Abstract
Background
Ureaplasma spp. are associated with various infectious diseases in females, but there is still limited evidence regarding whether they are related to nonspecific cervicitis. The aim of this study was to develop and evaluate a digital droplet PCR (ddPCR) assay for the detection and quantification of Ureaplasma spp. in cervical swabs.
Methods
A total of 267 non-specific cervicitis (NSC) patients and 195 asymptomatic females were included in this study. We produced standard curves for Ureaplasma spp. to evaluate the analytical performance of the ddPCR assay. Then, we detected and quantified the bacterial load of Ureaplasma spp. in cervical swabs.
Results
The prevalences of U. parvum were 37.8% (101/267) and 29.7% (58/195), U. urealyticum were 9.0% (24/267) and 8.7% (17/195) in the NSC group and control group, respectively. In addition, the median copy number of U. parvum was 2.5 × 104 copies/ml (n = 101) in the NSC group and 9.2 × 103 copies/ml (n = 58) in the control group. The U. parvum load in the NSC group was significantly higher than that in the asymptomatic individuals (P < 0.001). whereas the median load of U. urealyticum was 8.4 × 103 copies/ml (n = 24) and 1.4 × 103 (n = 17) copies/ml in the two groups, respectively, , the difference was not statistically significant (P = 0.450).
Conclusions
Our study is the first to develop a droplet digital PCR (ddPCR) method for the detection and quantification of Ureaplasma spp. in clinical samples, and the method has excellent analytical performance and a wide range of clinical application prospects.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-021-06355-6.
Keywords: Ureaplasma parvum, Ureaplasma urealyticum, qPCR, Droplet digital PCR, Absolute quantitation, Cervicitis
Introduction
Ureaplasma spp. has 14 known serotypes that were divided into two biovars: Ureaplasma parvum and Ureaplasma urealyticum. This pathogen is suspected to be the causative agents of non-gonococcal urethritis [1], bacterial vaginosis [2], cervicitis [3], and multiple adverse pregnancy outcomes such as spontaneous preterm birth [4], chorioamnionitis [5], and preterm premature rupture of membranes (PPROM) [6]. In addition, some case reports have described rare invasive infections from Ureaplasma spp., such as idiopathic hyperammonemia [7], spontaneous bacterial pericarditis [8], multifocal abscesses [9], and osteomyelitis [10], especially in immunocompromised patients.
However, the roles of U. parvum and U. urealyticum are sometimes controversial. A meta-analysis included seven eligible case-control studies, including 1507 NGU patients and 1223 controls. The results suggested that U. urealyticum was significantly more prevalent inNGU patients, but U. parvum was found more oftenin controls [11]. Nevertheless, several clinical and fundamental types of research havereported that U. parvum induces adverse outcomes by causing a severe inflammatory response [4, 12, 13]. The controversy has been well explainedby Viscardi RM et al. [14], attributing it partly due to the presence of potential confounding factors in specimens, such as U. parvum and U. urealyticum based on culture or PCR assays. These reasons make the interpretation of many previous studies extremely difficult. Moreover, some scholars have proposed that the host genetic background and bacteria load have an impacte on the disease outcomes with Ureaplasma spp. [15, 16].
Of note, The European STI Guidelines Editorial Board issued a position statement that only people with a high Ureaplasma spp. load should be considered for treatment [17]. The quantitative bacterial load in all previous studies has used qPCR technology with external references [18, 19]; undoubtedly, it is a challengingto generatea standard curve. Digital droplet PCR (ddPCR) is a method of absolute nucleic acid quantification without external references and robustness to variations in PCR efficiency [20, 21]. Several studies have showed that ddPCR has much higher sensitivity and is more resistant to PCR inhibition than qPCR [22, 23]. A recent study suggested that the copy numbers obtained by qPCR were 1 to 31 times higher than those obtained byddPCR (mean: 9.7; SD 7.7) [24], indicating that ddPCR has more accurate quantitative capabilities than qPCR.
Hence, the purpose of this study was to develop ddPCR for the detection and accurate quantification of Ureaplasma spp. in non-specific cervicitis patients and an asymptomatic physical examination population (control group).
Materials and methods
Study population
Patients attending the gynaecology outpatient or health clinic in Minhang Hospital, Fudan University were recruited for this study from July 2019 to November 2020. All women 1) were aged ≥18 years without co-infection with HIV or autoimmune diseases, 2) had exhibited symptoms of cervical discharge (purulent or mucopurulent endocervical exudate) or cervical bleeding [25], 3) had not received the gynaecologic intervention or antibiotics treatment within the preceding 3 months, 4) had not used an intrauterine contraceptive device and had been sexually active in last 3 months, 5) and were not currently pregnant or menstruating.
