Abstract
Background and Objective
Long non‐coding RNAs (lncRNAs) can act as competing endogenous RNAs (ceRNAs) to compete for micro‐RNAs (miRNAs) in regulation of downstream genes, various biological functions and diseases. Yet, the expression and regulation of lncRNAs in periodontitis are not fully understood. The objective of the study was to identify potential genes (lncRNA, messenger RNA [mRNA] and miRNA) involved in periodontitis, construct lncRNA‐miRNA‐mRNA ceRNA networks, explore gene functions and validate gene expressions.
Material and Methods
The data sets for the lncRNA, mRNA and miRNA expression profiles in gingival samples from periodontally healthy subjects and chronic periodontitis patients were obtained from the Gene Expression Omnibus. The differentially expressed lncRNAs (DElncRNAs), mRNAs (DEmRNAs) and miRNAs (DEmiRNAs) were identified, and ceRNA networks were then constructed. The expression of DElncRNAs and DEmRNAs was examined by quantitative real‐time polymerase chain reaction (qPCR). Moreover, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were performed for exploring the potential functions and biological pathways.
Results
The GSE80715 and GSE54710 data sets were retrieved. Subsequently, 26 DElncRNAs, 436 DEmRNAs and 12 DEmiRNAs were identified (|fold change| ≥2, adjusted p < 0.05). Further bioinformatics analysis contributed to establishment of the ceRNA networks, which consisted of 10 DElncRNAs, 11 DEmiRNAs and 83 DEmRNAs. Notably, the qPCR results showed a marked decrease in the expression of lncRNA H19 and two mRNAs (NOS1 and MAPT) which further supported the identified ceRNA network. The GO results revealed that the up‐regulated mRNAs were significantly enriched in inflammatory processes, whilst the down‐regulated mRNAs were enriched in cellular potentials.
Conclusion
Non‐coding RNAs are critically involved in the regulatory mechanisms in the pathogenesis of periodontitis. Further study is warranted to investigate the specific underlying genetic traits and networks.
Keywords: gene expression and regulatory networks, non‐protein‐coding RNA, pathogenesis, periodontitis
1. INTRODUCTION
Periodontitis is a common inflammatory disease of tooth‐supporting tissues worldwide that is associated with biofilms, and it leads to progressive destruction of periodontium and eventual tooth loss.1 Yet, periodontitis is closely associated with a variety of systemic diseases and disorders (eg, diabetes mellitus, cardiovascular disease and Alzheimer's disease) and thus poses serious risks to general health and well‐being.2, 3, 4
The gingiva plays an important role as a mechanical and biological barrier against invading pathogens and maintains effective innate immune responses for tissue homeostasis and periodontal health.5 Initially, the inflammation is limited to localised gingival tissues, whilst persistent and dysregulated immuno‐inflammatory responses crucially account for destruction of the entire periodontium.6 It is important to note that the pathogenesis of periodontitis can be significantly modulated by genetic and epigenetic factors via a pathogen‐induced host immune response.7
RNA is the most versatile biomolecule and is generally classified as protein‐coding (coding) or non‐protein‐coding (non‐coding) RNA.8 Non‐coding RNAs are not translated into proteins but typically regulate gene expression. They can be further subdivided into two major groups based on length: short non‐coding RNAs and long non‐coding RNAs (lncRNAs). MicroRNAs (miRNAs), with a length of 20 to 23 nucleotides, are short non‐coding RNAs and function as post‐transcriptional repressors by binding to messenger RNA (mRNA), which results in the silencing of a specific target gene.9 Several microarray studies have reported that miRNAs are differentially expressed in healthy gingiva and in gingiva with periodontitis, which indicates that miRNAs play critical roles in periodontal homeostasis and pathology of the gingival tissues.10, 11
LncRNAs, which have received increasing attention in recent years, are a large and diverse class of transcribed RNA molecules of more than 200 nucleotides in length. They are reported to be associated with the pathogenesis of various diseases and have the capacity to regulate gene expression at the transcriptional and post‐transcriptional levels.12 An accumulating body of evidence indicates that lncRNAs are differentially expressed in gingival tissues from periodontally healthy patients and from those with periodontitis, which indicates the crucial roles of lncRNAs in the pathogenesis of periodontitis.13, 14, 15, 16 It has also been reported that lncRNA can be a genetic risk factor for periodontitis.17 In addition to regulating mRNAs via independent mechanisms, lncRNAs can act as competing endogenous RNAs (ceRNAs) by competitively binding to miRNAs via miRNA response elements, thereby attenuating the down‐regulation of mRNA expression by miRNA and indirectly regulating mRNA expression.18 LncRNA‐mediated ceRNA interactions have been identified in various cancers and inflammatory diseases.18 However, similar studies on periodontitis are rare, and a comprehensive analysis of lncRNAs and miRNAs related to the pathogenesis of periodontitis is still lacking.
This bioinformatics study aimed to identify differentially expressed lncRNAs, mRNAs and miRNAs between healthy gingival tissues and those with chronic periodontitis, explore their functions, construct lncRNA‐miRNA‐mRNA ceRNA networks and validated genes of interest using quantitative real‐time polymerase chain reaction (qPCR).
2. MATERIALS AND METHODS
2.1. Acquisition and processing of data sets
To identify coding and non‐coding RNAs that potentially involved in the pathogenesis of periodontitis, the periodontitis data sets were searched within the data sets registered in the Gene Expression Omnibus (GEO) of the NCBI (www.ncbi.nlm.nih.gov/geo/). The data sets comprising the expression of mRNA and lncRNA or miRNA in gingival samples from patients with chronic periodontitis (the diseased group) and in gingival samples from healthy individuals (the control group) were obtained. RNA sequencing (RNA‐seq) is suggested to have advantages over traditional microarrays, as it provides more comprehensive information about the transcript characteristics and is not limited to the known genes on the microarray.19 Therefore, RNA‐seq data sets were prioritised for this study.
