Abstract
Background and objectives
During the ongoing pandemic of COVID‐19, SARS‐CoV‐2 RNA was detected in plasma and platelet products from asymptomatic blood donors, raising concerns about potential risk of transfusion transmission, also in the context of the current therapeutic approach utilizing plasma from convalescent donors. The objective of this study was to assess the efficacy of amotosalen/UVA light treatment to inactivate SARS‐CoV‐2 in human plasma to reduce the risk of potential transmission through blood transfusion.
Methods
Pools of three whole‐blood‐derived human plasma units (630–650 ml) were inoculated with a clinical SARS‐CoV‐2 isolate. Spiked units were treated with amotosalen/UVA light (INTERCEPT Blood System™) to inactivate SARS‐CoV‐2. Infectious titres and genomic viral load were assessed by plaque assay and real‐time quantitative PCR. Inactivated samples were subject to three successive passages on permissive tissue culture to exclude the presence of replication‐competent viral particles.
Results
Inactivation of infectious viral particles in spiked plasma units below the limit of detection was achieved by amotosalen/UVA light treatment with a mean log reduction of >3·32 ± 0·2. Passaging of inactivated samples on permissive tissue showed no viral replication even after 9 days of incubation and three passages, confirming complete inactivation. The treatment also inhibited NAT detection by nucleic acid modification with a mean log reduction of 2·92 ± 0·87 PFU genomic equivalents.
Conclusion
Amotosalen/UVA light treatment of SARS‐CoV‐2 spiked human plasma units efficiently and completely inactivated >3·32 ± 0·2 log of SARS‐CoV‐2 infectivity, showing that such treatment could minimize the risk of transfusion‐related SARS‐CoV‐2 transmission.
Keywords: amotosalen/UVA, pathogen inactivation, SARS‐CoV‐2, plasma
Introduction
A key element of blood safety is the prevention of transfusion‐transmitted infections. Multiple safety measures were introduced in the last decades, strongly accelerated by the recognition of HIV transfusion transmission in the early 1980s, significantly decreasing the risk for infectious adverse events. However, there is still a remaining risk for the recipients of labile blood components, considering the limitations of existing safety measures [1]. Newly emerging pathogens, particularly viruses, may pose a challenge to the safety of plasma transfusion, especially if subclinical infection occurs in combination with blood viremia. The majority of individuals infected by one of the recently emerging arboviruses develop no symptoms but may carry a high titre viral load in the blood [1, 2]. The emergence and risk for the blood supply caused by emerging infectious diseases are unpredictable [3], as demonstrated by the current COVID‐19 pandemic.
The first cases of COVID‐19 disease, an infection of the lower respiratory tract, were reported in late 2019, spreading rapidly in and from Wuhan, China [4]. The highly virulent pathogen causing the disease was identified as a beta‐Coronavirus (SARS‐CoV‐2) [5]. On 11 March 2020, the WHO declared a global COVID‐19 pandemic, up to date more than 47 million confirmed infections and more than 1·2 million disease‐related deaths have been reported globally, with highest disease burden in Europe and the Americas, followed by the Middle East and Asia. In Saudi Arabia, more than 348 000 confirmed cases and more than 5000 disease‐related deaths have been reported up to date according to the WHO COVID‐19 Dashboard [6]. Respiratory droplets were quickly identified as the main route of infection, other ways of transmission, especially smear infection/oral intake, may also play a role [7]. Not every infected individual develops the COVID‐19 disease, the prevalence of asymptomatic infected individuals has been reported by several studies [7, 8] including a study from South Korea that reported a prevalence of 19·2% [9].
