Abstract
Background
Hereditary breast cancer (BC), ovarian cancer (OC), and pancreatic cancer (PC) are the major BRCA-associated tumours. However, some BRCA1/2-wild-type (wt) patients with a strong personal and/or family history of cancer need a further genetic testing through a multi-gene panel containing other high- and moderate-risk susceptibility genes.
Patients and methods
Our study was aimed to assess if some BC, OC, or PC patients should be offered multi-gene panel testing, based on well-defined criteria concerning their personal and/or family history of cancer, such as earliness of cancer onset, occurrence of multiple tumours, or presence of at least two or more affected first-degree relatives. For this purpose, 205 out of 915 BC, OC, or PC patients, resulted negative for BRCA1/2 and with significant personal and/or family history of cancer, were genetically tested for germline pathogenic or likely pathogenic variants (PVs/LPVs) in genes different from BRCA1/2.
Results
Our investigation revealed that 31 (15.1%) out of 205 patients harboured germline PVs/LPVs in no-BRCA genes, including PALB2, CHEK2, ATM, MUTYH, MSH2, and RAD51C. Interestingly, in the absence of an analysis conducted through multi-gene panel, a considerable percentage (15.1%) of PVs/LPVs would have been lost.
Conclusions
Providing a multi-gene panel testing to BRCA1/2-wt BC/OC/PC patients with a strong personal and/or family history of cancer could significantly increase the detection rates of germline PVs/LPVs in other cancer predisposition genes beyond BRCA1/2. The use of a multi-gene panel testing could improve the inherited cancer risk estimation and clinical management of patients and unaffected family members.
Key words: breast cancer, germline pathogenic variants, multi-gene panel testing, ovarian cancer, pancreatic cancer
Highlights
-
•
Patients with significant personal and/or family history of BC, OC, or PC could benefit from a multi-gene panel testing.
-
•
A total of 205 out of 915 BRCA1/2-wt BC, OC, or PC patients were genetically tested for germline PVs/LPVs in other genes.
-
•
A total of 15.1% of 205 BC, OC, or PC patients harboured germline PVs/LPVs in cancer susceptibility genes different from BRCA1/2.
-
•
PALB2, CHEK2, ATM, and RAD51C have been shown to be the genes more frequently altered in BRCA1/2-wt patients.
-
•
Using a multi-gene panel testing could improve the clinical management of patients and their unaffected family members.
Introduction
The hereditary breast and ovarian cancer (HBOC) syndrome is an autosomal dominant inherited disorder that includes 5%-7% of all breast cancer (BC) cases and 10%-15% of all ovarian cancer (OC) cases.1 Although the majority of BC and OC cases are sporadic (75%-80%), ~15%-20% are considered familial type and 5%-10% are hereditary.2,3 Moreover, ~3%-10% of patients with pancreatic adenocarcinoma exhibit a family history of pancreatic cancer (PC) and ~10%-20% are due to a hereditary cause.4,5 In this context, BRCA1 and BRCA2 are the most common genes associated with hereditary BC, OC, and PC. BRCA1 and BRCA2 pathogenic variant (PV) carriers are characterized by a risk of 15%-45% and 10%-20%, respectively, of developing OC and 50%-85% of developing BC.2 The lifetime risk of developing PC is ~3% for BRCA1 PV carriers and 5%-10% for BRCA2 PV carriers.6,7 A BRCA1/2 genetic testing is recommended for patients with HBOC syndrome or PC.8 In spite of BRCA1 and BRCA2 being the two high-penetrance genes primarily associated with increased risk of hereditary BC/OC and PC, several studies have helped identify many other predisposition genes for these tumours.9,10 Other hereditary cancer syndromes could be associated with germline PVs in several high- and moderate-risk susceptibility genes such as CDH1, PALB2, PTEN, STK11, TP53, ATM, CHEK2, BARD1, BRIP1, RAD51C, and RAD51D.10,11
Recently, it has become necessary to study several genes in a short time and in an inexpensive way. This scenario has become possible with the advent of next-generation sequencing (NGS) technologies, which allow the simultaneous sequencing of multiple samples and genes.12 Because of the cost reduction, this approach offers a potential therapeutic application for patients with PV in other genes, beyond BRCA1/2.13
Kurian et al.14 showed an increased BC risk associated with several genes such as ATM, BARD1, BRCA1, BRCA2, CHEK2, PALB2, PTEN, and TP53, as well as an OC risk associated with various genes including ATM, BRIP1, RAD51C, RAD51D, MLH1, MSH6, MSH2, NBN, and STK11. Recently, Germani et al.9 carried out a multi-gene panel testing on 113 BRCA-negative patients with BC, OC, or PC, by identifying in 14 patients a PV or a likely pathogenic variant (LPV) beyond BRCA1/2 genes, such as CHEK2, RAD51C, ATM, MLH1, MSH2, and RECQL. Our recent study10 conducted on patients with bilateral breast cancer (BBC) showed that 14.4% of PVs in high- and moderate-penetrance BC susceptibility genes, such as PTEN, PALB2, CHEK2, ATM, and RAD51C, would have been lost in the absence of an analysis carried out via multi-gene panel.10
Based on a database of the Breast Cancer BRCA System retrospectively harvested at University Hospital Policlinico ‘P. Giaccone’ of Palermo, the aim of this work was to describe the typology and gene location of germline PVs detected in susceptibility genes to BC, OC, or PC, in order to investigate the prevalence of different inherited genetic variants in these patients and assess the utility of carrying out an NGS-based multi-gene panel testing in these individuals. This information could be useful and interesting in order to understand if the use of a multi-gene panel testing is recommended for BC, OC, or PC patients who fulfil specific criteria based on their personal and family history of cancer.
Patients and methods
Study population
A retrospective study was carried out from October 2016 to November 2020 at the ‘Sicilian Regional Center for the Prevention, Diagnosis and Treatment of Rare and Heredo-Familial Tumors’ of the Section of Medical Oncology of University Hospital Policlinico ‘P. Giaccone’ of Palermo. We collected and analysed all clinical information regarding 915 patients, 531 with primary BC, 345 affected by OC, and 39 with metastatic PC. Subsequently, we analysed 205 out of 915 patients with BC (165 individuals), OC (27 women), and PC (13 subjects) with negative test result for germline BRCA1/2 PVs who showed at least one of the following criteria: (i) at least other two first-degree relatives affected by BC, OC, and/or PC; (ii) early onset of cancer (age at diagnosis ≤36 years); or (iii) presence of synchronous/metachronous tumours (e.g. bilateral BC, BC and OC). The analysis was carried out by NGS-based multi-gene panel testing. The medical personal history of patients was acquired during genetic counselling in the presence of a multidisciplinary team constituted by an oncologist, a geneticist, and a psychologist. All patients provided an informed consent, and the information regarding personal and familial history of cancer, family geographical origin, age of cancer diagnosis, histological tumour subtype, molecular phenotype, and disease stages (I-IV) was anonymously recorded. The study (Protocol ‘G-Land 2017’) was approved by the ethical committee (Comitato Etico Palermo 1; approval number: 0103-2017) of the university-affiliated hospital AOUP ‘P. Giaccone’ of Palermo. All clinical information for each enrolled patient was anonymously recorded and coded.