The data set GSE80715, which is the only RNA‐seq data set that contains both mRNA and lncRNA expression profiles, was obtained. It comprised one mixed healthy sample from 10 healthy gingival tissues from nine periodontally healthy patients and one mixed diseased sample from 10 diseased periodontal tissues from seven patients with chronic periodontitis. The diseased sites had a probing depth (PD) ≥4 mm, a clinical attachment level (CAL) ≥4 mm and bleeding on probing (BOP). The expression profile was obtained from the Illumina HiSeq 2000 platform. The miRNA expression data were derived from GSE54710. The data set comprised 200 gingival tissue samples from 86 patients with aggressive or chronic periodontitis. Of the 200 samples, 128 (98 diseased and 30 healthy sites) were from patients with chronic periodontitis, and 72 (61 diseased and 11 healthy sites) were from patients with aggressive periodontitis. All diseased sites had a PD >4 mm, CAL ≥3 mm and BOP. The miRNA expression profile was obtained from the Agilent Human microRNA microarray (v16.0) platform.
2.2. Differential analysis
Differential gene analyses were carried out using the Linear Models for Microarray (limma) package of the R software. For the GSE80715 RNA‐seq data, the differentially expressed genes were re‐annotated and classified into protein‐coding genes (DEmRNAs) and non‐coding genes (DElncRNAs) according to the Genecode database (https://www.gencodegenes.org/). Only samples from patients with chronic periodontitis in the GSE54710 data set were included in the analysis. Differentially expressed miRNAs were identified between the diseased and healthy groups. MiRNAs with a |fold change| ≥2 and an adjusted P value <0.05 were considered to be significantly expressed (DEmiRNAs).
2.3. Prediction of lncRNA‐miRNA‐mRNA interactions and construction of ceRNA networks
The sequences of the DEmiRNAs were acquired from miRBase (www.mirbase.org). The interactions between mRNA‐miRNA and lncRNA‐miRNA were then predicted with miRanda (http://www.microrna.org/). The predicted mRNAs and lncRNAs that overlapped in the DEmRNAs and DElncRNAs were selected to construct the lncRNA‐miRNA‐mRNA ceRNA network with the Cytoscape software (v3.7.2; www.cytoscape.org).
2.4. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analyses
To explore the biological functions of DEmRNAs and DElncRNAs in the ceRNA network, GO and KEGG pathway analyses were performed using the R package ClusterProfiler.20 GO was performed to analyse the main function of the differentially expressed genes. Pathway analysis was performed to determine the significant biological pathways of the differential genes according to the KEGG database. A Benjamini‐Hochberg adjusted P value of 0.05 was set as the cut‐off value for significance.
2.5. Validation of DEmRNAs and DElncRNAs by qPCR
Human gingival tissue samples were collected from three healthy individuals and three patients with chronic periodontitis. All subjects were systemically healthy, non‐smoking adults (age ≥18 years). Healthy sites were defined as those with no periodontal inflammation, no BOP, no attachment loss and a PD ≤3 mm. The sites with chronic periodontitis had a PD ≥4 mm, CAL ≥3 mm and BOP. The gingival specimens (approximately 2 × 2 mm) were obtained from the marginal gingiva during periodontal flap surgery, rinsed with phosphate‐buffered saline solution, immediately stored in RNA‐later solution (Thermo Fisher Scientific, Waltham, USA) overnight at 4°C and thereafter stored at −80°C for subsequent RNA isolation. Total RNA was extracted using the RNeasy Cell Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions, and 1 μg of total RNA isolated from each sample was reverse‐transcribed into cDNA using Superscript III Reverse Transcriptase (Thermo Fisher Scientific). The primers of the target genes were designed with NCBI Primer‐BLAST and are listed in Table S1. qPCR was carried out using the Step One Real‐Time PCR System (Applied Biosystems, Grand Island, NY) with SYBR Green Master Mix (Applied Biosystems). The target gene copy numbers were normalised against the endogenous housekeeping gene glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH).
3. RESULTS
3.1. Identification of DElncRNAs, DEmRNAs and DEmiRNAs in periodontitis
A flow chart of the construction of the ceRNA network is shown in Figure 1. In data set GSE80715, 436 mRNAs and 26 lncRNAs were identified as being differentially expressed with a |fold change| ≥2 and an adjusted P value <0.05. Specifically, 379 mRNAs and 21 lncRNAs were up‐regulated, whilst 57 mRNAs and 5 lncRNAs were down‐regulated in gingival tissues with chronic periodontitis relative to normal tissues. The top five up‐regulated and top five down‐regulated mRNAs and lncRNAs are shown in Table 1. The full list of differentially expressed mRNAs and lncRNAs is shown in Tables S2 and S3.
FIGURE 1.
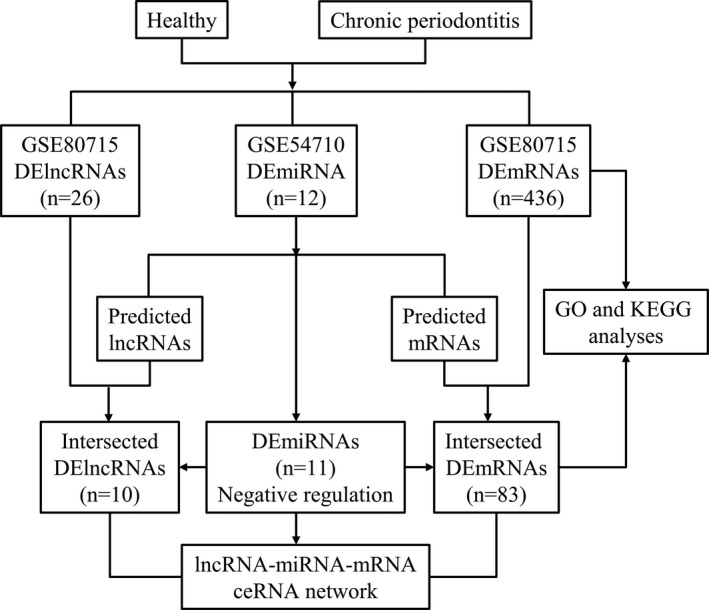
Flow chart of the construction of ceRNA networks
TABLE 1.