SARS‐CoV‐2 is closely related to other human coronaviruses, especially SARS‐CoV which caused an outbreak of the Severe Acute Respiratory Syndrome in Singapore and Korea in 2000–2002. By decision of the International Committee for the Taxonomy of Viruses, SARS‐CoV and SARS‐CoV‐2 are different strains of the same species [5]. Transmission by blood transfusion was not shown for SARS‐CoV, however, due to the detection of viral genomes in blood in the symptomatic phase, and the detection of SARS‐CoV genomes in leucocytes in the convalescent phase together with evidence for subclinical infection, transfusion transmission of SARS‐CoV is of concern and considered a theoretical risk [10]. Evidence for subclinical infection was also shown for SARS‐CoV‐2 [7]. Low viral load SARS‐CoV‐2 RNAemia in blood was detected frequently in symptomatic patients [11, 12]. A recent study from China reported SARS‐CoV RNAemia also in asymptomatic blood donors; viral nucleic acids were detected retrospectively in frozen plasma products, a platelet product and donor samples for pre‐screening [13]. Although transfusion transmission of SARS‐CoV‐2 is not documented to date, the detection of viral nucleic acids in blood products from asymptomatic donors raises concerns about potential transfusion transmission of SARS‐CoV‐2, especially because blood screening procedures for that pathogen are not established yet. Another potential risk became evident in light of the recent approaches to use convalescent donors’ plasma to treat COVID‐19 patients, an idea that seems to be one of the most promising therapeutic approach to date [14].
The safety of blood donations has been of major concern for transfusion medicine practitioners and researchers, this has led to the introduction of certain measures to reduce this risk including risk factor questionnaires, screening assay to detect transfusion‐transmitted pathogens in blood donations [15]. These measures have led to decreased prevalence of some of the transfusion‐transmitted infections such as HIV, HCV and HBV [16]. The increased improvement in the performance of screening assays was very helpful in reducing transmission risks. These assays are pathogen specific and are of no value to detect newly emerging and re‐emerging pathogens. Therefore, it is worthwhile to consider treating the donated plasma with a pathogen inactivation technology proven to be efficient against a large panel of pathogens including SARS‐CoV‐2 [17].
The INTERCEPT technology inactivates a broad spectrum of viruses, bacteria and protozoa in plasma prepared for transfusion [18]. During a photochemical reaction involving amotosalen (the photoactive compound) and UVA light, the pathogens’ genomes are modified in a targeted and specific manner by cross‐linking the genomic strands, preventing transcription and replication without affecting the clinical outcome of the plasma transfusion [19, 20]. This was also shown in a cohort of thrombotic thrombocytopenic purpura (TTP) patients requiring therapeutic plasma exchange [21, 22]. As an additional effect, residual white blood cells of the donor are inactivated more efficiently than by gamma‐irradiation [23], reducing the risk for immunological transfusion reactions and transfusion‐associated graft‐versus‐host disease (TA‐GvHD). Efficient inactivation by amotosalen/UVA treatment of the very closely related SARS‐CoV was reported in human plasma units and platelet concentrates [24, 25]. We also recently have shown that amotosalen/UVA treatment can efficiently inactivate the closely related Middle East Respiratory Syndrome Coronavirus (MERS‐CoV) in human plasma [26] and platelet concentrates [27], and now extending this work to evaluate the inactivation of SARS‐CoV‐2 in human therapeutic plasma units.
Materials and methods
Cell line and SARS‐CoV‐2 culture
Vero E6 cells were maintained in Dulbecco’s modified Eagle medium (DMEM) with 10% foetal bovine serum (FBS) as previously described (Al‐Amri et al, 2017). A human SARS‐CoV‐2 clinical patient isolate (SARS‐CoV‐2/human/SAU/85791C/2020, gene bank accession number: MT630432) was used in all experiments in the Special Infectious Agents Unit (SIAU) Biosafety level 3 facility at King Fahd Medical Research Center (KFMRC), King Abdulaziz University (KAU), Jeddah, Saudi Arabia.
Preparation of SARS‐CoV‐2 stock
Virus was inoculated on 90%–95% confluent Vero E6 cells (ATCC# CRL‐1586) in a T175 tissue culture flask at multiplicity of infection (MOI) of 1 and incubated in a humidified incubator at 5% CO2 and 37°C for one hour with gentle shaking every 15–20 min. Subsequently, the inoculum was replaced by 25 ml of viral inoculation medium (DMEM with 2% FBS, 1% penicillin/streptomycin and 10 mmol/l HEPES [pH 7·2]) and the cells were incubated in a humidified incubator at 5% CO2 and 37°C until 80%–90% of cells showed a cytopathic effect (CPE), typically three days post‐infection. Supernatant was collected and centrifuged to remove cellular debris for 5 min at 500× g at room temperature. Virus was then aliquoted and stored at –80°C, and the titre was determined by plaque assay.
