Abstract
Dysoxic marine waters (DMW, < 1 μM oxygen) are currently expanding in volume in the oceans, which has biogeochemical, ecological and societal consequences on a global scale. In these environments, distinct bacteria drive an active sulfur cycle, which has only recently been recognized for open‐ocean DMW. This review summarizes the current knowledge on these sulfur‐cycling bacteria. Critical bottlenecks and questions for future research are specifically addressed. Sulfate‐reducing bacteria (SRB) are core members of DMW. However, their roles are not entirely clear, and they remain largely uncultured. We found support for their remarkable diversity and taxonomic novelty by mining metagenome‐assembled genomes from the Black Sea as model ecosystem. We highlight recent insights into the metabolism of key sulfur‐oxidizing SUP05 and Sulfurimonas bacteria, and discuss the probable involvement of uncultivated SAR324 and BS‐GSO2 bacteria in sulfur oxidation. Uncultivated Marinimicrobia bacteria with a presumed organoheterotrophic metabolism are abundant in DMW. Like SRB, they may use specific molybdoenzymes to conserve energy from the oxidation, reduction or disproportionation of sulfur cycle intermediates such as S0 and thiosulfate, produced from the oxidation of sulfide. We expect that tailored sampling methods and a renewed focus on cultivation will yield deeper insight into sulfur‐cycling bacteria in DMW.
Introduction
Oxygen deficiency is a rather common phenomenon in marine waters caused by microbial aerobic respiration coupled to the degradation of organic matter, combined with insufficient supply of oxygen through water circulation or diffusion (Canfield et al., 2005). Oxygen‐minimum zones (OMZs) are waters in the open ocean containing < 20 μM oxygen occurring between 100 and 1,500 m depth. The largest OMZs are found in the Eastern Tropical North Pacific (ETNP), the Eastern Tropical South Pacific (ETSP) and the Arabian Sea (Fig. 1, Table S1). Together, OMZs amount to 10 million km3 or approximately 1% of the ocean's volume (Paulmier and Ruiz‐Pino, 2009). When there is sufficient input of sinking phytoplankton biomass, oxygen concentrations in OMZs can drop to below the common detection level of 1 μM and were considered anoxic and described as ‘anoxic marine zones’ (Ulloa et al., 2012). However, using a highly sensitive STOX oxygen sensor, Revsbech and colleagues (2009) and Thamdrup and colleagues (2012) showed that the supposedly anoxic OMZ waters, which will here be termed ‘OMZ core’, still may contain traces of oxygen (< 50 nM). Yet, these sensors have not yet been widely applied in marine sampling campaigns. Coastal waters can similarly experience oxygen deficiency and anoxia, for example in the Namibian Upwelling, Chesapeake Bay and the Pacific South‐American coastal waters (Fig. 1, Table S1).
Fig 1.
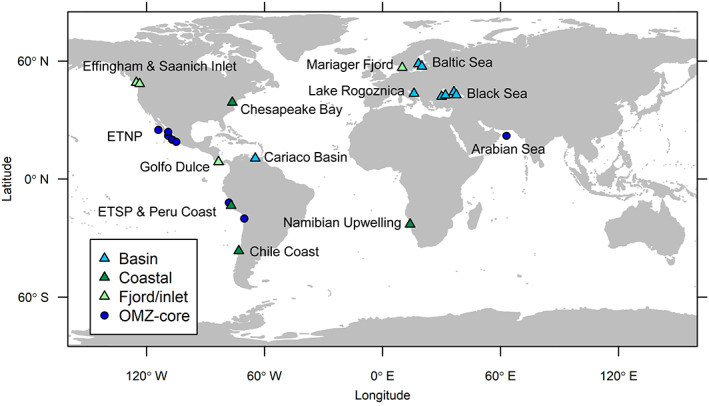
Dysoxic marine waters studied with respect to microorganisms driving the sulfur cycle. Triangles indicate locations that are permanently, seasonally or incidentally euxinic. For a list of studies per location, see Table S1.
ETNP, Eastern Tropical North Pacific; ETSP, Eastern Tropical South Pacific; OMZ, oxygen‐minimum zone. [Color figure can be viewed at wileyonlinelibrary.com]
In enclosed marine basins and fjords, as well as in coastal waters, stratification is a common factor in the development and persistence of oxygen deficiency (Canfield et al., 2005). Stratification can even lead to euxinia (Meyer and Kump, 2008), here defined as anoxic conditions with > 0.1 μM sulfide. Several euxinic marine basins, fjords and inlets have been studied with respect to their sulfur cycle and the associated microorganisms (Fig. 1, Table S1). The development of euxinia in OMZs is prevented by a relatively high advection of oxygenated water compared to enclosed environments, and by a negative feedback loop centered around nitrogen loss. Denitrifying and anaerobic ammonia‐oxidizing bacteria in the OMZ convert fixed forms of nitrogen such as ammonium and nitrate into N2 at such high rates that OMZs are responsible for 30% to 50% of the total loss of fixed nitrogen from the ocean (Lam and Kuypers, 2011). This causes surface phytoplankton to be limited in nitrogen, which in turn limits the input of organic matter to the OMZs, preventing the depletion of nitrate and nitrite (Canfield, 2006; Boyle et al., 2013). Hence, the ensuing development of euxinia is halted, as denitrifying bacteria outcompete sulfate‐reducing ones (Froelich et al., 1979; Chen et al., 2017).
OMZ core waters, despite being dominated by nitrogen cycling, can also harbor an active sulfur cycle, which has long been overlooked due to the absence of detectable sulfide (Canfield et al., 2010; Johnston et al., 2014; Carolan et al., 2015). Similar conditions are found in some stratified environments where the oxic and euxinic zones are separated by a suboxic zone (Murray et al., 1989; Lavik et al., 2009; Hawley et al., 2014; Findlay et al., 2017), here defined to contain no detectable oxygen (< 1 μM) or sulfide (< 0.1 μM) using standard methods. The exact concentration of oxygen in suboxic zones is still unclear. However, the detection of sulfur‐cycling microorganisms suggests an active sulfur cycle in suboxic zones, for instance in the Black Sea and Cariaco Basin (Neretin et al., 2007; Rodriguez‐Mora et al., 2016). For the purpose of this review, we use the term ‘dysoxic marine water’ (DMW, < 1 μM of oxygen) to describe all marine suboxic zones, OMZ core waters, anoxic waters and euxinic waters (Box 1).
Box 1. Glossary of definitions for marine oxygen‐deficient environments.
| Environmental terminology | Definition |
|---|---|
| Dysoxic water | < 1 μM oxygen |
| Anoxic water | Not containing oxygen |
| Euxinic water | Anoxic, > 0.1 μM sulfide |
| Oxygen‐minimum zone (OMZ) | Open ocean water with < 20 μM oxygen |
| OMZ core, also known as ‘anoxic marine zone’ | Open ocean water with < 50 nM oxygen |
| Suboxic zone | < 1 μM oxygen, < 0.1 μM sulfide, in between oxic and euxinic zones of stratified waters |
Over the last 60 years, DMW has expanded in volume more than fourfold (Schmidtko et al., 2017) because of oceanic warming – reducing oxygen solubility – and eutrophication (reviewed by Breitburg et al., 2018). This process is expected to continue. In addition, Ulloa and colleagues (2012) have predicted that the deposition of anthropogenically fixed nitrogen will cause OMZ cores to develop euxinia, since it counteracts the nitrogen‐loss‐based negative feedback loop. Potential long‐term, global consequences of expanding marine dysoxia and euxinia include changes in availability of key nutrients (iron, phosphorus, etc.) and trace metals (cadmium, copper, zinc, etc.), and loss of fishery stocks, affecting coastal economies and food security (Breitburg et al., 2018). Furthermore, since DMW environments are biogeochemical hotspots for microbial production of the greenhouse gas nitrous oxide (Naqvi et al., 2010), their expansion provides a feedback loop that in turn contributes to global warming. In the geological past, the rise of euxinic conditions has led to several mass extinction events such as during the end‐Permian (Meyer and Kump, 2008) and the mid‐Cretaceous (Kamyshny et al., 2009).
The biogeochemical sulfur cycle in DMW consists of abiotic and biologically mediated reactions (Fig. 2; Ehrlich et al., 2015), with the latter providing energy to many different microorganisms. Sulfate‐reducing bacteria (SRB) reduce sulfate () to sulfide (HS−), coupled to the oxidation of small organic compounds or H2 (Muyzer and Stams, 2008). Most of this sulfide is re‐oxidized by oxidized metals or sulfur‐oxidizing bacteria (SOB), either completely to sulfate (Jørgensen et al., 1991) or to different sulfur cycle intermediates (SCIs) including elemental sulfur (S0), polysulfides (), thiosulfate (), tetrathionate () and sulfite (; Zopfi et al., 2001; Kamyshny et al., 2011; Findlay, 2016). These SCIs can be used as electron donor or acceptor by various microorganisms including SRB and SOB (Rabus et al., 2013; Han and Perner, 2015; Dahl, 2017).
Fig 2.
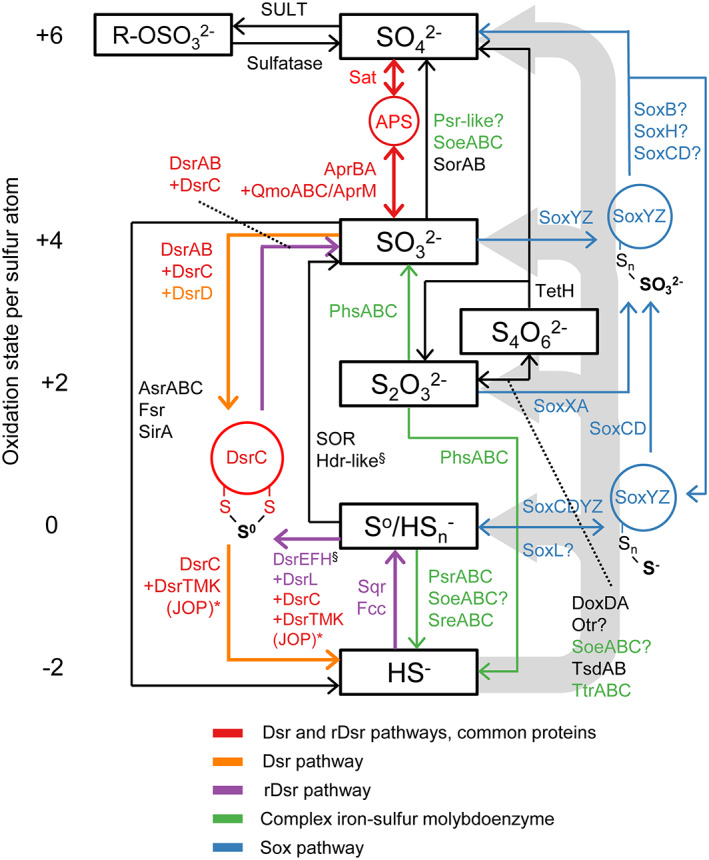
The dissimilatory conversions within the marine sulfur cycle. The oxidation state of the inorganic species is indicated at the left. Abiotic and assimilatory reactions are not indicated, except for the abiotic oxidation of sulfide which is illustrated by wide grey arrows. The S0 in DsrC‐trisulfide is considered zero‐valent (Santos et al., 2015). The sulfur atom in APS has an oxidation state of +6, and those in tetrathionate have an oxidation state of +2.5. A question mark symbol (?) shows that involvement is uncertain. The asterisk symbol (*) indicates that DsrT is required for sulfide oxidation in green sulfur bacteria (Holkenbrink et al., 2011), but is also found in SRB. Protein complexes other than DsrTMK(JOP) can also transfer electrons to DsrC to enable this reaction (Venceslau et al., 2014). The section symbol (§) indicates that the rhodanese sulfurtransferases Rhd‐TusA‐DsrE2 are also essential in the reaction mediated by this complex (Dahl, 2017).
Apr, APS reductase; Asr, anaerobic sulfite reductase; Dox, thiosulfate:quinone oxidoreductase; Dsr, dissimilatory sulfite reductase; Fcc, flavocytochrome c sulfide dehydrogenase; Fsr, F420‐dependent sulfite reductase; Hdr, heterodisulfide reductase; Otr, octaheme tetrathionate reductase; Phs, thiosulfate reductase; Psr, polysulfide reductase; Qmo, quinone‐interacting membrane‐bound oxidoreductase; Sat, sulfate adenylyltransferase; Sir, sulfite reductase; Soe, sulfite‐oxidizing enzyme; SOR, sulfur oxygenase/reductase; Sor, sulfite‐acceptor oxidoreductase; Sox, sulfur‐oxidizing multienzyme complex; Sqr, sulfide:quinone oxidoreductase; Sre, sulfur reductase; SULT, sulfotransferase; Tet, tetrathionate hydrolase; Tsd, thiosulfate dehydrogenase; Ttr, tetrathionate reductase. [Color figure can be viewed at wileyonlinelibrary.com]
The detection and analysis of sulfur‐cycling genes, transcripts and proteins in DMW yield a powerful perspective on the diversity and activity of sulfur‐cycling microorganisms (Fig. 2), and more so when applied to metagenome‐assembled genomes (MAGs) or single‐cell amplified genomes (SAGs). Various ‘omics’ studies have yielded insight into the dominant SOB in DMW (Lavik et al., 2009; Walsh et al., 2009; Callbeck et al., 2018; Plominsky et al., 2018). However, only few studies have addressed the broader diversity of sulfur‐cycling microorganisms (Canfield et al., 2010; Stewart et al., 2012; Schunck et al., 2013; Hawley et al., 2014), without tapping into the larger potential of genome‐centric metagenomics and the available metagenome data. To fill this knowledge gap, we screened MAGs from DMW environments for sulfur‐cycling marker genes (Supporting Information Methods). Part of the MAGs was assembled from metagenomes of the Arabian Sea and ETSP OMZ cores produced by Tara Oceans (Parks et al., 2017; Tully et al., 2018). Other MAGs were assembled from metagenomes of 15 different water depths of the Black Sea (Villanueva et al., 2020; Suominen et al., 2019; Supporting Information Methods), which served as model for enclosed DMW environments.
Despite the central role of the sulfur cycle in DMW, current biogeochemical and microbiological knowledge has not been comprehensively reviewed so far. Therefore, we herein provide an overview of sulfur cycle processes in DMW, and we discuss the diversity, metabolism and physiology of the bacteria involved in these processes. In the following section, we will discuss current knowledge on SRB, who form an essential part of the sulfur cycle through the production of sulfide. The subsequent section treats SOB, covering well‐studied groups such as SUP05 and Sulfurimonas, less explored groups such as BS‐GSO2, and putative sulfur oxidizers such as SAR324 members. The final section discusses which bacteria could be involved in sulfur reduction or disproportionation.
Sulfate‐reducing bacteria
The presence and activity of SRB in the dysoxic water column have been demonstrated through sulfate reduction rate measurements with isotopically labelled sulfate (35 ) performed in euxinic settings such as the Black Sea with sulfate reduction rates up to 36 nmol l−1 day−1 (Sorokin, 1972; Jørgensen et al., 1991; Albert et al., 1995; Pimenov et al., 2000) and Mariager Fjord with rates up to 140 nmol l−1 day−1 (Sørensen and Canfield, 2004), but also in the ETSP OMZ core with rates up to 16.9 nmol l−1 day−1 (Canfield et al., 2010). More extensively conducted taxonomic marker studies point to a universal presence of SRB in DMW since the 16S rRNA genes of canonical SRB lineages of the class Deltaproteobacteria have been widely detected (Madrid et al., 2001; Vetriani et al., 2003; Lin et al., 2006; Fuchsman et al., 2011; Wright et al., 2012; Schunck et al., 2013; Ganesh et al., 2014; Rodriguez‐Mora et al., 2015; Suter et al., 2018; Callbeck et al., 2019).
