Abstract
Salmonellosis is one of the most important bacterial diseases in pigeons. This study aimed to estimate the prevalence of Salmonella spp. in domestic pigeons (Columba livia f. domestica) in Poland, its antimicrobial susceptibility (both phenotypic and genotypic), and its capability for biofilm formation. The presence of selected virulence genes, nucleotide homology of selected genes, and susceptibility to bacteriophages were investigated as well. From the 585 pigeons tested, 5.47% turned out positive. All isolated strains were recognized as Salmonella enterica ser. Typhimurium. The asymptomatic pigeons were carriers of 37.5% of the isolates. The dominant variants were as follows: 1,4,[5],12,:i:1,2 (53.13%) and 1,4,[5],12,:‐:‐ (31.25%). Most of the strains analysed showed the ability to produce biofilm after 24 and 48 hr of incubation (59.38% and 53.13%, respectively). Over 90% of the strains were confirmed for lpfA, agafA, invA, sivH, and avrA virulence genes. Also, of the thirteen antimicrobial susceptibility genes, the following were confirmed: sul1, tet(A), blaTEM‐1 , floR, strA, and strB. The most common were the strB (18%) and tet(A) (12%) genes that are responsible for coding resistance to aminoglycosides and tetracyclines, respectively. Most of the strains were phenotypically resistant to oxytetracycline (46.88%), neomycin (53.13%) and tylosin (100%). The susceptibility of the investigated Salmonella strains to the bacteriophages was between 33% and 100%. MLST, PCR MP and ERIC PCR analyses indicated a very high genetic similarity of the investigated strains (over 99%). Results of our study indicate that Salmonella enterica ser. Typhimurium is still an important agent in domestic pigeons and that its antimicrobial resistance increases. Alarming is also the confirmation of a single‐phase variant 1,4,[5],12:i,‐, which could have increased virulence and multi‐drug resistance encoded on the plasmid. Most importantly, however, such strains have been isolated from humans with clinical symptoms of Salmonella infection.
Keywords: antimicrobial resistance, gene homology, genotyping, pigeons, Salmonella, virulence factors
1. INTRODUCTION
Salmonella is one of the members of the Enterobacteriaceae family and is an aetiologic agent of food‐borne poisoning—salmonellosis (Jahantigh & Nili, 2010). According to the World Health Organization (2017), salmonellosis is the prevailing zoonosis in Europe right after campylobacteriosis. In 2018, 91,857 cases of salmonellosis were detected in the European population, whereas in the United States, Salmonella spp. causes about 1.2 million infections every year (EFSA & ECDC, 2018, CDC).
In Poland, this bacterium still ranks first, and the number of cases of infections increases every year (Polański et al., 2019). Interestingly, domestic and urban pigeons are considered as one of the reservoirs of zoonotic agents, because of close contact with humans, domestic pigeons through the food chain or handling, whereas urban ones by contact with avian faeces (de Oliveira et al., 2018; Lawson et., 2014).
Infections of pigeons (Columba livia) with Salmonella enterica are usually caused by pigeon‐adapted strains of serovar Typhimurium which poses no serious health hazard to humans (Pasmans et al., 2004). However, a recent study conducted by Haesendonck et al. (2016) confirmed the presence of Salmonella Enteritidis phage type 4 (PT4), which is an important human pathogen, in a feral population of domestic pigeons in Brussels. The prevalence of Salmonella Enteritidis in the examined pigeon population was high (33%), which suggests that feral pigeons may pose a potential hazard to human health. Infections of pigeons caused by Salmonella spp. can lead to severe diseases in this bird species and are mainly induced by belonging to the serogroup B strains of Salmonella Typhimurium var. Copenhagen of the following phage types: DT2, PT4, PT46 and PT99 (Kriz et al., 2011; Pasmans et al., 2008; Teske et al., 2013). The clinical signs of pigeon salmonellosis may vary depending on the age, general condition, and immune status of individual birds. The greenish diarrhoea, loss of appetite, and ultimately death, affecting birds of a wide range of ages, are the most typical signs of this disease. Individual birds in the infected flocks can also show the following clinical symptoms: weight loss, arthritis, panophthalmitis, torticollis and other neurological signs. Embryo deaths and a high death rate of nestlings in the first 7 days of life can also be observed. The most common postmortem lesions are as follows: hepatomegalia, splenomegalia, haemorrhages in the liver, enteritis, and also abscesses that could be present in almost all internal organs. The capability of Salmonella for long‐term survival in the host macrophages causes the risk of chronical carriage. The subclinically infected pigeons become carriers of these bacteria and can shed it occasionally with faeces, which is the main route of infection in pigeon flocks (Pasmans et al., 2008).
Most of the epidemiological data concerning infections of pigeons with Salmonella spp. are based on investigations of feral pigeons (Dovc et al., 2004; Gargiulo et al., 2014; Haesebrouck et al., 2016; de Oliveira et al., 2018; de Sousa et al., 2010; Torres‐Mejía et al., 2018) and only a few derive from studies conducted with ornamental and racing pigeons (Stenzel et al., 2014; Teske et al., 2013). Pigeon breeding is still a popular and time‐consuming hobby (which leads to the long exposure of humans to pigeon pathogens), and given that these birds could be carriers of Salmonella, the human health hazard is real (Israili & Iqbal, 2017).
In the light of the above, this study aimed to estimate the prevalence of Salmonella spp. in domestic pigeons and to evaluate its antimicrobial susceptibility and virulence features (carriage of virulence genes and biofilm formation capacity), which could imply a worse outcome in the event of infection. Also, the study of nucleotide homology on selected genes and phage‐susceptibility testing were performed to analyse the genetic variability of the strains isolated.
2. MATERIALS AND METHODS
2.1. Ethical statement
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required as this study was carried out on swab samples collected from birds which were patients of the Department of Poultry Diseases, Faculty of Veterinary Medicine, University of Warmia and Mazury in Olsztyn (Poland) during standard diagnostic procedures and no other manipulations of live animals were done. Tissue samples were collected from naturally dead pigeons delivered for necropsy.
2.2. Samples
The field Salmonella strains used as an experimental material were collected in 2015 – 2017 (full years) and in the first two months of 2018 from domesticated pigeons (both carrier and ornamental birds) originating from different regions of Poland (Figure 1). The 585 pigeons used in this study were patients of the Department of Poultry Diseases, Faculty of Veterinary Medicine, University of Warmia and Mazury in Olsztyn, Poland. They were divided into 2 groups based on their health condition during veterinary examination. Pigeons not exhibiting clinical symptoms (415 birds) at the time of examination were marked with letter H (Healthy), whereas birds displaying one or more of the following symptoms: diarrhoea, arthritis, panophthalmitis, weight loss, torticollis, sudden death, and poor breeding results (embryo deaths, decreased fertility and laying), were marked with letter S (Sick, 170 birds). The sample material consisted of sterile swabs (Deltalab, Spain) collected from cloaca, crop and faeces of birds from group H and live birds from group S. Moreover, when deaths were recorded, samples of liver, kidneys, joints, ovary (when possible) and intestines were collected during the postmortem examination of dead pigeons. Information about the samples positive for Salmonella spp. is presented in Table S1.
Figure 1.
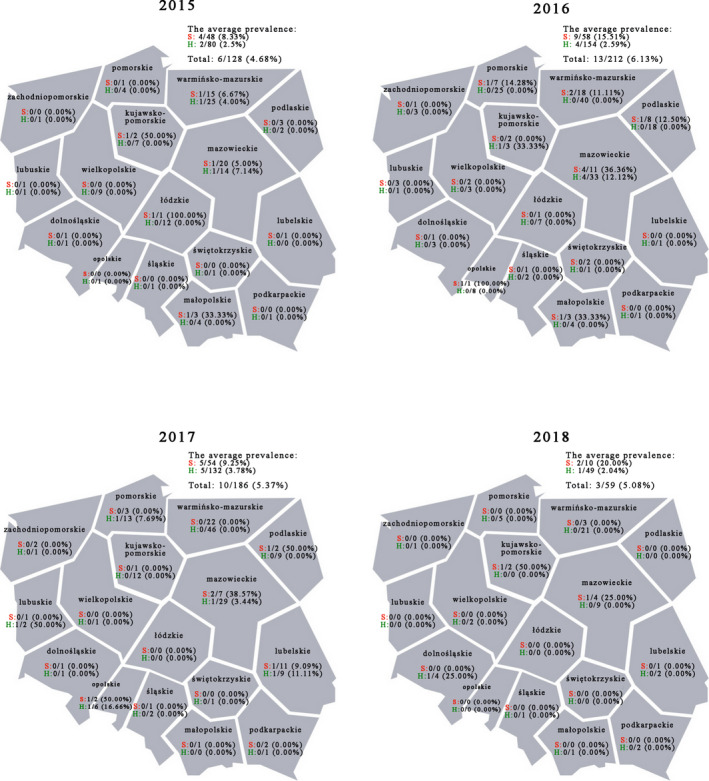
Prevalence of Salmonella enterica ser. Typhimurium isolated from pigeons with various health status in Poland. The samples were collected in the whole 2015–2017 years and in the first two months of the year 2018 from pigeons originating from various regions of Poland. Abbreviations: S—sick pigeons, H—pigeons with no clinical symptoms [Colour figure can be viewed at wileyonlinelibrary.com]
2.3. Microbiological screening for Salmonella
For pre‐incubation, the swabs were placed into non‐selective buffered peptone water (Merck) at 41.5°C for 24 hr. Afterwards, the samples were transferred into the Brilliance Salmonella medium (Oxoid). Plates were incubated at 41.5°C for 48h ± 2 hr under aerobic conditions. Afterwards, only purple colonies were considered for further examination. The final identification was performed using commercial latex agglutination tests: Salmonella test Kit (Oxoid) and API 20E (bioMérieux).
