Abstract
Aims:
Acute myeloid leukemia (AML) is an aggressive hematologic neoplasm, in which relapse due to drug resistance is the main cause for treatment failure and the disease progression. In this study, we aimed to investigate the molecular mechanism of KDM4C-dependent MALAT1/miR-328-3p/CCND2 axis in cytarabine (Ara-C) resistance in the context of AML.
Methods:
Bioinformatics analysis was performed to predict the targeting relationships among KDM4C, MALAT1, miR-328-3p, and CCND2 in AML, which were validated with chromatin immunoprecipitation and dual-luciferase reporter assay. Methylation-specific polymerase chain reaction was conducted to detect the methylation of MALAT1 promoter. After conducting gain- and loss-of-function assays, we investigated the effect of KDM4C on cell Ara-C resistance. A NOD/SCID mouse model was established to further investigate the roles of KDM4C/MALAT1/miR-328-3p/CCND2 in Ara-C resistant AML cells.
Results:
KDM4C expression was upregulated in AML. KDM4C upregulation promoted the demethylation in the promoter region of MALAT1 to increase its expression, MALAT1 targeted and inhibited miR-328-3p expression, enhancing the Ara-C resistance of HL-60/A. miR-328-3p targeted and suppressed the expression of CCND2 in HL-60/A to inhibit the Ara-C resistance. Mechanistically, KDM4C regulated miR-328-3p/CCND2 through MALAT1, resulting in Ara-C resistance in AML. Findings in an in vivo xenograft NOD/SCID mouse model further confirmed the contribution of KDM4C/MALAT1/miR-328-3p/CCND2 in the Ara-C resistant AML.
Conclusion:
Our study demonstrated that KDM4C may up-regulate MALAT1 expression, which decreases the expression of miR-328-3p. The downregulation of miR-328-3p increased the level of CCND2, which induced the Ara-C resistance in AML.
Keywords: acute myeloid leukemia, CCND2, cytarabine, KDM4C, MALAT1, miR-328-3p
Introduction
Acute myeloid leukemia (AML) is a very common heterogeneous blood disease featured by abnormal accumulation of immature myeloid cells in bone marrow, which can be classified into different subtypes in both children and adults.1,2 The annual incidence of AML is approximate three to five cases per 100,000 people in the USA.3 Treatment of AML includes allogeneic hematopoietic stem cell transplantation, chemotherapy, and targeted therapy.4–6 Cytarabine (Ara-C) has been among the standard induction chemotherapies in the treatment of AML for the past four decades.7 Although treatment with Ara-C alone or in combination with other agents can achieve 50–75% complete remission in AML patients, an important proportion of patients show relapse due to Ara-C resistance.8 In recent decades, targeted therapy has emerged as a promising strategy for the clinical treatment of AML to deal with the chemotherapy resistance.9
Epigenetics refers to inheritance of a phenotype without changes in the DNA sequence. For example, reversible epigenetic modifications of DNA or histones to open or close chromosomal territories can promote or suppress targeted gene expression, respectively.10 Histone demethylase KDM4C, which belongs to the Jumonji domain-containing protein 2C family, functions as a histone H3K9 demethylase.11 KDM4C has been documented as an oncogenic player in both solid tumors and AML,12–15 indicating KDM4C to be a potential candidate for the targeted therapy. However, the role of KDM4C in mediating Ara-C resistance in AML remains elusive. Pan et al.16 reported that KDM4C targeted and upregulated the expression of metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) in colorectal cancer. MALAT1 is a long non-coding RNA (lncRNA) that is highly expressed in the nucleus and regulates gene expression both at transcriptional and post-transcriptional levels,17 and has been reported to play roles in many cancers with respect to cancer cell proliferation, invasion, and metastasis.18 In AML, MALAT1 was upregulated and increased cell proliferation, which was correlated with poor prognosis.19 Wen et al.19 have shown that MALAT1 was notably upregulated in chronic myeloid leukemia (CML) and worked as a competitive endogenous RNA for microRNA-328 (miR-328-3p).20 Eiring et al.21 reported that miR-328-3p restoration induced differentiation and impeded blasts’ survival in CML.
Thus, we hypothesized that the KDM4C/MALAT1 axis mediates Ara-C resistance in AML, and therefore undertook an investigation of the downstream targets of MALAT1 and miR-328-3p. In this paper, we aimed to study the role of KDM4C in AML and investigate its mechanisms it the context of Ara-C resistance. In particular, we attempted to decipher the regulatory aspects of KDM4C and MALAT1, and also elucidate how the KDM4C/MALAT1/miR-328-3p/CCND2 axis affects Ara-C resistance in AML. Our work not only uncovers the molecular mechanism for resistance to Ara-C but also provides potential new therapeutic targets for the treatment of chemotherapy-resistant AML.
Material and methods
Bioinformatics analysis
A total of 173 AML samples from The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) and 70 normal bone marrow samples from the GTEx database (https://gtexportal.org) were downloaded. The downloaded data was integrated into a gene expression matrix, and the R software package DESeq2 was used for gene differential expression analysis. The miRNA-lncRNA targeting relationship database in the miRNA-Target function in the starBase database (http://starbase.sysu.edu.cn/) was used to predict the miRNA targeted and regulated by MALAT1, and the miRNA-mRNA in the miRNA-Target targeting relation database was also used to predict miRNAs that target the CCND2 gene. The non-coding RNA sequencing data (GSE128079) of 10 AML samples and nine control samples were obtained from the GEO database, and the R software package edgeR was used for miRNA differential expression analysis.
Clinical sample analysis
Thirty AML samples were collected and isolated from AML patients receiving several rounds of chemotherapy and 30 normal marrow samples were obtained from healthy people. Among the AML patients, there were 18 were males and 12 females, aged 14–77 years old, with a median age of 55 years. According to the French–American–British and World Health Organization classification systems, the AML samples were classified into three cases of M1, eight cases of M2, one case of M3, 10 cases of M4, five cases of M5, two cases of M6, and one case of M7 (Supplemental material Table 1 online).
Cell culture
AML HL-60 cell line and HL-60 Ara-C resistant cell line HL-60/A were purchased from the Institute of Hematology & Blood Diseases Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College (Tianjin, China). Cells were cultured with Roswell Park Memorial Institute1640 medium (Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin in a humidified incubator with 5% CO2 at 37°C.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
MTT assay was performed to test the effect of Ara-C on the cell viability. The concentration gradient was set to calculate the IC50. HL-60 and HL-60/A cells were seeded into a 96-well plate with a density of 1 × 104 cells/well and cultured in a 37°C with 5% CO2 incubator overnight until cells were adherent to the walls. Next, adherent cells were treated with Ara-C with concentrations of 0, 0.005, 0.01, 0.02, 0.05, 0.1, 0.2, 0.5, 1.0, and 2.0 μg/mL, with five duplicated wells of each concentration. After 48 h incubation, 10 μL of MTT was added to each well for another 3 h-incubation. Optical density (OD) was measured at 570 nm on a plate reader (Thermo, Waltham, MA, USA). Cell viability was analyzed with each Ara-C concentration using 0 μg/mL as control to present 100% cell viability. Experiments were repeated three times independently.
Cell infection
pLV-EGFP-N (overexpression vector; oe-vector) and pSIH1-H1-copGFP (shRNA knockdown vector; sh-vector) were purchased from GenePharma (Shanghai, China) and used for MALAT1 overexpression and KDM4C knockdown, respectively. Empty control vectors, oe-NC and sh-NC were also obtained from GenePharma (Shanghai, China). Lentiviral vectors were grouped as follows: pSIH1-H1-copGFP-sh-NC (sh-NC; shRNA negative control), pLV-EGFP-N (oe-NC; overexpression negative control), pLV-EGFP-CCND2 (oe-CCND2; overexpression of CCND2), and pSIH1-H1-copGFP-sh-KDM4C (sh-KDM4C; knockdown of KDM4C). The lentiviral transfer vector, packaging vector psPAX2, and envelop vector Pmd2g were mixed in a ratio of 3:3:1 (9 μg:9 μg:3 μg) in a sterile Eppendorf (EP) tube to prepare the packaging system, followed by addition of 50 μL of 2.5 M CaCl2. Then, double-distilled water was added to the system to a volume of 500 μL and added with 500 μL of 2 × HEPES. The sample was gently mixed and stood for 1 min at room temperature. Next, the lentiviral packaging system was added to a 10 cm diameter cell culture dish containing HEK293T cells for incubation for 48 h. Cell morphology and the percentage of cells expressing green fluorescence were observed under a fluorescence microscope. When the cells were in good condition and the percentage of cells expressing green fluorescence was above 50%, the cell supernatant was collected. Then, the supernatant was centrifuged in a low-temperature centrifuge at 3000 rev/min for 5 min to remove dead cells. Finally, the supernatant was filtered with a 0.45 μm sterile filter membrane and stored at −80°C.
