Abstract
Background
Quantification of myocardium scarring in late gadolinium enhanced (LGE) cardiac magnetic resonance imaging can be challenging due to low scar‐to‐background contrast and low image quality. To resolve ambiguous LGE regions, experienced readers often use conventional cine sequences to accurately identify the myocardium borders.
Purpose
To develop a deep learning model for combining LGE and cine images to improve the robustness and accuracy of LGE scar quantification.
Study Type
Retrospective.
Population
A total of 191 hypertrophic cardiomyopathy patients: 1) 162 patients from two sites randomly split into training (50%; 81 patients), validation (25%, 40 patients), and testing (25%; 41 patients); and 2) an external testing dataset (29 patients) from a third site.
Field Strength/Sequence
1.5T, inversion‐recovery segmented gradient‐echo LGE and balanced steady‐state free‐precession cine sequences
Assessment
Two convolutional neural networks (CNN) were trained for myocardium and scar segmentation, one with and one without LGE‐Cine fusion. For CNN with fusion, the input was two aligned LGE and cine images at matched cardiac phase and anatomical location. For CNN without fusion, only LGE images were used as input. Manual segmentation of the datasets was used as reference standard.
Statistical Tests
Manual and CNN‐based quantifications of LGE scar burden and of myocardial volume were assessed using Pearson linear correlation coefficients (r) and Bland–Altman analysis.
Results
Both CNN models showed strong agreement with manual quantification of LGE scar burden and myocardium volume. CNN with LGE‐Cine fusion was more robust than CNN without LGE‐Cine fusion, allowing for successful segmentation of significantly more slices (603 [95%] vs. 562 (89%) of 635 slices; P < 0.001). Also, CNN with LGE‐Cine fusion showed better agreement with manual quantification of LGE scar burden than CNN without LGE‐Cine fusion (%ScarLGE‐cine = 0.82 × %Scarmanual, r = 0.84 vs. %ScarLGE = 0.47 × %Scarmanual, r = 0.81) and myocardium volume (VolumeLGE‐cine = 1.03 × Volumemanual, r = 0.96 vs. VolumeLGE = 0.91 × Volumemanual, r = 0.91).
Data Conclusion
CNN based LGE‐Cine fusion can improve the robustness and accuracy of automated scar quantification.
Level of Evidence
3
Technical Efficacy
1
Keywords: Deep learning, image segmentation, image fusion, late gadolinium enhancement, myocardial scar, scar quantification
Myocardium scar quantified by late gadolinium enhanced (LGE) cardiac magnetic resonance imaging (MRI) has an important prognostic value in heart diseases and represents an important risk factor for ventricular arrhythmias,1, 2 sudden cardiac death,3, 4 and heart failure.5, 6 Currently, computer‐assisted manual segmentation of the myocardium borders and scarred regions is the reference method for LGE image analysis.3, 7 However, extensive manual intervention is a bottleneck in the image analysis workflow and is prone to reader variability.8, 9 Automatic image segmentation using deep convolutional neural networks (CNN) has been recently proposed to standardize LGE analysis and mitigate the time and effort of manual contouring.10, 11, 12 With CNN, large annotated datasets are used to automatically learn how to identify scarred and normal myocardium pixels in LGE images. One limitation of automatic methods is the unreliable identification of scars in the vicinity of hyperenhanced blood pool or adipose tissues.13 This limitation is accentuated further by low‐quality images and cases of uncommon myocardium shapes and scar patterns (e.g., hypertrophic cardiomyopathy [HCM]).14, 15 In practice, readers resolve this ambiguity by performing side‐by‐side reading of LGE and conventional cine image sequences to determine the correct borders of the scar and myocardium.14 However, this practice entails additional manual processing and delays an already prolonged image analysis workflow.
Automatic methods for facilitating the integration of cine and LGE image sequences can be classified into direct16, 17, 18 or indirect19, 20 image fusion approaches. In the direct approach, the myocardium is first delineated in the cine images. The resulting contours are then copied to the corresponding LGE image dataset to guide myocardium segmentation. This approach requires accurate registration of the cine and LGE images and involves heuristic design and careful selection of algorithmic parameters.16, 17 In the indirect approach, shape variations among the myocardium contours are modeled using an annotated set of cine images. Then, the modeled shapes are used to regularize (or constrain) the myocardium segmentation in LGE images not necessarily corresponding to the modeled set of cine images. Deep CNN based methods have been used to implement indirect integration of LGE and cine information.19, 20 Current methods of both approaches require a separate processing step to identify scars as hyperenhanced regions within the segmented myocardium. However, the accuracy of such intensity‐based identification of scars is limited and vulnerable to imaging artifacts and selection of algorithmic parameters. In this study, we present a deep CNN algorithm for robust segmentation of LGE scars in HCM patients by allowing fusion of LGE and cine images.
