Graphical abstract

Keywords: Tuberculosis, Multi-drug resistant tuberculosis, Pharmacometabolomics, Metabolomics, Antitubercular drugs, Drug discovery, Mechanisms of action
Abstract
Tuberculosis (TB), one of the oldest and deadliest bacterial diseases, continues to cause serious global economic, health, and social problems. Current TB treatments are lengthy, expensive, and routinely ineffective against emerging drug resistant strains. Thus, there is an urgent need for the identification and development of novel TB drugs possessing comprehensive and specific mechanisms of action (MoAs). Metabolomics is a valuable approach to elucidating the MoA, toxicity, and potency of promising chemical leads, which is a critical step of the drug discovery process. Recent advances in metabolomics methodologies for deciphering MoAs include high-throughput screening techniques, the integration of multiple omics methods, mass spectrometry imaging, and software for automated analysis. This review describes recently introduced metabolomics methodologies and techniques for drug discovery, highlighting specific applications to the discovery of new antitubercular drugs and the elucidation of their MoAs.
1. Introduction
Mycobacterium tuberculosis (M. tb.), the etiologic agent of human tuberculosis (TB), has ancient roots [1]. The genus Mycobacterium is hypothesized to have existed for 150 million years and an early ancestor of M. tb. was present in East Africa three million years ago [2]. The modern form of M. tb. has been present for at least 70,000 years [2]. TB is a global pandemic that mostly impacts low and middle income countries [3]. According to the World Health Organization (WHO) global tuberculosis report, 10 million people worldwide were diagnosed with TB in 2019, and 1.4 million people died from the disease [4]. TB is treatable with available antibiotics, but outcomes are often unsuccessful. In particular, the emergence of rifampicin resistant (RR-TB), multi-drug resistant (MDR-TB), or extensively drug-resistant (XDR-TB) forms of M. tb. has only exasperated the worldwide health crisis [5]. Globally, over two hundred thousand people tested positive for MDR-TB or RR-TB in 2019; this represents a 10% increase from 2018 [4]. Although cases of XDR-TB declined 6% from 2018 to 2019 according to the 2020 Global TB report [4], MDR-TB and XDR-TB remain a major concern in the fight to control TB [6]. MDR strains of M. tb., which are resistant to both first-line antituberculosis drugs (i.e., rifampicin and isoniazid), were responsible for approximately a quarter of all deaths caused by antimicrobial-resistant infections. Treatment regimens for MDR-TB have a low rate of success, and are unacceptably long (greater than 18 months), which decreases treatment compliance and increases the potential for side effects [7]. Limiting the occurrence of active MDR-TB incidences from the growing reservoir of latent MDR infections (i.e., latent tuberculosis caused by MDR M. tb. strains) is crucial to the successful management of TB [7].
The emergence of MDR and XDR strains has rapidly increased the need for new antitubercular drugs [8]. However, the rate of progress in the development of new TB drugs is hindered by a variety of technical challenges. A dangerous respiratory pathogen, M. tb. is a threat to laboratory personnel, which complicates the handling of the organism in a research setting. Biosafety Level 3 (BSL3) facilities are needed to safely investigate M. tb. [9], but these facilities are very expensive to maintain and add a significant burden to research [9]. In addition, M. tb. is a slow growing pathogen and requires long drug exposure times. The unique nature of the M. tb. cell wall, which is lipid-rich and includes an extremely thick peptidoglycan layer, prevents many small molecules from accessing M. tb internal targets [10]. Spontaneous chromosomal mutations selected under antibiotic pressure have led to target alterations or the overproduction of multiple efflux pumps, which are the most important mechanisms of resistance employed by M. tb. These mechanisms need to be better understood to combat resistance and recover drug efficacy [11]. Furthermore, M. tb. exists as heterogeneous populations within different microenvironments in infected hosts, which requires drug candidates to be active against multiple forms of M. tb. Finally, given that antitubercular therapies often involve a multi-drug regimen that may continue for more than eighteen months, any promising candidate should have minimum drug-drug interactions and should display minimal toxicity or side effects [8].
2. Antitubercular drugs
The standard care for TB patients involves two months of treatments with a four drug cocktail (isoniazid, rifampicin, pyrazinamide, and ethambutol) followed by four additional months of treatment with rifampicin and isoniazid [5], [12], [13]. This drug-treatment regimen is currently used for the majority of TB cases and has an 80–90% success rate for patients infected with drug-sensitive M. tb. Please see Table 1 for the current list of antituberculosis drugs used in the treatment of TB [5]. Conversely, an MDR-TB regimen requires at least 18 months of treatments with five or more TB drugs that include second-line and injectable drugs that are known to be more toxic, inefficient, and poorly tolerated. The current treatment of MDR-TB only has a cure rate of 60–70% [5], [14]. Unfortunately, treatment outcomes for pre-extensively drug resistant TB (Pre-XDR TB) and XDR-TB patients are still poor [15], [16]. The development of effective treatments for drug-resistant TB remains a major challenge.
Table 1.
| Name | MW | logP | HBD | HBA | PSA | ROTB | Ro5 | No. violation |
|---|---|---|---|---|---|---|---|---|
| cIsoniazid | 137.14 | −0.69 | 2 | 3 | 68.01 | 1 | Pass | 0 |
| cRifampicin | 822.95 | 2.95 | 6 | 14 | 220.15 | 5 | Fail | 3 |
| cPyrazinimide | 123.11 | −1.23 | 1 | 3 | 68.87 | 1 | Pass | 0 |
| cEthambutol | 204.31 | −0.06 | 4 | 4 | 64.52 | 9 | Pass | 0 |
| cStreptomycin | 581.58 | −7.06 | 12 | 19 | 336.43 | 9 | Fail | 3 |
| dAmikacin | 585.61 | −8.58 | 13 | 17 | 331.94 | 10 | Fail | 3 |
| dp-Amino salicylic acid | 153.14 | 0.83 | 3 | 4 | 83.55 | 1 | Pass | 0 |
| dCycloserine | 102.09 | −2.42 | 2 | 3 | 64.35 | 0 | Pass | 0 |
| dLevofloxacin | 361.37 | 0.51 | 1 | 7 | 73.32 | 2 | Pass | 0 |
| dMoxifloxacin | 401.44 | −0.50 | 2 | 7 | 82.11 | 4 | Pass | 0 |
| dEthionamide | 166.24 | 1.33 | 1 | 1 | 38.91 | 2 | Pass | 0 |
| dProthionamide | 180.27 | 1.77 | 1 | 1 | 38.91 | 3 | Pass | 0 |
| dTerizidone | 302.29 | −0.34 | 2 | 6 | 101.38 | 4 | Pass | 0 |
| dLinezolid | 337.35 | 0.64 | 1 | 5 | 71.11 | 4 | Pass | 0 |
| dClofazimine | 473.40 | 7.30 | 1 | 4 | 39.99 | 4 | Pass | 1 |
| dBedaquiline | 555.52 | 7.13 | 1 | 4 | 45.59 | 8 | Fail | 2 |
| dDelamanid | 534.49 | 6.14 | 0 | 8 | 101.12 | 9 | Fail | 2 |
| dMeropenem | 383.46 | −4.35 | 6 | 3 | 110.18 | 5 | Pass | 2 |
data derived from Motamen, S.; Quinn, R. J., Analysis of Approaches to Anti-tuberculosis Compounds. ACS Omega2020, 5 (44), 28529–28540. Copyright 2020 American Chemical Society.
Abbreviations correspond to: MW, molecular weight; log P: logarithm of partition coefficient of a molecule between aqueous and lipophilic phases, usually octanol and water; HBD, hydrogen bond donor; HBA, hydrogen bond acceptor; PSA, polar surface area; ROTB, rotatable bonds; Ro5, Lipinski Rule of Five; No. violations, number of quantitative estimate of drug-likeness that were violated: HBD ≤ 5, HBA ≤ 10, MW ≤ 500, log P ≤ 5, ROTBs ≤ 10, and PSA ≤ 140 Å.
First line TB drugs.
Second line TB drugs.
An often interacting collection of factors can contribute to drug resistance in TB [17]. Drug-resistant TB can be acquired by direct transmission but can also arise through unsuccessful treatment regimens, which may result from patient non-compliance, or misuse or extended use of first line drugs. Thus, despite extensive and ongoing efforts, major gaps still remain in achieving a successful treatment for the emerging problem of MDR-TB and XDR-TB. A notable hindrance to this effort is the inability to easily define the mechanism of action (MoA) of existing drugs or novel drug candidates. In this regard, untargeted metabolomics may prove to be a valuable approach [6].
3. Metabolomics to the rescue
Metabolomics is the quantitative and/or qualitative characterization of small molecular-weight molecules (i.e., metabolites) within biofluids, cells, organisms, or tissues [18], [19], [20]. NMR and mass spectrometry (MS) are the analytical platforms most commonly used to monitor metabolome changes [10], [18], [21], [22], [23]. Metabolomics has emerged as a tool to investigate the MoA of chemical leads and drug canidates [18]. Specifically, the metabolic impact of a drug treatment can be ascertained by comparing the metabolomes from cell extracts, biofluids (e.g., blood or urine) or tissues before and after a drug dose has been administered. Thus, metabolomics has increasingly been incorporated into all aspects of the antimicrobial drug discovery and development pipeline, from lead discovery to post-approval drug surveillance (Fig. 1) [24]. Proteomics, transcriptomics and genomics have also been used to investigate drug targets [25], [26], metabolomics may be used in combination with other omics techniques to accelerate the determination of MoAs. Overall, metabolomics is a critical tool for achieving new pharmaceutical developments [22], [26], [27], [28], [29]. In this review, several metabolomics techniques used in drug discovery will be described. We will also highlight the application of metabolomics to antitubercular drug discovery and the determination of drug candidate MoA.
Fig. 1.
Schematic of the drug discovery process with highlighted steps that may benefit from the incorporation of metabolomics methodology.
4. Metabolomics in drug discovery
Metabolomics has been applied to a variety of research areas from disease diagnosis and precision medicine to microbiology and environmental monitoring [18], [30], [31], [32]. The use of metabolomics in drug discovery research is often termed pharmacometabolomics, which is the study of how a subject reacts to a drug treatment [33]. Metabolomics profiles the small molecule or chemical signature of a biological system under a given set of conditions. These conditions may be a disease state or a genetic variant, or a response to a nutrient change, an exposure to a xenobiotic, or a drug treatment. Accordingly, metabolomics is useful for identifying new drug targets and understanding their MoA, and has beneficially impacted the discovery, validation and elucidation of the molecular mechanism of novel antimicrobials [34], [35]. A chemical lead or drug candidate that inhibits enzymes, signaling pathways, transporters, or other metabolic processes is likely to cause either an accumulation or depletion of specific substrates, products, nutrients or other metabolites. In this regard, it is possible to determine the lethal target of a drug by detecting the metabolic changes in response to a drug treatment [36]. In general, the global metabolomes of drug‐treated and untreated bacteria or M. tb. cells are compared in an unbiased manner, in which the levels of metabolite biomarkers provide a tool to identify potential drug failures, detect unexpected side effects or to inform pass/no-pass decisions in animal or clinical trials. Incorporating metabolomics into the drug discovery pipeline can significantly accelerate drug development by avoiding false-leads and wasted effort. The estimated cost of developing a new drug is upwards of $1.3 billion dollars [37] and the average time before FDA approval is at least twelve years [38]. Further exasperating this situation is the low (~10%) success rate of drug discovery [39]. Thus, there is a strong demand to find new technologies to make the research and development processes more rapid, more cost-effective, and more successful [40], [41]. Metabolomics or pharmacometabolomics is expected to dramatically facilitate the drug discovery process and reduce these overall costs [33], [42], [43]. In fact, technological advancements and the incorporation of high-throughput approaches are enabling metabolomics to contribute to all stages of the drug development pipeline, including target discovery, deciphering MoAs, or predicting toxicity (Fig. 1) [33].