Top up‐regulated and down‐regulated mRNAs and lncRNAs identified from GSE80715
| Ensemble ID | Gene symbol | Log2 (fold change) | Adjusted P value |
|---|---|---|---|
| Up‐regulated mRNAs | |||
| ENSG00000108342 | CSF3 | 7.50 | 5.85E‐21 |
| ENSG00000182759 | MAFA | 7.30 | 8.19E‐09 |
| ENSG00000117322 | CR2 | 6.12 | 1.53E‐07 |
| ENSG00000241566 | IGKV2D‐24 | 6.00 | 1.06E‐08 |
| ENSG00000211899 | IGHM | 5.68 | 2.18E‐15 |
| Down‐regulated mRNAs | |||
| ENSG00000165953 | SERPINA12 | −6.03 | 1.30E‐04 |
| ENSG00000102891 | MT4 | −5.75 | 5.45E‐04 |
| ENSG00000172867 | KRT2 | −5.45 | 5.41E‐15 |
| ENSG00000134765 | DSC1 | −4.48 | 3.61E‐11 |
| ENSG00000204538 | PSORS1C2 | −4.44 | 7.22E‐06 |
| Up‐regulated lncRNAs | |||
| ENSG00000240666 | MME‐AS1 | 38.83 | 2.05E‐09 |
| ENSG00000227619 | RP11‐492E3.2 | 19.91 | 2.24E‐04 |
| ENSG00000240040 | AC096579.13 | 19.87 | 6.25E‐11 |
| ENSG00000226777 | KIAA0125 | 15.46 | 2.21E‐04 |
| ENSG00000253364 | RP11‐731F5.2 | 14.96 | 1.50E‐09 |
| Down‐regulated lncRNAs | |||
| ENSG00000130600 | H19 | 0.02 | 3.95E‐15 |
| ENSG00000263698 | RP11‐408H20.3 | 0.04 | 1.09E‐09 |
| ENSG00000265888 | RP11‐408H20.2 | 0.05 | 3.47E‐06 |
| ENSG00000261786 | RP4‐555D20.2 | 0.11 | 3.55E‐04 |
| ENSG00000246100 | LINC00900 | 0.14 | 2.30E‐04 |
Abbreviations: lncRNA, long non‐coding RNA; mRNA, messenger RNA.
The data set GSE54710, which contained the miRNA expression profiles of healthy gingival tissue and gingival tissue with periodontitis, was obtained. Specifically, 71 gingival samples (18 healthy and 53 with chronic periodontitis) were included in the subsequent analysis. The results of differential analysis showed that six miRNAs (hsa‐miR‐451, hsa‐miR‐486‐5p, hsa‐miR‐575, hsa‐miR‐4299, hsa‐miR‐4306 and hsa‐miR‐3917) were up‐regulated and six miRNAs (hsa‐miR‐141, hsa‐miR‐210, hsa‐miR‐1246, hsa‐miR‐1260, hsa‐miR‐205 and hsa‐miR‐1260b) were down‐regulated in the diseased gingival tissues relative to the normal tissues (|fold change| ≥2 and adjusted P value <0.05) (Table 2). A volcano plot which visually demonstrate the distribution of differentially expressed miRNAs was shown in Figure S1.
TABLE 2.
Differentially expressed miRNAs identified from GSE54710 and their DElncRNA and DEmRNA targets in GSE80715
| DEmiRNAs | Log2(fold change) | Adjusted P value | DElncRNA targets | DEmRNA targets |
|---|---|---|---|---|
| Up‐regulation | ||||
| hsa‐miR−451 | 1.82 | 2.08E−04 | N/A | N/A |
| hsa‐miR−486‐5p | 1.60 | 1.78E−03 | H19 | NOS1 |
| hsa‐miR−575 | 1.13 | 6.80E−03 | RP4‐555D20.2 | ISL2, MAPT, NPR3, SHISA9, SFRP1 |
| hsa‐miR−4299 | 1.08 | 8.37E−04 |
H19 RP4‐555D20.2 |
FREM2, ISL2, MAPT, NOS1, SHISA9 |
| hsa‐miR−4306 | 1.07 | 2.67E−04 | RP4‐555D20.2 | FREM2, ISL2, NOS1, NPR3, SHISA9 |
| hsa‐miR−3917 | 1.13 | 7.93E−06 | RP4‐555D20.2 | MAPT, SFRP1 |
| Down‐regulation | ||||
| hsa‐miR−141 | −1.19 | 1.88E−06 |
KIAA0125 SLC7A11‐AS1 TRHDE‐AS1 |
ABCA13, ADAMTS20, BIRC3, C3, CSF3R, CXCL1, CXCL5, FCRL5, FCRLA, FHAD1, GALNT5, GPC6, HAS2, HS3ST1, HS6ST2, HSD11B1, HTR7, ICAM1, IL1A, IL24, IL7R, ITGB3, LAMC2, MME, NCEH1, NRCAM, OLR1, PHLDA1, PLAC8, PPP1R15A, RGS4, SELE, SERPINB9, SLC12A8, SLC16A3, SLC5A1, SOD2, ST3GAL1, STRIP2, TINAGL1, TNIP3, TREM1, TRHDE, TUSC3 |
| hsa‐miR−210 | −1.14 | 2.17E−08 |
KIAA0125 LIPE‐AS1 RP11‐270 M14.5 TRHDE‐AS1 |
C3, CCL18, CR1, GALNT5, HS3ST1, KYNU, LYN, POU2AF1, RGS4, SLC12A8, SLC16A3, TMPRSS4, TNIP3 |
| hsa‐miR−1246 | −1.18 | 2.57E−04 |
LIPE‐AS1 RP11‐66B24.4 SLC7A11‐AS1 TRHDE‐AS1 |