Presence and diversity
All known SRB reduce sulfate through the dissimilatory (bi)sulfite reductase (Dsr) pathway (Rabus et al., 2015). This consistency has facilitated functional marker investigations of the ecology of SRB. Such a functional marker approach is more appropriate than 16S rRNA gene surveys, as it does not require metabolic assumptions based on taxonomy. Although the core proteins of the Dsr pathway, Sat, AprBA and DsrAB, are also present in the reversed Dsr (rDsr) sulfur oxidation pathway (Dahl, 2017; Fig. 2), their reductive and oxidative versions are phylogenetically distinguishable (Meyer and Kuever, 2007; Loy et al., 2009; Müller et al., 2015; Pelikan et al., 2016). Reductive Dsr genes have been identified throughout suboxic and euxinic waters of the Black Sea (Neretin et al., 2007) and the Cariaco Basin (Rodriguez‐Mora et al., 2016), in sulfidic coastal waters off Peru (Schunck et al., 2013), in the core of the ETNP and ETSP OMZs (Canfield et al., 2010; Carolan et al., 2015), and in the Gdansk Deep within the Baltic Sea (Korneeva et al., 2015). Moreover, reductive Dsr genes were shown to be transcribed into mRNA (Stewart et al., 2012; Ulloa et al., 2012; Schunck et al., 2013; Rodriguez‐Mora et al., 2016; Saunders et al., 2019), providing evidence for activity of SRB. Although powerful, the results of Dsr markers should be cautiously interpreted (Anantharaman et al., 2018), as the Dsr pathway does not only facilitate dissimilatory sulfate reduction but can also mediate dissimilatory reduction and disproportionation of SCIs (Rabus et al., 2015; Florentino et al., 2017), and in some rare cases sulfur oxidation (Sigalevich and Cohen, 2000; Slobodkina et al., 2017; Thorup et al., 2017). Thus, although bacteria with reductive Dsr pathways are core members of the microbial community of DMW, their sulfur metabolism is not necessarily restricted to dissimilatory sulfate reduction. We therefore refer to them as putative SRB.
Surveys based on 16S rRNA or functional marker genes and metagenomic studies of DMW have revealed a high diversity of putative SRB, most of which are only distantly related to described species. The deltaproteobacterial putative SRB detected in 16S rRNA gene data sets are rarely affiliated with established genera (Fuchsman et al., 2011; Wright et al., 2012; Ganesh et al., 2014; Rodriguez‐Mora et al., 2015; Suter et al., 2018). Of all canonical SRB lineages, Desulfobacteraceae species are thought to be dominant due to the prevalence of their sequences in 16S rRNA data sets (Fuchsman et al., 2011; Wright et al., 2012; Rodriguez‐Mora et al., 2015; Suter et al., 2018) and metagenomic data sets (Canfield et al., 2010; Schunck et al., 2013). However, Desulfobulbaceae species have also been detected, specifically including the 16S rRNA genes of the genera Desulfocapsa and Desulforhopalus (Neretin et al., 2007; Canfield et al., 2010; Fuchsman et al., 2011; Fuchsman et al., 2012; Rodriguez‐Mora et al., 2015; Suter et al., 2018). Furthermore, bacteria related to the genus Desulfatiglans seem widespread in DMW since Desulfatiglans‐related sequences were retrieved from the Black Sea (16S rRNA genes; Vetriani et al., 2003; Neretin et al., 2007), coastal DMW off Peru (metagenomics; Schunck et al., 2013), the Gdansk Deep in the Baltic Sea (dsrB fragments; Korneeva et al., 2015) and the Cariaco Basin (dsrA fragments; Rodriguez‐Mora et al., 2016). Functional marker gene surveys indicated an even larger diversity of putative SRB beyond the Deltaproteobacteria, including Thermodesulfovibrio‐related bacteria in the ETSP OMZ core (Canfield et al., 2010) and diverse unknown putative SRB in euxinic basins (Korneeva et al., 2015; Rodriguez‐Mora et al., 2016).
The dsrD gene, encoding a small protein with a possible regulatory function (Mizuno et al., 2003; Venceslau et al., 2014), is an alternative functional marker gene for detection of SRB (Mussmann et al., 2005). It has been used to investigate bacterial genomes with dsrAB genes lacking a clear oxidative/reductive affiliation (Anantharaman et al., 2018), since dsrD forms a reliable marker for the reductive Dsr pathway when present together with other Dsr genes (Rabus et al., 2015). We detected the dsrD gene – in the context of other dsr genes – in metagenomes and MAGs from the Black Sea throughout the euxinic and suboxic zones (Fig. 3A,B), confirming previously reported distributions of putative SRB (Neretin et al., 2007). We could not obtain any assembled reductive dsrA genes from publicly available OMZ metagenomes (Canfield et al., 2010; Ganesh et al., 2014; Fuchsman et al., 2017; Tully et al., 2018; Saunders et al., 2019), likely due to sampling bias (see following subsection) and insufficient sequencing depth. However, many complete dsrA genes could be retrieved from the Black Sea metagenome (Supporting Information Methods). An analysis of these dsrA sequences supported the view emerging from previous studies: a large diversity of putative SRB, with a somewhat distant relationship to canonical SRB belonging to Desulfobacula, Desulfococcus, Desulfocapsa, Desulfatiglans and Thermodesulfovibrio, and to non‐canonical lineages other than the Deltaproteobacteria or Nitrospirae (Fig. 4). This view mirrors the overly large diversity of SRB that can generally be found in marine sediments (Muyzer and Stams, 2008; Müller et al., 2015). Despite the wealth of knowledge on the metabolism of SRB, the ecophysiological causes behind this diversity are currently poorly understood.
Fig 3.
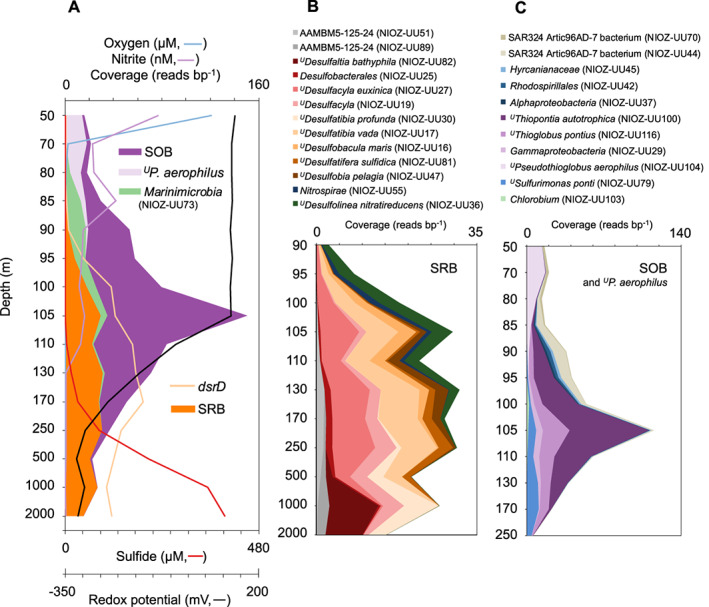
Black Sea water column distribution of metagenome‐assembled genomes (MAGs) of sulfur‐cycling bacteria based on their genetic capacity.
A. Physicochemical measurements and normalized cumulative metagenome coverage of all MAGs of putative sulfur‐oxidizing bacteria (SOB) combined, U P. aerophilus (NIOZ‐UU104), Marinimicrobia (NIOZ‐UU73), all dsrD genes combined and all MAGs of putative sulfate‐reducing bacteria (SRB) combined in samples of 15 different depths of the Black Sea. The oxygen, nitrite and sulfide data correspond to the PHOXY cruise of June–July 2013 (Sollai et al., 2019). The Black Sea metagenome was also constructed from samples taken during this cruise as detailed in Supporting Information Methods and Villanueva and colleagues (2020). Redox potential was measured during the 64PE408 NESSC/SIAM cruise of January–February 2016 from samples with a closely agreeing sulfide profile (Fig. S1).
B,C. Relative abundances of MAGs of (B) putative SRB and (C) putative SOB and U P. aerophilus were based on normalized metagenome coverage. See Supporting Information Methods for details on the methodology and data processing. [Color figure can be viewed at wileyonlinelibrary.com]
Fig 4.

Maximum‐likelihood phylogenetic reconstruction based on bacterial reductive DsrA proteins predicted from Black Sea MAGs and unbinned contigs (blue) and reference genomes (Anantharaman et al., 2018).
Black dots indicate support of > 95% out of 1,000 ultra‐fast bootstraps. The scale bar indicates substitutions per site. See Supporting Information Methods for methodology and Data S1 for the full phylogenetic tree in Newick format. [Color figure can be viewed at wileyonlinelibrary.com]
Physiology and metagenomics
We have a considerable understanding of the physiology of SRB from marine sediments, owing to a rich diversity of isolated SRB that are available for laboratory research (Muyzer and Stams, 2008; Rabus et al., 2015). In contrast, no SRB have been isolated from DMW, except for two subspecies of Desulfovibrio oceani from the ETSP OMZ (Finster and Kjeldsen, 2010). It can be reasonably assumed that most of the deltaproteobacterial putative SRB detected in DMW adhere to the general metabolism of dissimilatory reduction of sulfate and oxidation of small organic compounds or H2. However, the lack of closely related described SRB does not allow further constraining of metabolic niches based on taxonomic affiliation, nor does it allow hypotheses on the many other variable physiological aspects.
These challenges can be addressed by genome‐centric metagenomics, exemplified by recent explorations of the potential metabolism of Desulfatiglans‐related SAGs from marine sediments (Jochum et al., 2018) and of the identity and potential metabolism of non‐canonical putative SRB from various environments (Wasmund et al., 2016; Anantharaman et al., 2018; Hausmann et al., 2018; Thiel et al., 2018; Meier et al., 2019). With these aims, we mined metagenome data to obtain 13 MAGs of putative SRB from the Black Sea, encoding complete or incomplete reductive Dsr pathways (Figs 3A,B and 5, Table S2, Supporting Information Methods). In agreement with previous diversity studies, most of the putative SRB MAGs were affiliated with Desulfobacterales, Desulfobulbales and Nitrospirae but did not classify within established genera, except for U Desulfobacula maris. Four of the putative SRB MAGs from the Black Sea (52%–93% complete, 1%–3% contaminated) affiliated with the non‐canonical phyla Nitrospirae, Chloroflexi and candidate phylum AAMBM5‐125‐24. Other novel putative SRB within the phylum Nitrospirae, ‘Candidatus Sulfobium mesophilum’ (Zecchin et al., 2018) and ‘Candidatus Nitrobium versatile’ (Arshad et al., 2017) were only distantly related to Nitrospirae MAG NIOZ‐UU55 [< 51% amino acid identity (AAI), Fig. 5, Table S2]. Another phylogenetically related SAG containing sulfate‐reducing genes (AAMBM5‐125‐24) has been retrieved from the euxinic Zodletone Spring (Fig. 5; Youssef et al., 2019). Despite the novelty revealed by this genome‐centric approach, it should be noted that it did not encompass the complete diversity of putative SRB in the Black Sea detected according to the dsrD (Fig. 3A) and dsrA diversity (Fig. 4).
Fig 5.
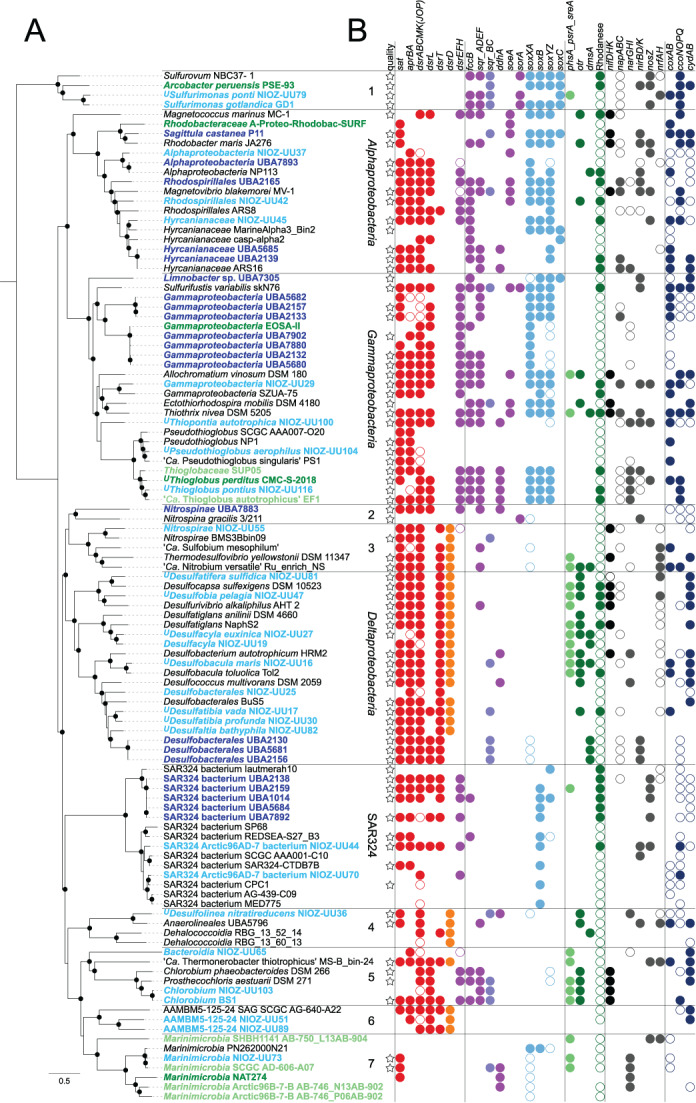
A phylogenomic and genetic overview of important microbial players in the marine sulfur cycle of dysoxic marine water (DMW) environments.
A. An unrooted phylogenomic maximum‐likelihood tree constructed from a concatenated alignment of 120 single‐copy household genes (Supporting Information Methods). Phylogenetic clades were identified, with numbers indicating the following lineages: 1, Campylobacterota; 2, Nitrospinae; 3, Nitrospirae; 4, Chloroflexi; 5, Bacteroidetes; 6, candidate phylum AAMBM5‐125‐24; 7, Marinimicrobia. Black dots indicate support by > 95% out of 1,000 ultra‐fast bootstraps. The scale bar indicates substitutions per site. The tree includes data from all available genomes (March 2020) from DMW sites (bold, coloured by environment following the colour code of Fig. 1) that contain dissimilatory sulfur genes, and relevant reference genomes (black). The superscript prefix ‘U’ indicates uncultured species for which a taxonomy has been proposed based on a high‐quality genome, functional annotation and environmental distribution (Konstantinidis et al., 2017) with the genome sequences as type material (Chuvochina et al., 2019; Murray et al., 2020; Supporting Information Protologue).
B. An overview of the presence of functional genes enabling conversions of sulfur, nitrogen and oxygen, following the colour scheme of Fig. 2. Shortly, red indicates core genes of the Dsr/rDsr pathways, orange indicates dsrD, purple indicates dsrEFH and various oxidative sulfur genes, light blue indicates sox genes, light‐green indicates phs/psr/sre genes, dark‐green indicates various (potentially) reductive sulfur genes, black/dark grey indicates nitrogen genes, dark blue indicates oxygen reduction genes. The presence of the indicated functional genes or gene clusters is shown with filled circles; open circles reveal incomplete gene clusters. For ‘Rhodanese’, filled circles indicate 10 or more rhodanese domains (Supporting Information Methods). Stars distinguish the high‐quality genomes (> 80% complete, < 5% contaminated) from the medium‐quality genomes (> 50% complete, < 10% contaminated) analysed. Only two low‐quality metagenome‐assembled genomes, that is, that of Dehalococcoidia RBG_13_52_14 (35% complete, 2% contaminated) and the population genome of Gammaproteobacteria EOSA‐II composed of multiple combined single‐cell amplified genomes (63% complete, 21% contaminated), were also included. A comprehensive overview of genome origin, quality, classification, annotation and average amino acid identity (AAI) between genomes can be found in Table S2. [Color figure can be viewed at wileyonlinelibrary.com]
More so than taxonomic affiliation, functional gene annotation offers insight into the possible energy metabolism(s) of these microorganisms. Metagenome mining yielded three Desulfobacterales MAGs from the Arabian Sea OMZ core (95%–96% complete, 0.7% contaminated) with a complete Dsr pathway but lacking the reductive marker dsrD (Fig. 5, Table S2, Supporting Information Methods). Moreover, they harbored oxidative instead of reductive dsrA genes possibly horizontally transferred from Chlorobia or SAR324 bacteria (Data S1). Together with the absence of dsrD and the presence of sqr (Fig. 5), this suggests a sulfur‐oxidizing rather than sulfate‐reducing metabolism. These MAGs thus question the common assumption that all Desulfobacterales reduce sulfate, and undermine taxonomy‐based physiological assumptions in general. Habitat profiling can further support metabolic hypotheses based on functional annotation of the genes in different MAGs. For instance, the SRB that reside exclusively in the deeper euxinic waters of the Black Sea (U Desulfatibia profunda, U Desulfaltia bathyphila, Desulfacyla NIOZ‐UU19; 95%–97% complete) apparently lack the genes to detoxify oxygen (cydAB) or hydrogen peroxide (catalase; Table S2) and to utilize alternative electron acceptors (Figs 3B and 5), reflecting their probably purely euxinic and strictly sulfate‐reducing lifestyle. In contrast, the MAGs of SRB relatively abundant in suboxic waters (U Desulfolinea nitratireducens, Nitrospirae NIOZ‐UU55, U Db. maris, U Desulfatibia vada, U Desulfacyla euxinica; 73%–93% complete, 1%–4% contaminated) encode a plethora of genes for the energy‐conserving reduction of alternative electron acceptors such as S0 or thiosulfate (psrA/phsA/sreA), tetrathionate (otr), nitrate (napABC, narGHI) and nitrite (otr, nirBD, nirK, nrfAH). They also encode terminal oxidases (coxAB, ccoNOPQ, cydAB; Figs 3B and 5), which could be part of complete oxygen respiratory chains. These metabolic potentials are in line with a complex sulfur cycle interlinked with nitrogen cycling and oxygen intrusions (see the following sections). Whether the same SRB species as herein detected in the Black Sea are also present in other DMW requires additional research. However, the metagenomic data from the Black Sea offer a basis for such investigations ranging from 16S rRNA surveys to genome‐centric genomics. Expression studies are required to investigate the metabolism of SRB in DMW, and whether they shift their metabolism in response to changing conditions. For instance, the expression of Desulfocapsa‐related nitrogen fixation (nif) genes in the suboxic and upper euxinic zones of the Black Sea (Kirkpatrick et al., 2018) suggests that the nif‐encoding SRB U Desulfatifera sulfidica (99% complete, 2% contaminated) and U Desulfobia pelagia (96% complete, no contamination) could be actively fixing nitrogen. These genes are also encoded by Nitrospirae NIOZ‐UU55 (73% complete, 1% contaminated). This supports a growing body of evidence for nitrogen fixation by putative SRB in DMW (Jayakumar et al., 2012; Bonnet et al., 2013; Loescher et al., 2014; Christiansen and Loescher, 2019).
