Abstract
Lipids are major and essential constituents of plant cells and provide energy for various metabolic processes. However, the function of the lipid signal in defence against Verticillium dahliae, a hemibiotrophic pathogen, remains unknown. Here, we characterized 19 conserved stearoyl‐ACP desaturase family proteins from upland cotton (Gossypium hirsutum). We further confirmed that GhSSI2 isoforms, including GhSSI2‐A, GhSSI2‐B, and GhSSI2‐C located on chromosomes A10, D10, and A12, respectively, played a dominant role to the cotton 18:1 (oleic acid) pool. Suppressing the expression of GhSSI2s reduced the 18:1 level, which autoactivated the hypersensitive response (HR) and enhanced cotton Verticillium wilt and Fusarium wilt resistance. We found that low 18:1 levels induced phenylalanine ammonia‐lyase‐mediated salicylic acid (SA) accumulation and activated a SA‐independent defence response in GhSSI2s‐silenced cotton, whereas suppressing expression of GhSSI2s affected PDF1.2‐dependent jasmonic acid (JA) perception but not the biosynthesis and signalling cascade of JA. Further investigation showed that structurally divergent resistance‐related genes and nitric oxide (NO) signal were activated in GhSSI2s‐silenced cotton. Taken together, these results indicate that SA‐independent defence response, multiple resistance‐related proteins, and elevated NO level play an important role in GhSSI2s‐regulated Verticillium wilt resistance. These findings broaden our knowledge regarding the lipid signal in disease resistance and provide novel insights into the molecular mechanism of cotton fungal disease resistance.
Keywords: GhSACPD, Gossypium hirsutum, lipid signal, molecular mechanism, Verticillium wilt resistance
GhSACPD gene family isoforms GhSSI2‐A, GhSSI2‐B, and GhSSI2‐C displayed an important role in regulating 18:1 fatty acid level and negatively regulated cotton disease resistance to Verticillium wilt.

1. INTRODUCTION
To cope with pathogen challenge, plants have developed a complex and multilayered immune system to recognize and combat the invading pathogens (Akira et al., 2006). There exist two types of defence responses that are triggered by the detection of different molecules secreted from pathogens. The first response occurs at the plant cell surface when pathogen‐associated molecular patterns (PAMPs) are detected by pattern‐recognition receptors (PRRs); this is known as PAMP‐triggered immunity (PTI). The other response is effector‐triggered immunity (ETI), which is triggered by effector molecules that are delivered from the pathogen directly into the plant cell and are recognized by the corresponding plant resistance (R) proteins (Nomura et al., 2011; Spoel & Dong, 2012). ETI is often accompanied by the hypersensitive response (HR), a form of programmed cell death (PCD), including generation of reactive oxygen species (ROS), activation of a string of pathogenesis‐related (PR) genes, and accumulation of secondary metabolites like phytoalexins, tannins, and phenolic compounds (Hammond‐Kosack & Jones, 1996; Muthamilarasan & Prasad, 2013).
Plants defend against pathogen infections by activating multiple resistance pathways in which the plant hormones salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) play major signalling roles. Studies of Arabidopsis and other plants showed that the rapid increase of endogenous SA levels after biotrophic or hemibiotrophic pathogen infection can induce a battery of pathogenesis‐related (PR) genes and the activation of systemic acquired resistance (SAR) (Durrant & Dong, 2004; Jiang et al., 2009; Vallad & Goodman, 2004). For example, Verticillium dahliae secretes isochorismatases (VdISC1) that hydrolyse isochorismate and suppress SA accumulation of cotton; VdISC1 deletion mutants produce significantly reduced disease symptoms in cotton (Liu et al., 2014). Cotton CBP60g, SARD1, and GhCBP60b were shown to serve as crucial components that positively regulate V. dahliae resistance by regulating the SA level (Qin et al., 2018). We also revealed that downregulation of GbEDS1, a key gene in SA synthesis, led to a decreased SA level and increased pathogen susceptibility in cotton (Zhang et al., 2017). Another phytohormone that is involved in plant defence is jasmonic acid ( JA), which plays crucial roles especially against necrotrophic pathogens, insects, and wounds (Ding et al., 2011). Not only do SA and JA activate defence pathways individually, they also interact synergistically or antagonistically to further fine‐turn downstream signalling (Kachroo et al., 2003). Previous studies proved that a transcriptional coactivator nonexpressor of pathogenesis‐related 1 (NPR1) plays a crucial role in SA signalling and in the crosstalk between SA and JA signalling in Arabidopsis (Dong, 2004; Pieterse & Van Loon, 2004; Spoel et al., 2003).
Aside from plant hormones, enzymes involved in lipid biosynthesis or degradation have also been linked to SAR (Gao et al., 2015; Lim et al., 2017). Fatty acids (FAs), a major and essential constituent of all plant cells, can provide structural integrity and energy for various metabolic processes, and can function as signal transduction mediators (Lim et al., 2017). For instance, accumulation of 18:2 and 18:3 FAs results in higher resistance in bean to Botrytis cinerea and in Arabidopsis to Pseudomonas syringae (Ongena et al., 2004; Yaeno et al., 2004). Glycerol‐3‐phosphate (G3P), a precursor for the biosynthesis of all plant glycerolipids, can also serve as a mobile inducer of SAR (Chanda et al., 2011). Azelaic acid, a C9 dicarboxylic acid generated from the oxidation of C18 FAs present on galactolipids, confers local and systemic resistance against P. syringae (Jung et al., 2009). An elevated level of monogalactosyl diacylglycerol (MGDG) and digalactosyl diacylglycerol (DGDG) in rice increases susceptibility to the bacterial blight pathogen Xanthomonas oryzae pv. oryzae, indicating the negative role they play in rice immunity (Gao et al., 2017). As the only soluble fatty acid desaturase, stearoyl‐ACP desaturase (SACPD) initiates the desaturation of FAs by introducing a cis double bond into the acyl‐ACPs at C9, and generates the monounsaturated FA oleic acid (18:1) in the plastids. Reduced SACPD activity is associated with enhanced resistance to multiple pathogens in Arabidopsis (Kachroo et al., 2001), rice (Jiang et al., 2009), soybean (Kachroo et al., 2008), and wheat (Song et al., 2013).
Defence signalling in cotton (Gossypium hirsutum), the most important fibre crop for humans worldwide, appears to differ in some aspects from the signalling in model plants or many monocotyledonous plants. One such difference is that the significant increase in SA levels in model plants on biotrophic and/or hemibiotrophic pathogen infection mainly depends on the induction of the ICS1 gene (Catinot et al., 2008; Uppalapati et al., 2007; Wildermuth et al., 2001), whereas in cotton the ICS1 gene or its homologous genes was not significantly induced on V. dahliae infection (our unpublished data). A similar phenomenon was observed in rice in which the SA levels were twice those in dicots and the SA levels did not increase further in response to pathogen infection (Silverman et al., 1995). This raises questions regarding the role of SA as a defence signal or pathway in cotton.
Several SACPD‐encoding genes have been isolated from various plant species, including Arabidopsis, rice, castor, rapeseed, and soybean (Byfield et al., 2006; Jiang et al., 2009; Kachroo et al., 2007; Shanklin & Somerville, 1991; Slocombe et al., 1992). However, the function of the SACPD gene family in the complex genome of allotetraploid cotton, the functional diversity among its subgroups, and the identity of the predominant member(s) functioning in response to pathogens are still unclear. As previously reported, the primary function of SACPD is regulating 18:1 FA levels. Low 18:1 FA levels can improve the resistance of plants to leaf disease (Jiang et al., 2009; Shekara et al., 2007), but it is not known whether it is effective against vascular disease. In this study, we examined the function of the GhSACPD gene family and discovered the predominant functioning member, revealing the molecular mechanism of GhSACPDs in cotton Verticillium wilt resistance, which provided novel insights into cotton immunity.
2. RESULTS
2.1. SACPD genes with high amino acid identity exist in upland cotton
To identify the SACPD isoforms in G. hirsutum, the full‐length amino acid sequence of AtSSI2 (At2g43710) was used as the query in a BLAST search against the G. hirsutum genome database (https://cottonfgd.org/) (Zhang, Maximova, et al., 2015). Then, the identified GhSACPDs were used as queries to search for further SACPD proteins in the G. hirsutum genome. All the candidate SACPD proteins were analysed using HMMER software to identify the FA_desaturase_2 domain with the pfam accession PF03405 (Tang et al., 2016). In this way, 19 SACPD protein‐encoding genes were identified in G. hirsutum and numbered according to genome location. The predicted peptides encoded by the 19 genes ranged from 387 to 397 amino acids, and most of them were predicted to localize in the plastids (Table S1). Of the 19 GhSACPD genes, six (GH_A10G1561, GH_A10G1562, GH_A10G1563, GH_D10G1329, GH_D10G1331, and GH_D10G1332) were located in tandem on chromosomes A10 and D10 with three as a group (Figure S1), which is similar to the Arabidopsis genome in which three out of seven stearoyl‐acyl carrier protein desaturase (SAD) genes (At3g02610, At3g02620, and At3g02630) are located in tandem on chromosome 3 (Kachroo et al., 2007). Another six genes (GH_A05G2715, GH_A05G3296, GH_A05G3299, GH_D05G2733, GH_D05G3475, and GH_D05G3478) were located on chromosomes A05 and D05 without a tandem relationship. All the other members were distributed on chromosomes A02, A07, A12, D02, D03, and D07 (Figure S1).
To better understand the evolutionary relationships between different members of the GhSACPD gene family, we constructed anunrooted phylogenetic tree with the GhSACPD protein sequences and performed a comparative analysis of exon‐intron structure (Figure S2). In general, the GhSACPD gene family presented a relatively simple genetic stucture. With three exceptions (GH_A07G1333, GH_D07G1317, and GH_A07G1333), the GhSACPD isoforms possess one or two introns (Figure S2). As in Arabidopsis and cacao (Kachroo et al., 2007; Zhang, Maximova, et al., 2015), the GhSACPDs shared high identity in amino acid sequence similarity in the FA_desaturase_2 domain (Figure S3, Table S2). To gain insights into potential catalytic activities of the cotton SACPD isozymes based on the three‐dimensional crystal structure of SAD proteins in castor (Cahoon et al., 1997; Ylva et al., 1996), we performed an amino acid sequence alignment using RcSAD1, AtSSI2 from Arabidopsis, and the 19 cotton GhSACPD sequences. The cotton GhSACPDs possessed 11 highly conserved α‐helices previously identified as important for Δ9 stearoyl‐acyl carrier protein desaturase activity (Figure S4).
2.2. The group represented by GH_A10G1563 play a dominant role on C18:1 fatty acid pool
To investigate the evolutionary relationships between the GhSACPD genes, the SACPD sequences from Arabidopsis and G. hirsutum were used to generate an unrooted phylogenetic tree. As shown in Figure 1a, the GhSACPDs were divided into seven groups: six groups, consisting of 15 GhSACPD genes, were gathered into the same clade as AtSSI2; group VII, containing the remaining four GhSACPD genes, was in the same clade as AtSAD6. None of the cotton Gh SACPD genes were in the same clade as other Arabidopsis SADs. Interestingly, SAD1, SAD2, SAD3, SAD4, and SAD5 were present in Arabidopsis but were absent from G. hirsutum, suggesting that these genes may have been acquired in Arabidopsis after divergence from the common ancestor of Arabidopsis and Gossypium.
FIGURE 1.
