Abstract
The hereditary disease Retinitis pigmentosa results in severe vision loss due to photoreceptor degeneration by unclear mechanisms. In several disease models, the second messenger cGMP accumulates in the degenerating photoreceptors, where it may over‐activate specific cGMP‐interacting proteins, like cGMP‐dependent protein kinase. Moreover, interventions that counteract the activity of these proteins lead to reduced photoreceptor cell death. Yet there is little or no information whether other than such regular cGMP‐interactors are present in the retina, which we, therefore, investigated in wild‐type and retinal degeneration (rd1, rd10, and rd2) mouse models. An affinity chromatography based proteomics approach that utilized immobilized cGMP analogs was applied to enrich and select for regular and potentially new cGMP‐interacting proteins as identified by mass spectrometry. This approach revealed 12 regular and 10 potentially new retinal cGMP‐interacting proteins (e.g., EPAC2 and CaMKIIα). Several of the latter were found to be expressed in the photoreceptors and to have proximity to cGMP and may thus be of interest when defining prospective therapeutic targets or biomarkers for retinal degeneration.

Keywords: cGMP, cGMP‐interacting proteins, Chemical proteomics, Photoreceptors, Retinal degeneration
The involvement of cGMP (cyclic guanosyl monophosphate) in the retinal degeneration mechanism(s) is elusive, so we applied a proteomics approach to look for cGMP‐binding proteins in healthy and degenerating retinas. This approach revealed known binding proteins (PKG, PKAs, and PDEs) as well as potential new cGMP‐interacting proteins (EPAC2 (RAPGEF4), p‐CaMKIIα, p‐GSK3β, and MAPK1/3). The potential new cGMP‐interacting proteins showed proximity with cGMP in situ in the retina. While an indirect or direct binding of cGMP to these proteins is not yet verified, the results indicate them as important proteins to decipher the cGMP signaling pathway and hence the degeneration mechanisms. Illustration created with https://biorender.com/.

Abbreviations
- ACN
acetonitrile
- ADP
adenosine 5′‐diphosphate
- AGC
automatic gain control
- BLAST
basic local alignment search tool
- C18
18 carbon chains
- CaMKIIγ
Ca2+/calmodulin‐dependent protein kinase II gamma
- CaMKIIδ
Ca2+/calmodulin‐dependent protein kinase II delta
- cAMP
adenosine 3’, 5’‐cyclic monophosphate
- cGMP
guanosine 3’, 5’‐cyclic monophosphate
- CNB
cyclic nucleotide binding
- CNG
cyclic nucleotide gated
- ctrl
control
- DAPI
4′,6‐diamidino‐2‐phenylindole
- DDA
data‐dependent acquisition
- DTT
DL‐Dithiothreitol
- EPAC2
Rap guanine nucleotide exchange factor
- E‐value
expect value
- FA
formic acid
- GDP
guanosine 5′‐diphosphate
- GSK3α
glycogen synthase kinase‐3 isoform alpha
- GSK3β
glycogen synthase kinase‐3 isoform beta
- HCD
high‐energy collision dissociation
- IAA
Iodoacetamide
- INL
inner nuclear layer
- LC
Liquid chromatography
- MAPKI
mitogen‐activated protein kinase I
- MS
mass spectrometry
- NCE
normalized collision energy
- ONL
outer nuclear layer
- PDE
phosphodiesterase
- PFA
paraformaldehyde
- PKA
cAMP‐dependent protein kinase
- PKG
cGMP‐dependent protein kinase
- PLA
proximity ligation assay
- PN
postnatal day
- RD
retinal degeneration
- RP
retinitis pigmentosa
- RRID
Research Resource Identifier (see scicrunch.org)
- RT
room temperature
- TFA
trifluoroacetic acid
- wt
wild‐type
1. INTRODUCTION
Retinitis pigmentosa (RP) is hereditary and leads to degeneration of retinal photoreceptors (i.e., rods and cones) (Fahim et al. 1993; Chang et al., 2011), and retinal remodeling (Pfeiffer et al., 2020). While it usually starts with night blindness, RP often leads to complete blindness (Hanna et al., 2017; Petit et al., 2012). The prevalence of RP is reported as 1 in 3,000 to 7,000 people worldwide (Haim, 2002; Fahim et al. 1993), and over 60 genes have been identified to cause the disease (“RetNet, https://sph.uth.edu/RetNet/”, (Daiger et al. 1998), albeit the degeneration mechanisms remain unknown. Some gene therapy studies have been promising and entered the clinical stage, with currently one approval for a defect in the RPE65 gene, which concerns Leber's Congenital Amaurosis, an RP subtype (Petit et al., 2012; Pichard et al., 2016; Russell et al., 2017), but otherwise, RP therapies are lacking. A significant reason for this is likely the poor understanding of its pathology.
To study the RP mechanisms, several human homolog animal models are available which carry similar mutations as certain RP patient cohorts, such as the rd1, rd10, and rd2 mice (Arango‐Gonzalez et al., 2014; Chang et al., 2002). The rd1 mutation affects the beta subunit of the rod‐specific phosphodiesterase 6 (PDE6b) (Arango‐Gonzalez et al., 2014; Bowes et al., 1990; Sahaboglu et al., 2013), leading to a lack of hydrolyzation of guanosine 3’, 5’‐cyclic monophosphate (cGMP) in these cells. The rd1 degeneration in the photoreceptors of the outer nuclear layer (ONL) is both early, with onset at about postnatal day (PN) 7–8, and rapid, peaking at around PN13 with very few rods left at PN28 (Arango‐Gonzalez et al., 2014; Sahaboglu et al., 2013). The rd10 likewise has a Pde6b mutation, but different from rd1 and with a later onset of its rod degeneration, which becomes detectable and peaks in the third week and continues until about PN60 (Arango‐Gonzalez et al., 2014; Chang et al., 2002, 2007). The rd2 model concerns a peripherin 2 gene mutation, with degeneration onset and peak after two to three weeks, and with continued degeneration for up to a year (Arango‐Gonzalez et al., 2014; Chang et al., 2002). Interestingly, also rd10 and rd2 display cGMP accumulation in photoreceptors, and this feature is seen in yet other models as well (Arango‐Gonzalez et al., 2014; Power et al., 2020). A key role for cGMP in at least several forms of RP, therefore, seems likely (Power et al., 2020), although its downstream interactors remain elusive, except for cGMP‐dependent protein kinase (PKG) (Paquet‐Durand et al., 2009; Vighi et al., 2018) and cyclic nucleotide‐gated (CNG) channels (Paquet‐Durand et al., 2011).
The presence of cGMP‐interacting proteins can be studied using, for example, proteomics, and previous such investigations have indeed resulted in the discovery of such proteins in multiple tissues (Kim & Park, 2003; Scholten et al., 2006, 2007). Regular cGMP‐interacting proteins include PKG (Corradini et al., 2015; Francis et al., 2005; Francis & Corbin, 2013; Paquet‐Durand et al., 2009), and cAMP‐dependent protein kinase (PKA), which shares homology with PKG (Francis & Corbin, 2013) and which can be cross‐activated by cGMP, as well as CNG‐channels, phosphodiesterases, and cGMP transport proteins (Francis et al., 2005). However, seemingly unrelated proteins have also been reported as potential interactors, like the mitogen‐activated protein kinase I (MAPKI) (Kim & Park, 2003). It is thus reasonable to assume that yet other cGMP‐targets exist in the retina, including in RP, but to our knowledge, such information is not available. This needs to be studied, particularly since it could reveal possible detrimental cGMP signaling pathway(s) and identify new molecular targets for RP therapy. We have therefore taken on a proteomic approach, using affinity chromatography and mass spectrometry (MS) in combination with the mouse models mentioned above, including healthy wild type (wt) animals, to elucidate cGMP’s retinal interactors and possible disease‐related pathway(s).
2. MATERIALS AND METHODS
2.1. Materials
All chemicals were commercially available. Protease inhibitor (cat no. P5726) and phosphatase inhibitor (cat no. P3840), adenosine 5′‐diphosphate (ADP, cat. no. 01905), guanosine 5′‐diphosphate (GDP, cat. no. G7127), ProteoSilver™ silver stain kit (cat. no. PROTSIL1‐1KT), iodoacetamide (IAA, cat. no. I6125), trifluoroacetic acid (TFA, cat. no. 299537) and ammonium bicarbonate (cat. no A6141) were obtained from Sigma. The Bio‐Rad Protein Assay Kit (cat. no. 5000112) and DL‐dithiothreitol (DTT, cat. no. 1610610) were from Bio‐Rad. Sequencing Grade Modified Trypsin, Part No. V511A was from Promega. PageBlue protein staining solution (cat. no. 24620) and Nupage Gels 4%–12% (cat. no. NP0322PK2), NuPage sample buffer (cat. no. NP0008), and MOPS buffer (cat. no. B0001) were purchased from Thermo Scientific. Acetonitrile (ACN, cat. no. 1.59002) LiChrosolv and formic acid (FA, cat. no. 1.59013) LiChrosolv were from Merck. Free cGMP (cat. no. G001), cAMP (cat. no. A001), and cyclic nucleotide affinity beads: 8‐(2‐Aminoethylthio)guanosine‐3’, 5’‐cyclic monophosphate (8‐AET‐cGMP, Cat. No: A 019), N2‐(6‐Aminohexyl)guanosine‐3’, 5’‐cyclic monophosphate (2‐AH‐cGMP, Cat. No. A 056), 2’‐O‐(6‐ Aminohexylcarbamoyl)guanosine‐ 3’, 5’‐cyclic monophosphate (2’‐AHC‐cGMP, Cat. No. A 059), and EtOH‐NH‐Agarose (Cat. No. A 010) were provided by BIOLOG Life Science Institute (Bremen, Germany). Eppendorf tubes, Axygen Maximum Recovery (cat. no. MCT‐150‐L‐C), were purchased from Fischer Scientific. Ultra Micro spin Silica C18 column (Part # SUM SS18V, 3–30 μg capacity) was purchased from The Nest Group Inc. South Borough, MA, USA. All utilized antibodies are listed in Table 1.
TABLE 1.
