Abstract
Immunologists have recently realized that there is more to the classic innate immune sensor systems than just mere protection against invading pathogens. It is becoming increasingly clear that such sensors, including the inflammasomes, toll‐like receptors, and the complement system, are heavily involved in the regulation of basic cell physiological processes and particularly those of metabolic nature. In fact, their “non‐canonical” activities make sense as no system directing immune cell activity can perform such task without the need for energy. Further, many of these ancient immune sensors appeared early and concurrently during evolution, particularly during the developmental leap from the single‐cell organisms to multicellularity, and therefore crosstalk heavily with each other. Here, we will review the current knowledge about the emerging cooperation between the major inter‐cell communicators, integrins, and the cell‐autonomous intracellularly and autocrine‐active complement, the complosome, during the regulation of single‐cell metabolism.
Linked Articles
This article is part of a themed issue on Canonical and non‐canonical functions of the complement system in health and disease. To view the other articles in this section visit http://onlinelibrary.wiley.com/doi/10.1111/bph.v178.14/issuetoc
Abbreviations
- α2M
α‐2 macroglobulin
- AA
amino acid
- ADMIDAS
adjacent to MIDAS
- ANA
anaphylatoxin
- AP
alternative pathway
- AP‐1
activator protein 1
- APC
antigen‐presenting cell
- BCL2
B cell lymphoma 2
- C3aR
C3a receptor
- C5aR
C5a receptor
- CP
classical pathway
- LP
lectin pathway
- CR
complement receptor
- CTL
cytotoxic T lymphocytes
- CTSL
cathepsin L
- ECM
extracellular matrix
- ER
endoplasmic reticulum
- FAK
focal adhesion kinase
- FB
factor B
- FH
factor H
- GLUT1
glucose transporter 1
- ICAM
intercellular adhesion molecule
- ILK
integrin‐linked kinase
- LAD‐1
leukocyte adhesion deficiency 1
- LAMTOR
late endosomal/lysosomal adaptor, MAPK, and mTOR activator 5
- LAT1
large neutral amino acid transporter
- LFA‐1
lymphocyte function‐associated antigen 1
- MAC
membrane attack complex
- MASP
mannose‐associated serine protease
- MCT4
monocarboxylate transporter 4
- MIDAS
metal ion‐dependent adhesion site
- mTORC1
mammalian target of rapamycin complex 1
- MYA
million years ago
- NLR
nod‐like receptor
- NLRP3
NLR family pyrin domain containing 3 protein
- OXPHOS
oxidative phosphorylation
- PINCH
Cys‐His‐rich protein
- PZP
pregnancy zone protein
- RA
rheumatoid arthritis
- Src
proto‐oncogene TK sarcoma
- SyMBS
synergistically active metal ion‐binding sites
- TCR
T‐cell receptor
- TEP
thioester‐containing protein
- Th1
T helper 1
- VCAM‐1
vascular cell adhesion molecule 1
- vWF
von Willebrand factor
1. INTRODUCTION
Evolutionary genetics is a powerful tool to classify species and their common ancestor(s) based on genome structure, genetic changes, and adaptations within populations. Evolution from single‐cell organisms into complex life and ultimately Homo sapiens was driven by genes supporting multicellularity and can be traced to the common ancestor of Choanozoa, appearing more than 750 million years ago (MYA; Brunet & King, 2017). Among key multicellularity‐driving genes, those encoding specific cell surface molecules such as cadherins, extracellular matrix (ECM) domain proteins, and integrins played a crucial role in mediating the interactions between cells and their environment. Over time, developing organisms increased in structural complexity, and particularly, the advent of body cavities and a vascular system was accompanied by the emergence of transcription factors and signalling molecules and the capability to discriminate between self and altered/non‐self and the defence against invading pathogens—in short, immunity. The complement system is among the first of such immune defence mechanisms that evolved, and it has been traced back to the common ancestor of the Eumetazoa, more than 650 MYA (Nonaka & Kimura, 2006). The normal activity of all living organisms, of the cell as a whole and of any cellular sensor or effector system, is critically dependent on metabolism. The metabolic activity of a given cell or organism is considered the root of life as catabolic and anabolic reactions together deliver the energy and the molecular key precursor molecules to build biomass (proteins, lipids, and DNA; Ralser, 2018).
Thus, metabolic pathways, integrins, and the complement system share about 500 million years of co‐evolution. Their structural and functional co‐conservation during evolution not only underpins their single biological values for the host but also promoted interactions between these three systems. Work over the last few years indeed demonstrated a close functional relationship between the complement system and single‐cell metabolism. Further, integrins and complement are traditionally connected as some complement components form close partnerships with selected single integrin chains to generate receptors with novel and/or unique ligand specificities. Excitingly, the recent identification of the integrin leukocyte adhesion molecule (leukocyte function‐associated antigen‐1 [LFA‐1]) as a key upstream regulator of tissue complement activity and downstream complement‐mediated cell metabolic events (Kolev et al., 2020) now amalgamates integrins, complement, and metabolism into a new triangular functional relationship.
Here, we will summarize what is known about the (co)evolutionary journeys of integrins (Figure 1a) and complement (Figure 1b) and discuss their functional cooperation over time with a specific eye on cell metabolism. As this may only be the “tip of an iceberg” with regard to this new functional network, we will also point out the methodological difficulties that we face to systematically detangle its complex and interconnected contributions to normal cell physiology.
FIGURE 1.
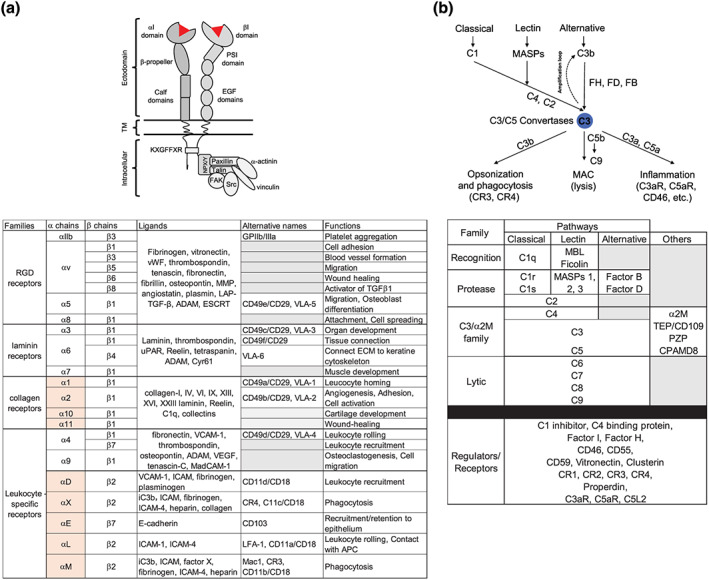
Integrins and complement—core structures, components, and functions. (a) Top: schematic of the general structure of integrin pairs with defining domains and the most common intracellular signalling scaffold engaged upon activation depicted. Bottom: overview of the 24 heterodimers formed by integrins in humans with their main ligands and biological functions. Boxes underlaid in light red mark the I domain‐containing α chains. (b) Top: simplified schematic of the three complement activation pathways and canonical complement system functions. Bottom: overview of the main proteins constituting the classic complement system cascade, categorized into five families: recognition molecules, proteases, TEP proteins of the terminal lytic pathway, and regulators/receptors
2. INTEGRINS: PAVING THE ROAD TOWARDS MULTICELLULARITY
2.1. Integrin structure
Due to the importance of their involvement in basically all types of cellular adhesion events, integrins are conserved across most of the metazoan phyla (Calderwood et al., 2003), including Porifera (Pancer, Kruse, Muller, & Muller, 1997) and Cnidaria (Brower, Brower, Hayward, & Ball, 1997). Integrins are normally formed as functional pairs of two distinct chains, specifically α and β chain combinations (Figure 1a). In invertebrates, at least 26 genes encoding distinct α chains and 17 genes encoding distinct β chains have been deposited in public databases.
Both α and β subunits of integrins are all composed of a cytoplasmic tail (generally 20–75 amino acids [AAs], except for the β4 cytoplasmic subunit, which exceeds 1,000 AAs in length), a transmembrane domain, and an ectodomain (750–1,000 AAs). All integrin α chain ectodomains contain a 7‐bladed β propeller with EF‐hand domains capable of binding Ca2+ to modulate ligand‐binding affinity (Oxvig & Springer, 1998; Figure 1a). Nine of the 18 α integrins are composed of a von Willebrand factor type A (vWF‐A) domain (also called I domain) inserted within the β propeller (Larson, Corbi, Berman, & Springer, 1989) and a metal ion‐dependent adhesion site (MIDAS) motif responsible for ligand binding (Johnson & Chouhan, 2014; Lee, Bankston, Arnaout, & Liddington, 1995). The cytoplasmic tail, though highly variable among different species, contains a preserved KXGFFXR motif that is responsible for the successful αβ heterodimer integrin formation. Structurally, β integrin ectodomains are defined by the presence of a vWF‐A domain, a plexin‐semaphorin‐integrin domain, and four EGF domain repeats (Figure 1a). Further, β integrins also display a MIDAS region, and this is flanked by synergistically active metal ion‐binding sites (SYMBS) and adjacent to MIDAS (ADMIDAS) domain. Each sequence binds to a divalent cation (Ca2+, Mg2+, or Mn2+), and together, they are the driving forces behind the switch from an inactivated (low affinity) to an activated (high affinity) conformation that allows ligand binding (Tiwari, Askari, Humphries, & Bulleid, 2011) in the integrin pairings lacking the I domain.
2.2. Integrins in cell–cell communication
Because integrin activity induces signalling events including particularly those instructing inter‐cell communication, their involvement in early developmental pathways of organisms is well acknowledged and studied (Gettner, Kenyon, & Reichardt, 1995; Stark et al., 1997; Wilcox, DiAntonio, & Leptin, 1989; Yee & Hynes, 1993). Integrins serve as main molecular link between cells and the ECM (i.e., laminins, fibronectins, and collagens), adhesion molecules, and plasma proteins (Harris, McIntyre, Prescott, & Zimmerman, 2000; Hynes, 2002; Pierschbacher & Ruoslahti, 1984) and are critical for normal tissue generation, maintenance, and repair (Anderson, Owens, & Naylor, 2014). They signal in response to extracellular stimuli (outside‐in signalling) or to certain intracellular events (inside‐out; Giancotti & Ruoslahti, 1999; Harburger & Calderwood, 2009; Hynes, 2002). Such inside‐out signals involve several cytoskeletal proteins that directly or indirectly bind the intracellular domain of the β subunits. Key proteins that can interact with several different β subunits include talin, kindlin, paxillin, α‐actinin, vinculin, integrin‐linked kinase (ILK), Cys‐His‐rich protein (PINCH), and parvin (Anderson et al., 2014). The interaction of these proteins with the integrin β chain most often culminates in the phosphorylation of proto‐oncogene TK sarcoma (Src) and focal adhesion kinase (FAK) family protein kinases and induction of cytoskeletal rearrangements (Bolos, Gasent, Lopez‐Tarruella, & Grande, 2010). The β‐integrin cytoplasmic domain is highly conserved, and the common NPX/Y motif functions as hub for the interaction with intracellular adaptor proteins (Figure 1a). In vertebrates, integrins are involved in both developmental and immunological processes, such as early embryogenesis, regulation of cell proliferation and differentiation, leukocyte migration, and complement receptor‐dependent phagocytosis. Mutations in integrins or the major effectors of integrin signalling pathways cause defective organ development, immunodeficiency, cancer, or autoimmune disease.
2.3. Evolutionary origin of integrins
Because of the central role of the integrins in many basic biological processes, it is not surprising that integrin genes can be traced back to the early common ancestors of Choanozoa (Sebe‐Pedros, Roger, Lang, King, & Ruiz‐Trillo, 2010; Figure 2). For example, the β chain ectodomains, MIDAS, ADMIDAS, and SYMBS, as well as the intracellular tail NPX/Y motif, are found in protist organisms. De facto, three to four α and four β subunits were already expressed by Capsaspora owczarzaki, the eukaryotic unicellular ancestors of animals. Moreover, the intracellular adaptor proteins talin, paxillin, α‐actinin, vinculin, parvin, ILK, and PINCH and the TKs c‐Src and FAK are present already in more ancient organisms, such as Unikonts (Amoebozoa) even before the emergence of integrin chains (Sebe‐Pedros et al., 2010).
FIGURE 2.