5. Metabolomic methods used in drug discovery
5.1. Mass spectrometry imaging
Mass spectrometry imaging (MSI) has recently been employed to understand the mode of action of drugs in vivo and in situ by spatially characterizing metabolic changes in tissue samples [44], [45]. The MSI protocol consists of (1) sample preparation, (2) analyte desorption and ionization, (3) mass analysis, and (4) image registration (Fig. 2A) [46]. The ionization techniques commonly used in MSI are secondary ionization mass spectrometry (SIMS), desorption electrospray ionization (DESI) and matrix assisted laser desorption ionization (MALDI) [47], [48], [49]. These ionization techniques are used to record the spatial distribution of molecules by scanning across a tissue sample and measuring a mass spectrum at each x, y grid coordinate [49]. MSI can achieve a spatial resolution from 100 μm to as low as 20 μm [50]. To obtain an image, a cryosection of a tissue sample with a 10 to 20 μm thickness is typically mounted on a glass slide coated with a conductive material and the surface sprayed with a matrix enabling ionization; 1,5-diaminonapthalene and 9-aminoacridine are two common matrix components. The sample is placed into the MS instrument and the tissue surface is rastered with a laser, with analytes desorbed and ionized from the surface at each site of laser ablation to yield a complete mass spectrum [44]. The MSI dataset is commonly presented as a two-dimensional distribution of ions superimposed onto an original image of the tissue sample [44].
Fig. 2.
(A) Strategy of Imaging Metabolomics. Reprinted with permission from He, J.; Luo, Z.; Huang, L.; He, J.; Chen, Y.; Rong, X.; Jia, S.; Tang, F.; Wang, X.; Zhang, R.; Zhang, J.; Shi, J.; Abliz, Z., Ambient mass spectrometry imaging metabolomics method provides novel insights into the action mechanism of drug candidates. Analytical Chemistry2015,87 (10), 5372-9. Copyright 2015 American Chemical Society. (B) Optical images of whole-body tissue sections at different time points after administration of NHBA, and spatiotemporal visualization of NHBA and six endogenous metabolites in rat whole-body tissue sections. Rats were given NHBA (40 mg/kg via intraperitoneal injection) and euthanized 10 min, 20 min, 30 min, or 1 h later. MSI data were acquired by AFADESI-MSI (MRM, NHBA: m/z 374.2 → 242.0; GABA: m/z 104.1 → 69.0; choline:m/z 104.1 → 60.0;valine: m/z 118.1 → 72.0;creatine: m/z 132.1 → 90.0;glycerophosphocholine: m/z 258.1 → 104.0; adenosine: m/z 268.0 → 136.0). Spatial resolution = 300 μm Å~ 500 μm. Reprinted with permission from He, J.; Luo, Z.; Huang, L.; He, J.; Chen, Y.; Rong, X.; Jia, S.; Tang, F.; Wang, X.; Zhang, R.; Zhang, J.; Shi, J.; Abliz, Z., Ambient mass spectrometry imaging metabolomics method provides novel insights into the action mechanism of drug candidates. Analytical Chemistry2015,87 (10), 5372–9. Copyright 2015 American Chemical Society.
MSI imaging is faster than conventional MS-based metabolomics; in particular, it does not require sample extraction or chromatography [51]. Notably, MSI can be performed under ambient conditions, where the ionization and desorption of analytes occurs in an open environment. In this regard, the direct analysis of metabolites from a tissue sample with minimal preparation or handling avoids biologically-irrelevant perturbations in the metabolome and erroneous scientific conclusions [49], [52]. The preparation protocol is sample dependent since animal tissues need to be sectioned and dried but plant components (e.g., leaves, stems, roots, etc.) can often be directly analyzed [49]. In contrast to the analysis of homogenized samples, which will only yield an average view of the composition of the metabolome and may provide an incomplete or incorrect view of a metabolic response due to a disease state or drug treatment, MSI can detect spatially dependent variations in metabolites. For example, any discrepancy in the spatial distribution of metabolites in heterogeneous cancer tissues is frequently lost with conventional liquid chromatography-mass spectrometry (LC-MS) analysis of tissue homogenates [51]. Simply, the metabolism and transport of metabolites in tissues are dynamic and spatially resolved. MSI has been used to image anticancer drugs and their metabolites within tumors to characterize drug distribution and to identify secondary metabolites and toxicity sources [53], [54]. Overall, imaging the metabolome provides a better view of the pathophysiology [55], where the spatial distribution of molecules is also valuable for biomarker and drug discovery [51]. MSI does have limitations, including slower throughput, less coverage of the metabolome (fewer detected metabolites), and lower sensitivity [55].
He et al. (2011) recently developed an MS method for imaging the metabolome based on air-flow assisted desorption electrospray ionization mass spectrometry imaging (AFADESI-MSI) under in situ conditions [56]. AFADESI-MSI is essentially an air flow assisted desorption MS electrospray ionization in which analytes are ionized and the resulting ions lifted from the surface by an ionizing stream containing one or more solvents. This technique has been used to map drugs and drug metabolites in whole-body tissue sections under ambient conditions. AFADESI-MSI is particularly useful for the analysis of these large tissue samples because the high efficiency of extraction improves ion collection and remote transport efficiency while also promoting charged droplet desolvation [47], [56], [57]. In comparison, whole-body MALDI imaging requires the subdividing of large tissue sections due to limited vacuum sample space [57]. In addition, imaging atop a MALDI matrix can cause sample movement and localization errors; a matrix is not required for AFADESI-MSI [57]. The AFADESI-MSI approach was illustrated by profiling the metabolome of the whole-body sagittal plane (i.e. longitudinal plane) of rats as cryosections harvested after administration of an anti-insomnia drug candidate, N6-(4-hydroxybenzyl)-adenosine (NHBA). NHBA acts within the brain and is of interest as a novel neuroprotective compound [47]. AFADESI-MSI was used to spatially map the distribution of NHBA within whole-body rat tissues. NHBA was observed to accumulate in the intestines with very little of the drug distributed to the brain. To understand the basis for the sedative and hypnotic effect of NHBA given the trace levels found in brain tissue, AFADESI-MSI in a full scan mode was next used to profile whole-body metabolome changes following administration of normal and high doses of NHBA compared to controls. A principal component analysis (PCA) of a hyperspectral MSI data matrix revealed molecular alterations in response to stimulation by NHBA. Specifically, six endogenous metabolites corresponding to adenosine, choline, creatine, GABA, glycerophosphocholine, and valine changed after drug administration (Fig. 2B). These metabolites were then analyzed at various time points to acquire a spatiotemporal response to NHBA, which revealed a pharmacological network of multiple mechanistic pathways associated with the sedative and hypnotic effects of NHBA [47].
MSI may accelerate drug development by facilitating the determination of MoAs, by identifying impacted metabolic pathways, and by validating therapeutic targets. MSI may also detect possible side effects or in situ toxicities. Overall, these outcomes can guide the evolution of chemical leads by identifying compounds that produced the spatially desired metabolic response. Simply, MSI can identify compounds that lead to metabolic changes consistent with a maintenance or enhancement of efficacy while avoiding side effects. Thus, MSI is anticipated to be very useful during the early preclinical stages of drug development as a tool for enhancing the identification and prioritization of drug candidates [47].
5.2. High-throughput metabolomics profiling
The lack of fast and efficient methods for deciphering MoAs for chemical leads is a current bottleneck in the drug discovery pipeline. HTS is an established and important component of the drug discovery process [58], [59], [60], [61]. HTS is commonly used during the early stages of drug discovery for the rapid evaluation of millions of compounds for potential inhibitors. While primarily used at present to identify chemical leads, robotic HTS may also help to improve the throughput of other steps of the drug discovery process. Simply, faster and robust high-throughput metabolomics assays may be the key to elucidating the MoAs of drug candidates. A methodology previously described by Halouska et al. (2012) provides a general approach to assign MoAs to a chemical lead (Fig. 3A) [62]. Simply, the MoA for a compound without an established MoA can be inferred on the basis of a relative similarity to the metabolic profiles observed after treatment with drugs with known MoAs.
Fig. 3.
(A) 2D OPLS-DA scores plot demonstrating the clustering pattern for 12 antibiotics with known biological targets and three compounds of unknown in vivo activity: untreated M. smegmatis cells, and chloramphenicol, ciprofloxacin, gentamicin, kanamycin, rifampicin, streptomycin, ethambutol, ethionamide, isoniazid, ampicillin, D-cycloserine, vancomycin, amiodorone, chlorprothixene, or clofazimine treated M. smegmatis cells. The ellipses correspond to the 95% confidence limits from a normal distribution for each cluster. The untreated M. smegmatis cells (■) was designated the control class and the remainder of the cells were designated as treated. The OPLS-DA used one predictive component and six orthogonal components to yield a R2X of 0.715, R2Y of 0.803 and Q2 of 0.671. Reprinted with permission from Halouska, S.; Fenton, R. J.; Barletta, R. G.; Powers, R., Predicting the in vivo mechanism of action for drug leads using NMR metabolomics. ACS Chem Biol2012,7 (1), 166–71. Copyright 2012 American Chemical Society. (B) Annotated Compound Activity Map. An expanded view of the Compound Activity Map with the extracts and m/z features separated into subclusters and colored coded using the Gephi modularity function. Each bioactive subcluster is composed of extracts containing a family of compounds with a defined biological activity. The Compound Activity Map is annotated with a representative molecule from each of the families of compounds that have been independently confirmed by purification and chemical analysis. Reprinted with permission from Kurita, K. L.; Glassey, E.; Linington, R. G., Integration of high-content screening and untargeted metabolomics for comprehensive functional annotation of natural product libraries. Proc Natl Acad Sci U S A2015,112 (39), 11999–2004. Copyright 2015 National Academy of Sciences.
Tiziani et al. (2011) introduced an NMR-based method for screening the metabolic response of drug-treated mammalian cells using a 96-well format [63]. Briefly, the method was validated using CCRF-CEM, SKOV-3 or HeLa cells seeded at a density of 1 × 104 to 1.5 × 105 cells per well and then treated with either dichloroacetate (20 μM), dexamethasone (50 nM), rapamycin (100 nM), or vincristine (1.0 nM) for 24 h. The 96-well plates were then treated with 0.5% w/v of sodium dodecyl sulfate, sealed and placed in a 4 °C ultra-sonicating water bath for 5 min to lyse the cells. The contents of each well were then transferred to 3 mm NMR tubes containing a phosphate buffer, D2O and TMSP for NMR data collection [e.g., 1D 1H NMR, Carr-Purcell-Meiboom-Gill (CPMG) spin echo, two-dimensional (2-D) 1H J-resolved (JRES)]. A multichannel electronic pipette was used to automate the liquid transfer steps. PCA models of the NMR datasets demonstrated clear group separation based on the type of drug treatment or the identity of the cell line. The CPMG spin echo or 2D JRES pulse sequence was observed to efficiently remove background signals from cell debris and biomolecules. The results are interesting given that the application of the CPMG experiment to metabolomics samples has previously yielded erroneous results due to protein-metabolite interactions [64]. The NMR-based metabolomics screen was tested using a small library of 56 known drugs with defined MoAs (i.e., kinase inhibitors). CCRF-CEM cells were treated with 1.0 μM of each drug, after which the cells were characterized by both ATP bioluminescence measurements and metabolome changes. Changes in the concentrations of several metabolites were correlated with both drug treatment and cell survivability; four of these hits were further validated using a clinical metabolic parameter (i.e., lactate/pyruvate ratio). Specifically, an eEF-2 kinase inhibitor and an NF-kB activation inhibitor increased the lactate/pyruvate ratio; whereas MK2 and PKA, PKC, and PKG inhibitors induced a decrease in the ratio of lactate/pyruvate. Accordingly, the NMR-based metabolomics screen may enable personalized medicine based upon rapid pre-screening of potential drug treatments against patient-derived cells and monitoring whether the metabolic profile of the treated cells predict an efficacious outcome [63].
