Abstract
IL‐22 is an alpha‐helical cytokine which belongs to the IL‐10 family of cytokines. IL‐22 is produced by RORγt+ innate and adaptive lymphocytes, including ILC3, γδ T, iNKT, Th17 and Th22 cells and some granulocytes. IL‐22 receptor is expressed primarily by non‐haematopoietic cells. IL‐22 is critical for barrier immunity at the mucosal surfaces in the steady state and during infection. Although IL‐22 knockout mice were previously shown to develop experimental autoimmune encephalomyelitis (EAE), a murine model of multiple sclerosis (MS), how temporal IL‐22 manipulation in adult mice would affect EAE course has not been studied previously. In this study, we overexpressed IL‐22 via hydrodynamic gene delivery or blocked it via neutralizing antibodies in C57BL/6 mice to explore the therapeutic impact of IL‐22 modulation on the EAE course. IL‐22 overexpression significantly decreased EAE scores and demyelination, and reduced infiltration of IFN‐γ+IL‐17A+Th17 cells into the central nervous system (CNS). The neutralization of IL‐22 did not alter the EAE pathology significantly. We show that IL‐22‐mediated protection is independent of Reg3γ, an epithelial cell‐derived antimicrobial peptide induced by IL‐22. Thus, overexpression of Reg3γ significantly exacerbated EAE scores, demyelination and infiltration of IFN‐γ+IL‐17A+ and IL‐17A+GM‐CSF+Th17 cells to CNS. We also show that Reg3γ may inhibit IL‐2‐mediated STAT5 signalling and impair expansion of Treg cells in vivo and in vitro. Finally, Reg3γ overexpression dramatically impacted intestinal microbiota during EAE. Our results provide novel insight into the role of IL‐22 and IL‐22‐induced antimicrobial peptide Reg3γ in the pathogenesis of CNS inflammation in a murine model of MS.
Keywords: experimental autoimmune encephalomyelitis, IL‐22, microbiota, multiple sclerosis treatment, Reg3γ
IL‐22 overexpression provides mice partial protection from EAE, underlining a therapeutic potential in human MS disease. This protection appears to be independent of Reg3γ. Our results also reveal that Reg3γ may directly and negatively regulate the function of Treg cells via STAT5‐mediated pathway; thus, its overexpression resulted in reduced Treg cell numbers and a more severe EAE course.
Abbreviations
- AhR
Aryl hydrocarbon receptor
- CNS
Central nervous system
- DMEM
Dulbecco's modified Eagle's minimal essential medium
- EAE
Experimental autoimmune encephalomyelitis
- Erk
Extracellular signal‐regulated kinase
- Extl
Exostosin‐like
- FBS
Fetal bovine serum
- Foxp3
Forkhead box protein 3
- GM‐CSF
Granulocyte–macrophage colony‐stimulating factor
- IL
Interleukin
- ILC
Innate lymphoid cell
- IFN
Interferon
- Jak1
Janus kinase
- MOG
myelin oligodendrocyte glycoprotein
- MS
Multiple sclerosis
- NKT
natural killer T
- PAMP
Pathogen‐associated molecular pattern
- PMA
Phorbol 12‐myristate 13‐acetate
- Reg
Regenerating islet‐derived
- RORγt+
Retinoic acid receptor‐related orphan receptor gamma t
- STAT
Signal transducer and activator of transcription
- Th
T helper
- Tyk2
Tyrosine kinase
Highlights.
Temporal IL‐22 overexpression ameliorates EAE pathology
IL‐22 neutralization exacerbates EAE scores marginally
Reg3γ overexpression exacerbates EAE pathology
Reg3γ blocks Treg expansion in vivo and in vitro and inhibits IL‐2‐mediated STAT5 phosphorylation
Reg3γ overexpression also results in dramatic changes in the intestinal microbiota during EAE
INTRODUCTION
Interleukin (IL)‐22 is a helical cytokine classified under the IL‐10 family of cytokines [1, 2]. It is produced mainly by retinoic acid receptor‐related orphan receptor gamma t (RORγt)+ innate and adaptive immune cells. These include group 3 innate lymphoid cells (ILC3) [3, 4, 5, 6], neutrophils [7], mast cells [8, 9], γδ T cells [10], natural killer T (NKT) cells [11], T helper (Th)17 [12] and Th22 cells [13]. IL‐22 is more abundantly expressed in the mucosal tissues and skin where most of the aforementioned cells are prevalent. Production of IL‐22 is induced by IL‐23 and IL‐1β, which are also highly expressed at the mucosal sites by antigen‐presenting cells in response to pathogen‐associated molecular patterns or by aryl hydrocarbon receptor (AhR) ligands [2, 14, 15, 16, 17, 18]. IL‐22 affects mostly non‐haematopoietic lineage, such as epithelial cells and Muller cells through its membrane receptor [1, 19, 20, 21]. Membrane‐bound IL‐22 receptor is composed of IL‐22RA1 and IL‐10Rβ, which are coupled to Janus kinase (Jak1) and tyrosine kinase 2 (Tyk2). IL‐22 receptor activates various signalling pathways including signal transducer and activator of transcription (Stat)1/3/5, c‐Jun‐N‐terminal kinase (JNK), p38 and extracellular signal‐regulated kinase (Erk)1/2 [22, 23]. A soluble receptor for IL‐22 (IL‐2BP) is also produced by antigen‐presenting cells from its gene IL22RA2 [24, 25, 26]. IL‐22‐BP is believed to regulate bioavailability and block signalling of IL‐22. IL‐22 signalling events turn on several genes that regulate proliferation of epithelial cells and mucosal tissue repair; production of antimicrobial peptides including four regenerating protein (Reg) family members, S100A/B, etc., and mucus; and production of several chemokines, which recruit leucocytes including neutrophils [1, 27]. Antimicrobial peptides are critical for the containment of commensals on the luminal side of the intestines. Thus, IL‐22 is critical in both steady state for mucosal barrier homeostasis and mucosal protective immunity to enteropathogens during infection.
IL‐22 has been studied in the context of multiple sclerosis (MS) [28]. Importantly, many variants near the IL22RA2 gene are associated with increased risk for MS, coeliac disease and psoriasis [29, 30, 31]. IL‐22 knockout B6 mice, however, developed experimental autoimmune encephalomyelitis (EAE), suggesting that pathology can develop independently of IL‐22 [32]. More recent studies revealed a protective role for IL‐22 in the MS context in mice [33, 34]. These studies revealed that the genetic ablation of IL‐22BP resulted in less severe EAE pathology, possibly by increasing the availability of IL‐22 and that gain‐of‐function variants of IL‐22RA2 in humans might confer susceptibility to MS. Those studies also showed a correlation between elevated interferon‐gamma (IFN‐γ) /Th1 response and increased IL‐22BP. Moreover, recombinant IL‐22 injections have been shown to alleviate experimental uveitis and Muller cells in the retina have been shown to express IL‐22RA1 and respond to IL‐22 [21]. These data suggest that IL‐22 might have potential in the context of MS as a therapeutic cytokine. However, to our knowledge, recombinant IL‐22 administration or its temporal overexpression has not yet been reported in the context of EAE.
In this study, we aimed to test the therapeutic potential of temporal overexpression of IL‐22 in the murine myelin oligodendrocyte glycoprotein (MOG)35‐55‐induced EAE model. We show that temporal IL‐22 overexpression ameliorates EAE and that this protection is independent of Reg3γ. Conversely, Reg3γ overexpression exacerbated EAE pathology and impacted the intestinal microbial community. We also show that Reg3γ may reduce regulatory T (Treg)‐cell number and expansion in vivo and ex vivo and may partly explain the exacerbation of EAE pathology.
MATERIALS AND METHODS
Mice
C57BL/6 mice were bred in specific pathogen‐free conditions at Erciyes University Betul‐Ziya Eren Genome and Stem Cell Center (GENKOK) and Experimental Research and Application Center (DEKAM) mouse facilities. C57BL/6 Foxp3‐YFP mice were a gift from Mohamed Oukka and were bred at Erciyes University GENKOK and DEKAM mouse facilities as well.
Human samples
Five hundred µl cerebrospinal fluid (CSF) from the leftovers of each individual, who underwent lumbar puncture, was collected. Of those CSF, five samples came from RRMS patients. Five controls (n = 5) were included. CSF fluid was spun at 400 g for 10 min at 4°C. The pellet was lysed in TRIzol, and RNA was extracted.
Study approval
All methods were approved by Erciyes University Institutional Review Boards for animal and human studies. The approval number for mouse study is 15/141. The ethics number for the human studies is 2019/743. Consent was taken from volunteers. Both studies were conducted according to the relevant guidelines and regulations.
Hydrodynamic gene delivery and neutralization
IL‐22 and Reg3γ cDNAs were amplified as described previously [35] using primers from total cDNA generated from colonic total RNA that was reverse‐transcribed by iScript cDNA Synthesis Kit. Both cDNAs cloned into TOPO vectors were validated, amplified and blunt‐end‐cloned into pBSHCRHP‐A backbone, which was generated by Miao et al. [36] For hydrodynamic injections, 15 µg plasmid was resuspended in 2 ml of saline. Hydrodynamic injections were performed intravenously in 5–10 s. IL‐22 protein in serum was confirmed by ELISA. Reg3γ overexpression was confirmed by Citrobacter rodentium colony‐forming assay [35]. IL‐22 was kindly provided by Genentech and was injected every other day (150 µg/mouse) for the first 10 days post‐immunization with MOG35‐55 peptide.