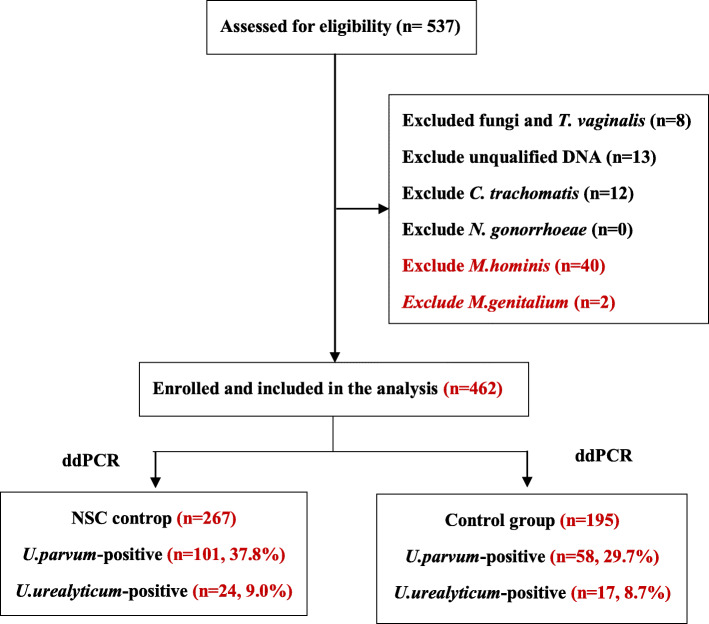
A total of 537 cervical samples were obtained according to protocols performed by experienced clinicians. All specimens were examined microscopically to rule out fungi and Trichomonas vaginalis and then subjected to HPV, C. trachomatis, N. gonorrhoea, M. hominis, and M. genitalium, U. parvum, and U. urealyticum testing by qPCR in septuplicate. Finally, a total of 462 samples were ultimately enrolled in this study, including 267 non-specific cervicitis patients [26, 27] and 195 asymptomatic individuals.
Specimen collection and DNA extraction
Total DNA from 200 μl fresh cervical swab samples was extracted using MiniBest Universal Genomic DNA Extraction Kit (Takara) following the manufacturer’s instructions. The extracts were resuspended in 100 μl of nuclease-free water and stored at − 20 °C or − 80 °C until use. The concentration of nucleic acids was determined with a NanoDrop 2000 Spectrophotometer (Thermo Scientific, USA). Human papillomavirus (HPV), C. trachomatis, N. gonorrhoea, M. hominis, and M. genitalium were confirmed by commercial kits based on TaqMan probes (Shanghai Kehua Bio-Engineering, or ZJ Bio-Tech Co., Ltd., Shanghai, China) according to the manufacturer’s instructions. U. parvum serovar 1 (ATCC 27813) and U. urealyticum serovar 4 (ATCC 27816) were used as standard strains.
Real-time quantitative (qPCR)
QPCR amplification was performed in a total volume of 20 μl, including the 10 μl of Probe qPCR Mix (with UNG) (Takara), 500 nM target/reference primers, 250 nM target/reference probes, and 9 μl of each template DNA. The qPCR conditions were as follows: 25 °C for 10 min, followed by 45 cycles of 15 s at 95 °C and 34 s at 60 °C. Fluorescent accumulation data were analysed using ABI 7500 Software Version 2.3 (Applied Biosystems). Threshold cycle (CT) values of < 40 were defined as a positive result for the U. parvum and U. urealyticum, respectively. In addition, the quality of PCR extraction was monitored by amplifying human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in cervical swabs. CT values of > 37 suggest the failure of DNA extraction or qPCR amplification. Standard curves were generated by plotting the CT of the qPCR performed on a ten fold dilution series of purified DNA from U. parvum and U. urealyticum.
The primers and probes were designed based on the ParC gene conserved regions of U. parvum and U. urealyticum, respectively (Additional file 1), which have previously been used for qPCR [28].
U. parvum, forward: 5′-TAGTTGCTCATAAAATCAC-3′,
Reverse: 5′-CGTTCCATATATAAACAGCTATAAC-3′,
Probe: 5′- FAM-CTATGCGTGAAAAGATG-BHQ1–3′;
U. urealyticum, forward: 5′- TAAGTGTTTTAGTATTAGTGAGC-3′;
Reverse: 5′- TGCTGCTAAAACGCTTTGTGC-3’C;
Probe: 5′- FAM-CACCATCACCACTTTATT-BHQ1–3′.
Digital droplet PCR (ddPCR)
The same primers and probes were used for ddPCR reactions as qPCR. ddPCR was performed using a QX200 Droplet Digital PCR system (Bio-Rad). The Mastermix for ddPCR included 1× ddPCR Supermix for Probes (no dUTP), 900 nM target/reference primers, 250 nM target/reference probes, and 9 μl sample DNA. The generation of droplets was performed by the QX200 Droplet Generator according to the manufacturer’s protocols. PCR amplification was carried out using an Applied Biosystems Veriti 96-Well Thermal Cycler with the following PCR conditions: 95 °C for 10 min followed by 40 cycles of denaturation at 94 °C for 30 s, 60 °C for 1 min and the enzyme was deactivated at 98 °C for 10 min. The plate was stored at 4 °C until the droplets were analysed with QX200 Droplet Reader and QuantaSoft software version 1.7.4 (Bio-Rad). The threshold between positive and negative droplet populations was set manually using per-plate positive and no-template controls as a guide.