List of antibodies used in this study
| Antibody | Provider | Cat. No. | Dilution IF/PLA |
|---|---|---|---|
| Rabbit anti‐PKGI | Abcam | Ab90502 | 1:500 |
| Sheep anti‐cGMP | Harry W.M. Steinbusch, Maastrich University, the Netherlands | Not applicable | 1:500 |
| Rabbit anti‐p‐CaMKII‐α (Thr 286) | Santa Cruz Biotechnology | SC‐12886‐R | 1:500 |
| Rabbit anti‐p‐GSK3β (Ser 9) | Cell Signaling Technology | 9,336 | 1:500 |
| Mouse anti‐MAPK1 | Santa Cruz Biotechnology | SC−1647 | 1:500 |
| Rabbit anti‐MAPK3 | Santa Cruz Biotechnology | SC−94 | 1:500 |
| Rabbit anti‐EPAC2 | Sigma | ABN492 | 1:500 |
2.2. Animals
The animals used were the following four different mouse lines: C3H rd1/rd1 (rd1, Sanyal & Bal, 1973), C57BL/6J rd10/rd10 (rd10, RRID:MGI:3581193, The Jackson Laboratory), C3H rd2/rd2 (rd2, Sanyal & Zeilmaker, 1984), and control C3H wild‐type (wt, Sanyal & Bal, 1973), that is, three retinal degeneration (RD) models and one healthy control line. All lines are kept and bred in house. No randomization of mice nor any sample calculation was performed in this study. The animals were housed under standard white cyclic lighting, had free access to food and water, and were used irrespective of sex. All the procedures were performed following the issued local animal ethics committee (permit #M92‐15) and the ARVO statement for the use of animals in ophthalmic and visual research. All efforts were made to minimize the number of animals used and their suffering. Prior to eye enucleation, the mice (PN11) were killed by decapitation, without prior anesthesia, as required by the permit. The different models display different degeneration characteristics, but for consistency, all animals were taken on PN11, as this would give the additional insight if the same cGMP‐interacting proteins were detected irrespective of the models’ mutations and progression.
The rd1 has the earliest onset of the three retinal degeneration (RD) models, yet at PN11, there is no reduction of its ONL thickness (Azadi et al., 2007; Hauck et al., 2006), and the risk for bias due to asymmetry in the retinal layer contribution to a sample is hence small. We, therefore, let these characteristics of the rd1 govern the time point for all analyses. The outcome from these RD models was compared to the one from the wt line. The study was not pre‐registered.
2.3. Sample preparation
A total number of 120 mice (i.e., 240 eyes, see Table 2) from the RD (60 retinas/RD model) and wt (60 retinas) mouse models were enucleated early morning. The retinas were dissected under a bright light in 4℃ homogenizing buffer [50 mM Tris‐HCl, 50 mM NaCl, 1 mM EDTA, 50 mM NaF, 5 mM NaH2PO4, 1 mM DTT, and protease‐ and phosphatase inhibitors] followed by homogenization using a sonicator. The homogenate was centrifuged 10.000 g for 30 min at 4℃. The protein concentration of the obtained soluble fraction was measured by the Bio‐Rad Protein Assay Kit, and the fraction was stored at −80℃ until use. The sample size, 60 retinas per mouse model, was optimized in preliminary experiments (here 108 mice were used to optimize the approach) where the used protein amount (i.e., 1.4 mg) showed to be sufficient to perform the proteomics approach and subsequently get an acceptable MS outcome. Furthermore, no exclusion criteria were pre‐determined.
TABLE 2.
Overview of mouse models and numbers of retinas used for this study. In all cases, the age of the animals was PN11
| Mouse model | Number of animals | Number of retinas used | Mutated gene |
|---|---|---|---|
| wt | 30 | 60 | – |
| rd1 | 30 | 60 | Pde6β |
| rd10 | 30 | 60 | Pde6β |
| rd2 | 30 | 60 | Peripherin 2 |
| Total number | 120 | 240 | – |
2.4. Affinity pull down
The soluble fraction [1.4 mg] was added to 60 µl of 8‐AET‐cGMP, 2‐AH‐cGMP, 2‐AHC‐cGMP, or EtOH‐NH beads, respectively. No blinding of the samples was used during the affinity chromatography. The lysate‐beads suspensions were left to incubate under agitation for 1 hr at 4℃, followed by washes in ice‐cold PBS. The beads were subsequently subjected to several elution steps to remove the unspecific binding and enrich for cGMP‐interacting proteins, as previously described (Scholten et al., 2006). The elution steps consisted of 2 × 50 µL of GDP [10 mM], ADP [10 mM], cAMP [10 mM], cGMP [10 mM], for 5 min at 4℃, followed by a short centrifugation at 10.000 g to pellet the beads. The protein concentration in the eluates was too low for determination, so eluted proteins (10 µl eluate) were instead visualized by silver staining after standard electrophoresis.
2.5. In‐solution sample preparation for liquid chromatography (LC)‐MS/MS
The rd2 eluates (no blinding of the samples was used in MS) were reduced with DTT to a final concentration of 10 mM and heated at 56°C for 30 min followed by alkylation with IAA to a final concentration of 20 mM for 30 min at 20°C in the dark. Digestion was performed by adding trypsin (0.1 µg, Sequencing Grade Modified Trypsin, Part No. V511A) followed by overnight incubation at 37°C, and terminated by the addition of 10% TFA. Each fraction underwent desalting by Ultra Micro spin Silica C18 column (SUM SS18V, 3–30 μg capacity, The Nest group Inc. South Borough) and was dried by a centrifugal evaporator before resuspension with solvent A (0.1% FA).
The peptide analyses were performed by MS on a Q Exactive HFX mass spectrometer (Thermo Scientific) connected to an Ultimate 3,000 ultra‐high‐performance LC system (Thermo Scientific). The digested peptides (10 µl) were loaded on a pre‐column (Thermo Scientific; ID 75 μm × 2 cm, column temperature 35°C) and then separated on an EASY‐Spray column (Thermo Scientific; Inner diameter 75 μm × 25 cm, column temperature 45°C). A non‐linear gradient of buffer B (80% ACN, 0.1% FA) in buffer A (aqueous 0.1% FA) was applied at a flow rate of 300 nl/min. For the gradient, the percentage of solvent B was maintained at 6% for 4 min, increased to 31% for 60 min, to 50% for 5 min, and to 90% for 13 min. Solvent B was then decreased to 6% and kept for 15 min.
One full MS scan (resolution 120.000 @ 200 m/z; mass range 375–1.500 m/z) was followed by inclusion‐list driven MS/MS scans (resolution 15.000 @ 200 m/z). The precursor ions were isolated with 1.2 m/z isolation width and fragmented using higher‐energy collisional‐induced dissociation at a normalized collision energy of 28. Charge state screening was enabled, and singly charged ions, as well as precursors with a charge state above 6, were rejected. The dynamic exclusion window was set to 40 s. The automatic gain control was set to 3e6 for MS and 1e5 for MS/MS with ion accumulation times of 50 ms and 19 ms, respectively. The intensity threshold for precursor ion selection was set to 1.6e5.
2.6. In‐gel digestion sample preparation for LC‐MS/MS
For wt, rd1, and rd10, the samples (used without blinding) were prepared by in‐gel digestion to avoid the presence of stray beads that risked to clog the liquid chromatography system, which was experienced with the in‐solution method used for the rd2 samples. The wt, rd1, and rd10 eluates were precipitated with 100% ethanol to a final concentration of 90% ethanol and allowed to stand at −20°C overnight. The pellets were dried and resuspended in 1 X NuPage sample buffer reduced with 0.5 M DTT and heated at 56°C for 30 min, followed by alkylation with IAA. The samples were applied to a 10% NuPage gel and run until the samples migrated out of the wells. The bands were then cut out and washed with MQ for 30 min on a shaker at 20°C, after which the liquid was removed (as it was after each of the following steps). The gel bands were then washed with 40% ACN in 25 mM ammonium bicarbonate for 30 min, followed by 100% ACN, and dried using a vacuum centrifuge, after which trypsin (0.01 µg/µl) was added for overnight incubation at 37°C. Peptides were then extracted by adding 1% FA in MQ, followed by 100% ACN for 5 min, and the liquid transferred to collection tubes. These two last steps were repeated twice in total. The extracted peptides were finally dried using a vacuum centrifuge and resolved in 2% ACN in 0.1% TFA.
2.7. Mass spectrometry acquisition
The LC‐MS detection was performed on a Tribrid mass spectrometer Fusion equipped with a Nanospray Flex ion source and coupled with an EASY‐nLC 1,000 ultra‐high‐pressure LC pump (Thermo Scientific). The peptides, 10 µl, were injected into the LC‐MS. First, the peptides were concentrated on an Acclaim PepMap 100 C18 precolumn (75 μm × 2 cm, Thermo Scientific) before being separated on an Acclaim PepMap RSLC column (75 μm × 25 cm, nanoViper, C18, 2 μm, 100 Å) at 40°C with a flow rate of 300 nl/min. Solvent A (0.1% FA in water) and solvent B (0.1% FA in ACN) were used to create a nonlinear gradient to elute the peptides.
The Orbitrap Fusion was operated in the positive data‐dependent acquisition (DDA) mode. The peptides were introduced into the LC‐MS via stainless steel Nano‐bore emitter (outer diameter 150 µm, inner diameter 30 µm) with the spray voltage of 2 kV, and the capillary temperature was set 275°C. Full MS survey scans from m/z 350–1350, with a resolution of 120.000, were performed in the Orbitrap detector. The automatic gain control (AGC) target was set to 4 × 105 with an injection time of 50 ms. The most intense ions (up to 20 ions) with charge states 2–5 from the full scan MS were selected for fragmentation in the Orbitrap. The MS2 precursors were isolated with a quadrupole mass filter set to a width of 1.2 m/z. Precursors were fragmented by high‐energy collision dissociation (HCD) at normalized collision energy (NCE) of 30%. The resolution was fixed at 30,000, and for the MS/MS scans, the values for the AGC target and injection time were 5 × 104 and 54 ms, respectively. The duration of dynamic exclusion was set to 45 s, and the mass tolerance window was 10 ppm.