EAE induction
Experimental autoimmune encephalomyelitis was induced as previously described [37] . Mice were immunized at the flanks with an emulsion of MOG35–55 peptide (100 μg) emulsified in CFA supplemented with 4 mg/ml Mycobacterium tuberculosis extract H37Ra (Difco) subcutaneously. The animals received 200 ng pertussis toxin (List Biological Laboratories) i.p. on days 0 and 2. Clinical signs of EAE were monitored daily. The disease was scored according to the following criteria [37]: 0, no signs of disease; 1, limp tail; 2, hind limb weakness; 3, hind limb paralysis; and 4, hind limb and forelimb paralysis.
Isolation of leucocytes from CNS, spleen and lymph nodes and intracellular cytokine staining
Brain and spinal cords were harvested and sliced into 1‐mm pieces in 2 ml Dulbecco's modified Eagle's minimal essential medium (DMEM) containing 10% fetal bovine serum (FBS) and 1 mg/ml Collagenase IV. The tissues were digested for 30 min at 37°C. The mix was spun at 300 g for 10 min and then resuspended in 70% Percoll, and overlayed with 37% Percoll. After 20‐min centrifugation at 500 g without brakes at room temperature, cells were collected from the interphase and after a wash with complete medium (300 g, 5 min) used for experiments. Lymph nodes (LNs) were mashed mechanically with the back of a plunge, cells were filtered through 70‐μm strainer and spun at 300 g for 5 min, and the pellet was resuspended in RPMI containing 10% FBS and used for experiments. Spleen was prepared similarly, then treated with ACK lysis buffer to rid cells of erythrocytes. To identify cytokine profile of lymphocytes, 0·5 to 1x 106 cells were stimulated in RPMI containing 10% FBS and Golgi plug (1 μl/ml), 50 ng/ml of phorbol 12‐myristate 13‐acetate (PMA) and 1 μg/ml of ionomycin for 4–5 h at 37°C. Cells were then fixed and stained using intracellular cytokine staining kit (BD Biosciences) according to the instructions from the manufacturer. Treg cells were determined after staining with CD4 and Forkhead box protein 3 (Foxp3). In some experiments Foxp3 Yellow Fluorescent Protein (YFP) reporter mice were used, thus surface staining with CD4/CD3 was sufficient [37].
Following antibodies were used for staining: Mice:CD4 (GK1·5, eBioscience), IFN‐γ (XMG1·2, BioLegend), IL‐17A (eBio17B7, eBioscience), Foxp3 (MF‐14, BioLegend), pSTAT5 (SRBCZX, eBioscience). IL‐22 (Poly5164, BioLegend). IL‐10 (JES5‐16E3, BioLegend) and GM‐CSF (MP1‐22E9, BioLegend).
Luxol fast blue staining and demyelination scoring
One‐centimetre spinal cord was fixed in 10% neutral‐buffered formalin and embedded in paraffin, and cryosectioned and mounted on slides. Luxol fast blue staining was performed. Demyelination was scored semi‐quantitatively based on Ref. 38: no demyelination, score 0; mild demyelination (10%), score 1; moderate demyelination (10–25%), score 2; and severe demyelination (>25%), score 3.
Real‐time QPCR
Tissues or cells were collected in 1 ml TRIzol and homogenized with the PRO200 Homogenizer (PRO Scientific), and total RNA was extracted. SYBR Green qPCR was performed with a 7500 Real‐Time PCR System per the instructions of the manufacturer (Roche 480). Expression of the tested genes was normalized to the housekeeping ribosomal protein L19 (rPL19) mRNA. Arbitrary relative expression units were calculated by division of expression of the gene of interest by rPL19 mRNA expression. Primer and probe sequences for each target are given in Figure S1A. For real‐time qPCR array, splenocytes from B6 mice were cultured for 24 h in Reg3γ‐conditioned or control media (from empty plasmid transfections) with the addition of CD3/CD28, 1 µg/ml of each. The cells were collected from the supernatant, as well as from the bottom of the plate by trypsin/EDTA, and lysed with TRIzol. A real‐time qPCR array was performed for a select set of genes using Qiagen‐PAMM 52Z‐Mouse.
Transfection
HEK293 T cells were cultured in DMEM supplemented with 10% FBS. Transfection was performed according to the manufacturer's protocol (Neon). Briefly, cells were detached before use and transfected with Reg3γ or empty control plasmid with Neon transfection kit and device. 1–5 × 106 cells and 30 µg plasmid per each 100 μL tip were used. A GFP expression plasmid was used as a control to assess transfection efficacy. The supernatant was collected 48 h and 96 h later and filtered through 0·22‐µm filters and frozen at −80°C for future use as Reg3γ‐conditioned or control medium, respectively.
In vivo TREG expansion
Six to eight‐week‐old Foxp3 YFP or B6 mice were injected with 2 ml of saline containing 15 µg/ml plasmid (IL‐22, Reg3γ or empty vector as control) on day −2 or −1. Then, on starting day 0, the mouse groups received 100 µl of intraperitoneal injections of 5 ng IL‐2 (BioLegend) per gram mouse (in PBS), daily for 7 days. The mice were killed on day 7, and LNs (inguinal and brachial) were mechanically disrupted by the end of a plunge, and the cells were stained for CD4 or and Foxp3 if B6 is used. Blood lymphocytes were prepared after lysing red blood cells, the same staining as in LN was performed.
In vitro TREG expansion
CD4+ Foxp3YFP+ Treg cells were sorted from LNs and spleens from Foxp3 YFP mice and were cultured in CD3/CD28‐coated (1 µg/ml) 96‐well plates with 2000 U IL‐2 for 3–5 days. After culture, cells were counted by Spherotech counting beads and for Foxp3 YFP on FACSAria III.
Microbiota analyses
Stool samples from ileum were collected either 6 days after hydrodynamic gene delivery for naïve non‐immunized mice or at the peak of EAE (~21 days after hydrodynamic gene delivery and MOG35‐55 immunization) in microcentrifuge tubes. The samples were buffered with 2 ml of DNA/RNA Shield Buffer (Puritan DNA/RNA Shield, Zymo Research) that protects the composition of stool from disruption, and samples were stored and transferred at room temperature until DNA isolation. The remaining stool samples were stored at −80°C until further processing. DNA isolation was performed using Qiagen PowerSoil DNA isolation kits according to the protocol of the manufacturer (http://www.mobio.com/files/protocol/12888.pdf). Before the DNA isolation step, samples were buffered in 5 ml MoBio lysis buffer, vortexed for 30–40 s and homogenized. Following homogenization, mixture of stool/cotton was centrifuged at 1500 rcf for 5 min and supernatant of 1 ml was put in Garnet Bead tubes containing 750 uL buffer. The sequencing of 16S rRNA was performed using Illumina MiSeq system generating at least 50 000 reads per sample, and all procedures before sequencing were performed according to the protocol of the manufacturer (16S Metagenomic Sequencing Library Preparation Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System). 16S rRNA gene amplification was performed targeting V3‐V4 regions. Kappa polymerase was used for polymerase chain reaction (PCR). The overhang adapters appropriate for the Illumina MiSeq system were appended to the primers creating a single amplicon of approximately 460 bp. The full length of the primers was as follows: forward 5′ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGC CTACGG GNGGCWGCAG3′ and reverse ′GTCTCGTGGGCTCGGAGATGTGTATAA GAGACAGGACTA CHVGGGTATCTAATCC3′ [39]. Sequencing data were analysed using the QIIME pipeline [40] after filtering and trimming the reads for Phred quality score 30 via the Trimmomatic tool [41], and merging the paired‐end reads into amplicons using FLASh tool [42]. Operational taxonomic units were determined using the Uclust method and GreenGenes database [43] with closed reference procedure.
Statistical analyses
Pairwise comparisons were performed by Student's t‐test or the Mann–Whitney U‐test. Multiple comparisons were performed by ANOVA and Dunn's multiple comparison test. p‐Value < 0·05 accepted as significant. Error bars indicate ±standard error of the mean (SEM).
For microbiota analysis, alpha‐ and beta‐diversity statistics were assessed accordingly by QIIME pipeline scripts. Statistical tests were performed by the Mann–Whitney U‐test, and false discovery rate correction on p‐values was done by the Benjamini–Hochberg procedure.