Furthermore, plasmids of miR-328-3p mimic, miR-328-3p inhibitor, and relevant NC (NC mimic and NC inhibitor) were provided by RiboBio (Guangzhou, China). Cells were transfected with miR-328-3p mimic, miR-328-3p inhibitor, short hairpin RNA targeting MALAT1 (sh-MALAT1), CCND2 overexpression vector (oe-CCND2) or relevant NC, alone or in combination, following the instructions of Lipofectamine-2000 (Invitrogen, Carlsbad, CA, USA).
Real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using Trizol (15596026, Invitrogen, Carlsbad, CA, USA). Complementary DNA was synthesized according to the protocol of PrimeScript RT reagent Kit (RR047A, Takara, Tokyo, Japan). qPCR was performed using a Fast SYBR Green PCR kit (Applied Biosystems, Foster City, CA, USA) and ABI PRISM 7300 RT-PCR machine (Applied Biosystems, Foster City, CA, USA) with three triplicated wells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as internal control. The 2−ΔΔCt method was applied to quantify the relative expression level of KDM4C, MALAT1, and CCND2. ΔΔCt = (Ctexperimental target − Ctinternal control) − (Ctcontrol target − Ctinternal control). RT-qPCR primers were designed using PubMed database and Primer Premier 5.0 and synthesized by BGI Genomics (Shenzhen, China) (Supplemental Table 2).
Western blot
Cells were washed with PBS and lysed with cell lysis buffer (C0481, Sigma, St. Louis, MO, USA) for 30 min at 4°C, followed by centrifugation in an EP tube at 12,000 g for 15 min at 4°C. The bicinchoninic acid method was used to quantify the protein concentration (Beyotime Biotechnology, Shanghai, China). Proteins were heated with loading buffer at 95°C for 5 min. Then, 20 μg portions of protein were separated via sodium dodecyl sulfate and polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA) with 0.3 A and 20 V. The PVDF membrane was blocked with 5% skimmed milk for 1 h at room temperature, followed by incubation with anti-KDM4C rabbit antibody (ab85454, 1: 200, Abcam, Cambridge, UK), anti-CCND2 rabbit antibody (ab207604, 1: 1000, Abcam), and anti-GAPDH rabbit antibody (D16H11, 1: 100, Cell Signaling Technology, Beverly, MA, USA) at 4°C overnight. The next day, the PVDF membrane was washed three times with TBST, followed by incubation with horseradish peroxidase (HRP)-conjugated mouse anti-rabbit Immunoglobulin G (IgG; ab9482, 1: 5000, Abcam) for 1 h at room temperature. The PVDF membrane was washed six times with TBST and developed using enhanced chemiluminescence reagent (Baomanbio, Shanghai, China). GAPDH served as loading control, and ImageJ was used to quantify the relative expression level of proteins as the ratio of target protein value/loading control value. Experiments were repeated three times.
Methylation-specific PCR (MS-PCR)
Genomic DNA was isolated from cells by a TIANamp Genomic DNA Kit (Tiangen, Biotech, Beijing, China). DNA purity and concentration were measured according to values of (optical density value) OD260 and OD280. DNA bisulfite modification was conducted with the EZ DNA Methylation Kit (Tiangen Biotech, Beijing, China). Here, bisulfite converted the unmethylated cytosine to thymine, while methylated cytosine remained intact. Two sets of MS primers (Table 1) were used to amplify each region of interest: one pair recognized a sequence in which CpG sites are unmethylated (bisulfite modified to CpG), and the other recognized a sequence in which CpG sites are methylated (unmodified by bisulfite treatment). The methylation status of MALAT1 promoter region was analyzed according to the MS-PCR protocol. One pair of primers was used to recognize the sequence where CpG sites were not methylated (modification by bisulfite), and the other pair recognized the sequence where CpG sites are methylated (no modification by bisulfite). The MS-PCR system was run in a volume of 25 μL, including 12.5 μL Hot-StarTaq Master Mix (Qiagen, Hilden, Germany), 1 μL bisulfite treated NDA template, 1.5 μL forward and reverse primers, respectively according to the following conditions: 95°C 12 min, 94°C 30 s, 62°C 30 s, 72°C 45 s, 40 cycles, and 72°C 10 min. The PCR product was analyzed via 1.5% agarose gel electrophoresis at 120 V, stained with ethidium bromide, and visualized under an ultraviolet illumination system. Experiments were repeated three times.
Table 1.
MALAT1 promoter primers for methylation-specific polymerase chain reaction.
| Genes | Sequences (5'–3') |
|---|---|
| MALAT1 M: | F:TTTTTGAGTAGTTGGGATTATAGGC |
| R:ACTTTAAAAAACTAAAACGAACGAA | |
| MALAT1 U: | F:TTTGAGTAGTTGGGATTATAGGTGT |
| R:ACTTTAAAAAACTAAAACAAACAAA |
M, methylated; U, unmethylated.
Chromatin immunoprecipitation (ChIP)
ChIP was performed using an EZ-Magna ChIP TMA kit (Millipore, Billerica, MA, USA). HL-60 cells in log phase were crosslinked with 1% formaldehyde for 10 min followed by de-crosslinking with 125 mM glycine for 5 min. Cells were washed two times with pre-chilled PBS and centrifuged at 2000 rev/min for 5 min. The cell pellet was resuspended in ChIP lysis buffer (150 mM NaCl, 50 mM Tris (pH 7.5), 5 mM EDTA, 0.005% NP40, 0.01% Triton X-100) supplemented with protease inhibitor complex (PIC) to a density of 2 × 106 cells/200 mL, followed by centrifugation at 5000 rev/min for 5 min. The pellet was resuspended with nucleus buffer and lysed on ice for 10 min. Chromatin was sonicated to 200–1000 bp fragments, followed by centrifugation at 14,000 g for 10 min at 4°C. The supernatant (100 μL) was added with 900 μL of ChIP dilution buffer and 20 μL of 50 × PIC. Next, 60 μL of Protein A Agarose/Salmon Sperm DNA was added to each sample, followed by incubation at 4°C for 1 h and centrifugation at 700 rev/min for 1 min. The supernatant was collected, from which 20 μL was aliquoted as the input. The remainder was incubated with either rabbit anti-KDM4C antibody or 1 μL of rabbit IgG control overnight. The next day, samples were incubated with 60 μL of Protein A Agarose/Salmon Sperm DNA for 2 h at 4°C, followed by centrifugation at 700 rev/min for 10 min. The agarose-antibody complex pellet was washed with 1 mL of low salt, high salt, LiCl, and TE buffer (two times) in sequence. DNA was eluted with two portions of 250 μL of ChIP elution buffer. Next, 20 μL of NaCl was used for de-crosslinking, followed by DNA purification. The relative level of MALAT1 promoter region was quantified by qPCR. The primers of MALAT1 promoter region are listed in Supplemental Table 2.
Dual-luciferase reporter assay
The dual luciferase reporter assay was performed to verify the targeting relationship between MALAT1 and miR-328-3p and that of miR-328-3p and CCND2. HEK293T cells were seeded in a 24-well plate and cultured for 24 h. Wild type (Wt) and mutant (Mut) CCND2 promoter luciferase reporter plasmid pGL4.10-hRluc Wt and pGL4.10-hRluc Mut were constructed, which were co-transfected with miR-328-3p into HEK293T cells. After 48 h transfection, the medium was removed, and cells were washed with PBS two times. Then, cells were lysed with 1 × PLB lysis buffer and subjected to the Nano-Glo® Dual-Luciferase® Reporter detection kit (Promega, Madison, WI, USA), followed by the measurement of luciferase activity according to the manufacturer’s protocols. Experiments were repeated three times. The interaction between MALAT1 and miR-328-3p was studied with the same procedures, except for the construction of Wt-miR-328-3p and Mut-miR-328-3p luciferase reporter vectors.
