Abstract
ABCG subfamily proteins are highly enriched in terrestrial plants. Many of these proteins secrete secondary metabolites that repel or inhibit pathogens. To establish why the ABCG subfamily proteins proliferated extensively during evolution, we constructed phylogenetic trees from a broad range of eukaryotic organisms. ABCG proteins were massively duplicated in land plants and in oomycetes, a group of agronomically important plant pathogens, which prompted us to hypothesize that plant and pathogen ABCGs coevolved. Supporting this hypothesis, full‐size ABCGs in host plants (Arabidopsis thaliana and Glycine max) and their pathogens (Hyaloperonospora arabidopsidis and Phytophthora sojae, respectively) had similar divergence times and patterns. Furthermore, generalist pathogens with broad ranges of host plants have diversified more ABCGs than their specialist counterparts. The hypothesis was further tested using an example pair of ABCGs that first diverged during multiplication in a host plant and its pathogen: AtABCG31 of A. thaliana and HpaP802307 of H. arabidopsidis. AtABCG31 expression was activated following infection with H. arabidopsidis, and disrupting AtABCG31 led to increased susceptibility to H. arabidopsidis. Together, our results suggest that ABCG genes in plants and their oomycete pathogens coevolved in an arms race, to extrude secondary metabolites involved in the plant's defense response against pathogens.
Abbreviations
- ABA
abscisic acid
- ABC
ATP‐binding cassette
- ABCG
G subfamily of ATP‐binding cassette proteins
- AVR proteins
avirulence proteins
- EDS1
Enhanced Disease Susceptibility 1
- Ka
non‐synonymous mutational rates
- Ks
synonymous mutational rates
- NBD
nucleotide‐binding domain
- PDR
pleiotropic drug resistance
- R genes
disease resistance genes
- TMD
transmembrane domain
- WBC
white‐brown complex
Introduction
ABC proteins contain ATP‐binding cassettes, which bind and hydrolyze ATP. They carry out diverse physiological functions by transporting a large number of different chemicals across biological membranes, and also by performing biochemical functions unrelated to transport (Theodoulou and Kerr 2015, Yu et al. 2015, Locher 2016). An interesting characteristic of ABC transporters is that, as a group, they transport vastly different types of chemicals, and some individual ABC proteins transport diverse substrates. Despite some progress in determining the structures of ABC proteins (Lee et al. 2016, Khunweeraphong et al. 2017, Taylor et al. 2017, Kim and Chen 2018, Manolaridis et al. 2018, Alam et al. 2019), the structural basis for substrate specificity is unclear in the proteins, and consequently, it is not possible to predict the substrates of ABC proteins that have not yet been functionally studied. Identifying the substrate and function of an ABC protein usually requires obtaining the corresponding mutants, followed by extensive functional studies. Thus, determining the functions of newly discovered ABC genes requires much time and effort. However, a systematic analysis of recent successes in determining the structures and functions of many ABC proteins (Thomas and Tampé 2018, Srikant and Gaudet 2019) might provide some clues as to the relationship between the structure and function of ABC proteins.
Another interesting aspect of ABC proteins is their evolution (Dean and Annilo 2005, Kovalchuk and Driessen 2010, Xiong et al. 2015, Ofori et al. 2018). ABC proteins are of ancient origin, since they exist in all organisms studied to date. They have undergone a wide range of changes throughout the history of life, as evidenced by the existence of many different types of ABC proteins with different structures and functions. Moreover, ABC protein domains were fused, modified and multiplied during evolution, and prokaryotes tend to have simpler and smaller ABC proteins than those found in eukaryotes (Xiong et al. 2015). Bioinformatic analysis of over a thousand plant transcriptomes found that simple organisms such as microalgae possess fewer unique ABC proteins than vascular land plants (Lane et al. 2016). Evolutionary studies of ABC genes are interesting not only because they reveal the evolutionary history of genes, but also because they imply the functions and substrates of many uncharacterized ABC proteins; it is reasonable to assume that ABC proteins that are closely related in the phylogenetic tree are more likely to perform similar functions than distantly related ones. Thus, we can speculate about the potential functions of unidentified ABC proteins by assessing their phylogenetic relationships with well‐characterized ABC proteins.
Terrestrial plants generally have more ABC transporters than animals or aquatic photosynthetic organisms (Andolfo et al. 2015, Hwang et al. 2016, Lane et al. 2016). A comparison of the different ABC protein subfamilies revealed that members of the G subfamily duplicated and proliferated substantially more than other subfamily members in plants (Andolfo et al. 2015, Hwang et al. 2016). We previously reported that many ABCG subfamily transporters allow plants to adapt to dry land environments, by secreting a surface coating, mediating a defense response, or facilitating the long‐distance hormonal control of growth and development (Hwang et al. 2016). In the same report, we presented evidence for the explosive multiplication of genes similar to the stress hormone ABA transporter, AtABCG40, during the evolution of terrestrial plants from unicellular green algae. Thus, the multiplication and diversification of ABCG transporters might have contributed substantially to the colonization of land by photosynthetic organisms.
To further examine this possibility and to better understand how these transporters evolved in the green plant lineage, we constructed phylogenetic trees of all ABCG subfamily genes of Arabidopsis thaliana and their homologs from a broad range of organisms, including Opistokonta (e.g. animals, fungi), Amoebozoa, Viridiplantae (e.g. green algae, land plants) and Chromalveolata (e.g. Stramenopiles, Alveolata, Haptophyta, Cryptophyta). Our phylogenetic analysis revealed several interesting insights. First, A. thaliana ABCG subfamily proteins and their homologs in land plants are clearly separated from those of other organisms, suggesting that they differentiated after the evolution of land plants. Second, A. thaliana ABCG subfamily proteins and their homologs in land plants diversified to seven different groups, with each group comprising multiple genes with similar physiological functions. Third, full‐size ABCGs rapidly diversified not only in land plants but also in oomycetes (e.g. Phytophthora). Since many oomycetes are plant pathogens (Kamoun et al. 2015) and some plant ABC transporters transport secondary metabolites important for pathogen defense (Stukkens et al. 2005, Yazaki 2006, Crouzet et al. 2013), we hypothesized that the ABCGs in plants and oomycetes might have coevolved in an evolutionary arms race. We report several lines of evidence that support this possibility.
Materials and methods
Phylogenetic tree analysis and collection of novel ABCGs
The protein sequences of ABCG family members were collected from NCBI (National Center of Biotechnology Information) and transcriptome database (e.g. MMESTP) to avoid a biased taxon sampling (Keeling et al. 2014). For Figs 1;2 and S1, AtABCG homologous genes were gathered using BLASTp (top matches count: 500, e‐value cutoff: 1e‐5). Instead of using raw data from a non‐redundant (nr) database, taxon selection in a local database (based on NCBI RefSeq database; summarized in Tables 1, S2 and S3) was performed to avoid saturation of certain lineages. In addition, Ginkgo and Chara ABCG genes were included in the phylogenetic analyses to broaden taxon sampling. CD‐Search (Marchler‐Bauer et al. 2011) and UniProtDB were used to predict the domain region of ABCGs.
FIGURE 1.
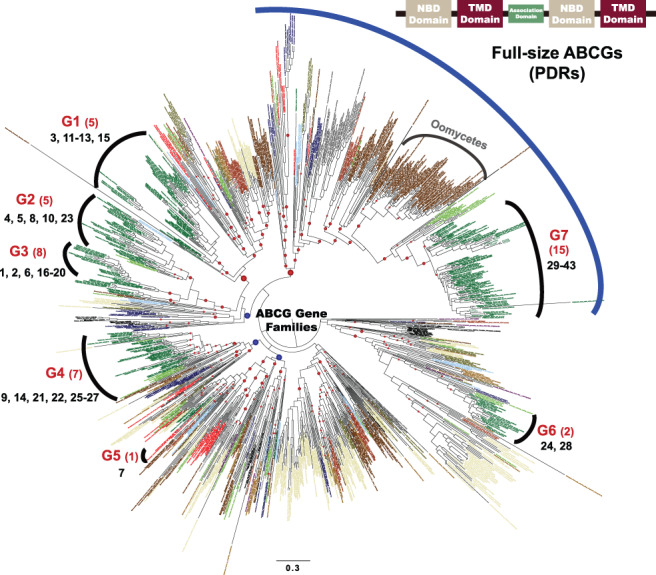
Phylogenetic tree of the G subfamily of the ATP‐Binding Cassette (ABCG) protein family. The phylogenetic tree was constructed using the maximum likelihood (ML) method. Taxon and gene IDs are summarized in Table S2. Blue and red circles indicate the bootstrap values between 70 and 80%, and >80%, respectively. ‘Group 1’ through ‘Group 7’ marked on the periphery are for the land plant lineage based on AtABCGs, and the numbers below the group numbers indicate the numbers of the A. thaliana ABCG proteins. The ABCGs under the blue curve on top are full‐size, while the others are half‐size. The scale bar indicates the branch length measured in the number of amino acid substitutions per site. NBD, nucleotide‐binding domain; TMD, transmembrane domain.
FIGURE 2.