Kurita et al. (2015) developed a compound activity mapping platform, which integrated an image based phenotypic screen with a high-resolution untargeted metabolomics analysis [65]. The platform can predict the identities and MoA of bioactive constituents from complex natural product extracts. The utility of the platform was demonstrated by treating HeLa cells in 384-well plates with a library of 234 natural product extracts. A total of 10,977 mass spectral features from untargeted metabolomics were combined with 58,032 biological measurements (e.g., size, shape, area, number of nuclei or mitotic cells, etc.) from cell images obtained for each well. The authors defined both activity scores and cluster scores to filter the mass spectral features that were correlated with the cytological profile for each natural product extract. From this analysis, 57 cytological profiles were predicted from 634 mass spectral features, which were then used to generate a network map. Bioactive clusters in the resulting network either contained a single known class of compounds, multiple known compound classes, or an unknown compound (Fig. 3B). Overall, 13 unique clusters were identified that contained 11 known and 4 new compound families. The new compounds included the quinocinnolinomycin family of natural products possessing a unique skeleton and set of m/z features that were correlated with endoplasmic reticulum stress. Accordingly, the compound activity mapping platform may expedite the discovery process by providing a HTS-compatible screening method which supports a rational approach to lead discovery.
Zampieri et al. (2018) recently developed a metabolome profiling strategy to classify the MoA of bioactive compounds targeting Mycolicibacterium (aka., Mycobacterium) smegmatis (https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=1866885), a commonly used non-pathogenic model for M. tb [27]. The method, which is both simple and rapid, is outlined in Fig. 4A. The metabolomics workflow uses 96-well plates to cultivate M. smegmatis, allowing the simultaneous comparison of multiple drug candidates. Cells were grown in 700 μl of a standard 7H9 media to an OD595 of approximately 0.4 and then treated with either 10 μM of a compound with an unknown MoA or a drug of a known MoA at a dosage already demonstrated to inhibit cell growth by 20% to 100% [27]. The chemical library consisted of 62 antibiotics with known MoAs and 212 antituberculosis compounds with unknown MoAs. After drug treatment, the cell cultures were tested at six different time points ranging from 5 to 600 min. A cold solvent extraction was used on the entire cell culture without any cell removal, where the supernatant was then directly injected into the mass spectrometer. The relative changes in metabolite intensities were measured to create a profile for each drug or compound. These metabolic profiles for known MoAs was then leveraged to classify the antimycobacterial compounds without an MoA by a direct comparison [27]. In this manner, 77% of the 212 antimycobacterial compounds induced metabolic responses in M. smegmatis consistent with the known MoAs. The MoAs for seven of these compounds were experimentally validated. 15% of the compounds did not show a significant metabolic response compared to controls. Only 8% of the compounds generated a unique metabolic response suggestive of a novel cellular target or MoA. The metabolic profile of six compounds suggested an interference with trehalose and lipid metabolism; the proposed MoA was supported by secondary assays. Thus, HTS metabolome profiling revealed new druggable pathways in M. tb. and enabled the discovery of drug candidates with potentially novel MoAs.
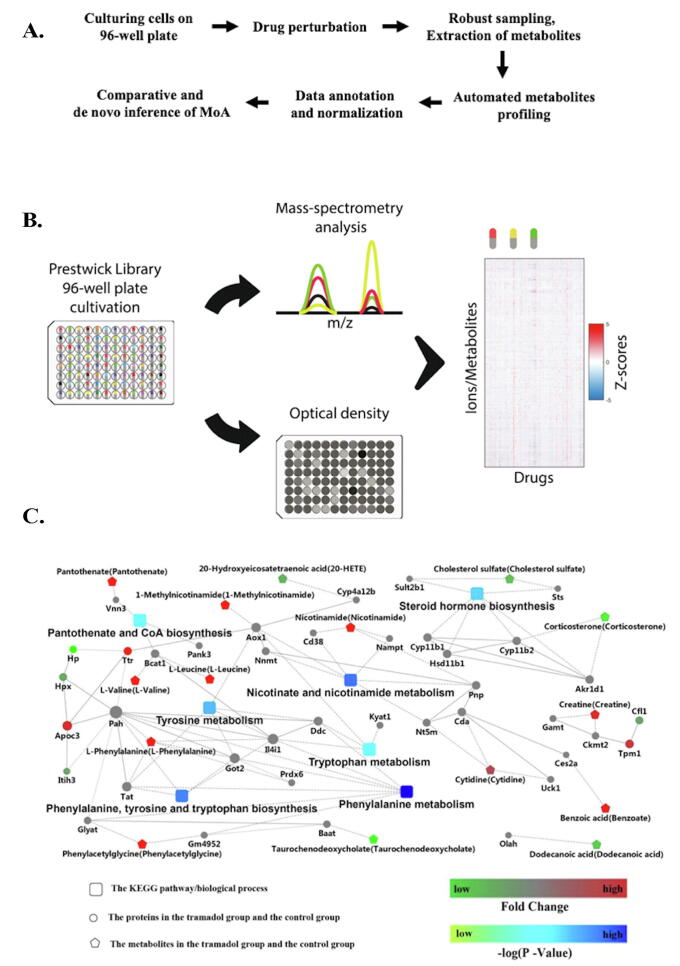
Fig. 4.
(A) Metabolomics workflow. Cells were grown in 700-µl volumes in 96-well plates to an OD595 of about 0.4, before addition of 10 µl of the antimicrobial compound. Cell culture (80 µl) was withdrawn from each well at each sampling time. Forty microliters were used to determine cell density, and the remaining 40 µl was added to cold extraction buffer. Supernatant was directly injected into a time-of-flight mass spectrometer, and relative changes in metabolite intensities were extrapolated from processing of the metabolome data. Adapted from Zampieri, M.; Szappanos, B.; Buchieri, M. V.; Trauner, A.; Piazza, I.; Picotti, P.; Gagneux, S.; Borrell, S.; Gicquel, B.; Lelievre, J.; Papp, B.; Sauer, U., High-throughput metabolomic analysis predicts mode of action of uncharacterized antimicrobial compounds. Sci Transl Med2018,10 (429). (B) Illustration of the metabolic drug profiling workflow. Growth is monitored using a plate reader up to 6 h after treatment, while metabolomics samples are collected after 2 h of treatment and analyzed by FIA-TOFMS. Reprinted with permission from Campos, A. I.; Zampieri, M., Metabolomics-Driven Exploration of the Chemical Drug Space to Predict Combination Antimicrobial Therapies. Mol Cell2019,74 (6), 1291–1303 e6. Copyright 2020 Elseiver. (C) Combination analyses with proteomics and metabolomics. Red represents up-regulated and green represents down-regulated. The rectangular node represents the KEGG pathway/biological process. The circular and pentagon nodes represent the proteins and the metabolites in the tramadol group and the control group, respectively. The P-value is represented by a blue color gradient; the deeper blue color indicates a larger P-value. Reprinted with permission from Jiang, S.; Liu, G.; Yuan, H.; Xu, E.; Xia, W.; Zhang, X.; Liu, J.; Gao, L., Changes on proteomic and metabolomic profile in serum of mice induced by chronic exposure to tramadol. Sci Rep2021,11 (1), 1454. Copyright 2021 Springer Nature. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
5.3. Integrating metabolomics with other omic techniques
Metabolomics has been shown to be a valuable stand-alone method for rapidly elucidating the MoA of drug candidates. Nevertheless, integrating metabolomics with other omics datasets is expected to improve the efficiency and accuracy of MoA elucidation while also enhancing understanding of the overall biological response to individual treatments [66], [67]. Campos et al. (2019) monitored the metabolic response of E. coli to 1279 chemically diverse drugs from the Prestwick chemical library using a rapid assay combining metabolomics and genomics data [68]. Only 11% of these drugs, all of which are FDA approved, have antimicrobial activity. As a result, the chemical library may be used to characterize drug-drug interactions to develop multi-drug therapies by combining drugs with distinct MoAs. The metabolic profile for the entire chemical library was acquired using the HTS method developed by Zampieri et al. as described above (Fig. 4B) [27]. The overall workflow enabled the rapid metabolic profiling of ~ 39,000 ions, with 969 of the spectral features putatively annotated to a metabolite. Seventy percent of the screened drugs induced strong and diverse metabolic responses in E. coli. Since only 15% of the compounds inhibited E. coli growth by more than 20%, the majority of the Prestwick drugs were likely targeting nonessential bacterial processes. The drug-specific metabolic changes were then mapped to the previously established genome-scale metabolic network for E. coli [69] categorizing the metabolic response associated with 3,807 single-gene deletion strains, each associated with the loss of a specific enzymatic function. In this regard, mapping the drug-induced metabolic response to the gene-induced metabolic response may identify a potential drug target (knockout gene) or an indirectly mediating drug response. This comparison resulted in 10,227 significant similarities that implied drug-gene associations. For one treatment, 70 different metabolic pathways were dysregulated, which hints at the potential diversity of metabolic responses across the entire screening library. Moreover, this approach also grouped-together antibiotics with similar MoAs regardless of the in vivo target or growth inhibition.
Predicting drug epistatic interactions remains challenging [70], [71]. To address this issue, the drug-induced metabolic profiles were compared to a previously obtained chemogenomic data set [72]. The E. coli gene-knockout strains were exposed to 82 chemicals to calculate a fitness score based on differences in colony size. In this regard, a Prestwick drug exhibiting a metabolic profile similar to a gene knockout, may epistatically interact with one of the 82 chemicals with a fitness score significantly above the average fitness score. Specifically, a positive fitness score would imply antibiotic resistance, while a negative fitness score would suggest susceptibility. The approach may discover non-antibiotic compounds that, when combined, exhibit promising antimicrobial properties [68].
Proteomics has also been used in combination with metabolomics [73], [74], [75]. An example of this approach is seen in a study of off-target effects of Tramadol, an opioid routinely used as an analgesic. The long-term side effects of Tramadol use are severe yet the basis for the toxicity is not very well understood [76]. Jiang et al. (2021) analyzed the proteomic and metabolomic profile of heart blood from tramadol-treated mice (oral, 50 mg/kg, 5 weeks). Proteomic analysis revealed 31 differentially expressed serum proteins, which included enzyme inhibitor-associated proteins (proteins that specifically bind to and inhibit an enzyme target; examples include plasma protease C1 inhibitor and inter-alpha-trypsin inhibitor heavy chain H3), mitochondria-related proteins, and cytoskeleton proteins. These dysregulated proteins were confirmed by ELISA analysis and formed a highly connected protein–protein interaction network. Some of the proteins were also previously observed in other studies of substance abuse.
An LC-MS metabolomics analysis of the tramadol-treated mouse serum samples revealed 29 significantly perturbed metabolites associated with a variety of KEGG pathways, including: biosynthesis of amino acids; protein digestion and absorption; valine, leucine and isoleucine biosynthesis; and valine, leucine and isoleucine degradation. A combination interaction network of the differentially expressed proteins and the dysregulated metabolites was also highly connected and enriched in KEGG pathways consisting of the biosynthesis of amino acids, steroid hormone biosynthesis, phenylalanine metabolism, tyrosine metabolism, nicotinate and nicotinamide metabolism, and focal adhesion (Fig. 4C). Overall, the combined proteomics and metabolomics results identified protein digestion and absorption as the most common enriched KEGG pathway from a combination of the proteomics and metabolomics results; this included the upregulation of isoleucine, valine, leucine, phenylalanine, glutamine and the down-regulation of carboxypeptidase B2 (CPB2). The down-regulation of CPB2 may offer a useful marker for Tramadol-associated toxicity since CPB2 may protect organs from oxidative damage.