Establishment of xenograft mice models
Non-obese diabetic/severe combined immunodeficient (NOD/SCID) mice were produced by the crossing of SCID mice and NOD/lt non-obese diabetic mice. NOD/SCID male mice (Beijing HFK Bioscience Co., Ltd, Beijing, China) aged 6–8 weeks were fed under specific pathogen free housing conditions. Five groups of NOD/SCID mice were injected intravenously with 0.2 mL of 1 × 107 cells, which were treated with sh-NC + oe-NC, sh-KDM4C + oe-NC, sh-NC + oe-MALAT1, sh-KDM4C + oe-MALAT1 or HL-60 as control, respectively (12 mice/group). Next, 2 μg/mL of Ara-C was prepared with saline solution and injected subcutaneously into the mice for 6 weeks. The survival of mice was recorded, and the number of leukocytes from peripheral blood obtained from the tail vein and retro-orbital vein was quantified and the ratio of leukocytes was calculated.22
Hematoxylin and eosin (H&E) staining
Samples were washed with distilled water (diH2O), rinsed thoroughly with PBS, 4% paraformaldehyde for 30–50 min, diH2O in orders, and subjected to dehydration, clearing, paraffin embedding, and sectioning. Sections were placed on slides and dried in a 45°C incubator, followed by dewaxing, rehydration with gradient ethanol from high to low concentration, and diH2O washing for 5 min. Next, sections were stained with hematoxylin for 5 min and washed with tap water for 3 s, 1% hydrochloric acid ethanol for 3 s, and 5% eosin for 3 min, followed by dehydration, clearing, and mounting. The morphology and cell infiltration in lymph were observed under a light microscope and recorded. ImageJ was used to record and analyze the data.
Immunohistochemistry (IHC)
Blood was collected from the mice, which were sacrificed by cervical dislocation. The blood samples were fixed in 4% paraformaldehyde, embedded in paraffin, sectioned to 4 μm, and dewaxed. Antigen retrieval was done by microwave heating. Then, sections were allowed to stand for 5 min, heated, washed with PBS, and blocked with normal goat serum. sh-NC + oe-NC was used as NC and HistostainTMSP-9000 kit (Zymed, CA, USA) was used for IHC. In brief, sections were incubated with anti-KDM4C or anti-CCND2 overnight at 4°C. The next day, sections were brought back to room temperature and washed with PBS, followed by incubation with HRP-conjugated anti-rabbit secondary antibody for 30 min at 37°C. Then, the sections were washed with PBS and developed with diaminobenzidine for 5–10 min under a microscope to adjust the development time. Sections were counterstained with hematoxylin for 1 min and mounted with balsam mounting medium. After that, sections were visualized and photographed. Five representative fields under high magnification (Upright optical microscope, NIKON, Japan) were selected and the number of positive cells with brown or yellow color was quantified.
Statistical analysis
All statistical analyses in this study were processed with SPSS 21.0 (IBM, USA). Data were presented as mean ± standard deviation. Data from two groups were compared by unpaired t-test. Data among multiple groups were analyzed by one-way analysis of variance (ANOVA). Data at different time points among multiple groups were analyzed by repeated measures ANOVA. p < 0.05 indicated statistically significant difference.
Results
KDM4C is upregulated in AML bone marrow and HL-60/A cell line
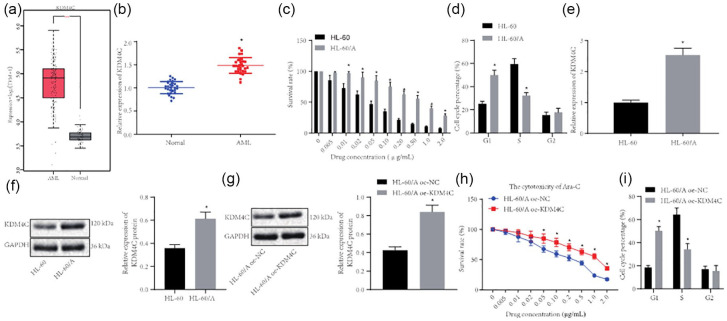
KDM4C is a subtype of the known demethylase KDM family, which plays important roles in the development of cancer by regulating lysine methylation of histone.15,23 Bioinformatics analysis of the TCGA and GTEx databases revealed that demethylase KDM4C expression was increased in AML [Figure 1(a)]. Thus, to determine whether KDM4C also plays important roles in the development of AML, we detected the expression of KDMC4 in 30 bone marrow tissue samples from AML patient and 30 normal bone marrow samples. RT-qPCR showed that KDM4C expression was significantly upregulated in bone marrow tissue samples from AML patients compared with normal bone marrow [Figure 1(b)]. Next, we selected HL-60 and HL-60/A cell lines and performed the MTT assay to test the effect of Ara-C on their proliferation. The data demonstrated that the IC50 of Ara-C for HL-60 was 0.044 μg/mL while the IC50 for HL-60/A was 0.268 μg/mL after Ara-C treatment for 48 h, indicating a 6.09 ratio of HL-60/A IC50 to HL-60 IC50, which indicated that HL-60/A was more resistant to Ara-C and thus suitable for the in vitro drug resistant model [Figure 1(c)]. To understand how HL-60 and HL-60/A responded differently to Ara-C, flow cytometry was applied to detect the cell cycle, which displayed that G1 phase of HL-60/A was notably higher than that of HL-60, while S phase was notably lower than that of HL-60, but G2 phase did not differ [Figure 1(d)]. Then, RT-qPCR and Western blot were performed to analyze the expression of KDM4C, which demonstrated that KDM4C expression was considerably higher in HL-60/A cells than that in HL-60 cells [Figure 1(e) and (f)]. We also overexpressed KNM4C in HL-60/A cells, whereupon MTT assay revealed the IC50 of Ara-C in control (oe-NC) was 0.158 μg/mL while the IC50 of Ara-C in oe-KDM4C cells was 0.428 μg/mL μg/mL, giving an IC50 ration of 2.71 [Figure 1(h)]. Flow cytometry data demonstrated that overexpression of KDM4C increased cells arrested at G1 phase, decreased cells arrested at S phase, but did not change the proportion of cells at G2 phase [Figure 1(i)]. These results suggested that KDM4C expression was upregulated in AML patients and the HL-60/A cell line and may be involved in Ara-C resistance in AML.
Figure 1.
KDM4C is upregulated in bone marrow tissue samples from AML patients and in the HL-60/A cell line. (a) Analysis of KDM4C expression level in AML based on TCGA (173 bone marrow tissue samples from AML patient) and GTEx (70 normal bone marrow samples) databases. (b) RT-qPCR detection of KDM4C expression in bone marrow tissue samples from 30 AML patients and 30 normal bone marrow samples, *comparison with normal group. (c) MTT assay to analyze HL-60 and HL-60/A viability after Ara-C treatment, *comparison with HL-60. (d) Flow cytometry to study the cell cycle of HL-60 and HL-60/A, *comparison with HL-60. (e) RT-qPCR to detect the expression of KDM4C in HL-60 and HL-60/A, *comparison with HL-60. (f) Western blot to detect the expression of KDM4C protein in HL-60 and HL-60/A cells, *comparison with HL-60. (g) Western blot to analyze the efficiency of KDM4C expression in HL-60/A, *comparison with HL-60/A oe-NC. (h) MTT assay to analyze HL-60/A cell viability after oe-KDM4C treatment, *p < 0.05 compared with oe-NC. (i) Flow cytometry to analyze the cell cycle after overexpression of KDM4C in HL-60/A, *comparison with HL-60/A oe-NC. Experiments were repeated three times. Data from two groups was compared by unpaired t-test.
