Summary
RNA-binding proteins (RBPs) are critical regulators of post-transcriptional gene expression and aberrant RBP-RNA interactions can promote cancer progression. Here, we interrogate the function of RBPs in cancer using pooled CRISPR-Cas9 screening and identify 57 RBP candidates with distinct roles in supporting MYC-driven oncogenic pathways. We find that disrupting YTHDF2-dependent mRNA degradation triggers apoptosis in triple-negative breast cancer (TNBC) cells and tumors. eCLIP- and m6A-sequencing reveal that YTHDF2 interacts with mRNAs encoding proteins in the MAPK pathway that, when stabilized, induce epithelial-to-mesenchymal transition and increase global translation rates. scRibo-STAMP profiling of translating mRNAs reveals unique alterations in the translatome of single cells within YTHDF2-depleted solid tumors, which selectively contribute to endoplasmic reticulum stress-induced apoptosis in TNBC cells. Thus, our work highlights the therapeutic potential of RBPs by uncovering a critical role for YTHDF2 in counteracting the global increase of mRNA synthesis in MYC-driven breast cancers.
Keywords: CRISPR screening, N6-methyladenosine, YTHDF2, MYC-driven cancer, scRNA-seq
eTOC Blurb
RNA-binding proteins (RBPs) are aberrantly expressed in cancer, but they remain largely unexplored as drug targets. Using RBP-focused CRISPR/Cas9 screening, Einstein et al. uncover an essential and selective role for YTHDF2 in MYC-driven breast cancer, highlighting the utility of discovering RBPs as safe and effective therapeutic targets.
Graphical Abstract

Introduction
Changes in cellular growth rate and identity that occur during cancer progression are driven by specific gene expression signatures programmed by the activity of DNA-binding transcription factors (TFs) and RNA-binding proteins (RBPs). To support high oncogenic growth rates, cancer cells generally require increased levels of transcription and global pre-RNA synthesis controlled by TFs, consequently increasing the cell’s dependence on post-transcriptional processing by RBPs. While TF mutations have been studied for decades, RBPs have been overlooked as drivers of disease and as therapeutically relevant targets. RBPs determine the fate of transcribed RNAs by regulating their splicing, polyadenylation, translation, subcellular localization and turnover (Hentze et al., 2018). Somatic mutations as well as epigenetic and post translational modifications can cause aberrant RBP expression in cancer cells, which can promote cell division, cell survival, metastasis, angiogenesis and host immune evasion (Pereira et al., 2017). However, RBPs remain largely unexplored as drug targets because their systematic evaluation has been limited by the lack of sensitive and efficient assays for phenotypic interrogation of individual RBPs (Pereira et al., 2017).
C-MYC (MYC) is the primary oncogenic driver of cancer gene expression programs in a broad spectrum of cancer types where cells become “addicted” to and dependent on MYC for survival (Zuber et al., 2011). Although small molecules with the potential to inhibit MYC directly have recently emerged as clinical candidates (Han et al., 2019b; Struntz et al., 2019), they are likely to incur systemic toxic effects with long term treatment due to inhibition of MYC’s physiological functions. Therefore, we employed an orthogonal method by identifying RBPs that are selectively required to sustain MYC’s oncogenic gene regulatory program using a synthetic lethality paradigm.
Here, we employed human mammary epithelial cells expressing a MYC estrogen receptor fusion (MYC-ER HMECs) to test the hypothesis that the mutation of two genes (MYC and an RBP candidate) causes cell death, while the mutation of either gene individually is tolerated (Kessler et al., 2012). Using a CRISPR-Cas9 library targeting over one thousand RBPs in the human genome, we identified 57 RBPs that are required for the survival of MYC-hyperactive cells (Wheeler et al., 2020). Proteins known to mediate RNA catabolism were overrepresented among RBP candidates and we specifically found that depletion of YTHDF2 induced apoptosis in human triple-negative breast cancer (TNBC) cell lines and impeded xenografted tumor growth in vivo. Integrated analyses identified a role for YTHDF2 in the turnover of RNAs that contribute to epithelial to mesenchymal transition (EMT) initiation and tumorigenesis to limit the endoplasmic reticulum (ER)-stress response that accompanies this process. Altogether, our study clarifies the molecular mechanisms underlying m6A-mediated gene regulation in MYC-driven breast cancer and highlights YTHDF2 as a safe and effective therapeutic target.
Results
Identification of critical RBPs in MYC-driven breast cancer
To screen for RBP dependencies in cancer, we used a CRISPR-Cas9 lentiviral library containing 10 single-guide (sg)RNAs for each of 1,078 RBPs, 628 sgRNAs targeting essential genes as positive controls and 1,058 non-targeting sgRNAs as negative controls (Wheeler et al., 2020). The small library size and specific focus on RBPs provides higher-confidence hits and therefore, higher reproducibility than whole-genome approaches typically using more shallow coverage of 3–4 sgRNAs per gene (Shalem et al., 2014). High-throughput sequencing confirmed that the plasmid library maintained sgRNA coverage and aliquots were tightly correlated (less than 0.065% of sgRNAs received undetectable normalized read counts; Figure S1A).
We transduced MYC-ER HMECs with the CRISPR library in biological duplicate, selected for transduced cells with puromycin, treated half of the cells with tamoxifen (TAM) to induce MYC activity and isolated genomic DNA (gDNA) 8 and 16 days after puromycin removal (Figure 1A). The representation of randomly integrated sgRNAs was tightly correlated with their frequencies in the original plasmid library and replicate libraries were highly correlated (Figure S1B and S1C). We observed a significant reduction in sgRNA diversity in surviving cells on days 8 and 16 in both MYC-induced and uninduced HMECs, indicating dropout of cells transduced with sgRNAs targeting essential genes (positive controls) compared to cells transduced with non-targeting sgRNAs (negative controls) (Figures 1B, S1D and S1E).
