Abstract
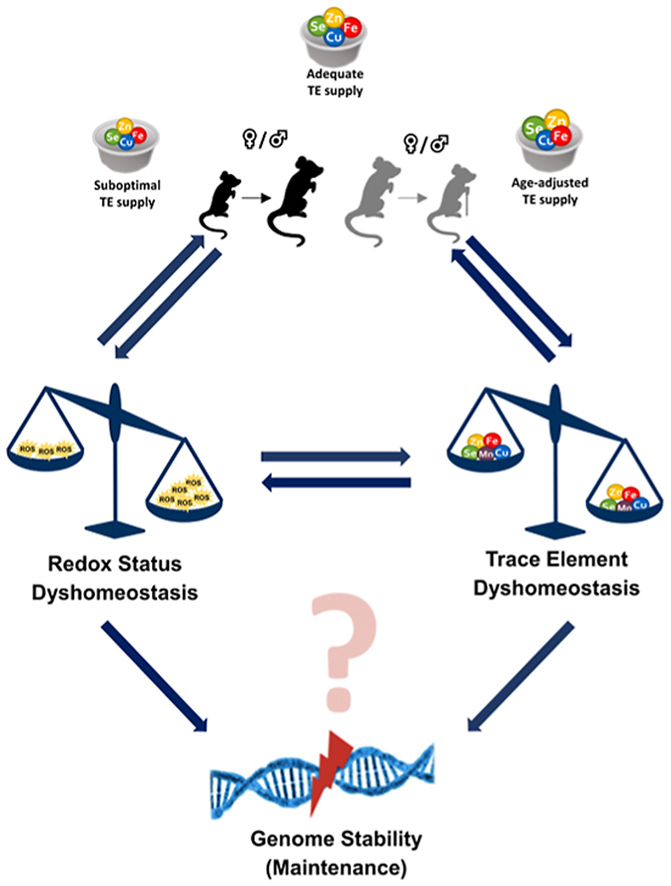
Trace elements (TEs) are essential for diverse processes maintaining body function and health status. The complex regulation of the TE homeostasis depends among others on age, sex, and nutritional status. If the TE homeostasis is disturbed, negative health consequences can result, e.g., caused by impaired redox homeostasis and genome stability maintenance. Based on age-related shifts in TEs which have been described in mice well-supplied with TEs, we aimed to understand effects of a long-term feeding with adequate or suboptimal amounts of four TEs in parallel. As an additional intervention, we studied mice which received an age-adapted diet with higher concentrations of selenium and zinc to counteract the age-related decline of both TEs. We conducted comprehensive analysis of diverse endpoints indicative for the TE and redox status, complemented by analysis of DNA (hydroxy)methylation and markers denoting genomic stability maintenance. TE concentrations showed age-specific alterations which were relatively stable and independent of their nutritional supply. In addition, hepatic DNA hydroxymethylation was significantly increased in the elderly mice and markers indicative for the redox status were modulated. The reduced nutritional supply with TEs inconsistently affected their status, with most severe effects regarding Fe deficiency. This may have contributed to the sex-specific differences observed in the alterations related to the redox status and DNA repair activity.
Overall, our results highlight the complexity of factors impacting on the TE status and its physiological consequences. Alterations in TE supply, age, and sex proved to be important determinants that need to be taken into account when considering TE interventions for improving general health and supporting convalescence in the clinics.
Keywords: Trace elements, Ageing, Sex, Redox status, Genome stability (maintenance)
Graphical abstract
Highlights
-
•
Trace element profiles differ by age and sex under moderately modulated TE supply.
-
•
Maintenance of age-related trace element shifts through all feeding groups.
-
•
Cu/Zn ratio and DNA hydroxymethylation emerge as appropriate murine ageing markers.
1. Introduction
Essential trace elements (TEs) are micronutrients and as such they fulfill a multitude of vital cellular key functions although they only account for a very small fraction of the total body weight [1]. Usually, TE concentrations are well-balanced and can, to a certain degree, be adapted to a varying physiological need or available amounts of TEs [[2], [3], [4]]. However, drastic alterations of the dietary intake can result in deficiency or intoxication with concomitant symptoms. Further, if the TE homeostasis is only marginally modulated, small changes can accumulate over time and result in health deprivations such as an increased risk for certain diseases. From a nutritional perspective, iron (Fe), zinc (Zn), and selenium (Se) are most often supplied in dosages below the reference intake and thus many people worldwide are at risk for a suboptimal status [[5], [6], [7]]. In contrast, copper (Cu) is more prone to accumulating in the body even if there is no excessive nutritional intake [8]. A common characteristic of all four of the above-mentioned TEs is their redox modulatory capacity. Se and Zn indirectly exert redox modulation by their functional, structural or catalytic involvement in redox regulating or antioxidant enzymes, in form of seleno- or Zn-containing proteins [9,10]. Cu and Fe are intrinsically redox active, thus directly impacting on redox homeostasis, but they are also part of antioxidant enzymes [8,11].
Ageing as one of the greatest present health challenges is associated with an altered TE homeostasis [12,13] and changes in the maintenance of cellular redox homeostasis. Several hypotheses addressing the underlying mechanisms of ageing focus on an age-dependent increase in oxidative stress leading to an accumulation of oxidized proteins, lipids, and DNA damage, while stress response and repair pathways are presumed to gradually decline with increasing age [[14], [15], [16], [17]]. One of those is the transcription factor nuclear factor erythroid 2-like 2 (Nrf2), which is considered as the master regulator of cellular redox homeostasis [18,19]. Yet, in ageing the transcriptional induction of Nrf2-regulated genes involved in antioxidant defence, redox homeostasis, DNA repair, cell fate decisions, and Fe metabolism is reduced [20]. In addition, the Nrf2 signalling pathway itself is susceptible to alterations of cellular TE concentrations [21], which has been shown for all four investigated in the present study [22].
Another important process modulated by both, ageing and TE dyshomeostasis, is DNA damage response (DDR) and repair [23,24]. This includes different key enzymes of the DDR machinery as well as the base excision repair (BER) pathway, which is responsible for the repair of non-bulky DNA lesions [[25], [26], [27], [28]]. One of the most frequently studied TEs in this respect is Zn as Zn-finger motives are important structural prerequisites for binding of proteins to DNA or RNA. For example, poly(ADP-ribose) polymerase 1 (PARP1) and p53 are well-known representatives of Zn-finger proteins, involved in safeguarding genome integrity [[29], [30], [31]]. Indeed, a recent study could determine the importance of Zn (signaling) in PARP1 activation finally resulting in genome stability maintenance [32].
Several studies reported age-dependent shifts of individual TE concentrations in human elderly which are e.g., well-established for increasing Cu levels and decreasing Zn concentrations in serum [13,33,34]. Only recently, age-dependent alterations of several TEs were analysed in parallel to establish age-dependent TE profiles. While serum Se and Zn concentrations declined with age, serum Cu and Fe concentrations showed an age-dependent increase [13,[33], [34], [35]]. Interestingly, we could recapitulate these human age-specific TE profiles in old mice (>110 weeks of age) in comparison to young adult mice (24 weeks of age) [36]. The old mice showed typical characteristics of ageing that manifested in increased hepatic DNA hydroxymethylation, markers for low grade inflammation, and changes in markers indicative for the redox status such as NAD(P)H quinone dehydrogenase 1 (NQO1) and glutathione S-transferase (GST) activity. However, in contrast to humans, mice were fed a standard chow diet throughout their whole life which contained high concentrations of TEs (1.5–10-fold above nutritional requirements). Based on these results, the question arises how age-related changes of the TE status develop in mice receiving an adequate or even suboptimal TE supply which resembles the nutritional supply of humans. Mice, young at age (three weeks), fed a combined suboptimal supply of the TEs Fe, Zn, Cu, Se, and iodine (I) developed splenomegaly and cardiomegaly as well as growth retardation together with signs of a low-grade inflammation within nine weeks [37]. In addition, markers for hepatic genomic instability were increased which appears to be mainly driven by an impaired Fe homeostasis.
Based on these results, we aimed to understand effects of varying TE supply starting i) after weaning or ii) in mice at an age of 11 months and lasting for six months. We hypothesised that age-related changes of the TE homeostasis would differ under conditions of a limited TE supply compared to a supply meeting the nutritional requirements. Furthermore, we were wondering whether we could counteract the age-related TE changes by including an additional intervention group receiving so-called age-adapted TE concentrations with a higher nutritional intake of Zn and Se to potentially counteract the decline of these two TEs during the ageing process, as observed in human studies [13,33,35].
Therefore, three and 40 weeks old C57BL/6Jrj mice of both sexes were divided into three different intervention groups receiving either a suboptimal (-TE), adequate (+TE), or age-adjusted (+TEaa) TE supply. After six months of intervention, effects on TE status markers, Nrf2 activity as well as genomic stability were investigated.
2. Materials and methods
2.1. Animal husbandry
Animal experiment was approved by the Ministry of Environment, Health and Consumer Protection of the federal state of Brandenburg, Germany (2347-44-2017) and conducted following national guidelines and institutional guidelines of the German Institute of Human Nutrition Potsdam-Rehbruecke, Germany. C57BL/6Jrj mice were housed in polycarbonate cages on a 12:12 h light/dark cycle at 22°C and constant humidity at 55 %. With the age of three (after weaning) and 40 weeks, mice received a diet low in five TEs (modified AIN93M, Cargill, Granovit, Kaiseraugst, Switzerland; 9.6, 0.75, 4.7, 0.44, 0.07 mg/kg Fe, Mn, Zn, Cu, and Se, respectively), but also low in Mg (40 mg/kg) and I (0.1 mg/kg), as reported before [37]. The animals were divided in three groups of eight to ten animals each and supplied with either suboptimal (-TE), adequate (+TE), or age-adjusted (+TEaa) amounts of TEs.
Drinking water of all mice was fortified with 500 mg Mg/kg (MgSO4, Merck (Sigma-Aldrich), Darmstadt, Germany) and 9.25 mg Mn/kg (MnCl2, Merck (Sigma-Aldrich)), whereas the intake of Fe (FeCl2, Merck), Zn (ZnSO4, Merck (Sigma-Aldrich)), Cu (CuSO4, Merck (Sigma-Aldrich)), and Se (Na2SeO3, Thermo Fisher Scientific, Waltham, USA) differed. While the -TE mice did not receive any other additives via the drinking water besides Mn and Mg, animals of the +TE group received 25.4, 5.56, 25.3, and 0.08 mg/kg Fe, Cu, Zn, and Se, respectively, according to feeding recommendations for an adequate supply [38]. The +TEaa group received identical concentrations of Fe and Cu compared to animals of the +TE group; contents of Zn and Se were administered at supplemented concentrations of 175.3 and 0.53 mg/kg, respectively. Both, food and water were offered ad libitum. Detailed information on feeding recommendations, TE content of feed and herein supplied concentrations in the drinking water are listed in Table 1.
Table 1.
TE supply of mice. Feeding recommendations [38] and content of the trace elements iron (Fe), copper (Cu), zinc (Zn), selenium (Se), manganese (Mn), and magnesium (Mg) in feed and water within the animal experiment. Feed TE contents were determined by ICP-MS/MS [37].
| TE | Feeding recommendations [mg/kg] | TE content of feed [mg/kg] | TE in drinking water [mg/kg] |
Total supply [mg/kg] |
Fold changes compared to feeding recommendation |
Fold changes |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -TE | +TE | +TEaa | -TE | +TE | +TEaa | -TE | +TE | +TEaa | -TE vs. +TE |
+TEaa vs. +TE | |||
| Cu | 6.00 | 0.44 | - | 5.56 | 5.56 | 0.44 | 6.00 | 6.00 | 0.07 | 0 | 0 | 0.07 | 0 |
| Fe | 35.0 | 9.62 | - | 25.4 | 25.4 | 9.62 | 35.0 | 35.0 | 0.3 | 0 | 0 | 0.3 | 0 |
| Mg | 500 | 40.0 | 500 | 500 | 500 | 540 | 540 | 540 | 1.1 | 1.1 | 1.1 | 0 | 0 |
| Mn | 10.0 | 0.75 | 9.25 | 9.25 | 9.25 | 10.0 | 10.0 | 10.0 | 0 | 0 | 0 | 0 | 0 |
| Se | 0.15 | 0.07 | - | 0.08 | 0.53 | 0.07 | 0.15 | 0.60 | 0.5 | 0 | 4 | 0.5 | 4 |
| Zn | 10.0 | 4.75 | - | 25.3 | 175.3 | 4.75 | 30.0 | 180 | 0.5 | 3 | 18 | 0.15 | 6 |
After 26 weeks, at the age of either 30 (adult) or 66 weeks (old), mice were anesthetised with isoflurane (Cp-pharma, Burgdorf, Germany). After blood withdrawal by heart puncture, serum was obtained after full coagulation at room temperature (RT) and centrifugation for 10 min (3000×g, 4°C). Liver was surgically dissected and aliquoted, snap-frozen in liquid nitrogen and stored at -80 °C until further use. For inductively coupled plasma tandem mass spectrometry (ICP-MS/MS), free Zn, gene expression, and global DNA (hydroxy)methylation analyses, hepatic tissue was pulverised under liquid nitrogen prior to analysis. Whenever possible, analyses were conducted with fully blinded samples from the point of tissue harvesting.