Materials and Methods
MRI Dataset
MRI datasets from three different sites were used to develop and test the proposed CNN model. All patients signed statements approved by the Investigational Review boards of the participating institutions, agreeing to the use of their medical information for research. The dataset was a subset of cases from a multicenter HCM study,3 in which patients with implantable cardioverter defibrillators, sustained ventricular tachycardia or ventricular fibrillation, myocardial infarction, or septal reduction procedures were excluded. Among these patients, we only included cases if both short‐axis LGE and cine sequences planned using the same reference scan were available. A set of MRI scans of 162 HCM patients acquired from two medical centers (Tufts Medical Center and Beth Israel Deaconess Medical Center) were combined and randomly split (as discussed below) to train, optimize, and test the proposed model. We refer to this image set as the development dataset. To increase the robustness of testing, we used a set of MRI scans of 29 patients from a third site for testing only. We refer to this image set as external testing dataset.
All acquisitions were performed on 1.5 T scanners (Philips Healthcare, Best, The Netherlands). Each patient dataset included electrocardiogram gated breath‐hold inversion‐recovery segmented gradient echo LGE and balanced steady‐state free precession (bSSFP) cine scans. LGE images were acquired 10–20 minutes after intravenous administration of 0.2 mmol/kg gadolinium‐diethylenetriamine penta‐acetic acid (Magnevist; Schering, Berlin, Germany).
The typical LGE imaging parameters of the development dataset were: repetition time (TR) = 3.4–4.9 msec, echo time (TE) = 1.1–2.9 msec, flip angle (α) = 15°–20°, field of view (FOV) = 360–400 × 360–400 mm2, pixel size = 1.0–1.25 × 1.0–1.25 mm2, number of slices = 7–24, slice thickness = 8–10 mm, and trigger delay = 332–1040 msec. The imaging parameters for the bSSFP cine sequences were as follows: TR = 2.6–3.6, TE = 1.2–1.7 msec, α = 15–60°, FOV = 360–400 × 360–400 mm2, pixel size = 0.97–1.25 × 1.0–1.25 mm2, and slice thickness = 5–10 mm.
The external testing dataset consisted of 29 patients from a third site using a 1.5 T scanner (Philips Healthcare, Best, The Netherlands). The acquisition parameters for the external dataset were as follows. LGE acquisition with TR = 3.3–8.8 msec, TE = 1.2–2.0 msec, α = 15°‐60°, FOV = 360–400 × 360–400 mm2, pixel size = 0.7–1.6 × 0.7–1.6 mm2, number of slices = 7–25, slice thickness = 8–12 mm, and trigger delay = 255–838 msec. The imaging parameters for bSSFP cine sequences were: TR = 3.0–4.2 msec, TE = 1.5–2.1 msec, α = 50–70°, FOV = 360–400 × 360–400 mm2, pixel size = 0.97–1.25 × 1.0–1.25 mm2, and slice thickness = 8–10 mm.
Data Splitting and Preprocessing
The development dataset was split into training (50%, 81 patients), validation (25%, 40 patients), and testing (25%, 41 patients) subsets (Fig. 1a). A stratified (patient‐wise) random splitting approach was used such that a similar ratio of cases with different LGE scar burden (<1%, 1–10%, and >10%) was maintained in each subset. The external dataset (29 patients) was used only for testing. For each LGE slice, a matched cine slice at the same cardiac phase and closest anatomical location was selected using the trigger delay and slice location information stored in the dicom file (Fig. 1b). In‐plane image misalignment due to different breath‐hold levels and/or patient motion between cine and LGE scans was reduced using in‐plane image translation. First, the center of the left ventricle (LV) was manually selected in the LGE and cine images. Then, in‐plane image translation was used to align the selected centers. Finally, the operator was able to overlay the translated LGE and cine images to visually check their alignment and repeat the process if needed. All images were normalized to intensity range from 0 to 1 and spatial resolution of 1.2 mm, and cropped to 160 × 160 matrix.
FIGURE 1.
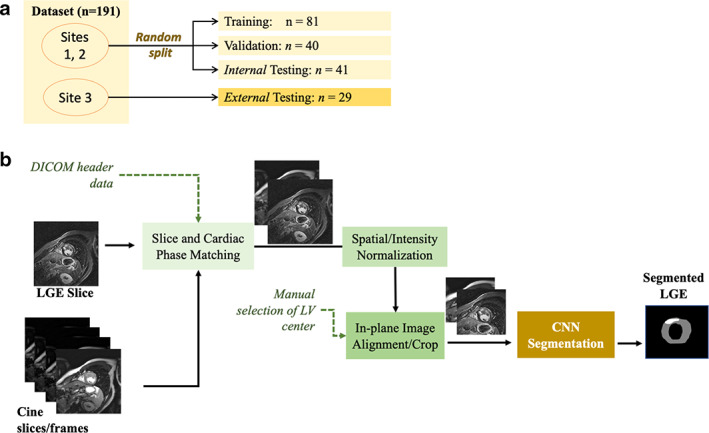
Flow chart of dataset splitting (a), and convolutional neural network (CNN) based fusion of late gadolinium enhancement (LGE) and balanced steady‐state free precession (bSSFP) cine sequences for myocardium and scar segmentation (b). DICOM = digital imaging and communications in medicine file format; LV = left ventricle.
