Abstract
Enhancer binding proteins (EBPs) are key players of σ54‐regulation that control transcription in response to environmental signals. In the anaerobic microorganism Desulfovibrio vulgaris Hildenborough (DvH), orp operons have been previously shown to be coregulated by σ54‐RNA polymerase, the integration host factor IHF and a cognate EBP, OrpR. In this study, ChIP‐seq experiments indicated that the OrpR regulon consists of only the two divergent orp operons. In vivo data revealed that (i) OrpR is absolutely required for orp operons transcription, (ii) under anaerobic conditions, OrpR binds on the two dedicated DNA binding sites and leads to high expression levels of the orp operons, (iii) increasing the redox potential of the medium leads to a drastic down‐regulation of the orp operons expression. Moreover, combining functional and biophysical studies on the anaerobically purified OrpR leads us to propose that OrpR senses redox potential variations via a redox‐sensitive [4Fe–4S]2+ cluster in the sensory PAS domain. Overall, the study herein presents the first characterization of a new Fe–S redox regulator belonging to the σ54‐dependent transcriptional regulator family probably advantageously selected by cells adapted to the anaerobic lifestyle to monitor redox stress conditions.
Keywords: anaerobes, Desulfovibrio, enhancer binding protein, iron‐sulfur protein, redox sensor, transcriptional regulator
Enhancer Binding Proteins (EBPs) are key players of σ54‐regulation that control transcription of genes expression in response to environmental signals. OrpR is a novel type of EBP that presents an unprecedented Fe–S cluster in the PAS sensory domain and detects the redox potential, not O2. In anaerobes, OrpR regulates the expression of genes crucial for growth in anaerobic conditions as a function of the redox potential of the environment.

1. INTRODUCTION
Transcription initiation is a highly regulated step of gene expression and is essential for the cells response to environmental changes (Werner & Grohmann, 2011). Bacterial transcription initiation is regulated by a complex network of cell signaling pathways, which culminates in the recruitment of RNA polymerase (RNAP) to specific promoter regions by σ factors and the formation of open promoter complexes (Zhang et al., 2002). The σ factors are directly responsible for promoter recognition and they are the targets of transcription activator proteins that are required for DNA melting to make a transcription competent open promoter complex. There are two families of σ factor, based on sequence homology and the mechanism of action: σ70 and σ54. Upon binding to the promoter, while the RNAP‐σ70 holoenzyme spontaneously forms an open promoter complex, the RNAP‐σ54 holoenzyme rarely spontaneously converts to an open complex (Glyde et al., 2017). Transcriptionally competent open complex formation by the σ54 holoenzyme requires the action of activators bound remotely upstream from the transcription starting site. These activators, also called bacterial enhancer‐binding proteins (bEBPs), belong to the AAA+ ATPase family whose members are associated with a diverse range of cellular activities. ATP hydrolysis by bEBPs is then required for the isomerization from the closed complex to the open complex (Bose et al., 2008). The σ54 subunit of bacterial RNA polymerases recognizes and binds regulatory conserved elements composed of GG ‐24 and TGC ‐12 elements upstream from the transcription starting site and bEBPs usually bind to a specific conserved upstream activating sequence (UAS) (Buck et al., 2000). bEBPs generally consist of three domains: an N‐terminal sensory domain that perceives the signal, a central AAA+ domain responsible for ATP hydrolysis, and a C‐terminal DNA binding domain (Bush & Dixon, 2012). Since bEBPs bind relatively far upstream of the transcriptional start site, DNA bends in order for the central AAA+ domain to interact with the RNA polymerase using ATPase activity to activate transcription (Gao et al., 2020). While the AAA+ and the DNA binding domains are well conserved in bEBPs, the sensing domain exhibits a great diversity. Sensory domains belonging to different domain families such as RR (Response Regulators), PAS (Per, ARNT, and Sim), XylR‐N, V4R (Vinyl 4 Reductase), and GAF (cGMP‐specific phosphodiesterases, adenylyl cyclases, and FhlA) have been described (Bush & Dixon, 2012). This variety of sensory domains allow bEBPs to respond to a wide range of signals through different mechanisms that regulate negatively or positively bEBPs activity by modulating the DNA binding, the ATPase activity, or the interaction with RNA polymerase (Shingler, 2011).
The highest number of bEBPs with relation to genome size is observed in anaerobic sulfate‐reducing Deltaproteobacteria and this is probably linked to the variety of their ecological niches (Kazakov et al., 2015). They are ubiquitous in anoxic habitats and are characterized by the ability to gain energy for biosynthesis and growth by coupling oxidation of organic compounds or molecular hydrogen to the reduction of sulfate into sulfide (Zhou et al., 2011).
Desulfovibrio vulgaris Hildenborough (DvH) is a well‐established model to study the complex physiology and stress responses of anaerobic sulfate‐reducing microbes (Zhou et al., 2011). The adaptative responses of Desulfovibrio species to various environmental conditions and stresses (oxidative, metals, salts, heat shock, starvation, alkaline pH, pressure, and syntrophy), as revealed by a variety of integrated systems biology approaches, led to the conclusion that these microorganisms use several strategies to cope with adverse environmental conditions such as shifting energy metabolism, set up of oxidative‐stress responses and activation of stress‐specific pathways (Amrani et al., 2016; Benomar et al., 2015; Cadby et al., 2016; 2017; da Silva et al., 2015; Figueiredo et al., 2012; Korte et al., 2014; Li et al., 2014; Nair et al., 2015; Rajeev et al., 2015; Ramel et al., 2013; Varela‐Raposo et al., 2013; Wilkins et al., 2014; Yurkiw et al., 2012; Zhou et al., 2013, 2017, 2011). The adaptative responses can be specific and fast, thanks to the transcriptional control.
Characterization of the σ54 regulome in DvH showed the presence of 37 genes encoding putative σ54‐associated bEBPs. Despite their high number in DvH and other Desulfovibrio species, only a few bEBPs have been functionally studied. This includes the two‐component regulatory system NrfS‐NrfR that regulates nitrite and nitrate reduction (Cadby et al., 2017; Rajeev et al., 2015), NorR involved in nitrosative stress response (Varela‐Raposo et al., 2013), DVU2956 controlling biofilm formation (Zhu et al., 2019), and OrpR (also called DVU2106) that, under anaerobic conditions, activates the transcription of genes putatively involved in cell division (Fiévet et al., 2011). The gene encoding OrpR is located between two divergent operons, orp3‐orp4‐orp5 and orp7‐orp8‐orp9 and collaborates with σ54‐RNA polymerase and the integration host factor (IHF) to orchestrate the simultaneous expression of the divergent orp operons (Fiévet et al., 2011, 2014) (Figure 1). OrpR is composed of the typical three domains found in bEBPs, the N‐terminal sensory domain, the central AAA+ domain, and the C‐terminal DNA binding domain. Interestingly, the sensory domain of OrpR belongs to the ubiquitous PAS domain family (Möglich et al., 2009) that has been described to detect signals via a bound cofactor (Taylor & Zhulin, 1999) and regulates processes as diverse as nitrogen fixation in rhizobium (David et al., 1988), phototropism in plants (Christie et al., 1998), circadian behavior in insects (Nambu et al., 1991) and gating of ion channels in vertebrates (Morais Cabral et al., 1998).
FIGURE 1.