2.2. ICP-MS/MS analysis of TEs in serum, liver, and feed
Feed samples were homogenised at RT using mortar and pestle. 20–50 mg of pulverised liver tissue or feed sample were subjected to acidic microwave digestion as addressed in detail before [21]. In short, microwave (CEM-Mars 6 or CEM-Discover SP-D, Kamp-Lintfort, Germany) digestion was conducted with HNO3 (65 % Suprapur, Merck) and H2O2 (30 %, Merck) in PTFE or glass microwave vessels, with addition of internal and isotope-dilution standards. By following 1:4 dilution of liver samples, a final concentration of 2.93 % HNO3 resulted. Serum samples needed no further sample preparation and were diluted 1:10 with a solution containing EDTA, ammonia, non-ionic surfactant (Triton X-100), and butanol. Analysis of diluted sample solutions was conducted by ICP-MS/MS (8800 ICP-QQQ-MS, Agilent Technologies, Waldbronn, Germany) as previously described [39,40]. For quality assurance, certified reference materials (ERM BB-422 (fish muscle) and ERM BB-186 (pig kidney) (both Merck) for liver tissue and feed samples as well as ClinChek lyophilised serum control, Ref. 8880–8882, LOT 1497 (RECIPE, Munich, Germany) for serum samples) were analysed with the samples according to the described protocol.
2.3. ELISA-based analysis of transferrin concentrations in serum
Transferrin levels in serum were analysed applying an enzyme-linked immunosorbent assay (ELISA) kit (ALPCO, Salem, USA). In brief, murine serum was diluted 1:200,000 and treated following manufacturer's instructions. Finally, the average absorbance at 450 nm was measured with an Infinite 200 Pro microplate reader (Tecan, Männedorf, Switzerland). Calculation of concentrations was performed based on standard curves.
2.4. Selenoprotein P analysis in serum by affinity-HPLC-ICP-MS/MS
The separation was performed by affinity high performance liquid chromatography coupled to inductively coupled plasma mass spectrometry (affinity-HPLC-ICP-MS/MS) according to the method published by Heitland and Köster [41] with minor adjustments. 5 μL murine serum were diluted 1:20. Separation was performed with a HPLC system (Infinity 1200, Agilent), using a GE Healthcare HiTrap Heparin column (material: Heparin-Sepharose, volume: 1 mL). Injection volume was 75 μL. Separation of selenoprotein P (Selenop) is achieved by the following elution program, using 2 different NH4OAc (Merck (Sigma-Aldrich)) buffers (A: 0.17 M, B: 1.3 M) with 2 % (v/v) EtOH (Merck (Sigma-Aldrich)) adjusted to pH 7 using 25 % (m/v) NH4OH in H2O (Honeywell (Fluka), Charlotte, USA): 0.0–1.0 min 100 % buffer A; 1.0–4.5 min 100 % buffer B; 4.5–5.5 min 100 % buffer A. Flow rate was 1 mL/min. The eluent was mixed online via t-piece with a solution containing 10 μg/L Ge (diluted from 1000 mg/L Ge ICP-standard solution, Carl Roth, Karlsruhe, Germany) in 2 % (v/v) EtOH as internal standard. This mixture was then directly introduced to an ICP-MS/MS (8800 ICP-QQQ-MS, Agilent) equipped with a MicroMist™-nebuliser and Scott-type spraychamber. Calibration was performed species-independently in the range of 0.1–50 μg/L Se in H2O, freshly prepared on a daily basis from commercially available Se ICP-standard solution (1000 mg/L, Carl Roth). Calculation of Selenop concentrations was performed based on the assumption that Selenop contains an average of 5 Se atoms per molecule and a mass of M = 40.69 kDa [42]. For quality control, the certified reference material NIST SRM 1950 “Metabolites in human plasma” was purchased via Merck (Sigma-Aldrich) and used to determine Selenop content of reference material ClinChek lyophilised serum, levels 1 and 2 (Ref. 8880-8882, LOT 1497, RECIPE, Munich, Germany). In each following measurement, at least one of the aforementioned reference materials was prepared as human serum and carried along to confirm correctness of results.
2.5. Analysis of free Zn in serum and liver
Free Zn was determined as described previously [43], using the low molecular weight fluorescent probe Zinpyr-1 and the formula by Grynkiewicz et al. [44]. For murine serum samples, the assay was adjusted as follows: 20 μL of pre-diluted serum (1:10 in assay buffer for improved storage at -80 °C) were mixed with 80 μL pre-warmed assay buffer containing Zinpyr-1 (final concentration 0.05 μM). The incubation times for F, Fmin, and Fmax were set to 60, 30, and 60 min, respectively. In order to induce Fmin and Fmax, 15 μL EDTA solution (800 μM) and ZnSO4 solution (25 mM) per well were added, resulting in final concentrations of 100 μM EDTA and 2.8 mM ZnSO4, respectively.
The determination of free Zn in murine liver tissue follows the principles of serum analysis. Assay buffer was added to approximately 10 mg aliquots of homogenised liver tissue to yield a final concentration of 40 mg liver tissue per mL. The slurry was mixed for 10 s using a vortex-shaker, incubated for 30 min at 37°C, mixed again for 10 s and then centrifuged for 4 min at 160×g. 50 μL of the supernatant and an equal volume of Zinpyr-1 solution (0.1 μM in assay buffer) were added to the wells of a 96-well-plate and incubated for 20 min before read-out of F, Fmin, and Fmax were measured subsequently after addition of 15 μL/well EDTA solution (7.8 mM, final concentration 1 mM) and incubation for 30 min (Fmin), followed by addition of 15 μL/well ZnSO4 solution (35 mM, final concentration 4 mM) and incubation for 20 min (Fmax). All incubations were carried out using a shaker at RT and in the dark. Fluorescence intensities were measured at RT (λex 507 nm, λem 527 nm, bandwidth 5 nm) using a SPARK fluorescence plate reader (Tecan).
2.6. RNA isolation, reverse transcription, and qRT-PCR in liver
Total liver RNA isolation was conducted using Trizol Reagent (Thermo Fisher Scientific (Invitrogen)) following manufacturer's instructions. After elimination of genomic DNA utilising PerfeCTa DNase I (Quanta BioSciences, Beverly, USA), 8 μg RNA were applied for reverse transcription in a final volume of 20 μL (qScript cDNA synthesis, Quanta BioSciences) to generate complementary DNA (cDNA). For cDNA amplification, 1x PerfeCTa SYBR Green Supermix (Quanta BioSciences), cDNA, and 250 nM primer (sequences are listed in Supplementary Table 2) were mixed in a total volume of 10 μL. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the CFX Connect Real-time PCR Detection System (Bio-Rad Laboratories, Munich, Germany). The thermal cycling program consists of the following steps: 3 min at 95°C, followed by 41 cycles of 15 s at 95°C, 20 s at 60°C, and 30 s at 72°C. Possible deviations in the melting temperatures can be found in Supplementary Information Table 2. Copy numbers were calculated based on standard curves. Expression levels were normalised to a composition factor based on the house keeper genes hypoxanthine phosphoribosyltransferase 1 (Hprt) and ribosomal protein L13a (Rpl13a).
2.7. Analysis of enzyme activities in serum and liver
For measurement of enzymatic activities, frozen liver samples were homogenised in Tris buffer (100 mM Tris (Carl Roth), 300 mM KCl (Applichem, Darmstadt, Germany), 0.1 % (v/v) Triton X-100, (Serva, Heidelberg, Germany)) with protease inhibitor (1 μL/mg (v/v); Merck Milipore, Burlington, USA), pH 7.6). To remove cellular debris, samples were centrifuged (10 min, 14,000×g, 4°C) followed by measurement of NQO1 [45], glutathione peroxidase (GPX) [46], thioredoxin reductase (TXNRD) [47], and GST [48] as previously described. Briefly, NQO1 was measured by a menadione-mediated reduction of 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide. GPX and TXNRD activity measurements were conducted using a NADPH-consuming glutathione reductase (GR)-coupled assay or with a NADPH-dependent reduction of 5,5′-dithio-bis-(2-nitrobenzoic acid), respectively. GST activity was measured using 1-chloro-2,4-dinitrobenzene as substrate in the presence of reduced glutathione (GSH). All measurements were performed in triplicates using a microplate reader (Synergy H1, BioTek, Bad Friedrichshall, Germany). Enzymatic activities were normalised to protein content measured by Bradford analysis (Bio-Rad Laboratories).
2.8. Measurement of GSH and GSSG via (HPLC-qTOF)
Sample preparation was based on a method published before [49], adapted to frozen liver aliquots of 40–60 mg. After carefully thawing the previously at -80°C stored samples, ice was used to work on to avoid oxidation of GSH to GSSG during sample preparation. In addition, N-ethylmaleimide (NEM, ≥ 98 %, Merck (Sigma-Aldrich)) was used in excess for complex formation with GSH, reducing the free thiol groups for GSSG formation and therefore allowing the quantitative measurement of both, GSH (as GSH-NEM) and GSSG in each sample. Tissue disruption was started by three freeze-thaw cycles using liquid nitrogen and a 37°C water bath. After adding zirconia beads (biolab, Bebensee, Germany) and 300 μL ice-cold extraction buffer (1 % Triton X-100, 0.6 % sulfosalicylic acid, 1 mM NEM, 1 % protease inhibitor in KPE buffer (0.1 M potassium phosphate buffer, 5 mM EDTA), and the stable isotope-labelled internal standard (Glutathione-[glycin-13C2,15N])), samples were homogenised three times for 20 s in a bead ruptor. After centrifugation, the supernatants were diluted 1:200 for GSH and 1:50 for GSSG measurements via high performance liquid chromatography quadrupole time of flight (HPLC-qTOF) analysis (HPLC: 1200 Infinite, Agilent; qTOF-MS: 6538, Agilent). The separation was conducted on an Atlantis T3 LC column and appropriate pre-column using dH2O + 0.01 % formic acid and acetonitrile +0.01 % formic acid as mobile phases in a gradient elution and 0.3 mL/min flow. For ionisation, electrospray ionisation in positive mode (ESI+) was applied. ESI+ parameters were set as following: gas temperature: 320°C, drying gas 12 L/min, nebuliser 37 psig, VCap 2750 V, fragmentor 175 V, skimmer 65 V and OCT 1 RF Vpp 750 V. For GSH-NEM m/z 304.0970 (14 eV collision energy (CE)) was used as quantifier and m/z 201.0700 and m/z 287.0693 (CE 19 eV; 14 eV) as qualifiers. The results were normalised to protein content, which was determined using a standard BCA (Merck (Sigma-Aldrich)) assay.
2.9. Alkaline comet assay in liver
To detect single- and double-strand breaks as well as alkali-labile sites, the alkaline comet assay was performed [50]. Cell isolation from 40 mg of frozen liver tissue, experimental procedure, and final analysis of 200 randomly selected cells (100 per technical replicate) by semi-automated image analysis software (Comet IV, Perceptive Instruments, Stone, UK) were carried out as previously published [51], following the OECD guideline 489 and the recommendations for minimum information for reporting on the comet assay (MICRA) [[51], [52], [53]].
2.10. ELISA-based analysis of oxidative DNA damage in liver
For preparation of tissue extracts, 200 μL ATL-buffer (Qiagen DNeasy Kit (Qiagen, Hilden, Germany)), 6.5 mM butylated hydroxytoluene (BHT) (Merck (Sigma-Aldrich)), and zirconia beads (biolab) were added to 20–30 mg frozen liver tissue, which was disrupted using a bead ruptor 12 (biolab) subsequently. Tissue debris and beads were removed by centrifugation (10 min, 10,000×g, 4°C). Supernatant was collected and DNA isolation was performed using a Qiagen DNeasy Kit followed by DNA content measurement via Nanodrop One (Thermo Fisher Scientific). To reduce potential occurring oxidation during sample preparation to a minimum, 6.5 mM BHT were additionally added to every existing volume employed in further sample preparation. DNA was digested by 1 U/μL benzonase (Merck) for 90 min at 37°C followed by a 30-min treatment with 1 U/μL alkaline phosphatase (Thermo Fisher Scientific) at 37°C. Samples were heated up to 95°C for 10 min and placed on ice until further processing. Eventually, samples were diluted to a DNA concentration of 100 ng/mL. 5 μg of DNA were added per well in triplicate and prepared according to the DNA/RNA Oxidative Damage ELISA Kit (Cayman Chemicals, Ann Arbor, USA) instructions. Absorbance was measured at 405 nm with an Infinite 200 Pro microplate reader (Tecan) after 90 min. Calculation of concentrations was performed based on standard curves. Finally, variances are expressed as fold changes compared to +TE male adults (mean +TE male adult = 1).