Particles as microhabitat
Sulfate is thermodynamically an inferior electron acceptor to nitrate and nitrite (Table S3), implying that denitrifying microorganisms will outcompete SRB for electron donors in suboxic waters and OMZ cores. How then is dissimilatory sulfate reduction sustained, especially in nitrite‐ and nitrate‐rich OMZ cores? It has been postulated that SRB occupy microhabitats inside organic particles, in which nitrate and nitrite have already been depleted (Fuchsman et al., 2011; Wright et al., 2012). These organic particles or ‘marine snow’ (Alldredge and Silver, 1988) are particularly abundant in suboxic waters (Karl and Knauer, 1991; Taylor et al., 2001; Sorokin, 2002) and OMZs (Whitmire et al., 2009; Roullier et al., 2014), compared to other regions of the oceanic water column. When oxygen levels drop below ~25 μM, particles of the predominant size range (100–200 μM in diameter; Roullier et al., 2014) develop inner anoxic microhabitats due to limitation of oxygen diffusion (Shanks and Reeder, 1993; Klawonn et al., 2015; Ploug and Bergkvist, 2015). This implies that in nitrate‐rich dysoxic waters, particles will develop a nitrate‐depleted core, as nitrate rarely exceeds a concentration of 25 μM in DMW and diffuses slower than oxygen (Fuchsman et al., 2019). Thus, sulfate‐reducing microhabitats may be abundant in non‐sulfidic DMW.
SRB in seawater seem to be more abundant in particles than in free suspension. The 16S rRNA gene sequences of canonical deltaproteobacterial SRB were found to be predominantly particle‐associated (> 30 μm) in marine suboxic zones (Fuchsman et al., 2011; Suter et al., 2017, 2018) and the ETNP OMZ core (Fuchsman et al., 2017). Moreover, reductive dsrA genes in the ETNP OMZ core were almost exclusively detected in the particle (> 30 μm) fraction, whereas oxidative dsrA genes showed no specific particle association (Saunders et al., 2019). This particle‐bound lifestyle causes SRB to be significantly underrepresented in some molecular ecological studies. It is common practice to employ a pre‐filter step for the collection of biomass to remove eukaryotes (1.6–10 μM pore size cut‐off), thus also removing particles and particle‐associated SRB. This methodology has been applied for investigations in DMW (Canfield et al., 2010; Stewart et al., 2012; Ulloa et al., 2012; Ganesh et al., 2014; Hawley et al., 2014), which may explain why SRB sequences were present in low abundance or absent. In contrast, studies omitting a pre‐filter have identified SRB sequences in substantial proportion (Neretin et al., 2007; Fuchsman et al., 2011; Carolan et al., 2015; Rodriguez‐Mora et al., 2016; Saunders et al., 2019). Particle sinking in standard Niskin sampling bottles also caused bias against particles and SRB sequences (Suter et al., 2017). Thus, complete circumvention of these potential biases requires in situ filtration devices, which have occasionally been used in DMW (Lavik et al., 2009; Marschall et al., 2010; Sollai et al., 2019). In situ filtration was also applied for obtaining the Black Sea MAGs presented herein, indeed resulting in higher estimated relative abundances of SRB than found by Neretin and colleagues (2007) in both the suboxic zone (< 13% vs. < 2% of all bacteria) and the euxinic zone (< 20% vs. < 5%, Supporting Information Methods). Since these estimated fractional abundances may still be biased by DNA extraction methods, ideally an extraction‐independent method should also be applied such as fluorescence in situ hybridization of functional genes (Barrero‐Canosa et al., 2017). To achieve comparability between different studies, similar sampling methodology with minimal bias is essential and should be carefully evaluated.
Sulfur‐oxidizing bacteria
The oxidative part of the sulfur cycle starts with the competition between SOB and abiotic reactions for sulfide (Luther et al., 2011). Depending on the sulfide oxidation route, a range of possible sulfide oxidation products can be formed (Fig. 2), having a significant effect on the rest of the biogeochemistry in DMW. Oxidized metals such as manganese oxide (MnO2) or ferric (oxy)hydroxides are so efficient in catalyzing sulfide oxidation (Yao and Millero, 1993; Ma et al., 2006) that even at micromolar concentrations MnO2 is thought to be abiotically responsible for the bulk of the sulfide oxidation in systems with broad stable chemoclines and low oxygen flux such as the Black Sea (Jørgensen et al., 1991; Konovalov et al., 2003; Trouwborst et al., 2006; Stanev et al., 2018) and Cariaco Basin (Ho et al., 2004). Chemical oxidation of sulfide produces SCIs such as S0 and thiosulfate, which are commonly detected in euxinic marine waters (Jørgensen and Bak, 1991; Zopfi et al., 2001; Li et al., 2008; Kamyshny et al., 2013; Findlay et al., 2014). Like sulfide, SCIs can be converted by SOB to sulfate as energy source.
Microorganisms that oxidize sulfur compounds possess widely differing metabolisms, including autotrophy or heterotrophy, and chemotrophy or phototrophy (Dahl, 2017). Some SOB oxidize a wide range of sulfur compounds as primary energy source, whereas others facultatively oxidize specific sulfur compounds such as thiosulfate as supplementary energy source (Sorokin, 2003). Photolithotrophic SOB – green or purple sulfur bacteria – can become dominant if euxinic marine waters overlap with the photic zone in shallow waters (Findlay et al., 2015; Pjevac et al., 2015; Findlay et al., 2017; Pjevac et al., 2019). The deeper the chemocline, the smaller the population and role of phototrophic SOB, exemplified by low‐light‐adapted Chlorobium bacteria in the Black Sea (approximately 100 m depth; Overmann et al., 1992; Manske et al., 2005; Marschall et al., 2010). They are outnumbered by chemolithotrophic SOB, with gammaproteobacterial SUP05 bacteria (Lavik et al., 2009; Canfield et al., 2010; Glaubitz et al., 2013) and sulfur‐oxidizing Campylobacterales bacteria such as Sulfurimonas species (Grote et al., 2008; Schunck et al., 2013; Callbeck et al., 2019) as foremost examples. However, the biogeochemical impact on element cycling is not always related to cellular abundance (Pester et al., 2012; Hausmann et al., 2019), exemplified by magnetotactic Magnetococcus‐related bacteria that shuttle the scarcely available phosphate from the Black Sea chemocline into the euxinic zone (Schulz‐Vogt et al., 2019), which suggests a sulfur‐oxidizing physiology akin to other Magnetococcus species (Bazylinski et al., 2013). This underscores the importance of using multiple approaches when studying functional groups of microorganisms, including sulfur‐cycling bacteria. Here, we will mainly discuss well‐studied chemolithotrophic SOB specifically abundant in OMZ core waters and other deep DMW.
SUP05 bacteria
Based on 16S rRNA gene surveys, specific gammaproteobacterial bacteria belonging to the SUP05 clade and closely related to known sulfur‐oxidizing symbionts have been identified as abundant putative SOB in DMW (reviewed by Wright et al., 2012). The capacity of these bacteria for chemolithoautotrophic nitrate reduction – most probably coupled to the oxidation of sulfide and/or SCIs – in DMW was strongly indicated by stable isotope probing experiments with labelled inorganic carbon (Grote et al., 2008; Glaubitz et al., 2010) and correlations with rate measurements of nitrate reduction and dark carbon fixation (Lavik et al., 2009; Schunck et al., 2013). Direct cell counts with fluorescent probes showed that SUP05 bacteria may form a dominant group of the microbial community; they comprised up to 50% of the microbial population in euxinic shelf waters off Namibia and Peru (< 3 × 106 cells ml−1; Lavik et al., 2009; Callbeck et al., 2018), up to 17% in the ETSP OMZ core (5 × 105 cells ml−1; Callbeck et al., 2018), up to 30% at an oxic–euxinic interface in the Baltic Sea (4 × 105 cells ml−1, Landsort Deep) and up to 10% and 13% in the suboxic and euxinic zone of the Black Sea respectively (< 7 × 104 cells ml−1; Glaubitz et al., 2013).
The presumed physiology of SUP05 bacteria was supported by the presence in their MAGs of genes for sulfide oxidation (sqr, fccAB), the ‘Sox’ thiosulfate oxidation pathway (soxXABYZ), the rDsr sulfur oxidation pathway (sat, aprBA, dsrABCMK, dsrEFH; Figs 2 and 5), nitrate reduction (narGHIJ) and inorganic carbon fixation through the Calvin–Benson–Bassham cycle (Walsh et al., 2009; Canfield et al., 2010; Murillo et al., 2014; Callbeck et al., 2018). This physiology has been confirmed by cultivation experiments of the only current SUP05 isolate, 'Candidatus Thioglobus autotrophicus’ (Shah et al., 2017), which showed growth with sulfide, thiosulfate, thiotaurine and stored S0 as energy source (Shah et al., 2019). The four available SUP05 genomes from DMW show variation in the presence of other nitrogen‐respiration genes (nirK, nirS, nirBD, norCB, nosZ; Fig. 5) and oxidative phosphorylation genes (coxBAC, ccoNOPQ, cytochrome bc 1 complex genes; Fig. 5), indicating metabolic diversification of the strains within this clade. Corresponding with their high abundance, SUP05 bacteria generally dominate the detection of rDsr pathway genes, transcripts and proteins in DMW (Canfield et al., 2010; Stewart et al., 2012; Hawley et al., 2014). The sister clade ARCTIC96BD‐19 is represented by ‘Candidatus Thioglobus singularis’ (Marshall and Morris, 2013), but ARCTIC96BD‐19 genomes are too divergent from SUP05 genomes to consider them the same genus (63%–66% AAI, Table S2). ‘Ca. T. singularis’ has an organoheterotrophic aerobic lifestyle and does not oxidize sulfur (Spietz et al., 2019). These features are probably representative of all ARCTIC96BD‐19 bacteria, based on the absence of most sulfur oxidation genes in their genomes (Swan et al., 2011; Fig. 5) and a preference for oxic waters (Wright et al., 2012; Fig. 3A; UP. aerophilus). To reflect their distinct taxonomy and physiology, we suggest to rename the ARCTIC96BD‐19 clade to Pseudothioglobus (Supporting Information Protologue).
The affinity of SUP05 bacteria for sulfide is higher than reported for any other bacterium or substrate (Crowe et al., 2018), demanding a re‐evaluation of the existing definition of euxinia. Currently, the sulfide concentration threshold to distinguish non‐sulfidic from euxinic conditions commonly falls in the range of 0.5–1 μM. This threshold is similar to the in vitro Michaelis–Menten half‐saturation constant (K m) of 2 μM of purified high‐affinity Sqr proteins (Schutz et al., 1997; Brito et al., 2009) and the K m found for phototrophic SOB (> 0.8 μM; Van Gemerden, 1984). However, the estimated K m of SUP05 bacteria is much lower (25–340 nM; Crowe et al., 2018). The most widely used method for determining sulfide concentrations has a sulfide detection limit of 0.1 μM (Cline, 1969; Jørgensen et al., 1991; Zopfi et al., 2001), hence falling within this estimated range. Thus, we suggest it is biologically sound and practically feasible to use a sulfide threshold of at most 0.1 μM to define euxinia, at least in marine environments. However, an even lower threshold would be more accurate, as SUP05 bacteria consume sulfide at < 5 nM (Crowe et al., 2018). Such low concentrations can be detected and quantified with sensitive voltammetric sensors (Luther et al., 1991; Luther et al., 2008). The findings of Crowe and colleagues (2018) illustrate the value of studies that quantify properties such as substrate affinity and should motivate further investigation, for instance with respect to sulfide toxicity or substrate affinity for sulfide of other key SOB. Such studies are required to advance biogeochemical models that explicitly take microbial community composition and function into account by integrating omics data (Reed et al., 2014; Louca et al., 2016).
Campylobacterota
The SOB of the phylum Campylobacterota (formerly Epsilonproteobacteria; Waite et al., 2017, 2018) are more diverse and environment‐specific than the common SUP05 bacteria. The most widespread Campylobacterota genus in DMW is Sulfurimonas. Members of this genus dominate the upper euxinic zone of the Black Sea and Baltic Sea at a count of 15% to 30% of all microorganisms (< 2 × 105 cells ml−1) and outnumber SUP05 bacteria (Brettar et al., 2006; Grote et al., 2008; Glaubitz et al., 2010). Members of another Campylobacterota genus, Arcobacter, are generally less abundant in euxinic basins (Glaubitz et al., 2008; Fuchsman et al., 2012; Rodriguez‐Mora et al., 2013), but proliferate in euxinic shelf waters during sulfidic events (< 25% of all cells; < 1 × 106 cells ml−1; Lavik et al., 2009; Schunck et al., 2013; Callbeck et al., 2019). Four Campylobacterota isolates have thus far been obtained from DMW, all facultative anaerobes capable of sulfur oxidation and nitrate reduction: Sulfurimonas gotlandica (Grote et al., 2012) and ‘Candidatus Sulfurimonas baltica’ (Henkel, 2019), both from a redoxcline in Gotland Basin; ‘Candidatus Sulfurimonas marisnigri’ from the Black Sea euxinic zone (Henkel et al., 2019); and Arcobacter peruensis from euxinic coastal waters off Peru (Callbeck et al., 2019). A. peruensis was only demonstrated to use sulfide as energy source (Callbeck et al., 2019), whereas the Sulfurimonas species were grown with sulfide, SCIs and H2 (Labrenz et al., 2013; Henkel, 2019). As is common in Campylobacterota members, the genomes of S. gotlandica and A. peruensis lack rDsr genes and these SOB are presumed to oxidize sulfur compounds through a variant of the Sox pathway encoded by two operons (soxXY 1 Z 1 AB and soxCDY 2 Z 2; Meier et al., 2017; Pjevac et al., 2018; Götz et al., 2019; Figs 2 and 5). We recovered a Sulfurimonas MAG from the Black Sea (U Sulfurimonas ponti), which also lacks most Sox genes despite being virtually complete (97% completeness, 4% contamination, Fig. 5). U S. ponti may oxidize sulfide incompletely to S0, or use an alternative route such as the Hdr‐like sulfur oxidation pathway (Boughanemi et al., 2016).
The heterotrophic A. peruensis was not capable of fixing inorganic carbon and requires an organic carbon source such as acetate (Callbeck et al., 2019), whereas the autotrophic Sulfurimonas species fixed inorganic carbon for growth, presumably through the reverse tricarboxylic acid cycle (Grote et al., 2008; Henkel, 2019). In euxinic coastal waters, sufficient organic carbon can be available to allow A. peruensis to successfully compete for sulfur substrate with autotrophic SOB by achieving a higher carbon assimilation rate and therefore probably also a higher growth rate (Callbeck et al., 2019). Sulfurovum species of the Campylobacterota phylum were highly abundant during sulfidic events in coastal waters (Lavik et al., 2009; Schunck et al., 2013; Callbeck et al., 2019) and possibly outnumber Sulfurimonas in Cariaco Basin (Rodriguez‐Mora et al., 2013; Rodriguez‐Mora et al., 2016; Taylor et al., 2018). Previous cultivation‐ and metagenomics‐based studies of Sulfurovum members have primarily addressed hydrothermal vent habitats. They revealed metabolic similarity to Sulfurimonas species with respect to sulfur oxidation, carbon fixation, and nitrate reduction (Yamamoto et al., 2010; Giovannelli et al., 2016; Jeon et al., 2017; Meier et al., 2017; Mori et al., 2018). However, one notable exception is Sulfurovum aggregans, which cannot oxidize sulfur but instead reduces it (Mino et al., 2014). As such, multiple biochemical roles are possible for Sulfurovum species in DMW.