2.4. Serotyping Salmonella strains
The differentiation into serovars was performed with the plate agglutination method using an SSI® Salmonella ‘Big Five’ Serotyping Kit (bioMérieux). All analyses were performed according to the manufacturer's protocols. Additionally, to confirm Salmonella enterica ser. Typhimurium and Copenhagen variants, presence or absence of STM4495 and oafA genes were tested by PCR according to the method described by He et al. (2016) (primer sequences, product size, and annealing temperatures are summarized in Table S2).
2.5. Biofilm assay
The experiment was performed using polystyrene microtitre plates with flat bottoms based on the techniques described by Ebrahimi et al. (2013) with slight modification. All strains were incubated in TSB with the addition of 1% glucose. Results were read after 24 and 48 hr. Biofilm production was interpreted according to the criteria described by Stepanović et al. (2007). The mean optical density (OD) of the negative control + 3 standard deviations of negative control was considered the cut‐off (ODc = 0.15), and biofilm producers were therefore categorized as follows:
not a biofilm producer: OD ≤ ODc (all strains which OD values were below 0.15),
weak biofilm producer ODc: < OD ≤ 2 × ODc (all strains which OD values were above 0.15 and below 0.3),
moderate biofilm producer: 2 × ODc < OD ≤ 4 × ODc (all strains which OD values were above 0.3 and below 0.6),
strong biofilm producer: OD > 4 × ODc (all strains which OD values were above 0.6).
2.6. Antimicrobial susceptibility
The sensitivity of the isolated strains to antibiotics, that is, to 13 antimicrobials commonly used in pigeon treatment: amoxicillin 10 μg (AML), amoxicillin with clavulanic acid 30 μg (AMC), doxycycline 30 μg (DO), enrofloxacin 5 μg (ENR), florfenicol 30 μg (FFC), flumequine 30 μg (UB), lincomycin/spectinomycin 109μg (LS), marbofloxacin 5 μg (MAR), neomycin 30μg (N), norfloxacin 10μg (NOR), oxytetracycline 30 μg (OT), sulphamethoxazole/trimetoprim 19:1 (SXT), and tylosin 30μg (TY), was evaluated with the disc diffusion method using Oxoid discs (UK). The resistance to antibiotics was assessed according to the CLSI (Clinical and Laboratory Standards Institute, USA, 2013) guidelines using the quality control strain Escherichia coli ATCC 25,922. All strains were categorized as sensitive (S), intermediate (I) or resistant (R) to antimicrobials.
2.7. Bacterial DNA extraction for gene analysis
Bacterial DNA was extracted using an ExtractMe DNA bacterial kit (Blirt, Poland) according to the manufacturer's protocols. Eluted DNA concentrations and quality were measured using a BioSpectrometer® (Eppendorf). Afterwards, eluted DNA was stored at −20ºC for further analyses.
2.8. Amplification of genes related to antimicrobial resistance and virulence factors
All PCR reactions were carried out with a HotStarTaq Plus Master Mix Kit (Qiagen) according to the manufacturer's protocols in a nexus gradient thermocycler (Eppendorf). The following genes were selected for the antimicrobial resistance tests: sul1, sul2, sul3, tet(A), tet(B), tet(G), blaTEM‐1 , floR, cat1, cat2, aac6, strA, and strB. In case of virulence, the investigated genes were: lpf, sivH, invA, agaF, and avaR. The list of investigated genes, primer sequences, product size, and annealing temperature are summarized in Table S2. All primers were synthesized by Genomed S.A. (Poland). Ten microlitres of PCR products were electrophoresed on a 2% agarose gel in the presence of Midori Green Advance (Nippon Genetics, Germany), at 120 V for 60 min. The results were read using the Quantum ST5 Gel Documentation System (Vilber). To confirm the specificity of the amplicons obtained, some PCR products of interest were randomly selected and purified using a CleanUp kit (A&A Biotechnology) for sequencing (Genomed).
2.9. Gene homology of selected isolates
One isolate representative for each serotype and feature: ability for biofilm formation, was selected for the amplification of seven genes used for the multilocus sequence typing (MLST) according to the Achtman scheme (Achtman et al., 2012). The analysis included seven housekeeping genes (Table S3).
PCR was performed with a HotStarTaq Plus Master Mix Kit (Qiagen) and the composition of the reaction mixture was as follows: 10 µl of HotStarTaq Plus Master Mix, 0.1 µl of each of the primers (concentration 100 µM), 2 µl of Coralload 10× loading dye, and 2 µl of eluted DNA. The reaction volume was made up to 20 µl with ribonuclease‐free water. The primer sequences were described by Kidgell et al. (2002) and annealing temperature for amplification of internal fragment of analysed genes and for sequencing were 55°C and 50°C respectively.
Next, the PCR products were purified from the reaction mixture residues with the use of a Clean‐Up Concentrator kit (A&A Biotechnology) and sent for Sanger sequencing to Genomed (Poland). The resulting sequences were assembled in the SeqMan Pro application (DNASTAR) and checked manually for sequencing errors using the same software. The obtained sequences of all genes were aligned using the MUSCLE (Edgar, 2004) method and MegAlign Pro software (DNASTAR). All aligned sequences were trimmed at the same 5' and 3' ends and used for further analyses. The sequences of each gene amplified for selected isolates were concatenated into one sequence with the use of Megalign Pro Software (DNASTAR). The sequences prepared in this way were analysed for sequence type (ST) with a Salmonella typing tool available on the PubMLST website (https://pubmlst.org/bigsdb?db=pubmlst_salmonella_seqdef&page=batchSequenceQuery).
2.10. Accession numbers
Sequences of concatenated Salmonella enterica ser. Typhimurium genes obtained in this study were submitted to the GenBank database to generate accession numbers (MT856703‐MT856744).
2.11. PCR MP
The PCR MP (PCR melting profile) procedure was conducted according to a modified protocol by Zaczek et al. (2015). In this method, restriction enzymes are used for genomic DNA digestion, and the DNA restriction fragments obtained are ligated with an oligonucleotide adaptor followed by PCR amplification with a reduction of temperature during denaturation stage in each cycle.
In this study, genomic DNA (about 1 µg) was digested during incubation of the mixture with a Fast Digest BamHI enzyme (1.0 µl) (Thermo Fisher Scientific) and 2.5 µl of a reaction buffer in a total volume of 25 µl, at 37°C for 45 min. Inactivation of the enzyme was performed at 80°C for 15 min. Next, the restriction fragments of genomic DNA (25 µl) were ligated to the adaptor using 5U/µL of T4 ligase (ThermoFisher Scientific) and 2.8 µl of 10 x T4 DNA ligase buffer (ThermoFisher Scientific) in a total volume of 32.2 µl with incubation at 16°C for 60 min. The purification of product ligation was performed at 65°C for 10 min. The adaptor was prepared by mixing equimolar amounts of two oligonucleotides: pcr/mp‐oligo‐lig and oli‐pom‐BamHI. Oligonucleotides were used in a final concentration of 10 µM and incubated at 60°C for 2 min. A volume of 4 µl of ligation product was amplified by PCR (Mastercycler nexus gradient, Eppendorf) in a reaction mixture with 0.25 µl of primer MP BamHI, 2.5 µl of PCR reaction buffer Shark 10× (DNA Gdańsk), 17.75 µl of water, 1 µl of 2.5 mM dNTPs (nzytech, Portugal), 1 µl of MgCl2 (DNA Gdańsk), and 0.5 µl of Hypernova polymerase (Blirt, Poland) in a total volume of 25 µl. The PCR was performed as follows: 7 min at 72°C; an initial denaturation step for 90 s; 24 cycles of denaturation for 1 min followed by annealing at 72°C for 2 min and elongation at 72°C for 2 min 15s, and a final elongation step at 72°C for 5 min. The denaturation temperature was determined during the optimization experiments for five randomly chosen strains using a range of temperatures (83.5°C, 84.4°C, 85.7°C, 86.9°C, 88°C, 89°C). Primer sequences are summarized in Table S4.
