munshi, S. K. (2020). Microbiological quality and antimicrobial potential of extracts of different spices. Food Research, 4(2), 375–379. 10.26656/fr.2017.4(2).303 [DOI] [Google Scholar]
- Clemenson, Steve (2019). Herbs and spices. In Swainson M. (Ed.), Swainson's Handbook of Technical and Quality Management for the Food Manufacturing Sector (pp. 433–455). Woodhead Publishing Series in Food Science, Technology and Nutrition. [Google Scholar]
- Costa, M. D. C., Cruz, A. I. C., Bispo, A. S. D. R., Ferreira, M. A., Costa, J. A., & Evangelista‐Barreto, N. S. (2020). Occurrence and antimicrobial resistance of bacteria in retail market spices/Ocorrencia e resistencia antimicrobiana de bacterias em especiarias comercializadas no varejo. Ciência Rural, 50(4), 10.1590/0103-8478cr20190775 [DOI] [Google Scholar]
- Drăgan‐Bularda, M. (2000). Microbiologie generală (General Microbiology), 3rd ed. (pp. 222–227). Babeș‐Bolyai University. [Google Scholar]
- Dutta, S., & Ramamurthy, T. (2020). Influence of abiotic factors in the emergence of antibiotic resistance. In Thomas S. (Ed.), Antimicrobial resistance, global challenges and future interventions (pp. 81–100). Springer. [Google Scholar]
- Fei, P., Xie, Q., Jiang, Y., Feng, H., Chang, Y., Kang, H., Xing, M., & Chen, J. (2020). Genotyping, antimicrobial susceptibility and biofilm formation of Bacillus cereus isolated from powdered food products in China. Foodborne Pathogens and Disease, 17, 8–15. 10.1089/fpd.2020.2802 [DOI] [PubMed] [Google Scholar]
- Fiedler, G., Schneider, C., Igbinosa, E. O., Kabisch, J., Brinks, E., Becker, B., Stoll, D. A., Cho, G.‐S., Huch, M., & Franz, C. M. A. P. (2019). Antibiotics resistance and toxin profiles of Bacillus cereus‐group isolates from fresh vegetables from German retail markets. BMC Microbiology, 19, 250. 10.1186/s12866-019-1632-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogele, B., Granta, R., Valciņa, O., & Bērziņš, A. (2018). Occurrence and diversity of Bacillus cereus and moulds in spices and herbs. Food Control, 83, 69–74. 10.1016/j.foodcont.2017.05.038 [DOI] [Google Scholar]
- Frentzel, H., Thanh, M. D., Krause, G., Appel, B., & Mader, A. (2018). Quantification and differentiation of Bacillus cereus group species in spices and herbs by real‐time PCR. Food Control, 83, 99–108. 10.1016/j.foodcont.2016.11.028 [DOI] [Google Scholar]
- György, É., Laslo, É., Kuzman, I. H., & Dezső András, C. (2020). The effect of essential oils and their combinations on bacteria from the surface of fresh vegetables. Food Science & Nutrition, 8(10), 5601–5611. 10.1002/fsn3.1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariram, U., & Labbé, R. (2015). Spore prevalence and toxigenicity of Bacillus cereus and Bacillus thuringiensis isolates from U.S. retail spices. Journal of Food Protection, 78(3), 590–596. 10.4315/0362-028X.JFP-14-380 [DOI] [PubMed] [Google Scholar]
- Gold, H. S., & Moellering, R. C. (1996). Antimicrobial‐drug resistance. The New England Journal of Medicine, 335(19), 1445–1453. [DOI] [PubMed] [Google Scholar]
- Jolliffe, I. T., & Cadima, J. (2016). Principal component analysis: A review and recent developments. Philosophical Transactions of the Royal Society A, 374, 20150202. 10.1098/rsta.2015.0202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, G., Saigal, S., & Elongavan, A. (2017). Action and resistance mechanisms of antibiotics: A guide for clinicians. Journal of Anaesthesiology Clinical Pharmacology, 33, 300–305. 10.4103/joacp.JOACP_349_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara, A. (2017). Practical guide to principal component methods in R, 1st ed. (pp. 12–42). [Google Scholar]
