Abstract
Carboxysomes are protein‐based organelles essential for carbon fixation in cyanobacteria and proteobacteria. Previously, we showed that the cyanobacterial nucleoid is used to equally space out β‐carboxysomes across cell lengths by a two‐component system (McdAB) in the model cyanobacterium Synechococcus elongatus PCC 7942. More recently, we found that McdAB systems are widespread among β‐cyanobacteria, which possess β‐carboxysomes, but are absent in α‐cyanobacteria, which possess structurally and phyletically distinct α‐carboxysomes. Cyanobacterial α‐carboxysomes are thought to have arisen in proteobacteria and then horizontally transferred into cyanobacteria, which suggests that α‐carboxysomes in proteobacteria may also lack the McdAB system. Here, using the model chemoautotrophic proteobacterium Halothiobacillus neapolitanus, we show that a McdAB system distinct from that of β‐cyanobacteria operates to position α‐carboxysomes across cell lengths. We further show that this system is widespread among α‐carboxysome‐containing proteobacteria and that cyanobacteria likely inherited an α‐carboxysome operon from a proteobacterium lacking the mcdAB locus. These results demonstrate that McdAB is a cross‐phylum two‐component system necessary for positioning both α‐ and β‐carboxysomes. The findings have further implications for understanding the positioning of other protein‐based bacterial organelles involved in diverse metabolic processes.
Plain language summary
Cyanobacteria are well known to fix atmospheric CO2 into sugars using the enzyme Rubisco. Less appreciated are the carbon‐fixing abilities of proteobacteria with diverse metabolisms. Bacterial Rubisco is housed within organelles called carboxysomes that increase enzymatic efficiency. Here we show that proteobacterial carboxysomes are distributed in the cell by two proteins, McdA and McdB. McdA on the nucleoid interacts with McdB on carboxysomes to equidistantly space carboxysomes from one another, ensuring metabolic homeostasis and a proper inheritance of carboxysomes following cell division. This study illuminates how widespread carboxysome positioning systems are among diverse bacteria. Carboxysomes significantly contribute to global carbon fixation; therefore, understanding the spatial organization mechanism shared across the bacterial world is of great interest.
The McdAB system distributes β‐carboxysomes across the nucleoid in β‐cyanobacteria to ensure proper inheritance following cell division. Here we show the McdAB system also functions to distribute the structurally‐ and phyletically‐distinct α‐carboxysomes of proteobacteria. We propose that McdAB proteins function in bacteria as anti‐aggregation systems for protein‐based organelles that have the capacity to self‐associate.
1. INTRODUCTION
Bacterial Microcompartments (BMCs) are large cytosolic protein‐based organelles that encapsulate sensitive metabolic processes to provide microbes with a distinct environmental growth advantage (Kerfeld et al., 2018). Diverse in structure and function, BMCs have been identified across 29 bacterial phyla and ~20% of all sequenced bacterial genomes (Axen et al., 2014), so their functions are of great ecological, evolutionary, biotechnological, and medical interest. Despite their prevalence and importance in bacterial metabolism, little is known about how BMCs are spatially regulated in the cell. The model BMC is the carboxysome, which is named for its involvement in carbon fixation. Carboxysomes are classified as either α or β depending on the type of Ribulose‐1,5‐bisphosphate carboxylase/oxygenase (Rubisco) they encapsulate; α‐carboxysomes contain Rubisco form 1A, β‐carboxysomes contain Rubisco form 1B, and both carboxysome types possess a phyletically distinct set of protein components necessary for encapsulation (Figure 1a,b) (Rae et al., 2013). Since Rubisco can utilize either CO2 or O2 as a substrate, driving either the Calvin–Benson–Bassham cycle (CO2) or the wasteful process of photorespiration (O2), encapsulation of Rubisco and carbonic anhydrase within a selectively permeable protein shell generates a high internal CO2 environment within carboxysomes that minimizes photorespiration (Badger et al., 1998; Kaplan et al., 1991; Price & Badger, 1989; Tcherkez et al., 2006). Through this mechanism, carboxysomes contribute to greater than 35% of global carbon fixation through atmospheric CO2 assimilation (Cohen & Gurevitz, 2006; Kerfeld & Melnicki, 2016).
FIGURE 1.
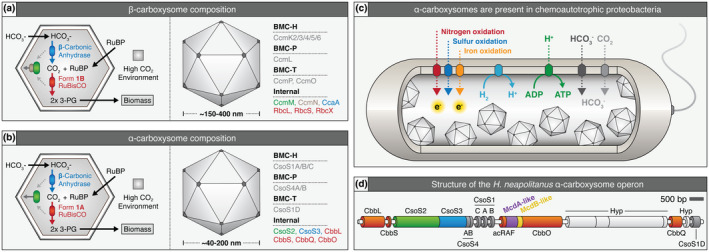
Overview of α‐ and β‐carboxysome composition, operon structure, and prevalence in proteobacteria. (a) Cartoon illustration of internal reactions (left) and known components (right) of β‐carboxysomes and (b) α‐carboxysomes. (c) α‐carboxysome‐containing proteobacteria display diverse metabolic capacities. (d) H. neapolitanus α‐carboxysome operon. Red = Rubisco or Rubisco associated, green = CsoS2 which mediates Rubisco/shell interaction, blue = carbonic anhydrase, purple = putative mcdA gene, yellow = putative mcdB gene, dark grey = shell component, light grey = hypothetical protein (Hyp). Gene colors are matched to the proteins shown in panel B and functionally equivalent proteins for β‐carboxysomes in panel A [Colour figure can be viewed at wileyonlinelibrary.com]
In our previous study, we identified and characterized the two‐component McdAB system (Maintenance of Carboxysome Distribution protein A and B) required for maintaining the equidistant positioning of multiple carboxysomes along the nucleoid in the model rod‐shaped cyanobacterium Synechococcus elongatus PCC 7942 (Maccready et al., 2018). We found that McdA, a ParA‐type ATPase, non‐specifically bound DNA in the presence of ATP. McdB, a novel small disordered protein, localized to carboxysomes and stimulated the ATPase activity of McdA, removing McdA from the nucleoid. Once removed, McdA then recycled its nucleotide and rebound the nucleoid at locations with the lowest concentration of McdB. The local removal of McdA created depletion zones in the vicinity of McdB‐bound carboxysomes that subsequently evolved into emergent global oscillations of McdA. McdB‐bound carboxysomes, therefore, generate and exploit dynamic McdA gradients to move in a directed and persistent manner toward increased concentrations of McdA on the nucleoid. Since we found that carboxysomes clustered in the absence of the McdAB system, we concluded that its Brownian‐ratchet‐based distribution mechanism functioned primarily as an “anti‐aggregation” system, while simultaneously ensuring proper inheritance of carboxysomes.
More recently, we found that two distinct types of McdAB systems (Type 1 and Type 2) exist among β‐cyanobacteria, which possess β‐carboxysomes (MacCready et al., 2020). Surprisingly, however, we also found that α‐cyanobacteria, which possess α‐carboxysomes, completely lack the McdAB system. We previously reported that many anoxygenic photosynthetic proteobacteria as well as chemoautotrophic bacteria (Figure 1c), also possess α‐carboxysomes and potentially encode for McdA and McdB within their carboxysome (cso) operon (Figure 1d) (Maccready et al., 2018). Interestingly, α‐carboxysomes are thought to have arisen in proteobacteria and were subsequently horizontally transferred into cyanobacteria, resulting in the distinct α‐cyanobacterial lineage. Thus, it remains unclear whether proteobacterial α‐carboxysomes are positioned by the McdAB system and whether this system evolved prior to or after horizontal transfer into cyanobacteria.
Here, we identify a novel McdAB system that positions α‐carboxysomes in proteobacteria. In the sulfur‐oxidizing chemoautotrophic proteobacterium Halothiobacillus neapolitanus c2, we show that uniform distributions of α‐carboxysomes are lost in the absence of either McdA or McdB. As shown for the β‐McdAB system in S. elongatus, we found that H. neapolitanus α‐McdA and α‐McdB interact with each other, and that α‐McdB co‐localizes with α‐carboxysomes in vivo. Furthermore, we found that α‐McdAB systems are widespread among α‐carboxysome‐containing proteobacteria, some possessing two distinct copies of α‐McdB. As we showed previously for β‐McdB proteins, purified H. neapolitanus α‐McdB undergoes liquid‐liquid phase separation (LLPS) in vitro. Despite this shared LLPS activity, we found that different McdB types had different minimal oligomeric units in vitro. β‐McdB Type 1 forms a hexamer, β‐McdB Type 2 forms a dimer, and α‐McdB remains a monomer. These results further our understanding of how different McdB proteins interface with their cognate McdA and carboxysome cargo. Collectively, we reveal a widespread McdAB system required for positioning structurally and phyletically distinct α‐carboxysomes in proteobacteria and provide insights into the evolution of α‐carboxysomes between cyanobacteria and proteobacteria. The findings have broad implications for understanding the subcellular organization of protein‐based organelles in bacteria.
2. RESULTS AND DISCUSSION
2.1. McdA‐ and McdB‐like proteins distribute carboxysomes in H. neapolitanus
We began this study by testing the hypothesis that the McdA‐like coding sequence that we previously identified within proteobacterial cso operons functioned to distribute α‐carboxysomes in the cell (Figure 1d) (Maccready et al., 2018). We also tested whether the small coding sequence following the putative mcdA gene encoded a functional homolog of cyanobacterial McdB. To explore this, we used the sulfur‐oxidizing chemoautotroph Halothiobacillus neapolitanus c2 (hereafter H. neapolitanus), which has been established as a model system for studying α‐carboxysomes in proteobacteria (Cai et al., 2015; Cannon & Shively, 1983; Dou et al., 2008; Holthuijzen et al., 1987; Oltrogge et al., 2020; Rae et al., 2013; Schmid et al., 2006; Shively et al., 1973). First, in H. neapolitanus, we fused the native gene that encodes the small subunit of Rubisco (CbbS) with the fluorescent protein mTurquoise2 to form CbbS‐mTQ (Goedhart et al., 2012). The CbbS‐mTQ signal appeared as several dynamic and overlapping foci (Video 1), which from a single image, looked like a nearly homogeneous signal across the length of individual H. neapolitanus cells (Figure 2a). In S. elongatus, fluorescent‐labeled carboxysomes are sufficiently separated from one another to be resolved (Cameron et al., 2013; Maccready et al., 2018; Savage et al., 2010). In general, H. neapolitanus carboxysomes are greater in number (4 to 18) and smaller (40 to 200 nm diameter) compared to the fewer (3 to 5) and larger (150 to 400 nm) β‐carboxysomes of S. elongatus (Cannon et al., 2001; Maccready et al., 2018; Rae et al., 2013; Savage et al., 2010; Schmid et al., 2006; Sun et al., 2019). Therefore, we interpret this data as α‐carboxysomes being distributed across the cell length (Figure 2a), but the smaller size of α‐carboxysomes combined with their high‐copy number precludes single carboxysome resolution using traditional fluorescence microscopy imaging of H. neapolitanus cells, which are also relatively small (0.5 × 1.5 µm). We, therefore, performed Transmission Electron Microscopy (TEM) to determine if this CbbS‐mTQ signal in wild‐type cells, indeed, represented assembled and distributed carboxysomes. Consistent with our interpretation of the fluorescent imaging, we frequently found that α‐carboxysomes were distributed over the nucleoid region of the cell and down the medial axis (Figure 2b). The distribution of fluorescent‐labeled carboxysomes in our TEM images are very similar to those shown previously by other groups imaging native carboxysomes in H. neapolitanus cells (Cannon & Shively, 1983; Dou et al., 2008; Rae et al., 2013; Schmid et al., 2006; Shively et al., 1973).
FIGURE 2.
