Summary
An anaerobic enrichment with CO from sediments of hypersaline soda lakes resulted in a methane‐forming binary culture, whereby CO was utilized by a bacterium and not the methanogenic partner. The bacterial isolate ANCO1 forms a deep‐branching phylogenetic lineage at the level of a new family within the class ‘Natranaerobiia’. It is an extreme haloalkaliphilic and moderate thermophilic acetogen utilizing CO, formate, pyruvate and lactate as electron donors and thiosulfate, nitrate (reduced to ammonia) and fumarate as electron acceptors. The genome of ANCO1 encodes a full Wood–Ljungdahl pathway allowing for CO oxidation and acetogenic conversion of pyruvate. A locus encoding Nap nitrate reductase/NrfA ammonifying nitrite reductase is also present. Thiosulfate respiration is encoded by a Phs/Psr‐like operon. The organism obviously relies on Na‐based bioenergetics, since the genome encodes for the Na+‐Rnf complex, Na+‐F1F0 ATPase and Na+‐translocating decarboxylase. Glycine betaine serves as a compatible solute. ANCO1 has an unusual membrane polar lipid composition dominated by diethers, more common among archaea, probably a result of adaptation to multiple extremophilic conditions. Overall, ANCO1 represents a unique example of a triple extremophilic CO‐oxidizing anaerobe and is classified as a novel genus and species Natranaerofaba carboxydovora in a novel family Natranaerofabacea.
Introduction
In view of the possibility of an early soda ocean (created by CO2 weathering of Na/K‐rich basalt crust of volcanic origin) in the geological history of Earth and Mars and a probability of the same in the current ocean on Europa, there is an interest in astrobiology of the existing haloalkaline environments on Earth, such as terrestrial soda lakes and deep sea serpentinization area (Kempe and Kazmierczak, 2002; Herschy et al., 2014; Fox‐Pawel et al., 2016). One of the potential common substrates for extraterrestrial microbes would be CO, which can form by photolysis of CO2 under strong UV radiation (King, 2015).
Modern saline‐alkaline soda lakes represent rare examples of stable highly alkaline natural habitats due to the presence of high concentrations of soluble sodium carbonates, which can reach molar concentrations in hypersaline soda lakes. Such lakes are located in arid areas characterized by a hot climate with long periods of evaporative salt concentration (Schagerl, 2016). Thus, prokaryotes thriving in soda lakes often exhibit triple extremophilic circumstances, i.e. halo‐alkalo‐thermo‐phily (Mesbah et al. 2007, 2009; Sorokin et al., 2017).
Extensive knowledge has been accumulated in the past three decades on the functionally important microbial communities in soda lakes, both by culture‐dependent (Sorokin et al., 2015; Grant and Jones, 2016; Sorokin, 2017) and culture‐independent molecular approaches (Vauvorakis et al., 2016; 2018; Zorz et al., 2019). However, several pieces of the puzzle are still missing. In particular, whether anaerobic carbon monoxide oxidation (carboxydotrophy) is possible under extremely haloalkaline conditions. Aerobic carboxydotrophy has been shown for a group of highly salt‐tolerant soda lake alkaliphiles belonging to two closely related genera Alkalispirillum/Alkalilimnicola of the Gammaproteobacteria (Hoeft et al., 2007; Sorokin et al., 2010). The only anaerobic isolate reported as capable of CO oxidation at high pH is the acetogenic Alkalibaculum bacchii, a nonhalophilic, alkalitolerant member of the Eubacteriaceae family (Firmicutes), isolated from soil (Allen et al., 2010; Liu et al., 2012).
Three major types of anaerobic carboxydotrophy have been established: by acetogens via the Wood–Ljungdahl pathway, resulting in CO2, acetate and/or ethanol; by methanogens, resulting in methane or acetate; and by hydrogenogens, resulting in H2 and CO2 (Diender et al., 2015). Since protons are the final electron acceptors of hydrogenogens, the hydrogenogenic metabolism is highly improbable under soda lake conditions, while both acetogenic and methanogenic CO utilization are possible. There is a fourth (marginal) pathway of anaerobic carboxydotrophy, the direct anaerobic oxidation of CO in presence of a suitable electron acceptor, but it has not yet been well studied, except for sulfate‐reducing conditions, exemplified by Desulfotomaculum species (Parshina et al., 2010) and Archaeoglobus fulgidus (Henstra et al., 2007).
Our previous attempts to directly enrich either acetogenic or methanogenic CO‐utilizing haloalkaliphiles from soda lakes at mesophilic conditions and low salinity (0.6 M total Na+, pH 10, 30°C and 0.2 atm CO in the gas phase) resulted in a positive acetogenic enrichment consisting of five clostridia members distantly related to the genera Anaerobranca and Dethiobacter. However, efforts to further enrich the consortium failed and the culture was eventually lost. Recently, another trial was undertaken to find out whether CO could replace formate/H2 as the electron donor for triple extremophilic methyl‐reducing methanogens of the class Methanonatronarchaeia found in hypersaline soda lakes (Sorokin et al., 2017, 2018). Methane was indeed formed in such enrichments but not directly by methanogens. Instead, a triple extremophilic anaerobic bacterium capable of utilizing CO as the electron donor was responsible for the initial CO conversion. Its phenotypic, phylogenetic and genomic properties are described in this article.
Results
Enrichment and isolation of a haloalkaliphilic carboxydotrophic bacterium
In the primary enrichment, inoculated with anaerobic sediment from a hypersaline soda lake, methane formation at 50°C in the presence of CO/MeOH as substrates was only observed at the minimal CO concentration of 0.05 atm in the gas phase. The onset of methane accumulation occurred after a prolonged lag period (0.3% in the gas phase after 53 days) and reached a maximum of 3.2% at day 85. Interestingly, all CO had already been consumed at day 65, indicating that the CO consumption and methane formation were not synchronized. However, no methane formation occurred in control incubations lacking CO. In the second 1:100 transfer of this culture, two major cell morphotypes were observed: bean‐shaped motile cells and tiny angular cocci typical for the genus Methanonatronarchaeum. This assumption was later confirmed by isolating this methyl‐reducing methanogen from the consortium using formate instead of CO as the electron donor. It had 100% 16S‐rRNA gene identity to one of the previously isolated strains of M. thermophilum (Sorokin et al., 2017, 2018) and it was unable to use CO as the electron donor in pure culture. Therefore, it was concluded that the bean‐shaped bacterium was the primary CO‐utilizing organism in this consortium, which might have converted part of CO into formate utilized by the methyl‐reducing methanogen. The CO‐utilizing bacterium was finally separated from the methanogen in serial dilutions using higher CO concentration (10%–20% in the gas phase) and omitting MeOH from the medium. Furthermore, we repeated the enrichment at these modified conditions with the inoculum diluted from the start and the same bacterium was selected up to a 10−8 dilution, indicating that this was the only carboxydotrophic anaerobe capable of growing at the used triple extremophilic conditions. The isolated pure culture strain was designated ANCO1.
Phylogeny of strain ANCO1
The 16S rRNA‐based phylogenetic analysis placed strain ANCO1 into the Firmicutes order Natranaerobiales (elevated to the level of a class ‘Natranaerobiia’ according to the phylogenomic taxonomy, https://gtdb.ecogenomic.org) as a deep‐branching lineage (Fig. 1A). The closest relative of ANCO1 (90% sequence identity) is the moderately salt‐tolerant alkaliphilic peptidolytic anaerobe Natranaerobaculum magadiense, isolated from hypersaline soda lake Magadi in Kenya (Zavarzina et al., 2013). A more advanced phylogenomic analysis based on 120 conserved single copy marker genes confirmed the independent position of strain ANCO1 within the class ‘Natranaerobiia’ at the level of a new family (Fig. 1B): the relative evolutionary divergence (RED) value of the node of N. carboxydovora offshoot calculated using GTDBtk was 0.663 (values of 0.773 and 0.629 considered as family and order thresholds, respectively) (Parks et al. 2018). AAI values between ANCO1 and members of the type genus Natranaerobius did not exceed 48%.
Fig 1.
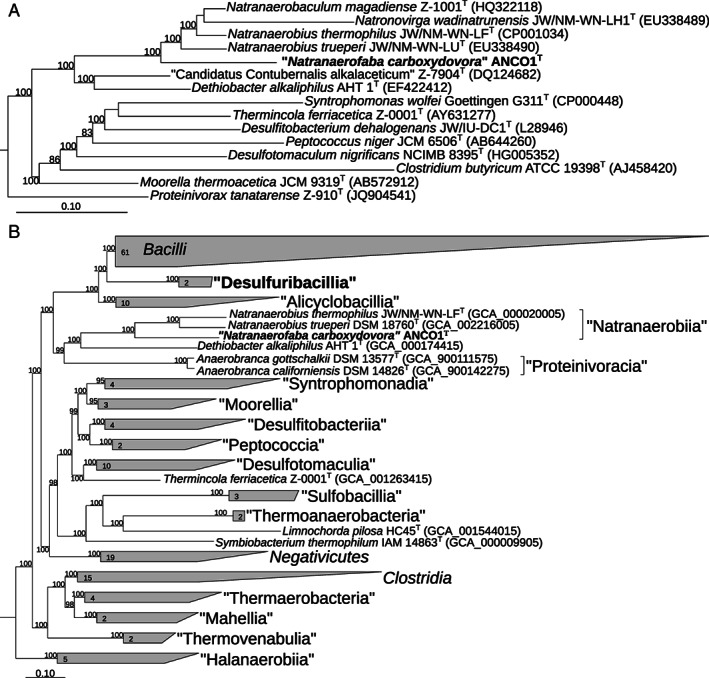
Phylogeny of ANCO1 based on (A) 16S rRNA gene sequence analysis and (B) the phylogenomic analysis of 120 bacterial conserved single‐copy protein coding genes (taxonomic designations correspond to the genome taxonomy database) (Parks et al., 2018). Only sequences from the type species were used. The trees were built using IQ‐TREE (Nguyen et al., 2015). Bootstrap values above 80% are shown at the nodes. Bar, 0.10 changes per position.
