ISG products are believed to directly block viral replication. We propose an alternative mechanism based on emerging evidence. We discuss that the metabolic products of two ISG proteins, namely RSAD2 and NOS, inhibit activity of GAPDH to regulate central carbon metabolism and support a broad‐spectrum antiviral immune response via at least four mechanisms: eicosanoids storm, antigen cross‐presentation via MHC‐I, modulating NFAT and NF‐κB pathways, and S‐nitrosylation of viral proteins.
Keywords: GAPDH, immunometabolism, ISG, viperin, viruses
Abstract
In response to viral infections, the innate immune system rapidly activates expression of several interferon‐stimulated genes (ISGs), whose protein and metabolic products are believed to directly interfere with the viral life cycle. Here, we argue that biochemical reactions performed by two specific protein products of ISGs modulate central carbon metabolism to support a broad‐spectrum antiviral response. We demonstrate that the metabolites generated by metalloenzymes nitric oxide synthase and the radical S‐adenosylmethionine (SAM) enzyme RSAD2 inhibit the activity of the housekeeping and glycolytic enzyme glyceraldehyde 3‐phosphate dehydrogenase (GAPDH). We discuss that this inhibition is likely to stimulate a range of metabolic and signalling processes to support a broad‐spectrum immune response. Based on these analyses, we propose that inhibiting GAPDH in individuals with deteriorated cellular innate immune response like elderly might help in treating viral diseases such as COVID‐19.
Abbreviations
- 1,3‐BPG
1,3‐biphosphoglycerate
- COVID‐19
coronavirus disease 2019
- cADPR
cyclic ADP‐ribose
- ddhCTP
3'‐deoxy‐3',4'‐didehydro analogue
- GAPDH
glyceraldehyde 3‐phosphate dehydrogenase
- G3P
glyceraldehyde 3‐phosphate
- hiPSCs
human induced pluripotent stem cells
- ISG
interferon‐stimulated gene
- NFAT
nuclear factor of activated T cells
- NOS
nitric oxide synthase
- RSAD2
radical S‐adenosylmethionine domain‐containing protein 2
- RdRps
RNA‐dependent RNA polymerases
- SAM
S‐adenosylmethionine
Introduction
Central carbon metabolism converts sugars into a range of metabolic precursors that are used to generate biomass and energy required for the cellular function [1] (Fig. 1A). Consequently, remodelling of central carbon metabolism occurs in many human diseases such as cancer [2] and is at the forefront of the host–pathogen interactions. Pathogens like bacteria or viruses are dependent on host cellular metabolites and proteins to support their reproduction. To fight viral infections, all cells are equipped with a nonspecific response consisting of the expression of several proteins and enzymes, induced by different types of interferons, and thus, are referred to as interferon‐stimulated gene (ISG) products. Most previous studies have led to the conclusion that the protein products of these genes directly act on the viral life cycles to restrict their replication [3, 4]. On the contrary, we propose a new model based on available data in the literature and an analogy from a system engineering perspective (Fig. 1B): A cell can be considered as a factory and central carbon metabolism as the main process for converting a raw material to products and energy for the factory to function (Fig. 1B). When an infectious agent enters the factory, it will highjack the main process and use the products for its reproduction. Under this circumstance, the first response of the control room would be to use some of the available products in a second reaction (analogous to the function of ISGs) to directly block viral replication and to inhibit the main process. This would limit the nutrients for the reproduction of the infectious agent, while redirecting the materials and energy to a third process that can eliminate the invading agent. Accordingly, we suggest that the metabolites generated by some ISG proteins contribute to the remodelling of the central carbon metabolism in support of a broad‐spectrum antiviral immune response. We discuss emerging evidence that supports this model. We show how the early metabolites generated by the biochemical reactions of two ISG metalloenzymes, namely nitric oxide synthase (NOS) and the radical‐SAM enzyme RSAD2, inhibit the glycolytic enzyme glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) and how this inhibition is likely to support a broad‐spectrum antiviral immune response.
Fig. 1.
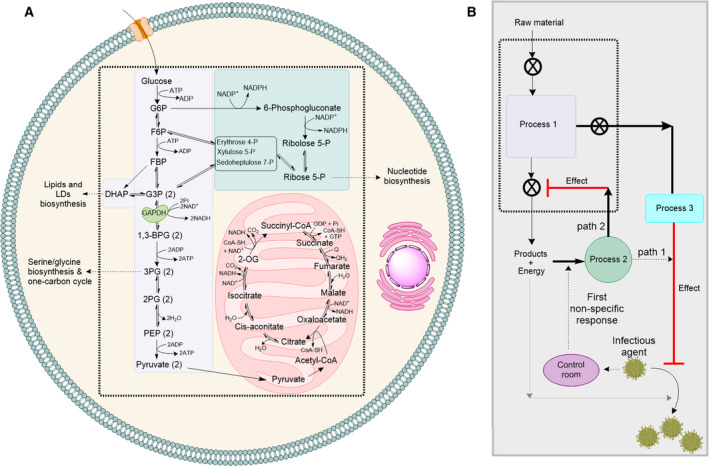
Central carbon metabolism and viral infection. (a) Central carbon metabolism (glycolysis, pentose phosphate pathway and TCA cycle) converts sugars to the building blocks of DNA and RNA, proteins and lipids. Additionally, it generates energy in the form of ATP and redox cofactors NAD+/NADH and NADP+/NADPH. Abbreviations: LDs, lipid droplets; G6P, glucose 6‐phosphate; F6P, fructose 6‐phosphate; FBP, fructose 1,6‐biphosphate; G3P, glyceraldehyde 3‐phosphate; DHAP, dihydroxyacetone phosphate; 1,3‐BPG, 1,3‐biphosphoglycerate; 3PG, 3‐phosphoglycerate; 2PG, 2‐phosphoglycerate; PEP, phosphoenolpyruvate; 2‐OG, 2‐oxoglutarate. (b) A systems engineering analogy describing function of the protein products of ISGs as (1) direct effectors of viral replication and (2) in the remodelling of central carbon metabolism to support broad‐spectrum immune response. The cell is like a factory and central carbon metabolism is the process 1. If an infectious agent enters the factory, the first response of the control room would be to use some of the available products and energy (process 2) to either directly inhibit viral replication (path 1) or to inhibit the production process (path 2). The outcomes of path 2 will be (i) reduction in formation of products and energy to limit access of pathogen to these resources and (ii) support of process 3, which restricts replication of the infectious agent.
Mechanisms adopted by cells to inhibit GAPDH
GAPDH is a housekeeping protein catalysing a critical step in glycolysis, with additional functions in DNA repair [5], cytoskeletal dynamics and vesicular trafficking between cellular compartments [6] and redox signalling and apoptosis [7]. As a glycolytic enzyme, it catalyses the NAD+‐dependent transformation of glyceraldehyde 3‐phosphate (G3P or GAP) to 1,3‐biphosphoglycerate (1,3‐BPG) (Fig. 1A). Metabolomic analysis together with computational studies have revealed that flux through GAPDH is a rate‐limiting step in glycolysis [8]. Here, we discuss mechanisms adopted by the cellular innate immune response to inhibit NAD+‐dependent conversion of G3P by GAPDH, focussing on those associated with ISG products specifically rather than other mechanisms such as malonylation [9].