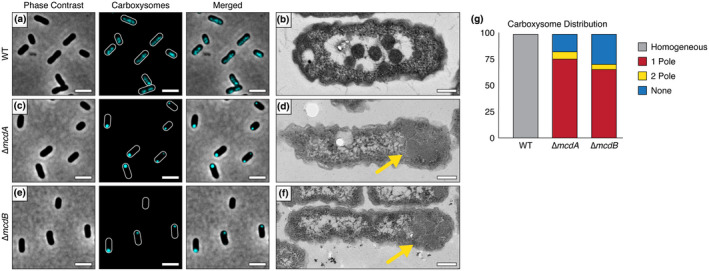
An McdAB system positions α‐carboxysomes. (a) The carboxysome reporter CbbS‐mTQ is distributed in WT H. neapolitanus cells. Scale bar: 2 µm. (b) Electron micrograph showing α‐carboxysomes confined to the nucleoid in WT cells. Scale bar: 200 nm. (c) Homogenous distribution of α‐carboxysomes is lost in the absence of McdA. Scale bar: 2 µm. (d) Polar localization of assembled α‐carboxysomes in the absence of McdA (yellow arrow). Scale bar: 200 nm. (e) Homogenous distribution of α‐carboxysomes is lost in the absence of McdB. Scale bar: 2 µm. (f) Polar localization of assembled α‐carboxysomes in the absence of McdB (yellow arrow). Scale bar: 200 nm. (g) Quantification of carboxysome distributions. In ∆mcdA (n = 210 cells), 76% of cells had a single polar focus, 7% displayed two foci at opposing poles, and 17% lacked a focus. In ∆mcdB (n = 220 cells), 66% of cells had a single polar focus, 5% displayed two foci at opposing poles, and 29% lacked a focus [Colour figure can be viewed at wileyonlinelibrary.com]
Next, using this CbbS‐mTQ α‐carboxysome reporter, we individually deleted our candidate mcdA or mcdB genes. In the absence of either mcdA or mcdB, the homogenous distribution of α‐carboxysomes down the cell length was lost (Figure 2c,e and Video 1). Instead, the majority of cells had a single high‐intensity CbbS‐mTQ focus at one pole (~76% of n = 210 cells for ∆mcdA and ~66% of n = 220 cells for ∆mcdB) (Figure 2g). Additionally, 7% of ∆mcdA and 5% of ∆mcdB cells displayed two foci at opposing cell poles. The remaining cells (17% of ∆mcdA and 29% of ∆mcdB) lacked a CbbS‐mTQ focus entirely, suggesting that these cells did not inherit α‐carboxysomes after division and that the carboxysomes have yet to be assembled de novo. We performed TEM on the ∆mcdA and ∆mcdB strains to determine if these massive foci of CbbS‐mTQ represented assembled carboxysomes or amorphous protein aggregates of carboxysome components. In the absence of either mcdA or mcdB, the vast majority of cells (~75% of n = 214 cells) had a single cluster of multiple assembled carboxysomes (Figure 1d,f). Once again, the TEM assisted in our interpretation of the fluorescent imaging. Now that we have identified the McdAB systems of α and β carboxysomes, from this point forward, we distinguish the two systems according to the type of carboxysome being distributed: α‐McdAB systems position proteobacterial α‐carboxysomes and β‐McdAB systems position cyanobacterial β‐carboxysomes.
Our identification of an α‐McdAB system for the positioning of proteobacterial α‐carboxysomes is intriguing on multiple fronts. First, α‐carboxysomes are typically much smaller and more numerous than β‐carboxysomes in cells (Cannon et al., 2001; Rae et al., 2013; Schmid et al., 2006; Sun et al., 2019). Therefore, given that H. neapolitanus cells are also smaller (0.5 × 1.5 microns) than S. elongatus cells (1.3 × 3 microns), it could have been reasoned that α‐carboxysomes would rely on random diffusion for their inheritance and distribution in the cell; whereas the larger size and lower copy number of cyanobacterial β‐carboxysomes necessitate an active positioning system. However, since we found that assembled α‐carboxysomes aggregated toward the polar region of cells in the absence of α‐McdA or α‐McdB, reminiscent of β‐carboxysomes in the absence of β‐McdA or β‐McdB (Maccready et al., 2018), we conclude that all McdAB systems actively position carboxysomes to not only ensure proper inheritance following cell division, but to primarily function as an anti‐aggregation system for protein‐based organelles that have the capacity to self‐associate. Our findings provide an explanation as to why α‐ and β‐carboxysomes, when heterologously expressed in the absence of their cognate McdAB systems, undergo extreme aggregation in other bacterial species or in plant chloroplasts (Baumgart et al., 2017; Bonacci et al., 2012; Lin et al., 2014; Long et al., 2018). It is very likely that adding the two‐component McdAB system will solve the aggregation issue that arises when carboxysome components are expressed in these heterologous hosts.
In addition to α‐ and β‐carboxysome size and quantity being significantly different between S. elongatus and H. neapolitanus, so is their nucleoid biology. While newborn H. neapolitanus cells are monoploid (Desmarais et al., 2019), S. elongatus cells are polypoid; harboring as many as 10 copies of their chromosome (Chen et al., 2012). Some β‐cyanobacteria that we have previously shown to encode a McdAB system, such as Synechocystis sp. PCC 6803, can have over 50 chromosome copies (Zerulla et al., 2016). Since McdA uses the nucleoid as a matrix for positioning McdB‐bound carboxysomes, our findings show that McdAB systems can function on nucleoids with drastically different chromosome contents. It remains to be determined how chromosome content influences McdA dynamics and carboxysome positioning on the nucleoid. It has been shown that the copy numbers of both carboxysomes and chromosomes are regulated in cyanobacteria by environmental growth conditions (Ohbayashi et al., 2019; Sun et al., 2019). It is possible that expression levels of the McdAB system may also be regulated by environmental cues, such as nutrient availability.
2.2. Without the McdAB system, clustered carboxysomes are nucleoid excluded to the cell poles
Macromolecular crowding of the cytoplasm in combination with the nucleoid acting as a formidable diffusion barrier have been shown to be responsible for the polar localization of several large intracellular bodies in bacteria (Straight et al., 2007; Winkler et al., 2010), such as plasmids lacking an active positioning system (Erdmann et al., 1999; Ringgaard et al., 2009). Without a ParA‐system, plasmids that were once distributed equally along the nucleoid length, become nucleoid “excluded”—the passive effect of the nucleoid as a diffusion barrier to mesoscale bodies in the cytoplasm (Planchenault et al., 2020). Therefore, it seems that ParA‐based positioning systems, such as McdAB, not only overcome the nucleoid as a diffusion barrier, but exploit it as a matrix for the movement and positioning of their mesoscale intracellular cargos.
H. neapolitanus is a halophile capable of growing under extreme hyperosmotic conditions where the cell membrane pushes against the compacted nucleoid (see Figure 2d,f), so that the nucleoid forms a barrier in the cell that can even restrict the diffusion of GFP (Konopka et al., 2009; Van Den Bogaart et al., 2007). We, therefore, asked if the polar localization of carboxysome clusters in the ∆mcdA and ∆mcdB strains is the result of nucleoid exclusion. To explore this question, we used 4′,6‐diamidino‐2‐phenylindole (DAPI) to stain the nucleoids of WT, ∆α‐mcdA, and ∆α‐mcdB H. neapolitanus strains encoding the CbbS‐mTQ α‐carboxysome reporter. In wild‐type cells, carboxysome foci were confined within the nucleoid signal (PCC = 0.80, n = 128 cells) (Figure 3a). The finding is consistent with our previous report that the McdAB system uses the nucleoid as a matrix to position carboxysomes (Maccready et al., 2018). In ∆α‐mcdA (PCC = 0.36, n = 436 cells) or ∆α‐mcdB (PCC = 0.33, n = 135 cells) strains, α‐carboxysome foci were nucleoid excluded at the cell poles (Figure 3b,c). Intriguingly, the nucleoids of both the ∆mcdA and ∆mcdB strains are shifted away from midcell in the opposing direction of the carboxysome cluster. The data suggest that the polar cluster of carboxysomes is large and dense enough to interfere with nucleoid placement in the cell. In some instances, the carboxysome clusters grew large enough to locally expand and deform the rod‐shaped morphology of the cell itself (Figure S1).
FIGURE 3.
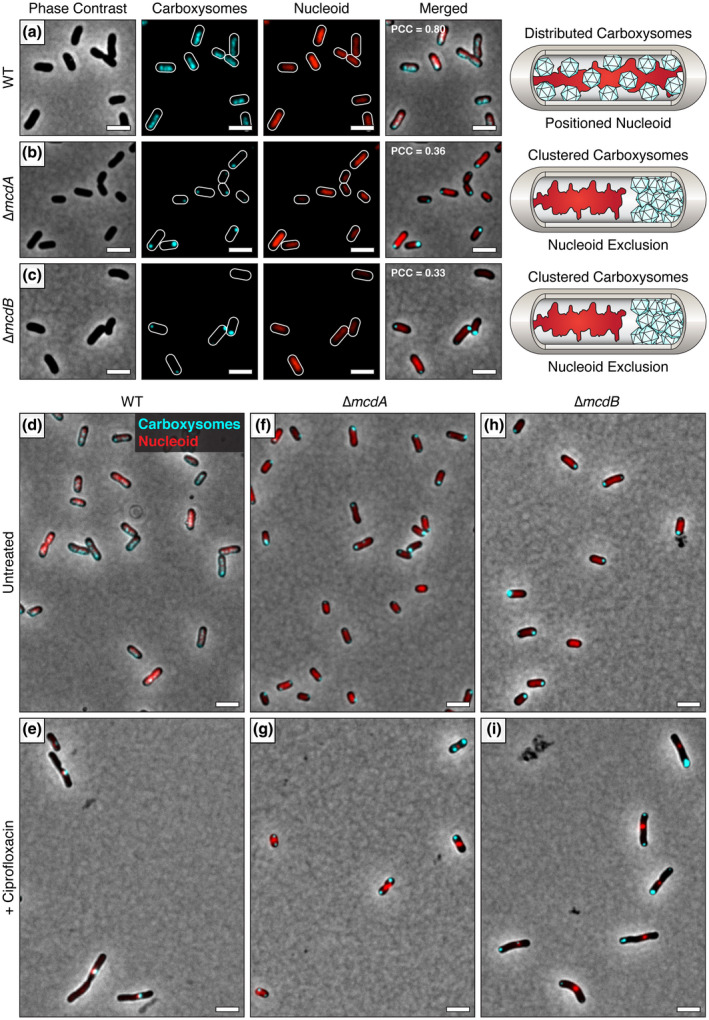
Aggregated α‐carboxysomes are nucleoid excluded. (a) The carboxysome reporter CbbS‐mTQ colocalizes with the DAPI‐stained nucleoid in WT H. neapolitanus cells. In (b) ∆mcdA and (c) ∆mcdB mutants, aggregated α‐carboxysomes are nucleoid excluded. PCC values were calculated from n ≥ 100 cells per cell population. (d) In WT cells, carboxysomes colocalize with the DAPI‐stained nucleoid. (e) After treatment with ciprofloxacin, the nucleoid condenses and carboxysomes remain colocalized with the DAPI‐stained nucleoid. In ∆mcdA (f) and ∆mcdB (h) strains, aggregated carboxysomes are excluded from the DAPI‐stained nucleoid. After ciprofloxacin treatment of ∆mcdA (g) and ∆mcdB (i) strains, the nucleoid condenses and carboxysomes remain aggregated despite the increased cytoplasmic space. Scale bar: 2 µm [Colour figure can be viewed at wileyonlinelibrary.com]
In the deletion strains, it was unclear whether the nucleoid‐excluded carboxysomes interacted to form aggregates at the cell poles or whether they simply accumulated at the poles, but without actually sticking to each other. To address this question, we sought to increase the cytoplasmic space by elongating cells with cephalexin treatment. However, the additional chromosomal content in these elongated cells prevented us from determining if carboxysomes aggregated in a manner that was independent of nucleoid exclusion (Figure S2). We next tried the gyrase‐inhibitor ciprofloxacin to condense the nucleoid, which significantly increased the cytoplasmic space in the cell (Figure 3d‐i). In WT cells, carboxysomes remained colocalized with the compacted nucleoid after ciprofloxacin treatment (Figure 3d‐e). In the ∆mcdA and ∆mcdB strains, carboxysomes remained aggregated despite the significant increase in diffusible cytoplasmic space (Figure 3f‐i). Altogether, these data show that the McdAB system functions to both position carboxysomes on the nucleoid and to prevent their aggregation.