ADAM28, ADAMTS20, ALDH1A3, BIRC3, C3, CLCA4, CLMP, CYP24A1, HS3ST1, HSD11B1, HTR7, INHBA, LAX1, MME, NNMT, OLR1, PHLDA1, POU2AF1, SAA2, SERPINB9, SLC12A8, SLC13A5, SLC7A11, SOD2, STRIP2, TINAGL1, TRHDE, TUSC3, XDH |
| hsa‐miR−1260 | −1.21 | 7.50E−03 |
CTD−2369P2.8 KIAA0125 RP11‐66B24.4 RP3‐467 K16.2 SLC7A11‐AS1 TRHDE‐AS1 |
ALDH1A3, C3, CCDC88B, CD79A, CLMP, CR1, CSF3R, CXCL5, CYP4X1, DERL3, FCGR3B, FCRLA, FHAD1, FPR1, GPR110, GRAMD1B, HAS2, HS3ST1, ICAM1, IL24, INHBA, LAMC2, LAX1, LTF, LYN, MZB1, OSM, PLAC8, POU2AF1, PPP1R15A, RGS4, SAA1, SAA2, SERPINB9, SLC13A5, SLC16A3, SOCS3, SOD2, STRIP2, TNFRSF10C, TRHDE |
| hsa‐miR−205* | −1.08 | 9.34E−09 |
LIPE‐AS1 RP11‐270 M14.5 RP3‐467 K16.2 SLC7A11‐AS1 TRHDE‐AS1 |
ABCA13, ADAM28, ADAMTS20, ALDH1A3, BIRC3, C3, CCL18, CLCA4, CLMP, CXCL1, CXCL5, CYP24A1, FCRL5, FPR1, GPC6, HS3ST1, HS6ST2, HSD11B1, HTR7, IL1A, IL7R, INHBA, ITGB3, KYNU, LAX1, LTF, MZB1, NCEH1, NNMT, NRCAM, OLR1, PHLDA1, PLAC8, SELE, SLC16A3, SLC5A1, SLC7A11, SOD2, ST3GAL1, STRIP2, TMPRSS4, TNFRSF10C, TNIP3, TREM1, TRHDE, XDH |
| hsa‐miR−1260b | −1.22 | 3.71E−03 |
CTD−2369P2.8 KIAA0125 SLC7A11‐AS1 TRHDE‐AS1 |
ALDH1A3, C3, CCDC88B, CD79A, CLMP, CR1, CSF3R, CXCL5, CYP4X1, DERL3, FCGR3B, FCRLA, FHAD1, FPR1, GPR110, GRAMD1B, HAS2, HS3ST1, ICAM1, IL24, INHBA, LAMC2, LAX1, LTF, LYN, MZB1, OSM, PLAC8, POU2AF1, PPP1R15A, RGS4, SAA1, SAA2, SLC13A5, SLC16A3, SOCS3, SOD2, STRIP2, TNFRSF10C, TRHDE |
Abbreviations: DElncRNA, differentially expressed long non‐coding RNA; DEmiRNA, differentially expressed microRNA; DEmRNA, differentially expressed messenger RNA.
3.2. Construction of ceRNA networks
Interactions between mRNA‐miRNA and lncRNA‐miRNA were obtained with miRanda. Target mRNAs or lncRNAs that overlapped with the 436 DEmRNAs or 26 DElncRNAs were selected. Table 2 shows the predicted DElncRNA‐DEmiRNA‐DEmRNA relationships. Finally, the lncRNA‐miRNA‐mRNA ceRNA networks consisted of 11 miRNAs (6 down‐regulated and 5 up‐regulated), 10 lncRNAs (2 down‐regulated and 8 up‐regulated) and 83 mRNAs (76 up‐regulated and 7 down‐regulated) (Figure 2).
FIGURE 2.
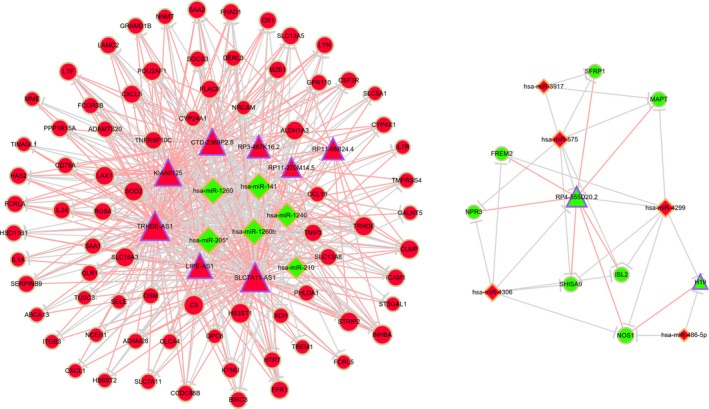
The lncRNA‐miRNA‐mRNA ceRNA networks. Red represents up‐regulation, and green represents down‐regulation. The diamonds represent miRNAs, circles represent mRNAs, and triangles represent lncRNAs. LncRNA‐mRNA pairs with ≥2 shared miRNAs were connected by red lines
3.3. GO and KEGG pathway enrichment analyses of DEmRNAs
3.3.1. DEmRNAs in GSE 80715
GO and KEGG pathway enrichment analyses were performed on 436 DEmRNAs in the GSE80715 data set. The GO results showed that DEmRNAs were enriched in all three aspects: biological process (BP), molecular function (MF) and cellular component (CC). In total, 126 BP, 11 CC and 22 MF items showed significant enrichment (P adjusted <0.05). In these three GO function domains, the humoral immune responses mediated by circulating immunoglobulin (GO:0002455, P adjusted = 8.37 × 10−119) and complement activation (GO:0006956, P adjusted = 4.47 × 10−118) were the most enriched terms of BP; the immunoglobulin complex (GO:0019814, P adjusted = 1.87 × 10−171) was the most enriched term of CC; and antigen binding (GO:0003823, P adjusted = 3.29 × 10−105) was the most enriched term of MF. In the KEGG pathway analysis, 22 pathways were found to be significantly enriched (P adjusted <0.05). Specifically, cytokine‐cytokine receptor interaction (hsa04060), tumour necrosis factor (TNF) signalling pathway (hsa04668) and interleukin (IL)‐17 signalling pathway (hsa04657) were the most enriched pathways. The top enriched GO and top 10 KEGG pathways are shown in Figure 3A,B.
FIGURE 3.