PCR MP products (10 µl) were run on a 2% agarose (EURx, Poland) gel at 30V for 2.5 hr, and the amplification patterns were determined by examination on Simply Safe (EURx, Poland) – stained gels illuminated by UV light (Proxima AQ‐4.2 2,650, Isogen Life Science, The Netherlands). Amplicon sizes were determined by comparing the bands with a Gene Ruler 100‐bp Plus DNA mass ladder (ThermoFisher Scientific).
2.12. ERIC PCR
ERIC PCR (Enterobacterial Repetitive Intergenic Consensus PCR) was conducted according to the protocol published by Anjay et al. (2015). The method uses long primers that are complementary to the conservative fragments of the bacterial genome which are differently distributed depending on the strain. The PCR reaction was standardized with 2.5 µl of 10x Shark buffer (DNA Gdańsk), 2.5 µl of MgCl2 (25mM) (DNA Gdańsk), 3 µl of dNTP (2 mM) (nzytech, Portugal), 30 pmol of each primer ERIC1R and ERIC2, 1U of Hypernova DNA polymerase (Blirt), 3 µl of template DNA, and nuclease‐free water up to a final volume of 25 µl. The amplification cycles included initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 60 s, annealing at 40°C for 90 s and extension at 72°C for 60 s with a single cycle for final extension at 72°C for 7 min. Primer sequences are summarized in Table S4.
Ten microlitres of PCR products were electrophoresed on a 1.5% agarose gel in the presence of Midori Green Advance (Nippon Genetics), at 120 V for 120 min. The results were read using the Quantum ST5 Gel Documentation System (Vilber).
2.13. Phage sensitivity test
The phage sensitivity test was conducted according to a modified protocol of the spot test. A series of 6 consequent tenfold dilutions of BAFASAL® (Proteon Pharmaceuticals) preparation (an equivalent mixture of four bacteriophages against Salmonella spp.: 3Sent1, 8Sent65, 8Sent1748, and 5Sent1) was prepared. Bacterial lawns of each strain were made using the double agar overlay method, on which 20 μL droplets of the tested dilutions of the phage (10–2–10–6) were applied. The degree of lysis of the lawns was determined for each tested dilution after overnight incubation. The following spot evaluation system was used: ‘cl’ when droplets showed complete lysis, ‘+’ when uncomplete lysis or separate plaques were present, and ‘‐’ when no plaques were observed. Strains susceptibility was calculated based on the comparison of absolute values of decimal logarithm of the highest dilution of BAFASAL® where plaques/lysis were present on plates of the tested strains and BAFASAL® host strain (Salmonella enterica ser. Gallinarum). The calculated values were shown as a percentage of susceptibility.
3. RESULTS
3.1. Prevalence of Salmonella spp. in domestic pigeons
The total number of pigeons positive for Salmonella spp. was 32, which accounted for 5.47% of the 585 birds examined. All detected strains belonged to the serovar Typhimurium. The average prevalence of this bacterium in pigeons varied from 4.68% to 6.13% depending on the year. However, it was higher in group S birds (8.33 to 20% depending on the year) than in the pigeons classified into group H (2.04 to 3.78% depending on the year) (Figure 1). From all the positive samples, 12 (37.50%) originated from pigeons with no clinical symptoms (Table S1).
3.2. Serotyping
All analysed strains were confirmed to be Salmonella enterica ser. Typhimurium. The dominant variants among the analysed strains were as follows: 1,4,[5],12,:i:1,2 (16 strains ‐ 50%) and 1,4,[5],12,:‐:‐ (10 strains ‐ 31.25%). The other three variants (1,4,[5],12,:‐:1,2; 1,4,[5],12,:i:‐ and Copenhagen) appeared sporadically in 3 isolates ‐ 9.38%, 2 isolates ‐ 6.25% and one isolate – 3.12% (Table 1).
Table 1.
Characterization of Salmonella enterica ser
| Sample ID | Biofilm | Serotype | Phage susceptibility | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BAFASAL® dilutions | BAFASAL® efficiency | ||||||||
| 24 hr | 48 hr | 10–2 | 10–3 | 10–4 | 10–5 | 10–6 | |||
| PL_245 | + | ++ | 1,4,[5],12:i:1,2 | cl | cl | + | + | + | 100% |
| PL_323 | + | ++ | 1,4,[5],12:i:1,2 | cl | cl | cl | + | − | 83% |
| PL_332 | − | − | 1,4,[5],12:i:1,2 | cl | + | + | − | − | 67% |
| PL_387 | + | − | 1,4,[5],12: −:− | cl | cl | + | + | − | 83% |
| PL_427 | − | − | 1,4,[5],12:i:1,2 | cl | + | + | − | − | 67% |
| PL_445 | + | + | 1,4,[5],12: −:− | cl | cl | + | + | − | 83% |
| PL_480 | − | − | 1,4,[5],12: −:− | cl | cl | cl | + | − | 83% |
| PL_481* | +++ | +++ | 1,4,[5],12: −:− | cl | cl | + | + | − | 83% |
| PL_490 | − | − | 1,4,[5],12:i:1,2 | cl | cl | + | − | − | 67% |
| PL_491* | + | + | 1,4,[5],12: −:1,2 | cl | cl | + | − | − | 67% |
| PL_511 | + | + | 1,4,[5],12:i:1,2 | cl | cl | cl | + | − | 83% |
| PL_519 | + | + | 1,4,[5],12: −:1,2 | cl | cl | + | − | − | 67% |
| PL_547 | − | + | 1,4,[5],12:i:1,2 | cl | cl | cl | + | − | 83% |
| PL_548 | + | − | 1,4,[5],12: −:− | cl | cl | + | − | − | 67% |
| PL_549 | ++ | + | 1,4,[5],12:i:1,2 | cl | cl | + | − | − | 67% |
| PL_550 | + | + | 1,4,[5],12:i:1,2 | cl | cl | − | − | − | 50% |
| PL_559 | ++ | ++ | 1,4,[5],12: −:− | cl | cl | cl | − | − | 67% |
| PL_575 | − | − | 1,4,[5],12:i:1,2 | cl | cl | cl | cl | + | 100% |
| PL_576* | − | − | 1,4,[5],12:i:1,2 | cl | cl | cl | − | − | 67% |
| PL_577 | − | + | 1,4,[5],12:i: − | cl | + | − | − | − | 50% |
| PL_580* | + | − | 1,4,[5],12:i:1,2 | + | − | − | − | − | 33% |
| PL_587 | ++ | +++ | 1,4,[5],12: −:1,2 | cl | cl | + | − | − | 67% |
| PL_605 | + | ++ | 1,4,[5],12: −:− | cl | + | − | − | − | 50% |
| PL_613 | + | − | 1,4,[5],12: −:− | cl | + | − | − | − | 50% |
| PL_614* | − | − | 1,4,[5],12: −:− | cl | + | − | − | − | 50% |
| PL_616* | + | + | 1,4,[5],12:i: − | cl | + | − | − | − | 50% |
| PL_619 | − | − | 1,4,[5],12: −:− | cl | cl | + | − | − | 67% |
| PL_620 | − | − | 1,4,[5],12:i:1,2 | cl | cl | + | − | − | 67% |
| PL_621 | − | − | 1,4,[5],12:i:1,2 | cl | + | + | − | − | 67% |
| PL_626 | − | − | 1,4,[5],12:i:1,2 | cl | cl | + | − | − | 67% |
| PL_628 | ++ | +++ | 1,4,[5],12:i:1,2 | cl | cl | cl | − | − | 67% |
| PL_629 | + | + | var. Copenhagen | cl | + | − | − | − | 50% |
Typhimurium isolates investigated in the study. Symbols: biofilm: +++ strong, ++ medium, + weak, − none; phage susceptibility: cl complete lysis, + uncomplete lysis or separate plaques were present, − lack of plaques. The isolates used for molecular analyses (sequencing) are marked with asterisk.