Plasma preparation
Whole blood units (450 ml ± 10%) were collected and prepared at King Abdulaziz University Hospital, Transfusion Services, Jeddah, Kingdom of Saudi Arabia, from voluntary donors. Briefly, blood units were centrifuged at 544× g for 10 min to separate the platelet (PLT)‐rich plasma. The PLT‐rich plasma was then centrifuged at 230× g at 20°C for 10 min. The plasma was then transferred into the plasma bag and kept at −30°C. All blood units were screened routinely for HCV antibody, HBsAg, HBc antibody, HIV (1/2) antibody, HTLV (1/2) antibody, Syphilis as well as HCV, HBV and HIV by NAT.
SARS‐CoV‐2 inactivation
Pools of three single‐donor plasma units were used in this study (volume 630–650 ml). All plasma pools were tested for the presence of anti‐SARS‐CoV‐2 neutralizing antibodies using an in‐house neutralization assay. Each plasma pool was inoculated with SARS‐CoV‐2 stock in a 1:100 dilution. Plasma units were treated with amotosalen/UVA using the INTERCEPT Processing Set for Plasma and an INTERCEPT Illuminator INT‐100 (Cerus Corporation, U.S.A.). The following samples were collected for testing: a positive control sample from the virus stock; a negative control sample from the plasma before inoculation with the virus; a pretreatment sample after the addition of amotosalen and a sample from the treated plasma units after INTERCEPT illumination (Fig. 1). All samples were stored at −80°C until testing.
Figure 1.
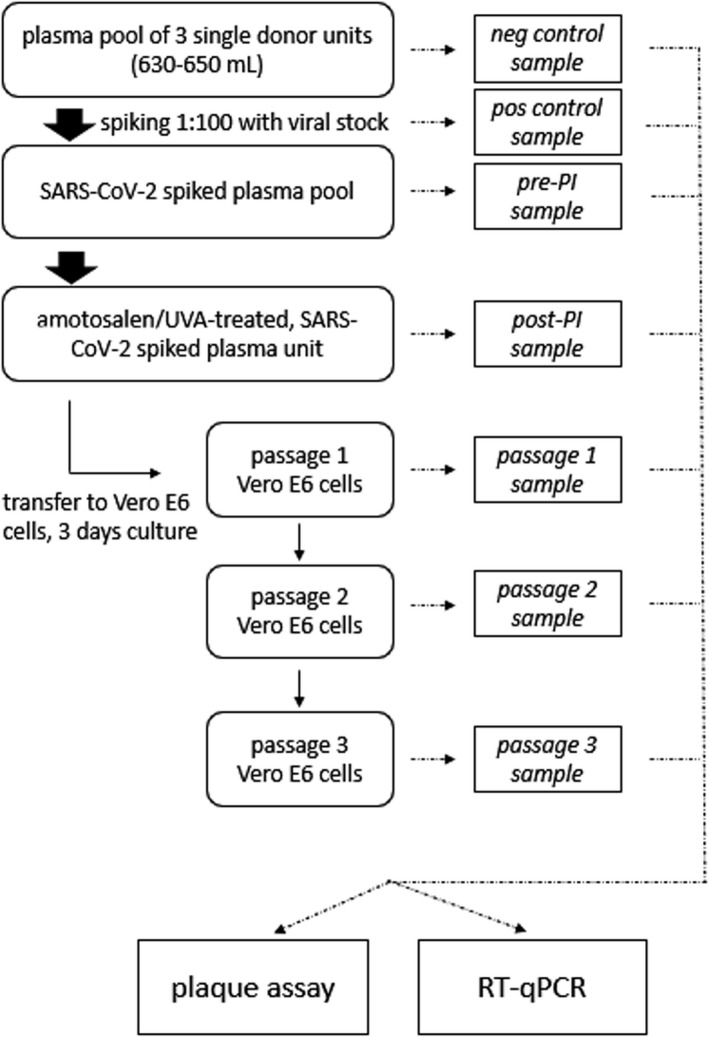
Experimental design. Schematic view of the experimental design. Briefly, pools of 3 plasma units were spiked in a 1:100 dilution with a SARS‐CoV‐2 viral stock. The pools were treated with amotosalen/UVA pathogen inactivation and post‐inactivation subject to 3 consecutive passages of 3 days respectively on Vero E6 cells. Samples were taken at various stages and assessed for replicating virus by plaque assay and viral genomes by RT‐qPCR.