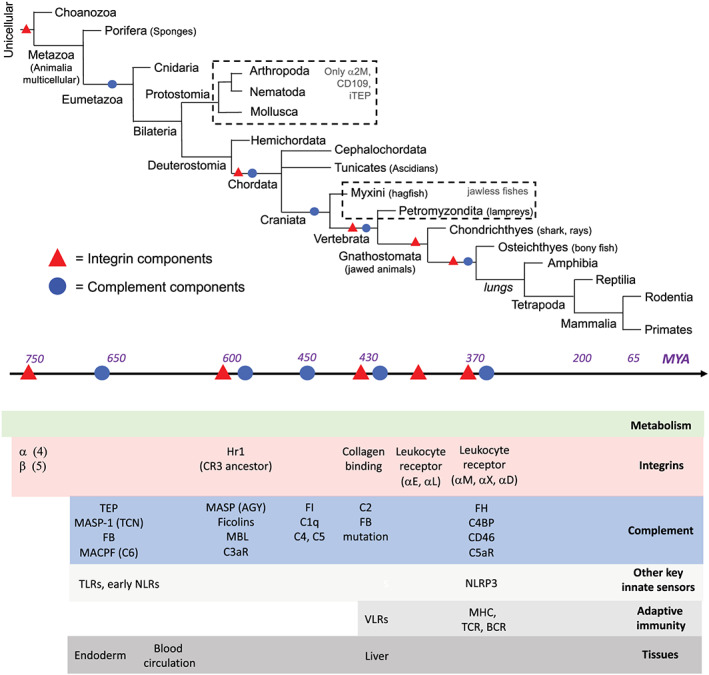
Evolution of integrins and complement. Upper part: simplified phylogenic tree of species with the evolutionary timeline of appearance in MYA aligned below. Bottom part: emergence of integrin and complement components during evolution. The appearance of key innate and adaptive immune system components and significant “leaps” in tissue development towards body cavities, the vascular system, and the liver are also shown. Timeline based on PMID: 27918074, 28405393, and 16598240
Even though the exact function of integrins in protist organisms has not been clearly identified on the molecular level, it is accepted that they are forerunners of classic integrin signalling (Baade, Paone, Baldrich, & Hauck, 2019; Sebe‐Pedros & Ruiz‐Trillo, 2010) and part of the protein machinery that allowed for the transition from unicellular to multicellular organisms (Meller et al., 2015). The first marked expansion and diversification of integrin genes during evolution after their emergence seems to have occurred ~450 MYA. Indeed, predictive models suggest expression of eight α chains with an I domain insertion (among 11 α chains in total) and five β integrin‐chains in the Ascidian Ciona intestinalis, indicating that that insertion of the I domain and defined ligand‐binding activity within the α chain(s) occurred in Chordata (Ewan et al., 2005). Two later studies confirmed expression of five α (α2, α3, α6, α9, α10, and α11) and two β‐paired distinct integrins by members of this phylum (Christiaen et al., 2008; Woznica et al., 2012).
Furthermore, sequencing comparison revealed a single αI domain in another Ascidian, Halocynthia roretzi. This protein was identified as Hr1, an ancestor of complement receptors (CRs) 3 and 4, which mediate C3b/iC3b‐dependent phagocytosis of yeast cells (see below; Figure 2; Miyazawa, Azumi, & Nonaka, 2001).
From that evolutionary time on, αI integrins were detected in all vertebrates (and potentially one αI domain detected in hagfish), with the first orthologue of human collagen‐binding integrin αI found in lampreys as this integrin protein can recognize human ligand epitopes (Chouhan et al., 2014). Finally, with the emergence of cartilaginous fish (Chondrichthyes), about 350 MYA, the first “modern” leukocyte integrin receptors (i.e., αE and αL, involved in intercellular adhesion molecule [ICAM]‐1 and E‐cadherin recognition, respectively) were detected, while sub‐specialization into αM, αX pairings (recognizing inactivated C3b [iC3b]), and αD (recognizing vascular cell adhesion molecule [VCAM‐1]) seems to have occurred with the later divergence of bony fish (Osteichthyes). It is interesting to note that this timeline indeed coincides with the emergence of the adaptive immune system, signified by the presence of the variable lymphocyte receptor diversity region in hagfish and later the occurrence of T‐cell receptors (TCRs) and B cell receptors and the major histocompatibility complex molecules in cartilaginous fish (Figure 2; Flajnik & Kasahara, 2010). Thus, the increasing sophistication of the integrin system with its growing ligand detection and signalling capabilities paralleled the evolution of specialized cells and tissues to support phagocytosis, activation, and migration of immune cells in more and more complex organisms.
3. COMPLEMENT: PROTECTING THE HEALTH OF THE DEVELOPING ANIMAL KINGDOM
The complement system is among the oldest part of immunity and can show for a timeline similar to those of integrins during evolution. In 1888, Jules Bordet first discovered antibacterial properties of plasma components that were later called complement by Paul Ehrlich, because these components complemented the action of antibodies. Ever since, complement is described as a protein‐based system acting not only as first line of host defence through opsonization and lytic activity against and noxious self but also as a central element for immune cell recruitment and activation (Merle, Noe, Halbwachs‐Mecarelli, Fremeaux‐Bacchi, & Roumenina, 2015).
Starting from a handful of key effector molecules, through 500 million years of evolution, complement developed into a complex system with about 50 circulating and/or cell membrane‐bound components in humans (Merle, Church, Fremeaux‐Bacchi, & Roumenina, 2015). In the absence of danger signals, the classic complement system idles in a mostly inactive form in blood and interstitial fluids. It can be activated in a cascade‐like fashion through three distinct pathways: the classical pathway (CP), lectin pathway (LP), or alternative pathway (AP), which all converge at the formation of C3 convertases that activate the central C3 component by cleavage into C3a and C3b. This triggers subsequent proteolytic activation of C5 into C5a and C5b. C3b is a major opsonin, and C5b seeds the terminal pathway (TP) with subsequent formation of the pore‐forming membrane attack complex (MAC; Figure 1b). The anaphylatoxins (ANAs) C3a and C5a mediate scavenger cell influx to the site of infection and the general inflammatory reaction via activating their respective GPCRs on vascular and immune cells (Klos et al., 2009). A number of complement regulators, either in circulation or expressed by cells, protect host tissue from unwanted or uncontrolled complement attack. Thus, complement members are classified into five families: recognition components, activating proteases, thioester‐containing proteins (TEP), lytic components, and receptors and regulators (Figure 1b).
The evolutionary origin of complement is generally considered to be the TEP superfamily (Figure 2). TEPs are defined by the canonical thioester motif GCGEQ that allows for the fast reaction of the TEP with cell surface macromolecules, antibodies, and so forth, and that is critically conserved among species. The TEP superfamily is divided into two families: the C3 family that contains the complement core components C3, C4, and C5 and the α‐2 macroglobulin (α2M) family that consists of α2M, CD109, pregnancy zone protein (PZP), and C3‐ and PZP‐like α2M domain‐containing 8. Family‐specific features include the presence of a so‐called C345C and an ANA domain in all C3 family proteins (Sekiguchi, Fujito, & Nonaka, 2012), while α2M family are characterized by a specific “protease bait domain.” TEP/C3 and α2M, along with the complement proteases Factor B (FB) and mannose‐associated serine protease 1 (MASP‐1) and other precursor proteins with lytic activity such as the C6‐like MAC/perforin protein, were detected in most of the studied Cnidarians (Figure 2). This indicates that eumetazoan ancestors were already capable of ensuring complement‐mediated danger recognition, complement activation, and target opsonization and possibly developing lytic activity (Fujito, Sugimoto, & Nonaka, 2010; Kimura, Sakaguchi, & Nonaka, 2009; Miller et al., 2007). Among the three complement pathways, the LP is the most ancient. Key LP recognition molecules such as the first ficolin and mannan‐binding lectin can be found in the humoral fluid of Tunicates where these sensors interact with MASP‐1 and then direct C3 activation to protect the organism's body cavities (Endo, Matsushita, & Fujita, 2015; Huang et al., 2011; Roberts et al., 2007). MASPs then diversified and increased their activity repertoire/specificity in Cephalochordata when the active site serine from a TCN type in MASP‐1 changed towards an AGY type in MASP‐3 and the MASPs developed the ability to ultimately activate both the LP and the AP, while the “younger” MASP‐2 seems to be more specific for the CP, which also appeared later in evolution (Sekine, Takahashi, Iwaki, & Fujita, 2013). Thus, the first complement sensor and activator molecules appeared early in evolution and remained functional in all vertebrates (Endo et al., 2003). With regard to the receptors, expression of a structural and functional homologue of the C3a receptor (C3aR) is found on haemocyte (early immune cells) membranes of Ciona intestinalis, indicating that complement‐mediated chemotaxis may have appeared early in evolution (Franchi & Ballarin, 2017). For more details on the advent of the first complement opsonin receptors, please see below.
The next major change in the composition of the early complement system occurred around 460 MYA: with the emergence of the first orthologous C1q gene (BjC1q) in Branchiostoma japonicum (BjC1q), a Cephalochordate (Gao, Li, Ma, & Zhang, 2014). Soon after, C4 and C5 (Ishiguro et al., 1992; Krisinger et al., 2012), as well as the first inhibitory protein, Factor I (Kimura, Ikeo, & Nonaka, 2009), appeared with the evolution of jawless fishes. In cartilaginous fish, FB structurally changed due to a single mutation that allowed emergence of the C2 component protein (Nonaka, 2014) and a fully functional CP as we know it today. A quickly following time period was marked by the birth of new complement activation fragment receptors such as the ANA receptor C5a receptor (C5aR), CR2, CR3, CR4, and the C3b receptor/regulator CD46 (Kaidoh & Gigli, 1989; Kumar, Bhandari, Sarde, & Goswami, 2014; Li, Sui, & Sun, 2017; Smith, 1998; Sun, Li, Wang, Zhang, & Zhang, 2010; Terado et al., 2003). The concurrent appearance of a battery of complement regulators such as C1 inhibitor, Factor H (FH) and FH‐related proteins, and C4b‐binding protein not only demonstrated the urgent need to tightly control this powerful immunological tool but also completed then in core the composition of the modern mammalian complement system (Figure 2; for an excellent in‐depth overview of complement evolution, please see Nonaka, 2014).
Of note, a “full‐blown” complement system was already in place when the adaptive immune system finally appeared in cartilaginous fish (Flajnik & Kasahara, 2010). Thus, it would be expected that the developing adaptive immune system arms integrated existing complement activities into their functional repertoire—and this notion is indeed reflected in the broadly acknowledged central role for complement in the instruction of normal B and T‐cell biology (Killick, Morisse, Sieger, & Astier, 2018).
4. COMPLEMENT AND INTEGRINS: PARTNERS MADE BY CO‐EVOLUTION
As delineated above, complement and integrins share more than 600 million years of co‐evolution. They were both there early on and, when organisms moved from single cells to more sophisticated multilayer and multi‐organ organisms, both systems accompanied this developmental progress by contributing specific independent activities but also by starting to cooperate with each other. Such cooperation is particularly clear on the level of the complement receptors, CR3 and CR4. CR3 (also known as Mac‐1 or integrin αMβ2) is composed of the integrin chains CD11b (gene ITGAM) and CD18 (gene ITGB2), and CR4 is composed of the integrin chains CD11c (gene ITGAX) and CD18; thus, the receptors belong to the β2 integrin family. Both, CR3 and CR4, share about 87% homology with each other and bind a complement regulator‐processed form of C3b, iC3b, and also C3d/dg (Erdei et al., 2019). CR3 and CR4 are mostly expressed on scavenger cells, and their interaction with iC3b bound to the surface of microbes or stressed/dying host cells induces the phagocytic uptake and safe removal of these tagged targets (Merle, Noe, et al., 2015). Fittingly, the emergence of complement regulators that can generate the iC3b (or C3c and C3d) protein occurred during a major diversification phase of α integrins, and notably at the same time as CR3 and CR4, in Chordata ~500 MYA (Miyazawa et al., 2001; Figure 2). This suggests that C3‐dependent phagocytosis of targets by haemocytes may have represented a critical selective advantage. In light of this, it is surprising that Protostomes have taken a different path with regard to their composition of integrin and complement proteins: They lost (or did not evolve) the C3 protein family and lack CR3 and CR4 expression, as well as some integrin types such as αL and αD (Hughes, 2001). Therefore, complement proteins may have exerted selective pressure on certain integrin genes (or vice versa) during evolution towards Deuterostome. Of course, this is speculative as differences in animal body structures (e.g., the open vascular system in Protostomes vs. the move towards a closed vascular system in Deuterostomes) or simply exposure to distinct environments or pathogens, and so forth, have likely also been driving forces. The ancient complement and integrin co‐operation extended quickly beyond orchestrating phagocytosis towards more extensive regulation of cell activity. It appears that the mutation of FB that led to the generation of the complement C2 component (and hence CP emergence) occurred at a similar time when collagen‐binding integrins evolved. The ability of integrins to particularly engage with ECM components such as collagens, laminins, cadherins, and MMPs is at the heart of their key role in cell–cell communication, migration, and tissue development and (re)modelling (Figure 1a; Hynes, 2002). The α2β1 integrin is a major collagen binder and also heavily involved in the activity of innate immune cells (Adorno‐Cruz & Liu, 2019). It is also a receptor for C1q and multiple collectins (Figure 1a; Edelson et al., 2006; Zutter & Edelson, 2007) and the binding of C1q to integrin α2β1 induces cytokine production and general activation in mast cells, indicating early cooperation in the broader control of innate immune cell activity between integrins and complement. Importantly, this alliance also carried into adaptive immunity because the α2β1 integrin represents a key co‐stimulatory molecule for effector T‐cell activation in arthritis pathogenesis and can be triggered by C1q (Boisvert, Chetoui, Gendron, & Aoudjit, 2010; Sasaki et al., 2003). Moreover, in‐depth studies into the role of complement in T‐cell control then led to the discovery of a novel functional crosstalk between complement, integrins, and cell metabolism (see below).