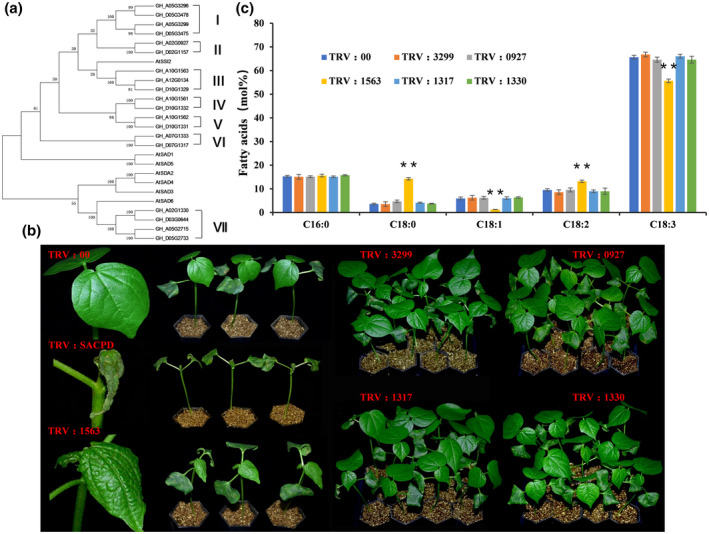
Characterization of different GhSACPD genes via phylogenetic tree analysis and virus‐induced gene silencing (VIGS) experiment. (a) Phylogenetic tree of SACPD proteins from Gossypium hirsutum (Gh) and Arabidopsis thaliana (At) plants. The neighbour‐joining tree was generated using the MEGA 5 program. According to the phylogenetic tree, GhSACPDs were divided into groups I to VII. (b) Spontaneous lesion formation on the leaves of TRV:SACPD and TRV:1563 plants. TRV:3299, TRV:0927, TRV:1317, and TRV:1330 plants showed no difference compared with TRV:00. Photographs were taken 2 weeks after VIGS. (c) Detection of fatty acid content in the silenced cotton lines. The values are the mean ± SE for three biological replicates (**p < 0.01, Student's t test)
To determine whether there is functional differentiation or redundancy in the GhSACPD gene family, the tobacco rattle virus (TRV)‐based virus‐induced gene silencing (VIGS) strategy was used to knock down the transcripts of GhSACPDs in cotton. Considering the sequence similarity between the At subgenome (where “t” stands for tetraploid) and the Dt subgenome and amino acid identity (95%–99%) in different groups of GhSACPDs, isoforms from one group were considered as a single unit throughout this work. For each group, one isoform was selected as the template; the fragment of 5′ untranslated region and the transit peptide sequence, which has lower sequence similarity than the conserved FA_desaturase_2 domain, was used for VIGS. Isoforms of GH_A05G3299, GH_A02G0927, GH_A10G1563, GH_A10G1561, GH_A10G1562, GH_D07G1317, and GH_A02G1330 were chosen to represent groups I to VII, respectively. Due to the low expression level, we failed to obtain fragments of GH_A10G1561 and GH_A10G1562, which represent groups IV and group V, respectively. We constructed common pTRV2 vectors that consisted of conserved coding sequences of GhSACPD members, and gene‐specific pTRV2 vectors (named as TRV:SACPD, TRV:3299, TRV:0927, TRV:1563, TRV:1317, and TRV:1330) based on the lower sequence similarity of the 5′ untranslated region (Figure S5). The pTRV2 vector without sequence insertion (TRV:00) or with a fragment of GhCLA1 (TRV:CLA1) was used as a control or a VIGS efficiency indicator (Figure S6a). The 7‐day‐old cotton seedlings were treated with recombinant virus according to a previous study (Gao, Wheeler, et al., 2011). Two weeks after treatment, the decreased expression levels of different GhSACPD genes were confirmed through quantitative reverse transcription PCR (RT‐qPCR) analysis (Figure S6b). Extremely low transcript levels of GH_A05G3299, GH_A02G0927, GH_A10G1563, GH_D07G1317, and GH_A02G1330 were detected in TRV:SACPD plants, while plants silenced for all the other five gene groups showed extremely decreased expression levels only on the target gene (Figure S7), which suggests that the whole GhSACPD gene family and specific gene members were silenced in cotton (Figure S7). To our surprise, a lesion mimic phenotype occurred on the leaves of TRV:SACPD and TRV:1563 plants specifically (Figure 1b), with curled leaves and spontaneous necrotic lesions along the leaf vein appearing in these plants (Figure 1b). In addition, necrotic lesions were also observed in the stem of TRV:SACPD plants (Figure S6c). No obvious phenotypic differences were observed between the TRV:3299, TRV:0927, TRV:1317, TRV:1330, and TRV:00 plants (Figure 1b). These results indicate that the group represented by GH_A10G1563 displayed a crucial role on the C18:1 pool.
To reveal the member that was responsible for the conversion of 18:0 to 18:1 in cotton, we determined the fatty acid level in each type of VIGS plant. As shown in Figure 1c, the TRV:1563 plants showed a significantly reduced oleic acid (18:1) level and consequently an increase in stearic acid (18:0) content. As the downstream products of oleic acid (18:1), the contents of linoleic acid (18:2) and α‐linolenic acid (18:3) were dramatically affected, being upregulated and downregulated, respectively. The downregulation of the other isoforms caused no obvious changes in fatty acid content (Figure 1c). These data indicate that group III isoforms (GH_A10G1563, GH_D10G1329, GH_A12G0134) play a dominant role in determining the 18:1 content in cotton. Thus, the gene group was named GhSSI2, and the three members GH_A10G1563, GH_D10G1329, and GH_A12G0134 were renamed GhSSI2‐A, GhSSI2‐B, and GhSSI2‐C, respectively.
2.3. Silencing of GhSSI2s leads to an HR‐like phenotype in cotton
HR is a form of PCD in plants, which results in necrotic lesion formation, sealing the pathogen in a tomb of dead cells (Coll et al., 2011). Because the GhSSI2‐silenced plants exhibited necrosis in the leaf vein (Figure 2a), we performed histochemical staining using trypan blue and confirmed the cell death phenotype in the leaf (Figure 2b). HR triggers rapid production of ROS and induces numerous PR genes. To evaluate whether the lesion mimic in GhSSI2‐silenced plants was due to HR‐like cell death, the H2O2 level and PR genes that areassociated with HR were analysed. Staining of the leaf cells with 3,3′‐diaminobezidine (DAB) for H2O2 revealed that the GhSSI2‐silenced plants produced much higher amounts of H2O2 than TRV:00, resulting in darker staining (Figure 2c). Furthermore, PR genes PR1, PR2, PR3, PR4, PR5, PR6, and PR10 were significantly upregulated in the GhSSI2‐silenced plant leaves (Figure 2d). Because SA is a sensor of the HR network that executes a tight control in HR lesion formation (Alvarez, 2000), we examined the SA level and found a significantly higher SA accumulation in the GhSSI2‐silenced plants than the control plants (Figure 2e). These data suggest that the low level of C18:1 led to constitutive activation of the HR‐like cell death phenotype in cotton seedlings.
FIGURE 2.
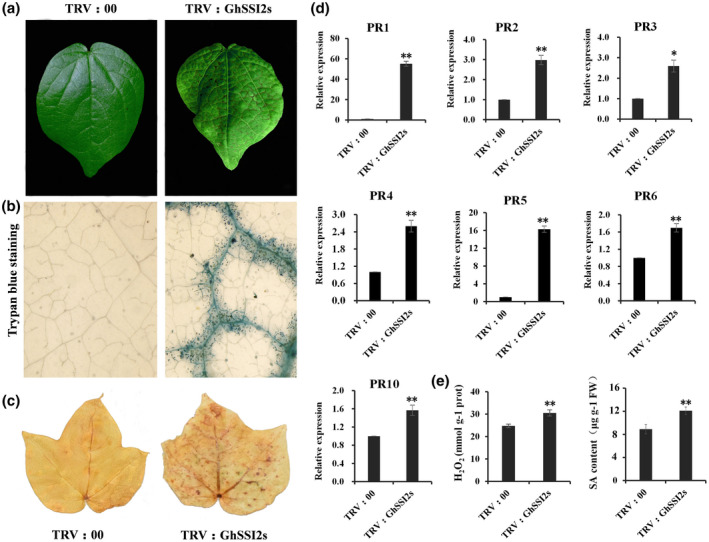
Knockdown of GhSSI2s triggered a hypersensitive response (HR)‐like phenotype. (a) Spontaneous lesions were observed on the leaves of TRV:GhSSI2s, especially along the leaf vein. (b) Trypan blue staining of TRV:GhSSI2 and TRV:00 leaves, showing obvious cell death phenotype in the TRV:GhSSI2 leaves. (c) H2O2 accumulation at the site of lesion formation in TRV:GhSSI2 leaves was visualized through 3,3′‐diaminobenzidine (DAB) staining. (d) Quantitative reverse transcription PCR analysis of PR gene expression in TRV:00 and TRV:GhSSI2 plants. The values are the mean ± SE for three biological replicates (*p < 0.05, ** p< 0.01, Student's t test). (e) Measurements of the H2O2 and salicylic acid (SA) contents in the silenced cotton leaves. The values are the mean ± SE for three biological replicates (**p < 0.01, Student's t test)
2.4. GhSSI2s negatively regulate disease resistance to V. dahliae and F usarium oxysporum
The lesion mimic phenotype often confers resistance to pathogen attacks, particularly to biotrophs (Molina et al., 1999; Zhang et al., 2016). To examine the response of GhSSI2 under V. dahliae infection, we monitored its expression changes within 72 hr postinoculation and found that GhSSI2 expression was significantly suppressed in nearly all the time points (Figure 3d). Then we silenced GhSSI2 genes in the susceptible cotton variety CCRI8. Two weeks after VIGS treatment of CCRI8, the seedlings were inoculated with highly virulent V. dahliae strain Linxi2‐1 (Yang et al., 2019). Three weeks after infection, the GhSSI2‐silenced plants displayed dramatically improved Verticillium wilt resistance, with less leaf chlorosis, less wilting, and lower disease index compared to TRV:00 plants (Figure 3a,c). Moreover, the fungal recovery assay indicated that a smaller number of fungal colonies were found in the GhSSI2‐silenced plants compared to TRV:00 plants, which further supported these results (Figure 3b). In contrast, we found that all the other GhSACPD isoforms (except the GhSSI2 group) showed a similar phenotype to the TRV:00 plants, suggesting that these genes did not participate in the defence response against V. dahliae (Figure S8). Fusarium wilt is another fungal disease that poses a great threat to cotton (Wang et al., 2018). We therefore investigated the function of GhSSI2s in defence against F. oxysporum infection. We silenced GhSSI2s in cotton variety Jimain11 (susceptible to F. oxysporum), then the plants were inoculated with the highly aggressive defoliating isolate FOV7. The results indicated that the silenced plants displayed improved Fusarium wilt resistance at 25 days postinoculation (dpi) (Figure S9a), whereas serious symptoms appeared in the control (CK) plants, as indicated by disease index results (Figure S9b). Thus, these results suggest that GhSSI2s negatively regulated vascular fungal disease resistance in cotton.
FIGURE 3.