RESULTS
IL‐22 overexpression alleviates EAE pathology by reducing CNS‐infiltrating lymphocytes
To test the therapeutic potential of IL‐22 overexpression, one group of B6 mice was hydrodynamically injected with IL‐22 expression plasmid 2 days before immunization with MOG35‐55. The plasmid backbone was initially engineered by Miao et al. for hydrodynamic injections and contains a liver tissue‐specific promoter [36]. We cloned IL‐22 into this vector and previously validated its expression in vivo in infection and colitis models [35, 44]. Another group of mice was injected with empty plasmid and was used as control (Figure S1B). Such injections were shown to provide systemic IL‐22 for at least 10 days up to ng levels (Figure S2A). Also, Il22 expression in the liver was detectable (Figure S2B). Hydrodynamic IL‐22 plasmid injection also increased some of the antimicrobial peptides in the small intestines such as S100A8 (Figure S2B). The mean EAE scores of the IL‐22‐injected group of mice were significantly lower compared with the empty plasmid‐injected group (Figure 1a). Importantly, demyelination scores of spinal cord sections after Luxol fast blue staining also revealed significantly lower demyelination, supporting a protective role for IL‐22 in this model of EAE (Figure 1b,c). Consistently, lymphocytes from the brains and spinal cords of IL‐22EAE group showed significantly less infiltration of lymphocytes, CD4+ T cells, IL‐17+ cells and IFN‐γ+ cells, suggesting that IL‐22 overexpression might reduce pathogenic T‐cell generation or infiltration into the CNS (Figure 1d, Fig S3A). Although the absolute number of Foxp3+ Treg cells in the brain was lower and the mean fluorescent intensity of Foxp3 in Treg cells on a per‐cell basis was comparable, Foxp3+/CD4+ ratio was higher in the IL‐22EAE group (Figure 1e). The production of IL‐17, IFN‐γ, IL‐10 and IL‐22 in the draining inguinal LNs and the spleen by lymphocytes was examined in the priming stage, on the 7th day of MOG35‐55 immunization, and revealed no differences between the IL‐22EAE and EmptyEAE control groups (Figure 1f). Foxp3+ Treg cell number and percentages were also unremarkable between the two groups (Figure 1g). Collectively, these data suggest that IL‐22 overexpression, possibly recombinant IL‐22, may provide partial protection against EAE, and this protection is associated with reduced lymphocyte infiltration into CNS.
FIGURE 1.
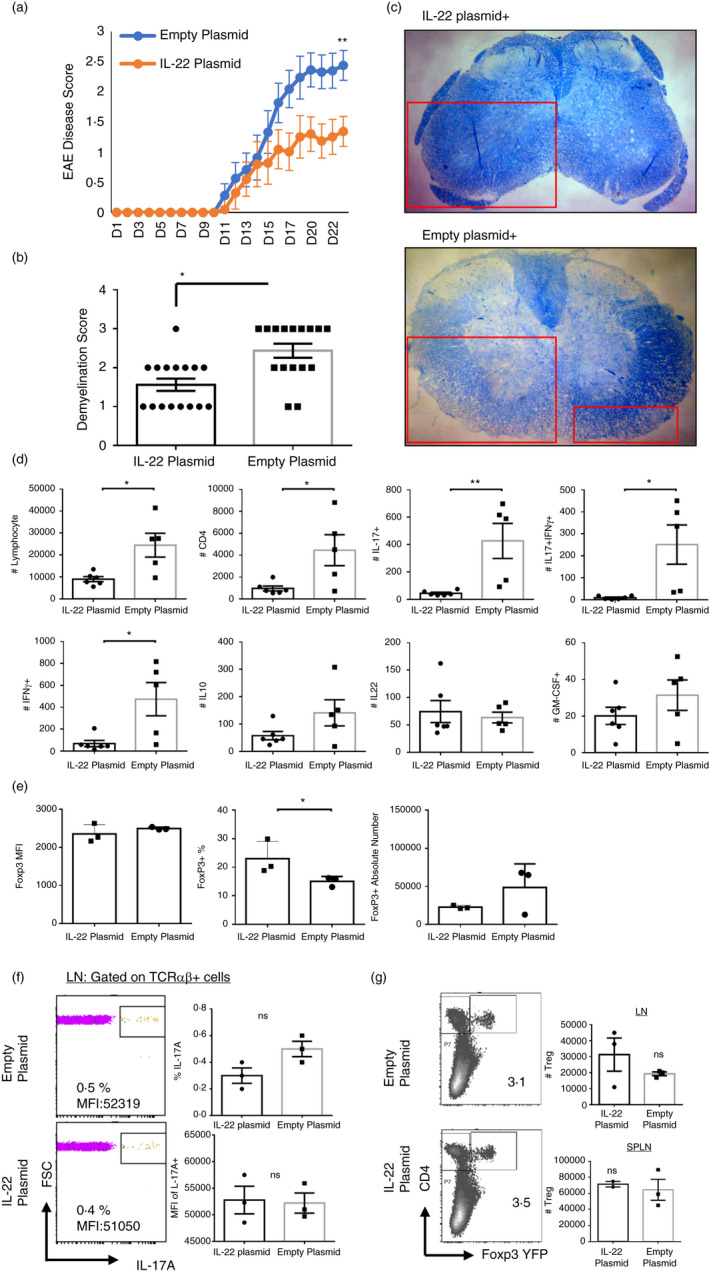
Temporal IL‐22 overexpression via hydrodynamic gene delivery ameliorates experimental autoimmune encephalomyelitis (EAE) pathology and disease scores. (a) IL‐22 plasmid or empty plasmid was injected on day −2, and EAE was induced by MOG35‐55 immunization on day 0. Mice were monitored for 3–4 weeks, and EAE disease was scored. (b) Spinal cords were removed and stained with Luxol fast blue to quantify demyelination 3 weeks after EAE induction (n = 11 mice for both groups). (c) A representative picture of Luxol fast blue staining. (d) Spinal cords and brain tissues from IL‐22 plasmid‐ or empty plasmid‐injected mouse groups were harvested 3 weeks after EAE induction, and infiltrating lymphocytes were quantified based on IFN‐γ, IL‐17A and GM‐CSF production. Absolute number or percentages of indicated cytokine‐producing CD4+ T cells were quantified (n = 11 mice (control), n = 11 mice). (e) Mean fluorescent intensity (MFI) of Foxp3 protein in Treg cells in the brain at 3 weeks after EAE induction, or percentages of Treg cells among CD4+ T cells, or their absolute numbers. (f) Lymph node or splenic IL‐17A production by T cells in IL‐22 plasmid‐ or empty plasmid‐injected mice on day 7 of EAE induction (priming phase of the disease). (g) Treg cell numbers in LN and spleen on day 7 of EAE induction. The experiments were repeated at least three times. (*) indicates p‐value <0·05, (**), p < 0·01
IL‐22 neutralization does not alter EAE pathology significantly
We next tested whether neutralization of IL‐22 would show an opposite effect compared with the overexpression system in the EAE model. One group of B6 mice was injected every other day with anti‐IL‐22 (150 µg/mice dose) for the first 10 days post‐immunization with MOG35‐55 peptide. The control group received isotype IgG. Such injections slightly increased the EAE disease score (Figure 2a); however, it did not result in significant differences between the groups with respect to demyelination scores (Figure 2b), or quantity and profile of leucocyte infiltrate into the CNS (Figure 2c). Importantly, when the (anti‐IL‐22) injections were applied every day, the disease scores were comparable (Figure S3B). The results suggest that temporal IL‐22 neutralization may not be beneficial in the clinical setting of MS. A recent study by Mattapallil et al. with IL‐22 KO mice showed higher EAE disease scores in the IL‐22 KO mice; however, the further details of the pathology (demyelination, lymphocyte infiltration) were not provided as the main model studied in the paper was experimental autoimmune uveitis (EAU) [21]. More importantly, the neutralizing antibody was not tested in the EAE setting in that study. However, the investigators tested IL‐22 neutralization by intraperitoneal injection, systemically, and intravitreal injections, locally, and provided evidence for protection in the EAU setting. These divergent results may be due to differences in the accessibility of anti‐IL‐22 to different organs or perhaps differences in the vivarium and commensals the mice are harbouring.
FIGURE 2.