Sulfur‐oxidizing autotrophs such as Sulfurimonas and SUP05 bacteria compete for very similar niches through different strategies. The metabolically specialized, streamlined (< 1.5 Mbp genomes) and non‐motile SUP05 bacteria prefer stable conditions, while the motile and more adaptable Sulfurimonas species benefit from a less stable chemocline with more mixing of sulfide, nitrate and oxygen (Rogge et al., 2017; Taylor et al., 2018). Furthermore, SUP05 bacteria are most abundant at low‐sulfidic conditions (< 5 μM; Glaubitz et al., 2013; Rogge et al., 2017), which may be due to their unparalleled high affinity for sulfide (Crowe et al., 2018) and their capability to store S0 for later usage when external substrates are absent (Shah et al., 2019). In euxinic basins, Sulfurimonas species can thrive simultaneously with SUP05 bacteria, but have a relative abundance peak in slightly deeper, more sulfidic waters (median 17 μM; Fig. 3C; Rogge et al., 2017). Here, the electron acceptors oxygen and nitrate are irregularly available (Konovalov et al., 2003; Glaubitz et al., 2010; Glaubitz et al., 2013). Sulfurimonas species have adapted to these conditions through motility and chemotaxis towards nitrate‐rich conditions (Grote et al., 2012), which is sustained by energy storage in the form of polyphosphate (Möller et al., 2019). Furthermore, Sulfurimonas species probably conserve more energy from nitrate than SUP05 bacteria, since instead of partial denitrification to nitrite (Shah et al., 2017) or possibly nitrous oxide (Walsh et al., 2009; Hawley et al., 2014), S. gotlandica can perform complete denitrification to nitrogen gas (Labrenz et al., 2013) and U S. ponti could perform ammonification (nrfAH, Fig. 5).
Intriguingly, ‘Ca. S. marisnigri’ is the first bacterium demonstrated to couple sulfur oxidation to the reduction of MnO2 to Mn2+ for growth (Henkel et al., 2019). This trait could be highly beneficial in euxinic basins such as the Black Sea since in contrast to oxygen and nitrate, MnO2 is in constant supply – albeit at low concentrations – since it is particulate and sinks (Tebo, 1991; Konovalov et al., 2004; Trouwborst et al., 2006). This metabolic capacity could also answer the long‐pending question of how high carbon fixation rates are sustained in euxinic waters without sufficient nitrate, nitrite or oxygen (Jørgensen et al., 1991; Taylor et al., 2001; Ho et al., 2004; Jost et al., 2010; Kirkpatrick et al., 2018). Indeed, a reaction–diffusion model by Yakushev and colleagues (2007) required coupling of MnO2 reduction to carbon fixation to reproduce the observed chemical profiles. There are indications that the reaction rates of abiotic and microbial sulfide oxidation by Mn in euxinic basins are in the same order of magnitude (Jørgensen et al., 1991; Sorokin et al., 1995; Henkel, 2019). However, there is currently no insight into the in situ abundance of ‘Ca. S. marisnigri’. Like S. gotlandica (Grote et al., 2012), it does not affiliate with the locally abundant Sulfurimonas GD17 subclade (95%–96% 16S rRNA gene sequence similarity). Further investigation through molecular studies is currently challenging, as the enzymatic pathway allowing MnO2 reduction is unknown. Nevertheless, these findings have large consequences for our view on euxinic biogeochemistry, as sulfide‐driven denitrification and nitrogen loss may effectively be bypassed. Mn‐dependent sulfide oxidation could even result in a fixed nitrogen gain, since both S. marisnigri and S. baltica can apparently fix nitrogen (Henkel, 2019), confirming a recently published hypothesis (Kirkpatrick et al., 2018).
Other sulfur‐oxidizing lineages
As described above, the physiology of some of the key SOB has been explored in some detail, but other microbial players are waiting to be described. Predominantly genomic studies point to a wide phylogenetic diversity of poorly studied SOB for which important ecological or biogeochemical roles in DMW have been demonstrated or are strongly indicated. The class Gammaproteobacteria probably contains relevant SOB that do not affiliate with the SUP05 clade, notwithstanding their key role. Firstly, genomes of the EOSA‐II lineage were retrieved from coastal waters and the OMZ in Southern Pacific waters (Fig. 5), actively expressing sulfur oxidation genes in the ETSP OMZ core (Plominsky et al., 2018). Secondly, the gammaproteobacterial BS‐GSO2 clade was detected in the Black Sea as autotrophic lineage with a peak in relative abundance at the euxinic interface together with SUP05, suggesting it uses sulfur as energy source (Glaubitz et al., 2010). This clade is especially noteworthy since sequencing studies indicate that BS‐GSO2 bacteria may outnumber SUP05 bacteria in the Black Sea (Fuchsman et al., 2011; Kirkpatrick et al., 2018; Fig. 3C) and Cariaco Basin (Suter et al., 2018; Taylor et al., 2018). The only currently available BS‐GSO2 MAG is that of U Thiopontia autotrophica obtained from the Black Sea metagenomes analyzed herein (NIOZ‐UU100, 93% complete, 0.1% contamination) with 99% 16S rRNA gene identity with the original BS‐GSO2 sequence reported by Glaubitz and colleagues (2010). Indeed, U T. autotrophica possesses the genes for sulfur oxidation (rDsr) and the Calvin–Benson–Bassham cycle, but it differs from SUP05 bacteria in lacking most Sox genes and encoding a complete denitrification pathway (Fig. 5). It thus seems the BS‐GSO2 clade has been overshadowed by SUP05, yet may successfully compete for the same niche.
Bacteria of the uncultivated SAR324 candidate phylum have been abundantly and ubiquitously detected in DMW (Fuchsman et al., 2011; Wright et al., 2012; Beman and Carolan, 2013; Lüke et al., 2016; Suter et al., 2018; Fig. 3C). These SAR324 bacteria encode the rDsr sulfur oxidation pathway, which could enable them to oxidize sulfur for energy (Swan et al., 2011; Sheik et al., 2014; Fig. 5). Notably, they may couple this process to the reduction of the greenhouse gas nitrous oxide as they encode nitrous oxide reductase genes (nosZ, Fig. 5). The uncultured alphaproteobacterial family Hyrcanianaceae also harbors putative SOB with genomes retrieved from hydrothermal vent plumes (Zhou et al., 2020), the Arabian Sea OMZ core, and the Black Sea (Fig. 5). Low‐abundance SOB could still significantly alter their environment, for instance through diazotrophy or N2 fixation. Examples of such SOB are the heterotrophic alphaproteobacterium Sagitulla castanea isolated from euxinic Peru shelf waters (Martínez‐Pérez et al., 2018) or photolithoautotrophic Chlorobium strains (Overmann et al., 1992; Manske et al., 2005; Marschall et al., 2010; nifDHK; Figs 3C, 5). Finally, marine Nitrospinae members have been shown to oxidize nitrite (Lücker et al., 2013; Sun et al., 2019; Kitzinger et al., 2020), but the presence of a complete rDsr pathway in a Nitrospinae MAG from the ETSP OMZ core (UBA7883, 97% complete, 1% contaminated) opens up the possibility that some members may use sulfur as additional or alternative energy source (Fig. 5).
Sulfur‐reducing and sulfur‐disproportionating bacteria
The SCIs formed or introduced in DMW could form the substrate for further oxidation by SOB, but could also be used as electron acceptor by sulfur‐reducing microorganisms, or as substrate for disproportionation (Fig. 2), thus shortcutting the sulfur cycle as has been suggested for other aquatic ecosystems (Tonolla et al., 2004; Wilbanks et al., 2014; Bhatnagar et al., 2020). Similar to abiotic sulfide oxidation, SOB may introduce SCIs in DMW through oxidation of sulfide to S0 (Dahl, 2017), which is stored intracellularly by the abundant SUP05 bacteria (Shah et al., 2019). Part of this S0 may be released into the environment due to grazing of SUP05 bacteria by protists (Lin et al., 2007; Glaubitz et al., 2008; Anderson et al., 2013) or due to lysis by SUP05‐infecting viruses (Cassman et al., 2012; Anantharaman et al., 2014; Roux et al., 2014; Roux et al., 2016). Additionally, S0 has been observed to be introduced into OMZs through the drifting off of S0 produced in coastal waters experiencing sulfidic events (Callbeck et al., 2018). These findings highlight the importance of considering full models of the sulfur cycle and avoiding simplified two‐reaction representations consisting only of dissimilatory sulfate reduction and chemolithotrophic re‐oxidation of sulfide to sulfate (Ulloa et al., 2012; Hawley et al., 2014). The extent of the other fluxes is currently a major unknown factor in DMW, with a large impact on the routes of sulfur‐driven carbon fixation and on the occurrence of other linkages with the carbon and nitrogen cycles.
Sulfur‐reducing bacteria and Marinimicrobia
The consumption of SCIs in DMW is thought to proceed through a combination of oxidation, reduction and disproportionation (Sorokin et al., 1995; Zopfi et al., 2001; Sørensen and Canfield, 2004). The relative importance of these consumption routes is currently unknown. Sulfur isotope fractionation studies offer little insight, since the measurements in various euxinic marine waters can be explained by sulfate reduction as well as sulfur reduction or disproportionation (Li et al., 2010; Kamyshny et al., 2011). The reduction or disproportionation of S0 and thiosulfate is more exergonic than sulfate reduction under the conditions found in DMW (Supporting Information Methods, Table S3), implying that SRB could gain more energy through these reactions. Many cultured SRB are able to reduce or disproportionate thiosulfate (Rabus et al., 2015) through the Dsr pathway and thiosulfate reductase (PhsABC; Fig. 2; Burns and DiChristina, 2009) and may prefer this electron acceptor over sulfate (Jørgensen, 1990). Many anaerobic microorganisms use S0 as electron acceptor (Rabus et al., 2013) mediated by polysulfide reductase (PsrABC) or sulfur reductase (SreABC; Fig. 2; Laska et al., 2003; Sorokin et al., 2015). These three protein complexes (Phs, Psr, Sre) are complex iron–sulfur molybdoenzymes with such a close phylogenetic relationship and with so few characterized representatives, that distinction based on sequence is currently impossible (Hedderich et al., 1998; Hinsley and Berks, 2002; Laska et al., 2003; Duval et al., 2008; Burns and DiChristina, 2009). Furthermore, various SOB also encode genes with similarity to psrABC (Wright et al., 2014), of which the resulting enzymes may well act in reverse (Eddie and Hanson, 2013; Weissgerber et al., 2013). Thus, the presence of psr‐like genes in a genome suggests the capability of some form of dissimilatory sulfur conversion, but this requires further investigation.
The reduction of SCIs in DMW was used as energy metabolism by an organoheterotrophic Shewanella strain isolated from the Black Sea (Perry et al., 1993). However, related microorganisms are unlikely to play a big role in DMW, as they have not been detected in microbial ecology studies. SRB remain probable candidates for mediating sulfur reduction, as several genomes of putative SRB retrieved from the Black Sea encoded psr‐like genes and a tetrathionate reductase gene (otr) in addition to their sulfate‐reducing genes (Fig. 5). Other MAGs from DMW metagenomes also showed possibly reductive psr‐like genes, such as Bacteroidia NIOZ‐UU65 from the Black Sea, a MAG of uncultivated clade SAR324 from the Arabian Sea OMZ core and three Marinimicrobia genomes (Fig. 5). Bacteria of the uncultivated candidate phylum Marinimicrobia (formerly known as Marine Group A and clade SAR406) are prevalent in DMW and contain genomic signatures of organoheterotrophy (Wright et al., 2014; Bertagnolli et al., 2017; Hawley et al., 2017), a metabolism supported by DNA‐based stable isotope probing incubations from the Black Sea (Suominen et al., 2019). It has been suggested that Marinimicrobia may reduce S0 based on the presence of psrABC genes (Wright et al., 2014; Hawley et al., 2017). As explained, we think it would be more accurate and unambiguous to broaden the hypothesis to Marinimicrobia having an unspecified dissimilatory sulfur metabolism. Furthermore, the preference for shallow waters with relatively oxidizing conditions by Marinimicrobia NIOZ‐UU73 (Fig. 3A; 90% complete, no contamination) suggests that for this specific member, a facultative sulfur‐oxidizing lifestyle is more likely. Another Marinimicrobia MAG (PN262000N21, 98% complete, no contamination) encodes an almost‐complete ‘Sox’ pathway conferring the potential for dissimilatory thiosulfate oxidation (Figs 2 and 5). The most straightforward path to revealing the energy metabolism of these uncultivated bacteria would be cultivation, isolation and characterization. Like genomes of SAR11 and SUP05 bacteria, Marinimicrobia genomes are extensively streamlined (Hawley et al., 2017) implying that these bacteria are highly adapted to in situ conditions. Hence, cultivation may require natural seawater as medium, or recently designed synthetic alternatives (Henson et al., 2016).
Sulfur‐disproportionating bacteria
Sulfur disproportionation is the simultaneous oxidation and reduction of an SCI, typically leading to the production of both sulfate and sulfide, which is an uncommon microbial trait (Finster, 2008; Slobodkin and Slobodkina, 2019). The biochemistry of sulfur disproportionation is unresolved and may involve the Dsr pathway in Deltaproteobacteria such as Desulfurivibrio alkaliphilus (Thorup et al., 2017) and Desulfocapsa sulfexigens (Finster, 2008; Finster et al., 2013), and Psr‐like molybdoenzymes and rhodanese sulfurtransferases in other microorganisms such as Desulfurella amilsii (Florentino et al., 2019). Although uncommon, disproportionation is probably influential in euxinic marine waters, as microorganisms growing through disproportionation of S0 or thiosulfate could be cultivated from the euxinic Mariager Fjord (Sørensen and Canfield, 2004), and a diffusion–reaction model of Chesapeake Bay required the inclusion of S0 disproportionation or reduction to explain the observed S0 concentration profiles (Findlay et al., 2017). In euxinic basins Desulfocapsa species could be involved, as their 16S rRNA genes were detected in the Cariaco Basin (Rodriguez‐Mora et al., 2015) as well as the Black Sea (Neretin et al., 2007; Fuchsman et al., 2011; Fuchsman et al., 2012). This hypothesis is difficult to test with genomic data due to the unclear biochemistry behind S0 disproportionation. Out of the MAGs obtained from the Black Sea, U Df. sulfidica is the most closely related to Desulfocapsa. Yet, it has a markedly different gene repertoire than Dc. sulfexigens, without molybdoenzymes or high numbers of rhodanese genes. However, plenty other sulfur‐disproportionating bacterial candidates with Dsr pathways, molybdoenzymes and high numbers of rhodanese genes remain (Fig. 5). Like Dv. alkaliphilus (Thorup et al., 2017), some might oxidize sulfide in a disproportionation‐dependent pathway including sulfide oxidation by Sqr (U Db. maris, U Db. vada, U Dl. nitratireducens, Arabian Sea OMZ core Desulfobacterales).
Since most characterized sulfur‐disproportionating microorganisms can grow autotrophically (Finster et al., 1998; Florentino et al., 2016; Mardanov et al., 2016; Slobodkin and Slobodkina, 2019), their presence could spell a role in the high rates of carbon fixation that are generally observed within euxinic marine waters just below the euxinic interface (Jørgensen et al., 1991; Taylor et al., 2001). This phenomenon is commonly attributed solely to sulfur oxidation by chemolithoautotrophic SOB (Grote et al., 2008; Glaubitz et al., 2010). Our hypothesis is in agreement with the stimulation of carbon fixation measured upon the addition of thiosulfate or polysulfide to euxinic samples from the Baltic Sea (Labrenz et al., 2005; Jost et al., 2010). However, these findings could be influenced by artificial introduction of oxygen (De Brabandere et al., 2012) and are in need of further testing. In general, more dedicated experimental work is needed to quantitatively constrain SCI‐consuming reactions, such as was done for a freshwater lake (Findlay and Kamyshny, 2017). Another unexplored factor in the marine sulfur cycle is the cycling of organic sulfur compounds, which has been highlighted for marine sediments (Wasmund et al., 2017). Such processes may be important in DMW, as dimethylsulfide oxidation genes (ddhA) were detected in genomes of SUP05 bacteria, U T. autotrophica and Marinimicrobia, and dimethylsulfoxide reductase genes (dmsA) in MAGs of Desulfacyla species, U Db. maris and OMZ Desulfobacterales. Future studies should address to what extent reactions of SCIs and organic sulfur compounds contribute to the overall sulfur cycle, and whether this is affected by environmental conditions.