AML, acute myeloid leukemia; Ara-C, cytarabine; NC, negative control; oe, overexpression; RT-qPCR, real-time quantitative polymerase chain reaction; TCGA, The Cancer Genome Atlas; GTEx, Genotype-Tissue Expression; MTT, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
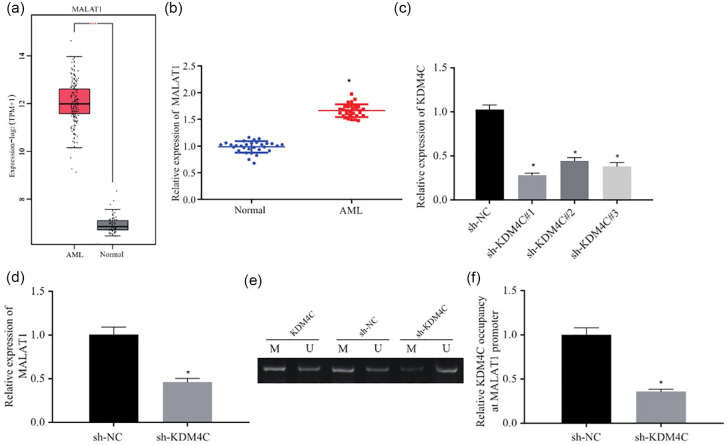
KDM4C elevates MALAT1 expression by demethylating the promoter of MALAT1
It has been reported that KDM4C could bind to MALAT1 promoter.24 In order to further investigate the mechanism by which KDM4C causes drug resistance, we tested the hypothesis that KDM4C influences the drug resistance via MALAT1. MALAT1 was predicted to be upregulated in AML in the TCGA and GTEx databases we used previously [Figure 2(a)]. We detected the expression of MALAT1 in bone marrow tissue samples from 30 AML patients and 30 normal bone marrow samples. RT-qPCR results showed that MALAT1 expression was significantly higher in bone marrow tissue samples from AML patients compared with normal bone marrow samples [Figure 2(b)]. To study the regulatory relationship between KDM4C and MALAT1, we knocked down KDM4C expression in HL-60/A cells and analyzed the expression of KDM4C. The data demonstrated that after knockdown by shRNA, KDM4C expression was remarkably downregulated in HL-60/A cells, among which sh-KDM4C#1 displayed the best knockdown effect, and was thus selected for the subsequent experiments [Figure 2(c)]. Next, RT-qPCR results demonstrated that MALAT1 expression was notably decreased in HL-60/A cells after shRNA KDM4C treatment [Figure 2(d)]. MS-PCR was then performed to detect the methylation of MALAT1 promoter region after silencing KDM4C in HL-60/A cells, which uncovered that the methylation level in MALAT1 promoter was considerably increased [Figure 2(e)]. ChIP results also demonstrated that KDM4C was enriched in the promoter region of MALAT1, while the enrichment of KDM4C was remarkably decreased after KDMC4 knockdown in HL-60/A cells [Figure 2(f)]. These results revealed that KDM4C upregulation in HL-60/A induced the expression of MALAT1 via demethylation in the promoter region.
Figure 2.
KDM4C increases MALAT1 expression by demethylating the promoter of MALAT1. (a) MALAT1 expression level in AML according to TCGA (bone marrow tissue samples from 173 AML patients) and GTEx (70 normal bone marrow samples) databases, *p < 0.05 compared with normal group. (b) RT-qPCR to detect the expression of MALAT1 in bone marrow tissue samples from 30 AML patients and 30 normal bone marrow samples, *p < 0.05 compared with normal group. (c) RT-qPCR to detect the expression of KDM4C after transfection with KDM4C shRNA in HL-60/A, *p < 0.05 compared with sh-NC. (d) RT-qPCR to detect the expression of MALAT1 after knockdown of KDM4C in HL-60/A, *p < 0.05 compared with sh-NC. (e) MS-PCR to analyze the methylation in MALAT1 promoter after knockdown of KDM4C in HL-60/A cells. (f) ChIP to detect the enrichment of KDM4C in the promoter region of MALAT1 in HL-60/A cells, *p < 0.05 compared with sh-NC. Experiments were repeated three times. Data from two groups was compared by unpaired t-test.
AML, acute myeloid leukemia; ChIP, chromatin immunoprecipitation; MS-PCR, methylation-specific polymerase chain reaction; NC, negative control; RT-qPCR, real-time quantitative polymerase chain reaction; sh, short hairpin; TCGA, The Cancer Genome Atlas; GTEx, Genotype-Tissue Expression.
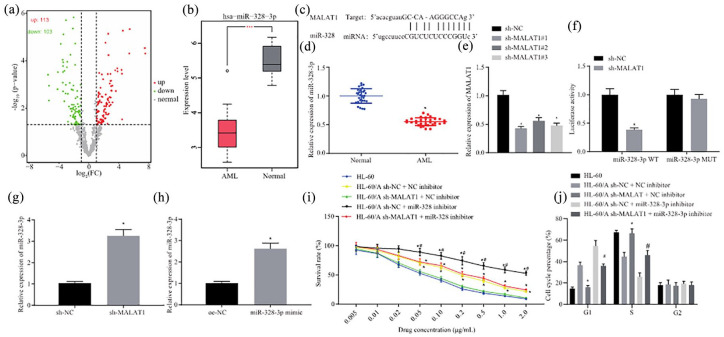
MALAT1 enhances AML cells resistant to Ara-C by suppressing miR-328-3p
First, starBase was applied to predict the downstream target of MALAT1. The analysis uncovered that there were many differentially expressed miRNAs downstream of MALAT1 targets, among which miR-328-3p expression was notably downregulated [Figure 3(a)]. The GSE128079 database revealed that miR-328-3p expression was considerably decreased in AML samples [Figure 3(b)]. To determine whether miR-328-3p is regulated by MALAT1 in AML, the interaction between MALAT1 and miR-328-3p was predicted by starBase, which demonstrated that MALAT1 could bind to miR-328-3p [Figure 3(c)]. RT-qPCR was applied to detect the expression of miR-328-3p in bone marrow tissue samples from AML patients and normal bone marrow samples, which uncovered that miR-328-3p expression was decreased in the patient bone marrow tissue samples [Figure 3(d)]. After knockdown of MALAT1 by shRNA in HL-60/A, RT-qPCR showed that MALAT1 expression was considerably decreased [Figure 3(e)]. Next, the dual-luciferase reporter assay was conducted, which showed that MALAT1 targeted miR-328-3p [Figure 3(f)]. RT-qPCR results showed that miR-328-3p expression was significantly upregulated in HL-60/A cells after silencing MALAT1 [Figure 3(g)].
Figure 3.
MALAT1 enhances resistance of AML cells to Ara-C by suppressing miR-328-3p. (a) Volcano analysis of differentially expressed miRNAs in AML by whole blood NGS (GSE128079). (b) miR-328-3p expression in AML based on whole blood NGS (GSE128079). (c) starBase prediction of the binding between MALAT1 and miR-328-3p. (d) RT-qPCR to detect the expression of miR-328-3p in bone marrow tissue samples from AML patients and normal bone marrow samples, *p < 0.05 compared with normal group. (e) RT-qPCR to detect the expression of MALAT1 after transfection with MALAT1 shRNA in HL-60/A, *p < 0.05 compared with normal group. (f) Dual-luciferase reporter assay to analyze the relationship between MALAT1 and miR-328-3p, *p < 0.05 compared with sh-NC. (g) RT-qPCR to detect the expression of miR-328-3p after knockdown of MALAT1 in HL-60/A, *p < 0.05 compared with sh-NC. (h) RT-qPCR to detect the overexpression of miR-328-3p in HL-60 and HL-60/A cells, *p < 0.05 compared with oe-NC. (i) MTT assay to analyze the viability of HL-60/A cells after knockdown of MALAT1 and overexpression of miR-328-3p, *comparison with sh-MALAT1 + NC inhibitor, #comparison with sh-NC + miR-328-3p inhibitor, p < 0.05. (j) Flow cytometry to analyze the cell cycle of HL-60/A after knockdown of MALAT1 and overexpression of miR-328-3p, *comparison with HL-60/sh-MALAT1 + NC inhibitor, #comparison with sh-NC + miR-328-3p inhibitor, p < 0.05. Experiments were repeated three times. Data from two groups were compared by unpaired t-test. Data among multiple groups were analyzed by one-way analysis of variance.
