Abstract
Endogenous homeostasis and peripheral tissue metabolism are disrupted by irregular fluctuations in activation, movement, feeding and temperature, which can accelerate negative biological processes and lead to immune reactions, such as rheumatoid arthritis (RA) and osteoarthritis (OA). This review summarizes abnormal phenotypes in articular joint components such as cartilage, bone and the synovium, attributed to the deletion or overexpression of clock genes in cartilage or chondrocytes. Understanding the functional mechanisms of different genes, the differentiation of mouse phenotypes and the prevention of joint ageing and disease will facilitate future research.
Keywords: cartilage, circadian clock, gene, osteoarthritis, phenotype
1. INTRODUCTION
Osteoarthritis (OA) is a highly prevalent rheumatic musculoskeletal disorder, encompassing progressive, inflammatory and immunological changes affecting joint structures.1 The clinical features of OA include cartilage loss, increased subchondral bone thickness, tidemark replication, decreased subchondral trabecular bone mass and bone marrow lesions (BML).2, 3 Endogenous biological clocks determine daily, monthly or annual rhythms in biological processes. Cells with endogenous self‐excited oscillations, caused by molecular fluctuations produced by a series of clock genes, form the basis of circadian rhythms. The classical biological clock pathway is comprised of three transcription‐translation feedback loops (TTFL) in mammals (Figure 1). A heterodimer is the core element of TTFLs, which is formed by BMAL1 (brain and muscle ARNT‐like 1/Arntl) and NPAS2 (Neuronal PAS domain protein 2) and/or CLOCK (Circadian locomotor output cycles kaput).4, 5 Additional proteins in the loops such as Cryptochrome (CRYs), Period (PERs), REV‐ERBs, retinoic acid receptor‐related orphan receptors (RORs) and D‐box binding protein (DBP) are synthesized at specific times of the day, accumulate and degrade in the cytoplasm.
FIGURE 1.
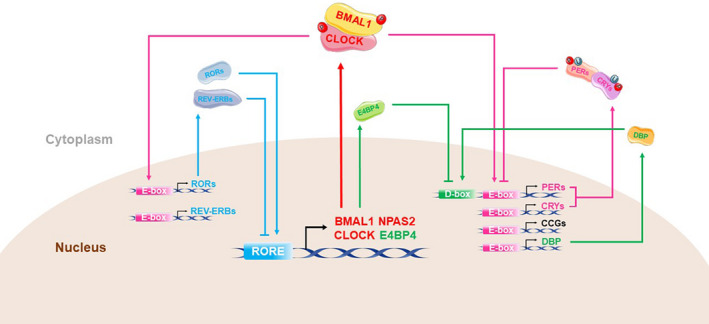
Circadian clock transcription‐translation feedback loops. In primary TTFL, phosphorylated BMAL1:CLOCK/NPAS2 is translocated into the nucleus and initiates the transcription of related genes by binding to the promoter regions E‐box (5′‐CACGTG‐3′) of several genes, including the core clock genes Cryptochrome (CRYs), Period (PERs), REV‐ERBs and retinoic acid receptor‐related orphan receptors (RORs). The PER and CRY proteins accumulate in the cytoplasm, enter the nucleus and inhibit the transcription and phosphorylation of BMAL1 and CLOCK. Consequently, further production of clock proteins in inhibited. In another feedback pathway, RORs and REV‐ERBs proteins competitively bind to ROR elements (ROREs) near the BMAL1 promoter. While REV‐ERBs inhibit the transcription of BMAL1, RORs promote the process. As such, REV‐ERBs show a strong circadian rhythm that is aligned with the rhythm of BMAL1. Additionally, D‐box binding protein (DBP), regulated by E‐box and the mammalian transcription factor E4 binding protein 4 (E4BP4; regulated by ROREs), accommodate the expression of PER gene through D‐box and, in turn, affect E‐box activity.6 Overall, these three cis‐acting elements coordinate a circadian cycle over approximately 24 h
A vast array of studies has elucidated specific genes or external factors that disrupt the body's natural clock. The systemic or specific deletion of classic clock genes in the biological clock's TTFLs and the artificial disruption of the sleep‐wake cycle, causes significant alterations in the human body, including changes in body mass and composition, impaired mental status, growth retardation, premature ageing and cardiovascular disease.7, 8, 9, 10 In this review, we summarize the integral articular phenotypic variations and the disordered expression of molecular oscillations caused by the disruption of representative clock genes throughout the whole body or within cartilage. These insights provide valuable clues regarding the biological clock's involvement in maintaining cartilage and joint developmental, ageing, and metabolic processes, and healthy matrix homeostasis.