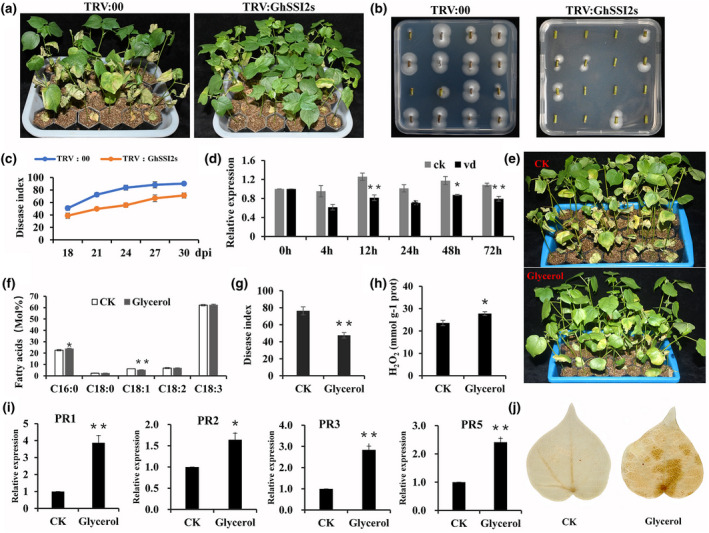
GhSSI2s‐silenced plants conferred enhanced resistance to V erticillium dahliae. (a) Disease symptoms of TRV:00 and TRV:GhSSI2s plants inoculation with V. dahliae at 24 days post inoculation (dpi). (b) Fungal recovery experiments in TRV:GhSSI2s and control plants 4 days after recovery. (c) The disease index of the TRV:00 and TRV:GhSSI2s plants. The values were the means ± SE for three independent experiments (30 seedlings in each experiment). (D) Quantitative reverse transcription PCR (RT‐qPCR) analysisof expression of GhSSI2s following inoculation with V. dahliae in resistant ND601. GhUBQ14 was used as an internal control. The values were the means ± SE for three biological replicates (** p< 0.01, Student’s t test). (e) Disease symptoms of cotton plants treated with 100 mM glycerol following inoculation with V. dahliae. Photographs were taken at 20 dpi.(f) Detection of fatty acid composition in the glycerol‐treated plants. The values were the means ± SE for three biological replicates (*p < 0.05, **p < 0.01, Student’s t test). (g) Disease index of cotton plants after glycerol treatment at 20 dpi. Error bars were SE calculated from threebiological replicates, each containing 35 plants. (h) Measurements of the H2O2 contents in the glycerol‐treated plants. The values were the means ± SE for three biological replicates (* p< 0.05, Student’s t test). (I) RT‐qPCR analysis of PR gene expressionin control and glycerol‐treated plants. The values were the means ± SE for three biological replicates (*p < 0.05, **p < 0.01, Student’s t test). (J) Detection of H2O2 accumulation in the control and glycerol treatment cottons via 3,3′‐diaminobenzidine staining
Glycerol can be phosphorylated to G3P, which subsequently can be acylated with C18:1‐ACP (stearoyl‐acyl carrier protein) to yield lysophosphatidic acid (LPA), leading to a decreased level of oleic acid (18:1) (Jiang et al., 2009; Kachroo et al., 2004; Zhang, Smith, et al., 2015). To determine whether reduction of endogenous oleic acid (18:1) levels via exogenous application of glycerol has a similar effect to the silenced GhSSI2s, we conducted foliar spraying with glycerol on cotton seedlings, and found that glycerol application mimicked the GhSSI2‐silencing treatment in reducing the C18:1 level, elevating PR expression, accumulation of H2O2, and improving disease resistance (Figure 3e–j). However, the HR‐like phenotype was not observed in the seedlings treated by glycerol. This could be a result of the different 18:1 levels between GhSSI2‐silenced and glycerol‐treated cotton plants, indicating a dosage effect of 18:1 in the cotton defence system.
2.5. Suppressing GhSSI2s activates an SA‐independent defence response in cotton
The increased SA level in GhSSI2‐silenced plants prompted us to investigate whether the SA pathway is required in the regulation of disease resistance by GhSSI2s. We determined the expression levels of genes essential to the SA signal pathway (Ding et al., 2018; Feys et al., 2001; Garcion et al., 2008; Rietz et al., 2011). Unexpectedly, the expression levels of EDS1, PAD4, NPR1, CBP60, SARD, and ICS1 did not significantly differ between the TRV:GhSSI2s and TRV:00 plants (Figure 4a), indicating the nonactivation of the SA pathway. Plant SA is derived from chorismate either by isochorismate synthase (ICS) or phenylalanine ammonia‐lyase (PAL) catalysed steps (Shine et al., 2016). To further verify our result, we generated double‐gene‐silenced cotton lines with simultaneously downregulated gene combinations of GhSSI2s & GhPAL (TRV:GhSP) and GhSSI2s & GhICS1 (TRV:GhSI) (Figures 4b and S10). Both of the TRV:GhSP and TRV:GhSI cotton lines showed a reduced SA level compared with the TRV:GhSSI2s plants (Figure 4c). Even so, we still observed elevated H2O2 content and upregulated PR expression, which were consistent with those in the TRV:GhSSI2s plants (Figures 3 and 4d,e,f). These data, together with the nonactivated SA signal pathway components (Figure 4a), indicate SA‐independent activation of the defence response in the GhSSI2‐silenced plants.
FIGURE 4.
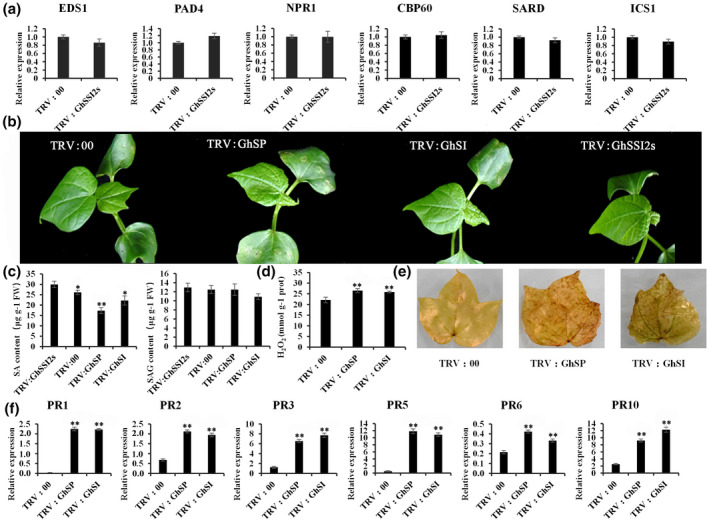
Salicylic acid (SA) is not essential for the GhSSI2‐conferred activation of defence phenotypes. (a) The transcript levels of SA signal transduction pathway genes. The values are the mean ± SE for three biological replicates (**p < 0.01, Student's t test). (b) Morphological features of double‐gene‐silenced plants by virus‐induced gene silencing (VIGS). Co‐suppressed plants had similar phenotypes to TRV:GhSSI2 plants. TRV:SP (TRV:GhSSI2 & Gh PAL) and TRV:SI (TRV:GhSSI2 & Gh ICS1) represented cosuppressed plants with different gene combinations. Photographs were obtained 2 weeks after VIGS. (c) Measurements of the SA and SA glucoside (SAG) in the cosuppressed cotton leaves. TRV:SP and TRV:SI had lower SA levels than the TRV:GhSSI2 plants. The values are the mean ± SE for three biological replicates (* p< 0.05, **p < 0.01, Student's t test). (d, e) The content of H2O2 was elevated in the cosuppressed cotton leaves. (f) Expression patterns of PR genes in the TRV:SP and TRV:SI plants
Inconsistent with the elevated SA level in TRV:GhSSI2s, the ICS1 gene, contributing up to 90% of pathogen‐induced SA synthesis in Arabidopsis or several other species (Garcion et al., 2008), did not show obvious expression change compared to TRV:00 plants (Figure 4a). This result demonstrated that the enhanced SA level in TRV:GhSSI2s was independent of ICS1 involvement in SA synthesis. Thus, we speculated that the enhanced SA level could mainly derive from the PAL pathway. To obtain supporting evidence for this hypothesis, we identified 13 GhPAL isoforms in the G. hirsutum genome. We then selected three (GhPAL1, GhPAL2, and GhPAL3) out of the 13 GhPAL members based on the high basal expressions in a transcriptome database of plants inoculated by V. dahliae (Wang et al., 2021) for further study (Table S3). The expression of GhPAL1 and GhPAL2 was significantly higher in the TRV:GhSSI2s plants compared to the TRV:00 plants (Figure 5b), whereas the GhPAL3 transcript level was not significantly altered. Higher PAL enzyme activity was also detected in the GhSSI2‐silenced plants (Figure 5c), indicating that the high level of SA in the low 18:1 system in TRV:GhSSI2 plants was probably mainly generated by PAL‐catalysed reactions. To gain more evidence for this, we examined the transcript levels of GhICS1 and GhPALs following V. dahliae infection, and discovered significant upregulation of PAL1 and PAL2 but not ICS1 (Figure 5a). Moreover, we examined the SA level when ICS1 and PALs were silenced. As shown in Figure 5d, the TRV:GhPAL plants exhibited dark green and wrinkled leaves whereas the TRV:GhICS1 plants showed no obvious difference from the TRV:00 plants. The TRV:GhPAL plants showed reduced levels of both free SA and SA glucoside (SAG) (bound conditions) content while the TRV:GhICS1 plants showed only SAG content decreased compared to TRV:00 plants (Figure 5f). Together, these results demonstrate that the PAL pathway plays a leading role in SA synthesis in cotton.
FIGURE 5.
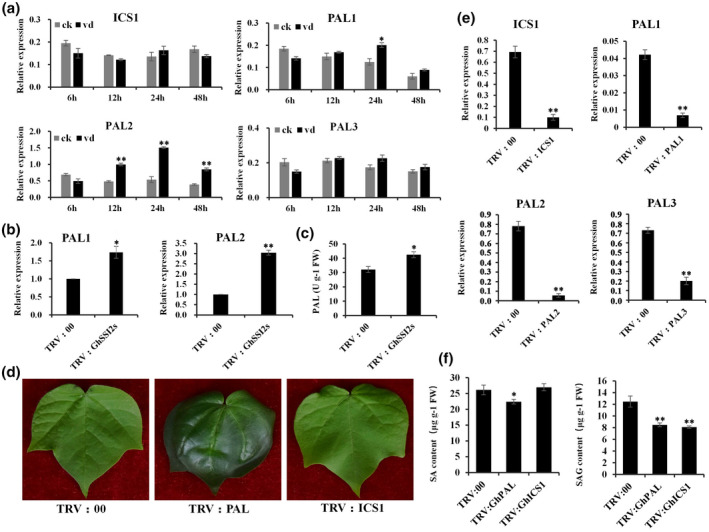
The phenylalanine ammonia‐lyase (PAL) pathway is important for salicylic acid (SA) synthesis in cotton. (a) Quantitative reverse transcription PCR (RT‐qPCR) analysis of GhICS1 and GhPAL expression at different time points following inoculation with Verticillium dahliae. The values are the mean ± SE for three biological replicates (**p < 0.01, Student's t test). (b) The expression of GhPALs in TRV:00 and TRV:GhSSI2 plants. The values are the mean ± SE for three biological replicates (*p < 0.05, **p < 0.01, Student's t test). (c) Analysis of PAL enzyme activity in TRV:00 and TRV:GhSSI2 leaves. The values are the mean ± SE for three biological replicates. (*p < 0.05, Student's t test). (d) Phenotypes of TRV:00, TRV:GhPAL, and TRV:GhICS1 plants. The TRV:GhPAL leaves exhibited a dark green colour. (e) RT‐qPCR analysis indicated that the transcripts of GhICS1, GhPAL1, GhPAL2, and GhPAL3 were reduced in the corresponding gene‐silenced plants. (f) The contents of SA and SA glucoside (SAG) in TRV:00, TRV:GhPAL, and TRV:GhICS1 cotton leaves
2.6. Suppressing GhSSI2s affects the PDF1.2‐dependent JA signalling pathway
The fatty acid‐derived phytohormone JA is particularly well known for its role in orchestrating the environmental responsiveness of biotic and abiotic stress (Wasternack & Hause, 2013). JA biosynthesis is initiated by the release of α‐linolenic acid (18:3) through hydrolysis of the chloroplast membranes by phospholipases. The low level of C18:1 in GhSSI2‐silenced plants led to the decrease in the α‐linolenic acid (18:3) content (Figure 1c), which encouraged us to examine whether JA biosynthesis was affected. We discovered that the JA level was significantly decreased in GhSSI2‐silenced plants compared to the control (Figure 6c), as a result of the lack of the substrate (C18:3) for JA biosynthesis. Subsequently, we assessed the ability of GhSSI2‐silenced plants to activate JA‐dependent defence responses. RT‐qPCR analysis showed that the JA response gene PDF1.2 could not be induced under JA treatment (Figure 6d). However, we observed the upregulation of JA biosynthesis‐related genes (LOX1, LOX3, LOX5, AOS, AOC1, and OPR3) and JA signalling was activated, as indicated by expression of JA signalling‐related genes (MYC, JAZ1, JAZ10, COI1, and VSP1) in the GhSSI2‐silenced plants (Figure 6a,b). Furthermore, root growth was inhibited inTRV:GhSSI2s and TRV:00 plants on JA treatment (Figure 6b,e). The induced expression of GhVSP (another JA‐responsive gene) combined with the similar phenotype of TRV:GhSSI2s and TRV:00 plants under JA treatment suggested that GhSSI2‐silenced plants could perceive JA just like in TRV:00 plants. Taken together, these results indicate that suppressing the expression of GhSSI2s affects the PDF1.2‐dependent JA signalling pathway in cotton.