2.8. Data analysis
The raw DDA data were analyzed with Proteome Discoverer™ 2.2 (PD 2.2) Software (Thermo Scientific). Peptides were identified using SEQUEST HT against UniProtKB Mouse + isoforms database (release 20190617). The search was performed with the following parameters applied: Static modification: cysteine carbamidomethylation and Dynamic modifications: N‐terminal acetylation and phosphorylation. The precursor tolerance was set to 10 ppm, and fragment tolerance was set to 0.02 ppm for the HFX instrument and 0.05 for the Fusion. Up to 2 missed cleavages were allowed, and Percolator was used for peptide validation at a q‐value of a maximum of 0.01. Filter settings at the protein level: Master is equal to Master.
2.9. Proximity ligation assay (PLA)
PLA was performed on cryosections of 4% paraformaldehyde (PFA) fixed retinas, using Detection Reagents Red kit, the PLA Probe anti‐rabbit or anti‐goat MINUS, and a cGMP antibody, which had a PLUS probe attached using Duolink® In Situ Probemaker PLUS (all Duolink, Olink Bioscience). The procedure followed the manufacturer's instructions. In short, the retinal sections were permeabilized and blocked with blocking solution (1% BSA, 0.25% Triton X‐100 in PBS) for 45 min at 20°C, and primary antibodies were incubated overnight at 4°C. PLA Probe anti‐rabbit MINUS incubated for 1 hr at 37°C. The ligation and amplification steps were performed using the Detection Reagents Red, followed by the addition of mounting medium with 4′,6‐diamidino‐2‐phenylindole (DAPI, Abcam, Cambridge, England). To visualize the PLA punctuations (i.e., reaction products from the cGMP and protein of interest when maximally 40 nm apart), pictures were taken of the full retinal cross‐sections at 20x magnification using the Zeiss Axio Imager.M2 microscope. The total number of PLA‐punctuations was counted in images from samples from independent retinas (three retinas/ RD model and nine retinas/wt model), respectively, with the aid of Zen (2.5) blue edition software (Zeiss Zen software). The PLA images had first been made unidentifiable, and the punctuations counted manually by a person, thus unaware of the samples' identity. After counting, the identity of the pictures was disclosed and the results were compiled and depicted with histograms as the ratio of the ONL over the inner nuclear layer (INL), i.e., ONL/INL, using GraphPad Prism 8.1.
2.10. Statistical analysis
To quantify the number of PLA punctuations on the respective sections (i.e., wt, rd1, rd10, and rd2) the nonparametric statistical method Kruskal‐Wallis was applied. The test was performed using GraphPad Prism 8. The figure (i.e., Figure 3) displays mean and error bars representing SD and level of significance was set to p < .05. The probability of finding statistically significant differences between the RD and wt models per staining condition (i.e., PKGI, EPAC2, p‐CaMKIIα, p‐GSK3β, and MAPK1/3) was only high in the EPAC2:cGMP staining between the rd10 and rd2 model.
FIGURE 3.
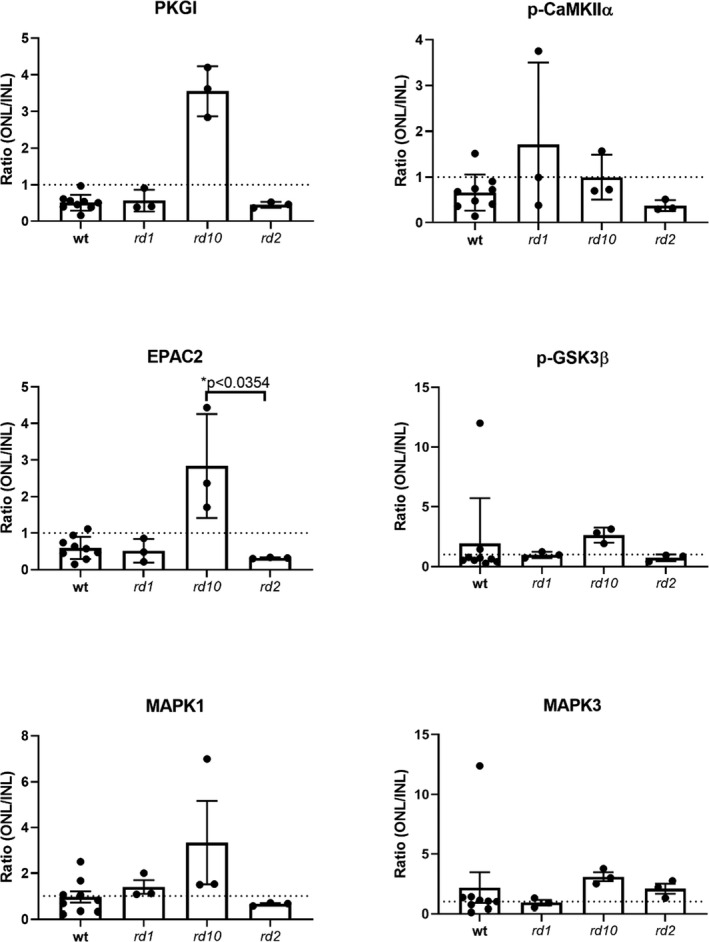
Overview of the ratios of the counted PLA punctuations in the retinas. The PLA punctuations were counted in a blinded fashion and the ratio of ONL/INL displayed in histograms. The statistical method Kruskal‐Wallis was applied to calculate whether some of the proteins had more proximity with cGMP in the respective RD and wt models and revealed that the probability to have more proximity between the cGMP:EPAC2 was higher in the rd10 compared to the rd2 model. Data is based on three biologically independent RD retinas (n = 3) and nine biologically independent wt retinas (n = 9). Error bars indicate standard deviation. Note the different scales of the y‐axes
3. RESULTS
Here we employed an optimized affinity chromatography method (summarized in Figure 1a) to evaluate RD and wt retinas for cGMP‐interacting proteins. The sample size, 60 retinas per mouse model, was optimized in preliminary experiments, using different amounts of protein, where the highest amount tested (i.e., 1.4 mg) was found to be sufficient to perform the affinity step and subsequently get a good MS outcome. Isolation of cGMP‐interacting proteins was achieved using affinity beads with synthetic cGMP linked at different sites, to increase the potential to enrich proteins that may bind to cGMP at different positions (Figure 1b). Since the aim was to study cGMP interactors as broadly as possible, we combined the identified proteins from the various beads in one table.
FIGURE 1.
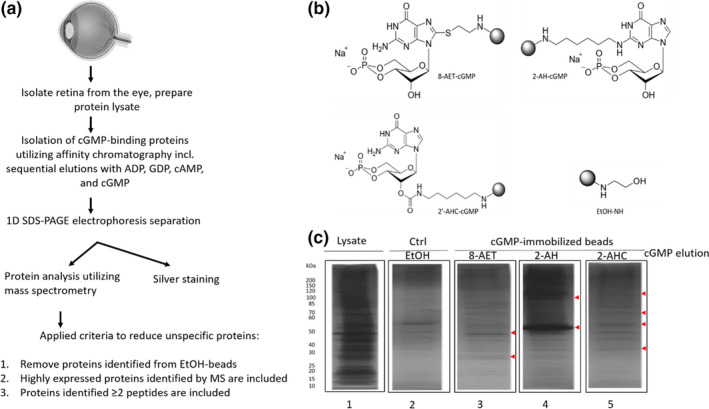
Enriched cGMP‐interacting proteins in the various RD models and wt. (a) Schematic overview of the sequential elution protocol used for the enrichment of potential cGMP binding‐proteins, (eye from BioRender.com) (b) Chemical structures of different cGMP‐immobilized beads used in this study, (c) Silver stained SDS‐PAGE indicating a successful decrease in unspecific proteins. From left, lane 1: Raw retinal wt tissue (1 µg). Lane 2: Proteins eluted with cGMP from control (Ctrl) beads (EtOH). Lane 3–5: cGMP‐interacting proteins eluted with cGMP from cGMP‐immobilized beads (8‐AET, 2‐AH, 2‐AHC). Red arrowheads indicate protein bands at various sizes, which are unique for the cGMP‐immobilized beads compared to Ctrl beads
3.1. Sequential separation reveals proteins having an affinity for cGMP
To avoid the analysis of proteins that bound unspecifically to the immobilized cGMP or the beads as such (8‐AET, 2‐AH, and 2‐AHC, see overview in Figure 1a) proteins were eluted by sequential separation steps with related analogs (GDP, ADP, cAMP) before elution with cGMP as the final step (Scholten et al., 2006). The cGMP elution would thus contain enriched cGMP‐interacting proteins, although the proteins eluted by cAMP were also analyzed since it is well‐known that cAMP may cross‐activate some cGMP‐interactors (Jiang et al., 1992; Kim et al., 2011; Pavlaki & Nikolaev, 2018; Scholten et al., 2006, 2007; Shabb, 2010). As another measure to reveal false positives, we performed similar sequential elutions of proteins bound to the control (Ctrl; EtOH) beads. The initial SDS‐PAGE gel analysis of the affinity chromatography revealed a difference in the pattern of wt retinal proteins eluted by free cGMP from the cGMP beads (8‐AET, 2‐AH, and 2‐AHC, see Figure 1c; see Figure S1 for rd1, rd10, and rd2 silver stainings) when compared to the raw lysate. This indicated that the beads captured specific proteins and not merely reflected the raw lysate's protein composition. Moreover, the cGMP elution patterns differed between the cGMP beads and the Ctrl beads, suggesting that some bands were unique to the cGMP beads (examples shown with arrowheads in Figure 1c). These results indicate that there appear to be proteins whose binding shows specificity for cGMP and, conversely, that some proteins may bind to the beads regardless if they carry an immobilized cGMP analog or not.