Data concerning the histological type and cancer diagnosis were obtained by medical pathology reports in diagnostic core biopsies or tumour resections. After genetic counselling, patients were evaluated for germline BRCA1/2 genetic test based on the probability rate of carrying PV assessed by BRCAPRO genetic risk prediction model15 and according to the family and personal criteria established by guidelines of the Italian Association of Medical Oncology (AIOM) (https://www.aiom.it/linee-guida-aiom-neoplasie-della-mammella-2019/ and https://www.aiom.it/wp-content/uploads/2019/10/2019_Racc_BRCA_pancreas.pdf).16
Sample collection and next-generation sequencing analysis
Peripheral blood was collected from BC, OC, or PC patients. Genomic DNA was extracted from the peripheral blood using the DNeasy® Blood Kit (QIAGEN, Hilden, Germany) and quantified by Qubit®3.0 fluorometer (Thermo Fisher Scientific, Waltham, MA). Its quality was evaluated using 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The genetic analysis for BRCA1/2 was carried out as previously described.2,17,18 Sequencing analysis was carried out using Ion 520 Chip (Thermo Fisher Scientific) and Ion Torrent S5 (Thermo Fisher Scientific) instrument. The obtained data were processed with two different software called Amplicon Suite (SmartSeq s.r.l., Novara, Italy) and Ion Reporter Software v.5.14 (Thermo Fisher Scientific).
The genetic analysis by multi-gene panel, which included 22 genes involved in risk of hereditary BC, OC, PC, and colorectal cancer, and other inherited cancer syndromes (ATM, APC, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, EPCAM, MLH1, MSH2, MSH6, MUTYH, NBN, PALB2, PMS2, PTEN, RAD50, RAD51C, RAD51D, STK11, and TP53), was carried out as previously described.10
Sanger sequencing
PVs and LPVs identified with NGS were validated by Sanger sequencing using SeqStudio (Thermo Fisher Scientific) and BigDye Terminator 3.1 Cycle Sequencing Kit (Life Technologies, Carlsbad, CA), according to the manufacturers’ protocols.
Genetic variant classification
The detected genetic variants were categorized according to criteria developed by the Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA) consortium (https://enigmaconsortium.org/) and International Agency for Research on Cancer recommendations,19 and classified into five classes: benign (class I), likely benign (class II), variant of uncertain significance (VUS, class III), likely pathogenic (class IV), and pathogenic (class V). Several databases were used for the identification and classification of genetic variants, such as ClinVar, BRCA Exchange, LOVD, and Varsome.
The detected variants have been named based on the recommendations for the description of sequence variants supplied by the Human Genome Variation Society.20
Statistical analysis
The clinico-pathological characteristics of patients and prevalence of PVs in BRCA1/2 and other genes were assessed for each subgroup of patients. The differences between subgroups were evaluated by Fisher’s exact test. P values <0.05 were considered statistically significant. Statistical analysis was carried out using IBM SPSS Statistics for Windows Version 23.0 (IBM Corporation, Armonk, NY).
Results
Detection of germline variants in cancer susceptibility genes by multi-gene panel testing
Nine hundred and fifteen patients with BC, OC, or PC, enrolled from October 2016 to November 2020 at our institute, who met the criteria concerning personal and family history of cancer recommended by the AIOM national guidelines, were genetically tested for germline BRCA1/2 LPVs/PVs. Subsequently, 205 out of 915 patients, including 165 individuals with BC, 27 with OC, and 13 with PC, who resulted negative to germline BRCA1/2 testing and with a strong personal and/or family history of BC, OC, and/or PC, were selected for a further analysis by NGS-based multi-gene panel testing, in order to detect the presence of germline LPVs/PVs in different cancer susceptibility genes, beyond BRCA1/2 (Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2021.100235).
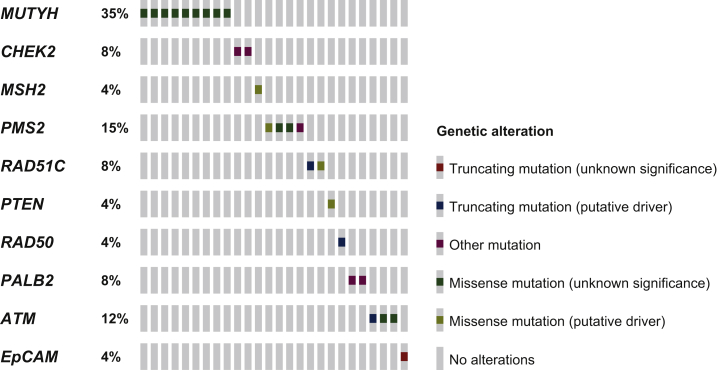
The indications for the selection of these probands were provided by the multidisciplinary team. This further mutational screening revealed that 31 (15.1%) out of 205 genetically tested patients with BC, OC, and PC harboured germline PVs/LPVs (classes IV and V) in other cancer susceptibility genes different from BRCA1/2 (Figure 1). In particular, our analysis showed that 24 out of 31 individuals positively tested in no-BRCA genes by multi-gene panel had BC, whereas 4 were affected by OC and only 3 had PC.
Figure 1.
Distribution of PVs/LPVs detected in 31 BC, OC, or PC patients analysed with multi-gene panel testing.
The OncoPrint, showing the identified PVs/LPVs by heatmap, was obtained by the informatics tool Mutation Mapper (cBioPortal for Cancer Genomics). The intronic variant sequences (IVS) are not shown. BC, breast cancer; LPV, likely pathogenic variant; OC, ovarian cancer; PC, pancreatic cancer; PV, pathogenic variant.
Therefore, 24 (14.5%) out of 165 BC patients (160 women and 5 men) analysed by multi-gene panel approach showed PVs/LPVs in other cancer susceptibility genes (no-BRCA) (Table 1).
Table 1.