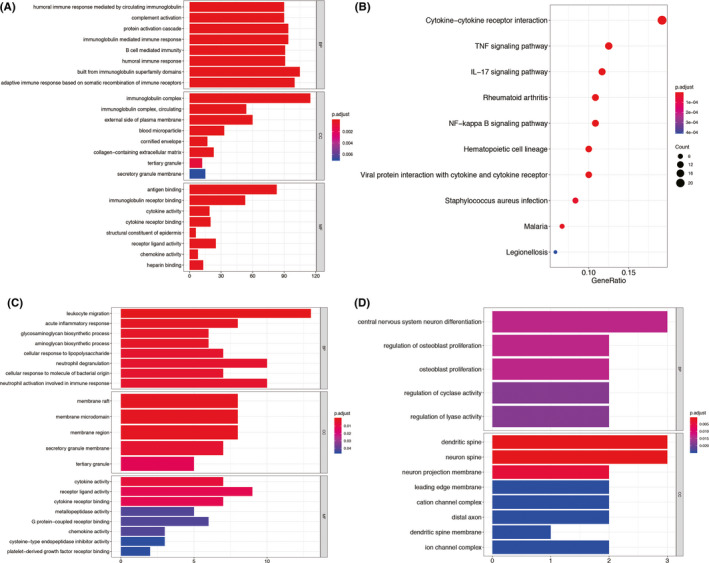
Top enriched GO annotations and KEGG pathways of DEmRNAs identified from GSE80715 and the ceRNA network. (A) Top enriched GO terms of 436 DEmRNAs in GSE80715. (B) Top 10 enriched KEGG pathways of 436 DEmRNAs in GSE80715. (C) Top enriched GO terms of 76 up‐regulated mRNAs in the ceRNA network. (D) Top enriched GO terms of 7 down‐regulated mRNAs in the ceRNA network. The horizontal axes of Figure 3A,C,D indicate the number of genes (gene count) enriched in the GO term. BP, biological process; CC, cellular component; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and genomes; MF, molecular function; NF‐kappa B, Nuclear factor‐kappa B; TNF, tumour necrosis factor
3.3.2. DEmRNAs in the ceRNA network
To further clarify the dysregulated ceRNA networks and reveal the potential role of DElncRNAs in patients with periodontitis, GO and KEGG pathway enrichment analyses were performed on the 83 DEmRNAs (76 up‐regulated and 7 down‐regulated) in the networks.
The 76 up‐regulated mRNAs were significantly enriched in 53 BP, 5 CC and 9 MF GO terms. Specifically, leukocyte migration (GO:0050900, P adjusted <0.001) and acute inflammatory responses (GO:0002526, P adjusted =0.002) were the most enriched terms of BP, membrane raft (GO:0045121, P adjusted =0.002) was the most enriched term of CC, and cytokine activity (GO:0005125, P adjusted =0.007) was the most enriched term of MF. The 7 down‐regulated mRNAs were significantly enriched in 5 BP and 45 CC GO terms. Specifically, central nervous system neuron differentiation (GO:0021953, P adjusted =0.014), regulation of osteoblast proliferation (GO:0033688, P adjusted =0.014) and osteoblast proliferation (GO:0033687, P adjusted =0.014) were the most enriched terms of BP, and dendritic spine (GO:0043197, P adjusted <0.001) was the most enriched term of CC. The top enriched GO terms are shown in Figure 3C,D. In the KEGG pathway analysis, the 76 up‐regulated mRNAs were found to be enriched in 10 pathways, which are shown in Figure S2. The down‐regulated mRNAs were only enriched in Alzheimer disease (hsa05010).
3.4. Expression levels of DElncRNAs and DEmRNAs
Gingival tissue specimens were obtained from three healthy subjects (one male and two female, mean age, 31.33 ± 4.64 years) and three patients with chronic periodontitis (one male and two female, mean age, 33.67 ± 6.80 years). We selected three DEmRNAs [nitric oxide synthase 1 (NOS1), microtubule‐associated protein tau (MAPT) and ISL LIM homeobox 2 (ISL2)] and six DElncRNAs (H19, RP11‐290L1.3, RP11‐492E3.2, RP11‐731F5.2, KIAA0125 and AC096579.13) of interest and examined their expression with qPCR. The expressions of NOS1 and MAPT were significantly down‐regulated in the periodontitis group, which was consistent with the RNA‐seq results of GSE80715. Regarding the lncRNAs, our results showed that the expression of H19 was significantly down‐regulated, and the expressions of RP11‐290L1.3 and RP11‐492E3.2 were up‐regulated in the periodontitis group, which was consistent with the RNA‐seq results of GSE80715. However, no statistically significant difference was found in the expressions of ISL2, AC096579.13, KIAA0125 and RP11‐731F5.2 between the healthy group and the periodontitis group. The qPCR results were shown in Figure 4.
FIGURE 4.
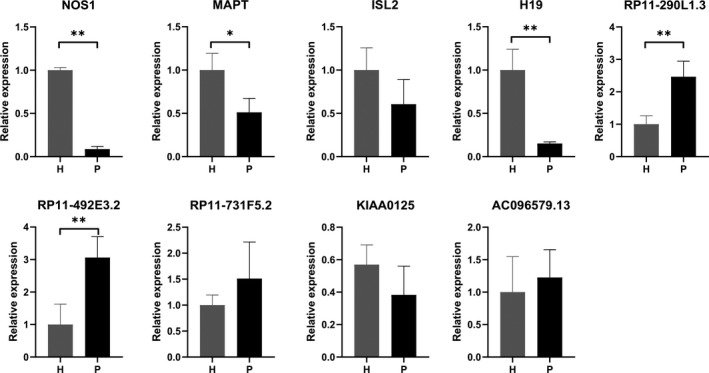
The expression levels of three DEmRNAs [nitric oxide synthase 1 (NOS1), microtubule‐associated protein tau (MAPT) and ISL LIM homeobox 2 (ISL2)] and six DElncRNAs (H19, RP11‐290L1.3, RP11‐492E3.2, RP11‐731F5.2, KIAA0125 and AC096579.13) were examined by quantitative real‐time polymerase chain reaction. Results were normalised to glyceraldehyde 3‐phosphate dehydrogenase (GAPDH). H, healthy group; P, periodontitis group. (*p < 0.05, **p < 0.01)
4. DISCUSSION
Periodontitis is a multifactorial and complex inflammatory disease characterised by progressive destruction of tooth‐supporting tissues, including gingiva, periodontal ligament, cementum and alveolar bone. Evidence suggests that epigenetic processes participate in the pathogenesis and progression of periodontitis via regulation of the pathogen‐induced host immune response, because microbiome infection alters the expression of non‐coding RNAs.7, 13, 21 Non‐coding RNAs, such as lncRNAs and miRNAs, function to regulate gene expression at transcriptional, post‐transcriptional and epigenetic levels.8, 9, 12 To better understand the potential pathogenic mechanisms of periodontitis, it is crucial to elucidate the role of non‐coding RNAs in periodontitis. This study analysed differentially expressed lncRNAs, mRNAs and miRNAs in gingival tissue from patients with chronic periodontitis, explored their functions, elucidated their interactions by constructing ceRNA networks and validated several DElncRNAs and DEmRNAs of interest by qPCR.