Inhibition of GAPDH by ddhCTP ribonucleotide analogue generated by RSAD2 (viperin)
Radical S‐adenosylmethionine (SAM) domain‐containing protein 2 (RSAD2) also known as viperin is a member of the radical‐SAM superfamily of enzymes [10]. The RSAD2 gene is an interferon‐stimulated gene (ISG) whose expression is induced by type‐I, type‐II and type‐III interferons, directly by viruses and by LPS [11, 12, 13, 14, 15, 16, 17, 18]. It is known that expression of RSAD2 restricts replication of a wide range of RNA and DNA viruses in different cells [19, 20] and this effect is proposed to result from an altered metabolic state [21]. Biochemical and cell biological studies revealed that RSAD2 can catalyse the transformation of CTP to its 3'‐deoxy‐3',4'‐didehydro analogue (ddhCTP) [22]. Isotope labelling experiments [22], structural analysis [23] and biochemical studies [24] have also shed light on the enzymatic mechanism. Biochemical experiments showed that ddhCTP may act as a chain terminator of viral RNA‐dependent RNA polymerases (RdRps) (IC50> 20,000 µM) [22]. It should be noted that the reported IC50 values of ddhCTP as a chain terminator of viral RdRps were not corrected for the observed background effect of CTP on biochemical assays used to measure chain‐termination activity [25]. Nevertheless, if ddhCTP acts as a chain terminator of viral RdRps, the question that arises is why does then the cellular activity of RSAD2 affects many processes like glucose homeostasis [26] and expression of immune‐related genes [27]? To answer this fundamental question, metabolomic experiments using HEK293T cells and macrophages derived from human induced pluripotent stem cells (hiPSCs) were used. It was discovered that cellular activity of RSAD2 diminishes activity of NAD+‐dependent enzymes including that of GAPDH inside cells (Fig. 2) increasing intracellular levels of G3P and metabolites of pentose phosphate pathway [25, 28]. Subsequent biochemical studies confirmed that ddhCTP inhibits activity of GAPDH in a test tube with an IC50 value of 55.8 ± 0.2 µM [25]. This value is bout 400‐fold less than the reported IC50 value of ddhCTP as chain terminator and is less than the reported cellular concentration of ddhCTP (100–300 µM) [22]. These data suggest that under physiological conditions ddhCTP is more efficient in inhibiting GAPDH than acting as a chain terminator of RdRps.
Fig. 2.
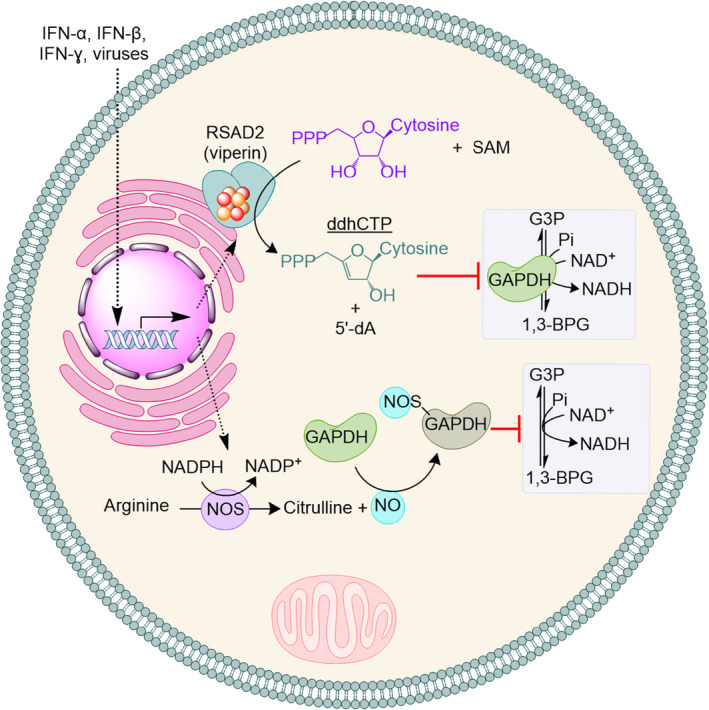
The early response of the cellular innate immune system inhibits NAD+‐dependent activity of GAPDH. In response to interferons, viruses or bacteria, the cells express metalloenzymes RSAD2 (viperin) and/or nitric oxide synthase (NOS). RSAD2 uses S‐adenosylmethionine (SAM) to catalyse transformation of CTP to ddhCTP, which inhibit activity of GAPDH. 5´‐deoxyadenosine (5´‐dA) is formed as a by‐product. On the other hand, NOS generates NO, which induces S‐nitrosylation of GAPDH and inhibits its activity.
S‐nitrosylation
Nitric oxide (NO) has emerged as a key player in innate immune response to bacterial and viral pathogens [29, 30]. It is synthesized from L‐arginine by the catalytic function of the metalloenzyme nitric oxide synthase (NOS). In humans at least three isoforms of NOS have been reported (NOS‐I, NOS‐II and NOS‐III) [31, 32]. These metalloenzymes have binding sites for NADPH, FMN, FAD and calmodulin (CaM). The active site of all three isoforms has a haem cofactor and catalyses conversion of L‐arginine to NO and L‐citrulline in two steps [33]. Several reports have shown that NO induces S‐nitrosylation of GPADH, which inhibits its activity (Fig. 2) [34, 35, 36, 37, 38]. It is suggested that S‐nitrosylation of the active site thiol leads to nonenzymatic ADP‐ribosylation, which inactivates the protein [37].
Inhibition of GAPDH and a broad‐spectrum antiviral response
Inhibition of GAPDH by ddhCTP or S‐nitrosylation will likely result in an increase in the cellular availability of NAD+. This will support protein ADP‐ribosylation and biosynthesis of cyclic ADP‐ribose (cADPR) [39, 40] (Fig. 3), both of which require NAD+ as a substrate. Consistently, it is shown that S‐nitrosylation of GAPDH and inhibition of its activity increases endogenous protein ADP‐ribosylation [34]. ADP‐ribosylation is shown to increase proteasomal activity [41, 42]. cADPR on the other hand, is a second messenger metabolite involved in modulation of Ca2+ signalling and homeostasis [43, 44, 45] (Fig. 3A). cADPR binds to ryanodine receptor (RyRs), which is expressed in many cell types including macrophages and T cells [46], and initiates the release of Ca2+ from the intracellular store (Fig. 3) [47, 48, 49]. Aligned with these observations, it has been shown that the cellular level of NAD+ controls Ca2+ store and release [50].
Fig. 3.