Detection of replicating SARS‐CoV‐2
Detection of replicating SARS‐CoV‐2 in amotosalen/UVA‐treated plasma was performed as previously described for SARS‐CoV‐1 and MERS‐CoV with minor modifications [25, 26]. Briefly, collected pretreatment and inactivated samples were diluted at 1:10 dilution in DMEM with 10% FBS, inoculated on Vero E6 cells in 6 well plates in duplicates and incubated for 1 h at 37°C. Inoculum was then removed and replaced with 2 ml DMEM with 10% FBS and incubated for 3 days at 37°C. Then, supernatants were collected, diluted 1:10 in DMEM with 10% FBS and re‐inoculated on Vero E6 cells for two more successive passages. Supernatants collected from each passage were also used for viral load quantification by RT‐qPCR.
Plaque assay
Plaque assays were performed as previously described [26] with minor modifications. Briefly, samples were serially diluted in DMEM with 10% FBS starting from 1:10 and 1 ml from each dilution was inoculated on confluent Vero E6 cell monolayers and incubated for 1 h at 37°C. Then, the inoculum was removed and overlaid with DMEM containing 0·8% agarose and incubated for 3 days at 37°C. Cells were then stained with crystal violet for 4 h at 37°C, and plaques were counted to determine the viral titre as plaque forming unit (PFU)/ml.
RT‐qPCR quantitation
Viral RNA was extracted from all samples collected directly from the plasma units (positive, negative, pretreatment and post‐treatment samples) using the QIAmp Viral RNA Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. Relative quantification of the SARS‐CoV‐2 viral load was performed by one‐step dual‐target real‐time RT‐PCR (RealStar SARS‐CoV‐2 RT‐PCR Kit 1.0, Altona Diagnostics, Germany) according to the manufacturer’s instructions using a 7500 Fast Real‐Time PCR System (Applied Biosystems, U.S.A.). The PCR detects a beta‐coronavirus specific target (E‐gene), a SARS‐CoV‐2 specific target (S‐gene) and an internal control. The decrease in viral load was expressed by comparing the cycle threshold (CT) values from each sample relative to the CT values of the pretreatment inoculated sample (with the SARS‐CoV‐2 specific primers). The SARS‐CoV‐2 titres were expressed as PFU equivalents per ml (PEq/ml) using a standard curve (standard: serial dilutions of the viral stock) and choosing dilutions of the original sample (10−1 to 10−8) with CT values in the exponential phase. Each run included a positive viral template control and no‐template negative control. Each sample was tested in duplicate, and the mean is reported as PEq/ml.
IRB approval
The study was approved by the Unit of Biomedical Ethics of the King Abdulaziz University Hospital (approval # 285‐20).
Results
Inactivation of SARS‐CoV‐2 in human plasma units
Five pools (A‐E) of human plasma collected from healthy donors were spiked with SARS‐CoV‐2. Spiked units were then treated with amotosalen and UVA light. The mean infectious viral titre in pretreatment samples was 3·32 ± 0·19 log10 PFU/ml (range: 3·1–3·6 log10 PFU/ml) (Table 1). Treatment of spiked units resulted in a mean reduction of >3·32 ± 0·19 log10 PFU/ml as no infectious virus was detected by plaque assay (Table 1). Figure 2 shows a representative plaque assay result for all tested units. As expected, testing of negative control samples collected before spiking with SARS‐CoV‐2 showed no replication‐competent virus. Of note, the viral infectivity titre in viral stock was 5·6 ± 0·2 log10 PFU/ml and it was reduced to a mean infectivity of 3·32 ± 0·19 log10 PFU/ml after spiking in a 1:100 dilution, which is close to an expected post‐dilution titre of 3·6 log10 PFU/ml. Considering additional dilution of the spiked plasma by the addition of amotosalen solution, the dilution of the viral stock in plasma did not lead to unexpected loss of viral infectivity.
Table 1.