Upon their discovery, it was frequently noted that the carboxysomes of cyanobacteria and chemoautotrophs are commonly associated with the nucleoid region of the cell (Gantt & Conti, 1969; Shively et al., 1970, 1973; Wolk, 1973). Our work shows that, for both groups of bacteria, it is the McdAB system that is responsible for this association. We also find that in the absence of the α‐McdAB system, nucleoid positioning and cell morphology are altered by the carboxysome aggregate, which likely has drastic physiological consequences. While α‐ and β‐carboxysome aggregation does not result in a high CO2‐requiring phenotype (Desmarais et al., 2019; Maccready et al., 2018), we recently found that β‐carboxysome aggregation in S. elongatus results in slower growth, cell elongation, asymmetric cell division, and altered cellular levels of Rubisco (Rillema et al., 2020). It was also recently found that inactive carboxysomes are degraded at polar regions of a cyanobacterial cell (Hill et al., 2020). How the polar localization of carboxysome aggregates, from a lack of a McdAB system, perturbs carboxysome function and turnover remains unclear. A future direction will be the study of α‐ and β‐McdAB systems under varying biotic and abiotic conditions to understand the impact of carboxysome mispositioning and aggregation on cellular physiology.
2.3. α‐McdB is targeted to α‐carboxysomes and interacts with α‐McdA
We recently showed that β‐McdB acts as an adaptor between β‐McdA on the nucleoid and β‐carboxysomes in the cyanobacterium S. elongatus. To determine whether H. neapolitanus α‐McdB is targeted to α‐carboxysomes, we expressed a second copy of α‐McdB as a fusion to the fluorescent protein mNeonGreen (mNG) (Shaner et al., 2013), producing mNG‐α‐McdB, in our native CbbS‐mTQ strain. Consistent with mNG‐β‐McdB loading onto β‐carboxysomes in S. elongatus (Maccready et al., 2018), we found that mNG‐α‐McdB colocalized with the CbbS‐mTQ signal of H. neapolitanus α‐carboxysomes (PCC = 0.81; n = 173 cells) (Figure 4a and Video 2). Due to the diffuse nature of the α‐carboxysome fluorescence signal, as described above, we took advantage of our finding that α‐carboxysomes cluster to form bright and clearly resolved puncta at the cell poles without α‐McdA present. In our Δα‐mcdA deletion strain, mNG‐α‐McdB clearly colocalized with α‐carboxysome aggregates (PCC = 0.90; n = 355 cells) (Figure 4b and Video 3). The data show that α‐McdB associates with α‐carboxysomes, and α‐McdA is not required for this association.
FIGURE 4.
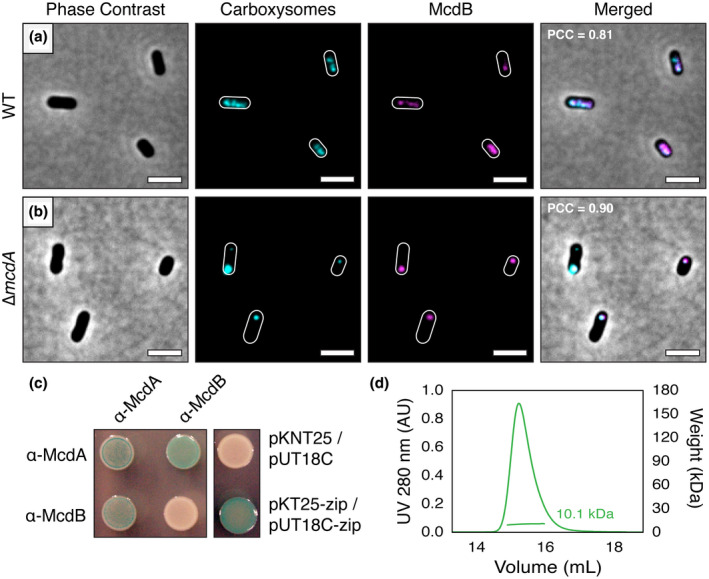
α‐McdB loads onto α‐carboxysomes and interacts with α‐McdA. (a) mNG‐McdB (magenta) colocalizes with the carboxysome reporter CbbS‐mTQ (cyan) in WT H. neapolitanus. (b) In the absence of McdA, McdB strongly colocalizes with carboxysome aggregates. PCC values were calculated from n ≥ 100 cells per cell population. Scale bar: 2 µm. (c) Bacterial‐2‐Hybrid (B2H) analysis of α‐McdA and α‐McdB. α‐McdA was positive for self‐association. α‐McdA directly interacts with α‐McdB. α‐McdB did not self‐associate. B2H image is representative of three independent trials. (d) SEC‐MALS plot for H. neapolitanus α‐McdB; monomer MW = 10 kDa [Colour figure can be viewed at wileyonlinelibrary.com]
Finally, as we did for the β‐McdAB system in S. elongatus, we sought to determine if α‐McdA and α‐McdB of H. neapolitanus self‐associate and directly interact with each other by performing a Bacterial‐2‐Hybrid assay (B2H) in E. coli. Consistent with ParA‐family members forming dimers (Schumacher, 2007), including a β‐McdA‐like homolog (Schumacher et al., 2019), α‐McdA was positive for self‐association (Figure 4c). Also, like the β‐McdAB system of S. elongatus, α‐McdA directly interacts with α‐McdB. Surprisingly, however, we found that H. neapolitanus α‐McdB did not self‐associate in our B2H assay, whereas S. elongatus β‐McdB strongly self‐associates (Maccready et al., 2018). It is difficult to draw firm conclusions on the lack of McdB self‐interaction because the T18/T25 domains may sterically hinder the interaction, or influence protein stability. Therefore, we subsequently performed Size Exclusion Chromatography—Multiple Angle Laser Light Scattering (SEC‐MALS) and found that purified α‐McdB, indeed, remains a monomer in solution (Figure 4d). This difference in α‐ and β‐McdB self‐association has implications for understanding how McdB proteins are recruited to structurally distinct α‐ and β‐carboxysomes as well as how McdBs interact with their cognate McdA ATPase on the nucleoid.
2.4. Defining the conserved features of α‐McdAB proteins
We recently found that McdAB systems are widespread across β‐cyanobacteria, which possess β‐carboxysomes (MacCready et al., 2020). A surprising finding in this search was that McdAB systems were absent in α‐cyanobacteria, which possess α‐carboxysomes. Thus, our finding here of an α‐McdAB system that positions α‐carboxysomes in H. neapolitanus was unexpected. We next sought to determine how widespread this system was among α‐carboxysome‐containing proteobacteria. Given the amino acid sequence diversity, we previously found among cyanobacterial β‐McdA proteins, and especially for β‐McdB proteins, we first searched for α‐McdAB systems by performing neighborhood analyses within and around proteobacterial α‐carboxysome operons. Using BlastP, we identified α‐carboxysome‐containing proteobacteria within NCBI and JGI IMG databases using α‐carboxysome components CsoS2, CsoS4A, or CsoS4B as queries (see Figure 1d). Next, we searched around each α‐carboxysome operon in these organisms to identify α‐mcdAB‐like genes. We classified positive hits for α‐mcdA as proteins containing the deviant‐Walker‐A box, which defines the ParA family of ATPases (Koonin, 1993), as well as Walker A’ and Walker B boxes. We classified positive hits for α‐mcdB as a small coding sequence immediately following the α‐mcdA gene.
Alignment of the amino acid sequences of all α‐McdA hits identified through our analyses revealed a high percentage of similarity (~53% pairwise identity), but also revealed three regions of conservation among α‐McdA proteins not present among classical ParA‐type proteins (Figures 5a and S3). While α‐McdA proteins were largely conserved, α‐McdB proteins displayed extreme diversity. This was also true for cyanobacterial β‐McdAB systems (MacCready et al., 2020). All α‐McdB hits also shared many of the general sequence features present in β‐McdB proteins, including: (i) a well‐conserved charged N‐terminus, (ii) an invariant C‐terminal tryptophan residue (all ended with the amino acid sequence V(I)WPD), (iii) low hydrophobicity, (iv) biased amino acid composition, and (v) intrinsic disorder (Figures 5b and S4).
FIGURE 5.
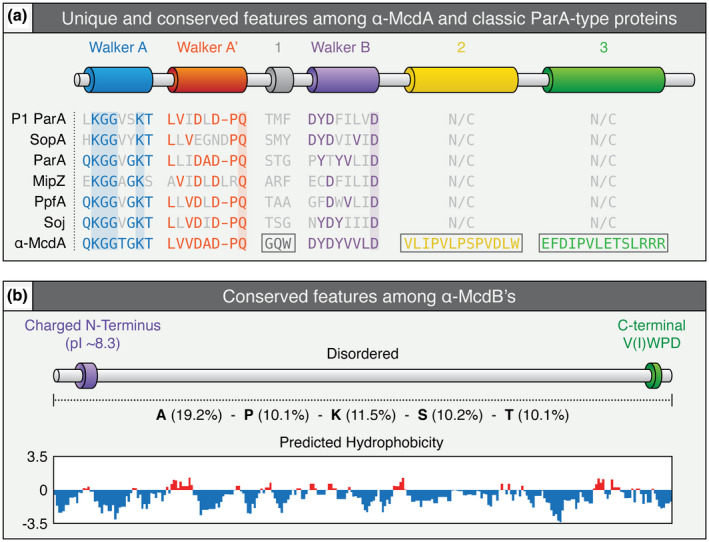
Conserved features among α‐McdAB proteins within cso operons. (a) Features conserved or distinct (boxed) among identified α‐McdA proteins encoded within the cso operon of α‐carboxysome‐containing proteobacteria compared to classical ParA‐type proteins involved in the positioning of diverse cargoes. The deviant‐Walker A (blue), A’ (red), and B (purple) boxes, are conserved among all ParA family proteins. ParA‐type proteins shown: Escherichia coli phage P1 ParA (plasmid partitioning—YP_006528) (Abeles et al., 1985), Escherichia coli F plasmid SopA (plasmid partitioning—NP_061425) (Mori et al., 1986), Caulobacter crescentus ParA (chromosome segregation—AAB51267) (Mohl & Gober, 1997), Caulobacter crescentus MipZ (cell‐division positioning—NP_420968) (Thanbichler & Shapiro, 2006), Rhodobacter sphaeroides PpfA (chemotaxis cluster distribution—EGJ21499) (Roberts et al., 2012), and Bacillus subtilis Soj (chromosome segregation—NP_391977) (Marston & Errington, 1999). (b) General features of α‐McdB proteins encoded within the cso operon of α‐carboxysome‐containing proteobacteria. Percent composition of the amino acids alanine (A), proline (P), lysine (K), serine (S), and threonine (T) are presented to illustrate the strong bias for these amino acids (center). All α‐McdB proteins identified are highly hydrophilic across the entire primary sequence (bottom) [Colour figure can be viewed at wileyonlinelibrary.com]
2.5. α‐McdAB systems are widespread among α‐carboxysome‐containing proteobacteria
We previously found that many β‐cyanobacteria had mcdA and mcdB genes at distant loci from the operons encoding carboxysome components (MacCready et al., 2020). Our identification of highly conserved regions that are specific to α‐McdA proteins, along with the conserved features among α‐McdB proteins, allowed us to re‐search proteobacterial genomes to identify more α‐McdA and α‐McdB genes that did not fall within or near the α‐carboxysome operon. Across 250 α‐carboxysome‐containing proteobacterial genomes, we identified 228 α‐McdA sequences (~91% of genomes analyzed) and 278 α‐McdB sequences (100% of genomes analyzed) (Figure 6a). These results present a similar trend to what we previously found in several β‐cyanobacteria—β‐McdB can exist without the presence of β‐McdA (“orphaned” McdBs) and β‐McdB proteins greatly varied in length (51–169 amino acids) (Figure 6a). Interestingly, 12% of the genomes analyzed contained a second α‐mcdB gene (see McdB sequences labeled as “Special Orphan” in Figure S4). Orphaned McdB paralogs lack the charged N‐terminus conserved in McdB proteins that are encoded adjacent to the mcdA gene. Aside from this distinction, both McdB paralogs are predicted to be completely disordered and both have the invariant V(I)WPD sequence present across all α‐McdB proteins identified thus far. To our knowledge, this is the first example of two cognate ParA partner protein paralogs potentially involved in the same process.
FIGURE 6.