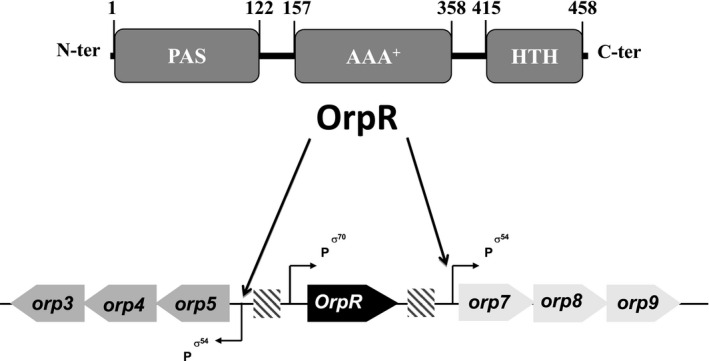
OrpR collaborates with σ54‐RNA polymerase to orchestrate the simultaneous expression of the divergent orp operons. OrpR is composed of three domains: an N‐terminal PAS, a central AAA+‐type ATPase, and a C‐terminal HTH DNA‐binding domains. Schematic representation of the orp operons (orp5‐orp4‐orp3 and orp7‐orp8‐orp9) and the OrpR encoding gene is shown. The position of σ54 and σ70 promoters is indicated and the OrpR‐binding regions are indicated by hatched rectangles
The signal sensed by OrpR and how it is perceived are important missing points to be addressed. In this study, we showed that OrpR is absolutely required for the expression of both orp operons under anaerobic conditions. We further demonstrated that OrpR undergoes a switch from active to inactive state when the redox potential of the medium increases and we identified that the signal sensed by OrpR as the redox potential rather than the O2 molecule itself. This switch is controlled by the PAS domain and requires three conserved cysteine residues. Spectroscopic studies revealed that the PAS domain binds a redox‐sensitive [4Fe–4S]2+ cluster which can be converted into a [3Fe–4S]1+ cluster when a mild oxidant is added. Finally, we reported ChIP‐seq analysis to define the DvH OrpR regulon.
Altogether, these results lead us to propose a mechanistic model in which the bEBP OrpR of DvH senses redox potential variations to cope with environmental changes encountered by anaerobic bacteria.
2. RESULTS
2.1. OrpR is a redox sensor
We previously showed, using Escherichia coli as a host, that OrpR collaborates with σ54‐RNA polymerase to activate expression of both orp5‐orp4‐orp3 and orp7‐orp8‐orp9 operons under anaerobiosis (Fiévet et al., 2011). Here, we further confirm such conclusion in vivo in DvH. Hence, when grown under anaerobiosis, a DvH mutant in which orpR was inactivated by transposon insertion (TnOrpR) did not show the presence of the Orp4 and Orp9 proteins while both proteins were produced in the wild‐type strain (Figure 2). In order to identify the signal sensed by the OrpR regulator, we monitored the expression of two OrpR regulated genes, orp4 and orp9, in different growth conditions. We then determined by qRT‐PCR the expression of orp4 and orp9 genes when cells were exposed to continuous sparging of oxygen (0.02%), an environmental condition in which DvH was still able to grow (Fievet et al., 2015). Figure 3a showed that when cells were cultured under continuous 0.02% O2 sparging, expression of orp4 and orp9 genes was drastically decreased when compared with anaerobic growth conditions. Western blot experiments using antibodies against Orp9 further confirmed that cells grown under continuous 0.02% O2 sparging exhibited a strong decrease in Orp9 production (Figure 3c). These results indicate that OrpR was inactive under microaerobic condition. Surprisingly, we found that when cells were exposed to continuous sparging of nitrogen (100%), OrpR was also inactive. Hence, the level of transcription of orp4 and orp9 genes and the production of Orp9 protein were drastically reduced when compared to the levels in cells grown under anaerobic conditions without sparging of nitrogen (Figure 3b,c). As control, we checked that under N2 and O2 sparging conditions, the amount of orpR transcripts and OrpR protein was not decreased (Figures 3a,b and S1). We then measured the redox potentials of the medium of DvH cultures grown in the three conditions described above. Redox potential (E, values given vs SHE) was −425 mV for anaerobic conditions, while it was −280 mV and −290 mV under continuous 0.02% O2 and N2 sparging, respectively. This suggests that the increase of the redox potential of the medium is sufficient to inactivate OrpR. During anaerobic syntrophic co‐culture of DvH with Methanosarcina barkeri, a growth condition during which the orp genes are not expressed (Scholten et al., 2007) and the Orp proteins are not produced (Figure 3d), the redox potential of the growth medium was −260 mV. This is consistent with our results, and suggests that OrpR is inactivated by an increase in redox potential of the medium and not solely by the O2 molecule.
FIGURE 2.
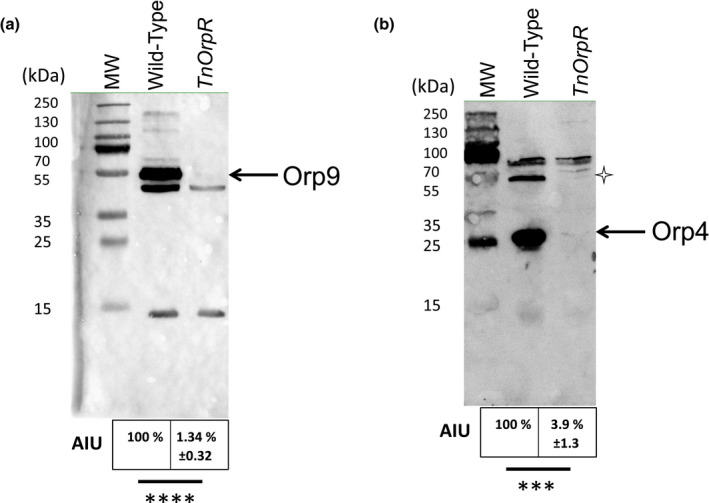
OrpR is required for the production of Orp4 and Orp9 proteins. 25 μg of total protein extract prepared from wild‐type and TnOrpR null mutant strains were analyzed by Western blot using antibodies against Orp9 (A) and Orp4 (B). The proteins Orp4 (31 kDa) and Orp9 (50 kDa) are indicated by arrows. The star indicates a dimeric form of Orp4. Quantification was carried out with Fiji (arbitrary units AIU) and statistical analysis was performed with Student's test, ****p < 0.0001 and ***p < 0.001. Data represent mean values of three biological replicates
FIGURE 3.

Increase of the medium redox potential leads to strong down‐regulation of orp operons expression. (A) Relative gene expression by qRT‐PCR of orp4 (grey), orpR (light grey) and orp9 (black) after growth of the wild‐type strain in the presence of 0.02% oxygen sparging vs anaerobic conditions. (B) Relative gene expression by qRT‐PCR of orp4 (grey), orpR (light grey) and orp9 (black) after growth of the wild‐type strain in the presence of nitrogen sparging vs anaerobic conditions. The average of three independent biological samples is shown, with standard deviations calculated from biological replicates. The number of transcripts under anaerobic condition was taken as a reference for each sample. (C) Twenty‐five micrograms of total protein extract prepared from wild‐type strains grown under anaerobic conditions or in the presence of N2 and oxygen sparging (0.02%) (measured redox potential Eh = −280 mV) were analyzed by Western blot using antibodies against Orp9. (D) Twenty‐five micrograms of total protein extract prepared from wild‐type strains grown under pure anaerobic culture (measured redox potential Eh = −455 mV) or under syntrophic growth of DvH growth with Methanosarcina barkeri (measured redox potential Eh = −260 mV) were analyzed by Western blot using antibodies against Orp9. Quantification was carried out with Fiji arbitrary units (AIU) and statistical analysis was performed with Student's test, ****p < 0.000, ***p < 0.001 and **p < 0.01. Data represent mean values ± standard deviation of three biological replicates
Altogether, these data allowed us to suggest that (1) OrpR senses the redox potential of the medium, (2) increasing redox potential abolishes OrpR activity.
2.2. The N‐terminal PAS domain senses the signal and is required to control OrpR activity
To investigate the role of the OrpR PAS domain, we constructed a truncated version of OrpR in which the PAS domain was deleted (strain OrpRΔPAS). The production of Orp9 in the DvH OrpRΔPAS mutant strain grown under anaerobic conditions or in the presence of 0.02% O2 was monitored. Figure 4 shows that in cells grown under anaerobic conditions, the DvH OrpRΔPAS and the OrpR WT strain produced the Orp9 protein (however with a 30% lower production in the OrpRΔPAS compared to the WT strain, Figure 4b), indicating that the PAS domain of OrpR is not required to activate σ54‐RNA polymerase. In cells grown under 0.02% oxygen sparging, the OrpRΔPAS strain still produced a large amount of Orp9 protein which contrasts with the WT situation (Figure 4). These results indicate that in absence of the PAS domain the expression of orp operons is constitutive whatever the redox potential of the medium. These data also suggest that DNA binding seems to occur independently of the PAS domain. We conclude that the PAS domain perceives the signal and acts negatively to control the activation of σ54‐dependent transcription.
FIGURE 4.