Reduction of infectious SARS‐CoV‐2 titres in human plasma units after amotosalen/UVA treatment
| Experiment | Viral infectivity titre, log10 PFU/ml | Log reduction | |||
|---|---|---|---|---|---|
| Positive control | Negative control | Pretreatment sample* | Inactivated sample | ||
| A | 5·5 | ND | 3·4 | ND | >3·4 |
| B | 5·7 | ND | 3·2 | ND | >3·2 |
| C | 5·6 | ND | 3·6 | ND | >3·6 |
| D | 5·9 | ND | 3·3 | ND | >3·3 |
| E | 5·3 | ND | 3·1 | ND | >3·1 |
| Mean ± SD | 5·6 ± 0·20 | ND | 3·32 ± 0·19 | ND | >3·32 ± 0·19$ |
ND indicates not detected.
After addition of amotosalen.
This indicates complete inactivation of the virus.
Figure 2.
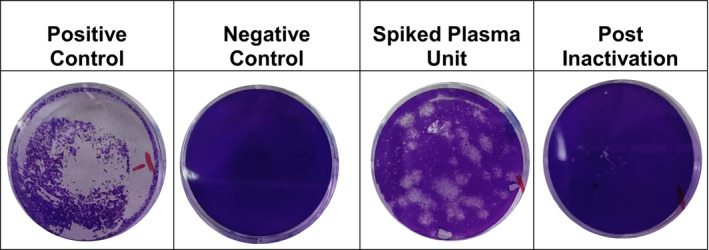
Inactivation of SARS‐CoV‐2 in plasma by amotosalen and UVA treatment assessed by a plaque assay. Vero E6 cells were inoculated for 1 h with the following samples in a 1:10 dilution in DMEM: the SARS‐CoV‐2 viral stock (Positive Control), human plasma (Negative Control), plasma from a SARS‐CoV‐2 spiked pretreatment sample (Spiked Plasma Unit) and amotosalen/UVA‐treated, SARS‐CoV‐2 spikes plasma (Post‐iInactivation). The cells were overlaid with agarose, incubated for three more days followed by neutral red staining. Experiments were conducted in serial dilutions. Photographs (4×) are shown from one of five representative experiments.
The impact of pathogen inactivation treatment on the genomic viral load
To further confirm these results, the viral genomic viral load was determined for all collected samples. The median CT preinactivation was 25·6 (18·3–29·1). The pretreatment samples mean genomic viral load was 2·92 ± 0·87 log10 PEq/ml, which was in the same range as the infectious titre (3·32 ± 0·19 log10 PFU/ml). After treatment, no viral genomes were detectable by PCR (Table 2). The internal PCR control was always positive indicating no PCR inhibition and confirming the decrease in the signal from the SARS‐CoV‐2 specific primers is due to a mean minimum inactivation of 2·92 ± 0·87 log10 PEq/ml.
Table 2.
| Experiment | Positive control | Negative control | Pretreatment sample | Inactivated sample |
|---|---|---|---|---|
| A | 5·99 | ND | 3·60 | ND |
| B | 5·88 | ND | 2·82 | ND |
| C | 5·53 | ND | 3·96 | ND |
| D | 5·36 | ND | 2·44 | ND |
| E | 5·18 | ND | 1·79 | 0·24 |
| Mean ± SD | 5·59 ± 0·34 | ND | 2·92 ± 0·87 | 0·04 ± 0·11 |
ND indicates not detected.
Data are shown as log10 PEq/ml.
Titres were determined from the same samples used in Table 1.
Passaging of INTERCEPT treated plasma to confirm complete inactivation
To exclude the possibility of any remaining replicating SARS‐CoV‐2 associated with the presence of viral genomic load in the treated plasma units (Table 2), we inoculated the collected samples on Vero E6 cells and evaluated infectivity over three successive passages. While culture of all pretreatment samples showed viral replication and complete CPE within 3 days post‐inoculation similar to that of positive control, neither viral replication nor CPE was observed in cells inoculated with inactivated samples similar to negative controls (Fig. 3) even after 9 days of incubation in all three passages. For further confirmation, we determined the genomic viral load from supernatants collected from all passages inoculated with either pretreatment or post‐treatment samples. As shown in Table 3, passaging of pretreatment samples showed viral replication as evident by CPE. On the other hand, viral genomes in cells inoculated with inactivated samples in culture supernatants were not detectable. Together, these data confirm the complete inactivation of SARS‐CoV‐2 in the tested platelets units and the absence of replication‐competent virus post‐inactivation.
Figure 3.