Figure 1. RBP CRISPR screen identifies mRNA decay as a vulnerability in MYC-driven cancer.
(A) Schematic describing the generation and implementation of the RBP-targeted CRISPR screen in MYC-ER HMECs.
(B) Cumulative distribution of normalized read counts for sgRNAs in each sample. Each condition is representative of 2 independent replicates. Two-sided Kolmogorov-Smirnov tests compared to day 0.
(C) Comparison of β score replicates on day 16. Inset highlights synthetic lethal candidate RBPs and genes enriched for “negative regulation of macromolecule metabolic process” are labeled.
(D) Schematic describing the function of RBP candidates identified in (C). RBPs in bold are significantly upregulated in basal-like TNBC tumors compared with ER/PR/HER2 positive tumors from TCGA Data Portal (Liu et al., 2018). p < 0.05, two-sided Student’s T-test.
(E) Box plot comparing mRNA expression levels for MYC and YTHDF2 between TNBC and ER/PR/HER2 positive tumors from TCGA data portal (Liu et al., 2018). p*** < 0.001, two-sided Student’s T-test.
(F) Western blot confirming shRNA-mediated knockdown in MYC-ER HMECs.
(G) Quantification of Annexin-V staining in MYC-ER HMECs with indicated knockdowns with (blue) and without (red) 24 hours of MYC-induction. p* < 0.05, p*** < 0.001, two-tailed Student’s T-test. Bars are mean ± SD, n = 3 independent replicates.
(H) Quantification of propidium iodide (PI) staining in MYC-ER HMECs with indicated knockdowns with (blue) and without (red) 24 hours of MYC-induction. p* < 0.05, p*** < 0.001, two-tailed Student’s T-test. Bars are mean ± SD, n = 3 independent replicates.
(I) Quantification of cell cycle phase using PI in MYC-ER HMECs with indicated knockdowns with and without 3 days of MYC-induction. ***p < 0.001, ****p < 0.0001, two-way ANOVA test with Dunnett’s post-hoc test for multiple comparisons. Bars are mean ± SD. n = 3 independent replicates.
(J) Cell proliferation of shYTHDF2 cells compared with NTC over time in TNBC and HR+ breast cancer cell lines. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, two-way ANOVA test with Dunnett’s post-hoc test for multiple comparisons. Values normalized to initial average confluence. Bars are mean ± SD, n = 6 independent replicates.
See also Figure S1.
sgRNAs targeting 57 RBP candidates (p < 0.05) were depleted in MYC-induced cell populations compared to uninduced control cells (Figure 1C and Table S1). Our analysis revealed RBPs that have been implicated in gene regulation downstream of MYC, including PTBP1 (He et al., 2014), PCBP2 (Wan et al., 2016), ELAVL1 (Lafon et al., 1998) and TRA2B (Park et al., 2019) and identified ~40 RBPs previously unknown in this context. We observed enrichment for genes that negatively regulate macromolecule assembly, including several genes that repress exon inclusion (HNRNPA2B1, PTBP1, SRSF9), repress transcription (ZGPAT, SLTM), promote mRNA turnover (YTHDF2, UPF3B, UPF3A, XRN1, DCP2, AGO2), and prevent translation initiation (EIF4ENIF1, EIF4E2) (Figure 1C). This suggests vulnerabilities in MYC-hyperactive cells by RBPs that reduce the stability of mRNAs for translation, or RBPs that reduce protein translation rates.
We next cross referenced public data from The Cancer Genome Atlas (TCGA) pan-cancer clinical data resource (Liu et al., 2018) and found that 67% of our RBP candidates are highly expressed in typically MYC-amplified basal-like, TNBC tumors, compared to luminal A/luminal B, hormone receptor positive (HR+; estrogen, progesterone, or human epidermal growth factor receptor 2 [ER/PR/HER2] positive) breast tumors (Green et al., 2016) (Figure 1D). Together, our results indicate that several RBPs controlling various stages of the RNA life cycle are aberrantly expressed in TNBC. Since MYC-amplified cells typically contain increased quantities of mRNA, caused in part by 3’UTR shortening (Mayr and Bartel, 2009), we further examined RBP candidates that regulate RNA stability and turnover. We found that the overall survival of patients with TNBC tumors containing above median MYC expression levels was significantly improved when tumors contained below median expression levels of mRNA decay factors AGO2, EXOSC7, FUS, YTHDF2 and ZFP36L2 (Figure S1F).
Depleting YTHDF2 induces apoptosis of TNBC tumors in vivo
Consistent with previous studies implicating aberrant mRNA methylation (Lan et al., 2019) and expression of m6A methyltransferases, demethylases and m6A binding proteins in various cancers (Huang et al., 2018), we found that the m6A reader protein YTHDF2 is significantly upregulated in TNBC compared to HR+ breast cancers (Figure 1E). YTHDF2 mediates mRNA turnover by recruiting the CCR4-NOT deadenylase complex to initiate deadenylation and decay of m6A-containing transcripts before localizing with bound targets to processing (P) bodies for committed degradation (Du et al., 2016; Wang et al., 2014). However, the role of YTHDF2-mediated turnover of methylated RNA in cancer is not clear due to conflicting findings regarding its function (Paris et al., 2019; Zhong et al., 2019). Moreover, the direct and functionally relevant YTHDF2 target RNAs have yet to be defined in the mammary epithelium or in human breast cancer.