A general common scheme among integrins and complement receptors/sensors is that they are all relatively promiscuous; thus, none of them has a single exclusive ligand that triggers a single specific signalling event (Humphries, Byron, & Humphries, 2006). For example, integrins bind to insoluble ECM proteins (e.g., fibronectins, laminins, and collagens), matricellular proteins (e.g., cysteine‐rich angiogenic protein 61/CTGF/NOV [CCN]), cell surface ligands (e.g., ICAMs and VCAM‐1) and soluble (e.g., fibrinogen, complement proteins, VEGF, FGF2, angipoietin‐1, or TGFβ) ligands (Lau, 2016, Ruegg & Alghisi, 2010). Complement CR3 and CR4 bind iC3b (C3c and C3d) but also ICAM‐1, fibrinogen, and factor X (Figure 2; Erdei et al., 2019). C1q and the collectins and ficolins interact each with numerous proteins, and novel binding partners are still discovered (Ghebrehiwet, Hosszu, & Peerschke, 2017). Further, complement activation regulates the expression of integrin ligands in the vasculature (Skeie, Fingert, Russell, Stone, & Mullins, 2010), and integrins emerge as master regulators of extrahepatic core complement component expression (see below), indicating cross‐regulation between these systems. Overall, it seems that by joining forces early during evolution, these two old systems have created an extensive network that can sense and integrate a vast repertoire of cellular/environmental signals and dangers and together translate these effectively into accurate cellular responses from development to immunity. We therefore have likely only scratched the surface of this multifaceted relationship, and there are many more discoveries to be made.
5. THE COMPLOSOME: ORCHESTRATOR OF NUTRIENT USAGE AND METABOLIC REPROGRAMMING
Connections between complement and whole‐body metabolism have been made early on: It is long known that serum complement activity partakes in the regulation of lipid metabolism via the des‐Arginated form of C3a, C3a‐desArg (initially identified as acylation stimulation protein), which induces triglyceride accumulation and glucose transport in adipocytes (Cui et al., 2007). Furthermore, perturbations in serum complement activation beyond the homeostatic level are connected with obesity, insulin resistance, and diabetes (Cianflone, Xia, & Chen, 2003; Hajishengallis, Reis, Mastellos, Ricklin, & Lambris, 2017; Kahn & Flier, 2000). For an excellent review on the roles of serum complement in metabolic disease, please see Phieler, Garcia‐Martin, Lambris, and Chavakis (2013). However, it was the observation of a cell‐intrinsic and intracellularly active complement system that led to the discovery of complement‐mediated control of single‐cell metabolic activity. We have coined cell‐autonomous and autocrine‐active complement “the complosome” to set it apart from the liver‐derived complement and to signify that it heavily engages with other assembly‐prone intracellular sensor systems such as the inflammasomes (see below).
This new non‐canonical role of complement was observed first in human T‐cells, and a key mediator of this activity is the human‐specific complement receptor and regulator CD46 (membrane cofactor protein; Liszewski & Kemper, 2019). CD46 is absent in rodent somatic tissue in its “modern” form, and a functional homologue that fully recapitulates its activity has not yet been defined (Riley, Kemper, Leung, & Atkinson, 2002; Tsujimura et al., 1998; Yamamoto, Fara, Dasgupta, & Kemper, 2013). We will therefore focus in the following on human T‐cells and only succinctly summarize the connection between the complosome and single‐cell metabolism as more comprehensive reviews on this subject have recently been published (Hess & Kemper, 2016; West & Kemper, 2019; West, Kunz, & Kemper, 2020).
Human circulating CD4+ T‐cells express constantly low basal levels of the C3 gene and thus contain intracellular stores of C3. They also express an intracellular C3aR on lysosomes and contain pools of the ancient protease cathepsin L (CTSL) in lysosomes and the endoplasmic reticulum (ER). CTSL continuously processes intracellular C3 (which is also found in lysosomes and the ER) into bioactive C3a and C3b in resting T‐cells, and the engagement of lysosomal C3aR by intracellular C3a mediates low‐level activation of the nutrient sensor mammalian target of rapamycin (mTOR). This, in turn, sustains basal levels of glycolysis and thus cell survival (Figure 3a; Kolev, Le Friec, & Kemper, 2014). CD46 also contributes to maintaining T‐cell homeostasis during the quiescent T‐cell state by binding to the Notch‐1 ligand Jagged‐1 on the T‐cell surface and via this, prevents unwanted Notch‐1 activation that would normally lead to T‐cell activation (Le Friec et al., 2012).
FIGURE 3.
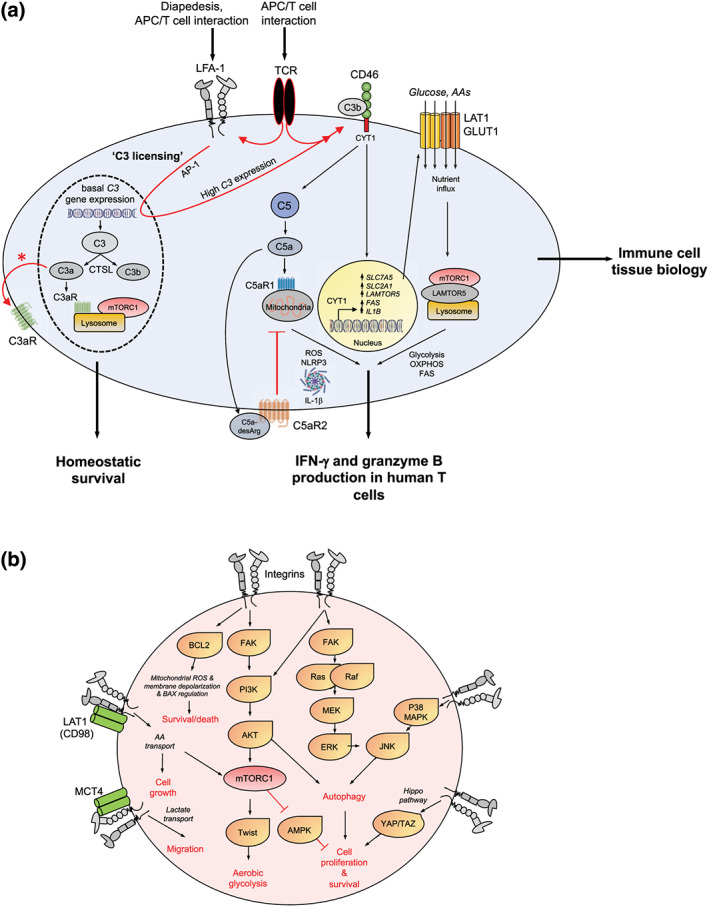
LFA‐1‐induced C3 licensing drives metabolic reprogramming in activated T‐cells. (a) Resting human CD4+ and CD8+ T‐cells sustain basal levels of C3 gene expression and generate continuously C3a and C3b via CTSL cleavage. Cell‐autonomous C3a stimulates the lysosomal C3aR that in turn sustains tonic mTORC1 activation needed for homeostatic survival. Diapedesis or the cognate APC/T‐cell interaction engages T‐cell expressed LFA‐1 (via ICAM‐1) and induces increased C3 gene expression and protein generation—C3 licensing. Timely incoming TCR signals are required to mediate increased intrinsic C3 activation (TCR signals also further activate LFA‐1), rapid surface translocation of C3a and the C3aR (denoted by an asterisk) and of C3b that then induces autocrine CD46 stimulation. CD46 downstream signals include the cleavage and nuclear translocation of its intracellular CYT‐1 domain that drives expression of nutrient transporters GLUT1 (glucose), LAT1 (amino acids, AA), fatty acid synthase or fatty acid synthesis (FAS), and the lysosomal scaffolder LAMTOR5. Together, these events support mTORC1 assembly at the lysosomes and high glycolysis, fatty acid synthesis, OXPHOS, and tricarboxylic acid cycle. CD46 also induces activation of intracellular C5 with C5a engaging the intracellular C5aR1 to produce ROS, NLRP3 inflammasome activation, and intrinsic active IL‐1β that sustains Th1 responses. Surface C5aR2 is a negative regulator of Th1 induction. The CD46/IL‐2R‐driven signals that co‐induce IL‐10 and Th1 contraction are not depicted here. Transmigration‐induced expression of a cell‐autonomous complosome is a defining feature of immune cells in tissue and also needed for normal monocyte/macrophage tissue activity (not shown). (b) Cell metabolic events known to be engaged by different integrins in a range of cells and their outcome on cell survival, proliferation, and migration (see text for details)
Thus, an intracellularly active and cell‐autonomous complement C3 system sustains T‐cell homeostasis via tonic mTOR activity. However, and somewhat reminiscent of the hydrolysis‐driven “idling” AP of complement in serum, this system can be engaged rapidly in the presence of danger to support effector T‐cell activity. Specifically, upon TCR engagement, intracellular CTSL‐mediated C3a and C3b generation is dramatically increased and also, with the C3aR, shuttled to the cell surface. T‐cell‐derived C3b now engages CD46 (the C3b and Jagged‐1 binding sites within CD46 do not overlap), and such engagement induces CD46 signalling and shedding of CD46 from the T‐cell membrane via metalloproteinases (Figure 3; Jorcano & Ruiz‐Carrillo, 1979; Le Friec et al., 2012). Further, the intracellular signalling domains of CD46, CYT‐1 and CYT‐2 (the CD46 isoforms CD46CYT‐1 and CD46CYT‐2), that arise through alternative splicing of a single gene (Johnstone, Russell, Loveland, & McKenzie, 1993) are cleaved intracellularly by γ‐secretase and translocate to the nucleus. There, they induce the increased expression of the glucose and AA transporters, glucose transporter 1 (GLUT1; SLC2A1) and large neutral amino acid transporter (LAT1; SLC7A5), respectively, thereby mediating the nutrient influx needed for T‐cell activation (Figure 3a). In addition, CYT‐1 of CD46 also up‐regulates the late endosomal/lysosomal adaptor, MAPK, and MTOR activator 5 (LAMTOR5), which then drives mammalian target of rapamycin complex 1 (mTORC1) assembly at the lysosomes with subsequent induction of high levels of glycolysis and oxidative phosphorylation (OXPHOS; Figure 3). In parallel to the direct impact on the cell metabolic machinery, autocrine CD46 signalling also results in increased expression of IL‐2Rα (CD25) and assembly of the high‐affinity IL‐2 receptor (Liao, Lin, & Leonard, 2011; Liao, Lin, & Leonard, 2013; West, Kolev, & Kemper, 2018). These are all events specifically required for IFN‐γ production and T helper type 1 (Th1) effector responses (Chang et al., 2013; Kolev et al., 2015). The central role of the cell autonomous CD46‐C3 axis in Th1 induction is underpinned by the findings that patients deficient in either CD46 or its ligand (C3/C3b) are unable to mount productive Th1 responses (at least early in life) and suffer from recurrent infections. Of note, CD4+ T‐cells from these patients proliferate normally and have no defect in Th2 responses, and serum C3b cannot rescue lack of T‐cell‐derived C3b, suggesting a specific functional connection between the T‐cell‐intrinsic complosome and IFN‐γ generation (Ghannam, Fauquert, Thomas, Kemper, & Drouet, 2014; Le Friec et al., 2012; Liszewski et al., 2013).