2. EVIDENCE OF CIRCADIAN CLOCK IN CARTILAGE
The circadian rhythm of peripheral tissues can be regulated by specific inputs from external factors, such as eating, temperature, light and physical exercise. These factors provide coordinated input to circadian rhythmicity through crosstalk between metabolic process and neural circuits.11, 12, 13 In 1962, Simmons described diurnal variations in cell proliferation and bone growth plate metabolism.14, 15 Soon after, the metabolic rhythms of the epiphysis and condylar cartilage were also studied in detail.16, 17, 18, 19, 20, 21, 22 Additionally, research reported that, in the absence of alterations in environmental factors, the thickness and volume of articular cartilage also vary from day to night.23, 24 Collectively, these findings suggest the possibility of an autonomously controlled clock in cartilage. A recent series of PER2::Luc clock reporter mouse model studies more directly demonstrated the autonomic functioning of the circadian clock in cartilage and cartilage explants in vivo.25, 26, 27 With the emergence of DNA microarrays, mass spectrometry and other advanced molecular biological technologies, more clock genes have been detected in cartilage. More than 600 genes and 145 proteins expressed in cartilage have periodic rhythmicity controlled by clock genes.28, 29, 30, 31
3. CARTILAGE AND BONE PHENOTYPES OF MOUSE CLOCK GENE MUTATIONS
3.1. BMAL1
3.1.1. BMAL1 may not be necessary for embryonic limb cartilage development and bone growth
Many studies have shown that BMAL1 deficiency significantly reduces early embryonic development and the implantation potential of female mice.32, 33, 34 However, in normal mouse embryo development, BMAL1 might not be required for cartilage and long bone development. Yu et al using Alcian blue and Alizarin red staining analysis found insufficient cranial cartilage calcification and smaller and shorter mandibular condyle phenotypes in E14.5‐E18.5 embryos of global BMAL1 deletion and BMAL1fl/fl Twist‐Cre mice.35 This finding suggests that BMAL1 occupies a crucial position in the chondrogenesis and entochondrostosis processes of the mandibular condyles. However, the abnormal progression of endochondral ossification in the limbs was not mentioned. To further understand its role, Ma et al specifically ablated BMAL1 from the cartilage of E13.5‐E18 mice. They found no abnormalities in body mass, bone length (femur, tibia and humerus) and growth plates compared to wide‐type (WT) mice,36 suggesting that BMAL1 is not necessary for the elongation of long bones (Table 1). The sharp contrast between the two studies may be attributed to the different stages of endochondral osteogenesis between the mandibular condyle and limb bones.
TABLE 1.
Summary of phenotypes of genome‐editing mouse models in cartilage circadian clock disruption. Description of mouse models in the foetus, adolescence and adult stage in terms of alterations of body height/weight, cartilage/growth plate and bone compared with age‐matched wide‐type mice
| References | Mice | Detection of the Δallele | Age | General | Location | Visual observation and pathological | Protein | |
|---|---|---|---|---|---|---|---|---|
| Cartilage tissue/growth plate | Bone and other tissues | Cartilage tissue | ||||||
| Foetus stage | ||||||||
| Yu, et al35 | Bmal1 −/− | Exon 5 and part of exon 4 | E14.5‐18.5 | Body size decreased | Mandibular condyle | Chondrogenesis decreased | Alizarin red staining decreased | COL2A1, COL10A1, ACAN, SOX9, CyclinD1 and Ki67 decreased; TUNEL‐positive cells increased |
| Bmal1fl/fl; Twist2‐Cre | ||||||||
| Ma, et al36 | TamCart Bmal1fl/fl | Not mentioned | E13.5‐18.5 | No abnormal | ||||
| Adolescence stage | ||||||||
| Bunger,MK., et al 46 | Bmal1 −/− | Exon 5 and part of exon 4 | W6‐11 | No abnormal | ||||
| Yu, et al 35 | Bmal1 −/− | Exon 5 and part of exon 4 | W4‐20 | —— | Mandibular condyle | Shorter PZ and HZ; number of chondrocytes per column in PZ and HZ decreased | BMD, BV/TV, Tb. Th and Tb.N decreased; Tb. Sp increased | COL2A1, ACAN, COL10A1 and Ki67 decreased; TUNEL‐positive cells increased |
| Suyama,K., et al 47 | Bmal1 −/− | Exon 5 and part of exon 4 | W10 | —— | Lumbar spine | Disc height and DHI decreased; lower matrix‐to‐cell ratio in NP; hyperplasia in AF | Vertebral bone BV, TV, BV/TV, Tb.N and Tb. Th decreased; Tb. Sp increased | —— |
| Ma, et al 36 | TamCart Bmal1fl/fl | Not mentioned | W4‐6 | Body weight and length decreased | Hindlimb | Shorter PZ; larger HZ | Shorter | BrdU‐positive cells, COL10A1, VEGF, HIF1α, and Bcl‐2 decreased; TUNEL‐positive cells, Caspase‐3, MMP13 and Runx2 increased |
| Dudek, et al 44 | Col2α1‐Bmal1 −/− | Exon 8 | W4‐12 | No abnormal | Knee joint | Cell apoptosis near the tidemark; loss of chondrocytes and voids in ECM | No abnormal | —— |
| Takarada, et al 45 | Bmal1 −/− | Exon 6‐8 | W3‐15 | Body length/weight decreased | Rib | —— | —— | IHH decreased; no abnormal with Col II, Col X, Sox9, Runx2 |
| α1(II)‐collagen‐Cre; Bmal1fl/fl | Hindlimb | —— | Shorter | IHH decreased | ||||
| Adult stage | ||||||||
| Bunger, et al46 | Bmal1 −/− | Exon 5 and part of exon 4 | W20‐40 | Body weight decreased; abnormal gait and posture | Tarsal joint | —— | Calcified nodules; calcaneal tendon calcification; osteocalcin increased; no abnormal in BMD | —— |
| Vertebrae | Proliferative bone bridging from the costal cartilage to the sternum; increased matrix deposition and bone proliferation in perichondrium | Bridging ankylosis and osteophyte in intervertebral and lumbar joints; no abnormal in BMD | —— | |||||
| Knee joint | No abnormal | Tendons and ligaments ectopic calcification/ossification; no abnormal in BMD | —— | |||||
| Schroder, et al 54 | Bmal1 −/− | Exon 8 | W20‐22 | —— | Hindlimb and Ribcage | Alcian Blue decreased | Distal tibia thicker; calcification of the calcaneal tendon | —— |
| iMS‐Bmal1 −/− | W75‐79 | Hair loss; dermatitis increased; kyphosis | ||||||
| Dudek, et al52 | Col2α1‐Bmal1 −/− | Exon 8 | W24‐48 | —— | Intervertebral disc | Thinner; gradual disappearance of CEP; bone bridges; fibrosis | Ectopic ossification | Adamts1, Adamts5, Adamts15 and Follistatin increased |
| Hand, et al56 | Col6α1‐Bmal1 −/− | Exon 8 | W12‐36 | —— | Ankle joint | —— | Density increased; calcaneal spur; calcification between the metatarsal and phalangeal bones; chondroid metaplasia at the junction of the synovium, joint capsule and enthesis repair tissue | —— |
| Intervertebral disc | Calcification | —— | —— | |||||
| Yuan, et al 58 | ClockΔ19 | Exon 19 | W24 | Knee joint | —— | Safranin‐O staining decreased; cartilage and meniscus damage histologic scores increased | —— | —— |
| Kc, et al 11 | ClockΔ19/Δ19 | Exon 19 | W7‐9 | Knee, shoulder, spine disc and facet joints | No abnormal | |||
PZ: Proliferative zone; HZ: Hypertrophic zone; BMD: Bone mineral density; BV/TV: bone volume/total volume; Tb.Th: Trabecular thickness; Tb.N: Trabecular number; Tb.Sp: Trabecular separation; CEP: Cartilaginous end plate; IVD: Intervertebral disc; DHI: Disc height index; NP: Nucleus pulposus; AF: Annulus fibrosus; ECM: Extracellular matrix.