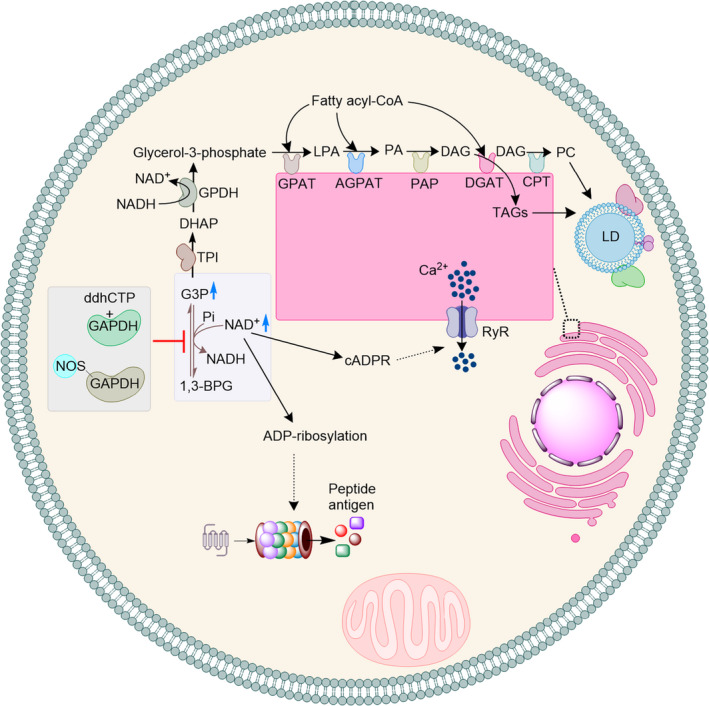
Inhibition of GAPDH increases the intracellular availability (blue arrow) of glyceraldehyde 3‐phosphate (G3P) and NAD+. An increase in the availability of G3P will support biosynthesis of TAGs and PC, which are the building blocks of lipid droplets (LDs). Increase in the availability of NAD+ will support synthesis of cADPR and ADP‐ribosylation. cADPR activates RyR receptor and induces release of Ca2+ from the cellular stores. ADP‐ribosylation can increase proteasomal activity and formation of peptide antigens.
Inhibition of GAPDH can also increase the cellular availability of G3P. Consistently, macrophages expressing RSAD2, which can produce ddhCTP, show a higher intracellular level of G3P as compared to RSAD2‐KO macrophages [25]. Increase in the cellular availability of G3P supports biosynthesis of triacylglycerols (TAGs) and phosphatidylcholine (PC), which are the building blocks of lipid droplets (LDs) [51, 52]. G3P is converted to dihydroxyacetone phosphate (DHAP) by the catalytic activity of triosephosphate isomerase (Fig. 3). Subsequently, DHAP is converted by the NADH‐dependent activity of glycerol‐3‐phosphate dehydrogenase (GPDH) to glycerol‐3‐phosphate (Fig. 3). Next, in a series of enzymatic reactions [52], which have been studied since early 1950s and are localized at the cytosolic face of the endoplasmic reticulum (ER), glycerol‐3‐phosphate and fatty acyl‐CoA are combined to generate TAGs and PC. Consistently, using 13C‐labelling experiments it was found that upon formation of classically activated macrophages (M1 macrophages), in which expression of RSAD2 is highly induced [53], formation of LD increases and the carbon for the synthesis of LDs originates from G3P [54].
Stimulation of proteasome activity by ADP‐ribosylation, cADPR‐dependent stimulation of Ca2+ release from the cellular stores, and an increase in biosynthesis of TAGs and PC, is likely to support a systemic immune response in cells in at least four ways (Fig. 4):
Fig. 4.
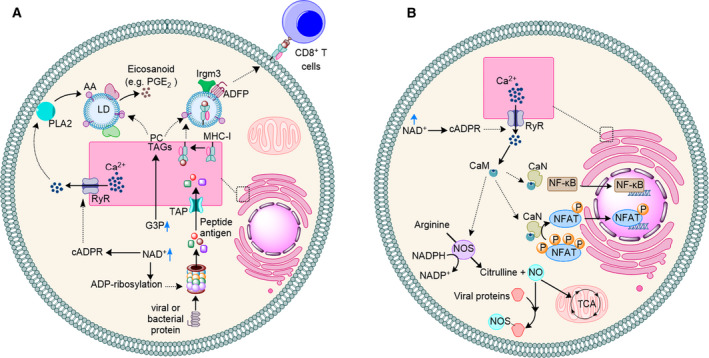
Inhibition of GAPDH and increase in the cellular availability of G3P and NAD+ support a broad‐spectrum immune response via at least four mechanisms. (a) Increase in the cellular availability of G3P and NAD+ supports eicosanoid synthesis and antigen cross‐presentation via MHC‐I. (b) Increase in the cellular availability of NAD+ will induce release of Ca2+ from the cellular stores and CaM‐dependent activation of NFAT and NF‐κB. On the other hand, CaM induces synthesis of NO by iNOS, which promotes S‐nitrosylation of viral proteins to restrict viral replication in immune cells like macrophages or modulates TCA cycle to induce formation of inflammatory macrophages.
Eicosanoids storm: Eicosanoids have a wide range of functions in inflammation and immune response to pathogens. Overall, available data suggest that eicosanoids can have both pro‐inflammatory and anti‐inflammatory activities depending on context and thus, may contribute to a balanced immune response [55]. It has been established that LDs are not just fat‐storing organelles and that they are important mediators of the innate immune response to pathogens [56]. LDs are shown to be a site for biosynthesis of eicosanoids [57]. Formation of eicosanoids occurs via a complex and highly regulated process [57] starting with liberation of arachidonic acid (AA) from phospholipids by Ca2+‐dependent phospholipases (PL)A2 [58, 59]. In different cells including innate immune cells like macrophages, the LDs are rich in AA [60, 61, 62, 63, 64]. Characterization of lipid droplets in different cells has revealed that enzymes involved in catalytic conversion of AA to eicosanoid like PGE2, namely cyclooxygenase 1 and 2 (COX1 and COX2), are localized to the LDs [65]. These data strongly suggest that LDs are at least partially involved in synthesis of inflammatory eicosanoids from AA and their downstream signalling pathways. Inhibition of GAPDH is likely to support LDs‐mediated eicosanoids biosynthesis in at least two ways: (i) increase in the cellular availability of G3P for biosynthesis of TAGs and PC and formation of LDs (Fig. 4A) and (ii) increase in the cellular availability of NAD+ and induction of cADPR‐dependent release of Ca2+ from cellular stores to support activity of the Ca2+‐dependent phospholipases PLA2 and liberation of AA (Fig. 4A).
Antigen cross‐presentation via major histocompatibility class I: This process requires proteasomal activity and LDs. An increase in proteasomal activity increases the rate of formation of peptide antigens for cross‐presentation via major histocompatibility class I (MHC‐I) [66, 67, 68]. In many cell types including macrophages and DCs, a major path of antigen cross‐presentation involves transfer of peptide antigen into the endoplasmic reticulum (ER) lumen by the ATP‐dependent function of the TAP system (Fig. 4A) [69, 70]. In the ER lumen, peptide antigens bind to MHC‐1 and the complex is transported to the cell surface for presentation to CD8+ T cells. A mechanism of transportation to the cell surface is through LDs [56]. In dendritic cells (DCs), the immune‐related GTPs protein, namely Irgm3, localizes to the LDs [71]. When the gene expressing Irgm3 or adipose differentiation‐related protein (ADRP, also known as ADFP), which regulates LD biogenesis and dynamics [72, 73], was inactivated, formation of LDs was impaired and cross‐presentation of antigen to CD8+ T cells was abrogated [71]. Additionally, saponin‐based adjuvants (SBAs), which are used in cancer vaccines, induce formation of LDs in CD11b+ DCs and this increase causes a saponin‐dependent increase in antigen cross‐presentation and T‐cell activation [74]. Therefore, a concomitant increase in the cellular availability of G3P and NAD+ due to inhibition of GAPDH will ensure formation of LDs as carriers of the MHC‐I/antigen complex, and increase proteasomal activity via an ADP‐ribosylation pathway to provide the peptide antigens (Fig. 4A). Consistent with this mechanism, the cellular activity of RSAD2 (viperin), which generates the ddhCTP metabolite and inhibits GAPDH, stimulates degradation of Zika virus and tick‐borne encephalitis virus nonstructural protein NS3 via a proteasome‐dependent manner [75].