Diagnostic performance of ddPCR for Ureaplasma spp.
First, the specificity of the primers and probes for U. parvum and U. urealyticum were evaluated with 12 kinds of common microorganisms of the genitourinary tract, including M. hominis, M. genitalium, C. trachomatis, Human papillomavirus (HPV), Herpes simplex virus (HSV), N. gonorrhoea, S. agalactiae, S. aureus, E. faecium, E. coli, P. aeruginosa, C. albicans. Second, the linear dynamic range of the ddPCR assay was assessed using the serial 10-fold dilutions of the standard DNA containing the target region as described previously [18]. Each dilution in the series was tested in three technical replicate wells. DNase/RNase-free water was used as the negative control (NC). The analytical limit of the assay was determined to be the concentration of the maximum dilution factor of the series in which all 10 replicates tested positive. To compare the dynamic range of ddPCR and qPCR, 10-fold serial dilutions of U. parvum and U. urealyticum were tested using the same primers/probe sets for both ddPCR and qPCR. Octuplicate results were used for intra-assay and inter-assay calculations.
Statistical analysis
Continuous variables are expressed as “the mean ± standard deviation”. Intra- and interassay coefficients of variability (CV) were calculated for methodological evaluation. Continuous variables between the two groups were compared using Student's t-test. The medians were compared using Wilcoxon rank sum test. The Pearson correlation coefficient was performed to estimate the correlation betweenqPCR and ddPCR results of clinical samples. A P-value < 0.05 was considered a significant difference. The data were calculated in Stata 13.0 (StataCorp, TX, USA) and plots of linear regression were generated with GraphPad Prism 8.0 Software, Inc.
Results
Analytical performance of the ddPCR assay
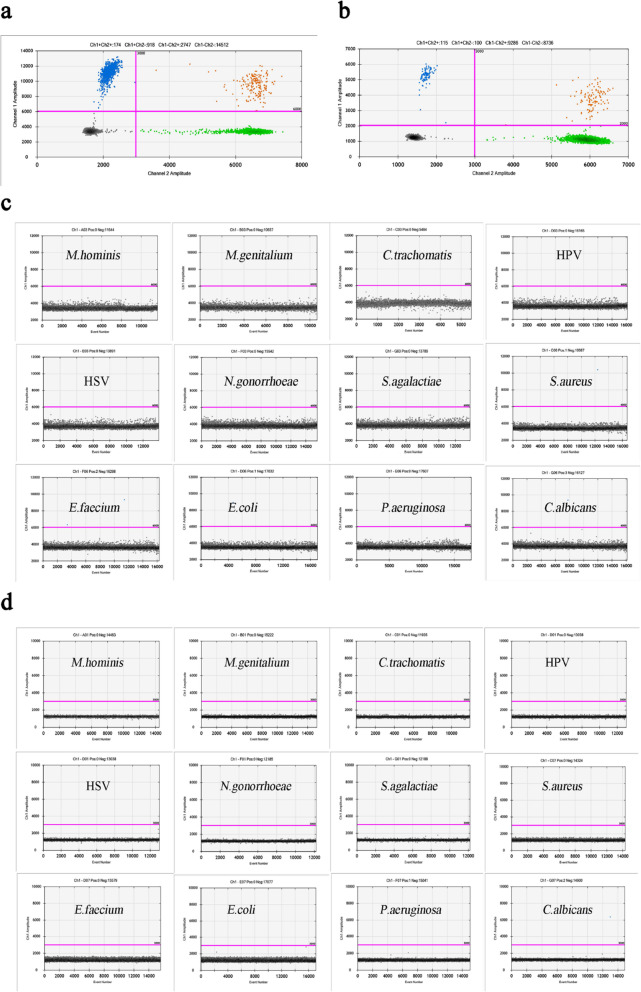
We found positive droplets of U. parvum and U. urealyticum with channel amplitude signals above 6000 and 2000, respectively (Fig. 1). The ddPCR assay showed excellent specificity with no cross-reaction with twelve kinds of urogenital microorganisms. In addition, the ddPCR assay exhibited excellent intra- and inter assay coefficients of variability (CV). ddPCR has a lower intra-assay CV (0.36–0.42%) and a lower inter-assay CV (0.78–1.59%) for U. parvum, as well as a lower intra-assay CV (0.31–0.81%) and a lower inter-assay CV (1.43–1.48%) for the U. urealyticum, respectively (Table 1).
Fig. 1.
Specificity of Primers and probes for U. parvum and U. urealyticum. a Positive droplets of U. parvum with channel amplitude signals above 6000. b Positive droplets of U. urealyticum with channel amplitude signals above 2000. c and d The primers and probes for U. parvum (c) and U. urealyticum (d) showed excellent specificity, without cross-reaction to 12 kinds of common microorganisms of the genitourinary tract, including M. hominis, M.genitalium, C. trachomatis, Human papilloma virus, Herpes simplex virus, N. gonorrhoeae, S.agalactiae, S. aureus, E. faecium, E. coli, P. aeruginosa, and C. albicans
Table 1.