The primary synovial joint articular cartilage grows from chondrocytes in the central layer of the epiphyseal plate, gradually differentiated from E11.5. Its shape and biosynthetic properties remain invariable throughout its lifetime.37, 38 Inversely, condyle cartilage, as secondary cartilage, does not develop significantly until E15. Growth starts from mesenchymal tissue covering the prenatal or postnatal condyle, and it easily adapts to alterations in the environment.39, 40 In contrast to the significant effect of BMAL1 on the postnatal endochondral ossification of bone (detailed below), studies have shown that these gene products may not yet form a functional circadian feedback loop during the early stages of development.41, 42, 43 However, more research is needed to support this proposal.
3.1.2. BMAL1 significantly affects adolescent cartilage and bone development
Although the expression of BMAL1 in cartilage and bone gradually decreases with body development, its inverse role is indispensable.44 Genome‐wide RNA sequencing shows the differential expression of chondrogenesis‐related genes, and Hedgehog pathway‐related proteins were highest during adolescence (4‐6 weeks) compared to postpubescence (8‐10 weeks).35 BMAL1 and CLOCK were highly expressed by proliferous zone (PZ) or prehypertrophic to hypertrophic zone (HZ) chondrocytes in the growth plates of WT mice,35, 45 which alluded that BMAL1 was closely related to chondrogenesis.
The study of mice with global BMAL1 deletion was pioneered by Bunger et al.46 No significant abnormal changes were observed in body mass, body length, degenerated bone and joint pathology before 15 weeks of age in MOP3−/− mice. Alternatively, Bunger et al suggested that MOP3 was critical for maintaining joint homeostasis in adulthood. In another study, body mass and length, as well as the longitudinal length of the tibia and femur, of BMAL1−/− mice were significantly reduced from three weeks of age, indicating that BMAL1 influences the elongation of long bones, but it does not indicate obvious cartilage pathology.45 However, in a study of the mandibular condyle, Yu et al 35 reported that cartilage thickness and matrix were significantly reduced across the whole‐body BMAL1‐deleted mice, accompanied by reduced chondrocyte proliferation and increased apoptosis. These findings are completely contrary to the previous conclusions, possibly due to the reasons proposed above. Using microcomputed tomography to evaluate intervertebral disc height in mice, the annulus fibrosus (AF) tissue of BMAL1−/− mice was shown to be hyperplasic, the lumbar vertebrae and intervertebral discs were significantly smaller and thinner, and the vertebral bone parameters were also significantly reduced.47 A possible explanation for the distinct cartilage pathology at these different anatomical sites may be that articular cartilage maintains a low‐level of metabolic synthesis and proliferation after birth. In contrast, secondary cartilage or intervertebral discs are sensitive to external stimuli and mechanical pressures and can deform flexibly.