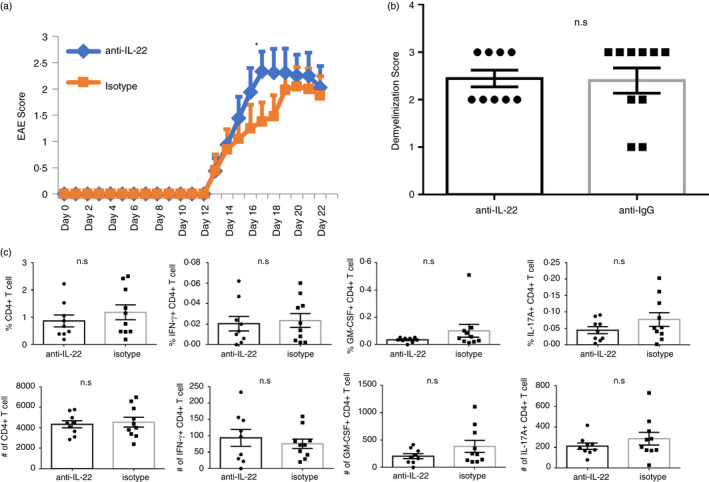
Neutralization of IL‐22 exacerbates experimental autoimmune encephalomyelitis (EAE) disease scores (a) After EAE was induced by MOG35‐55 immunization, one group of mice received every other day 150 µg anti‐mouse IL‐22, and the other group received isotype antibody until day 10 post‐immunization. The EAE was scored daily. (b) On day 22, spinal cords were removed and stained with Luxol fast blue and scored to quantify demyelination (n = 9–10 per group). (c) Spinal cords and brain tissues from anti‐IL‐22 and isotype antibody (control) groups were harvested 3 weeks after EAE induction, and infiltrating lymphocytes were quantified based on IFN‐γ, IL‐17A and GM‐CSF production. Absolute number or percentages of indicated cytokine‐producing CD4+ T cells were quantified (n = 9–10 per group). The experiments were repeated 3 times. (*) indicates p‐value <0·05
Reg3γ overexpression does not recapitulate IL‐22 overexpression and exacerbates EAE
IL‐22 induces production of four groups of Reg family proteins both in humans and in mice. These include Reg1, Reg2, Reg3a, Reg3b, Reg3 g, Reg3d and Reg4 in mice and REG1A, REG1B, REG3A, REG3B and REG4 in humans [27]. Reg proteins bind bacterial sugars and peptidoglycans through their C‐type lectin domains (CTLD) via Ca2+‐independent interactions and exert their bactericidal effects. Several receptors on other cells of the host have also been defined for Reg proteins (Reg1a‐Exostosin‐like 3 (Extl3); REG3A‐EXTL3, Reg3γ‐Extl3; REG3A‐EGFR; Reg3β‐gp130; Reg4‐GPR37) though some are controversial [45]. Anti‐inflammatory effects on epithelial cells of REG3A (human ortholog of murine Reg3γ) have been described [46]. Dose‐dependent SOCS induction or inhibition of NFκB translocation into nucleus by REG3A has been shown [47]. Therefore, we put to test whether protective effects of IL‐22, when overexpressed, might have been mediated by Reg3γ. To this end, hydrodynamic injections were performed with Reg3γ plasmid or empty vector, 2 days prior to EAE induction by MOG35‐55 immunizations. Reg3γ overexpression was confirmed by real‐time qPCR in the liver tissues and small intestine (Figure 3a). The hydrodynamic gene delivery‐based plasmid Reg3γ expression has been previously validated and did not augment IL‐17, GM‐CSF or IL‐22 in the serum of naïve (non‐EAE) mice (Figure S2C) [35]. Interestingly, EAE disease scores were significantly higher in the Reg3γ PlasmidEAE group compared with Empty PlasmidEAE group (Figure 3b). In line with this, demyelination scores (Figure 3c), and the number of lymphocytes, IL‐17+IFN‐γ+ cells, IL‐17+GM‐CSF+ cells, IL‐17A+ cells, IL‐22+ cells and IFN‐γ+ CD4+ T cells have all been elevated in the Reg3γ PlasmidEAE group compared with the Empty PlasmidEAE group at the peak of the disease (Figure 3d, Figure S3C). The number of CD4+Foxp3+ Treg cells between Reg3γ PlasmidEAE and Empty PlasmidEAE group mice was not significantly different at the peak of the disease (Day 19) (Figure 3d). When the draining LNs were collected at the peak the disease, IL‐17A, IL‐22 and IL‐17+IFN‐γ+ lymphocytes were significantly elevated in the Reg3γ PlasmidEAE group compared with the Empty PlasmidEAE group (Figure 3e). These data suggested that Reg3γ might have directly or indirectly affected, and augmented proliferation and/or infiltration of pathogenic T cells into CNS.
FIGURE 3.
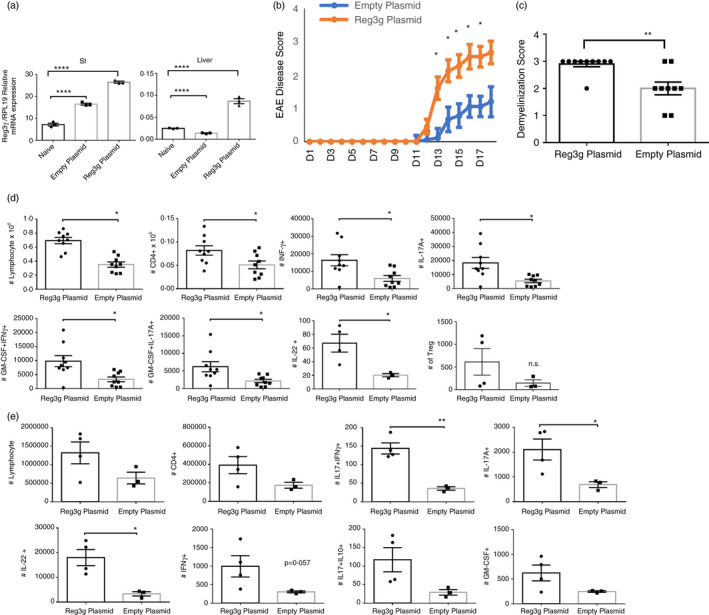
Temporal Reg3γ overexpression via hydrodynamic gene delivery exacerbates experimental autoimmune encephalomyelitis (EAE) pathology and disease scores (a) Reg3γ plasmid or empty plasmid were injected on day −2, and 3 days later, Reg3γ message was quantified by real‐time qPCR in the liver and small intestine tissue (n = 3). (b) Reg3γ plasmid or empty vector was injected on day −2, and EAE was induced by MOG35‐55 immunization on day 0. Mice were monitored for 3‐4 weeks, and EAE disease was scored (n = 9–10 per group). (c) Spinal cords were removed and stained with Luxol fast blue to quantify demyelination 3 weeks after EAE induction (n = 9–10 per group). (d) Spinal cords and brain tissues from Reg3γ plasmid‐ or empty vector‐injected mouse groups were harvested 3 weeks after EAE induction, and infiltrating lymphocytes were quantified based on IFN‐γ, IL‐17A and GM‐CSF production. Absolute number of indicated cytokine‐producing CD4+ T cells or Treg cells (last panel) was quantified (n = 9–10 mice per group). (e) Absolute number of indicated cytokine‐producing CD4+ T cells in the draining LNs of Reg3γ plasmid‐ or empty plasmid‐injected mouse groups at 3 weeks post‐immunization of EAE. (n = 4 mice per group). (*) indicates p‐value <0·05; (**), p < 0·01; (***), p < 0·001; and (****), p < 0·0001
Reg3γ blocks treg expansion in vivo and ex vivo
Because Treg cells are crucial in EAE and other autoimmune context, and Reg3γ or its human ortholog REG3A has been implicated in STAT1/3/5‐mediated pathways and NFκB signalling, we wanted to further pursue whether Reg3γ would have an impact on the expansion of Treg cells at the steady state in vivo. We injected hydrodynamically one group of mice (n = 5) with Reg3γ plasmid and another group with IL‐22 plasmid, and the control group received empty plasmid. Each group then received low‐dose IL‐2 daily, for 7‐days, to expand Treg cells in vivo, as previously described. As a control, another group of mice who received empty plasmid was injected with PBS. Such low‐dose IL‐2 injections indeed expanded Treg cell numbers in mice compared with PBS recipients (Figure 4a). More importantly, Treg cell numbers were significantly lower in the LNs of the Reg3γ overexpression group, suggesting that Reg3γ overexpression may block IL‐2‐dependent Treg expansion (Figure 4a). We further confirmed this ex vivo. To do this, HEK293 T cells were transfected with either empty or Reg3γ plasmid and the supernatants were collected within 3 days post‐transfection (Figure 4b). Then, Treg cells were sorted from naïve B6 Foxp3‐YFP mice and the cells were cultured in IL‐2/CD3/CD28 in the presence of Reg3γ‐conditioned or control media (from empty plasmid transfections). In such conditions, Treg cells expanded less in Reg3γ‐conditioned media (Figure 4c,d). These results led us to explore whether IL‐2 signalling may be impacted by Reg3γ. Indeed, when sorted Treg cells were pretreated for 2 h with Reg3γ‐conditioned media and then stimulated with IL‐2, STAT5 phosphorylation was significantly reduced compared with Treg cells pretreated with empty vector‐conditioned media (Figure 4e). Phospho‐STAT5 levels in Treg cells were also significantly reduced without exogenous IL‐2 stimulation. These results suggest that reduced Treg expansion in vivo and ex vivo may partly be due to Reg3γ‐mediated inhibition of IL‐2 signalling. To gain more insight into the transcriptional changes instigated by Reg3γ in immune cells, splenocytes from B6 mice were cultured for 24 h in Reg3γ‐conditioned or control media (from empty plasmid transfections) with the addition of CD3/CD28, 1 µg/ml of each. A real‐time qPCR array was performed for a select set of genes (Figure 4f). These experiments revealed that Reg3γ‐conditioned medium significantly blocked Foxp3 transcription along with Nfkbia, which supports our in vivo and ex vivo data that Reg3γ may inhibit Treg expansion (Figure 4f). These data collectively suggest that exacerbation of EAE after Reg3γ overexpression may partly be explained by the impact of Reg3γ on Treg cell numbers owing to its inhibitory effect on IL‐2 signalling and Foxp3 transcription.
FIGURE 4.