The diagnostic performance of ddPCR for Ureaplasma spp.
| Analytical performance | U. parvum | U. urealyticum |
|---|---|---|
| Specificity (%) | 100 | 100 |
| Liner range (copies/μl) | 3.20 ~ 3200 | 1.50–1500 |
| The limit of detection | 0.64 | 1.1 |
| Intra-assay CV (%) | 0.36–0.42 | 0.31–0.81 |
| Inter-assay CV (%) | 0.78–1.59 | 1.43–1.48 |
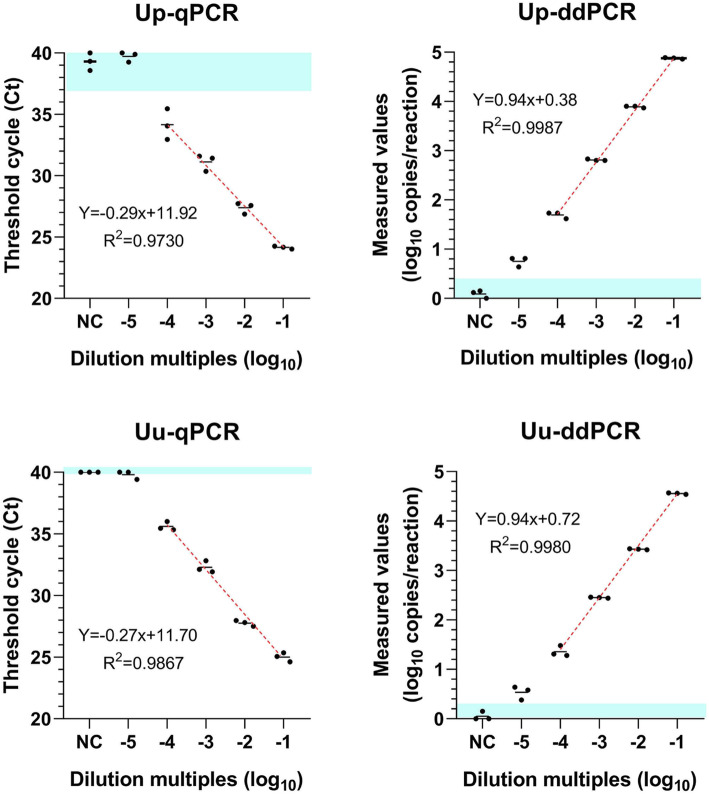
As shown in Fig. 2, the liner range of ddPCR was 3.2–3200 copies/μl with R2 = 0.9987 for U. parvum and was 1.5–1500 copies/μl with R2 = 0.9980 for U. urealyticum, respectively. The limit of detection of qPCR is ten times for that of ddPCR for U. parvum and U. urealyticum (Fig. 2).
Fig. 2.
Comparison of the dynamic range of ddPCR and qPCR. A plot of the results from a linearity experiment to compare the correlation coefficient of ddPCR and qPCR. Dilution multiples are plotted on the X-axis measured values of qPCR (A and C) and ddPCR (converted to log10) are plotted on the Y-axis using GraphPad Prism 8.0. The data represent three independent experiments, with each concentration repeated 3 times (mean ± SD). Linear regression of the ddPCR assay for ten fold serially diluted DNA of Ureaplasma spp.
Prevalence of Ureaplasma spp. in the NSC and control groups
There were 267 females with non-specific cervicitis in the case group, with a mean age of 40.56 (range 20–74 years; SD ± 11.06 years), and 195 asymptomatic females in the control group, with a mean age of 43.37 (range 23–68 years; SD ± 8.03 years). There were no differences in age between the two groups (P > 0.05). The prevalence of U. parvum was 37.8% (101/267) in the NSC group and 29.7% (58/195) in the control group, respectively. The prevalence of U. urealyticum was 9.0%24/267) in the NSC group and 8.7%17/195) in the control group (Fig. 3).
Fig. 3.