Several transgenic mice variants with specific BMAL1 deletion from the cartilage have been constructed to investigate its effect on cartilage homeostasis. Dudek et al established adult chondro‐specific BMAL1‐KO mice44 and discovered significant degeneration of knee cartilage, extracellular matrix (ECM), cell damage, and tidemark loss, commencing from the eighth week; the observations were restricted to articular cartilage and did not include the synovium or its affiliated ligaments. However, no distinct decreases in body mass and length were observed, which was inconsistent with another study of αⅠ(Ⅱ)‐collagen‐Cre; Bmal1fl/fl mice.45 The conspicuous discrepancy may be explained by differences in the type II collagen a1 (COL2A1) promoter targeting strategies of the two groups of COL2A1‐CRE transgenic deletion strains.48 Ma et al also reported improvements in skeletal lengths 36 and suggested the increase in apoptotic cells, with reductions of collagen (COL), SRY‐Box transcription factor 9 (SOX9), vascular endothelial growth factor (VEGF), other basic components of the cartilage matrix and development related proteins, results in the disappearance of the circadian rhythm. These observations revealed that the deletion of BMAL1 hindered the mineralization and calcification of normal hypertrophic chondrocytes and delayed angiogenic‐osteogenic coupling. These outcomes appear to be closely linked with the precise regulation of the Indian hedgehog (IHH) protein, a pivotal member of the developmental regulatory Hedgehog family.35, 45 The BMAL1:CLOCK heterodimer complex could bind to the E‐box (CACGTG) in the promoter region of IHH and hedgehog ligand 1, binding to Patched (PTCH1), a key receptor of hedgehog pathway. This would regulate the sequential differentiation of chondrocytes, which could be influenced by PER1 and parathyroid hormone (PTH).45 Early in the mouse lifecycle, the hedgehog pathway activator, SAG, partially saves the mandible shortness phenotype caused by BMAL1 deficiency rather than in adulthood.35 This is consistent with the rate of cartilage repair during development being much higher in the young compared to adults and the elderly, suggesting the reversibility of cartilage formation. Transforming growth factor‐β (TGF‐β) and the nuclear factor of activated T cells (NFAT) signalling also play a non‐negligible role in maintaining stable growth and ageing of articular cartilage 49, 50 by RNA sequencing in cKO mice. Consistently, TGF‐β signalling has also been significantly altered by an in vitro RNA‐seq of knock‐down BMAL1 and NR1D1.51
3.1.3. Lack of BMAL1 accelerates ageing and inflammation of limbs and joints in adult mice
Internal body factors, such as ageing, alter the circadian rhythm in cartilage manifested by classical CLOCK, BMAL1, NR1D1 expression, causing changes in amplitude inversely proportional to age (rather than other clock proteins).25, 35, 44, 51, 52 As mentioned above, Bunger et al found that after twenty weeks of age, Mop3−/− mice showed abnormal phenotypes, including reduced body mass, anorexia, dehydration and abnormal gait.46 Typical non‐inflammatory joint diseases were found through radiological and histological analysis, and there was enhanced heterotopic ossification of hind limb joints, but there were no significant changes in global bone density. Inversely, the articular cartilage of the tibia and femur, and other soft structures (eg muscles), seemed to remain intact and uneroded. Similarly, degeneration of the lumbar intervertebral disc (IVD) was observed in Col2a1‐BMAL1 cKO mice after six months, and extensive denaturation, ecstatic ossification and increased fibrosis at twelve months.52 These outcomes suggest that BMAL1 significantly disturbs bone homeostasis during the ageing process.
Consistent with previous research,53 phenotypic changes in inducible skeletal muscle‐specific deletion of BMAL1 in adult mice aged 16‐20 weeks were not limited to the muscle transition to an ageing phenotype.54 They reported some phenotypes similar to those demonstrated by Bunger et al,46 including a limited range of movement, spinal and tail deformity, and increased tendon and ligament calcifications. Complementarily, bone calcification and decreased cartilage staining, similar to the OA‐like joint phenotypes induced by systemic BMAL1‐KO mice, were also found in this study. An additional viewpoint was thus proposed that skeletal muscle systems might share a set of peripheral clock regulators enabling synchronization.55 Dudek et al provided further evidence for this hypothesis by comparing the rhythm gene sequence of IVD, cartilage and tendon and identified eight core clock genes (including ARTNL, PER2 and Dbp) among the rare number of overlapping genes.52
By ablating BMAL1 from articular mesenchymal cells expressing COL6A1,56 overall joint alterations further encompassed changes in fibroblast‐like synoviocytes (FLS) and MHCⅡ macrophages in the synovium, owing to FLS and chondrocytes originated from the progenitor cells. Similar to Bunger et al, significant thickening of the ankle joint synovium, cartilage and bone hyperplasia and chondroid metaplasia were interspersed at the tendon and ligament insertion sites.46 However, no further variations in the knee joint were recorded. These phenotypes provide additional evidence of the biological clock's central role in maintaining healthy joints, as the FLS can secrete a myriad of inflammatory factors leading to RA. Likewise, the synovial membrane is the only tissue in the joint cavity filled with blood vessels. As such, inflammatory factors may damage adjacent tissues through the circulatory system.
3.2. Other clock genes
As another pivotal member of the BMAL1:CLOCK heterodimer, CLOCK also plays a noteworthy role in maintaining normal articular cartilage. Large‐scale DNA microarray analysis showed that CLOCK might be a mechanically sensitive clock gene, as it was significantly up‐regulated following mechanical stress.30 According to Safranin‐O staining, abnormal weight loss and cartilage degeneration phenotypes, similar to BMAL1 deletion, were found in CLOCKΔ19 mice, where the nuclear factor κB (NF‐κB) signalling was activated to trigger chronic inflammation.57, 58 This was inconsistent with the vacant study of Kc et al,11 who found no significant pathological differences in Safranin‐O fast green staining in the cartilage of CLOCK and Tau‐mutant mice. This might be due to age differences, as the genetic background was the same between the studies. The loss of CLOCK gene fragments may alter cartilage phenotypes in adult mice. In addition to cartilage degradation, shorter tibiae and lower bone mass could also be induced by CLOCK mutation.59 The latest research demonstrated that the motor ability of 18‐month‐old mice, promoted by injecting lentiviral vectors encoding CLOCK, to enhance cartilage regeneration, decreased ageing markers (Cdkn1a and Cdkn2a), as well as activating genes involved in cartilage development.60 These results imply that the CLOCK gene supports cartilage rejuvenation and alleviates ageing. However, the importance of other clock genes, such as CRYs and PERs, in maintaining joint phenotypes has not been studied.