NFAT‐ and NF‐κB‐mediated immune regulation. In innate immune cells like macrophages or T cells release of Ca2+ activates a range of immune defence mechanisms (Fig. 3B). Ca2+ binds to calmodulin (CaM) and the complex activates the phosphatase calcineurin (CaN) [76, 77]. In turn, CaN dephosphorylates and activates nuclear factor of activated T cells (NFAT) [78]. Additionally, CaN plays a role in LPS‐induced nuclear factor‐κB (NF‐κB) activation in macrophages [79, 80, 81]. NFAT‐ and NF‐κB regulate expression of several genes involved in immune cell response and function including IL‐10, IL‐6, IL‐8, IFN‐1, IFN‐2, TNF‐α and multiple TLR‐inducible genes including iNOS [82, 83, 84, 85, 86, 87, 88, 89, 90]. Thus, inhibition of GAPDH by ddhCTP or NO, and the likely increase in the cellular availability of NAD+, will modulate NFAT‐ or NF‐κB‐dependent expression of inflammatory genes. In summary, inhibition of GAPDH and an increase in the cellular level of NAD+ are likely to induce stimulation of cADPR and release of Ca2+. This will modulate activity of NFAT‐ and NF‐κB for a balanced and effective antiviral immune response. There is growing evidence in support of this mechanism. Firstly, when RSAD2 (viperin) gene was knocked out, thereby abrogating the inhibition of GAPDH by ddhCTP, the mRNA level of genes whose expression is regulated by NFAT or NF‐κB including iNOS and TNF‐α was affected in macrophages [91]. Secondly, NFAT and NF‐κB regulate Th2 response [92] and cellular activity of RSAD2 modulates activity of NF‐κB and AP1, which interact with NFAT, for optimal Th2 response [93]. Finally, overexpression of viperin upregulates expression of a wide range of immune‐related genes including IL‐8, IFN‐1 and IFN‐2 [27], whose expression is regulated by NFAT.
Nitric oxide and immune response. In many cells, nitric oxide (NO) is an important product of the innate immune response and has broad‐spectrum antiviral and antibacterial activity [94, 95, 96, 97]. It can contribute to viral restriction via different mechanisms. NO causes S‐nitrosylation of different viral proteins, abolishes their activity and reduces viral replication [98] (Fig. 4B). Viral components, including proteases [99, 100, 101, 102], RNA‐dependent RNA polymerases (RdRps) [103], and transcriptase [104], are known to be inhibited by NO‐mediated S‐nitrosylation. Replication of a number of viruses is restricted by NO including herpes simplex virus type 1 [105], Japanese encephalitis virus [106], coxsackievirus [101], dengue virus type 2 (DNGV‐2) [103], influenza virus [107] and HIV‐1 [102, 104]. Additionally, NO can modulate mitochondrial metabolism to induce formation of inflammatory macrophages [108]. Therefore, inhibition of GAPDH and an increase in the cellular availability of NAD+, which will induce cADPR‐dependent Ca2+ release, will induce activity of iNOS via CaM binding (Fig. 4B). The resulting NO can support a broad‐spectrum antiviral response via S‐nitrosylation of viral protein or by further modulating the metabolism in immune cells like macrophages.
Concluding remarks
In summary, we demonstrate that the glycolytic and housekeeping enzyme GAPDH is inhibited or modified by the metabolites, namely ddhCTP and NO, produced by two ISG protein products, RSAD2 and nitric oxide synthase, respectively. Inhibition of the NAD+‐dependent conversion of G3P by GAPDH supports several downstream metabolic and signalling pathways, specifically biosynthesis of TAGs and PC, which are precursors of LDs, protein ADP‐ribosylation and synthesis of cADPR. Together, these metabolites stimulate a balanced immune response via inflammatory eicosanoids, antigen cross‐presentation, activation of NFAT and NF‐κB and stimulation of formation of NO. This immunometabolic regulation of central carbon metabolism to stimulate a broad‐spectrum immune response provides an explanation for the wide range of effects observed due to expression of RSAD2 (viperin) in many cell types: these include the broad‐spectrum antiviral response [19], optimal Th2 cytokine production [93], which requires NFAT function [109], modulation of cellular lipid metabolism during human cytomegalovirus and influenza virus infections [110, 111], induction of type‐1 interferon production in plasmacytoid dendritic cells via a Toll‐like receptor‐mediated mechanism [112], interference with glucose homeostasis [26] and regulation of macrophage polarization [91].
Our analyses suggest that inhibition of GAPDH by the cellular innate immune response primes a broad‐spectrum immune response to viral infection. This is in opposed to recent reports [113, 114] suggesting that inhibition of GAPDH reduces immune response and thus, is a potential therapeutic approach for treating inflammatory diseases. These studies are based on use of small molecules such as the drug dimethyl fumarate (DMF) [113], which is used to treat autoimmune diseases, or 4‐octyl itaconate [114]. These molecules were suggested to directly modify Cys150 or Cys22, respectively, in GAPDH, and inhibit its activity. This inhibition was linked to a reduction in synthesis of inflammatory cytokines such as TNF‐α in T cells and macrophages with the assumption that no other protein in the cell was modified [113, 114]. In contrast to this assumption, analysis of global proteome in T cells reveals more than 2400 cysteine residues that could potentially be modified by DMF [115]. It was shown that two cysteine residues in protein kinase Cθ are target of modification by DMF and these modifications interfere with T‐cell activation [115].
Because the cellular innate immune response adopts mechanism that leads to inhibition of GAPDH, which as we discussed is likely to induce a broad‐spectrum immune response, we propose that in individuals with weakened cellular innate immune system inhibition of GAPDH might be a therapeutic approach to help prime the innate immune response via at least four mechanisms: (i) supporting formation of eicosanoids, (ii) assisting antigen cross‐presentation via MHC‐I, (iii) mediating immune response via NFAT and NF‐κB and (iv) stimulating synthesis of NO. Hence, we speculate that inhibition of GAPDH might help in the treatment of infection with viruses such as SARS‐CoV‐2. Future works should test the validity of this proposal.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
KHE conceived the study and wrote the manuscript with contribution from all the authors.