5.4. Software for dose–response metabolomics
Metabolomics may be used to decipher a mechanism of toxicity, identify drug side-effects, determine a toxic dosage, elucidate drug metabolism, and investigate drug-microbiota interactions [31], [77], [78]. However, a simple pairwise comparison of metabolic changes (treated versus untreated) for a single drug-dosage is not sufficient to understand the effects of disrupting two or more independent protein targets [79]. Yao et al. (2020) illustrated a dose–response metabolomics methodology that includes their TOXcms software (http://pattilab.wustl.edu/software/toxcms) to evaluate compounds that differentially target multiple proteins.
Briefly, mammalian cells were treated with multiple doses of the drug or toxicant to generate dose–response curves. In general, a 10-fold range of concentrations was used, centered on the effective dose at median (ED50) of the drug of interest. Metabolites were then extracted from the lysed cells and analyzed with LC-MS based metabolomics. The raw data files from each dose–response experiment were converted to an mzXML format. Peak detection and correspondence determination were performed by the XCMS R packages [80]. The resulting peak tables were then analyzed by TOXcms to generate dose–response curves (i.e., relative peak intensity versus drug concentration) for each spectral feature. TOXcms organizes the dose–response curves based on statistical or similar trends. Plots are grouped based on the directional change in peak intensity resulting from the drug treatment: 1) peak intensity decreases with increasing drug concentration (decrease group), 2) peak intensity increases with increasing drug concentration (increase group), 3) both an increase and decrease in peak intensity (inflection group), 4) features without a clear dosage-dependent trend (no difference group). An ED50 is calculated for only the spectral features assigned to either the increase or decrease group by fitting the data to a logistic dose–response model [81]. A PCA model is created from all features, which is color-coded by the calculated ED50 values. In this regard, the PCA scores plot allows for the identification of metabolic features with similar dose–response trends and ED50 values. Distinct dose–response patterns signify a metabolic response to a different protein target with a unique drug binding affinity.
The capability of TOXcms was evaluated on BT549 breast cancer cells treated with etomoxir. TOXcms identified 908 features, from a total of 26,920 features, demonstrating a statistically significant monotonic increase or decrease in peak intensity as a function of etomoxir dosage. The 3D PCA scores plot (Fig. 5A) showed a bimodal distribution of ED50 values indicating that etomoxir has at least two in vivo targets. Etomoxir is known to inhibit the enzyme carnitine palmitoyl-transferase I (CPT1), which transforms acyl-CoAs to acyl-carnitines (ACs). Features from the PCA scores plot with ED50 values less than 2 μM included an increase in carnitine (Fig. 5B) and a decrease in many acyl chains (Fig. 5C–F), observations which were consistent with an altered CPT1 activity. Conversely, a second set of features from the PCA scores plot had high 200 μM ED50 values associated with a change in central carbon metabolism among other unassigned features. Notably, there was no difference in central carbon metabolism at the lower doses. The large difference in ED50 values implied the alterations in central carbon metabolism were not dependent on CPT1 inhibition and more likely resulted from an off-target effect. Thus, TOXcms revealed a possible second protein target with a lower affinity for etomoxir, which was recently identified as complex I of the electron transport chain and adenine nucleotide translocase [82]. The dose–response trends of acylcarnitine were not correlated with cell proliferation (ED50 94 μM), but TOXcms reported a high correlation between central carbon metabolism (among others) and cell proliferation (Fig. 5H) and mitochondrial respiration (Fig. 5I). Overall, a metabolomic dose–response analysis of etomoxir by TOXcms provided clear evidence that the drug has multiple targets and at high concentrations inhibits both CPT1 activity and mitochondrial respiration. Notably, the analysis also points to the need to understand ED50 as a means of limiting off-target effects.
Fig. 5.
TOXcms reveals that etomoxir has a second protein target at 200 μM in BT549 breast cancer cells. (A) PCA plot shows distribution of ED50 values calculated by TOXcms. Each dot on the plot represents a distinct feature. Features with increasing dose–response trends are shown on the left of the plot, and features with decreasing dose–response trends are shown on the right. ED50 values are color-coded. Features in the data that do not show a statistically significant trend are colored gray. Data from etomoxir-treated samples show a bimodal distribution of ED50 values (purple and green circles). (B-F) Features in the purple circles were identified as substrates and products of the primary target of etomoxir (i.e., CPT1). (G) Features in the green circles were identified to be in pathways distinct from CPT1, and hence were annotated as off-target. (H) At hundreds of micromolar, etomoxir decreases proliferation of BT549 cells. The dose–response curve of this phenotype does not correlate with the dose–response curve of the substrates and products of CPT1. Instead, the dose–response curve of proliferation correlates with the off-target effect of etomoxir. (I) Similarly, the dose–response curve of mitochondrial respiration correlates with the dose–response curve of etomoxir’s off-target effect. Taken together, these data demonstrate that etomoxir inhibits CPT1 at low concentrations but inhibits mitochondrial respiration via an off-target effect at high concentrations. Reprinted with permission from Yao, C. H.; Wang, L.; Stancliffe, E.; Sindelar, M.; Cho, K.; Yin, W.; Wang, Y.; Patti, G. J., Dose-Response Metabolomics To Understand Biochemical Mechanisms and Off-Target Drug Effects with the TOXcms Software. Anal Chem2020,92 (2), 1856–1864. Copyright 2020 American Chemical Society. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
6. Examples of metabolomics in antitubercular drug discovery
Metabolomics delivers unique insights into the composition, organization, activity and the regulation of M. tb. cellular networks [32], [75], [83], [84], [85]. In this regard, the application of metabolomics to M. tb. is facilitating the discovery of new antitubercular drugs, and, accordingly, is a rapidly growing endeavour [32], [83], [84], [85]. NMR metabolomics has been previously used to predict the in vivo MoA for chemical leads by combining 1D 1H NMR data sets with multivariate statistics like PCA or orthogonal projections to latent structures (OPLS) [62]. M. smegmatis cell cultures were treated with twelve antitubercular drugs corresponding to three general MoA classes (e.g., inhibitors of biosynthesis of cell wall, mycolic acids, or proteins). The drugs were dosed at a concentration that induced a 50% decrease in cell growth to avoid cell death and metabolic changes unrelated to the drug’s MoA. The cell extracts were then analyzed by NMR with the scores plot from a PCA or OPLS model used to characterize the relative similarities in metabolic profiles (Fig. 3A). The basic hypothesis is that drugs with similar MoAs or biological activities will induce similar changes to the M. smegmatis metabolome and cluster together in a PCA or OPLS scores plot. M. smegmatis was similarly treated with three chemical leads from the Tuberculosis Antimicrobial Acquisition and Coordinating Facility (TAACF) library, each possessing demonstrated activity but unknown MoA: amiodorone, chlorprothixene, and clofazimine. The three TAACF compounds clustered together with vancomycin and D-cycloserine in the OPLS scores plot. This suggests the three TAACF compounds have a similar MoA with vancomycin and D-cycloserine and are likely inhibitors of cell wall synthesis [62].
The complete characterization of the MoAs for a TB drug may enable the development of a combination therapy to increase treatment efficacy and to suppress drug resistance. One approach is to identify the metabolic response correlated with the known antimicrobial activity of the drug and a secondary metabolic response associated with a previously unknown MoA. The new MoA can then be leveraged as a target for a second drug treatment. Wang et al. (2019) demonstrated this approach by investigating the metabolic impact of bedaquiline (i.e., ATP synthase inhibitor), the first new FDA approved TB drug in over 40 years [86]. Unfortunately, bedaquiline has exhibited serious side effects and resistant strains were quickly reported. Thus, a multidrug treatment that specifically targets the metabolic response to bedaquiline may reverse drug resistance while simultaneously minimizing toxicity by allowing for a lower dose of bedaquiline. The authors had previously described an LC-MS metabolomic profiling platform for M. tb [32], which was used to identify M. tb. metabolite changes due to bedaquiline treatments. Not surprisingly, bedaquiline induced a two-fold decrease in cellular ATP levels. A total of 130 other metabolites were monitored for 48 h following a range of drug doses (e.g., 0x to 50x MIC). A Pearson correlation between these 130 metabolites and ATP identified glutamine as being the most highly correlated (r = 0.809) metabolite. Glutamine synthetase, which is the primary source of glutamine and requires ATP, was identified as a likely secondary target of bedaquiline. The activity of bedaquiline was potentiated upwards of 20-fold by the addition of glutamine synthetase inhibitors (i.e., 2,4,5-trisubstituted imidazoles, methionine sulfoximine), which could be reversed by the addition of glutamine. Thus, the synergistic combination of bedaquiline with a glutamine synthetase inhibitor would require a significantly lower dose of bedaquiline to achieve the same cellular response, and potentially avoiding its toxic side effects. The development of drug resistance may equally be avoided since two enzymes that are part of the same metabolic network were inhibited.
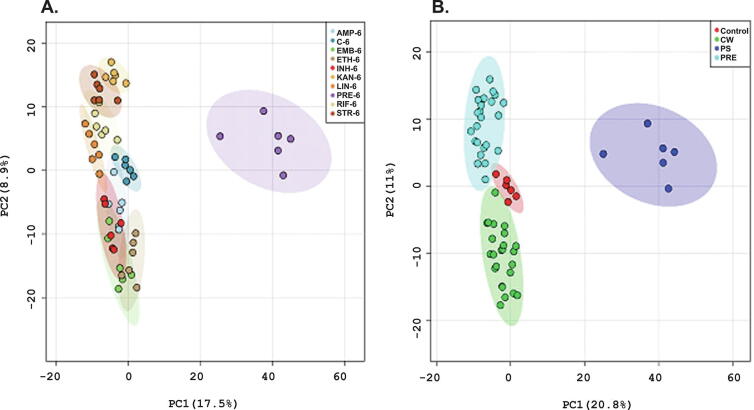
Another example involves Pretomanid, a candidate TB drug in clinical phase III trials, but with a poorly defined MoA [23]. Pretomanid has been extensively studied as a candidate for the treatment of both drug-susceptible and drug-resistant TB [87], [88]. Baptista et al. (2018) used untargeted metabolomics to compare Pretomanid with eight different antitubercular drugs with a range of known MoAs. M. smegmatis was used as a TB model and the metabolome was characterized with flow infusion electrospray ion high resolution mass spectrometry (FIE-HRMS) and gas-chromatography time-of flight mass spectrometry (GC-tofMS). The resulting PCA models from the mass spectrometry data sets showed that Pretomanid treatment generated a unique metabolic profile (Fig. 6) in M. smegmatis cell extracts when compared to the other tested antibiotics. Alterations in the pentose phosphate pathway was the most prominent metabolic change due to Pretomanid treatment. An increase in phosphate sugars was previously linked to the accumulation of the toxic metabolite, methylglyoxal, which showed significant antimicrobial activity against M. smegmatis [23]. Methylglyoxal was detected 6 h following treatment with Pretomanid, which suggests a potential MoA for the TB drug candidate.
Fig. 6.
Demonstrative principal component analysis (PCA) score plots of FIE-MS data of 2285 normalized m/z intensity values (P < 0.05) in both negative and positive ionization mode, 6 h after antibiotic treatment: (A) AMP-ampicillin, C-control group, EMB-ethambutol, ETH-ethionamide, INH-isoniazid, KAN-kanamycin, LIN-linezolid, PRE-pretomanid, RIF-rifampicin, STR-streptomycin; (B) samples were clustered based on their mechanism of action, CW-cell wall synthesis, PS-protein synthesis, PRE-pretomanid treatment. Colored regions display 95% confidence. Reprinted with permission Baptista, R.; Fazakerley, D. M.; Beckmann, M.; Baillie, L.; Mur, L. A. J., Untargeted metabolomics reveals a new mode of action of pretomanid (PA-824). Scientific Reports2018,8 (1), 5084. Copyright 2018 Springer Nature.