3.3. Biofilm production
The biofilm capacity test was performed in two time periods. In both cases (24 and 48 hr), most of the strains analysed showed the ability to produce biofilm, that is 59.38% and 53.13%, respectively (Table 1).
3.4. Antimicrobial susceptibility—phenotypic analysis
The greatest percentage of the isolates was susceptible to amoxicillin with clavulanic acid (71.88%), florfenicol (68.75%) and quinolones (enrofloxacin 71.88%, marbofloxacin 75.00%, norfloxacin 71.85%). Among all analysed strains, the highest resistance was noted for neomycin (53.13%), oxytetracycline (46.88%) and tylosin (100.00%) (Figure 2).
Figure 2.
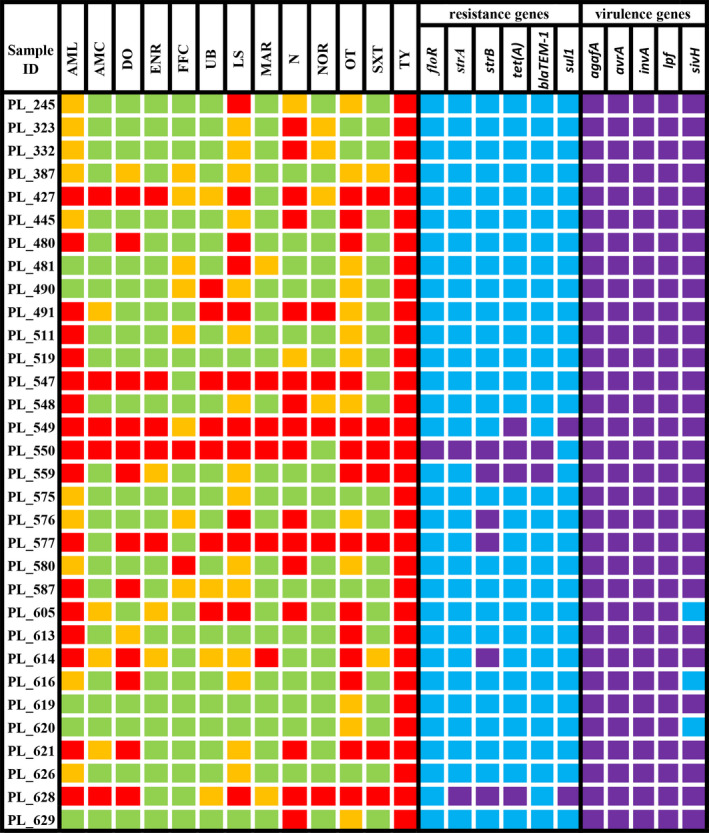
Antimicrobial susceptibility, frequency of chosen antimicrobial resistance genes, and virulence genes distributions among Salmonella enterica ser. Typhimurium isolated from domestic pigeons in Poland. The susceptibility to certain antimicrobials is marked with the following colours: red—resistant strain, orange—intermediate resistant strain, and green—susceptible strain. The presence of a certain gene is marked with purple, whereas the absence of a certain gene is marked with blue. Abbreviations: (amoxycillin 10μg (AML), amoxicillin with clavulanic acid 30μg (AMC), doxycycline 30μg (DO), enrofloxacin 5μg (ENR), florfenicol 30μg (FFC), flumequine 30μg (UB), lincomycin/spectinomycin 109μg (LS), marbofloxacin 5 μg (MAR), neomycin 30μg (N), norfloxacin 10μg (NOR), oxytetracycline 30μg (OT), sulphamethoxazole/trimetoprim 19:1 (SXT), tylosin 30μg (TY)) [Colour figure can be viewed at wileyonlinelibrary.com]
3.5. PCR detection of antimicrobial resistance genes and virulence genes
In the performed experiment, we analysed the presence of genes associated with antibiotic resistance to sulfonamides, tetracyclines, β‐lactams, florfenicol, chloramphenicol, and aminoglycosides. Of the thirteen genes tested, six were confirmed. The most common were the strB and tet(A) genes responsible for coding resistance to aminoglycosides and tetracyclines, respectively. The floR gene was found in one strain only (Figure 2).
In most of the strains, we did not confirm any of the analysed antibiotic resistance genes (78.13%). In the other strains, the number of confirmed genes ranged from 1 to 5 (Figure 2).
All five analysed virulence genes were confirmed in over 90% strains of Salmonella enterica ser. Typhimurium (Figure 2).
3.6. Nucleotide identity of selected isolates
The seven‐locus genotyping revealed that all examined isolates were characterized by high gene homology in the analysed genes—among 3,336 nt of concatenated sequences only 1 nucleotide substitution was detected The non‐synonymous substitution of G with C was observed in all strains except PL_580 in 478 position of the analysed sucA gene fragment and contributed to a change in the amino acid sequence (substitution of valine with leucine in position 160 of the amino acid alignment). It allowed dividing our strains into two STs: 128 (strains no PL_481, PL_491, PL_576, PL_614, PL_616) and 19 (strain PL_580). The alleles detected for sucA gene were 55 (all strains except PL_580) and 9. The blast analysis of this gene revealed 100% homology of 128 STs with Salmonella enterica ser. Typhimurium strain DT2 (Acc. No. HG326213). The details of MLST for all investigated strains are presented in Table 2. The blasted sequences of all strains revealed that they were 99.8%–100% homologous with the Salmonella enterica ser. Typhimurium sequences available in the GenBank.
Table 2.
Allelic profiles and MLST sequence types (STs) of selected Salmonella enterica ser. Typhimurium isolated in this study. The allele and ST numbers were assigned with the Salmonella typing tool available on the website: https://pubmlst.org/bigsdb?db=pubmlst_salmonella_seqdef&page=batchSequenceQuery
| Sample ID | MLST allelic profile | ST | SNP | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| aroC | dnaN | hemD | hisD | purE | sucA | thrA | Gene | mutation | positiona | ||
| PL_481 | 10 | 7 | 12 | 9 | 5 | 55 | 2 | 128 | sucA | G > C | 478 |
| PL_491 | 10 | 7 | 12 | 9 | 5 | 55 | 2 | 128 | sucA | G > C | 478 |
| PL_576 | 10 | 7 | 12 | 9 | 5 | 55 | 2 | 128 | sucA | G > C | 478 |
| PL_580 | 10 | 7 | 12 | 9 | 5 | 9 | 2 | 19 | none | none | none |
| PL_614 | 10 | 7 | 12 | 9 | 5 | 55 | 2 | 128 | sucA | G > C | 478 |
| PL_616 | 10 | 7 | 12 | 9 | 5 | 55 | 2 | 128 | sucA | G > C | 478 |
The nucleotide position in the analysed gene fragment.
3.7. PCR MP and ERIC PCR
At 88°C used as a denaturation temperature, the band profiles were obtained for each of the analysed strains. However, they were the same for all tested bacteria (Supplementary Figure 1). The described collection could not be differentiated also in the case of ERIC PCR profiles (Figure S2).
3.8. Phage susceptibility test
The performed test showed that all strains were susceptible to BAFASAL®. Considering the BAFASAL®’s host strain (Salmonella enterica ser. Gallinarum) susceptibility to the preparation as a reference, the susceptibility of S. Typhimurium strains was between 33% and 100%. The highest (100%) efficacy of BAFASAL® was observed against two strains numbered: 245 and 575, while the lowest one (33%) against one strain marked: PL_580. The other bacterial strains showed susceptibility over or equal to 50% (Table 1).
4. DISCUSSION
Although known for years, Salmonella is still an important animal and human health issue across the world. Every year, it causes enormous economic losses in both food and veterinary industry. While prevention programmes have been introduced for poultry that have successfully reduced the number of salmonellosis cases on farms, there are no adequate procedures for other birds, which can also be a reservoir of Salmonella for humans (Haesendonck et al., 2016). Noteworthy is also that both small rodents and wildlife birds can spread this pathogen into poultry farms, especially when none biosecurity procedures are strictly followed (Żebrowska et al., 2017). In many countries, domestic pigeons are kept for pigeon sport (carrier/ racing pigeons) or as ornamental pet birds, but they are also delicacy in the Asian and European cuisine. Although, only a few cases of this bacterium transmission from pigeon to human are known, this route of infection should not be ignored (Osman et al., 2014).