Acknowledgements
Due to limitations, we have cited most relevant studies. We apologize to all our colleagues whose work we have not been able to cite. KHE is grateful to Professor Fraser Armstrong (University of Oxford, UK) and Professor Wilfred R. Hagen (TU Delft, the Netherlands) for generous support of his scientific endeavour and to the European Molecular Biology Organization (EMBO). The research on RSAD2 (viperin) was supported by an EMBO long‐term fellowship to Dr Kourosh H. Ebrahimi (ALTF 157‐2015) and an EPA Abraham Research Fund (RF259) to Professor William S. James and Dr Kourosh H. Ebrahimi. The research in the group of Professor James McCullagh is supported by Biotechnology and Biological Sciences Research Council (BBSRC) (BB/R013829/1) and Wellcome Institutional Strategic Support Fund (204826/Z/16/Z).
References
- 1.Noor E, Eden E, Milo R & Alon U (2010) Central carbon metabolism as a minimal biochemical walk between precursors for biomass and energy. Mol Cell 39, 809–820. [DOI] [PubMed] [Google Scholar]
- 2.Warburg O (1956) On the origin of cancer cells. Science (80‐ ) 123, 309–314. [DOI] [PubMed] [Google Scholar]
- 3.Schoggins JW & Rice CM (2011) Available online at www.sciencedirect.comInterferon‐stimulated genes and their antiviral effector functions. Curr Opin Virol 1, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider WJ, Chevillotte MD & Rice CM (2014) Interferon‐stimulated genes: a complex web of host defenses. Annu Rev Immunol 32, 513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer‐Siegler K, Mauro DJ, Seal G, Wurzer J, DeRiel JK & Sirover MA (1991) A human nuclear uracil DNA glycosylase is the 37‐kDa subunit of glyceraldehyde‐3‐phosphate dehydrogenase. Proc Natl Acad Sci 88, 8460–8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zala D, Hinchelmann MV, Yu H, da Gunha MML, Liot G, Cordelieres FP, Marco S & Saudou F (2013) Vesicular glycolysis provides on‐board energy for fast axonal transport. Cell 152, 479–491. [DOI] [PubMed] [Google Scholar]
- 7.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LDet al, (2005) S‐nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol 7, 665–674. [DOI] [PubMed] [Google Scholar]
- 8.Shestov AA, Liu X, Ser Z, Cluntun AA, Hung YP, Huang L, Kim D, Le A, Yellen G, Albeck JG & et al, (2014) Quantitative determinants of aerobic glycolysis identify flux through the enzyme GAPDH as a limiting step. Elife 3, e03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galvan‐Pena S, Carroll RG, Newman C, Hinchy EC, Palsson‐McDermott E, Robinson EK, Covarrubias S, Nadin A, James AM, Haneklaus Met al, (2019) Malonylation of GAPDH is an inflammatory signal in macrophages. Nat Commun 10, 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duschene KS & Broderick JB (2010) The antiviral protein viperin is a radical SAM enzyme. FEBS Lett 584, 1263–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin K‐C & Cresswell P (2001) Viperin (cig5), an IFN‐inducible antiviral protein directly induced by human cytomegalovirus. Proc Natl Acad Sci USA 98, 15125–15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Severa M, Coccia EM & Fitzgerald KA (2006) Toll‐like receptor‐dependent and ‐independent viperin gene expression and counter‐regulation by PRDI‐binding factor‐1/BLIMP1. J Biol Chem 281, 26188–26195. [DOI] [PubMed] [Google Scholar]
- 13.Mikulecky P, Andreeva E, Amara P, Weissenhorn W, Nicolet Y & Macheboeuf P (2018) Human viperin catalyzes the modification of GPP and FPP potentially affecting cholesterol synthesis. FEBS Lett 592, 199–208. [DOI] [PubMed] [Google Scholar]
- 14.Seo J‐Y, Yaneva R & Cresswell P (2011) Viperin: a multifunctional, interferon‐inducible protein that regulates virus replication. Cell Host Microbe 10, 534–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helbig KJ & Beard MR (2014) The role of viperin in the innate antiviral response. J Mol Biol 426, 1210–1219. [DOI] [PubMed] [Google Scholar]
- 16.Seo J‐Y, Yaneva R, Hinson ER & Cresswell P (2011) Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science (80‐ ) 332, 1093–1097. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL & Hartmann R (2007) Type III interferon (IFN) induces a type I IFN‐like response in a restricted subset of cells through signaling pathways involving both the Jak‐STAT pathway and the mitogen‐activated protein kinases. J Virol 81, 7749–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang C, Huang M, Xiong T, Rong F, Li L, Liu DX & Chen RA (2020) Transcriptomic analysis and functional characterization reveal the duck interferon regulatory factor 1 as an important restriction factor in the replication of Tembusu virus. Front Microbiol 11, 2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh S & Marsh ENG (2020) Viperin: an ancient radical‐SAM enzyme finds its place in modern cellular metabolism and innate immunity. J Biol Chem. 10.1074/jbc.REV120.012784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattijssen S & Pruijn GJM (2012) Viperin, a key player in the antiviral response. Microbes Infect 14, 419–426. [DOI] [PubMed] [Google Scholar]
- 21.Honarmand Ebrahimi K (2018) A unifying view of the broad‐spectrum antiviral activity of RSAD2 (viperin) based on its radical‐SAM chemistry. Metallomics 10, 539–552. [DOI] [PubMed] [Google Scholar]
- 22.Gizzi AS, Grove TL, Arnold JJ, Jose J, Jangra RK, Garforth SJ, Du Q, Cahill SM, Dulyaninova NG, Love JDet al, (2018) A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature 558, 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenwick MK, Su D, Dong M, Lin H & Ealick SE (2020) Structural basis of the substrate selectivity of viperin. Biochemistry 59, 652–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honarmand Ebrahimi K, Rowbotham J, McCullagh J & James WS (2020) Mechanism of diol dehydration by a promiscuous radical‐SAM enzyme homologue of the antiviral enzyme viperin (RSAD2). ChemBioChem 11, 1605–1612. [DOI] [PubMed] [Google Scholar]
- 25.Ebrahimi KH, Vowles J, Browne C, McCullagh J & James WS (2020) ddhCTP produced by the radical‐SAM activity of RSAD2 (viperin) inhibits the NAD+‐dependent activity of enzymes to modulate metabolism. FEBS Lett 10, 1631–1644. [DOI] [PubMed] [Google Scholar]