We first verified that loss of YTHDF2 expression causes cell death in MYC-induced HMECs using two independent short hairpin (sh)RNA-mediated knockdowns of YTHDF2 (shYTHDF2–1 and shYTHDF2–2) (Figures 1F and S1G). After 24 hours of MYC-induction, we observed a significant increase in apoptotic, Annexin V-positive cells and early cell death in MYC-induced HMECs compared to uninduced controls (Figures 1G, 1H, and S1H). We also confirmed epistasis between YTHDF2 and m6A-modified RNA targets using shRNA-mediated knockdown of m6A writer, METTL3 (shMETTL3) (Figure 1F), which similarly triggered apoptotic cell death (Figures 1G, 1H and S1H). In addition, using PI staining we found that after three days of MYC-induction significantly more YTHDF2-depleted HMECs were assigned to sub-G1 cell cycle phase and significantly less were assigned to G0/G1 phase, suggesting G1 checkpoint arrest. Uninduced HMECs did not show significant changes in fractions of cells assigned to G0/G1, S or G2/M phases following YTHDF2 depletion (Figure 1I, Figure S1I). We also found that proliferation rates of TNBC cell lines (MDA-MB-231 and secondary lung metastatic MDA-MB-231-LM2) were significantly reduced following YTHDF2 depletion while HR+ breast cancer cell lines (MCF-7 and SKBR3) were not (Figure 1J).
To determine the importance of genes encoding regulators of mRNA turnover on MYC-driven tumor growth in vivo we generated a pool of MDA-MB-231-LM2 cells transduced with various doxycycline (DOX) inducible, TurboRFP-tagged shRNAs targeting several proteins mediating RNA stability, three of which target YTHDF2, and assessed dropout of shRNAs both in vitro cultured cells and in vivo by subcutaneous mouse xenograft (Figure 2A and Table S2). We identified a YTHDF2-targeting hairpin that was significantly depleted in resected tumors and in cells cultured in vitro (p < 0.001), indicating a growth disadvantage in TNBC cells upon silencing of YTHDF2 (Figures 2B-2D). To verify that YTHDF2 inhibition negatively affects tumor growth in vivo we generated stable MDA-MB-231-LM2 cells transduced with the DOX inducible shRNA identified by the screen (Figures S2A and S2B). Following DOX treatment, we confirmed that cells expressing the highest levels of the YTHDF2-targeting shRNA became depleted over time when cultured in vitro, suggesting death of cells with sufficient YTHDF2 depletion (Figure S2C). After initial tumor engraftment, we observed reduced growth rates and significantly smaller final tumor volumes from DOX-treated mice compared with tumors from vehicle-treated mice (Figure 2E, 2F and 2G). In addition to fewer proliferating cells, we observed increased caspase-3 cleavage and a reduction in host angiogenic vascular endothelial cell markers in tumors from DOX-treated mice (Figures 2H, 2I and S2D).
Figure 2. Depletion of YTHDF2 in TNBC cells suppresses tumor growth in vivo.
(A) Schematic of pooled shRNA screen conducted in MDA-MB-231-LM2 cells in vitro and in vivo.
(B-D) Dot plots quantifying shRNA normalized abundance following doxycycline (DOX)-induced knockdown of MDA-MB-231-LM2 cells (B) in vitro (n = 4 independent replicates) and (C, D) in vivo. Dots represent individual tumor samples. Bars are median ± quartile. T0 is the tumor composition when DOX was introduced. −DOX=16 mice, +DOX=10 mice.
(E) Average tumor volume over time of DOX-induced, YTHDF2-depleted xenografted mice compared to vehicle controls. p*** < 0.001, p**** < 0.0001, two-way ANOVA test with Dunnett’s post-hoc test for multiple comparisons. Bars are mean ± SEM, −DOX = 9 mice, +DOX = 6 mice.
(F) Images of mice 35 days following DOX-induction compared to vehicle controls. Scale bars = 1 cm.
(G) Tumor volumes 35 days following DOX-induction compared to vehicle controls. −DOX = 9 mice, +DOX = 6 mice. P-values calculated by two-sided Student’s t-test.
(H) RT-qPCR analysis relative to Gapdh of mRNA extracted from final tumors, 35 days following DOX-induction. **p < 0.01, n.s. = not significant, two-sided Student’s T-test. Bars are mean ± SEM, −DOX = 9 vehicle mice, +DOX = 6 mice.
(I) Immunohistochemical staining of DOX-treated and vehicle control tumor sections, 35 days following DOX-induction. Scale bars = 100 μm.
See also Figure S2.
Next, we generated inducible multi-tissue CAG-CreERT;Ythdf2fl/fl mice by crossing CAG-CreERT mice with previously generated Ythdf2fl/fl mice (Li et al., 2018b) (Figure S2E) to expose any effects on the viability of healthy cells in other organs. Systemic genetic depletion of Ythdf2 resulted in no gross physiological abnormalities or changes in body weight for at least four weeks following TAM administration (Figures S2F and S2G), nor did it induce programmed cell death in female reproductive tissues known to rely on m6A regulation by YTHDF2 (Ivanova et al., 2017) (Figure S2H). This suggests that inhibition of Ythdf2 in an intact organism has no adverse effects in non-cancerous somatic tissues and that YTHDF2 depletion safely and specifically inhibits growth of cells predisposed to MYC addiction.
YTHDF2 targets are enriched for genes regulating growth factor signaling
To identify transcript-specific functions of YTHDF2 targets in MYC-driven cancer, we performed enhanced Crosslinking and Immunoprecipitation (eCLIP) (Van Nostrand et al., 2017) and m6A-sequencing (m6A-seq) (Dominissini et al., 2013) in MYC-induced and uninduced HMECs, MDA-MB-231-LM2 cells, MDA-MB-231 cells, MCF-7 cells and SKBR3 cells in biological duplicate (Figure S3A and Table S3). In agreement with previous YTHDF2 CLIP and RNA immunoprecipitation (RIP)-seq experiments, we found that a majority of high confidence YTHDF2 and m6A peaks map to the 3’end of transcripts in the coding sequence (CDS) and 3’ untranslated region (3’UTR) (Figure 3A) and contain the DRACH (D=A, G or U; R=A or G; H=A, C or U) sequence motif (Figure 3B). We confirmed that YTHDF2 binding sites generally overlap with m6A modified sites using pairwise comparisons between eCLIP and m6A-seq IP reads at each m6A site and that eCLIP IP reads were enriched at m6A sites over size-matched (SM)Inputs (Figures S3B and S3C). Collectively our epitranscriptomic analyses confirm binding of YTHDF2 at m6A modified sites on RNAs containing the consensus sequence.