Schematic flow chart showing the inclusion specimens in this study
Correlation analysis of ddPCR and qPCR
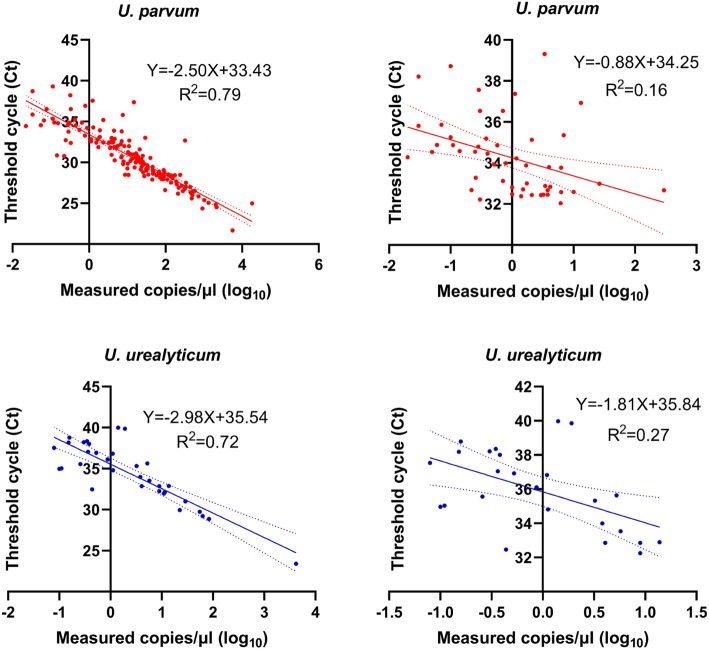
U. parvum and U. urealyticum positive specimens were tested by ddPCR and qPCR simultaneously. The correlation coefficients between ddPCR and qPCR were 0.79 and 0.72 for U. parvum and U. urealyticum, respectively. However, it is worth noting that the Spearman correlation coefficient was very low when the Ct value was greater than 32 (R2 = 0.16 for U. parvum and R2 = 0.27 for U. urealyticum) (Fig. 4).
Fig. 4.
Correlation of U. parvum and U. urealyticum between qPCR and ddPCR assays. Correlation and regression analysis of ddPCR and qPCR. Measured copies are plotted on the X-axis (converted to log10) and Ct values of qPCR on the Y-axis using GraphPad Prism 8.0
Absolute quantitation of U. parvum and U. urealyticum
Based on ddPCR, the median bacterial load for U. parvum was 2.5 × 104 copies/ml in the NSC group (n = 101) and 9.2 × 103 copies/ml in the control group (n = 58). However, the median load for U. urealyticum was 8.44 × 103 copies/ml in the NSC group (n = 24) and 1.38 × 103 copies/ml in the control group (n =17 ) (Table 2). The load of U. parvum was significantly different between the NSC and control groups, whereas, the load of U. urealyticum was not significantly different between the two groups (P > 0.05).
Table 2.
Copy numbers of U. parvum and U. urealyticum by ddPCR in the NSC and control groups
| Group | U. parvum | U. urealyticum | ||||
|---|---|---|---|---|---|---|
| N | Median copies/ml (IQR) | P | N | Median copies/ml (IQR) | P | |
| NSC | 101 |
2.5 × 104 (6.0 × 103 -1.1 × 105) |
0.000 | 19 |
8.4 × 103 (1.2 × 103 -3.1 × 104) |
0.450 |
| Control | 58 |
9.2 × 103 (5.9 × 102 -3.4 × 104) |
14 |
1.4 × 103 (5.2 × 102–2.5 × 104) |
||
Discussion
The purpose of this study was to develop a ddPCR assay for the detection and quantification of Ureaplasma spp.. According to the standard curve of the ddPCR assay for DNA from ATCC reference strains of Ureaplasma spp., the assays showed excellent analytical performance. Our data suggested that the precision of ddPCR is higher than that of qPCR, especially for the low-copy specimens, similar to the report of Mahendran P[29]. It can contribute to the results that the correlation of Ureaplasma spp. was higher between ddPCR and qPCR methods, (Fig. 4) but very poor when the Ct value was larger than 32 in clinical samples. Thus, ddPCR would more accurately quantify the load of Ureaplasma spp. than qPCR assay, Nevertheless, the Ureaplasma spp. load was assessed by qPCR assay in previous studies [18, 30]. According to Table 2, the absolutely copy number in the non-specific cervicitis group was higher than that in the control group (P = 0.003). In contrast, the difference in U. urealyticum copy number between the two groups was not statistically significant (P = 0.450).
Ureaplasma spp. can be isolated from blood, amniotic fluid, cerebrospinal fluid, sputum, bronchoalveolar lavage, pleural fluid, and semen [14]. Here, we for the first time quantify the precise bacterial load of U. parvum and U. urealyticum using ddPCR technology. It is generally considered that Ureaplasma spp. load ≥104 CCU/ml is a sign of active infection and may require treatment with antibiotics. The establishment of this reference value is liquid culture based. However, traditional liquid culture is only a qualitative method with relatively poor sensitivity, with false positive and false negative, as well as the inability to distinguish U. parvum and U. urealyticum [31]. Real-time PCR has become a common technique in molecular diagnosis and overcomes the above-mentioned shortcomings of liquid culture, with relative quantification through a standard curve, though the construction of a standard curve is complicated and time-consuming. Moreover, the quantitative accuracy of qPCR is easily affected by amplification efficiency and PCR inhibitors.