FIGURE 6.
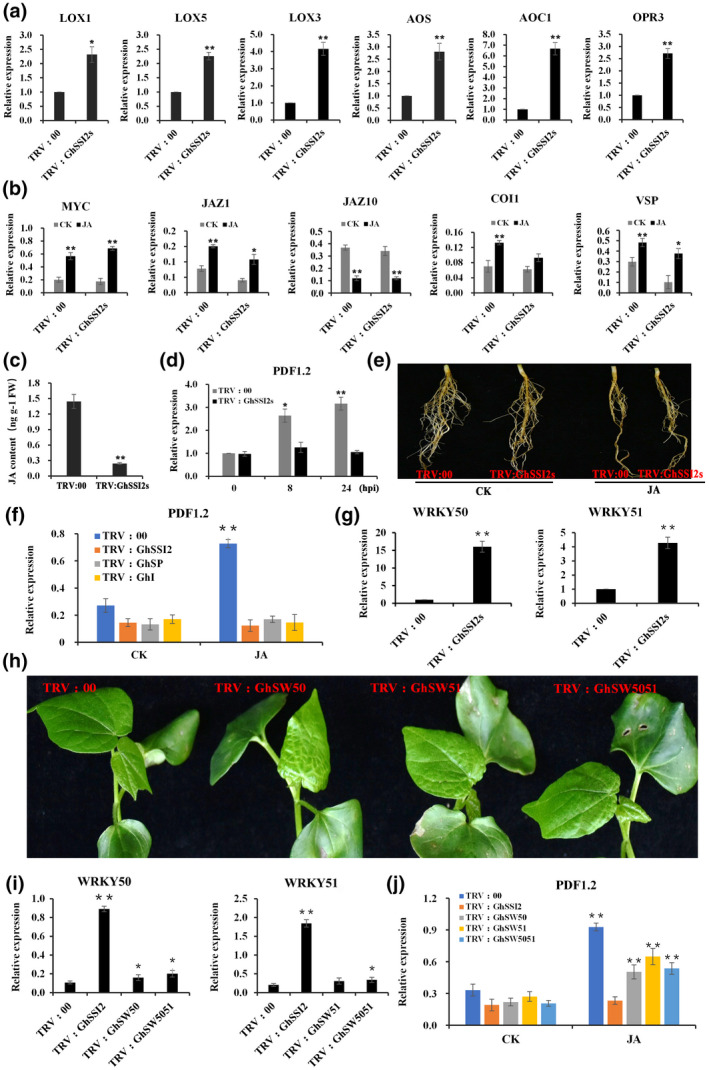
The impaired induction of PDF1.2 in the GhSSI2s‐silenced plants due to the elevated expression of GhWRKY50 and GhWRKY51. (a) Quantitative reverse transcription PCR (RT‐qPCR) analysis of the expression pattern of genes involved in jasmonic acid (JA) synthesis in the GhSSI2s‐silenced plants. (b) The transcript levels of JA signal transduction pathway genes on JA treatment of the TRV:00 and TRV:GhSSI2 plants. The values are the mean ± SE for three biological replicates (*p < 0.05, **p < 0.01, Student's t test). (c) Measurements of JA content in cotton leaves. The values are the mean ± SE for three biological replicates (**p <0 .01, Student's t test). (d) Detection of PDF1.2 gene expression under treatment with JA at different time points. (e) JA treatment affected the root growth in TRV:00 and TRV:GhSSI2 plants. (f) Expression patterns of PDF1.2 in the double‐gene‐silenced TRV:GhSI and TRV:GhSP plants after JA treatment. (g) Expression levels of WRKY genes in TRV:00 and TRV:GhSSI2 plants. All the values are the mean ± SE for three biological replicates (*p < 0.05, **p < 0.01, Student's t test). (h) Morphological features of double‐ or triple‐gene‐silenced plants by virus‐induced gene silencing (VIGS). Cosuppressed plants had similar phenotypes to TRV:GhSSI2 plants. TRV:SW50, TRV:SW51, and TRV:SW5051 represent cosuppressed plants with different combinations. Photographs were obtained 2 weeks after VIGS. (i) RT‐qPCR analysis indicated that the transcripts of GhWRKY50 and GhWRKY51 were reduced in the corresponding gene‐silenced plants. (j) Downregulation of WRKY50 and WRKY51 in the low C18 system restored partial JA‐inducible PDF1.2 expression (**p < 0.01, Student's t test)
Because SA and JA have been reported to antagonize the activation of each other's defence responses (Kachroo et al., 2001), we tested the possibility of the elevated levels of SA in GhSSI2‐silenced plants inhibiting the JA signalling pathway. As shown in Figure 6f, when treated with JA, the expression of PDF1.2 could be induced only in TRV:00 plants. Despite a depressed SA level occurring in TRV:GhSP and TRV:GhSI plants, the JA‐responsive gene PDF1.2 was not activated in either TRV:GhSP or TRV:GhSI plants. Based on these results, we ruled out the possibility that the impaired JA response in the GhSSI2‐silenced plants was due to the elevated SA levels. Arabidopsis ssi2 mutant plants can restore PDF1.2 expression by mutations of transcription factors AtWRKY50 and AtWRKY51 (Gao, Venugopal, et al., 2011). Therefore, we hypothesized that the blocked JA pathway in cotton TRV:GhSSI2 plants might due to the induction of the cotton genes homologous to AtWRKY50 and AtWRKY51. We observed elevated expression of GhWRKY50 and GhWRKY51 in TRV:GhSSI2 plants compared to TRV:00 plants (Figure 6g). We therefore carried out VIGS with combinational vectors to generate double‐ or triple‐gene‐silenced plants with simultaneously silenced genes of GhSSI2s & GhWRKY50 (TRV:GhSW50), GhSSI2s & GhWRKY51 (TRV:GhSW51), and GhSSI2s & GhWRKY50 & GhWRKY50 (TRV:GhSW5051). The cosuppressed seedlings exhibited similar phenotypes to TRV:GhSSI2s (Figure 6h,i). In addition, PDF1.2 expression displayed upregulation in TRV:GhSW50, TRV:GhSW51, and TRV:GhSW5051 plants, which was consistent with that in TRV:00 plants (Figure 6j). These results demonstrate that the impaired C18 system in cotton blocks the PDF1.2‐dependent JA response, whereas the canonical JA signalling cascade is not impaired.
2.7. Knockdown GhSSI2s up‐regulates expression of structurally divergent resistance‐related genes
The major mediators of plant defence signalling pathways include the phytohormones SA, JA, ethylene (ET), abscisic acid (ABA), and auxin, which not only induce defence responses individually, but also interact synergistically or antagonistically to further orchestrate downstream signalling (Berens et al., 2017; Kachroo & Kachroo, 2009; Robert‐Seilaniantz et al., 2011). However, in this study, the GhSSI2‐silenced cotton displayed significant disease resistance even though the SA and JA signalling pathways were not activated (Figures 4 and 5). An interesting question is, what is the molecular mechanism that GhSSI2‐silenced cotton uses to resist pathogen infection? A genome‐wide transcriptome analysis from GhSSI2‐silenced cotton leaves revealed that as many as 17,654 genes were significantly differentially expressed compared to TRV:00, comprising 8,663 upregulated and 8,991 downregulated genes (p < 0.05, fold change >2; Table S4). Gene ontology (GO) analysis of the biological processes showed that categories including systemic acquired resistance, response to stress, response to fungus, defence response, and response to stimulus were enriched among the upregulated genes (Figure 6a and Table S5), which indicates the activation of the defence‐related pathway in the TRV:GhSSI2 plants. Strikingly, 79 PR genes were significantly upregulated in TRV:GhSSI2 plants, demonstrating a 2.0‐ to 2,272.5‐fold increase in FPKM value (Table S6) that overlapped with that of Ghlmm mutant plants containing a nonsense mutation in the 5‐aminolevulinic acid dehydratase gene and exhibiting necrotic leaf damage and enhanced disease resistance to V. dahliae (Chai et al., 2017). Of the 48 PR genes upregulated in Ghlmm mutant plants, 36 were also induced in TRV:GhSSI2 plants (Table S6), which suggests a correlation between elevated V. dahliae resistance in TRV:GhSSI2 plants and in Ghlmm mutant plants (Figure 7a).
FIGURE 7.