Cell morphology
ANCO1 has bean‐shaped cells of variable length, from 2 to 5 μm, depending on the growth rate, motile with a few flagella located on the concave side of the cell (Fig. 2). Although endospore formation was not observed at the growth conditions used, the genome does contain several extended loci encoding this capacity.
Fig 2.
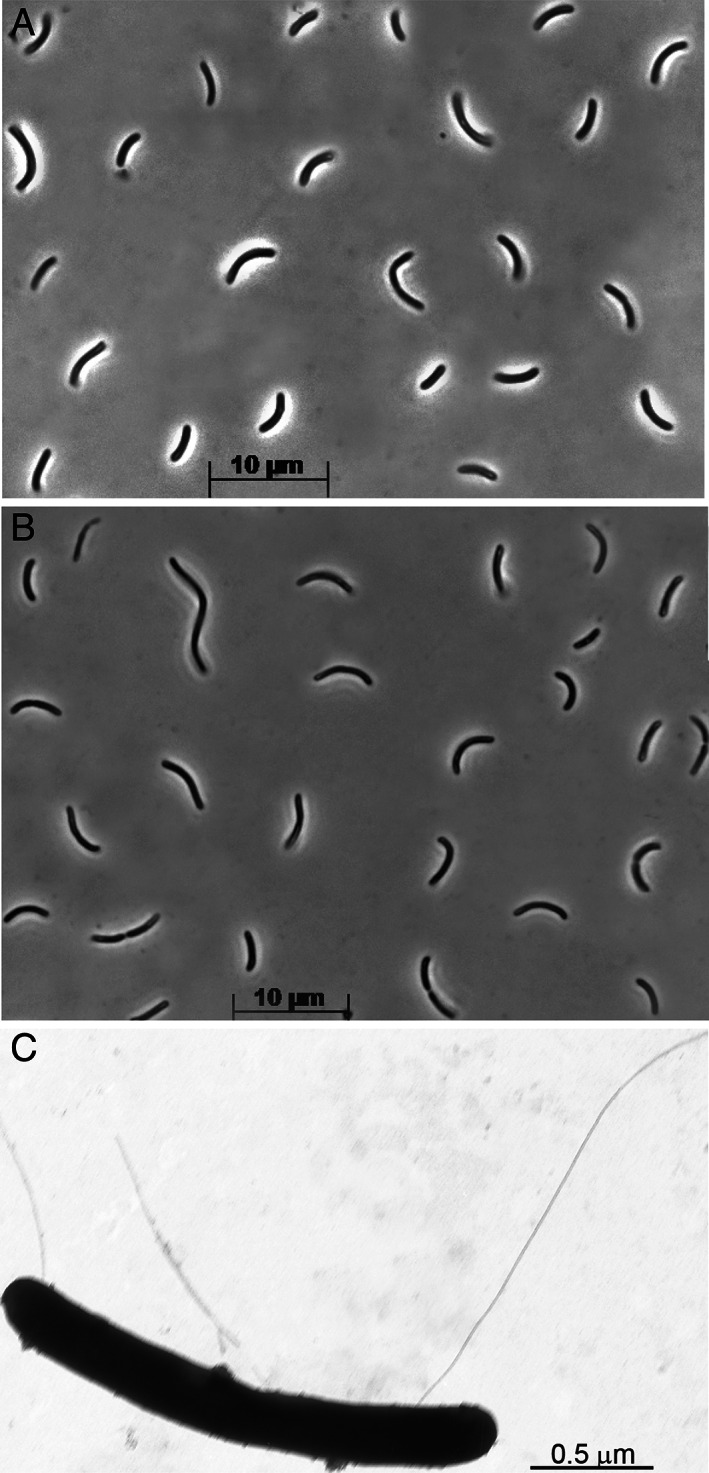
Cell morphology of strain ANCO1 grown at 4 M total Na+, pH 9.7 and 50°C. A,B. Phase‐contrast microphotographs of cells grown with CO and pyruvate, respectively. C. Electron microphotograph of a cell showing flagellation.
Membrane polar lipids
ANCO1 contains a wide range of intact polar lipids (IPLs) (Table 2). These all had either a phosphocholine (PC) or phosphoglycerol (PG) polar head group and the PCs were dominant over the PGs. The apolar cores of the IPLs comprised a distinctive range of different structures. The alkyl chains (containing between 14 and 19 carbon atoms) were bound to the glycerol backbone via either ester or ether bonds giving rise to diacyl lipids (two ester‐bonded chains), diether lipids and mixed acyl/ether lipids (including plasmalogen lipids). Mixed acyl/ether lipids have been reported in anaerobic bacteria, with some exceptions in aerobic bacteria, and are thought to improve cell resistance to extreme external conditions (Grossi et al., 2015). The majority (≤80%) of the lipids in ANCO1 were diethers, which are rare in bacteria but are characteristic of archaeal membranes. A number of the bacterial species that have been reported to produce diether lipids belong to thermophiles (e.g. Langworthy et al., 1983; Huber et al., 1992; Hamilton‐Brehm et al., 2013). Macrocyclic diethers were also detected in ANCO1 as a minor component. These have not previously been described in pure bacterial cultures but have been detected in environments including authigenic carbonates from marine evaporites, probably formed at high temperatures and high salinity (Baudrand et al., 2010; Ziegenbalg et al., 2012), iron sulfide nodules formed during the anaerobic oxidation of methane (van Dongen et al., 2007), geothermal hot springs (Pancost et al., 2006) and iron sulfides from a Mid‐Atlantic hydrothermal field (Blumenberg et al., 2007). The archaeal equivalent of this lipid, an isoprenoidal macrocyclic diether (known as macrocyclic archaeol) has been shown to augment membrane impermeability and stability in thermophilic methanogenic archaea (Koga et al., 1998), which explains its accumulation in extremophilic archaea (Sprott et al., 1991; Bale et al., 2019b).
Table 2.
Relative abundance (% of total) of core lipids released after acid hydrolysis of cells of ANCO1.
| Alkyl carbon number | Alkyl component | Individual % | Carbon number sum % |
|---|---|---|---|
| 14 | C14:0 FAa | 1.6 | 15.5 |
| C14:0 MGEb | 13.9 | ||
| 15 | iso C15:0 MGEb | 0.3 | 0.5 |
| C15:0 MGEb | 0.2 | ||
| 16 | C16:0 FAa | 2.9 | 5.8 |
| C16:0 DAc | 0.3 | ||
| C16:0 MGEb | 2.6 | ||
| 17 | 10‐Me C16:0 FAa | 3.9 | 23.3 |
| iso + anteiso C17:0 DAc | 17.9 | ||
| iso C17:0 MGEb | 0.7 | ||
| anteiso C17:0 MGEb | 0.6 | ||
| C17:0 MGEb | 0.2 | ||
| 18 | C18:0 FAa | 2.4 | 3.1 |
| C18:0 DAc | 0.2 | ||
| 1‐OH C18:0 alcohol | 0.5 | ||
| 19 | iso C19:0 FAa | 0.3 | 1.4 |
| iso C19:0 DAc | 1.1 | ||
| Dialkyls | C29:0 DGEd | 0.5 | |
| C31:0 DGEd | 21.1 | ||
| C31:0 Macrocyclic DGEd | 1.3 | ||
| C32:0 DGEd | 3.1 | ||
| C33:0 DGEd | 4.4 | ||
| C34:0 DGEd | 12.8 |
Components formed during acid hydrolysis.
Fatty acid derived from ester‐bound alkyl chains.
Monoalkyl glycerol ethers, not including glycerol carbons.
Dimethyl acetals derived from alk‐1‐enyl ether substituents of plasmalogen lipids.
Dialkyl glycerol ethers (sum of two alkyl chains, not including glycerol carbons). Trace level components (≤ 0.1%) are not given.
The individual constituents of the core lipids were also analysed directly in more detail, after hydrolysis of the biomass to cleave off the polar head groups (see Supporting Information Fig. S2, e.g., of structures and theoretical hydrolysis schemes). This procedure released fatty acids (FA; formed from ester bound alkyl chains), dimethyl acetals [DA; formed from plasmalogen lipids; van Gelder et al. (2014)] and mono and dialkyl glycerol ethers (MGEs and DGEs; formed from ether‐bound alkyl chains). Both straight chain and branched alkyl chains were present. As the DGEs contain two alkyl chains (Supporting Information Fig. S2), their total number of alkyl carbon atoms range between 29 and 34. Overall conclusion of the lipid analysis is that its unusual composition most probably reflects the triple extremophilic nature of strain ANCO1.
The respiratory lipoquinoines were not detectable in ANCO1, either because they were present at a very low level or because they are not produced. The latter is likely since the genome does not contain the genes encoding the enzymes required for menaquinone biosynthesis.
Growth physiology of strain ANCO1
Growth with CO as the electron donor (and other electron donors) was only possible in the presence of at least 100 mg l−1 of yeast extract. After gradual adaptation, ANCO1 was able to tolerate up to 50% CO in the gas phase. However, the maximum biomass yield reached only 10 mg cell protein l−1 even at highest CO concentration, and the full CO consumption was observed only at 5%–10% CO. The only gaseous product detected was CO2, although most of it was apparently absorbed into the highly alkaline culture medium (Supporting Information Fig. S1). Liquid analysis at the end point in the incubation with 5% CO (11.5 mM) showed formation of two organic products – acetate as a major and formate as a minor component (2.5 and 0.8 mM, respectively), indicating that the mode of anaerobic CO conversion in ANCO1 is homoacetogenic (4 mol CO > 1 mol acetate). Both formate and acetate can be utilized by Methanonatroarchaeum (as electron donor and C‐source, respectively) which might explain why the initial enrichment supported growth of this methyl‐reducing methanogen.
An enzymatic assay confirmed the presence of anaerobic CODH activity in strain ANCO1 (1.77 ± 0.22 μmol min−1 of CO conversion per mg protein in cell free extract). The only another compound supporting acetogenic growth from a range of substrates tested (H2, formate, MeOH, EtOH, PrOH, BuOH, lactate, and sugars) was pyruvate, which was converted into acetate and lactate with a minor formation of H2 (0.2–0.3 mM).
The ability of ANCO1 to use various electron acceptors was first tested with the two positive electron donors, i.e. CO and pyruvate. A definite growth stimulation was observed with fumarate and thiosulfate. The latter was partially reduced to sulfite and sulfide, while succinate formation from fumarate was very weak, which is difficult to explain, especially because fumarate reductase genes are present in the genome (Table 1).