Conclusions and future perspectives
This review has presented a compendium of the current insights into sulfur‐cycling bacteria in DMW (Fig. 6). Based on current experimental evidence, it is difficult to investigate in situ sulfur reactions beyond sulfide oxidation and sulfate reduction. Euxinic marine waters are thought to host a complex network of reactions, whereas this remains more uncertain for suboxic waters and OMZ cores. Molecular studies have revealed a high diversity of putative SRB and SOB, which we expect to be explored further and consolidated over the coming years, specifically with the use of improved genome‐centric metagenomics. Sampling methods without bias against particle‐associated microorganisms can give an accurate and intercomparable view on diversity and abundance across DMW, specifically of SRB. With this in mind, it could be evaluated whether the community of novel putative SRB genomically revealed by us in the Black Sea is representative of euxinic marine basins and perhaps DMW in general. Together, the diverse sulfur‐cycling bacteria form a myriad of connections with other elemental cycles. SUP05 bacteria fix inorganic carbon with energy from very low sulfide concentrations, warranting a biologically meaningful reshaping of our concept of euxinia. These insights into sulfide affinity are crucial building blocks for biogeochemical modelling efforts, which could be further improved by estimations of critical biological parameters including biomass yield and sulfide tolerance. Metatranscriptomics and metaproteomics experiments might play a role in testing under which conditions SRB use their genomic potential for respiration of oxygen and diverse nitrogen compounds. Notably, genomic classifications of some bacteria as ‘SRB’ are for now putative, as the Dsr pathway on which this is based could also confer other forms of sulfur metabolism, such as sulfur reduction, disproportionation or oxidation. Similarly, the lack of fundamental insight into the relation of sequence and function of prevalent Psr‐related molybdoenzymes hinders metabolic predictions. Thus, genomic‐based research can find strong support in cultivation experiments and the in vitro study of heterologously expressed sulfur enzymes. The power of cultivation has been showcased by the isolation of SUP05, Sulfurimonas and Arcobacter bacteria, and specifically that of the Mn‐reducing and probably nitrogen‐fixing ‘Ca. S. marisnigri’. The cultivation and isolation of SRB from DMW is also feasible (Teske et al., 1996; Zopfi et al., 2001; Sørensen and Canfield, 2004; Finster and Kjeldsen, 2010), but more challenging than cultivation from sediment due to rapid oxidation of sampled water. These efforts could be facilitated by genome‐guided cultivation (Gutleben et al., 2018) or a reverse genomic approach (Cross et al., 2019). Finally, the factors controlling nitrogen fixation by SOB and SRB require further investigation, as this may be an important factor in the expected development of open‐ocean euxinia (Ulloa et al., 2012). The advances as summarized and predicted herein will enable the construction of biogeochemical models of the sulfur cycle from meta‐omics data, as has been done for the nitrogen cycle in the Arabian Sea (Reed et al., 2014) and in the Saanich Inlet (Louca et al., 2016). In the future, such endeavors could aid in predicting the biogeochemical response to expanding dysoxia and euxinia.
Fig 6.
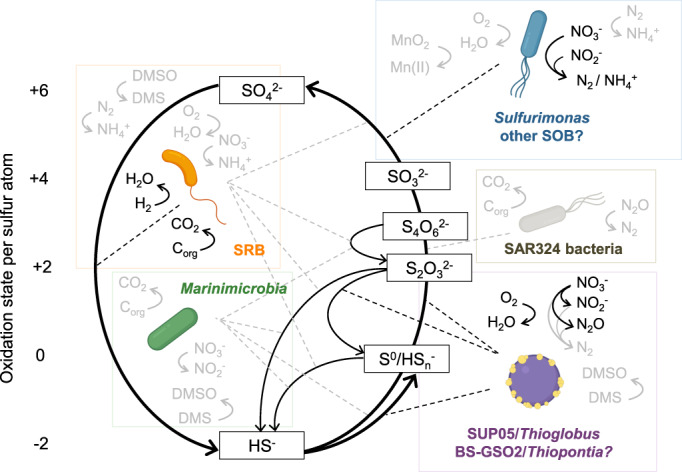
Conceptual ecophysiological model of the sulfur cycle and the involved microorganisms in DMW.
Question marks and the light gray colour are used when there are indications for involvement of specific microorganisms and/or processes but definitive proof is lacking. Bacterial images were created with BioRender. [Color figure can be viewed at wileyonlinelibrary.com]
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Data S1. Maximum‐likelihood phylogenetic reconstruction of dsrA genes in newick format. See Supporting Information Methods for methodology.
Table S1. Microbiological and biogeochemical studies of the sulfur cycle in dysoxic marine waters, grouped by environment. Volumes based on the work of Paulmier and Ruiz‐Pino (2009) correspond to the estimated volume of waters containing > 0.5 μM nitrite. The maximum volume of anoxic water off the Namibian coast was calculated from the largest observed extent of sulfidic bottom waters (7000 km2; Lavik et al., 2009) and an assumed sulfidic layer thickness of 10 m.
Table S2. Origin, quality, classification, annotation, and average amino acid identity (AAI) of the analysed genomes in Fig. 5. Methods are described in Supporting Information Methods. The AAI values were calculated with an enveomics script using Diamond because of computational limitations, which could lead to significantly overestimated AAI values between 50% and 60% (https://rodriguez-r.com/blog/aai-blast-vs-diamond/). Following the thresholds proposed by Konstantinidis and colleagues (2017), values exceeding the 65% genus‐level lower threshold are coloured green, and values exceeding the 45% family‐level lower threshold are coloured yellow.
Table S3. Gibbs free energies [ΔG (kJ e−)] of common dissimilatory conversions mediated by anaerobic microorganisms under conditions representative of the upper euxinic zone of the Black Sea and the core of the ETSP OMZ. Calculation methodology and variables used can be found in Supporting Information Methods.
Appendix S1. Supplementary Information Methods.
Appendix S2. Supplementary Information Protologue.
Acknowledgements
We would like to thank all crew and scientific party of the 64PE371 and 64PE408 Black Sea cruises aboard R/V Pelagia for sampling, the sulfur thesis ring of the Wageningen University Laboratory of Microbiology and Department of Environmental Technology for constructive proofreading, Dr. Karthik Anantharaman for freely sharing his HMMs before peer‐reviewed publication, and prof. Friedrich Widdel for helpful discussion on the sulfur cycle and autotrophy. This research was supported through SIAM Gravitation grant 024.002.002 to AJMS and JSSD of the Netherlands Ministry of Education, Culture and Science and the Netherlands Organisation for Scientific Research (NWO). BED and FABvM were supported by the NWO Vidi grant 864.14.004. BED was supported by the European Research Council (ERC) Consolidator grant 865694: DiversiPHI.
References
- Albert, D.B., Taylor, C., and Martens, C.S. (1995) Sulfate reduction rates and low‐molecular‐weight fatty‐acid concentrations in the water column and surficial sediments of the Black Sea. Deep Sea Res Part I Oceanogr Res Pap 42: 1239–1260. [Google Scholar]
- Alldredge, A.L., and Silver, M.W. (1988) Characteristics, dynamics and significance of marine snow. Prog Oceanogr 20: 41–82. [Google Scholar]
- Anantharaman, K., Duhaime, M.B., Breier, J.A., Wendt, K.A., Toner, B.M., and Dick, G.J. (2014) Sulfur oxidation genes in diverse deep‐sea viruses. Science 344: 757–760. [DOI] [PubMed] [Google Scholar]
- Anantharaman, K., Hausmann, B., Jungbluth, S.P., Kantor, R.S., Lavy, A., Warren, L.A., et al. (2018) Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J 12: 1715–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, R., Wylezich, C., Glaubitz, S., Labrenz, M., and Jurgens, K. (2013) Impact of protist grazing on a key bacterial group for biogeochemical cycling in Baltic Sea pelagic oxic/anoxic interfaces. Environ Microbiol 15: 1580–1594. [DOI] [PubMed] [Google Scholar]
- Arshad, A., Dalcin Martins, P., Frank, J., Jetten, M.S.M., Op den Camp, H.J.M., and Welte, C.U. (2017) Mimicking microbial interactions under nitrate‐reducing conditions in an anoxic bioreactor: enrichment of novel Nitrospirae bacteria distantly related to Thermodesulfovibrio . Environ Microbiol 19: 4965–4977. [DOI] [PubMed] [Google Scholar]
- Barrero‐Canosa, J., Moraru, C., Zeugner, L., Fuchs, B.M., and Amann, R. (2017) Direct‐geneFISH: a simplified protocol for the simultaneous detection and quantification of genes and rRNA in microorganisms. Environ Microbiol 19: 70–82. [DOI] [PubMed] [Google Scholar]
- Bazylinski, D.A., Williams, T.J., Lefevre, C.T., Berg, R.J., Zhang, C.L., Bowser, S.S., et al. (2013) Magnetococcus marinus gen. nov., sp. nov., a marine, magnetotactic bacterium that represents a novel lineage (Magnetococcaceae fam. nov., Magnetococcales ord. nov.) at the base of the Alphaproteobacteria . Int J Syst Evol Microbiol 63: 801–808. [DOI] [PubMed] [Google Scholar]
- Beman, J.M., and Carolan, M.T. (2013) Deoxygenation alters bacterial diversity and community composition in the ocean's largest oxygen minimum zone. Nat Commun 4: 2705. [DOI] [PubMed] [Google Scholar]
- Bertagnolli, A.D., Padilla, C.C., Glass, J.B., Thamdrup, B., and Stewart, F.J. (2017) Metabolic potential and in situ activity of marine Marinimicrobia bacteria in an anoxic water column. Environ Microbiol 19: 4392–4416. [DOI] [PubMed] [Google Scholar]
- Bhatnagar, S., Cowley, E.S., Kopf, S.H., Pérez Castro, S., Kearney, S., Dawson, S.C., et al. (2020) Microbial community dynamics and coexistence in a sulfide‐driven phototrophic bloom. Environ Microbiome 15: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet, S., Dekaezemacker, J., Turk‐Kubo, K.A., Moutin, T., Hamersley, R.M., Grosso, O., et al. (2013) Aphotic N2 fixation in the Eastern Tropical South Pacific Ocean. PLoS One 8: e81265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughanemi, S., Lyonnet, J., Infossi, P., Bauzan, M., Kosta, A., Lignon, S., et al. (2016) Microbial oxidative sulfur metabolism: biochemical evidence of the membrane‐bound heterodisulfide reductase‐like complex of the bacterium Aquifex aeolicus . FEMS Microbiol Lett 363: fnw156. [DOI] [PubMed] [Google Scholar]
- Boyle, R.A., Clark, J.R., Poulton, S.W., Shields‐Zhou, G., Canfield, D.E., and Lenton, T.M. (2013) Nitrogen cycle feedbacks as a control on euxinia in the mid‐Proterozoic Ocean. Nat Commun 4: 1533. [DOI] [PubMed] [Google Scholar]
- Breitburg, D., Levin, L.A., Oschlies, A., Gregoire, M., Chavez, F.P., Conley, D.J., et al. (2018) Declining oxygen in the global ocean and coastal waters. Science 359: eaam7240. [DOI] [PubMed] [Google Scholar]
- Brettar, I., Labrenz, M., Flavier, S., Botel, J., Kuosa, H., Christen, R., and Hofle, M.G. (2006) Identification of a Thiomicrospira denitrificans‐like epsilonproteobacterium as a catalyst for autotrophic denitrification in the Central Baltic Sea. Appl Environ Microbiol 72: 1364–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito, J.A., Sousa, F.L., Stelter, M., Bandeiras, T.M., Vonrhein, C., Teixeira, M., et al. (2009) Structural and functional insights into sulfide:quinone oxidoreductase. Biochemistry 48: 5613–5622. [DOI] [PubMed] [Google Scholar]
- Burns, J.L., and DiChristina, T.J. (2009) Anaerobic respiration of elemental sulfur and thiosulfate by Shewanella oneidensis MR‐1 requires psrA, a homolog of the phsA gene of Salmonella enterica serovar typhimurium LT2. Appl Environ Microbiol 75: 5209–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callbeck, C.M., Lavik, G., Ferdelman, T.G., Fuchs, B., Gruber‐Vodicka, H.R., Hach, P.F., et al. (2018) Oxygen minimum zone cryptic sulfur cycling sustained by offshore transport of key sulfur oxidizing bacteria. Nat Commun 9: 1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callbeck, C.M., Pelzer, C., Lavik, G., Ferdelman, T.G., Graf, J.S., Vekeman, B., et al. (2019) Arcobacter peruensis sp. nov., a chemolithoheterotroph isolated from sulfide‐and organic‐rich coastal waters off Peru. Appl Environ Microbiol 85: e01344–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield, D.E. (2006) Models of oxic respiration, denitrification and sulfate reduction in zones of coastal upwelling. Geochim Cosmochim Acta 70: 5753–5765. [Google Scholar]
- Canfield, D.E., Kristensen, E., and Thamdrup, B. (2005) Aquatic Geomicrobiology. Oxford, UK: Gulf Professional Publishing. [DOI] [PubMed] [Google Scholar]
- Canfield, D.E., Stewart, F.J., Thamdrup, B., De Brabandere, L., Dalsgaard, T., Delong, E.F., et al. (2010) A cryptic sulfur cycle in oxygen‐minimum‐zone waters off the Chilean coast. Science 330: 1375–1378. [DOI] [PubMed] [Google Scholar]
- Carolan, M.T., Smith, J.M., and Beman, J.M. (2015) Transcriptomic evidence for microbial sulfur cycling in the eastern tropical North Pacific oxygen minimum zone. Front Microbiol 6: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassman, N., Prieto‐Davo, A., Walsh, K., Silva, G.G., Angly, F., Akhter, S., et al. (2012) Oxygen minimum zones harbour novel viral communities with low diversity. Environ Microbiol 14: 3043–3065. [DOI] [PubMed] [Google Scholar]
- Chen, J., Hanke, A., Tegetmeyer, H.E., Kattelmann, I., Sharma, R., Hamann, E., et al. (2017) Impacts of chemical gradients on microbial community structure. ISME J 11: 920–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen, C.F., and Loescher, C.R. (2019) Facets of diazotrophy in the OMZ off Peru revisited: what we could not see from a single marker gene approach. bioRxiv: 558072. [Google Scholar]
- Chuvochina, M., Rinke, C., Parks, D.H., Rappe, M.S., Tyson, G.W., Yilmaz, P., et al. (2019) The importance of designating type material for uncultured taxa. Syst Appl Microbiol 42: 15–21. [DOI] [PubMed] [Google Scholar]
- Cline, J.D. (1969) Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr 14: 454–458. [Google Scholar]
- Cross, K.L., Campbell, J.H., Balachandran, M., Campbell, A.G., Cooper, S.J., Griffen, A., et al. (2019) Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat Biotechnol 37: 1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe, S.A., Cox, R.P., Jones, C., Fowle, D.A., Santibanez‐Bustos, J.F., Ulloa, O., and Canfield, D.E. (2018) Decrypting the sulfur cycle in oceanic oxygen minimum zones. ISME J 12: 2322–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl, C. (2017) Sulfur metabolism in phototrophic bacteria. In Modern Topics in the Phototrophic Prokaryotes: Metabolism, Bioenergetics, and Omics, Hallenbeck, P.C. (ed). Cham, Switzerland: Springer International Publishing, pp. 27–66. [Google Scholar]
- De Brabandere, L., Thamdrup, B., Revsbech, N.P., and Foadi, R. (2012) A critical assessment of the occurrence and extend of oxygen contamination during anaerobic incubations utilizing commercially available vials. J Microbiol Methods 88: 147–154. [DOI] [PubMed] [Google Scholar]
- Duval, S., Ducluzeau, A.L., Nitschke, W., and Schoepp‐Cothenet, B. (2008) Enzyme phylogenies as markers for the oxidation state of the environment: the case of respiratory arsenate reductase and related enzymes. BMC Evol Biol 8: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddie, B.J., and Hanson, T.E. (2013) Chlorobaculum tepidum TLS displays a complex transcriptional response to sulfide addition. J Bacteriol 195: 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich, H.L., Newman, D.K., and Kappler, A. (2015) Ehrlich's Geomicrobiology. Boca Raton, FL: CRC Press. [Google Scholar]
- Findlay, A.J. (2016) Microbial impact on polysulfide dynamics in the environment. FEMS Microbiol Lett 363: fnw103. [DOI] [PubMed] [Google Scholar]