Reference Segmentation
The reference standard segmentations for all LGE images were obtained as a part of core laboratory analyses (PERFUSE, Boston, MA) using manual analysis by a cardiologist (R.H.C., with 3‐year experience in LGE scar analysis in HCM patients). Both datasets were analyzed by the same reader using a commercial software (QMASS version 7.4, Medis Inc., Raleigh, NC). First, the endocardium and epicardium boundaries were manually delineated. Then, the reader manually adjusted a gray‐scale threshold to identify all visually apparent hyper‐enhanced areas within the myocardium. If necessary, the reader used manual drawing to include LGE scars not identified by the intensity threshold and/or exclude hyper‐enhanced areas representing noise or artifacts.3 These areas were then summed to generate a total volume of LGE and expressed as a proportion of the total LV myocardium (%Scar).
Network Architecture
We developed two 2D‐CNN models for LGE segmentation (one with and one without LGE‐Cine fusion) based on U‐Net architecture.21 The models included four multiresolution processing levels (image down‐sampling factor = 2 per level), 3 × 3 convolutional kernels with maximum pooling at each resolution level, ReLU activation, batch normalization, and dropout layers. Long and short skip‐connections, typically used for U‐Net, were used to improve network performance.22 The input to the CNN model was an LGE image (size = 160 × 160) or a stack of two matched LGE‐Cine images (size = 160 × 160 × 2) for CNN without or with fusion, respectively. A softmax layer was used as the output layer of the network to produce four maps representing the probability of each pixel to belong to scar, normal‐myocardium, blood, or background regions. Background and blood pool regions were then merged into one background region. During model training, a cross‐entropy loss function was used to measure the error between the CNN based segmentation and the manual segmentation.21 Model training was done for a fixed number of epochs (N = 250) to minimize the loss function and the best performing model (i.e., that with highest segmentation accuracy in the validation subset) was selected as the final model. Image augmentation (using translation, rotation, and up–down/left–right flipping) was used to increase the training dataset size and avoid over‐fitting.23
Optimization of CNN Model Parameters
Training of each model was repeated using different sets of hyper‐parameters arbitrarily selected from the following ranges: number of channels at the first processing layer (16, 24, 32, 48, 64), dynamic learning rate (initial = 0.01 or 0.005; minimum = 0.0005, reduction factor = 0.8, plateau interval = 30 epochs), dropout probabilities (0.25 and 0.5), and batch size (2 and 4 patients ~20–40 images). We used the validation dataset to evaluate the performance of each trained model and the final model was selected as the one with the smallest number of parameters that yielded the highest myocardium segmentation accuracy. The architecture of the optimal models with and without fusion was comprised of 0.58 × 106 and 1.3 × 106 parameters, respectively, with learning rate = 0.005, dropout = 0.25, and batch size = 4 patients. The number of channels in the four multiresolution (from high to low) levels were 32/64/128/128 and 48/96/196/196 for the CNN with and without fusion, respectively. The networks were implemented using Python‐V3.6 (Python Software Foundation, www.python.org) and Tensorflow‐V2.1 (Google Inc., Mountain View, CA). Implementation was performed on both Nvidia DGX‐1 workstation and the cloud‐based Precision Medicine Platform (PMP) of the American Heart Association (AHA). Network implementation, model parameters, and sample datasets are publicly available at https://doi.org/10.7910/dvn/w22fya.
Image Postprocessing
The outputs of the CNN models were processed to create categorical images with each pixel labeled normal myocardium, scar, and background (including blood pool). The segmented myocardium was first processed to fill small gaps within the myocardium (using a morphological closing operation, disk with radius = 3 pixels). If multiple disjoint myocardium regions were segmented, only the largest one was selected and all others deleted. Morphological dilation with 1 pixel was used to account for eroded boundaries observed in the segmented images.
Statistical Analysis
Linear correlation (Pearson correlation coefficient, r) and Bland–Altman analyses were used to evaluate the agreement between automatic and manual quantification of LGE scar burden and myocardium volume in the testing datasets. Chi‐square test was used to compare sample proportions. LGE scar burden, %Scar (defined as the ratio of scar volume to the total LV myocardium volume) and presence of scar (defined as %Scar >1%) were analyzed. Slices with substantial errors such that <50% of myocardium area was correctly segmented, were identified and removed from data analysis. Data analyses were performed using Statistics Toolbox of Matlab‐R2018b (Mathworks Inc., Natick, MA).