Figure 3. eCLIP and m6A-seq identify MAPK/ERK pathway transcripts that are regulated by YTHDF2.
(A) Metagene profiles of filtered and unfiltered eCLIP (blue) and m6A-seq (red) peak enrichment.
(B) Motif analysis of unfiltered eCLIP (top) and m6A-seq (bottom) data. Representative of 2 independent replicates.
(C) Venn diagram overlaps of high-confidence m6A-seq peak and gene enrichment.
(D) Four-way Venn diagram of overlapping, high-confidence YTHDF2 target genes among TNBC and HR+ breast cancer cell lines. Genes overlapping in TNBC cell lines are outlined.
(E) Hierarchical cluster map illustrating expression levels of overlapping YTHDF2 target genes outlined in (D). Gene clusters are depicted in the dendrogram.
(F) Gene ontology (GO) enrichment of genes in dendrogram clusters from (E).
See also Figure S3.
To determine if MYC activity influences the m6A landscape we compared high-confidence m6A sites among MYC-induced and uninduced HMECs and among TNBC and HR+ breast cancer cell lines. Interestingly, we found that the secondary lung metastatic MDA-MB-231-LM2 tumor cells appeared to acquire new m6A modifications since ~72% of m6A sites (14,873 peaks) and ~46% modified genes (4,512 genes) were unique from the parental MDA-MB-231 cell line (Figure 3C). Consistent with previous studies (Lin et al., 2016), this suggests that the m6A landscape is modified during tumor evolution and metastasis, indicating that cancer cells become increasingly reliant on m6A-dependent RNA regulation as they become more invasive. However, MYC-induced HMECs did not display robust differences in m6A modified sites compared to uninduced HMECs in either direction, neither did HR+ breast cancer cell lines compared to MDA-MB-231 cells, suggesting that MYC activity does not determine the m6A landscape in mammary tumors (Figure 3C).
Since RBP binding affinity is driven largely by RNA sequence and secondary structure, which are conserved across cell types and states (Lambert et al., 2014), we expect that differences in YTHDF2 binding targets among cancer cell lines result from differences in target RNA expression level and alterations in m6A status. To understand YTHDF2’s role in promoting MYC-driven cancer cell growth and survival and to pinpoint the targets that uniquely contribute to apoptosis, we analyzed RNA expression data for overlapping YTHDF2 targets in TNBC cell lines (Figure 3D). Using hierarchical clustering of z-scores, we identified two clusters (red and blue) where YTHDF2 targets are highly expressed in TNBC cells compared to HR+ cells (Figure 3E). These clusters were enriched for genes that regulate wound healing, cell adhesion, ERK1/2 signaling, and EMT, while clusters including genes that are also highly expressed in MCF-7 cells (yellow and green) generally lack enrichment for genes belonging to these ontology terms (Figure 3F).
Depletion of YTHDF2 upregulates EMT-specific pathways
We next verified that TNBC cells undergo expression changes resembling EMT following YTHDF2 depletion by performing RNA-sequencing (RNA-seq) in YTHDF2-depleted, MYC-induced HMECs in biological duplicate. We found that transcripts that are bound by YTHDF2 are upregulated compared to transcripts lacking binding sites (Figure 4A), however, inspection of the individual upregulated mRNAs revealed that >50% of upregulated transcripts were not direct YTHDF2 targets. These include CPA4, HMOX1, and MMP3, which are known to contribute to or are transcribed in response to cell migration, wound healing, and metastatic phenotypes (Handa et al., 2019; Zhu et al., 2017; Zhu et al., 2019) (Figure 4B). These genes exhibited very low or undetectable RNA expression (transcripts per million (TPM) ≤ 1) in NTC cells yet were dramatically increased (~30–85 TPM) in YTHDF2-depleted cells. 4.8% of expressed genes (Bonferroni adjusted p-value (padj) < 0.001, log2FoldChange > 1) were differentially upregulated in YTHDF2-depleted, MYC-induced HMECs compared to NTC (Table S4) and were enriched for ontologies associated with mesenchymal cell transition (i.e. tissue morphogenesis, TGF-β receptor signaling, and signaling responses to growth factor stimuli), while downregulated transcripts were enriched for ontologies associated with epithelial cell processes (i.e. adhesion and junction organization, keratinization, and epidermis development) (Figure 4C). In addition, we found that upregulated genes overlapping with those in YTHDF2-depleted MYC-induced HMECs over YTHDF2-depleted uninduced HMECs were enriched for ontologies associated with inflammatory and stress response, but not EMT-specific pathways (Figure S4A), indicating that general inhibition of YTHDF2 promotes EMT and growth factor signaling, however, only in conjunction with elevated MYC activity does YTHDF2 inhibition lead to apoptosis.
Figure 4. Depletion of YTHDF2 triggers activation of EMT.
(A) Cumulative distribution of the fold change in mRNA expression of shYTHDF2 cells over NTC. Two-sided Kolmogorov-Smirnov test compared to non-targets. n = 4 replicates (2 hairpins x 2 biological replicates).
(B) Volcano plot describing the upregulated (red) and downregulated (blue) genes in shYTHDF2 cells compared to NTC in MYC-induced HMECs. Direct YTHDF2 eCLIP targets are labeled in bold, green text.
(C) Gene ontology (GO) enrichment of differentially expressed genes in shYTHDF2 cells compared to NTC in MYC-induced HMECs.
(D) Immunofluorescent staining in TNBC cell lines. Arrow heads indicate cell projections. Scale bar = 50 μm.
(E) Schematic displaying upstream signaling pathways that induce ERK1/2 signaling and EMT in breast cancer. Overlapping YTHDF2 targets in TNBC cell lines are listed.