- Findlay, A.J., Bennett, A.J., Hanson, T.E., and Luther, G.W., 3rd. (2015) Light‐dependent sulfide oxidation in the anoxic zone of the Chesapeake Bay can be explained by small populations of phototrophic bacteria. Appl Environ Microbiol 81: 7560–7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay, A.J., Di Toro, D.M., and Luther, G.W. (2017) A model of phototrophic sulfide oxidation in a stratified estuary. Limnol Oceanogr 62: 1853–1867. [Google Scholar]
- Findlay, A.J., Gartman, A., MacDonald, D.J., Hanson, T.E., Shaw, T.J., and Luther, G.W. (2014) Distribution and size fractionation of elemental sulfur in aqueous environments: the Chesapeake Bay and Mid‐Atlantic Ridge. Geochim Cosmochim Acta 142: 334–348. [Google Scholar]
- Findlay, A.J., and Kamyshny, A. (2017) Turnover rates of intermediate sulfur species in anoxic freshwater and sediments. Front Microbiol 8: 2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finster, K. (2008) Microbiological disproportionation of inorganic sulfur compounds. J Sulfur Chem 29: 281–292. [Google Scholar]
- Finster, K., Liesack, W., and Thamdrup, B. (1998) Elemental sulfur and thiosulfate disproportionation by Desulfocapsa sulfoexigens sp. nov., a new anaerobic bacterium isolated from marine surface sediment. Appl Environ Microbiol 64: 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finster, K.W., and Kjeldsen, K.U. (2010) Desulfovibrio oceani subsp. oceani sp. nov., subsp. nov. and Desulfovibrio oceani subsp. galateae subsp. nov., novel sulfate‐reducing bacteria isolated from the oxygen minimum zone off the coast of Peru. Antonie Van Leeuwenhoek 97: 221–229. [DOI] [PubMed] [Google Scholar]
- Finster, K.W., Kjeldsen, K.U., Kube, M., Reinhardt, R., Mussmann, M., Amann, R., and Schreiber, L. (2013) Complete genome sequence of Desulfocapsa sulfexigens, a marine deltaproteobacterium specialized in disproportionating inorganic sulfur compounds. Stand Genomic Sci 8: 58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentino, A.P., Brienza, C., Stams, A.J.M., and Sanchez‐Andrea, I. (2016) Desulfurella amilsii sp. nov., a novel acidotolerant sulfur‐respiring bacterium isolated from acidic river sediments. Int J Syst Evol Microbiol 66: 1249–1253. [DOI] [PubMed] [Google Scholar]
- Florentino, A.P., Pereira, I.A.C., Boeren, S., van den Born, M., Stams, A.J.M., and Sanchez‐Andrea, I. (2019) Insight into the sulfur metabolism of Desulfurella amilsii by differential proteomics. Environ Microbiol 21: 209–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentino, A.P., Stams, A.J., and Sanchez‐Andrea, I. (2017) Genome sequence of Desulfurella amilsii strain TR1 and comparative genomics of Desulfurellaceae family. Front Microbiol 8: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froelich, P.N., Klinkhammer, G.P., Bender, M.L., Luedtke, N.A., Heath, G.R., Cullen, D., et al. (1979) Early oxidation of organic matter in pelagic sediments of the eastern equatorial Atlantic: suboxic diagenesis. Geochim Cosmochim Acta 43: 1075–1090. [Google Scholar]
- Fuchsman, C.A., Devol, A.H., Saunders, J.K., McKay, C., and Rocap, G. (2017) Niche partitioning of the N cycling microbial community of an offshore oxygen deficient zone. Front Microbiol 8: 2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsman, C.A., Kirkpatrick, J.B., Brazelton, W.J., Murray, J.W., and Staley, J.T. (2011) Metabolic strategies of free‐living and aggregate‐associated bacterial communities inferred from biologic and chemical profiles in the Black Sea suboxic zone. FEMS Microbiol Ecol 78: 586–603. [DOI] [PubMed] [Google Scholar]
- Fuchsman, C.A., Murray, J.W., and Staley, J.T. (2012) Stimulation of autotrophic denitrification by intrusions of the bosporus plume into the anoxic black sea. Front Microbiol 3: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsman, C.A., Paul, B., Staley, J.T., Yakushev, E.V., and Murray, J.W. (2019) Detection of transient denitrification during a high organic matter event in the Black Sea. Global Biogeochem Cycles. 80: 402–416. [Google Scholar]
- Ganesh, S., Parris, D.J., DeLong, E.F., and Stewart, F.J. (2014) Metagenomic analysis of size‐fractionated picoplankton in a marine oxygen minimum zone. ISME J 8: 187–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannelli, D., Chung, M., Staley, J., Starovoytov, V., Le Bris, N., and Vetriani, C. (2016) Sulfurovum riftiae sp. nov., a mesophilic, thiosulfate‐oxidizing, nitrate‐reducing chemolithoautotrophic epsilonproteobacterium isolated from the tube of the deep‐sea hydrothermal vent polychaete Riftia pachyptila . Int J Syst Evol Microbiol 66: 2697–2701. [DOI] [PubMed] [Google Scholar]
- Glaubitz, S., Kiesslich, K., Meeske, C., Labrenz, M., and Jurgens, K. (2013) SUP05 dominates the Gammaproteobacterial sulfur oxidizer assemblages in pelagic redoxclines of the Central Baltic and Black Seas. Appl Environ Microbiol 79: 2767–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaubitz, S., Labrenz, M., Jost, G., and Jurgens, K. (2010) Diversity of active chemolithoautotrophic prokaryotes in the sulfidic zone of a Black Sea pelagic redoxcline as determined by rRNA‐based stable isotope probing. FEMS Microbiol Ecol 74: 32–41. [DOI] [PubMed] [Google Scholar]
- Glaubitz, S., Lueders, T., Abraham, W.R., Jost, G., Jürgens, K., and Labrenz, M. (2008) 13C‐isotope analyses reveal that chemolithoautotrophic Gamma‐and Epsilonproteobacteria feed a microbial food web in a pelagic redoxcline of the Central Baltic Sea. Environ Microbiol 11: 326–337. [DOI] [PubMed] [Google Scholar]
- Götz, F., Pjevac, P., Markert, S., McNichol, J., Becher, D., Schweder, T., et al. (2019) Transcriptomic and proteomic insight into the mechanism of cyclooctasulfur‐ versus thiosulfate‐oxidation by the chemolithoautotroph Sulfurimonas denitrificans . Environ Microbiol 21: 244–258. [DOI] [PubMed] [Google Scholar]
- Grote, J., Jost, G., Labrenz, M., Herndl, G.J., and Jurgens, K. (2008) Epsilonproteobacteria represent the major portion of chemoautotrophic bacteria in sulfidic waters of pelagic redoxclines of the Baltic and Black Seas. Appl Environ Microbiol 74: 7546–7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote, J., Schott, T., Bruckner, C.G., Glockner, F.O., Jost, G., Teeling, H., et al. (2012) Genome and physiology of a model Epsilonproteobacterium responsible for sulfide detoxification in marine oxygen depletion zones. Proc Natl Acad Sci USA 109: 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutleben, J., Chaib De Mares, M., van Elsas, J.D., Smidt, H., Overmann, J., and Sipkema, D. (2018) The multi‐omics promise in context: from sequence to microbial isolate. Crit Rev Microbiol 44: 212–229. [DOI] [PubMed] [Google Scholar]
- Han, Y., and Perner, M. (2015) The globally widespread genus Sulfurimonas: versatile energy metabolisms and adaptations to redox clines. Front Microbiol 6: 989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann, B., Pelikan, C., Herbold, C.W., Kostlbacher, S., Albertsen, M., Eichorst, S.A., et al. (2018) Peatland Acidobacteria with a dissimilatory sulfur metabolism. ISME J 12: 1729–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann, B., Pelikan, C., Rattei, T., Loy, A., and Pester, M. (2019) Long‐term transcriptional activity at zero growth of a cosmopolitan rare biosphere member. MBio 10: e02189–e02118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley, A.K., Brewer, H.M., Norbeck, A.D., Pasa‐Tolic, L., and Hallam, S.J. (2014) Metaproteomics reveals differential modes of metabolic coupling among ubiquitous oxygen minimum zone microbes. Proc Natl Acad Sci USA 111: 11395–11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley, A.K., Nobu, M.K., Wright, J.J., Durno, W.E., Morgan‐Lang, C., Sage, B., et al. (2017) Diverse Marinimicrobia bacteria may mediate coupled biogeochemical cycles along eco‐thermodynamic gradients. Nat Commun 8: 1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedderich, R., Klimmek, O., Kroger, A., Dirmeier, R., Keller, M., and Stetter, K.O. (1998) Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol Rev 22: 353–381. [Google Scholar]
- Henkel, J.V. (2019) Bacterial H2S oxidation coupled to the reduction of MnO2 studied on new isolated species of the genus Sulfurimonas . In Leibniz‐Institut für Ostseeforschung Warnemünde (IOW). Rostock, Germany: Universität Rostock, p. 109. [Google Scholar]
- Henkel, J.V., Dellwig, O., Pollehne, F., Herlemann, D.P.R., Leipe, T., and Schulz‐Vogt, H.N. (2019) A bacterial isolate from the Black Sea oxidizes sulfide with manganese(IV) oxide. Proc Natl Acad Sci USA 116: 12153–12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson, M.W., Pitre, D.M., Weckhorst, J.L., Lanclos, V.C., Webber, A.T., and Thrash, J.C. (2016) Artificial seawater media facilitate cultivating members of the microbial majority from the Gulf of Mexico. mSphere 1: e00028–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsley, A.P., and Berks, B.C. (2002) Specificity of respiratory pathways involved in the reduction of sulfur compounds by Salmonella enterica . Microbiology 148: 3631–3638. [DOI] [PubMed] [Google Scholar]
- Ho, T.Y., Taylor, G.T., Astor, Y., Varela, R., Muller‐Karger, F., and Scranton, M.I. (2004) Vertical and temporal variability of redox zonation in the water column of the Cariaco Basin: implications for organic carbon oxidation pathways. Mar Chem 86: 89–104. [Google Scholar]
- Holkenbrink, C., Barbas, S.O., Mellerup, A., Otaki, H., and Frigaard, N.‐U. (2011) Sulfur globule oxidation in green sulfur bacteria is dependent on the dissimilatory sulfite reductase system. Microbiology 157: 1229–1239. [DOI] [PubMed] [Google Scholar]
- Jayakumar, A., Al‐Rshaidat, M.M., Ward, B.B., and Mulholland, M.R. (2012) Diversity, distribution, and expression of diazotroph nifH genes in oxygen‐deficient waters of the Arabian Sea. FEMS Microbiol Ecol 82: 597–606. [DOI] [PubMed] [Google Scholar]
- Jeon, W., Priscilla, L., Park, G., Lee, H., Lee, N., Lee, D., et al. (2017) Complete genome sequence of the sulfur‐oxidizing chemolithoautotrophic Sulfurovum lithotrophicum 42BKT(T). Stand Genomic Sci 12: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochum, L.M., Schreiber, L., Marshall, I.P.G., Jorgensen, B.B., Schramm, A., and Kjeldsen, K.U. (2018) Single‐cell genomics reveals a diverse metabolic potential of uncultivated Desulfatiglans‐related Deltaproteobacteria widely distributed in marine sediment. Front Microbiol 9: 2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, D.T., Gill, B.C., Masterson, A., Beirne, E., Casciotti, K.L., Knapp, A.N., and Berelson, W. (2014) Placing an upper limit on cryptic marine sulphur cycling. Nature 513: 530–533. [DOI] [PubMed] [Google Scholar]
- Jørgensen, B.B. (1990) A thiosulfate shunt in the sulfur cycle of marine sediments. Science 249: 152–154. [DOI] [PubMed] [Google Scholar]
- Jørgensen, B.B., and Bak, F. (1991) Pathways and microbiology of thiosulfate transformations and sulfate reduction in a marine sediment (Kattegat, Denmark). Appl Environ Microbiol 57: 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen, B.B., Fossing, H., Wirsen, C.O., and Jannasch, H.W. (1991) Sulfide oxidation in the anoxic Black Sea chemocline. Deep Sea Res Part A Oceanogr Res Pap 38: S1083–S1103. [Google Scholar]
- Jost, G., Martens‐Habbena, W., Pollehne, F., Schnetger, B., and Labrenz, M. (2010) Anaerobic sulfur oxidation in the absence of nitrate dominates microbial chemoautotrophy beneath the pelagic chemocline of the eastern Gotland Basin, Baltic Sea. FEMS Microbiol Ecol 71: 226–236. [DOI] [PubMed] [Google Scholar]
- Kamyshny, A., Borkenstein, C.G., and Ferdelman, T.G. (2009) Protocol for quantitative detection of elemental sulfur and polysulfide zero‐valent sulfur distribution in natural aquatic samples. Geostand Geoanal Res 33: 415–435. [Google Scholar]
- Kamyshny, A., Yakushev, E.V., Jost, G., and Podymov, O.I. (2013) Role of sulfide oxidation intermediates in the redox balance of the oxic–anoxic interface of the Gotland Deep, Baltic Sea. In Chemical Structure of Pelagic Redox Interfaces: Observation and Modeling. Berlin/Heidelberg, Germany: Springer, pp. 95–119. [Google Scholar]
- Kamyshny, A., Zerkle, A.L., Mansaray, Z.F., Ciglenecki, I., Bura‐Nakic, E., Farquhar, J., and Ferdelman, T.G. (2011) Biogeochemical sulfur cycling in the water column of a shallow stratified sea‐water lake: speciation and quadruple sulfur isotope composition. Mar Chem 127: 144–154. [Google Scholar]
- Karl, D.M., and Knauer, G.A. (1991) Microbial production and particle flux in the upper 350 m of the Black Sea. Deep Sea Res Part A Oceanogr Res Pap 38: S921–S942. [Google Scholar]
- Kirkpatrick, J.B., Fuchsman, C.A., Yakushev, E.V., Egorov, A.V., Staley, J.T., and Murray, J.W. (2018) Dark N2 fixation: nifH expression in the redoxcline of the Black Sea. Aquat Microb Ecol 82: 43–58. [Google Scholar]
- Kitzinger, K., Marchant, H.K., Bristow, L.A., Herbold, C.W., Padilla, C.C., Kidane, A.T., et al. (2020) Single cell analyses reveal contrasting life strategies of the two main nitrifiers in the ocean. Nat Commun 11: 767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klawonn, I., Bonaglia, S., Bruchert, V., and Ploug, H. (2015) Aerobic and anaerobic nitrogen transformation processes in N2‐fixing cyanobacterial aggregates. ISME J 9: 1456–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konovalov, S., Samodurov, A., Oguz, T., and Ivanov, L. (2004) Parameterization of iron and manganese cycling in the Black Sea suboxic and anoxic environment. Deep Sea Res Part I Oceanogr Res Pap 51: 2027–2045. [Google Scholar]
- Konovalov, S.K., Luther, G.W., Friederich, G.E., Nuzzio, D.B., Tebo, B.M., Murray, J.W., et al. (2003) Lateral injection of oxygen with the Bosporus plume – fingers of oxidizing potential in the Black Sea. Limnol Oceanogr 48: 2369–2376. [Google Scholar]
- Konstantinidis, K.T., Rosselló‐Móra, R., and Amann, R. (2017) Uncultivated microbes in need of their own taxonomy. ISME J 11: 2399–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneeva, V.A., Pimenov, N.V., Krek, A.V., Tourova, T.P., and Bryukhanov, A.L. (2015) Sulfate‐reducing bacterial communities in the water column of the Gdansk Deep (Baltic Sea). Mikrobiologiia 84: 250–260. [PubMed] [Google Scholar]
- Labrenz, M., Grote, J., Mammitzsch, K., Boschker, H.T., Laue, M., Jost, G., et al. (2013) Sulfurimonas gotlandica sp. nov., a chemoautotrophic and psychrotolerant epsilonproteobacterium isolated from a pelagic redoxcline, and an emended description of the genus Sulfurimonas . Int J Syst Evol Microbiol 63: 4141–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrenz, M., Jost, G., Pohl, C., Beckmann, S., Martens‐Habbena, W., and Jurgens, K. (2005) Impact of different in vitro electron donor/acceptor conditions on potential chemolithoautotrophic communities from marine pelagic redoxclines. Appl Environ Microbiol 71: 6664–6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, P., and Kuypers, M.M. (2011) Microbial nitrogen cycling processes in oxygen minimum zones. Ann Rev Mar Sci 3: 317–345. [DOI] [PubMed] [Google Scholar]
- Laska, S., Lottspeich, F., and Kletzin, A. (2003) Membrane‐bound hydrogenase and sulfur reductase of the hyperthermophilic and acidophilic archaeon Acidianus ambivalens . Microbiology 149: 2357–2371. [DOI] [PubMed] [Google Scholar]
- Lavik, G., Stuhrmann, T., Bruchert, V., Van der Plas, A., Mohrholz, V., Lam, P., et al. (2009) Detoxification of sulphidic African shelf waters by blooming chemolithotrophs. Nature 457: 581–584. [DOI] [PubMed] [Google Scholar]