Results
LGE scar was present in 10 patients (26% of 41) in the internal testing datasets and 15 patients (52% of 29) in the external testing datasets, with average burden, %Scar, of 7.1% ± 6.4% (median = 5.3%) and 7.8% ± 7.5% (median = 5.2%), respectively (Table 1). The number of patients with scar in the combined testing dataset (25% of the development dataset + external dataset) was 25 (36% of 70) patients with average burden 6% ± 5% (median = 5%). Both CNN models (with and without fusion) successfully segmented the myocardium in 66 patients (94% of 70 patients of the combined testing dataset) and showed strong correlation with manual quantification in the internal (r > 0.91, N = 40), external (r > 0.73, N = 26), and combined (r > 0.81, N = 66) testing datasets (Table 2). CNN analysis time was less than 0.05 seconds/slice in both models while image postprocessing was less than 0.10 seconds/slice. In four cases, low image quality (N = 1, internal dataset) and severe anomaly in myocardium shape (N = 3 in external dataset) lead to substantial errors in both models and thus four datasets were excluded from further analysis (Fig. S1 in the Supplemental Material). CNN with LGE‐Cine fusion allowed segmentation of significantly more slices compared to CNN without fusion (603 [95%] vs. 562 [89%] of 635 slices, P < 0.001) (Fig. 2 and Table 3). Quantification by CNN with LGE‐Cine fusion showed strong agreement with manual quantification of %Scar (%ScarLGE‐cine = 0.82 × %Scarmanual, r = 0.84; 66 patients, 603 slices) (Fig. 3) and myocardium volume (VolumeLGE‐cine = 1.03 × Volumemanual, r = 0.96; 66 patients, 603 slices) (Fig. 4). Strong correlation was also observed for 2‐parameter regression line (i.e., with an intercept) for both %Scar (%ScarLGE‐cine = 0.12 + 0.81 × %Scarmanual, r = 0.84; 66 patients, 603 slices) and myocardium volume (VolumeLGE‐cine = 18 + 0.91 × Volumemanual, r = 0.96; 66 patients, 603 slices). The estimation bias in Bland–Altman graph was −0.3% for %Scar and 7 mL for myocardium volume with >90% of measurements within the limits of agreement for %Scar (±5%) and myocardium volume (±41 mL) (Fig. 3). In comparison to CNN with LGE‐Cine fusion, in all testing datasets, CNN without fusion showed greater underestimation of manual quantification of %Scar (%ScarLGE = 0.47 × %Scarmanual, r = 0.81; 66 patients, 562 slices) (Fig. 3) and myocardium volume (VolumeLGE = 0.91 × Volumemanual, r = 0.91; 66 patients, 562 slices) (Fig. 4). Also, compared to CNN with LGE‐Cine fusion, CNN without LGE‐Cine fusion showed lower accuracy (80% vs. 89%, P = 0.15, N = 66), specificity (93% vs. 96%, P = 0.54, N = 44), and sensitivity (57% vs. 77%, P = 0.14, N = 22) of identifying patients with scar.
TABLE 1.
Patients' Demographics in Training, Validation, and Testing Subsets
| Training (N = 81) | Validation (N = 40) | Internal Testing (N = 41) | External Testing (N = 29) | All (N = 191) | |
|---|---|---|---|---|---|
| Age, years | 42 ± 17 (46) | 47 ± 16 (50) | 50 ± 16 (49) | 40 ± 16 (41) | 45 ± 17 (46) |
| Male, N (%) | 49(60%) | 32(80%) | 29(71%) | 9(31%) | 119 (62%) |
| Body surface areaa, m2 | 1.86 ± 0.26 (1.89) | 2.0 ± 0.25 (2.02) | 2.0 ± 0.22 (2.0) | 1.82 ± 0.25 (1.85) | 1.92 ± 0.26 (1.91) |
| %Scar | 6.4 ± 7.1% (3.9%) | 6.6 ± 7.2% (3.4%) | 7.1 ± 6.4% (5.3%) | 7.8 ± 7.5% (5.2%) | 6.8 ± 7.0% (4.3%) |
| %Scar > 1% | 27 (33%) | 13 (33%) | 10 (26%) | 15 (52%) | 65 (32%) |
| Atrial fibrillationa, N (%) | 12 (15%) | 5 (13%) | 9 (22%) | 2 (7%) | 28 (15%) |
| NSVTa, N (%) | 8 (10%) | 6 (15%) | 5 (12%) | 4 (14%) | 23 (12%) |
| CADa, N (%) | 4 (5%) | 4 (10%) | 2 (5%) | 0 (0%) | 10 (5%) |
| NYHA classa, N (%) | |||||
| I | 39 (48%) | 14 (35%) | 18 (44%) | 12 (41%) | 83 (43%) |
| II | 26 (32%) | 16 (40%) | 4 (10%) | 9 (31%) | 55 (29%) |
| III | 11 (14%) | 7 (18%) | 15 (37%) | 5 (17%) | 38 (20%) |
| IV | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
Data are given as mean ± standard deviation (median) or number of patients (%).
%Scar = percentage of left ventricular scar to myocardium volume; CAD = coronary artery disease; NSVT = nonsustained ventricular tachycardia; NYHA = New York Heart Association.
Data was missing for 15 patients (training, 5; validation, 3; internal testing, 4; and external testing, 3).
TABLE 2.