2.11. BER incision activity assay in liver
BER incision activity was assessed as previously described [51]. In brief, 50 mg of snap-frozen liver tissue were employed for tissue extract preparation. Tissue extracts were incubated with three different fluorescently labelled DNA lesion containing hairpin-oligonucleotides (DNA lesions incorporated: 8-oxo-2'-deoxyguanine (8-oxodG), 5-hydroxy-2'-deoxyguanine (5-OHdU), apurinic/apyrimidinic site (AP site) analogue). Via denaturing polyacrylamide gel electrophoresis (PAGE), intact and incised oligonucleotides were separated and quantitated via fluorescence measurement. The percentage of incised oligonucleotide in relation to the intact oligonucleotide is indicative for incision activity of BER glycosylases.
2.12. Analysis of poly(ADP-ribosyl)ation status in liver via HPLC-MS/MS
Analysis of PARylation levels was performed via HPLC-MS/MS (HPLC: 1260 Infinity, Agilent; MS/MS: 6495A, Agilent) after extraction of poly(ADP-ribose) (PAR) from murine liver tissue according to previously published protocols [51,54]. Briefly, 10–20 mg of liver tissue were applied for PAR analysis. The frozen tissue was disrupted in trichloroacetic acid resulting in precipitation. As internal standard, 13C, 15N-labelled PAR was added. Via alkaline treatment, protein-bound PAR was detached and subsequently, DNA, RNA, and proteins were removed by enzymatic digestion. For extraction of PAR, the sample solution was subjected to solid-phase extraction. The extracted PAR was enzymatically hydrolysed to its monomeric units for final quantification via HPLC-MS/MS. Results were normalised to DNA concentration, which was measured using Hoechst 33258.
2.13. Western blot analysis in liver
Protein extracts were prepared as previously described [51] for BER incision activity assay, diluted 1:3 with 4x Laemmli sample buffer (0.25 M Tris pH 6.8, 8 % SDS, 40 % Glycerol, 0.03 % bromophenol blue) followed by denaturation for 5 min at 95°C, and stored at -80°C until further use [51]. Proteins were loaded and separated on 15 % polyacrylamide gels and transferred to a 0.45 μm nitrocellulose membrane (Amersham Protran, Merck (Sigma-Aldrich)) via tank blotting (Bio-Rad Laboratories). Subsequently, membranes were blocked in 3 % (w/v) not-fat dry milk Tris-buffered saline containing 0.1 % (v/v) Tween 20 (T-TBS) for 1 h at RT. Primary antibodies were diluted in blocking solution and were incubated overnight at 4°C. Anti-DNA polymerase beta (Polβ) (1:1000, ab175197, abcam, Cambridge, UK) and anti-β-actin (1:10,000, ab8227, abcam) were used as primary antibodies for immunoblot detection. The secondary antibody, horseradish peroxidase-conjugated goat anti-rabbit IgG (1:50,000, 7074S, Cell Signaling, Danvers, US), was incubated for 1 h in 3 % (w/v) non-fat dry milk in T-TBS at RT. Finally, for protein detection, SuperSignalTM West Dura (Thermo Fisher Scientific) was used. Band intensities were quantified by ChemiDoc MP Imaging System and the appropriate software Image Lab (Bio-Rad Laboratories). Protein expression was normalised to housekeeping gene β-actin.
Several antibodies for DNA ligase (Lig) 1 and 3 quantification in murine liver samples were tested, however none of the tested antibodies showed appropriate specificity with regard to the proteins of interest.
2.14. Measurement of global DNA (hydroxy)methylation via HPLC-MS/MS in liver
Sample preparation, enzymatic hydrolysis and analytical method were conducted as described earlier and are based on measurement of methylated (mdC) and hydroxymethylated (hmdC) cytidine in total DNA [37,[55], [56], [57]]. Briefly, 30 mg of frozen, pulverised liver tissue were lysed and homogenised. RNA and proteins were enzymatically removed, and DNA was extracted via phenol/chloroform/isoamylalcohol extraction. Following addition of the internal standard, containing [15N2, 13C]-dC, mdC-D3, and hmdC-D3, 12 μg of DNA were subjected to enzymatic hydrolysis yielding the respective nucleosides for analysis. Samples were measured in a final dilution of 1:20 for mdC and dC, and 1:6 for hmdC. Quantitative analytical measurement was performed via HPLC-MS/MS (HPLC: 1260 Infinity, Agilent; MS/MS: 6495A, Agilent). Results are presented in relation to total cytidine (dC) in DNA (mdC/dC [%] and hmdC/dC [%]).
2.15. Statistical analysis
Statistical evaluation was performed in GraphPad Prism 9 (GraphPad Software, La Jolla, USA). After removal of statistical outliers by Gubbs test, normal distribution (Shapiro-Wilk test) and variance (Levene test) were tested. Three-Way analysis of variance (ANOVA) followed by Bonferroni's post-test was performed for statistical analysis. While p-values are summarised in Table 2, an overview of mean values ± standard deviation, as well as p-values is provided in Supplementary Information Table 1. Calculation of correlation coefficient was performed according to Spearman (referred to as rs). Statistical significances are shown as *p < 0.05, **p < 0.01, ***p < 0.001.
Table 2.
Statistical analysis overview of selected TE profiles and biomarkers in serum and liver, hepatic relative mRNA expression level, as well as hepatic redox and genomic stability markers. Statistical analysis based on Three-Way ANOVA followed by Bonferroni's post-test. Statistical significances are shown as *p < 0.05, **p < 0.01, ***p < 0.001.
| parameters | diet | age | sex | diet x age | diet x sex | sex x age | diet x sex x age | ||
|---|---|---|---|---|---|---|---|---|---|
| Zn status as well as related biomarkers and genes in murine serum and liver | |||||||||
| serum | Zn | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | ** | |
| free Zink | n.s. | n.s. | *** | ** | n.s. | n.s. | n.s. | ||
| liver | Zn | n.s. | 0.069 | n.s. | n.s. | n.s. | n.s. | n.s. | |
| free Zn | n.s. | n.s. | ** | 0.078 | n.s. | n.s. | n.s. | ||
| rel. Mt1 mRNA | n.s. | n.s. | *** | n.s. | * | n.s. | 0.067 | ||
| rel. Mt2 mRNA | n.s. | n.s. | ** | n.s. | ** | 0.071 | 0.098 | ||
| rel. Zip3 mRNA | n.s. | ** | *** | n.s. | * | n.s. | n.s. | ||
| rel. Zip5 mRNA | * | *** | *** | * | * | *** | * | ||
| rel. Znt10 mRNA | n.s. | * | n.s. | n.s. | n.s. | n.s. | n.s. | ||
| Se status as well as related biomarkers and genesinmurine serum and liver | |||||||||
| serum | Se | ** | *** | * | n.s. | n.s. | n.s. | n.s. | |
| GPX acitivity | n.s. | n.s. | n.s. | n.s. | * | n.s. | n.s. | ||
| Selenop concentration | * | *** | ** | n.s. | n.s. | n.s. | n.s. | ||
| liver | Se | * | *** | * | n.s. | n.s. | n.s. | 0.056 | |
| GPX acitivity | ** | ** | n.s. | n.s. | n.s. | n.s. | n.s. | ||
| rel. Selenop mRNA | n.s. | n.s. | *** | n.s. | n.s. | n.s. | n.s. | ||
| Cu status as well as related biomarkers and genesinmurine serum and liver | |||||||||
| serum | Cu | ** | *** | *** | 0.065 | n.s. | n.s. | n.s. | |
| Cu/Se ratio | ** | *** | * | ** | * | n.s. | 0.097 | ||
| Cu/Zn ratio | n.s. | *** | ** | n.s. | n.s. | n.s. | * | ||
| liver | Cu | n.s. | *** | *** | n.s. | n.s. | n.s. | n.s. | |
| rel. Atp7b mRNA | n.s. | n.s. | *** | n.s. | n.s. | n.s. | n.s. | ||
| rel. Ctr1 mRNA | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | ||
| Fe and Mn status as well as related biomarkers and genesinmurine serum and liver | |||||||||
| serum | transferrin | *** | ** | ** | * | * | n.s. | n.s. | |
| Mn | n.s. | n.s. | * | 0.055 | n.s. | n.s. | 0.060 | ||
| liver | Fe | *** | *** | *** | n.s. | *** | *** | n.s. | |
| rel. Dmt1 mRNA | n.s. | * | *** | n.s. | n.s. | n.s. | n.s. | ||
| rel. Fech mRNA | n.s. | n.s. | * | n.s. | n.s. | n.s. | n.s. | ||
| rel. Fpn mRNA | * | n.s. | ** | n.s. | n.s. | n.s. | * | ||
| rel. Hamp mRNA | *** | ** | *** | * | n.s. | n.s. | n.s. | ||
| rel. Hmox1 mRNA | n.s. | ** | ** | n.s. | * | ** | n.s. | ||
| Mn | *** | * | 0.091 | * | ** | n.s. | ** | ||
| Markers indicative for redox status in murine liver | |||||||||
| liver | GST activity | ** | n.s. | *** | n.s. | n.s. | *** | n.s. | |
| rel. Nqo1 mRNA | n.s. | ** | *** | n.s. | n.s. | n.s. | n.s. | ||
| NQO1 activity | n.s. | *** | *** | n.s. | n.s. | n.s. | n.s. | ||
| rel. Txnrd1 mRNA | * | n.s. | n.s. | 0.087 | * | n.s. | n.s. | ||
| TXNRD activity | n.s. | ** | * | n.s. | n.s. | n.s. | n.s. | ||
| Genomic stability markers and global DNA (hydroxy)methylation in murine liver | |||||||||
| liver | DNA in tail | 0.074 | 0.088 | n.s. | n.s. | n.s. | n.s. | n.s. | |
| rel. 8-oxodG level | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | ||
| PAR | 0.051 | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | ||
| incision AP site | n.s. | n.s. | * | n.s. | n.s. | ** | n.s. | ||
| incision 8-oxodG | n.s. | n.s. | *** | n.s. | n.s. | n.s. | n.s. | ||
| incision 5 OHdU | n.s. | n.s. | 0.068 | n.s. | n.s. | n.s. | n.s. | ||
| rel. Polß mRNA | * | n.s. | *** | n.s. | n.s. | n.s. | n.s. | ||
| Polß protein | ** | * | n.s. | n.s. | n.s. | n.s. | n.s. | ||
| rel. Aptx mRNA | 0.099 | n.s. | * | n.s. | n.s. | n.s. | n.s. | ||
| rel. Lig1 mRNA | n.s. | ** | *** | n.s. | n.s. | n.s. | n.s. | ||
| mdC/dC | n.s. | n.s. | * | n.s. | n.s. | n.s. | n.s. | ||
| hmdC/dC | * | *** | n.s. | n.s. | n.s. | n.s. | n.s. | ||
3. Results
To study diet-, age-, and sex-dependent changes in the TE status, mice at the age of 30 (adult) or 66 weeks (old) of both sexes were sacrificed after receiving either a suboptimal (-TE), adequate (+TE) or age-adjusted (+TEaa) TE supply for 26 weeks via diet and drinking water. TE concentrations of Cu, Fe, Mn, Se, and Zn, as well as functional biomarkers for Fe, Se, and Zn were analysed in serum and liver. For mechanistic insights, mRNA expression analysis for various transporters and binding proteins was performed in murine liver (Fig. 1, Fig. 2, Fig. 3, Fig. 4, Supplementary Information Fig. 2).
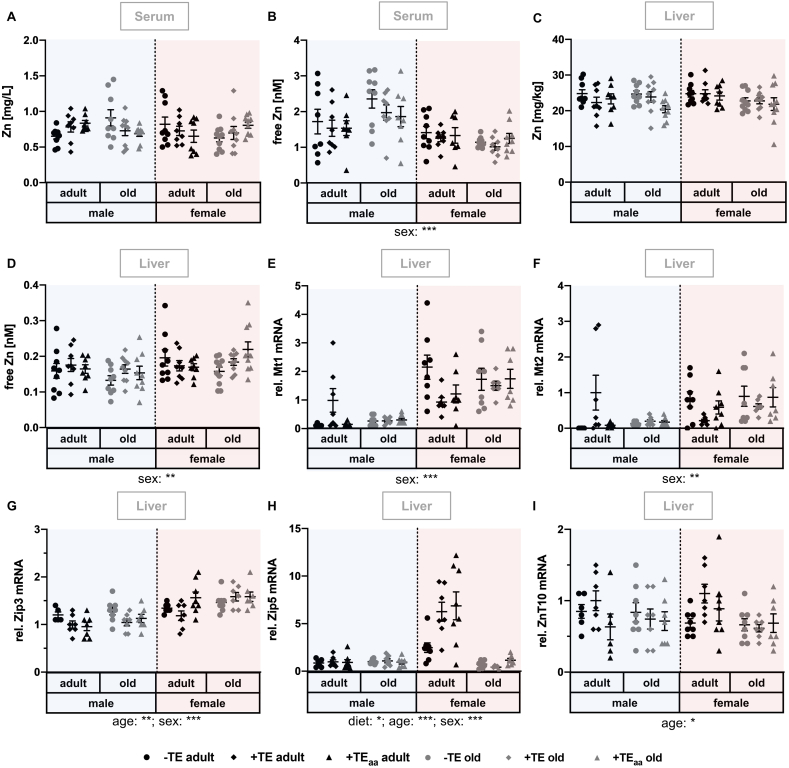
Fig. 1.