PVs/LPVs harboured by patients with BC, OC, or PC analysed by multi-gene panel testing
| Gene | Variant type | HGVS nomenclature | Protein change | Variant interpretation | Patients n (%) |
|---|---|---|---|---|---|
| Breast cancer patients | |||||
| MUTYH | M | c.1145G>A | p.Gly382Asp | PV | 5 (20.6) |
| CHEK2 | fs | c.1229del | p.Thr410fs | PV | 2 (8.2) |
| CHEK2 | IVS | c.721+3A>T | — | CIP/PV | 2 (8.2) |
| CDH1 | IVS | c.2164+2T>C | — | PV | 1 (4.2) |
| MSH2 | M | c.1045C>G | p.Pro349Ala | CIP/PV | 1 (4.2) |
| PMS2 | fs | c.2182_2184delinsG | p.Thr728Alafs | CIP/PV | 1 (4.2) |
| PMS2 | M | c.137G>T | p.Ser46Ile | LPV | 1 (4.2) |
| PMS2 | M | C.2T>C | p.Met1Thr | PV | 1 (4.2) |
| RAD51C | IVS | c.1026+5_1026+7del | — | LPV | 1 (4.2) |
| RAD51C | NS | c.224dup | p.Tyr75Ter | PV | 1 (4.2) |
| RAD51C | M | c.773G>A | p.Arg258His | LPV | 1 (4.2) |
| PTEN | M | c.284C>A | p.Pro95Gln | PV | 1 (4.2) |
| RAD50 | NS | c.3598C>T | p.Arg1200Ter | PV | 1 (4.2) |
| MUTYH | M | c.494A>G | p.Tyr165Cys | PV | 1 (4.2) |
| PALB2 | fs | c.758dup | p.Ser254fs | PV | 1 (4.2) |
| PALB2 | fs | c.1050_1053del | p.Thr351fs | PV | 1 (4.2) |
| ATM | M | c.8147T>C | p.Val2716Ala | LPV | 1 (4.2) |
| ATM | NS | c.8818_8821dup | p.Ser2941Ter | PV | 1 (4.2) |
| Ovarian cancer patients | |||||
| MUTYH | M | c.1145G>A | p.Gly382Asp | PV | 2 (50) |
| ATM | IVS | c.4776+1G>T | — | LPV | 1 (25) |
| PMS2 | M | c.2249G>A | p.Gly750Asp | LPV | 1 (25) |
| Pancreatic cancer patients | |||||
| MUTYH | M | c.1145G>A | p.Gly382Asp | PV | 1 (33.3) |
| EPCAM | NS | c.227C>G | p.Ser76Ter | PV | 1 (33.3) |
| ATM | M | c.8558C>T | p.Thr2853Met | LPV | 1 (33.3) |
BC, breast cancer; CIP, conflicting interpretations of pathogenicity; fs, frameshift; IVS, intronic variant sequences; LPV, likely pathogenic variant; M, missense; NS, nonsense; OC, ovarian cancer; PC, pancreatic cancer; PV, pathogenic variant.
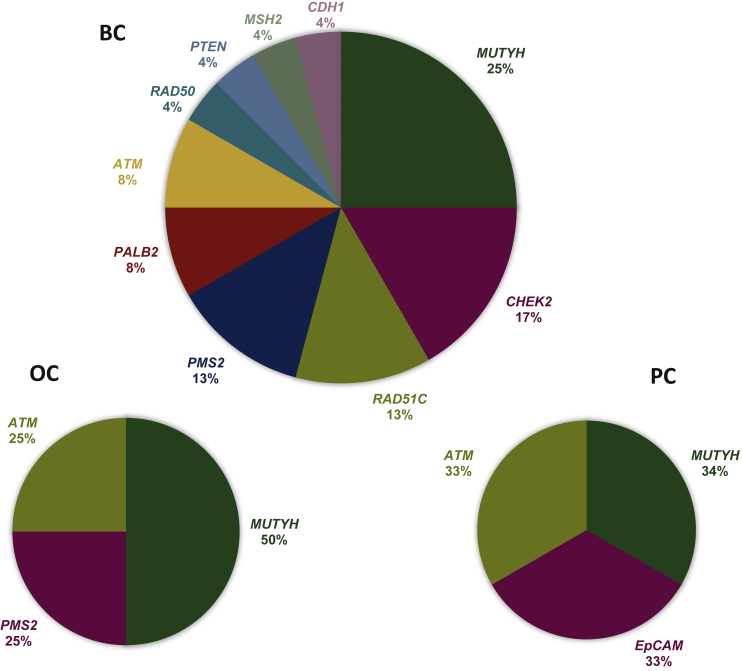
Specifically, six (25%) probands have been shown to harbour PVs in MUTYH gene, four (17%) patients in CHEK2, three (13%) subjects in RAD51C and PMS2 genes, respectively, and two (8%) individuals in PALB2 and ATM genes, respectively. In addition, single PVs in RAD50, PTEN, MSH2, and CDH1 genes were detected in four patients, respectively (Figure 2).
Figure 2.
Percentage distribution of no-BRCA genes altered in BC, OC, or PC patients detected by multi-gene panel testing.
BC, breast cancer; OC, ovarian cancer; PC, pancreatic cancer.
Our analysis revealed that 11 (45.8%) out of 24 no-BRCA patients were affected by BBC. Specifically, the PVs were distributed in 11 probands as follows: 2 in CHEK2, 2 in PALB2, 2 in ATM, 2 in RAD51C, 2 in MUTYH, and 1 in MSH2 genes.
The most frequent PV, observed in five probands (20.6%) of our retrospective analysis, resulted to be a missense mutation in a heterozygous condition in MUTYH gene named c. 1145G>A, which, if present in a biallelic form, is usually linked to MUTYH-associated colon polyposis syndrome and colorectal cancer.21
The second most recurrent PV observed in two (8.3%) individuals affected by BBC was detected in CHEK2 gene and is named c.1229del. This result is coherent with the previously published data in our work.10 In fact, this variant has been shown to be correlated with BBC and, in particular, with luminal A/B molecular phenotype, estrogen receptor positivity >60%, and progesterone receptor positivity between 20% and 60%.10
Another interesting variant detected in CHEK2 gene was an intronic variant sequence called c.721+3A>T. The nature of this variant is still to be defined. In fact, the ClinVar database includes it among the alterations with conflicting interpretations of pathogenicity (CIP), although it could be probably considered a PV being a splice-site mutation.
Despite the fact that PALB2 has been shown to be one of the most frequently altered genes in BC patients,22 in our analysis only two patients (8%) exhibited an alteration in this gene. However, both these patients showed BBC whereby a correlation between this PV and BBC could be hypothesized.
Among 27 OC patients, 4 (14.9%) showed a germline PV/LPV in no-BRCA genes (Table 1). In particular, two patients (50%) carried a monoallelic PV in MUTYH gene, whereas one in ATM and one in PMS2 (Figure 2). Like BC patients, even those with OC harboured the monoallelic MUTHY PV named c.1145G>A. One patient showed a variant in ATM gene called c.4776+1G>T, acting as a donor splice site in intron 31 of the gene. Considering results from in silico analysis, this variant has been classified as LPV, because it is expected to disrupt RNA splicing, resulting in the absence of protein product. Finally, one OC patient resulted to harbour a germline biallelic PMS2 PV named c.2249G>A. This variant involves a substitution of glycine, a highly conserved residue, with aspartic acid at codon 750 and has been observed in patients with constitutional mismatch repair (MMR) deficiency syndrome.