The prevalence of Salmonella enterica ser. Typhimurium in the examined pigeons was low (~ 5.5%), which is consistent with previous findings by Teske et al. (2013) and Stenzel et al. (2013) who revealed 0.9–3.7 and 7% prevalence of Salmonella spp. in domestic pigeons, respectively. The comparison of our results with those available in literature shows that the prevalence of this bacterium in the populations of feral/urban pigeons is similar to those observed in domesticated pigeons and oscillates around 0.9%–6.34% (Gargiulo et al., 2014; de Oliviera et al., 2018; de Sousa et al., 2010). Noteworthy is that ca. 40% of Salmonella enterica ser. Typhimurium strains obtained in this study were isolated from clinically healthy pigeons, which corresponds with results of the investigation conducted in Germany (Teske et al., 2013). The subclinical infections with Salmonella in domestic pigeons could result from improperly performed treatment, which was observed in the case of florfenicol (Pasmans et al., 2008). Subclinically infected pigeons are carriers of this bacterium and they could shed it with the faeces periodically; this in turn could promote infection spreading in the flock and increase the hazard posed to human health.
One of the key tools in Salmonella determination is the serotyping, which is used to check which serotypes dominate across the analysed area and whether there are new variants of a given strain (Bugarel et al., 2012; Madajczak & Szych, 2010). In our research, the dominant variants were 1,4,[5],12,:i:1,2 (50%) and 1,4,[5],12,:‐,‐ (~31%). The other three variants occurred sporadically (below 10%). It is difficult to refer to these results, because there are no data on the prevalence of different variants of Salmonella in pigeons. However, we are concerned about two facts. The first is that the presence of all four variants was confirmed in the analysed area. This phenomenon seems alarming and may pose a challenge in the development/selection of targeted prevention programmes. The second fact is the confirmation of a single‐phase variant 1,4,[5],12:i,‐. This variant has been raising concerns and interest around the world for many years, because of its increased virulence. Most reports indicate its multi‐drug resistance encoded on the plasmid (including active substances used to treat pigeons such as tetracyclines and sulfonamides). In addition, it was isolated from individuals with a much severe course of the disease (Bugarel et al., 2012; Echeita et al., 2001; Hoszowski & Wasyl, 2011). In our study, one of the strains qualified for this variant also showed high phenotypic resistance (resistant to almost all active substances—except florfenicol and amoxicillin with clavulanic acid) and confirmed one gene associated with antibiotic resistance strB (data not shown). Nevertheless, in our study, we performed the determination only by serotyping and according to Zając et al. (2015). This method is not perfect and has some limitations. To definitely check whether a given strain is monophasic, it is recommended to use PCR for fljB, fliC, insertion element IS200 genes and the gene responsible for the expression of antigens of the H:E complex (e, n, x; e, n, z15; e, h) (Bugarel et al., 2012; Hoszowski & Wasyl, 2011). Therefore, more detailed research is needed in this respect in the analysed area.
The antibiotic therapy is one of the most popular ways to treat sick pigeons. However, sometimes it fails to bring satisfactory effects, mainly due to: improperly selected active substance, too short period of antibiotic administration or incorrect dose of the drug, ignorance of breeders, and also Salmonella ability for intracellular presence and high resistance to environmental factors. It should also be remembered that after the treatment a part of individuals can become carriers and sowers of bacteria, thus posing an epidemiological threat to other birds in the flock (Majer‐Dziedzic et al., 2014). Our results on the analysis of Salmonella enterica ser. Typhimurium antimicrobial susceptibility correspond only partially with results obtained by Yousef and Mamdouh (2016). Our results differ from those obtained in Egypt, where pigeon Salmonella strains were susceptible to norfloxacin and mainly resistant to lincomycin and trimethoprim/ sulfamethoxozole (Osman et al., 2014). They differ also from those described earlier in Poland by Stenzel et al. (2014), who showed that 100% of the isolates were susceptible to amoxicillin, amoxicillin with clavulanic acid, enrofloxacin, flumequine, florfenicol, and trimethoprim/sulfamethoxazole. In our current study, higher antimicrobial resistance was noted, especially in the case of amoxicillin. A similar trend was also noted by Ledwoń et al., who detected increasing resistance to amoxicillin, amoxicillin with clavulanic acid, gentamicin, tetracycline, and neomycin in field Salmonella strains isolated in 2007–2012 compared to those isolated between the years 2013 to 2017 (Ledwoń et al., 2019).
Noteworthy is also that pigeon Salmonella strains isolated at the beginning of the XXI century were usually susceptible to all antimicrobials tested (Kimpe et al., 2002). The comparative analysis of results obtained in this study and previous findings points to the increasing antimicrobial resistance in the field pigeon Salmonella strains, which could be due to the overuse of antibiotics by pigeon breeders. The above is related to the fact that in Poland, likewise in most of the European countries, domestic pigeons are used as racing birds (carrier pigeons) or as ornamental birds (fancy pigeons), and not for meat production, hence their treatment is not subject to strict veterinary inspection control.
In addition to the phenotypic method, genotypic determinations were performed for the antibiotic resistance mechanisms for various active substances. Only six of the thirteen selected genes were confirmed in the analysed Salmonella enterica ser. Typhimurium strains (sul1, tet(A), blaTEM‐1 , floR and strB). On the one hand, the minimal presence of genes associated with antibiotic resistance is not a surprise to us. As reported by Samah and Shalaby (2013), the phenomenon of multi‐drug resistance among Salmonella strains is rather sporadic and these strains show high sensitivity to the majority of active substances tested. However, the relatively high resistance of these strains in vitro to the tested antibiotics suggests that there may be other genes associated with this phenomenon in the area we analysed. Tet(A) and strB were the most common genes and these results are consistent with those obtained by other research groups (Adesiji et al., 2014; Frech et al., 2003; Samah & Shalaby, 2013). The presence of these genes may be due to the abuse of both tetracyclines and aminoglycosides both in non‐targeted therapies and in the agricultural industry in Europe (Pezzella et al., 2004). Interestingly, results of phenotypic and genotypic studies of antibiotic resistance were not always consistent considering the analysed strains, likely due to the occurrence of ‘silent genes’ (Enne et al., 2006; Kime et al., 2019). In other words, these genes may have no proper connection to the promoter, which results in the lack of their expression. The second reason may be that there are no point mutations necessary to activate them. Nevertheless, their appearance among the analysed strains is disturbing (possibility of horizontal and vertical gene transfer), thus further research is needed in this respect (Gao et al., 2012; Palmer et al., 2018).
Due to the antibiotic resistance phenomenon, the world is looking for alternative therapies. One of them are bacteriophages. The use of host‐specific phages is increasingly often considered in controlling zoonotic pathogens (Atterburry et al., 2020). In the case of S. Typhimurium, there are already some reports on the effectiveness of bacteriophages in eradicating this microorganism. However, these studies concern pigs or poultry (Abhisingha et al., 2020; Atterburry et al., 2020; Hooton et al., 2011). Our in vitro preliminary research shows that the bacteriophages used proved to be highly effective (average more than 60%) against strains of Salmonella enterica ser. Typhimurium obtained from pigeons. However, these studies require an in vivo experiment to confirm BAFASAL® efficacy.
Biofilm formation is one of the important virulence factors. On the one hand, it helps the bacteria survive in unfavourable conditions, while on the other hand, it facilitates colonization of a host organism (Merino et al., 2019). This structure is particularly important to the veterinary and human medicine because bacteria able to form biofilms are far more resistant to antibiotics (Kaczorek et al., 2017). Olson et al. (2002) showed that Salmonella strains capable of producing a ‘biological membrane’ were significantly less sensitive to the selected active substances compared to the bacteria growing in the planktonic form. Interestingly, the researchers used in their study, among others, such active substances as amoxicillin, oxytetracycline, and enrofloxacin, which are used in Poland for pigeon treatment. In our experiment, over 50% of the analysed strains showed the ability to form biofilm. Similar results have been noted among Salmonella strains isolated from poultry farms in Spain (Marin et al., 2009). The fact that numerous analysed serotypes produce biofilm may indicate a difficult course of veterinary treatment in the future. Therefore, research aimed to monitor the biofilm and prevent its production in the pigeon industry should be conducted.
Additionally, we analysed Salmonella enterica ser. Typhimurium strains in search for the presence of the selected virulence genes. All of the analysed genes were confirmed in our Salmonella strains. These results were unsurprising; the high incidence rate of virulence genes was previously observed in Salmonella strains isolated from animals, humans, and in the food industry (Hertwig et al., 2019). The presence of the invA gene in all strains we analysed is consistent with the results reported by other authors, which confirms that this is a conserved gene in the whole Salmonella genus (Dashan et al., 2010; Oliviera et al., 2003; Salehi et al., 2005). For this reason, this gene is already used as a tool for the development of rapid diagnostic methods, and it could be used as a targeted prevention programme for this bacterium in the future (Chiu & Ou, 1996). The lpfA and agafA genes are associated with cell adhesion and thus encode the ability to produce biofilm (Campos‐Galvão et al., 2016). Their presence in all of the analysed Salmonella strains may suggest that this virulence factor may be crucial in the pathogenesis of the disease in pigeons in the future.