AML, acute myeloid leukemia; Ara-C, cytarabine; Mut, mutant; NC, negative control; NGS, next-generation sequencing; oe, overexpression; RT-qPCR, real-time quantitative polymerase chain reaction; sh, short hairpin; WT, wild type; MTT, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; miR-328-3p, microRNA-328-3p.
The effect of miR-328-3p on the Ara-C resistance was then investigated. HL-60/A and HL-60 cells were transfected with miR-328-3p-mimic, and subsequent RT-qPCR results demonstrated that miR-328-3p was remarkably upregulated after transfection with miR-328-3p-mimic in HL-60/A and HL-60 cells, thus validating the successful transfection of miR-328-3p [Figure 3(h)]. The MTT assay was then performed to detect the effect of knockdown of MALAT1 and/or inhibition of miR-328-3p on the resistance to Ara-C. The results uncovered that down-regulation of MALAT1 suppressed HL-60/A cells resistance to Ara-C, while suppression of miR-328-3p induced greater resistance of HL-60/A cells to Ara-C [Figure 3(i)]. Flow cytometry was also performed to evaluate the cell cycle distribution after knockdown of MALAT1 and/or inhibition of miR-328-3p. Results revealed that after knockdown of MALAT1, Ara-C treated HL-60/A cells exhibited a similar increase in the proportion of S-phase cells and a decrease in G1 cells, while suppression of miR-328-3p in HL-60/A cells led to more cells arrested in the G1 phase and fewer cells in S phase [Figure 3(j)]. Coherently, MALAT1 enhanced AML cells resistance to Ara-C by suppressing miR-328-3p.
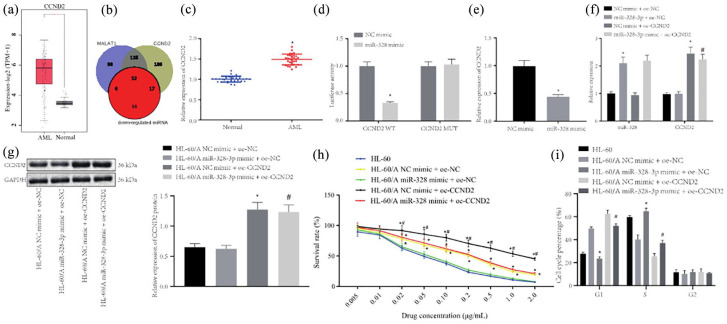
miR-328-3p inhibits AML cells resistant to Ara-C by targeting CCND2
starBase predicted that miR-328-3p targeted CCND2, which belongs to the cyclin family and has been shown to correlate with drug resistance in cancer.25 CCND2 was found to be upregulated in AML through analysis of the TCGA and GTEx databases [Figure 4(a)]. The Venn diagram was plotted, which revealed 12 differentially expressed miRNAs, including miR-328-3p [Figure 4(b)]. We made a preliminary determination of the targeting effect of miR-328-3p on AML in CCND2, and speculated that CCND2 may be regulated by miR-328-3p to participate in AML Ara-C resistance. Next, RT-qPCR was used to detect the expression of CCND2, which demonstrated that CCND2 expression was notably increased in bone marrow tissue samples from AML patients compared with normal bone marrow samples [Figure 4(c)]. Verification through dual-luciferase reporter assay also uncovered that miR-328-3p inhibited CCND2 expression [Figure 4(d)].
Figure 4.
miR-328-3p inhibits AML cells resistant to Ara-C by targeting CCND2. (a) CCND2 expression in AML according to TCGA (bone marrow tissue samples from 173 AML patients) and GTEx (70 normal bone marrow samples) databases, *p < 0.05 compared with normal group. (b) Venn diagram to show the miRNA targets of MALAT1 and CCND2 according to the analysis from starBase. (c) RT-qPCR to detect the expression of MALAT1 in bone marrow tissue samples from AML patients and normal bone marrow samples, *p < 0.05 compared with normal group. (d) Dual-luciferase reporter assay to detect the relationship between miR-328-3p and CCND2, *p < 0.05 compared with NC mimi. (e) RT-qPCR to detect the expression of CCND2 after overexpression of miR-328-3p in HL-60/A cells, *p < 0.05 compared with NC mimic. (f) RT-qPCR to detect the expression of miR-328-3p and CCND2 after overexpression of miR-328-3p and CCND2 in HL-60/A cells, *p < 0.05 compared with NC mimic + oe-NC, #p < 0.05 compared with miR-328-3p mimic + oe-NC. (g) Western blot of the expression of CCND2 after overexpression of miR-328-3p and CCND2 in HL-60/A cells, *p < 0.05 compared with NC mimic + oe-NC, #p < 0.05 compared with miR-328-3p mimic + oe-NC. (h) MTT assay to detect the viability of HL-60/A cells after overexpression of miR-328-3p and CCND2, *p < 0.05 compared with HL-60/miR-328-3p mimic + oe-NC, #p < 0.05 compared with NC mimic + oe-NC/miR-328-3p mimic + oe-CCND2. (i) Flow cytometry to analyze the cell cycle of HL-60/A after overexpression of miR-328-3p and CCND2, *p < 0.05 compared with NC mimic + oe-NC, #p < 0.05 compared with NC mimic + oe-CCND2. Experiments were repeated three times. Data from two groups were compared by unpaired t-test. Data among multiple groups were analyzed by one-way analysis of variance.
AML, acute myeloid leukemia; Ara-C, cytarabine; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Mut, mutant; NC, negative control; oe, overexpression; RT-qPCR, real-time quantitative polymerase chain reaction; TCGA, The Cancer Genome Atlas; Wt, wild type; GTEx, Genotype-Tissue Expression; miRNA, microRNA.
To verify whether miR-328-3p can activate CCND2 expression in HL-60/A cells, HL-60/A cells were overexpressed with miR-328-3p and CCND2 or respective controls. RT-qPCR and Western blot were conducted to analyze the expression of miR-328-3p and CCND2 in these cells. The results demonstrated that CCND2 expression was reduced while miR-328-3p expression was enhanced in cells transfected with miR-328-3p mimic. However, after cells were co-treated with miR-328-3p mimic and oe-CCND2, miR-328-3p expression was not significantly altered, while CCND2 expression was restored [Figure 4(e) to (g)].
The effect of miR-328-3p targeting CCND2 on AML resistance to Ara-C was subsequently studied. HL-60/A cells were transfected as previously described, and the MTT assay was performed to calculate cell resistance and IC50. The results displayed that the IC50 values of Ara-C for these cells were 0.041 μg/mL (HL-60), 0.126 μg/mL (NC-mimic + oe-NC), 0.045 μg/mL (miR-328 mimic + oe-NC); 0.233 μg/mL (NC-mimic + oe-CCND2); 0.132 μg/mL (miR-328 mimic + oe-CCND2), which indicated that the cell resistance of HL-60/A cells infected with NC-mimic + oe-CCND2 > those with mimic + oe-NC ≈ those with miR-328 mimic + oe-CCND2 > those with miR-328 mimic + oe-NC with HL-60 cells serving as an NC [Figure 4(h)]. Then, flow cytometry was conducted to evaluate the cell cycle, which revealed that overexpression of miR-328-3p decreased cells at G1 phase, while increased cells at S phase, and unchanged cells at G2 phase, and similar findings for cell cycle in HL-60 cells, which suggested that miR-328-3p promoted the sensitivity of HL-60/A to Ara-C. However, overexpression of CCND2 caused increased cells at G1 phase, while it decreased cells at S phase, with no change in G2 phase cells, which indicated that CCND2 overexpression inhibited HL-60/A sensitivity to Ara-C [Figure 4(i)]. The above results indicated that miR-328-3p inhibited AML resistant to Ara-C by targeting CCND2.