Among 13 PC patients who underwent multi-gene panel testing, 3 (23%) showed heterozygous PVs/LPVs in genes different from BRCA1/2, such as MUTYH, EPCAM, and ATM (Table 1, Figure 2). In particular, the same MUTYH variant c.1145G>A, observed in BC and OC patients, was detected also in one PC subject, suggesting that this alteration is relatively recurrent in Sicilian individuals with BC, OC, and PC (8/205 probands; ~4%). The missense variant c.227C>G in EPCAM gene causes an amino acid substitution of cytosine with a guanine which determines the premature stop codon formation, resulting in the loss of normal protein function. This alteration, reported as PV by LOVD, though in a homozygous state, was described by Thoeni et al.23 in one patient affected by congenital tufting enteropathy. Lastly, one patient revealed a missense variant named c.8558C>T in ATM gene. The nature of this variant, which determines an amino acid substitution of threonine with methionine at codon 2853, is still to be defined. In fact, the ClinVar database reported it as VUS, because the threonine residue is highly conserved and exhibits a moderate physicochemical difference with methionine. Conversely, the Varsome database classified this alteration as LPV, because it is located in a mutational hotspot.
Lastly, we investigated the presence of VUS and variants with CIP in genes different from BRCA1/2, by identifying 70 VUS/CIP in BC patients, 18 in OC women, and 4 in PC subjects (Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2021.100235). The most frequent VUS named c.663A>C and detected in MSH6 gene was observed in three BC patients and one with OC. The total number of patients harbouring VUS/CIP was 102, since some subjects were carriers of two or three VUS/CIP. BC patients have been shown to harbour a higher number of VUS/CIP mainly in ATM (six VUS and six CIP), APC (six VUS and two CIP), RAD50 (four VUS and four CIP), and CHEK2 (five VUS and two CIP) genes, whereas OC patients carried VUS/CIP mainly in CHEK2 (three VUS and one CIP) and MSH6 (two VUS and one CIP) genes.
Association between germline PVs/LPVs and clinical factors
Our analysis highlighted that the median age at diagnosis of BC patients with PVs/LPVs in cancer susceptibility genes different from BRCA1/2 was lower than that of BRCA1/2-positive patients and all wild-type (wt) individuals (median age: 32 versus 41 years, P = 0.8; 32 versus 44 years, P = 0.9, respectively), whereas the mean age instead was higher than that of other groups (mean age: 45.6 versus 42.6 versus 44.5 years, respectively). Most of the patients with PVs/LPVs either in BRCA1/2 or in other susceptibility genes had an age at diagnosis ≤40 years. Significant clinico-pathological differences among three subgroups of BC were observed (Table 2).
Table 2.
Comparison between baseline characteristics and clinico-pathological parameters of BC patients analysed by germline BRCA1/2 and multi-gene panel testing
| BRCA1/2 mut | Multi-gene panel mut | All wt | P valuea | P valueb | |
|---|---|---|---|---|---|
| Number of patients | 83 | 24 | 589 | — | — |
| Age at diagnosis (years) | |||||
| Median | 41 | 32 | 44 | ||
| Mean | 42.6 | 45.6 | 44.5 | ||
| Range | 28-80 | 27-71 | 21-84 | 0.32 | 0.24 |
| Age groups (years), n (%) | |||||
| ≤40 | 40 (48.2) | 9 (37.5) | 241 (40.9) | ||
| 41-50 | 26 (31.3) | 7 (29.2) | 194 (32.9) | ||
| 51-60 | 13 (15.7) | 5 (20.8) | 105 (17.9) | ||
| >60 | 4 (4.8) | 3 (12.5) | 49 (8.3) | 0.48 | 0.86 |
| Histological subtype, n (%) | |||||
| Ductal | 72 (86.8) | 13 (54.2) | 456 (77.4) | ||
| Lobular | 7 (8.4) | 2 (8.3) | 47 (8) | ||
| Others | 4 (4.8) | 2 (8.3) | 49 (8.3) | ||
| Unknown | — | 7 (29.2) | 37 (6.3) | <0.0001 | <0.0001 |
| Molecular subtype, n (%) | |||||
| Luminal | 46 (55.4) | 15 (62.5) | 397 (67.4) | ||
| HER2E | 3 (3.7) | 1 (4.2) | 33 (5.6) | ||
| TNBC | 34 (40.9) | 3 (12.5) | 149 (25.3) | ||
| Unknown | — | 5 (20.8) | 10 (1.7) | 0.0001 | <0.00001 |
| ER, n (%) | |||||
| ≤20 | 8 (9.7) | — | 19 (3.2) | ||
| >20 | 34 (40.9) | 16 (66.6) | 355 (60.3) | ||
| Negative | 37 (44.6) | 4 (16.7) | 165 (28) | ||
| Unknown | 4 (4.8) | 4 (16.7) | 50 (8.5) | 0.014 | 0.41 |
| PR, n (%) | |||||
| ≤20 | 19 (22.9) | 2 (8.4) | 76 (12.9) | ||
| >20 | 23 (27.7) | 12 (50) | 286 (48.6) | ||
| Negative | 37 (44.6) | 5 (20.8) | 165 (28) | ||
| Unknown | 4 (4.8) | 5 (20.8) | 62 (10.5) | 0.004 | 0.38 |
| Ki-67, n (%) | |||||
| <20 | 8 (9.7) | 5 (20.8) | 158 (26.9) | ||
| 20-50 | 31 (37.3) | 8 (33.3) | 211 (35.8) | ||
| >50 | 39 (47) | — | 134 (22.7) | ||
| Unknown | 5 (6) | 11 (45.9) | 86 (14.6) | <0.00001 | 0.0006 |
| Histological grade, n (%) | |||||
| G1 | 2 (2.4) | 2 (8.4) | 62 (10.5) | ||
| G2 | 18 (21.7) | 8 (33.3) | 212 (36) | ||
| G3 | 58 (69.9) | 3 (12.5) | 226 (38.4) | ||
| Unknown | 5 (6) | 11 (45.8) | 89 (15.1) | <0.00001 | 0.0005 |
| Tumour size (T), n (%) | |||||
| T1 | 38 (45.8) | 8 (33.3) | 261 (44.4) | ||
| T2 | 23 (27.7) | 4 (16.7) | 145 (24.6) | ||
| T3 | 3 (3.6) | — | 18 (3) | ||
| T4 | 2 (2.4) | — | 6 (1) | ||
| unknown | 17 (20.5) | 12 (50) | 159 (27) | 0.12 | 0.16 |
| Axillary nodal involvement (N), n (%) | |||||
| N0 | 30 (36.1) | 2 (8.3) | 257 (43.7) | ||
| N1 | 24 (28.9) | 6 (25) | 108 (18.3) | ||
| N2 | 8 (9.6) | 3 (12.5) | 23 (3.9) | ||
| N3 | 5 (6) | — | 9 (1.5) | ||
| unknown | 16 (19.4) | 13 (54.2) | 192 (32.6) | 0.009 | 0.005 |
| Bilateral, n (%) | |||||
| Yes | 19 (22.9) | 11 (45.8) | 121 (20.5) | ||
| No | 64 (77.1) | 13 (54.2) | 468 (79.5) | 0.04 | 0.008 |
| Median age at diagnosis (years) | |||||
| Primary tumour | 40 | 53 | 46 | ||
| Secondary tumour | 50 | 63 | 52 | 0.86 | 0.85 |
| Time between first and second tumours (years) | |||||
| Median | 8 | 7 | 3 |
ER, estrogen receptor; PR, progesterone receptor; mut, mutated; TNBC, triple-negative breast cancer; wt, wild type.