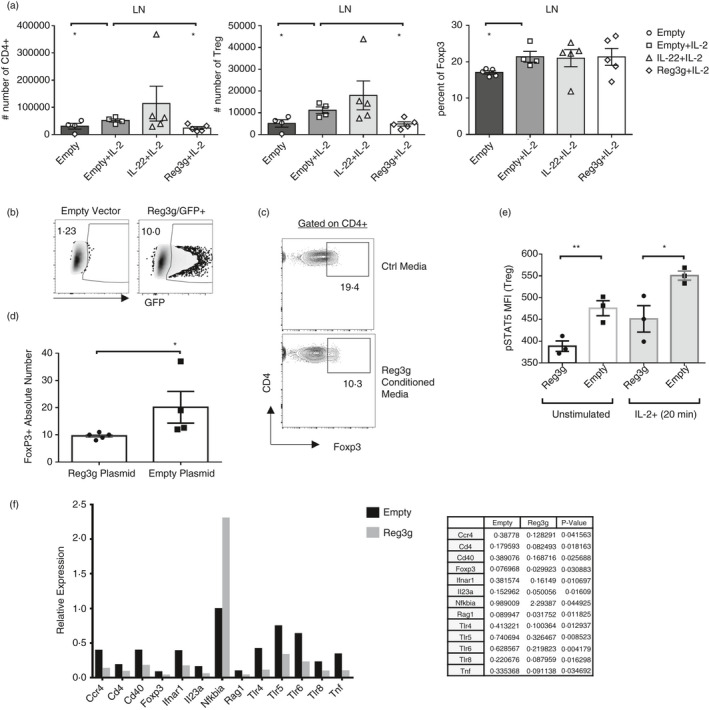
Reg3γ blocks Treg expansion in vivo and ex vivo (a) Foxp3‐YFP or B6 mice were injected with 2 ml of saline containing 15 µg/ml plasmid (IL‐22, Reg3γ or empty plasmid as control) on day −2 or −1. Then, on starting day 0, the mouse groups received 100 µl of intraperitoneal injections of 5 ng IL‐2 per gram mouse (in PBS), daily for 7 days. The mice were killed on day 7, and the absolute number of CD4+ T cells and Treg cells and Treg cell percentage in LNs (inguinal and brachial) (a) were quantified. (b) HEK293 T cells were transfected with Reg3γ or empty plasmid or as control GFP vector. The supernatant (conditioned media) was collected on days 2 and 5. (c) Filtered antibiotic‐added conditioned media were added into CD3/CD28‐activated sorted splenocytes obtained from Foxp3‐YFP reporter mice and cultured for 3 days. A representative flow plot of Treg cell expansion. (d) Quantification of the absolute number of Treg cells in ‘C’. (e) Sorted CD4+ Foxp3‐YFP+ Treg cells were cultured for 2 h in Reg3γ or empty plasmid‐conditioned media from HEK293 T cell transfections. Then, cells were stimulated with IL‐2 and STAT5 phosphorylation was quantified by nuclear staining. MFI indicates mean fluorescent intensity. (f) Splenocytes from B6 mice were cultured for 24 h in Reg3γ or empty plasmid‐conditioned media from HEK293 T cell transfections with the addition of CD3/CD28 1 µg/ml each. Both the adherent and suspension cells were collected and used for real‐time qPCR array (Qiagen, PAMM 52Z‐Mouse). A bar graph (top) and list (bottom) of differentially regulated genes (p < 0·05 for all) are given. (*) indicates p‐value <0·05; and (**), p < 0·01. Experiments in (a–e) were repeated three times
Reg3γ and its putative receptor Extl3’s expression kinetics during EAE
Next, we explored the expression of Reg3γ and its putative receptor Extl3 in the small intestine and spinal cord tissues of mice (Figure 5a). Previous studies have shown the expression of members of Reg family proteins in neurons in various species [27]. The nomenclature has been somewhat confusing across different species in earlier reports [48]. Nevertheless, in rat motor neurons, following injury, Reg1, PAP1 (also known as Reg2/Reg3β), PAPII (Reg3α), PAPIII (Reg3γ), and Reg4 were shown to be increased [48]. In the kainic acid‐induced seizure model of rats, Reg3β and Reg3γ expressions in hippocampal and parahippocampal areas were detected [49]. Moreover, rat PAP‐III (Reg3γ) was shown to promote neurite growth [50]. Similarly, Reg1α was reported to stimulate neurite outgrowth by binding EXTL‐3 receptor in rats and in mice [51]. Despite the abovementioned data, Reg family members have not been assessed in EAE models until now. In the spinal cord of mice, Reg3γ expression was significantly downregulated following EAE induction on day 7 and after EAE symptoms developed at the peak of disease (Figure 5a). In contrast, its putative receptor Extl3 was upregulated significantly following EAE induction on day 7th and beyond at the peak of disease. Reg1α on the other hand was expressed significantly more at the peak of the disease compared with the naïve mice and the priming phase of EAE both in the spinal cord and in the small intestine. Reg3γ expression in the small intestine followed an opposite trend compared with the spinal cord, and the mRNA level of Reg3γ was elevated after immunization on day 7th and at the peak of the disease, while the receptor Extl3 showed a downward trend as the disease progressed. These data reveal that Reg3γ and Extl3 are differentially regulated during EAE in the spinal cord and the small intestinal tissue.
FIGURE 5.
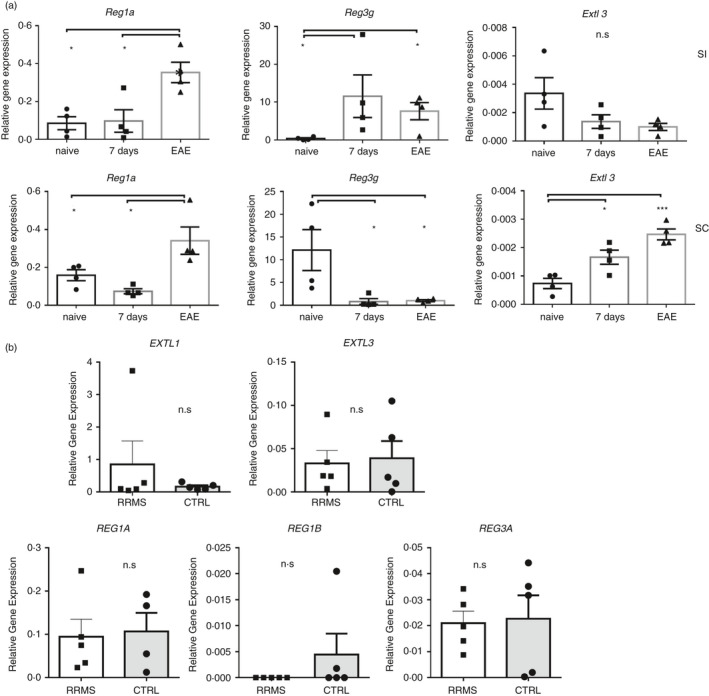
Reg3γ and its putative receptor Extl3’s expression kinetics in the spinal cord and intestinal tissues during experimental autoimmune encephalomyelitis (EAE) and in MS patient CSF cells (a) Spinal cord (SC) and small intestine (SI) tissues from naïve, 7‐ and 20‐day post‐immunization from EAE mice were collected, and real‐time qPCR was performed for Reg1α, Reg3γ and Extl3 (n = 3). (b) The cerebrospinal fluid (CSF) from MS patients or controls was spun, and the cell pellet was used for real‐time qPCR for EXTL1, EXTL3, REG1A, REG1B and REG3A (n = 5 for each group). (*) indicates p‐value <0·05
The human ortholog of murine Reg3γ is REG3A due to higher sequence similarity despite the presence of another ortholog named REG3G in humans [27]. REG1 and REG3 expressions were reported in plaques of Alzheimer's patients and CSF [52]. REG3A has also been reported to be overexpressed in central neurons of traumatic brain injury patients [49] but not in MS patients. Therefore, to test their expression in MS patients, CSF from 5 RRMS patients and 5 controls who had undergone lumbar puncture but returned negative for MS was spun, and cells were collected and tested for REG1A, REG1B, REG3A and EXTL3 (Figure 5b). REG1A and EXTL3 expressions were higher compared with REG3A and REG1B; however, there was no difference between MS patients and controls with respect to any of the tested genes (Figure 5b).
Reg3γ overexpression changes microbiota in EAE mice
Lastly, to gain insight into the changes in the intestinal microbiota instigated by Reg3γ overexpression, we compared ileal microbiota of Reg3γ plasmid‐injected mice with those of empty plasmid‐injected control mice. In the naïve (non‐EAE) mice, Reg3γ overexpression did not induce major changes within 6 days of hydrodynamic gene delivery compared with empty plasmid‐injected mice. Principal co‐ordinate analyses did not show well separation between Reg3γ and empty plasmid overexpression groups. The significant changes were detected only in the Mycoplasma and unclassified Clostridiales taxon (Figure S5A–D). On the other hand, in the EAE mice 3 weeks after MOG35‐55 immunizations and hydrodynamic gene delivery, principal co‐ordinate analysis ordinated based on the Unweighted UniFrac beta‐diversity metric revealed that Reg3γ overexpression brought dramatic changes to microbiota and that individual mouse in each group clustered together and was distanced away from the members of the other group (Figure 6a). Exacerbation of EAE following Reg3γ overexpression was associated with a total reduction in gut microbiota sequencing reads and microbiota biodiversity (Figure 6b,c). Importantly, the most significant shifts appeared as the disappearance of several taxa such as Actinobacteria and Deferribacteres phyla; Blautia, Chlamydia, Sutterella, Staphylococcus and Aerococcus genera; and Lachnospiraceae family, as well as a significant loss of Verrucomicrobia phylum and Planococcaceae family (Figure 6d). Interestingly, the Bacteroides genus and a specific class of Alphaproteobacteria, that is unclassified Rickettsiales, are significantly enriched in the Reg3γ‐overexpressed EAE group (Figure 6e). In the literature, Bacteroides fragilis has been described as an important commensal, which impacted EAE severity through its polysaccharide A by modulation of Treg cells [53]. Similarly, another Bacteroidetes member Prevotella histicola has also been shown to provide protection in EAE models through Treg induction [54]. On the other hand, Rickettsiales, which are highly enriched after Reg3γ overexpression, are known to be potential manipulators of eukaryotic cells [55], and some members of the taxa can act as the drivers of autoimmunity via structural mimicry [56]. It is also noteworthy that Lachnospiraceae family, which is depleted in Reg3γ‐overexpressed EAE mice, includes butyrate‐producing genera, which was shown to promote Treg cell generation in the intestines [57, 58]. Thus, exacerbation of EAE following Reg3γ overexpression appears to take place despite an enrichment in some of the potentially protective commensals Bacteroidetes but is associated with dysbiosis, Rickettsiales enrichment and depletion of butyrate‐producing Lachnospiraceae family. It should be noted that the microbiota analysis was conducted at higher clade levels, and further studies are required to determine the involvement of specific genera.