Table 1.
Utilization of various electron donors/acceptors by strain ANCO1 during anaerobic growth at 48°C in a medium containing 4M total Na+, pH 9.7, supplemented with 500 mg l−1 of yeast extract as the carbon source.
| e‐donor | e‐acceptor | Growth | Biomass yield increase (times)a | Products from donor | Products from acceptor |
|---|---|---|---|---|---|
| CO | − | + | Acetate/formate | − | |
| Fumarate | + | 1.5 | Acetate | Succinate (0.5 mM) | |
| Thiosulfate | + | 1.2 | nd | HS− (up to 8 mM) | |
| Pyruvate | − | + | Acetate/lactate/H2 | − | |
| Fumarate | + | 2.2 | Acetate/lactate | Succinate 0.5 (mM) | |
| Thiosulfate | + | 1.6 | nd | HS− (up to 11 mM) | |
| Lactate | − | − | |||
| Fumarate | w | − | Acetate | Succinate (0.5 mM) | |
| Thiosulfate | + | − | Acetate | HS− (up to 12 mM) | |
| Formate | − | − | |||
| Fumarate | + | − | CO2 | Succinate (5.7 mM) | |
| Thiosulfate | + | − | HS− (up to 4 mM) | ||
| Nitrate/nitrite | + | − | NH3 (up to 5 mM) |
In comparison to growth without acceptor; nd, not determined;
The genomic data (see below) suggested that ANCO1 has a potential for dissimilatory reduction of nitrate to ammonia (DNRA). However, tests with CO and pyruvate showed neither growth stimulation nor nitrate (5 mM) consumption, and addition of even 2 mM nitrite inhibited growth. Therefore, other potential electron donors (suggested by the genome composition) were tested: formate, H2 and lactate. The latter two were negative, while anaerobic growth with formate as the electron donor was possible with thiosulfate, fumarate and nitrate/nitrite. The ammonifying growth of ANCO1 was not easy to prove. When nitrate was added at 5 mM from the start, it was converted only to nitrite. At 2 mM initial concentration, however, ammonifying growth with formate as donor was possible up to two further additions of 2 mM portions of nitrate. In total, 6 mM of nitrate were consumed without formation of intermediate nitrite and 4 mM NH3 was detected in the end of incubation. Nitrite could also support ammonifying growth with formate, but only when it was added at low concentration of 2 mM together with formate (50 mM) to a culture, pre‐grown with pyruvate or to a culture started to grow with formate + fumarate. It was possible to add in total up to 6 mM nitrite in three portions and the final ammonia production reached 5.3 mM with a prominent biomass increase in comparison to the control without nitrite addition. A direct addition of even 2 mM nitrite to the formate culture from the start revealed the complete absence of growth. Furthermore, formate was the only electron donor for which significant amount of succinate was detected during growth with fumarate as the electron acceptor. On the other hand, sulfidogenic growth on formate with thiosulfate produced the least sulfide from other positive cases (Table 1).
Lactate as the potential electron donor was also tested with fumarate and thiosulfate as the electron acceptors. In both cases, growth was observed, with very active sulfide formation from thiosulfate, but only trace amount of succinate was formed during growth with fumarate. No growth was detected with H2 at any tested conditions (Table 1 ).
Influence of salt and pH on growth of ANCO1 was tested with pyruvate + fumarate. Both profiles showed growth only at a limited range, indicating an extreme specialization of this organism to live in hypersaline soda lake conditions (Fig. 3). The salt profile (obtained at pH 9.5) showed that ANCO1 is an extreme halophile, growing within a range of total Na+ from 2.5 to 4.5 M (optimum at 4 M). It is an obligate alkaliphile, growing within a narrow pH range above 9 and up to 10.5 (optimum at 9.5; the final pH measured at 30°C). On its temperature dependence, ANCO1 can be qualified as a moderate thermophile with the growth range from 35 to 56°C and an optimum at 48–50°C.
Fig 3.
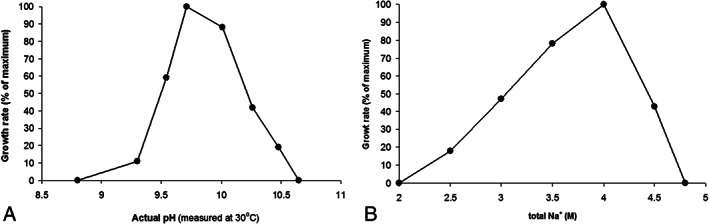
Influence of pH (at 4 M total Na+) (A) and salinity (at pH 9.5) (B) on growth of strain ANCO1 with pyruvate + fumarate at 48°C. The data are mean from parallel duplicate cultures.
Functional genomic and proteomic analyses
The genome of ANCO1 (accession number CP054394) was assembled into a single contig of 3.26 MB, which is predicted to be circular and has a GC content of 35.3 mol%. Plasmids were not detected. Annotation produced 3120 coding sequences. The key functional pathways found in the genome of ANCO1 are summarized in Fig. 4 and the identified genes/proteins are listed in the Supporting Information Table S1. The key expressed functional proteins are shown in the Supporting Information Fig. S3 and listed in the Supporting Information Table S2. The full range of expressed proteins is provided in the extended proteomic data file.
Fig 4.
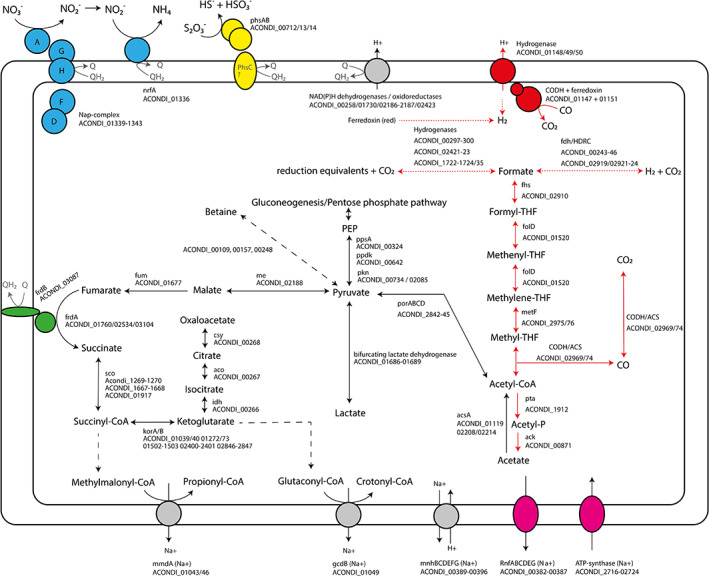
Overview of the metabolism of ANCO1, including associated locus tags. Dashed lines represent multiple reactions. Dotted lines represent putative hydrogenase reactions taking place in the metabolism. Subunits of complexes that are shadowed in grey could not be found. Colours indicate specific pathways: CO‐utilization pathways (red), Rnf‐complex and ATP‐synthase (pink), fumarate respiratory complex (green), nitrate respiration complex (blue), thiosulfate respiration complex (yellow). CODH: carbon monoxide dehydrogenase; ACS: acetyl‐CoA synthase; fdh/HDRC: formate dehydrogenase; formyl‐THF synthase; metF: methylene‐THF reductase; folD: formyl‐THF cyclohydrolase/ methylene‐THF dehydrogenase; pta: phospho‐acetyltransferase; ack: acetate kinase; acsA: acetyl‐CoA synthase; por: pyruvate oxidoreductase; ppd: phosphopyruvate decarboxylase; pps: phosphoenolpyruvate synthase; pkn: pyruvate dehydrogenase; me: malic enzyme; fum: fumarate hydratase; frd: fumarate reductase; sco: succinyl‐CoA synthetase; kor: 2‐oxoglutarate oxidoreductase; idh: isocitrate dehydrogenase; aco: cis‐aconitase; csy: citrate synthase; mmd: methylmalonyl‐CoA decarboxylase; gcd: glutaconyl‐CoA decarboxylase; mnh: Na+/H+ antiporter; nrfA: nitrite reductase; phs: thiosulfate reductase. [Color figure can be viewed at wileyonlinelibrary.com]
The ANCO1 genome contains all the genes of the Wood–Ljungdahl pathway and corresponding enzymes were indeed detected in the proteome of ANCO1 grown on CO and pyruvate. The presence of three CODH‐encoding genes in the genome and the detection of CODH activity in cell free extracts confirms the ability of the bacterium to utilize CO as electron donor. ANCO1 encodes for one CODH/ACS cluster, likely involved in CO‐oxidation as well as CO2 fixation. When the organization of this gene cluster is compared to corresponding CODH/ACS clusters of other carboxydotroph genomes, most similarity is observed with the thermophilic acetogens Moorella thermoacetica and Carboxydothermus hydrogenoformans (Fig. 5). Next to a CODH/ACS cluster, ANCO1 encodes for two other CODHs that are likely monofunctional. One lies adjacent to a hydrogenase complex (ACONDI_01147‐1151). However, carboxydotrophic hydrogenogenic activity was not apparent from the physiological tests (no H2 formation from CO), which means that this hydrogenase is not linked with the CODH, like, for example, Ech is.
Fig 5.