4. MOLECULAR EXPRESSION DISORDER CAUSED BY DELETION OF SPECIFIC CLOCK GENES IN CARTILAGE AND CHONDROCYTES
4.1. BMAL1
As a core clock factor in the circadian rhythm feedback loop, BMAL1 is crucial for governing the prosodic expression of other rhythm genes and internal or external destructing factors influencing cartilage. RNA‐seq and PER2::Luc bioluminescence imaging confirmed the loss of circadian rhythm in the cartilage of BMAL1‐KO/cKO mice.44, 52, 56 Systemic or cartilage‐specific elimination of BMAL1 impaired the circadian rhythm of postpartum mice, showing significant loss of prosodic expression of several genes associated with cartilage, along with a distinct elevation in the expression of Matrix metalloproteinase 3 (MMP3), MMP13, VEGF and Runt‐related transcription factor 2 (Runx2).9, 36 Of note, BMAL1 was expressed irregularly in chondrocytes cultured in normal and BMAL1‐cKO mice 36 compared to other studies.28, 51 These two opposing manifestations may be due to a discrepancy in the way the chondrocytes were treated or because different species of organisms were used. siBMAL1 enhanced the reduction of proteoglycan and COL2A1 caused by IL‐1β and increased chondrogenic degrading enzymes,61 which was consistent with the trends observed in BMAL1−/− mouse chondrocytes.45 Meanwhile, Sirt1, NAD, and NAMPT were significantly dampened by knocking down BMAL1 in light of the combination of BMAL1 and Sirt1,61 providing an effective basis for the disorders of circadian rhythm seen with ageing.
A significant decrease in PER1 and REV‐ERBα expression was observed in Bmal1−/− mice, or chondrocytes and FLS, with elevated levels of PER2 and CRY1, but not CRY2.36, 45, 56 BMAL1 and PER2/CRY1 were consistently expressed inversely,62 evidenced by robust oscillations in the expression of BMAL1::Luc reporter in SW‐1353 over several days, in antiphase to PER2::Luc reporter.25, 29 Remarkably, BMAL1 excision deleted both BMAL1 and BMAL2, which could not substitute BMAL1 to maintain the expression oscillation of most genes in the tissues.63 These results allude to the consistent regulatory role of BMAL1 on other clock genes. In contrast, other studies have shown that BMAL1 was incapable of mediating RORA, RORC and RBX1.51
4.2. Other clock genes
4.2.1. CLOCK
There have been no obvious rhythmic fluctuations in the CLOCK gene detected in mouse cartilage tissues, such as rib growth plates 45 and intervertebral discs.64 It is possible that NPAS2 is combining with BMAL1 instead of the non‐oscillating CLOCK. The oscillations of protein expression in xiphoid tissue cultures disappeared after CLOCK mutation,25 leading to the nuclear translocation of NF‐κB, acetylation inhibition and phosphorylation activation.58 These observations shed light on the role clock genes play in the probability of developing OA.
4.2.2. PERs
Overexpressing PER1 attenuates ALP, COLⅡ, COLⅩ, SOX6 and IHH expression in ATDC5 cell lines,45, 65 which are pivotal to chondrogenesis. Thus, PER1 could inhibit the biological processes of chondrocyte differentiation to osteoblasts or hypertrophic chondrocytes. Of note, PER1 was not sensitive to these genes in primary costal chondrocytes,45 which explains the diverse roles of clock genes in the different stages of cartilage formation. Rong et al found that SOX9 reduction and overexpression of chondrodegradation‐related genes MMP13 and ADAMTS5 caused by IL‐1β (but not BMAL1) were significantly neutralized by PER2 knock‐down in human chondrocytes.66 This reflects the irreplaceability of BMAL1 in the biological clock pathway.
4.2.3. REV‐ERBs
Although they are members of the secondary circadian clock pathway feedback loop, REV‐ERBs in chondrocytes are infrequently studied. After suppressing REV‐ERBα in chondrocytes, Yang et al demonstrated the significant activation of BMAL1 and Sirt1, produces a similar inflammation‐neutralizing effect as the overexpression of BMAL1 (mentioned above).61 As such, the reduction of REV‐ERBα results in the retention of BMAL1 in the cytoplasm in the clock pathway feedback loop. However, in the treatment of specific clock genes, drugs targeted at REV‐ERBs and RORs have shown remarkable efficacy against diabetes, atherosclerosis, autoimmune diseases and cancer.67 Das et al confirmed that REV‐ERB agonist SR9009 alleviates cartilage matrix loss due to OA surgery and significantly reduces pain through intra‐articular injection compared with partial medial meniscectomy (PMM) after 12 weeks of drug administration.68 In addition, SR9009 and CRY1/2 agonist KL001 reduce abnormal collagen fibre synthesis and collagen accumulation.69 These findings have proved the importance of the circadian rhythm in balancing collagen fibre synthesis and degradation.
5. FUTURE RESEARCH DIRECTIONS
Currently, most clock genes are involved in maintaining cartilage health, yet several new challenges have emerged that require further research. First of all, a limitation of the experimental identification and verification methods to elucidate gene function exists; the pathological phenotypes of model mice cannot fully reflect the variations of in vivo function. This is because the level of aspartate aminotransferase (AST), alanine transaminase (ALT) and urea nitrogen (BUN) in the blood of MOP3−/− mice aged about four months was significantly higher than that of their MOP3+/‐ or MOP3+/+ siblings. In contrast, no remarkable pathological changes were observed in the livers, hearts, lungs and kidneys.70 Therefore, further studies into the cellular and molecular mechanisms of temporal and spatial gene expression profiles, along with predictions of their functions, need to be explored. Additionally, the biological clock has cellular specificity. Many researchers have proposed variant ways (differing between cells) in clock genes interact with other genes.36, 47, 71 Furthermore, recent research has identified that some genes expressed in periodic 24‐27 h oscillations after synchronization in isolated liver and skin tissues of BMAL1−/− might be due to ETS transcription factor and PRDX protein.63 These observations highlight the complex and sophisticated regulatory role of BMAL1 function in transcription, post‐transcription, translation and post‐translation of protein expression in different tissues. But it has not yet been reported in cartilage. Our subsequent work will focus on the decisive role of specific core clock genes in inflammatory or degenerative arthropathies or in combination with other proteins to maintain homeostasis in healthy cartilage.