Evaluation of Convolutional Neural Network (CNN) Based Quantification of LGE Scar Burden, %Scar, and Myocardium Volume With and Without Image Fusion
| Dataset | CNN Model | Successful Segmentationa | %Scar (Slope/R)b | Myocardium Volume (Slope/R) | Scar Detectionc |
|---|---|---|---|---|---|
| All testing (66 patients) | Fusion | 603 (95% of 635) | 0.82/0.84 | 1.03/0.96 | 89/96/77% (N = 22) |
| No fusion | 562 (89% of 635; P < 0.001) | 0.47/0.81 | 0.91/0.91 | 80/93/57% (N = 22) | |
| Internal testing (40 patients) | Fusion | 298 (95% of 313) | 0.80/0.91 | 1.04/0.92 | 90/93/80% (N = 10) |
| No fusion | 273 (87% of 313; P < 0.001) | 0.40/0.93 | 0.92/0.90 | 83/97/46% (N = 10) | |
| External testing (26 patients) | Fusion | 305 (95% of 322) | 0.85/0.73 | 1.02/0.96 | 88/100/75% (N = 12) |
| No fusion | 289 (90% of 322; P = 0.015) | 0.65/0.75 | 0.90/0.88 | 77/86/67% (N = 12) |
Data are given as number of slices (% of total number of slices).
Data are given as Unitless linear regression slope and coefficient of determination (R).
Data are given as Accuracy/specificity/sensitivity (number of patients with LGE scar burden >1%).
FIGURE 2.
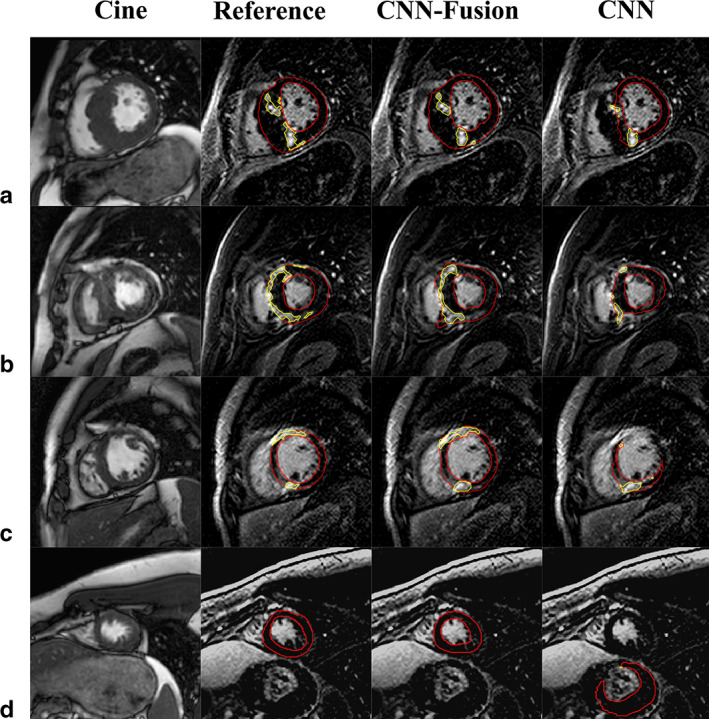
Segmentation of late gadolinium enhancement (LGE) in short‐axis slices for four different patients (a–d). Myocardium and scar contours (red and yellow, respectively) overlaid on the LGE slices (columns 2–4) indicate: manual segmentation (Reference, column 2), convolutional neural network (CNN) with fusion (CNN‐Fusion, column 3), and CNN without fusion (CNN, column 4). Column 1 displays the cine images corresponding to each LGE slice (matched location and cardiac phase).
TABLE 3.
Success Rate of Convolutional Neural Network (CNN) Based Segmentation Categorized by Slice Location
| Dataset | CNN Model | Apical Slices | Mid‐Cavity Slices | Basal Slices | All Slices |
|---|---|---|---|---|---|
| All (66 patients) | Fusion | 177 (89% of 198) | 318 (99% of 320) | 108 (92% of 117) | 603 (95% of 635) |
| No fusion | 156 (79% of 198; P = 0.006) | 307 (96% of 320; P = 0.015) | 99 (85% of 117; P = 0.091) | 562 (89% of 635; P < 0.001) | |
| Internal testing (40 patients) | Fusion | 98 (92% of 107) | 150 (99% of 151) | 50 (91% of 55) | 298 (95% of 313) |
| No fusion | 84 (79% of 107; P = 0.006) | 145 (96% of 151; P = 0.094) | 44 (80% of 55; P = 0.097) | 273 (87% of 313; P < 0.001) | |
| External testing (26 patients) | Fusion | 79 (87% of 91) | 168 (99% of 169) | 58 (94% of 62) | 305 (95% of 322) |
| No fusion | 72 (79% of 91; P = 0.149) | 162 (96% of 169; P = 0.076) | 55 (89% of 62; P = 0.316) | 289 (90% of 322; P = 0.015) |
Data are given as number of slices successfully segmented (% of total number of slices). P‐values are reported for the difference between successful segmentation using fusion versus no fusion.
FIGURE 3.