Changes in Zn status as well as related biomarkers and genes. Markers for the evaluation of the Zn status were determined in serum and liver of adult (30 weeks) and old (66 weeks) male and female C57BL/6Jrj mice (n = 8–10) receiving a -TE, +TE, or +TEaa diet for 26 weeks. Total Zn and free Zn concentrations were analysed in serum (A, B) and liver (C, D) by ICP-MS/MS and via fluorescent probes, respectively. To further evaluate the Zn status, relative expression levels of the Zn regulators Mt1 (E) and Mt2 (F) as well as the Zn transporters Zip3 (G), Zip5 (H), and ZnT10 (I) were examined via qRT-PCR analysis. Hepatic transcription levels were normalised to a composite factor based on the house keeper genes Hprt and Rpl13a. Variances are expressed as fold changes compared to +TE male adults (mean + TE male adult = 1). Statistical testing based on Three-Way ANOVA and Bonferroni's post-test with *p < 0.05, **p < 0.01, ***p < 0.001. Detailed results of statistical testing are summarised in Table 2.
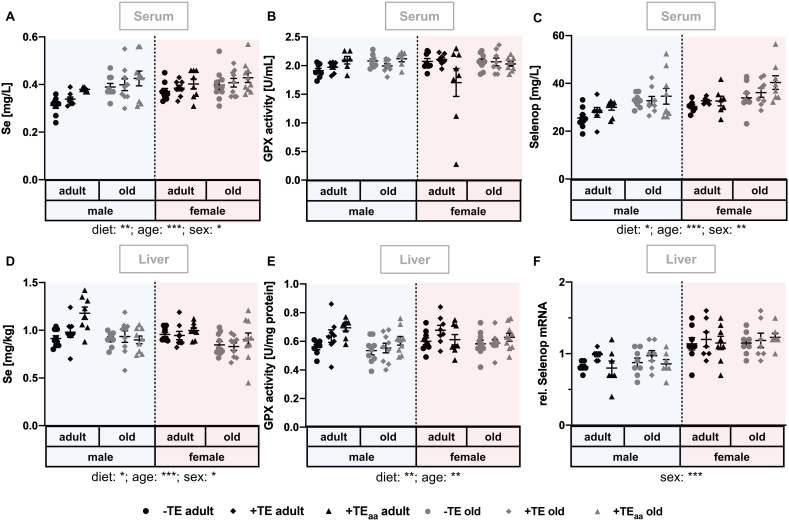
Fig. 2.
Changes in Se status as well as related biomarkers and genes. Markers for the evaluation of the Se status were determined in serum and liver of adult (30 weeks) and old (66 weeks) male and female C57BL/6Jrj mice (n = 8–10) receiving a -TE, +TE, or +TEaa diet for 26 weeks. Se concentrations were analysed in serum (A) and liver (D) by ICP-MS/MS. To further evaluate the Se status, GPX enzyme activity was analysed by a NADPH-consuming assay in serum (B) and liver (E). Additionally, serum Selenop concentrations (C) were analysed by affinity-HPLC-ICP-MS/MS and hepatic Selenop transcription level was examined by qRT-PCR analysis, whereby hepatic transcription levels were normalised to a composite factor based on the house keeper genes Hprt and Rpl13a. Variances are expressed as fold changes compared to +TE male adults (mean +TE male adult = 1). Statistical testing based on Three-Way ANOVA and Bonferroni's post-test with *p < 0.05, **p < 0.01, ***p < 0.001. Detailed results of statistical testing are summarised in Table 2.
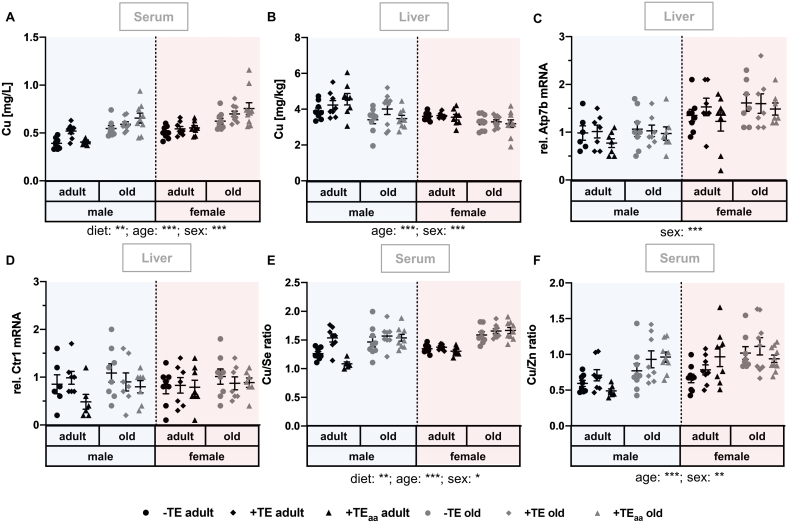
Fig. 3.
Changes in Cu status as well as related biomarkers and genes. Markers for the evaluation of the Cu status were determined in serum and liver of adult (30 weeks) and old (66 weeks) male and female C57BL/6Jrj mice (n = 8–10) receiving a -TE, +TE, or +TEaa diet for 26 weeks. Cu concentrations were analysed in serum (A) and liver (B) by ICP-MS/MS. On the basis of the analysed TE concentrations, serum Cu/Se (E) and serum Cu/Zn ratio (F) were calculated. To further evaluate the Cu status, relative hepatic expression levels of Cu transporters Ctr1 (C) and Atp7b (D) were examined by qRT-PCR analysis. Hepatic transcription levels were normalised to a composite factor based on the house keeper genes Hprt and Rpl13a. Variances are expressed as fold changes compared to +TE male adults (mean +TE male adult = 1). Statistical testing based on Three-Way ANOVA and Bonferroni's post-test with *p < 0.05, **p < 0.01, ***p < 0.001. Detailed results of statistical testing are summarised in Table 2.
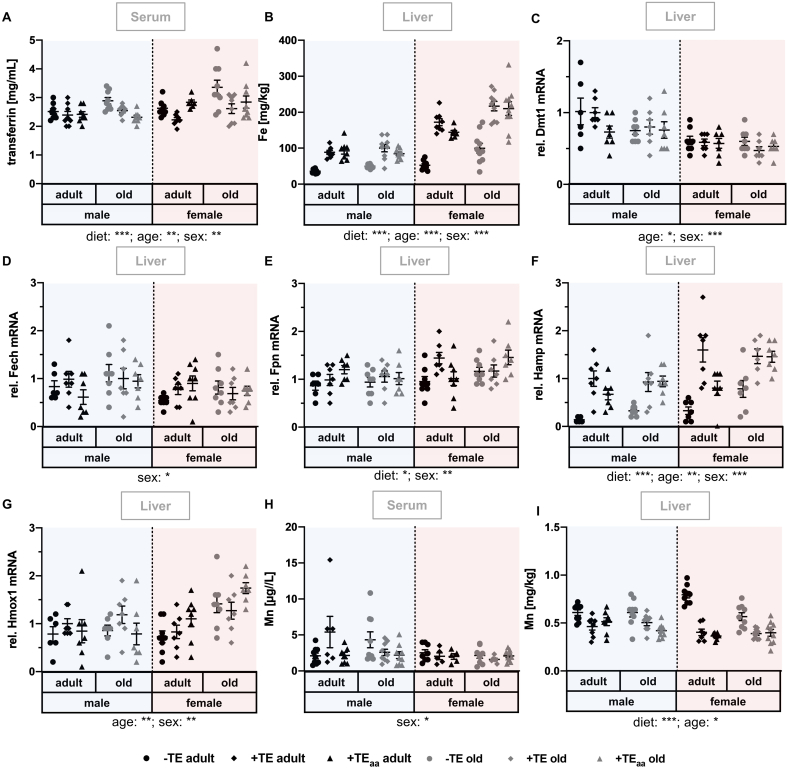
Fig. 4.
Changes in Fe and Mn status, as well as related biomarkers and genes. Markers for the evaluation of the Fe and Mn status were determined in serum and liver of adult (30 weeks) and old (66 weeks) male and female C57BL/6Jrj mice (n = 8–10) receiving a -TE, +TE, or +TEaa diet for 26 weeks. Hepatic Fe (B) as well as serum (H) and hepatic (I) Mn concentrations were determined via ICP-MS/MS. To further evaluate the Fe status, serum transferrin (A) concentrations were analysed via ELISA and relative hepatic expression levels of genes involved in Fe metabolism including Dmt1 (C), Fech (D), Fpn (E), Hamp (F), and Hmox1 (G) were examined by qRT-PCR analysis. Hepatic transcription levels were normalised to a composite factor based on the house keeper genes Hprt and Rpl13a. Variances are expressed as fold changes compared to +TE male adults (mean +TE male adult = 1). Statistical testing based on Three-Way ANOVA and Bonferroni's post-test with *p < 0.05, **p < 0.01, ***p < 0.001. Detailed results of statistical testing are summarised in Table 2.
3.1. Changes in Zn status as well as related biomarkers and genes
The nutritional Zn supply was modulated from 4.75 mg/kg in the -TE group to 30.0 mg/kg in the +TE group to 180 mg/kg in the +TEaa group (Table 1). Irrespective of that, total serum and hepatic Zn concentrations (Fig. 1A, C) were not modulated by the dietary intervention. Hepatic Zn concentrations showed a trend to decrease with increasing age (age: p = 0.069). Free Zn was unaffected by diet and age but it was modulated by sex in both liver and serum (Fig. 1B, D). Female mice showed decreased serum and increased hepatic free Zn concentrations compared to male mice.
mRNA expression of the metal-binding metallothioneins 1 (Mt1) and 2 (Mt2) were significantly increased in female mice (Fig. 1E, F). Zrt- and Irt-like protein (Zip) 5 showed the opposite diet-induced effect in adult female mice being upregulated in both +TE groups in comparison to the -TE group (-TE vs. +TE: p = 0.0015; -TE vs. +TEaa: p < 0.001) (Fig. 1H). mRNA expression of Zip 3 was significantly increased in female mice and also in old mice but differences were rather small (Fig. 1G). Zinc transporter (ZnT) 10 was not modulated by diet, but decreased with increasing age (Fig. 1I).
3.2. Changes in Se status as well as related biomarkers and genes
The nutritional Se supply was modulated from 0.07 mg/kg in the -TE group to 0.15 mg/kg in the +TE group to 0.60 mg/kg in the +TEaa group (Table 1). In serum, Se concentrations were increased with increasing dietary supply, but only to a small extent (Fig. 2A). Unexpectedly, Se concentrations were higher in old than in adult mice and were higher in females than in males. Comparable results were obtained for serum levels of Selenop with regard to diet, age, and sex (Fig. 2C). In contrast, serum GPX activity was constant throughout all groups (Fig. 2B). In liver, for both Se concentration and GPX activity, a diet-induced increase was observed, which had a fold-change of 1.3 at maximum for both (Fig. 2D, E). In contrast to the serum Se concentration, both hepatic Se levels and GPX activity were decreased in old mice. Additionally, a sex-dependent alteration in Selenop mRNA expression was observed with female mice showing increased mRNA expression as compared to male mice (Fig. 2F).
3.3. Changes in Cu status as well as related biomarkers and genes
The nutritional Cu supply was modulated from 0.44 mg/kg in the -TE group to 6.00 mg/kg in the +TE and the +TEaa group (Table 1). Serum Cu concentrations were also very moderately, yet significantly, altered by diet (Fig. 3A). Effects were mainly observed when comparing the -TE groups with either of the respective +TE groups. In addition, Cu levels were increased in an age-dependent manner with higher levels in old mice. Also, female mice had higher Cu levels than male mice. In contrast, hepatic Cu concentrations (Fig. 3B) showed inverse effects with significantly decreased Cu concentrations in females in comparison to males and old in comparison to adult animals. While mRNA expression of high affinity copper uptake protein 1 (Ctr1) was not affected by any factor investigated (Fig. 3D), mRNA expression of copper-transporting ATPase 2 (Atp7b) was significantly increased in female as compared to male mice (Fig. 3C).
Both the serum Cu/Zn and the Cu/Se ratio were increased in old in comparison to adult mice. In line with the serum Cu concentrations, both ratios were also increased in females as compared to males. Male adult +TE mice showed a significantly higher Cu/Se ratio as compared to both, male adult -TE mice (p < 0.05) and male adult +TEaa mice (p < 0.001). These specific diet-dependent effects were limited to male adult animals.