It’s becoming gradually accepted that role of Ureaplasma spp. as a pathogen in multiple diseases with increasing clinical observation studies [32, 33]. In addition, several in vitro studis have demonstrated that Ureaplasma spp. modulate cytokine [34, 35] and pro-inflammatory responses in a dose-dependent manner [36]. Interestingly, one study indicated that a high load of U. parvum (107 CFU/ml) can significantly increase prothrombin/thrombin mRNA expression to promote the onset of PPROM but that a lower load (105 CFU/ml) had no significant effect [6]. It indicated that there may exist a threshold effect in various association diseases with U. parvum infection. Therefore, it is vital to detect the load of Ureaplasma spp. through more precisely quantitative technology. At this time, ddPCR, an emerging technology, is partitioned the PCR reaction into about 20,000 nm-sized droplets. Then, the relative concentration of targets, primers and probes are higher, while the inhibitor is relatively lower, which perfectly overcoming the shortcomings of conventional qPCR [23].
Our study had several limitations. First, due to the specialized instruments and consumables, ddPCR has a higher cost than qPCR, which limits its clinical application to a large extent. In addition, the prevalence of U. urealyticum is relatively lower and the number of populations recruited for the study was relatively small. Finally, this was a single-centre cross-sectional study, and multicenter prospective studies with a large population should be carried out to provide more evidence.
The ddPCR assay that we established has great application prospects. It can more accurately quantify the burden of Ureaplasma spp. than qPCR in both asymptomatic and symptomatic individuals, and monitor dynamic changes in bacterial load after the use of antibiotics. Moreover, based on its high sensitivity and high tolerance to PCR inhibitors of ddPCR, it can be used for detection of samples with low bacterial content that are readily available (urine, amniotic fluid, semen, peripheral blood, etc). Further, it can be used with chorionic amniotic tissue or lung tissue which may contain potential PCR inhibitors, promoting analysis of multiple potential diseases, such as infertility, miscarriage, chorioamnionitis, bronchopulmonary dysplasia, bacteremia, and rare invasive infections of Ureaplasma spp.
In conclusion, this study is the first to report the absolute quantitative analysis of Ureaplasma spp. with excellent analytical performance, which may be a promising technology.
Supplementary Information
Additional file 1. Multiple sequence alignment of the ParC genes of 14 serotypes of Ureaplasma spp. (a) Sequence of the U. parvum primes/probe; (b) Sequence of the U. ureaplasma primers/probe.
Acknowledgements
We are sincerely obliged to Biomed-Union Co. Ltd. (Shanghai, China) for excellent technical assistance with ddPCR technology.
Abbreviations
- ddPCR
Digital droplet PCR
- U. parvum
Ureaplasma parvum
- U. urealyticum
Ureaplasma urealyticum
- NSC
Non-specific cervicitis
- PPROM
Preterm premature rupture of membranes
- HPV
Human papillomavirus
- CT
Threshold cycle
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
Authors’ contributions
Yanfang Huang designed the experiments, collected clinical specimens, performed the experiments and analysed the data, and drafted the manuscript. Huifen Pan designed and performed the experiments, analysed the data and obtained funding. Xiaoqin Xu and Panpan Lv collected the clinical specimens and performed the experiments. Xinxin Wang performed the experiments. Zhen Zhao designed the experiments, analysed the data, obtained funding, supervised the study, and drafted the manuscript. All authors participated in revising of the manuscript for important intellectual content. All authors read and approved the final manuscript.
Funding
This work was supported by the Minhang District Science and Technology Committee (Grant numbers 2017MHZ82).
Availability of data and materials
The data generated during the current study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
The study was approved by the Ethical Committee of the Minhang Hospital, Fudan University.
The study abides by the ethical norms and principles for research as established by the Declaration of Helsinki. Informed consent was obtained from all subjects. All patients gave permission for the collection of their cervical swabs and for their medical records to be accessed and recorded.
Consent for publication
No.
Competing interests
The authors have declared that no conflicts of interest exist. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yanfang Huang and Huifen Pan contributed equally to this work.