FIGURE 6.
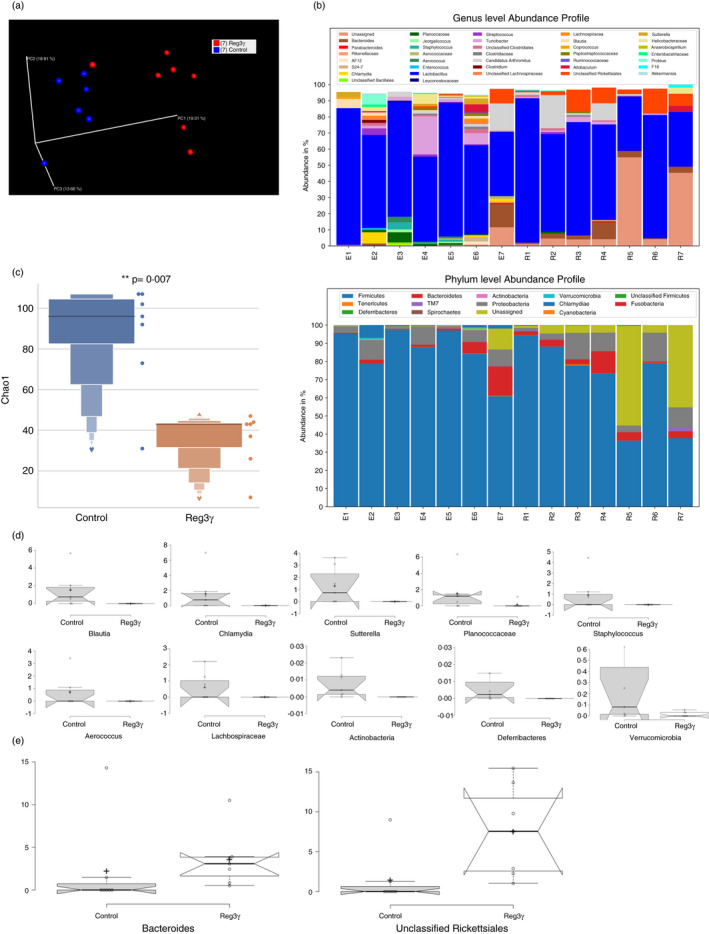
Reg3γ overexpression causes dysbiosis in the intestinal microbiota in experimental autoimmune encephalomyelitis (EAE) mice (a) Reg3γ plasmid or empty vector was injected on day −2, and EAE was induced by MOG35‐55 immunization on day 0. Mice were killed on day 20, and ileal faeces were collected for microbiota analyses. Principal co‐ordinate analysis ordinated based on the Unweighted UniFrac beta‐diversity metric plot (n = 7 per group). (b) Genus‐ and phylum‐level abundance profile of ileal microbiota in Reg3γ plasmid‐ or empty vector‐injected EAE mice at the peak of the disease (n = 7 per group). (c) Total microbiota abundance of ileal microbiota in Reg3γ plasmid‐ or empty vector (control)‐injected EAE mice at the peak of the disease (n = 7 per group), p‐value <0·05. (d) Significantly reduced genera of microbes in ileal microbiota of Reg3γ plasmid‐injected compared with empty vector‐injected EAE mice at the peak of the disease (n = 7 per group). All genera plots shown in ‘D’ are significantly altered (p‐value <0·05). (e) Significantly enriched (all plots shown in ‘E’ have p‐value <0·05) genera of microbes in ileal microbiota in Reg3γ plasmid‐injected compared with empty vector‐injected EAE mice at the peak of the disease (n = 7 per group), (*) indicates p‐value <0·05
DISCUSSION
In this study, we show that temporal overexpression of murine IL‐22 has a protective effect on the severity of MOG35‐55 peptide‐induced EAE, a murine model of MS. This was demonstrated by reduced EAE disease scores, reduced demyelination. IL‐22 overexpression also resulted in reduced lymphocyte infiltration to CNS. Thus, pathogenic IFN‐γ+IL‐17+ and GM‐CSF+ T cells were reduced in the CNS after IL‐22 expression. Conversely, IL‐22 neutralization slightly exacerbated the EAE disease scores. These results argue that IL‐22 may provide some protection from the symptoms of EAE when prophylactically given, at least in the B6 strain and MOG35‐55‐induced setting. To our knowledge, the therapeutic effects of injections of recombinant IL‐22 on EAE have not been reported in the literature. IL‐22 transgenic mice have been shown to develop psoriasis‐like phenotype [59], the EAE model has not yet been described in these transgenic mice either. In our study, we chose hydrodynamic gene delivery, and thus a systemic expression of IL‐22, owing to its low cost compared with recombinant protein injections. Our results support the findings of Laaksonen et al. and Lindahl et al. that IL‐22BP, by neutralizing IL‐22, promotes disease progression which they showed by knocking out IL‐22BP [33, 34]. Our results are also in line with Mattapallil et al.’s recent findings that local (intravitreal) or systemic administration of IL‐22 has protective effects on the experimental autoimmune immune uveitis model (EAU) [21]. Although IL‐22 KO mice were used in both reports, IL‐22 injections (as recombinant protein or gene delivery) have not been assessed or reported in the EAE model. Therefore, our results provide evidence for the protective effects of IL‐22 in EAE by temporal overexpression of IL‐22 gene.
Reduced lymphocyte infiltration, particularly Th1 and Th17 cells into the brain and spinal cord, suggests that IL‐22 blocks pathogenic T‐cell generation in the periphery or their migration into the CNS or their expansion. Examination of draining LNs or spleen, seven days following MOG35‐55 immunization, did not reveal significant differences between IL‐22 recipient or control mice with respect to Th1 and Th17 cells. Changes in chemokine receptors’ expression for such T cells in the IL‐22 overexpression group are likely and have not been explored in this study. The effects of IL‐22 on immune cells are expected to be indirect as IL‐22R subunits are not expressed by T cells.
To explore the mechanisms of IL‐22‐mediated protection from EAE, we assessed the potential involvement of a Reg protein family member Reg3γ. A study by Xia et al. showed beneficial role for Reg3γ, when overexpressed, in type 1 diabetes of non‐obese diabetic mice [60]. Although a recent report by Zhou et al. has recently studied REGγ/ PA28γ/ Psme3 in the context of EAE, the gene of interest in that study was not the antimicrobial Reg3ɣ, but encoded a proteasomal protein [61]. Therefore, to our knowledge Reg3γ has not been investigated in the context EAE. In our experiments, surprisingly, Reg3γ overexpression exacerbated EAE scores and demyelination and was associated with elevated infiltration of lymphocytes and Th1/Th17 cells into the brain and spinal cord suggesting that Reg3γ directly or indirectly may impact lymphocyte functions. Anti‐apoptotic, pro‐proliferative and anti‐inflammatory roles have been described for several Reg family members; however, never in the context of lymphocytes have such effects of Reg proteins been described in detail before [27]. Treg cells are crucial in restraining inflammation; thus, we first tested the impact of Reg3γ on murine Treg cells. Our ex vivo and in vivo results indeed point to an inhibitory role of Reg3γ on IL‐2‐mediated STAT5 phosphorylation and Foxp3 transcription. Negative regulation of both might explain the reduced Treg expansion in vivo in the Reg3γ‐overexpressed mice and ex vivo in the Reg3γ exposure cultures. Vaishnava et al. previously reported elevated IFN‐γ+ CD4+ T‐cell frequency in the Reg3γ‐/‐ mouse lamina propria [62]. Although the authors showed that the frequency of Th17 and Treg cells was unchanged, the results were given only as per cent of cells in this paper. The absolute number of IFN‐γ‐ or IL‐17A‐positive cells has not been quantified, nor have the other organs been explored. More importantly, direct effects of Reg3γ in ex vivo cultures on T cells have not been explored. On the other hand, Reg3γ knockout hosts were shown to be more susceptible to graft‐versus‐host disease (GVHD) due in part to Reg3γ‐mediated protection of intestinal stem cells from apoptosis [63]. A more recent study revealed that local or systemic human REG3A delivery in mice provided some protection to mice from dextran sulphate sodium or 2,4,6‐trinitrobenzenesulphonic acid (TNBS)‐induced colitis [64]. This study does not investigate REG3A’s impact on Th17 or Treg, and focuses on the microbiome; at the same time, it reports reduced IL‐1β levels after REG3A treatment in the intestine. We believe that in both the colitis models and the GVHD model, the anti‐apoptotic role of REG3A and murine Reg3γ may become more important as compared to the potential negative impact of REG3A or Reg3γ on Treg cells. Importantly, the negative impact of human REG3A, ortholog of murine Reg3γ, is yet to be confirmed. We believe that Reg3γ‐mediated Treg regulation is plausible and may serve as a negative feedback mechanism in the presence of infection to temporally block suppressive mechanisms. It is conceivable that when Reg3γ is expressed in abundance, possibly due to an infection, it restrains regulatory mechanisms such as Treg cells to allow the immune system to function until the pathogen is cleared. During the revision of our manuscript, You et al. reported that REG3A inhibition by miRNA (MiR‐10a‐3p) in human T cells is able to shift the balance between Th17/Treg in favour of the latter [65]. The role for murine Reg3γ is unclear from this work; nevertheless, this study underlines that Reg family members may have direct impact on CD4+ T‐cell lineage choices.