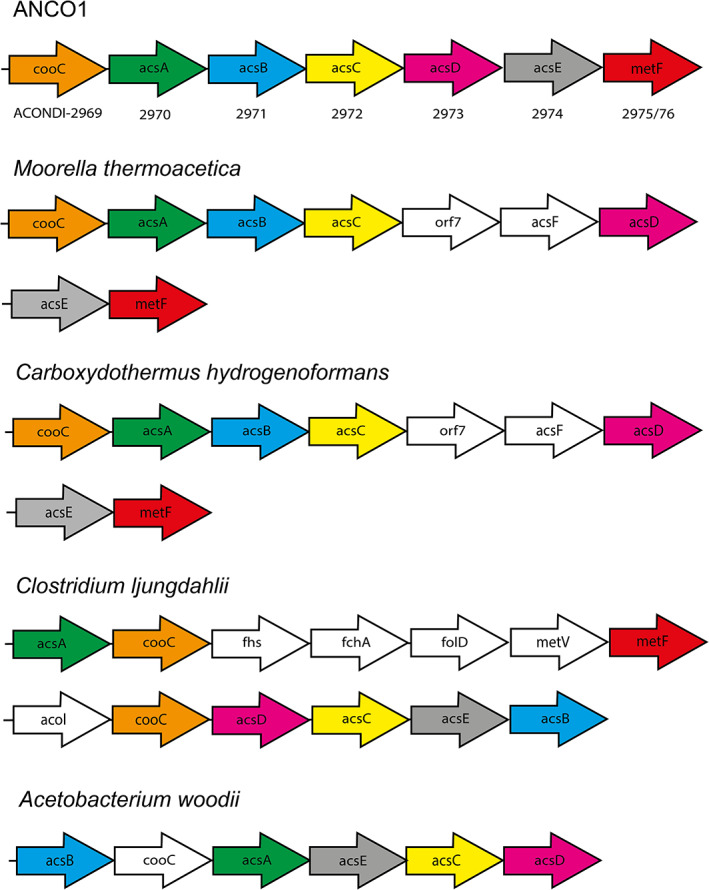
Organization of the CODH/acetyl‐CoA synthase (ACS) operon in ANCO1 compared to other carboxydotrophs. Gene abbreviations: cooC: CODH chaperone; acsA: CODH; acs B: ACS; acsC: corrinoid iron–sulfur protein large subunit; orf7: unknown; acsF: ACS chaperone; acsD: corrinoid iron–sulfur protein small subunit; acsE: methyltransferase A; fhs: formyl‐THF synthase; metF: methylene‐THF reductase; acoL: dihydrolipoamide dehydrogenase; folD: formyl‐THF cyclohydrolase/methylene‐THF dehydrogenase. [Color figure can be viewed at wileyonlinelibrary.com]
The annotated cytoplasmic Fe‐only hydrogenases exhibit similarity to bifurcating NADPH‐dependent hydrogenases potentially involved in redox distribution, in particular to recycle NAD(P)H and ferredoxin pools used as reductants for biosynthesis, building membrane potential and anaerobic respiration (Buckel and Thauer, 2013). Several formate dehydrogenases are also present, of which two (ACONDI 00243‐00246 and 02919‐02924) show similarity with the CO‐reactive hydrogenase of Acetobacterium woodii (HDCR type, Schuchmann and Müller, 2013), and may be used to link hydrogen oxidation to formate production. Genes encoding acetate kinase and acetyl‐phosphotransferase are present, enabling acetate production from acetyl‐CoA and energy conservation.
Comparative proteomic analysis of cells grown acetogenically either with CO or with pyruvate was performed in order to understand more about the CO‐specific metabolism of ANCO1. However, it appears that the same set of enzymes involved mostly in the Wood–Ljungdahl pathway and pyruvate oxidation were equally expressed (Supporting Information Fig. S3 and Table S2). This might indicate that both CO and pyruvate are funnelled into the same pathway.
Regarding key energy‐related reactions, ANCO1 encodes for a Rnf‐complex and F1F0 ATP synthase (both annotated as sodium‐translocating). In addition, two membrane bound primary sodium‐translocating decarboxylases are present: glutaconyl‐CoA and methylmalonyl‐CoA decarboxylase, which are potentially involved in the sodium‐based bioenergetics and a control of sodium levels and osmoregulation in anaerobic alkaliphiles (Dimroth and Schink, 1998; Buckel, 2001). Further fate of the thioester products (propionyl‐CoA and crotonyl‐CoA) of these two putative reactions is difficult to predict, but, likely, they might be channelled into anabolic routs, such as amino acid or lipid biosynthesis.
Several proton/sodium antiporters are also encoded, including two multi‐subunit Mnh complexes, one independent (ACONDI 02179‐02187) and another one in a large locus together with the Na+‐Rnf complex (ACONDI 00389–00396), enabling linkage of the proton and sodium gradients and maintenance of pH homeostasis. Also, a secondary sodium pump ‐ potassium‐stimulated Na+‐pyrophosphatase, involved the ATP‐dependent Na+ removal from the cytoplasm is encoded by ACONDI 00187.
For osmoprotection, the genome predicts two potential pathways of glycine betaine biosynthesis. The first is sequential glycine methylation (ACONDI 00109/00157), including glycine/sarcosine and sarcosine/N,N‐dimethylglycine methyltransferases (Roberts, 2005). The second is the oxidative pathway from carnitin (ACONDI 01812/01814), including l‐carnithine dehydrogenase LcdH and 3‐keto‐5‐aminohexanoate cleavage enzyme Kce (Rebouche, 1998). In addition, a presence in the ANCO1 genome of genes coding for multiple types of transporters for betaine/carnitin/choline import (Angelidis and Smith, 2003; Pflüger and Müller, 2004; Ongagna‐Yhombi et al., 2015) strongly suggest that active transport of betaine and its precursors is active in the ANCO1 as well. These include the following families: BCCT, Gbu, BetS, OpuAC, OpuAB, OpuE and OpuD (the full list with the locus tags is present in the Supporting Information Table S1). A possible source of betaine for import might be yeast extract required by ANCO1 for growth at any conditions. Furthermore, potassium may play a significant role in osmoprotection too, judging from the presence of a large repertoire of genes encoding potassium import/export (KefC, TrkA, TrkAH, KtrAB, NhaP2, Kch) (Supporting Information Table S1). Calculation of the total proteome acidity using the IPC calculator (http://isoelectric.org/calculate.php) showed a bimodal distribution of the pI value, with a major moderately acidic peak between pH 4 and 6 and a smaller basic peak between pH 8 and 10 (Supporting Information Fig. S4), indicating that ANCO1 is most probably using the ‘salt‐out’ strategy as the dominant osmoprotection mechanism with potassium import as a secondary mechanism. But for the definite conclusion, both intracellular betaine and potassium accumulation needs to be determined.
Despite the inability to grow on sugars, the genome of ANCO1 encodes for the complete pentose‐phosphate pathway and glycolysis. However, it lacks sugar transporters, suggesting that these pathways are most likely used for gluconeogenesis. The presence of pyruvate oxidoreductase enables conversion of acetyl‐CoA into pyruvate for assimilatory metabolism. An incomplete TCA‐cycle lacking malate dehydrogenase is encoded by the genome of ANCO1.
Despite the absence of lipoquinones (both by direct analysis and predicted by the lack of genes encoding menaquinone biosynthesis), the genome analysis confirms the potential of ANCO1 to obtain energy by nitrate/nitrite ammonifcation and thiosulfate‐ and fumarate‐dependent respiration found in growth experiments. For DNRA, ANCO1 potentially uses an unusual module encoded in a single locus consisting of an extracellular truncated Nap type respiratory nitrate reductase NapAGHDF (ACONDI 01139‐01143) and an exported ammonifying nitrite reductase NrfA (ACONDI 01136). The locus, however, appears to lack NrfH, a membrane tetraheme c involved in electron transport to NrfA from quinones. As the NrfA present in the genome is predicted to have a single transmembrane helix and lies in the same operon as the Nap nitrate reductase, this might indicate a possibility for direct electron transfer from a membrane electron donor (which is still unknown) and an interaction between the two complexes. On the other hand, the genome also contains another locus that encodes a mosaic redox complex with an uncertain (to predict) function (ACONDI 02948‐02952/02956). It includes a membrane diheme b556 cytochrome of the PhsC/NarI type (might be a missing subunit of the Phs operon, see below), followed by a putative NarG (catalytic subunit of the membrane respiratory nitrate reductase). Next, a tetraheme c cytochrome of the NapC/CymA type containing a single transmembrane loop is encoded. While not homologous to the integral membrane NrfH, it still might potentially be involved in the DNRA of ANCO1. The last part of this locus codes for the electron transfer complex FixXCA.
The genome of ANCO1 includes an operon coding for PhsAB complex (both subunits are exported) potentially involved in a 2‐electron reduction of thiosulfate, but apparently lacking its membrane electron‐transferring anchor PhsC, which might be encoded in another genomic locus (see above). Subunits of fumarate reductase (FrdAB) potentially involved in fumarate respiration are also present. A membrane associated NADPH oxidoreductases and a formate dehydrogenase (ACONDI‐00243‐00246) might be involved in donation of electrons to the respiration metabolism. However, the absence of respiratory lipoquinones still questions how those are linked to the electron‐accepting respiratory complexes. The only candidate for the electron transfer encoded in the genome is a low‐potential diheme c split‐Soret cytochrome (ACONDI‐02684), which is exported and attached to membrane. Such cytochromes serve in electron transport of sulfate‐reducing bacteria (Devreese et al., 1997).
Despite the observed lactate production and utilization by the strain, lactate dehydrogenase could not be clearly annotated. However, the genome encodes for an operon encoding for a bifurcating lactate dehydrogenase, similar to an earlier reported bifurcating lactate dehydrogenase complex (Weghoff et al., 2015). This complex might be involved in the utilization of lactate for respiration and could additionally be involved in lactate production when grown on pyruvate, which is in line with the proteomics data showed relatively high abundance of this complex in cell grown on pyruvate.
Discussion
Strain ANCO1 represents the first example of an extremely haloalkaliphilic CO‐utilizing anaerobe. It is a member of a deep‐branching phylogenetic lineage in the phylum Firmicutes, class ‘Natraanaerobiia’, which currently consists of only three genera found in soda lakes: the extremely salt‐tolerant genera Natranaerobius and Natronovirga (Mesbah et al., 2007; Mesbah and Wiegel, 2009) and a moderately salt‐tolerant genus Natranaerobaculum (Zavarzina et al., 2013). All of them are moderately thermophilic, obligately anaerobic fermentative heterotrophs with potential for anaerobic respiration, and in these the new member of this branch, strain ANCO1, is most similar to Natranaerobius thermophilus, which is also evident from the genome analysis (see Supporting Information Table S1). However, the ability to utilize CO as electron donor has not been reported for any of the ANCO1 relatives and the available genomes from the Natranaerobius species lack the respective genetic potential for such metabolism. The CODH/ACS cluster in ANCO1 is similar to that of other thermophilic carboxydotrophs, like Moorella thermoacetica and Carboxydothermus hydrogenoformans (Fig. 5). The main product of CO conversion by strain ANCO1 is acetate, similar to what is observed in M. thermoacetica (Drake and Daniel, 2004). Thermophilic carboxydotrophs M. thermoacetica and C. hydrogenoformans lack the RnF‐complex and employ energy converting hydrogenases (Ech) in order to generate a cation gradient and produce ATP (Wu et al., 2005; Schuchmann and Müller, 2014). Although a CODH‐hydrogenase like gene‐cluster is found in the genome of ANCO1 (Fig. 4), the absence of clear hydrogen production suggests it is not employing it. In addition, this cluster does not show the typical Ech structure, indicating that in contrast to other CO‐utilizing thermophiles, ANCO1 uses the Rnf‐complex for its energy metabolism.