3.4. Changes in Fe and Mn status as well as related biomarkers and genes
The nutritional Fe supply was modulated from 9.62 mg/kg in the -TE group to 35.0 mg/kg in the +TE and the +TEaa group (Table 1). Both biomarkers for the Fe status, serum transferrin (Fig. 4A) and hepatic Fe concentrations (Fig. 4B), were modulated in a diet-dependent manner. Within all age- and sex-specific subgroups, transferrin concentrations were highest in mice receiving the -TE diet as compared to +TE or +TEaa supply. Hepatic Fe concentrations were always lower in mice with -TE supply in comparison to either of the respective +TE groups. Further, serum transferrin and hepatic Fe concentrations were significantly increased in female compared to male and old compared to adult mice. Regarding genes involved in Fe-metabolism, hepcidin antimicrobial peptide (Hamp) and hepatic Fe-exporter ferroportin-1 (Fpn) mRNA expression were modulated by diet, being suppressed in the -TE groups compared to +TE- and +TEaa-supplied mice (Fig. 4C–G). These effects were much more pronounced for Hamp than for Fpn. mRNA expression of ferrochelatase (Fech) and divalent metal transporter 1 (Dmt1) were decreased in female mice (Fig. 4C, D). In contrast, Hamp, Fpn, and heme oxygenase 1 (Hmox1) showed increased expression levels in female mice (Fig. 4E–G). Furthermore, increased Hamp and Hmox1 mRNA expression was observed in old compared to adult mice (Fig. 4F, G), while Dmt1 mRNA expression was decreased in old as compared to adult mice (Fig. 4C).
The nutritional Mn supply was not modulated in any intervention group, but met the recommendation for mice (Table 1). Serum Mn concentration (Fig. 4H) was not affected by diet or age, but was moderately reduced in female compared to male mice. In contrast to serum, hepatic Mn concentrations (Fig. 4I) showed a clear diet-dependency despite constant Mn supply over all groups. The hepatic Mn concentration was increased in mice receiving the -TE supply compared to +TE and +TEaa supply. These effects were most pronounced in female mice independent of their age.
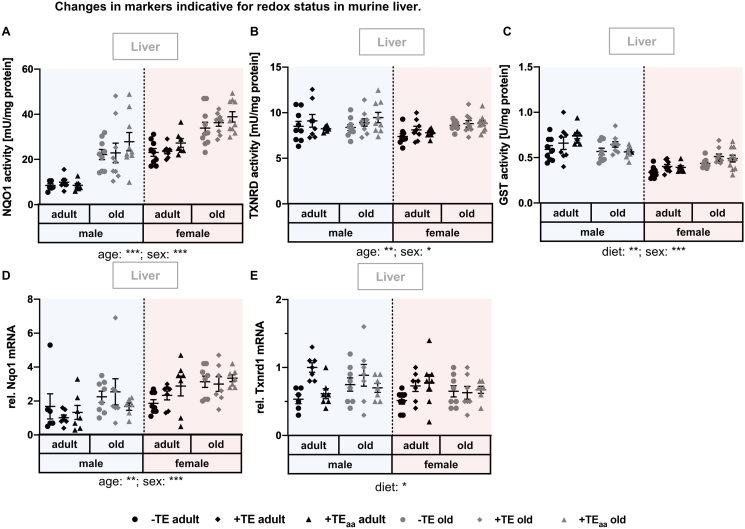
3.5. Changes in hepatic markers indicative for the redox status
Different genes, in part Nrf2 target genes, were analysed in liver, including NQO1, TXNRD, and GST activity, as well as Txnrd1 and Nqo1 mRNA expression (Fig. 5). Only GST activity was significantly affected by the diet. The -TE supply reduced enzyme activity compared to +TE or +TEaa supply (Fig. 5C). In +TE mice, Txnrd1 mRNA expression was specifically increased in adult and old males, as well as adult females (Fig. 5E). Both NQO1 activity and mRNA expression as well as TXNRD activity were increased in old compared to adult mice (Fig. 5A, B, D). Furthermore, GST activity was higher in males than in females. Comparable sex effects (increase in male vs. female mice) could be determined for TXNRD enzyme activity (Fig. 5B). In contrast, Nqo1 mRNA expression as well as NQO1 enzyme activity increased in female in comparison to male mice (Fig. 5A, D). Also, hepatic GSH concentrations showed a sex-dependent alteration, with increased GSH concentration in male compared to female mice (Supplementary Information Fig. 3).
Fig. 5.
Changes in markers indicative for the redox status in murine liver. Various markers indicative for the redox status were analysed in liver tissue of adult (30 weeks) and old (66 weeks) male and female C57BL/6Jrj mice (n = 8–10) receiving a -TE, +TE, or + TEaa diet for 26 weeks. Enzyme activities of the Nrf2 targets NQO1 (A), TXNRD (B), and GST (C) were determined by activity assays. Relative expression levels of Nqo1 (D) and Txnrd1 (E) mRNA were examined by qRT-PCR analysis. Hepatic transcription levels were normalised to a composite factor based on the house keeper genes Hprt and Rpl13a. Variances are expressed as fold changes compared to + TE male adults (mean + TE male adult = 1). Statistical testing based on Three-Way ANOVA and Bonferroni's post-test with * p < 0.05, **p < 0.01, ***p < 0.001. Detailed results of statistical testing are summarised in Table 2.
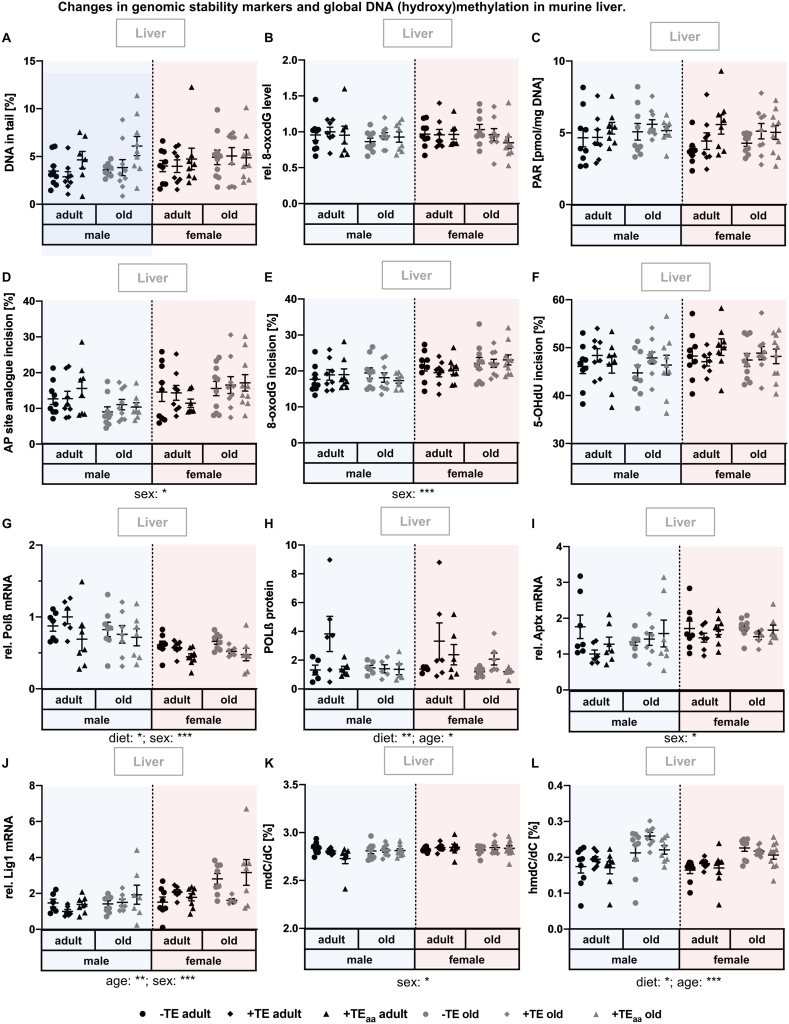
3.6. Changes in genomic stability markers and global DNA (hydroxy)methylation
Neither DNA damage nor relative 8-oxodG level, as well as PARylation status were affected by diet, age, or sex (Fig. 6A–C). In contrast, AP site analogue and 8-oxodG incision activity showed a sex-dependent increase in incision activity in female mice (Fig. 6D, E), while for 5-OHdU this was only observed as a trend (p = 0.0678) (Fig. 6F). Polβ mRNA expression showed a diet-dependent effect with highest expression level in -TE mice and lowest in +TEaa mice (Fig. 6G). While an increased Polβ mRNA expression was observed in male mice, this sex-dependent effect could not be confirmed for Polβ protein expression level. In addition, Polβ protein levels were increased in adult in comparison to old and in +TE-supplied mice (Fig. 6H). Furthermore, mRNA expression level of aprataxin (Aptx) and Lig1 were increased in female compared to male mice (Fig. 6I and J). Besides, Lig1 mRNA expression levels were age-dependently increased in old compared to adult mice.
Fig. 6.
Changes in genomic stability markers and global DNA (hydroxy)methylation in murine liver. Genomic stability markers were analysed in liver tissue of adult (30 weeks) and old (66 weeks) male and female C57BL/6Jrj mice (n = 8–10) receiving a -TE, +TE, or + TEaa diet for 26 weeks. Regarding DNA damage, the total amount of DNA strand breaks as well as alkali-labile sites (A), and relative 8-oxodG levels (B) were analysed by alkaline comet assay and applying an ELISA kit, respectively. PARylation levels were determined via HPLC-MS/MS (C), and BER incision activity was determined towards an AP site analogue (D), an 8-oxodG (E), and a 5-OHdU (F) containing oligonucleotide by a non-radioactive incision activity assay. Relative expression levels of Polβ (G), Aptx (I), and Lig1 (J) mRNA were examined by qRT-PCR analysis and hepatic transcription levels were normalised to a composite factor based on the house keeper genes Hprt and Rpl13a. Variances are expressed as fold changes compared to + TE male adults (mean + TE male adult = 1). Polβ protein quantification was performed via western blot analysis using β-actin for normalisation of determined protein levels. DNA (hydroxy)methylation was quantified via HPLC-MS/MS (K, L). Statistical testing based on Three-Way ANOVA and Bonferroni's post-test with * p < 0.05, **p < 0.01, ***p < 0.001. Detailed results of statistical testing are summarised in Table 2.
While global DNA methylation in the liver was only significantly increased in female compared to male mice (Fig. 6K), global DNA hydroxymethylation was significantly increased in old as compared to adult mice (Fig. 6L). Furthermore, global DNA hydroxymethylation was increased in +TE-supplied mice compared to both -TE- and +TEaa-supplied mice.
4. Discussion
This study aimed to analyse the effects of a long-term feeding period with suboptimal supply of the essential TEs Fe, Cu, Zn, and Se, starting either at young or older age in comparison to mice receiving adequate TE supply. As a third intervention group, we included mice which were fed a so-called age-adjusted diet with a higher supply of Se and Zn above adequate levels. This intervention aimed to compensate for the anticipated decline in the Zn and Se status during ageing. Age-dependent shifts in serum TE concentrations have been described before for single TEs [[58], [59], [60], [61]], but lately have been published also for whole panels of TEs [36,62]. In contrast to these published data, Zn concentrations were almost unchanged by ageing in the mice analysed herein (Fig. 1). As expected, serum Cu concentration was upregulated in old mice (Fig. 3A) [13,63]. Additionally, the hepatic Cu concentration showed a trend to decrease with increasing age (Fig. 3B), an effect that has been observed in humans before [64]. In contrast to our previous results, serum Se and Selenop concentrations were increased in old mice (Fig. 2A, B). Cao et al. also found an increase in serum Se and Selenop concentrations in male mice with increasing age [61]. In parallel, hepatic Se and GPX activity decreased with age (Fig. 2D, E) indicating that the hepatic Se pool is deprived for increasing the Se transport to peripheral organs. Also, hepatic Fe concentrations and biomarkers increased with age (Fig. 4). These differences could be due to the younger age of the old mice studied herein which were sacrificed at an age of 66 weeks. In our previous study, the old mouse cohort had an age of more than 100 weeks [36]. Yet, the older mice suffered from much more age-related diseases, mostly tumours, which was avoided by choosing younger mice for our current old mouse cohort. Another possibly contributing factor to be considered is that the amount of the life-long TE supply could be important for determining the age-related shifts in TE profiles, which was lower than in almost all other published studies in which usually a standard mouse chow was used which is very high in TEs.
Neither serum nor hepatic Zn concentrations or Zn-related biomarkers were modulated by the dietary intervention (Fig. 1), leading to the conclusion that the Zn status is well maintained independent of the herein studied nutritional supply of the mice. In comparison to other studies focusing on Zn deficiency in mice, the supplied Zn level in -TE animals was approximately 2-fold higher. Thus, it was not expected that symptoms of severe Zn deficiency would develop in the -TE mice [65,66]. But, also in +TEaa-supplied mice receiving 6-fold higher Zn concentrations than recommended, no alterations of the Zn status were observed. Interestingly, free Zn concentrations in serum and liver varied in female compared to male mice in an opposite manner (Fig. 1B, D). While sex-dependent differences have been reported for human serum before [43], to the best of our knowledge, an increase in hepatic free Zn concentrations of female mice has not been reported, so far. In line with this, hepatic mRNA expression of transporters of the Zip-family as well as Mts (Fig. 1E–H), which are essential for the maintenance of cellular Zn homeostasis, was increased in female mice confirming the known sexual dimorphisms for Mts [67,68].