Our data also provide information regarding select Reg family member's expression during EAE in a murine model. Reg3γ level in the spinal cord tissue appears to go down as the disease progresses and its putative receptor Extl3 follows an opposite trend. In human MS patient's CSF, cells after centrifugation have also been tested for expression of REG3A and EXTL3. The mRNA message levels of these two molecules did not differ between cells of MS and control non‐MS group CSF. Although the sample size for the human study was small, both human and murine data are the first in the literature to assess Reg3γ, Extl3 and REG3A, EXTL3 expression in the EAE and MS context, respectively.
Finally, the data presented here also provide novel insight into the murine Reg3γ‐induced changes in the intestinal microbiota of MOG35‐55‐induced EAE mice, a murine model of MS, exclusively at the peak of the disease. Reg3γ binds to peptidoglycan, chitin and mannan but not to dextran, which thus has bactericidal effects on Gram‐positive bacteria [27, 66]. Human REG3A has similar properties. Darnaud et al. have recently overexpressed human REG3A in mice and investigated its effects on chemically induced colitis models and microbiota at the basal and inflammatory states [64]. Human REG3A overexpression resulted in the enrichment of Clostridiales (Ruminococcaceae, Lachnospiraceae) and depletion of Sutterellaceae (phylum Proteobacteria), Prevotellaceae, Porphyromonadaceae and Bacteroidaceae (phylum Bacteroidetes) and Bacteroidetes (Prevotellaceae) in the basal state. In the colitic mice, compared with the basal state, WT mice displayed enrichment of Prevotellaceae and unclassified Bacteroidales, and depletion of Porphyromonadaceae, Sutterellaceae and unclassified Desulfovibrionales, whereas REG3A‐TG mice displayed an increase in Prevotellaceae, unclassified Bacteroidales, Bacteroidaceae and Verrucomicrobiaceae and a decrease in Lactobacillaceae, unclassified Clostridia and Lachnospiraceae [64]. In our model, the overexpression of murine Reg3γ by hydrodynamic gene delivery resulted in the disappearance of several taxa such as Actinobacteria and Deferribacteres phyla; Blautia, Chlamydia, Sutterella, Staphylococcus and Aerococcus genera; and Lachnospiraceae family, as well as a significant loss of Verrucomicrobia phylum and Planococcaceae family at the peak of EAE. Interestingly, the Bacteroides genus and a specific class of Alphaproteobacteria, that is unclassified Rickettsiales, are significantly enriched in the Reg3γ‐overexpressed EAE group. Enrichment of some Bacteroides members and depletion of Sutterellaceae and Lachnospiraceae appear to be a recurring theme in Reg3γ and REG3A‐overexpressed mice, particularly in the inflammatory state despite the difference in EAE and colitis models.
In conclusion, the data presented here collectively reveal that IL‐22 overexpression provides mice partial protection from EAE, and points to therapeutic potential in human MS disease. This protection appears to be independent of Reg3γ. Our results also reveal that Reg3γ may directly and negatively regulate the function of Treg cells via STAT5‐mediated pathway; thus, its overexpression resulted in reduced Treg cell numbers and perhaps a more severe EAE course.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
AE conceptualized the entire study. MM, HC, HAD, MO and MFY helped in the design. AE, SE, YH, MC, FZO, ZBA, TNG, KA, AB, MOK, MH, OUN and AG performed the experiments. AE wrote the manuscript. MFY, MA and MM provided the human patient and control samples. All authors read the manuscript and contributed to the data interpretation, revision and discussions intellectually.
Supporting information
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
ACKNOWLEDGEMENT
We thank Prof. Vehbi Gunes, all DEKAM Mouse facility members and Omer Kilic. This work was supported partly by the Erciyes University BAP grant, TDK‐2019‐9220; and The Scientific and Technological Research Council of Turkey (TUBITAK) grants 315S315 and 215S725 to AE.
Funding information
This work was supported partly by the Erciyes University BAP grant, TDK‐2019‐9220; and grants 315S315 and 215S725 to AE.
REFERENCES
- 1.Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL‐22. Nat Immunol. 2011;12:383–90. [DOI] [PubMed] [Google Scholar]
- 2.Zenewicz LA. IL‐22: There is a gap in our knowledge. ImmunoHorizons. 2018;2:198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satoh‐Takayama N, Vosshenrich CAJ, Lesjean‐Pottier S, Sawa S, Lochner M, Rattis F, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–70. [DOI] [PubMed] [Google Scholar]
- 4.Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA. Innate and adaptive interleukin‐22 protects mice from inflammatory bowel disease. Immunity. 2008;29:947–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, et al. Interleukin‐22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–9. [DOI] [PubMed] [Google Scholar]
- 6.Spits H, Di Santo JP. The expanding family of innate lymphoid cells: Regulators and effectors of immunity and tissue remodeling. Nat. Immunol. 2011;12:21–7. [DOI] [PubMed] [Google Scholar]
- 7.Zindl CL, Lai J‐F, Lee YK, Maynard CL, Harbour SN, Ouyang W, et al. IL‐22‐producing neutrophils contribute to antimicrobial defense and restitution of colonic epithelial integrity during colitis. Proc Natl Acad Sci U S A. 2013;110:12768–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumoutier L, Louahed J, Renauld J‐C. Cloning and characterization of IL‐10‐related T Cell‐derived inducible factor (IL‐TIF), a novel cytokine structurally related to IL‐10 and inducible by IL‐9. J Immunol. 2000;164:1814–9. [DOI] [PubMed] [Google Scholar]
- 9.Mashiko S, Bouguermouh S, Rubio M, Baba N, Bissonnette R, Sarfati M. Human mast cells are major IL‐22 producers in patients with psoriasis and atopic dermatitis. J Allergy Clin Immunol. 2015;136(351–359):e1. [DOI] [PubMed] [Google Scholar]
- 10.Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, Mills KHG. Interleukin‐1 and IL‐23 Induce Innate IL‐17 Production from γδ T Cells, Amplifying Th17 Responses and Autoimmunity. Immunity. 2009;31:331–41. [DOI] [PubMed] [Google Scholar]
- 11.Goto M, Murakawa M, Kadoshima‐Yamaoka K, Tanaka Y, Nagahira K, Fukuda Y, et al. Murine NKT cells produce Th17 cytokine interleukin‐22. Cell Immunol. 2009;254:81–4. [DOI] [PubMed] [Google Scholar]
- 12.Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi‐Joannopoulos K, Collins M, et al. Interleukin (IL)‐22 and IL‐17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eyerich S, Eyerich K, Pennino D, Carbone T, Nasorri F, Pallotta S, et al. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J Clin Invest. 2009;119:3573–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veldhoen M, Hirota K, Christensen J, O’Garra A, Stockinger B. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med. 2009;206:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–40. [DOI] [PubMed] [Google Scholar]
- 16.Crellin NK, Trifari S, Kaplan CD, Satoh‐Takayama N, Di Santo JP, Spits H. Regulation of cytokine secretion in human CD127+ LTi‐like innate lymphoid cells by toll‐like receptor 2. Immunity. 2010;33:752–64. [DOI] [PubMed] [Google Scholar]
- 17.Qiu J, Heller JJ, Guo X, Chen ZME, Fish K, Fu YX, et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glatzer T, Killig M, Meisig J, Ommert I, Luetke‐Eversloh M, Babic M, et al. RORγt+ innate lymphoid cells acquire a proinflammatory program upon engagement of the activating receptor NKp44. Immunity. 2013;38:1223–35. [DOI] [PubMed] [Google Scholar]
- 19.Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R. IL‐22 increases the innate immunity of tissues. Immunity. 2004;21:241–54. [DOI] [PubMed] [Google Scholar]
- 20.Tachiiri A, Imamura R, Wang Y, Fukui M, Umemura M, Suda T. Genomic structure and inducible expression of the IL‐22 receptor α chain in mice. Genes Immun. 2003;4:153–9. [DOI] [PubMed] [Google Scholar]
- 21.Mattapallil MJ, Kielczewski JL, Zárate‐Bladés CR, St Leger AJ, Raychaudhuri K, Silver PB, et al. Interleukin 22 ameliorates neuropathology and protects from central nervous system autoimmunity. J Autoimmun. 2019;102:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie MH, Aggarwal S, Ho WH, Foster J, Zhang Z, Stinson J, et al. Interleukin (IL)‐22, a novel human cytokine that signals through the interferon receptor‐related proteins CRF2‐4 and IL‐22R. J Biol Chem. 2000;275:31335–9. [DOI] [PubMed] [Google Scholar]
- 23.Lejeune D, Dumoutier L, Constantinescu S, Kruijer W, Schuringa JJ, Renauld JC. Interleukin‐22 (IL‐22) activates the JAK/STAT, ERK, JNK, and p38 MAP kinase pathways in a rat hepatoma cell line: Pathways that are shared with and distinct from IL‐10. J Biol Chem. 2002;277:33676–82. [DOI] [PubMed] [Google Scholar]