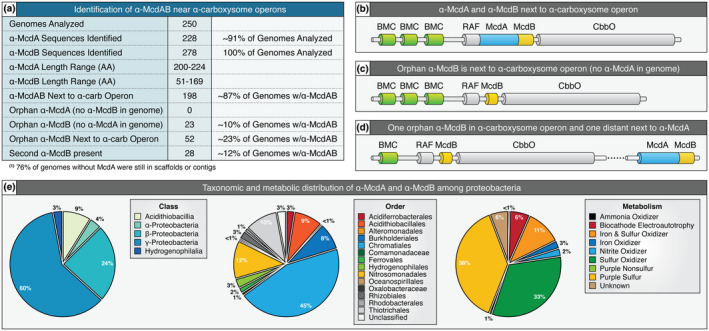
McdAB systems are widespread among carboxysome‐containing proteobacteria. (a) Table highlighting the prevalence and genomic context of all identified α‐McdAB sequences within α‐carboxysome‐containing proteobacteria. (b) Genomic arrangement when α‐McdAB are encoded within the cso operon. (c) Genomic arrangement when only α‐McdB is encoded within the cso operon. (d) Genomic arrangement when one copy of α‐McdB is encoded within the cso operon and a second copy of α‐McdB is encoded next to α‐mcdA at a distant locus. (e) α‐McdAB systems are widely distributed among proteobacterial taxonomic classes (left), orders (center), and metabolisms (right) [Colour figure can be viewed at wileyonlinelibrary.com]
In total, we found three possible genomic arrangements of α‐mcdA and α‐mcdB genes: (i) α‐mcdA and α‐mcdB were both within the α‐carboxysome operon (Figure 6b), (ii) only α‐mcdB was found within the operon (Figure 6c), or (iii) one orphan α‐mcdB gene was within the operon, and a second α‐mcdB gene was present at a distant locus but next to α‐mcdA (Figure 6d). Consistent with our identification of orphan McdB proteins, we have recently found that McdB in S. elongatus plays an important, but currently unclear, role in carboxysome function outside of its role in positioning carboxysomes with McdA (Rillema et al., 2020). Indeed, ParA‐partner proteins involved in the positioning of other protein‐based cargos, such as chemotaxis clusters (Alvarado et al., 2017; Ringgaard et al., 2011; Roberts et al., 2012), are also key functional components of the cargo itself. We note, however, that 76% of genomes without α‐mcdA were still incomplete; therefore, it is still possible that an unidentified α‐mcdA gene is present and next to the additional α‐mcdB gene in these genomes.
Overall, we found that α‐McdAB systems are present across most major taxonomic classes and orders of proteobacteria; largely present in the class γ‐proteobacteria (60% of genomes) and order Chromatiales (45% of genomes) (Figure 6e). Unlike cyanobacteria, which perform oxygenic photosynthesis, the metabolisms of α‐carboxysome‐containing proteobacteria can greatly vary (see Figure 1c). We found α‐McdAB systems in nitrite, ammonia, and iron utilizers, as well as biocathode electroautotrophs that can acquire energy by taking up electrons from electrodes, while using CO2 as an inorganic carbon source and terminal electron acceptor (Nevin et al., 2010) (Figure 6e). However, α‐McdAB systems were primarily found in purple sulfur bacteria (38% of genomes), which perform anoxygenic photosynthesis, and sulfur‐oxidizing chemoautotrophs, such as H. neapolitanus (33% of genomes). Purple sulfur bacteria use sulfide and hydrogen as electron donors, whereas purple non‐sulfur bacteria use organic compounds (Madigan & Jung, 2009). Interestingly, the size and quantity of β‐carboxysomes in cyanobacteria are directly linked to not only CO2 availability, but also light intensity and quality (Rillema et al., 2020; Rohnke et al., 2018, 2020; Sun et al., 2016, 2019). Given the prevalence of McdAB systems in sulfur utilizers, it is intriguing to speculate that sulfur‐limitation will also alter α‐carboxysome size, number, and/or distribution in H. neapolitanus.
2.6. Cyanobacterial α‐carboxysomes likely originated from a proteobacterium lacking α‐mcdA within their cso operon
It is largely believed that α‐carboxysomes emerged in proteobacteria and then were horizontally transferred into cyanobacteria; creating the distinct phylogenetic clade of α‐cyanobacteria (Badger & Bek, 2008; Badger et al., 2002; Badger & Price, 2003; Marin et al., 2007; Rae et al., 2013). However, following our recent study (MacCready et al., 2020), why α‐cyanobacteria lack the McdAB system remained an outstanding question. In an attempt to better understand α‐carboxysome evolution among α‐cyanobacteria and proteobacteria, we constructed a Maximum Likelihood phylogenetic tree inferred using a concatenation of the major α‐carboxysome components CbbL, CbbS, CsoS3, CsoS4A, and CsoS4B (see Figure 1d). We note that the major carboxysome component CsoS2 was omitted from this analysis due to its intrinsically disordered nature (Oltrogge et al., 2020), which has relaxed selection due to the lack of structural constraint.
Recall that we found α‐carboxysome‐containing proteobacteria encoding two α‐McdB genes, where one α‐mcdB gene is orphaned within the cso operon without an adjacent α‐mcdA gene, and the other is encoded next to α‐mcdA at a distant locus away from the cso operon (see Figure 6d). Intriguingly, we found that α‐carboxysomes from proteobacteria with two copies of α‐mcdB formed a single clade (Figure 7a). This clade was more closely related to α‐cyanobacterial α‐carboxysomes than to proteobacterial α‐carboxysomes that have both α‐mcdA and α‐mcdB encoded within the cso operon. Indeed, while H. neapolitanus possess both α‐mcdA and α‐mcdB within their cso operon (Figure 7b), many other proteobacterial species, such as Thiohalospira halophila DSM 15071, only possess an orphaned α‐mcdB within their cso operon; α‐mcdA and the second α‐mcdB gene are encoded at a distant locus (Figure 7c). Thus, our phylogenetic tree suggests that α‐cyanobacteria, such as Cyanobium gracile PCC 6307, likely horizontally inherited a cso operon from a proteobacterium that encoded α‐mcdA and a second α‐mcdB gene away from the cso operon (Figure 7d). While it is possible that α‐cyanobacteria inherited the orphan α‐mcdB gene present within the proteobacterial cso operon, it is likely that this protein no longer provided a fitness advantage in the absence of α‐McdA, or the second α‐McdB protein, and was, therefore, lost.
FIGURE 7.
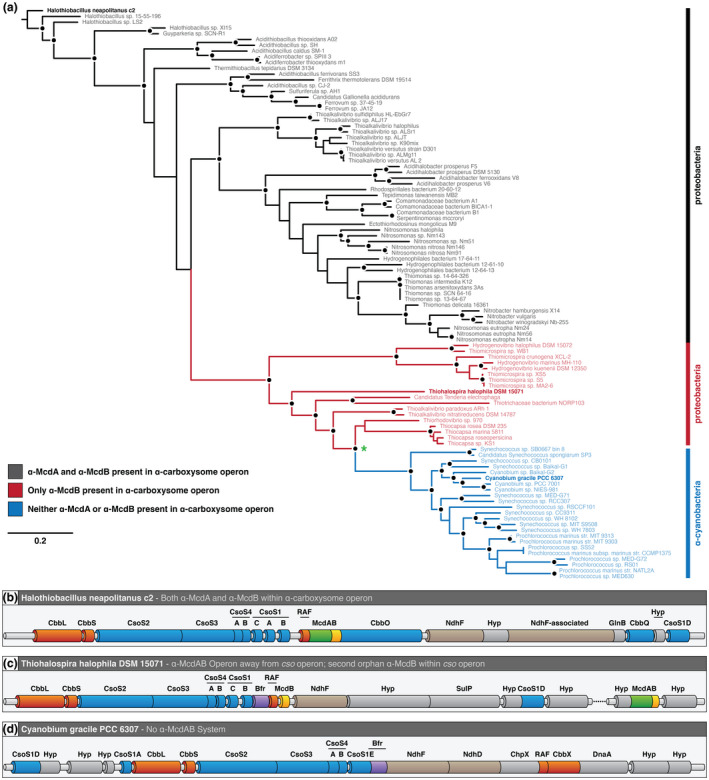
α‐carboxysome evolution among proteobacteria and α‐cyanobacteria. (a) Inferred phylogeny of α‐carboxysome‐containing proteobacteria and α‐cyanobacteria. Line colors: α‐McdAB found within cso operon (black), only α‐McdB found within cso operon (red), and neither α‐McdAB found within cso operon (blue). Black dot represents >70% bootstrap support (500 replicates). Green asterisk represents a shared cso operon ancestor among α‐cyanobacteria and proteobacteria that lacks the α‐mcdA gene. (b) Genomic arrangement of the H. neapolitanus cso operon. (c) Genomic arrangement of the T. halophila cso operon. (D) Genomic arrangement of the C. gracile cso operon, which lacks a McdAB system [Colour figure can be viewed at wileyonlinelibrary.com]
Our finding that McdAB systems are widespread in β‐cyanobacteria and proteobacteria, but completely absent in α‐cyanobacteria raises the question: What is different about α‐carboxysomes in α‐cyanobacteria that makes active distribution by the McdAB system unnecessary? In general, β‐carboxysomes are significantly larger in diameter (β‐carboxysome of Spirulina platensis is > 500 nm) than proteobacterial α‐carboxysomes (α‐carboxysome of proteobacterium Nitrobacter agilis ~ 120 nm), and of the limited number of α‐carboxysomes studied in α‐cyanobacteria, these are typically the smallest (Prochlorococcus marinus ~ 90 nm) (Rae et al., 2013). α‐carboxysome copy number in the cell is also typically much higher. This inverse correlation of carboxysome size and copy number is also true for plasmids. High‐copy plasmids are typically smaller, whereas low‐copy plasmids are larger (Planchenault et al., 2020). Intriguingly, it was recently found across the sequenced genomes of enterobacteria that ParA‐based plasmid partition systems are present in larger plasmids (>25 kb) and lacking in smaller plasmids (Planchenault et al., 2020). All plasmids larger than 180 kb encoded a ParA‐based partition system without exception. Therefore, it is attractive to speculate that, like for small plasmids, the α‐carboxysomes of α‐cyanobacteria may rely on their high‐copy number and smaller size for inheritance, which would make the McdAB system dispensable in these organisms. In line with this reasoning, not a single active distribution system for encapsulin nanocompartments that are even smaller in diameter (~30 nm) has yet to be identified.
2.7. α‐McdAB systems are distinct from the β‐McdAB systems of cyanobacteria
Given the quantity of α‐ and β‐McdAB systems that we have identified across cyanobacteria and proteobacteria, we next sought to identify similarities and differences among these subtypes. We previously identified two distinct β‐McdAB systems exclusive to β‐cyanobacteria, with Type 2 systems being more ancestral and more abundant than Type 1 (MacCready et al., 2020). Here, we found that α‐McdA proteins share more features with β‐McdA Type 2 proteins, including: (i) the presence of the signature lysine residue in the deviant‐Walker A box, which defines the ParA family but is absent in β‐McdA Type 1, (ii) a conserved central tryptophan residue in region 1, (iii) a highly similar Walker B box, and (iv) greater conservation in region 4 (Figure 8a). This further demonstrates the divergence of Type 1 β‐McdA proteins (present in S. elongatus), which lack the signature lysine residue in the deviant‐Walker A box and possesses a large mid‐protein extension (MacCready et al., 2020).
FIGURE 8.