The OrpR PAS domain and cysteines residues are required for sensing cellular redox variations. (a) Western blot analysis of wild‐type, TnOrpR, OrpRΔPAS and OrpRmutCys mutant strains grown under anaerobic conditions (‐) or in the presence of 0.02% oxygen sparging (+) using antibodies against Orp9. 25 μg of total protein extract was loaded onto the gel. (b) Quantification of the Orp9 protein (50 kDa indicated by an arrow) in the different strains under anaerobic conditions or in the presence of 0.02% oxygen sparging. Errors bars indicate ± SD of three independent experiments. Statistical analysis was carried out with Tukey's test, ****p < 0.0001 and ns no significant
2.3. The conserved cysteines are required for the function of the PAS sensor domain
Sequence alignment of OrpR homologs from various anaerobic Deltaproteobacteria was performed and allowed us to identify C52, C61, and C65 as fully conserved residues in the PAS domain of the DvH OrpR (Figure S2). To determine whether the conserved cysteine residues are required for the function of the PAS domain, we constructed a DvH mutant strain, OrpR_mutCys, in which the three conserved cysteine residues (C52, C61, and C65) were substituted by serine residues. Production of the Orp9 protein in the OrpR_mutCys strain was observed whatever the media's redox state, as it was described for the OrpRΔPAS strain (Figure 4). It should be noted that the higher and lower amounts of OrpRΔPAS and OrpR_mutCys proteins, respectively, do not affect the Orp9 production (Figures S3 and 4). These results indicate that the conserved cysteine residues of the PAS domain are important for the function of the PAS domain.
2.4. The sensing PAS domain binds a Fe–S cluster
To investigate how the PAS domain senses the redox potential variations, OrpR was overproduced with a histidine tag either homologously in DvH or heterologously in E. coli. Protein was purified under anaerobic conditions to homogeneity and analyzed by spectroscopic methods. Anaerobically purified samples displayed a straw brown color which was lost upon exposure to air, after buffer‐exchange or sample concentration steps. Both OrpR samples produced in DvH or E. coli exhibited a similar absorption visible spectrum with absorption bands around 410 nm and 320 nm suggesting the presence of either a [4Fe–4S] or a [3Fe–4S] cluster (Figures 5a and 6a). Fe–S clusters can exist on several oxidation states and can be either diamagnetic or paramagnetic, the latter being visible by EPR spectroscopy. Anaerobically purified OrpR samples were EPR‐silent, which together with the visible absorption spectrum supports the presence of a diamagnetic (S = 0) [4Fe–4S]2+ cluster in the PAS domain (Figures 5b and 6b). We estimated the amount of Fe–S center incorporated into OrpR from the absorbance at 410 nm using an extinction coefficient of 15 000 M–1.cm–1, a typical average for values found for [4Fe–4S] clusters (Lippard & Berg, 1994). The anaerobically purified samples contained between 0.3 and 0.5 [4Fe–4S]2+ cluster per polypeptide chain (average of five preparations). Furthermore, after reduction with dithionite, anaerobically purified OrpR overproduced in E. coli showed a drastic absorbance decrease at 410 nm (Figure 6a), which could be interpreted as either a reduction of the [4Fe–4S]2+ into [4Fe–4S]1+ cluster and/or as cluster degradation. By EPR spectroscopy, a dithionite‐reduced sample produced a weak anisotropic signal (g‐values = 2.05, 1.91, and 1.89) consistent with a reduced (S = 1/2) [4Fe–4S]1+ center (Guerrini et al., 2008) (Figures 6b and S4). However, this signal accounted for only 10% of the initial [4Fe‐4Fe]2+ center, thus suggesting that large absorbance decay at 410 nm was mainly due to Fe–S cluster instability in the presence of dithionite. This behavior has been already reported for other [4Fe–4S] transcriptional factors such as FNR and NreB (Esbelin et al., 2012; Müllner et al., 2008). Thus, all these results suggest that as‐purified OrpR produced in DvH or E. coli likely harbors a diamagnetic (S = 0) [4Fe–4S]2+ cluster.
FIGURE 5.

OrpR binds a Fe–S cluster. UV‐Visible and EPR signatures of OrpR produced in DvH. (a) UV‐visible absorption spectrum of anaerobically purified OrpR homologously produced in DvH. Cuvette pathlength 1 mm contained 90 μM of protein sample (concentration determined using a standard colorimetric BCA assay) in 100 mM Tris, 500 mM NaCl, 350 mM imidazole, 10% glycerol, pH 7.5 (elution buffer). (b) EPR spectra of anaerobically purified OrpR homologously produced in DvH, as‐isolated (green line), exposed to 2 molar equivalents O2 (blue trace) or to 2 equivalents DCPIP (black line). Inserts: Zoom on the g ~ 4.3 signal (S = 5/2 Fe3+species) and on the g ~ 2.01 signal (S = 1/2 [3Fe–4S]1+). The number of molar equivalents of oxidant is given according to the concentration of the [4Fe–4S]2+ cluster (80 μM) present in the purified OrpR sample (200 μM). The [4Fe–4S]2+ concentration was estimated from the absorbance at 410 nm in a freshly purified sample. The sample was frozen at 1 min after addition of air‐saturated buffer or DCPIP solution. Recording conditions: Microwave frequency, 9.48 GHz, microwave power, 10mW (or 0.4 mWfor the g = 2.01 signal), modulation amplitude, 1 mT (Inserts: 2mT and 0.5mT for the g = 4.3 and g = 2.01 signals, respectively), modulation frequency, 100 kHz, temperature 15 K, number of accumulations: 10–25. Spectra were normalized by considering dilution factor of the sample and the number of accumulations to allow direct comparison of EPR signal amplitude [Colour figure can be viewed at wileyonlinelibrary.com]
FIGURE 6.

UV‐Visible and EPR signatures of purified OrpR produced in Escherichia coli. (A) UV‐visible absorption spectra of as‐purified (black line) and dithionite‐reduced (dot line) OrpR. The asterisk (*) marks the absorption band at 320 nm due to a slight excess of dithionite. (B) EPR spectra of as‐isolated (anaerobic) and dithionite‐reduced (8 molar equivalents) OrpR. The weak peak at 340 mT (g ~ 2.1) in the as‐purified sample represents less than 1% of protein and may correspond to traces of [3Fe–4S]1+ cluster coming from Fe–S cluster degradation. OrpR concentration was 90 µM in 50 mM Hepes, 500 mM NaCl, 500 mM imidazole, 10% glycerol pH 7.5. EPR recording conditions: 9.48 GHz, microwave power 10mW, modulation amplitude 1 mT, modulation frequency 100 kHz, temperature 15 K
The behavior of the [4Fe–4S]2+ cluster exposed to O2 was evaluated by EPR and UV‐visible spectroscopies. EPR‐spectrum of purified OrpR sample exposed to air is shown in Figures 5b and S4. Only a derivate peak at g ~ 4.3 was observed, consistent with high spin (S = 5/2) Fe3+ species likely coming from dismantling of the [4Fe–4S]2+ cluster (Figures 5b and S4). Furthermore, the absorbance decay at 410 nm observed on the absorption spectra suggests full degradation of the Fe–S cluster upon exposure to air with a half‐life constant of about 10 min under these conditions (Figure S5). Next, the behavior of the OrpR [4Fe–4S]2+ cluster of OrpR was investigated when treated with a mild oxidant such as DCPIP (E°’pH7.5 = + 220 mV against + 840 mV for O2). In the DCPIP‐treated sample, an intense pseudo‐axial signal developed at g = 2.015 and 2.005 which can be attributed to an oxidized (S = 1/2) [3Fe–4S]1+ species (Figures 5b and S4). The [3Fe–4S]1+ species was also observed after OrpR oxidation with ferricyanide (E°’pH7.5 = + 420 mV) (Figure S6) but was clearly absent in the O2‐treated sample (Figures 5b and S4). Also, the derivate peak at g ~ 4.3 coming from high spin (S = 5/2) Fe3+ species, with additional EPR lines at 9.5, 6.8, 4.6, 4.27, and 4.16, was also observed in the DCPIP‐treated sample and was more intense than in the O2‐treated sample (Figure 5b). All these results indicate that anaerobically purified OrpR likely contains a redox‐sensitive [4Fe–4S]2+ cluster, which dissembles in vitro into [3Fe–4S]1+ and Fe3+ species when treated with mild oxidants such as DCPIP or ferricyanide, while O2 induces further degradation of Fe–S cluster.