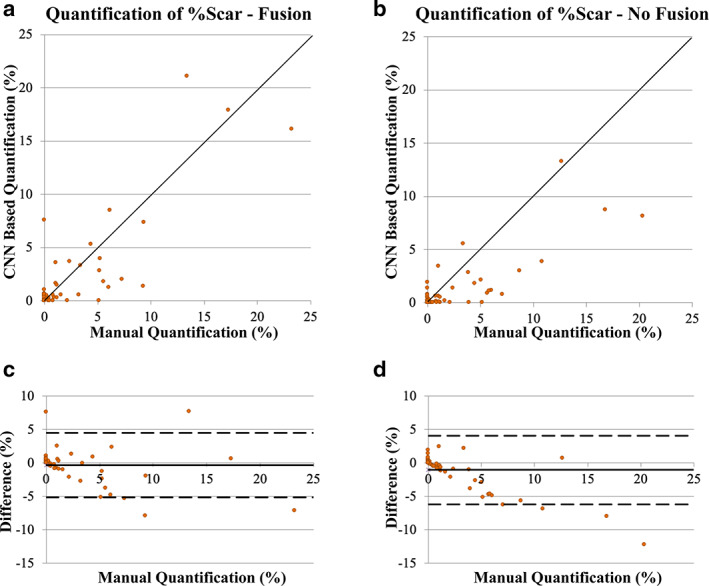
Assessment of convolutional neural network (CNN) based quantification of LGE scar burden (%Scar) with (a, c) and without (b, d) LGE‐Cine fusion versus manual quantification. The solid line in the scatter plots (a, b) represents unity‐slope regression line. The solid and dashed horizontal lines in Bland–Altman plots (c, d) represent bias and limit of agreement, respectively.
FIGURE 4.
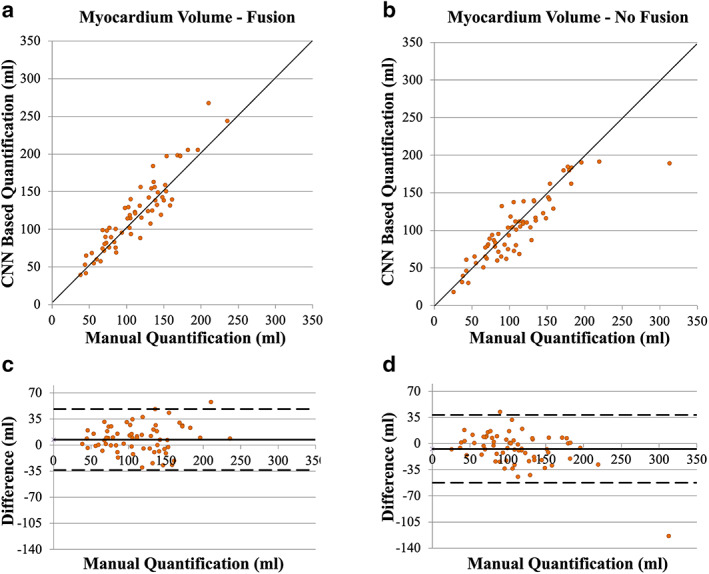
Assessment of convolutional neural network (CNN) based quantification of myocardium volume with (a, c) and without (b, d) LGE‐Cine fusion versus manual quantification. The solid line in the scatter plots (a, b) represents unity‐slope regression line. The solid and dashed horizontal lines in Bland–Altman plots (c, d) represent bias and limit of agreement, respectively.
Discussion
In this study, we have presented a CNN model for improved LGE scar quantification using automated LGE‐Cine fusion. The developed model attempts to mimic the common clinical practice of reading LGE and cine sequences. Our results demonstrated that CNN with LGE‐Cine fusion improved the quantification accuracy of LGE scar burden and myocardium volume and allowed better detection of LGE scars compared to CNN without fusion. The results also showed that CNN with LGE‐Cine fusion enabled segmentation of LGE slices in cases where conventional CNN without fusion failed.
Model Development
The CNN models in our study were trained using a dataset from two medical centers implementing the same imaging protocol on 1.5 T scanners from a single vendor. This relative homogeneity of the dataset allowed effective training of smaller CNN models compared to what has been presented in previous studies.10, 11 Also, we noted that a more efficient representation of LGE patterns and image contrast may be achieved by incorporating cine images into the model. This was indicated by the smaller size (<50%) of the optimal CNN based fusion model compared to that of the CNN model without fusion. To avoid overfitting the model to the training dataset, we employed three techniques: dropout layers, image augmentation, and selecting the optimal model based on the accuracy of segmenting a validation dataset.