Various molecular methods targeting different genes have been used to replace the conventional serotyping method for Salmonella enterica serovars. MLST is one of the suitable methods for typing Salmonella serovars because of its high discriminative ability (Achtman et al., 2012; Sukhnanand et al., 2005). Its disadvantage, however, is its high costs due to the necessity of performing multiple PCR assays. In the present study, we amplified 7 housekeeping genes of selected Salmonella enterica ser. Typhimurium isolates, used in MLST, to investigate the nucleotide homology. The nucleotide sequences of six of the analysed genes were identical in all investigated isolates. Our analysis confirms a low degree of genetic diversity between the analysed Salmonella enterica ser. Typhimurium isolates (>99.8%), thus indicating limited usefulness of MLST gene analysis in the context of nucleotide identity investigation in different isolates belonging to the same serotype. According to Achtman MLST scheme, the majority of S. Typhimurium isolates are sequence type 19 which is an and ancestral genotype (Achtman et al., 2012).Numerous single‐ or double‐locus MLST variants radiate from the ST19 central hub. One of them is single‐locus variant ST128, which is a host restricted to Columba livia, but closely related to S. Typhimurium isolates that circulates in livestock and can cause zoonotic infections in humans. For this reason, the fact that most of the strains examined in MLST belonged to this ST was not surprising. However, one of the isolates (PL_580) belonged to typical ST19.
Two additional molecular approaches: ERIC‐PCR and PCR‐MP, were used to evaluate the genetic diversity of the isolated strains. These two independent analytical methods were chosen on the basis of their discriminatory power described previously for Salmonella bacteria (Anjay et al., 2015; Zaczek et al., 2015). However, our results were surprising. Even though the strains were isolated from different regions, at different seasons and different years, all analysed strains obtained the same genetic profiles. These results suggest that there is an endemic clone in Poland (Hellmuth et al., 2017). Further monitoring studies should be undertaken in this, especially for the areas of Poland which were not included in our research. It might appear that only the sequencing of the whole genomic DNA will allow for precise description of bacterial strains investigated in this study.
5. CONCLUSIONS
Even though the prevalence of Salmonella among domestic pigeons in Poland is relatively low, it should not be ignored. These birds are considered a potential reservoir of Salmonella enterica ser. Typhimurium. The increasing resistance to antibiotics and the occurrence of strains capable of producing biofilm and carrying the virulence gene are disturbing phenomena. Therefore, research should be continued to both develop and monitor respective prevention programmes.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Fig S1
Fig S2
Table S1
Table S2
Table S3
Table S4
ACKNOWLEDGEMENTS
The authors would like to thank Ewa Szczucińska for her assistance during microbiological examination. Project financially co‐supported by Minister of Science and Higher Education in the range of the programme entitled ‘Regional initiative of Excellence' for the years 2019‐2022, Project No. 010/RID/2018/19, amount of funding 12.000.000 PLN
Kaczorek‐Łukowska E, Sowińska P, Franaszek A, Dziewulska D, Małaczewska J, Stenzel T. Can domestic pigeon be a potential carrier of zoonotic Salmonella? Transbound Emerg Dis.2021;68:2321–2333. 10.1111/tbed.13891
Contributor Information
Edyta Kaczorek‐Łukowska, Email: edyta.kaczorek@uwm.edu.pl.
Tomasz Stenzel, Email: tomasz.stenzel@uwm.edu.pl.
DATA AVAILABILITY STATEMENT
The part of the data that support the findings of this study are available in the supplementary material of this article. The rest of the data are available from the corresponding author upon reasonable request.
REFERENCES
- Abhisingha, M., Dumnil, J., & Pitaksutheepong, C. (2020). Efficiency of phage cocktail to reduce Salmonella Typhimurium on chicken meat during low temperature storage. LWT, 129, 109580. 10.1016/j.lwt.2020.109580 [DOI] [Google Scholar]
- Achtman, M., Wain, J., Weill, F. X., Nair, S., Zhou, Z., Sangal, V., Krauland, M. G., Hale, J. L., Harbottle, H., Uesbeck, A., Dougan, G., Harrison, L. H., Brisse, S., & S.E.M.S. Group (2012). Multilocus sequence typing as a replacement for serotyping in Salmonella enterica . PLoS Path, 8(6), e1002776. 10.1371/journal.ppat.1002776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adesiji, Y. O., Deekshit, V. K., & Karunasagar, I. (2014). Antimicrobial‐resistant genes associated with Salmonella spp. isolated from human, poultry, and seafood sources. Food Science & Nutrition, 2(4), 436–442. 10.1002/fsn3.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjay, A. K., Agarwal, R. K., Ramees, T. P., Dubal, Z. B., Kaushik, P., Kumar, M. S., Dudhe, N. C., Milton, A. A. P., Abhishek, B. K., & Shagufta, B. (2015). Molecular Typing of Salmonella Typhimurium and S. Enteritidis Serovars from Diverse Origin by ERIC‐PCR. Journal of Pure and Applied. Microbiology, 9(3), 2627–2634. 10.1111/jam.12805 [DOI] [Google Scholar]
- Atterbury, R. J., Gigante, A. M., Rubio Lozano, M., Méndez Medina, R. D., Robinson, G., Alloush, H., Barrow, P. A., & Allen, V. A. (2020). Reduction of Salmonella contamination on the surface of chicken skin using bacteriophage. Virology Journal, 17(98), 10.1186/s12985-020-01368-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugarel, M., Granier, S. A., Bonin, E., Vignaud, M. L., Roussel, S., Fach, P., & Brisabois, A. (2012). Genetic diversity in monophasic (1,4,[5],12:I:‐ and 1,4,[5],12:‐:1,2) and in non‐motile (1,4,[5],12:‐:‐) variants of Salmonella enterica S. Typhimurium. Food Research International, 45(2), 1016–1024. 10.1016/j.foodres.2011.06.057- [DOI] [Google Scholar]
- Campos‐Galvão, M. E., Ribon, A. O., Araújo, E. F., & Vanetti, M. C. (2016). Changes in the Salmonella enterica Enteritidis phenotypes in presence of acyl homoserine lactone quorum sensing signals. Journal of Basic Microbiology, 56(5), 493–501. 10.1002/jobm.201500471 [DOI] [PubMed] [Google Scholar]
- Centers of Disease Control and Prevention . Retrieved from https://www.cdc.gov/nationalsurveillance/pdfs/NationalSalmSurveillOverview_508.pdf.