2.5. Oxidized forms of OrpR are able to bind DNA
To investigate whether the redox status of OrpR might influence its OrpR DNA binding ability, we performed electrophoretic mobility shift assay (EMSA) experiments. Biotin‐labeled DNA fragment containing the OrpR DNA binding site previously determined (Fiévet et al., 2011) was incubated in anaerobiosis with increasing concentrations of anaerobically as‐prepared [4Fe–4S]2+ OrpR form or treated with either DCPIP or air. Figures 7a,b, and S7a show that as‐prepared OrpR and oxidant‐treated OrpR were both able to bind their DNA target (Figures 7a,b, and S7a). It should be noted that at high OrpR concentration and whatever the OrpR redox status, the unbound DNA fraction was unchanged and a slower rate DNA migration is observed (Figures 7a,b and S7a). As a control, BSA protein was not able to bind on the OrpR DNA target (Figures 7a,b and S7a, lanes 1). The affinity of the interaction between OrpR and DNA was then determined in anaerobically as‐prepared [4Fe–4S]2+ OrpR form untreated, or treated with either DCPIP or air (Figures 7c,d and S7b). KD values of 227 ± 57 nM for holo‐OrpR, 111 ± 38 nM for air‐treated OrpR and 162 ± 62 nM were obtained and are within the same order of magnitude (Figures 7c,d and S7b) whatever the redox state of the Fe–S cluster. Altogether these data suggest that the OrpR redox status does not influence the interaction between OrpR and DNA.
FIGURE 7.

OrpR DNA binding ability. (a) Electrophoretic Mobility Shift Assay (EMSA) of 5 fmoles of biotin‐labeled DNA fragments containing the OrpR binding site incubated with 5 μM of BSA (lane 1), without OrpR protein (lane 2) and with increasing concentrations (nM) of OrpR purified under anaerobic conditions (lanes 3 to 11). (b) Electrophoretic Mobility Shift Assay (EMSA) of 5 fmoles of biotin‐labeled DNA fragments containing the OrpR binding site incubated with 5 μM of BSA (lane 1), without OrpR protein (lane 2) and with increasing concentrations (nM) of O2‐treated OrpR (lanes 3 to 13). (C and D) Affinity of as prepared‐ or O2‐treated OrpR to DNA. The quantification of band intensities was determined by measuring the fluorescent signal of the unbound DNA (100% corresponding to the DNA fluorescence without protein, lanes 2). The fraction of DNA bound was then determined using the following expression: 100%—unbound DNA. The fraction of DNA bound was plotted vs the concentration of OrpR and the values of the KD of 227 ± 57 nM for as prepared‐OrpR (C) and 111 ± 38 nM for O2‐treated OrpR (D) were obtained by fitting the data with the “one site binding” equation in OriginPro 8,5. Data correspond to two or three biological replicates
2.6. Identification of OrpR target genes in vivo
Last, we investigated whether OrpR regulates other targets than the two orp operons. In order to study in vivo the genes that compose the OrpR regulon, ChIP‐seq analysis was carried out on exponential grown DvH cells using polyclonal anti‐OrpR antibodies. Immunoprecipitated DNA was then sequenced by Illumina MiSeq technology as previously described (Fioravanti et al., 2013; Pini et al., 2015). Using an arbitrary 76 bp‐long paired‐end sequencing reads, we identified three main enriched DNA sequences (Figure 8a). Visualization of these 3 peaks using Integrative Genomic Viewer (Fioravanti et al., 2013) showed that the highest DNA enrichment (second peak surrounded by a dashed black line in Figure 8a) was split into two regions on both sides of the gene encoding OrpR, mapping to the promoter regions of the two orp operons (Figure 8b). These two enriched DNA sequences contained the conserved 17‐bp imperfect palindromic site (GGGCGYRTTTTGCGCCC) previously shown as target of OrpR in vitro (Fiévet et al., 2011). The two other minor peaks mapped to the promoter region of DVU1258 encoding a glutamine synthetase and to the intergenic region between DVU2956 and DVU2958 encoding a σ54‐dependent transcriptional regulator and a putative membrane protein, respectively (third peak surrounded by the dashed line in Figure 8a). σ54‐dependent promoters were identified upstream DVU1258 and DVU2958 (Kazakov et al., 2015) as well. We then measured by qRT‐PCR the expression of DVU1258, DVU2958, and both orp genes orp5 and orp7 as positive controls. Expression of DVU595, which is under the control of a σ70‐dependent promoter, was measured as a negative control both in wild‐type and TnOrpR null mutant strains (Figure 8c). We found that the expression of DVU0595, DVU1258, and DVU2958 genes were similar in wild‐type and TnOrpR null mutant which did not produce OrpR while, as control, the expression of the two orp genes was strongly downregulated in the TnOrpR null mutant (Figure 8c).
FIGURE 8.

The orp operons are the only targets of OrpR. (a) Whole genome ChIP‐Seq analysis. The 750 kb DvH genome containing the three main enriched peaks are in dashed lines. (b) The enriched peak corresponding to the orp operons expression sequences is zoomed. (c) Relative quantification of gene expression by real‐time PCR of DVU0595, DVU1258, Orp5, Orp7, and DVU2958 transcripts in the wild ‐type (white box) and TnOrpR null mutant (grey bars) strains. The average of three independent biological samples are shown. The amount of each transcript in the wild‐type strain was taken as reference
Altogether, these results showed that OrpR directly binds in vivo to the two orp operon promoter regions which are the sole genes that it regulates.
3. DISCUSSION
bEBPs tightly regulate the σ54‐dependent transcription in response to environmental signals (Bush & Dixon, 2012). Recent works have shed light on the diversity of mechanisms used by bEBPs to activate σ54‐dependent in response to the signals perceived. Interestingly anaerobic bacteria such as DvH are relying on many different bEBPs for their adaptation. The present study expands the diversity of bEBPs since we propose that OrpR is a new type of σ54‐dependent regulator using an Fe–S cluster‐containing PAS domain for redox‐sensing in anaerobic bacteria.
Previous studies using E. coli as host organism described that OrpR is required to initiate transcription of the orp genes (Fiévet et al., 2011). Here, we show that OrpR is active in vivo under anaerobic standard growth conditions because a DvH mutant strain lacking OrpR (TnOrpR) was not able to express the orp genes in anaerobiosis. Also, ChIP seq experiments validated that the two orp operons were the direct and specific targets of OrpR. bEBPs are known to modulate the expression of their target depending on environmental conditions, thus we aimed at determining the signal perceived by OrpR. Here, we provide data indicating that OrpR senses redox potential increase rather that the oxygen concentration since flushing DvH strains with 0.02% oxygen, but also with pure N2, abolished the expression of the OrpR regulated genes. Measuring the redox potential of the medium under 0.02% oxygen and pure N2 sparging, revealed that inactivation of OrpR was linked to the increase of the redox potential by about +140 mV (such redox change might be a consequence of the H2S elimination by the gas flow used in sparging conditions). Altogether, OrpR can be viewed as a bEBPs that is able to activate σ54‐dependent transcription at low redox potential in physiological conditions and which is inactive when the redox potential of the medium increases.
OrpR obeys the general observation that many N‐terminal domains of bEBPs are sensory domains (Shingler, 2011). Hence, we showed that the mutant DvH strain, OrpRΔPAS, possessing OrpR lacking its N‐terminal domain had lost the conditional expression of the orp genes, since Orp9 was produced in all the conditions tested, that is, constitutively.