Exploring the metabolism of M. tb. is important to understand its ability to survive in a host and disease pathogenesis. Agapova et al. (2019) investigated how M. tb. utilized amino acids as a nitrogen source by studying bacterial physiology and by using stable isotope tracing methods [89]. M. tb. cells were cultured in the presence of each individual 15N-labeled amino acid as the sole nitrogen source. Liquid chromatography-mass spectrometry (LC-MS) was then used to identify the metabolites derived from the 15N-labeled amino acid and the metabolic pathways enriched in 15N-nitrogens. M. tb. took up all amino acids, but had a growth preference and rapid utilization of glutamic acid, aspartic acid, asparagine, and glutamine (i.e., Glu > Asp > Asn > Gln). Notably, some amino acids were homeostatically controlled regardless of the supplied amino acid in the culture medium; whereas, the cellular concentration of other amino acids depended on the extracellular availability of amino acids. Similarly, the rate of 15N-labeling of the metabolome was dependent on the supplied 15N-labeled amino acid and the position of the 15N-label. Overall, alanine and alanine dehydrogenase were identified as critical nodes in nitrogen metabolism, with alanine dehydrogenase playing an obvious role in alanine utilization. In fact, M. tb. lacking alanine dehydrogenase activity did not grow with Ala as the sole nitrogen source, but grew normally with either Gln or NH4+. Taken together, M. tb. revealed a flexible metabolic network with characteristics that are likely a product of evolution in a human host [89].
Fatty acid and lipid synthesis and utilization are critical to the production and stability of the M. tb. cell wall. Accordingly, Sharma et al. (2020) illustrated the importance of lipidomics to characterizing the MoAs of TB drugs. Ultra performance liquid chromatography-electrospray ionization and mass spectrometry (UPLC-ESI-MS) was used to characterize the impact of the antimycobacterial vanillin on various lipid extracts from M. smegmatis. The treatment of M. smegmatis cells with vanillin changed the composition of specific lipid classes, causing a disruption in cell membrane homeostasis [90]. Specifically, vanillin caused an overall increase in total lipids, but a decrease in keto-, α-, and methoxy-mycolic acids, which are abundant in the M. tb. cell wall and essential for its survival in the host. These results suggested vanillin disrupted the synthesis of mycolic acids as evident by the accumulation of mycolipanolic acid. These drug-induced lipid changes highlight the importance of individual components to the integrity of the thick lipid-rich cell wall that enables M. tb. survival under adverse conditions. It also demonstrated the importance of combining lipidomics with metabolomics for profiling TB drugs that target cell walls and mycolic acid biosynthesis.
M. tb. is a persistent intracellular pathogen that is intrinsically tolerant to some antibiotics. An untargeted metabolomics approach was successfully used to identify an enzyme that mediates the broad antibiotic tolerance in M. tb [91], [92]. Metabolic profiling of three different antitubercular drugs, isoniazid, rifampicin, and streptomycin, showed that each targeted different cellular processes but all three triggered the activation of isocitrate lyase (ICL). ICL deficient M. tb. strains were found to be more susceptible to the three antibiotics compared to WT. Thus, ICL may play a role in adaptive antibiotic resistance [91].
The failure of first line TB drugs to treat MDR-TB highlights the need for new antibiotics with novel MoAs. Koen et al. (2018) used 2D gas chromatography time-of-flight mass spectrometry (GCxGC-TOFMS) metabolomics to understand the mechanism of action of colistin sulfate (CS), a polymyxin antibiotic [93]. CS is already established as a last resort antibiotic for treating multidrug-resistant Gram-negative infections such as Pseudomonas aeruginosa, Acinetobacter baumannii, and Klebsiella pneumonia. CS acts as a detergent and the MOA is based upon disruption of the integrity of the bacterial membrane. CS also has a proven activity against mycobacteria and is a lead candidate for treating MDR-TB [93]. The metabolic response to CS treatment, an increase in both fatty acid synthesis and glycolysis, is consistent with the MoA observed for other bacteria, suggesting CS-treated mycobacteria must tap an additional energy source to support cell wall repair.
Ballinger et al. (2019) illustrated the use of LC-MS-based metabolic and lipidomic profiling in combination with an in-depth crystallography study to understand the MoA of HTS chemical leads [94]. An HTS cell-based assay identified the amidino-urea 1-[(2,6-diethylphenyl)-3-N-ethylcarbamimodoyl]urea (referred to as compound 8918), which killed M. tb. cells and drug-resistant clinical isolates with MIC90 values ranging from 0.56 to 3.1 μM. Compound 8918 was as effective as rifampin in treating BALB/c mice infected with M. tb., but with an unknown MoA. The metabolic and lipidomic changes caused by the amidino-urea differed from those seen with other TB drugs (e.g., proguanil, isoniazid, streptomycin, rifampin, paraaminosalicylic acid). Notably, treatment with compound 8918 dysregulated 4′-phosphopantetheine (Ppt), coenzyme A (CoA), and precursors in the metabolic pathway. The amidino-urea also inhibited the synthesis of trehalose dimycolates, trehalose monomycolates, and phthiocerol dimycoserosates. M. tb. strains resistant to compound 8918 had a point mutation in phosphopantetheimnyl transferase (PptT), which synthesizes cell wall lipids critical for virulence. PptT transfers Ppt from CoA to acyl carrier proteins (ACPs), which are involved in the synthesis of cell wall mycolic acids. Ppt hydrolase (PptH) releases Ppt to recycle the ACP. Thus, synergy between PptT and PptH may enhance the lethality of compound 8918. A 1.8 Å crystal structure of PptT with compound 8918 bound within the Ppt binding site supports the hypothesis that compound 8918 targets this transferase. Overall, CoA metabolism was identified as a potentially new target for the development of TB drugs with novel MoAs.
Metabolomics may also be used to reveal toxic side-effects of drug candidates in clinical and clinical toxicology studies. Cao et al. (2018) used the UPLC-MS metabolomics analysis of patient urine samples to study the hepatoxicty of antitubercular drugs. Newly diagnosed TB patients were treated with isoniazid, rifampicin, ethambutol, pyrazinimide and streptomycin for at least one month. The resulting metabolic profiles were assigned to one of three different groups: (i) a preliminary TB diagnosis group without drug treatment, (ii) a drug-induced liver injury (DILI) group, and (iii) a non-DILI group. Urine metabolites affected by the first-line TB treatments were associated with several pathways: tricarboxylic acid circulation, arginine and proline metabolism, and purine metabolism. Specifically, the levels of pyroglutamate, isocitrate, citrate, and xanthine were significantly decreased after the administration of TB drugs. These metabolic perturbations were also different between DILI and non-DILI patients. Urate and cis-4-octenedioic acid levels were significantly increased in DILI compared to non-DILI, while cis-aconitate and hypoxanthine levels were significantly decreased. Overall, the metabolic results suggest superoxide generation may aggravate the hepatotoxic effects of first-line TB drugs [95].
7. Conclusions and future directions
In this review, we focused on recent metabolomics-based techniques and methods that have been applied to the elucidation of MoAs for drug candidates, and to the identification of new druggable targets. We also highlighted recent advancements in the application of metabolomics to the development of antitubercular drugs, and the elucidation of MoAs and processes associated with undesired toxicity. The multiple TB drug candidates that are now in clinical trials have also relied on the incorporation of metabolomics into the drug discovery pipeline. Metabolomics techniques have advanced the development of multi-drug therapies, providing insights into M. tb. physiology, drug efficacy, antibiotic resistance, and the MoA of novel lead candidates.
Nevertheless, there are still significant gaps in the application of metabolomics to the drug discovery process that need to be addressed such as the integration of the various omics platforms, the further development of new screening platforms, and the introduction of various isotopomeric labels. The implementation of machine learning technologies would be particularly useful to improve screening platforms that integrate bactericidal and host toxicity data. Isotopomers may improve the elucidation of precursor-product relationships with the potential to discover new druggable targets and pathways. This should be applied to both in vitro studies of M. tb. metabolic routes and to the eukaryotic host pathways impacted by the pharmacokinetic and pharmacodynamics of developmental drugs. To this end, a more extensive use of rodent models should be used prior to large animals such as swine and primate M. tb. models that better mimic the human host. Finally, a direct link of metabolomics technology to click chemistry approaches should allow for target identification and the subsequent development of novel compounds.
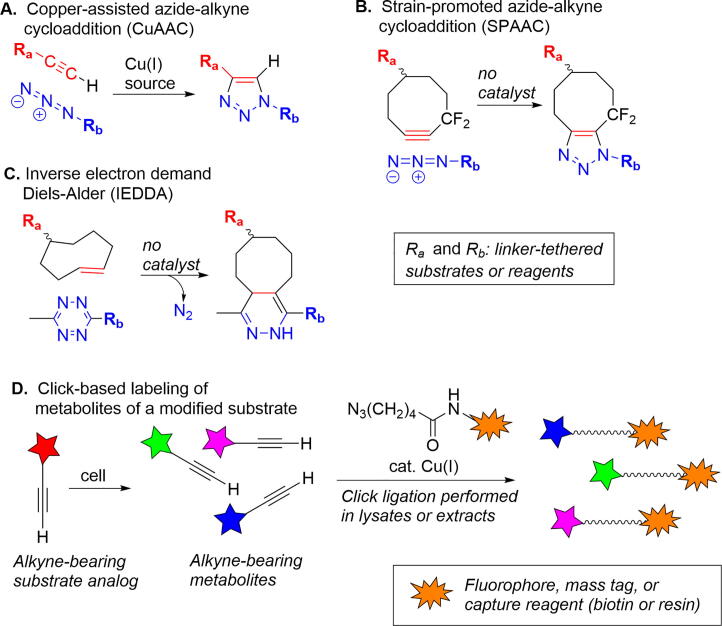
The challenges faced by metabolomics investigations often include the need to efficiently extract and concentrate low abundant metabolites, and to discriminate those metabolites associated with a particular pathway of interest. Click reactions employing minimally modified substrate analogs are emerging as powerful tools for interrogation of metabolic processes. “Click” in this context describes efficient and highly specific reactions between pairs of otherwise unreactive functional groups to form stable linkages (Fig. 7A–C) [96]. While the best known click process remains the copper assisted cycloaddition of azides and terminal alkynes [97], [98], there is increasing interest in “biorthogonal” click reactions unaffected by biological media and compatible with living systems. Most significant among this latter group are metal-free cycloadditions of azides with strained alkynes and inverse electron-demand Diels-Alder cycloadditions of strained alkenes and 1,2,4,5-tetrazines [99], [100], [101].
Fig. 7.
Overview of click chemistry and illustration of application to metabolomics. (A) Copper-assisted azide-alkyne cycloaddition (CuAAC). (B) Strain-promoted azide-alkyne cycloaddition (SPAAC). (C) Inverse electron demand Diels-Alder reaction (IEDDA). (D) Identifying metabolites using alkyne-containing analogs of substrates.
In an example of the application of click methodology to metabolomics, alkyne-terminated analogs of oleic (18:1) and palmitic (16:0) acids were followed through metabolic processes using a post-extraction Cu-promoted reaction with an azide-functionalized fluorophore [102], [103]. A complimentary version of this strategy involves azide derivatives of fatty acids and other natural products [104]; the authors noted that application of this approach for metabolite tracking and discovery is facilitated by the high tolerance of bacterial secondary metabolic pathways for clickable analogs of natural products. A recent report illustrates a click-based strategy for targeted metabolomics based upon incubation with alkyne-containing analogs of natural substrates, followed by selective capture of alkyne-containing species using an azide-containing solid phase resin [105]. The technology, termed DIMEN (deep interrogation of metabolism via enrichment), was applied to tracing the metabolic fate of hydroxy acid glycosides present as signaling molecules in C. elegans. Replacement of a methyl (CH3) sidechain with an ethynyl (–C≡CH, alkyne) group enabled selective capture of downstream species by an azide-containing resin; the characteristic fragmentation pattern of ions derived from the clicked species provided a ready means for their discrimination within complex mixtures. Application of the DIMEN methodology to a single alkyne-containing fatty acid analog revealed a large number of novel metabolites. An overview of the application of click chemistry within metabolomics profiling is shown in Fig. 7D; note that the example could as easily have included azide-modified substrates and capture by an alkyne-containing reagent.