Comparison between BRCA1/2 mut versus multi-gene panel mut patients.
Comparison between multi-gene panel mut versus all wt patients.
Also, we observed that patients with germline alterations in BRCA1/2 had more frequently a ductal histotype of BC (86.8%) compared to patients with alterations in other predisposition genes different from BRCA1/2 (54.2%).
Interestingly, only 3 patients (12.5%) with PVs/LPVs in no-BRCA genes showed triple-negative breast cancer (TNBC) compared to 34 BRCA1/2-positive patients (40.9%; P = 0.12) and 149 (25.3%; P < 0.00001) subjects without any alteration in analysed genes.
Furthermore, it has been observed that breast tumours harboured by PV carriers in no-BRCA genes showed low proliferation rates and lower histological grade than BRCA1/2 PV carriers (P = 0.003) and wt subjects (P = 0.24). Additionally, a significant axillary nodal involvement was not detected compared to other two groups (P = 0.07; P = 0.002).
As mentioned earlier, 11 patients who showed gene alterations by multi-gene panel analysis had BBC. This result was statistically significant either compared to BRCA1/2 PV carriers (P = 0.04) or to wt individuals (P = 0.008).
Comparing between them only patients analysed by multi-gene panel, no statistically significant correlation was shown between mutated and wt patients. However, our analysis showed that mutated patients had a median age at diagnosis considerably lower than wt subjects (median age: 32 versus 43.5 years; P = 0.24). As regards BBC patients, the median time between first and second tumour was longer in mutated than in patients without alteration.
A comparison among OC patients with PVs/LPVs in BRCA1/2 and no-BRCA genes and all wt subjects was carried out (Table 3). We observed that women with PVs/LPVs in susceptibility genes different from BRCA1/2 tend to develop OC before than the other two groups (median age: 52.5 versus 56 versus 58 years). Also, it was observed that most of the BRCA-positive and wt women developed OC over the fifth decade of life.
Table 3.
Comparison between baseline characteristics and clinico-pathological parameters of OC patients analysed by germline BRCA1/2 and multi-gene panel testing
| BRCA1/2 mut | Multi-gene panel mut | All wt | P valuea | P valueb | |
|---|---|---|---|---|---|
| Number of patients | 85 | 4 | 283 | — | — |
| Age at diagnosis (years) | |||||
| Median | 56 | 52.5 | 58 | ||
| Mean | 57.9 | 55.5 | 58 | ||
| Range | 37-81 | 38-79 | 28-84 | 0.9 | 0.8 |
| Age groups (years), n (%) | |||||
| ≤40 | 3 (3.5) | 1 (25) | 22 (7.8) | ||
| 41-50 | 19 (22.4) | 1 (25) | 33 (11.6) | ||
| 51-60 | 31 (36.5) | — | 112 (39.6) | ||
| 61-70 | 22 (25.8) | 1 (25) | 72 (25.4) | ||
| >70 | 10 (11.8) | 1 (25) | 44 (15.6) | 0.45 | 0.77 |
| Cancer site, n (%) | |||||
| Monolateral Ovarian carcinoma | 54 (63.5) | 4 (100) | 236 (83.4) | ||
| Bilateral Ovarian carcinoma | 29 (34.1) | — | 21 (7.4) | ||
| Fallopian tube carcinoma | 0 (0) | — | 2 (0.7) | ||
| Primary peritoneal carcinoma | 2 (2.4) | — | 24 (8.5) | 0.24 | 0.01 |
| FIGO stage, n (%) | |||||
| I | 3 (3.5) | — | 12 (4.2) | ||
| II | 4 (4.7) | 1 (25) | 10 (3.5) | ||
| III | 49 (57.7) | 2 (50) | 39 (13.8) | ||
| IV | 9 (10.6) | — | 11 (3.9) | ||
| Unknown | 20 (23.5) | 1 (25) | 211 (74.6) | 0.43 | 0.03 |
| Histological subtype, n (%) | |||||
| HGSC | 68 (80) | 3 (75) | 194 (68.6) | ||
| Clear cell | 2 (2.4) | — | 6 (2.1) | ||
| Endometrioid | 6 (7.1) | — | 25 (8.8) | ||
| LGSC | 3 (3.5) | — | 2 (0.7) | ||
| Papillary | 3 (3.5) | 1 (25) | 2 (0.7) | ||
| Unknown | 3 (3.5) | — | 54 (19.1) | 0.2 | 0.0004 |
| Personal cancer history before EOC, n (%) | |||||
| Breast cancer history | 15 (17.6) | 2 (50) | 20 (7) | ||
| Other cancers | 1 (12) | 1 (25) | 21 (7.4) | ||
| No cancer history | 69 (81.2) | 1 (25) | 242 (85.6) | 0.001 | 0.001 |
| Surgery, n (%) | |||||
| Staging | 11 (12.9) | 2 (50) | 34 (12) | ||
| Primary cytoreductive | 32 (37.6) | 1 (25) | 126 (44.5) | ||
| Unknown | 42 (49.4) | 1 (25) | 123 (43.5) | 0.1 | 0.07 |
EOC, epithelial ovarian cancer; HGSC, high-grade serous carcinoma; LGSC, low-grade serous carcinoma; mut, mutated; wt, wild type.
Comparison between BRCA1/2 mut versus multi-gene panel mut patients.
Comparison between multi-gene panel mut versus all wt patients.
Based on tumour characteristics, the most represented OC histological subtype in all three patient groups (BRCA-positive, no-BRCA PV carriers, and wt) was the high-grade serous carcinoma.
Interestingly, three out of four patients harbouring PVs/LPVs in susceptibility genes different from BRCA1/2 showed a personal history of BC or other cancers. The clinico-pathological features of analysed OC patients are reported in Table 3. Obviously, some of these data are not statistically significant due to the low number of patients analysed by multi-gene panel.
Concerning the PC patients genetically tested by multi-gene panels, all had metastatic disease, but it was not possible to carry out a comparative analysis due to the very low number of individuals.