Biochemical and biophysical analyses of OrpR allowed us to bring some clues on how the signal is perceived by the OrpR PAS domain. Hence, UV‐visible and EPR spectra of the anaerobically purified OrpR showed that it likely binds a [4Fe–4S]2+ cluster. In almost all Fe–S proteins, Fe–S clusters are covalently bound to the protein via the ligands which are thiolates from cysteinyl residues. Interestingly, sequence alignment of OrpR homologs from various anaerobic Deltaproteobacteria points out C52, C61, and C65 as fully conserved residues in the PAS domain suggesting that the Fe–S center is located in the sensory domain. A structural model of the PAS domain shows that these three conserved cysteine residues C52, C61, and C65 are located within 5–8 Å from each other which is compatible with the binding of a Fe–S cluster (Figure S8). In the case of other redox sensor protein families characterized so far, the [4Fe–4S]2+ cluster is coordinated by four cysteines and occasionally by three cysteines and an aspartate (Barth et al., 2018). It should be noted that an aspartate or a glutamate residue in position 54 in DvH OrpR sequence is also conserved in OrpR homologues and it is in proximity for binding the Fe–S cluster (within 6 Å distance of the cysteines triad) (Figure S8). Further structural, biochemical, and spectroscopic characterizations of the purified mutants will be necessary to unambiguously assess the coordination mode of the [4Fe–4S]2+ cluster in the PAS domain. The fact that the Fe–S cluster is important for OrpR signal perception was strongly suggested, thanks to the construction of the OrpR mutant, OrpR_mutCys, in which the three conserved cysteines have been replaced by serine residues. Indeed, the OrpR_mutCys mutant DvH strain did not perceive the signal and thus constitutively produced Orp9. Therefore, this work is the first report of a bEBP using a Fe–S cluster‐containing PAS sensory domain. This is also the first illustration that like aerobes, anaerobic microorganisms can use Fe–S binding proteins as redox sensors.
Further characterization of the OrpR Fe–S cluster by spectroscopic methods indicated that the cluster is sensitive to O2. Interestingly, using DCPIP or ferricyanide as milder oxidants, the [4Fe–4S]2+ cluster underwent an oxidation process and dissociated into [3Fe–4S]1+ and Fe3+ species. Preliminary experiments indicate that the [3Fe–4S]1+ species is transient, contrary to the Fe3+ species that is stable over time in the conditions tested (not shown). Whether the appearance of a [3Fe–4S]1+ intermediate results from a degradative oxidative process due to the use of chemical oxidants, or if it is a physiological intermediate in the redox stress sensing mechanism, remains unknown so far. Further in vivo and in vitro studies will address this issue.
In this study, we showed that OrpR can be ranked among bEBP, such as NorR or DmpR, whose sensory domain prevents σ54‐dependent transcription (D’Autréaux et al., 2005; Shingler & Pavel, 1995). Hence, truncation of the PAS domain of OrpR allowed Orp9 to be expressed in conditions under which the full length OrpR did not. Such negative control of the PAS domain leading to an inactive bEBP can arise by modulating the activity of the other two domains, the AAA+ ATPase domain and the DNA binding domain, via structural rearrangements (D’Autréaux et al., 2005; Lazazzera et al., 1996). The EMSA experiments showed that the affinity of the interaction between OrpR and its DNA target is unchanged even after oxidative treatment (which changed the redox status and/or on the stability of the OrpR Fe–S cluster). Thus, we ruled out that perception of the signal alters drastically DNA binding activity. Functional and structural consequences of the signal sensing on OrpR protein remain to be determined.
Altogether, the present data enabled the proposal of a working model for the OrpR‐dependent regulatory mechanism. OrpR binds in vivo to the dedicated DNA binding sites. Under anaerobic growth conditions generating low redox potential of the medium, OrpR is under a reduced form that binds to dedicated DNA binding sites, has a [4Fe–4S]2+ cluster in the PAS domain and is able to activate σ54‐RNA polymerase. Increase of medium redox potential induces an OrpR oxidized form, very likely affecting the redox state and/or stability of its Fe–S cluster, which leads to the inability of OrpR to remodel the σ54‐RNA polymerase‐promoter complex and to its incapacity to activate transcription.
In this study, together with our previous results (Fiévet et al., 2011, 2014), we demonstrate that OrpR is absolutely required for the transcription of two operons, flanking the OrpR coding gene, under anaerobic conditions by directly binding to their promoter regions. These results are in agreement with the previous σ54‐dependent regulome analysis in DvH showing that in most cases the bEBP‐encoding genes are co‐localized with their cognate σ54‐dependent promoter (Kazakov et al., 2015). In the case of OrpR, the ChIP‐seq analysis performed here clearly demonstrates that OrpR binds in vivo on the imperfect palindrome motif previously determined in vitro (GGGCGYRTTTTGCGCCC) located upstream the two orp operons (Fiévet et al., 2011). These results bring experimental validation of our previous in silico analysis showing that this conserved palindromic binding sequence of orp operons are the two sole targets of OrpR (Fiévet et al., 2011). Thus, the morphological defects observed in DvH cells when OrpR is inactivated (Fiévet et al., 2011) are only linked to the decrease in the amount of the Orp proteins. The role of the Orp proteins is however still unclear, a hypothesis being that they intervene directly or indirectly in a cell division process (Fiévet et al., 2011). As increasing the medium redox potential switches off orp transcription, we can assume that the Orp proteins are mainly associated with the anaerobic lifestyle. Considering that most of the Orp proteins are Fe–S proteins, it is tempting to propose that OrpR participates in an adaptative response developed by anaerobic bacteria to save energy and avoiding biosynthesis of fragile Fe–S clusters‐containing proteins under oxidative stress conditions. The regulator OrpR seems to be restricted to species harboring Orp proteins which belong to δ‐Proteobacteria exhibiting various metabolisms such as the sulfate‐ and sulfur‐reducing bacteria and some syntrophic bacteria. As suggested before, the ORP system may have originated from Archaea and later radiated into Bacteria through horizontal gene transfer (Scholten et al., 2007). The increase in the O2 concentration of the Earth's atmosphere about 2.5 billion years ago confronts anaerobic microorganisms with deleterious oxidation processes and increasing redox potential. These conditions forced anaerobes to develop mechanisms to detect and cope with O2 and redox stress conditions. It is probably in this context that the δ‐Proteobacteria advantageously added the regulatory OrpR system totally linked to the anaerobic lifestyle.
4. EXPERIMENTAL PROCEDURES
4.1. Strains and growth conditions
Strains used in this study are listed in Table S1. E. coli TG1 strains were grown in Luria‐Bertani (LB) medium at 37°C with the appropriate antibiotic when required (0.27 mM for ampicillin, 0.15 mM for chloramphenicol). Cultures of DvH were grown in medium PC (Postgate et al., 1984) supplemented with 0.17 mM kanamycin or 0.15 mM thiamphenicol when required, at 33°C under anaerobic conditions. Anaerobic work was performed in an anaerobic chamber (COY Laboratory Products or MBraun) filled with a 10% H2‐90% N2 mixed‐gas atmosphere. Solutions were made anoxic by flushing with N2 for removal of O2. Solutions, glass, and plastic materials were equilibrated for at least 12 hr inside the anaerobic chamber before use. Syntrophic co‐cultures of DvH and M. barkeri were performed in Hungate tubes in the dark at 33°C under shaking (200 rpm) in CCM medium (Walker et al., 2009) amended with 30 mM lactate. The headspace of the Hungate tube was filled with 80% H2‐20% CO2. Co‐cultures were established by inoculating 10% (vol/vol) of an exponentially growing culture of DvH and 10% (vol/vol) of an exponentially growing culture of M. barkeri.
4.2. Plasmid construction for protein production in DvH
Plasmids pBMC6PC3HisOrpR, pBMC6PC3OrpRΔPAS, and pBMC6PC3OrpRmut‐cys were constructed by first PCR amplification of the 250bp of the dvu3171 promoter from DvH chromsomal DNA using promcyc_HindIII and promcyc_SalINdeI. The corresponding PCR fragment was digested by HindIII/SalI enzymes and subcloned into the HindIII and SalI sites of the plasmid pBMC6 to obtain pBMC6PC3. The appropriated primers described in Table S2 were used to amplify orpR and orpRΔPAS genes from DvH genomic DNA. The obtained PCR products and the pBMC6PC3 plasmid were digested with NdeI and SacI restriction enzymes and ligated into the multiple cloning site of the plasmid to get pBMC6PC3HisOrpR and pBMC6PC3OrpRΔPAS, respectively. For all constructs, successful ligations were confirmed via DNA sequencing and subsequently introduced in DvH cells by electroporation.
4.3. Site‐directed mutagenesis
Simultaneous mutations of Cys52, Cys61 and Cys65 residues from OrpR were generated by using pBMC6PC3HisOrpR as the PCR template and the Q5 site‐directed mutagenesis kit from Biolabs. All the primers used for mutagenesis (Table S2) were designed using the online NEB primer design software NEBaseChangerTM, mutations were confirmed by DNA sequencing and subsequently electroporated into DvH cells.