KDM4C induces Ara-C resistance through regulation of MALAT1/miR-328-3p/CCND2
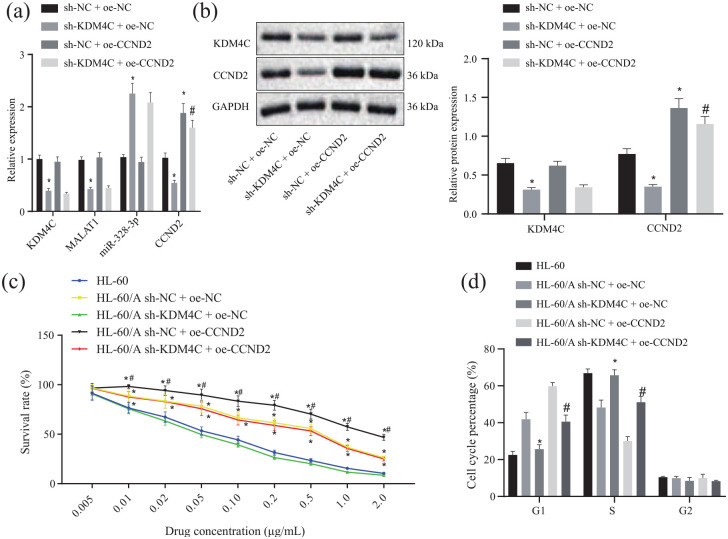
To confirm that KDM4C participates in Ara-C resistance in AML by regulating miR-328-3p/CCND2 through MALAT1, HL-60/A cells were transfected with sh-KDM4C, oe-CCND2, or corresponding controls. RT-qPCR and Western blot were conducted to detect the expression of KDM4C, MALAT1, miR-328-3p, and CCND2. The results showed that, after knockdown of KDM4C in HL-60/A cells, MALAT1 expression was down-regulated, miR-328-3p expression was upregulated, and CCND2 was also partially down-regulated upon overexpression of miR-328-3p. Overexpression of CCND2 could rescue the CCND2 expression [Figure 5(a) and (b)]. These data indicated that KDM4C regulated miR-328-3p/CCND2 through MALAT1.
Figure 5.
KDM4C regulates miR-328-3p/CCND2 through MALAT1 leading to Ara-C resistance in AML. (a) RT-qPCR to detect the expression of KDM4C, MALAT1, miR-328-3p, and CCND2 after knockdown of KDM4C or overexpression of CCND2 in HL-60/A cells, *p < 0.05 compared with sh-NC + oe-NC, #p < 0.05 compared with sh-KDM4C + oe-NC. (b) Western blot to detect the expression of KDM4C and CCND2 after knockdown of KDM4C and overexpression of CCND2 in HL-60/A cells, *p < 0.05 compared with sh-NC + oe-NC, #p < 0.05 compared with sh-KDM4C + oe-NC. (c) MTT assay to analyze the viability of HL-60/A cells after knockdown of KDM4C and overexpression of CCND2 in HL-60/A cells, *p < 0.05 compared with HL-60/sh-KDM4C + oe-NC, #p < 0.05 compared with sh-NC + oe-NC/sh-KDM4C + oe-CCND2. (d) Flow cytometry to analyze the cell cycle of HL-60 and HL-60/A cells, *p < 0.05 compared with sh-NC + oe-NC, #p < 0.05 compared with sh-NC + oe-CCND2. Experiments were repeated three times. Data among multiple groups were analyzed by one-way analysis of variance.
AML, acute myeloid leukemia; Ara-C, cytarabine; G1, G1-phase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; NC, negative control; oe, overexpression; RT-qPCR, real-time quantitative polymerase chain reaction; S, S-phase; sh, short hairpin; MTT, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; miR-328-3p, microRNA-328-3p.
Then, we considered whether KDM4C/MALAT1/miR-328-3p/CCND2 axis regulates the sensitivity of AML cells to Ara-C. Our results from the MTT assay demonstrated that the IC50 of the sh-NC + oe-NC + Ara-C, sh-KDM4C + oe-NC + Ara-C, sh-NC + oe-CCND2 + Ara-C, and sh-KDM4C + oe-CCND2 + Ara-C groups were 0.268 μg/mL, 0.044 μg/mL, 0.553 μg/mL, and 0.235 μg/mL with HL-60 as control (0.057 μg/mL) [Figure 5(c)], which revealed that knockdown of KDM4C inhibited HL-60/A resistance to Ara-C, while overexpression of CCND2 could recover the resistance to Ara-C in HL-60/A cells. Moreover, flow cytometry showed that after knockdown of KDM4C in HL-60/A, cells at G1 phase were decreased, cells at S phase were increased, but cells at G2 phase were unchanged, which showed a similar trend for HL-60 cells. However, overexpression of CCND2 reversed the distribution of G1 and S phases [Figure 5(d)].
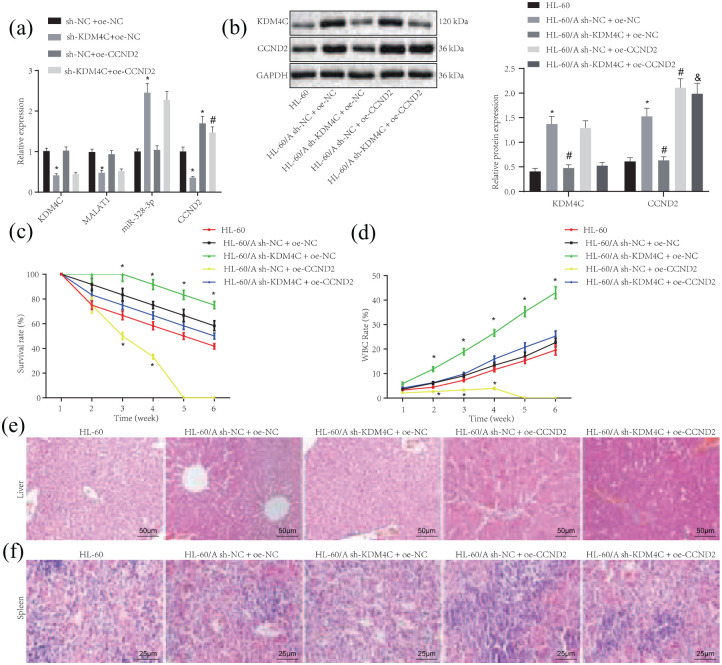
KDM4C/MALAT1/miR-328-3p/CCND2 axis regulates in vivo Ara-C resistance in AML
To further elucidate the contribution of the KDM4C/MALAT1/miR-328-3p/CCND2 axis to Ara-C resistance in AML, a xenograft tumor model was established. RT-qPCR determination revealed that, compared with sh-NC + oe-NC group, the expression of KDM4C, MALAT1, and CCND2 was reduced while miR-328-3p expression was increased in sh-KDM4C + oe-NC group. Moreover, compared with sh-NC + oe-CCND2 group, the expression of KDM4C and MALAT1 was reduced but miR-328-3p expression was increased while CCND2 expression was not significantly changed [Figure 6(a)].
Figure 6.
KDM4C/MALAT1/miR-328-3p/CCND2 axis regulates in vivo Ara-C resistance in AML. (a) RT-qPCR to detect the expression of KDM4C, MALAT1, miR-328-3p, and CCND2 in corresponding stably transfected HL-60/A cells, *p < 0.05 compared with sh-NC + oe-NC, #p < 0.05 compared with sh-KDM4C + oe-NC. (b) Western blot analysis to detect the expression of KDM4C, MALAT1, miR-328-3p, and CCND2 in NOD/SCID mice under light microscope (left); statistical chart of Western blot analysis (right), *p < 0.05 compared with HL-60, # p < 0.05 compared with HL-60/A sh-NC + oe-NC, & HL-60/A sh-KDM4C + oe-CCND2. (c) The survival of mice in each group, *p < 0.05 compared with HL-60/HL-60/A sh-NC + oe-NC/HL-60/A sh-KDM4C + oe-CCND2. (d) The quantification of the ratio of leukocytes, *p < 0.05 compared with HL-60/HL-60/A sh-NC + oe-NC/HL-60/A sh-KDM4C + oe-CCND2. (e) H&E staining to show the lesion and tumor infiltration in liver tissues of grouped mice under light microscope, × 200. (f) H&E staining to show the lesion and tumor infiltration in spleen tissues of grouped mice under light microscope, × 400. Experiments were repeated three times. Data among multiple groups were analyzed by one-way analysis of variance (ANOVA). Data at different time points among multiple groups were analyzed by repeated measures ANOVA. p < 0.05 indicated statistically significant difference.