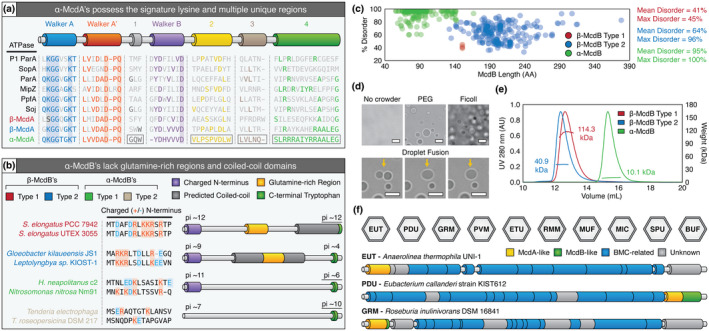
Similarities and differences among all known McdA and McdB proteins. (a) Features conserved or distinct among α‐McdA (green), β‐McdA Type 1 (red), β‐ McdA Type 2 (blue), and diverse classical ParA‐type proteins. All Walker‐boxes are well‐conserved among all classic ParA family proteins. ParA‐type proteins shown: Escherichia coli phage P1 ParA (plasmid partitioning—YP_006528) (Abeles et al., 1985), Escherichia coli F plasmid SopA (plasmid partitioning—NP_061425) (Mori et al., 1986), Caulobacter crescentus ParA (chromosome segregation—AAB51267) (Mohl & Gober, 1997), Caulobacter crescentus MipZ (cell‐division positioning—NP_420968) (Thanbichler & Shapiro, 2006), Rhodobacter sphaeroides PpfA (chemotaxis cluster distribution—EGJ21499) (Roberts et al., 2012), and Bacillus subtilis Soj (chromosome segregation—NP_391977) (Marston & Errington, 1999). (b) α‐McdB proteins lack the predicted central coiled‐coil and glutamine‐rich regions found in β‐McdB proteins. α‐McdB Type 2 proteins lack the charged N‐terminus conserved in all other McdB types. (c) PONDR disorder scatter plot for all McdB protein types. (d) DIC microscopy images showing purified H. neapolitanus McdB undergoes LLPS in vitro in the presence of the crowders PEG or Ficoll. Droplets exhibit liquid‐like properties such as fusion (yellow arrows). (e) SEC‐MALS plot for a representative β‐McdB Type 1 (S. elongatus McdB; monomer MW = 17 kDa), β‐McdB Type 2 (Synechococcus sp. PCC 7002 McdB; monomer MW = 21 kDa), and α‐McdB (H. neapolitanus McdB; monomer MW = 10 kDa). (f) McdA/B‐like sequences genomically neighbor BMC components across diverse microbes [Colour figure can be viewed at wileyonlinelibrary.com]
Next, we examined the similarities and differences among all McdB amino acid sequences identified. ParB proteins that connect plasmid or chromosomal cargo to their cognate ParA ATPase use a charged N‐terminal region to interact with ParA and stimulate its ATPase activity (Badrinarayanan et al., 2015; Baxter & Funnell, 2014). We found that all McdB types encoded next to their cognate mcdA gene also possess conserved highly charged N‐termini (Figure 8b). But in proteobacteria where the second α‐mcdB gene is orphaned in the cso operon, this highly charged N‐terminus is absent (Figure 8b). Given this observation, we designate α‐McdB proteins with the charged N‐terminal region and encoded downstream of the α‐mcdA gene as Type 1, and α‐McdB proteins lacking the charged N‐terminus and orphaned in the cso operon as Type 2. It remains to be determined if the charged N‐terminus of McdB proteins is, indeed, responsible for interaction with McdA. Further dividing α‐ and β‐McdB proteins, we find that α‐McdB proteins lack the predicted coiled‐coil region and mid‐protein glutamine‐rich stretches conserved among β‐McdB proteins (Figure 8b). Despite these differences among subtypes, all McdB proteins possess an invariant C‐terminal tryptophan residue. This amino acid is intriguing because many proteins involved in the assembly of viral‐ or phage‐capsids also contain a tryptophan at their C‐terminus (Deeb, 1973; Johnson et al., 2020; Komla‐Soukha & Sureau, 2006; Marintcheva et al., 2006; Skoging & Liljeström, 1998; Tsuboi et al., 2003). Given the capsid‐like icosahedral structure of the carboxysome, it is attractive to speculate that the C‐terminal tryptophan residue plays a role in McdB recruitment to carboxysomes.
Our finding that many proteobacteria possess an additional distinct copy of α‐McdB was intriguing. We have yet to identify a cyanobacterial species with two mcdB genes (MacCready et al., 2020). The need for two α‐McdB proteins is not clear, but an absence of the charged N‐terminus that is conserved in the McdB paralog suggests a role in modulating carboxysome interactions with McdA on the nucleoid. These α‐McdB paralogs may work together in modulating interactions with α‐McdA, possibly tuning the stimulation of McdA ATPase activity and release from the nucleoid. For instance, both McdB copies likely associate with carboxysomes, but only the paralog with the charged N‐terminus would interact with α‐McdA. Therefore, the relative ratio of these McdB paralogs would dictate the extent to which carboxysomes associate with McdA on the nucleoid. Paralog interactions regulating each other's function are not uncommon. One such example is FtsZ paralogs (FtsZ1 and FtsZ2) involved in archaeal cell division (Liao et al., 2020) and chloroplast division (Chen et al., 2018). In the case of chloroplasts, these paralogs assemble into heteropolymers at mid‐chloroplast to define the division site. FtsZ1 increases FtsZ2 turnover, which is thought to facilitate FtsZ2 remodeling during chloroplast constriction (TerBush & Osteryoung, 2012). In this way, FtsZ1 functions as a regulator of FtsZ2 activity and is a highly conserved feature throughout photosynthetic lineages (TerBush et al., 2018). Elucidating how α‐McdB paralogs influence carboxysome function and organization is an exciting avenue of future research.
2.8. All McdB types show LLPS activity, but with different minimal oligomeric units
A conserved feature across all McdB types we identified is intrinsic disorder, but to varying degrees (Figure 8c). While β‐McdB Type 1 proteins are on average 41% disordered and β‐McdB Type 2 proteins are on average 64% disordered, α‐McdB proteins are significantly more disordered at ~95%. This dramatic difference in disorder for α‐McdB proteins is likely due to the lack of the predicted coiled‐coil found in both β‐McdB types (see Figure 8b). In addition to intrinsic disorder, all McdB proteins share low complexity regions with repetitive and biased amino acid compositions, and low hydrophobicity. All these features are hallmarks of proteins that can undergo Liquid‐Liquid Phase Separation (LLPS) (Alberti et al., 2019). LLPS refers to the ability of biomolecules, such as protein, to separate in solution to form a condensed liquid phase with material properties distinct from those of the surrounding dilute phase. We have previously shown that purified representatives from both types of β‐McdB proteins (β‐McdB Type 1 from S. elongatus and β‐McdB Type 2 from Synechococcus sp. PCC 7002) can undergo LLPS in vitro (MacCready et al., 2020). We purified H. neapolitanus α‐McdB and found that it too has LLPS activity, forming droplets with liquid‐like behaviors such as the fusion of two adjacent droplets into one (Figure 8d, Video 4). We conclude that LLPS is a conserved activity across all McdB proteins, but it remains to be determined how McdB LLPS activity plays a role in carboxysome positioning and function.
The fact that β‐McdB proteins have a predicted coiled‐coil, while α‐McdB proteins are almost completely disordered, suggests that β‐McdB proteins form oligomers. We performed Size Exclusion Chromatography ‐ Multiple Angle Laser Light Scattering (SEC‐MALS) to determine the oligomeric state of purified α‐ and β‐McdB proteins. We found that S. elongatus Type 1 β‐McdB formed a hexamer, Synechococcus sp. PCC 7002 Type 2 β‐McdB formed a dimer, and consistent with lacking a predicted coiled‐coil, H. neapolitanus α‐McdB remained a monomer (Figure 8e). These findings are consistent with our B2H data, where β‐McdB of S. elongatus strongly self‐associates (Maccready et al., 2018), whereas H. neapolitanus α‐McdB showed no self‐association (see Figure 4c). We conclude that the predicted coiled‐coil domains exclusive to β‐McdB proteins are likely required for oligomerization and are important for β‐carboxysome positioning and function, whereas α‐McdB proteins function as monomers.
A future study will determine how differences in oligomeric state influence the ability of McdB proteins to undergo LLPS.
2.9. McdAB systems are not restricted to carboxysome BMCs
Last, an outstanding question is whether the McdAB system is restricted to carboxysome BMCs. While carboxysomes are the only known anabolic BMC, several other catabolic BMCs exist (Kerfeld et al., 2018), including: (i) EUT (Ethanolamine UTilization microcompartment), (ii) PDU (1,2‐Propanediol Utilization microcompartment), (iii) GRM (Glycyl Radical enzyme‐containing Microcompartment), (iv) PVM (Planctomycetes and Verrucomicrobia Microcompartment), (v) ETU (EThanol‐Utilizing microcompartment), (vi) RMM (Rhodococcus and Mycobacterium Microcompartment), (vii) MUF (Metabolosome of Unknown Function microcompartment), (viii) MIC (Metabolosome with Incomplete Core microcompartment), (ix) SPU (Sugar Phosphate Utilizing microcompartment), and (x) BUF (BMC of Unknown Function) (Figure 8f). In four such examples, we find McdB‐ and/or McdA‐like sequences within or neighboring these BMC operons (Figure 8f). In cases where McdB‐like sequences were observed, all possess a C‐terminal aromatic residue (tyrosine instead of tryptophan) within the last four amino acids. As we detailed above, a C‐terminal tryptophan residue is invariant across all carboxysome‐associated McdB proteins we have identified to date (Figure 8b), and may play a role in McdB association with their cognate BMC.
Collectively, our results show that α‐McdAB systems are widespread among α‐carboxysome containing proteobacteria and function as an anti‐aggregation mechanism to ensure proper distribution and inheritance of α‐carboxysomes following cell division. Our results have important implications for understanding the evolution and function of diverse McdA and McdB proteins as it relates to structurally and phyletically distinct α‐ and β‐carboxysomes, and also have much broader implications for understanding the equidistant positioning of diverse catabolic BMCs across the bacterial domain.
3. METHODS
3.1. Strains and growth conditions
All mutants described in this study were constructed using wild‐type Halothiobacillus neapolitanus (Parker) Kelly and Wood (ATCC® 23641™) purchased from ATCC. Cultures were grown in ATCC® Medium 290: S‐6 medium for Thiobacilli (Hutchinson et al., 1965) and incubated at 30°C, while shaken at 130 RPM in air supplemented with 5% CO2. Strains were preserved frozen at −80°C in 10% DMSO.
3.2. Construct designs and cloning
All constructs were generated using Gibson Assembly and verified by sequencing. Fragments for assembly were synthesized by PCR or ordered as a gBlock (IDT). Constructs contained flanking DNA that ranged from 750 to 1,100 bp in length upstream and downstream of the targeted insertion site to promote homologous recombination into target genomic loci. Cloning of plasmids was performed in chemically competent E. coli Top10 or Stellar cells (Takara Bio).
3.3. Transformation in H. neapolitanus
Competent cells of H. neapolitanus were generated by growing 1 L of log culture in 2.8 L baffled flasks. Cultures were harvested by centrifugation at 3,000g for 45 min. Cell pellets were resuspended and washed twice with 0.5 volumes of ice‐cold Millipore water. The resulting pellet was resuspended in 1 × 10–3 volumes (1 ml) of ice‐cold Millipore water. These competent cells were used immediately or frozen at −80°C for future use. Frozen competent cells were thawed on ice for 4 hr or overnight at 4°C. Competent cells were mixed with 5 µl plasmid DNA (1.5–5 µg) and incubated on ice for 5 min. This mixture was then transferred to a tube containing 5 ml of ice‐cold S6 medium without antibiotics and incubated on ice for 5 min. Cells were then incubated for 16–36 hr at 30°C, while shaken at 130 RPM in air supplemented with 5% CO2. Transformants were selected by plating on selective S6 medium with antibiotics. Colonies were restreaked. Restreaked colonies were verified for mutation by PCR.
3.4. Native fluorescent fusions
For the native CbbS fluorescent fusion, the sequences encoding the fluorescent protein mTurquoise (mTq) were attached to the 3ʹ region of the native cbbS coding sequence, separated by a GSGSGS linker. A kanamycin resistance cassette was inserted immediately downstream of the cbbS coding sequence. The mutant was selected by plating on S6 agar plates supplemented with 50 µg/ml of kanamycin. The CbbS‐mTQ fusion was verified by PCR.
3.5. Single gene deletions of mcdA and mcdB
mcdA (Hn0912) and mcdB (Hn0911) are the second and third genes in their operon. The deletion construct of mcdA (Hn0912) was created by replacing the mcdA coding sequence with a spectinomycin resistance cassette. The promoter upstream of Hn0913 was duplicated and inserted downstream of the antibiotic cassette to minimize disruption of downstream genes. The deletion construct of mcdB was created by removing mcdB (Hn0911), along with the two upstream genes (Hn0913 and Hn0912). These genes were replaced with a spectinomycin resistance cassette, followed by the promoter upstream of Hn0913 and the codon‐optimized genes Hn0913 and Hn0912. This allowed for a clean deletion of the mcdB coding sequence. Mutants were selected by plating on S6 agar plates supplemented with 50 µg/ml of spectinomycin. The mutations were verified by PCR.