Serum as well as hepatic Se concentrations and biomarkers were very moderately modulated by feeding three different Se concentrations (Fig. 2). Both serum Se as well as Selenop concentrations increased with an increasing dietary Se supply (Fig. 2A, C), but fold changes were much smaller than the ones described for the classical torula yeast-based diet which is fed to induce Se deficiency [21]. Furthermore, higher serum Se and Selenop concentrations were detected in female than in male mice, which is in line with the literature (Fig. 2A, C, F) [[69], [70], [71]]. As also hepatic Selenop mRNA expression was increased in female mice, an estrogen-induced increase of Selenop transcription could be the reason for this effect [72].
The Cu supply of the +TE intervention groups was 13.6-times higher than that of the -TE groups. However, there was only a marginal difference in serum Cu concentrations between the -TE and +TE groups. In old mice additionally supplied with Zn and Se (+TEaa), serum Cu concentrations were slightly higher than in mice of the respective -TE and +TE intervention groups. In accordance with our previous findings [36], female mice showed higher serum Cu concentrations than male mice, while in liver the Cu concentration tended to be lower in female than in male mice. The increased Atp7b mRNA expression in female in comparison to male mice (Fig. 3C) could be the underlying mechanism for this observation, by facilitating higher Cu export from the liver into the blood. In line with higher serum Cu concentrations, hepatic Mt mRNA expression was increased in female mice (Fig. 1E, F). mRNA expression of the metal-binding Mt1 and Mt2 is regulated via the transcription factor metal regulatory transcription factor 1 which is mainly activated in response to increased Zn but also Cu concentrations [73,74]. Furthermore, Mts are connected to the overall redox status, as Mt2 is known to be a Nrf2 target gene [75]. In contrast, hepatic Ctr1 mRNA expression remained unaffected (Fig. 3D), thus not reflecting the observed changes in Cu concentrations. An inverse relationship of serum and liver Cu concentrations was mathematically substantiated by a moderate negative correlation revealed by Spearman correlation analysis (rs = -0.576, p < 0.001) (Supplementary Information Fig. 4).
Out of the four TEs, the strongest modulation by diet was observed for the Fe status indicated by significantly decreased hepatic Fe concentrations and Hamp mRNA expression (Fig. 4B, F). Hamp encodes for the Fe-homeostasis regulating hormone hepcidin, which upon downregulation facilitates intestinal Fe absorption and release of Fe from other cell types to balance the Fe status [11]. Accordingly, serum transferrin concentrations were increased in -TE mice (Fig. 4A). This indicates impairment of the Fe status, because enhanced transferrin synthesis by the liver ameliorates the capacity of Fe binding and distribution within the body to ensure adequate Fe supply of peripheral tissues [76]. Besides dietary effects, the hepatic Fe concentrations were increased in female mice (Fig. 4B). This could be explained by the observed higher Hmox1 mRNA expression in females (Fig. 4G), fostering an increased release of Fe from heme [77], while at the same time decreased hepatic mRNA expression of Fech might lower incorporation of Fe into heme (Fig. 4D) [78].
Although the Mn supply was constant in all intervention groups, liver Mn concentrations were significantly increased in -TE mice (Fig. 4I). This can be explained by the inverse interrelation of Fe and Mn homeostases [79], which is based on the fact that Dmt1 becomes upregulated upon Fe deficiency, not only transporting Fe but also Mn [80]. Fe deficiency can result in an increase in Mn concentrations in peripheral tissues, which is dependent on the availability of Fe relative to Mn [79,81]. The reduced Fe supply in -TE mice resulted in a decrease in hepatic Fe concentration, while hepatic Mn concentration increased, overall confirming the antagonistic nature of Mn and Fe regulation.
Nowadays, rather than individual serum TE concentrations, composite TE biomarkers are used as estimate of the TE and health status. An increased serum Cu/Zn ratio has become a robust predictive nutritional biomarker of age, age-related decline, in particular frailty, and all-cause mortality in the elderly human population [34,[82], [83], [84]]. In line with serum Cu concentrations, mice of the -TE groups showed decreased Cu/Zn ratios compared to mice of the respective +TE intervention groups. Further, as we observed previously [36], also in this study the Cu/Zn ratio was higher in old mice (Fig. 3F). The serum Cu/Se ratio is another composite biomarker related to the redox status and upon increase indicative for a state of (infectious) disease and poor health in both humans and mice [[85], [86], [87], [88]]. The serum Cu/Se ratio was lower in the +TEaa groups in comparison to the -TE and +TE groups, which is plausible as within the +TEaa intervention, mice received increased amounts of Se (Fig. 3E). In addition, in male adult but not old or female mice, the Cu/Se ratio could be modulated by diet. These results reflect the altered Cu and Se supply in the three intervention groups, but limited to adult male mice. Additionally, the Cu/Se ratio was increased in both, old as compared to adult, and female as compared to male mice, likely driven by the increased serum Cu concentrations in these groups. In this context, it needs to be considered that we modulated the supply of both Zn and Se, yet this had no, in case of Zn, and only marginal, in case of Se, effect on the analysed status markers allowing to consider the ratios with Cu in both cases as reasonable biomarkers.
Overall, the TE concentrations were only very mildly modulated by our long-term dietary intervention, as was the cellular redox status, which was addressed by analysing Nrf2 activity via target gene expression analysis (Fig. 5D,E). Yet, ageing impacted on the cellular redox status as indicated by higher mRNA expression and enzyme activity of redox-regulating NQO1 (Fig. 5A, D) and antioxidant TXNRD (Fig. 5B, E) [[89], [90], [91]]. Accordingly, an age-related increase in hepatic NQO1 levels in C57BL/6 mice has previously been reported by us [36], and others [[92], [93], [94]]. Alterations in the redox balance could also be related to the age-dependent increase in global hepatic DNA hydroxymethylation we observed, which has been shown to increase in a redox-dependent manner [95]. Global hepatic DNA hydroxymethylation showed increased concentrations in old as well as -TE-supplied mice (Fig. 6L). The latter might be related to the observed Fe deficiency via the dependency of enzymes of the ten-eleven translocation family on Fe, responsible for the conversion of mdC to hmdC.
In female as compared to male mice, hepatic mRNA expression and enzyme activity of NQO1 were increased, while hepatic GST activity was decreased (Fig. 5A, C, D). This has been reported before [36]. Possibly, increased Nqo1 mRNA expression and enzyme activity in females could be related to the higher levels of estrogen in female mice [96], which are in turn causal to formation of DNA damaging estrogen quinones. These can be counteracted by NQO1, although a key role for NQO1 in this detoxification is controversially discussed [97,98]. Thus, in females the increased NQO1 together with reduced GST and TXNRD activity (Fig. 5B, C) imply an impaired redox status [99,100], that is likely related to the observed sex-dependent differences in the redox-active TEs Fe and Cu. Via activation of Nrf2, these TEs can initiate upregulation of Nrf2 targets such as Nqo1, Hmox1, Hamp, and Mt2 [[101], [102], [103]], all of which showed elevated hepatic mRNA expression in female mice.
Despite observed characteristics of ageing in the old mice as well as intervention-induced effects especially in -TE-supplied mice, we found only marginal effects on the analysed endpoints related to genomic stability (Fig. 6). These results are contradicting several studies that indicated an effect of both TE imbalances [26,27,104] as well as ageing [105,106] on genome stability maintenance. Especially with regard to the widespread theory of impaired DNA repair and accumulation of DNA damage as key factors causing and promoting ageing, this was unexpected [105,106]. Nevertheless, several human and rodent studies revealed comparable results with regard to DNA damage showing a missing correlation between increased DNA damage levels and age [51,107,108]. In line with the unaffected DNA damage levels, hepatic PARylation, key to DDR and BER [109], several mRNA expression levels of central BER proteins, as well as incision activities towards 8-oxodG, 5-OHdU, and an AP site analogue were not affected by diet and age (Fig. 6C–J). In literature, for both gene expression analysis and incision activity heterogenous results are reported [[110], [111], [112], [113], [114], [115]]. Conclusively, these results strengthen the hypothesis that the initial key players in the well-regulated BER system are not affected by the TE supply studied or the age of the mice [51].
Interestingly, female mice showed significantly increased incision activity towards the AP site analogue and 8-oxodG, while (oxidative) DNA damage level as well as PARylation status were not affected (Fig. 6A–E). In this context, the increased activity of analysed key BER glycosylases in female mice could represent one potential mechanism of female mice to counteract altered redox status associated (oxidative) DNA damage accumulation resulting in comparable DNA damage levels as in male mice. Furthermore, sex-dependent changes in mRNA and protein expression levels of BER genes and proteins involved in successful strand polymerisation as well as ligation indicate that sex-dependent alterations in the DNA repair mechanism occur additionally at different steps apart from initial incision (Fig. 6G–J).
In contrast to marginally affected global hepatic DNA methylation, global hepatic DNA hydroxymethylation was increased in old mice as previously observed (Fig. 6K and L) [36,116]. This is in line with the common hypothesis that aberrant epigenetic changes might be a common driver of ageing, highlighting global DNA hydroxymethylation as a potential suitable ageing marker. The aforementioned interrelation with alterations in the redox status, as observed in ageing [117], corroborates this assumption.
5. Conclusion
The dietary intervention used in this study modulated the TE supply of mice only mildly, which reflects changes that can also occur in humans in developed countries with an appropriate nutritional status. Regardless of these physiological variations in the dietary TE supply, age-related shifts in TE concentrations were maintained throughout all feeding groups indicating that these effects are driven by other mechanisms which could not be modulated by the dietary supply. Based thereon, these data indicate that just increasing the nutritional supply of critical TEs such as Zn and Se in older individuals will not help to maintain or even to improve their TE status and to overcome ageing-induced effects on TE homeostasis. Thus, it is essential to better understand the molecular mechanisms driving these age-related changes of the TE homeostasis and to think about putative (pharmacological) interventions, e.g., by modulating transporter or binding protein expression to improve the TE status of the elderly.
Funding
This research was funded by the German Research Foundation (DFG) Research Unit TraceAge (FOR 2558) and by the German Federal Ministry of Education and Research for the Competence Cluster NutriAct – Nutrition Research Berlin-Potsdam (FKS: 01EA1806B).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102083.
Contributor Information
Viktoria K. Wandt, Email: vwandt@uni-potsdam.de.
Nicola Winkelbeiner, Email: winkelbeiner@uni-potsdam.de.
Kristina Lossow, Email: kristina.lossow@uni-jena.de.
Johannes F. Kopp, Email: research.jfkopp@posteo.de.
Maria Schwarz, Email: schwarz.maria@uni-jena.de.
Wiebke Alker, Email: alker@tu-berlin.de.
Merle M. Nicolai, Email: merle.nicolai@uni-wuppertal.de.
Luise Simon, Email: Luisesimon@gmx.net.
Caroline Dietzel, Email: carolinedietzel@gmx.de.
Barbara Hertel, Email: bhertel@uni-potsdam.de.
Gabriele Pohl, Email: gpohl@uni-potsdam.de.
Franziska Ebert, Email: fraebert@uni-potsdam.de.
Lutz Schomburg, Email: lutz.schomburg@charite.de.
Julia Bornhorst, Email: bornhorst@uni-wuppertal.de.
Hajo Haase, Email: haase@tu-berlin.de.
Anna P. Kipp, Email: anna.kipp@uni-jena.de.
Tanja Schwerdtle, Email: tanja.schwerdtle@uni-potsdam.de.