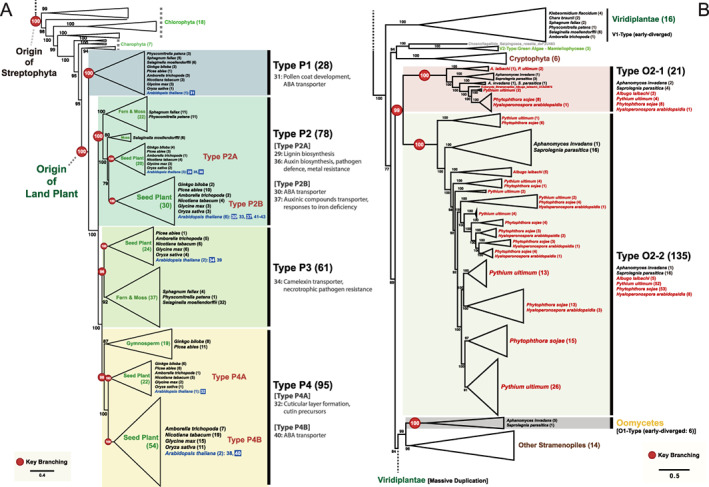
Phylogeny of the full‐size ABCGs representing the major duplication events in land plants and oomycetes. (A) A subtree of full‐size ABCGs of Chlorophyta and land plants. The AtABCG transporters, which were identified in previous studies (listed in Table S1), are on a blue background and their functions are summarized in the figure. (B) A subtree of full‐size ABCGs of Stramenopiles rooted with Viridiplantae. The plant pathogenic oomycetes are indicated with red letters. The subtrees in (A) and (B) are derived from the full tree presented in Fig. S1. The number next to the name of the species indicates the number of ABCGs. Branch support is shown as the ultrafast bootstrap support value and the red circles represent key branching points. Some clades were merged to improve readability. The scale bar indicates the branch length measured in the number of amino acid substitutions per site.
TABLE 1.
Taxon list of oomycete full‐size ABCG transporters used in the phylogenetic analysis shown in Fig. S2.
| Taxonomic group | Host infection type | Species name | Number of full‐size ABCGs | References |
|---|---|---|---|---|
| Oomycetes – Saprolegniales – Saprolegniaceae | Animal pathogen | Saprolegnia diclina | 21 | Diéguez‐Uribeondo et al. 2007 |
| Oomycetes – Saprolegniales – Saprolegniaceae | Animal pathogen | Saprolegnia parasitica | 21 | Diéguez‐Uribeondo et al. 2007 |
| Oomycetes – Saprolegniales – Leptolegniaceae | Animal pathogen | Aphanomyces invadans | 8 | Diéguez‐Uribeondo et al. 2009 |
| Oomycetes – Saprolegniales – Leptolegniaceae | Animal pathogen | Aphanomyces astaci | 7 | Makkonen et al. 2012 |
| Oomycetes – Peronosporales – Peronosporaceae | Generalist pathogen (Plant) | Phytophthora infestans | 49 | Kamoun et al. 2015 |
| Oomycetes – Peronosporales – Peronosporaceae | Generalist pathogen (Plant) | Phytophthora parasitica | 60 | Meng et al. 2014 |
| Oomycetes – Peronosporales – Peronosporaceae | Generalist pathogen (Plant) | Phytophthora sojae | 61 | Erwin and Ribeiro 1996 |
| Oomycetes – Pythiales | Generalist pathogen (Plant) | Pythium aphanidermatum | 42 | Chen et al. 1992 |
| Oomycetes – Pythiales | Generalist pathogen (Plant) | Pythium ultimum | 56 | Kamoun et al. 2015 |
| Oomycetes – Albuginales | Generalist pathogen (Plant) | Albugo candida | 10 | McMullan et al. 2015 |
| Oomycetes – Peronosporales – Peronosporaceae | Specialist pathogen (Plant) | Hyaloperonospora arabidopsidis | 9 | Thines et al. 2009 |
| Oomycetes – Albuginales | Specialist pathogen (Plant) | Albugo laibachii | 7 | Kamoun et al. 2015 |
The ABCG sequences used in Figs 1;2 and S1–S3 were basically aligned using MAFFT v7 (Katoh and Standley 2013). The resulting alignment of entire ABCGs (Fig. 1) was trimmed to remove gaps using automated trimming options in trimAl (Capella‐Gutierrez et al. 2009). The maximum likelihood (ML) analysis was conducted in IQ‐TREE (Nguyen et al. 2015) with 1000 ultrafast bootstrap replications. The evolutionary model used in the phylogenetic analyses was selected by ModelFinder, which is incorporated in IQ‐TREE (Kalyaanamoorthy et al. 2017). The phylogeneic analysis results from IQ‐TREE are visualized by FigTree v1.4.4 (https://github.com/rambaut/figtree/) and bootstrap supporting values of less than 50% were removed.
Unannotated ABCGs from published genomes in an available database (e.g. Ginkgo genome from GIGAn DB [http://gigadb.org/dataset/100209]) were collected based on annotated ABCGs by homologous search and chosen as putative ABCGs. The ABCGs were confirmed by phylogenetic analysis using previously known ABCGs. To filter out ABCG paralogs from putative ABCGs, only ABCGs that formed a monophyletic clade with other ABCGs were selected. In addition, short proteins (<100 amino acids) were removed from the analysis.
Relative molecular clock analysis
The full‐size ABCG transporters used in constructing the phylogenetic trees (Table S2) were included in the divergence time estimation. To calculate relative divergence time, a Markov Chain Monte Carlo (MCMC) Bayesian approach implemented on the program Bayesian Evolutionary Analysis Sampling Trees (BEAST) v1.10.4 (Drummond et al. 2012) was used under LG + G4 + I and a strict clock model. Based on the tree topology of full‐size ABCG transporters (Fig. S1), the most widely used Yule process in speciation model and the simple birth–death process for speciation (Gernhard 2008) was applied for the tree prior as an operator option. More detailed options are available in Data S1, which was generated by Bayesian Evolutionary Analysis Utility (BEAUti) program. MCMC chains were run with 10 million generations, and 10% of the total number of iterations (1 million states) was used as the burn‐in for an analysis.
Ks and Ka distributions
Full‐size ABCG transporter sequences of four species (two host species: A. thaliana and Glycine max; two pathogen species: P. sojae and Hyaloperonospora arabidopsidis) were collected from their genome data. The number of nonsynonymous substitutions per non‐synonymous site (Ka) of gene pairs from each species was esitimated using ‘KaKs_Calculator’ implemented in ParaAT (Parallel Alignment and back‐Translation; Zhang et al. 2006, Zhang et al. 2012). Synonymous substitutions per synonymous site (Ks) were calculated in the same way.
Comparative analysis of evolutionary rates between plants and pathogens
To quantify the difference of evolutionary rates, log‐fold changes of non‐synonymous over synonymous alterations (dN/dS) between pairs of full‐size ABCGs in plant hosts and those in plant pathogenic oomycetes were calculated using the equation logFC(dN/dS) =|log(dNdS Ath − Gmax ) − log(dNdS Hpa − Psojae )|. To test the statistical significance of the evolutionary‐rate difference, logFC(dN/dS) of the ABCG31 homologs from the two plant–pathogen pairs (Plants: AtABCG31 of A. thaliana and dxK7M810 of G. max; Pathogens: HpaP802307 of H. arabidopsidis and dxG5ADX0 of P. sojae) were compared with those from random pairs. Sequences of random pairs (n = 1225) were taken from ABCG31 homologs in 50 different land plants in representative genomic databases (Goodstein et al. 2011). The P‐value was calculated with the percentage of random pairs that have lower logFC(dN/dS) than the observed value.
The difference of divergence time, logFC(dS) = |log(dS Ath − Gmax ) − log(dS Hpa − Psojae )|, was used as an independent variable to control the intrinsic difference in divergence time‐scales between plant and pathogen pairs. To calculate P‐value taken account for the divergence time‐scale, we tested the random pairs whose logFC(dS) values approximate to that of the observed value. Multiple tests by differentiating the criteria for the dispersion of logFC(dS) were conducted and the minimum p‐value was chosen. Evolutionary rate (dN/dS) and divergence time (dS) were calculated using codeml implemented in the PAML package (Yang 1997).
Plant growth and pathogen inoculation
Arabidopsis thaliana ecotype Col‐0 and mutants derived from the Col‐0 background, eds1‐2, abcg31‐2 (GK064C11), abcg31‐3 (GK242G07), were used. Seeds were sown in pots (diameter 10 cm, height 8.7 cm) of soil and grown for 10 days (22/18°C, 14/10 h day–night cycles). Hyaloperonospora arabidopsidis isolate Noco2 was maintained on the A. thaliana Col‐0. For inoculation, infected cotyledons were harvested and suspended in sterilized water containing 0.05% (w/v) Tween‐20. The resuspension was filtered using Miracloth (Millipore Corp.), and conidiosporangia that passed through the cloth were diluted with water containing 0.05% (w/v) Tween‐20 to a final concentration of 5 × 104 spores ml−1.
The 10‐day‐old Arabidopsis seedlings were inoculated with the conidiosporangia of H. arabidopsidis following the method described previously (Asai et al. 2015). The inoculated seedlings were then covered with a transparent cover (Cleanwrap) to maintain high humidity (above 80%), and kept at 17°C, 14/10 h day–night conditions. The degree of infection was estimated by counting the number of spores per cotyledon.
RNA extraction and assay of gene expression level
Total RNA was extracted using homemade Trizol reagent. To obtain cDNA to use as the template for quantitative RT‐PCR (qRT‐PCR), 4 μg of RNA was subjected to reverse transcription with SuperiorScriptIII Reverse Transcriptase (Enzynomics). To determine the gene expression level, qRT‐PCR was conducted using RbTaq qPCR 2X PreMIX (Enzynomics). The primer sets used to detect AtABCG31 expression were RTABCG31_F, 5′‐TCCAAAAGCCTTTGATTCCAGT‐3′ and RTABCG31_R, 5′‐TCCCACAGGACTTTTTTTATCTTCTC‐3′ (Kang et al. 2015). The qRT‐PCR results were normalized by detecting the expression level of a housekeeping gene AtUbiquitin11, using the primer sets of AtUbiquitin11, Ubi11_F, 5′‐ GAACCAAGTTCATGTATCGT‐3′ and Ubi_R, 5′‐ ACACTCATCAAACTAAGCAC‐3′.