Compared to the other omics techniques, metabolomics is still a young and progressing discipline. Thus, there are extensive opportunities to improve the accuracy, coverage, reproducibility, sensitivity, and throughput of metabolomics data through the development of new methods and tools. Open access data and software repositories is one critical means to facilitate the technological advancements needed by the field. The ready availability of metabolomics data sets with the associated processing protocols, software, and manually curated results would provide an invaluable framework to evaluate and disseminate best practices to the community. In the same manner that the availability of multiple, large scale, HTS results may provide new chemical leads against a diversity of diseases or therapeutic targets, a similar collection of drug-induced metabolic profiles of M. tb. could be leveraged to infer MoAs for novel antitubercular drugs. Drug discovery databases and repositories are still under development, especially regarding mycobacterial research, but their continued growth and development will contribute to advancements in M. tb. drug development.
HTS and various secondary screens often rely on cell-based assays to identify and validate potential leads. The resulting minimal inhibitory concentrations (MICs) are then used to prioritize the chemical leads for further development. But, the outcomes from these assays, including the application of metabolomics as described above, are inherently biased by the assay conditions and/or cell culture media [106]. Drug efficacies and the associated MoAs can drastically change with differences in cell growth conditions [107]. In essence, most cell culture media and growth conditions are designed to promote cell growth instead of mimicking the in vivo conditions of a host. This issue is well illustrated by the MoA of bedaquiline in both M. smegmatis and M. tb. Bedaquiline is an inhibitor of the ATP synthase complex that results in lower ATP levels and a compromised energy state for the bacterium. This mechanism is possible because Mycobacterium must use oxidative phosphorylation to generate ATP from non-fermentable lipids, the preferred carbon source for intracellular pathogenic mycobacteria [108], [109]. As substrate-level phosphorylation is inadequate to supply sufficient ATP, ATP synthase becomes essential for in vivo survival of both replicating and dormant mycobacteria as both states require to a greater or lesser extent a level of ATP for survival. In contrast, in standard culture media with high concentrations of fermentable carbon sources, the effect of bedaquiline is masked, illustrating how different environmental conditions modulate the MoA and perturb drug efficacies. Thus, obtaining reliable cell-based metabolomics data to investigate the MoAs of promising antitubercular drugs will require employing culture conditions that mimic essential details of the host environment, such as the limited availability of critical nutrients or oxygen, the addition of blood, serum or other key components from the host, or the co-culturing with other cell-types to simulate a heterogeneous system.
As exemplified throughout this review, M. smegmatis has often been used as an omics model for M. tb. and as a target in cell-based assays to screen for TB drugs. This raises a serious question – is M. smegmatis a valid surrogate for M. tb.? M. smegmatis has been and will continue to be an excellent model for M. tb [110]. This does not mean that the results of an omics analysis in M. smegmatis can be directly extrapolated to M. tb to elucidate the MoA of a given drug. M. smegmatis strain mc2155 genome is 6.68 Mb (NCBI Reference Sequence: NC_008596.1) in comparison to 4.41 Mb for M. tb. H37Rv strain (NCBI Reference Sequence: NC_000962.3). The main advantages of the M. smegmatis model are the low pathogenicity and the faster growth rate. In addition, there is significant homology at the amino acid level for 60% of M. tb. genes that have M. smegmatis orthologs [111]. Nonetheless, M. tb. is a host-adapted human pathogen that contrasts with the environmental characteristic of M. smegmatis that uses most of the extra genetic repertoire for greater redundancy and encoding of functions involved in the uptake of extracellular nutrients [112]. In contrast, M. tb. possesses several specific genes involved in pathogenesis and cell wall structure, which includes the ability to synthesize longer mycolic acid chains with cyclopropane rings [113]. Nonetheless, the core, essential metabolic pathways that encode drug targets are conserved in both species. The main findings in M. smegmatis are, in many cases, directly translatable to M. tb. In our previous studies to determine the lethal target of D-cycloserine (DCS), we showed that the metabolic impact of DCS was directly linked to the D-alanine pathway, which identified D-alanine ligase as the lethal drug target in both M. tb. and M. smegmatis [114]. However, DCS also had profound effects on the rest of the metabolome where there were significant differences: the M. tb. metabolome showed a higher concentration of carbohydrates and amino sugars that differed from the case of M. smegmatis, where the metabolites being synthesized from D-alanine were well dispersed throughout the metabolome. Nonetheless, even these metabolome differences may yield insights into the pathogenic adaptation.
Overall, metabolomics is a valuable and important component of the drug discovery process for TB. It is critical to understanding the mechanisms of drug resistance, for identifying new druggable targets, and for advancing the field of precision medicine. Metabolomics is currently assisting in elucidating the MoA, toxicity, and potency of promising new antitubercular drugs; and will be critical for the successful delivery of new and desperately needed TB therapies to the world.
CRediT authorship contribution statement
Isin T. Sakallioglu: Conceptualization, Writing - original draft, Writing - review & editing. Raúl G. Barletta: Writing - review & editing, Funding acquisition. Patrick H. Dussault: Writing - review & editing, Funding acquisition. Robert Powers: Conceptualization, Writing - original draft, Writing - review & editing, Funding acquisition, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported in part by funding from the National Science Foundation under Grant Number 1660921, the Redox Biology Center (P30 GM103335, NIGMS), the Nebraska Center for Integrated Biomolecular Communication (P20 GM113126, NIGMS), and the Nebraska Agricultural Experiment Station with funding from the USDA National Institute of Food and Agriculture (NEB-39-179 and NEB-39-178), the School of Veterinary Medicine and Biomedical Sciences, and the University of Nebraska-Lincoln College of Arts and Sciences.
References
- 1.Gutierrez M.C., Brisse S., Brosch R., Fabre M., Omais B., Marmiesse M. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 2005;1(1):e5. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barberis I., Bragazzi N.L., Galluzzo L., Martini M. The history of tuberculosis: from the first historical records to the isolation of Koch's bacillus. J Prev Med Hyg. 2017;58(1):E9–E12. [PMC free article] [PubMed] [Google Scholar]
- 3.Delogu G., Sali M., Fadda G. The biology of mycobacterium tuberculosis infection. Mediterr J Hematol Infect Dis. 2013;5(1):e2013070. doi: 10.4084/MJHID.2013.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. Global tuberculosis report. World Health Organization, Geneva; 2020. www.who.int/news-room/fact-sheets/detail/tuberculosis.
- 5.Motamen S., Quinn R.J. Analysis of Approaches to Anti-tuberculosis Compounds. ACS Omega. 2020;5(44):28529–28540. doi: 10.1021/acsomega.0c03177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuyiringire N., Tusubira D., Munyampundu J.P., Tolo C.U., Muvunyi C.M., Ogwang P.E. Application of metabolomics to drug discovery and understanding the mechanisms of action of medicinal plants with anti-tuberculosis activity. Clin Transl Med. 2018;7(1):29. doi: 10.1186/s40169-018-0208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knight G.M., McQuaid C.F., Dodd P.J., Houben R. Global burden of latent multidrug-resistant tuberculosis: trends and estimates based on mathematical modelling. Lancet Infect Dis. 2019;19(8):903–912. doi: 10.1016/S1473-3099(19)30307-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grzelak E.M., Choules M.P., Gao W., Cai G., Wan B., Wang Y. Strategies in anti-Mycobacterium tuberculosis drug discovery based on phenotypic screening. J Antibiot (Tokyo) 2019;72(10):719–728. doi: 10.1038/s41429-019-0205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Soolingen D., Wisselink H.J., Lumb R., Anthony R., van der Zanden A., Gilpin C. Practical biosafety in the tuberculosis laboratory: containment at the source is what truly counts. Int J Tuberc Lung Dis. 2014;18(8):885–889. doi: 10.5588/ijtld.13.0629. [DOI] [PubMed] [Google Scholar]
- 10.Fatima Z., Nandan S., Hameed S. Understanding Mass Spectrometry-based Global Mycobacterial Lipidomics. Curr Mol Med. 2020;20(8):607–623. doi: 10.2174/1566524020666200206120840. [DOI] [PubMed] [Google Scholar]
- 11.Balganesh M., Dinesh N., Sharma S., Kuruppath S., Nair A.V., Sharma U. Efflux pumps of Mycobacterium tuberculosis play a significant role in antituberculosis activity of potential drug candidates. Antimicrob Agents Chemother. 2012;56(5):2643–2651. doi: 10.1128/AAC.06003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Awasthi D., Freundlich J.S. Antimycobacterial Metabolism: Illuminating Mycobacterium tuberculosis Biology and Drug Discovery. Trends Microbiol. 2017;25(9):756–767. doi: 10.1016/j.tim.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh V., Chibale K. Strategies to Combat Multi-Drug Resistance in Tuberculosis. Acc Chem Res. 2021;54(10):2361–2376. doi: 10.1021/acs.accounts.0c00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibabaw A., Gelaw B., Gebreyes W., Robinson R., Wang S.H., Tessema B. The burden of pre-extensively and extensively drug-resistant tuberculosis among MDR-TB patients in the Amhara region, Ethiopia. PLoS ONE. 2020;15(2):e0229040. doi: 10.1371/journal.pone.0229040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmero D., Gonzalez Montaner P., Cufre M., Garcia A., Vescovo M., Poggi S. First series of patients with XDR and pre-XDR TB treated with regimens that included meropenen-clavulanate in Argentina. Arch Bronconeumol. 2015;51(10):e49–e52. doi: 10.1016/j.arbres.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 16.WHO announces updated definitions of extensively drug-resistant tuberculosis. World Health Organization; 2021.