Human CD4+ T‐cells also contain storages of intracellular C5 and constantly generate low‐level C5a in the resting state. The enzyme that cleaves and activates intracellular C5 into C5a is currently not defined (Arbore et al., 2016) but could include an intracellular C5 convertase or a distinct C5‐cleaving protease. Autocrine engagement of CD46 driven by TCR activation amplifies intracellular C5a generation which then supports the production of ROS that are critical to normal Th1 induction via intrinsic IL‐2 induction (Sena et al., 2013). Furthermore, intracellular ROS production triggers the downstream assembly of the canonical nod‐like receptor (NLR) family pyrin domain containing 3 protein (NLRP3) inflammasome and subsequent secretion of mature IL‐1β, which sustains Th1 responses specifically in mucosal tissues (Figure 3a; Arbore et al., 2016). Similar to CD4+ T‐cells, human cytotoxic CD8+ T‐cells (CTLs) also express stores of activated C3 and C5 and their cognate complement receptors C3aR1, C5aR1 and C5aR2, and CD46. TCR and CD28 stimulation increases intracellular C3 and C5 activation and leads to autocrine engagement of CD46 also in these T‐cells. Furthermore, CD46 co‐stimulation augments specifically IFN‐γ production and granzyme B expression, therefore endowing human CTLs with optimal effector/cytotoxic activity. This effect is also mediated by metabolic reprogramming as CD46 signalling enhances nutrient influx, glycolysis, and fatty acid synthesis in these lymphocytes (Figure 3a). Of note, although intracellular C5 and NLRP3 are present in circulating human CD8+ T‐cells, canonical NLPR3 inflammasome activity and intrinsic IL‐1β production seem to not be required for their normal IFN‐γ secretion and CTL activity (Arbore et al., 2018. Further, the role of intrinsic C5 in human CTL biology has not been explored yet.
Importantly, and in line with a now broadly acknowledged role of complement in the resolution phase of immune responses and tissue repair, the complosome is also instrumental in inducing the metabolic changes that underly Th1 contraction to limit the pathological consequences of an over‐exuberant or prolonged T‐cell response (O'Garra & Vieira, 2007; Stummvoll et al., 2008; Trinchieri, 2007). CD46, together with incoming signals from the IL‐2R, orchestrates Th1 contraction via the co‐induction of IL‐10 in Th1 cells once sufficient IFN‐γ production and Th1‐derived IL‐2 levels are established. The exact signals downstream of the IL‐2R or CD46 that mediate IL‐10 production are currently ill‐defined. They seem, however, to depend mostly on the CYT‐2 domain of CD46 and operate via reduction of CD25 expression, limiting of nutrient influx, down‐regulation of GLUT1, LAT1, and LAMTOR5, reduced mTORC1 activity, increase of cholesterol flux, and the general return of the cell to a metabolically resting state (Cardone et al., 2010; Hess & Kemper, 2016; Kolev et al., 2015; Liszewski et al., 2013; Perucha et al., 2019). Thus, failures in the normal activity of the complosome during Th1 contraction are connected with T‐cell‐driven disease states: For example, uncontrolled intracellular C3 expression and activation in CD4+ T‐cells contributes to hyper‐Th1 responses observed in rheumatoid arthritis (RA) and in systemic lupus erythematosus (Cardone et al., 2010; Ellinghaus et al., 2017; Kolev et al., 2020), and a reduction in CD46‐driven IL‐10 co‐production is connected with progression of multiple sclerosis (Astier, Meiffren, Freeman, & Hafler, 2006). Intrinsic C5 activity also contributes to the negative control of human Th1 responses as increased C5a‐desArg production by T‐cells observed during Th1 expansion engages the inhibitory C5aR2 in an autocrine fashion and leads to a cessation in ROS generation and NLRP3 inflammasome activation (Figure 3a; Arbore et al., 2016). The exact mechanism, as to how C5aR2‐triggered signalling pathways lead to its suppressive effects, is currently not known.
In sum, a cell‐autonomous and in part intracellularly active complosome exists in human T‐cells. Further, this system is an integral part of the T‐cell homeostatic and effector function induction/contraction life cycle phases via direct impact on nutrient influx and metabolism.
6. INTEGRIN LFA‐1: NOVEL KEY CONTROLLER OF COMPLOSOME C3 EXPRESSION
As outlined above, the complosome is integral to normal human Th1 and cytotoxic CD8+ T‐cell responses, and increased or decreased intracellular C3 contributes to autoimmunity and infections respectively (West et al., 2018). Importantly, we had previously shown that pharmacological targeting of pathologically augmented intracellular C3 activity can normalize hyperactive Th1 responses in RA (Liszewski et al., 2013). Therefore, controlled modulation of intracellular complement may be a novel means to ameliorate autoimmunity and/or chronic infections. Until recently, the mechanisms that regulate C3 gene expression in T‐cells were undefined as TCR and/or CD28 engagement induce proteolytic activation of existing C3 pools but do not increase C3 gene transcription per se significantly (Liszewski et al., 2013). In culture, T‐cells can endocytose the hydrolyzed form of C3, C3(H2O), from serum supplemented into media that may help sustaining homeostatic survival. In tissues and lymph nodes, however, where T‐cell activation occurs and effector function develops (Verma et al., 2012, Masopust & Schenkel, 2013, Verma et al., 2016a), a source of high levels of exogenous C3 is not available. We therefore argued that induction of high C3 gene expression needed to support Th1/CTL effector function may occur during endothelial diapedesis of T‐cells from the blood into tissue. T‐cells express at least 12 of the 24 known integrin heterodimers, with a specific expression pattern dictated by the nature (CD4+, CD8+, regulatory T‐cell, etc.) and activation state of the cell (von Andrian & Mackay, 2000). The integrin αLβ2 (LFA‐1; CD11a [ITGAL gene]/CD18 [ITGB2 gene]) is the most abundant and widespread in expression with regard to T‐cells. LFA‐1 and very late antigen 4 (α4β1 integrin, VLA‐4; CD49d [ITGA4 gene]/CD29 [ITGB1] gene) play key roles throughout the T‐cell life cycle and specifically during lymphocyte extravasation. Indeed, our assessment of a range of integrins and selectins mediating T‐cell diapedesis for their ability to induce C3 transcription by human CD4+ T‐cells identified LFA‐1 as a novel key inducer of C3 gene activation (Kolev et al., 2020). LFA‐1 stimulation triggers the activation of the transcription factor activator protein 1 (AP‐1) that in turn augments C3 gene transcription (Figure 3a). Moreover, signals driven by LFA‐1 after its engagement by ICAM‐1 during endothelial transmigration are also critically required to induce intrinsic C3 expression in human CD8+ T‐cells and in monocytes (the dependency of LFA‐1‐induced C3 gene transcription on AP‐1 in these cells has not been explored). Importantly, such LFA‐1 mediated “C3 licensing” is not sufficient to induce effector function in T‐cells and monocytes on its own. IFN‐γ and granzyme B production in T‐cells also requires a timely incoming TCR signal (Figure 3a), while IL‐1β secretion by monocytes needs toll‐like receptor engagement as second trigger. Thus, LFA‐1‐mediated C3 licensing seems a broadly applicable novel pathway and a prerequisite for effector immune cell function upon their (antigen)‐specific activation. In consequence, patients with mutations in the ITGB2 gene and that suffer from leukocyte adhesion deficiency 1 (LAD‐1) cannot increase intracellular C3 in immune cells and display specifically reduced Th1 and CTL responses as well as diminished IL‐1β production by monocytes when these cells are stimulated (Kolev et al., 2020). Conversely, T‐cells that have transmigrated in an LFA‐1‐dependent fashion into the inflamed joints of patients with RA have significantly increased intracellular steady‐state C3 expression. Furthermore, C3 levels associate with disease severity and may represent a novel biomarker distinguishing inflamed versus uninflamed RA.
Thus, the integrin network emerges as a novel and central regulator of the complosome. Another exciting outcome of this work is the finding that cell‐intrinsic transcription of complement, including C3, is one of the most significantly enriched biological pathway immune cell populations in the tissue (Kolev et al., 2020). Therefore, a common cardinal feature of T‐cells and monocytes when transitioning from the blood into the tissue is the expression induction of basically the complete repertoire of complement components including effector molecules, activators, receptors, and regulators. This is an additional strong indication of the central role of the complosome, independently of liver‐derived and circulating complement, in the regulation of cellular activity of cells particularly once they moved into tissues.
7. SHARED INTERESTS: THE COMPLOSOME AND INTEGRINS IN THE REGULATION OF METABOLISM
Immune cells from patients with LAD‐1 have defective CD18 activity, and their immune cells are compromised in their ability to migrate into tissues as LFA‐1 is required for this process. However, immune cells in these patients are also impaired in normal effector activity. The perturbed ability of CD4+ T‐cells from patients with LAD‐1 to generate Th1 responses was thought exclusively to be rooted in reduced TCR signalling (Abraham, Griffith, & Miller, 1999; Chirathaworn et al., 2002; Hogg, Patzak, & Willenbrock, 2011; Varga et al., 2010), suboptimal peripheral supramolecular activation cluster formation in the immunological synapse during T‐cell priming (Monks, Freiberg, Kupfer, Sciaky, & Kupfer, 1998), and/or reduction in Notch activity on CD4+ T‐cells (Verma et al., 2016b), events which are all regulated by LFA‐1. The new finding that LFA‐1 is upstream of the C3‐CD46 axis links the inability of LAD‐1 T‐cells to generate sufficient intracellular C3 and engage CD46‐driven metabolic reprogramming with their reduced Th1 immunity. This idea is supported by the fact that CD46 activity per se mimics the core functions of LFA‐1 during T‐cell stimulation: CD46‐mediated signals synergize with the TCR to activate cellular kinases (Astier, Trescol‐Biemont, Azocar, Lamouille, & Rabourdin‐Combe, 2000; Kemper & Atkinson, 2007; Kolev et al., 2015) and CD46 is required for Notch‐1 and Jagged‐1 up‐regulation in human CD4+ T‐cells (Le Friec et al., 2012). In addition, CD4+ T‐cells from CD46‐deficient patients, similar to those from patients that suffer from LAD‐1, proliferate normally and are able to produce Th2 and Th17 cytokines normally (Le Friec et al., 2012). Importantly, LFA‐1 stimulation on CD4+ T‐cells parallels the metabolic changes previously observed upon direct CD46 co‐stimulation, including activation of mTORC1, glycolysis, and OXPHOS. Thus, this novel integrin–complement connection converges on the level of cell metabolism.
This is of little surprise as integrins have their own long history of drivers of cell metabolic activities via a number of mechanisms (Figure 3b). For example, as CD46, they are also key in the regulation of nutrient influx into cells: The β1 integrin interacts directly with CD98 (Rintoul et al., 2002; Zent et al., 2000), a constituent of the AA transporters LAT‐1 and LAT‐2, and can increase nutrient‐driven cell growth via this interaction (Nicklin et al., 2009). Further, β1 integrin also interacts with the monocarboxylate transporter 4 (MCT4), which functions as a lactate transporter (Gallagher, Castorino, & Philp, 2009). In epithelial cells, β1 integrin and MCT4 cooperate at the leading edge of migrating cells, and perturbation of MCT4 slows lactate‐dependent cell migration (Gallagher et al., 2009). Increased nutrient flux or availability is a general signal to turn the key that engages the cellular nutrient sensor machinery, that is, mTORC1 activation (Nicklin et al., 2009). In addition to nutrient‐induced mTOR activation, integrins can also trigger signals that directly induce mTORC1. This includes activating FAK and via this the PI3K‐Akt pathway, a master inducer of mTORC1 activity (Xia, Nho, Kahm, Kleidon, & Henke, 2004). Integrin β1‐mediated signals can also directly impact on how cells engage glycolysis. For example, β2 integrin‐induced FAK‐PI3K‐Akt‐mTOR axis activation increases expression of the transcription factor Twist in epithelial cells, which in turn mediates a shift from anaerobic to aerobic glycolysis (Yang et al., 2015). In all cases, cellular mTORC1 activation simultaneously inhibits the activity of the AMP‐activated protein kinase, which is the key negative controller of cell proliferation (Ling et al., 2020), explaining the strong general mitotic activity of integrins. In addition, integrins also regulate the induction of cell survival promoting autophagy (which is modulated by nutrient availability) by engagement of PI3K‐Akt and MAPK signalling upon ECM binding and control the nutrient‐sensitive Hippo signalling pathway. Hippo operates through expression induction of genes controlling cell survival and death via activation of the transcriptional co‐activators yes‐associated protein and transcriptional coactivator with PDZ‐binding motif (WWTR1; Moroishi, Hansen, & Guan, 2015; Piccolo, Dupont, & Cordenonsi, 2014; Santinon, Pocaterra, & Dupont, 2016).