3.6. Exogenous expression of mNG‐McdB
The sequence encoding the fluorescent protein mNeonGreen (mNG) was attached to the 5ʹ region of the mcdB coding sequence, separated by a GSGSGS linker. The gene was codon‐optimized, placed under the expression of a Ptrc promoter, and inserted into a neutral site, located between genes Hn0933 and Hn0934. Mutants were selected by plating on S6 agar plates supplemented with 25 µg/ml of chloramphenicol. The insertion was verified by PCR.
Exogenous expression of mNG‐McdB was not induced. All images obtained resulted from leaky expression of the Ptrc promoter. Exogenous expression images of mNG‐McdB were compared back to empty vector control, which showed no fluorescence signal.
3.7. Nucleoid staining
Cells were harvested by centrifugation at 5,000g for 5 min. Following centrifugation, the cells were washed in PBS. The resulting cell pellet was resuspended in 50 µl PBS and stained with DAPI at a final concentration of 25 µM for 5–10 min immediately prior to imaging.
3.8. Ciprofloxacin and cephalexin treatments
Cells were collected at an OD of 0.05–0.1 and treated with 50 µM ciprofloxacin or with 8 µM cephalexin for 24 hr. Cells were then stained with 2 µg/ml of DAPI for 15 min and imaged.
3.9. Fluorescence imaging
All live‐cell microscopy was performed using exponentially growing cells. Three microliters of cells were dropped onto a square of 2% agarose + S6 pad and imaged on a glass‐bottom dish (MatTek Life Sciences). All fluorescence and phase contrast imaging were performed using a Nikon Ti2‐E motorized inverted microscope controlled by NIS Elements software with a SOLA 365 LED light source, a 100X Objective lens (Oil CFI Plan Apochromat DM Lambda Series for Phase Contrast), and a Photometrics Prime 95B Back‐illuminated sCMOS camera or a Hamamatsu Orca Flash 4.0 LT + sCMOS camera. mNG‐McdB was imaged using a “YFP” filter set (C‐FL YFP, Hard Coat, High Signal‐to‐Noise, Zero Shift, Excitation: 500/20nm [490‐510 nm], Emission: 535/30 nm [520‐550 nm], Dichroic Mirror: 515 nm). CbbS‐mTQ labeled carboxysomes were imaged using a “CFP” filter set (C‐FL CFP, Hard Coat, High Signal‐to‐Noise, Zero Shift, Excitation: 436/20 nm [426‐446 nm], Emission: 480/40 nm [460‐500 nm], Dichroic Mirror: 455 nm). DAPI fluorescence was imaged using a standard “DAPI” filter set (C‐FL DAPI, Hard Coat, High Signal‐to‐Noise, Zero Shift, Excitation: 350/50 nm [325‐375 nm], Emission: 460/50 nm [435‐485 nm], Dichroic Mirror: 400 nm). All videos were taken at one frame per minute for a duration of 15 min.
3.10. Transmission electron microscopy
Log cultures of H. neapolitanus were pelleted and fixed overnight at 4°C in 2.5% glutaraldehyde in 0.1 M Sorensen's buffer (pH 7.4). After several washes in 0.1 M Sorensen's buffer, cells were post‐fixed overnight at 4°C in 1% osmium tetroxide in 0.1 M Sorensen's buffer. Cells were washed until the solution became clear. Cells were then immobilized in 1% agarose and sliced thinly into 0.5 mm slices. Slices were dehydrated in an increasing series of ethanol or acetone (30%–100%) washes for 10 min each. Slices were infiltrated with Embed812 resin (25% increments for 1–16 hr each at room temperature). A final incubation in full strength resin was performed at room temperature under vacuum. Cells were then embedded in blocks, incubated under vacuum overnight, and transferred to a 60°C oven to polymerize for 1–2 days. Thin sections of approximately 50 nm were obtained by a Leica UC7 Ultramicrotome. Sections were post‐stained with 7% uranyl acetate for 10 min, Reynold's lead citrate for 5 min, and visualized on a JEOL 1400‐plus transmission electron microscope equipped with an XR401 AMT sCMOS camera.
3.11. Bacterial two‐hybrid analysis
N‐terminal T18 and T25 fusions of McdA and McdB were constructed using plasmids pKT25, pKNT25, pUT18C, and pUT18, sequence‐verified, and co‐transformed into E. coli BTH101 (Karimova et al., 1998). Several colonies of T18/T25 cotransformants were isolated and grown in LB medium with 100 μg/ml of ampicillin, 50 μg/ml of kanamycin, and 0.5 mM IPTG overnight at 30°C, while shaking at 225 RPM. Overnight cultures were spotted on indicator Xgal plates supplemented with 100 μg/ml of ampicillin, 50 μg/ml of kanamycin and 0.5 mM IPTG. Plates were incubated at 30°C up to 48 hr before imaging.
3.12. α‐McdAB homolog search and neighborhood analysis
Identification of all α‐carboxysome containing proteobacteria within the NCBI and JGI databases was performed via BlastP using the α‐carboxysome components CsoS2, CsoS4A, or CsoS4B as queries. Neighborhood analyses for McdA‐ and McdB‐like sequences were then carried out within these resulting genomes by manually searching 4,000 bp upstream and downstream of the cso operon. Multiple sequence alignment for identified putative McdA proteins was performed using MAFFT 1.3.7 under the G‐INS‐I algorithm (Katoh & Standley, 2013), whereas the E‐INS‐I algorithm was used for McdB proteins due to long gaps caused by intrinsic disorder. Subsequent identification of McdA‐ and McdB‐like sequences away from the cso operon was performed via BlastP using highly conserved regions/features identified from the multiple sequence alignments.
3.13. α‐McdB sequence analysis
Coiled‐coil predictions for α‐McdB proteins were performed using DeepCoil (Ludwiczak et al., 2019). α‐McdB protein disorder predictions were conducted using PONDR with the VL‐XT algorithm (Li et al., 1999; Romero et al., 1997, 2001). Analysis of α‐McdB hydrophobicity was performed using ProtScale using the Kyte and Doolittle scale (Gasteiger et al., 2005; Kyte & Doolittle, 1982).
3.14. Phylogenetic inference
Ortholog sequences for H. neapolitanus CbbL (Hneap_0922), CbbS (Hneap_0921), CsoS3 (Hneap_0919), CsoS4A (Hneap_0918), and CsoS4B (Hneap_0917) were obtained via BlastP for each proteobacterium and cyanobacterium. Multiple sequence alignments for each protein sequence were performed using MAFFT 1.3.7 (Katoh & Standley, 2013) under the G‐INS‐I algorithm and BLOSUM62 scoring matrix. The five resulting alignments were then concatenated into one alignment using Geneious 11.1.5 (https://www.geneious.com). Regions of low conservation within the resulting alignment were removed using gBlocks 0.91 b (Castresana, 2000; Talavera & Castresana, 2007). A phylogenetic tree was then estimated with maximum likelihood analyses using RAxML 8.2.11 (Stamatakis, 2014) under the LG + Gamma scoring model of amino acid substitution. Bootstrap values were calculated from 500 replicates.
3.15. Expression and purification of McdB homologs
All McdB proteins were expressed with an N‐terminal His‐SUMO tag off a pET11b plasmid. The expression plasmids were transformed into competent BL21‐AI cells. Expression cultures were grown in 2 L of LB + carbenicillin (100 µg/ml) at 37°C to an OD600 of 0.6. Cultures were then induced with final concentrations of IPTG at 1 mM and L‐arabinose at 0.2% (w/v) and allowed to grow for 4 hr at 37°C. Cultures were pelleted and resuspended in 80 ml of lysis buffer (50 mM Tris‐HCl pH 8.5, 300 mM KCl, 5 mM 2‐Mercaptoethanol (BME), 0.05 mg/ml of lysozyme, 0.05 µl/ml of Benzonase, protease inhibitor) on ice. Cells were lysed using a tip sonicator at 50% power with 10s on/20s off cycles for 6 min at 4°C. Lysates were spun down at 35,000g at 4°C, and supernatants loaded onto 5 ml HP HIS‐TRAP columns (GE Healthcare Life Sciences) equilibrated in buffer A (50 mM Tris‐HCl pH 8.5, 300 mM KCl, 5 mM BME, 20 mM imidazole). Proteins were eluted using a 5%–100% gradient of buffer B (50 mM Tris‐HCl pH 8.5, 300 mM KCl, 5 mM BME, 500 mM imidazole) via an AKTA pure system (GE Healthcare Life Sciences). Peak fractions were pooled and diluted in buffer A to a final imidazole concentration < 100 mM. Purified His‐Ulp1 (300 µl) was added and reactions were incubated at 30°C to cleave the His‐SUMO tag. Reactions were then passed over 5 ml HP HIS‐TRAP columns to remove both the His‐SUMO tag and His‐Ulp1 enzyme. Flow‐through was concentrated and passed over an SEC column (HiLoad 16/600 Superdex 200 pg; GE Healthcare Life Sciences) equilibrated in buffer C (50 mM Tris‐HCl pH 8.5, 150 mM KCl, 5 mM BME, 10% glycerol) using an AKTA pure system. Peak fractions were pooled, flash‐frozen with liquid N2, and stored at −80°C.
3.16. Microscopy of McdB phase separation in vitro
Images were taken of 450 µM McdB in a buffer consisting of 20 mM HEPES (pH 7.0) and 100 mM KCl, with or without the addition of either 15% (w/v) PEG‐8000 or Ficoll‐400 as indicated. All samples were left to incubate for 2 hr prior to imaging at room temperature. Imaging was performed using 16 well CultureWells (Grace BioLabs). Wells were passivated by overnight incubation in 5% (w/v) Pluronic acid (Thermo‐Fischer), and washed thoroughly with the corresponding buffer prior to use. Imaging of McdB droplet formation was performed using a Nikon Ti2‐E motorized inverted microscope (60 × DIC objective and DIC analyzer cube) with a Transmitted LED Lamp house and a Photometrics Prime 95B Back‐illuminated sCMOS Camera. Image and video analyses were performed using Fiji v 1.0.
3.17. Size‐exclusion chromatography with multi‐angle light scattering (SEC–MALS)
For each McdB homolog analyzed, 500 µl of the sample at 1.5 mg/ml was passed over an SEC column (Superdex 200 Increase 10/300 GL; GE Healthcare Life Sciences) at a flow rate of 0.15 ml/min in buffer (50 mM Tris‐HCl pH 8.5, 150 mM KCl, 5 mM BME) at 4°C. Following SEC, the samples were analyzed using an A280 UV detector (AKTA pure; GE Healthcare Life Sciences), the DAWN HELEOS‐II MALS detector (Wyatt Technology), and the Optilab rEX refractive index detector (Wyatt Technology). The data were analyzed to calculate mass using ASTRA software (Wyatt Technology). Bovine serum albumin was used as the standard for calibration.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Supporting information
Table S1
Video S1
Video S2
Video S3
Video S4
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Dr. JK Nandakumar for assistance in the SEC‐MALS experiments. This work was supported by the National Science Foundation to A.G.V. (Award No. 1817478 and CAREER Award No. 1941966), NSF GRFP Award to L.T. (DGE 1841052), Rackham Graduate Student Research Grant to L.T., and from research initiation funds provided by the MCDB Department to A.G.V., University of Michigan and the Michigan Life Sciences Fellows Program to J.S.M. We also thank Jeffrey Harrison from the UM Microscopy Core for assistance with TEM preparation and imaging.
MacCready JS, Tran L, Basalla JL, Hakim P, Vecchiarelli AG. The McdAB system positions α‐carboxysomes in proteobacteria. Mol Microbiol. 2021;116:277–297. 10.1111/mmi.14708
Joshua S. MacCready and Lisa Tran are co‐first authorship.
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article.