Despite the presence of soluble hydrogenases encoded in the genome, strain ANCO1 was unable to grow acetogenically on H2/CO2 or use H2 as electron donor for anoxic respiration. The strain did produce minor amounts of H2 when grown on pyruvate. This raises the question why strain ANCO1 is able to use CO but not H2 to drive acetogenesis. A possible reason are the thermodynamic constrains imposed during H2‐dependent acetogenesis – the fact that the standard redox potential of the pair 2H++2e−/H2 (≈ −400 mV) is not sufficiently negative to reduce ferredoxin, demands for bifurcation mechanisms to perform CO2 reduction in the WLP (Schuchmann and Müller, 2012; Buckel and Thauer, 2013). Utilization of CO (with a substantially lower standard redox potential, i.e. E0' (CO2/CO) ≈ −520 mV) circumvents this bottleneck, and ferredoxin can be directly reduced via CODH. If ANCO1 is unable to link H2‐oxidation to ferredoxin reduction via a bifurcating hydrogenase, this would explain the absence of acetogenic growth with H2/CO2. The H2 that is produced during growth on pyruvate might originate from Fe‐only hydrogenases using ferredoxin as donor, and favouring the hydrogen producing reaction direction (Adams 1990).
As ANCO1 encodes for a HDCR‐like complex (ACONDI 00243‐00246 and 02919‐02924), the WLP appears to be H2‐dependent in this strain. Hydrogen production via Fe‐only hydrogenases with ferredoxin (derived from CO oxidation) might play a role in introducing the reduction equivalents via the HDCR‐like complex into the WLP. Such internal cycling of hydrogen by acetogens has been observed before from organic substrates such as fructose (ref. Wiechmann et al., 2020). To note that activity of HDCR‐like complex in ANCO1 was not confirmed experimentally in this study, it is also possible that formate alternatively originates from bifurcating cytoplasmic enzymes that require NAD(P)H and ferredoxin, similar to what is observed in Clostridium autoethanogenum (Wang et al., 2013). In this case, H2 would not be required to drive the WLP, allowing operation on CO without intermediate H2 production. The potential roles for hydrogenases and their role in acetogenesis are depicted in Fig. 4 (dotted red lines). Overall, with current information, we can only speculate on the H2 metabolism of strain ANCO1 and more biochemical research is required to further elucidate this.
Regarding anaerobic respiration, the ability to utilize fumarate and thiosulfate as the electron acceptors is reported for Natranaerobius thermophilus and Natranaerobaculum magadiense, although succinate formation from fumarate was not measured. But the genome of N. thermophilus does contain the genes coding for fumarate reductase and thiosulfate reductase (Nther_2664‐2665 and Nther_0643‐0644, respectively) with the highest similarity to the ones present in ANCO1 genome. What is questionable, however, is the universal ability for the DNRA in the whole ‘Natranaerobiia’ members described previously. The reasons for doubts are the following: (i) formation of ammonia as the final product was not analysed in Natranaerobius truperii and Natronovirga and (ii) genes encoding for respiratory nitrate reductases (Nar/Nap) and the NrfAH‐type multiheme cytochromes c are not present in the genomes of both Natranaerobius species (genomes of Natronovirga and Natranaerobaculum are not available). Therefore, this important mode of anaerobic respiration currently cannot be assumed as a common trait? in this group. It is also questionable that as much as 20 mM nitrate was used in the tests with the ‘Natranaerobiia’ members. Our experience with ANCO1 showed that the complete ammonification is inhibited at far lower nitrate concentrations. Furthermore, the inability of ANCO1 to directly utilize nitrite for DNRA is also intriguing, pointing out that there is a problem. The root of this ‘sluggishness’ might lay in the unusual modularity of the DNRA system in this Gram‐positive bacterium. The classical ammonifying nitrite reductase consists of the periplasmic catalytic pentaheme c NrfA and its electron donor tetraheme c NrfH, which is an integral membrane quionol‐dehydrogenase (Simon and Klotz, 2013). In ANCO1, only the NrfA subunit is present, which is also a part of a locus containing a truncated Nap nitrate reductase lacking its cytochrome c containing electron donating NapB and NapC. Such deviations from the classical structures might be related to the facts that (i) ANCO1 does not have a periplasm, and (ii) that it does not have the respiratory quinones, which normally serve as the immediate electron donors for the periplasmically located respiratory complexes. Therefore, the only available low‐redox potential electron donors in ANCO1 appears to be produced in the cytoplasm [ferredoxins and NAD(P)H]. This could explain the difficulties with the DNRA growth we observed in ANCO1, particularly that the nitrite reduction was only possible after initiation of growth either by pyruvate fermentation or at fumarate‐reducing conditions. This might have been necessary to build up the required low redox potential electron donor pool for nitrate/nitrite reduction. Regarding the ability for DNRA reported previously in the H2/CO‐utilizing acetogens (REFS), it looks like that this is rather complex. At least in two documented cases (Moorella thermoacetica and Clostridium ljungdahlii) the ammonification of nitrate/nitrite does happen, however, no evidence for genes encoding multicytochrome c type of ammonifying Nir (NrfA) are present in the genomes. Instead, the cytoplasmic assimilatory NADH‐dependent NirB seems to operate as the electron think rather resembling ‘dump of electrons’ in fermentative prokaryotes (Seifritz et al., 2003; Pierce et al., 2008; Emerson et al., 2018). Therefore, such process is fundamentally different from the respiratory DNRA.
Taxonomy
Based on the phylogenomic analysis and unique phenotypic properties, we suggest to classify strain ANCO1 into a new genus and species Natranaerofaba carboxydovora, which forms a new family Natranaerofabaceae within the class ‘Natranaerobiia’.
Natranaerofabaceae fam. nov
Natr.an.ae.ro.fa.ba.ce'ae. N.L. fem. n. Natranaerofaba a bacterial genus; −aceae ending to denote a family; N.L. fem. pl. n. Natranaerofabaceae, the family of Natranaerofaba.
The family includes obligately anaerobic, acetogenic, extremely haloalkaliphilic bacteria living in hypersaline soda lakes. It is a member of class ‘Natranarobiia’, phylum Firmicutes. The family consists of a single genus and species Natranaerofaba carboxydovora. The type genus is Natranaerofaba.
Natranaerofaba gen. nov
Natr.an.ae.ro.fa'ba N.L. neut. n. natron, derived from Arabic natrun soda (sodium carbonate); Gr. pref. an‐, not; Gr. masc. n. aer, air; L. fem. n. faba, bean; N.L. fem. n. Natranaerofaba, bean‐shaped soda loving anaerobe.
The genus includes obligately anaerobic acetogenic bacteria with bean‐shaped cells. The polar lipids are dominated by diethers with phosphocholine and phosphoglycerol polar heads. They are extremely halophilic and obligately alkaliphilic moderate thermophiles capable of anaerobic respiration. They can utilize CO and pyruvate during acetogenic growth and formate and lactate in presence of thiosulfate, fumarate or nitrate as electron acceptors. Form a deep‐branching lineage within the class ‘Natranarobiia’, phylum Firmicutes. The type species is Natranaerofaba carboxydovora. Family classification: Natranaerofabaceae.
Natranaerofaba carboxydovora sp. nov
car.bo.xy.do.vo'ra L. pref. carboxydum, carbon monoxide; L. v. voro, to devour, consume; L. fem. adj. carboxydovora, consuming carbon monoxide.
Cells possess a Gram‐positive type of cell wall and are bean‐shaped rods, 0.4 × 3–6 μm, motile by peritrichous flagella. Endospore formation is not observed, but the potential is present in the genome. Polar lipids include phosphocholines and phsphoglycerols, with the most common core components being mostly present as ether‐bound C14:0 and plasmalogen‐derived aiC17:0. Respiratory lipoquinones are not present. The species is strictly anaerobic and heterotrophic acetogen capable of anaerobic respiration. Acetogenic growth is possible with pyruvate and CO as the electron donors with production of acetate/lactate and acetate/formate as products respectively. Anaerobic respiration is possible with CO, pyruvate, formate and lactate as donors and fumarate and thiosulfate (2‐electron reduction) as the electron acceptors. Lactate is converted to acetate. Formate also supports anaerobic growth with nitrate as acceptor resulting in formation of ammonia. Yeast extract is utilized as a C‐source. Obligately alkaliphilic with a pH range for growth between 9 and 10.5 and an optimum at pH 9.5–9.7. Extremely salt‐tolerant with a total Na+ range for growth from 2.5 to 4.5 M (optimum 3.5–4 M). Moderately thermophilic with a temperature range of 35–56°C and an optimum at 48–50°C. The type strain was obtained from anaerobic sediments of a hypersaline soda lake in Kulunda Steppe (Altai region, Russia). DNA G + C is 35.3 mol% (genome). Type strain is ANCO1T (DSM 108926). The EMBL/GenBank genome accession number is CP054394. [Correction added on 24 March 2021, after first online publication: The DSM collection number has been corrected in this version for accuracy.]