(F) Western blot analysis of cell lysates from NTC and shYTHDF2 TNBC and HR+ breast cancer cell lines of EMT transcription factor protein expression.
(G) Western blot analysis of cell lysates from NTC and shYTHDF2 TNBC and HR+ breast cancer cell lines of ERK1/2 pathway activation and downstream effectors.
(H) RT-qPCR analysis relative to GAPDH of mRNA extracted from shYTHDF2 cells compared to NTC in TNBC and HR+ breast cancer cell lines. ****p < 0.0001, ***p < 0.001, **p < 0.01, two-way ANOVA test with Dunnett’s post hoc test for multiple comparisons. Bars are mean ± SD, n = 3 independent replicates.
See also Figure S4.
Supporting this hypothesis, we observed that YTHDF2-depleted, TNBC cells displayed a spindle-shape morphology denoted by high levels of vimentin localized to cell projections (Figure 4D). Additionally, we found that several upstream regulators of EMT are YTHDF2 targets (Figure 4E) and that depletion of YTHDF2 resulted in upregulation of downstream EMT transcription factors, ZEB-1 and SNAIL (Figure 4F). EMT is executed transcriptionally in response to the activation of the MAPK/ERK cascades, often leading to upregulation of MYC itself and increased expression of several translation initiation factors known to contribute to cancer progression (Chen et al., 2013; Truitt et al., 2015; Yin et al., 2017). Following YTHDF2-depletion, we detected ERK1/2 phosphorylation, increased expression of translation initiation factors and elevated MYC protein expression (Figure 4G). To determine if YTHDF2 limits the activation of signaling pathways operating upstream or in parallel to MYC, we next tested if YTHDF2 regulates MYC’s transcriptional targets or the MYC transcript, itself. We found that YTHDF2 binding did not always correspond with MYC transcript abundance (Figures S4B and S4C) and although MYC targets were significantly upregulated in YTHDF2-depleted MYC-induced HMECs (Figure S4D), very few significantly upregulated MYC targets were bound by YTHDF2 (Figure S4E). Furthermore, the adaptive ER stress pathway is typically engaged during EMT to alleviate metabolic and oxidative stress that accompanies cancer cell transformation and growth by facilitating protein folding and averting cell death (Dey et al., 2013; Feng et al., 2014; Rutkowski et al., 2006). Consistently, we detected activation of the adaptive unfolded protein response (UPR) in all breast cancer subtypes including upregulation of genes downstream of the serine/threonine PERK kinase, ATF4, GADD34 and CHOP and splicing of XBP1 (sXBP1) downstream of the serine/threonine IRE1 kinase (Figure 4H). In conclusion, we found that depletion of YTHDF2 generally increases the expression of EMT pathway genes in breast cancer cells, however, only TNBC cells were highly sensitive to upregulation of these pathways.
Depletion of YTHDF2 sensitizes TNBC cells to proteotoxicity
While it is well-established that MYC can actuate both cell proliferation and apoptosis, evidence suggests that rather than inducing apoptosis directly, high levels of MYC expression may be responsible for sensitizing cells to apoptotic triggers (Hart et al., 2012). Since depletion of YTHDF2 has recently been shown to sensitize MYC-amplified acute myeloid leukemia (AML) cells to TNF-induced apoptosis (Paris et al., 2019), we next dissected the extrinsic and intrinsic apoptotic pathways driving this phenotype (Figure 5A). Of the most significantly upregulated YTHDF2 target transcripts in YTHDF2-depleted, MYC-induced HMECs, with strong YTHDF2 binding signal centered on m6A sites we first focused on TNF Receptor Superfamily Member 10b (TNFRSF10B i.e., DR5) (Figure S5A) by probing DR5-induced apoptotic activity through co-depletion of YTHDF2 and DR5. While we did observe a reduction in caspase-8 cleavage with co-depletion compared to YTHDF2-only depleted cells, we surprisingly did not observe reduced cleavage of downstream caspase-3, indicating that DR5 knockdown did not halt the apoptotic cascade in YTHDF2-depleted cells (Figure S5B).
Figure 5. YTHDF2-depleted, TNBC cells initiate apoptosis from intrinsic mitochondrial stress.
(A) Schematic describing the pathways that contribute to extrinsic vs. intrinsic apoptosis.
(B) Western blot analysis of cell lysates from NTC and shYTHDF2 TNBC and HR+ breast cancer cell lines assessing activation of the terminal unfolded protein response pathway.
(C) Genome browser tracks (hg19) depicting YTHDF2 eCLIP peaks (light blue) over size-matched (SM)Input (dark blue) and m6A methylation peaks (light red) over input (dark red) on the PRSS23 transcript.
(D) Box plot displaying mRNA expression levels for MYC and PRSS23 in TNBC and ER/PR/HER2 positive tumors from TCGA data portal (Liu et al., 2018). p*** < 0.001, two-sided Student’s T-test.
(E) Cell proliferation of shYTHDF2 and YTHDF2/PRSS23 co-depleted TNBC cells. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, two-way ANOVA test with Dunnett’s post-hoc test for multiple comparisons. Values normalized to initial average confluence. Bars are mean ± SD, n = 6 independent replicates.
(F) Western blot analysis of cell lysates from shYTHDF2 and YTHDF2/PRSS23 co-depleted TNBC cells assessing protein expression of UPR pathway genes and translation initiation factors.
(G) Cumulative distribution of the fold change in mRNA expression between shYTHDF2 compared with NTC in MYC-induced HMECs. Describes direct TCF12 targets (ENCSR000BUN) (red) or non-targets (blue). Two-sided Kolmogorov-Smirnov test compared to non-targets, n = 4 replicates (2 hairpins x 2 biological replicates).
(H) Venn diagram describing the overlap of TCF12 targets determined in (G) with high confidence YTHDF2 target genes and differentially upregulated genes in shYTHDF2 MYC-induced HMECs.