REFERENCES
- Abeles, A.L., Friedman, S.A. & Austin, S.J. (1985) Partition of unit‐copy miniplasmids to daughter cells. III. The DNA sequence and functional organization of the P1 partition region. Journal of Molecular Biology, 185, 261–272. Available from: 10.1016/0022-2836(85)90402-4 [DOI] [PubMed] [Google Scholar]
- Alberti, S., Gladfelter, A. & Mittag, T. (2019) Considerations and challenges in studying liquid‐liquid phase separation and biomolecular condensates. Cell, 176(3), 419–434. Available from: 10.1016/j.cell.2018.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado, A., Kjæ, A., Yang, W., Mann, P., Briegel, A., Waldor, M.K. et al. (2017) Coupling chemosensory array formation and localization. eLife, 6. Available from: https://elifesciences.org/articles/31058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axen, S.D., Erbilgin, O. & Kerfeld, C.A. (2014) A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Computational Biology, 10, e1003898. Available from: 10.1371/journal.pcbi.1003898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger, M.R., Andrews, T.J., Whitney, S.M., Ludwig, M., Yellowlees, D.C., Leggat, W. et al. (1998) The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast‐based CO2‐concentrating mechanisms in algae. Canadian Journal of Botany, 76, 1052–1071. Available from: 10.1139/b98-074 [DOI] [Google Scholar]
- Badger, M.R. & Bek, E.J. (2008) Multiple Rubisco forms in proteobacteria: Their functional significance in relation to CO2 acquisition by the CBB cycle. Journal of Experimental Botany, 59, 1525–1541. Available from: 10.1093/jxb/erm297 [DOI] [PubMed] [Google Scholar]
- Badger, M.R., Hanson, D. & Price, G.D. (2002) Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Functional Plant Biology, 29, 161–173. Available from: 10.1071/PP01213 [DOI] [PubMed] [Google Scholar]
- Badger, M.R. & Price, G.D. (2003) CO2 concentrating mechanisms in cyanobacteria: Molecular components, their diversity and evolution. Journal of Experimental Botany, 54, 609–622. Available from: 10.1093/jxb/erg076 [DOI] [PubMed] [Google Scholar]
- Badrinarayanan, A., Le, T.B.K. & Laub, M.T. (2015) Bacterial chromosome organization and segregation. Annual Review of Cell and Developmental Biology, 31, 171–199. Available from: 10.1146/annurev-cellbio-100814-125211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart, M., Huber, I., Abdollahzadeh, I., Gensch, T. & Frunzke, J. (2017) Heterologous expression of the Halothiobacillus neapolitanus carboxysomal gene cluster in Corynebacterium glutamicum. Journal of Biotechnology, 258, 126–135. Available from: https://www.sciencedirect.com/science/article/pii/S0168165617301244?via%3Dihub. 10.1016/j.jbiotec.2017.03.019 [DOI] [PubMed] [Google Scholar]
- Baxter, J.C. & Funnell, B.E. (2014) Plasmid partition mechanisms. Microbiology Spectrum, 2. Available from: http://www.asmscience.org/content/journal/microbiolspec/10.1128/microbiolspec.PLAS‐0023‐2014 [DOI] [PubMed] [Google Scholar]
- Bonacci, W., Teng, P.K., Afonso, B., Niederholtmeyer, H., Grob, P., Silver, P.A. et al. (2012) Modularity of a carbon‐fixing protein organelle. Proceedings of the National Academy of Sciences of the United States of America, 109, 478–483. Available from: 10.1073/pnas.1108557109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, F., Sutter, M., Bernstein, S.L., Kinney, J.N. & Kerfeld, C.A. (2015) Engineering bacterial microcompartment shells: Chimeric shell proteins and chimeric carboxysome shells. ACS Synthetic Biology, 4, 444–453. Available from: 10.1021/sb500226j [DOI] [PubMed] [Google Scholar]
- Cameron, J.C., Wilson, S.C., Bernstein, S.L. & Kerfeld, C.A. (2013) Biogenesis of a bacterial organelle: The carboxysome assembly pathway. Cell, 155, 1131–1140 Available from: http://www.sciencedirect.com/science/article/pii/S0092867413013627. 10.1016/j.cell.2013.10.044 [DOI] [PubMed] [Google Scholar]
- Cannon, G.C., Bradburne, C.E., Aldrich, H.C., Baker, S.H., Heinhorst, S. & Shively, J.M. (2001) Microcompartments in prokaryotes: Carboxysomes and related polyhedra. Applied and Environment Microbiology, 67, 5351–5361 Available from: http://www.ncbi.nlm.nih.gov/pubmed/11722879. 10.1128/AEM.67.12.5351-5361.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon, G.C. & Shively, J.M. (1983) Characterization of a homogenous preparation of carboxysomes from Thiobacillus neapolitanus . Archives of Microbiology, 134, 52–59. Available from: http://link.springer.com/10.1007/BF00429407. 10.1007/BF00429407 [DOI] [Google Scholar]
- Castresana, J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution, 17, 540–552. Available from: 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Chen, A.H., Afonso, B., Silver, P.A. & Savage, D.F. (2012) Spatial and temporal organization of chromosome duplication and segregation in the cyanobacterium Synechococcus elongatus PCC 7942. PLoS One, 7, e47837. Available from: 10.1371/journal.pone.0047837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., MacCready, J.S., Ducat, D.C. & Osteryoung, K.W. (2018) The molecular machinery of chloroplast division. Plant Physiology, 176, 138–151. Available from: http://www.plantphysiol.org/content/plantphysiol/176/1/138.full.pdf. 10.1104/pp.17.01272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, Y. & Gurevitz, M. (2006) The cyanobacteria—Ecology, physiology and molecular genetics BT. In: Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.‐H., and Stackebrandt, E. (Eds.) The prokaryotes: Volume 4: Bacteria: firmicutes, cyanobacteria. : Springer US, pp. 1074–1098. Available from: 10.1007/0-387-30744-3_39 [DOI] [Google Scholar]
- Deeb, S.S. (1973) Involvement of a tryptophan residue in the assembly of bacteriophages 80 and lambda. Journal of Virology, 11, 353–358. Available from: https://pubmed.ncbi.nlm.nih.gov/4570923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmarais, J.J., Flamholz, A.I., Blikstad, C., Dugan, E.J., Laughlin, T.G., Oltrogge, L.M. et al. (2019) DABs are inorganic carbon pumps found throughout prokaryotic phyla. Nature Microbiology, 4, 2204–2215. Available from: 10.1038/s41564-019-0520-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou, Z., Heinhorst, S., Williams, E.B., Murin, C.D., Shively, J.M. & Cannon, G.C. (2008) CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. Journal of Biological Chemistry, 283, 10377–10384. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18258595. [DOI] [PubMed] [Google Scholar]
- Erdmann, N., Petroff, T. & Funnell, B.E. (1999) Intracellular Localization of P1 ParB protein depends on ParA and parS. Proceedings of the National Academy of Sciences of the United States of America, 96, 14905–14910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt, E. & Conti, S.F. (1969) Ultrastructure of blue‐green algae. Journal of Bacteriology, 97, 1486–1493. Available from: http://jb.asm.org/content/97/3/1486.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M.R., Appel, R.D. et al. (2005) Protein identification and analysis tools on the ExPASy Server BT. In: Walker, J.M. (Ed.) The proteomics protocols handbook. Humana Press, pp. 571–607. Available from: 10.1385/1-59259-890-0:571 [DOI] [Google Scholar]
- Goedhart, J., von Stetten, D., Noirclerc‐Savoye, M., Lelimousin, M., Joosen, L., Hink, M.A. et al. (2012) Structure‐guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nature Communications, 3, 751. Available from: 10.1038/ncomms1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, N.C., Tay, J.W., Altus, S., Bortz, D.M. & Cameron, J.C. (2020) Life cycle of a cyanobacterial carboxysome. Science Advances, 6, eaba1269. Available from: http://advances.sciencemag.org/content/6/19/eaba1269.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuijzen, Y.A., Kuenen, J.G. & Konings, W.N. (1987) Activity of ribulose‐1,5‐bisphosphate carboxylase in intact and disrupted carboxysomes of Thiobacillus neapolitanus . FEMS Microbiology Letters, 42, 121–124. Available from: http://www.sciencedirect.com/science/article/pii/0378109787904587 [Google Scholar]
- Hutchinson, M., Johnstone, K.I. & White, D. (1965) The taxonomy of certain thiobacilli. Journal of General Microbiology, 41, 357–366. https://www.microbiologyresearch.org/content/journal/micro/10.1099/00221287‐41‐3‐357 [DOI] [PubMed] [Google Scholar]
- Johnson, K.A., Pokhrel, R., Budicini, M.R., Gerstman, B.S., Chapagain, P.P. & Stahelin, R.V. (2020) A conserved tryptophan in the Ebola virus matrix protein C‐terminal domain is required for efficient virus‐like particle formation. Pathogens, 9, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, A., Schwarz, R., Lieman‐Hurwitz, J., & Reinhold, L. (1991) Physiological and molecular aspects of the inorganic carbon‐concentrating mechanism in cyanobacteria. Plant Physiology, 97, 851–855. Available from: http://www.plantphysiol.org/content/97/3/851.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova, G., Pidoux, J., Ullmann, A. & Ladant, D. (1998) A bacterial two‐hybrid system based on a reconstituted signal transduction pathway. Proceedings of the National Academy of Sciences of the United States of America, 95(10), 5752–5756. Available from: 10.1073/pnas.95.10.5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment Software Version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780. Available from: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld, C.A., Aussignargues, C., Zarzycki, J., Cai, F. & Sutter, M. (2018) Bacterial microcompartments. Nature Reviews Microbiology, 16, 277–290 Available from: http://www.nature.com/doifinder/10.1038/nrmicro.2018.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld, C.A. & Melnicki, M.R. (2016) Assembly, function and evolution of cyanobacterial carboxysomes. Current Opinion in Plant Biology, 31, 66–75 Available from: https://www.sciencedirect.com/science/article/pii/S136952661630036X?via%3Dihub [DOI] [PubMed] [Google Scholar]
- Komla‐Soukha, I. & Sureau, C. (2006) A tryptophan‐rich motif in the carboxyl terminus of the small envelope protein of hepatitis B virus is central to the assembly of hepatitis delta virus particles. Journal of Virology, 80, 4648–4655. Available from: http://jvi.asm.org/content/80/10/4648.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka, M.C., Sochacki, K.A., Bratton, B.P., Shkel, I.A., Record, M.T. & Weisshaar, J.C. (2009) Cytoplasmic protein mobility in osmotically stressed Escherichia coli . Journal of Bacteriology, 191, 231–237. Available from: http://jb.asm.org/content/191/1/231.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin, E.V. (1993) A superfamily of ATPases with diverse functions containing either classical or deviant ATP‐binding Motif. Journal of Molecular Biology, 229, 1165–1174. Available from: http://www.sciencedirect.com/science/article/B6WK7‐45PV5P8‐8Y/2/3d3e0e9a3a714c549a715f2c4e22502e [DOI] [PubMed] [Google Scholar]
- Kyte, J. & Doolittle, R.F. (1982) A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology, 157, 105–132. Available from: http://www.sciencedirect.com/science/article/pii/0022283682905150 [DOI] [PubMed] [Google Scholar]
- Li, X., Romero, P., Rani, M., Dunker, A.K. & Obradovic, Z. (1999) Predicting protein disorder for N‐, C‐, and internal regions. Genome Inform Ser Workshop Genome Inform, 10, 30–40. Available from: http://europepmc.org/abstract/MED/11072340 [PubMed] [Google Scholar]
- Liao, Y., Ithurbide, S., Löwe, J., & Duggin, I.G. (2020) Two FtsZ proteins orchestrate archaeal cell division through distinct functions in ring assembly and constriction. bioRxiv 2020.06.04.133736. Available from: http://biorxiv.org/content/early/2020/06/05/2020.06.04.133736.abstract
- Lin, M.T., Occhialini, A., Andralojc, P.J., Devonshire, J., Hines, K.M., Parry, M.A.J. et al. (2014) β‐Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. The Plant Journal, 79, 1–12. Available from: 10.1111/tpj.12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, B.M., Hee, W.Y., Sharwood, R.E., Rae, B.D., Kaines, S., Lim, Y.‐L. et al. (2018) Carboxysome encapsulation of the CO2‐fixing enzyme Rubisco in tobacco chloroplasts. Nature Communications, 9, 3570. Available from: http://www.nature.com/articles/s41467‐018‐06044‐0. 10.1038/s41467-018-06044-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwiczak, J., Winski, A., Szczepaniak, K., Alva, V. & Dunin‐Horkawicz, S. (2019) DeepCoil—A fast and accurate prediction of coiled‐coil domains in protein sequences. Bioinformatics, 35, 2790–2795. Available from: 10.1093/bioinformatics/bty1062 [DOI] [PubMed] [Google Scholar]