AML, acute myeloid leukemia; Ara-C, cytarabine; H&E, hematoxylin and eosin; NC, negative control; oe, overexpression; RTqPCR, real-time quantitative polymerase chain reaction; sh, short hairpin; miR-328-3p, microRNA-328-3p.
Next, the expression of KDM4C and CCND2 in the peripheral blood of modeled mice in each group was determined by Western blot assay, with the results showing that the expression of KDM4C and CCND2 was significantly increased in the HL-60 cells compared with HL-60/A cells, whereas the expression of KDM4C and CCND2 decreased in the HL-60/A cells after silencing KDM4C; compared with sh-KDM4C + oe-NC group, CCND2 expression increased but no significant change was found in KDM4C expression in sh-KDM4C + oe-CCND2 group [Figure 6(b)], indicating that the cells were stably transduced. Subsequent observations showed that the xenograft mice treated with the HL-60/A sh-KDM4C + oe-NC had the best survival rate, while those with HL-60/A sh-NC + oe-CCND2 had the worst survival. Moreover, compared with the HL-60/A sh-KDM4C + oe-NC group, the HL-60/A sh-KDM4C + oe-CCND2 group displayed worse survival [Figure 6(c)]. Similar results were found in the peripheral blood leukocytes [Figure 6(d)]. The hepatic and splenic injury of mice in each group was observed by H&E staining, and the results showed lowered cellular infiltration of spleen with multiple focal necrosis in the liver of mice injected with HL-60/A cells with CCND2 silencing, which was increased in the HL-60/A cells overexpressing CCND2. Furthermore, the liver injury and splenic injury were aggravated in the mice injected with HL-60/A cells expressing sh-KDM4C + oe-CCND2 compared with mice treated with the HL-60/A cells expressing sh-KDM4C + oe-NC [Figure 6(e) and (f)]. These results further validated the role of KDM4C in regulating the sensitivity of AML cells to Ara-C by modulating the MALAT1/miR-328-3P/CCND2 axis.
Discussion
The abnormal activation or overexpression of epigenetic regulators like KDM4C histone demethylase has been reported to change gene expression patterns so as to increase the cell survival and cause drug resistance in tumor lines.26–28 In our study, we applied bioinformatics analysis, gain- and loss-of-function studies, in vitro Ara-C resistant cell line, and in vivo AML mice to elucidate how the KDM4C/MALAT1/miR-328-3p/CCND2 axis contributes to Ara-C resistance in AML. We found that KDM4C was significantly upregulated in AML patient samples and in the HL-60/A cell line; MALAT1 expression was notably increased in AML and positively correlated with KDM4C expression. Furthermore, miR-328-3p was the target of MALAT1 and was downregulated in AML, to an extent correlating negatively with Ara-C resistance. CCND2 proved to be the downstream target of miR-328-3p and induced resistance to Ara-C. Finally, KDM4C knockdown could increase the survival of mice and benefit the treatment response to Ara-C in AML.
There are limited findings about the function of KDM4C in the development of AML and Ara-C resistance in AML. Cheung et al.15 revealed that KDM4C in cooperation with PRMT1 remodeled epigenetic programs and deregulated transcriptional patterns by removing the repressive effect of H3K9me3 on HOXA9 in AML. Agger et al.29 uncovered that KDM4C is required for the survival of AML cells. Our study demonstrated the upregulation of KDM4C in AML patient samples and Ara-C resistant cell line, and showed that KDM4C conferred resistance to Ara-C by targeting MALAT1, consistent with results of previous studies indicating the induction of growth advantage of AML cells by KDM4. lncRNAs participate in diverse regulatory networks regarding gene transcription and translation and have been implicated in the etiology of various cancers.30,31 In CML, MALAT1 serves as a sponge of miR-328-3p, and Huang et al.20 reported that MALAT1 can benefit cell proliferation in AML and was associated with poor prognosis.19 Our present data reveal that MALAT1 is targeted and positively regulated by KDM4C via demethylation, while miR-328-3p-38 is the downstream target of MALAT1, and that upregulation of MALAT1 decreases the expression of miR-328-3p, which imparts Ara-C resistance in AML. MiR-328 has been shown to undergo downregulation in primary glioblastoma, non-small cell lung cancer, and colorectal cancer, which is all consistent with the anti-tumor effect of miR-328-3p.32–34 Our results and previous work inspired us to investigate the downstream target of miR-328-3p that is responsible for Ara-C resistance in AML. The bioinformatics analysis indicated CCND2 as the target of miR-328-3p, which has also been shown to be upregulated in AML patient samples and to induce Ara-C resistance. CCND2 is a member of the cyclin family, which regulates cyclin dependent kinase across the cell cycle.35 CCND2 is frequently found to be mutated in AML patients.36 In bladder cancer, Yu et al.25 demonstrated that CCND2 mRNA and protein were down-regulated by miR-194-5p, which resulted in chemoresistance to cisplatin. Our data uncover that CCND2 is targeted and downregulated by miR-328-3p, which increases the proportion of cells in the G1 phase and inhibits the sensitivity of HL-60/A to Ara-C. Finally, we undertook corroborative studies in AML model mice, which revealed that knockdown of KDM4C significantly increased survival of AML mice and promoted the effectiveness of Ara-C therapy.
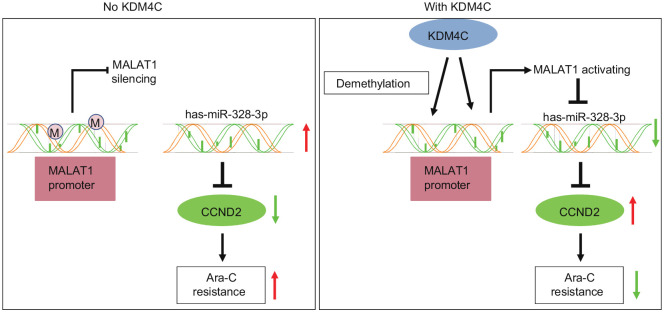
Conclusion
Taken together, results of this paper decipher how the KDM4C/MALAT1/miR-328-3p/CCND2 axis contributes to Ara-C resistance in AML. KDM4C demethylates the MALAT1 promoter region to upregulate its expression, which in turn downregulates miR-328-3p expression, causing an increased level of CCND2, finally leading to Ara-C resistance in AML (Figure 7). Our work provides a novel mechanism of an epigenetic regulator that induces Ara-C resistance in AML, and sheds light on a new strategy to develop targeted treatment of Ara-C resistant AML. It is a matter for future studies to determine how demethylation on H3K9 affects chromosomal accessibility at the genome wide level and how other target genes may influence Ara-C resistance in AML due to this epigenetic change.
Figure 7.
KDM4C demethylates MALAT1 promoter region to upregulate its expression, which in turn downregulates miR-328-3p expression, causing the increased level of CCND2, finally leading to Ara-C resistance in acute myeloid leukemia.
Ara-C, cytarabine.
Supplemental Material
Supplemental material, sj-pdf-1-taj-10.1177_2040622321997259 for KDM4C contributes to cytarabine resistance in acute myeloid leukemia via regulating the miR-328-3p/CCND2 axis through MALAT1 by Lu Xue, Chunhuai Li, Jin Ren and Yue Wang in Therapeutic Advances in Chronic Disease
Acknowledgments
We would like to give our sincere appreciation to the reviewers for their helpful comments on this article.
Footnotes
Author contributions: Lu Xue, and Yue Wang wrote the paper and conceived and designed the experiments; Chunhuai Li and Jin Ren analyzed the data; Lu Xue and Jin Ren collected and provided the sample for this study. All authors have read and approved the final submitted manuscript.
Availability of data and materials: All data generated or analyzed during this study are included in this published article.
Conflict of interest statement: The authors declare that there is no conflict of interest.
Ethics statement: All experiments were approved by the Ethics Committee of the First Hospital of Jilin University (institutional review board approval number: 2020-640) and conforming to principles originating in the Declaration of Helsinki. Animal experiments were performed in accordance with the standard of The guide for the care and use of laboratory animals published by the National Institutes of Health. The animal experimental procedures were ratified by the Animal Ethics Committee of the First Hospital of Jilin University (approval number: 2020-0186).