4.4. ChIP‐Seq experiments
DvH cells were grown until OD600nm of approximately 0.4 to 0.5 in medium C. Protocol was performed as previously published (Fioravanti et al., 2013; Pini et al., 2015) except for the sonication step that was performed using a Bioruptor (Diagenode) for 10 min (Interval 30 sec + 30 sec; Intensity HIGH).
4.5. Protein production and purification
Homologous productions of proteins in DvH strains were performed in 20 L of PC medium. Cells were cultured until the optical density at 600 nm reached 0.8 to 0.9. Heterologous productions of proteins in E. coli were obtained as follows: 1 mM isopropyl β‐d‐thiogalactoside (IPTG) was added to an exponentially growing culture at 37°C of recombinant E. coli strains in 2 L of LB medium containing 100 µg/ml ampicillin and grown for additional 4 hr at 37°C. Proteins were purified anaerobically in an anaerobic chamber with identical purification protocols. The bacterial pellet of OrpR was resuspended in buffer A (50 mM Tris‐HCl, pH 7.5, 500 mM NaCl, 10% glycerol) and containing 40 mM imidazole, DNase I (Roche), and cOmpleteTM protease inhibitor EDTA free (from Roche Applied Science). Cell suspensions were disrupted two times in a chilled (4°C) French press (Thermo‐FA‐080A) at 1,200 psi. Cell lysates were clarified by ultracentrifugation (45,000g for 80 min at 4°C). After filtration of the supernatant across a 0.2 µm filter, the soluble proteins were loaded onto a 5 ml Ni‐Sepharose affinity column (His Trap HP, GE Healthcare) equilibrated with buffer A containing 40 mM imidazole. OrpR poly‐histidine tagged proteins were eluted with buffer A containing 350 mM imidazole. Protein concentration was determined using a BCA PierceTM 660 nm Protein Assay (Thermo) colorimetric kit, using bovine serum albumin as standard. Purity of the fraction was checked by SDS‐PAGE. Protein concentration was also estimated from absorbance measurement at 280nm using a calculated extinction coefficient of 16 000 M–1.cm–1 based on the protein sequence (https://web.expasy.org/protparam/). Values found were in good agreement with the protein concentration deduced from BCA protein assays.
4.6. Western blotting experiments
DvH cells were grown in PC medium until the OD600 reached 0.4–0.6. To prepare samples, 0.5 OD600 units of DvH cultures were centrifuged and the pellets were resuspended in 50 µl of 2X loading buffer (120 mM Tris‐HCl pH 6.8, 20% of glycerol and 0.2% of bromophenol blue) supplemented with 0.69 mM SDS and 10 mM DTT. Then, samples were boiled for 10 min. After separation by electrophoresis in 12.5% SDS polyacrylamide gel, proteins were transferred onto a nitrocellulose membrane followed by blocking of the membrane with PBS containing 3% BSA and 0.5% Tween‐20 for 1 hr at room temperature. After three washes in PBS buffer containing 0.1% of Tween‐20, the membrane was incubated overnight with polyclonal rabbit anti‐Orp4 diluted at 1:2,500 and anti‐Orp6 and anti‐Orp9 serums used at 1:25,000. All rabbit polyclonal antibodies have been generated by Agro‐Bio from purified proteins. The membrane was then washed three times in PBS buffer supplemented with 0.1% of Tween‐20 and incubated 1 hr at room temperature with an HRP‐conjugated anti‐rabbit secondary antibody from Thermo Scientific (1:2,500). After two washes in PBS buffer supplemented with 0.1% of Tween‐20 followed by two washes in PBS buffer, SuperSignal® West Pico Chemiluminescent Substrate kit (Thermo Scientific) was used for detection according to the manufacturers’ instructions. The signal detection was realized by using ImageQuant LAS 4000 mini from GE Healthcare.
4.7. Electrophoretic mobility shift assays for DNA binding
DNA fragment containing the OrpR binding site was amplified by PCR with the biotin‐labeled primers prom2107b and prom2107comp shown in Table S2. EMSAs were performed using lightShift Chemiluminescent EMSA Kit (Thermo Scientific). The different redox states of OrpR were incubated at room temperature in 10 mM Tris pH 7.5, 100 mM KCl, 50 ng/µl poly(dI‐dC), and 0.05% Nonidet P‐40 binding buffer with 5 fmoles of the biotin‐labeled DNA fragment for 30 min. Reactions were then resolved by a pre‐run electrophoresis in a 5% agarose gel in TBE buffer (450 mM Tris, 450 mM borate, and 0.01 mM EDTA). The samples were blotted onto a 0.45‐µm Biodyne B nylon membrane and then cross‐linked to the membrane using a 312 nm UV Transilluminator (Uvitec) for 1 min. Membranes were processed as recommended in the Chemiluminescent Nucleic Acid Detection Module Kit (ThermoScientific). For EMSAs in presence of as‐purified OrpR with or without DCPIP, all steps described previously were performed in an anaerobic chamber (COY Laboratory Products or MBraun). O2‐treated OrpR was obtained by incubating as‐prepared OrpR 30 min under aerobic conditions before incubation. EMSAs experiments with O2‐treated OrpR were performed in aerobic conditions.
4.8. Biophysical experiments
EPR spectroscopy experiments were performed on a Bruker Elexsys E500 spectrometer fitted with an Oxford Instruments ESR‐900 helium gas flow for cryogenic temperature studies. UV‐Visible absorption spectra were recorded on a Lambda‐25 Perkin‐Elmer and Uvikon (Kontron Instruments) spectrophotometers. All experiments were performed on freshly purified samples under anaerobic conditions (JACOMEX glovebox, pO2 < 2 ppm). The purified protein was kept in the elution buffer and conserved at 4°C under anaerobic conditions for a few days when necessary. Depending on the purification, elution buffer was 50 mM Tris‐HCl pH7.5, 500 mM NaCl, 350 mM imidazole, 10% glycerol for the recombinant His‐tagged (N‐terminus) OrpR. Purified OrpR samples were reduced by adding small aliquots of fresh stock solution of dithionite (at about 100 mM dissolved in a pH 8‐buffered solution) or oxidized anaerobically by adding small aliquots of 2, 6‐dichlorophenolindophenol (DCPIP; E°’pH7.5 = +220 mV vs SHE), potassium hexacyanoferrate (III) (K3[Fe(CN)6]; E°’pH7.5 = 420 mV vs SHE) solutions. DCPIP and ferricyanide stock solutions were prepared at 10 mM in deionized water. Air‐saturated buffer solution at 20°C (the concentration of O2 dissolved in the buffer solution was estimated to 220 μM according to reference (Crack et al., 2004) was added directly to the sample in a sealed EPR tube and quickly frozen in cold ethanol inside the glovebox prior to storage in liquid nitrogen.
AUTHOR CONTRIBUTIONS
B.B. and C.A. contributed to the conception or design of the study. A.F., M.M., G.B., D.E., E.G.B., O.V., S.R.P., A.D., Z.D., B.B. and C.A. contributed to the acquisition of the data, analysis, or interpretation of the data and A.F., G.B., E.G.B., S.R.P., A.D., Z.D., B.B. and C.A. contributed to the writing of the manuscript.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the contribution of Marielle Bauzan for the cultures in syntrophy and Yann Denis for transcriptomic facilities and Grant Zane (Judy Wall laboratory) for providing us the GZ3621Orp ::mini‐Tn5 strain. The authors are also grateful to Emilien Etienne and the EPR facilities available at the national EPR network RENARD (IR CNRS 3443) and the Aix‐Marseille University EPR center. We also thank Martine Company for technical assistance, Katia Villion for her help on the project during its school program, Béatrice Py and Bruno Guigliarelli for insightful discussions. The French National Research Agency (ANR) funded this research project (ANR‐12‐ISV8‐0003‐01). The project leading to this publication has received funding from the Excellence Initiative of Aix‐Marseille University—A*MIDEX, a French “Investissements d'Avenir” programme and is part of the Institute of Microbiologies and Biotechnology—IM2B (AMX‐19‐IET‐006). This work was supported by Fundação para a Ciência e Tecnologia (FCT) (grant to SRP, FCT‐ANR/BBB‐MET/0023/2012) and by the Belgian Federal Science Policy Office (Belspo) (IAP7/44, iPROS project). SRP was also supported by the Applied Molecular Biosciences Unit‐UCIBIO financed by national funds from FCT (UIDP/04378/2020 and UIDB/04378/2020). This article is based upon work from COST Action CA15133, supported by COST (European Cooperation in Science and Technology).