- Chiu, C. H., & Ou, J. T. (1996). Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture‐multiplex PCR combination assay. Journal of Clinical Microbiology, 34(10), 2619–2622. 10.1128/JCM.34.10.2619-2622.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2013). Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. 4th. Approved standard, CLSI document VET01‐A4. Clinical and Laboratory Standards Institute. [Google Scholar]
- Dahshan, H., Shahada, F., Chuma, T., Moriki, H., & Okamoto, K. (2010). Genetic analysis of multidrug‐resistant Salmonella enterica serovars Stanley and Typhimurium from cattle. Veterinary Microbiology, 145(1–2), 76–83. 10.1016/j.vetmic.2010.02.035 [DOI] [PubMed] [Google Scholar]
- de Oliveira, M. C. V., Camargo, B. Q., Cunha, M. P. V., Saidenberg, A. B., Teixeira, R. H. F., Matajira, C. E. C., Moreno, L. Z., Gomes, V. T. M., Christ, A. P. G., Barbosa, M. R. F., Sato, M. I. Z., Moreno, A. M., & Knöbl, T. (2018). Free‐ranging synanthropic birds (Ardea alba and Columba livia domestica) as Carriers of Salmonella spp. and Diarrheagenic Escherichia coli in the Vicinity of an Urban Zoo. Vector‐Borne and Zoonotic Diseases, 18(1), 65–69. 10.1089/vbz.2017.2174 [DOI] [PubMed] [Google Scholar]
- de Sousa, E., Berchieri, A. J., Pinto, A. A., Machado, R. Z., de Carrasco, A. O., Marciano, J. A., & Werther, K. (2010). Prevalence of Salmonella spp. antibodies to Toxoplasma gondii, and Newcastle disease virus in feral pigeons (Columba livia) in the city of Jaboticabal, Brazil. Journal of Zoo and Wildlife Medicine, 41(4), 603–607. 10.1638/2008-0166.1 [DOI] [PubMed] [Google Scholar]
- Dovc, A., Zorman‐Rojs, O., Vergles Rataj, A., Bole‐Hribovsek, V., Krapez, U., & Dobeic, M. (2004). Health status of free‐living pigeons (Columba livia domestica) in the city of Ljubljana. Acta Veterinaria Hungarica, 52(2), 219–226. 10.1556/AVet.52.2004.2.10 [DOI] [PubMed] [Google Scholar]
- Ebrahimi, A., Moatamedi, A., Lotfalian, S., & Mirshokraei, P. (2013). Biofilm formation, hemolysin production and antimicrobial susceptibilities of Streptococcus agalactiae isolated from the mastitis milk of dairy cows in Shahrekord district, Iran. Veterinary Research Forum, 4(4), 269–272. PMID: 25568683. [PMC free article] [PubMed] [Google Scholar]
- Echeita, M. A., Herrera, S., & Usera, M. A. (2001). Atypical, fljB‐negative Salmonella enterica subsp. enterica strain of serovar 4,5,12:I:‐ appears to be a monophasic variant of serovar Typhimurium. Journal of Clinical. Microbiology, 39(8), 2981–2983. 10.1128/JCM.39.8.2981-2983.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research., 32(5), 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA, and ECDC (2018). The European Union One Health 2018 Zoonoses Report EFSA Journal, 17(12). [DOI] [PMC free article] [PubMed]
- Enne, V. I., Delsol, A. A., Roe, J. M., & Bennett, P. M. (2006). Evidence of antibiotic resistance gene silencing in Escherichia coli . Antimicrobial Agents and Chemotherapy, 50(9), 3003–3010. 10.1128/AAC.00137-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frech, G., Kehrenberg, C., & Schwarz, S. (2003). Resistance phenotypes and genotypes of multiresistant Salmonella enterica subsp. enterica serovar Typhimurium var. Copenhagen isolates from animal sources. Journal of Antimicrobial Chemotherapy, 51(1), 180–182. 10.1093/jac/dkg058 [DOI] [PubMed] [Google Scholar]
- Gao, J., Yu, F. Q., Luo, L. P., He, J. Z., Hou, R. G., Zhang, H. Q., Li, S. M., Su, J. L., & Han, B. (2012). Antibiotic resistance of Streptococcus agalactiae from cows with mastitis . The Veterinary Journal, 194(3), 423–424. 10.1016/j.tvjl.2012.04.020 [DOI] [PubMed] [Google Scholar]
- Gargiulo, A., Russo, T. P., Schettini, R., Mallardo, K., Calabria, M., Menna, L. F., Raia, P., Pagnini, U., Caputo, V., Fioretti, A., & Dipineto, L. (2014). Occurrence of enteropathogenic bacteria in urban pigeons (Columba livia) in Italy. Vector‐Borne Zoonotic Diseases, 14(4), 251–255. 10.1089/vbz.2011.0943 [DOI] [PubMed] [Google Scholar]
- Haesendonck, R., Rasschaert, G., Martel, A., Verbrugghe, E., Heyndrickx, M., Haesebrouck, F., & Pasmans, F. (2016). Feral pigeons: A reservoir of zoonotic Salmonella Enteritidis strains? Veterinary Microbiology, 15(195), 101–103. 10.1016/j.vetmic.2016.09.017 [DOI] [PubMed] [Google Scholar]
- He, X. H., Xu, X. B., Li, K., Liu, B., & Yue, T. L. (2016). Identification of Salmonella enterica Typhimurium and variants using a novel multiplex PCR assay. Food Control, 65, 152–159. 10.1016/j.foodcont.2016.01.015 [DOI] [Google Scholar]
- Hellmuth, J. E., Hitzeroth, A. C., Bragg, R. R., & Boucher, C. A. (2017). Evaluation of the ERIC‐PCR as a probable method to differentiate Avibacterium paragallinarum serovars. Avian Pathology, 46(3), 272–277. 10.1080/03079457.2016.1259610 [DOI] [PubMed] [Google Scholar]
- Hooton, S. P., Atterbury, R. J., & Connerton, I. F. (2011). Application of a bacteriophage cocktail to reduce Salmonella Typhimurium U288 contamination on pig skin. International Journal of Food Microbiology, 151(2), 157–163. 10.1016/j.ijfoodmicro.2011.08.015 [DOI] [PubMed] [Google Scholar]
- Hoszowski, A., & Wasyl, D. (2011). Epidemiological importance and identification of the monophasic strains of Salmonella Typhimurium 1,4,[5],12:I:‐. Medycyna Weterynaryjna, 67(9), 589–593. [Google Scholar]
- Israili, Z. H., & Iqbal, N. J. (2017). Effect of pigeon keeping on health and family life. International Journal of Advances in Social Science and Humanities, 5(09), 5–14. 10.4172/2471-9846.1000190 [DOI] [Google Scholar]
- Jahantigh, M., & Nili, H. (2010). Drug resistance of Salmonella spp. isolated from pigeon eggs. Comparative Clinical Pathology, 19(4), 437–439. 10.1007/s00580-010-1005-6 [DOI] [Google Scholar]
- Kaczorek, E., Małaczewska, J., Wójcik, R., & Siwicki, A. K. (2017). Biofilm production and other virulence factors in Streptococcus spp. isolated from clinical cases of bovine mastitis in Poland. BMC Veterinary Research, 13, 398. 10.1186/s12917-017-1322-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidgell, C., Reichard, U., Wain, J., Linz, B., Torpdahl, M., Dougan, G., & Achtman, M. (2002). Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infection, Genetics and Evolution, 2(1), 39–45. 10.1016/s1567-1348(02)00089-8 [DOI] [PubMed] [Google Scholar]
- Kime, L., Randall, C. P., Banda, F. I., Coll, F., Wright, J., Richardson, J., Empel, J., Parkhill, J., & O'Neill, A. J. (2019). Transient silencing of antibiotic resistance by mutation represents a significant potential source of unanticipated therapeutic failure. MBio, 10(5), e01755, 19. 10.1128/mBio.01755-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpe, A., Decostere, A., Martel, A., Haesebrouck, F., & Devriese, L. A. (2002). Prevalence of antimicrobial resistance among pigeon isolates of Streptococcus gallolyticus, Escherichia coli and Salmonella enterica serotype Typhimurium. Avian Pathology, 31(4), 393–397. 10.1080/03079450220141679 [DOI] [PubMed] [Google Scholar]
- Kriz, P., Sisak, F., Slana, I., Karpiskova, R., Docekal, J., Skoric, M., Fictum, P., Babak, V., & Pavlik, I. (2011). Mycobacterium avium Subsp. avium and Salmonella enterica serotype Typhimurium var. Copenhagen phage type DT2 in pigeons. Foodborne Pathogens and Disease, 8(10), 1135–1137. 10.1089/fpd.2010.0836 [DOI] [PubMed] [Google Scholar]
- Lawson, B., de Pinna, E., Horton, R. A., Macgregor, S. K., John, S. K., Chantrey, J., Duff, J. P., Kirkwood, J. K., Simpson, V. R., Robinson, R. A., Wain, J., & Cunningham, A. A. (2014). Epidemiological evidence that garden birds are a source of human salmonellosis in England and Wales. PLoS One, 9(2), e88968. 10.1371/journal.pone.0088968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledwoń, A., Rzewuska, M., Czopowicz, M., Kizerwetter‐Świda, M., Chrobak‐Chmiel, D., & Szeleszczuk, P. (2019). Occurrence and antimicrobial susceptibility of Salmonella spp. isolated from domestic pigeons Columba livia var. domestica in 2007–2017 in Poland. Medycyna Weterynaryjna, 75(12), 735–737. 10.21521/mw.6280 [DOI] [Google Scholar]