Experimental procedures
Cultivation conditions
The basic medium used for enrichment and routine cultivation of pure culture was obtained by 1:1 mixing of a NaCl and a sodium carbonate bases both containing 4 M total Na+ and sterilized separately at 120°C for 40 min. The NaCl base contained (g l−1): NaCl 240, KCl 5, K2HPO4 2.5, NH4Cl 0.5. The pH was adjusted to 7 by titration with 10% KHPO4 before sterilization. The carbonate base contained (g l−1): Na2CO3 190, NaHCO3 30, NaCl 16, K2HPO4 1 (pH 10). After sterilization, both bases were supplemented with 1 ml l−1 each of filter‐sterilized vitamin and acidic trace metal stock solutions (Pfennig and Lippert, 1966), 1 ml l−1 of filter‐sterilized alkaline W‐Se solution (Plugge, 2005) and 1 mM MgSO4. The alkaline base was also supplemented with 4 mM NH4Cl from 2 M sterile stock solution. Finally, the two bases were mixed 1:1 in a sterilized flask, resulting in a 4 M Na+ Cl‐carbonate alkaline medium with a final of pH 9.7. For enrichment of the potential CO‐dependent methyl‐reducing methanogens, this medium was amended with 50 mM MeOH, 2 mM sodium acetate, 100 mg l−1 yeast extract and 1% (v/v) of autoclaved 1:1 sediment:brine suspension from Bitter‐1 soda lake (according to Sorokin et al., 2017). The 40 ml complete medium was dispensed into 115 ml serum bottles, supplemented with 0.5 mM of sodium filter‐sterilized Na2S, closed with butyl rubber stoppers and made anoxic by three cycles of vacuum boiling/sterile argon flushing. Finally, the medium was reduced by adding 20 μl of sterile dithionite solution in 1 M sodium bicarbonate. Sterile CO gas was added at 5%–50% in the gas phase on the top of argon. The bottles were inoculated with 1 cm3 of 1:1 sediment‐brine suspension and incubated statically with periodic hand shaking at 50°C.
Cultivation of pure culture was mostly done with the same basic medium, whereby CO and MeOH were replaced by various electron donors with or without electron acceptors. Tests for the Cl− dependence were done by either decreasing the proportion of NaCl base to 0 or increasing it up to a maximum of 3.9 M (retaining carbonates at a minimal concentration which allowed sufficient alkalinity for growth). Test for total Na+ range for growth was done using 1:1 mixture (based of Na+ molarity) of NaCl/Na‐carbonates, creating a range from 1 to 4.5 M Na+ at pH 9.5. The pH range for growth was tested in three buffer systems containing 4 M total Na+: for the pH from 7 to 8, 50 mM HEPES‐50 mM K2HPO4/4 M NaCl; for the pH 8–8.5, 4 M NaCl/0.5 M filter‐sterilized NaHCO3; for the pH 9–11, 2 M NaCl/2 M Na‐carbonate buffer with different proportions of bicarbonate and carbonate.
Analyses
CO, CO2, CH4 and H2 in the gas phase were quantified by the GC equipped with a methanator and flame ionization and thermal conductivity detectors [Chromateck Crystall 5000 (Ufa, Russia); column Hayesep 80–100 mesh, 2 m ×3 mm, 40°C; carrier gas Ar]. Nitrite and sulfide were quantified by colorimetric methods (Trüper and Schlegel, 1964; Eck, 1966). Nitrate was monitored qualitatively using Merck sticks after neutralizing the culture supernatant to pH 7. The organic acids in the liquid phase were analysed by HPLC. The 0.4 ml of cell‐free culture supernatant was added to 0.6 ml 0.05 M H2SO4 solution containing 10 mM DMSO as and internal standard. Samples were run on a Shimazu iSeries Plus (Kioto, Japan), equipped with a MetaCarb 67H column (Agilent technologies, Santa Clara, CA), operated either at 30 or 45°C with 0.005 M H2SO4 as eluent at a flow rate of 0.9 ml min−1. Both, a UV and RI detector were used for detection of components. Concentrations could be accurately determined down to 0.1 mM.
Activity of anaerobic carbon monoxide dehydrogenase (CODH) was measured via enzymatic assays. Cells grown with CO were harvested from a 50 ml late exponential phase culture by centrifugation in anaerobic chamber, the cell pellet was washed with anoxic 0.8 M NaHCO3 (pH 8.5), cooled on ice and resuspended in 0.5 M NaHCO3 (pH 8.5) containing 2 mM DTT. The cells were disrupted by sonication with a Bandelin Sonopuls (Berlin, Germany), under anaerobic conditions, and the resulting cell free extract was transferred to rubber stoppered glass vials and kept on ice. Enzymatic assays were performed in quartz cuvettes sealed with a rubber stopper. After placing the rubber stopper, the gas phase was flushed twice with nitrogen to remove oxygen, and a final time with CO. Cuvettes were filled with 0.9 ml 0.8 M NaHCO3 via syringe and methyl viologen was added at 10 mM final concentration. Before start of the assay the absorbance in the cuvette was set between 0.1 and 0.3 at 578 nm using drops of 100 mM dithionite. The sample was placed in a pre‐heated (50°C) socket in a U‐2010 spectrophotometer (Hitachi, Tokyo, Japan). The reaction was started by adding 100, 50 or 25 μl cell free extract. The CO‐dependent methyl viologen reduction activity was measured at 578 nm, while temperature was kept constant at 50°C. The background activity was determined with N2 in the headspace instead of CO. Overall background activity was 10 times lower as measured in the samples fed with CO and activity ceased fast (<1 min). Protein concentrations in the cell free extract was analysed by Roti‐Nanoquant protein assay (CarlRoth, Karlsruhe, Germany), according to manufacturer instructions.
For the lipid analyses, the cells of pure culture of strain ANCO1 were grown with pyruvate at 50°C until late exponential phase, harvested by centrifugation, washed once with 2 M NaCl and freeze dried. The extracts for core lipid analysis were hydrolyzed in HCl/MeOH (1.5 N) by refluxing for 3 h. The hydrolysate was adjusted with aqueous KOH to pH 5, extracted three times with dichloromethane, and dried over Na2SO4. Derivatization of the hydrolyzed extract and its analysis were carried out as described previously (Bale et al., 2019b). Briefly, core lipid quantification was carried out on an Agilent Technologies 7890B GC with a silica column (CP Sil‐5, 25 × 0.32 mm) using the method described by Bale et al. (2019b). Compound identification was carried out on an Agilent Technologies 7890A gas chromatograph (GC) coupled to an Agilent Technologies 5975C VL MSD mass spectrometer (MS), operated at 70 eV, with a mass range m/z 50–800 and a scan rate of 3 scans s−1. The column and oven settings were the same as for the quantification GC analysis. Compounds were identified based on literature data and library mass spectra. Intact polar lipids (IPLs) were extracted from freeze‐dried biomass using a modified Bligh–Dyer procedure and analysed by ultra‐high‐pressure liquid chromatography–high‐resolution mass spectrometry (UHPLC‐HRMS) as described previously (Bale et al., 2019a). The presence of respiratory menaquinones was tested as described previously (Sorokin et al., 2019).
For electron microscopy, cells from 1 ml of pure culture grown with pyruvate were harvested by centrifugation, washed once and resuspended in 0.2 ml 2 M NaCl, pH 7 and stained for 1 min in 1% (w/v) uranyl acetate. The preparations were examined in JEOL 100 model transmission electron microscope (Japan).
Genome sequencing and phylogenomic analysis
Genomic DNA was extracted from cells grown with pyruvate + fumarate using the MasterPure Gram positive DNA extraction kit (Middleton, Wisconsin, USA), according to manufacturer instructions. Genomic DNA was commercially sequenced at Novogene (Beijing, China) using the PacBio technology. Genome was assembled using Flye (version 2.7.1) and checked for completeness and contamination using CheckM (version 1.1.2). The assembled genome was annotated using Prokka (version 1.14.0). The genome was deposited in the GenBank under accession number CP054394. The identity of selected functional proteins (including mostly catabolic and pH‐salt homeostasis functions, see Supporting Information Table S1) from the automatic annotation was verified by using manual Blast in UniProt.
For 16S rRNA gene‐based phylogeny, sequences were aligned using MAFFT v7.427 (GINS‐i strategy) (Nakamura et al., 2018). For phylogenomic analysis, the list of 120 bacterial core genes was taken from Genome Taxonomy Database (GTDB) (Parks et al., 2018). These marker genes were identified in selected genomes, aligned and concatenated using GTDBtk v1.2.0 (Chaumeil et al., 2019). Alignment was automatically trimmed using trimAl 1.2rev59 by using automated1 and gt 0.95 options (Capella‐Gutiérrez et al., 2009). The resulting alignment consisted of 21 189 amino acid residues. In both cases, phylogenetic tree was built using IQ‐TREE 1.6.12 program (Nguyen et al., 2015) with SH‐aLRT test (Anisimova et al., 2011) as well as ultrafast bootstrap with 1000 replicates (Hoang et al., 2018) and ModelFinder to determine the best‐fit model (Kalyaanamoorthy et al., 2017).
Proteomic analysis
For the shot‐gun proteomic analysis, strain ANCO1 was grown either with pyruvate or CO as the electron donors and yeast extract as the C‐source. The 20 mg of pelleted cell biomass each (wet weight) were lysed using B‐PER reagent/TEAB buffer and bead beating followed by centrifugation at 14.000g under cooling to collect protein supernatant. The proteins were precipitated with TCA (20% v/v final) at 4°C followed by washing twice using ice cold acetone. The protein pellet was redissolved in 200 mM ammonium bicarbonate containing 6 M urea, reduced in a 10 mM dithiothreitol solution and further alkylated using 20 mM iodoacetamide. The solution was diluted to below 1 M urea and digested using Trypsin at a ratio protease to protein of 1:50. Before analysis, peptides were desalted using an Oasis HLB solid phase extraction sorbent (Waters) according to the manufacturer protocols. An aliquot corresponding to approximately 100 ng of protein digest was analysed in duplicates using an one‐dimensional shot‐gun proteomics approach (Köcher et al., 2012). For this, 1 μl of samples were analysed using a nano‐liquid‐chromatography system consisting of an ESAY nano LC 1200, equipped with an Acclaim PepMap RSLC RP C18 separation column (50 μm × 150 mm, 2 μm), and an QE plus Orbitrap mass spectrometer (Thermo). The flow rate was maintained at 300 nl min−1 over a linear gradient from 5% to 30% solvent B over 90 min and finally to 75% B over 25 min. Solvent A was H2O containing 0.1% formic acid, and solvent B consisted of 80% acetonitrile in H2O and 0.1% formic acid. The Orbitrap was operated in data‐depended acquisition mode acquiring peptide signals from m/z 350 to 1400, where the top 10 signals were isolated at a window of m/z 2.0 and fragmented using an NCE of 30. The collected data were analysed against the established proteome database, using PEAKS Studio 8.5 (Bioinformatics Solutions) allowing 20 ppm parent ion mass error tolerance, three missed cleavages, carbamidomethylation as fixed and methionine oxidation and N/Q deamidation as variable modifications. Peptide spectrum matches were filtered against 1% false discovery rate (FDR) and protein identifications were accepted as being significant when having two unique peptides minimum. Relative protein abundances were correlated with protein molecular weight normalized spectral counts (Hongbin et al., 2004; Keiji and Itoh, 2008). The obtained dataset was deposited at the proteomics identification database (PRIDE Archive) and are openly available through the proteome exchange server (http://www.proteomexchange.org, PXD020223; http://www.ebi.ac.uk/pride, reviewer account details: reviewer26358@ebi.ac.uk/phJgNecx).