Funding: This research was supported by talents reserve project of the First Hospital of Jilin University (xuelu) a.
Informed consent: Written informed consent was obtained from patients.
ORCID iD: Yue Wang  https://orcid.org/0000-0001-7956-106X
https://orcid.org/0000-0001-7956-106X
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Lu Xue, Department of Pediatrics Hematology, The First Hospital of Jilin University, Changchun, P.R. China.
Chunhuai Li, Department of Pediatrics Hematology, The First Hospital of Jilin University, Changchun, P.R. China.
Jin Ren, Department of Respiratory Medicine, The Second Hospital of Jilin University, Changchun, P.R. China.
Yue Wang, Department of Pediatrics Hematology, The First Hospital of Jilin University, No. 1, Xinmin Street, Chaoyang District, Changchun, Jilin Province 130021, P.R. China.
Reference
- 1.Bonnet D, Dick JE.Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997; 3: 730–737. [DOI] [PubMed] [Google Scholar]
- 2.Wang G, Li S, Xue K, et al. PFKFB4 is critical for the survival of acute monocytic leukemia cells. Biochem Biophys Res Commun 2020; 526: 978–985. [DOI] [PubMed] [Google Scholar]
- 3.De Kouchkovsky I, Abdul-Hay M. Acute myeloid leukemia: a comprehensive review and 2016 update. Blood Cancer J 2016; 6: e441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koreth J, Schlenk R, Kopecky KJ, et al. Allogeneic stem cell transplantation for acute myeloid leukemia in first complete remission: systematic review and meta-analysis of prospective clinical trials. JAMA 2009; 301: 2349–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perl AE.The role of targeted therapy in the management of patients with AML. Blood Adv 2017; 1: 2281–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robak T, Wierzbowska A.Current and emerging therapies for acute myeloid leukemia. Clin Ther 2009; 31 Pt2: 2349–2370. [DOI] [PubMed] [Google Scholar]
- 7.Lowenberg B, Pabst T, Vellenga E, et al. Cytarabine dose for acute myeloid leukemia. N Engl J Med 2011; 364: 1027–1036. [DOI] [PubMed] [Google Scholar]
- 8.Cai J, Damaraju VL, Groulx N, et al. Two distinct molecular mechanisms underlying cytarabine resistance in human leukemic cells. Cancer Res 2008; 68: 2349–2357. [DOI] [PubMed] [Google Scholar]
- 9.van Dijk AD, de Bont E, Kornblau SM.Targeted therapy in acute myeloid leukemia: current status and new insights from a proteomic perspective. Expert Rev Proteomics 2020; 17: 1–10. [DOI] [PubMed] [Google Scholar]
- 10.Nebbioso A, Tambaro FP, Dell’Aversana C, et al. Cancer epigenetics: moving forward. PLoS Genet 2018; 14: e1007362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das PP, Shao Z, Beyaz S, et al. Distinct and combinatorial functions of Jmjd2b/Kdm4b and Jmjd2c/Kdm4c in mouse embryonic stem cell identity. Mol Cell 2014; 53: 32–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Fang R, Yue C, et al. Wnt-induced stabilization of KDM4C is required for Wnt/beta-catenin target gene expression and glioblastoma tumorigenesis. Cancer Res 2020; 80: 1049–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin C-Y, Wang B-J, Chen B-C, et al. Histone demethylase KDM4C stimulates the proliferation of prostate cancer cells via activation of AKT and c-Myc. Cancers (Basel) 2019; 11: 1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia J, Lizcano F.KDM4C activity modulates cell proliferation and chromosome segregation in triple-negative breast cancer. Breast Cancer (Auckl) 2016; 10: 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung N, Fung TK, Zeisig BB, et al. Targeting aberrant epigenetic networks mediated by PRMT1 and KDM4C in acute myeloid leukemia. Cancer Cell 2016; 29: 32–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan T, Song Z, Wu L, et al. USP49 potently stabilizes APOBEC3G protein by removing ubiquitin and inhibits HIV-1 replication. Elife 2019; 8: e48318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutschner T, Hämmerle M, Diederichs S.MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013; 91: 791–801. [DOI] [PubMed] [Google Scholar]
- 18.Shi X, Sun M, Liu H, et al. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett 2013; 339: 159–166. [DOI] [PubMed] [Google Scholar]
- 19.Wen F, Cao Y-X, Luo Z-Y, et al. LncRNA MALAT1 promotes cell proliferation and imatinib resistance by sponging miR-328 in chronic myelogenous leukemia. Biochem Biophys Res Commun 2018; 507: 1–8. [DOI] [PubMed] [Google Scholar]
- 20.Huang JL, Liu W, Tian LH, et al. Upregulation of long non-coding RNA MALAT-1 confers poor prognosis and influences cell proliferation and apoptosis in acute monocytic leukemia. Oncol Rep 2017; 38: 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eiring AM, Harb JG, Neviani P, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 2010; 140: 652–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zorzoli A, Di Carlo E, Cocco C, et al. Interleukin-27 inhibits the growth of pediatric acute myeloid leukemia in NOD/SCID/Il2rg-/- mice. Clin Cancer Res 2012; 18: 1630–1640. [DOI] [PubMed] [Google Scholar]
- 23.Soini Y, Kosma VM, Pirinen R.KDM4A, KDM4B and KDM4C in non-small cell lung cancer. Int J Clin Exp Pathol 2015; 8: 12922–12928. [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X, Li R, Song Q, et al. JMJD2C promotes colorectal cancer metastasis via regulating histone methylation of MALAT1 promoter and enhancing beta-catenin signaling pathway. J Exp Clin Cancer Res 2019; 38: 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu G, Zhou H, Yao W, et al. lncRNA TUG1 promotes cisplatin resistance by regulating CCND2 via epigenetically silencing miR-194-5p in bladder cancer. Mol Ther Nucleic Acids 2019; 16: 257–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berry WL, Janknecht R.KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res 2013; 73: 2936–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregory BL, Cheung VG.Natural variation in the histone demethylase, KDM4C, influences expression levels of specific genes including those that affect cell growth. Genome Res 2014; 24: 52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai CT, So CW.Epigenetic therapies by targeting aberrant histone methylome in AML: molecular mechanisms, current preclinical and clinical development. Oncogene 2017; 36: 1753–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agger K, Miyagi S, Pedersen MT, et al. Jmjd2/Kdm4 demethylases are required for expression of Il3ra and survival of acute myeloid leukemia cells. Genes Dev 2016; 30: 1278–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huarte M.The emerging role of lncRNAs in cancer. Nat Med 2015; 21: 1253–1261. [DOI] [PubMed] [Google Scholar]
- 31.Long Y, Wang X, Youmans DT, et al. How do lncRNAs regulate transcription? Sci Adv 2017; 3: eaao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Z, Sun L, Wang H, et al. MiR-328 expression is decreased in high-grade gliomas and is associated with worse survival in primary glioblastoma. PLoS One 2012; 7: e47270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma W, Ma C-N, Zhou N-N, et al. Up- regulation of miR-328-3p sensitizes non-small cell lung cancer to radiotherapy. Sci Rep 2016; 6: 31651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu XT, Xu Q, Tong JL, et al. MicroRNA expression profiling identifies miR-328 regulates cancer stem cell-like SP cells in colorectal cancer. Br J Cancer 2012; 106: 1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiong Y, Menninger J, Beach D, et al. Molecular cloning and chromosomal mapping of CCND genes encoding human D-type cyclins. Genomics 1992; 13: 575–584. [DOI] [PubMed] [Google Scholar]
- 36.Eisfeld A-K, Kohlschmidt J, Schwind S, et al. Mutations in the CCND1 and CCND2 genes are frequent events in adult patients with t(8;21)(q22;q22) acute myeloid leukemia. Leukemia 2017; 31: 1278–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-taj-10.1177_2040622321997259 for KDM4C contributes to cytarabine resistance in acute myeloid leukemia via regulating the miR-328-3p/CCND2 axis through MALAT1 by Lu Xue, Chunhuai Li, Jin Ren and Yue Wang in Therapeutic Advances in Chronic Disease