Model Performance
The developed models were tested using a diverse dataset from 70 HCM patients including an external dataset from a different imaging center (which used the same scanner and imaging protocol but with different imaging parameters). Scar quantification in HCM patients is highly challenging due to the patchy multifoci appearance of scars.14, 15 Additionally, in HCM, myocardium hypertrophy and changed LV geometry limit the ability to differentiate hyperenhanced myocardium from the blood pool.13, 14 In both the internal and external test datasets, CNN with LGE‐Cine fusion outperformed CNN without fusion in terms of segmentation robustness (i.e., number of segmented slices) and %Scar quantification accuracy (i.e., linear regression slope closer to 1). Also, in the combined dataset, but not the internal or the external datasets separately, CNN with LGE‐Cine fusion showed higher correlation coefficient. The robustness and accuracy of segmenting the myocardium by both models were comparable in the internal and external datasets. However, a lower correlation coefficient between manual and CNN based quantification of LGE scar burden was observed in the external dataset. Additionally, most failed segmentations (three out of four cases) belonged to the external dataset. A performance drop of pretrained DL models is usually expected when applied to datasets different from those used during model development. Although both the internal and external datasets were from HCM patients and acquired using 1.5 T scanner from the same vendor, there were differences among the two datasets that contributed to performance drop in the external dataset. This discrepancy included differences in patient characteristics (e.g., scar prevalence), imaging parameters (e.g., spatial resolution), and other implicit differences in implementing the imaging protocol (e.g., subjective setting of inversion delay and injection‐to‐imaging delay). Our results highlight the need to develop new methods for improving the generalizability of deep learning based LGE image analyses.
Image Matching
In our study, LGE and cine image datasets were acquired with breath‐holding and automatically matched based on slice location and cardiac phase parameters stored in the DICOM files. However, different levels of breath‐holding and voluntary patient motion had to be compensated especially with the relatively long time interval between the cine scans and the LGE scans. To compensate patient motion, we used a simple manual image translation (to match the manually selected centers of the LV in each image) to compensate in‐plane image shift and improve the alignment of the matched LGE and cine slices. Also, the multiresolution processing architecture adopted in our CNN model provided more robustness to residual misalignment of LGE and cine images.
Limitations
Only one reader did manual segmentation of the LGE images and we did not compare CNN quantification errors to interobserver variability. However, in our study, CNN versus manual scar quantification variability (−0.7 ± 7.4 g corresponding to −0.3 ± 5.0%) is comparable to the previously reported variability among expert readers (inter‐observer: −1.3 ± 6.5 g and intra‐observer: 0.3 ± 7.8 g).13 Also, we did not employ advanced transfer learning techniques to improve the model performance in the external dataset. A dedicated study may be needed to evaluate the potential advantage of including cine images in LGE segmentation models when applied to external datasets. A further limitation is that image mismatching caused by errors in cardiac gating or through‐plane motion are not corrected for by the in‐plane translation implemented to match LGE and cine images. Although automated nonrigid image registration has been previously proposed to improve LGE‐Cine image alignment,17 it has several limitations. First, it is very challenging to automatically achieve accurate registration of LGE and cine sequences given their substantial differences in image contrast and characteristics. Also, boundary and shape deformation introduced by nonrigid registration algorithms can distort the anatomical information embedded in the cine images and lead to segmentation errors.
Conclusion
We have presented a CNN based method for LGE‐Cine image fusion that allows robust and accurate quantification of myocardium LGE scar burden and enhances LGE image analysis workflow. The developed CNN model outperformed a conventional CNN model that analyzed only LGE images without incorporating cine sequences.
Supporting information
Fig S1 Sample of three slices (columns) from the four cases with failed automatic segmentation. Cases 1–3 (external testing dataset) have severe hypertrophy with large scars, a pattern that did not present in the patient cohort used for training the model. Case 4 (local testing dataset) shows no contrast between myocardium and background.
Acknowledgments
The project described was supported in part by American Heart Association (AHA) research grants (15EIA22710040) (Dallas, TX, USA), AHA Institute for Precision Cardiovascular Medicine (19AIML34850090) (Dallas, TX, USA) and National Institutes of Health 1R01HL129157‐01A1 and 5R01HL129185 (Bethesda, MD, USA).
The copyright line for this article was changed on 26th May 2021 after original online publication.