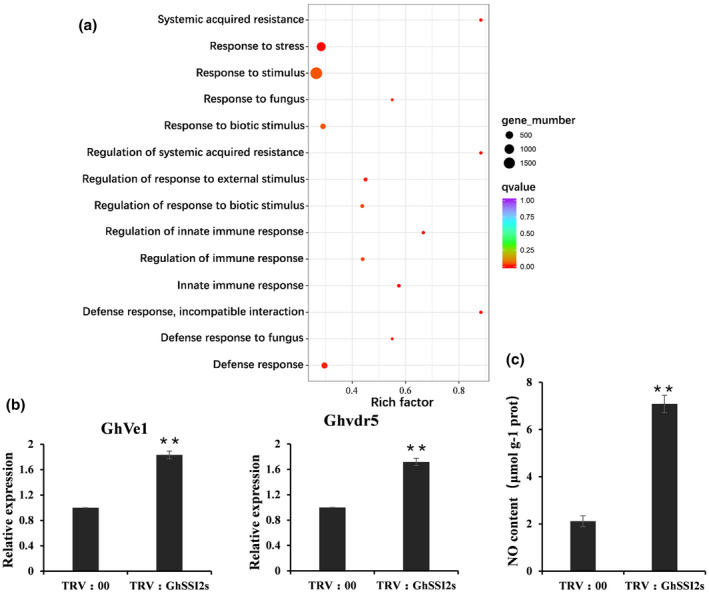
Downregulation of GhSSI2s activated the cotton defence system and elevated NO accumulation. (a) Gene ontology (GO) enrichment analysis of the differentially expressed genes between TRV:00 and TRV:GhSSI2 plants. RNA isolated from leaves of three individual plants was used for RNA sequencing. The q value is the p value after multiple hypothesis test correction. Rich factor: percentage of enriched genes compared with background in the corresponding GO term. (b) Quantitative reverse transcription PCR analysis of GhVe1 and Ghvdr5 expression levels in TRV:00 and TRV:GhSSI2 plants. (c) Measurements of the NO content in cotton leaves. The values are the mean ± SE for three biological replicates (**p < 0.01, Student's t test)
To examine whether silencing of GhSSI2 affected resistance‐related proteins, we analysed the expression of different resistance‐related genes in TRV:GhSSI2 plants. Based on the NB‐ARC domain (pfam accession PF00931) screening with HMMER 3.0 across the G. hirsutum genome, 533 nucleotide‐binding site (NBS) genes were identified (Table S7), among which 111 genes were upregulated in the TRV:GhSSI2 plants (Table S8), including CN (19), CNL (16), N (32), NL (7), RN (7), TN (23), and 7 TNL types. As a member of the extracellular leucine‐rich repeat receptor‐like proteins (eLRR‐RLP) type of disease resistance proteins, Ve1 provides resistance against V. dahliae in tomato (Fradin et al., 2009). We identified 124 eLRR‐RLP type genes (annotated as receptor‐like protein 12) in the G. hirsutum genome (Table S9) and as many as 50 of these displayed elevated transcript levels in TRV:GhSSI2 plants (Table S10). The Ve1 homologous gene GhVe1 (Ghir_D09G017000) and Ghvdr5 (Ghir_A01G022030), conferring resistance to V. dahliae in cotton (Yang et al., 2015; Zhang et al., 2011, 2012), were also included (Figure 7b).
Because nitric oxide (NO) is a well‐known regulator of disease physiologies, and the accumulation of NO can trigger nuclear gene expression in the low 18:1 system in Arabidopsis (Mandal et al., 2012), we examined the NO level in TRV:GhSSI2s cotton and the results showed a significant elevation of the endogenous NO content in the TRV:GhSSI2 plants (Figure 7c).
Taken together, these results indicate that suppressing GhSSI2 expression can activate multiple resistance‐related proteins and an elevated NO signal in cotton.
3. DISCUSSION
S‐ACP‐DES is an archetypical member of a family of soluble FA desaturases that mediates conversion of stearic acid (18:0) to oleic acid (18:1) and regulates levels of unsaturated FAs in cells (Kachroo et al., 2001, 2004). Moreover, oleic acid (18:1) also functions as signalling mediator in plant resistance against several pathogens (Walley et al., 2013). It has been demonstrated that mutation in the major SACPD isoform (SSI2) in Arabidopsis caused improved resistance to bacterial and oomycete pathogens, and turnip crinkle virus but hypersusceptibility to necrotrophic pathogens (Gao, Venugopal, et al., 2011; Kachroo et al., 2004; Shah et al., 2001; Shekara et al., 2007). In addition, rice OsSSI2‐knockdown plants showed markedly enhanced resistance to leaf diseases, such as the blast fungus Magnaporthe grisea and leaf blight bacterium X. oryzae pv. oryzae (Jiang et al., 2009). In this study, we characterized the cotton SACPD gene family and revealed the different response and function of each SACPD isoform in a vascular fungal disease inoculated via root tip or wound root. Despite the high sequence similarity of SACPD members between G. hirsutum and other plant species, including Arabidopsis (Kachroo et al., 2007), cacao (Zhang, Maximova, et al., 2015), and tobacco (Zhang et al., 2014), the isoforms located on chromosomes A10, D10, and A12 (designated as GhSSI2‐A, GhSSI2‐B, GhSSI2‐C) play a dominant role in the C18:1 pool, which is different from the previous report that the Gossypium barbadense GbSSI2 gene made little contribution to the C18:1 pool (Gao et al., 2013). Our results further demonstrated that the GhSSI2‐silenced plants showed enhanced resistance to V. dahliae, with a significant reduction of C18:1 and C18:3. In addition, exogenous spraying of glycerol significantly improved cotton disease resistance in this study, mimicking the downregulation of GhSSI2s in vivo, which is consistent with the fact that glycerol could be phosphorylated to G3P by glycerol kinase and subsequently acylated with C18:1‐ACP (stearoyl‐acyl carrier protein), resulting in a decreased level of 18:1 (Jiang et al., 2009; Kachroo et al., 2004; Zhang, Smith, et al., 2015). Taken together, our results revealed that GbSSI2s, in the cotton SACPD gene family, function in regulating the oleic acid (18:1) level and contribute to Verticillium wilt resistance.
PCD, a kind of cellular suicide phenomenon, generally occurs when plants encounter biotic or abiotic stress (Brodersen et al., 2002). As one of the best characterized PCDs in plants, HR is an innate immune response to protect hosts from pathogen attacks via inducing cell death at the sites of infection to further inhibit pathogen spread (Heath, 2000; Lam et al., 2001). Lesion mimic mutants (LMMs) spontaneously produce necrotic lesions and activate defence responses without mechanical damage and/or pathogen infection, and are considered an effective tool for understanding the cellular mechanisms governing PCD in plants (Bruggeman et al., 2015; Chai et al., 2017). Except for the P450 gene GhCYP82D, tryptophan synthase GbTSA1/GbTSB1, and 5‐aminolevulinic acid dehydratase GhLMMD (Chai et al., 2017; Miao et al., 2019; Sun et al., 2014), other genes that could induce a lesion mimic phenotype have seldom been exposed in the complicex allotetraploid cotton genome. In the present study, we revealed that suppressing the expression of cotton GhSSI2s caused an obvious PCD phenotype, which is a rare and exciting phenomenon. Many lesion mimic mutants are SA‐dependent, such as the GbTSA1 and GbTSB1 knockdown mutants and the Ghlmm mutant (Chai et al., 2017; Miao et al., 2019). However, the lesion mimic in GhCYP82D‐RNAi plants is independent of SA synthesis and SA signalling (Sun et al., 2014). We found that knockdown of GhSSI2s triggered a lesion mimic phenotype with an elevated SA level but without activating the SA signalling pathway, which was not the same as either GbTSA1/GbTSB1 or the GhCYP82D genes. This finding also differed from the SA immunity system in rice, in which the SA marker genes were highly expressed but SA levels were not upregulated towards the hemibiotrophic pathogen Rhizoctonia solani (Kouzai et al., 2018). Our findings expose a novel aspect in dicotyledonous plants where the accumulation of free SA might not be always associated with activation of SA signalling (Noutoshi et al., 2012).
C18 unsaturated FAs play a vital role in maintaining the structural integrity and normal physiological function of cells (Lim et al., 2017; Yang et al., 2016). Plant proteins, encoded by resistance (R) genes, in the cell surface also play a crucial role by specific recognition of pathogen‐derived molecules from avirulent pathogens, leading to cell death at the sites of pathogen entry (Shirasu & Schulze‐Lefert, 2000). In the present study, on the one hand, downregulation of GhSSI2s altered the C18 metabolism of cotton and adversely affected the functioning of the plasmalemma, which might release multiple signals that mimic pathogen invasion and result in being recognized by the a receptor on the surface of the membrane. Recognition is followed by local accumulation of ROS, SA, and upregulation of pathogenesis‐related genes (PRs), as indicated in Figure 2a–e. On the other hand, 18:1 also functions as a kind of defence signalling and low 18:1 could directly upregulate the expression of different resistance genes in an SA‐independent manner (Shekara et al., 2007; Venugopal et al., 2009). We found that as many as 79 PR genes, 111 NBS‐LRR genes, and 50 receptor‐like protein 12 genes were upregulated in the GhSSI2s‐silenced cotton. In contrast, the SA signalling pathway and the biosynthesis and perception of JA were powerless in GhSSI2‐silenced cotton. These results indicate that knockdown of GhSSI2s directly activates the R gene‐dependent defence system, which is independent of the SA and/or JA pathways.
There is also a branch of SAR consisting of NO‐ROS‐AzA‐G3P that operates in parallel with SA, and both branches are essential for the activation of SAR (Gao et al., 2014). As shown in Figure 6c, the NO level significantly increased, which might be responsible for activating the above branch. Thus, we deduced that the fatty acids and lipids might use a different signalling mechanism from the SA pathway in defence against hemibiotrophic pathogen challenge.
Once a plant has been attacked by pathogens, it must reallocate energy from growth to defence, resulting in a fitness penalty (Brown, 2002). Thus, immunity signalling must be precisely regulated to ensure that the plant allocates resources to growth in the absence of pathogen infection (Ning et al., 2017). A low level of 18:1 can result in automatic activation of defence and, simultaneously, severely retard growth in Arabidopsis (Shah et al., 2001). As the only enzyme that catalyses the conversion of 18:0 to 18:1 in plants, SACPD must be precisely controlled. In this study, the TRV:GhSSI2s plants did not display a lesion phenotype upon V. dahliae infection (data not shown). However, suppressing GhSSI2s via VIGS made the seedlings display an HR‐like phenotype, differing from the performance of the inoculated plants. We deduce it could result from a dosage effect induced by the 18:1 level, which is influenced by expression of GhSSI2s. Generally, plants can modulate trade‐offs between immunity and normal growth by fine‐tuning the compounds that induce plant immunity, such as oleic acid (18:1). Only when the content of the oleic acid is under a certain threshold can a necrotic phenotype be induced, while fluctuations of GhSSI2 expression within a certain range cannot induce the necrotic phenotype.
4. EXPERIMENTAL PROCEDURES
4.1. Plant materials and growth conditions
Cotton plants G. hirsutum 'CCRI 8' (susceptible cultivar), and G. hirsutum 'Nongda 601' (resistant cultivar) were grown in an environmentally controlled chamber under a 16‐hr light/8‐hr dark cycle at 30 ± 2 °C/25 ± 2 °C (day/night) conditions. Nongda 601 was used for treatment and sampling, while CCRI 8 was used for disease index calculation.
4.2. Treatment with V. dahliae, methyl jasmonate, and glycerol
Uniform cotton seedlings (2 weeks old) were sprayed with methyl jasmonate (MeJA)
(100 μM) and glycerol (100 mM) or inoculated with V. dahliae strain Linxi 2‐1 conidial suspensions (107 conidia/ml). Glycerol solution was prepared in sterile water containing 0.04% Silwett L‐77. Water containing 0.04% Silwett L‐77 was used as the control treatment.
The leaves of nine individual seedlings were collected from each treatment at each time point. Three plants were mixed as a sample and each time point contained three biological replicates.
4.3. RT‐qPCR and RT‐PCR
The total RNA was extracted from cotton tissues using an EASYspin Plus Plant RNA Kit (Aidlab Biotech) according to the manufacturer's instructions. First‐strand cDNA was generated from 2 μg of total RNA using a PrimeScript II First Strand cDNA Synthesis Kit (TaKaRa). The RT‐qPCR was performed using a CFX96 Real‐time Detection System (Bio‐Rad) with an SYBR Premix Dimer Eraser (Perfect Real Time) (TaKaRa). Three biological replicates were used for each analysis with three technical replicates each. Gh UBQ14 was used as an internal control. The primers used in our study are listed in Table S11.