CONFLICT OF INTEREST
The authors confirm that there are no conflicts of interest.
AUTHOR CONTRIBUTIONS
Xiaopeng Song: Conceptualization (lead); Investigation (lead); Resources (lead); Writing‐original draft (lead); Writing‐review & editing (lead). Hui Bai: Formal analysis (equal); Resources (equal); Writing‐review & editing (equal). Xinghua Meng: Software (lead); Writing‐review & editing (equal). Jianhua Xiao: Supervision (equal); Validation (equal). Li Gao: Funding acquisition (lead); Supervision (lead); Validation (lead).
ACKNOWLEDGEMENTS
This work was supported by funding from The National Key Research and Development Program of China (Project No. 2017YFD0502200) and Applied Technology Research and Development Plan of Heilongjiang, China (GA18B203).
Song X, Bai H, Meng X, Xiao J, Gao L. Drivers of phenotypic variation in cartilage: Circadian clock genes. J Cell Mol Med. 2021;25:7593–7601. 10.1111/jcmm.16768
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article.
REFERENCES
- 1.Morris JL, Letson HL, Gillman R, et al. The CNS theory of osteoarthritis: Opportunities beyond the joint. Semin Arthritis Rheu. 2019;49(3):331‐336. [DOI] [PubMed] [Google Scholar]
- 2.Yuan XL, Meng HY, Wang YC, et al. Bone‐cartilage interface crosstalk in osteoarthritis: Potential pathways and future therapeutic strategies. Osteoarthr Cartilage. 2014;22:1077‐1089. [DOI] [PubMed] [Google Scholar]
- 3.Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum. 2012;64:1697‐1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gekakis N, Staknis D, Nguyen HB, et al. Role of the clock protein in the mammalian circadian mechanism. Science (New York, NY). 1998;280:1564‐1569. [DOI] [PubMed] [Google Scholar]
- 5.Hogenesch JB, Gu YZ, Moran SM, Shimomura K, Bradfield CA. The basic helix‐loop‐helix‐pas protein MOP9 is a brain‐specific heterodimeric partner of circadian and hypoxia factors. J Neurosci 2000;20:RC83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowell IG. E4BP4/NFIL3, a par‐related bZIP factor with many roles. BioEssays. 2002;24:1023‐1029. [DOI] [PubMed] [Google Scholar]
- 7.Doi M. Circadian clock‐deficient mice as a tool for exploring disease etiology. Biol Pharm Bull 2012;35:1385‐1391. [DOI] [PubMed] [Google Scholar]
- 8.Yang G, Zhang J, Jiang T, et al. Bmal1 deletion in myeloid cells attenuates atherosclerotic lesion development and restrains abdominal aortic aneurysm formation in hyperlipidemic mice. Arterioscler Thromb Vasc Biol. 2020;40:1523‐1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J, Zhou X, Tang Q, et al. Bmal1 deficiency contributes to mandibular dysplasia by upregulating mmp3. Stem Cell Rep. 2018;10:180‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubrovsky YV, Samsa WE, Kondratov RV. Deficiency of circadian protein clock reduces lifespan and increases age‐related cataract development in mice. Aging (Albany NY). 2010;2:936‐944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kc R, Li X, Voigt RM, et al. Environmental disruption of circadian rhythm predisposes mice to osteoarthritis‐like changes in knee joint. J Cell Physiol. 2015;230:2174‐2183. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Berenbaum F. Osteoarthritis: When chondrocytes don't wake up on time. Arthritis Rheum. 2013;65:2233‐2235. [DOI] [PubMed] [Google Scholar]
- 13.Tahara Y, Shibata S. Entrainment of the mouse circadian clock: Effects of stress, exercise, and nutrition. Free Radic Biol Med. 2018;119:129‐138. [DOI] [PubMed] [Google Scholar]
- 14.Simmons DJ. Diurnal periodicity in epiphyseal growth cartilage. Nature. 1962;195:82‐83. [DOI] [PubMed] [Google Scholar]
- 15.Simmons DJ, Nichols G Jr. Diurnal periodicity in the metabolic activity of bone tissue. Am J Physiol. 1966;210:411‐418. [DOI] [PubMed] [Google Scholar]
- 16.Simmons DJ. Periodicity of S35 uptake in rat femurs. Experientia. 1964;20:137‐138. [DOI] [PubMed] [Google Scholar]
- 17.Simmons DJ. Daily rhythm of S32 incorporation into epiphyseal cartilage in mice. Experientia. 1968;24:363‐364. [DOI] [PubMed] [Google Scholar]
- 18.Bahorsky MS, Bernardis LL. Circadian fluctuations in tibia cartilage. Assayable pituitary homogenates of fed and starved weanling female rats. Experientia. 1969;25:755‐756. [DOI] [PubMed] [Google Scholar]
- 19.Oudet C, Petrovic A. Effect of active retrodisplacement on the rate of growth of the condylar cartilage in young rats in circadian and annual growth: Significance of changes in the direction of condylar growth. Orthod Fr. 1976;47:15‐26. [PubMed] [Google Scholar]
- 20.Oudet C, Petrovic A. Growth rhythms of the cartilage of the mandibular condyle: Effects of orthopaedic appliances. Int J Chronobiol. 1978;5:545‐564. [PubMed] [Google Scholar]
- 21.Russell JE, Grazman B, Simmons DJ. Mineralization in rat metaphyseal bone exhibits a circadian stage dependency. Proc Soc Exp Biol Med. 1984;176:342‐345. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Z, Wang Z, Lei X, Yang X, Chen Y. Circadian rhythm of PGE2 level in plasma and condylar cartilage of young growing rats. Hua Xi Yi Ke Da Xue Xue Bao. 1997;28:23. [PubMed] [Google Scholar]