Fiévet A, Merrouch M, Brasseur G, et al. OrpR is a σ54‐dependent activator using an iron‐sulfur cluster for redox sensing in Desulfovibrio vulgaris Hildenborough. Mol Microbiol. 2021;116:231–244. 10.1111/mmi.14705
[Correction added on 04 June 2021, after first online publication: The copyright line was changed.]
Contributor Information
Bénédicte Burlat, Email: bburlat@imm.cnrs.fr.
Corinne Aubert, Email: aubert@imm.cnrs.fr.
REFERENCES
- Amrani, A., van Helden, J., Bergon, A., Aouane, A., Ben Hania, W., Tamburini, C. et al. (2016) Deciphering the adaptation strategies of Desulfovibrio piezophilus to hydrostatic pressure through metabolic and transcriptional analyses. Environmental Microbiology Reports, 8, 520–526. [DOI] [PubMed] [Google Scholar]
- Barth, C., Weiss, M.C., Roettger, M., Martin, W.F. & Unden, G. (2018) Origin and phylogenetic relationships of [4Fe‐4S]‐containing O2 sensors of bacteria. Environmental Microbiology, 20, 4567–4586. [DOI] [PubMed] [Google Scholar]
- Benomar, S., Ranava, D., Cárdenas, M.L., Trably, E., Rafrafi, Y., Ducret, A. et al. (2015) Nutritional stress induces exchange of cell material and energetic coupling between bacterial species. Nature Communications, 6, 6283. Available from: 10.1038/ncomms7283. [DOI] [PubMed] [Google Scholar]
- Bose, D., Joly, N., Pape, T., Rappas, M., Schumacher, J., Buck, M. et al. (2008) Dissecting the ATP hydrolysis pathway of bacterial enhancer‐binding proteins. Biochemical Society Transactions, 36, 83–88. Available from: 10.1042/BST0360083. [DOI] [PubMed] [Google Scholar]
- Buck, M., Gallegos, M.T., Studholme, D.J., Guo, Y. & Gralla, J.D. (2000) The bacterial enhancer‐dependent sigma(54) (sigma(N)) transcription factor. Journal of Bacteriology, 182, 4129–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, M. & Dixon, R. (2012) The role of bacterial enhancer binding proteins as specialized activators of σ54‐dependent transcription. Microbiology and Molecular Biology Reviews, 76, 497–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadby, I.T., Faulkner, M., Cheneby, J., Long, J., van Helden, J., Dolla, A. & et al. (2017) Coordinated response of the Desulfovibrio desulfuricans 27774 transcriptome to nitrate, nitrite and nitric oxide. Scientific Reports, 7, 16228. Available from: 10.1038/s41598-017-16403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadby, I.T., Ibrahim, S.A., Faulkner, M., Lee, D.J., Browning, D., Busby, S.J. et al. (2016) Regulation, sensory domains and roles of two Desulfovibrio desulfuricans ATCC27774 Crp family transcription factors, HcpR1 and HcpR2, in response to nitrosative stress. Molecular Microbiology, 102, 1120–1137. [DOI] [PubMed] [Google Scholar]
- Christie, J.M., Reymond, P., Powell, G.K., Bernasconi, P., Raibekas, A.A., Liscum, E. et al. (1998) Arabidopsis NPH1: A flavoprotein with the properties of a photoreceptor for phototropism. Science, 282, 1698–1701. Available from: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- Crack, J., Green, J. & Thomson, A.J. (2004) Mechanism of oxygen sensing by the bacterial transcription factor fumarate‐nitrate reduction (FNR). Journal of Biological Chemistry, 279, 9278–9286. Available from: 10.1074/jbc.M309878200. [DOI] [PubMed] [Google Scholar]
- D’Autréaux, B., Tucker, N.P., Dixon, R. & Spiro, S. (2005) A non‐haem iron centre in the transcription factor NorR senses nitric oxide. Nature, 437, 769–772. Available from: 10.1038/nature03953. [DOI] [PubMed] [Google Scholar]
- da Silva, S.M., Amaral, C., Neves, S.S., Santos, C., Pimentel, C. & Rodrigues‐Pousada, C. (2015) An HcpR paralog of Desulfovibrio gigas provides protection against nitrosative stress. FEBS Open Bio, 5, 594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, M., Daveran, M.‐L., Batut, J., Dedieu, A., Domergue, O., Ghai, J. et al. (1988) Cascade regulation of nif gene expression in Rhizobium meliloti . Cell, 54, 671–683. Available from: 10.1016/S0092-8674(88)80012-6. [DOI] [PubMed] [Google Scholar]
- Esbelin, J., Jouanneau, Y. & Duport, C. (2012) Bacillus cereus Fnr binds a [4Fe‐4S] cluster and forms a ternary complex with ResD and PlcR. BMC Microbiology, 12, 125. Available from: 10.1186/1471-2180-12-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiévet, A., Cascales, E., Valette, O., Dolla, A. & Aubert, C. (2014) IHF Is required for the transcriptional regulation of the Desulfovibrio vulgaris Hildenborough orp Operons. PLoS One, 9, e86507. Available from: 10.1371/journal.pone.0086507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fievet, A., Ducret, A., Mignot, T., Valette, O., Robert, L., Pardoux, R. et al. (2015) Single‐cell analysis of growth and cell division of the anaerobe Desulfovibrio vulgaris Hildenborough. Frontiers in Microbiology, 6, 1378. Available from: 10.3389/fmicb.2015.01378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiévet, A., My, L., Cascales, E., Ansaldi, M., Pauleta, S.R., Moura, I. et al. (2011) The anaerobe‐specific orange protein complex of Desulfovibrio vulgaris hildenborough is encoded by two divergent operons coregulated by σ54 and a cognate transcriptional regulator. Journal of Bacteriology, 193, 3207–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo, M.C.O., Lobo, S.A.L., Carita, J.N., Nobre, L.S. & Saraiva, L.M. (2012) Bacterioferritin protects the anaerobe Desulfovibrio vulgaris Hildenborough against oxygen. Anaerobe, 18, 454–458. Available from: 10.1016/j.anaerobe.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Fioravanti, A., Fumeaux, C., Mohapatra, S.S., Bompard, C., Brilli, M., Frandi, A. et al. (2013a) DNA binding of the cell cycle transcriptional regulator GcrA depends on N6‐adenosine methylation in Caulobacter crescentus and other Alphaproteobacteria. PLoS Genetics, 9, e1003541. Available from: 10.1371/journal.pgen.1003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, F., Danson, A.E., Ye, F., Jovanovic, M., Buck, M. & Zhang, X. (2020) Bacterial enhancer binding proteins—AAA+ proteins in transcription activation. Biomolecules, 10. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7175178/ [Accessed 11 September 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glyde, R., Ye, F., Darbari, V.C., Zhang, N., Buck, M. & Zhang, X. (2017) Structures of RNA polymerase closed and intermediate complexes reveal mechanisms of dna opening and transcription initiation. Molecular Cell, 67, 106–116.e4. Available from: 10.1016/j.molcel.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrini, O., Burlat, B., Léger, C., Guigliarelli, B., Soucaille, P. & Girbal, L. (2008) Characterization of two 2[4Fe4S] ferredoxins from Clostridium acetobutylicum . Current Microbiology, 56, 261–267. Available from: 10.1007/s00284-007-9072-x. [DOI] [PubMed] [Google Scholar]
- Kazakov, A.E., Rajeev, L., Chen, A., Luning, E.G., Dubchak, I., Mukhopadhyay, A. et al. (2015) σ54‐dependent regulome in Desulfovibrio vulgaris Hildenborough. BMC Genomics, 16, 919. Available from: 10.1186/s12864-015-2176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte, H.L., Fels, S.R., Christensen, G.A., Price, M.N., Kuehl, J.V., Zane, G.M. et al. (2014) Genetic basis for nitrate resistance in Desulfovibrio strains. Frontiers in Microbiology, 5, 153. Available from: 10.3389/fmicb.2014.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazazzera, B.A., Beinert, H., Khoroshilova, N., Kennedy, M.C. & Kiley, P.J. (1996) DNA binding and dimerization of the Fe‐S‐containing FNR protein from Escherichia coli are regulated by oxygen. Journal of Biological Chemistry, 271, 2762–2768. Available from: 10.1074/jbc.271.5.2762. [DOI] [PubMed] [Google Scholar]