Abbreviations
- 5-OHdU
5-hydroxy-2'-deoxyguanine
- 8-oxodG
8-oxo-2'-deoxyguanine
- affinity-HPLC-ICP-MS/MS
affinity high performance liquid chromatography coupled to inductively coupled plasma mass spectrometry
- ANOVA
analysis of variance
- AP site
apurinic/apyrimidinic site
- Aptx
aprataxin
- Atp7b
copper-transporting ATPase 2
- BER
base excision repair
- BHT
butylated hydroxytoluene
- cDNA
complementary DNA
- CE
collision energy
- Ctr1
high affinity copper uptake protein 1
- Cu
copper
- DDR
DNA damage response
- Dmt1
divalent metal transporter 1
- ELISA
enzyme-linked immunosorbent assay
- ESI+
electrospray ionisation in positive mode
- Fe
iron
- Fech
ferrochelatase
- Fpn
ferroportin-1
- GPX
glutathione peroxidase
- GR
glutathione reductase
- GSH
glutathione
- GSSG
glutathione disulfide
- GST
glutathione S-transferase
- Hamp
hepcidin antimicrobial peptide
- hmdC
hydroxymethylated cytidine
- Hmox1
heme oxygenase 1
- HPLC-MS/MS
high performance liquid chromatography tandem mass spectrometry
- Hprt
hypoxanthine phosphoribosyltransferase 1
- I
iodine
- Lig
DNA ligase
- mdC
methylated cytidine
- Mn
manganese
- Mt
metallothionein
- NEM
N-ethylmaleimide
- NQO1
NAD(P)H quinone dehydrogenase 1
- Nrf2
nuclear factor erythroid 2-like 2
- PAGE
polyacrylamide gel electrophoresis
- PAR
poly(ADP-ribose)
- PARP1
poly(ADP-ribose) polymerase 1
- PARylation
poly(ADP-ribosyl)ation
- PCR
polymerase chain reaction
- Polβ
polymerase beta
- qRT-PCR
quantitative real-time polymerase chain reaction
- qTOF
quadrupole time of flight
- Rpl13a
ribosomal protein L13a
- RT
room temperature
- Se
selenium
- Selenop
selenoprotein P
- TE(s)
trace element(s)
- TXNRD
thioredoxin reductase
- Zip
Zrt- and Irt-like protein
- Zn
zinc
- ZnT
zinc transporter
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bornhorst J., Kipp A.P., Haase H., Meyer S., Schwerdtle T. The crux of inept biomarkers for risks and benefits of trace elements. Trends Anal. Chem. 2018;104:183–190. [Google Scholar]
- 2.WHO Trace elements in human nutrition. Report of a WHO expert committee. World Health Organ Tech Rep Ser. 1973;532:1–65. [PubMed] [Google Scholar]
- 3.Malbohan I.M., Fialova L. [Trace elements, human nutrition and health] Cas. Lek. Cesk. 1997;136(11):356–359. [PubMed] [Google Scholar]
- 4.Mertz W. The essential trace elements. Science. 1981;213(4514):1332–1338. doi: 10.1126/science.7022654. [DOI] [PubMed] [Google Scholar]
- 5.Livingstone C. Zinc: physiology, deficiency, and parenteral nutrition. Nutr. Clin. Pract. 2015;30(3):371–382. doi: 10.1177/0884533615570376. [DOI] [PubMed] [Google Scholar]
- 6.WHO . 2015. The Global Prevalence of Anaemia in 2011. [Google Scholar]
- 7.Combs G.F., Jr. Selenium in global food systems. Br. J. Nutr. 2001;85(5):517–547. doi: 10.1079/bjn2000280. [DOI] [PubMed] [Google Scholar]
- 8.Chen J. The molecular mechanisms of copper metabolism and its roles in human diseases. Pflügers Archiv. 2020;472(10):1415–1429. doi: 10.1007/s00424-020-02412-2. [DOI] [PubMed] [Google Scholar]
- 9.Fairweather-Tait S.J. Selenium in human health and disease. Antioxidants Redox Signal. 2011;14(7):1337–1383. doi: 10.1089/ars.2010.3275. [DOI] [PubMed] [Google Scholar]
- 10.Maret W. Zinc biochemistry: from a single zinc enzyme to a key element of life. Adv Nutr. 2013;4(1):82–91. doi: 10.3945/an.112.003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yiannikourides A., Latunde-Dada G.O. A short review of iron metabolism and pathophysiology of iron disorders. Medicine. 2019;6(3) doi: 10.3390/medicines6030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meplan C. Trace elements and ageing, a genomic perspective using selenium as an example. J. Trace Elem. Med. Biol. 2011;25(Suppl 1):S11–S16. doi: 10.1016/j.jtemb.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Baudry J. Changes of trace element status during aging: results of the EPIC-Potsdam cohort study. Eur. J. Nutr. 2020;59(7):3045–3058. doi: 10.1007/s00394-019-02143-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minelli A. Oxidative stress-related aging: a role for prostate cancer? Biochim. Biophys. Acta. 2009;1795(2):83–91. doi: 10.1016/j.bbcan.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Finkel T., Holbrook N.J., Oxidants Oxidative stress and the biology of ageing. Nature. 2000;408(6809):239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 16.Haigis M.C., Yankner B.A. The aging stress response. Mol. Cell. 2010;40(2):333–344. doi: 10.1016/j.molcel.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoeijmakers J.H. DNA damage, aging, and cancer. N. Engl. J. Med. 2009;361(15):1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 18.Schmidlin C.J. Redox regulation by NRF2 in aging and disease. Free Radic. Biol. Med. 2019;134:702–707. doi: 10.1016/j.freeradbiomed.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou L. Aging-related decline in the induction of Nrf2-regulated antioxidant genes in human bronchial epithelial cells. Redox Biol. 2018;14:35–40. doi: 10.1016/j.redox.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39(4):199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Schwarz M. Crosstalk of Nrf2 with the trace elements selenium, iron, zinc, and copper. Nutrients. 2019;11(9) doi: 10.3390/nu11092112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brigelius-Flohe R., Kipp A.P. Selenium in the redox regulation of the Nrf2 and the Wnt pathway. Methods Enzymol. 2013;527:65–86. doi: 10.1016/B978-0-12-405882-8.00004-0. [DOI] [PubMed] [Google Scholar]
- 23.Cai Z., Zhang J., Li H. Selenium, aging and aging-related diseases. Aging Clin. Exp. Res. 2019;31(8):1035–1047. doi: 10.1007/s40520-018-1086-7. [DOI] [PubMed] [Google Scholar]
- 24.Aslan M. Lymphocyte DNA damage and oxidative stress in patients with iron deficiency anemia. Mutat. Res. 2006;601(1–2):144–149. doi: 10.1016/j.mrfmmm.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Wandt V.K. A matter of concern - trace element dyshomeostasis and genomic stability in neurons. Redox Biol. 2021;41:101877. doi: 10.1016/j.redox.2021.101877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linder M.C. Copper and genomic stability in mammals. Mutat. Res. 2001;475(1–2):141–152. doi: 10.1016/s0027-5107(01)00076-8. [DOI] [PubMed] [Google Scholar]
- 27.Sharif R. The role of zinc in genomic stability. Mutat. Res. 2012;733(1–2):111–121. doi: 10.1016/j.mrfmmm.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Schwerdtle T. Impact of copper on the induction and repair of oxidative DNA damage, poly(ADP-ribosyl)ation and PARP-1 activity. Mol. Nutr. Food Res. 2007;51(2):201–210. doi: 10.1002/mnfr.200600107. [DOI] [PubMed] [Google Scholar]
- 29.Cassandri M. Zinc-finger proteins in health and disease. Cell Death Dis. 2017;3:17071. doi: 10.1038/cddiscovery.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burkle A. Poly(ADP-ribosyl)ation, genomic instability, and longevity. Ann. N. Y. Acad. Sci. 2000;908:126–132. doi: 10.1111/j.1749-6632.2000.tb06641.x. [DOI] [PubMed] [Google Scholar]
- 31.Ou H.L., Schumacher B. DNA damage responses and p53 in the aging process. Blood. 2018;131(5):488–495. doi: 10.1182/blood-2017-07-746396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wedler N. Impact of the cellular zinc status on PARP-1 activity and genomic stability in HeLa S3 cells. Chem. Res. Toxicol. 2021 Mar 15;34(3):839–848. doi: 10.1021/acs.chemrestox.0c00452. Epub 2021 Mar 1. PMID: 33645215. [DOI] [PubMed] [Google Scholar]
- 33.Mocchegiani E. Zinc: dietary intake and impact of supplementation on immune function in elderly. Age. 2013;35(3):839–860. doi: 10.1007/s11357-011-9377-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mocchegiani E. Cu to Zn ratio, physical function, disability, and mortality risk in older elderly (ilSIRENTE study) Age. 2012;34(3):539–552. doi: 10.1007/s11357-011-9252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lymbury R. Selenium status of the Australian population: effect of age, gender and cardiovascular disease. Biol. Trace Elem. Res. 2008;126(Suppl 1):S1–S10. doi: 10.1007/s12011-008-8208-6. [DOI] [PubMed] [Google Scholar]
- 36.Lossow K. Aging affects sex- and organ-specific trace element profiles in mice. Aging (N Y) 2020;12(13):13762–13790. doi: 10.18632/aging.103572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finke H. Effects of a cumulative, suboptimal supply of multiple trace elements in mice: trace element status, genomic stability, inflammation, and epigenetics. Mol Nutr Food Res. 2020 doi: 10.1002/mnfr.202000325. [DOI] [PubMed] [Google Scholar]
- 38.Hedrich H. second ed. 2012. p. 868. (The Laboratory Mouse). [Google Scholar]
- 39.Kopp J.F. A quick and simple method for the determination of six trace elements in mammalian serum samples using ICP-MS/MS. J. Trace Elem. Med. Biol. 2019;54:221–225. doi: 10.1016/j.jtemb.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 40.Meyer S. Development, validation and application of an ICP-MS/MS method to quantify minerals and (ultra-)trace elements in human serum. J. Trace Elem. Med. Biol. 2018;49:157–163. doi: 10.1016/j.jtemb.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 41.Heitland P., Koster H.D. Biomonitoring of selenoprotein P in human serum by fast affinity chromatography coupled to ICP-MS. Int. J. Hyg Environ. Health. 2018;221(3):564–568. doi: 10.1016/j.ijheh.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 42.Hill K.E. The selenium-rich C-terminal domain of mouse selenoprotein P is necessary for the supply of selenium to brain and testis but not for the maintenance of whole body selenium. J. Biol. Chem. 2007;282(15):10972–10980. doi: 10.1074/jbc.M700436200. [DOI] [PubMed] [Google Scholar]
- 43.Alker W. A zinpyr-1-based fluorimetric microassay for free zinc in human serum. Int. J. Mol. Sci. 2019;20(16) doi: 10.3390/ijms20164006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grynkiewicz G., Poenie M., Tsien R.Y. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 1985;260(6):3440–3450. [PubMed] [Google Scholar]
- 45.Muller M. Nrf2 target genes are induced under marginal selenium-deficiency. Genes Nutr. 2010;5(4):297–307. doi: 10.1007/s12263-010-0168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Florian S. Loss of GPx2 increases apoptosis, mitosis, and GPx1 expression in the intestine of mice. Free Radic. Biol. Med. 2010;49(11):1694–1702. doi: 10.1016/j.freeradbiomed.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krehl S. Glutathione peroxidase-2 and selenium decreased inflammation and tumors in a mouse model of inflammation-associated carcinogenesis whereas sulforaphane effects differed with selenium supply. Carcinogenesis. 2012;33(3):620–628. doi: 10.1093/carcin/bgr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Habig W.H., Pabst M.J., Jakoby W.B., S-transferases Glutathione. The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 1974;249(22):7130–7139. [PubMed] [Google Scholar]
- 49.Neumann C. The role of poly(ADP-ribose) polymerases in manganese exposed Caenorhabditis elegans. J. Trace Elem. Med. Biol. 2020;57:21–27. doi: 10.1016/j.jtemb.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azqueta A., Collins A.R. The essential comet assay: a comprehensive guide to measuring DNA damage and repair. Arch. Toxicol. 2013;87(6):949–968. doi: 10.1007/s00204-013-1070-0. [DOI] [PubMed] [Google Scholar]
- 51.Winkelbeiner N. A multi-endpoint Approach to base excision repair incision activity augmented by PARylation and DNA damage levels in mice: impact of sex and age. Int. J. Mol. Sci. 2020;21(18) doi: 10.3390/ijms21186600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moller P. Minimum Information for Reporting on the Comet Assay (MIRCA): recommendations for describing comet assay procedures and results. Nat. Protoc. 2020;15(12):3817–3826. doi: 10.1038/s41596-020-0398-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.OECD, Test No. 489 . OECD Publishing; Paris, France: 2016. Vivo Mammalian Alkaline Comet Assay, OECD Guidelines for the Testing of Chemicals, Section 4: Health Effects. [Google Scholar]
- 54.Martello R. Quantification of cellular poly(ADP-ribosyl)ation by stable isotope dilution mass spectrometry reveals tissue- and drug-dependent stress response dynamics. ACS Chem. Biol. 2013;8(7):1567–1575. doi: 10.1021/cb400170b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Speckmann B. Selenium increases hepatic DNA methylation and modulates one-carbon metabolism in the liver of mice. J. Nutr. Biochem. 2017;48:112–119. doi: 10.1016/j.jnutbio.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 56.Muller S.M. Arsenic-containing hydrocarbons: effects on gene expression, epigenetics, and biotransformation in HepG2 cells. Arch. Toxicol. 2018;92(5):1751–1765. doi: 10.1007/s00204-018-2194-z. [DOI] [PubMed] [Google Scholar]
- 57.Schiesser S. Deamination, oxidation, and C-C bond cleavage reactivity of 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxycytosine. J. Am. Chem. Soc. 2013;135(39):14593–14599. doi: 10.1021/ja403229y. [DOI] [PubMed] [Google Scholar]
- 58.Quinn J.F. Gender effects on plasma and brain copper. Int. J. Alzheimer's Dis. 2011;2011:150916. doi: 10.4061/2011/150916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woodward W.D., Filteau S.M., Allen O.B. Decline in serum zinc level throughout adult life in the laboratory mouse. J. Gerontol. 1984;39(5):521–524. doi: 10.1093/geronj/39.5.521. [DOI] [PubMed] [Google Scholar]
- 60.Wong C.P., Magnusson K.R., Ho E. Increased inflammatory response in aged mice is associated with age-related zinc deficiency and zinc transporter dysregulation. J. Nutr. Biochem. 2013;24(1):353–359. doi: 10.1016/j.jnutbio.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cao L. Analyses of selenotranscriptomes and selenium concentrations in response to dietary selenium deficiency and age reveal common and distinct patterns by tissue and sex in telomere-dysfunctional mice. J. Nutr. 2017;147(10):1858–1866. doi: 10.3945/jn.117.247775. [DOI] [PubMed] [Google Scholar]
- 62.Ma S. Organization of the mammalian ionome according to organ origin, lineage specialization, and longevity. Cell Rep. 2015;13(7):1319–1326. doi: 10.1016/j.celrep.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Massie, H., Colacicco, J., Alello, V., Changes With Age in Copper and Ceruloplasmin in Serum from Humans and C57BL/6J Mice 1979.