Identified proteins eluted with cAMP or cGMP from the cGMP‐immobilized beads were merged (from now on referred to as cGMP‐bead‐eluates), as was the cAMP and cGMP eluates from the Ctrl beads (from now on referred to as Ctrl‐bead‐eluates). The MS analyses of the cGMP‐bead‐eluates resulted in a high number (>100) of different proteins (see Table [Link], [Link], [Link], [Link] for wt, rd1, rd10, and rd2 MS data, respectively). To focus on cGMP‐interacting proteins and to avoid unspecific proteins, strict and defined criteria for accepting bona fide cGMP‐interacting proteins were employed: Proteins identified in the Ctrl‐bead‐eluates were excluded from further analyses, and an acceptance threshold of a minimum of two unique peptides identified for each protein was applied for the remaining proteins (Table [Link], [Link], [Link], [Link]). Utilizing these criteria resulted in several regular cGMP‐interacting proteins from all the RD and wt models. Some of these were kinases like PKGI and PKAs (PKAIa, PKAIIa, PKAIb, and PKAIIb, Table 3), which due to their known cGMP and or cAMP binding (Kim et al., 2011; Lorenz et al., 2017; Osborne et al., 2011) supported that the experimental set‐up works. Another group of proteins that interact with cGMP and or cAMP was likewise detected, namely six members/subunits of the PDE family: Pde1a, Pde1b, Pde1c, Pde2a, Pde5a, and Pde6a (Table 3). It was noted that the identified protein profile was not similar for all the models.
TABLE 3.
Regular cGMP‐interacting proteins in the retina of the mouse models
| Accession | Gene | Protein | wt | rd1 | rd10 | rd2 |
|---|---|---|---|---|---|---|
| Q61481 | Pde1a** | Calcium/calmodulin‐dependent 3',5'‐cyclic nucleotide phosphodiesterase 1A | X | |||
| Q01065 | Pde1b | Calcium/calmodulin‐dependent 3',5'‐cyclic nucleotide phosphodiesterase 1B | X | X | X | X |
| Q64338 | Pde1c* | Calcium/calmodulin‐dependent 3',5'‐cyclic nucleotide phosphodiesterase 1C | X | X | X | |
| Q922S4 | Pde2a | cGMP‐dependent 3',5'‐cyclic phosphodiesterase | X | X | X | |
| Q8CG03 | Pde5a** | cGMP‐specific 3',5'‐cyclic phosphodiesterase | X | X | ||
| P27664 | Pde6a | Rod cGMP‐specific 3',5'‐cyclic phosphodiesterase subunit alpha | X | X | ||
| P0C605 | PKGI | cGMP‐dependent protein kinase 1 | X | X | X | X |
| Q9DBC7 | PKAIa | cAMP‐dependent protein kinase type I‐alpha regulatory subunit | X | X | X | X |
| P12367 | PKAIIa | cAMP‐dependent protein kinase type II‐alpha regulatory subunit | X | X | X | X |
| P12849 | PKAIb | cAMP‐dependent protein kinase type I‐beta regulatory subunit | X | X | X | |
| P31324 | PKAIIb | cAMP‐dependent protein kinase type II‐beta regulatory subunit | X | X | X | X |
| P68181 | Prkacb | cAMP‐dependent protein kinase catalytic subunit beta | X |
Table 3 Overview of eluted and selected regular cGMP‐interacting proteins in the retina. The table shows regular cGMP‐interacting proteins enriched from the mouse models (wt, rd1, rd10, and rd2). An X indicates which protein was identified in which mouse model. The superscripts +, * and ** indicate that the corresponding protein was detected by only 1 unique peptide (see MS data in Supporting information) and hence not included in this list for wt+, rd1*, and rd10**, respectively.
The BLAST (Basic Local Alignment Search Tool) search, which identifies regions of similarities between proteins, was then applied to help identifying a protein as a potential novel cGMP‐interactor. We here used PKGI as the query in the search algorithm for the identified proteins of all the models (Table [Link], [Link], [Link], [Link]), since PKGI is known to bind cGMP in the cyclic nucleotide‐binding (CNB) domain (Huang et al., 2014), which revealed numerous proteins having sequence similarities with PKGI. We moreover applied a threshold to include only proteins with a negative Expect value (E‐value), since this signifies high sequence similarity with PKGI. The BLAST outcome identified the expected PKAs (already listed in Table 3), which can bind cGMP (Lorenz et al., 2017), which thus supported the BLAST search's ability to identify cGMP‐interacting proteins. Nine other kinases (with sequence similarities with PKGI in the catalytic region) and one exchange protein (the Rap guanine nucleotide exchange factor; EPAC2, with sequence similarity in the CNB domain) were further identified as potential new cGMP‐interacting proteins in the retina (Table 4). These proteins were all found in the cGMP‐bead‐eluates of the wt, rd10, and rd2 models. However, in the rd1 model, only three of them were detected, namely Ca2+/calmodulin‐dependent protein kinase II gamma and delta (CaMKIIγ and CaMKIIδ) and MAPKI, and this discrepancy, or inconsistency, thus resembles the situation above concerning Table 3. Other BLAST searches with the queries Pde5, CNG, and PKA (i.e., other proteins known to interact with cGMP), were correspondingly tested. These identified a few proteins with high sequence similarity, but which all were already included in the PKGI BLAST results and therefore not specified here in detail.
TABLE 4.
Potential retinal cGMP‐interacting proteins based on high sequence similarity with PKGI
| Accession | Gene | Description | wt | rd1 | rd10 | rd2 | Previously detected in the retina | Previously shown photoreceptor protective effect |
|---|---|---|---|---|---|---|---|---|
| P28028 | Braf | Serine/threonine‐protein kinase B‐raf | X | X | X |
Yes (Xie et al., 2017) |
||
| P11798 | CamkIIa | Calcium/calmodulin‐dependent protein kinase type II subunit alpha | X | X | X | Yes (Bronstein et al., 1988) |
Yes (Laabich & Cooper, 2000) |
|
| P28652 | Camk2IIb | Calcium/calmodulin‐dependent protein kinase type II subunit beta | X | X | X |
Yes |
||
| Q923T9 | CamkIIg | Calcium/calmodulin‐dependent protein kinase type II subunit gamma | X | X | X | X |
Yes (Bronstein et al., 1988) |
|
| Q6PHZ2 | CamkIId | Calcium/calmodulin‐dependent protein kinase type II subunit delta | X | X | X | X |
Yes (Bronstein et al., 1988) |
|
| Q9EQZ6 | EPAC2 | Rap guanine nucleotide exchange factor 4 | X | X | X |
Yes (Whitaker & Cooper, 2010a) |
||
| Q2NL51 | Gsk3a | Glycogen synthase kinase−3 alpha | X | X | X |
Yes (Marchena et al., 2017; Pérezleón et al., 2013; Sánchez‐Cruz et al., 2018) |
Yes |
|
| Q9WV60 | Gsk3b | Glycogen synthase kinase−3 beta | X | X | X |
Yes (Marchena et al., 2017; Pérezleón et al., 2013; Sánchez‐Cruz et al., 2018) |
Yes |
|
| P63085 | Mapk1 | Mitogen‐activated protein kinase 1 | X | X | X | X |
Yes (Shinde et al., 2012) |
Yes |
Table 4 Overview of BLAST results. Potential cGMP‐interacting partners fulfilling the requirements described in the text, including a high sequence similarity to the known cGMP‐interacting protein PKGI when compared using a BLAST search. CaMKIIa, GSK3, and MAPK1/3 have previously been shown to have a photoreceptor protective effect (i.e., reduced photoreceptor cell death) when intervention against these proteins was applied in relevant situations; see text for more information. An X indicates which protein was identified in which mouse model.
We thus conclude that employing defined criteria and performing focused searches brought to light both expected and new cGMP interactors. The next question was if such interactions with cGMP, particularly those of the novel cGMP‐interacting proteins, are taking place in situ, or are just a result of the opportunity to interact directly or indirectly (e.g., in a complex) in the homogenate.
3.2. Potential retinal cGMP‐interacting proteins indicate interaction in situ
To understand if the potentially new cGMP binding proteins have relevant interactions in the retina we selected five of these (EPAC2, CamKIIα, GSK3β, MAPK1, and MAPK3), based on if the proteins previously had been detected in retinal photoreceptors, and if interventions against these proteins had shown a photoreceptor protective effect, which hence would tie them to the well‐being of these cells (Table 4). Their proximity to cGMP was hence tested by PLA in situ on retinal cross‐sections from wt and RD models.
3.3. PKGI
The direct interaction between cGMP and PKGI has previously been inferred (Huang et al., 2014) and hence chosen as a positive control. For negative controls, we used the omission of antibodies (see Figure 2 and Figure S2). The PLA assay with anti‐cGMP and anti‐PKGI (cGMP:PKGI) showed clear punctuations in the various layers of the retina (see Figure 2a for a section of the wt retina, Figure S2 for magnified PLA snapshots from wt and RD models, and Table S5 for average PLA counts for each model). The ratio from the cGMP:PKGI showed relatively more punctuations in the INL than in the ONL, giving a low ONL/INL ratio in wt as well as rd1 and rd2 (but not rd10; Figure 3). However, the average number of cGMP:PKGI punctuations in the ONL was nonetheless considerable in the rd1 retinas when compared with wt, as well as with rd10 and rd2 (Table S5).
FIGURE 2.
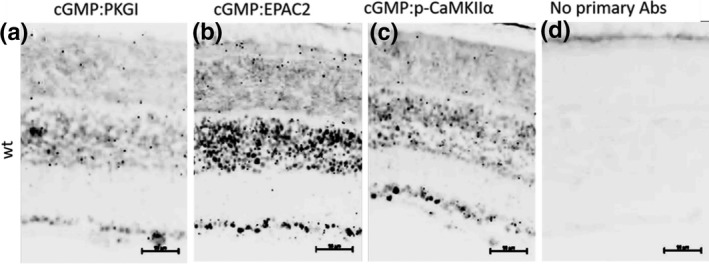
cGMP interactions in the retina. Proximity ligation assay showing (a) cGMP proximity to PKG, a known cGMP interactor. (b and c) cGMP proximity to EPAC2 and p‐CaMKIIα, respectively, either via direct interaction, or indirectly in a complex. (d) The negative control, where no primary antibodies were added to the sections, showing background staining from the PLA kit which is virtually non‐existing. Black (inverted pictures) punctuations are a positive outcome from the PLA, suggesting interaction. Scale bars: 50 µm. Biologically independent wt retinas n = 9
3.4. EPAC2 and P‐CaMKIIα
The exchange protein EPAC2 is an accepted cGMP‐interacting protein, at least in vitro (Rehmann et al., 2008), but EPAC2 has, to the best of our knowledge, never been shown to interact with cGMP in the retina (i.e., at the in situ level). PLA with EPAC2 revealed proximity to cGMP in the entire retina in all models (see Figure 2 for the situation in wt and Figure S2 for RD models). The rd10 model again had a numerically higher ONL/INL ratio value than any of the other retinas. The PLA with the cGMP:p‐CaMKIIα combination likewise gave numerous punctuations in all the layers in the retina, similar to the cGMP:PKGI PLA. The ONL/INL ratios displayed quite extensive variations within and between the different groups (Figure 3).