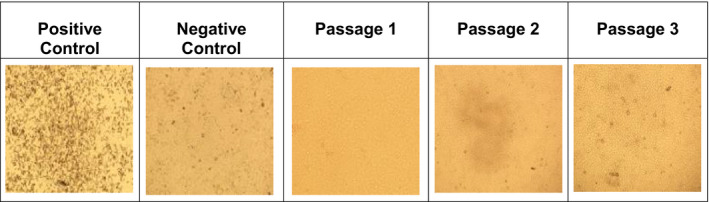
Assessment of complete inactivation of replicative SARS‐CoV‐2 post‐amotosalen/UVA treatment by passaging experiments. Vero E6 cells were inoculated for 1 h with the following samples in a 1:10 dilution in DMEM: plasma from a SARS‐CoV‐2 spiked pretreatment sample (Positive Control), human plasma (Negative Control) and amotosalen/UVA‐treated, SARS‐CoV‐2 spikes plasma (Passage 1–3) and passaged for three consecutive passages. Both the positive control and the pretreatment sample caused extensive CPE by day 3 post‐inoculation in all three passages. Negative control and inactivated sample did not show any CPE in Vero E6 cells. Photographs (4×) are shown from one of five representative experiments on day 3 post‐inoculation in each passage.
Table 3.
Results of passaging experiments of SARS‐CoV‐2 in Vero E6 cells before and after inactivation of spiked plasma*, †
| Experiment | Passage 1 | Passage 2 | Passage 3 | |
|---|---|---|---|---|
| A | Pretreatment sample | 5·05 | 4·54 | 4·14 |
| Inactivated sample | ND | ND | ND | |
| B | Pretreatment sample | 5·12 | 4·61 | 4·31 |
| Inactivated sample | ND | ND | ND | |
| C | Pretreatment sample | 5·24 | 4·05 | 4·01 |
| Inactivated sample | ND | ND | ND | |
| D | Pretreatment sample | 4·96 | 4·01 | 3·95 |
| Inactivated sample | ND | ND | ND | |
| E | Pretreatment sample | 5·16 | 4·21 | 3·97 |
| Inactivated sample | ND | ND | ND | |
ND indicates not detected.
Data are shown as log10 PEq/ml.
Samples in Table 1 were used in this experiment. Samples were used at 1:10 dilution, and titre was determined on day 3 post‐inoculation.
Discussion
Newly emerging pathogens are an ongoing potential threat for safety of the blood supply. Pathogen inactivation technology may provide immediate enhanced safety in an outbreak setting, in contrast to diagnostic tools which, even if they already exist, need to be developed and validated for multiple weeks to months [28].
We were able to show complete inactivation of >3·32 ± 0·19 log PFU/ml and 2·92 ± 0·87 log PEq/ml SARS‐CoV‐2 in human plasma pools of 630–650 ml in the present study, corresponding to volumes usually collected by plasmapheresis for the production of convalescent plasma (CP). Since a clinical isolate was used, the maximum titre obtained in vitro was not very high. Higher in vitro infectivity titres could likely show higher minimum inactivation efficacy. No CPE and no genomic viral load were detectable after 3 consecutive rounds of passaging on Vero E6 cells, pointing towards complete inactivation. The reported genomic viral load in the blood of symptomatic patients and asymptomatic donors is relatively low, often close to the limit of detection [11, 12, 13, 29], pointing towards sufficient inactivation efficacy of amotosalen/UVA to prevent potential transfusion transmission (Table 2).
Recently, the inactivation of a different SARS‐CoV‐2 isolate (USA‐WA1‐2020) from the USA with Riboflavin/UVB in single‐donor plasma units (>3·4 log PFU/ml) and platelets (>4·53 log PFU/ml) was reported [30]. Another study using Riboflavin/UVB technology with the same isolate reported an inactivation efficacy of >4·59 ± 0·15 log/PFU in single‐donor plasma units and >3·30 ± 0·26 log PFU/ml in whole blood [31]. Both Riboflavin/UVB studies did not report passaging experiments or used sensitive NAT testing to confirm the findings of the plaque assay used in these studies.
The current study also supports the use of amotosalen/UVA for mitigating the risk of potential superinfection with SARS‐CoV‐2 through convalescent plasma.