Discussion
Hereditary BC, OC, and PC, together with prostate cancer and melanoma, are the major BRCA-associated cancers.24,25 In fact, some of these tumours have been shown to be associated with the HBOC syndrome, because the risk of developing them significantly enhances in germline BRCA1/2 PV carriers.26, 27, 28 However, it has been observed that many BC, OC, or PC patients with a strong personal and/or family history of cancer are negative for germline BRCA1/2 PVs/LPVs and, thus, a further genetic testing via a broader gene panel should be requested. Indeed, some BRCA1/2-wt patients may carry a still unidentified PV/LPV in high- and moderate-risk susceptibility genes, including PALB2, CHEK2, ATM, TP53, and PTEN, involved in several hereditary cancer syndromes.29, 30, 31 Today, the use of multi-gene panels, which include several susceptibility genes related to hereditary cancer syndromes, is becoming progressively recurrent, thanks also to the advancement acquired by NGS technology which transformed the clinical approach to genetic testing.32, 33, 34
Recent studies highlighted that, due to overlapping phenotypes, a considerable number of PVs/LPVs, which, could be missed if independently tested for the HBOC, Li–Fraumeni, Peutz–Jeghers, Cowden, Lynch syndromes, and other hereditary tumour syndromes, may be more easily identified by multi-gene panel testing. For this reason, multi-gene panels able to cover all these syndromes rather than panels specific for a single syndrome were produced.35 Certainly, clinicians may obtain more information about one or more inherited cancer syndromes in a single test by means of the use of a multi-gene panel.36 Several studies have recently evaluated the importance of complete multi-gene panels in the clinical management of hereditary BC, OC, or PC patients.32,37,38 Today, however, the specific selection of high-risk individuals who should be tested by multi-gene panel as well as the choice of genes to comprise in testing for each hereditary syndrome still remain an open question. In fact, until now there are no precise guidelines which allow to establish specific criteria for identifying the most appropriate subjects to undergo multi-gene panel testing.10 A relevant clinical implication is attributed to the choice of genes to include in a multi-gene panel.
In the present study, 205 out of 915 patients affected by BC, OC, or PC, with previous BRCA1/2 negative test result, and significant personal and/or family history of BC, OC, and/or PC were selected by our multidisciplinary team and genetically tested for germline PVs/LPVs in different cancer susceptibility genes beyond BRCA1 and BRCA2, by NGS-based multi-gene panel testing. This investigation was aimed to assess if some BC, OC, or PC patients should be offered multi-gene panel testing, based on well-defined criteria concerning their personal and family history of cancer, such as earliness of cancer onset, occurrence of multiple tumours, and presence of two or more affected relatives.
Our study showed that 31 (15.1%) out of 205 patients genetically tested by multi-gene panel were carriers of a PV/LPV in high- and moderate-risk cancer susceptibility genes different from BRCA1/2, including PALB2, CHEK2, ATM, MUTYH, MSH2, and RAD51C. Among 31 carriers of PVs/LPVs in no-BRCA genes, 24 out of 165 (14.5%) were BC probands, 4 out of 27 (14.9%) had OC, and 3 out of 13 (23%) were PC subjects. These data reveal how a noteworthy percentage of BC, OC, and PC patients showed deleterious alterations in other cancer predisposition genes beyond BRCA1 and BRCA2, using a multi-gene panel approach.
Heterozygous alterations in the moderate-risk susceptibility gene CHEK2 were detected only in BC patients, as already previously reported by other authors.39,40
Interestingly, 11 (45.8%) out of 24 BC patients positively tested by multi-gene panel in genes beyond BRCA1/2 have been shown to have BBC. Besides the founder mutation c.1100delC detected in the moderate-risk susceptibility gene CHEK2, further deleterious alterations in BBC patients were found in other different genes such as PALB2, ATM, and RAD51C, suggesting a potential association between BBC and HRD.
In our investigation, a low rate of PALB2 alterations was detected probably due to the selection criteria used to recruit patients which led to the exclusion by analysis of TNBC individuals >60 years of age and no-TNBC subjects (in the absence of multiple tumours and/or family history) >36 years of age.
As regards the analysed OC patients, interestingly, one out of four women harbouring alterations in no-BRCA genes was carrier of a germline homozygous PMS2 LPV (c.2249G>A) and, therefore, her OC was associated with constitutional MMR deficiency syndrome rather than with HBOC syndrome. Instead, the EPCAM PV (c.227C>G) detected in one out of three PC patients is not associated with Lynch syndrome as expected, since this variant does not involve a 3′ deletion within gene.
Since, in the last years, several studies investigated also the impact of germline monoallelic MUTYH PVs in genetic susceptibility to the development of BC, OC, or PC, showing increased risk rates associated with heterozygosity condition,41, 42, 43, 44, 45, 46 we have decided to report these data concerning the heterozygous MUTYH alterations detected in our study cohort. Interestingly, nine patients (six of whom were affected by BC, two with OC, and one with PC) showed a germline monoallelic MUTYH PV. Although the association between monoallelic MUTYH PVs and risk of BC, OC, or PC remains controversial, this information could represent, in the future, an important step in defining the risk and cancer types associated with these alterations, beyond colorectal cancer.
Additionally, we have observed that 12 (50%) out of 24 germline PVs/PVLs detected in 31 tested carrier subjects were missense mutations (Table 1).
Our study also showed that carriers of PVs/LPVs in cancer susceptibility genes different from BRCA1/2 had a lower probability of developing a TNBC in comparison to BRCA-positive carriers and wt individuals who, instead, harboured often a luminal-like BC histotype. Furthermore, the presence of tumours with reduced proliferation rate, lower histological grade, and without axillary nodal involvement in patients carrying germline PVs/LPVs in no-BRCA genes suggested that these carriers show a greater tendency to develop less aggressive BCs.
However, comparing the clinico-pathological features of three groups of analysed patients, in some cases no statistically significant differences were detected, probably due to the low number of patients genetically tested by multi-gene panel. Therefore, a future perspective of this work will be to increase the size of our study population.
In this study, we investigated the potential clinical impact of NGS-based multi-gene panel testing in patients affected by BC, OC, or PC, in order to evaluate the usefulness of carrying out a most comprehensive genetic investigation in individuals who had at least other two first-degree family members with BC, OC, and/or PC, or early onset of cancer (age at diagnosis ≤36 years), or presence of synchronous/metachronous tumours (e.g. bilateral BC, BC and OC). Our work highlighted that an improvement of detection rates of germline deleterious variants in patients with BC, OC, and PC could be achieved through the adoption of an NGS-based multiple-gene panel testing, because we have observed that, in the absence of a genetic analysis carried out by means of a multi-gene panel, a significant percentage (15.1%) of PVs/LPVs would have been lost.
Lastly, our investigation showed results coherent with the recommendations of the current National Comprehensive Cancer Network (NCCN) guidelines, offering a more cost-effective cancer risk assessment compared with a gene-by-gene approach and avoiding underestimating the number of individuals affected by a hereditary tumour syndrome.