After re‐annotation of GSE80715, 436 mRNAs and 26 lncRNAs were identified as being differentially expressed. Among the DEmRNAs, 379 were up‐regulated and 57 were down‐regulated in diseased gingival tissues relative to normal tissues. The top five up‐regulated mRNAs were CSF3, MAFA, complement receptor (CR) 2, immunoglobulin kappa variable 2D‐24 (IGKV2D‐24) and immunoglobulin heavy constant mu (IGHM). CSF3, also known as granulocyte‐colony stimulating factor, is a key regulator of granulopoiesis and can be generated by many cell types upon pro‐inflammatory stimuli such as TNF or lipopolysaccharide. It plays a critical role in the host response to infection, which has potential implications for inflammatory and autoimmune diseases.22 Offenbacher et al.23 reported that CSF3 was associated with host‐bacteria interaction, and its expression increased significantly during induction of gingivitis. MAFA, a member of the large MAF family of transcription factors that contain a basic‐leucine zipper (b‐Zip) motif, has been reported to have great significance in the development and maturation of beta‐cells and glucose‐responsive insulin secretion.24 CR2 can be activated and modulated by bacteria in the biofilm of the periodontal pocket. The increased activation of CR2 on B cells or follicular dendritic cells can stimulate the production of inflammatory mediators and up‐regulate the immune response, which may contribute to local tissue destruction and the pathogenesis of periodontitis.25 IGKV2D‐24 and IGHM belong to the immunoglobulin family, which has anti‐inflammatory effects and exerts a wide range of immunoregulatory functions in autoimmune and inflammatory diseases.26
Based on the results of our analysis, we selected three DEmRNAs and six DElncRNAs of interests for further validation via qPCR. The expression of two mRNAs (NOS1 and MAPT) was significantly down‐regulated, in consistence with the RNA‐seq results of GSE80715. Nitric oxide (NO) is a free radical produced from l‐arginine via a family of NO synthases (NOS). Unlike NOS2, or inducible NOS (iNOS), which is involved in inflammatory processes, whereas NOS1, or neuronal NOS, is a constitutive NOS isoform that produces a low concentration of NO to function in physiological homeostatic processes.27 MAPT, a neural phosphoprotein member of the MAP family, is an important structural element with functions in assembly and stabilisation of microtubules in healthy neurons. It has been reported to be associated with neurodegenerative diseases, such as Parkinson's disease and Alzheimer's disease.28 Similarly, Guzeldemi‐Akcakanat et al.29 used microarray to detect a significant decrease in the expression of MAPT in the gingival tissue of patients with aggressive periodontitis. Yu et al.30 reported MAPT as a crosstalk gene associated with peri‐implantitis and type‐2 diabetes mellitus.
The expression of six DElncRNAs (H19, RP11‐290L1.3, RP11‐492E3.2, AC096579.13, KIAA0125 and RP11‐731F5.2) was further examined by qPCR. The results suggested that the expression of H19 significantly decreased, whilst the expressions of RP11‐290L1.3 and RP11‐492E3.2 increased, in line with the RNA‐seq results of GSE80715.
In recent years, an increasing number of studies have reported that lncRNAs function as ceRNAs (miRNA ‘sponges’) via competitive binding to miRNAs. These lncRNAs share miRNA response elements with coding mRNAs, protecting them from miRNA‐mediated degradation. Recently, Li et al.31 reported a ceRNA network on pathogenesis of periodontitis, which involved three lncRNAs (MALAT1, TUG1 and FGD5‐AS1). These differences between the results of their and our studies may have arisen from differences in the datasets, analysis methods and periodontitis samples. Li et al.31did not report lncRNA expression data, and the identified target lncRNAs in the ceRNA network were based purely on predictions. In contrast, we retrieved lncRNA expression data from GSE80715. Moreover, Li et al.31 included samples of both aggressive and chronic periodontitis as the periodontitis group in their dataset. In contrast, we only included samples from patients diagnosed with chronic periodontitis to avoid heterogeneity among the samples.
The top down‐regulated DElncRNA, H19, which is an imprinted maternally expressed transcript, has been implicated in the pathological processes of both cancers and inflammatory diseases.32, 33 Recently, accumulating evidence has highlighted the role of H19 as a ceRNA in inflammation and osteogenesis. Fang et al.34 found that H19 functioned as a ceRNA in the regulation of miR‐874 and restoration of dysregulated inflammatory responses in lipopolysaccharide‐induced sepsis. Liang et al.35 reported that H19 could enhance osteogenesis in vivo and in vitro by acting as a ceRNA to compete for binding with miR‐141 and miR‐22. Both of these miRNAs are negative regulators of osteogenesis targeting β‐catenin; hence, by competitively binding with these two miRNAs, H19 ultimately activated the Wnt/β‐catenin pathway and promoted osteogenesis. In addition, research suggests that H19 could enhance the osteogenic differentiation of human bone marrow mesenchymal stem cell and bone formation via the miR‐675‐TGF‐β1‐Smad3‐HDAC signalling pathway, whereas knockdown of H19 suppresses these effects.36 Recently, Guo et al.37 found that H19 participates in periodontal inflammation by activating autophagy. However, the potential role of H19 as a ceRNA in periodontitis has rarely been reported. We obtained the expression profile of lncRNAs from existing data set and predicted that H19 could bind miRNAs (miR‐486‐5p and miR‐4299) to form ceRNA pairs with NOS1 and MAPT. The qPCR validation results of reduced H19, NOS1 and MAPT expression in the periodontitis group could provide further support to our prediction. Regarding the other two validated differentially expressed lncRNAs, RP11‐290L1.3 and RP11‐492E3.2, existing studies suggest that these are differentially expressed in cancer, adenovirus infection and inflammatory bowel disease and might have implications in autoimmune diseases such as multiple sclerosis and rheumatoid arthritis.38, 39, 40, 41 However, no study has reported the relationships of these lncRNAs with periodontal disease.