- Kumar, S. B., Arnipalli, S. R., & Ziouzenkova, O. (2020). Antibiotics in food chain: The consequences for antibiotic resistance. Antibiotics, 9(10), 6889. 10.3390/antibiotics9100688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leboffe, M. J., & Pierce, B. E. (2011). A Photographic atlas for the microbiology laboratory, 4th ed. (pp. 5–7). Morton Publishing Company. [Google Scholar]
- Lobova, T. I., Yemelyanova, E., Andreeva, I. S., Puchkova, L. I., & Repin, V. Y. (2015). Antimicrobial resistance and plasmid profile of bacterial strains isolated from the urbanized Eltsovka‐1 River (Russia). Microbial. Drug Resistance, 21, 477–490. 10.1089/mdr.2014.0203 [DOI] [PubMed] [Google Scholar]
- Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., Harbarth, S., Hindler, J. F., Kahlmeter, G., Olsson‐Liljequist, B., Paterson, D. L., Rice, L. B., Stelling, J., Struelens, M. J., Vatopoulos, A., Weber, J. T., & Monnet, D. L. (2012). Multidrug‐resistant, extensively drug‐resistant and pandrug‐resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection, 18(3), 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Mancilla‐Becerra, L. M., Lías‐Macías, T., Ramírez‐Jiménez, C. L., & León, J. B. (2019). Multidrug‐resistant bacterial foodborne pathogens: impact on human health and economy (pp. 1–17). Pathogenic Bacteria; IntechOpen. [Google Scholar]
- Mandal, M., Das, S. N., & Mandal, S. (2020). Principal component analysis exploring the association between antibiotic resistance and heavy metal tolerance of plasmid‐bearing sewage wastewater bacteria of clinical relevance. Access. Microbiology, 2(3). 10.1099/acmi.0.000095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meidong, R., Doolgindachbaporn, S., Jamjan, W., Sakai, K., Tashiro, Y., Okugawa, Y., & Tongpim, S. (2017). A novel probiotic Bacillus siamensis B44v isolated from Thai pickled vegetables (Phak‐dong) for potential use as a feed supplement in aquaculture. The Journal of General Applied Microbiology, 63, 246–253. 10.2323/jgam.2016.12.002 [DOI] [PubMed] [Google Scholar]
- Melo González, M. G., Romero, S. M., Arjona, M., Larumbe, A. G., & Vaamonde, G. (2017). Microbiological quality of Argentinian paprika. Revista Argentina De Microbiología, 49(4), 339–346. 10.1016/j.ram.2017.02.006 [DOI] [PubMed] [Google Scholar]
- Mercimek Takci, H. A., Bakirhan, P., Ozdemir, E., & Yalcin, A. (2020). Antibiotic susceptibility patterns of biofilm producing gram negative bacilli isolated from Kilis local cheese (Food‐related antibiotic resistance). Banat's Journal of Biotechnology, XI, 21, 58–63. 10.7904/2068-4738-XI(21)-58 [DOI] [Google Scholar]
- Moore, R. E., Millar, B. C., Panickar, J. R., & Moore, J. E. (2019). Microbiological safety of spices and their interaction with antibiotics: Implications for antimicrobial resistance and their role as potential antibiotic adjuncts. Food Quality and Safety, 3, 93–97. 10.1093/fqsafe/fyz008 [DOI] [Google Scholar]
- Muriuki, S. W., Neondo, J. O., & Budambula, N. L. M. (2020). Detection and profiling of antibiotic resistance among culturable bacterial isolates in vended food and soil samples. International Journal of Microbiology, 2020, 1–12. 10.1155/2020/6572693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaneethan, Y., & Effarizah, M. E. (2021). Prevalence, toxigenic profiles, multidrug resistance, and biofilm formation of Bacillus cereus isolated from ready‐to eat cooked rice in Penang. Malaysia. Food Control, 121, 107553. 10.1016/j.foodcont.2020.107553 [DOI] [Google Scholar]