The use of pathogen inactivation technology for CP also raises the question of the impact on product quality, hence the neutralizing activity. A study with Ebolavirus CP (EBOV CP) showed no significant impact on the neutralizing activity by amotosalen/UVA treatment [32]. A single case study showed a loss of 2–4% total IgG after amotosalen/UVA treatment of EBOV CP without assessing the consequences for the neutralizing activity [33]. Recently, no significant impact of amotosalen/UVA treatment on the neutralizing activity and neutralizing antibody quantity of COVID‐19 convalescent plasma (CCP) was reported [34]. These reports point towards the preservation of neutralizing activity during amotosalen/UVA treatment.
Conclusion
In the present study, we showed efficient, complete inactivation of SARS‐CoV‐2 in large human plasma units by amotosalen/UVA using a local clinical isolate, which may serve as an additional layer of safety in plasma transfusion during the COVID‐19 pandemic. These finding are in line with former findings of efficient inactivation of human coronaviruses in plasma by amotosalen/UVA pathogen inactivation [24, 26].
Conflict of interest
There is no conflict of interest identified.
Funding
The study was funded by the Ministry of Health of the Kingdom of Saudi Arabia (grant number 540).
Acknowledgements
The authors acknowledge the support by Cerus Corporation in the form of Equipment, reagents and solutions.
Contributor Information
Esam I. Azhar, Email: eazhar@kau.edu.sa.
Salwa I. Hindawi, Email: sihindawi@yahoo.com.
References
- 1.Busch MP, Bloch EM, Kleinman S: Prevention of transfusion‐transmitted infections. Blood 2019; 133:1854–64 [DOI] [PubMed] [Google Scholar]
- 2.Kleinman S: Pathogen inactivation: emerging indications. Curr Opin Hematol 2015; 22:547–53 [DOI] [PubMed] [Google Scholar]
- 3.Glynn SA, Busch MP, Dodd RY, et al.: Emerging infectious agents and the nation's blood supply: responding to potential threats in the 21st century. Transfusion 2013; 53:438–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N, Zhang D, Wang W, et al.: A Novel Coronavirus from Patients with Pneumonia in China, 2019. New England Journal of Medicine 2020; 382(8):727–33. 10.1056/nejmoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coronaviridae Study Group of the International Committee on Taxonomy of V : The species Severe acute respiratory syndrome‐related coronavirus: classifying 2019‐nCoV and naming it SARS‐CoV‐2. Nat Microbiol 2020; 5:536–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO : WHO COVID‐19 Dashboard . WHO, 2020. (Last accessed November 5 2020).
- 7.Lotfi M, Hamblin MR, Rezaei N: COVID‐19: Transmission, prevention, and potential therapeutic opportunities. Clin Chim Acta 2020; 508:254–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Z, Xu Y, Sun C, et al.: A systematic review of asymptomatic infections with COVID‐19. J Microbiol Immunol Infect = Wei mian yu gan ran za zhi 2020: 10.1016/j.jmii.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim GU, Kim MJ, Ra SH, et al.: Clinical characteristics of asymptomatic and symptomatic patients with mild COVID‐19. Clin microbiol infect 2020; 26:948.e1‐e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang L, Yan Y, Wang L: Coronavirus Disease 2019: Coronaviruses and Blood Safety. Transfus Med Rev 2020; 34(2):75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C, Wang Y, Li X, et al.: Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395(10223):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W, Lan Y, Yuan X, et al.: Detectable 2019‐nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg Microbes Infect 2020; 9:469–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang L, Zhao L, Gong H, et al.: Severe acute respiratory syndrome Coronavirus 2 RNA detected in blood donations. Emerg Infect Dis 2020; 26(7):1631–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casadevall A, Pirofski LA: The convalescent sera option for containing COVID‐19. J Clin Invest 2020; 130:1545–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.WHO : WHO Guidelines Approved by the Guidelines Review Committee; Blood Donor Counselling: Implementation Guidelines. Geneva, World Health Organization Copyright © World Health Organization 2014., 2014.