Finally, there is strong connection between integrin signalling and control of mitochondrial activity and integrity. In fibroblast, integrin‐mediated signalling induces mitochondrial ROS production in an B cell lymphoma 2 (BCL2)‐dependent manner and concomitant integrin‐dependent mitochondrial membrane depolarization (Werner & Werb, 2002). Several integrins can trigger BCL2 activation on a broad range of cells, which points towards a key role of the integrin network in counter‐acting stress‐induced mitochondrial dysfunction (Figure 3b). For example, PI3K‐Akt‐induced signals activate phosphoinositide‐dependent kinases that ultimately lead to the displacement of the pro‐apoptotic protein Bad from mitochondria and its proteasomal destruction (Stupack & Cheresh, 2002). Furthermore, LFA‐1‐mediated signals, engaged during the cognate T‐cell and antigen‐presenting cells (APCs) interaction, “move” these organelles as a whole towards the immunological synapse, likely to provide local energy for this intense cell–cell interaction (Contento et al., 2010).
The overwhelming net outcome of integrin signalling on cell metabolism is that of supporting an activated, proliferative, and migratory phenotype. It is therefore not surprising that perturbations in integrins signalling pathways—or in the mechanisms that restrain integrin activity—are connected with human metabolic diseases such as cancer and fibrosis. (Winograd‐Katz, Fassler, Geiger, & Legate, 2014). Importantly, cell metabolism itself emerges as key controller of integrin activity (reviewed in Ata & Antonescu, 2017), implying the existence of positive and negative feedback loops between metabolites and integrin activity. The integrin field is starting to look now more deeply into this new crosstalk to better define the molecular mechanisms underlying associated disease states. We suggest that it may be of interest to also include the integrated complement–metabolism axis into these studies and considerations (Figure 4).
FIGURE 4.

The integrin–complosome–metabolism network in the regulation of T‐cell effector function. Summary model depiction of the known cooperative actions of LFA‐1/integrins, the complosome, and key metabolic events that underly successful effector function induction in human CD4+ and CD8+ T‐cells. T‐cell receptor, growth factor receptor, and intrinsic inflammasome signals as well as negative/positive feedback loops (e.g., between generated metabolites and LFA‐1) have been omitted for simplicity
8. INTRACELLULAR COMPLEMENT/THE COMPLOSOME: DID IT ALL START HERE?
Although cell‐intrinsic, intracellularly activated, and functioning complement has been best defined in human T‐cells so far, intrinsic intracellular C3a can be found in a range of cell types (Liszewski et al., 2013). Moreover, with the discovery that expression induction of the complosome is a defining feature of immune cells in tissue (Kolev, West et al., 2020), our early suggestion that this system is operative in principle in all cells and hence of broad physiological significance is gathering support. Indeed, cell‐intrinsic C3 is critical in regulating autophagy in pancreatic β cells during time of glycolytic stress (King et al., 2019). Thus, the complosome may be directly connected to the classic metabolic disease diabetes (see contribution by Blom and colleagues in this Special Issue).
We also champion an evolutionary path of complement that deviates from the commonly accepted one in which the liver‐derived complement system was “first.” Based on the location‐driven activity of complement, its functionally important cross‐activity with other intracellular danger sensor systems, and the emerging key role for all of these systems in cell physiology, we suggest that complement may have originally appeared in single‐cell organisms as an intracellular sensor and rectifier of metabolic stress. With the evolution of into multi‐organ organisms—and particularly with the development of a vascular system—complement took a bifurcated path: Part of the original C3 remained functional mostly within cells to control cell physiology, while part of C3 segregated into a secreted form, mostly produced by hepatocytes and assumed the role of the systemic pattern recognition system (Elvington, Liszewski, & Atkinson, 2016; Hess & Kemper, 2016; Kolev & Kemper, 2017; West et al., 2020; West & Kemper, 2019). We suggest that the structure and the post‐translational modifications of intra‐ and extra‐cellular C3 is different: liver‐derived C3 is folded into the classically known structure that can generate the opsonin C3b as well as the backbone for C3/C5 convertase formation. Intracellular C3, on the other hand, may exist in several different forms of which some may be folded “correctly” but some may not as those are rather processed by cell‐specific proteases to release the domains mediating its cell metabolic activities (e.g., C3a). The structure of intracellular C3 is likely dictated by the specific environment (acidity, salt concentration, etc.) of the subcellular compartment that it is shunted into. This model could explain why processed/activated C3 analysed in the lysate of immune cells show a distinct fragment pattern when compared with liver‐derived activated C3 (Elvington, Liszewski, Bertram, Kulkarni, & Atkinson, 2017; Liszewski et al., 2013). In addition, it would also explain why immune cells in culture only utilize about 10% of the C3(H2O) that they can take up from serum for intracellular CTSL‐mediated C3a generation and expel the remaining non‐cleaved 90% (Elvington et al., 2017): the latter likely indicates that cell‐expressed C3 is transported to and activated within specific cellular sub‐compartments, which are distinct from those of the C3 “uptake route.” Such notion also aligns with our inability to fully rescue Th1 responses in T‐cells from LAD‐1 (or some C3 deficient) patients with C3 or C3a by viral delivery or electroporation (Ghannam et al., 2014; Kolev et al., 2020)—C3/C3a was likely not as efficiently targeted to its intracellular receptor in lysosomal/ER compartments as it would have been via the cell‐autonomous “natural” pathway.
An originally intracellular activity of complement sits also well with the recent finding that many biologically important protein families that have classically been viewed as “cell surface only” molecules have also central intracellular roles. For example, intracellular pools of LFA‐1 regulate asymmetric T‐cell differentiation (Capece et al., 2017), intracellular complement GPCRs drive ROS production and possibly gene regulation (Arbore et al., 2016; Kremlitzka et al., 2019), and intracellularly active MMPs are critical in the regulation of gene transcriptional control (Jobin, Butler, & Overall, 2017).
9. INTEGRINS, THE COMPLOSOME AND CELL METABOLISM: A TANGLED SPECIES‐SPECIFIC WEB
The modulation of cellular metabolic pathways for therapeutic application is currently a major scientific and clinical focus, particularly in cancer where application of antimetabolites to inhibit expansion of neoplastic cells has yielded success (Luengo, Gui, & Vander Heiden, 2017). Thus, understanding the signals that modulate cell metabolism including those provided by integrins and the complosome (Figure 4) could advance the development of such therapeutics.
This, however, will not be an easy undertaking. Firstly, most cellular sensor and effector systems engage into extensive crosstalk and cross‐regulation with each other. To pinpoint function, however, researchers tend to study their system of choice most often independently—without controlling the effect of loss/overactivity of one system on the others. The “everything is connected problem” is even more an obstacle when studying cell metabolism. For example, the analysis of immune cell populations ex vivo under restricted or controlled nutrient conditions is an important first approach to gauge their biological effects. However, cells in culture respond differently to metabolic modulations as compared to their in vivo behaviour as cells in tissues are embedded within an active network of surrounding cells that all modulate environmental nutrient availability and affect each other's activity (Ma et al., 2019). In vivo animal models, on the other hand, do not allow to control for the compensatory metabolic pathways that cells tend to engage in when encountering engineered restrictions in key metabolic pathways (Biancur et al., 2017; Johnson et al., 2018).
With regard to the complosome, small animal models have their own issues. CD46 activity is integral to human Th1 and CTL responses but does not exist in this form in rodents. Thus, further studying the biological significance of CD46 in depth—for example, during T‐cell memory and/or tissue residency—is currently hampered by a lack of a suitable in vivo model. Also, even complosome components that are present in both mice and men do not always follow the same expression pattern or mediate comparable functions. For example, there is an ongoing discussion in the field about the presence of ANA receptors on mouse T‐cells, while their activity on human T‐cells has been broadly observed (Laumonnier, Karsten, & Kohl, 2017). Similarly, human T‐cells rely on CTSL‐cleaved intrinsic C3 activation for IFN‐γ production, while CTSL is not the C3‐activating protease in mouse T‐cells and their T‐cells can also utilize exogenous C3a for Th1 induction. Finally, almost all human/mouse complement receptors function not only in a cell‐specific fashion but several of them also in a subcellular‐specific and/or biphasic mode during cell homeostasis and activation. A similar situation exists when comparing mouse and human integrins, where species specific expression patterns and ligands specificities are also observed (Holmes & Rout, 2011). Thus, it will likely take the generation of new cell‐specific inducible integrin and complement models in combination with comparative, integrated, advanced, and more accessible metabolic profiling approaches of patients with inborn errors in these components to detangle this intricate integrin–complement–metabolism web.
9.1. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20 (Alexander, Christopoulos, et al., 2019, Alexander, Fabbro, et al., 2019; Alexander, Kelly, et al., 2019).
AUTHOR CONTRIBUTIONS
N.M., P.S., and J.R. contributed equally.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGEMENTS
Work in the Complement and Inflammation Research Section (CIRS) is financed by the Division of Intramural Research, National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health (NIH). We further acknowledge the work of the many scientists, particularly working on integrins and T‐cell metabolism, that we were unable to cite here due to focus on the emerging connection between intracellular complement, integrins, and metabolism.
Merle NS, Singh P, Rahman J, Kemper C. Integrins meet complement: The evolutionary tip of an iceberg orchestrating metabolism and immunity. Br J Pharmacol. 2021;178:2754–2770. 10.1111/bph.15168
REFERENCES
- Abraham, C., Griffith, J., & Miller, J. (1999). The dependence for leukocyte function‐associated antigen‐1/ICAM‐1 interactions in T cell activation cannot be overcome by expression of high density TCR ligand. Journal of Immunology, 162(8), 4399–4405. [PubMed] [Google Scholar]
- Adorno‐Cruz, V., & Liu, H. (2019). Regulation and functions of integrin alpha2 in cell adhesion and disease. Genes & Diseases, 6(1), 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H., Christopoulos, A., Davenport, A. P., Kelly, E., Mathie, A., Peters, J. A., … Collaborators . (2019). The Concise Guide to PHARMACOLOGY 2019/20: G protein‐coupled receptors. British Journal of Pharmacology, 176(Suppl 1), S21–S141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H., Fabbro, D., Kelly, E., Mathie, A., Peters, J. A., Veale, E. L., … Collaborators . (2019). The Concise Guide to PHARMACOLOGY 2019/20: Catalytic receptors. British Journal of Pharmacology, 176(Suppl 1), S247–S296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H., Fabbro, D., Kelly, E., Mathie, A., Peters, J. A., Veale, E. L., … Collaborators . (2019). The Concise Guide to PHARMACOLOGY 2019/20: Enzymes. British Journal of Pharmacology, 176(Suppl 1), S297–S396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H., Kelly, E., Mathie, A., Peters, J. A., Veale, E. L., Armstrong, J. F., … Collaborators . (2019a). The Concise Guide to PHARMACOLOGY 2019/20: Introduction and other protein targets. British Journal of Pharmacology, 176(Suppl 1), S1–S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H., Kelly, E., Mathie, A., Peters, J. A., Veale, E. L., Armstrong, J. F., … Collaborators . (2019b). The Concise Guide to PHARMACOLOGY 2019/20: Transporters. British Journal of Pharmacology, 176(Suppl 1), S397–S493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, L. R., Owens, T. W., & Naylor, M. J. (2014). Structural and mechanical functions of integrins. Biophysical Reviews, 6(2), 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Andrian, U. H., & Mackay, C. R. (2000). T‐cell function and migration. Two sides of the same coin. The New England Journal of Medicine, 343(14), 1020–1034. 10.1056/NEJM200010053431407 [DOI] [PubMed] [Google Scholar]