- 17.Iacobino A., Fattorini L., Giannoni F. Drug-Resistant Tuberculosis 2020: Where We Stand. Appl Sci. 2020;10(6):2153. [Google Scholar]
- 18.Powers R. NMR metabolomics and drug discovery. Magn Reson Chem. 2009;47(Suppl 1):S2–S11. doi: 10.1002/mrc.2461. [DOI] [PubMed] [Google Scholar]
- 19.du Preez I., Luies L., Loots D.T. The application of metabolomics toward pulmonary tuberculosis research. Tuberculosis (Edinb) 2019;115:126–139. doi: 10.1016/j.tube.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Drapal M., Fraser P.D. Metabolite Profiling: A Tool for the Biochemical Characterisation of Mycobacterium sp. Microorganisms. 2019;7(5) doi: 10.3390/microorganisms7050148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leenders J., Frederich M., de Tullio P. Nuclear magnetic resonance: a key metabolomics platform in the drug discovery process. Drug Discov Today Technol. 2015;13:39–46. doi: 10.1016/j.ddtec.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Bhinderwala F., Powers R. NMR Metabolomics Protocols for Drug Discovery. Methods Mol Biol. 2019;2037:265–311. doi: 10.1007/978-1-4939-9690-2_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baptista R., Fazakerley D.M., Beckmann M., Baillie L., Mur L.A.J. Untargeted metabolomics reveals a new mode of action of pretomanid (PA-824) Sci Rep. 2018;8(1):5084. doi: 10.1038/s41598-018-23110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minias Alina Ż.L., Ewelina Lechowicz, Filip Gąsior, Agnieszka Knast, Sabina Podlewska, Daria Zygała, Jarosław Dziadek. Early Drug Development and Evaluation of Putative Antitubercular Compounds in the -Omics Era. Front Microbiol. 2021;11:3640. doi: 10.3389/fmicb.2020.618168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powers R. The current state of drug discovery and a potential role for NMR metabolomics. J Med Chem. 2014;57(14):5860–5870. doi: 10.1021/jm401803b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russell C., Rahman A., Mohammed A.R. Application of genomics, proteomics and metabolomics in drug discovery, development and clinic. Ther Deliv. 2013;4(3):395–413. doi: 10.4155/tde.13.4. [DOI] [PubMed] [Google Scholar]
- 27.Zampieri M., Szappanos B., Buchieri M.V., Trauner A., Piazza I., Picotti P. High-throughput metabolomic analysis predicts mode of action of uncharacterized antimicrobial compounds. Sci Transl Med. 2018;10(429) doi: 10.1126/scitranslmed.aal3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilcoxen K.M., Uehara T., Myint K.T., Sato Y., Oda Y. Practical metabolomics in drug discovery. Expert Opin Drug Discov. 2010;5(3):249–263. doi: 10.1517/17460441003631854. [DOI] [PubMed] [Google Scholar]
- 29.Rabinowitz J.D., Purdy J.G., Vastag L., Shenk T., Koyuncu E. Metabolomics in drug target discovery. Cold Spring Harb Symp Quant Biol. 2011;76:235–246. doi: 10.1101/sqb.2011.76.010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wishart D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov. 2016;15(7):473–484. doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- 31.Ramirez T., Daneshian M., Kamp H., Bois F.Y., Clench M.R., Coen M. Metabolomics in toxicology and preclinical research. ALTEX. 2013;30(2):209–225. doi: 10.14573/altex.2013.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nandakumar M., Prosser G.A., de Carvalho L.P., Rhee K. Metabolomics of Mycobacterium tuberculosis. Methods Mol Biol. 2015;1285:105–115. doi: 10.1007/978-1-4939-2450-9_6. [DOI] [PubMed] [Google Scholar]
- 33.Beger R.D., Schmidt M.A., Kaddurah-Daouk R. Current Concepts in Pharmacometabolomics, Biomarker Discovery, and Precision Medicine. Metabolites. 2020;10(4) doi: 10.3390/metabo10040129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maldonado-Carmona N., Vazquez-Hernandez M., Patino Chavez O.J., Rodriguez-Luna S.D., Jimenez Rodriguez O., Sanchez S. Impact of approximately omics in the detection and validation of potential anti-infective drugs. Curr Opin Pharmacol. 2019;48:1–7. doi: 10.1016/j.coph.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Li S.G., Vilcheze C., Chakraborty S., Wang X., Kim H., Anisetti M. Evolution of a thienopyrimidine antitubercular relying on medicinal chemistry and metabolomics insights. Tetrahedron Lett. 2015;56(23):3246–3250. doi: 10.1016/j.tetlet.2015.02.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tulloch L.B., Menzies S.K., Coron R.P., Roberts M.D., Florence G.J., Smith T.K. Direct and indirect approaches to identify drug modes of action. IUBMB Life. 2018;70(1):9–22. doi: 10.1002/iub.1697. [DOI] [PubMed] [Google Scholar]
- 37.Wouters OJ, M M, Luyten J. Estimated Research and Development Investment Needed to Bring a New Medicine to Market, 2009-2018. JAMA. 2020;323(9):844–53. [DOI] [PMC free article] [PubMed]
- 38.Van Norman G.A. Drugs, Devices, and the FDA: Part 1: An Overview of Approval Processes for Drugs. JACC Basic Transl Sci. 2016;1(3):170–179. doi: 10.1016/j.jacbts.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takebe T., Imai R., Ono S. The Current Status of Drug Discovery and Development as Originated in United States Academia: The Influence of Industrial and Academic Collaboration on Drug Discovery and Development. Clin Transl Sci. 2018;11(6):597–606. doi: 10.1111/cts.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wishart D.S. Applications of metabolomics in drug discovery and development. Drugs R D. 2008;9(5):307–322. doi: 10.2165/00126839-200809050-00002. [DOI] [PubMed] [Google Scholar]
- 41.Mikami T., Aoki M., Kimura T. The application of mass spectrometry to proteomics and metabolomics in biomarker discovery and drug development. Curr Mol Pharmacol. 2012;5(2):301–316. doi: 10.2174/1874467211205020301. [DOI] [PubMed] [Google Scholar]
- 42.Mastrangelo A., Armitage E.G., Garcia A., Barbas C. Metabolomics as a tool for drug discovery and personalised medicine. A review. Curr Top Med Chem. 2014;14(23):2627–2636. doi: 10.2174/1568026614666141215124956. [DOI] [PubMed] [Google Scholar]
- 43.Han J., Xia Y., Lin L., Zhang Z., Tian H., Li K. Next-generation Metabolomics in the Development of New Antidepressants: Using Albiflorin as an Example. Curr Pharm Des. 2018;24(22):2530–2540. doi: 10.2174/1381612824666180727114134. [DOI] [PubMed] [Google Scholar]
- 44.Cobice D.F., Goodwin R.J., Andren P.E., Nilsson A., Mackay C.L., Andrew R. Future technology insight: mass spectrometry imaging as a tool in drug research and development. Br J Pharmacol. 2015;172(13):3266–3283. doi: 10.1111/bph.13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen G., Fan M., Liu Y., Sun B., Liu M., Wu J. Advances in MS Based Strategies for Probing Ligand-Target Interactions: Focus on Soft Ionization Mass Spectrometric Techniques. Front Chem. 2019;7:703. doi: 10.3389/fchem.2019.00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDonnell L.A., Rompp A., Balluff B., Heeren R.M., Albar J.P., Andren P.E. Discussion point: reporting guidelines for mass spectrometry imaging. Anal Bioanal Chem. 2015;407(8):2035–2045. doi: 10.1007/s00216-014-8322-6. [DOI] [PubMed] [Google Scholar]
- 47.He J., Luo Z., Huang L., He J., Chen Y., Rong X. Ambient mass spectrometry imaging metabolomics method provides novel insights into the action mechanism of drug candidates. Anal Chem. 2015;87(10):5372–5379. doi: 10.1021/acs.analchem.5b00680. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y., Ma X., Lin Z., He M., Han G., Yang C. Imaging mass spectrometry with a low-temperature plasma probe for the analysis of works of art. Angew Chem Int Ed Engl. 2010;49(26):4435–4437. doi: 10.1002/anie.200906975. [DOI] [PubMed] [Google Scholar]
- 49.Wu C., Dill A.L., Eberlin L.S., Cooks R.G., Ifa D.R. Mass spectrometry imaging under ambient conditions. Mass Spectrom Rev. 2013;32(3):218–243. doi: 10.1002/mas.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Q., Comi T.J., Li B., Rubakhin S.S., Sweedler J.V. On-Tissue Derivatization via Electrospray Deposition for Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging of Endogenous Fatty Acids in Rat Brain Tissues. Anal Chem. 2016;88(11):5988–5995. doi: 10.1021/acs.analchem.6b01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun C., Wang F., Zhang Y., Yu J., Wang X. Mass spectrometry imaging-based metabolomics to visualize the spatially resolved reprogramming of carnitine metabolism in breast cancer. Theranostics. 2020;10(16):7070–7082. doi: 10.7150/thno.45543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perez C.J., Bagga A.K., Prova S.S., Yousefi Taemeh M., Ifa D.R. Review and perspectives on the applications of mass spectrometry imaging under ambient conditions. Rapid Commun Mass Spectrom. 2019;33(Suppl 3):27–53. doi: 10.1002/rcm.8145. [DOI] [PubMed] [Google Scholar]
- 53.Torok S., Vegvari A., Rezeli M., Fehniger T.E., Tovari J., Paku S. Localization of sunitinib, its metabolites and its target receptors in tumour-bearing mice: a MALDI-MS imaging study. Br J Pharmacol. 2015;172(4):1148–1163. doi: 10.1111/bph.12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jove M., Spencer J., Clench M., Loadman P.M., Twelves C. Precision pharmacology: Mass spectrometry imaging and pharmacokinetic drug resistance. Crit Rev Oncol Hematol. 2019;141:153–162. doi: 10.1016/j.critrevonc.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 55.Miura D., Fujimura Y., Wariishi H. In situ metabolomic mass spectrometry imaging: recent advances and difficulties. J Proteomics. 2012;75(16):5052–5060. doi: 10.1016/j.jprot.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 56.He J., Tang F., Luo Z., Chen Y., Xu J., Zhang R. Air flow assisted ionization for remote sampling of ambient mass spectrometry and its application. Rapid Commun Mass Spectrom. 2011;25(7):843–850. doi: 10.1002/rcm.4920. [DOI] [PubMed] [Google Scholar]
- 57.Luo Z., He J., Chen Y., He J., Gong T., Tang F. Air flow-assisted ionization imaging mass spectrometry method for easy whole-body molecular imaging under ambient conditions. Anal Chem. 2013;85(5):2977–2982. doi: 10.1021/ac400009s. [DOI] [PubMed] [Google Scholar]
- 58.Broach J.R., Thorner J. High-throughput screening for drug discovery. Nature. 1996;384(6604 Suppl):14–16. [PubMed] [Google Scholar]
- 59.Harrigan G.G., Yates L.A. High-throughput screening, metabolomics and drug discovery. IDrugs. 2006;9(3):188–192. [PubMed] [Google Scholar]
- 60.Bajorath J. Integration of virtual and high-throughput screening. Nat Rev Drug Discov. 2002;1(11):882–894. doi: 10.1038/nrd941. [DOI] [PubMed] [Google Scholar]