- Li, X., Gilhooly, W.P., Zerkle, A.L., Lyons, T.W., Farquhar, J., Werne, J.P., et al. (2010) Stable sulfur isotopes in the water column of the Cariaco Basin. Geochim Cosmochim Acta 74: 6764–6778. [Google Scholar]
- Li, X.N., Taylor, G.T., Astor, Y., and Scranton, M.I. (2008) Relationship of sulfur speciation to hydrographic conditions and chemoautotrophic production in the Cariaco Basin. Mar Chem 112: 53–64. [Google Scholar]
- Lin, X., Scranton, M.I., Varela, R., Chistoserdov, A., and Taylor, G.T. (2007) Compositional responses of bacterial communities to redox gradients and grazing in the anoxic Cariaco Basin. Aquat Microb Ecol 47: 57–72. [Google Scholar]
- Lin, X., Wakeham, S.G., Putnam, I.F., Astor, Y.M., Scranton, M.I., Chistoserdov, A.Y., and Taylor, G.T. (2006) Comparison of vertical distributions of prokaryotic assemblages in the anoxic Cariaco Basin and Black Sea by use of fluorescence in situ hybridization. Appl Environ Microbiol 72: 2679–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loescher, C.R., Groskopf, T., Desai, F.D., Gill, D., Schunck, H., Croot, P.L., et al. (2014) Facets of diazotrophy in the oxygen minimum zone waters off Peru. ISME J 8: 2180–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louca, S., Hawley, A.K., Katsev, S., Torres‐Beltran, M., Bhatia, M.P., Kheirandish, S., et al. (2016) Integrating biogeochemistry with multiomic sequence information in a model oxygen minimum zone. Proc Natl Acad Sci USA 113: E5925–E5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy, A., Duller, S., Baranyi, C., Mußmann, M., Ott, J., Sharon, I., et al. (2009) Reverse dissimilatory sulfite reductase as phylogenetic marker for a subgroup of sulfur‐oxidizing prokaryotes. Environ Microbiol 11: 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücker, S., Nowka, B., Rattei, T., Spieck, E., and Daims, H. (2013) The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front Microbiol 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüke, C., Speth, D.R., Kox, M.A.R., Villanueva, L., and Jetten, M.S.M. (2016) Metagenomic analysis of nitrogen and methane cycling in the Arabian Sea oxygen minimum zone. PeerJ 4: e1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther, G.W., 3rd, Findlay, A.J., Macdonald, D.J., Owings, S.M., Hanson, T.E., Beinart, R.A., and Girguis, P.R. (2011) Thermodynamics and kinetics of sulfide oxidation by oxygen: a look at inorganically controlled reactions and biologically mediated processes in the environment. Front Microbiol 2: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther, G.W., Church, T.M., and Powell, D. (1991) Sulfur speciation and sulfide oxidation in the water column of the Black Sea. Deep‐Sea Res Part A Oceanogr Res Pap 38: S1121–S1137. [Google Scholar]
- Luther, G.W., Glazer, B.T., Ma, S.F., Trouwborst, R.E., Moore, T.S., Metzger, E., et al. (2008) Use of voltammetric solid‐state (micro)electrodes for studying biogeochemical processes: laboratory measurements to real time measurements with an in situ electrochemical analyzer (ISEA). Mar Chem 108: 221–235. [Google Scholar]
- Ma, S., Noble, A., Butcher, D., Trouwborst, R.E., and Luther, G.W. (2006) Removal of H2S via an iron catalytic cycle and iron sulfide precipitation in the water column of dead end tributaries. Estuar Coast Shelf Sci 70: 461–472. [Google Scholar]
- Madrid, V.M., Taylor, G.T., Scranton, M.I., and Chistoserdov, A.Y. (2001) Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the Cariaco Basin. Appl Environ Microbiol 67: 1663–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manske, A.K., Glaeser, J., Kuypers, M.M., and Overmann, J. (2005) Physiology and phylogeny of green sulfur bacteria forming a monospecific phototrophic assemblage at a depth of 100 meters in the Black Sea. Appl Environ Microbiol 71: 8049–8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardanov, A.V., Beletsky, A.V., Kadnikov, V.V., Slobodkin, A.I., and Ravin, N.V. (2016) Genome analysis of Thermosulfurimonas dismutans, the first thermophilic sulfur‐disproportionating bacterium of the phylum Thermodesulfobacteria . Front Microbiol 7: 950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall, E., Jogler, M., Hessge, U., and Overmann, J. (2010) Large‐scale distribution and activity patterns of an extremely low‐light‐adapted population of green sulfur bacteria in the Black Sea. Environ Microbiol 12: 1348–1362. [DOI] [PubMed] [Google Scholar]
- Marshall, K.T., and Morris, R.M. (2013) Isolation of an aerobic sulfur oxidizer from the SUP05/Arctic96BD‐19 clade. ISME J 7: 452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Pérez, C., Mohr, W., Schwedt, A., Dürschlag, J., Callbeck, C.M., Schunck, H., et al. (2018) Metabolic versatility of a novel N2‐fixing Alphaproteobacterium isolated from a marine oxygen minimum zone. Environ Microbiol 20: 755–768. [DOI] [PubMed] [Google Scholar]
- Meier, D.V., Pjevac, P., Bach, W., Hourdez, S., Girguis, P.R., Vidoudez, C., et al. (2017) Niche partitioning of diverse sulfur‐oxidizing bacteria at hydrothermal vents. ISME J 11: 1545–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, D.V., Pjevac, P., Bach, W., Markert, S., Schweder, T., Jamieson, J., et al. (2019) Microbial metal‐sulfide oxidation in inactive hydrothermal vent chimneys suggested by metagenomic and metaproteomic analyses. Environ Microbiol 21: 682–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, B., and Kuever, J. (2007) Phylogeny of the alpha and beta subunits of the dissimilatory adenosine‐5′‐phosphosulfate (APS) reductase from sulfate‐reducing prokaryotes–origin and evolution of the dissimilatory sulfate‐reduction pathway. Microbiology 153: 2026–2044. [DOI] [PubMed] [Google Scholar]
- Meyer, K.M., and Kump, L.R. (2008) Oceanic Euxinia in earth history: causes and consequences. Annu Rev Earth Planet Sci 36: 251–288. [Google Scholar]
- Mino, S., Kudo, H., Arai, T., Sawabe, T., Takai, K., and Nakagawa, S. (2014) Sulfurovum aggregans sp. nov., a hydrogen‐oxidizing, thiosulfate‐reducing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep‐sea hydrothermal vent chimney, and an emended description of the genus Sulfurovum . Int J Syst Evol Microbiol 64: 3195–3201. [DOI] [PubMed] [Google Scholar]
- Mizuno, N., Voordouw, G., Miki, K., Sarai, A., and Higuchi, Y. (2003) Crystal structure of dissimilatory sulfite reductase D (DsrD) protein – possible interaction with B‐ and Z‐DNA by its winged‐helix motif. Structure 11: 1133–1140. [DOI] [PubMed] [Google Scholar]
- Möller, L., Laas, P., Rogge, A., Goetz, F., Bahlo, R., Leipe, T., and Labrenz, M. (2019) Sulfurimonas subgroup GD17 cells accumulate polyphosphate under fluctuating redox conditions in the Baltic Sea: possible implications for their ecology. ISME J 13: 482–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, K., Yamaguchi, K., and Hanada, S. (2018) Sulfurovum denitrificans sp. nov., an obligately chemolithoautotrophic sulfur‐oxidizing epsilonproteobacterium isolated from a hydrothermal field. Int J Syst Evol Microbiol 68: 2183–2187. [DOI] [PubMed] [Google Scholar]
- Müller, A.L., Kjeldsen, K.U., Rattei, T., Pester, M., and Loy, A. (2015) Phylogenetic and environmental diversity of DsrAB‐type dissimilatory (bi)sulfite reductases. ISME J 9: 1152–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo, A.A., Ramírez‐Flandes, S., DeLong, E.F., and Ulloa, O. (2014) Enhanced metabolic versatility of planktonic sulfur‐oxidizing γ‐proteobacteria in an oxygen‐deficient coastal ecosystem. Front Mar Sci 1: 18. [Google Scholar]
- Murray, A.E., Freudenstein, J., Gribaldo, S., Hatzenpichler, R., Hugenholtz, P., Kampfer, P., et al. (2020) Roadmap for naming uncultivated Archaea and Bacteria . Nat Microbiol, 5: 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, J.W., Jannasch, H.W., Honjo, S., Anderson, R.F., Reeburgh, W.S., Top, Z., et al. (1989) Unexpected changes in the oxic anoxic interface in the Black Sea. Nature 338: 411–413. [Google Scholar]
- Mussmann, M., Richter, M., Lombardot, T., Meyerdierks, A., Kuever, J., Kube, M., et al. (2005) Clustered genes related to sulfate respiration in uncultured prokaryotes support the theory of their concomitant horizontal transfer. J Bacteriol 187: 7126–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer, G., and Stams, A.J. (2008) The ecology and biotechnology of sulphate‐reducing bacteria. Nat Rev Microbiol 6: 441–454. [DOI] [PubMed] [Google Scholar]
- Naqvi, S.W.A., Bange, H.W., Farías, L., Monteiro, P.M.S., Scranton, M.I., and Zhang, J. (2010) Marine hypoxia/anoxia as a source of CH4 and N2O. Biogeosciences 7: 2159–2190. [Google Scholar]
- Neretin, L.N., Abed, R.M., Schippers, A., Schubert, C.J., Kohls, K., and Kuypers, M.M. (2007) Inorganic carbon fixation by sulfate‐reducing bacteria in the Black Sea water column. Environ Microbiol 9: 3019–3024. [DOI] [PubMed] [Google Scholar]
- Overmann, J., Cypionka, H., and Pfennig, N. (1992) An extremely low‐light‐adapted phototrophic sulfur bacterium from the Black Sea. Limnol Oceanogr 37: 150–155. [Google Scholar]
- Parks, D.H., Rinke, C., Chuvochina, M., Chaumeil, P.‐A., Woodcroft, B.J., Evans, P.N., et al. (2017) Recovery of nearly 8,000 metagenome‐assembled genomes substantially expands the tree of life. Nat Microbiol 2: 1533–1542. [DOI] [PubMed] [Google Scholar]
- Paulmier, A., and Ruiz‐Pino, D. (2009) Oxygen minimum zones (OMZs) in the modern ocean. Prog Oceanogr 80: 113–128. [Google Scholar]
- Pelikan, C., Herbold, C.W., Hausmann, B., Müller, A.L., Pester, M., and Loy, A. (2016) Diversity analysis of sulfite‐and sulfate‐reducing microorganisms by multiplex dsrA and dsrB amplicon sequencing using new primers and mock community‐optimized bioinformatics. Environ Microbiol 18: 2994–3009. [DOI] [PubMed] [Google Scholar]
- Perry, K.A., Kostka, J.E., Luther, G.W., 3rd, and Nealson, K.H. (1993) Mediation of sulfur speciation by a black sea facultative anaerobe. Science 259: 801–803. [DOI] [PubMed] [Google Scholar]
- Pester, M., Knorr, K.H., Friedrich, M.W., Wagner, M., and Loy, A. (2012) Sulfate‐reducing microorganisms in wetlands ‐ fameless actors in carbon cycling and climate change. Front Microbiol 3: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenov, N.V., Rusanov, I.I., Yusupov, S.K., Fridrich, J., Lein, A.Y., Wehrli, B., and Ivanov, M.V. (2000) Microbial processes at the aerobic‐anaerobic interface in the deep‐water zone of the Black Sea. Microbiology 69: 436–448. [PubMed] [Google Scholar]
- Pjevac, P., Dyksma, S., Goldhammer, T., Mujakić, I., Koblížek, M., Mußmann, M., et al. (2019) In situ abundance and carbon fixation activity of distinct anoxygenic phototrophs in the stratified seawater lake Rogoznica. Environ Microbiol 21: 3896–3908. [DOI] [PubMed] [Google Scholar]
- Pjevac, P., Korlevic, M., Berg, J.S., Bura‐Nakic, E., Ciglenecki, I., Amann, R., and Orlic, S. (2015) Community shift from phototrophic to chemotrophic sulfide oxidation following anoxic holomixis in a stratified seawater lake. Appl Environ Microbiol 81: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pjevac, P., Meier, D.V., Markert, S., Hentschker, C., Schweder, T., Becher, D., et al. (2018) Metaproteogenomic profiling of microbial communities colonizing actively venting hydrothermal chimneys. Front Microbiol 9: 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plominsky, A.M., Trefault, N., Podell, S., Blanton, J.M., De la Iglesia, R., Allen, E.E., et al. (2018) Metabolic potential and in situ transcriptomic profiles of previously uncharacterized key microbial groups involved in coupled carbon, nitrogen and sulfur cycling in anoxic marine zones. Environ Microbiol 20: 2727–2742. [DOI] [PubMed] [Google Scholar]
- Ploug, H., and Bergkvist, J. (2015) Oxygen diffusion limitation and ammonium production within sinking diatom aggregates under hypoxic and anoxic conditions. Mar Chem 176: 142–149. [Google Scholar]
- Rabus, R., Hansen, T.A., and Widdel, F. (2013) Dissimilatory sulfate‐and sulfur‐reducing prokaryotes. In The Prokaryotes, Berlin, Heidelberg: Springer, pp. 309–404. [Google Scholar]
- Rabus, R., Venceslau, S.S., Wohlbrand, L., Voordouw, G., Wall, J.D., and Pereira, I.A. (2015) A post‐genomic view of the ecophysiology, catabolism and biotechnological relevance of sulphate‐reducing prokaryotes. Adv Microb Physiol 66: 55–321. [DOI] [PubMed] [Google Scholar]
- Reed, D.C., Algar, C.K., Huber, J.A., and Dick, G.J. (2014) Gene‐centric approach to integrating environmental genomics and biogeochemical models. Proc Natl Acad Sci USA 111: 1879–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revsbech, N.P., Larsen, L.H., Gundersen, J., Dalsgaard, T., Ulloa, O., and Thamdrup, B. (2009) Determination of ultra‐low oxygen concentrations in oxygen minimum zones by the STOX sensor. Limnol Oceanogr Methods 7: 371–381. [Google Scholar]
- Rodriguez‐Mora, M.J., Edgcomb, V.P., Taylor, C., Scranton, M.I., Taylor, G.T., and Chistoserdov, A.Y. (2016) The diversity of sulfide oxidation and sulfate reduction genes expressed by the bacterial communities of the Cariaco Basin, Venezuela. Open Microbiol J 10: 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Mora, M.J., Scranton, M.I., Taylor, G.T., and Chistoserdov, A.Y. (2013) Bacterial community composition in a large marine anoxic basin: a Cariaco Basin time‐series survey. FEMS Microbiol Ecol 84: 625–639. [DOI] [PubMed] [Google Scholar]
- Rodriguez‐Mora, M.J., Scranton, M.I., Taylor, G.T., and Chistoserdov, A.Y. (2015) The dynamics of the bacterial diversity in the redox transition and anoxic zones of the Cariaco Basin assessed by parallel tag sequencing. FEMS Microbiol Ecol 91: fiv088. [DOI] [PubMed] [Google Scholar]
- Rogge, A., Vogts, A., Voss, M., Jurgens, K., Jost, G., and Labrenz, M. (2017) Success of chemolithoautotrophic SUP05 and Sulfurimonas GD17 cells in pelagic Baltic Sea redox zones is facilitated by their lifestyles as K‐ and r‐strategists. Environ Microbiol 19: 2495–2506. [DOI] [PubMed] [Google Scholar]
- Roullier, F., Berline, L., Guidi, L., Durrieu De Madron, X., Picheral, M., Sciandra, A., et al. (2014) Particle size distribution and estimated carbon flux across the Arabian Sea oxygen minimum zone. Biogeosciences 11: 4541–4557. [Google Scholar]
- Roux, S., Brum, J.R., Dutilh, B.E., Sunagawa, S., Duhaime, M.B., Loy, A., et al. (2016) Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537: 689–693. [DOI] [PubMed] [Google Scholar]
- Roux, S., Hawley, A.K., Torres Beltran, M., Scofield, M., Schwientek, P., Stepanauskas, R., et al. (2014) Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single‐cell‐ and meta‐genomics. Elife 3: e03125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, A.A., Venceslau, S.S., Grein, F., Leavitt, W.D., Dahl, C., Johnston, D.T., and Pereira, I.A. (2015) A protein trisulfide couples dissimilatory sulfate reduction to energy conservation. Science 350: 1541–1545. [DOI] [PubMed] [Google Scholar]
- Saunders, J.K., Fuchsman, C.A., McKay, C., and Rocap, G. (2019) Complete arsenic‐based respiratory cycle in the marine microbial communities of pelagic oxygen‐deficient zones. Proc Natl Acad Sci USA 116: 9925–9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidtko, S., Stramma, L., and Visbeck, M. (2017) Decline in global oceanic oxygen content during the past five decades. Nature 542: 335–339. [DOI] [PubMed] [Google Scholar]