References
- 1.Beeton ML, Payne MS, Jones L. The Role of Ureaplasma spp. in the Development of Nongonococcal Urethritis and Infertility among Men. Clin Microbiol Rev. 2019;32(4):e00137–18. [DOI] [PMC free article] [PubMed]
- 2.Xiao B, Wu C, Song W, Niu X, Qin N, Liu Z, et al. Association analysis on recurrence of bacterial vaginosis revealed Microbes and clinical variables important for treatment outcome. Front Cell Infect Mi. 2019;9:189. [DOI] [PMC free article] [PubMed]
- 3.Lusk MJ, Garden FL, Rawlinson WD, Naing ZW, Cumming RG, Konecny P. Cervicitis aetiology and case definition: a study in Australian women attending sexually transmitted infection clinics. Sex Transm Infect. 2016;92(3):175–181. doi: 10.1136/sextrans-2015-052332. [DOI] [PubMed] [Google Scholar]
- 4.Rittenschober-Böhm J, Waldhoer T, Schulz SM, Pimpel B, Goeral K, Kasper DC, Witt A, Berger A. Vaginal Ureaplasma parvum serovars and spontaneous preterm birth. Am J Obstet Gynecol. 2019;220(6):591–594. doi: 10.1016/j.ajog.2019.01.237. [DOI] [PubMed] [Google Scholar]
- 5.Sweeney EL, Dando SJ, Kallapur SG, Knox CL. The human Ureaplasma species as causative agents of Chorioamnionitis. Clin Microbiol Rev. 2017;30(1):349–379. doi: 10.1128/CMR.00091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng L, Allen TK, Marinello WP, Murtha AP. Infection-induced thrombin production: a potential novel mechanism for preterm premature rupture of membranes (PPROM) Am J Obstet Gynecol. 2018;219(1):101. doi: 10.1016/j.ajog.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith M, Crews JD, Cheek N, Srivastava R, Appachi E. Hyperammonemic encephalopathy due to Ureaplasma parvum infection in an immunocompromised child. Pediatrics. 2019;144(2):e20190601. doi: 10.1542/peds.2019-0601. [DOI] [PubMed] [Google Scholar]
- 8.Contini C, Rotondo JC, Magagnoli F, Maritati M, Seraceni S, Graziano A, Poggi A, Capucci R, Vesce F, Tognon M, Martini F. Investigation on silent bacterial infections in specimens from pregnant women affected by spontaneous miscarriage. J Cell Physiol. 2018;234(1):100–107. doi: 10.1002/jcp.26952. [DOI] [PubMed] [Google Scholar]
- 9.Diaz Pallares C, Griener T, Vaughan S. Ureaplasma urealyticum disseminated multifocal abscesses in an immunocompromised adult patient: a case report. BMC Infect Dis. 2020;20(1):47. [DOI] [PMC free article] [PubMed]
- 10.Frangogiannis NG, Cate TR. Endocarditis and Ureaplasma urealyticum osteomyelitis in a hypogammaglobulinemic patient. A case report and review of the literature. J Infect. 1998;37(2):181–184. doi: 10.1016/S0163-4453(98)80174-6. [DOI] [PubMed] [Google Scholar]
- 11.Zhang N, Wang R, Li X, Liu X, Tang Z, Liu Y. Are Ureaplasma spp. a Cause of Nongonococcal Urethritis? A Systematic Review and Meta-Analysis. Plos One. 2014;9(12):e113771. doi: 10.1371/journal.pone.0113771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Payne MS, Ireland DJ, Watts R, Nathan EA, Furfaro LL, Kemp MW, et al. Ureaplasma parvum genotype, combined vaginal colonisation with Candida albicans, and spontaneous preterm birth in an Australian cohort of pregnant women. BMC Pregnancy Childb. 2016;16(1):312. [DOI] [PMC free article] [PubMed]
- 13.Motomura K, Romero R, Xu Y, Theis KR, Galaz J, Winters AD, Slutsky R, Garcia-Flores V, Zou C, Levenson D, et al. Intra-amniotic infection with Ureaplasma parvum causes preterm birth and neonatal mortality that are prevented by treatment with clarithromycin. MBIO. 2020;11(3):e720–e797. doi: 10.1128/mBio.00797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viscardi RM. Ureaplasma species: role in diseases of prematurity. Clin Perinatol. 2010;37(2):393–409. doi: 10.1016/j.clp.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Chamier M, Allam A, Brown MB, Reinhard MK, Reyes L. Host genetic background impacts disease outcome during intrauterine infection with Ureaplasma parvum. PLoS One. 2012;7(8):e44047. doi: 10.1371/journal.pone.0044047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kacerovsky M, Pliskova L, Bolehovska R, Musilova I, Hornychova H, Tambor V, Jacobsson B. The microbial load with genital mycoplasmas correlates with the degree of histologic chorioamnionitis in preterm PROM. Am J Obstet Gynecol. 2011;205(3):211–213. doi: 10.1016/j.ajog.2011.04.028. [DOI] [PubMed] [Google Scholar]
- 17.Horner P, Donders G, Cusini M, Gomberg M, Jensen JS, Unemo M. Should we be testing for urogenital Mycoplasma hominis, Ureaplasma parvum and Ureaplasma urealyticum in men and women?-a position statement from the European STI guidelines editorial board. J Eur Acad Dermatol. 2018;32(11):1845–1851. doi: 10.1111/jdv.15146. [DOI] [PubMed] [Google Scholar]
- 18.Liu L, Cao G, Zhao Z, Zhao F, Huang Y. High bacterial loads of Ureaplasma may be associated with non-specific cervicitis. Scand J Infect Dis. 2014;46(9):637–641. doi: 10.3109/00365548.2014.922696. [DOI] [PubMed] [Google Scholar]