Results
Phylogeny and evolution of ABCG members in a wide range of organisms
Functions, and hence putative substrates, of ABCG transporters are tremendously diverse, including transport of hormones, surface coating materials, materials for structural support and secondary metabolites for pathogen defense. As such a broad range of substrates is unusual among transporter groups, we were curious how such diversity evolved. As a first step to elucidating the phylogenetic relationships of ABCG proteins of plants, we gathered information of all ABCG genes of Arabidopsis, including their putative substrates (Table S1). These ABCG genes encode proteins with either 577–1107 amino acids (aa) for half‐size white‐brown complex (WBC)‐type or 1382–1469 aa for full‐size pleiotropic drug resistance (PDR)‐type transporters. These genes have 1–25 exons and are spread over many different chromosomes.
We next searched for sequences homologous to AtABCG subfamily members in a broad range of eukaryotic organisms, including those belonging to Viridiplantae, Metazoa, Fungi, Stramenopiles, Alveolata, Haptophyta and Amoebozoa (Table S2). Notably, we did not detect any bacterial homologs, suggesting that ABCGs originated independently at an early stage of eukaryotic evolution. Next, we reconstructed a phylogenetic tree based on aligned ABCG protein sequences using the maximum likelihood method (Fig. 1). Since the origin of ABCG transporter is unknown, we used midpoint rooting method to root the tree. In the tree, most homologs from all taxonomic groups (listed in Table S2) are widely distributed, but some homologs are clustered in specific lineages (i.e. Viridiplantae‐specific, Alveolata‐specific, Stramenopiles‐specific), suggesting that, in these specific lineages, ABCG proteins diversified soon after each lineage emerged. Among them, we focused on the ABCG transporters of land plant, especially on the 43 Arabiodpsis ABCG subfamily proteins (AtABCGs), because these AtABCGs were well characterized (Hwang et al. 2016). The 43 AtABCG transporters are clustered into seven clades (G1–G7), with other plant homologs (Fig. 1, Table S1). In each group, the green algal ABCGs diverged first in most cases and the land plant ABCGs followed, suggesting that ABCG proteins of land plants diversified from the pre‐existing ancestral green algal homologs. This result is consistent with the widely accepted notion that the land plant lineage evolved from green algal lineage (Leliaert et al. 2012). The half‐size ABCG transporters, known as WBC‐type (AtABCG genes 1–28) are paraphyletically distributed in six major groups (i.e. G1–G6), whereas full‐size ABCG transproters, known as PDR‐type (AtABCG genes 29–43) are grouped together in the clade G7. Interestingly, many oomycete ABCGs (full‐size) are positioned right next to clade G7, suggesting that they have a common origin with full‐size ABCGs of plants (Fig. 1).
Functional similarities and differences among Arabidopsis ABCG proteins
Since proteins of the same ancestral origin might share some functional similarities, we suspected that some Arabidopsis ABCG protein members belonging to the same group described above have similar functions. Indeed, a comparison of Fig. 1 and Table S1 revealed that similar functions were shared by some members of the same group. Group 1 (G1) includes AtABCG11, AtABCG12 and AtABCG13, which transport surface coating chemicals, such as wax and cutin (Bird et al. 2007). Group 3 (G3) includes AtABCG1, AtABCG2, AtABCG6 and AtABCG16, which transport materials needed for the synthesis of a suberin barrier on the seed coat (AtABCG2, AtABCG6) and/or construction of the intact pollen wall (AtABCG1, AtABCG16) (Yadav et al. 2014, Yim et al. 2016). This group may also include drug efflux transporters, since AtABCG19 overexpression increased kanamycin resistance (Mentewab and Stewart Jr. 2005). Group 4 (G4) contains AtABCG9 and AtABCG26, which transport pollen surface coating materials (steryl glucoside for ABCG9 and sporopollenin lipid precursors for AtABCG26; Quilichini et al. 2010, Choi et al. 2014) and plant hormone transporters (cytokinin for AtABCG14 and ABA for AtABCG25; Kuromori et al. 2010, Ko et al. 2014). Group 7 (G7) has the highest functional diversity among the Arabidopsis ABCG groups, with substrates including abscisic acid (AtABCG30, AtABCG31, AtABCG40), lignin precursors (AtABCG29), cutin precursors (AtABCG32) and disease response factors (AtABCG34, AtABCG36; Strader and Bartel 2009, Kang et al. 2010, Ruzicka et al. 2010, Choi et al. 2014, Khare et al. 2017). This feature of multiple substrates of ABCGs is likely evolved by acquisition of new functions. Why and how the G7 members multiplied to such diverse functions are intriguing questions.
Evolution of G7 ABCG proteins in land plants
To determine the origins and phylogenetic relationships among G7 ABCG proteins of land plants, we included ABCGs from additional taxa (Table S3), and then classified the G7 members in more detail. At the macro‐scale, the full‐size ABCGs of Viridiplantae appeared to be a result of paraphyletic evolution and could be divided into three different types: (1) V1‐Type; (2) V2‐Type and (3) V3‐Type (Fig. S1). The V1‐Type is the early‐diverged full‐size ABCGs that could not be found in chlorophyceaen algae but appeared in Charophyta (Chara and Klebsormidium), Bryophyta (moss; Physcomitrella and Sphagnum), Lycopodiophyta (fern ally; Selaginella) and in the basal angiosperm species Amborella trichopoda. However, this type could not be detected in any other angiosperm. The V2‐Type is Mamiellophyceae‐specific, found only in a class of Chlorophyta, Mamiellophyceae. The V3‐Type included most of the ABCG transporters present in seed plants (Fig. S1). Thus, the tree suggests that there were three different types of full‐size ABCGs in the early evolutionary stage of Viridiplantae. Later on, the V3‐Type massively multiplied and diversified only in land plants.
To better understand the evolution of ABCGs of Viridiplantae, we further analyzed the G7 members of land plants (Fig. 2A). The ABCG proteins of the Charophyta (Klebsomidium) share the most recent common ancestor with the G7 members, which suggested that the G7 land plant members were derived from the ABCG proteins of the Charophyta. The G7 members of land plants clustered into four types: Types P1–P3 including moss, ferns and seed plants, and Type P4 including only gymnosperms and angiosperms. This suggests that the G7 members of land plant ABCGs diversifed to three types in moss (Physcomitrella) and fern (Selaginella), and greatly expanded in seed plants, especially in flowering plants; there were 78 genes encoding Type P2 G7 ABCG proteins, 50 of which were in seed plants, including 9 in Arabidopsis (i.e. AtABCGs 29, 30, 33, 35, 36, 37, 41, 42, 43), and 95 G7 transporters from diverse seed plants in Type P4. Thus, G7 ABCG proteins seem to have evolved gradually from green algae (Chlorophyta) through Charophyta to land plants, where they diversified explosively (Fig. 2A).
Evolution of full‐size ABCG proteins in oomycetes
Strikingly, we observed that full‐size ABCGs underwent massive diversification in oomycetes also (Fig. S1). This is consistent with the observation that full‐size ABCGs genes (G7) in both land plants and oomycetes expanded explosively, and share a common ancestor (Fig. 1). To better understand the relationship of these intensive diversifications between the two groups, we further analyzed full‐size ABCG members of oomycetes (Fig. 2B).
Oomycetes exhibit hyphal growth and reproduce by dispersing spores, but are phylogenetically closer to diatoms and brown algae than to fungi (Dick 2001, Thines and Kamoun 2010). More than 60% of oomycetes are known plant pathogens, and some cause substantial damage to important crop species (Kamoun 2003, Thines and Kamoun 2010). Oomycetes have a high number of full‐size ABCG copies that clustered into two major clades: O1‐Type and O2‐Type (Fig. 2B). Oomycetes early‐diverged O1‐Type formed a monophyletic group with those of other stramenopiles (i.e. brown algae, diatoms), indicating that oomycete O1‐Type ABCGs originated from a common ancestor of the stramenopiles. The rest of oomycetes ABCGs were grouped in the O2‐Type, which subdivied into two different clades, Type O2‐1 and Type O2‐2. Among them, we found that there were many more Type O2‐2 members (135 copies) than other types (21 copies of Type O2‐1 and 6 copies of early‐diverged O1‐Type). This analysis also revealed that certain species (i.e. those belonging to the genera Phytophthora and Pythium) have extremely high numbers of full‐size ABCGs (Fig. 2B and Table S3). For instance, in P. sojae, a causal agent of root rot in soybean (G. max), there were 8 copies of Type O2‐1, and 53 copies of Type O2‐2 full‐size ABCGs.
To determine whether such multiplication occurred in other oomycete species as well, we analyzed full‐size ABCGs of six other oomycete species whose genomes have been sequenced, including two Phytophthora species (Phytophthora infestans and Phytophthora parasitica), Pythium aphanidermatum, Albugo candida, Saprolegnia diclina and Aphanomyces astaci (Fig. S2 and Table S3). The early‐diverged O1‐Type was found only in Saprolegniales, and it had much less copy numbers (16 copies) than the O2‐Type (47 copies in Type O2‐1 and 293 copies in O2‐2; Fig. S2). Two major types of O2‐Type (O2‐1 and O2‐2) included ABCGs from all six species used in this analysis. Interestingly, Type O2‐2 contained extremely high copy numbers of ABCG genes (Fig. S2), especially in P. sojae (53 genes) and Pythium ultimum (52 genes), suggesting that massive gene duplications had occurred in these plant pathogenic species. Taken together, this analysis demonstrated that the ABCGs in plants and oomycetes have common evolutionary patterns; both include the early‐diverged but less duplicated genes (i.e. V1‐Type vs O1‐Type) and lately‐diverged and massively duplicatred ABCGs (i.e. V3‐Type vs O2‐Type; see Figs 2 and S2).
Coevolution of full‐size ABCGs of land plants and plant pathogenic oomycetes
Based on the phylogenetic analysis described above, we hypothesized that Phytophthora might have developed ABCGs with functional similarities to G7 ABCGs of land plants, and that these protected the pathogen from the secondary metabolites secreted by the infected plants. Thus, our hypothesis predicted that the two groups of ABCGs coevolved through their specific interactions as drug pumps of plants and their pathogens.