- 61.Zampieri M., Zimmermann M., Claassen M., Sauer U. Nontargeted Metabolomics Reveals the Multilevel Response to Antibiotic Perturbations. Cell Rep. 2017;19(6):1214–1228. doi: 10.1016/j.celrep.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 62.Halouska S., Fenton R.J., Barletta R.G., Powers R. Predicting the in vivo mechanism of action for drug leads using NMR metabolomics. ACS Chem Biol. 2012;7(1):166–171. doi: 10.1021/cb200348m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tiziani S., Kang Y., Choi J.S., Roberts W., Paternostro G. Metabolomic high-content nuclear magnetic resonance-based drug screening of a kinase inhibitor library. Nat Commun. 2011;2:545. doi: 10.1038/ncomms1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagana Gowda G.A., Raftery D. Quantitating metabolites in protein precipitated serum using NMR spectroscopy. Anal Chem. 2014;86(11):5433–5440. doi: 10.1021/ac5005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kurita K.L., Glassey E., Linington R.G. Integration of high-content screening and untargeted metabolomics for comprehensive functional annotation of natural product libraries. Proc Natl Acad Sci U S A. 2015;112(39):11999–12004. doi: 10.1073/pnas.1507743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Selevsek N., Caiment F., Nudischer R., Gmuender H., Agarkova I., Atkinson F.L. Network integration and modelling of dynamic drug responses at multi-omics levels. Commun Biol. 2020;3(1):573. doi: 10.1038/s42003-020-01302-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ebrahim A., Brunk E., Tan J., O'Brien E.J., Kim D., Szubin R. Multi-omic data integration enables discovery of hidden biological regularities. Nat Commun. 2016;7:13091. doi: 10.1038/ncomms13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Campos A.I., Zampieri M. Metabolomics-Driven Exploration of the Chemical Drug Space to Predict Combination Antimicrobial Therapies. Mol Cell. 2019;74(6):1291–1303 e6. doi: 10.1016/j.molcel.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Orth J.D., Conrad T.M., Na J., Lerman J.A., Nam H., Feist A.M. A comprehensive genome-scale reconstruction of Escherichia coli metabolism–2011. Mol Syst Biol. 2011;7:535. doi: 10.1038/msb.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Facchetti G., Zampieri M., Altafini C. Predicting and characterizing selective multiple drug treatments for metabolic diseases and cancer. BMC Syst Biol. 2012;6:115. doi: 10.1186/1752-0509-6-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chandrasekaran S., Cokol-Cakmak M., Sahin N., Yilancioglu K., Kazan H., Collins J.J. Chemogenomics and orthology-based design of antibiotic combination therapies. Mol Syst Biol. 2016;12(5):872. doi: 10.15252/msb.20156777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nichols R.J., Sen S., Choo Y.J., Beltrao P., Zietek M., Chaba R. Phenotypic landscape of a bacterial cell. Cell. 2011;144(1):143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X., Zhang A., Yan G., Sun W., Han Y., Sun H. Metabolomics and proteomics annotate therapeutic properties of geniposide: targeting and regulating multiple perturbed pathways. PLoS ONE. 2013;8(8):e71403. doi: 10.1371/journal.pone.0071403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gioria S., Lobo Vicente J., Barboro P., La Spina R., Tomasi G., Urban P. A combined proteomics and metabolomics approach to assess the effects of gold nanoparticles in vitro. Nanotoxicology. 2016;10(6):736–748. doi: 10.3109/17435390.2015.1121412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang X., Zhang A., Wang P., Sun H., Wu G., Sun W. Metabolomics coupled with proteomics advancing drug discovery toward more agile development of targeted combination therapies. Mol Cell Proteomics. 2013;12(5):1226–1238. doi: 10.1074/mcp.M112.021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jiang S., Liu G., Yuan H., Xu E., Xia W., Zhang X. Changes on proteomic and metabolomic profile in serum of mice induced by chronic exposure to tramadol. Sci Rep. 2021;11(1):1454. doi: 10.1038/s41598-021-81109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duan L., Guo L., Wang L., Yin Q., Zhang C.M., Zheng Y.G. Application of metabolomics in toxicity evaluation of traditional Chinese medicines. Chin Med. 2018;13:60. doi: 10.1186/s13020-018-0218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang P., Shehu A.I., Ma X. The Opportunities of Metabolomics in Drug Safety Evaluation. Curr Pharmacol Rep. 2017;3(1):10–15. doi: 10.1007/s40495-016-0079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yao C.H., Wang L., Stancliffe E., Sindelar M., Cho K., Yin W. Dose-Response Metabolomics To Understand Biochemical Mechanisms and Off-Target Drug Effects with the TOXcms Software. Anal Chem. 2020;92(2):1856–1864. doi: 10.1021/acs.analchem.9b03811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mahieu N.G., Genenbacher J.L., Patti G.J. A roadmap for the XCMS family of software solutions in metabolomics. Curr Opin Chem Biol. 2016;30:87–93. doi: 10.1016/j.cbpa.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ritz C., Baty F., Streibig J.C., Gerhard D. Dose-Response Analysis Using R. PLoS ONE. 2015;10(12):e0146021. doi: 10.1371/journal.pone.0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raud B., Roy D.G., Divakaruni A.S., Tarasenko T.N., Franke R., Ma E.H. Etomoxir Actions on Regulatory and Memory T Cells Are Independent of Cpt1a-Mediated Fatty Acid Oxidation. Cell Metab. 2018;28(3):504–515 e7. doi: 10.1016/j.cmet.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rhee K. Minding the gaps: metabolomics mends functional genomics. EMBO Rep. 2013;14(11):949–950. doi: 10.1038/embor.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baughn A.D., Rhee K.Y. Metabolomics of Central Carbon Metabolism in Mycobacterium tuberculosis. Microbiol Spectr. 2014;2(3) doi: 10.1128/microbiolspec.MGM2-0026-2013. [DOI] [PubMed] [Google Scholar]
- 85.Rhee K.Y., de Carvalho L.P., Bryk R., Ehrt S., Marrero J., Park S.W. Central carbon metabolism in Mycobacterium tuberculosis: an unexpected frontier. Trends Microbiol. 2011;19(7):307–314. doi: 10.1016/j.tim.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Z, Soni V, Marriner G, Kaneko T, Boshoff HIM, Barry CE, Rhee KY. Mode-of-action profiling reveals glutamine synthetase as a collateral metabolic vulnerability of M. tuberculosis to bedaquiline. Proc Natl Acad Sci 2019;116(39):19646–19651. [DOI] [PMC free article] [PubMed]
- 87.Mirzayev F., Viney K., Linh N.N., Gonzalez-Angulo L., Gegia M., Jaramillo E. World Health Organization recommendations on the treatment of drug-resistant tuberculosis, 2020 update. Eur Respir J. 2020 doi: 10.1183/13993003.03300-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Conradie F., Everitt D., Crook A.M. Treatment of Highly Drug-Resistant Pulmonary Tuberculosis. Reply. N Engl J Med. 2020;382(24):2377. doi: 10.1056/NEJMc2009939. [DOI] [PubMed] [Google Scholar]
- 89.Agapova A., Serafini A., Petridis M., Hunt D.M., Garza-Garcia A., Sohaskey C.D. Flexible nitrogen utilisation by the metabolic generalist pathogen Mycobacterium tuberculosis. Elife. 2019;8 doi: 10.7554/eLife.41129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sharma S., Hameed S., Fatima Z. Lipidomic insights to understand membrane dynamics in response to vanillin in Mycobacterium smegmatis. Int Microbiol. 2020;23(2):263–276. doi: 10.1007/s10123-019-00099-9. [DOI] [PubMed] [Google Scholar]
- 91.Nandakumar M., Nathan C., Rhee K.Y. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat Commun. 2014;5:4306. doi: 10.1038/ncomms5306. [DOI] [PubMed] [Google Scholar]
- 92.Nieto R.L., Mehaffy C., Islam M.N., Fitzgerald B., Belisle J., Prenni J. Biochemical Characterization of Isoniazid-resistant Mycobacterium tuberculosis: Can the Analysis of Clonal Strains Reveal Novel Targetable Pathways? Mol Cell Proteomics. 2018;17(9):1685–1701. doi: 10.1074/mcp.RA118.000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koen N., van Breda S.V., Loots D.T. Elucidating the antimicrobial mechanisms of colistin sulfate on Mycobacterium tuberculosis using metabolomics. Tuberculosis (Edinb) 2018;111:14–19. doi: 10.1016/j.tube.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 94.Ballinger E., Mosior J., Hartman T., Burns-Huang K., Gold B., Morris R. Opposing reactions in coenzyme A metabolism sensitize Mycobacterium tuberculosis to enzyme inhibition. Science. 2019;363(6426) doi: 10.1126/science.aau8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cao J., Mi Y., Shi C., Bian Y., Huang C., Ye Z. First-line anti-tuberculosis drugs induce hepatotoxicity: A novel mechanism based on a urinary metabolomics platform. Biochem Biophys Res Commun. 2018;497(2):485–491. doi: 10.1016/j.bbrc.2018.02.030. [DOI] [PubMed] [Google Scholar]
- 96.McKay CS, Finn MG. Click chemistry in complex mixtures: bioorthogonal bioconjugation. Chem Biol (Oxford, UK) 2014;21(9):1075–1101. [DOI] [PMC free article] [PubMed]
- 97.Meldal M, Tornoe CW, Cu-catalyzed azide-alkyne cycloaddition. Chem Rev (Washington, DC, US) 2008;108(8):2952–3015. [DOI] [PubMed]
- 98.Hein J.E., Fokin V.V. Copper-catalyzed azide-alkyne cycloaddition (CuAAC) and beyond: new reactivity of copper(i) acetylides. Chem Soc Rev. 2010;39(4):1302–1315. doi: 10.1039/b904091a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim E., Koo H. Biomedical applications of copper-free click chemistry: in vitro, in vivo, and ex vivo. Chem Sci. 2019;10(34):7835–7851. doi: 10.1039/c9sc03368h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Knall A.-C., Slugovc C. Inverse electron demand Diels-Alder (iEDDA)-initiated conjugation: a (high) potential click chemistry scheme. Chem Soc Rev. 2013;42(12):5131–5142. doi: 10.1039/c3cs60049a. [DOI] [PubMed] [Google Scholar]
- 101.Deb T, Tu J, Franzini RM. Mechanisms and substituent effects of metal-free bioorthogonal reactions. Chem Rev (Washington, DC, US) 2021:121(12):6850–914. [DOI] [PubMed]
- 102.Thiele C., Papan C., Hoelper D., Kusserow K., Gaebler A., Schoene M. Tracing Fatty Acid Metabolism by Click Chemistry. ACS Chem Biol. 2012;7(12):2004–2011. doi: 10.1021/cb300414v. [DOI] [PubMed] [Google Scholar]
- 103.Thiele C., Wunderling K., Leyendecker P. Multiplexed and single cell tracing of lipid metabolism. Nat Methods. 2019;16(11):1123–1130. doi: 10.1038/s41592-019-0593-6. [DOI] [PubMed] [Google Scholar]
- 104.Perez A.J., Wesche F., Adihou H., Bode H.B. Solid-Phase Enrichment and Analysis of Azide-Labeled Natural Products: Fishing Downstream of Biochemical Pathways. Chem - Eur J. 2016;22(2):639–645. doi: 10.1002/chem.201503781. [DOI] [PubMed] [Google Scholar]
- 105.Hoki J.S., Le H.H., Mellott K.E., Zhang Y.K., Fox B.W., Rodrigues P.R. Deep Interrogation of Metabolism Using a Pathway-Targeted Click-Chemistry Approach. J Am Chem Soc. 2020;142(43):18449–18459. doi: 10.1021/jacs.0c06877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Crabbe A., Ledesma M.A., Nickerson C.A. Mimicking the host and its microenvironment in vitro for studying mucosal infections by Pseudomonas aeruginosa. Pathog Dis. 2014;71(1):1–19. doi: 10.1111/2049-632X.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Postnikova E., Cong Y., DeWald L.E., Dyall J., Yu S., Hart B.J. Testing therapeutics in cell-based assays: factors that influence the apparent potency of drugs. PLoS ONE. 2018;13(3):e0194880/1. doi: 10.1371/journal.pone.0194880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Patil V., Jain V. Insights into the physiology and metabolism of a mycobacterial cell in an energy-compromised state. J Bacteriol. 2019;201(19):e00210. doi: 10.1128/JB.00210-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Koul A., Vranckx L., Dhar N., Gohlmann H.W.H., Ozdemir E., Neefs J.-M. Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nat Commun. 2014;5:4369/1. doi: 10.1038/ncomms4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lelovic N., Mitachi K., Yang J., Lemieux M.R., Ji Y., Kurosu M. Application of Mycobacterium smegmatis as a surrogate to evaluate drug leads against Mycobacterium tuberculosis. J Antibiot. 2020;73(11):780–789. doi: 10.1038/s41429-020-0320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Judd J., Boucher N., Van Roey E., Gray T.A., Derbyshire K.M. Application of distributive conjugal DNA transfer in Mycobacterium smegmatis to establish a genome-wide synthetic genetic array. J Bacteriol. 2017;199(20):e00410. doi: 10.1128/JB.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wee W.Y., Dutta A., Choo S.W. Comparative genome analyses of mycobacteria give better insights into their evolution. PLoS ONE. 2017;12(3):e0172831/1. doi: 10.1371/journal.pone.0172831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Barkan D, Liu Z, Sacchettini JC, Glickman MS. Mycolic acid cyclopropanation is essential for viability, drug resistance, and cell wall integrity of Mycobacterium tuberculosis. Chem Biol (Cambridge, MA, US) 2009;16(5):499–509. [DOI] [PMC free article] [PubMed]
- 114.Halouska S., Fenton R.J., Zinniel D.K., Marshall D.D., Barletta R.G., Powers R. Metabolomics analysis identifies D-Alanine-D-Alanine ligase as the primary lethal target of D-cycloserine in mycobacteria. J Proteome Res. 2014;13(2):1065–1076. doi: 10.1021/pr4010579. [DOI] [PMC free article] [PubMed] [Google Scholar]