3.5. Phosphorylated‐GSK3β AND MAPK1/3
The PLA with cGMP:p‐GSK3βSer9 revealed proximity (see Figure 2 for wt and Figure S2 for RD models) with variable ONL/INL ratio outcomes between the groups (Figure 3). PLA with MAPK1 and MAPK3 also confirmed proximity to cGMP (Figure 2 and Figure S2). The ONL/INL ratios of the MAPKs likewise displayed variations within and between groups (Figure 3).
The PLA profiles for the cGMP: p‐GSK3βSer9 and cGMP:MAPK1/3 in general revealed only very few PLA punctuations in situ in the entire wt retina (PLA counts for cGMP:p‐GSK3βSer9 and cGMP:MAPK1/3 revealed between 30 to 350 punctuations, whereas, for example, cGMP:PKGI revealed >1,000 punctuations, see Table S5) when compared with the other PLA profiles in Figure 2. The proximity of these two proteins to cGMP may in quantitative terms thus be limited. Overall the statistical method Kruskal‐Wallis did calculate that there is a higher probability for more proximity between cGMP and EPAC2 in the rd10 than the rd2 model.
4. DISCUSSION
Here we successfully pulled‐down and identified cGMP‐interacting proteins from retinal tissue. Together with the PLA assay, this revealed that retinal cGMP is in proximity to several previously undescribed retinal cGMP‐interacting proteins (e.g., EPAC2, MAPK1/3, p‐GSK3β, and p‐CaMKIIα), which is information that may help elucidate the cGMP mechanism(s) during retinal degeneration (see Figure 4 for proposed pathway interactions).
FIGURE 4.
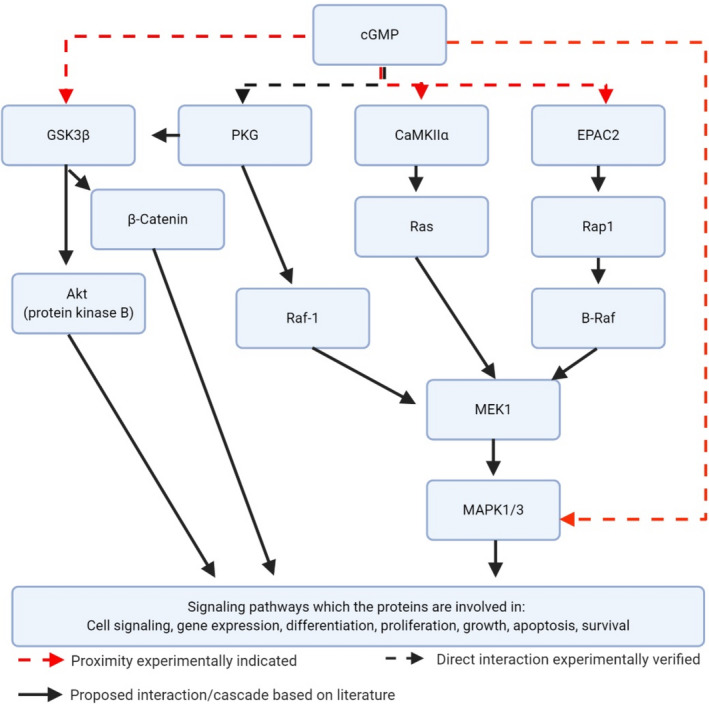
Illustration of the potential cGMP interaction proteins, and corresponding pathways, identified by the proteomics approach. cGMP has previously been experimentally verified to interact directly with PKG (Huang et al., 2014), whereas p‐GSK3β has previously been indicated to interact with PKG (Das et al., 2008; Hong et al., 2012). cGMP is an accepted EPAC2 interactor (Rehmann et al., 2008) and is suggested to be involved in the MAPK1/3 cascade (Maymó et al., 2012) as well is CaMKIIα (Caunt et al., 2006; McKay & Morrison, 2007). The present study revealed that EPAC2, MAPK1/3, p‐CaMKIIα and p‐GSK3β are in proximity to cGMP in situ in the retina. Created with Biorender.com
4.1. Identification of cGMP‐interacting proteins from retinas using a proteomics approach
Our strategies to reduce and refine the high number of the MS identified proteins resulted in two more limited sets, representing either expected or potentially new cGMP‐interacting proteins, respectively. The explanation for the originally high number of proteins in the cGMP eluates could include, 1) that some proteins still, even after washes and pre‐elutions, for yet unknown reasons remained bound to the beads in an unspecific way until released by cAMP or cGMP, and/or 2) that they represent proteins that themselves were bound to “true” in vivo cGMP‐interacting proteins and thus co‐eluted by cGMP in a complex.
When comparing regular as well as potentially new cGMP‐interacting proteins identified by MS, several of these showed up in all the models studied, whereas some were lacking from one or more of these. Altogether, of the four models rd1 had the fewest hits, which may be related to the aggressiveness of its degeneration, that perhaps affects metabolic or other aspects to prevent the proteins in question from being separated and identified. This possibility is strengthened by that EPAC2, GSK3β, and MAPK3 still appeared in the rd1 retina in the in situ PLA studies, despite not appearing in the proteomics approach. Furthermore, despite their different mutations, all retina models were taken at PN11 where the actual extent of degeneration, and therefore possibly the protein expression profiles, differs between the models, which in turn could have influenced the outcome (Liu et al., 2016; Punzo & Cepko, 2007). Finally, although less likely, proteins could have been lost during the sample preparation (Scholten et al., 2007).
The membrane‐bound CNG‐channels are clearly expressed in the retina (Fesenko et al., 1985; Nache et al., 2016) but were not identified in our samples. Since no strong detergents were used in our experimental set‐up the extraction of membrane proteins was probably not extensive (Carpenter et al., 2008), and these proteins may hence have been absent in the initial supernatant fractions. Moreover, analyzing hydrophobic membrane proteins, such as in CNG channels (Giorgetti et al., 2005; Weitz et al., 2002), by MS is more complicated than the analysis of hydrophilic proteins. The commonly used desalting steps, including the one used here, are based on the peptides' hydrophobicity and would, therefore, discard highly hydrophilic and highly hydrophobic peptides (Carroll et al., 2007; Kim et al., 2016). Our results may thus be restricted by that membrane‐bound cGMP‐interactors could have escaped detection. Likewise, the small sample size (1.4 mg initial load) for the stepwise elution from the cGMP‐immobilized beads, may in combination with the applied criteria have limited the detection of yet other regular or new potential cGMP‐interacting proteins.
4.2. EPAC2 and p‐CAMKIIα proximity to cGMP in situ in the retina
To verify that the selected retinal proteins would be in proximity to cGMP in situ, we applied the PLA assay on retinal sections and established that cGMP was in proximity to the known cGMP‐interactor PKGI. This added validity to the assay and it was also in agreement with other studies, reporting that cGMP and PKG interact directly (Huang et al., 2014), as well as that PKGI mRNA (Gamm et al., 2000) and phosphorylated PKG‐substrates (Paquet‐Durand et al., 2009) are found throughout the mouse retina. However, the low cGMP:PKGI ONL/INL ratio in rd1 and rd2 was unexpected. As cGMP is increased in the ONL in all the models used (Arango‐Gonzalez et al., 2014; Paquet‐Durand et al., 2009), one would predict a higher ONL/INL ratio of cGMP:PKGI interactions in the RD situations compared to wt, but this was only suggested for rd10. While we currently do not know the reason for this, yet undefined metabolic or other changes may occur due to the RD, and these might differ between the RD‐models. In turn, this may affect the cGMP:PKGI interaction (or any of the other studied interactions) in the retina, and hence resolve in different ratios, as seen here. Moreover, the PLA only provides a snapshot of the existing interactions at the time of fixation and will not describe their number or persistence over a longer period.
It is known that cAMP can interact with and activate the exchange protein EPAC2 (Das et al., 2009; Rehmann et al., 2003; Schwede et al., 2015), and in the retina, EPAC2 has been localized to the inner and outer segments of cone photoreceptors (Whitaker & Cooper, 2010b). However, cGMP is also an accepted EPAC2 interactor (albeit with no or very weak ability to activate EPAC2 in vitro (Rehmann et al., 2008)), meaning that retinal cGMP:EPAC2 interactions may occur in situ. This was corroborated here by PLA in the entire retina in all models, and given the previous observations on cGMP‐EPAC2 interactions (Das et al., 2009; Rehmann et al., 2003, 2008; Schwede et al., 2015), EPAC2 may thus very well be a retinal cGMP‐interacting protein.
Amongst the potentially new cGMP‐interacting proteins found were also four isoforms of the multifunctional serine/threonine‐protein kinase CaMKII, that is, α, β, γ, and δ, which regulate various neuronal functions (Gao et al., 2012). These CaMKII isoforms are activated by Ca2+/calmodulin, and the calmodulin‐binding initiates a conformational change that allows autophosphorylation, turning a CaMKII isoform (e.g., CaMKIIα) into a constitutively active form (e.g., p‐CaMKIIα) (Migues et al., 2006). Some CaMKII isoforms (CaMKIIα, CaMKIIβ, and CaMKIIδ) have been investigated for possible roles in the retina, including their potential as therapeutic targets against stress and cell death (Kim et al., 2010; Fan et al., 2007; Tetenborg et al., 2017; Aulia Jusuf et al. 2015; Laabich & Cooper, 2000). Moreover, a previous study found CaMKII and p‐CaMKII to be present in the photoreceptors and suggested CaMKII to be involved in their degeneration in the rd1 model, but no discrimination was made between the various CaMKII isoforms (Hauck et al., 2006). Our results, therefore, add that the phosphorylated variant of CaMKIIα is present in and has proximity to cGMP in the mouse retina and that it thus may be important for cGMP‐dependent mechanisms, including cGMP dependent cell death.