Our study highlighted that a deeper genetic investigation by NGS-based multi-gene panel testing could help us to identify PVs/LPVs in high- and moderate-risk cancer susceptibility genes different from most common BRCA1/2 genes, allowing to stratify a significant percentage of inherited BC, OC, or PC patients who could take advantage from the screening programs, intensive surveillance pathways, and/or risk-reducing surgical strategies, if necessary. Additionally, this investigation could allow the identification of family members with a higher risk of developing BC, OC, and/or PC, in order to put into practice prevention and surveillance strategies for these individuals. According to the recent NCCN guidelines (version 2.2021),47 helpful surveillance programs for the BC and/or OC prevention are recommended to LPV/PV carriers in no-BRCA1/2 genes, depending on the specific altered gene (Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2021.100235). For carriers of germline monoallelic alterations in CHEK2, PALB2, ATM, PTEN, and CDH1 genes, the BC risk management involves an annual mammogram with eventual tomosynthesis and breast magnetic resonance imaging with contrast (starting from age 30 years for PALB2 and CDH1, from 30 to 35 years for PTEN, and from 40 years for CHEK2 and ATM), whereas no surveillance is recommended for OC risk. However, for PALB2 and ATM LPV/PV carriers, a risk-reducing salpingo-oophorectomy (RRSO) may be suggested based on family history. For carriers of PALB2 and PTEN alterations, a risk-reducing mastectomy may be discussed, whereas this option may be suggested only based on family history for CHEK2, ATM, and CDH1 LPV/PV carriers. For RAD51C LPV/PV carriers, the RRSO may be considered starting from age 45 to 50 years, whereas the surveillance for BC risk is managed only based on family history. For carriers of germline monoallelic MSH2, PMS2, and EPCAM LPVs/PVs, the BC risk management is based on family history only, whereas surveillance for OC risk involves annually the transvaginal ultrasound and assessment of CA-125 serum levels. No recommendation for BC/OC risk management is suggested for carriers of heterozygous RAD50 and MUTYH LPVs/PVs (Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2021.100235).
However, larger study cohorts are required to more accurately detect the rates of PVs/PVLs in BC, OC, or PC patients with previously negative BRCA genetic testing result. In addition, the approach via multi-gene panel shows the disadvantage of determining an increase in detection rates of germline VUS and PVs/LPVs in moderate-/low-penetrance genes or with limited clinical relevance. This drawback could cause not only a risk overestimation but also a poor clinical management of the patient, resulting in an increase in the number of prophylactic surgical interventions without benefits as well as unnecessary health care costs.
Acknowledgements
All authors thank Dr Chiara Drago for the revision of the English language.
Funding
None declared.
Disclosure
The authors have declared no conflicts of interest.
Supplementary data
References
- 1.El Ansari F.Z., Jouali F., Marchoudi N. Screening of BRCA1/2 genes mutations and copy number variations in patients with high risk for hereditary breast and ovarian cancer syndrome (HBOC) BMC Cancer. 2020;20(1):747. doi: 10.1186/s12885-020-07250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Incorvaia L., Fanale D., Badalamenti G. Hereditary breast and ovarian cancer in families from Southern Italy (Sicily)—prevalence and geographic distribution of pathogenic variants in BRCA1/2 genes. Cancers. 2020;12(5):1158. doi: 10.3390/cancers12051158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russo A., Calò V., Bruno L. Is BRCA1-5083del19, identified in breast cancer patients of Sicilian origin, a Calabrian founder mutation? Breast Cancer Res Treat. 2008;113(1):67–70. doi: 10.1007/s10549-008-9906-7. [DOI] [PubMed] [Google Scholar]
- 4.Pilarski R. The role of BRCA testing in hereditary pancreatic and prostate cancer families. Am Soc Clin Oncol Educ Book. 2019;39:79–86. doi: 10.1200/EDBK_238977. [DOI] [PubMed] [Google Scholar]
- 5.Mocci E., Milne R.L., Méndez-Villamil E.Y. Risk of pancreatic cancer in breast cancer families from the Breast Cancer Family Registry. Cancer Epidemiol Biomarkers Prev. 2013;22(5):803–811. doi: 10.1158/1055-9965.EPI-12-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhen D.B., Rabe K.G., Gallinger S. BRCA1, BRCA2, PALB2, and CDKN2A mutations in familial pancreatic cancer: a PACGENE study. Genet Med. 2014;17(7):569–577. doi: 10.1038/gim.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Asperen C.J., Brohet R.M., Meijers-Heijboer E.J. Cancer risks in BRCA2 families: estimates for sites other than breast and ovary. J Med Genet. 2005;42(9):711–719. doi: 10.1136/jmg.2004.028829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daly M.B., Pilarski R., Yurgelun M.B. NCCN Guidelines Insights: genetic/familial high-risk assessment: breast, ovarian, and pancreatic, version 1.2020. J Natl Compr Canc Netw. 2020;18(4):380–391. doi: 10.6004/jnccn.2020.0017. [DOI] [PubMed] [Google Scholar]
- 9.Germani A., Petrucci S., De Marchis L. Beyond BRCA1 and BRCA2: deleterious variants in DNA repair pathway genes in Italian families with breast/ovarian and pancreatic cancers. J Clin Med. 2020;9(9):3003. doi: 10.3390/jcm9093003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fanale D., Incorvaia L., Filorizzo C. Detection of germline mutations in a cohort of 139 patients with bilateral breast cancer by multi-gene panel testing: impact of pathogenic variants in other genes beyond BRCA1/2. Cancers. 2020;12(9):2415. doi: 10.3390/cancers12092415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah P.D., Patil S., Dickler M.N., Offit K., Hudis C.A., Robson M.E. Twenty-one-gene recurrence score assay in BRCA-associated versus sporadic breast cancers: differences based on germline mutation status. Cancer. 2016;122(8):1178–1184. doi: 10.1002/cncr.29903. [DOI] [PubMed] [Google Scholar]
- 12.Fountzilas C., Kaklamani V.G. Multi-gene panel testing in breast cancer management. Cancer Treat Res. 2018;173:121–140. doi: 10.1007/978-3-319-70197-4_8. [DOI] [PubMed] [Google Scholar]
- 13.Neben C.L., Zimmer A.D., Stedden W. Multi-gene panel testing of 23,179 individuals for hereditary cancer risk identifies pathogenic variant carriers missed by current genetic testing guidelines. J Mol Diagn. 2019;21(4):646–657. doi: 10.1016/j.jmoldx.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Kurian A.W., Hughes E., Handorf E.A. Breast and ovarian cancer penetrance estimates derived from germline multiple-gene sequencing results in women. JCO Precis Oncol. 2017;1:1–12. doi: 10.1200/PO.16.00066. [DOI] [PubMed] [Google Scholar]
- 15.Mazzola E., Blackford A., Parmigiani G., Biswas S. Recent enhancements to the genetic risk prediction model BRCAPRO. Cancer Inform. 2015;14(suppl 2):147–157. doi: 10.4137/CIN.S17292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gori S., Barberis M., Bella M.A. Recommendations for the implementation of BRCA testing in ovarian cancer patients and their relatives. Crit Rev Oncol Hematol. 2019;140:67–72. doi: 10.1016/j.critrevonc.2019.05.012. [DOI] [PubMed] [Google Scholar]