- Madajczak, G., & Szych, J. (2010). Evaluation of PREMI®TEST Salmonella kit for identification of not‐typable by conventional methods Salmonella. Medycyna Doświadczalna I Mikrobiologia, 62(1), 29–36. [PubMed] [Google Scholar]
- Majer‐Dziedzic, B., Pochodyła, M., Tokarzewski, S., & Pochodyła, A. (2014). Isolation of Salmonella Typhimurium var. Copenhagen strains from dead pigeons and evaluation of their immunogenicity. Medycyna Weterynaryjna, 70(10), 620–625. [Google Scholar]
- Marin, C., Hernandez, A., & Lainez, M. (2009). Biofilm development capacity of Salmonella strains isolated in poultry risk factors and their resistance against disinfectants. Poultry Science, 88(2), 424–431. 10.3382/ps.2008-00241 [DOI] [PubMed] [Google Scholar]
- Merino, L., Procura, F., Trejo, F. M., Bueno, D. J., & Golowczyc, M. A. (2019). Biofilm formation by Salmonella sp. in the poultry industry: Detection, control and eradication strategies. Food Research International, 119, 530–540. 10.1016/j.foodres.2017.11.024 [DOI] [PubMed] [Google Scholar]
- Oliveira, S. D., Rodenbusch, C. R., Michael, G. B., Cardoso, M. I. R., Canal, C. W., & Brandelli, A. (2003). Detection of virulence genes in Salmonella Enteritidis isolates from different sources. Brazilian Journal of Microbiology, 34(1), 123–124. 10.1590/S1517-83822003000500042 [DOI] [Google Scholar]
- Olson, M., Ceri, H., Morck, D., Buret, A., & Read, R. (2002). Biofilm bacteria: Formation and comparative susceptibility to antibiotics. Canadian Journal of Veterinary Research, 66(2), 86–92. PMID: 11989739. [PMC free article] [PubMed] [Google Scholar]
- Osman, K. M., Marouf, S. H., Mehana, O. A., & AlAtfeehy, N. (2014). Salmonella enterica serotypes isolated from squabs reveal multidrug resistance and a distinct pathogenicity gene repertoire. Revue Scientifique Et Technique (International Office of Epizootics), 33(3), 997–1006. 10.20506/rst.33.3.2336 [DOI] [PubMed] [Google Scholar]
- Palmer, A. C., Chait, R., & Kishony, R. (2018). Nonoptimal gene expression creates latent potential for antibiotic resistance. Molecular Biology and Evolution, 35(11), 2669–2684. 10.1093/molbev/msy163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmans, F., Baert, K., Martel, A., Bousquet‐Melou, A., Lanckriet, R., De Boever, S., Van Immerseel, F., Eeckhaut, V., De Backer, P., & Haesbrouck, F. (2008). Induction of the carrier state in pigeons infected with Salmonella enterica subspecies enterica serovar typhimurium PT99 by treatment with florfenicol: A matter of pharmacokinetics. Antimicrobial Agents and Chemotherapy, 52(3), 954–961. 10.1128/AAC.00575-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmans, F., Van Immerseel, F., Hermans, K., Heyndrickx, M., Collard, J. M., Ducatelle, R., & Haesebrouck, F. (2004). Assessment of virulence of pigeon isolates of Salmonella enterica subsp. enterica serovar typhimurium variant copenhagen for humans. Journal of Clinical Microbiology, 42(5), 2000–2002. 10.1128/jcm.42.5.2000-2002.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzella, C., Ricci, A., DiGiannatale, E., Luzzi, I., & Carattoli, A. (2004). Tetracycline and Streptomycin Resistance Genes, Transposons, and Plasmids in Salmonella enterica Isolates from Animals in Italy. Antimicrobial Agents and Chemotherapy, 48(3), 903–908. 10.1128/AAC.48.3.903-908.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polański, P., Radziszewski, F., Księżak, E., Wiktor, A., & Sadkowska‐Todys, M. (2019). Foodborne infections and intoxications in Poland in 2017. Przegląd Epidemiologiczny, 73, 451–462. 10.32394/pe.73.43 [DOI] [PubMed] [Google Scholar]
- Salehi, T. Z., Mahzounieh, M., & Saeedzadeh, A. (2005). Detection of invA gene in isolated Salmonella from broilers by PCR Method. International Journal of Poultry Science, 4(8), 557–559. 10.3923/ijps.2005.557.559 [DOI] [Google Scholar]
- Samah, E. A., & Shalaby, A. G. (2013). Molecular characterization of Salmonella species isolated from pigeon. Animal Health Research Journal, 1(4), 1–13. [Google Scholar]
- Stenzel, T., Bancerz‐Kisiel, A., Tykałowski, B., Smiałek, M., Pestka, D., & Koncicki, A. (2014). Antimicrobial resistance in bacteria isolated from pigeons in Poland. Polish Journal of Veterinary Sciences, 17(1), 169–171. 10.2478/pjvs-2014-0023 [DOI] [PubMed] [Google Scholar]
- Stenzel, T., Szulia, G., Fordońska, J., Górska, J., Śmiałek, M., Tykałowski, B., Pestka, D., & Koncicki, A. (2013). Antimicrobial resistance of selected bacteria isolated from pigeons in Poland in the years 2010‐2013. Conference proceedings: Actual problems in poultry pathology . 27‐28.06.2013, , 174‐181
- Stepanović, S., Vuković, D., Hola, V., Di Bonaventura, G., Djukić, S., Cirković, I., & Ruzicka, F. (2007). Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS, 115(8), 891–899. 10.1111/j.1600-0463.2007.apm_630.x [DOI] [PubMed] [Google Scholar]
- Sukhnanand, S., Alcaine, S., Warnick, L. D., Su, W. L., Hof, J., Craver, M. P. J., McDonough, P., Boor, K. J., & Wiedmann, M. (2005). DNA sequence‐based subtyping and evolutionary analysis of selected Salmonella enterica serotypes. Journal of Clinical Microbiology, 43(8), 3688–3698. 10.1128/JCM.43.8.3688-3698.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske, L., Ryll, M., Rubbenstroth, D., Hänel, I., Hartmann, M., Kreienbrock, L., & Rautenschlein, S. (2013). Epidemiological investigations on the possible risk of distribution of zoonotic bacteria through apparently healthy homing pigeons. Avian Pathology, 42(5), 397–407. 10.1080/03079457.2013.822468 [DOI] [PubMed] [Google Scholar]
- Torres‐Mejía, A. M., Blanco‐Peña, K., Rodríguez, C., Duarte, F., Jiménez‐Soto, M., & Esperón, F. (2018). Zoonotic Agents in Feral Pigeons (Columba livia) from Costa Rica: Possible Improvements to Diminish Contagion Risks. Vector‐Borne and Zoonotic Diseases, 18(1), 49–54. 10.1089/vbz.2017.2131 [DOI] [PubMed] [Google Scholar]
- von Hertwig, A. M., Amorim Neto, D. P., de Almeida, E. A., Casas, M. R. T., & Nascimento, M. D. S. D. (2019). Genetic diversity, antimicrobial resistance and virulence profile of Salmonella isolated from the peanut supply chain. International Journal of Food Microbiology, 294, 50–54. 10.1016/j.ijfoodmicro.2019.02.005 [DOI] [PubMed] [Google Scholar]
- World Health Organisation (2017). The burden of foodborne diseases in the who European region. : World Health Organisation. Retrieved from https://www.euro.who.int/__data/assets/pdf_file/0005/402989/50607‐WHO‐Food‐Safety‐publicationV4_Web.pdf [Google Scholar]
- Yousef, S., & Mamdouh, R. (2016). Class I Integron and β‐lactamase encoding genes of multidrug resistance Salmonella isolated from pigeons and their environments. Cellular and Molecular Biology (Noisy‐le‐grand), 62(14), 48–54. 10.14715/cmb/2016.62.14.8 [DOI] [PubMed] [Google Scholar]
- Zaczek, A., Wojtasik, A., Izdebski, R., Gorecka, E., Wojcik, E. A., Nowak, T., Kwiecinski, P., & Dziadek, J. (2015). PCR melting profile as a tool for outbreak studies of Salmonella enterica in chickens. BMC Veterinary Research, 11(137). 10.1186/s12917-015-0451-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zając, M., Hoszowski, A., Skarżyńska, M., Lalak, A., Samcik, I., & Wasyl, D. (2015). Assessment of the efficacy of phase inversion methods in Salmonella diagnostics. Medycyna Weterynaryjna, 71(12), 796–800. [Google Scholar]
- Żebrowska, J. P., Witkowska, D., Sobczak, J., Mituniewicz, T., & Sowińska, J. (2017). Occurrence of Salmonella on poultry farms, in flocks of pigeons and feed monitored by the Department of Veterinary Hygiene in Olsztyn in the years 2014–2015. Medycyna Weterynaryjna, 73(2), 111–117. 10.21521/mw.5636 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Table S1
Table S2
Table S3
Table S4
Data Availability Statement
The part of the data that support the findings of this study are available in the supplementary material of this article. The rest of the data are available from the corresponding author upon reasonable request.