To test this hypothesis, we first compared the relative molecular divergence of full‐size ABCGs in plants and their pathogens, because interacting proteins are likely to have similar divergence times (Edger et al. 2015). For this purpose, we selected two pairs of host–pathogen species: A. thaliana–H. arabidopsidis and G. max–P. sojae. The estimated divergence times of their full‐size ABCGs ranged between 2.65 and 144.15 (relative values without real time scale; Figs 3A and S3). The median value of divergence times was 35.36 (Q1–Q3: 22.07–55.28, standard error: 4.87) among the 14 divergence points of A. thaliana full‐size ABCGs, while that of the pathogen H. arabidopsidis was 49.16 (Q1–Q3: 42.54–60.76, standard error: 9.11) from 9 divergence points. Wilcoxon rank sum statistical tests were applied to test a significant difference of divergence times between the two groups; a null hypothesis ‘there are no differences between full‐size ABCGs in the host and the pathogen in relative divergence times’ was tested. The distribution of these two divergence times was not significantly different (P‐value: 0.3054; Fig. 3A). Similar to this case, the distribution of divergence times of G. max–P. sojae was not significantly different to reject the null hypothesis (P‐value: 0.4152; Fig. 3A). In case of G. max, the median value was 27.93 (Q1–Q3: 20.55–38.74, standard error: 2.79) from 30 divergence points, whereas in the case of P. sojae, it was 32.89 (Q1–Q3: 22.47–42.54, standard error: 2.31) from 61 divergence points (Fig. 3A). This molecular clock analysis suggests that the divergence times of full‐size ABCG genes in plants and their pathogens did not differ significantly. Thus, duplications of ABCG genes in plant hosts and pathogenic oomycetes likely occurred at a similar time.
FIGURE 3.
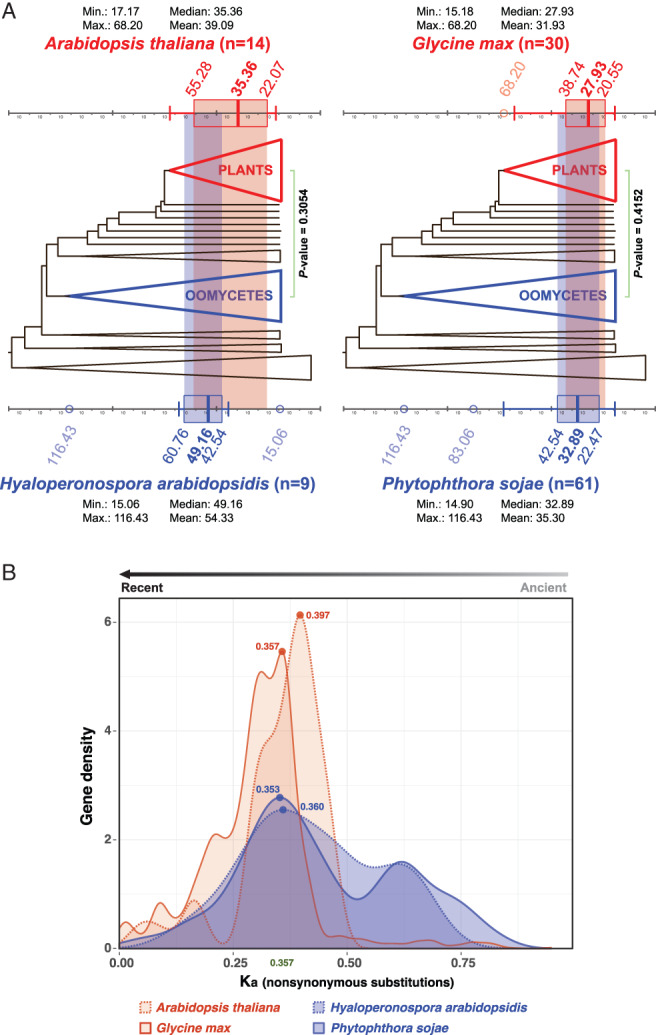
Concurrent diversification of full‐size ABCGs in land plants and plant pathogenic oomycetes. (A) Concurrent relative molecular divergence times of full‐size ABCGs in plant and plant pathogenic oomycetes (details in Fig. S3). The host plants (A. thaliana and G. max) are indicated in red, and plant pathogenic oomycetes (H. arabidopsidis and P. sojae) are indicated in blue. (n; the number of divergence points) (B) Density distribution of non‐synonymous (Ka) mutational rates of full‐size ABCGs in the four host/pathogen species. The major peaks of four species are in similar positions.
The second method we used to test the hypothesis was to compare non‐synonymous mutational rates (Ka) and synonymous mutational rates (Ks) of full‐size ABCGs in the same two plant–pathogen pairs: A. thaliana–H. arabidopsidis and G. max–P. sojae. The Ka distribution is used to study gene duplication in distantly related taxonomic groups (Albà and Castresana 2004), since nonsynonymous substitutions change amino acid sequences, which is directly related to gene selection in an evolutionary process. Major peaks (i.e. the highest gene density with the same Ka value) of A. thaliana, H. arabidopsidis, G. max and P. sojae were 0.397 (red dot), 0.360 (blue dot), 0.357 (red line) and 0.353 (blue line), respectively (Fig. 3B). The densities of genes with similar mutation rates between the host–pathogen pairs largely overlapped, suggesting that major gene diversifications of ABCGs in land plants and plant pathogenic oomycetes coincided (Fig. 3B), although the mutation rates of individual genes could differ. We also calculated the pairwise Ks distribution of gene pairs (total: 5332 pairs; Fig. S4), but the distribution was too broad to yield any conclusion, likely due to the saturated rates of synonymous mutations.
Third, we compared the number of full‐size ABCGs in generalist plant pathogenic oomycetes and specialist oomycetes. Since different plant hosts secrete different secondary metabolites (Parker et al. 2009, Sana et al. 2010, Kushalappa and Gunnaiah 2013, Arbona and Gomez‐Cadenas 2016), a generalist pathogen, which infects many plant species, might need many different ABCGs to remove the diverse secondary metabolites from their cytosol. By contrast, a specialist pathogen, which infects only a few specific species, might need fewer full‐size ABCGs to pump out the few secondary metabolites secreted from its hosts. Phytophthora sojae (Peronosporales) is a generalist plant pathogenic oomycete that infects various fabacean and other species (Erwin and Ribeiro 1996). Similarly, P. ultimum (Pythiales) is a generalist parasitic oomycete that can infect hundreds of diverse plant hosts (Kamoun et al. 2015). By contrast, Albugo laibachii (Albuginales) and H. arabidopsidis (Peronosporales) are specialist pathogens that infect a specific host, i.e. A. thaliana (Thines et al. 2009, Kamoun et al. 2015). Our phylogenetic analysis of ABCG transporters revealed that generalist parasitic oomycetes have more full‐size ABCG transporters than do specialists (Fig. 2 and Table 1). The generalist parasites P. sojae and P. ultimum have 56–61 copies of full‐size ABCG transporters, while the specialist pathogens A. laibachii and H. arabidopsidis have 7–9 copies. Interestingly, P. sojae and H. arabidopsidis are close relatives in the same family (Peronosporaceae), but the generalist P. sojae (61 copies) has about seven times more full‐size ABCG transporters than does the specialist H. arabidopsidis (9 copies).
Finally, to examine the possibility of coevolution in a specific case, we obtained ABCG sequences from two plant–pathogen pairs (i.e. A. thaliana–H. arabidopsidis and G. max–P. sojae) and compared the phylogenetic trees of their full‐size ABCGs (Fig. 4A). Type P1 ABCGs in host plants and Type O2‐1 ABCGs in pathogens clustered alone in a monophyletic clade (Fig. 4A) and they first diverged during the massive diversification of ABCGs (Fig. S3), suggesting the possibility of coevolution. To test the possibility of coevolution further, we selected two pairs of genes that belong to the Type P1 and Type O2‐1 ABCGs: ABCG31 of A. thaliana and dxK7M810 of G. max and HpaP802307 of H. arabidopsidis and dxG5ADX0 of P. sojae. We then calculated the evolutionary rates of the full‐size ABCGs, (logFC(dN/dS)) between random pairs of ABCG31 homologs in land plants and a pair of ABCGs in pathogenic oomycetes (HpaP802307 and dxG5ADX0) (Fig. 4B). The AtABCG31 and dxK7M810 pair (observed value) had a higher similarity of evolutionary rate with the pathogen pair than random pairs of ABCG31 homologs in land plants. Specifically, the similarity of the observed evolutionary rates was statistically significant when the difference of divergence time (logFC(dS)) was taken into account (minimal P = 0.012; Fig. 4B inset). This result suggests that ABCG31 homologs from two plant–pathogen pairs evolved at similar rates.
FIGURE 4.