4.4. VIGS procedure
The fragments containing part of the 5′ untranslated region (UTR) and the coding sequence, the conserved regions of GhSACPD family members, and the conserved regions of GhPAL, GhICS1, GhWRKY50, and GhWRKY51 were amplified from the cDNA of cv. Nongda 601 using the corresponding primer pairs. The primer sequences are listed in Table S2. The fragments representing a conserved region within this group and differing from the other group were then cloned into pTRV2 (Gao, Wheeler, et al., 2011), constructing vectors named TRV:3299, TRV:0927, TRV:1563, TRV:1317, TRV:1330, TRV:SACPD, TRV:PAL, TRV:ICS1, TRV:WRKY50, and TRV:WRKY51. These vectors and pTRV1 were introduced into Agrobacterium tumefaciens GV3101 through the freeze thawing method. The Agrobacterium strains containing different pTRV2 vectors were mixed in equal volumes with Agrobacterium strains containing pTRV1 at a ratio of 1:1. For the cosuppression of two or more genes, A. tumefaciens samples with different TRV vectors were mixed in equal volumes to suppress two or more genes. These mixtures were infiltrated in cotton cotyledons, as described previously (Gao, Wheeler, et al., 2011). Two weeks after injection, RNA was extracted from newly grown leaves to detect the expression of target genes. More than 30 seedlings were used for each analysis. The TRV:CLA1 construct served as a positive marker for evaluating VIGS efficiency. Plants that were successfully silenced were selected for the subsequent V. dahliae infection assay and other assays. All VIGS assays were performed three times independently.
4.5. Pathogen preparation, infection disease assay, and recovery assay
The highly aggressive defoliating V. dahliae strains Linxi2‐1 and F. oxysporum f. sp. vasinfectum FOV7 were cultured on potato dextrose agar (PDA) at 25 °C for 7 days. Next, the fungal colonies were inoculated into potato dextrose broth (200 g/L potato, 12.5 g/L glucose, pH 7) for 5 days at 25 °C until the spore concentration reached c.107 spores/ml. The spores were adjusted to a concentration of 107 conidia/ml with sterile distilled water for inoculation. Seedlings of Nongda 601 inoculated with V. dahliae were used for RNA extraction and seedlings of CCRI 8 inoculated with V. dahliae after agroinfiltration were used for observation of the disease symptoms. Seedlings of Jimian 11 inoculated with F. oxysporum after agroinfiltration were used for observation of the disease symptoms. The disease index was scored using at least 30 plants per treatment and repeated at least three times. The severity of disease symptoms on each cotton seedling was scored according to previous descriptions (Xu et al., 2012). The fungal recovery assay was performed based on the methods described previously (Zhang et al., 2011). The colonies of V. dahliae were observed after 4 days’ incubation.
4.6. Hormone determination
SA and SAG were measured according to the method described by Chai et al. (2017). For JA analysis, the ground samples (400 mg) were extracted in 10 times the sample volume with acetonitrile overnight at 4 °C. After centrifugation, the pellet was extracted with five times the volume of acetonitrile. The supernatants were combined and added to 16 mg of C18 QuEChERS to purify the samples. The supernatant was dried by evaporation with nitrogen gas. The residues were dissolved in 200 μl methanol. The supernatants were analysed using an HPLC‐MS/MS system (AB SCIEX QTRAP6500LC/MS/MS). The content of SA and JA was calculated according to the standard curve that was compiled from commercial SA and JA purchased from Sigma.
4.7. Histochemical assay
For DAB staining, leaves of TRV:GhSSI2 and TRV:00 plants were submerged in DAB‐HCl solution (1 mg/ml, pH 3.8; Sigma‐Aldrich) and kept under vacuum for 15 min then placed in the dark for 12 hr. After washing with distilled water, the leaves were then put into ethanol (95%) for decolourization until they were free of chlorophyll. The appearance of reddish spots was used as evidence of H2O2. For trypan blue staining, leaves of TRV:GhSSI2 and TRV:00 plants were incubated in trypan blue solution (25% phenol, 25% glycerol, 25% lactic acid, trypan blue 3.75 mg/ml) and kept under vacuum for 15 min. The leaves were then boiled for 8 min, kept at room temperature for 6–8 hr, and then destained with chloral hydrate (2.5 g/ml).
4.8. Lipid analysis
FAs were extracted from 0.1 g of leaf tissue of the gene‐silenced and glycerol‐treated plants as described previously (Zhang et al., 2005). Briefly, 1 ml of 2.5% H2SO4 in methanol was added to each tissue sample and incubated without shaking at 80 °C for 1.5 hr. Fatty acid methyl esters were re‐extracted by adding 1.5 ml of NaCl 0.9% (wt/vol) and 1 ml hexane. Heptadecanoic acid (C17:0) (Sigma) was added to serve as an internal standard throughout the whole procedure. FAs were separated and quantified by gas chromatography (Agilent; 7890B) and identified according to their retention times against the commercial standard.
4.9. Measurement of H2O2 and NO content and PAL activity
The levels of H2O2 and NO in plant tissues were determined using the spectrophotometric method with an H2O2 assay kit (Nanjing Jiancheng). The PAL enzyme activity in leaves was determined with a PAL detection kit (Nanjing Jiancheng).
4.10. Transcriptome sequencing
The leaves of TRV:GhSSI2 and TRV:00 plants were collected 2 weeks after VIGS injection, and the total RNA was extracted using an EASYspin Plus Plant RNA Kit (Aidlab Biotech). Sequencing was performed on the Illumina HiSeq 2000 platform in the Beijing Novogene Genomics Company. The differentially expressed genes were identified using the DESeq package with a negative binomial distribution (FDR < 0.05). All samples contained three biological repeats.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Supporting information
FIGURE S1 Distribution of the GhSACPD gene family in the Gossypium hirsutum genome. There were 19 copies of GhSACPD distributed across different cotton chromosomes and the horizontal lines indicate the relative position of each copy on the chromosomes
FIGURE S2 Phylogenetic relationship and gene structure of GhSACPD genes. Exons are represented by yellow boxes and introns by black lines
FIGURE S3 Weblogo graph of the SACPD gene family in cotton. Amino acid sequences between the two red triangles indicate the FA_desaturase_2 domain
FIGURE S4 Multiple amino acid sequence alignment of GhSACPD with SACPD from castor bean (RcSAD1) and Arabidopsis (AtSSI2). Secondary protein structures of SACPD are annotated according to the crystal structure of RcSAD1. Backgrounds indicate percentage of amino acid similarity: black, 100%; dark grey, 75%; light grey, 50%
FIGURE S5 Construction of pTRV2 vectors based on diversity sequence. (a) The fragments using for each grouped silence were shaded. The nucleotides highlighted in red indicate the initiation codon. According to the gene ID the vectors are marked TRV:3299, TRV:0927, TRV:1563, TRV:1317, and TRV:1330. (b) Fragment using for suppressing the whole SACPD gene family
FIGURE S6 Morphology of the cotton virus‐induced gene silencing (VIGS) experiment and expression analysis of GhSACPDs in GhSACPD KD plants. (a) Photobleaching phenotype of the plants inoculated with TRV:CLA for 2 weeks. (b) Reverse transcription PCR analysis indicated that the transcripts of GhSACPDs were reduced 14 days after infiltration. Photographs were obtained 2 weeks after VIGS. (c) The lesion phenotype on stems of TRV:SACPD plants. Photographs were obtained 3 weeks after VIGS
FIGURE S7 Off‐target examination in virus‐induced gene silencing plants. Quantitative reverse transcription PCR analyses of the target gene and other GhSACPD homologs in TRV:SACPD, TRV:1563, TRV:3299, TRV:0927, TRV:1317, and TRV:1330 plants. Error bars are SE calculated from three biological replicates
FIGURE S8 Disease symptoms of virus‐induced gene silencing plants on Verticillium dahliae inoculation. (a) Disease symptoms of the TRV:3299, TRV:0927, TRV:1317, TRV:1330, TRV:GhSSI2 (TRV:1563), and TRV:00 plants. Photographs were taken 24 days after V. dahliae inoculation. (b) Disease index of TRV:3299, TRV:0927, TRV:1317, TRV:1330, TRV:GhSSI2 (TRV:1563), and TRV:00 plants. Error bars are SE calculated from three biological replicates, with 30 plants in each replicate
FIGURE S9 Disease symptoms of virus‐induced gene silencing plants on Fusarium oxysporum inoculation. (a) Disease symptoms of the TRV:GhSSI2 and TRV:00 plants. Photographs were taken 30 days after F. oxysporum inoculation. (b) Disease index of TRV:GhSSI2 and TRV:00 plants. Error bars are SE calculated from three biological replicates, with 30 plants in each replicate
FIGURE S10 Quantitative reverse transcription PCR analysis indicates that the transcripts of GhICS1, GhPAL1, GhPAL2, and GhPAL3 were reduced in the corresponding double‐gene‐silenced plants. Error bars are SE calculated from three biological replicates (**p < .01, Student’s t test)
TABLE S1SACPD genes identified in Gossypium hirsutum
TABLE S2 Comparison of homology between SACPD family genes in Gossypium hirsutum
TABLE S3 PAL genes identified in Gossypium hirsutum
TABLE S4 Information of 8,663 up‐regulated genes identified by transcriptome comparative analysis between TRV:GhSSI2 and TRV:00
TABLE S5 Gene ontology analysis in biological processes
TABLE S6 PR genes significantly up‐regulated in TRV:GhSSI2s plants
TABLE S7 NB‐LRR genes identified in Gossypium hirsutum
TABLE S8 NBS‐LRR genes significantly up‐regulated in TRV:GhSSI2 plants
TABLE S9 Receptor‐like protein 12 genes identified in Gossypium hirsutum
TABLE S10 RLP genes significantly up‐regulated in TRV:GhSSI2 plants
TABLE S11 Primers used in this study
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (no. 31871672), the China Agriculture Research System (CARS‐15‐03), the Outstanding Youth Fund of Hebei Province (C2019204365), and the Talents Support Program of Hebei Province.
Mo S, Zhang Y, Wang X, et al. Cotton GhSSI2 isoforms from the stearoyl acyl carrier protein fatty acid desaturase family regulate Verticillium wilt resistance. Mol Plant Pathol. 2021;22:1041–1056. 10.1111/mpp.13093
Shaojing Mo, Yan Zhang, and Xingfen Wang contributed equally to this work.
DATA AVAILABILITY STATEMENT
All the data generated or analysed during this study were included in this article and its additional data files.