- 24.Dumoutier L, Lejeune D, Colau D, Renauld J‐C. Cloning and characterization of IL‐22 binding protein, a natural antagonist of IL‐10‐related T cell‐derived inducible factor/IL‐22. J Immunol. 2001;166:7090–5. [DOI] [PubMed] [Google Scholar]
- 25.Kotenko SV, Izotova LS, Mirochnitchenko OV, Esterova E, Dickensheets H, Donnelly RP, et al. Identification, cloning, and characterization of a novel soluble receptor that binds IL‐22 and neutralizes its activity. J Immunol. 2001;166:7096–103. [DOI] [PubMed] [Google Scholar]
- 26.Xu W, Presnell SR, Parrish‐Novak J, Kindsvogel W, Jaspers S, Chen Z, et al. A soluble class II cytokine receptor, IL‐22RA2, is a naturally occurring IL‐22 antagonist. Proc Natl Acad Sci U S A. 2001;98:9511–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z, Downing S, Tzanakakis ES. Four decades after the discovery of regenerating Islet‐derived (Reg) proteins: current understanding and challenges. Front. Cell Dev. Biol. 2019;7:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perriard G, Mathias A, Enz L, Canales M, Schluep M, Gentner M, et al. Interleukin‐22 is increased in multiple sclerosis patients and targets astrocytes. J Neuroinflammation. 2015;12:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawcer S, Hellenthal G, Pirinen M, Spencer CCA, Patsopoulos NA, Moutsianas L, et al. Genetic risk and a primary role for cell‐mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, et al. Analysis of immune‐related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beyeen AD, Adzemovic MZ, Öckinger J, Stridh P, Becanovic K, Laaksonen H, et al. IL‐22RA2 associates with multiple sclerosis and macrophage effector mechanisms in experimental neuroinflammation. J Immunol. 2010;185:6883–90. [DOI] [PubMed] [Google Scholar]
- 32.Kreymborg K, Etzensperger R, Dumoutier L, Haak S, Rebollo A, Buch T, et al. IL‐22 is expressed by Th17 cells in an IL‐23‐dependent fashion, but not required for the development of autoimmune encephalomyelitis. J Immunol. 2007;179:8098–104. [DOI] [PubMed] [Google Scholar]
- 33.Laaksonen H, Guerreiro‐Cacais AO, Adzemovic MZ, Parsa R, Zeitelhofer M, Jagodic M, et al. The multiple sclerosis risk gene IL22RA2 contributes to a more severe murine autoimmune neuroinflammation. Genes Immun. 2014;15:457–65. [DOI] [PubMed] [Google Scholar]
- 34.Lindahl H, Guerreiro‐Cacais AO, Bedri SK, Linnerbauer M, Lindén M, Abdelmagid N, et al. IL‐22 binding protein promotes the disease process in multiple sclerosis. J Immunol. 2019;203:888–98. [DOI] [PubMed] [Google Scholar]
- 35.Eken A, Singh AK, Treuting PM, Oukka M. IL‐23R+ innate lymphoid cells induce colitis via interleukin‐22‐dependent mechanism. Mucosal Immunol. 2014;7:143–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miao CH, Ye X, Thompson AR. High‐level factor VIII gene expression in vivo achieved by nonviral liver‐specific gene therapy vectors. Hum Gene Ther. 2003;14:1297–305. [DOI] [PubMed] [Google Scholar]
- 37.Eken A, Duhen R, Singh AK, Fry M, Buckner JH, Kita M, et al. S1P1 deletion differentially affects TH17 and Regulatory T cells. Sci Rep. 2017;7:12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandevelde M, Higgins RJ, Kristensen B, Kristensen F, Steck AJ, Kihm U. Demyelination in experimental canine distemper virus infection: Immunological, pathologic, and immunohistological studies. Acta Neuropathol. 1982;56:285–93. [DOI] [PubMed] [Google Scholar]
- 39.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next‐generation sequencing‐based diversity studies. Nucleic Acids Res. 2013;41: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high‐throughput community sequencing data. Nat. Methods. 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera‐checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh AK, Eken A, Fry M, Bettelli E, Oukka M. DOCK8 regulates protective immunity by controlling the function and survival of RORγt+ ILCs. Nat Commun. 2014;5:4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shin JH, Seeley RJ. REG3 proteins as gut hormones? Endocrinology. 2019;160:1506–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gironella M, Iovanna JL, Sans M, Gil F, Peñalva M, Closa D, et al. Anti‐inflammatory effects of pancreatitis associated protein in inflammatory bowel disease. Gut. 2005;54:1244–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Zhou H, Han Y, Liu X, Wang M, Wang X, et al. SOCS3 methylation in synergy with Reg3A overexpression promotes cell growth in pancreatic cancer. J Mol Med. 2014;92:1257–69. [DOI] [PubMed] [Google Scholar]
- 48.Namikawa K, Fukushima M, Murakami K, Suzuki A, Takasawa S, Okamoto H, et al. Expression of Reg/PAP family members during motor nerve regeneration in rat. Biochem Biophys Res Commun. 2005;332:126–34. [DOI] [PubMed] [Google Scholar]
- 49.März‐Weiss P, Kunz D, Bimmler D, Berkemeier C, Özbek S, Dimitriades‐Schmutz B, et al. Expression of pancreatitis‐associated protein after traumatic brain injury: A mechanism potentially contributing to neuroprotection in human brain. Cell Mol Neurobiol. 2011;31:1141–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Konishi H, Matsumoto S, Namikawa K, Kiyama H. N‐terminal cleaved pancreatitis‐associated protein‐III (PAP‐III) serves as a scaffold for neurites and promotes neurite outgrowth. J Biol Chem. 2013;288:10205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Ba IAT, Marchal S, François F, Silhol M, Lleres C, Michel B, et al. Regenerating islet‐derived 1α (Reg‐1α) protein is new neuronal secreted factor that stimulates neurite outgrowth via exostosin tumor‐like 3 (EXTL3) receptor. J Biol Chem. 2012;287:4726–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duplan L, Michel B, Boucraut J, Barthellémy S, Desplat‐Jego S, Marin V, et al. Lithostathine and pancreatitis‐associated protein are involved in the very early stages of Alzheimer’s disease. Neurobiol Aging. 2001;22:79–88. [DOI] [PubMed] [Google Scholar]
- 53.Ochoa‐Repáraz J, Mielcarz DW, Wang Y, Begum‐Haque S, Dasgupta S, Kasper DL, et al. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol. 2010;3:487–95. [DOI] [PubMed] [Google Scholar]
- 54.Shahi SK, Freedman SN, Murra AC, Zarei K, Sompallae R, Gibson‐Corley KN, et al. Prevotella histicola, A human gut commensal, Is as potent as COPAXONE® in an animal model of multiple sclerosis. Front Immunol. 2019;10:462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Renvoisé A, Merhej V, Georgiades K, Raoult D. Intracellular rickettsiales: insights into manipulators of eukaryotic cells. Trends Mol. Med. 2011;17:573–83. [DOI] [PubMed] [Google Scholar]
- 56.Paiardini A, Pascarella S. Structural mimicry between SLA/LP and Rickettsia surface antigens as a driver of autoimmune hepatitis: Insights from an in silico study. Theor Biol Med Model. 2013;10:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meehan CJ, Beiko RG. A phylogenomic view of ecological specialization in the lachnospiraceae, a family of digestive tract‐associated bacteria. Genome Biol Evol. 2014;6:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe‐derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–50. [DOI] [PubMed] [Google Scholar]
- 59.Park O, Wang H, Weng H, Feigenbaum L, Li H, Yin S, et al. In vivo consequences of liver‐specific interleukin‐22 expression in mice: Implications for human liver disease progression. Hepatology. 2011;54:252–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xia F, Cao H, Du J, Liu X, Liu Y, Xiang M. Reg3g overexpression promotes β cell regeneration and induces immune tolerance in nonobese‐diabetic mouse model. J Leukoc Biol. 2016;99:1131–40. [DOI] [PubMed] [Google Scholar]
- 61.Zhou L, Yao L, Zhang Q, Xie W, Wang X, Zhang H, et al. REGγ controls Th17 cell differentiation and autoimmune inflammation by regulating dendritic cells. Cell Mol Immunol. 2020;17:1136–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science (80‐) 2011; 334: 255–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao D, Kim YH, Jeong S, Greenson JK, Chaudhry MS, Hoepting M, et al. Survival signal REG3α prevents crypt apoptosis to control acute gastrointestinal graft‐versus‐host disease. J Clin Invest. 2018;128:4970–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Darnaud M, Dos Santos A, Gonzalez P, Augui S, Lacoste C, Desterke C, et al. Enteric delivery of regenerating family member 3 alpha alters the intestinal microbiota and controls inflammation in mice with colitis. Gastroenterology. 2018;154:1009–1023.e14. [DOI] [PubMed] [Google Scholar]
- 65.You G, Cao H, Yan L, He P, Wang Y, Liu B, et al. MicroRNA‐10a‐3p mediates Th17/Treg cell balance and improves renal injury by inhibiting REG3A in lupus nephritis. Int Immunopharmacol. 2020;88:106891. [DOI] [PubMed] [Google Scholar]
- 66.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5