- 26.Qi Z, Xia J, Xue X, Liu J, Liu W & Zhuzhe D (2017) Targeting viperin improves diet‐induced glucose intolerance but not adipose tissue inflammation. Oncotarget 8, 101418–101436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang B, Zhang J, Xiao ZZ & Sun L (2014) Rock bream (Oplegnathus fasciatus) viperin is a virus‐responsive protein that modulates innate immunity and promotes resistance against megalocytivirus infection. Developmental Comp Immunol 45, 35–42. [DOI] [PubMed] [Google Scholar]
- 28.Honarmand Ebrahimi K, Howie D, Rowbotham J, McCullagh J, Armstrong F & James WS (2020) Viperin, through its radical‐SAM activity, depletes cellular nucleotide pools and interferes with mitochondrial metabolism to inhibit viral replication. FEBS Lett 10, 1624–1630. [DOI] [PubMed] [Google Scholar]
- 29.Chakravortty D & Hensel M (2003) Inducible nitric oxide synthase and control of intracellular bacterial pathogens. Microbes Infect 5, 621–627. [DOI] [PubMed] [Google Scholar]
- 30.Akaike T & Maeda H (2000) Nitric oxide and virus infection. Immunology 101, 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forstermann U, Schmidt HHHW, Pollock JS, Sheng H, Mitchell JA, Warner TD, Nakane M & Murad F (1991) Isoforms of nitric oxide synthase characterization and purification from different cell types. Biochem Pharmacol 42, 1849–1857. [DOI] [PubMed] [Google Scholar]
- 32.Forstermann U, Gath I, Schwarz P, Closs EI & Kleinert H (1995) Isoforms of nitric oxide synthase: Properties, cellular distribution and expressional control. Biochem Pharmacol 50, 1321–1332. [DOI] [PubMed] [Google Scholar]
- 33.Rosen GM, Tsai P & Pou S (2002) Mechanism of free‐radical generation by nitric oxide synthase. Chem Rev 102, 1191–1200. [DOI] [PubMed] [Google Scholar]
- 34.Vedia LMY, McDonald B, Reep B, Brune B, Silvio MD, Billiar TR & Lapetina EG (1992) Nitric oxide‐induced S‐nitrosylation of glyceraldehyde‐3‐phosphate dehydrogenase inhibits enzymatic activity and increases endogenous ADP‐ribosylation. J Biol Chem 267, 24929–24932. [PubMed] [Google Scholar]
- 35.Dimmeler S, Lottspeich F & Brune B (1992) Nitric oxide causes ADP‐ribosylation and inhibition of glyceraldehyde‐3‐phosphate dehydrogenase. J Biol Chem 267, 16771–16774. [PubMed] [Google Scholar]
- 36.Mohr S, Hallak H, de Boitte A, Lapetina EG & Brune B (1999) Nitric oxide‐induced S‐glutathionylation and inactivation of glyceraldehyde‐3‐phosphate dehydrogenase. J Biol Chem 274, 9427–9430. [DOI] [PubMed] [Google Scholar]
- 37.Mohr S, Stamler JS & Brune B (1994) Mechanism of covalent modification of glyceraldehyde‐3‐phosphate dehydrogenase at its active site thiol by nitric oxide, peroxynitrite and related nitrosating agents. FEBS Lett 348, 223–227. [DOI] [PubMed] [Google Scholar]
- 38.Ishii T, Sunami O, Nakajima H, Nishio H, Takeuchi T & Hata F (1999) Critical role of sulfenic acid formation of thiols in the inactivation of glyceraldehyde‐3‐phosphate dehydrogenase by nitric oxide. Biochem Pharmacol 58, 133–143. [DOI] [PubMed] [Google Scholar]
- 39.Kim H, Jacobson EL & Jacobson MK (1993) Synthesis and degradation of cyclic ADP‐ribose by NAD glycohydrolases. Science (80‐ ) 261, 1330–1333. [DOI] [PubMed] [Google Scholar]
- 40.Lee HC (1994) Cyclic ADP‐ribose: a calcium mobilizing metabolite of NAD+. Mol Cell Biochem 138, 229–235. [DOI] [PubMed] [Google Scholar]
- 41.Ullrich O, Ciftci O & Hass R (2000) Proteasome activation by poly‐ADP‐ribose‐polymerase in human myelomonocytic cells after oxidative stress. Free Radic Biol Med 29, 995–1004. [DOI] [PubMed] [Google Scholar]
- 42.Ullrich O, Reinheckel T, Sitte N, Hass R, Grune T & Davies KJA (1999) Poly‐ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones. Proc Natl Acad Sci USA 96, 6223–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berridge MJ, Bootman MD & Roderick HL (2003) Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Cell Biol 4, 517–529. [DOI] [PubMed] [Google Scholar]
- 44.Krebs J, Agellon LB & Michalak M (2015) Ca2+ homeostasis and endoplasmic reticulum (ER) stress: an integrated view of calcium signaling. Biochem Biophys Res Commun 460, 114–121. [DOI] [PubMed] [Google Scholar]
- 45.Trebak M & Kinet JP (2019) Calcium signalling in T cells. Nat Rev Immunol 19, 154–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hosoi E, Nishizaki C, Gallgher KL, Wyre HW, Matsuo Y & Sei Y (2001) Expression of the Ryanodine receptor isoforms in immune cells. J Immunol 167, 4887–4894. [DOI] [PubMed] [Google Scholar]
- 47.Meszaros LG, Bak J & Chu A (1993) Cyclic ADP‐ribose as an endogenous regulator of the non‐skeletal type ryanodine receptor Ca2+ channel. Nature 364, 76–79. [DOI] [PubMed] [Google Scholar]
- 48.Sonnleitner A, Conti A, Bertocchini F, Schindler H & Sorrentino V (1998) Functional properties of the ryanodine receptor type 3 (RyR3) Ca2+ release channel. EMBO J 17, 2790–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee HC (2012) Cyclic ADP‐ribose and Nicotinic Acid Adenine Dinucleotide Phosphate (NAADP) as messengers for calcium mobilization. J Biol Chem 287, 31633–31640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magnone M, Bauer I, Poggi A, Mannino E, Sturla L, Brini M, Zocchi E, De Flora A, Nencioni A & Bruzzone S (2012) NAD+ levels control Ca2+ store replenishment and mitogen‐induced increase of cytosolic Ca2+ by cyclic ADP‐ribose‐dependent TRPM2 channel gating in human T lymphocytes. J Biol Chem 287, 21067–21081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pol A, Gross SP & Parton RG (2013) Biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J Cell Biol 204, 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo Y, Cordes KR, Farese RV & Walther TC (2009) Lipid droplets at a glance. J Cell Sci 122, 749–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satoh JI (2018) Gene expression profiles of M1 and M2 microglia characterized by comparative analysis of public datasets. Neuroimmunology 9, 124–138. [Google Scholar]
- 54.Rossa‐Ballina M, Guan XL, Schmidt A & Bumann D (2020) Classical activation of macrophages leads to lipid droplet formation without de novo fatty acid synthesis. Front Immunol 11, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dennis EA & Norris PC (2015) Eicosanoid storm in infection and inflammation. Nat Rev Immunol 15, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saka HA & Valdivia R (2012) Emerging roles for lipid droplets in immunity and host‐pathogen interactions. Annu Rev Cell Dev Biol 28, 411–437. [DOI] [PubMed] [Google Scholar]
- 57.Jarc E & Petan T (2020) A twist of FATe: Lipid droplets and inflammatory lipid mediators. Biochimie 169, 69–87. [DOI] [PubMed] [Google Scholar]