REFERENCES
- Akira, S., Uematsu, S. & Takeuchi, O. (2006) Pathogen recognition and innate immunity. Cell, 124, 783–801. [DOI] [PubMed] [Google Scholar]
- Alvarez, M.E. (2000) Salicylic acid in the machinery of hypersensitive cell death and disease resistance. Plant Molecular Biology, 44, 429–442. [DOI] [PubMed] [Google Scholar]
- Berens, M.L., Berry, H.M., Mine, A., Argueso, C.T. & Tsuda, K. (2017) Evolution of hormone signaling networks in plant defense. Annual Review of Phytopathology, 55, 401–425. [DOI] [PubMed] [Google Scholar]
- Brodersen, P., Petersen, M., Pike, H.M., Olszak, B., Brian, O., Skov, S. et al. (2002) Knockout of Arabidopsis ACCELERATED‐CELL‐DEATH11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes & Development, 16, 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.K.M. (2002) Yield penalties of disease resistance in crops. Current Opinion in Plant Biology, 5, 339–344. [DOI] [PubMed] [Google Scholar]
- Bruggeman, Q., Raynaud, C., Benhamed, M. & Delarue, M. (2015) To die or not to die? Lessons from lesion mimic mutants. Frontiers in Plant Science, 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byfield, G.E., Xue, H. & Upchurch, R.G. (2006) Two genes from soybean encoding soluble Δ9 stearoyl‐ACP desaturases. Crop Science, 46, 840–846. [Google Scholar]
- Cahoon, E.B., Lindqvist, Y., Schneider, G. & Shanklin, J. (1997) Redesign of soluble fatty acid desaturases from plants for altered substrate specificity and double bond position. Proceedings of the National Academy of Sciences of the United States of America, 94, 4872–4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catinot, J., Buchala, A., Abou‐Mansour, E. & Métraux, J.‐P. (2008) Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana . FEBS Letters, 582, 473–478. [DOI] [PubMed] [Google Scholar]
- Chai, Q., Shang, X., Wu, S., Zhu, G., Cheng, C., Cai, C. et al. (2017) 5‐aminolevulinic acid dehydratase gene dosage affects programmed cell death and immunity. Plant Physiology, 175, 511–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda, B., Xia, Y.E., Mandal, M.K., Yu, K., Sekine, K.‐T., Gao, Q.‐M. et al. (2011) Glycerol‐3‐phosphate is a critical mobile inducer of systemic immunity in plants. Nature Genetics, 43, 421–427. [DOI] [PubMed] [Google Scholar]
- Coll, N.S., Epple, P. & Dangl, J.L. (2011) Programmed cell death in the plant immune system. Cell Death and Differentiation, 18, 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, L., Xu, H., Yi, H., Yang, L., Kong, Z., Zhang, L. et al. (2011) Resistance to hemi‐biotrophic F. graminearum infection is associated with coordinated and ordered expression of diverse defense signaling pathways. PLoS One, 6, e19008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Y., Sun, T., Ao, K., Peng, Y., Zhang, Y., Li, X. et al. (2018) Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell, 173, 1454–1467. [DOI] [PubMed] [Google Scholar]
- Dong, X. (2004) NPR1, all things considered. Current Opinion in Plant Biology, 7, 547–552. [DOI] [PubMed] [Google Scholar]
- Durrant, W.E. & Dong, X. (2004) Systemic acquired resistance. Annual Review of Phytopathology, 42, 185–209. [DOI] [PubMed] [Google Scholar]
- Feys, B., Moisan, L., Newman, M. & Parker, J. (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. The EMBO Journal, 20, 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin, E.F., Zhang, Z., Juarez Ayala, J.C., Castroverde, C.D., Nazar, R.N., Robb, J. et al. (2009) Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1 . Plant Physiology, 150, 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M., Yin, X., Yang, W., Lam, S.M., Tong, X., Liu, J. et al. (2017) GDSL lipases modulate immunity through lipid homeostasis in rice. PLoS Pathogens, 13, e1006724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Q.M., Venugopal, S., Navarre, D. & Kachroo, A. (2011) Low oleic acid‐derived repression of jasmonic acid‐inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiology, 155, 464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Q.‐M., Yu, K., Xia, Y.E., Shine, M.B., Wang, C., Navarre, D. et al. (2014) Mono‐ and digalactosyldiacylglycerol lipids function nonredundantly to regulate systemic acquired resistance in plants. Cell Reports, 9, 1681–1691. [DOI] [PubMed] [Google Scholar]
- Gao, Q.M., Zhu, S., Kachroo, P. & Kachroo, A. (2015) Signal regulators of systemic acquired resistance. Frontiers in Plant Science, 6, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, W., Long, L.u., Zhu, L.‐F., Xu, L.i., Gao, W.‐H., Sun, L.‐Q. et al. (2013) Proteomic and virus‐induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae . Molecular & Cellular Proteomics, 12, 3690–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X., Wheeler, T., Li, Z., Kenerley, C.M., He, P. & Shan, L. (2011) Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. The Plant Journal, 66, 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcion, C., Lohmann, A., Lamodiere, E., Catinot, J., Buchala, A., Doermann, P. et al. (2008) Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis . Plant Physiology, 147, 1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond‐Kosack, K.E. & Jones, J.D. (1996) Resistance gene‐dependent plant defense responses. The Plant Cell, 8, 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, M. (2000) Hypersensitive response‐related death. Plant Molecular Biology, 44, 321–334. [DOI] [PubMed] [Google Scholar]
- Jiang, C.‐J., Hasegawa, M., Shimono, M., Sugano, S., Maeda, S., Inoue, H. et al. (2009) Suppression of the rice fatty‐acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Molecular Plant‐Microbe Interactions, 22, 820–829. [DOI] [PubMed] [Google Scholar]
- Jung, H.W., Tschaplinski, T.J., Wang, L., Glazebrook, J. & Greenberg, J.T. (2009) Priming in systemic plant immunity. Science, 324, 89–91. [DOI] [PubMed] [Google Scholar]
- Kachroo, A., Fu, D., Havens, W., Navarre, D., Kachroo, P. & Ghabrial, S. (2008) An oleic acid‐mediated pathway induces constitutive defense signaling and enhanced resistance to multiple pathogens in soybean. Molecular Plant‐Microbe Interactions, 21, 564–575. [DOI] [PubMed] [Google Scholar]
- Kachroo, A. & Kachroo, P. (2009) Fatty Acid‐derived signals in plant defense. Annual Review of Phytopathology, 47, 153–176. [DOI] [PubMed] [Google Scholar]
- Kachroo, A., Shanklin, J., Whittle, E., Lapchyk, L., Hildebrand, D. & Kachroo, P. (2007) The Arabidopsis stearoyl‐acyl carrier protein‐desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Molecular Biology, 63, 257–271. [DOI] [PubMed] [Google Scholar]
- Kachroo, A., Venugopal, S.C., Lapchyk, L., Falcone, D., Hildebrand, D. & Kachroo, P. (2004) Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis . Proceedings of the National Academy of Sciences of the United States of America, 101, 5152–5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo, P., Kachroo, A., Lapchyk, L., Hildebrand, D. & Klessig, D.F. (2003) Restoration of defective cross talk in ssi2 mutants: role of salicylic acid, jasmonic acid, and fatty acids in SSI2‐mediated signaling. Molecular Plant‐Microbe Interactions, 16, 1022–1029. [DOI] [PubMed] [Google Scholar]
- Kachroo, P., Shanklin, J., Shah, J., Whittle, E.J. & Klessig, D.F. (2001) A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proceedings of the National Academy of Sciences of the United States of America, 98, 9448–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzai, Y., Kimura, M., Watanabe, M., Kusunoki, K., Osaka, D., Suzuki, T. et al. (2018) Salicylic acid‐dependent immunity contributes to resistance against Rhizoctonia solani, a necrotrophic fungal agent of sheath blight, in rice and Brachypodium distachyon . New Phytologist, 217, 771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E., Kato, N. & Lawton, M. (2001) Programmed cell death, mitochondria and the plant hypersensitive response. Nature, 411, 848–853. [DOI] [PubMed] [Google Scholar]
- Lim, G.‐H., Singhal, R., Kachroo, A. & Kachroo, P. (2017) Fatty acid‐ and lipid‐mediated signaling in plant defense. Annual Review of Phytopathology, 55, 505–536. [DOI] [PubMed] [Google Scholar]
- Liu, T., Song, T., Zhang, X., Yuan, H., Su, L., Li, W. et al. (2014) Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nature Communications, 5, 4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, M.K., Chandra‐Shekara, A.C., Jeong, R.D., Yu, K., Zhu, S., Chanda, B. et al. (2012) Oleic acid‐dependent modulation of NITRIC OXIDE ASSOCIATED1 protein levels regulates nitric oxide‐mediated defense signaling in Arabidopsis . The Plant Cell, 24, 1654–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, Y., Xu, L., He, X., Zhang, L., Shaban, M., Zhang, X. et al. (2019) Suppression of tryptophan synthase activates cotton immunity by triggering cell death via promoting SA synthesis. The Plant Journal, 98, 329–345. [DOI] [PubMed] [Google Scholar]
- Molina, A., Volrath, S., Guyer, D., Maleck, K., Ryals, J. & Ward, E. (1999) Inhibition of protoporphyrinogen oxidase expression in Arabidopsis causes a lesion‐mimic phenotype that induces systemic acquired resistance. The Plant Journal, 17, 667–678. [DOI] [PubMed] [Google Scholar]
- Muthamilarasan, M. & Prasad, M. (2013) Plant innate immunity: an updated insight into defense mechanism. Journal of Biosciences, 38, 433–449. [DOI] [PubMed] [Google Scholar]
- Ning, Y., Liu, W. & Wang, G.L. (2017) Balancing immunity and yield in crop plants. Trends in Plant Science, 22, 1069–1079. [DOI] [PubMed] [Google Scholar]
- Nomura, K., Mecey, C., Lee, Y.N., Imboden, L.A., Chang, J.H. & He, S.Y. (2011) Effector‐triggered immunity blocks pathogen degradation of an immunity‐associated vesicle traffic regulator in Arabidopsis . Proceedings of the National Academy of Sciences of the United States of America, 108, 10774–10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noutoshi, Y., Okazaki, M., Kida, T., Nishina, Y., Morishita, Y., Ogawa, T. et al. (2012) Novel plant immune‐priming compounds identified via high‐throughput chemical screening target salicylic acid glucosyltransferases in Arabidopsis . The Plant Cell, 24, 3795–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongena, M., Duby, F., Rossignol, F., Fauconnier, M., Dommes, J. & Thonart, P. (2004) Stimulation of the lipoxygenase pathway is associated with systemic resistance induced in bean by a nonpathogenic Pseudomonas strain. Molecular Plant‐Microbe Interactions, 17, 1009–1018. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J. & Van Loon, L.C. (2004) NPR1: the spider in the web of induced resistance signaling pathways. Current Opinion in Plant Biology, 7, 456–464. [DOI] [PubMed] [Google Scholar]
- Qin, J., Wang, K., Sun, L., Xing, H., Wang, S., Li, L. et al. (2018) The plant‐specific transcription factors CBP60g and SARD1 are targeted by a Verticillium secretory protein VdSCP41 to modulate immunity. eLife, 7, e34902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietz, S., Stamm, A., Malonek, S., Wagner, S., Becker, D., Medina‐Escobar, N. et al. (2011) Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytologist, 191, 107–119. [DOI] [PubMed] [Google Scholar]
- Robert‐Seilaniantz, A., Grant, M. & Jones, J.D.G. (2011) Hormone crosstalk in plant disease and defense: more than just JASMONATE‐SALICYLATE antagonism. Annual Review of Phytopathology, 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., Nandi, A. & Klessig, D.F. (2001) A recessive mutation in the Arabidopsis SSI2 gene confers SA‐ and NPR1‐independent expression of PR genes and resistance against bacterial and oomycete pathogens. The Plant Journal, 25, 563–574. [DOI] [PubMed] [Google Scholar]