Supporting information
Appendix S1: Supporting information.
Acknowledgements
D.Y.S., M.P., M.D., D.Z.S., and J.S.S.D. acknowledge support from the SIAM‐Gravitation Program of the Dutch Ministry of Education, Culture and Science (24002002). D.Y.S. was also supported by the Russian Foundation of Basic Research (19‐04‐00401). A.M. was supported by the Russian Science Foundation (17‐74‐30025). N.B. and J.S.S.D. were supported by the under the European Union's Horizon 2020 research and innovation programme (grant agreement No. 694569).
References
- Adams, M. (1990) Structure and mechanism of iron‐only hydrogenases. Biochim Biophys Acta 1020: 115–145. [DOI] [PubMed] [Google Scholar]
- Allen, T.D., Caldwell, M.E., Lawson, P.A., Huhnke, R.L., and Tanner, R.S. (2010) Alkalibaculum bacchi gen.nov., sp.nov., a CO‐oxidizing, ethanol‐producing acetogen isolated from livestock‐impacted soil. Int J Syst Evol Microbiol 60: 2483–2489. [DOI] [PubMed] [Google Scholar]
- Angelidis, A.S., and Smith, G.M. (2003) Three transporters mediate uptake of glycine betaine and carnitine by Listeria monocytogenes in response to hyperosmotic stress. Appl Environ Microbiol 69: 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova, M., Gil, M., Dufayard, J.F., Dessimoz, C., and Gascuel, O. (2011) Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood‐based approximation schemes. Syst Biol 60: 685–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale, N.J., Sorokin, D.Y., Hopmans, E.C., Koenen, M., Rijpstra, W.I.C., Villanueva, L., et al. (2019a) New insights into the polar lipid composition of extremely halo(alkali)philic euryarchaea from hypersaline lakes. Front Microbiol 10: 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale, N.J., Rijpstra, W.I.C., Oshkin, I.Y., Belova, S.E., Dedysh, S.N., and Sinninghe Damsté, J.S. (2019b) Fatty acid and hopanoid adaption to cold in the methanotroph Methylovulum psychrotolerans . Front Microbiol 10: 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudrand, M., Grossi, V., Pancost, R., and Aloisi, G. (2010) Non‐isoprenoid macrocyclic glycerol diethers associated with authigenic carbonates. Org Geochem 41: 1341–1344. [Google Scholar]
- Blumenberg, M., Seifert, R., Petersen, S., and Michaelis, W. (2007) Biosignatures present in a hydrothermal massive sulfide from the mid‐Atlantic ridge. Geobiology 5: 435–450. [Google Scholar]
- Buckel, W. (2001) Sodium ion‐translocating decarboxylases. Biochim Biophys Acta 1527: 15–25. [DOI] [PubMed] [Google Scholar]
- Buckel, W., and Thauer, R.K. (2013) Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim Biophys Acta 1827: 94–113. [DOI] [PubMed] [Google Scholar]
- Capella‐Gutiérrez, S., Silla‐Martínez, J.M., and Gabaldón, T. (2009) trimAl: a tool for automated alignment trimming in large‐scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil, P.A., Mussig, A.J., Hugenholtz, P., and Parks, D.H. (2019) GTDB‐Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36: 1525–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreese, B., Costa, C., Demol, H., Papaefthymiou, V., Moura, I., Moura, J.J.G., et al. (1997) The primary structure of the split‐Soret cytochrome c from Desulfovibrio desulfuricans ATCC 27774 reveals an unusual type of diheme cytochrome c . Eur J Biochem 248: 445–451. [DOI] [PubMed] [Google Scholar]
- Diender, M., Stams, A.J.M., and Sousa, D.Z. (2015). Pathways and bioenergetics of anaerobic carbon monoxide fermentation. Front. Microbiol 6: 1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimroth, P., and Schink, B. (1998) Energy conservation in the decarboxylation of dicarboxylic acids by fermenting bacteria. Arch Microbiol 170: 69–77. [DOI] [PubMed] [Google Scholar]
- Drake, H.L., and Daniel, S.L. (2004) Physiology of the thermophilic acetogen Moorella thermoacetica . Res Microbiol 155: 869–883. [DOI] [PubMed] [Google Scholar]
- Emerson, D.F., Woolston, B.M., Liu, N., Donnelly, M., Currie, D.H., and Stephanopoulos, G. (2018) Enhancing hydrogen‐dependent growth of and carbon dioxide fixation by Clostridium ljungdahlii through nitrate supplementation. Biotechnol Bioeng 116: 294–306. [DOI] [PubMed] [Google Scholar]
- Fox‐Powell, M.G., Hallsworth, J.E., Cousins, C.R., and Cockell, C.S. (2016) Ionic strength is a barrier to the habitability of Mars. Astrobiology 16: 427–442. [DOI] [PubMed] [Google Scholar]
- Eck, G.‐R.‐v. (1966) Physiological and Chemical Test for Drinking Water. NEN 1056, IY‐2. Rijswijk, The Netherland: Nederlandse Normalisatie Instituut. [Google Scholar]
- Grant, W.D., and Jones, B.E. (2016). Bacteria, archaea and viruses of soda lakes. In Schagerl (ed). Soda lakes of East Africa. M. (97–147). Switzerland: Springer International Publishing, pp. 97‐147. [Google Scholar]
- Grossi, V., Mollex, D., Vinçon‐Laugier, A., Hakil, F., Pacton, M., and Cravo‐Laureau, C. (2015) Mono‐ and dialkyl glycerol ether lipids in anaerobic bacteria: biosynthetic insights from the mesophilic sulfate reducer Desulfatibacillum alkenivorans PF2803T. Appl Environ Microbiol 81: 3157–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton‐Brehm, S.D., Gibson, R.A., Green, S.J., Hopmans, E.C., Schouten, S., van der Meer, M.T.J., et al. (2013) Thermodesulfobacterium geofontis sp. nov., a hyperthermophilic, sulfate‐reducing bacterium isolated from obsidian Pool, Yellowstone national park. Extremophiles 17: 251–263. [DOI] [PubMed] [Google Scholar]
- Henstra, A.M., Dijkema, C., and Stams, A.J. (2007) Archaeoglobus fulgidus couples CO oxidation to sulfate reduction and acetogenesis with transient formate accumulation. Environ Microbiol 9: 1836–1841. [DOI] [PubMed] [Google Scholar]
- Herschy, B., Whicher, A., Camprubi, E., Watson, C., Dartnell, L., Ward, J., et al. (2014) An origin‐of‐life reactor to simulate alkaline hydrothermal vents. J Mol Evol 79: 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang, D.T., Chernomor, O., von Haeseler, A., Minh, B.Q., and Vinh, L.S. (2018) UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35: 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeft, S.E., Switzer, B.J., Stolz, J.F., Tabita, F.R., Witte, B., King, G.M., et al. (2007) Alkalilimnicola ehrlichii sp. nov., a novel arsenite‐oxidizing, haloalkaliphilic gammaproteobacterium capable of chemoautotrophic or heterotrophic growth with nitrate or oxygen as the electron acceptor. Int J Syst Evol Microbiol 57: 504–512. [DOI] [PubMed] [Google Scholar]
- Hongbin, L., Sadygov, R.G., and Yates, J.R. (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Сhem 76: 4193–4201. [DOI] [PubMed] [Google Scholar]
- Huber, R., Wilharm, T., Huber, D., Trincone, A., Burggraf, S., König, H., et al. (1992) Aquifex pyrophilus gen. nov. sp. nov., represents a novel group of marine hyperthermophilic hydrogen‐oxidizing bacteria. Syst Appl Microbiol 15: 340–351. [Google Scholar]
- Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K.F., von Haeseler, A., and Jermiin, L.S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14: 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiji, K., and Itoh, T. (2008) Mass spectrometry‐based approaches toward absolute quantitative proteomics. Curr Genomics 9: 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempe, S., and Kazmierczak, J. (2002) Biogenesis and early life on Earth and Europa: favored by an alkaline ocean? Astrobiology 2: 123–130. [DOI] [PubMed] [Google Scholar]
- King, G.M. (2015) Carbon monoxide as a metabolic energy source for extremely halophilic microbes: implications for microbial activity in Mars regolith. Proc Nat Ac USA (PNAS) 112: 4465–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga, Y., Morii, H., Akagawa‐Matsushita, M., and Ohga, M. (1998) Correlation of polar lipid composition with 16S rRNA phylogeny in methanogens. Further analysis of lipid component parts. Biosci Biotechnol Biochem 62: 230–236. [DOI] [PubMed] [Google Scholar]
- Köcher, T., Pichler, P., Swart, R., and Mechtler, K. (2012) Analysis of protein mixtures from whole‐cell extracts by single‐run nanoLC‐MS/MS using ultralong gradients. Nat Protocols 7: 882–890. [DOI] [PubMed] [Google Scholar]
- Langworthy, T.A., Holzer, G., Zeikus, J.G., and Tornabene, T.G. (1983) Iso‐ and anteiso‐branched glycerol diethers of the thermophilic anaerobe Thermodesulfotobacterium commune . Syst Appl Microbiol 4: 1–17. [DOI] [PubMed] [Google Scholar]
- Liu, K., Atiyeh, H.K., Tanner, R.S., Wilkins, M.R., and Huhnke, R.L. (2012) Fermentative production of ethanol from syngas using moderately alkaliphilic strains of Alkalibaculum bacchi . Bioresour Technol 104: 336–341. [DOI] [PubMed] [Google Scholar]
- Mesbah, N.M., Hedrick, D.B., Peacock, A.D., Rohde, M., and Wiegel, J. (2007) Natranaerobius thermophilus gen. nov. sp. nov., a halophilic, alkalithermophilic bacterium from soda lakes of the Wadi an Natrun, Egypt, and proposal of Natranaerobiaceae fam. nov. and Natranaerobiales ord. nov. Int J Syst Evol Microbiol 57: 2507–2512. [DOI] [PubMed] [Google Scholar]