- Arbore, G., West, E. E., Rahman, J., Le Friec, G., Niyonzima, N., Pirooznia, M., … Kemper, C. (2018). Complement receptor CD46 co‐stimulates optimal human CD8(+) T cell effector function via fatty acid metabolism. Nature Communications, 9(1), 1–15. 10.1038/s41467-018-06706-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbore, G., West, E. E., Spolski, R., Robertson, A. A. B., Klos, A., Rheinheimer, C., … Kemper, C. (2016). T helper 1 immunity requires complement‐driven NLRP3 inflammasome activity in CD4(+) T cells. Science, 352(6292), 1424–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astier, A., Trescol‐Biemont, M. C., Azocar, O., Lamouille, B., & Rabourdin‐Combe, C. (2000). Cutting edge: CD46, a new costimulatory molecule for T cells, that induces p120CBL and LAT phosphorylation. Journal of Immunology, 164(12), 6091–6095. 10.4049/jimmunol.164.12.6091 [DOI] [PubMed] [Google Scholar]
- Astier, A. L., Meiffren, G., Freeman, S., & Hafler, D. A. (2006). Alterations in CD46‐mediated Tr1 regulatory T cells in patients with multiple sclerosis. The Journal of Clinical Investigation, 116(12), 3252–3257. 10.1172/JCI29251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ata, R., & Antonescu, C. N. (2017). Integrins and cell metabolism: An intimate relationship impacting cancer. International Journal of Molecular Sciences, 18(1), 189–220. 10.3390/ijms18010189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baade, T., Paone, C., Baldrich, A., & Hauck, C. R. (2019). Clustering of integrin beta cytoplasmic domains triggers nascent adhesion formation and reveals a protozoan origin of the integrin‐talin interaction. Scientific Reports, 9(1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biancur, D. E., Paulo, J. A., Malachowska, B., Del Rey, M. Q., Sousa, C. M., Wang, X. X., … Kimmelman, A. C. (2017). Compensatory metabolic networks in pancreatic cancers upon perturbation of glutamine metabolism. Nature Communications, 8, 1–15. 10.1038/ncomms15965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert, M., Chetoui, N., Gendron, S., & Aoudjit, F. (2010). Alpha2beta1 integrin is the major collagen‐binding integrin expressed on human Th17 cells. European Journal of Immunology, 40(10), 2710–2719. 10.1002/eji.201040307 [DOI] [PubMed] [Google Scholar]
- Bolos, V., Gasent, J. M., Lopez‐Tarruella, S., & Grande, E. (2010). The dual kinase complex FAK‐Src as a promising therapeutic target in cancer. Onco Targets and Therapy, 3, 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower, D. L., Brower, S. M., Hayward, D. C., & Ball, E. E. (1997). Molecular evolution of integrins: Genes encoding integrin beta subunits from a coral and a sponge. Proceedings of the National Academy of Sciences of the United States of America, 94(17), 9182–9187. 10.1073/pnas.94.17.9182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet, T., & King, N. (2017). The origin of animal multicellularity and cell differentiation. Developmental Cell, 43(2), 124–140. 10.1016/j.devcel.2017.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood, D. A., Fujioka, Y., de Pereda, J. M., Garcia‐Alvarez, B., Nakamoto, T., Margolis, B., … Ginsberg, M. H. (2003). Integrin beta cytoplasmic domain interactions with phosphotyrosine‐binding domains: A structural prototype for diversity in integrin signaling. Proceedings of the National Academy of Sciences of the United States of America, 100(5), 2272–2277. 10.1073/pnas.262791999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capece, T., Walling, B. L., Lim, K., Kim, K. D., Bae, S., Chung, H. L., … Kim, M. (2017). A novel intracellular pool of LFA‐1 is critical for asymmetric CD8(+) T cell activation and differentiation. The Journal of Cell Biology, 216(11), 3817–3829. 10.1083/jcb.201609072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone, J., Le Friec, G., Vantourout, P., Roberts, A., Fuchs, A., Jackson, I., … Kemper, C. (2010). Complement regulator CD46 temporally regulates cytokine production by conventional and unconventional T cells. Nature Immunology, 11(9), 862–871. 10.1038/ni.1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C. H., Curtis, J. D., Maggi, L. B.Jr., Faubert, B., Villarino, A. V., O'Sullivan, D., … Pearce, E. L. (2013). Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell, 153(6), 1239–1251. 10.1016/j.cell.2013.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirathaworn, C., Kohlmeier, J. E., Tibbetts, S. A., Rumsey, L. M., Chan, M. A., & Benedict, S. H. (2002). Stimulation through intercellular adhesion molecule‐1 provides a second signal for T cell activation. Journal of Immunology, 168(11), 5530–5537. 10.4049/jimmunol.168.11.5530 [DOI] [PubMed] [Google Scholar]
- Chouhan, B. S., Kapyla, J., Denessiouk, K., Denesyuk, A., Heino, J., & Johnson, M. S. (2014). Early chordate origin of the vertebrate integrin alphaI domains. PLoS ONE, 9(11), 1–16, e112064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiaen, L., Davidson, B., Kawashima, T., Powell, W., Nolla, H., Vranizan, K., & Levine, M. (2008). The transcription/migration interface in heart precursors of Ciona intestinalis. Science, 320(5881), 1349–1352. 10.1126/science.1158170 [DOI] [PubMed] [Google Scholar]
- Cianflone, K., Xia, Z., & Chen, L. Y. (2003). Critical review of acylation‐stimulating protein physiology in humans and rodents. Biochimica et Biophysica Acta, 1609(2), 127–143. 10.1016/S0005-2736(02)00686-7 [DOI] [PubMed] [Google Scholar]
- Contento, R. L., Campello, S., Trovato, A. E., Magrini, E., Anselmi, F., & Viola, A. (2010). Adhesion shapes T cells for prompt and sustained T‐cell receptor signalling. The EMBO Journal, 29(23), 4035–4047. 10.1038/emboj.2010.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, W., Paglialunga, S., Kalant, D., Lu, H., Roy, C., Laplante, M., … Cianflone, K. (2007). Acylation‐stimulating protein/C5L2‐neutralizing antibodies alter triglyceride metabolism in vitro and in vivo. American Journal of Physiology. Endocrinology and Metabolism, 293(6), E1482–E1491. 10.1152/ajpendo.00565.2006 [DOI] [PubMed] [Google Scholar]
- Edelson, B. T., Stricker, T. P., Li, Z., Dickeson, S. K., Shepherd, V. L., Santoro, S. A., & Zutter, M. M. (2006). Novel collectin/C1q receptor mediates mast cell activation and innate immunity. Blood, 107(1), 143–150. 10.1182/blood-2005-06-2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus, U., Cortini, A., Pinder, C. L., Le Friec, G., Kemper, C., & Vyse, T. J. (2017). Dysregulated CD46 shedding interferes with Th1‐contraction in systemic lupus erythematosus. European Journal of Immunology, 47(7), 1200–1210. 10.1002/eji.201646822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvington, M., Liszewski, M. K., & Atkinson, J. P. (2016). Evolution of the complement system: From defense of the single cell to guardian of the intravascular space. Immunological Reviews, 274(1), 9–15. 10.1111/imr.12474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvington, M., Liszewski, M. K., Bertram, P., Kulkarni, H. S., & Atkinson, J. P. (2017). A C3(H20) recycling pathway is a component of the intracellular complement system. The Journal of Clinical Investigation, 127(3), 970–981. 10.1172/JCI89412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, Y., Matsushita, M., & Fujita, T. (2015). New insights into the role of ficolins in the lectin pathway of innate immunity. International Review of Cell and Molecular Biology, 316, 49–110. [DOI] [PubMed] [Google Scholar]
- Endo, Y., Nonaka, M., Saiga, H., Kakinuma, Y., Matsushita, A., Takahashi, M., … Fujita, T. (2003). Origin of mannose‐binding lectin‐associated serine protease (MASP)‐1 and MASP‐3 involved in the lectin complement pathway traced back to the invertebrate, amphioxus. Journal of Immunology, 170(9), 4701–4707. 10.4049/jimmunol.170.9.4701 [DOI] [PubMed] [Google Scholar]
- Erdei, A., Lukacsi, S., Macsik‐Valent, B., Nagy‐Balo, Z., Kurucz, I., & Bajtay, Z. (2019). Non‐identical twins: Different faces of CR3 and CR4 in myeloid and lymphoid cells of mice and men. Seminars in Cell & Developmental Biology, 85, 110–121. 10.1016/j.semcdb.2017.11.025 [DOI] [PubMed] [Google Scholar]
- Ewan, R., Huxley‐Jones, J., Mould, A. P., Humphries, M. J., Robertson, D. L., & Boot‐Handford, R. P. (2005). The integrins of the urochordate Ciona intestinalis provide novel insights into the molecular evolution of the vertebrate integrin family. BMC Evolutionary Biology, 5, 1–18. 10.1186/1471-2148-5-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik, M. F., & Kasahara, M. (2010). Origin and evolution of the adaptive immune system: Genetic events and selective pressures. Nature Reviews. Genetics, 11(1), 47–59. 10.1038/nrg2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi, N., & Ballarin, L. (2017). Immunity in protochordates: The tunicate perspective. Frontiers in Immunology, 8, 1–16. 10.3389/fimmu.2017.00674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujito, N. T., Sugimoto, S., & Nonaka, M. (2010). Evolution of thioester‐containing proteins revealed by cloning and characterization of their genes from a cnidarian sea anemone, Haliplanella lineate. Developmental and Comparative Immunology, 34(7), 775–784. 10.1016/j.dci.2010.02.011 [DOI] [PubMed] [Google Scholar]
- Gallagher, S. M., Castorino, J. J., & Philp, N. J. (2009). Interaction of monocarboxylate transporter 4 with beta1‐integrin and its role in cell migration. American Journal of Physiology. Cell Physiology, 296(3), C414–C421. 10.1152/ajpcell.00430.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Z., Li, M., Ma, J., & Zhang, S. (2014). An amphioxus gC1q protein binds human IgG and initiates the classical pathway: Implications for a C1q‐mediated complement system in the basal chordate. European Journal of Immunology, 44(12), 3680–3695. 10.1002/eji.201444734 [DOI] [PubMed] [Google Scholar]
- Gettner, S. N., Kenyon, C., & Reichardt, L. F. (1995). Characterization of beta pat‐3 heterodimers, a family of essential integrin receptors in C. elegans. The Journal of Cell Biology, 129(4), 1127–1141. 10.1083/jcb.129.4.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannam, A., Fauquert, J. L., Thomas, C., Kemper, C., & Drouet, C. (2014). Human complement C3 deficiency: Th1 induction requires T cell‐derived complement C3a and CD46 activation. Molecular Immunology, 58(1), 98–107. 10.1016/j.molimm.2013.11.010 [DOI] [PubMed] [Google Scholar]
- Ghebrehiwet, B., Hosszu, K. H., & Peerschke, E. I. (2017). C1q as an autocrine and paracrine regulator of cellular functions. Molecular Immunology, 84, 26–33. 10.1016/j.molimm.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancotti, F. G., & Ruoslahti, E. (1999). Integrin signaling. Science, 285(5430), 1028–1032. 10.1126/science.285.5430.1028 [DOI] [PubMed] [Google Scholar]
- Hajishengallis, G., Reis, E. S., Mastellos, D. C., Ricklin, D., & Lambris, J. D. (2017). Novel mechanisms and functions of complement. Nature Immunology, 18(12), 1288–1298. 10.1038/ni.3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harburger, D. S., & Calderwood, D. A. (2009). Integrin signalling at a glance. Journal of Cell Science, 122(Pt 2), 159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, S. D., Sharman, J. L., Faccenda, E., Southan, C., Pawson, A. J., Ireland, S., … Nc, I. (2018). The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: Updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Research, 46(D1), D1091–D1106. 10.1093/nar/gkx1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E. S., McIntyre, T. M., Prescott, S. M., & Zimmerman, G. A. (2000). The leukocyte integrins. The Journal of Biological Chemistry, 275(31), 23409–23412. 10.1074/jbc.R000004200 [DOI] [PubMed] [Google Scholar]
- Hess, C., & Kemper, C. (2016). Complement‐mediated regulation of metabolism and basic cellular processes. Immunity, 45(2), 240–254. 10.1016/j.immuni.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg, N., Patzak, I., & Willenbrock, F. (2011). The insider's guide to leukocyte integrin signalling and function. Nature Reviews. Immunology, 11(6), 416–426. [DOI] [PubMed] [Google Scholar]