- 17.Incorvaia L., Fanale D., Bono M. BRCA1/2 pathogenic variants in triple-negative versus luminal-like breast cancers: genotype–phenotype correlation in a cohort of 531 patients. Ther Adv Med Oncol. 2020;12 doi: 10.1177/1758835920975326. 175883592097532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fanale D., Fiorino A., Incorvaia L. Prevalence and spectrum of germline BRCA1 and BRCA2 variants of uncertain significance in breast/ovarian cancer: mysterious signals from the genome. Front Oncol. 2021;11:682445. doi: 10.3389/fonc.2021.682445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plon S.E., Eccles D.M., Easton D. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29(11):1282–1291. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.den Dunnen J.T., Dalgleish R., Maglott D.R. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37(6):564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 21.Stjepanovic N., Moreira L., Carneiro F. Hereditary gastrointestinal cancers: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2019;30(10):1558–1571. doi: 10.1093/annonc/mdz233. [DOI] [PubMed] [Google Scholar]
- 22.Janatova M., Kleibl Z., Stribrna J. The PALB2 gene is a strong candidate for clinical testing in BRCA1- and BRCA2-negative hereditary breast cancer. Cancer Epidemiol Biomarkers Prev. 2013;22:2323–2332. doi: 10.1158/1055-9965.EPI-13-0745-T. [DOI] [PubMed] [Google Scholar]
- 23.Thoeni C., Amir A., Guo C. A novel nonsense mutation in the EpCAM gene in a patient with congenital tufting enteropathy. J Pediatr Gastroenterol Nutr. 2014;58(1):18–21. doi: 10.1097/MPG.0000000000000106. [DOI] [PubMed] [Google Scholar]
- 24.Mersch J., Jackson M.A., Park M. Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer. 2015;121(2):269–275. doi: 10.1002/cncr.29041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Lorenzo S., Fanale D., Corradino B. Absence of germline CDKN2A mutation in Sicilian patients with familial malignant melanoma: could it be a population-specific genetic signature? Cancer Biol Ther. 2015;17(1):83–90. doi: 10.1080/15384047.2015.1108494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen F.C., van Overeem Hansen T., Sørensen C.S. Hereditary breast and ovarian cancer: new genes in confined pathways. Nat Rev Cancer. 2016;16(9):599–612. doi: 10.1038/nrc.2016.72. [DOI] [PubMed] [Google Scholar]
- 27.Dudley B., Karloski E., Monzon F.A. Germline mutation prevalence in individuals with pancreatic cancer and a history of previous malignancy. Cancer. 2018;124(8):1691–1700. doi: 10.1002/cncr.31242. [DOI] [PubMed] [Google Scholar]
- 28.Beebe-Dimmer J.L., Kapron A.L., Fraser A.M., Smith K.R., Cooney K.A. Risk of prostate cancer associated with familial and hereditary cancer syndromes. J Clin Oncol. 2020;38(16):1807–1813. doi: 10.1200/JCO.19.02808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desmond A., Kurian A.W., Gabree M. Clinical actionability of multigene panel testing for hereditary breast and ovarian cancer risk assessment. JAMA Oncol. 2015;1(7):943–951. doi: 10.1001/jamaoncol.2015.2690. [DOI] [PubMed] [Google Scholar]
- 30.Tsaousis G.N., Papadopoulou E., Apessos A. Analysis of hereditary cancer syndromes by using a panel of genes: novel and multiple pathogenic mutations. BMC Cancer. 2019;19(1):535. doi: 10.1186/s12885-019-5756-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tung N., Battelli C., Allen B. Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer. 2015;121:25–33. doi: 10.1002/cncr.29010. [DOI] [PubMed] [Google Scholar]
- 32.Easton D.F., Pharoah P.D., Antoniou A.C. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372(23):2243–2257. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin H.-C., Lee H.B., Yoo T.K. Detection of germline mutations in breast cancer patients with clinical features of hereditary cancer syndrome using a multi-gene panel test. Cancer Res Treat. 2020;52(3):697–713. doi: 10.4143/crt.2019.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corredor J., Woodson A.H., Gutierrez Barrera A., Arun B. Multigene panel testing results in patients with multiple breast cancer primaries. Breast J. 2020;26:1337–1342. doi: 10.1111/tbj.13762. [DOI] [PubMed] [Google Scholar]
- 35.Crawford B., Adams S.B., Sittler T. Multi-gene panel testing for hereditary cancer predisposition in unsolved high-risk breast and ovarian cancer patients. Breast Cancer Res Treat. 2017;163(2):383–390. doi: 10.1007/s10549-017-4181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurian A.W., Hare E.E., Mills M.A. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol. 2014;32(19):2001–2009. doi: 10.1200/JCO.2013.53.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LaDuca H., Stuenkel A.J., Dolinsky J.S. Utilization of multigene panels in hereditary cancer predisposition testing: analysis of more than 2,000 patients. Genet Med. 2014;16(11):830–837. doi: 10.1038/gim.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu C., LaDuca H., Shimelis H. Multigene hereditary cancer panels reveal high-risk pancreatic cancer susceptibility genes. JCO Precis Oncol. 2018:1–28. doi: 10.1200/PO.17.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding D., Zhang Y., He X., Meng W., Ma W., Zheng W. Frequency of the CHEK2 1100delC mutation among women with early-onset and bilateral breast cancer. Breast Cancer Res. 2012;14(2):401. doi: 10.1186/bcr3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weischer M., Nordestgaard B.G., Pharoah P. CHEK2∗1100delC heterozygosity in women with breast cancer associated with early death, breast cancer–specific death, and increased risk of a second breast cancer. J Clin Oncol. 2012;30(35):4308–4316. doi: 10.1200/JCO.2012.42.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rizzolo P., Silvestri V., Bucalo A. Contribution of MUTYH variants to male breast cancer risk: results from a multicenter study in Italy. Front Oncol. 2018;8:583. doi: 10.3389/fonc.2018.00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Win A.K., Reece J.C., Dowty J.G. Risk of extracolonic cancers for people with biallelic and monoallelic mutations in MUTYH. Int J Cancer. 2016;139(7):1557–1563. doi: 10.1002/ijc.30197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wasielewski M., Out A.A., Vermeulen J. Increased MUTYH mutation frequency among Dutch families with breast cancer and colorectal cancer. Breast Cancer Res Treat. 2010;124(3):635–641. doi: 10.1007/s10549-010-0801-7. [DOI] [PubMed] [Google Scholar]
- 44.Rennert G, Lejbkowicz F, Cohen I, et al. MutYH mutation carriers have increased breast cancer risk. Cancer. 118(8):1989-1993. [DOI] [PubMed]
- 45.Hutchcraft M.L., Gallion H.H., Kolesar J.M. MUTYH as an emerging predictive biomarker in ovarian cancer. Diagnostics. 2021;11(1):84. doi: 10.3390/diagnostics11010084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaffee K.G., Oberg A.L., McWilliams R.R. Prevalence of germ-line mutations in cancer genes among pancreatic cancer patients with a positive family history. Genet Med. 2017;20(1):119–127. doi: 10.1038/gim.2017.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daly M.B., Pal T., Berry M.P. Genetic/familial high-risk assessment: breast, ovarian, and pancreatic, version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2021;19(1):77–102. doi: 10.6004/jnccn.2021.0001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.