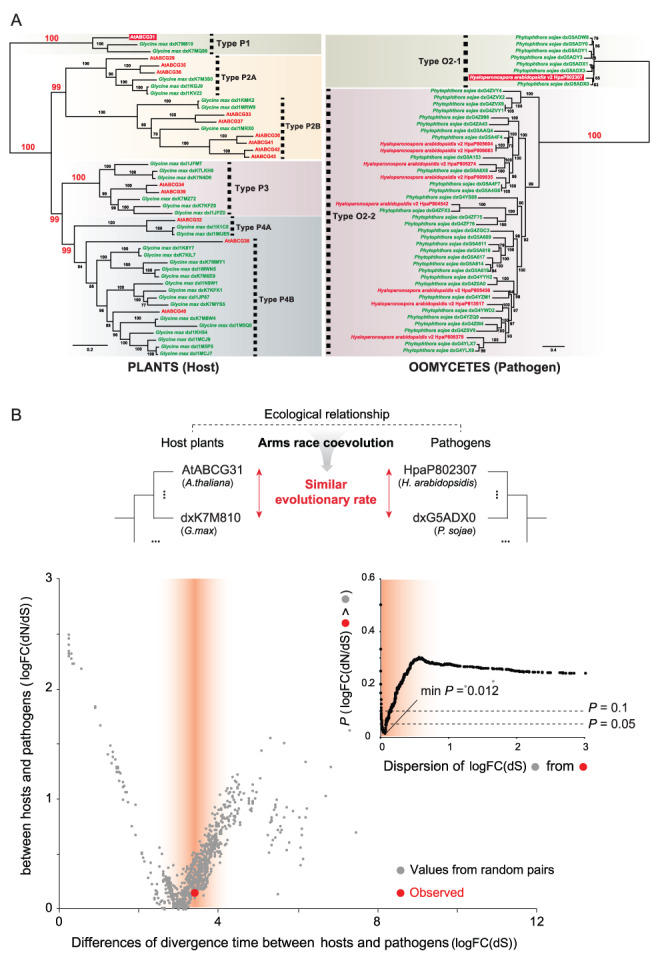
Phylogeny of the full‐size ABCGs and evolutionary rate differences between pairs of full‐size ABCGs in plant hosts and those in their pathogens. (A) The phylogeny was constructed using full‐size ABCG genes in host plants (Arabidopsis thaliana, G. max) and their pathogens (Hyaloperonospora arabidopsidis, P. sojae). Each host–pathogen pair is indicated with the same color (red: A. thaliana–H. arabidopsidis, green: G. max–P. sojae). The scale bar indicates the branch length measured in the number of amino acid substitutions per site. (B) The large graph shows the differences in evolutionary rates logFC(dN/dS) of the ABCG31 homologs from the two plant–pathogen pairs (observed value; red dot) compared to those from random pairs (gray dots). The observed value is from AtABCG31 of A. thaliana and dxK7M810 of G. max pair. Values from random pairs were obtained from pairs (n = 1225) of ABCG31 homologs in 50 different land plants. The difference of divergence time, logFC(dS), is used as an independent variable to control the intrinsic difference in divergence time‐scales between plant and pathogen pairs. The red gradient displays the dispersion of logFC(dS) centered from the observed value. Inset graph shows the P‐value of statistical tests calculated with the percentage of gray dots that have lower logFC(dN/dS) values than the red dot in the large graph.
We next used molecular biological techniques to test the possibility that AtABCG31 and HpaP802307 coevolved. We chose this pair of genes based on three reasons. First, H. arabidopsidis is a specialist pathogen that infects only A. thaliana (Thines et al. 2009). Second, AtABCG31 and HpaP802307 first diverged during the massive diversification of full‐size ABCGs (Fig. S3). Third, their evolutionary rates are similar (Fig. 4B). If AtABCG31 renders Arabidopsis plants more resistant to H. arabidopsidis infection, its expression may increase upon pathogen infection. Indeed, AtABCG31 expression increased more than twofold in the 4 days following H. arabidopsidis inoculation, whereas its expression decreased after the control water treatment (Fig. 5A). Generally the expression level of ABC transporters does not change dramatically in reponse to stimuli, but only around two‐ to three‐fold (Kang et al. 2010, Ko et al. 2014).
FIGURE 5.
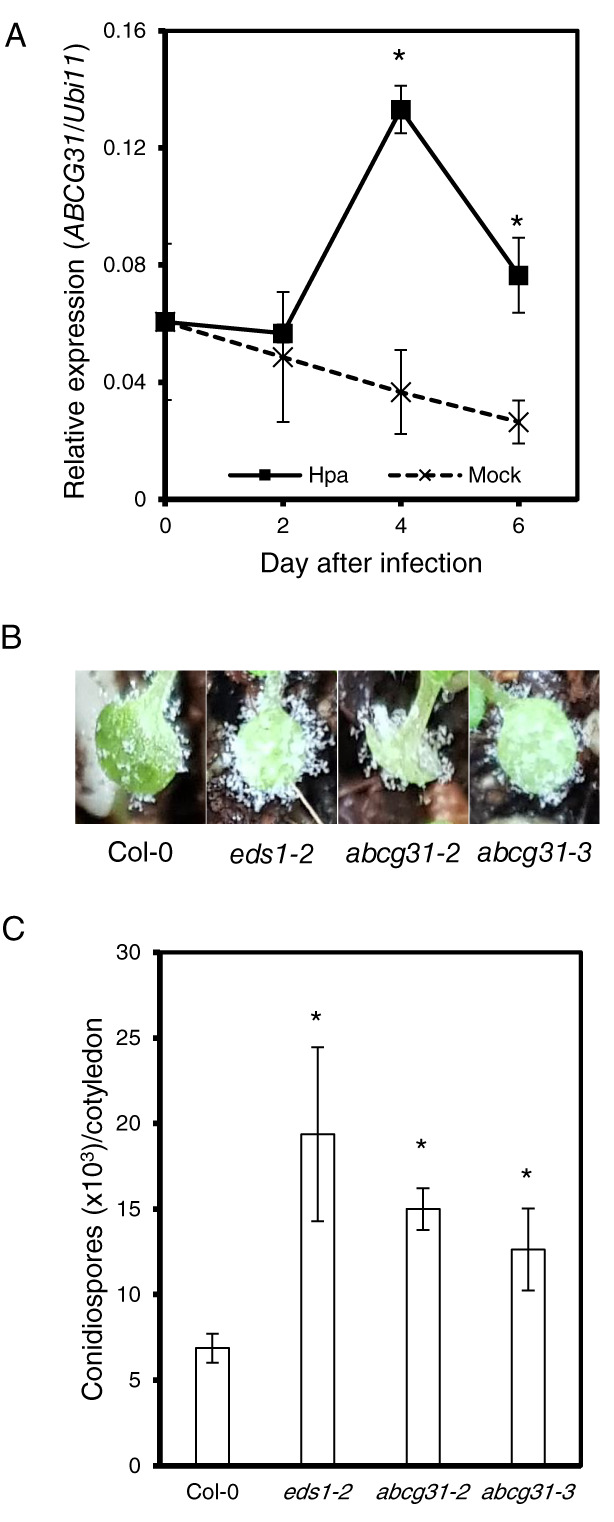
Responses of Arabidopsis abcg31 mutants to Hyaloperonospora arabidopsidis infection. (A) Expression level of AtABCG31 in Col‐0 wild‐type plants after infection with H. arabidopsidis isolate Noco2. Quantitative real time PCR was performed using total RNA extracted from Hpa or mock‐treated Col‐0 wild‐type cotyledons. (B) Photographs of diseased cotyledons 7 days after inoculation. (C) Numbers of conidiospores produced on 20 cotyledons 7 days after inoculation. *P < 0.05, Student's t‐test. Error bars represent standard errors from four biological replicates.
Next, we infected wild‐type Arabidopsis plants and two independent knockout alleles of atabcg31 (abcg31‐2, abcg31‐3; Choi et al. 2014) with H. arabidopsidis, and compared their disease susceptibility. For this purpose, we measured the asexual sporulation frequency of H. arabidopsidis on the cotyledons of Arabidopsis plants by counting the number of conidiosporangia. The Col‐0 eds1‐2 Arabidopsis mutant, which is highly susceptible to many pathogens (Aarts et al. 1998) due to disruption of EDS1 (ENHANCED DISEASE SUSCEPTIBILITY 1), was used as a positive control, and indeed the cotyledons of this plant were covered with many more H. arabidopsidis spores than were wild‐type Col‐0 plants (Fig. 5B,C). The abcg31‐2 and abcg31‐3 mutant cotyledons were also covered with more spores than those of the wild‐type (Fig. 5B,C), suggesting that AtABCG31 contributes to the resistance of Arabidopsis to H. arabidopsidis infection.
Finally, we constructed and compared 3D structural models of the two ABCG proteins. The structures of AtABCG31 and HpaP802307 were predicted using the structure of HsABCG2 (PDB ID: 6FFC) as template. Since HsABCG2 has only one transmembrane domain (TMD) and one nucleotide‐binding domain (NBD), which together form a homodimer as a functional unit, and because AtABCG31 and HpaP802307 have two units of TMD and NBD, we generated the first half and the second half TMD–NBD model of AtABCG31 and HpaP802307 independently, and then combined them to generate a full‐length model structure. AtABCG31 and HpaP802307 have 42.9 and 44.1% amino acid sequence similarity with HsABCG2, respectively. The 3D conformation predicted for AtABCG31 and HpaP802307 matched well at the TMD, where substrates are usually recognized (Fig. S5). Further structural studies might reveal similar substrate binding domains if AtABCG31 and HpaP802307 transport the same or similar secondary metabolites deployed during defense.
Discussion
In this study, we combined phylogenetic analyses with molecular genetic experiments, and obtained some interesting and consistent insights into ABCG evolution. First, we found evidence of an ancient origin of ABCG proteins in land plants followed by their explosive diversification (Fig. 2A). Second, we found that ABCGs of a large number of organisms clustered into seven distinct groups (Fig. 1), and many members belonging to the same group shared similar substrates. This is interesting, because ABCGs have been shown to transport such diverse substrates that it has not been possible to categorize these transporters based on their substrate specificities. Third, we analyzed the evolutionary relationships among ABCG genes of diverse eukaryotic phyla, and found that full‐size ABCGs explosively proliferated not only in land plants, but also in their destructive pathogens oomycetes (Figs 1;2 and S1, S2), and the phylogenetic patterns of these two clades mirrored each other. Lastly, we provided multiple lines of evidence from a range of other analyses, which suggested that ABCG genes in plants and oomycetes have coevolved (Figs 3, 4, 5 and Table 1). The substrates of these ABCG proteins are likely to be important for defense response of plants, although their identities remain mostly unknown.