4.3. GSK3β and MAPK1/3 proximity in situ in the retina
The GSK3β isoform of the constitutively active glycogen synthase kinase‐3 has been shown to be localized to inner segments of photoreceptors (Pérezleón et al., 2013), and the levels of GSK3β phosphorylated at serine 9 (GSK3βser9) have been reported to be elevated in the rd10 model compared to wt (Sánchez‐Cruz et al., 2018). It is also clear that interventions with small molecules that target and inhibit GSK3 can reduce photoreceptor cell death (Marchena et al., 2017), making GSK3 a promising therapeutic target for RD. Our PLA revealed proximity between cGMP and GSK3βser9 in situ, but since this appeared limited the cGMP association may primarily have been indirect, via PKG. PKGI has indeed previously been shown to phosphorylate GSK3βSer9 (Das et al., 2008; Hong et al., 2012), which inhibits the GSK3β activity, and this inhibition may thus reduce photoreceptor cell death. Consequently, a potential over‐phosphorylation of GSK3β by PKGI is likely not the mechanism by which cGMP contributes to RP cell death.
The last proteins selected for PLA consisted of the MAPK pathway proteins MAPK1 and MAPK3 (also known as ERK2 and ERK1, respectively). It has been reported that loss of MAPK1/3 function causes thinning of the retinal ONL and INL layers, suggesting that these MAPKs play a crucial role in photoreceptor survival (Pyakurel et al., 2017). The MAPK1/3 proteins showed limited proximity to cGMP, which, as reasoned above for p‐GSK3βSer9, could be because cGMP interacts with these proteins indirectly in a complex, perhaps including PKG. This fits with that when cGMP is elevated, MAPK1/3 becomes phosphorylated most likely by PKG (Ho et al., 1999; Zaragoza et al., 2002), although a direct binding between PKG and MAPK1/3 has, to our best knowledge, not been verified yet. Finally, it may after all also be that cGMP interacts directly with these kinases as previously suggested (Kim & Park, 2003), and if p‐GSK3βSer9 and MAPK1/3 are not the most favorable cGMP interactors this becomes reflected in the PLA outcome.
5. CONCLUSION
We successfully applied a proteomics approach to look for cGMP‐interacting proteins in healthy and degenerating retinas and employed several measures to exclude non‐specific binding proteins from the outcomes of the final cGMP‐eluates. The results revealed multiple regular cGMP‐interacting proteins, like PKGI, PKAs, and PDEs, which underlines the validity of the undertaking as well as reinforces the idea that RP therapies could target, for instance, PKG. Moreover, to reach new potential cGMP interactors in this context, a BLAST search of the eluted proteins provided information on ten other proteins showing high sequence similarity with PKGI. Five of these (p‐CaMKIIα, GSK3β, MAPK1/3, and EPAC2) were selected based on their relevance to photoreceptors and subjected to a PLA assay, which revealed that cGMP had proximity to them all in situ in the photoreceptor layer. While an indirect binding of cGMP to some of these (GSK3β and MAPK1/3) cannot yet be excluded, the results already indicate them as interesting proteins in the deciphering of the cGMP signaling pathway and hence the degeneration mechanisms.
CONFLICT OF INTEREST
FS is employed at Biolog Life Science Institute, which offers certain analogs and other products of this study on a commercial basis.
Supporting information
Table S1
Table S2
Table S3
Table S4
Supplementary Material
ACKNOWLEDGMENT
The authors thank Hodan Abdshill and Tamara Gehringer for technical assistance, and Patricia Veiga Crespo, and Sven Kjellström for pilot experimentation for this study. This project was funded by the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska‐Curie grant agreement No 765441 (transMed; H2020‐MSCA‐765441), Stiftelsen för Synskadade i f.d. Malmöhus län, Kronprinsessan Margaretas Arbetsnämnd för synskadade, and Ögonfonden.
All experiments were conducted in compliance with the ARRIVE guidelines.
Rasmussen M, Welinder C, Schwede F, Ekström P. The cGMP system in normal and degenerating mouse neuroretina: New proteins with cGMP interaction potential identified by a proteomics approach. J Neurochem.2021;157:2173–2186. 10.1111/jnc.15251
REFERENCES
- Arango‐Gonzalez, B., Trifunović, D., Sahaboglu, A., Kranz, K., Michalakis, S., Farinelli, P., Koch, S., Koch, F., Cottet, S., Janssen‐Bienhold, U., Dedek, K., Biel, M., Zrenner, E., Euler, T., Ekström, P., Ueffing, M., & Paquet‐Durand, F. (2014). Identification of a common non‐apoptotic cell death mechanism in hereditary retinal degeneration. PLoS One, 9, 1–11. 10.1371/journal.pone.0112142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulia, J. A., Sakagami, H., Kikkawa, S., & Terashima, T. (2015). Expression of beta subunit 2 of Ca 2+ /calmodulin‐dependent protein kinase I in the developing rat retina. Kobe Journal of Medical Sciences, 61, 115–123. [PubMed] [Google Scholar]
- Azadi, S., Johnson, L. E., Paquet‐Durand, F., Perez, M. T. R., Zhang, Y., Ekström, P. A. R., van Veen, T. (2007). CNTF + BDNF treatment and neuroprotective pathways in the rd1 mouse retina. Brain Research, 1129, 116–129. 10.1016/j.brainres.2006.10.031 [DOI] [PubMed] [Google Scholar]
- Bowes, C., Li, T., Danciger, M., Baxter, L. C., Applebury, M. L., & Farber, D. B. (1990). Retinal degeneration in the rd mouse is caused by a defect in the β subunit of rod cGMP‐phosphodiesterase. Nature, 347, 677–680. 10.1038/347677a0 [DOI] [PubMed] [Google Scholar]
- Bronstein, J. M., Wasterlain, C. G., Bok, D., Lasher, R., & Farber, D. B. (1988). Localization of retinal calmodulin kinase. Experimental Eye Research, 47, 391–402. 10.1016/0014-4835(88)90050-4 [DOI] [PubMed] [Google Scholar]
- Carpenter, E. P., Beis, K., Cameron, A. D., & Iwata, S. (2008). Overcoming the challenges of membrane protein crystallography. Current Opinion in Structural Biology, 18, 581–586. 10.1016/j.sbi.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, J., Altman, M. C., Fearnley, I. M., & Walker, J. E. (2007). Identification of membrane proteins by tandem mass spectrometry of protein ions. Proceedings of the National Academy of Sciences of the United States of America, 104, 14330–14335. 10.1073/pnas.0706817104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caunt, C. J., Finch, A. R., Sedgley, K. R., & McArdle, C. A. (2006). Seven‐transmembrane receptor signalling and ERK compartmentalization. Trends in Endocrinology and Metabolism, 17, 276–283. 10.1016/j.tem.2006.07.008 [DOI] [PubMed] [Google Scholar]
- Chang, B., Hawes, N. L., Hurd, R. E., Davisson, M. T., Nusinowitz, S., & Heckenlively, J. R. (2002). Retinal degeneration mutants in the mouse. Vision Research, 42, 517–525. 10.1016/S0042-6989(01)00146-8 [DOI] [PubMed] [Google Scholar]
- Chang, B., Hawes, N. L., Pardue, M. T., German, A. M., Hurd, R. E., Davisson, M. T., Nusinowitz, S., Rengarajan, K., Boyd, A. P., Sidney, S. S., Phillips, M. J., Stewart, R. E., Chaudhury, R., Nickerson, J. M., Heckenlively, J. R., & Boatright, J. H. (2007). Two mouse retinal degenerations caused by missense mutations in the β‐subunit of rod cGMP phosphodiesterase gene. Vision Research, 47, 624–633. 10.1016/j.visres.2006.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S., Vaccarella, L., Olatunji, S., Cebulla, C., & Christoforidis, J. (2011). Diagnostic challenges in retinitis pigmentosa: Genotypic multiplicity and phenotypic variability. Current Genomics, 12, 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradini, E., Burgers, P. P., Plank, M., Heck, A. J. R., & Scholten, A. (2015). Huntingtin‐associated protein 1 (HAP1) is a cGMP‐dependent Kinase Anchoring Protein (GKAP) specific for the cGMP‐dependent protein kinase Iβ isoform. Journal of Biological Chemistry, 290, 7887–7896. 10.1074/jbc.M114.622613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daiger, S., Rossiter, B., Greenberg, J., Christoffels, A., & Hide, W. (1998) Data services and software for identifying genes and mutations causing retinal degeneration. Investigative Ophthalmology & Visual Science 39, S295. University of Texas Houston Health Science Center. Houston, Texas. [Google Scholar]
- Das, A., Xi, L., & Kukreja, R. C. (2008). Protein kinase G‐dependent cardioprotective mechanism of phosphodiesterase‐5 inhibition involves phosphorylation of ERK and GSK3β. Journal of Biological Chemistry, 283, 29572–29585. 10.1074/jbc.M801547200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, R., Chowdhury, S., Mazhab‐Jafari, M. T., SilDas, S., Selvaratnam, R., & Melacini, G. (2009). Dynamically driven ligand selectivity in cyclic nucleotide binding domains. Journal of Biological Chemistry, 284, 23682–23696. 10.1074/jbc.M109.011700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahim, A. T., Daiger, S. P., & Weleber, R. G. (1993) Nonsyndromic Retinitis Pigmentosa Overview. (2000) Aug 4. In: Adam MP, Ardinger HH, Pagon RA, et al., editors. GeneReviews® [Internet]. [PubMed]