- Schulz‐Vogt, H.N., Pollehne, F., Jürgens, K., Arz, H.W., Beier, S., Bahlo, R., et al. (2019) Effect of large magnetotactic bacteria with polyphosphate inclusions on the phosphate profile of the suboxic zone in the Black Sea. ISME J 13: 1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schunck, H., Lavik, G., Desai, D.K., Grosskopf, T., Kalvelage, T., Loscher, C.R., et al. (2013) Giant hydrogen sulfide plume in the oxygen minimum zone off Peru supports chemolithoautotrophy. PLoS One 8: e68661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz, M., Shahak, Y., Padan, E., and Hauska, G. (1997) Sulfide‐quinone reductase from Rhodobacter capsulatus. Purification, cloning, and expression. J Biol Chem 272: 9890–9894. [DOI] [PubMed] [Google Scholar]
- Shah, V., Chang, B.X., and Morris, R.M. (2017) Cultivation of a chemoautotroph from the SUP05 clade of marine bacteria that produces nitrite and consumes ammonium. ISME J 11: 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, V., Zhao, X., Lundeen, R.A., Ingalls, A.E., Nicastro, D., and Morris, R.M. (2019) Morphological plasticity in a sulfur‐oxidizing marine bacterium from the SUP05 clade enhances dark carbon fixation. mBio 10: e00216–e00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks, A.L., and Reeder, M.L. (1993) Reducing microzones and sulfide production in marine snow. Mar Ecol Prog Ser 96: 43–47. [Google Scholar]
- Sheik, C.S., Jain, S., and Dick, G.J. (2014) Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ Microbiol 16: 304–317. [DOI] [PubMed] [Google Scholar]
- Sigalevich, P., and Cohen, Y. (2000) Oxygen‐dependent growth of the sulfate‐reducing bacterium Desulfovibrio oxyclinae in coculture with Marinobacter sp. strain MB in an aerated sulfate‐depleted chemostat. Appl Environ Microbiol 66: 5019–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobodkin, A.I., and Slobodkina, G.B. (2019) Diversity of sulfur‐disproportionating microorganisms. Microbiology 88: 509–522. [Google Scholar]
- Slobodkina, G.B., Mardanov, A.V., Ravin, N.V., Frolova, A.A., Chernyh, N.A., Bonch‐Osmolovskaya, E.A., and Slobodkin, A.I. (2017) Respiratory ammonification of nitrate coupled to anaerobic oxidation of elemental sulfur in deep‐sea autotrophic thermophilic bacteria. Front Microbiol 8: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollai, M., Villanueva, L., Hopmans, E.C., Reichart, G.J., and Sinninghe Damste, J.S. (2019) A combined lipidomic and 16S rRNA gene amplicon sequencing approach reveals archaeal sources of intact polar lipids in the stratified Black Sea water column. Geobiology 17: 91–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen, K.B., and Canfield, D.E. (2004) Annual fluctuations in sulfur isotope fractionation in the water column of a euxinic marine basin. Geochim Cosmochim Acta 68: 503–515. [Google Scholar]
- Sorokin, D.Y. (2003) Oxidation of inorganic sulfur compounds by obligately organotrophic bacteria. Microbiology 72: 641–653. [PubMed] [Google Scholar]
- Sorokin, D.Y., Kublanov, I.V., Gavrilov, S.N., Rojo, D., Roman, P., Golyshin, P.N., et al. (2015) Elemental sulfur and acetate can support life of a novel strictly anaerobic haloarchaeon. ISME J 10: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin, Y.I. (1972) The bacterial population and the processes of hydrogen sulphide oxidation in the Black Sea. Journal du Conseil 34: 423–454. [Google Scholar]
- Sorokin, Y.I. (2002) The Black Sea: Ecology and Oceanography. Amsterdam, The Netherlands: Backhuys Publishers. [Google Scholar]
- Sorokin, Y.I., Sorokin, P.Y., Avdeev, V.A., Sorokin, D.Y., and Ilchenko, S.V. (1995) Biomass, production and activity of bacteria in the Black Sea, with special reference to chemosynthesis and the sulfur cycle. Hydrobiologia 308: 61–76. [Google Scholar]
- Spietz, R.L., Lundeen, R.A., Zhao, X., Nicastro, D., Ingalls, A.E., and Morris, R.M. (2019) Heterotrophic carbon metabolism and energy acquisition in Candidatus Thioglobus singularis strain PS1, a member of the SUP05 clade of marine Gammaproteobacteria . Environ Microbiol 0: 2391–2401. [DOI] [PubMed] [Google Scholar]
- Stanev, E.V., Poulain, P.M., Grayek, S., Johnson, K.S., Claustre, H., and Murray, J.W. (2018) Understanding the dynamics of the oxic‐anoxic interface in the Black Sea. Geophys Res Lett 45: 864–871. [Google Scholar]
- Stewart, F.J., Ulloa, O., and DeLong, E.F. (2012) Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ Microbiol 14: 23–40. [DOI] [PubMed] [Google Scholar]
- Sun, X., Kop, L.F.M., Lau, M.C.Y., Frank, J., Jayakumar, A., Lücker, S., and Ward, B.B. (2019) Uncultured Nitrospina‐like species are major nitrite oxidizing bacteria in oxygen minimum zones. ISME J 13: 2391–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suominen, S., Dombrowski, N., Sinninghe Damste, J.S., and Villanueva, L. (2019) A diverse uncultivated microbial community is responsible for organic matter degradation in the Black Sea sulphidic zone. Environ Microbiol. https://doi.org/0.1111/1462‐2920.14902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter, E.A., Pachiadaki, M., Taylor, G.T., Astor, Y., and Edgcomb, V.P. (2018) Free‐living chemoautotrophic and particle‐attached heterotrophic prokaryotes dominate microbial assemblages along a pelagic redox gradient. Environ Microbiol 20: 693–712. [DOI] [PubMed] [Google Scholar]
- Suter, E.A., Scranton, M.I., Chow, S., Stinton, D., Medina Faull, L., and Taylor, G.T. (2017) Niskin bottle sample collection aliases microbial community composition and biogeochemical interpretation. Limnol Oceanogr 62: 606–617. [Google Scholar]
- Swan, B.K., Martinez‐Garcia, M., Preston, C.M., Sczyrba, A., Woyke, T., Lamy, D., et al. (2011) Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333: 1296–1300. [DOI] [PubMed] [Google Scholar]
- Taylor, G.T., Iabichella, M., Ho, T.Y., Scranton, M.I., Thunell, R.C., Muller‐Karger, F., and Varela, R. (2001) Chemoautotrophy in the redox transition zone of the Cariaco Basin: a significant midwater source of organic carbon production. Limnol Oceanogr 46: 148–163. [Google Scholar]
- Taylor, G.T., Suter, E.A., Pachiadaki, M.G., Astor, Y., Edgcomb, V.P., and Scranton, M.I. (2018) Temporal shifts in dominant sulfur‐oxidizing chemoautotrophic populations across the Cariaco Basin's redoxcline. Deep Sea Res Part II Top Stud Oceanogr 156: 80–96. [Google Scholar]
- Tebo, B.M. (1991) Manganese (II) oxidation in the suboxic zone of the Black Sea. Deep Sea Res Part A Oceanogr Res Pap 38: S883–S905. [Google Scholar]
- Teske, A., Wawer, C., Muyzer, G., and Ramsing, N.B. (1996) Distribution of sulfate‐reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most‐probable‐number counts and denaturing gradient gel electrophoresis of PCR‐amplified ribosomal DNA fragments. Appl Environ Microbiol 62: 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamdrup, B., Dalsgaard, T., and Revsbech, N.P. (2012) Widespread functional anoxia in the oxygen minimum zone of the Eastern South Pacific. Deep Sea Res Part I Oceanogr Res Pap 65: 36–45. [Google Scholar]
- Thiel, V., Garcia Costas, A.M., Fortney, N.W., Martinez, J.N., Tank, M., Roden, E.E., et al. (2018) ‘Candidatus Thermonerobacter thiotrophicus,’ a non‐phototrophic member of the Bacteroidetes/Chlorobi with dissimilatory sulfur metabolism in hot spring mat communities. Front Microbiol 9: 3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorup, C., Schramm, A., Findlay, A.J., Finster, K.W., and Schreiber, L. (2017) Disguised as a sulfate reducer: growth of the Deltaproteobacterium Desulfurivibrio alkaliphilus by sulfide oxidation with nitrate. mBio 8: e00671–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonolla, M., Peduzzi, S., Demarta, A., Peduzzi, R., and Hahn, D. (2004) Phototropic sulfur and sulfate‐reducing bacteria in the chemocline of meromictic Lake Cadagno, Switzerland. J Limnol 63: 161–170. [Google Scholar]
- Trouwborst, R.E., Clement, B.G., Tebo, B.M., Glazer, B.T., and Luther, G.W., 3rd. (2006) Soluble Mn(III) in suboxic zones. Science 313: 1955–1957. [DOI] [PubMed] [Google Scholar]
- Tully, B.J., Graham, E.D., and Heidelberg, J.F. (2018) The reconstruction of 2,631 draft metagenome‐assembled genomes from the global oceans. Sci Data 5: 170203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa, O., Canfield, D.E., DeLong, E.F., Letelier, R.M., and Stewart, F.J. (2012) Microbial oceanography of anoxic oxygen minimum zones. Proc Natl Acad Sci USA 109: 15996–16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gemerden, H. (1984) The sulfide affinity of phototrophic bacteria in relation to the location of elemental sulfur. Arch Microbiol 139: 289–294. [Google Scholar]
- Venceslau, S.S., Stockdreher, Y., Dahl, C., and Pereira, I.A. (2014) The ‘bacterial heterodisulfide’ DsrC is a key protein in dissimilatory sulfur metabolism. Biochim Biophys Acta 1837: 1148–1164. [DOI] [PubMed] [Google Scholar]
- Vetriani, C., Tran, H.V., and Kerkhof, L.J. (2003) Fingerprinting microbial assemblages from the oxic/anoxic chemocline of the Black Sea. Appl Environ Microbiol 69: 6481–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva, L., von Meijenfeldt, F.A.B., Westbye, A.B., Yadav, S., Hopmans, E.C., Dutilh, B.E., and Sinninghe Damste, J.S. (2020) Bridging the membrane lipid divide: bacteria of the FCB group superphylum have the potential to synthesize archaeal ether lipids. ISME J, 1–15. https://doi.org/10.1038/s41396‐020‐00772‐2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite, D.W., Vanwonterghem, I., Rinke, C., Parks, D.H., Zhang, Y., Takai, K., et al. (2017) Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front Microbiol 8: 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite, D.W., Vanwonterghem, I., Rinke, C., Parks, D.H., Zhang, Y., Takai, K., et al. (2018) Erratum: addendum: comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front Microbiol 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, D.A., Zaikova, E., Howes, C.G., Song, Y.C., Wright, J.J., Tringe, S.G., et al. (2009) Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science 326: 578–582. [DOI] [PubMed] [Google Scholar]
- Wasmund, K., Cooper, M., Schreiber, L., Lloyd, K.G., Baker, B.J., Petersen, D.G., et al. (2016) Single‐cell genome and group‐specific dsrAB sequencing implicate marine members of the class Dehalococcoidia (phylum Chloroflexi) in sulfur cycling. mBio 7: e00266–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmund, K., Mussmann, M., and Loy, A. (2017) The life sulfuric: microbial ecology of sulfur cycling in marine sediments. Environ Microbiol Rep 9: 323–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissgerber, T., Dobler, N., Polen, T., Latus, J., Stockdreher, Y., and Dahl, C. (2013) Genome‐wide transcriptional profiling of the purple sulfur bacterium Allochromatium vinosum DSM 180T during growth on different reduced sulfur compounds. J Bacteriol 195: 4231–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmire, A.L., Letelier, R.M., Villagran, V., and Ulloa, O. (2009) Autonomous observations of in vivo fluorescence and particle backscattering in an oceanic oxygen minimum zone. Opt Express 17: 21992–22004. [DOI] [PubMed] [Google Scholar]
- Wilbanks, E.G., Jaekel, U., Salman, V., Humphrey, P.T., Eisen, J.A., Facciotti, M.T., et al. (2014) Microscale sulfur cycling in the phototrophic pink berry consortia of the Sippewissett Salt Marsh. Environ Microbiol 16: 3398–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, J.J., Konwar, K.M., and Hallam, S.J. (2012) Microbial ecology of expanding oxygen minimum zones. Nat Rev Microbiol 10: 381–394. [DOI] [PubMed] [Google Scholar]
- Wright, J.J., Mewis, K., Hanson, N.W., Konwar, K.M., Maas, K.R., and Hallam, S.J. (2014) Genomic properties of Marine Group A bacteria indicate a role in the marine sulfur cycle. ISME J 8: 455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakushev, E., Pollehne, F., Jost, G., Kuznetsov, I., Schneider, B., and Umlauf, L. (2007) Analysis of the water column oxic/anoxic interface in the Black and Baltic seas with a numerical model. Mar Chem 107: 388–410. [Google Scholar]
- Yamamoto, M., Nakagawa, S., Shimamura, S., Takai, K., and Horikoshi, K. (2010) Molecular characterization of inorganic sulfur‐compound metabolism in the deep‐sea epsilonproteobacterium Sulfurovum sp. NBC37‐1. Environ Microbiol 12: 1144–1153. [DOI] [PubMed] [Google Scholar]
- Yao, W., and Millero, F.J. (1993) The rate of sulfide oxidation by δMnO2 in seawater. Geochim Cosmochim Acta 57: 3359–3365. [Google Scholar]
- Youssef, N.H., Farag, I.F., Hahn, C.R., Jarett, J., Becraft, E., Eloe‐Fadrosh, E., et al. (2019) Genomic characterization of candidate division LCP‐89 reveals an atypical cell wall structure, microcompartment production, and dual respiratory and fermentative capacities. Appl Environ Microbiol 85: e00110–e00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecchin, S., Mueller, R.C., Seifert, J., Stingl, U., Anantharaman, K., von Bergen, M., et al. (2018) Rice Paddy Nitrospirae carry and express genes related to Sulfate respiration: proposal of the new genus ‘Candidatus Sulfobium’. Appl Environ Microbiol 84: e02224‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Tran, P.Q., Kieft, K., and Anantharaman, K. (2020) Genome diversification in globally distributed novel marine Proteobacteria is linked to environmental adaptation. ISME J 14: 2060–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopfi, J., Ferdelman, T.G., Jorgensen, B.B., Teske, A., and Thamdrup, B. (2001) Influence of water column dynamics on sulfide oxidation and other major biogeochemical processes in the chemocline of Mariager Fjord (Denmark). Mar Chem 74: 29–51. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Maximum‐likelihood phylogenetic reconstruction of dsrA genes in newick format. See Supporting Information Methods for methodology.
Table S1. Microbiological and biogeochemical studies of the sulfur cycle in dysoxic marine waters, grouped by environment. Volumes based on the work of Paulmier and Ruiz‐Pino (2009) correspond to the estimated volume of waters containing > 0.5 μM nitrite. The maximum volume of anoxic water off the Namibian coast was calculated from the largest observed extent of sulfidic bottom waters (7000 km2; Lavik et al., 2009) and an assumed sulfidic layer thickness of 10 m.
Table S2. Origin, quality, classification, annotation, and average amino acid identity (AAI) of the analysed genomes in Fig. 5. Methods are described in Supporting Information Methods. The AAI values were calculated with an enveomics script using Diamond because of computational limitations, which could lead to significantly overestimated AAI values between 50% and 60% (https://rodriguez-r.com/blog/aai-blast-vs-diamond/). Following the thresholds proposed by Konstantinidis and colleagues (2017), values exceeding the 65% genus‐level lower threshold are coloured green, and values exceeding the 45% family‐level lower threshold are coloured yellow.
Table S3. Gibbs free energies [ΔG (kJ e−)] of common dissimilatory conversions mediated by anaerobic microorganisms under conditions representative of the upper euxinic zone of the Black Sea and the core of the ETSP OMZ. Calculation methodology and variables used can be found in Supporting Information Methods.
Appendix S1. Supplementary Information Methods.
Appendix S2. Supplementary Information Protologue.