No statistically significant difference was found in the expressions of ISL2, AC096579.13, KIAA0125 or RP11‐731F5.2 between the healthy and periodontitis groups. This lack of consistency with the RNA‐seq results of GSE80715 might be due to large individual variations and the small sample size. The gingival samples used for our PCR validation differed from those of RNA‐seq, and only three samples in each group were examined, which may not have bestowed enough power to detect the difference.
In terms of GO functional enrichment analysis, up‐regulated genes in the ceRNA network were enriched in various inflammatory processes, including leukocyte migration and the cellular response to lipopolysaccharide, which further explained their roles in periodontal inflammation. The down‐regulated genes in the ceRNA network were mainly enriched in processes related to cellular potentials, such as neuron differentiation and osteoblast proliferation, which were consistent with previous studies in which cells harvested from inflammatory periodontal tissues were suggested to show less differentiation potential than healthy cells.42, 43 Last but not least, KEGG pathway analysis identified significant enrichment of pathways in periodontitis‐affected gingival tissues, including cytokine‐cytokine receptor interaction, TNF signalling pathway, IL‐17 signalling pathway and NF‐kappa B signalling pathway, among others. Further validation of these pathways is needed to explore their regulation in the pathogenesis of chronic periodontitis.
5. CONCLUSIONS
The present novel findings on the differential expression of lncRNAs, miRNAs and mRNAs in the gingival tissue of patients with periodontitis suggest the essential involvement of non‐coding RNAs and related regulatory mechanisms in the pathogenesis of periodontitis. The significantly down‐regulated expression of lncRNA H19 and mRNAs (NOS1 and MAPT) provided further support to our prediction of the lncRNA‐miRNA‐mRNA ceRNA network. Each ceRNA pair identified might be a potential candidate regulator of the disease, and further investigation is warranted to elucidate the specific underlying genetic traits and interaction networks.
CONFLICT OF INTEREST
The authors report no conflicts of interest related to this study.
Supporting information
Fig S1
Fig S2
Table S1
Table S2
Table S3
ACKNOWLEDGEMENTS
The study was supported by the Hong Kong Research Grants Council (GRF No. 17106619), Hong Kong SAR, China.
Contributor Information
Yifan Lin, Email: yflin@hku.hk.
Yanqi Yang, Email: yangyanq@hku.hk.
REFERENCES
- 1.Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809‐1820. [DOI] [PubMed] [Google Scholar]
- 2.Bui FQ, Almeida‐da‐Silva CLC, Huynh B, et al. Association between periodontal pathogens and systemic disease. Biomed J. 2019;42:27‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin LJ, Lamster IB, Greenspan JS, Pitts NB, Scully C, Warnakulasuriya S. Global burden of oral diseases: emerging concepts, management and interplay with systemic health. Oral Dis. 2016;22:609‐619. [DOI] [PubMed] [Google Scholar]
- 4.Tonetti MS, Jepsen S, Jin L, Otomo‐Corgel J. Impact of the global burden of periodontal diseases on health, nutrition and wellbeing of mankind: A call for global action. J Clin Periodontol. 2017;44:456‐462. [DOI] [PubMed] [Google Scholar]
- 5.Bartold PM, Van Dyke TE. Periodontitis: a host‐mediated disruption of microbial homeostasis. Unlearning learned concepts. Periodontol. 2000;2013(62):203‐217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon DR, Bainbridge BW, Darveau RP. Modulation of the innate immune response within the periodontium. Periodontol. 2000;2004(35):53‐74. [DOI] [PubMed] [Google Scholar]
- 7.Meyle J, Chapple I. Molecular aspects of the pathogenesis of periodontitis. Periodontol. 2000;2015(69):7‐17. [DOI] [PubMed] [Google Scholar]
- 8.Chhabra R. The Epigenetics of Noncoding RNA. In: Handbook of Epigenetics. Amsterdam: Elsevier. 2017; 47‐59. [Google Scholar]
- 9.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post‐transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102‐114. [DOI] [PubMed] [Google Scholar]
- 10.Lee YH, Na HS, Jeong SY, Jeong SH, Park HR, Chung J. Comparison of inflammatory microRNA expression in healthy and periodontitis tissues. Biocell. 2011;35:43‐49. [PubMed] [Google Scholar]
- 11.Stoecklin‐Wasmer C, Guarnieri P, Celenti R, Demmer RT, Kebschull M, Papapanou P. MicroRNAs and their target genes in gingival tissues. J Dent Res. 2012;91:934‐940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629‐641. [DOI] [PubMed] [Google Scholar]
- 13.Larsson L, Castilho RM, Giannobile WV. Epigenetics and its role in periodontal diseases: a state‐of‐the‐art review. J Periodontol. 2015;86:556‐568. [DOI] [PubMed] [Google Scholar]
- 14.Sánchez‐Muñoz F, Martínez‐Coronilla G, Leija‐Montoya AG, et al. Periodontitis may modulate long‐non coding RNA expression. Arch Oral Biol. 2018;95:95‐99. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Wu F, Song Y, et al. Long noncoding RNA related to periodontitis interacts with miR‐182 to upregulate osteogenic differentiation in periodontal mesenchymal stem cells of periodontitis patients. Cell Death Dis. 2016;7:e2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou Y, Li C, Shu F, et al. lncRNA expression signatures in periodontitis revealed by microarray: the potential role of lncRNAs in periodontitis pathogenesis. J Cell Biochem. 2015;116:640‐647. [DOI] [PubMed] [Google Scholar]