- MacCready, J.S., Basalla, J.L. & Vecchiarelli, A.G. (2020) Origin and evolution of carboxysome positioning systems in cyanobacteria. Molecular Biology and Evolution, 37, 1434–1451. Available from: https://academic.oup.com/mbe/advance‐article/doi/10.1093/molbev/msz308/5695716. 10.1093/molbev/msz308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccready, J.S., Hakim, P., Young, E.J., Hu, L., Liu, J., Osteryoung, K.W. et al. (2018) Protein gradients on the nucleoid position the carbon‐fixing organelles of cyanobacteria. eLife, 7. Available from: https://elifesciences.org/articles/39723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan, M.T. & Jung, D.O. (2009) An overview of purple bacteria: Systematics, physiology, and habitats. In: Hunter, C.N., Daldal, F., Thurnauer, M.C. and Beatty, J.T. (Eds.) The purple phototrophic bacteria. Springer, pp. 1–15. Available from: 10.1007/978-1-4020-8815-5_1 [DOI] [Google Scholar]
- Marin, B., Nowack, E.C.M., Glöckner, G. & Melkonian, M. (2007) The ancestor of the Paulinella chromatophore obtained a carboxysomal operon by horizontal gene transfer from a Nitrococcus‐like γ‐proteobacterium. BMC Evolutionary Biology, 7, 85. Available from: 10.1186/1471-2148-7-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marintcheva, B., Hamdan, S.M., Lee, S.‐J. & Richardson, C.C. (2006) Essential residues in the C terminus of the bacteriophage T7 gene 2.5 single‐stranded DNA‐binding protein. Journal of Biological Chemistry, 281, 25831–25840. Available from: http://www.jbc.org/content/281/35/25831.abstract. 10.1074/jbc.M604601200. [DOI] [PubMed] [Google Scholar]
- Marston, A.L. & Errington, J. (1999) Dynamic movement of the ParA‐like Soj protein of B. subtilis and its dual role in nucleoid organization and developmental regulation. Molecular Cell, 4, 673–682. [DOI] [PubMed] [Google Scholar]
- Mohl, D.A. & Gober, J.W. (1997) Cell cycle dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus . Cell, 88, 675–684. Available from: 10.1016/S0092-8674(00)81910-8 [DOI] [PubMed] [Google Scholar]
- Mori, H., Kondo, A., Ohshima, A., Ogura, T. & Hiraga, S. (1986) Structure and function of the F plasmid genes essential for partitioning. Journal of Molecular Biology, 192, 1–15. [DOI] [PubMed] [Google Scholar]
- Nevin, K.P., Woodard, T.L., Franks, A.E., Summers, Z.M. & Lovley, D.R. (2010) Microbial electrosynthesis: Feeding microbes electricity to convert carbon dioxide and water to multicarbon extracellular organic compounds. mBio, 1, e00103‐10. Available from: http://mbio.asm.org/content/1/2/e00103‐10.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi, R., Nakamachi, A., Hatakeyama, T.S., Watanabe, S., Kanesaki, Y., Chibazakura, T. et al. (2019) Coordination of polyploid chromosome replication with cell size and growth in a cyanobacterium. mBio, 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltrogge, L.M., Chaijarasphong, T., Chen, A.W., Bolin, E.R., Marqusee, S. & Savage, D.F. (2020) Multivalent interactions between CsoS2 and Rubisco mediate α‐carboxysome formation. Nature Structural & Molecular Biology, 27, 281–287. Available from: 10.1038/s41594-020-0387-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchenault, C., Pons, M.C., Schiavon, C., Siguier, P., Rech, J., Guynet, C. et al. (2020) Intracellular positioning systems limit the entropic eviction of secondary replicons toward the nucleoid edges in bacterial cells. Journal of Molecular Biology, 432, 745–761. Available from: http://www.sciencedirect.com/science/article/pii/S002228362030036X [DOI] [PubMed] [Google Scholar]
- Price, G.D. & Badger, M.R. (1989) Isolation and characterization of high CO2‐requiring‐mutants of the cyanobacterium Synechococcus PCC7942. Plant Physiology, 91, 514–525. Available from: http://www.plantphysiol.org/content/91/2/514.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae, B.D., Long, B.M., Badger, M.R. & Price, G.D. (2013) Functions, compositions, and evolution of the two types of carboxysomes: Polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiology and Molecular Biology Reviews, 77, 357–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rillema, R., MacCready, J.S., and Vecchiarelli, A.G. (2020) Cyanobacterial growth and morphology are influenced by carboxysome positioning and temperature. bioRxiv 2020.06.01.127845. Available from: http://biorxiv.org/content/early/2020/06/01/2020.06.01.127845.abstract
- Ringgaard, S., Schirner, K., Davis, B.M. & Waldor, M.K. (2011) A family of ParA‐like ATPases promotes cell pole maturation by facilitating polar localization of chemotaxis proteins. Genes & Development, 25, 1544–1555. Available from: http://genesdev.cshlp.org/content/25/14/1544.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringgaard, S., van Zon, J., Howard, M. & Gerdes, K. (2009) Movement and equipositioning of plasmids by ParA filament disassembly. Proceedings of the National Academy of Sciences of the United States of America, 106, 19369–19374. Available from: http://www.pnas.org/content/106/46/19369.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, M.A.J., Wadhams, G.H., Hadfield, K.A., Tickner, S. & Armitage, J.P. (2012) ParA‐like protein uses nonspecific chromosomal DNA binding to partition protein complexes. Proceedings of the National Academy of Sciences of the United States of America, Available from: http://www.pnas.org/content/early/2012/04/09/1114000109.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohnke, B.A., Rodríguez Pérez, K.J. & Montgomery, B.L. (2020) Linking the dynamic response of the carbon dioxide‐concentrating mechanism to carbon assimilation behavior in Fremyella diplosiphon. mBio, 11, e01052‐20. Available from: http://mbio.asm.org/content/11/3/e01052‐20.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohnke, B.A., Singh, S.P., Pattanaik, B. & Montgomery, B.L. (2018) RcaE‐dependent regulation of carboxysome structural proteins has a central role in environmental determination of carboxysome morphology and abundance in Fremyella diplosiphon. mSphere, 3, e00617‐17. Available from: http://msphere.asm.org/content/3/1/e00617‐17.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, P., Obradovic, Z. & Dunker, A.K. (1997) Sequence data analysis for long disordered regions prediction in the Calcineurin family. Genome Informatics, 8, 110–124. [PubMed] [Google Scholar]
- Romero, P., Obradovic, Z., Li, X., Garner, E.C., Brown, C.J. & Dunker, A.K. (2001) Sequence complexity of disordered protein. Proteins: Structure, Function, and Bioinformatics, 42, 38–48. Available from: [DOI] [PubMed] [Google Scholar]
- Savage, D.F., Afonso, B., Chen, A.H. & Silver, P.A. (2010) Spatially ordered dynamics of the bacterial carbon fixation machinery. Science (80‐), 327, 1258–1261. Available from: http://www.sciencemag.org/cgi/content/abstract/327/5970/1258 [DOI] [PubMed] [Google Scholar]
- Schmid, M.F., Paredes, A.M., Khant, H.A., Soyer, F., Aldrich, H.C., Chiu, W. et al. (2006) Structure of Halothiobacillus neapolitanus carboxysomes by cryo‐electron tomography. Journal of Molecular Biology, 364, 526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, M.A. (2007) Structural biology of plasmid segregation proteins. Current Opinion in Structural Biology, 17, 103–109. Available from: http://www.sciencedirect.com/science/article/B6VS6‐4MJBTHM‐1/2/378b298b8361fd8789e4f21c4f951e22 [DOI] [PubMed] [Google Scholar]
- Schumacher, M.A., Henderson, M. & Zhang, H. (2019) Structures of maintenance of carboxysome distribution Walker‐box McdA and McdB adaptor homologs. Nucleic Acids Research, 47, 5950–5962. Available from: https://academic.oup.com/nar/article/47/11/5950/5491742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner, N.C., Lambert, G.G., Chammas, A., Ni, Y., Cranfill, P.J., Baird, M.A. et al. (2013) A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum . Nature Methods, 10, 407–409. Available from: 10.1038/nmeth.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively, J.M., Ball, F.L. & Kline, B.W. (1973) Electron microscopy of the carboxysomes (polyhedral bodies) of Thiobacillus neapolitanus . Journal of Bacteriology, 116, 1405–1411. Available from: http://www.ncbi.nlm.nih.gov/pubmed/4127632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively, J.M., Decker, G.L. & Greenawalt, J.W. (1970) Comparative ultrastructure of the thiobacilli. Journal of Bacteriology, 101, 618–627. Available from: http://jb.asm.org/content/101/2/618.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoging, U. & Liljeström, P. (1998) Role of the C‐terminal tryptophan residue for the structure‐function of the alphavirus capsid protein. Journal of Molecular Biology, 279, 865–872. Available from: http://www.sciencedirect.com/science/article/pii/S0022283698918174 [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2014) RAxML version 8: A tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics, 30, 1312–1313. Available from: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight, P.D., Fischbach, M.A., Walsh, C.T., Rudner, D.Z. & Kolter, R. (2007) A singular enzymatic megacomplex from Bacillus subtilis . Proceedings of the National Academy of Sciences of the United States of America, 104, 305–310. Available from: http://www.pnas.org/content/104/1/305.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Casella, S., Fang, Y., Huang, F., Faulkner, M., Barrett, S. et al. (2016) Light modulates the biosynthesis and organization of cyanobacterial carbon fixation machinery through photosynthetic electron flow. Plant Physiology, 171, 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Wollman, A.J.M., Huang, F., Leake, M.C. & Liu, L.N. (2019) Single‐organelle quantification reveals stoichiometric and structural variability of carboxysomes dependent on the environment. Plant Cell, 31, 1648–1664. Available from: www.plantcell.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera, G. & Castresana, J. (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology, 56, 564–577. Available from: 10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- Tcherkez, G.G.B., Farquhar, G.D., & Andrews, T.J. (2006) Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proceedings of the National Academy of Sciences of the United States of America, 103, 7246–7251. Available from: http://www.pnas.org/content/103/19/7246.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush, A.D., MacCready, J.S., Chen, C., Ducat, D.C., & Osteryoung, K.W. (2018) Conserved dynamics of chloroplast cytoskeletal FtsZ proteins across photosynthetic lineages. Plant Physiology, 176, 295–306. Available from: http://www.plantphysiol.org/content/176/1/295.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush, A.D. & Osteryoung, K.W. (2012) Distinct functions of chloroplast FtsZ1 and FtsZ2 in Z‐ring structure and remodeling. Journal of Cell Biology, 199, 623–637. Available from: 10.1083/jcb.201205114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler, M. & Shapiro, L. (2006) MipZ, a spatial regulator coordinating chromosome segregation with cell division in caulobacter. Cell, 126, 147–162. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867406007665 [DOI] [PubMed] [Google Scholar]
- Tsuboi, M., Overman, S.A., Nakamura, K., Rodriguez‐Casado, A. & Thomas, G.J. (2003) Orientation and interactions of an essential tryptophan (Trp‐38) in the capsid subunit of Pf3 filamentous virus. Biophysical Journal, 84, 1969–1976. Available from: http://www.sciencedirect.com/science/article/pii/S000634950375005X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Bogaart, G., Den, H.N., Krasnikov, V. & Poolman, B. (2007) Protein mobility and diffusive barriers in Escherichia coli: Consequences of osmotic stress. Molecular Microbiology, 64, 858. Available from: 10.1111/j.1365-2958.2007.05705.x [DOI] [PubMed] [Google Scholar]
- Winkler, J., Seybert, A., Konig, L., Pruggnaller, S., Haselmann, U., Sourjik, V. et al. (2010) Quantitative and spatio‐temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO Journal, 29, 910. Available from: 10.1038/emboj.2009.412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk, C.P. (1973) Physiology and cytological chemistry blue‐green algae. Bacteriology Reviews, 37, 32–101. Available from: https://pubmed.ncbi.nlm.nih.gov/4633592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerulla, K., Ludt, K. & Soppa, J. (2016) The ploidy level of synechocystis sp. PCC 6803 is highly variable and is influenced by growth phase and by chemical and physical external parameters. Microbiology (United Kingdom), 162, 730–739. Available from: https://www.microbiologyresearch.org/content/journal/micro/10.1099/mic.0.000264 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Video S1
Video S2
Video S3
Video S4
Supplementary Material
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.