- 64.Vuori E., Huunan-Seppala A., Kilpio J.O. Biologically active metals in human tissues. I. The effect of age and sex on the concentration of copper in aorta, heart, kidney, liver, lung, pancreas and skeletal muscle. Scand. J. Work. Environ. Health. 1978;4(2):167–175. doi: 10.5271/sjweh.2712. [DOI] [PubMed] [Google Scholar]
- 65.Chen Y. Zinc deficiency promotes testicular cell apoptosis in mice. Biol. Trace Elem. Res. 2020;195(1):142–149. doi: 10.1007/s12011-019-01821-4. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Q. Zinc deficiency induces oxidative damage and causes spleen fibrosis. Biol. Trace Elem. Res. 2020;194(1):203–209. doi: 10.1007/s12011-019-01762-y. [DOI] [PubMed] [Google Scholar]
- 67.Zhang D. Diurnal-and sex-related difference of metallothionein expression in mice. J. Circadian Rhythms. 2012;10(1):5. doi: 10.1186/1740-3391-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ljubojevic M. Sex-dependent expression of metallothioneins MT1 and MT2 and concentrations of trace elements in rat liver and kidney tissues: effect of gonadectomy. J. Trace Elem. Med. Biol. 2019;53:98–108. doi: 10.1016/j.jtemb.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 69.Schomburg L. Effect of age on sexually dimorphic selenoprotein expression in mice. Biol. Chem. 2007;388(10):1035–1041. doi: 10.1515/BC.2007.128. [DOI] [PubMed] [Google Scholar]
- 70.Seale L.A., Ogawa-Wong A.N., Berry M.J. Sexual dimorphism in selenium metabolism and selenoproteins. Free Radic. Biol. Med. 2018;127:198–205. doi: 10.1016/j.freeradbiomed.2018.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riese C. Selenium-dependent pre- and posttranscriptional mechanisms are responsible for sexual dimorphic expression of selenoproteins in murine tissues. Endocrinology. 2006;147(12):5883–5892. doi: 10.1210/en.2006-0689. [DOI] [PubMed] [Google Scholar]
- 72.Zhou X. Estrogen status alters tissue distribution and metabolism of selenium in female rats. J. Nutr. Biochem. 2012;23(6):532–538. doi: 10.1016/j.jnutbio.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takahashi S. Positive and negative regulators of the metallothionein gene (review) Mol. Med. Rep. 2015;12(1):795–799. doi: 10.3892/mmr.2015.3459. [DOI] [PubMed] [Google Scholar]
- 74.Krezel A., Maret W. The functions of metamorphic metallothioneins in zinc and copper metabolism. Int. J. Mol. Sci. 2017;18(6) doi: 10.3390/ijms18061237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu R. In vivo pharmacokinetics and regulation of gene expression profiles by isothiocyanate sulforaphane in the rat. J. Pharmacol. Exp. Therapeut. 2004;310(1):263–271. doi: 10.1124/jpet.103.064261. [DOI] [PubMed] [Google Scholar]
- 76.Gkouvatsos K., Papanikolaou G., Pantopoulos K. Regulation of iron transport and the role of transferrin. Biochim. Biophys. Acta. 2012;1820(3):188–202. doi: 10.1016/j.bbagen.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 77.Poss K.D., Tonegawa S. Heme oxygenase 1 is required for mammalian iron reutilization. Proc. Natl. Acad. Sci. U. S. A. 1997;94(20):10919–10924. doi: 10.1073/pnas.94.20.10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ferreira G.C. Ferrochelatase. Int J Biochem Cell Biol. 1999;31(10):995–1000. doi: 10.1016/s1357-2725(99)00066-7. [DOI] [PubMed] [Google Scholar]
- 79.Ye Q. Influence of iron metabolism on manganese transport and toxicity. Metall. 2017;9(8):1028–1046. doi: 10.1039/c7mt00079k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shawki A. Intestinal DMT1 is critical for iron absorption in the mouse but is not required for the absorption of copper or manganese. Am. J. Physiol. Gastrointest. Liver Physiol. 2015;309(8):G635–G647. doi: 10.1152/ajpgi.00160.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Seo Y.A., Li Y., Wessling-Resnick M. Iron depletion increases manganese uptake and potentiates apoptosis through ER stress. Neurotoxicology. 2013;38:67–73. doi: 10.1016/j.neuro.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Malavolta M. Plasma copper/zinc ratio: an inflammatory/nutritional biomarker as predictor of all-cause mortality in elderly population. Biogerontology. 2010;11(3):309–319. doi: 10.1007/s10522-009-9251-1. [DOI] [PubMed] [Google Scholar]
- 83.Gaier E.D. High serum Cu and Cu/Zn ratios correlate with impairments in bone density, physical performance and overall health in a population of elderly men with frailty characteristics. Exp. Gerontol. 2012;47(7):491–496. doi: 10.1016/j.exger.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giacconi R. Main biomarkers associated with age-related plasma zinc decrease and copper/zinc ratio in healthy elderly from ZincAge study. Eur. J. Nutr. 2017;56(8):2457–2466. doi: 10.1007/s00394-016-1281-2. [DOI] [PubMed] [Google Scholar]
- 85.Schwarz M. Copper interferes with selenoprotein synthesis and activity. Redox Biol. 2020;37:101746. doi: 10.1016/j.redox.2020.101746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ozturk P., Belge Kurutas E., Ataseven A. Copper/zinc and copper/selenium ratios, and oxidative stress as biochemical markers in recurrent aphthous stomatitis. J. Trace Elem. Med. Biol. 2013;27(4):312–316. doi: 10.1016/j.jtemb.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 87.Sun Q. Selenium and copper as biomarkers for pulmonary arterial hypertension in systemic sclerosis. Nutrients. 2020;12(6) doi: 10.3390/nu12061894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hackler J. Copper and selenium status as biomarkers of neonatal infections. J. Trace Elem. Med. Biol. 2020;58:126437. doi: 10.1016/j.jtemb.2019.126437. [DOI] [PubMed] [Google Scholar]
- 89.Ross D., Siegel D. Functions of NQO1 in cellular protection and CoQ10 metabolism and its potential role as a redox sensitive molecular switch. Front. Physiol. 2017;8:595. doi: 10.3389/fphys.2017.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Holmgren A. Antioxidant function of thioredoxin and glutaredoxin systems. Antioxidants Redox Signal. 2000;2(4):811–820. doi: 10.1089/ars.2000.2.4-811. [DOI] [PubMed] [Google Scholar]
- 91.Ross D., Siegel D. NQO1 in protection against oxidative stress. Current Opinion in Toxicology. 2018;7:67–72. [Google Scholar]
- 92.Chhetri J., King A.E., Gueven N. Alzheimer's disease and NQO1: is there a link? Curr. Alzheimer Res. 2018;15(1):56–66. doi: 10.2174/1567205014666170203095802. [DOI] [PubMed] [Google Scholar]
- 93.Zhang H. Nrf2-regulated phase II enzymes are induced by chronic ambient nanoparticle exposure in young mice with age-related impairments. Free Radic. Biol. Med. 2012;52(9):2038–2046. doi: 10.1016/j.freeradbiomed.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fu Z.D., Csanaky I.L., Klaassen C.D. Effects of aging on mRNA profiles for drug-metabolizing enzymes and transporters in livers of male and female mice. Drug Metab. Dispos. 2012;40(6):1216–1225. doi: 10.1124/dmd.111.044461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao B. Redox-active quinones induces genome-wide DNA methylation changes by an iron-mediated and Tet-dependent mechanism. Nucleic Acids Res. 2014;42(3):1593–1605. doi: 10.1093/nar/gkt1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nilsson M.E. Measurement of a comprehensive sex steroid profile in rodent serum by high-sensitive gas chromatography-tandem mass spectrometry. Endocrinology. 2015;156(7):2492–2502. doi: 10.1210/en.2014-1890. [DOI] [PubMed] [Google Scholar]
- 97.Chandrasena R.E. Problematic detoxification of estrogen quinones by NAD(P)H-dependent quinone oxidoreductase and glutathione-S-transferase. Chem. Res. Toxicol. 2008;21(7):1324–1329. doi: 10.1021/tx8000797. [DOI] [PubMed] [Google Scholar]
- 98.Dinkova-Kostova A.T., Talalay P. NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch. Biochem. Biophys. 2010;501(1):116–123. doi: 10.1016/j.abb.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lima C.F. The drinking of a Salvia officinalis infusion improves liver antioxidant status in mice and rats. J. Ethnopharmacol. 2005;97(2):383–389. doi: 10.1016/j.jep.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 100.Lovell M.A., Xie C., Markesbery W.R. Decreased glutathione transferase activity in brain and ventricular fluid in Alzheimer's disease. Neurology. 1998;51(6):1562–1566. doi: 10.1212/wnl.51.6.1562. [DOI] [PubMed] [Google Scholar]
- 101.Kerins M.J., Ooi A. The roles of NRF2 in modulating cellular iron homeostasis. Antioxidants Redox Signal. 2018;29(17):1756–1773. doi: 10.1089/ars.2017.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bayele H.K., Balesaria S., Srai S.K. Phytoestrogens modulate hepcidin expression by Nrf2: implications for dietary control of iron absorption. Free Radic. Biol. Med. 2015;89:1192–1202. doi: 10.1016/j.freeradbiomed.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gu J. Metallothionein is downstream of Nrf2 and partially mediates sulforaphane prevention of diabetic cardiomyopathy. Diabetes. 2017;66(2):529–542. doi: 10.2337/db15-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pra D. Iron and genome stability: an update. Mutat. Res. 2012;733(1–2):92–99. doi: 10.1016/j.mrfmmm.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 105.Lopez-Otin C. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vermeij W.P., Hoeijmakers J.H., Pothof J. Genome integrity in aging: human syndromes, mouse models, and therapeutic options. Annu. Rev. Pharmacol. Toxicol. 2016;56:427–445. doi: 10.1146/annurev-pharmtox-010814-124316. [DOI] [PubMed] [Google Scholar]
- 107.Jacob K.D. Markers of oxidant stress that are clinically relevant in aging and age-related disease. Mech. Ageing Dev. 2013;134(3–4):139–157. doi: 10.1016/j.mad.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maslov A.Y. DNA damage in normally and prematurely aged mice. Aging Cell. 2013;12(3):467–477. doi: 10.1111/acel.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mangerich A., Burkle A. Pleiotropic cellular functions of PARP1 in longevity and aging: genome maintenance meets inflammation. Oxid Med Cell Longev. 2012:321653. doi: 10.1155/2012/321653. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mikkelsen L. Aging and defense against generation of 8-oxo-7,8-dihydro-2'-deoxyguanosine in DNA. Free Radic. Biol. Med. 2009;47(5):608–615. doi: 10.1016/j.freeradbiomed.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 111.Tian F. Age-dependent down-regulation of mitochondrial 8-oxoguanine DNA glycosylase in SAM-P/8 mouse brain and its effect on brain aging. Rejuvenation Res. 2009;12(3):209–215. doi: 10.1089/rej.2009.0849. [DOI] [PubMed] [Google Scholar]
- 112.Cabelof D.C. Attenuation of DNA polymerase beta-dependent base excision repair and increased DMS-induced mutagenicity in aged mice. Mutat. Res. 2002;500(1–2):135–145. doi: 10.1016/s0027-5107(02)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Swain U., Subba Rao K. Study of DNA damage via the comet assay and base excision repair activities in rat brain neurons and astrocytes during aging. Mech. Ageing Dev. 2011;132(8–9):374–381. doi: 10.1016/j.mad.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 114.Gorniak J.P. Tissue differences in BER-related incision activity and non-specific nuclease activity as measured by the comet assay. Mutagenesis. 2013;28(6):673–681. doi: 10.1093/mutage/get047. [DOI] [PubMed] [Google Scholar]
- 115.Hamilton M.L. Does oxidative damage to DNA increase with age? Proc. Natl. Acad. Sci. U. S. A. 2001;98(18):10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tammen S.A. Aging alters hepatic DNA hydroxymethylation, as measured by liquid chromatography/mass spectrometry. J Cancer Prev. 2014;19(4):301–308. doi: 10.15430/JCP.2014.19.4.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jones D.P. Redox theory of aging. Redox Biol. 2015;5:71–79. doi: 10.1016/j.redox.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.