References
- 1.Di Marco A, Anguera I, Schmitt M, et al. Late gadolinium enhancement and the risk for ventricular arrhythmias or sudden death in dilated cardiomyopathy. JACC Heart Fail 2017;5(1):28‐38. [DOI] [PubMed] [Google Scholar]
- 2.Disertori M, Rigoni M, Pace N, et al. Myocardial fibrosis assessment by LGE is a powerful predictor of ventricular tachyarrhythmias in ischemic and nonischemic LV dysfunction: A meta‐analysis. JACC Cardiovasc Imaging 2016;9(9):1046‐1055. [DOI] [PubMed] [Google Scholar]
- 3.Chan RH, Maron BJ, Olivotto I, et al. Prognostic value of quantitative contrast‐enhanced cardiovascular magnetic resonance for the evaluation of sudden death risk in patients with hypertrophic cardiomyopathy. Circulation 2014;130(6):484‐495. [DOI] [PubMed] [Google Scholar]
- 4.Neilan TG, Coelho‐Filho OR, Danik SB, et al. CMR quantification of myocardial scar provides additive prognostic information in nonischemic cardiomyopathy. JACC Cardiovasc Imaging 2013;6(9):944‐954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shanbhag SM, Greve AM, Aspelund T, et al. Prevalence and prognosis of ischaemic and non‐ischaemic myocardial fibrosis in older adults. Eur Heart J 2019;40(6):529‐538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong TC, Piehler KM, Zareba KM, et al. Myocardial damage detected by late gadolinium enhancement cardiovascular magnetic resonance is associated with subsequent hospitalization for heart failure. J Am Heart Assoc 2013;2(6):e000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karim R, Bhagirath P, Claus P, et al. Evaluation of state‐of‐the‐art segmentation algorithms for left ventricle infarct from late gadolinium enhancement MR images. Med Image Anal 2016;30:95‐107. [DOI] [PubMed] [Google Scholar]
- 8.Klem I, Heiberg E, Van Assche L, et al. Sources of variability in quantification of cardiovascular magnetic resonance infarct size—Reproducibility among three core laboratories. J Cardiovasc Magn Reson 2017;19(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White SK, Flett AS, Moon JC. Automated scar quantification by CMR: A step in the right direction. J Thorac Dis 2013;5(4):381‐382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fahmy AS, Neisius U, Chan RH, et al. Three‐dimensional deep convolutional neural networks for automated myocardial scar quantification in hypertrophic cardiomyopathy: A multicenter multivendor study. Radiology 2019;294(1):52‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fahmy AS, Rausch J, Neisius U, et al. Automated cardiac MR scar quantification in hypertrophic cardiomyopathy using deep convolutional neural networks. JACC Cardiovasc Imaging 2018;11(12):1917‐1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jani Vivek P, Ostovaneh Mohammad R, Chamera E, Lima Joao A, Ambale‐Venkatesh B. Automatic segmentation of left ventricular myocardium and scar from LGE‐CMR images utilizing deep learning with weighted categorical cross entropy loss function weight initialization. Circulation 2019;140(Suppl 1):A15934. [Google Scholar]
- 13.Mikami Y, Kolman L, Joncas SX, et al. Accuracy and reproducibility of semi‐automated late gadolinium enhancement quantification techniques in patients with hypertrophic cardiomyopathy. J Cardiovasc Magn Reson 2014;16(1):85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turkbey EB, Nacif MS, Noureldin RA, et al. Differentiation of myocardial scar from potential pitfalls and artefacts in delayed enhancement MRI. Br J Radiol 2012;85(1019):e1145‐e1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vermes E, Carbone I, Friedrich MG, Merchant N. Patterns of myocardial late enhancement: Typical and atypical features. Arch Cardiovasc Dis 2012;105:300‐308. [DOI] [PubMed] [Google Scholar]
- 16.Tao Q, Piers SRD, Lamb HJ, van der Geest RJ. Automated left ventricle segmentation in late gadolinium‐enhanced MRI for objective myocardial scar assessment. J Magn Reson Imaging 2015;42(2):390‐399. [DOI] [PubMed] [Google Scholar]
- 17.Dikici E, O'Donnell T, Setser R, White RD, editors. Quantification of delayed enhancement MR images. Medical image computing and computer‐assisted intervention—MICCAI. Saint‐Malo: Springer; 2004. [DOI] [PubMed] [Google Scholar]
- 18.Leong CO, Lim E, Tan LK, et al. Segmentation of left ventricle in late gadolinium enhanced MRI through 2D‐4D registration for infarct localization in 3D patient‐specific left ventricular model. Magn Reson Med 2019;81(2):1385‐1398. [DOI] [PubMed] [Google Scholar]
- 19.Yue Q, Luo X, Ye Q, Xu L, Zhuang X, editors. Cardiac segmentation from LGE MRI using deep neural network incorporating shape and spatial priors. Medical image computing and computer assisted intervention—MICCAI. Shenzhen: Springer; 2019. [Google Scholar]
- 20.Tao X, Wei H, Xue W, Ni D, editors. Segmentation of multimodal myocardial images using shape‐transfer GAN. Statistical atlases and computational models of the heart multi‐sequence CMR segmentation, CRT‐EPiggy and LV full quantification challenges. Shenzhen: Springer; 2019. [Google Scholar]
- 21.Ronneberger O, Fischer P, Brox T, editors. U‐Net: convolutional networks for biomedical image segmentation. Medical image computing and computer‐assisted intervention—MICCAI. Munich: Springer; 2015. [Google Scholar]
- 22.Drozdzal M, Vorontsov E, Chartrand G, Kadoury S, Pal C, editors. The importance of skip connections in biomedical image segmentation. Deep learning and data labeling for medical applications. Athens: Springer; 2016. [Google Scholar]
- 23.Hussain Z, Gimenez F, Yi D, Rubin D, editors. Differential data augmentation techniques for medical imaging classification tasks. AMIA annual symposium proceedings. Washington, DC: American Medical Informatics Association; 2017. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1 Sample of three slices (columns) from the four cases with failed automatic segmentation. Cases 1–3 (external testing dataset) have severe hypertrophy with large scars, a pattern that did not present in the patient cohort used for training the model. Case 4 (local testing dataset) shows no contrast between myocardium and background.