(I) Quantification of reactive oxygen species (ROS) in shYTHDF2 and YTHDF2/PRSS23 co-depleted MDA-MB-231-LM2 cells compared to NTC. **p < 0.01, ***p < 0.001, one-way ANOVA test with Dunnett’s post-hoc test for multiple comparisons. Bars represent median values, n = 3 independent replicates.
See also Figure S5.
Since we detected activation of the adaptive UPR in all YTHDF2-depleted breast cancer cells, we hypothesized that the terminal UPR may be activated in YTHDF2-depleted TNBC cells leading to intrinsic apoptosis from excessive ER stress (Figure 5A). Indeed, we observed upregulated expression of ER chaperone and foldase proteins, indicating unfolded protein accumulation (Figure S4F) and we detected activation of the c-Jun N-terminal kinase (JNK), resulting in caspase-3 cleavage in TNBC cell lines, but not in HR+ cell lines (Figure 5B). Furthermore, unfolded protein accumulation and apoptosis were reversed following treatment with 4-phenylbutyric acid (4-PBA), an ER stress inhibitor (Figure S5C), confirming proteotoxicity as the underlying cause of cell death in YTHDF2-depleted TNBC cells. Additionally, cross-reference of public mass spectrometry data for 83 tumor samples provided by the Clinical Proteomic Tumor Analysis Consortium (CPTAC) from TCGA’s Cancer Proteome Study of Breast Tissue (Mertins et al., 2016) indicated that YTHDF2 expression is negatively correlated with MAPK activity and that tumor samples with lower YTHDF2 expression have higher levels of ASK1 (MAP3K5), JNK1/2 (MAPK8/9) and p38 (MAPK14) phosphorylation (Figure S5D).
To substantiate that proteotoxicity is a result of disrupting YTHDF2-mediated regulation of its target mRNAs, we identified and modulated the expression of the most significantly upregulated direct YTHDF2 target upon YTHDF2 depletion, Serine Protease 23 (PRSS23) (Figures 4B and 5C). PRSS23 is significantly downregulated in patients with TNBC tumors compared to patients with HR+ tumors (Figure 5D) and co-depletion of YTHDF2 and PRSS23 rescued cell proliferation rates in YTHDF2-depleted, TNBC cells (Figure 5E). Additionally, co-depletion of YTHDF2 and PRSS23 produced a substantial reduction in protein levels of translation initiation factors, eiF4E and eiF2α, MYC and caspase-3 cleavage compared to silencing of YTHDF2 alone (Figure 5F). Previous studies have implicated PRSS23 in translation control (Han et al., 2019a) and PRSS23 localizes to the nucleus and interacts with TCF12, a TF that regulates transcription of genes that promote MYC expression and metastatic phenotypes (Chen et al., 2013; Stelzl et al., 2005; Tang et al., 2019). In agreement with these previous reports, we found that TCF12 targets identified in publicly available ChIP-seq data (ENCODE; ENCSR000BUN) are upregulated in YTHDF2-depleted, MYC-induced HMECs, and that the majority of upregulated TCF12 targets are not YTHDF2 targets (Figure 5G and 5H). Moreover, we found that co-depletion of YTHDF2 and PRSS23 partially rescued BiP expression (Figure 5F), alleviated reactive oxygen species (ROS) levels by ~50% (Figure 5I) and dampened intrinsic apoptotic signaling (Figure S5B). Moreover, we identified a new facet of regulation by m6A in MYC-driven breast cancer where YTHDF2 binds to and targets several transcripts for degradation, including PRSS23, and in doing so safeguards cancer cells from proteotoxicity resulting from excessive ER stress.
Depletion of YTHDF2 boosts translation rates in single cells across tumors
To further test our hypothesis that apoptosis of YTHDF2-depleted, TNBC cells is driven by accumulated unfolded proteins, we measured rates of protein synthesis in these cells using puromycin incorporation as a surrogate measure of protein translation. We observed that YTHDF2-depleted cells contained higher levels of puromycin incorporation into nascent peptides, indicating increased levels of protein synthesis (Figure S6A). To substantiate this observation in vivo, we leveraged Ribosome-STAMP (Surveying Targets by APOBEC-Mediated Profiling), a method to assess translation by directing C-to-U base editing of ribosome-bound transcripts using the cytidine deaminase enzyme APOBEC1 fused to the C-terminus of the 40S ribosomal protein S2 (RPS2) (Brannan et al., 2021). We generated DOX inducible, HA-tagged, RPS2-APOBEC1 (RPS2-STAMP) and APOBEC1-only (Control-STAMP) expressing cell lines both in wild-type (WT) and in DOX-inducible shRNA-targeting YTHDF2 (TurboRFP-tagged) MDA-MB-231-LM2 cells (Figure S6B). After engraftment, mice were given DOX water for 3 days to induce both shRNA and STAMP transgene expression prior to tumor resection, dissociation and single-cell (sc)RNA-seq using 10X Chromium Single Cell capture. At this early timepoint, we uncovered cells harboring molecularly distinct translational signatures driven by both tumor heterogeneity and gene expression changes reflected by C-to-U sequence changes in mRNA (Figure 6A).
Figure 6. Analysis of single cells within tumors reveals increased translation rates in YTHDF2-depleted, TurboRFP-tagged cells.
(A) Schematic describing experimental workflow for generating cell lines, engrafting mice and dissociating tumors followed by single cell RNA-seq (scRNA) to profile the translatome within tumors.
(B) Uniform Manifold Approximation and Projection (UMAP) analysis of mRNA expression of merged cells from wild-type (WT) Control-STAMP cells (blue, n = 6,089), WT RPS2-STAMP cells (green, n = 5,886), shYTHDF2 Control-STAMP cells (brown, n = 6,851) and shYTHDF2 RPS2-STAMP cells (yellow, n = 6,630). Experiments performed in MDA-MB-231-LM2 cells.
(C) UMAP analysis of mRNA expression from merged cells from all samples colored by TurboRFP expression. Outline indicates the cluster of highest TurboRFP-expressing cells.