A correlation between the wide host range and the abundance of ABCGs in plant pathogenic oomycetes
Based on their host range, oomycete species can be classified into two parasitic types: generalist and specialist. Interestingly, our phylogenetic analysis of ABCG transporters revealed that generalist pathogenic oomycetes have more full‐size ABCG transporters than do specialist oomycetes (Fig. S2 and Table 1). Therefore, the numbers of full‐size ABCG transpoters do not simply align with the phylogenetic lineage, but seem to be enriched in generalist pathogens. One exception was the case of the genus Albugo (Albuginales). Albugo species are equipped with special mosaic‐like genomes, which facilitate infection of a broad range of host plants (Kamoun et al. 2015). Thus, diversified full‐size ABCG transporters might have made at least some generalist oomycetes, including Peronosporales (Phytophthora and Hyaloperonospora) species and Pythiales (Pythium), more resistant to the host defense system.
Host–parasite coevolution of ABCG transporters as an evolutionary arms race
ABCG members multiplied and differentiated independently in land plants and oomycetes, but they might have coevolved to transport similar secondary metabolites via convergent evolution. This hypothesis can be supported by Haldane's theory of balancing selection that posits that coevolution of a host and pathogen leads to the maintenance of many highly variable defense genes in the genomes of both organisms (Haldane 1949). This idea was experimentally supported in studies of host–pathogen interactions between the water flea Daphnia and its microparasites (Decaestecker et al. 2007), and the plant Linum and a rust fungus Melampsora (Thrall et al. 2012). Recently, it has been reported that ABCG genes function in the defense response in both host plants and their pathogens. On the plant side, many ABCG genes encode transporters of defense chemicals. AtABCG36 mediates the secretion of secondary metabolites that are effective in defending plants from virulent bacterial and eukaryotic pathogens (Kobae et al. 2006, Stein et al. 2006). AtABCG40 is important for Phytophthora resistance (Wang et al. 2015) and resistance to other pathogens (Campbell et al. 2003). AtABCG34 secretes camalexin to the leaf surface, which protects the plant from necrotrophic fungal infection (Khare et al. 2017). Nicotiana benthamiana full‐size ABCGs (Nb‐ABCG1 and Nb‐ABCG2) are involved in defense against P. infestans (Shibata et al. 2016). Nicotiana tabacum NtPDR1 transports diterpenes known as antimicrobial secondary metabolites (Crouzet et al. 2013). On the pathogen side also, Botrytis cinerea, a pathogen of Arabidopsis, has a full‐size ABCG transporter, BcatrB, which is a critical virulence factor, without which the pathogen fails to infect the host plant (Stefanato et al. 2009).
In this study, we found three lines of evidence that the ABCGs of plants and their Phytophthora pathogens coevolved (Fig. S6). First, the divergence times of ABCGs in host plant and their pathogens are similar (Fig. 3). Second, there is an association between the wide host range and the abundance of ABCGs in plant pathogenic oomycetes; generalist parasite oomycetes have more full‐size ABCGs transporters than do specialists (Fig. S2 and Table 1). Third, the absence of ABCG31 in the atabcg31 knockout (abcg31‐2, abcg31‐3) increased disease susceptibility to infection by H. arabidopsidis (Fig. 5B,C). It is interesting that both the hosts and their pathogens used the same ABCG sub‐family transporters for the arms race. These transporters were chosen for the race most likely because they are particularly suitable for chemical warfare, by transporting defense chemicals.
Significance of coevolution study
Coevolution between interacting species has most likely contributed to biological diversity. However, rigorous genetic/molecular proof for this coevolution is almost impossible at the present, since, very often, the organisms involved are not amenable to genetic modifications and their mutants are not readily available. For example, many plant pathogens including H. arabidopsidis are not yet genetically modifiable, and their mutants are not available. Despite this difficulty, it is important to identify the molecular genetic basis of plant–pathogen coevolution (e.g. Occhipinti 2013), since coevolution contributes to the emergence and fixation of new genetic traits.
Previous coevolution studies in the field of plant molecular pathology focused on the initial events of recognition/detection of pathogens by plants, and evasion of the detection by pathogens; plants developed disease resistance (R) genes whose products, either directly or indirectly, recognize corresponding pathogen effectors [termed avirulence (AVR) proteins] whose primary function is to downregulate defense responses (Karasov et al. 2014). Such coevolutionary mechanisms have been extensively studied using various plants and microbial species that interact with plants, including RPP13 R gene of A. thaliana and ATR13 AVR gene of Hyaloperonospora parasitica (Allen et al. 2004), RPM1 of A. thaliana and avrRpm1 of Pseudomonas syringae (Stahl et al. 1999), RPS2 of A. thaliana and avrRpt2 of P. syringae (Mauricio et al. 2003), and RPS5 of A. thaliana and avrPphB of P. syringae (Tian et al. 2002). According to an evolutionary analysis of plant R genes (Gao et al. 2018), plant–pathogen interactions triggered the adaptive evolution of the R genes of the basal‐branching streptophytes including Charophyta, as a strategy to overcome pathogen infection. Similarly, our phylogenetic analysis revealed that the full‐size ABCGs in land plants (V3‐Type) have massively diversified, and these genes were clustered closely together with those of Chlorophyta and Charophyta (Fig. S1).
Our results suggest that the coevolution of plants and their pathogens might have occurred at a late stage of interactions, at the stage of exchanging secondary metabolites through ABC transporters, in addition to the initial recognition/detection step. In future, it would be interesting to find out whether the coevolution of proteins occured more or less evenly at all steps of the interactions between partners of interactions, or if it is more abundant for proteins involved in initial recognition by receptors and the final exchange of materials through transporters. Our study provides perspectives for future research on the ABCGs in diverse organisms, including substrate identities, physiological functions, and coevolution with ABCGs in their interacting partner organisms.
Author contributions
Y.L., H.S.Y., K.H.S., M.P., S.J., S.K.H. and C.H.C. designed the experiments; C.H.C., H.S.Y., M.P. and D.H. constructed phylogenetic trees; S.J., D.C., S.C. and H.K. performed Hpa infection experiments; M.‐S.K. computed the structures of ABCG transporters; Y.L., C.H.C., B.Y.C., H.S.Y., M.‐S.K., K.H.S., S.K. and M.P. analyzed the data and wrote the manuscript. All the authors agreed on the contents of the paper and posted no conflicting interest.
Data availibility statement
Sequence data we used in phylogenetic and Ka/Ks analyses are available from the Data S2. Sequences were collected from public databases including NCBI (https://www.ncbi.nlm.nih.gov/), UniProtKB (https://www.uniprot.org/), MMESTP (https://www.imicrobe.us/) and GIGAn DB (http://gigadb.org/). The accession numbers of the sequence are shown in the phylogenetic trees.
Supporting information
Data S1. Bayesian Evolutionary Analysis Utility (BEAUti) file provides all parameters used for BEAST analysis (Fig. S3).
Data S2. Sequence data we used in phylogenetic and Ka/Ks analyses.
Fig. S1. Phylogenetic tree of the full‐size ABCGs (PDR‐type transporters) in broad taxon sampling listed in Table S2.
Fig. S2. Phylogenetic tree of the 351 oomycete full‐size ABCGs listed in Table S3.
Fig. S3. Relative molecular clock trees of full‐size ABCGs.
Fig. S4. Density distribution of synonymous (Ks) mutational rates of full‐size ABCGs in the four species.
Fig. S5. Comparison of predicted structures of AtABCG31, HpaP802307 and other ABCG transporters.
Fig. S6. Coevolution of full‐size ABCGs in host plant and pathogenic oomycetes is supported by three lines of evidence in this paper.
Table S1. General characteristics of Arabidopsis ABCGs.
Table S2. Taxon list of ABCGs used in the phylogenetic analysis shown in Fig. 1
Table S3. Taxon list of full‐size ABCGs used in the phylogenetic analysis shown in Figs 2 and S1.
Acknowledgements
We thank Professor Enrico Martinoia (University of Zurich, Zurich, Switzerland) for critical comments on the manuscript and Dr. Jane Parker (MPIZ, Cologne, Germany) for making H. arabidopsidis isolate Noco2 available to us. Y.L. was supported by the National Research Foundation of Korea (NRF) grant (2018R1A2A1A05018173) funded by the Korean government (Ministry of Science and ICT). H.S.Y was supported by grants from the Collaborative Genome Program of the Korea Institute of Marine Science and Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (MOF) (20180430), the National Research Foundation of Korea (NRF‐2017R1A2B3001923) and the Next‐generation BioGreen21 Program (PJ01389003) from the RDA (Rural Development Administration), Korea. M.P. was supported by grants from Innovation Fund Denmark (LESSISMORE) and the Carlsberg Foundation (CF18‐111). K.H.S was supported by National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2018R1A5A1023599, SRC).
Edited by J. Schjoerring
Contributor Information
Kee Hoon Sohn, Email: khsohn@postech.ac.kr.
Hwan Su Yoon, Email: hwansu@gmail.com.
Youngsook Lee, Email: ylee@postech.ac.kr.