- 23.Waterton JC, Solloway S, Foster JE, et al. Diurnal variation in the femoral articular cartilage of the knee in young adult humans. Magn Reson Med. 2000;43:126‐132. [DOI] [PubMed] [Google Scholar]
- 24.Coleman JL, Widmyer MR, Leddy HA, et al. Diurnal variations in articular cartilage thickness and strain in the human knee. J Biomech. 2013;46:541‐547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gossan N, Zeef L, Hensman J, et al. The circadian clock in murine chondrocytes regulates genes controlling key aspects of cartilage homeostasis. Arthritis Rheum. 2013;65:2334‐2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoo S‐H, Yamazaki S, Lowrey PL, et al. Period2: Luciferase real‐time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339‐5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okubo N, Minami Y, Fujiwara H, et al. Prolonged bioluminescence monitoring in mouse ex vivo bone culture revealed persistent circadian rhythms in articular cartilages and growth plates. PLoS One. 2013;8:e78306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinoi E, Ueshima T, Hojo H, Iemata M, Takarada T, Yoneda Y. Up‐regulation of per mrna expression by parathyroid hormone through a protein kinase a‐creb‐dependent mechanism in chondrocytes. J Biol Chem. 2006;281:23632‐23642. [DOI] [PubMed] [Google Scholar]
- 29.Honda KK, Kawamoto T, Ueda HR, et al. Different circadian expression of major matrix‐related genes in various types of cartilage: Modulation by light‐dark conditions. J Biochem. 2013;154:373‐381. [DOI] [PubMed] [Google Scholar]
- 30.Kanbe K, Inoue K, Xiang C, Chen Q. Identification of clock as a mechanosensitive gene by large‐scale DNA microarray analysis: Downregulation in osteoarthritic cartilage. Mod Rheumatol. 2006;16:131‐136. [DOI] [PubMed] [Google Scholar]
- 31.Dudek M, Angelucci C, Pathiranage D, et al. Circadian time series proteomics reveals daily dynamics in cartilage physiology. Osteoarthr Cartilage. 2021;29:739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boden MJ, Varcoe TJ, Voultsios A, Kennaway DJ. Reproductive biology of female BMAL1 null mice. Reproduction. 2010;139:1077‐1090. [DOI] [PubMed] [Google Scholar]
- 33.Xu J, Li Y, Wang Y, Xu Y, Zhou C. Loss of BMAL1 decreases oocyte fertilization, early embryo development and implantation potential in female mice. Zygote. 2016;24:760‐767. [DOI] [PubMed] [Google Scholar]
- 34.Ratajczak CK, Boehle KL, Muglia LJ. Impaired steroidogenesis and implantation failure in BMAL1‐/‐ mice. Endocrinology. 2009;150:1879‐1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu S, Tang Q, Xie M, et al. Circadian BMAL1 regulates mandibular condyle development by hedgehog pathway. Cell Prolif. 2020;53:e12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma Z, Jin X, Qian Z, et al. Deletion of clock gene BMAL1 impaired the chondrocyte function due to disruption of the HIF1alpha‐VEGF signaling pathway. Cell Cycle (Georgetown, Tex). 2019;18:1473‐1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song X, Hu H, Zhao M, Ma T, Gao L. Prospects of circadian clock in joint cartilage development. FASEB J. 2020;34: 14120‐14135. [DOI] [PubMed] [Google Scholar]
- 38.Martin P. Tissue patterning in the developing mouse limb. Int J Dev Biol. 1990;34:323‐336. [PubMed] [Google Scholar]
- 39.Shen G, Darendeliler MA. The adaptive remodeling of condylar cartilage – a transition from chondrogenesis to osteogenesis. J Dent Res. 2005;84:691‐699. [DOI] [PubMed] [Google Scholar]
- 40.Hirouchi H, Kitamura K, Yamamoto M, et al. Developmental characteristics of secondary cartilage in the mandibular condyle and sphenoid bone in mice. Arch Oral Biol. 2018;89:84‐92. [DOI] [PubMed] [Google Scholar]
- 41.Weger M, Weger BD, Diotel N, et al. Real‐time in vivo monitoring of circadian E‐box enhancer activity: A robust and sensitive zebrafish reporter line for developmental, chemical and neural biology of the circadian clock. Dev Biol. 2013;380:259‐273. [DOI] [PubMed] [Google Scholar]
- 42.Dekens MPS, Whitmore D. Autonomous onset of the circadian clock in the zebrafish embryo. EMBO J. 2008;27:2757‐2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dierickx P, Vermunt MW, Muraro MJ, et al. Circadian networks in human embryonic stem cell‐derived cardiomyocytes. EMBO Rep. 2017;18:1199‐1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dudek M, Gossan N, Yang N, et al. The chondrocyte clock gene Bmal1 controls cartilage homeostasis and integrity. J Clin Invest. 2016;126:365‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takarada T, Kodama A, Hotta S, et al. Clock genes influence gene expression in growth plate and endochondral ossification in mice. J Biol Chem 287:36081‐36095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bunger MK, Walisser JA, Sullivan R, et al. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal‐1 locus. Genesis. 2005;41:122‐132. [DOI] [PubMed] [Google Scholar]