- Shanklin, J. & Somerville, C. (1991) Stearoyl‐acyl‐carrier‐protein desaturase from higher plants is structurally unrelated to the animal and fungal homologs. Proceedings of the National Academy of Sciences of the United States of America, 88, 2510–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekara, A.C.C., Venugopal, S.C., Barman, S.R., Kachroo, A. & Kachroo, P. (2007) Plastidial fatty acid levels regulate resistance gene‐dependent defense signaling in Arabidopsis . Proceedings of the National Academy of Sciences of the United States of America, 104, 7277–7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine, M.B., Yang, J.‐W., El‐Habbak, M., Nagyabhyru, P., Fu, D.‐Q., Navarre, D. et al. (2016) Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytologist, 212, 627–636. [DOI] [PubMed] [Google Scholar]
- Shirasu, K. & Schulze‐Lefert, P. (2000) Regulators of cell death in disease resistance. Plant Molecular Biology, 44, 371–385. [DOI] [PubMed] [Google Scholar]
- Silverman, P., Seskar, M., Kanter, D., Schweizer, P., Metraux, J.‐P. & Raskin, I. (1995) Salicylic acid in rice (biosynthesis, conjugation, and possible role). Plant Physiology, 108, 633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocombe, S.P., Cummins, I., Jarvis, R.P. & Murphy, D.J. (1992) Nucleotide sequence and temporal regulation of a seed‐specific Brassica napus cDNA encoding a stearoyl‐acyl carrier protein (ACP) desaturase. Plant Molecular Biology, 20, 151–155. [DOI] [PubMed] [Google Scholar]
- Song, N.A., Hu, Z., Li, Y., Li, C., Peng, F., Yao, Y. et al. (2013) Overexpression of a wheat stearoyl‐ACP desaturase (SACPD) gene TaSSI2 in Arabidopsis ssi2 mutant compromise its resistance to powdery mildew. Gene, 524, 220–227. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H. & Dong, X. (2012) How do plants achieve immunity? Defence without specialized immune cells. Nature Reviews Immunology, 12, 89–100. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H., Koornneef, A., Claessens, S.M.C., Korzelius, J.P., Van Pelt, J.A., Mueller, M.J. et al. (2003) NPR1 modulates cross‐talk between salicylate‐ and jasmonate‐dependent defense pathways through a novel function in the cytosol. The Plant Cell, 15, 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L., Zhu, L., Xu, L., Yuan, D., Min, L. & Zhang, X. (2014) Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nature Communications, 5, 5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, K., Dong, C. & Liu, J. (2016) Genome‐wide analysis and expression profiling of the phospholipase D gene family in Gossypium arboreum . Science China Life Science, 59, 130–141. [DOI] [PubMed] [Google Scholar]
- Uppalapati, S.R., Ishiga, Y., Wangdi, T.W., Kunkel, B.N., Anand, A., Mysore, K.S. et al. (2007) The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Molecular Plant‐Microbe Interactions, 20, 955–965. [DOI] [PubMed] [Google Scholar]
- Vallad, G.E. & Goodman, R.M. (2004) Systemic acquired resistance and induced systemic resistance in conventional agriculture. Crop Science, 44, 1920–1934. [Google Scholar]
- Venugopal, S.C., Jeong, R.‐D., Mandal, M.K., Zhu, S., Chandra‐Shekara, A.C., Xia, Y.E. et al. (2009) Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene‐mediated signaling. PLoS Genetics, 5, e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley, J.W., Kliebenstein, D.J., Bostock, R.M. & Dehesh, K. (2013) Fatty acids and early detection of pathogens. Current Opinion in Plant Biology, 16, 520–526. [DOI] [PubMed] [Google Scholar]
- Wang, C., He, X., Li, Y., Wang, L., Guo, X. & Guo, X. (2018) The cotton MAPK kinase GhMPK20 negatively regulates resistance to Fusarium oxysporum by mediating the MKK4–MPK20–WRKY40 cascade. Molecular Plant Pathology, 19, 1624–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G., Wang, X., Zhang, Y., Yang, J., Li, Z., Wu, L. et al. (2021) Dynamic characteristics and functional analysis provide new insights into long non‐coding RNA responsive to Verticillium dahliae infection in Gossypium hirsutum . BMC Plant Biology, 21, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. & Hause, B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany, 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth, M.C., Dewdney, J., Wu, G. & Ausubel, F.M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Xu, F., Yang, L., Zhang, J., Guo, X., Zhang, X., & Li, G. (2012) Prevalence of the defoliating pathotype of Verticillium dahliae on cotton in central China and virulence on selected cotton cultivars. Journal of Phytopathology, 160, 369–376. [Google Scholar]
- Yaeno, T., Matsuda, O. & Iba, K. (2004) Role of chloroplast trienoic fatty acids in plant disease defense responses. The Plant Journal, 40, 931–941. [DOI] [PubMed] [Google Scholar]
- Yang, J., Wang, G., Ke, H., Zhang, Y., Ji, L., Huang, L. et al. (2019) Genome‐wide identification of cyclophilin genes in Gossypium hirsutum and functional characterization of a CYP with antifungal activity against Verticillium dahliae . BMC Plant Biology, 19, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W., Dong, R., Liu, L.i., Hu, Z., Li, J., Wang, Y. et al. (2016) A novel mutant allele of SSI2 confers a better balance between disease resistance and plant growth inhibition on Arabidopsis thaliana . BMC Plant Biology, 16, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Ling, X., Chen, T., Cai, L., Liu, T., Wang, J. et al. (2015) A cotton Gbvdr5 gene encoding a leucine‐rich‐repeat receptor‐like protein confers resistance to Verticillium dahliae in transgenic Arabidopsis and upland cotton. Plant Molecular Biology Reporter, 33, 987. [Google Scholar]
- Ylva, L., Weijun, H., Gunter, S. & John, S. (1996) Crystal structure of Δ9 stearoyl‐acyl carrier protein desaturase from castor seed and its relationship to other di‐iron proteins. The EMBO Journal, 15, 4081–4092. [PMC free article] [PubMed] [Google Scholar]
- Zhang, B., Yang, Y., Chen, T., Yu, W., Liu, T., Li, H. et al. (2012) Island cotton Gbve1 gene encoding a receptor‐like protein confers resistance to both defoliating and non‐defoliating isolates of Verticillium dahliae . PLoS One, 7, e51091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Li, J., Garcia‐Ruiz, H., Bates, P.D., Mirkov, T.E. & Wang, X. (2014) A stearoyl‐acyl carrier protein desaturase, NbSACPD‐C, is critical for ovule development in Nicotiana benthamiana . The Plant Journal, 80, 489–502. [DOI] [PubMed] [Google Scholar]
- Zhang, M., Barg, R., Yin, M., Gueta‐Dahan, Y., Leikin‐Frenkel, A., Salts, Y. et al. (2005) Modulated fatty acid desaturation via overexpression of two distinct ω‐3 desaturases differentially alters tolerance to various abiotic stresses in transgenic tobacco cells and plants. The Plant Journal, 44, 361–371. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Wang, F., Wu, W., Wang, D., Yang, W., Sun, J. et al. (2016) Characterization and genetic analysis of a novel light‐dependent lesion mimic mutant, lm3, showing adult‐plant resistance to powdery mildew in common wheat. PLoS One, 11, e0155358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Maximova, S.N. & Guiltinan, M.J. (2015) Characterization of a stearoyl‐acyl carrier protein desaturase gene family from chocolate tree, Theobroma cacao L. Frontiers in Plant Science, 6, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Smith, P., Maximova, S.N. & Guiltinan, M.J. (2015) Application of glycerol as a foliar spray activates the defence response and enhances disease resistance of Theobroma cacao . Molecular Plant Pathology, 16, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Wang, X., Rong, W., Yang, J., Li, Z., Wu, L. et al. (2017) Histochemical analyses reveal that stronger intrinsic defenses in Gossypium barbadense than in G. hirsutum are associated with resistance to Verticillium dahliae . Molecular Plant‐Microbe Interactions, 30, 984–996. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Wang, X., Yang, S., Chi, J., Zhang, G. & Ma, Z. (2011) Cloning and characterization of a Verticillium wilt resistance gene from Gossypium barbadense and functional analysis in Arabidopsis thaliana . Plant Cell Reports, 30, 2085–2096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Distribution of the GhSACPD gene family in the Gossypium hirsutum genome. There were 19 copies of GhSACPD distributed across different cotton chromosomes and the horizontal lines indicate the relative position of each copy on the chromosomes
FIGURE S2 Phylogenetic relationship and gene structure of GhSACPD genes. Exons are represented by yellow boxes and introns by black lines
FIGURE S3 Weblogo graph of the SACPD gene family in cotton. Amino acid sequences between the two red triangles indicate the FA_desaturase_2 domain
FIGURE S4 Multiple amino acid sequence alignment of GhSACPD with SACPD from castor bean (RcSAD1) and Arabidopsis (AtSSI2). Secondary protein structures of SACPD are annotated according to the crystal structure of RcSAD1. Backgrounds indicate percentage of amino acid similarity: black, 100%; dark grey, 75%; light grey, 50%
FIGURE S5 Construction of pTRV2 vectors based on diversity sequence. (a) The fragments using for each grouped silence were shaded. The nucleotides highlighted in red indicate the initiation codon. According to the gene ID the vectors are marked TRV:3299, TRV:0927, TRV:1563, TRV:1317, and TRV:1330. (b) Fragment using for suppressing the whole SACPD gene family
FIGURE S6 Morphology of the cotton virus‐induced gene silencing (VIGS) experiment and expression analysis of GhSACPDs in GhSACPD KD plants. (a) Photobleaching phenotype of the plants inoculated with TRV:CLA for 2 weeks. (b) Reverse transcription PCR analysis indicated that the transcripts of GhSACPDs were reduced 14 days after infiltration. Photographs were obtained 2 weeks after VIGS. (c) The lesion phenotype on stems of TRV:SACPD plants. Photographs were obtained 3 weeks after VIGS
FIGURE S7 Off‐target examination in virus‐induced gene silencing plants. Quantitative reverse transcription PCR analyses of the target gene and other GhSACPD homologs in TRV:SACPD, TRV:1563, TRV:3299, TRV:0927, TRV:1317, and TRV:1330 plants. Error bars are SE calculated from three biological replicates
FIGURE S8 Disease symptoms of virus‐induced gene silencing plants on Verticillium dahliae inoculation. (a) Disease symptoms of the TRV:3299, TRV:0927, TRV:1317, TRV:1330, TRV:GhSSI2 (TRV:1563), and TRV:00 plants. Photographs were taken 24 days after V. dahliae inoculation. (b) Disease index of TRV:3299, TRV:0927, TRV:1317, TRV:1330, TRV:GhSSI2 (TRV:1563), and TRV:00 plants. Error bars are SE calculated from three biological replicates, with 30 plants in each replicate
FIGURE S9 Disease symptoms of virus‐induced gene silencing plants on Fusarium oxysporum inoculation. (a) Disease symptoms of the TRV:GhSSI2 and TRV:00 plants. Photographs were taken 30 days after F. oxysporum inoculation. (b) Disease index of TRV:GhSSI2 and TRV:00 plants. Error bars are SE calculated from three biological replicates, with 30 plants in each replicate
FIGURE S10 Quantitative reverse transcription PCR analysis indicates that the transcripts of GhICS1, GhPAL1, GhPAL2, and GhPAL3 were reduced in the corresponding double‐gene‐silenced plants. Error bars are SE calculated from three biological replicates (**p < .01, Student’s t test)
TABLE S1SACPD genes identified in Gossypium hirsutum
TABLE S2 Comparison of homology between SACPD family genes in Gossypium hirsutum
TABLE S3 PAL genes identified in Gossypium hirsutum
TABLE S4 Information of 8,663 up‐regulated genes identified by transcriptome comparative analysis between TRV:GhSSI2 and TRV:00
TABLE S5 Gene ontology analysis in biological processes
TABLE S6 PR genes significantly up‐regulated in TRV:GhSSI2s plants
TABLE S7 NB‐LRR genes identified in Gossypium hirsutum
TABLE S8 NBS‐LRR genes significantly up‐regulated in TRV:GhSSI2 plants
TABLE S9 Receptor‐like protein 12 genes identified in Gossypium hirsutum
TABLE S10 RLP genes significantly up‐regulated in TRV:GhSSI2 plants
TABLE S11 Primers used in this study
Data Availability Statement
All the data generated or analysed during this study were included in this article and its additional data files.