- Holmes, R. S., & Rout, U. K. (2011). Comparative studies of vertebrate Beta integrin genes and proteins: Ancient genes in vertebrate evolution. Biomolecules, 1(1), 3–31. 10.3390/biom1010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H., Huang, S., Yu, Y., Yuan, S., Li, R., Wang, X., … Xu, A. (2011). Functional characterization of a ficolin‐mediated complement pathway in amphioxus. The Journal of Biological Chemistry, 286(42), 36739–36748. 10.1074/jbc.M111.245944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A. L. (2001). Evolution of the integrin alpha and beta protein families. Journal of Molecular Evolution, 52(1), 63–72. 10.1007/s002390010134 [DOI] [PubMed] [Google Scholar]
- Humphries, J. D., Byron, A., & Humphries, M. J. (2006). Integrin ligands at a glance. Journal of Cell Science, 119(Pt 19), 3901–3903. 10.1242/jcs.03098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes, R. O. (2002). Integrins. Cell, 110(6), 673–687. [DOI] [PubMed] [Google Scholar]
- Ishiguro, H., Kobayashi, K., Suzuki, M., Titani, K., Tomonaga, S., & Kurosawa, Y. (1992). Isolation of a hagfish gene that encodes a complement component. The EMBO Journal, 11(3), 829–837. 10.1002/j.1460-2075.1992.tb05120.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobin, P. G., Butler, G. S., & Overall, C. M. (2017). New intracellular activities of matrix metalloproteinases shine in the moonlight. Biochimica et Biophysica Acta, Molecular Cell Research, 1864(11 Pt A), 2043–2055. [DOI] [PubMed] [Google Scholar]
- Johnson, M. O., Wolf, M. M., Madden, M. Z., Andrejeva, G., Sugiura, A., Contreras, D. C., … Rathmell, J. C. (2018). Distinct regulation of Th17 and Th1 cell differentiation by glutaminase‐dependent metabolism. Cell, 175(7), 1780–1795e1719. 10.1016/j.cell.2018.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, M. S., & Chouhan, B. S. (2014). Evolution of integrin I domains. Advances in Experimental Medicine and Biology, 819, 1–19. [DOI] [PubMed] [Google Scholar]
- Johnstone, R. W., Russell, S. M., Loveland, B. E., & McKenzie, I. F. C. (1993). Polymorphic expression of CD46 protein isoforms due to tissue‐specific RNA splicing. Molecular Immunology, 30(14), 1231–1241. 10.1016/0161-5890(93)90038-D [DOI] [PubMed] [Google Scholar]
- Jorcano, J. L., & Ruiz‐Carrillo, A. (1979). H3.H4 tetramer directs DNA and core histone octamer assembly in the nucleosome core particle. Biochemistry, 18(5), 768–774. 10.1021/bi00572a005 [DOI] [PubMed] [Google Scholar]
- Kahn, B. B., & Flier, J. S. (2000). Obesity and insulin resistance. The Journal of Clinical Investigation, 106(4), 473–481. 10.1172/JCI10842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaidoh, T., & Gigli, I. (1989). Phylogeny of regulatory proteins of the complement system. Isolation and characterization of a C4b/C3b inhibitor and a cofactor from sand bass plasma. Journal of Immunology, 142(5), 1605–1613. [PubMed] [Google Scholar]
- Kemper, C., & Atkinson, J. P. (2007). T‐cell regulation: With complements from innate immunity. Nature Reviews Immunology, 7(1), 9–18. 10.1038/nri1994 [DOI] [PubMed] [Google Scholar]
- Killick, J., Morisse, G., Sieger, D., & Astier, A. L. (2018). Complement as a regulator of adaptive immunity. Seminars in Immunopathology, 40(1), 37–48. 10.1007/s00281-017-0644-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, A., Ikeo, K., & Nonaka, M. (2009). Evolutionary origin of the vertebrate blood complement and coagulation systems inferred from liver EST analysis of lamprey. Developmental and Comparative Immunology, 33(1), 77–87. 10.1016/j.dci.2008.07.005 [DOI] [PubMed] [Google Scholar]
- Kimura, A., Sakaguchi, E., & Nonaka, M. (2009). Multi‐component complement system of Cnidaria: C3, Bf, and MASP genes expressed in the endodermal tissues of a sea anemone, Nematostella vectensis. Immunobiology, 214(3), 165–178. 10.1016/j.imbio.2009.01.003 [DOI] [PubMed] [Google Scholar]
- King, B. C., Kulak, K., Krus, U., Rosberg, R., Golec, E., Wozniak, K., … Blom, A. M. (2019). Complement component C3 is highly expressed in human pancreatic islets and prevents beta cell death via ATG16L1 interaction and autophagy regulation. Cell Metabolism, 29(1), 202–210e206. 10.1016/j.cmet.2018.09.009 [DOI] [PubMed] [Google Scholar]
- Klos, A., Tenner, A. J., Johswich, K.‐O., Ager, R. R., Reis, E. S., & Köhl, J. (2009). The role of the anaphylatoxins in health and disease. Molecular Immunology, 46(14), 2753–2766. 10.1016/j.molimm.2009.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev, M., Dimeloe, S., Le Friec, G., Navarini, A., Arbore, G., Povoleri, G. A., … Kemper, C. (2015). Complement regulates nutrient influx and metabolic reprogramming during Th1 cell responses. Immunity, 42(6), 1033–1047. 10.1016/j.immuni.2015.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev, M., & Kemper, C. (2017). Keeping it all going—Complement meets metabolism. Frontiers in Immunology, 8, 1–18. https://www.frontiersin.org/articles/10.3389/fimmu.2017.00001/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev, M., Le Friec, G., & Kemper, C. (2014). Complement—Tapping into new sites and effector systems. Nature Reviews. Immunology, 14(12), 811–820. 10.1038/nri3761 [DOI] [PubMed] [Google Scholar]
- Kolev, M., West, E. E., Kunz, N., Chauss, D., Moseman, E. A., Rahman, J., … Kemper, C. (2020). Diapedesis‐induced integrin signaling via LFA‐1 facilitates tissue immunity by inducing intrinsic complement C3 expression in immune cells. Immunity, 52(3), 513–527e518. 10.1016/j.immuni.2020.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremlitzka, M., Nowacka, A. A., Mohlin, F. C., Bompada, P., De Marinis, Y., & Blom, A. M. (2019). Interaction of serum‐derived and internalized C3 with DNA in human B cells—A potential involvement in regulation of gene transcription. Frontiers in Immunology, 10, 493. 10.3389/fimmu.2019.00493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisinger, M. J., Goebeler, V., Lu, Z., Meixner, S. C., Myles, T., Pryzdial, E. L. G., & Conway, E. M. (2012). Thrombin generates previously unidentified C5 products that support the terminal complement activation pathway. Blood, 120(8), 1717–1725. 10.1182/blood-2012-02-412080 [DOI] [PubMed] [Google Scholar]
- Kumar, A., Bhandari, A., Sarde, S. J., & Goswami, C. (2014). Molecular phylogeny of C1 inhibitor depicts two immunoglobulin‐like domains fusion in fishes and ray‐finned fishes specific intron insertion after separation from zebrafish. Biochemical and Biophysical Research Communications, 450(1), 219–226. 10.1016/j.bbrc.2014.05.097 [DOI] [PubMed] [Google Scholar]
- Larson, R. S., Corbi, A. L., Berman, L., & Springer, T. (1989). Primary structure of the leukocyte function‐associated molecule‐1 alpha subunit: An integrin with an embedded domain defining a protein superfamily. The Journal of Cell Biology, 108(2), 703–712. 10.1083/jcb.108.2.703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, L. F. (2016). Cell surface receptors for CCN proteins. Journal of Cell Communication and Signaling, 10(2), 121–127. 10.1007/s12079-016-0324-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumonnier, Y., Karsten, C. M., & Kohl, J. (2017). Novel insights into the expression pattern of anaphylatoxin receptors in mice and men. Molecular Immunology, 89, 44–58. 10.1016/j.molimm.2017.05.019 [DOI] [PubMed] [Google Scholar]
- Le Friec, G., Sheppard, D., Whiteman, P., Karsten, C. M., Shamoun, S. A., Laing, A., … Kemper, C. (2012). The CD46‐Jagged1 interaction is critical for human TH1 immunity. Nature Immunology, 13(12), 1213–1221. 10.1038/ni.2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.‐O., Bankston, L. A., Arnaout, M. A., & Liddington, R. C. (1995). Two conformations of the integrin A‐domain (I‐domain): A pathway for activation? Structure, 3(12), 1333–1340. 10.1016/S0969-2126(01)00271-4 [DOI] [PubMed] [Google Scholar]
- Li, M. F., Sui, Z. H., & Sun, L. (2017). A teleost CD46 is involved in the regulation of complement activation and pathogen infection. Scientific Reports, 7(1), 1–13, 15028. 10.1038/s41598-017-15124-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, W., Lin, J. X., & Leonard, W. J. (2011). IL‐2 family cytokines: New insights into the complex roles of IL‐2 as a broad regulator of T helper cell differentiation. Current Opinion in Immunology, 23(5), 598–604. 10.1016/j.coi.2011.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, W., Lin, J. X., & Leonard, W. J. (2013). Interleukin‐2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity, 38(1), 13–25. 10.1016/j.immuni.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, N. X. Y., Kaczmarek, A., Hoque, A., Davie, E., Ngoei, K. R. W., Morrison, K. R., … Petersen, J. (2020). mTORC1 directly inhibits AMPK to promote cell proliferation under nutrient stress. Nature Metabolism, 2(1), 41–49. 10.1038/s42255-019-0157-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszewski, M. K., & Kemper, C. (2019). Complement in motion: The evolution of CD46 from a complement regulator to an orchestrator of normal cell physiology. Journal of Immunology, 203(1), 3–5. 10.4049/jimmunol.1900527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszewski, M. K., Kolev, M., Le Friec, G., Leung, M., Bertram, P. G., Fara, A. F., … Kemper, C. (2013). Intracellular complement activation sustains T cell homeostasis and mediates effector differentiation. Immunity, 39(6), 1143–1157. 10.1016/j.immuni.2013.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luengo, A., Gui, D. Y., & Vander Heiden, M. G. (2017). Targeting metabolism for cancer therapy. Cell Chemical Biology, 24(9), 1161–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, E. H., Verway, M. J., Johnson, R. M., Roy, D. G., Steadman, M., Hayes, S., … Jones, R. G. (2019). Metabolic profiling using stable isotope tracing reveals distinct patterns of glucose utilization by physiologically activated CD8(+) T cells. Immunity, 51(5), 856–870e855. 10.1016/j.immuni.2019.09.003 [DOI] [PubMed] [Google Scholar]
- Masopust, D., & Schenkel, J. M. (2013). The integration of T cell migration, differentiation and function. Nature Reviews. Immunology, 13(5), 309–320. 10.1038/nri3442 [DOI] [PubMed] [Google Scholar]
- Meller, J., Rogozin, I. B., Poliakov, E., Meller, N., Bedanov‐Pack, M., Plow, E. F., … Byzova, T. V. (2015). Emergence and subsequent functional specialization of kindlins during evolution of cell adhesiveness. Molecular Biology of the Cell, 26(4), 786–796. 10.1091/mbc.E14-08-1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merle, N. S., Church, S. E., Fremeaux‐Bacchi, V., & Roumenina, L. T. (2015). Complement system part I—Molecular mechanisms of activation and regulation. Frontiers in Immunology, 6, 262. 10.3389/fimmu.2015.00262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merle, N. S., Noe, R., Halbwachs‐Mecarelli, L., Fremeaux‐Bacchi, V., & Roumenina, L. T. (2015). Complement system part II: Role in immunity. Frontiers in Immunology, 6, 257. 10.3389/fimmu.2015.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, D. J., Hemmrich, G., Ball, E. E., Hayward, D. C., Khalturin, K., Funayama, N., … Bosch, T. C. (2007). The innate immune repertoire in cnidaria—Ancestral complexity and stochastic gene loss. Genome Biology, 8(4), 1–13, R59. 10.1186/gb-2007-8-4-r59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa, S., Azumi, K., & Nonaka, M. (2001). Cloning and characterization of integrin alpha subunits from the solitary ascidian, Halocynthia roretzi . Journal of Immunology, 166(3), 1710–1715. 10.4049/jimmunol.166.3.1710 [DOI] [PubMed] [Google Scholar]
- Monks, C. R., Freiberg, B. A., Kupfer, H., Sciaky, N., & Kupfer, A. (1998). Three‐dimensional segregation of supramolecular activation clusters in T cells. Nature, 395(6697), 82–86. 10.1038/25764 [DOI] [PubMed] [Google Scholar]
- Moroishi, T., Hansen, C. G., & Guan, K. L. (2015). The emerging roles of YAP and TAZ in cancer. Nature Reviews. Cancer, 15(2), 73–79. 10.1038/nrc3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin, P., Bergman, P., Zhang, B., Triantafellow, E., Wang, H., Nyfeler, B., … Murphy, L. O. (2009). Bidirectional transport of amino acids regulates mTOR and autophagy. Cell, 136(3), 521–534. 10.1016/j.cell.2008.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka, M. (2014). Evolution of the complement system. Sub‐Cellular Biochemistry, 80, 31–43. 10.1007/978-94-017-8881-6_3 [DOI] [PubMed] [Google Scholar]