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene‐mediated signaling pathways in Arabidopsis . Proc Natl Acad Sci USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam A, Kowal J, Broude E, Roninson I, Locher KP (2019) Structural insight into substrate and inhibitor discrimination by human P‐glycoprotein. Science 363: 753–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albà MM, Castresana J (2004) Inverse relationship between evolutionary rate and age of mammalian genes. Mol Biol Evol 22: 598–606 [DOI] [PubMed] [Google Scholar]
- Allen RL, Bittner‐Eddy PD, Grenville‐Briggs LJ, Meitz JC, Rehmany AP, Rose LE, Beynon JL (2004) Host‐parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306: 1957–1960 [DOI] [PubMed] [Google Scholar]
- Andolfo G, Ruocco M, Di Donato A, Frusciante L, Lorito M, Scala F, Ercolano MR (2015) Genetic variability and evolutionary diversification of membrane ABC transporters in plants. BMC Plant Biol 15: 51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbona V, Gomez‐Cadenas A (2016) Metabolomics of disease resistance in crops. Curr Issues Mol Biol 19: 13–30 [PubMed] [Google Scholar]
- Asai S, Shirasu K, Jones JDG (2015) Hyaloperonospora arabidopsidis (downy mildew) infection assay in Arabidopsis . Bio Protoc 5: e1627 [Google Scholar]
- Bird D, Beisson F, Brigham A, Shin J, Greer S, Jetter R, Kunst L, Wu X, Yephremov A, Samuels L (2007) Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J 52: 485–498 [DOI] [PubMed] [Google Scholar]
- Campbell EJ, Schenk PM, Kazan K, Penninckx IA, Anderson JP, Maclean DJ, Cammue BP, Ebert PR, Manners JM (2003) Pathogen‐responsive expression of a putative ATP‐binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis . Plant Physiol 133: 1272–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella‐Gutierrez S, Silla‐Martinez JM, Gabaldon T (2009) trimAl: a tool for automated alignment trimming in large‐scale phylogenetic analyses. Bioinformatics 25: 1972–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Schneider RW, Hoy JW (1992) Taxonomic and phylogenetic analyses of ten Pythium species using isozyme polymorphisms. Phytopathology 82: 1234–1234 [Google Scholar]
- Choi H, Ohyama K, Kim YY, Jin JY, Lee SB, Yamaoka Y, Muranaka T, Suh MC, Fujioka S, Lee Y (2014) The role of Arabidopsis ABCG9 and ABCG31 ATP binding cassette transporters in pollen fitness and the deposition of steryl glycosides on the pollen coat. Plant Cell 26: 310–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouzet J, Roland J, Peeters E, Trombik T, Ducos E, Nader J, Boutry M (2013) NtPDR1, a plasma membrane ABC transporter from Nicotiana tabacum, is involved in diterpene transport. Plant Mol Biol 82: 181–192 [DOI] [PubMed] [Google Scholar]
- Dean M, Annilo T (2005) Evolution of the ATP‐binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet 6: 123–142 [DOI] [PubMed] [Google Scholar]
- Decaestecker E, Gaba S, Raeymaekers JAM, Stoks R, Van Kerckhoven L, Ebert D, De Meester L (2007) Host–parasite ‘Red Queen’ dynamics archived in pond sediment. Nature 450: 870–873 [DOI] [PubMed] [Google Scholar]
- Dick MW (2001) Straminipilous Fungi: Systematics of the Peronosporomycetes Including Accounts of the Marine Straminipilous Protists, the Plasmodiophorids and Similar Organisms. Kluwer Academic Publishers, Dordrecht: [Google Scholar]
- Diéguez‐Uribeondo J, Fregeneda‐Grandes JM, Cerenius L, Pérez‐Iniesta E, Aller‐Gancedo JM, Tellería MT, Söderhäll K, Martín MP (2007) Re‐evaluation of the enigmatic species complex Saprolegnia diclina–Saprolegnia parasitica based on morphological, physiological and molecular data. Fungal Genet Biol 44: 585–601 [DOI] [PubMed] [Google Scholar]
- Diéguez‐Uribeondo J, García MA, Cerenius L, Kozubíková E, Ballesteros I, Windels C, Weiland J, Kator H, Söderhäll K, Martín MP (2009) Phylogenetic relationships among plant and animal parasites, and saprotrophs in Aphanomyces (Oomycetes). Fungal Genet Biol 46: 365–376 [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, Heidel‐Fischer HM, Bekaert M, Rota J, Glöckner G, Platts AE, Heckel DG, Der JP, Wafula EK, Tang M, Hofberger JA, Smithson A, Hall JC, Blanchette M, Bureau TE,Wright SI, dePamphilis CW, Eric Schranz M, Barker MS, Conant GC, Wahlberg N,Vogel H, Pires JC, Wheat CW (2015) The butterfly plant arms‐race escalated by gene and genome duplications. Proc Natl Acad Sci USA 112: 8362–8366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin DC, Ribeiro OK (1996) Phytophthora Diseases Worldwide. American Phytopathological Society Press, St. Paul: [Google Scholar]
- Gao Y, Wang W, Zhang T, Gong Z, Zhao H, Han G‐Z (2018) Out of water: the origin and early diversification of plant R genes. Plant Physiol 177: 82–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gernhard T (2008) The conditioned reconstructed process. J Theor Biol 253: 769–778 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2011) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS (1949) Disease and evolution. Ric Sci 19: 68–76 [Google Scholar]
- Hwang JU, Song WY, Hong D, Ko D, Yamaoka Y, Jang S, Yim S, Lee E, Khare D, Kim K, Palmgren M, Yoon Hwan S, Martinoia E, Lee Y (2016) Plant ABC transporters enable many unique aspects of a terrestrial plant's lifestyle. Mol Plant 9: 338–355 [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14: 587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S (2003) Molecular genetics of pathogenic oomycetes. Eukaryot Cell 2: 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S, Furzer O, Jones JDG, Judelson HS, Ali GS, Dalio RJD, Roy SG, Schena L, Zambounis A, Panabières F, Cahill D, Ruocco M, Figueiredo A, Chen X‐R, Hulvey J, Stam R,Lamour K, Gijzen M, Tyler BM, Grünwald NJ, Mukhtar MS, Tomé DFA, Tör M, Van DenAckerveken G, McDowell J, Daayf F, Fry WE, Lindqvist‐Kreuze H, Meijer HJG,Petre B, Ristaino J, Yoshida K, Birch PRJ, Govers F (2015) The top 10 oomycete pathogens in molecular plant pathology. Mol Plant Pathol 16: 413–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, Martinoia E, Lee Y (2010) PDR‐type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci USA 107: 2355–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Yim S, Choi H, Kim A, Lee KP, Lopez‐Molina L, Martinoia E, Lee Y (2015) Abscisic acid transporters cooperate to control seed germination. Nat Commun 6: 8113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov TL, Horton MW, Bergelson J (2014) Genomic variability as a driver of plant–pathogen coevolution? Curr Opin Plant Biol 18: 24–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral‐Zettler LA, Armbrust EV, Archibald JM, Bharti AK, Bell CJ, Beszteri B, Bidle KD, Cameron CT, Campbell L, Caron DA,Cattolico RA, Collier JL, Coyne K, Davy SK, Deschamps P, Dyhrman ST, Edvardsen B, Gates RD, Gobler CJ, Greenwood SJ, Guida SM, Jacobi JL, Jakobsen KS, James ER, Jenkins B, John U, Johnson MD, Juhl AR, Kamp A, Katz LA, Kiene R,Kudryavtsev A, Leander BS, Lin S, Lovejoy C, Lynn D, Marchetti A, McManus G,Nedelcu AM, Menden‐Deuer S, Miceli C, Mock T, Montresor M, Moran MA, Murray S,Nadathur G, Nagai S, Ngam PB, Palenik B, Pawlowski J, Petroni G, Piganeau G,Posewitz MC, Rengefors K, Romano G, Rumpho ME, Rynearson T, Schilling KB,Schroeder DC, Simpson AGB, Slamovits CH, Smith DR, Smith GJ, Smith SR, SosikHM, Stief P, Theriot E, Twary SN, Umale PE, Vaulot D, Wawrik B, Wheeler GL,Wilson WH, Xu Y, Zingone A, Worden AZ (2014) The marine microbial eukaryote transcriptome sequencing project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol 12: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare D, Choi H, Huh SU, Bassin B, Kim J, Martinoia E, Sohn KH, Paek KH, Lee Y (2017) Arabidopsis ABCG34 contributes to defense against necrotrophic pathogens by mediating the secretion of camalexin. Proc Natl Acad Sci USA 114: E5712–E5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khunweeraphong N, Stockner T, Kuchler K (2017) The structure of the human ABC transporter ABCG2 reveals a novel mechanism for drug extrusion. Sci Rep 7: 13767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Chen J (2018) Molecular structure of human P‐glycoprotein in the ATP‐bound, outward‐facing conformation. Science 359: 915–919 [DOI] [PubMed] [Google Scholar]
- Ko D, Kang J, Kiba T, Park J, Kojima M, Do J, Kim KY, Kwon M, Endler A, Song WY, Martinoia E, Sakakibara H, Lee Y (2014) Arabidopsis ABCG14 is essential for the root‐to‐shoot translocation of cytokinin. Proc Natl Acad Sci USA 111: 7150–7155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobae Y, Sekino T, Yoshioka H, Nakagawa T, Martinoia E, Maeshima M (2006) Loss of AtPDR8, a plasma membrane ABC transporter of Arabidopsis thaliana, causes hypersensitive cell death upon pathogen infection. Plant Cell Physiol 47: 309–318 [DOI] [PubMed] [Google Scholar]
- Kovalchuk A, Driessen AJ (2010) Phylogenetic analysis of fungal ABC transporters. BMC Genomics 11: 177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, Moriyama Y, Shinozaki K (2010) ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci USA 107: 2361–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushalappa AC, Gunnaiah R (2013) Metabolo‐proteomics to discover plant biotic stress resistance genes. Trends Plant Sci 18: 522–531 [DOI] [PubMed] [Google Scholar]
- Lane TS, Rempe CS, Davitt J, Staton ME, Peng Y, Soltis DE, Melkonian M, Deyholos M, Leebens‐Mack JH, Chase M, Rothfels CJ, Stevenson D, Graham SW, Yu J, Liu T, Pires JC, Edger PP, Zhang Y, Xie Y, Zhu Y, Carpenter E, Wong GK‐S, Stewart CN (2016) Diversity of ABC transporter genes across the plant kingdom and their potential utility in biotechnology. BMC Biotechnol 16: 47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Kinch LN, Borek DM, Wang J, Wang J, Urbatsch IL, Xie XS, Grishin NV, Cohen JC, Otwinowski Z, Hobbs HH, Rosenbaum DM (2016) Crystal structure of the human sterol transporter ABCG5/ABCG8. Nature 533: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, de Clerck O (2012) Phylogeny and molecular evolution of the green algae. CRC Crit Rev Plant Sci 31: 1–46 [Google Scholar]