- 58.Balsinde J, Barbour SE, Bianco ID & Dennis EA (1994) Arachidonic acid mobilization in P388D1 macrophages is controlled by two distinct Ca(2+)‐dependent phospholipase A2 enzymes. Proc Natl Acad Sci USA 91, 11060–11064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Channon JY & Leslie CC (1990) A calcium‐dependent mechanism for associating a soluble arachidonoyl‐hydrolyzing phospholipase A2 with membrane in the macrophage cell line RAW 264.7. J Biol Chem 265, 5409–5413. [PubMed] [Google Scholar]
- 60.Dvorak AM, Dvorak SP, Shulman ES, MacGlashan DW Jr, Pyne K, Harvey VS, Galli SJ & Lichtenstein LM (1983) Lipid bodies: cytoplasmic organelles important to arachidonate metabolism in macrophages and mast cells. J Immunol 131, 2965–2976. [PubMed] [Google Scholar]
- 61.Weller PF, Monahan‐Earley RA, Dvorak HF & Dvorak AM (1991) Cytoplasmic lipid bodies of human eosinophils. Subcellular isolation and analysis of arachidonate incorporation. Am J Pathol 138, 141–148. [PMC free article] [PubMed] [Google Scholar]
- 62.Dichlberger A, Schlager S, Kovanen PT & Schneider WJ (2016) Lipid droplets in activated mast cells – a significant source of triglyceride‐derived arachidonic acid for eicosanoid production. Eur J Pharmacol 785, 59–69. [DOI] [PubMed] [Google Scholar]
- 63.Pacia MZ, Majzner K, Czamara K, Sternak M, Chlopicki S & Baranska M (2020) Estimation of the content of lipids composing endothelial lipid droplets based on Raman imaging. Biochim Biophys Acta (BBA)‐Molecular Cell Biol Lipids 1865, 158758. [DOI] [PubMed] [Google Scholar]
- 64.Stiebing C, Matthaus C, Krafft C, Keller AA, Weber K, Lorkowski S & Popp J (2014) Complexity of fatty acid distribution inside human macrophages on single cell level using Raman micro‐spectroscopy. Anal Bioanal Chem 406, 7037–7046. [DOI] [PubMed] [Google Scholar]
- 65.Weller PF (2016) Leukocyte lipid bodies‐structure and function as “eicosasomes”. Trans Am Clin Climatol Asoc 127, 328–340. [PMC free article] [PubMed] [Google Scholar]
- 66.Kloetzel PM (2004) The proteasome and MHC class I antigen processing. Biochim Biophys Acta 1695, 217–225. [DOI] [PubMed] [Google Scholar]
- 67.Sijts EJAM & Kloetzel P‐M (2011) The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell Mol Life Sci 68, 1491–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grant EP, Michalek MT, Goldberg AL & Rock KL (1995) Rate of antigen degradation by the ubiquitin‐proteasome pathway influences MHC class I presentation. J Immunol 155, 3750–3758. [PubMed] [Google Scholar]
- 69.Neefjes JJ, Momburg F & Hammerling GJ (1993) Selective and ATP‐dependent translocation of peptides by the MHC‐encoded transporter. Science (80‐ ) 261, 769–771. [DOI] [PubMed] [Google Scholar]
- 70.Karttumen JT, Trowsdale J & Lehner PJ (1999) Antigen presentation: TAP dances with ATP. Curr Biol 9, R820–R824. [DOI] [PubMed] [Google Scholar]
- 71.Bougneres L, Helft J, Sangeeta T, Vargas P, Chang BHJ, Chan L, Campisi L, Lauvau G, Hugues S, Kumar Pet al, (2009) A role for lipid bodies in the cross‐presentation of phagocytosed antigens by MHC class I in dendritic cells. Immunity 31, 232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brasaemle DL, Barber T, Wolins NE, Serrero G, Blanchette‐Mackie EJ & Londos C (1997) Adipose differentiation‐related protein is an ubiquitously expressed lipid storage droplet‐associated protein. J Lipid Res 38, 2249–2263. [PubMed] [Google Scholar]
- 73.Nakamura N & Fujimoto T (2003) Adipose differentiation‐related protein has two independent domains for targeting to lipid droplets. Biochem Biophys Res Commun 306, 333–338. [DOI] [PubMed] [Google Scholar]
- 74.den Brok MH, Bull C, Wassink M, de Graaf AM, Wagenaars JA, Minderman M, Thakur M, Amigorena S, Rijke EO, Schrier CC & et al, (2016) Saponin‐based adjuvants induce cross‐presentation in dendritic cells by intracellular lipid body formation. Nat Commun 7, 13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Panayiotou C, Lindqvist R, Kurhade C, Vonderstein K, Pasto J, Edlund K, Upadhyay AS & Overby AK (2018) Viperin restricts Zika virus and tick‐borne encephalitis virus replication by targeting NS3 for proteasomal degradation. J Virol 92, e02054–e2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klee CB, Crouch TH & Krinks MH (1979) Calcineurin: a calcium‐ and calmodulin‐binding protein of the nervous system. Proc Natl Acad Sci 76, 6270–6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feske S, Okamura H, Hogan PG & Rao A (2003) Ca2+/calcineurin signalling in cells of the immune system. Biochem Biophys Res Commun 311, 1117–1132. [DOI] [PubMed] [Google Scholar]
- 78.Jain J, McCafffrey PG, Miner Z, Kerppola TK, Lambert JN, Verdine GL, Gurran T & Rao A (1993) The T‐cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature 365, 352–355. [DOI] [PubMed] [Google Scholar]
- 79.Jeon S, Kim SH, Shin SY & Lee YH (2018) Clozapine reduces Toll‐like receptor 4/NF‐κB‐mediated inflammatory responses through inhibition of calcium/calmodulin‐dependent Akt activation in microglia. Prog Neuro‐Psychopharmacology Biol Psychiatry 81, 477–487. [DOI] [PubMed] [Google Scholar]
- 80.Kim YH, Moon JS, Lee KS, Park SY, Cheong J, Kang HS, Lee HY & Kim HD (2004) Ca2+/calmodulin‐dependent protein phosphatase calcineurin mediates the expression of iNOS through IKK and NF‐κB activity in LPS‐stimulated mouse peritoneal macrophages and RAW 264.7 cells. Biochem Biophys Res Commun 314, 695–703. [DOI] [PubMed] [Google Scholar]
- 81.Hsuan SL, Kannan MS, Jeyaseelan S, Prakash YS, Malazdrewich C, Abrahamsen MS, Sieck GC & Maheswaran SK (1999) Pasteurella haemolyticaleukotoxin and endotoxin induced cytokine gene expression in bovine alveolar macrophages requires NF‐κB activation and calcium elevation. Microb Pathog 26, 263–273. [DOI] [PubMed] [Google Scholar]
- 82.Macian F (2005) NFAT proteins: key regulators of T‐cell development and function. Nat Rev Immunol 5, 472–484. [DOI] [PubMed] [Google Scholar]
- 83.Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SFet al, (2006) FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell 126, 375–387. [DOI] [PubMed] [Google Scholar]
- 84.Trama J, Lu Q, Hawley RG & Ho SN (2000) The NFAT‐related protein NFATL1 (TonEBP/NFAT5) is induced upon T cell activation in a calcineurin‐dependent manner. J Immunol 165, 4884–4894. [DOI] [PubMed] [Google Scholar]
- 85.Fric J, Zelante T, Wong AYW, Mertes A, Yu HB & Ricciardi‐Castagnoli P (2012) NFAT control of innate immunity. Blood 120, 1380–1389. [DOI] [PubMed] [Google Scholar]