- Fan, B., Wang, B. F., Che, L., Sun, Y. J., Liu, S. Y., Li, G. Y. (2017). Blockage of NOX2/MAPK/NF‐κB Pathway Protects Photoreceptors against Glucose Deprivation‐Induced Cell Death. Oxidative Medicine and Cellular Longevity, 2017, 1–14. 10.1155/2017/5093473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, W., Li, X., & Cooper, N. G. F. (2007). CaMKIIαB mediates a survival response in retinal ganglion cells subjected to a glutamate stimulus. Investigative Ophthalmology and Visual Science, 48, 3854–3863. 10.1167/iovs.06-1382 [DOI] [PubMed] [Google Scholar]
- Fesenko, E. E., Kolesnikov, S. S., & Lyubarsky, A. L. (1985). Induction by cyclic GMP of cationic conductance in plasma membrane of retinal rod outer segment. Nature, 313, 310–313. 10.1038/313310a0 [DOI] [PubMed] [Google Scholar]
- Francis, S. H., Blount, M. A., Zoraghi, R., & Corbin, J. D. (2005). Molecular properties of mammalian proteins that interact with cGMP: Protein kinases, cation channels, phosphodiesterases, and multi‐drug anion transporters. Frontiers in Bioscience: A Journal and Virtual Library, 10, 2097–2117. [DOI] [PubMed] [Google Scholar]
- Francis, S. H., & Corbin, J. D. (2013). Cyclic nucleotide‐dependent protein kinases. Encyclopedia of Biological Chemistry: Second Edition, 50, 574–579. [Google Scholar]
- Gamm, D. M., Barthel, L. K., Raymond, P. A., & Uhler, M. D. (2000). Localization of cGMP‐dependent protein kinase isoforms in mouse eye. Investigative Ophthalmology and Visual Science, 41, 2766–2773. [PubMed] [Google Scholar]
- Gao, N., Luo, J., Uray, K., Qian, A., Yin, S., Wang, G., Wang, X., Xia, Y., Wood, J. D., & Hu, H. (2012). CaMKII is essential for the function of the enteric nervous system. PLoS One, 7, e44426. 10.1371/journal.pone.0044426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti, A., Nair, A. V., Codega, P., Torre, V., & Carloni, P. (2005). Structural basis of gating of CNG channels. FEBS Letters, 579, 1968–1972. 10.1016/j.febslet.2005.01.086 [DOI] [PubMed] [Google Scholar]
- Haim, M. (2002). The epidemiology of retinitis pigmentosa in Denmark. Acta Ophthalmologica Scandinavica, Supplement, 80, 1–34. 10.1046/j.1395-3907.2002.00001.x [DOI] [PubMed] [Google Scholar]
- Hanna, J., Yücel, Y. H., Zhou, X., Mathieu, E., Paczka‐Giorgi, L. A., & Gupta, N. (2017). Progressive loss of retinal blood vessels in a live model of retinitis pigmentosa. Canadian Journal of Ophthalmology, 53, 391–401. 10.1016/j.jcjo.2017.10.014 [DOI] [PubMed] [Google Scholar]
- Hauck, S. M., Ekström, P. A. R., Ahuja‐Jensen, P., Suppmann, S., Paquet‐Durand, F., Veen, T., & van Ueffing M.(2006). Differential modification of phosducin protein in degenerating rd1 retina is associated with constitutively active Ca2+ /calmodulin kinase II in rod outer segments. Molecular and Cellular Proteomics, 5, 324–336. [DOI] [PubMed] [Google Scholar]
- Ho, A. K., Hashimoto, K., & Chik, C. L. (1999). 3’,5’‐cyclic guanosine monophosphate activates mitogen‐activated protein kinase in rat pinealocytes. Journal of Neurochemistry, 73, 598–604. [DOI] [PubMed] [Google Scholar]
- Hong, L., Xi, J., Zhang, Y., Tian, W., Xu, J., Cui, X., & Xu, Z. (2012). Atrial natriuretic peptide prevents the mitochondrial permeability transition pore opening by inactivating glycogen synthase kinase 3β via PKG and PI3K in cardiac H9c2 cells. European Journal of Pharmacology, 695, 13–19. 10.1016/j.ejphar.2012.07.053 [DOI] [PubMed] [Google Scholar]
- Huang, G. Y., Kim, J. J., Reger, A. S., Lorenz, R., Moon, E.‐W., Zhao, C., Casteel, D. E., Bertinetti, D., VanSchouwen, B., Selvaratnam, R., Pflugrath, J. W., Sankaran, B., Melacini, G., Herberg, F. W., & Kim, C. (2014). Structural basis for cyclic‐nucleotide selectivity and cGMP‐selective activation of PKG i. Structure, 22, 116–124. 10.1016/j.str.2013.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H., Shabb, J. B., & Corbin, J. D. (1992). Cross‐activation: Overriding cAMP/cGMP selectivities of protein kinases in tissues. Biochemistry and cell biology = . Biochimie Et Biologie Cellulaire, 70, 1283–1289. 10.1139/o92-175 [DOI] [PubMed] [Google Scholar]
- Kim, E.‐ E.‐K., & Park, J.‐M.‐ M. (2003). Identification of novel target proteins of cyclic GMP signaling pathways using chemical proteomics. BMB Reports, 36, 299–304. 10.5483/BMBRep.2003.36.3.299 [DOI] [PubMed] [Google Scholar]
- Kim, J. J., Casteel, D. E., Huang, G., Kwon, T. H., Ren, R. K., Zwart, P., Headd, J. J., Brown, N. G., Chow, D.‐C., Palzkill, T., & Kim, C. (2011). Co‐crystal structures of pkg iβ (92–227) with cgmp and camp reveal the molecular details of cyclic‐nucleotide binding. PLoS One, 6, e18413. 10.1371/journal.pone.0018413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. S., Zhong, J., & Pandey, A. (2016). Common errors in mass spectrometry‐based analysis of post‐translational modifications. Proteomics, 16, 700–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y. H., Kim, Y. S., Kang, S. S., Cho, G. J., & Choi, W. S. (2010). Resveratrol inhibits neuronal apoptosis and elevated Ca2+/ calmodulin‐dependent protein kinase II activity in diabetic mouse retina. Diabetes, 59, 1825–1835. 10.2337/db09-1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laabich, A., & Cooper, N. G. F. (2000). Neuroprotective effect of AIP on N‐methyl‐D‐aspartate‐induced cell death in retinal neurons. Molecular Brain Research, 85, 32–40. 10.1016/S0169-328X(00)00226-6 [DOI] [PubMed] [Google Scholar]
- Liu, Y., Beyer, A., & Aebersold, R. (2016). On the dependency of cellular protein levels on mRNA abundance. Cell, 165, 535–550. 10.1016/j.cell.2016.03.014 [DOI] [PubMed] [Google Scholar]
- Lorenz, R., Moon, E. W., Kim, J. J., Schmidt, S. H., Sankaran, B., Pavlidis, I. V., Kim, C., & Herberg, F. W. (2017). Mutations of PKA cyclic nucleotide‐binding domains reveal novel aspects of cyclic nucleotide selectivity. Biochemical Journal, 474, 2389–2403. 10.1042/BCJ20160969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchena, M., Villarejo‐Zori, B., Zaldivar‐Diez, J., Palomo, V., Gil, C., Hernández‐Sánchez, C., Martínez, A., de la Rosa, E. J. (2017). Small molecules targeting glycogen synthase kinase 3 as potential drug candidates for the treatment of retinitis pigmentosa. Journal of Enzyme Inhibition and Medicinal Chemistry, 32, 522–526. 10.1080/14756366.2016.1265522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maymó, J. L., Pérez, P. A., Maskin, B., Dueñas, J. L., Calvo, J. C., Sánchez, M. V., & Varone, C. L. (2012). The alternative Epac/cAMP pathway and the MAPK pathway mediate hCG induction of leptin in placental cells. PLoS One, 7, e46216. 10.1371/journal.pone.0046216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay, M. M., & Morrison, D. K. (2007). Integrating signals from RTKs to ERK/MAPK. Oncogene, 26, 3113–3121. 10.1038/sj.onc.1210394 [DOI] [PubMed] [Google Scholar]
- Migues, P. V., Lehmann, I. T., Fluechter, L., Cammarota, M., Gurd, J. W., Sim, A. T. R., Dickson, P. W., & Rostas, J. A. P. (2006). Phosphorylation of CaMKII at Thr253 occurs in vivo and enhances binding to isolated postsynaptic densities. Journal of Neurochemistry, 98, 289–299. 10.1111/j.1471-4159.2006.03876.x [DOI] [PubMed] [Google Scholar]
- Nache, V., Wongsamitkul, N., Kusch, J., Zimmer, T., Schwede, F., & Benndorf, K. (2016). Deciphering the function of the CNGB1b subunit in olfactory CNG channels. Scientific Reports, 6, 1–11. 10.1038/srep29378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne, B. W., Wu, J., McFarland, C. J., Nickl, C. K., Sankaran, B., Casteel, D. E., Woods, V. L., Kornev, A. P., Taylor, S. S., & Dostmann, W. R. (2011). Crystal structure of cGMP‐dependent protein kinase reveals novel site of interchain communication. Structure, 19, 1317–1327. 10.1016/j.str.2011.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet‐Durand, F., Beck, S., Michalakis, S., Goldmann, T., Huber, G., Mühlfriedel, R., Trifunović, D., Fischer, M. D., Fahl, E., Duetsch, G., Becirovic, E., Wolfrum, U., van Veen, T., Biel, M., Tanimoto, N., & Seeliger, M. W. (2011). A key role for cyclic nucleotide gated (CNG) channels in cGMP‐related retinitis pigmentosa. Human Molecular Genetics, 20, 941–947. 10.1093/hmg/ddq539 [DOI] [PubMed] [Google Scholar]
- Paquet‐Durand, F., Hauck, S. M., Veen, T., Van, U. M., & Ekström, P. (2009). PKG activity causes photoreceptor cell death in two retinitis pigmentosa models. Journal of Neurochemistry, 108, 796–810. 10.1111/j.1471-4159.2008.05822.x [DOI] [PubMed] [Google Scholar]
- Pavlaki, N., & Nikolaev, V. (2018). Imaging of PDE2‐ and PDE3‐mediated cGMP‐to‐cAMP cross‐talk in cardiomyocytes. Journal of Cardiovascular Development and Disease, 5, 4. 10.3390/jcdd5010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérezleón, J. A., Osorio‐Paz, I., Francois, L., & Salceda, R. (2013). Immunohistochemical localization of glycogen synthase and GSK3β: Control of glycogen content in retina. Neurochemical Research, 38, 1063–1069. 10.1007/s11064-013-1017-0 [DOI] [PubMed] [Google Scholar]