- Poretsky, R., Rodriguez‐R, L. M., Luo, C., Tsementzi, D., & Konstantinidis, K. T. (2014). Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One, 9(4), e93827. 10.1371/journal.pone.0093827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Säde, E., Lassila, E., & Björkroth, J. (2016). Lactic acid bacteria in dried vegetables and spices. Food Microbiology, 53, 110–114. 10.1016/j.fm.2015.09.005 [DOI] [PubMed] [Google Scholar]
- Sagoo, S. K., Little, C. L., Greenwood, M., Mithani, V., Grant, K. A., McLauchlin, J., de Pinna, E. , & Threlfall, E. J. (2009). Assessment of the microbiological safety of dried spices and herbs from production and retail premises in the United Kingdom. Food Microbiology, 26, 39–43. 10.1016/j.fm.2008.07.005 [DOI] [PubMed] [Google Scholar]
- Sanlier, N., & Gencerb, F. (2020). Role of spices in the treatment of diabetes mellitus: A minireview. Trends in Food Science & Technology, 99, 441–449. 10.1016/j.tifs.2020.03.018 [DOI] [Google Scholar]
- Sentausa, E., & Fournier, P. E. (2013). Advantages and limitations of genomics in prokaryotic taxonomy. Clinical Microbiology and Infection, 19(9), 790–795. 10.1111/1469-0691.12181 [DOI] [PubMed] [Google Scholar]
- Székács, A., Wilkinson, M., Mader, A., & Appel, B. (2018). Environmental and food safety of spices and herbs along global food chains. Food Control, 83, 1–6. 10.1016/j.foodcont.2017.06.033 [DOI] [Google Scholar]
- Thanh, M. D., Frentzel, H., Fetsch, A., Krause, G., Appel, B., & Mader, A. (2018). Tenacity of Bacillus cereus and Staphylococcus aureus in dried spices and herbs. Food Control, 83, 75–84. 10.1016/j.foodcont.2016.12.027 [DOI] [Google Scholar]
- Thapa, S. P., Shrestha, S., & Anal, A. K. (2020). Addressing the antibiotic resistance and improving the food safety in food supply chain (farm‐to‐fork) in Southeast Asia. Food Control, 108, 106809. 10.1016/j.foodcont.2019.106809 [DOI] [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing (2016). Breakpoint tables for interpretation of MICs and zone diameters (EUCAST). Retrieved from http://www.eucast.org. Accessed 10 September 2020 [Google Scholar]
- Wang, H., Zhou, Y., & Zhang, L. (2019). Antimicrobial resistance, gut microbiota, and health. In: Doyle M. P., Diez‐Gonzalez F., & Hill C. (Eds.), Food microbiology: fundamentals and frontiers (pp. 903‐926). : ASM Press. [Google Scholar]
- Wang, N., Yuan, L., Sadiq, F. A., & He, G. (2019). Inhibitory effect of Lactobacillus plantarum metabolites against biofilm formation by Bacillus licheniformis isolated from milk powder products. Food Control, 106, 106721. 10.1016/j.foodcont.2019.106721. https://www.ezbiocloud.net/taxonomy; https://www.megasoftware.net/ [DOI] [Google Scholar]
- Zarzecka, U., Zadernowska, A., & Chajęcka‐Wierzchowska, W. (2020). Starter cultures as a reservoir of antibiotic resistant microorganisms. LWT, 127, 109424. 10.1016/j.lwt.2020.109424 [DOI] [Google Scholar]
- Zhang, R. Q., Ying, G. G., Su, H. C., Zhou, L. J., & Liu, Y. S. (2013). Antibiotic resistance and genetic diversity of Escherichia coli isolates from traditional and integrated aquaculture in South China. Journal of Environmental Science and Health Part B, 48(11), 999–1013. 10.1080/03601234.2013.816611 [DOI] [PubMed] [Google Scholar]
- Zweifel, C., & Stephan, R. (2012). Spices and herbs as source of Salmonella‐related foodborne diseases. Food Research International, 45(2), 765–769. 10.1016/j.foodres.2011.02.024 [DOI] [Google Scholar]