- 17.Bochenek G, Hasler R, El Mokhtari NE, et al. The large non‐coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum Mol Genet. 2013;22:4516‐4527. [DOI] [PubMed] [Google Scholar]
- 18.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353‐358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Gerstein M, Snyder M. RNA‐Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S, Barros SP, Moretti AJ, et al. Epigenetic regulation of TNFA expression in periodontal disease. J Periodontol. 2013;84:1606‐1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamilton JA. Colony‐stimulating factors in inflammation and autoimmunity. Nat Rev Immunol. 2008;8:533‐544. [DOI] [PubMed] [Google Scholar]
- 23.Offenbacher S, Barros SP, Paquette DW, et al. Gingival transcriptome patterns during induction and resolution of experimental gingivitis in humans. J Periodontol. 2009;80:1963‐1982. [DOI] [PubMed] [Google Scholar]
- 24.Artner I, Hang Y, Guo M, Gu G, Stein R. MafA is a dedicated activator of the insulin gene in vivo. J Endocrinol. 2008;198:271‐279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Damgaard C, Holmstrup P, Van Dyke TE, Nielsen CH. The complement system and its role in the pathogenesis of periodontitis: current concepts. J Periodontal Res. 2015;50:283‐293. [DOI] [PubMed] [Google Scholar]
- 26.Bayry J, Thirion M, Misra N, et al. Mechanisms of action of intravenous immunoglobulin in autoimmune and inflammatory diseases. Neurol Sci. 2003;24(Suppl 4):S217‐221. [DOI] [PubMed] [Google Scholar]
- 27.Brennan PA, Thomas GJ, Langdon JD. The role of nitric oxide in oral diseases. Arch Oral Biol. 2003;48:93‐100. [DOI] [PubMed] [Google Scholar]
- 28.Lei P, Ayton S, Finkelstein DI, Adlard PA, Masters CL, Bush AI. Tau protein: relevance to Parkinson's disease. Int J Biochem Cell Biol. 2010;42:1775‐1778. [DOI] [PubMed] [Google Scholar]
- 29.Guzeldemir‐Akcakanat E, Sunnetci‐Akkoyunlu D, Orucguney B, et al. Gene‐Expression Profiles in Generalized Aggressive Periodontitis: A Gene Network‐Based Microarray Analysis. J Periodontol. 2016;87:58‐65. [DOI] [PubMed] [Google Scholar]
- 30.Yu T, Acharya A, Mattheos N, et al. Molecular mechanisms linking peri‐implantitis and type 2 diabetes mellitus revealed by transcriptomic analysis. PeerJ. 2019;7:e7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S, Liu X, Li H, et al. Integrated analysis of long noncoding RNA‐associated competing endogenous RNA network in periodontitis. J Periodontal Res. 2018;53:495‐505. [DOI] [PubMed] [Google Scholar]
- 32.Han Y, Ma J, Wang J, Wang L. Silencing of H19 inhibits the adipogenesis and inflammation response in ox‐LDL‐treated Raw264.7 cells by up‐regulating miR‐130b. Mol Immunol. 2018;93:107‐114. [DOI] [PubMed] [Google Scholar]
- 33.Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 Long non‐coding RNA in cancer initiation, progression and metastasis ‐ a proposed unifying theory. Mol Cancer. 2015;14:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang Y, Hu J, Wang Z, et al. LncRNA H19 functions as an Aquaporin 1 competitive endogenous RNA to regulate microRNA‐874 expression in LPS sepsis. Biomed Pharmacother. 2018;105:1183‐1191. [DOI] [PubMed] [Google Scholar]
- 35.Liang WC, Fu WM, Wang YB, et al. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci Rep. 2016;6:20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Y, Zheng Y, Jia L, Li W. Long Noncoding RNA H19 Promotes Osteoblast Differentiation Via TGF‐beta1/Smad3/HDAC Signaling Pathway by Deriving miR‐675. Stem Cells. 2015;33:3481‐3492. [DOI] [PubMed] [Google Scholar]
- 37.Guo R, Huang Y, Liu H, Zheng Y, Jia L, Li W. Long Non‐Coding RNA H19 Participates in Periodontal Inflammation via Activation of Autophagy. J Inflamm Res. 2020;13:635‐646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shang D, Zheng T, Zhang J, Tian Y, Liu Y. Profiling of mRNA and long non‐coding RNA of urothelial cancer in recipients after renal transplantation. Tumour Biol. 2016;37:12673‐12684. [DOI] [PubMed] [Google Scholar]
- 39.Zhao H, Chen M, Lind SB, Pettersson U. Distinct temporal changes in host cell lncRNA expression during the course of an adenovirus infection. Virology. 2016;492:242‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haberman Y, BenShoshan M, Di Segni A, et al. Long ncRNA landscape in the ileum of treatment‐naive early‐onset Crohn disease. Inflamm Bowel Dis. 2018;24:346‐360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teimuri S, Hosseini A, Rezaenasab A, et al. Integrative analysis of lncRNAs in Th17 cell lineage to discover new potential biomarkers and therapeutic targets in autoimmune diseases. Mol Ther Nucleic Acids. 2018;12:393‐404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park JC, Kim JM, Jung IH, et al. Isolation and characterization of human periodontal ligament (PDL) stem cells (PDLSCs) from the inflamed PDL tissue: in vitro and in vivo evaluations. J Clin Periodontol. 2011;38:721‐731. [DOI] [PubMed] [Google Scholar]
- 43.Tang HN, Xia Y, Yu Y, Wu RX, Gao LN, Chen FM. Stem cells derived from "inflamed" and healthy periodontal ligament tissues and their sheet functionalities: a patient‐matched comparison. J Clin Periodontol. 2016;43:72‐84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Table S1
Table S2
Table S3