- 47.Suyama K, Silagi ES, Choi H, et al. Circadian factors Bmal1 and RORalpha control HIF‐1alpha transcriptional activity in nucleus pulposus cells: Implications in maintenance of intervertebral disc health. Oncotarget. 2016;7:23056‐23071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doody KM, Bottini N. Chondrocyte clocks make cartilage time‐sensitive material. J Clin Invest. 2016;126:38‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Kraan PM. The changing role of TGFβ in healthy, ageing and osteoarthritic joints. Nat Rev Rheumatol. 2017;13:155‐163. [DOI] [PubMed] [Google Scholar]
- 50.Greenblatt MB, Ritter SY, Wright J, et al. NFATC1 and NFATC2 repress spontaneous osteoarthritis. Proc Natl Acad Sci USA. 2013;110:19914‐19919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akagi R, Fisch KM, Alvarez‐Garcia O, et al. Dysregulated circadian rhythm pathway in human osteoarthritis: NR1D1 and BMAL1 suppression alters TGF‐β and IL‐1β signaling in chondrocytes. Osteoarthr Cartilage. 2017;25(6):943‐951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dudek M, Yang N, Ruckshanthi JP, et al. The intervertebral disc contains intrinsic circadian clocks that are regulated by age and cytokines and linked to degeneration. Ann Rheum Dis. 2017;76:576‐584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age‐related pathologies in mice deficient in Bmal1, the core componentof the circadian clock. Genes Dev. 2006;20:1868‐1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schroder EA, Harfmann BD, Zhang X, et al. Intrinsic muscle clock is necessary for musculoskeletal health. J Physiol. 2015;593:5387‐5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeung C‐YC, Gossan N, Lu Y, et al. Gremlin‐2 is a BMP antagonist that is regulated by the circadian clock. Sci Rep 2014;4:5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hand LE, Dickson SH, Freemont AJ, Ray DW, Gibbs JE. The circadian regulator Bmal1 in joint mesenchymal cells regulates both joint development and inflammatory arthritis. Arthritis Res Ther. 2019;21:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alibhai FJ, Reitz CJ, Peppler WT, et al. Female clockΔ19/Δ19 mice are protected from the development of age‐dependent cardiomyopathy. Cardiovasc Res. 2018;114:259‐271. [DOI] [PubMed] [Google Scholar]
- 58.Yuan G, Xu L, Cai T, et al. Clock mutant promotes osteoarthritis by inhibiting the acetylation of NFkappab. Osteoarthr Cartilage. 2019;27:922‐931. [DOI] [PubMed] [Google Scholar]
- 59.Yuan G, Hua B, Yang Y, et al. The circadian gene clock regulates bone formation via PDIA3. J Bone Miner Res. 2017;32:861‐871. [DOI] [PubMed] [Google Scholar]
- 60.Liang C, Liu Z, Song M, et al. Stabilization of heterochromatin by clock promotes stem cell rejuvenation and cartilage regeneration. Cell Res. 2021;31:187‐205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang W, Kang X, Liu J, et al. Clock gene Bmal1 modulates human cartilage gene expression by crosstalk with Sirt1. Endocrinology. 2016;157:3096‐3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alagha MA, Vago J, Katona E, et al. A synchronized circadian clock enhances early chondrogenesis. Cartilage 2020:1947603520903425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ray S, Valekunja UK, Stangherlin A, et al. Circadian rhythms in the absence of the clock gene. Science (New York, NY). 2020;367:800‐806. [DOI] [PubMed] [Google Scholar]
- 64.Numaguchi S, Esumi M, Sakamoto M, et al. Passive cigarette smoking changes the circadian rhythm of clock genes in rat intervertebral discs. J Orthop Res. 2016;341:39‐47. [DOI] [PubMed] [Google Scholar]
- 65.Le NQ, Binh NT, Takarada T, Takarada‐Iemata M, Hinoi E, Yoneda Y. Negative correlation between PER1 and SOX6 expression during chondrogenic differentiation in pre‐chondrocytic ATDC5 cells. J Pharmacol Sci. 2013;122:318‐325. [DOI] [PubMed] [Google Scholar]
- 66.Rong J, Zhu M, Munro J, et al. Altered expression of the core circadian clock component period2 contributes to osteoarthritis‐like changes in chondrocyte activity. Chronobiol Int. 2019;36:319‐331. [DOI] [PubMed] [Google Scholar]
- 67.Kojetin DJ, Burris TP. REV‐ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov. 2014;13:197‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Das V, Kc R, Li X, et al. Pharmacological targeting of the mammalian clock reveals a novel analgesic for osteoarthritis‐induced pain. Gene. 2018;655:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chang J, Garva R, Pickard A, et al. Circadian control of the secretory pathway maintains collagen homeostasis. Nat Cell Biol. 2020;22:74‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun Y, Yang Z, Niu Z, et al. The mortality of MOP3 deficient mice with a systemic functional failure. J Biomed Sci. 2006;13:845‐851. [DOI] [PubMed] [Google Scholar]
- 71.Kim EJ, Yoo YG, Yang WK, et al. Transcriptional activation of HIF‐1 by RORalpha and its role in hypoxia signaling. Arterioscler Thromb Vasc Biol. 2008;28:1796‐1802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article.