- Li, X., Zhang, H., Ma, Y., Liu, P. & Krumholz, L.R. (2014) Genes required for alleviation of uranium toxicity in sulfate reducing bacterium Desulfovibrio alaskensis G20 [corrected]. Ecotoxicology, 23, 726–733. [DOI] [PubMed] [Google Scholar]
- Lippard, S.J. & Berg, J.M. (1994) Principles of bioinorganic chemistry. University Science Books. [Google Scholar]
- Möglich, A., Ayers, R.A. & Moffat, K. (1993) (2009) Structure and signaling mechanism of Per‐ARNT‐Sim domains. Structure, 17, 1282–1294. Available from: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais Cabral, J.H., Lee, A., Cohen, S.L., Chait, B.T., Li, M. & Mackinnon, R. (1998) Crystal structure and functional analysis of the HERG potassium channel N terminus: A eukaryotic PAS domain. Cell, 95, 649–655. Available from: 10.1016/S0092-8674(00)81635-9. [DOI] [PubMed] [Google Scholar]
- Müllner, M., Hammel, O., Mienert, B., Schlag, S., Bill, E. & Unden, G. (2008) A PAS domain with an oxygen labile [4Fe‐4S](2+) cluster in the oxygen sensor kinase NreB of Staphylococcus carnosus . Biochemistry, 47, 13921–13932. [DOI] [PubMed] [Google Scholar]
- Nair, R.R., Silveira, C.M., Diniz, M.S., Almeida, M.G., Moura, J.J.G. & Rivas, M.G. (2015) Changes in metabolic pathways of Desulfovibrio alaskensis G20 cells induced by molybdate excess. JBIC Journal of Biological Inorganic Chemistry, 20, 311–322. Available from: 10.1007/s00775-014-1224-4. [DOI] [PubMed] [Google Scholar]
- Nambu, J.R., Lewis, J.O., Wharton, K.A. & Crews, S.T. (1991) The Drosophila single‐minded gene encodes a helix‐loop‐helix protein that acts as a master regulator of CNS midline development. Cell, 67, 1157–1167. Available from: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- Pini, F., De Nisco, N.J., Ferri, L., Penterman, J., Fioravanti, A., Brilli, M. et al. (2015) Cell cycle control by the master regulator CtrA in Sinorhizobium meliloti . PLoS Genetics, 11, e1005232. Available from: 10.1371/journal.pgen.1005232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postgate, J.R., Kent, H.M., Robson, R.L. & Chesshyre, J.A. (1984) The genomes of Desulfovibrio gigas and D. vulgaris . Journal of General Microbiology, 130, 1597–1601. Available from: 10.1099/00221287-130-7-1597. [DOI] [PubMed] [Google Scholar]
- Rajeev, L., Chen, A., Kazakov, A.E., Luning, E.G., Zane, G.M., Novichkov, P.S. et al. (2015) Regulation of nitrite stress response in Desulfovibrio vulgaris Hildenborough, a model sulfate‐reducing bacterium. Journal of Bacteriology, 197, 3400–3408. Available from: 10.1128/JB.00319-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel, F., Amrani, A., Pieulle, L., Lamrabet, O., Voordouw, G., Seddiki, N. et al. (2013) Membrane‐bound oxygen reductases of the anaerobic sulfate‐reducing Desulfovibrio vulgaris Hildenborough: Roles in oxygen defence and electron link with periplasmic hydrogen oxidation. Microbiology, 159, 2663–2673. Available from: 10.1099/mic.0.071282-0. [DOI] [PubMed] [Google Scholar]
- Scholten, J.C., Culley, D.E., Brockman, F.J., Wu, G. & Zhang, W. (2007) Evolution of the syntrophic interaction between Desulfovibrio vulgaris and Methanosarcina barkeri: Involvement of an ancient horizontal gene transfer. Biochemical and Biophysical Research Communications, 352, 48–54. Available from: 10.1016/j.bbrc.2006.10.164. [DOI] [PubMed] [Google Scholar]
- Shingler, V. (2011) Signal sensory systems that impact σ54‐dependent transcription. FEMS Microbiology Reviews, 35, 425–440. [DOI] [PubMed] [Google Scholar]
- Shingler, V. & Pavel, H. (1995) Direct regulation of the ATPase activity of the transcriptional activator DmpR by aromatic compounds. Molecular Microbiology, 17, 505–513. Available from: 10.1111/j.1365-2958.1995.mmi_17030505.x. [DOI] [PubMed] [Google Scholar]
- Taylor, B.L. & Zhulin, I.B. (1999) PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiology and Molecular Biology Reviews, 63, 479–506. Available from: 10.1128/MMBR.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela‐Raposo, A., Pimentel, C., Morais‐Silva, F., Rezende, A., Ruiz, J.C. & Rodrigues‐Pousada, C. (2013) Role of NorR‐like transcriptional regulators under nitrosative stress of the δ‐proteobacterium, Desulfovibrio gigas . Biochemical and Biophysical Research Communications, 431, 590–596. Available from: 10.1016/j.bbrc.2012.12.130. [DOI] [PubMed] [Google Scholar]
- Walker, C.B., He, Z., Yang, Z.K., Ringbauer, J.A., He, Q., Zhou, J. et al. (2009) The electron transfer system of syntrophically grown Desulfovibrio vulgaris . Journal of Bacteriology, 191, 5793–5801. Available from: 10.1128/JB.00356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, F. & Grohmann, D. (2011) Evolution of multisubunit RNA polymerases in the three domains of life. Nature Reviews Microbiology, 9, 85–98. Available from: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- Wilkins, M.J., Hoyt, D.W., Marshall, M.J., Alderson, P.A., Plymale, A.E., Markillie, L.M. et al. (2014) CO2 exposure at pressure impacts metabolism and stress responses in the model sulfate‐reducing bacterium Desulfovibrio vulgaris strain Hildenborough. Frontiers in Microbiology, 5, 507. Available from: 10.3389/fmicb.2014.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkiw, M.A., Voordouw, J. & Voordouw, G. (2012) Contribution of rubredoxin:oxygen oxidoreductases and hybrid cluster proteins of Desulfovibrio vulgaris Hildenborough to survival under oxygen and nitrite stress. Environmental Microbiology, 14, 2711–2725. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Chaney, M., Wigneshweraraj, S.R., Schumacher, J., Bordes, P., Cannon, W. et al. (2002) Mechanochemical ATPases and transcriptional activation. Molecular Microbiology, 45, 895–903. Available from: 10.1046/j.1365-2958.2002.03065.x. [DOI] [PubMed] [Google Scholar]
- Zhou, A., Baidoo, E., He, Z., Mukhopadhyay, A., Baumohl, J.K., Benke, P. et al. (2013) Characterization of NaCl tolerance in Desulfovibrio vulgaris Hildenborough through experimental evolution. ISME Journal, 7, 1790–1802. Available from: 10.1038/ismej.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, A., Lau, R., Baran, R., Ma, J., von Netzer, F., Shi, W., et al. (2017) Key metabolites and mechanistic changes for salt tolerance in an experimentally evolved sulfate‐reducing bacterium, Desulfovibrio vulgaris . mBio, 8, e01780‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., He, Q., Hemme, C.L., Mukhopadhyay, A., Hillesland, K., Zhou, A. et al. (2011) How sulphate‐reducing microorganisms cope with stress: Lessons from systems biology. Nature Reviews Microbiology, 9, 452–466. Available from: 10.1038/nrmicro2575 [DOI] [PubMed] [Google Scholar]
- Zhu, L., Gong, T., Wood, T.L., Yamasaki, R. & Wood, T.K. (2019) σ54 ‐Dependent regulator DVU2956 switches Desulfovibrio vulgaris from biofilm formation to planktonic growth and regulates hydrogen sulfide production. Environmental Microbiology, 21, 3564–3576. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