(D) UMAP analysis of mRNA expression from merged cells from all samples colored by expression score of differentially upregulated genes from in vitro bulk RNA-seq in shYTHDF2 MYC-induced HMECs (Figure 4B; upregulated). Outline indicates the cluster of highest TurboRFP-expressing cells.
(E) UMAP analysis of mRNA expression from merged cells colored by mRNA expression Louvain clustering. Outline indicates the cluster of highest TurboRFP-expressing cells.
(F) Bar chart describing the fraction of each mRNA expression Louvain cluster in TurboRFP low- (n= 21,464) or high-expressing (n= 3,992) populations. TurboRFP high fraction defined as cells with TurboRFP z-score > 0.
(G) Clustered heatmaps of enriched gene ontology (GO) terms extracted from differentially expressed genes belonging to each mRNA expression Louvain cluster (p < 0.05).
(H) UMAP analysis of unfiltered EPKM of merged cells colored by sample.
(I) UMAP analysis of unfiltered EPKM of merged cells colored by average EPKM per cell.
(J) UMAP analysis of unfiltered EPKM of merged cells colored by EPKM Louvain clustering. Cells filtered based on assignment to Control- or RPS2-STAMP cluster. Bar chart describes the fraction of each cluster contained in each sample.
(K) Distribution of Control-STAMP filtered, average EPKM per cell for TurboRFP low- and high-expressing populations. TurboRFPHigh fraction defined as cells with TurboRFP z-score > 0. One-sided Kolmogorov-Smirnov test.
See also Figure S6.
Next, we tested if mRNA expression level changes in YTHDF2-depleted single cells resembled our bulk, in vitro observations. First, YTHDF2-depleted tumor cell populations (Control-STAMP and RPS2-STAMP) contained cells with high raw read coverage in mitochondrial genes compared to WT samples, indicating higher fractions of apoptotic or lysed cells (Figure S6C). Second, UMAP visualization of mRNA expression revealed clusters of YTHDF2-depleted tumor cells that were distinct from their WT counterpart (Figure 6B) and cells expressing the TurboRFP tag (TurboRFPHigh) generally had lower YTHDF2 mRNA expression on average compared to cells that did not (TurboRFPLow) (Figure S6D). Third, UMAP visualization defined a cell group containing the majority of TurboRFPHigh cells (Figure 6C), which also contained the highest expression of genes that were upregulated in bulk YTHDF2 depleted, MYC-induced HMECs (Figure 6D). This group of cells corresponded to three Louvain mRNA expression clusters (Figure 6E, clusters 1,2,3), with enrichment of TurboRFPHigh cells in cluster 2 (Figure 6F). Fourth, GO analysis on genes from cells assigned to the different mRNA expression Louvain clusters confirmed enrichment for ontologies related to EMT and stress response such as cytokine-mediated signaling pathway, response to wounding, blood vessel development, extracellular matrix (ECM) organization and positive regulation of cell death in clusters 1 and 2, while clusters 3 and 4 where highly enriched for genes involved in cell cycle progression (Figure 6G). Fifth, in agreement with our in vitro studies, we found that clusters 1 and 2 contained larger fractions of cells assigned to G1 phase of the cell cycle based on their transcriptomic expression profiles (Macosko et al., 2015), while cluster 3 was overrepresented for S phase and cluster 4 for G2M phase (Figure S6E). From these analyses we concluded that the majority of TurboRFPHigh cells undergoing G1 arrest exist in clusters 1 (Control-STAMP) and 2 (RPS2-STAMP) and that cluster 3 may contain fewer TurboRFPHigh cells that either have yet to undergo cell cycle arrest or have not undergone sufficient YTHDF2 depletion to initiate apoptosis since the mRNA expression profile in cluster 3 more closely resembles that of cluster 4 (Figure 6G), which contains cells expressing proliferative and tumorigenic markers.
Next, we analyzed the translatomes of single tumor cells (Brannan et al., 2021) by detecting C-to-U edits on transcripts and assigning site coverage (Edited-reads Per Kilobase of transcript, per Million mapped reads; EPKM). EPKM from CDS correlated strongly with values from both CDS and 3’UTR regions, consistent with the previous observation that 3’UTR signal is specific to RPS2-STAMP-mediated editing (Brannan et al., 2021) (Figure S6F). UMAP visualization of EPKM across all tumor samples revealed distinct clustering of Control-STAMP samples away from RPS2-STAMP samples (Figures 6H and 6I) and using Louvain clustering by EPKM, we filtered the dataset by excluding cells assigned to the Control-STAMP cluster (blue) that did not contain substantial editing (Figure 6J). Since transcriptomes and proteomes of individual tumor cells are known to be heterogenous (Ramon et al., 2018), we expected Ribo-STAMP to capture the full spectrum of translational states in tumors regardless of YTHDF2 modulation. Indeed, the distribution of EPKM values for filtered cells revealed a full range of edits throughout the TurboRFPLow population (Figure 6K). Generally, we found higher edits per gene in the YTHDF2 depleted, TurboRFPHigh population, suggesting increased rates of protein synthesis (Figure 6K).