- Nonaka, M., & Kimura, A. (2006). Genomic view of the evolution of the complement system. Immunogenetics, 58(9), 701–713. 10.1007/s00251-006-0142-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Garra, A., & Vieira, P. (2007). T(H)1 cells control themselves by producing interleukin‐10. Nature Reviews. Immunology, 7(6), 425–428. 10.1038/nri2097 [DOI] [PubMed] [Google Scholar]
- Oxvig, C., & Springer, T. A. (1998). Experimental support for a beta‐propeller domain in integrin alpha‐subunits and a calcium binding site on its lower surface. Proceedings of the National Academy of Sciences of the United States of America, 95(9), 4870–4875. 10.1073/pnas.95.9.4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancer, Z., Kruse, M., Muller, I., & Muller, W. E. (1997). On the origin of Metazoan adhesion receptors: Cloning of integrin alpha subunit from the sponge Geodia cydonium. Molecular Biology and Evolution, 14(4), 391–398. 10.1093/oxfordjournals.molbev.a025775 [DOI] [PubMed] [Google Scholar]
- Perucha, E., Melchiotti, R., Bibby, J. A., Wu, W., Frederiksen, K. S., Roberts, C. A., … Cope, A. P. (2019). The cholesterol biosynthesis pathway regulates IL‐10 expression in human Th1 cells. Nature Communications, 10(1), 1–13, 498. 10.1038/s41467-019-08332-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phieler, J., Garcia‐Martin, R., Lambris, J. D., & Chavakis, T. (2013). The role of the complement system in metabolic organs and metabolic diseases. Seminars in Immunology, 25(1), 47–53. 10.1016/j.smim.2013.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo, S., Dupont, S., & Cordenonsi, M. (2014). The biology of YAP/TAZ: Hippo signaling and beyond. Physiological Reviews, 94(4), 1287–1312. 10.1152/physrev.00005.2014 [DOI] [PubMed] [Google Scholar]
- Pierschbacher, M. D., & Ruoslahti, E. (1984). Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature, 309(5963), 30–33. 10.1038/309030a0 [DOI] [PubMed] [Google Scholar]
- Ralser, M. (2018). An appeal to magic? The discovery of a non‐enzymatic metabolism and its role in the origins of life. The Biochemical Journal, 475(16), 2577–2592. 10.1042/BCJ20160866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, R. C., Kemper, C., Leung, M., & Atkinson, J. P. (2002). Characterization of human membrane cofactor protein (MCP; CD46) on spermatozoa. Molecular Reproduction and Development, 62(4), 534–546. 10.1002/mrd.10144 [DOI] [PubMed] [Google Scholar]
- Rintoul, R. C., Buttery, R. C., Mackinnon, A. C., Wong, W. S., Mosher, D., Haslett, C., & Sethi, T. (2002). Cross‐linking CD98 promotes integrin‐like signaling and anchorage‐independent growth. Molecular Biology of the Cell, 13(8), 2841–2852. 10.1091/mbc.01-11-0530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, B., Davidson, B., MacMaster, G., Lockhart, V., Ma, E., Wallace, S. S., & Swalla, B. J. (2007). A complement response may activate metamorphosis in the ascidian Boltenia villosa . Development Genes and Evolution, 217(6), 449–458. 10.1007/s00427-007-0157-0 [DOI] [PubMed] [Google Scholar]
- Ruegg, C., & Alghisi, G. C. (2010). Vascular integrins: Therapeutic and imaging targets of tumor angiogenesis. Recent Results in Cancer Research, 180, 83–101. 10.1007/978-3-540-78281-0_6 [DOI] [PubMed] [Google Scholar]
- Santinon, G., Pocaterra, A., & Dupont, S. (2016). Control of YAP/TAZ activity by metabolic and nutrient‐sensing pathways. Trends in Cell Biology, 26(4), 289–299. 10.1016/j.tcb.2015.11.004 [DOI] [PubMed] [Google Scholar]
- Sasaki, K., Tsuji, T., Jinushi, T., Matsuzaki, J., Sato, T., Chamoto, K., … Nishimura, T. (2003). Differential regulation of VLA‐2 expression on Th1 and Th2 cells: A novel marker for the classification of Th subsets. International Immunology, 15(6), 701–710. 10.1093/intimm/dxg066 [DOI] [PubMed] [Google Scholar]
- Sebe‐Pedros, A., Roger, A. J., Lang, F. B., King, N., & Ruiz‐Trillo, I. (2010). Ancient origin of the integrin‐mediated adhesion and signaling machinery. Proceedings of the National Academy of Sciences of the United States of America, 107(22), 10142–10147. 10.1073/pnas.1002257107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebe‐Pedros, A., & Ruiz‐Trillo, I. (2010). Integrin‐mediated adhesion complex: Cooption of signaling systems at the dawn of Metazoa. Communicative & Integrative Biology, 3(5), 475–477. 10.4161/cib.3.5.12603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi, R., Fujito, N. T., & Nonaka, M. (2012). Evolution of the thioester‐containing proteins (TEPs) of the arthropoda, revealed by molecular cloning of TEP genes from a spider, Hasarius adansoni . Developmental and Comparative Immunology, 36(2), 483–489. 10.1016/j.dci.2011.05.003 [DOI] [PubMed] [Google Scholar]
- Sekine, H., Takahashi, M., Iwaki, D., & Fujita, T. (2013). The role of MASP‐1/3 in complement activation. Advances in Experimental Medicine and Biology, 735, 41–53. 10.1007/978-1-4614-4118-2_3 [DOI] [PubMed] [Google Scholar]
- Sena, L. A., Li, S., Jairaman, A., Prakriya, M., Ezponda, T., Hildeman, D. A., … Chandel, N. S. (2013). Mitochondria are required for antigen‐specific T cell activation through reactive oxygen species signaling. Immunity, 38(2), 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeie, J. M., Fingert, J. H., Russell, S. R., Stone, E. M., & Mullins, R. F. (2010). Complement component C5a activates ICAM‐1 expression on human choroidal endothelial cells. Investigative Ophthalmology & Visual Science, 51(10), 5336–5342. 10.1167/iovs.10-5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. L. (1998). Shark complement: An assessment. Immunological Reviews, 166, 67–78. [DOI] [PubMed] [Google Scholar]
- Stark, K. A., Yee, G. H., Roote, C. E., Williams, E. L., Zusman, S., & Hynes, R. O. (1997). A novel alpha integrin subunit associates with betaPS and functions in tissue morphogenesis and movement during Drosophila development. Development, 124(22), 4583–4594. [DOI] [PubMed] [Google Scholar]
- Stummvoll, G. H., DiPaolo, R. J., Huter, E. N., Davidson, T. S., Glass, D., Ward, J. M., & Shevach, E. M. (2008). Th1, Th2, and Th17 effector T cell‐induced autoimmune gastritis differs in pathological pattern and in susceptibility to suppression by regulatory T cells. Journal of Immunology, 181(3), 1908–1916. 10.4049/jimmunol.181.3.1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupack, D. G., & Cheresh, D. A. (2002). Get a ligand, get a life: Integrins, signaling and cell survival. Journal of Cell Science, 115(Pt 19), 3729–3738. 10.1242/jcs.00071 [DOI] [PubMed] [Google Scholar]
- Sun, G., Li, H., Wang, Y., Zhang, B., & Zhang, S. (2010). Zebrafish complement factor H and its related genes: Identification, evolution, and expression. Functional & Integrative Genomics, 10(4), 577–587. 10.1007/s10142-010-0182-3 [DOI] [PubMed] [Google Scholar]
- Terado, T., Okamura, K., Ohta, Y., Shin, D. H., Smith, S. L., Hashimoto, K., … Nonaka, M. (2003). Molecular cloning of C4 gene and identification of the class III complement region in the shark MHC. Journal of Immunology, 171(5), 2461–2466. 10.4049/jimmunol.171.5.2461 [DOI] [PubMed] [Google Scholar]
- Tiwari, S., Askari, J. A., Humphries, M. J., & Bulleid, N. J. (2011). Divalent cations regulate the folding and activation status of integrins during their intracellular trafficking. Journal of Cell Science, 124(Pt 10), 1672–1680. 10.1242/jcs.084483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchieri, G. (2007). Interleukin‐10 production by effector T cells: Th1 cells show self control. The Journal of Experimental Medicine, 204(2), 239–243. 10.1084/jem.20070104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura, A., Shida, K., Kitamura, M., Nomura, M., Takeda, J., Tanaka, H., … Seya, T. (1998). Molecular cloning of a murine homologue of membrane cofactor protein (CD46): Preferential expression in testicular germ cells. The Biochemical Journal, 330(Pt 1), 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga, G., Nippe, N., Balkow, S., Peters, T., Wild, M. K., Seeliger, S., … Grabbe, S. (2010). LFA‐1 contributes to signal I of T‐cell activation and to the production of T(h)1 cytokines. The Journal of Investigative Dermatology, 130(4), 1005–1012. 10.1038/jid.2009.398 [DOI] [PubMed] [Google Scholar]
- Verma, N. K., Dempsey, E., Long, A., Davies, A., Barry, S. P., Fallon, P. G., … Kelleher, D. (2012). Leukocyte function‐associated antigen‐1/intercellular adhesion molecule‐1 interaction induces a novel genetic signature resulting in T‐cells refractory to transforming growth factor‐beta signaling. The Journal of Biological Chemistry, 287(32), 27204–27216. 10.1074/jbc.M112.376616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, N. K., Fazil, M. H., Ong, S. T., Chalasani, M. L., Low, J. H., Kottaiswamy, A., … Kelleher, D. (2016a). Correction: LFA‐1/ICAM‐1 ligation in human T cells promotes Th1 polarization through a GSK3beta signaling‐dependent notch pathway. Journal of Immunology, 197(5), 2039–2040. 10.4049/jimmunol.1601160 [DOI] [PubMed] [Google Scholar]
- Verma, N. K., Fazil, M. H., Ong, S. T., Chalasani, M. L., Low, J. H., Kottaiswamy, A., … Kelleher, D. (2016b). LFA‐1/ICAM‐1 ligation in human T cells promotes Th1 polarization through a GSK3beta signaling‐dependent notch pathway. Journal of Immunology, 197(1), 108–118. 10.4049/jimmunol.1501264 [DOI] [PubMed] [Google Scholar]
- Werner, E., & Werb, Z. (2002). Integrins engage mitochondrial function for signal transduction by a mechanism dependent on Rho GTPases. The Journal of Cell Biology, 158(2), 357–368. 10.1083/jcb.200111028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, E. E., & Kemper, C. (2019). Complement and T cell metabolism: Food for thought. Immunometabolism, 1(T Cell Metabolic Reprogramming), 1–18, e190006. http://doi:10.20900/immunometab20190006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, E. E., Kolev, M., & Kemper, C. (2018). Complement and the regulation of T cell responses. Annual Review of Immunology, 36, 309–338. 10.1146/annurev-immunol-042617-053245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, E. E., Kunz, N., & Kemper, C. (2020). Complement and human T cell metabolism: Location, location, location. Immunological Reviews, 295, 68–81. 10.1111/imr.12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox, M., DiAntonio, A., & Leptin, M. (1989). The function of PS integrins in Drosophila wing morphogenesis. Development, 107(4), 891–897. [DOI] [PubMed] [Google Scholar]
- Winograd‐Katz, S. E., Fassler, R., Geiger, B., & Legate, K. R. (2014). The integrin adhesome: From genes and proteins to human disease. Nature Reviews. Molecular Cell Biology, 15(4), 273–288. [DOI] [PubMed] [Google Scholar]
- Woznica, A., Haeussler, M., Starobinska, E., Jemmett, J., Li, Y., Mount, D., & Davidson, B. (2012). Initial deployment of the cardiogenic gene regulatory network in the basal chordate, Ciona intestinalis . Developmental Biology, 368(1), 127–139. 10.1016/j.ydbio.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, H., Nho, R. S., Kahm, J., Kleidon, J., & Henke, C. A. (2004). Focal adhesion kinase is upstream of phosphatidylinositol 3‐kinase/Akt in regulating fibroblast survival in response to contraction of type I collagen matrices via a beta 1 integrin viability signaling pathway. The Journal of Biological Chemistry, 279(31), 33024–33034. 10.1074/jbc.M313265200 [DOI] [PubMed] [Google Scholar]
- Yamamoto, H., Fara, A. F., Dasgupta, P., & Kemper, C. (2013). CD46: The ‘multitasker’ of complement proteins. The International Journal of Biochemistry & Cell Biology, 45(12), 2808–2820. 10.1016/j.biocel.2013.09.016 [DOI] [PubMed] [Google Scholar]
- Yang, L., Hou, Y., Yuan, J., Tang, S., Zhang, H., Zhu, Q., … Liu, M. (2015). Twist promotes reprogramming of glucose metabolism in breast cancer cells through PI3K/AKT and p53 signaling pathways. Oncotarget, 6(28), 25755–25769. 10.18632/oncotarget.4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee, G. H., & Hynes, R. O. (1993). A novel, tissue‐specific integrin subunit, beta nu, expressed in the midgut of Drosophila melanogaster . Development, 118(3), 845–858. [DOI] [PubMed] [Google Scholar]
- Zent, R., Fenczik, C. A., Calderwood, D. A., Liu, S., Dellos, M., & Ginsberg, M. H. (2000). Class‐ and splice variant‐specific association of CD98 with integrin beta cytoplasmic domains. The Journal of Biological Chemistry, 275(7), 5059–5064. 10.1074/jbc.275.7.5059 [DOI] [PubMed] [Google Scholar]
- Zutter, M. M., & Edelson, B. T. (2007). The alpha2beta1 integrin: A novel collectin/C1q receptor. Immunobiology, 212(4–5), 343–353. 10.1016/j.imbio.2006.11.013 [DOI] [PubMed] [Google Scholar]