- Petit, L., Lhériteau, E., Weber, M., Le Meur, G., Deschamps, J.‐Y., Provost, N., Mendes‐Madeira, A., Libeau, L., Guihal, C., Colle, M.‐A., Moullier, P., & Rolling, F. (2012). Restoration of vision in the PDE6β‐deficient dog, a large animal model of rod‐cone dystrophy. Molecular Therapy, 20, 2019–2030. 10.1038/mt.2012.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer, R. L., Marc, R. E., & Jones, B. W. (2020). Persistent remodeling and neurodegeneration in late‐stage retinal degeneration. Progress in Retinal and Eye Research, 74, 100771. 10.1016/j.preteyeres.2019.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichard, V., Provost, N., Mendes‐Madeira, A., Libeau, L., Hulin, P., Tshilenge, K.‐T., Biget, M., Ameline, B., Deschamps, J.‐Y., Weber, M., Le Meur, G., Colle, M.‐A., Moullier, P., & Rolling, F. (2016). AAV‐mediated gene therapy halts retinal degeneration in PDE6β‐deficient dogs. Molecular Therapy, 24, 867–876. 10.1038/mt.2016.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, M., Das, S., Schütze, K., Marigo, V., Ekström, P., & Paquet‐Durand, F. (2020). Cellular mechanisms of hereditary photoreceptor degeneration – Focus on cGMP. Progress in Retinal and Eye Research, 74, 100772. 10.1016/j.preteyeres.2019.07.005 [DOI] [PubMed] [Google Scholar]
- Punzo, C., & Cepko, C. (2007). Cellular responses to photoreceptor death in the rd1 mouse model of retinal degeneration. Investigative Ophthalmology and Visual Science, 48, 849–857. [DOI] [PubMed] [Google Scholar]
- Pyakurel, A., Balmer, D., Saba‐El‐Leil, M. K., Kizilyaprak, C., Daraspe, J., Humbel, B. M., Voisin, L., Le, Y. Z., von Lintig, J., Meloche, S., & Roduit, R. (2017). Loss of extracellular signal‐regulated kinase 1/2 in the retinal pigment epithelium leads to RPE65 decrease and retinal degeneration. Molecular and Cellular Biology, 37, 295–312. 10.1128/MCB.00295-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmann, H., Arias‐Palomo, E., Hadders, M. A., Schwede, F., Llorca, O., & Bos, J. L. (2008). Structure of Epac2 in complex with a cyclic AMP analogue and RAP1B. Nature, 455, 124–127. 10.1038/nature07187 [DOI] [PubMed] [Google Scholar]
- Rehmann, H., Schwede, F., Doøskeland, S. O., Wittinghofer, A., & Bos, J. L. (2003). Ligand‐mediated activation of the cAMP‐responsive guanine nucleotide exchange factor Epac. Journal of Biological Chemistry, 278, 38548–38556. 10.1074/jbc.M306292200 [DOI] [PubMed] [Google Scholar]
- Roche, S. L., Kutsyr, O., Cuenca, N., & Cotter, T. G. (2019). Norgestrel, a progesterone analogue, promotes significant long‐term neuroprotection of cone photoreceptors in a mouse model of retinal disease. Investigative Ophthalmology and Visual Science, 60, 3221–3235. 10.1167/iovs.19-27246 [DOI] [PubMed] [Google Scholar]
- Russell, S., Bennett, J., Wellman, J. A., Chung, D. C., Yu, Z.‐F., Tillman, A., Wittes, J., Pappas, J., Elci, O., McCague, S., Cross, D., Marshall, K. A., Walshire, J., Kehoe, T. L., Reichert, H., Davis, M., Raffini, L., George, L. A., Hudson, F. P., … Maguire, A. M. (2017). Efficacy and safety of voretigene neparvovec (AAV2‐hRPE65v2) in patients with RPE65‐mediated inherited retinal dystrophy: A randomised, controlled, open‐label, phase 3 trial. The Lancet, 390, 849–860. 10.1016/S0140-6736(17)31868-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahaboglu, A., Paquet‐Durand, O., Dietter, J., Dengler, K., Bernhard‐Kurz, S., Ekström, P. A. R., Hitzmann, B., Ueffing, M., & Paquet‐Durand, F. (2013). Retinitis pigmentosa: Rapid neurodegeneration is governed by slow cell death mechanisms. Cell Death and Disease, 4, 1–8. 10.1038/cddis.2013.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Cruz, A., Villarejo‐Zori, B., Marchena, M., Zaldivar‐Díez, J., Palomo, V., Gil, C., Lizasoain, I., de la Villa, P., Martínez, A., de la Rosa, E. J., & Hernández‐Sánchez, C. (2018). Modulation of GSK‐3 provides cellular and functional neuroprotection in the rd10 mouse model of retinitis pigmentosa. Molecular Neurodegeneration, 13, 19. 10.1186/s13024-018-0251-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal, S., & Bal, A. K. (1973). Comparative light and electron microscopic study of retinal histogenesis in normal and rd mutant mice. Zeitschrift Für Anatomie Und Entwicklungsgeschichte, 142, 219–238. 10.1007/BF00519723 [DOI] [PubMed] [Google Scholar]
- Sanyal, S., & Zeilmaker, G. H. (1984). Development and degeneration of retina in rds mutant mice: Light and electron microscopic observations in experimental chimaeras. Experimental Eye Research, 39, 231–246. 10.1016/0014-4835(84)90011-3 [DOI] [PubMed] [Google Scholar]
- Scholten, A., Poh, M. K., Veen, T. A. B., Van, B. B., Van, V. M. A., & Heck, A. J. R. (2006). Analysis of the cGMP/cAMP interactome using a chemical proteomics approach in mammalian heart tissue validates sphingosine kinase type 1‐interacting protein as a genuine and highly abundant AKAP. Journal of Proteome Research, 5, 1435–1447. 10.1021/pr0600529 [DOI] [PubMed] [Google Scholar]
- Scholten, A., Veen, T. A. B., Van, V. M. A., & Heck, A. J. R. (2007). Diversity of cAMP‐dependent protein kinase isoforms and their anchoring proteins in mouse ventricular tissue. Journal of Proteome Research, 6, 1705–1717. 10.1021/pr060601a [DOI] [PubMed] [Google Scholar]
- Schwede, F., Bertinetti, D., Langerijs, C. N., Hadders, M. A., Wienk, H., Ellenbroek, J. H., de Koning, E. J. P., Bos, J. L., Herberg, F. W., Genieser, H.‐G., Janssen, R. A. J., & Rehmann, H. (2015). Structure‐guided design of selective Epac1 and Epac2 agonists. PLoS Biology, 13, 1–26. 10.1371/journal.pbio.1002038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabb, J. B. (2010). Cyclic nucleotide specificity and cross‐activation of cyclic nucleotide receptors, in Handbook of Cell Signaling, pp. 1549–1554. Elsevier Inc. [Google Scholar]
- Shinde, V. M., Sizova, O. S., Lin, J. H., LaVail, M. M., & Gorbatyuk, M. S. (2012). Er stress in retinal degeneration in S334ter Rho rats. PLoS One, 7, e33266. 10.1371/journal.pone.0033266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetenborg, S., Yadav, S. C., Brüggen, B., Zoidl, G. R., Hormuzdi, S. G., Monyer, H., van Woerden, G. M., Janssen‐Bienhold, U., & Dedek, K. (2019). Localization of Retinal Ca2+/Calmodulin‐Dependent Kinase II‐β (CaMKII‐β) at Bipolar Cell Gap Junctions and Cross‐Reactivity of a Monoclonal Anti‐CaMKII‐β Antibody With Connexin36. Frontiers in Molecular Neuroscience, 12, 1–13. 10.3389/fnmol.2019.00206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetenborg, S., Yadav, S. C., Hormuzdi, S. G., Monyer, H., Janssen‐Bienhold, U., & Dedek, K. (2017). Differential distribution of retinal Ca2+ /calmodulin‐dependent kinase II (CaMKII) isoforms indicates CaMKII‐β and ‐δ as specific elements of electrical synapses made of connexin36 (Cx36). Frontiers in Molecular Neuroscience, 10, 1–15. 10.3389/fnmol.2017.00425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vighi, E., Trifunović, D., Veiga‐Crespo, P., Rentsch, A., Hoffmann, D., Sahaboglu, A., Strasser, T., Kulkarni, M., Bertolotti, E., van den Heuvel, A., Peters, T., Reijerkerk, A., Euler, T., Ueffing, M., Schwede, F., Genieser, H.‐G., Gaillard, P., Marigo, V., Ekström, P., & Paquet‐Durand, F. (2018). Combination of cGMP analogue and drug delivery system provides functional protection in hereditary retinal degeneration. Proceedings of the National Academy of Sciences, 115(13), E2997–E3006. 10.1073/pnas.1718792115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitz, D., Ficek, N., Kremmer, E., Bauer, P. J., & Kaupp, U. B. (2002). Subunit stoichiometry of the CNG channel of rod photoreceptors. Neuron, 36, 881–889. 10.1016/S0896-6273(02)01098-X [DOI] [PubMed] [Google Scholar]
- Whitaker, C. M., & Cooper, N. G. F. (2010a). Differential distribution of epacs within the adult rat retina. Neuroscience, 165, 955–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker, C. M., & Cooper, N. G. F. (2010b). Differential distribution of exchange proteins directly activated by cyclic AMP within the adult rat retina. Neuroscience, 165, 955–967. 10.1016/j.neuroscience.2009.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, M. S., Zheng, Y. Z., Huang, L., & Bin, X. G. X. (2017). Infliximab relieves blood retinal barrier breakdown through the p38 MAPK pathway in a diabetic rat model. International Journal of Ophthalmology, 10, 1824–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza, C., Soria, E., López, E., Browning, D., Balbín, M., López‐Otín, C., & Lamas, S. (2002). Activation of the mitogen activated protein kinase extracellular signal‐regulated kinase 1 and 2 by the nitric oxide‐cGMP‐cGMP‐dependent protein kinase axis regulates the expression of matrix metalloproteinase 13 in vascular endothelial cells. Molecular Pharmacology, 62, 927–935. 10.1124/mol.62.4.927 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Table S3
Table S4
Supplementary Material