- Locher KP (2016) Mechanistic diversity in ATP‐binding cassette (ABC) transporters. Nat Struct Mol Biol 23: 487 [DOI] [PubMed] [Google Scholar]
- Makkonen J, Jussila J, Kokko H (2012) The diversity of the pathogenic oomycete (Aphanomyces astaci) chitinase genes within the genotypes indicate adaptation to its hosts. Fungal Genet Biol 49: 635–642 [DOI] [PubMed] [Google Scholar]
- Manolaridis I, Jackson SM, Taylor NMI, Kowal J, Stahlberg H, Locher KP (2018) Cryo‐EM structures of a human ABCG2 mutant trapped in ATP‐bound and substrate‐bound states. Nature 563: 426–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler‐Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese‐Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F,Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N,Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39: D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauricio R, Stahl EA, Korves T, Tian D, Kreitman M, Bergelson J (2003) Natural selection for polymorphism in the disease resistance gene Rps2 of Arabidopsis thaliana . Genetics 163: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan M, Gardiner A, Bailey K, Kemen E, Ward BJ, Cevik V, Robert‐Seilaniantz A, Schultz‐Larsen T, Balmuth A, Holub E, van Oosterhout C, Jones JDG (2015) Evidence for suppression of immunity as a driver for genomic introgressions and host range expansion in races of Albugo candida, a generalist parasite. Elife 4: e04550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Zhang Q, Ding W, Shan W (2014) Phytophthora parasitica: a model oomycete plant pathogen. Mycology 5: 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentewab A, Stewart CN Jr (2005) Overexpression of an Arabidopsis thaliana ABC transporter confers kanamycin resistance to transgenic plants. Nat Biotechnol 23: 1177–1180 [DOI] [PubMed] [Google Scholar]
- Nguyen L‐T, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ‐TREE: a fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Mol Biol Evol 32: 268–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occhipinti A (2013) Plant coevolution: evidences and new challenges. J Plant Interact 8: 188–196 [Google Scholar]
- Ofori PA, Mizuno A, Suzuki M, Martinoia E, Reuscher S, Aoki K, Shibata D, Otagaki S, Matsumoto S, Shiratake K (2018) Genome‐wide analysis of ATP binding cassette (ABC) transporters in tomato. PLoS One 13: e0200854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D, Beckmann M, Zubair H, Enot DP, Caracuel‐Rios Z, Overy DP, Snowdon S, Talbot NJ, Draper J (2009) Metabolomic analysis reveals a common pattern of metabolic re‐programming during invasion of three host plant species by Magnaporthe grisea . Plant J 59: 723–737 [DOI] [PubMed] [Google Scholar]
- Quilichini TD, Friedmann MC, Samuels AL, Douglas CJ (2010) ATP‐binding cassette transporter G26 (ABCG26) is required for male fertility and pollen exine formation in Arabidopsis thaliana . Plant Physiol 154: 678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K, Strader LC, Bailly A, Yang HB, Blakeslee J, Langowski L, Nejedla E, Fujita H, Itoh H, Syono K, Hejátko J, Gray WM, Martinoia E, Geisler M, Bartel B, Murphy AS, Friml J (2010) Arabidopsis PIS1 encodes the ABCG37 transporter of auxinic compounds including the auxin precursor indole‐3‐butyric acid. Proc Natl Acad Sci USA 107: 10749–10753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana TR, Fischer S, Wohlgemuth G, Katrekar A, Jung K, Ronald PC, Fiehn O (2010) Metabolomic and transcriptomic analysis of the rice response to the bacterial blight pathogen Xanthomonas oryzae pv. oryzae. Metabolomics 6: 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Ojika M, Sugiyama A, Yazaki K, Jones DA, Kawakita K, Takemoto D (2016) The full‐size ABCG transporters Nb‐ABCG1 and Nb‐ABCG2 function in pre‐ and post‐invasion defense against Phytophthora infestans in Nicotiana benthamiana . Plant Cell 28: 1163–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikant S, Gaudet R (2019) Mechanics and pharmacology of substrate selection and transport by eukaryotic ABC exporters. Nat Struct Mol Biol 26: 792–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J (1999) Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis . Nature 400: 667 [DOI] [PubMed] [Google Scholar]
- Stefanato FL, Abou‐Mansour E, Buchala A, Kretschmer M, Mosbach A, Hahn M, Bochet CG, Metraux JP, Schoonbeek HJ (2009) The ABC transporter BcatrB from Botrytis cinerea exports camalexin and is a virulence factor on Arabidopsis thaliana . Plant J 58: 499–510 [DOI] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sánchez‐Rodríguez C, Hou B‐H, Molina A, Schulze‐Lefert P, Lipka V, Somerville S (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Bartel B (2009) The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole‐3‐butyric acid. Plant Cell 21: 1992–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukkens Y, Bultreys A, Grec S, Trombik T, Vanham D, Boutry M (2005) NpPDR1, a pleiotropic drug resistance‐type ATP‐binding cassette transporter from Nicotiana plumbaginifolia, plays a major role in plant pathogen defense. Plant Physiol 139: 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NMI, Manolaridis I, Jackson SM, Kowal J, Stahlberg H, Locher KP (2017) Structure of the human multidrug transporter ABCG2. Nature 546: 504 [DOI] [PubMed] [Google Scholar]
- Theodoulou FL, Kerr ID (2015) ABC transporter research: going strong 40 years on. Biochem Soc Trans 43: 1033–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines M, Kamoun S (2010) Oomycete‐plant coevolution: recent advances and future prospects. Curr Opin Plant Biol 13: 427–433 [DOI] [PubMed] [Google Scholar]
- Thines M, Choi YJ, Kemen E, Ploch S, Holub EB, Shin HD, Jones JDG (2009) A new species of Albugo parasitic to Arabidopsis thaliana reveals new evolutionary patterns in white blister rusts (Albuginaceae). Persoonia 22: 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Tampé R (2018) Multifaceted structures and mechanisms of ABC transport systems in health and disease. Curr Opin Struct Biol 51: 116–128 [DOI] [PubMed] [Google Scholar]
- Thrall PH, Laine A‐L, Ravensdale M, Nemri A, Dodds PN, Barrett LG, Burdon JJ (2012) Rapid genetic change underpins antagonistic coevolution in a natural host‐pathogen metapopulation. Ecol Lett 15: 425–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Araki H, Stahl E, Bergelson J, Kreitman M (2002) Signature of balancing selection in Arabidopsis . Proc Natl Acad Sci USA 99: 11525–11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cordewener JH, America AH, Shan W, Bouwmeester K, Govers F (2015) Arabidopsis lectin receptor kinases LecRK‐IX.1 and LecRK‐IX.2 are functional analogs in regulating Phytophthora resistance and plant cell death. Mol Plant Microbe Interact 28: 1032–1048 [DOI] [PubMed] [Google Scholar]
- Xiong J, Feng J, Yuan D, Zhou J, Miao W (2015) Tracing the structural evolution of eukaryotic ATP binding cassette transporter superfamily. Sci Rep 5: 16724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Molina I, Ranathunge K, Castillo IQ, Rothstein SJ, Reed JW (2014) ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis . Plant Cell 26: 3569–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Bioinformatics 13: 555–556 [DOI] [PubMed] [Google Scholar]
- Yazaki K (2006) ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett 580: 1183–1191 [DOI] [PubMed] [Google Scholar]
- Yim S, Khare D, Kang J, Hwang J‐U, Liang W, Martinoia E, Zhang D, Kang B, Lee Y (2016) Postmeiotic development of pollen surface layers requires two Arabidopsis ABCG‐type transporters. Plant Cell Rep 35: 1863–1873 [DOI] [PubMed] [Google Scholar]
- Yu J, Ge J, Heuveling J, Schneider E, Yang M (2015) Structural basis for substrate specificity of an amino acid ABC transporter. Proc Natl Acad Sci USA 112: 5243–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li J, Zhao X‐Q, Wang J, Wong GK‐S, Yu J (2006) KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4: 259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xiao J, Wu J, Zhang H, Liu G, Wang X, Dai L (2012) ParaAT: a parallel tool for constructing multiple protein‐coding DNA alignments. Biochem Biophys Res Commun 419: 779–781 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Bayesian Evolutionary Analysis Utility (BEAUti) file provides all parameters used for BEAST analysis (Fig. S3).
Data S2. Sequence data we used in phylogenetic and Ka/Ks analyses.
Fig. S1. Phylogenetic tree of the full‐size ABCGs (PDR‐type transporters) in broad taxon sampling listed in Table S2.
Fig. S2. Phylogenetic tree of the 351 oomycete full‐size ABCGs listed in Table S3.
Fig. S3. Relative molecular clock trees of full‐size ABCGs.
Fig. S4. Density distribution of synonymous (Ks) mutational rates of full‐size ABCGs in the four species.
Fig. S5. Comparison of predicted structures of AtABCG31, HpaP802307 and other ABCG transporters.
Fig. S6. Coevolution of full‐size ABCGs in host plant and pathogenic oomycetes is supported by three lines of evidence in this paper.
Table S1. General characteristics of Arabidopsis ABCGs.
Table S2. Taxon list of ABCGs used in the phylogenetic analysis shown in Fig. 1
Table S3. Taxon list of full‐size ABCGs used in the phylogenetic analysis shown in Figs 2 and S1.