- 86.Mancini M & Toker A (2009) NFAT proteins: emerging roles in cancer progression. Nat Rev Cancer 9, 810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Crabtree GR & Schreiber SL (2009) SnapShot: Ca2+‐Calcineurin‐NFAT Signaling. Cell 138, 210–210.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Muller MR & Rao A (2010) NFAT, immunity and cancer: a transcription factor comes of age. Nat Rev Immunol 10, 645–656. [DOI] [PubMed] [Google Scholar]
- 89.Elloumi HZ, Maharshak N, Rao KN, Kobayashi T, Ryu HS, Muhlbauer M, Li F, Jobin C & Plevy SE (2012) A cell permeable peptide inhibitor of NFAT inhibits macrophage cytokine expression and ameliorates experimental colitis. PLoS One 7, e34172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu FX, Wu CL, Zhu ZA, Li MQ, Mao YQ, Liu M, Wang XQ, Yu DG & Tang TT (2013) Calcineurin/NFAT pathway mediates wear particle‐induced TNF‐α release and osteoclastogenesis from mice bone marrow macrophages in vitro. Acta Pharmacol Sin 34, 1457–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eom J, Yoo J, Kim JJ, Lee JB, Choi W, Park CG & Seo JY (2018) Viperin deficiency promotes polarization of macrophages and secretion of M1 and M2 cytokines. Immune Netw 18, e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ranger AM, Oukka M, Rengarajan J & Glimcher LH (1998) Inhibitory function of Two NFAT Family Members in Lymphoid Homeostasis and Th2 Development. Immunity 9, 627–635. [DOI] [PubMed] [Google Scholar]
- 93.Qiu L‐Q, Cresswell P & Chin K‐C (2009) Viperin is required for optimal Th2 responses and T‐cell receptor–mediated activation of NF‐kB and AP‐1. Blood 113, 3520–3529. [DOI] [PubMed] [Google Scholar]
- 94.Reiss CS & Komatsu T (1998) Does nitric oxide play a critical role in viral infections? J Virol 4547–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bogdan C, Rollinghoff M & Diefenbach A (2000) The role of nitric oxide in innate immunity. Immunol Rev 173, 17–26. [DOI] [PubMed] [Google Scholar]
- 96.Bogdan C (2015) Nitric oxide synthase in innate and adaptive immunity: an update. Trends Immunol 36, 161–178. [DOI] [PubMed] [Google Scholar]
- 97.Bogdan C (2001) Nitric oxide and the immune response. Nat Immunol 2, 907–916. [DOI] [PubMed] [Google Scholar]
- 98.Colasanti M, Persichini T, Venturini G & Ascenzi P (1999) S ‐Nitrosylation of viral proteins: molecular bases for antiviral effect of nitric oxide. IUBMB Life 48, 25–31. [DOI] [PubMed] [Google Scholar]
- 99.Badorff C, Fichtlscherer B, Rhoads RE, Zeiher AM, Muelsch A, Dimmeler S & Knowlton KU (2000) Nitric oxide inhibits dystrophin proteolysis by coxsackieviral protease 2A through S‐nitrosylation. Circulation 102, 2276–2281. [DOI] [PubMed] [Google Scholar]
- 100.Zell R, Markgraf R, Schmidtke M, Gorlach M, Stelzner A, Henke A, Sigusch HH & Gluck B (2004) Nitric oxide donors inhibit the coxsackievirus B3 proteinases 2A and 3C in vitro, virus production in cells, and signs of myocarditis in virus‐infected mice. Med Microbiol Immunol 193, 91–100. [DOI] [PubMed] [Google Scholar]
- 101.Saura M, Zaragoza C, McMillan A, Quick RA, Hohenadl C, Lowenstein JM & Lowenstein CJ (1999) An antiviral mechanism of nitric oxide: inhibition of a viral protease. Immunology 10, 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Persichini T, Colasanti M, Lauro GM & Ascenzi P (1998) Cysteine nitrosylation inactivates the HIV‐1 protease. Biochem Biophys Res Commun 250, 575–576. [DOI] [PubMed] [Google Scholar]
- 103.Tkhampunya R, Padmanabhan R & Ubol S (2006) Antiviral action of nitric oxide on dengue virus type 2 replication. J Gen Virol 87, 3003–3011. [DOI] [PubMed] [Google Scholar]
- 104.Persichini T, Colasanti M, Fraziano M, Colizzi V, Medana C, Polticelli F, Venturini G & Ascenzi P (1999) Nitric oxide inhibits the HIV‐1 reverse transcriptase activity. Biochem Biophys Res Commun 258, 624–627. [DOI] [PubMed] [Google Scholar]
- 105.Croen KD (1993) Evidence for antiviral effect of nitric oxide. Inhibition of herpes simplex virus type 1 replication. J Clin Invest 91, 2446–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lin YL, Huang YL, Ma SH, Yeh CT, Chiou SY, Chen LK & Liao CL (1997) Inhibition of Japanese encephalitis virus infection by nitric oxide: antiviral effect of nitric oxide on RNA virus replication. J Virol 71, 5227–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rimmelzwaan GF, Baars MMJW, de Lijster P, Fouchier RAM & Osterhaus ADME (1999) Inhibition of influenza virus replication by nitric oxide. J Virol 73, 8880–8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bailey JD, Diotallevi M, Nicol T, McNeill E, Shaw A, Chuaiphichai S, Hale A, Starr A, Nandi M, Stylianou Eet al, (2019) Nitric oxide modulates metabolic remodeling in inflammatory macrophages through TCA Cycle Regulation and Itaconate Accumulation. Cell Rep 28, 218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Trevillyan JM, Chiou XG, Chen YW, Ballaron SJ, Sheet MP, Smith ML, Wiedeman PE, Warrior U, Wilkins J, Gubbins EJet al, (2001) Potent inhibition of NFAT activation and T cell cytokine production by novel low molecular weight Pyrazole compounds. J Biol Chem 276, 48118–48126. [DOI] [PubMed] [Google Scholar]
- 110.Seo J‐Y & Cresswell P (2013) Viperin regulates cellular lipid metabolism during human cytomegalovirus infection. PloS Pathog 9, e1003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang X, Hinson ER & Cresswell P (2007) The interferon‐inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2, 96–105. [DOI] [PubMed] [Google Scholar]
- 112.Saitoh T, Satoh T, Yamamoto N, Uematsu S, Takeuchi O, Kawai T & Akira S (2011) Antiviral protein viperin promotes toll‐like receptor 7‐ and toll‐like receptor 9‐mediated type I interferon production in plasmacytoid dendritic cells. Immunity 34, 352–363. [DOI] [PubMed] [Google Scholar]
- 113.Kornberg MD, Bhargava P, Kim PM, Putluri V, Snowman AM, Putluri N, Calabresi PA & Snyder SH (2018) Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science (80‐ ) 360, 449–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liao ST, Han C, Xu DQ, Fu XW, Wang JS & Kong LY (2019) 4‐Octyl itaconate inhibits aerobic glycolysis by targeting GAPDH to exert anti‐inflammatory effects. Nat Commun 10, 5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Blewell MM, Xie J, Zaro BW, Backus KM, Altman A, Teijaro JR & Cravatt BF (2016) Chemical proteomic map of dimethyl fumarate–sensitive cysteines in primary human T cells. Sci Signal 9, rs10. [DOI] [PMC free article] [PubMed] [Google Scholar]