- Mesbah, N.M., Cook, G.M., and Wiegel, J. (2009) The halophilic alkalithermophile Natranaerobius thermophilus adapts to multiple environmental extremes using a large repertoire of Na+(K+)/H+ antiporters. Mol Microbiol 74: 270–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesbah, N.M., and Wiegel, J. (2009) Natronovirga wadinatrunensis gen. nov., sp. nov. and Natranaerobius trueperi sp. nov., halophilic, alkalithermophilic micro‐organisms from soda lakes of the Wadi an Natrun, Egypt. Int J Syst Evol Microbiol 59: 2042–2048. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Yamada, K.D., Tomii, K., and Katoh, K. (2018) Parallelization of MAFFT for large‐scale multiple sequence alignments. Bioinformatics 34: 2490–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, L.T., Schmidt, H.A., von Haeseler, A., and Minh, B.Q. (2015) IQ‐TREE: a fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Mol Biol Evol 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongagna‐Yhombi, S.Y., McDonald, N.D., and Boyd, E.F. (2015) Deciphering the role of multiple betaine‐carnitine‐choline transporters in the halophile Vibrio parahaemolyticus . Appl Environ Microbiol 81: 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancost, R.D., Pressley, S., Coleman, J.M., Talbot, H.M., Kelly, S.P., Farrimond, P., et al. (2006) Composition and implications of diverse lipids in New Zealand geothermal sinters. Geobiology 4: 71–92. [Google Scholar]
- Parks, D.H., Chuvochina, M., Waite, D.W., Rinke, C., Skarshewski, A., Chaumeil, P.‐A., and Hugenholtz, P. (2018) A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36: 996–1004. [DOI] [PubMed] [Google Scholar]
- Parshina, S.N., Sipma, J., Henstra, A.M., and Stams, A.J.M. (2010) Carbon monoxide as an electron donor for the biological reduction of sulphate. Int J Microbiol 2010: 319527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfennig, N., and Lippert, K.D. (1966) Uber das Vitamin B12–bedurfnis phototropher Schwefel Bakterien. Arch Microbiol 55: 245–256. [Google Scholar]
- Pflüger, K., and Müller, V. (2004) Transport of compatible solutes in extremophiles. J Bioenerg Biomembr 36: 17–24. [DOI] [PubMed] [Google Scholar]
- Pierce, E., Xie, G., Barabote, R.D., Saunders, E., Han, C.S., Detter, J.C., et al. (2008) The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum). Environ Microbiol 10: 2550–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plugge, C.M. (2005) Anoxic media design, preparation and considerations. Methods Enzymol 397: 3–16. [DOI] [PubMed] [Google Scholar]
- Rebouche, C.J. (1998) Carnitine metabolism and its regulation in microorganisms and mammals. Ann Rev Nutr 18: 39–61. [DOI] [PubMed] [Google Scholar]
- Roberts, M.F. (2005) Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst 1: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagerl, M. (ed). (2016) Soda lakes of East Africa. Switzerland: Springer International Publishing AG, p. 408. [Google Scholar]
- Schuchmann, K., and Müller, V. (2012) A bacterial electron‐bifurcating hydrogenase. J Biol Chem 287: 31165–31171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchmann, K., and Müller, V. (2013) Direct and reversible hydrogenation of CO2 to formate by a bacterial carbon dioxide reductase. Science 342: 1382–1385. [DOI] [PubMed] [Google Scholar]
- Schuchmann, K., and Müller, V. (2014) Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat Rev Microbiol 12: 809–821. [DOI] [PubMed] [Google Scholar]
- Seifritz, C., Drake, H.L., and Daniel, S.L. (2003) Nitrite as an energy‐conserving electron sink for the acetogenic bacterium Moorella thermoacetica . Curr Microbiol 46: 329–333. [DOI] [PubMed] [Google Scholar]
- Simon, J., and Klotz, M.G. (2013) Diversity and evolution of bioenergetic systems involved in microbial nitrogen compound transformations. Biochim Biophys Acta 1827: 114–135. [DOI] [PubMed] [Google Scholar]
- Sorokin, D.Y. (2017) Anaerobic haloalkaliphiles. In Encyclopedia Life Science. Chichester: John Wiley & Sons, Ltd. 10.1002/9780470015902.a0027654. [DOI] [Google Scholar]
- Sorokin, D.Y., Banciu, H.L., and Muyzer, G. (2015) Functional microbiology of soda lakes. Curr Opin Microbiol 25: 88–96. [DOI] [PubMed] [Google Scholar]
- Sorokin, D.Y., Makarova, K.S., Abbas, B., Ferrer, M., Golyshin, P.N., Galinski, E.A., et al. (2017) Discovery of extremely halophilic, methyl‐reducing euryarchaea provides insights into the evolutionary origin of methanogenesis. Nat Microbiol 2: 17081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin, D.Y., Merkel, A.Y., Abbas, B., Makarova, K., Rijpstra, W.I.C., Koenen, M., et al. (2018) Methanonatronarchaeum thermophilum gen. nov., sp. nov, and 'Candidatus Methanohalarchaeum thermophilum' ‐ extremely halo(natrono)philic methyl‐reducing methanogens from hypersaline lakes representing a novel euryarchaeal class Methanonatronarchaeia classis nov. Int J Syst Evol Microbiol 68: 2199–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin, D.Y., Yakimov, M.M., Messina, E., Merkel, A.Y., Bale, N.J., and Sinninghe Damsté, J.S. (2019) Natronolimnobius sulfurireducens sp. nov., and Halalkaliarchaeum desulfuricum gen. nov., sp. nov., the first sulfur‐respiring alkaliphilic haloarchaea from hypersaline alkaline lakes. Int J Syst Evol Microbiol 69: 2662–2673. [DOI] [PubMed] [Google Scholar]
- Sprott, G.D., Meloche, M., and Richards, J.C. (1991). Proportions of diether, macrocyclic diether, and tetraether lipids in Methanococcus jannaschii grown at different temperatures. J Bacteriol 173: 3907–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trüper, H.G., and Schlegel, H.G. (1964) Sulfur metabolism in Thiorhodaceae. 1. Quantitative measurements on growing cells of Chromatium okenii . Antonie Van Leeuwenhoek 30: 225–238. [DOI] [PubMed] [Google Scholar]
- van Dongen, B.E., Roberts, A.P., Schouten, S., Jiang, W.‐T., Florindo, F., and Pancost, R.D. (2007) Formation of iron sulfide nodules during anaerobic oxidation of methane. Geochim Cosmochim Acta 71: 5155–5167. [Google Scholar]
- van Gelder, A.H., Sousa, D.Z., Rijpstra, W.I.C., Sinninghe Damsté, J.S., Stams, A.J.M., and Sánchez‐Andrea, I. (2014) Ercella succinigenes gen. nov., sp. nov., an anaerobic succinate‐producing bacterium. Int J Syst Evol Microbiol 64: 2449–2454. [DOI] [PubMed] [Google Scholar]
- Vavourakis, C.D., Andrei, A.S., Mehrshad, M., Ghai, R., Sorokin, D.Y., and Muyzer, G. (2018) A metagenomics roadmap to the uncultured genome diversity in hypersaline soda lake sediments. Microbiome 6: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavourakis, C.D., Ghai, R., Rodriguez‐Valera, F., Sorokin, D.Y., Tringe, S.G., Hugenholtz, P., et al. (2016) Metagenomic insights into the uncultured diversity and physiology of microbes in four hypersaline soda lake brines. Front Microbiol 7: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Huang, H., Kahnt, J., Müller, A.P., Köpke, M., and Thauer, R.K. (2013) NADP‐specific electron‐bifurcating [fefe]‐hydrogenase in a functional complex with formate dehydrogenase in Clostridium autoethanogenum grown on CO. J Bacteriol 195: 4373–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weghoff, M.C., Bertsch, W.J., and Müller, V. (2015) A novel mode of lactate metabolism in strictly anaerobic bacteria. Environ Microbiol 17: 670–677. [DOI] [PubMed] [Google Scholar]
- Wiechmann, A., Ciurus, S., Oswald, F., Seiler, V.N., and Müller, V. (2020) It does not always take two to tango: “Syntrophy” via hydrogen cycling in one bacterial cell. ISME J 14: 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M., Ren, Q., Durkin, A.S., Daugherty, S.C., Brinkac, L.M., Dodson, R.J., et al. (2005) Life in hot carbon monoxide: the complete genome sequence of Carboxydothermus hydrogenoformans Z‐2901. PLoS Genet 1: e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavarzina, D.G., Zhilina, T.N., Kuznetsov, B.B., Kolganova, T.V., Osipov, G.A., Kotelev, M.S., and Zavarzin, G.A. (2013) Natranaerobaculum magadiense gen. nov., sp. nov., an anaerobic, alkalithermophilic bacterium from soda lake sediment. Int J Syst Evol Microbiol 63: 4456–4461. [DOI] [PubMed] [Google Scholar]
- Ziegenbalg, S.B., Birgel, D., Hoffmann‐Sell, L., Pierre, C., Rouchy, J.M., and Peckmann, J. (2012) Anaerobic oxidation of methane in hypersaline Messinian environments revealed by 13C‐depleted molecular fossils. Chem Geol 292–293: 140–148. [Google Scholar]
- Zorz, J.K., Sharp, C., Kleiner, M., Gordon, P.M.K., Pon, R.T., Dong, X., and Strous, M. (2019) A shared core microbiome in soda lakes separated by large distances. Nat Comm 10: 4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting information.