- 19.Frølund M, Lidbrink P, Wikström A, Cowan S, Ahrens P, Jensen J. Urethritis-associated pathogens in urine from men with non-gonococcal urethritis: a case-control study. Acta Dermato Venereologica. 2016;96(5):689–694. doi: 10.2340/00015555-2314. [DOI] [PubMed] [Google Scholar]
- 20.Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10(10):1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuypers J, Jerome KR. Applications of digital PCR for clinical microbiology. J Clin Microbiol. 2017;55(6):1621–1628. doi: 10.1128/JCM.00211-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song N, Tan Y, Zhang L, Luo W, Guan Q, Yan M, Zuo R, Liu W, Luo F, Zhang X. Detection of circulating Mycobacterium tuberculosis-specific DNA by droplet digital PCR for vaccine evaluation in challenged monkeys and TB diagnosis. Emerg Microbes Infec. 2018;7(1):1–9. doi: 10.1038/s41426-018-0076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poh TY, Ali NATB, Chan LLY, Tiew PY, Chotirmall SH. Evaluation of droplet digital polymerase chain reaction (ddPCR) for the absolute quantification of Aspergillus species in the human airway. Int J Mol Sci. 2020;21(9):3043. doi: 10.3390/ijms21093043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lillsunde Larsson G, Helenius G. Digital droplet PCR (ddPCR) for the detection and quantification of HPV 16, 18, 33 and 45 - a short report. Cell Oncol. 2017;40(5):521–527. doi: 10.1007/s13402-017-0331-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Workowski KA. Sexually transmitted diseases treatment guidelines, 2015. Ann Emerg Med. 2015;66(5):526–528. doi: 10.1016/j.annemergmed.2015.07.526. [DOI] [Google Scholar]
- 26.Nyirjesy P. Nongonococcal and nonchlamydial cervicitis. Curr Infect Dis Rep. 2001;3(6):540–545. doi: 10.1007/s11908-001-0092-6. [DOI] [PubMed] [Google Scholar]
- 27.Lus MJ, Konecny P. Cervicitis: a review. Curr Opin Infect Dis. 2008;21:49–55. doi: 10.1097/QCO.0b013e3282f3d988. [DOI] [PubMed] [Google Scholar]
- 28.Lu L, Yu-hua JI, Zhen Z, Fang Z, Guo-jun C, Fu-zhi X, Wei-hong Z. Clinical application of genotyping detection of human mycoplasma urealyticum based on parC gene sequences. Chin J Microbiol Immunol. 2011;31(9):843–846. [Google Scholar]
- 29.Mahendran P, Liew JWK, Amir A, Ching X, Lau Y. Droplet digital polymerase chain reaction (ddPCR) for the detection of Plasmodium knowlesi and Plasmodium vivax. Malaria J. 2020;19(1):241. [DOI] [PMC free article] [PubMed]
- 30.Contini C, Rotondo JC, Magagnoli F, Maritati M, Seraceni S, Graziano A, Poggi A, Capucci R, Vesce F, Tognon M, Martini F. Investigation on silent bacterial infections in specimens from pregnant women affected by spontaneous miscarriage. J Cell Physiol. 2019;234(1):100–107. doi: 10.1002/jcp.26952. [DOI] [PubMed] [Google Scholar]
- 31.Zhao F, Feng X, Lv P, Xu X, Zhao Z. Real-time PCR assay may be used to verify suspicious test results of Ureaplasmas spp from the liquid culture method. J Microbiol Meth. 2020;169:105831. doi: 10.1016/j.mimet.2020.105831. [DOI] [PubMed] [Google Scholar]
- 32.Saha SK, Schrag SJ, El Arifeen S, Mullany LC, Shahidul Islam M, Shang N, Qazi SA, Zaidi AKM, Bhutta ZA, Bose A, et al. Causes and incidence of community-acquired serious infections among young children in South Asia (ANISA): an observational cohort study. Lancet. 2018;392(10142):145–159. doi: 10.1016/S0140-6736(18)31127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bharat A, Cunningham SA, Scott Budinger GR, Kreisel D, DeWet CJ, Gelman AE, Waites K, Crabb D, Xiao L, Bhorade S, et al. Disseminated Ureaplasma infection as a cause of fatal hyperammonemia in humans. Sci Transl Med. 2015;7(284):283r–284r. doi: 10.1126/scitranslmed.aaa8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glaser K, Silwedel C, Fehrholz M, Waaga-Gasser AM, Henrich B, Claus H, et al. Ureaplasma species differentially modulate pro- and anti-inflammatory cytokine responses in newborn and adult human monocytes pushing the state toward pro-inflammation. Front Cell Infect Mi. 2017;7:484. [DOI] [PMC free article] [PubMed]
- 35.Pavlidis I, Spiller OB, Sammut Demarco G, MacPherson H, Howie SEM, Norman JE, et al. Cervical epithelial damage promotes Ureaplasma parvum ascending infection, intrauterine inflammation and preterm birth induction in mice. Nat Commun. 2020;11(1):199. [DOI] [PMC free article] [PubMed]
- 36.Noh EJ, Kim DJ, Lee JY, Park JH, Kim J, Han JW, et al. Ureaplasma Urealyticum infection contributes to the development of pelvic endometriosis through toll-like receptor 2. Front Immunol. 2019;10. 10.3389/fimmu.2019.02373. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Multiple sequence alignment of the ParC genes of 14 serotypes of Ureaplasma spp. (a) Sequence of the U. parvum primes/probe; (b) Sequence of the U. ureaplasma primers/probe.
Data Availability Statement
The data generated during the current study are available from the corresponding author upon reasonable request.