- 16.Epstein JS: Alternative strategies in assuring blood safety: An overview. Biologicals 2010; 38:31–5 [DOI] [PubMed] [Google Scholar]
- 17.Epstein J, Burnouf T: Points to consider in the preparation and transfusion of COVID‐19 convalescent plasma. Vox Sang 2020; 115:485–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanteri MC, Santa‐Maria F, Laughhunn A, et al.: Inactivation of a broad spectrum of viruses and parasites by photochemical treatment of plasma and platelets using amotosalen and ultraviolet A light. Transfusion 2020; 60:1319–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prowse CV: Component pathogen inactivation: a critical review. Vox Sang 2013; 104:183–99 [DOI] [PubMed] [Google Scholar]
- 20.Mintz PD, Bass NM, Petz LD, et al.: Photochemically treated fresh frozen plasma for transfusion of patients with acquired coagulopathy of liver disease. Blood 2006; 107:3753–60 [DOI] [PubMed] [Google Scholar]
- 21.Bost V, Chavarin P, Boussoulade F, et al.: Independent evaluation of tolerance of therapeutic plasma inactivated by amotosalen‐HCl‐UVA (Intercept) over a 5‐year period of extensive delivery. Vox Sang 2015; 109:414–6 [DOI] [PubMed] [Google Scholar]
- 22.Guignier C, Benamara A, Oriol P, et al.: Amotosalen‐inactivated plasma is as equally well tolerated as quarantine plasma in patients undergoing large volume therapeutic plasma exchange. Transfus Clin Biol 2018; 25:73–7 [DOI] [PubMed] [Google Scholar]
- 23.Castro G, Merkel PA, Giclas HE, et al.: Amotosalen/UVA treatment inactivates T cells more effectively than the recommended gamma dose for prevention of transfusion‐associated graft‐versus‐host disease. Transfusion 2018; 58:1506–15 [DOI] [PubMed] [Google Scholar]
- 24.Singh Y, Sawyer LS, Pinkoski LS, et al.: Photochemical treatment of plasma with amotosalen and long‐wavelength ultraviolet light inactivates pathogens while retaining coagulation function. Transfusion 2006; 46:1168–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinna D, Sampson‐Johannes A, Clementi M, et al.: Amotosalen photochemical inactivation of severe acute respiratory syndrome coronavirus in human platelet concentrates. Transfus Med 2005; 15:269–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hindawi SI, Hashem AM, Damanhouri GA, et al.: Inactivation of Middle East respiratory syndrome‐coronavirus in human plasma using amotosalen and ultraviolet A light. Transfusion 2018; 58:52–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashem AM, Hassan AM, Tolah AM, et al.: Amotosalen and ultraviolet A light efficiently inactivate MERS‐coronavirus in human platelet concentrates. Transfus Med 2019; 29:434–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Domanovic D, Ushiro‐Lumb I, Compernolle V, et al.: Pathogen reduction of blood components during outbreaks of infectious diseases in the European Union: an expert opinion from the European Centre for Disease Prevention and Control consultation meeting. Blood Transfus = Trasfusione del sangue 2019; 17:433‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young BE, Ong SWX, Kalimuddin S, et al.: Epidemiologic features and clinical course of patients infected with SARS‐CoV‐2 in Singapore. JAMA 2020; 323(15):1488–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keil SD, Ragan I, Yonemura S, et al.: Inactivation of severe acute respiratory syndrome coronavirus 2 in plasma and platelet products using a riboflavin and ultraviolet light‐based photochemical treatment. Vox Sanguinis 2020; 115(6):495–501. 10.1111/vox.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ragan I, Hartson L, Pidcoke H, et al.: Pathogen reduction of SARS‐CoV‐2 virus in plasma and whole blood using riboflavin and UV light. PLoS One 2020; 15:e0233947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dean CL, Hooper JW, Dye JM, et al.: Characterization of Ebola convalescent plasma donor immune response and psoralen treated plasma in the United States. Transfusion 2020; 60:1024–31 [DOI] [PubMed] [Google Scholar]
- 33.Geisen C, Kann G, Strecker T, et al.: Pathogen‐reduced Ebola virus convalescent plasma: first steps towards standardization of manufacturing and quality control including assessment of Ebola‐specific neutralizing antibodies. Vox Sang 2016; 110:329–35 [DOI] [PubMed] [Google Scholar]
- 34.Tonn T, Corman VM, Johnsen M, et al.: Stability and neutralising capacity of SARS‐CoV‐2‐specific antibodies in convalescent plasma. The Lancet Microbe 2020; 1(2):e63. 10.1016/s2666-5247(20)30037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]