scRibo-STAMP identifies unique translation profiles for single YTHDF2-depleted cells within heterogenous tumors
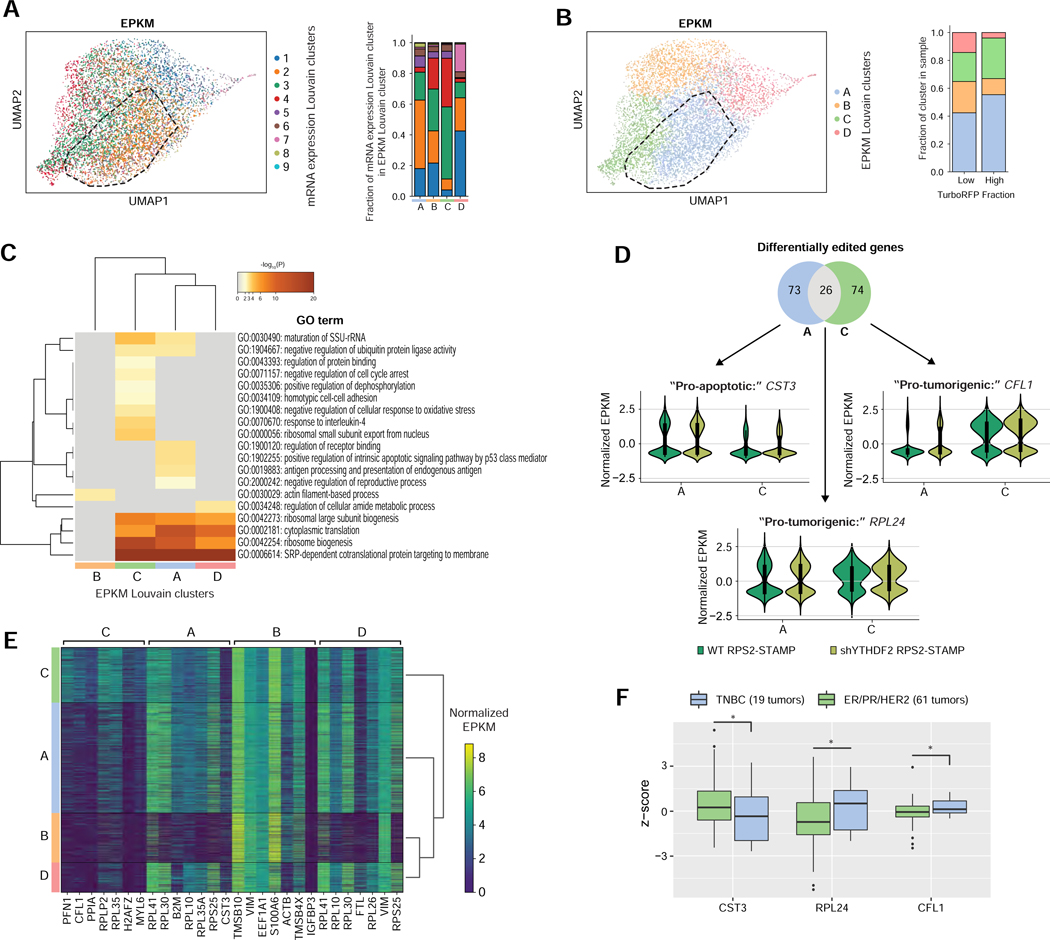
Tumors are composed of subpopulations of cells that differ at both the genomic and proteomic level (Ramon et al., 2018). Interestingly, we found that in addition to increased rates of protein synthesis, TurboRFPHigh cells also contained edits on more genes per cell on average (Figure S7A), suggesting that YTHDF2-depletion may initiate distinct translation programs. To identify differentially translated mRNAs, we focused on a subpopulation of cells that comprise of the most TurboRFPHigh cells (Figure S7B) with high EPKM (Figure S7C). This group contained enrichment for cells assigned to cluster 2 (Figure 7A), confirming that many of these cells originated from the shYTHDF2 RPS2-STAMP tumor. Re-visualizing the UMAP space by Louvain clustering on EPKM produced very similar clusters with substantial overlap of mRNA expression-defined cluster 2 with EPKM-defined cluster A (Figures 7A and 7B). This supports our observation that modulation of m6A modified transcripts on the RNA level incites widespread mRNA translation changes. Next, we performed GO analysis on genes from cells assigned to the different EPKM Louvain clusters. Clusters A and C contained the highest number of differentially edited transcripts (≥99 genes) and enrichment for TurboRFPHigh cells (Figure 7B) and each cluster respectively corresponded to mRNA expression-defined cluster 2 and clusters 3 and 4 (Figure 7A). Both clusters had high levels of editing within transcripts encoding genes known to contribute globally to translation (Figure 7C). Uniquely, cluster A (“Pro-apoptotic” cluster) was enriched for edits within genes involved in apoptosis and antigen presentation while cluster C (“Pro-tumorigenic” cluster) was enriched for edits within genes involved in preventing cell cycle arrest, homotypic cell adhesion and dampening oxidative stress, all of which are all associated with tumor progression (Figure 7C). 26% of the top 100 differentially edited genes (p < 0.05) between EPKM clusters A and C overlapped, while ~74% were unique to each cluster (Figure 7D). The most highly edited genes in each cluster included Cystatin C (CST3) in cluster A (“Pro-apoptotic” cluster), Cofilin 1 (CFL1) in cluster C (“Pro-tumorigenic” cluster) and 60S Ribosomal Protein L24 (RPL24) among overlapping genes (Figures 7D, 7E and Table S5). CST3 is a cysteine protease inhibitor that is secreted in response to hypoxia (Rosenow et al., 2013). While being an attractive biomarker in several types of cancers, CST3 is also involved in caspase-mediated cell death (Malone et al., 2020). Conversely, RPL24 is required for polysome assembly and is known to be upregulated in MYC-driven cancers (Wilson-Edell et al., 2014), and CFL1 is known to drive invasiveness in basal breast cancers (Quintela-Fandino et al., 2010). Accordingly, we found that TNBC tumors generally express significantly higher levels of CFL1 and RPL24 protein and less CST3 protein compared to HR+ tumors (Mertins et al., 2016), supporting a role for CST3 in promoting MYC-driven apoptosis (Figure 7F). In conclusion, Ribo-STAMP data corroborated our findings that YTHDF2-depleted tumor cells contain unique translatomes displaying increased translation of tumorigenic and apoptotic transcripts while lacking translation of cell cycle regulators important for maintaining oncogenic proliferation.
Figure 7. scRibo-STAMP identifies unique changes in the translatome of single cells clustering with TurboRFP-expressing populations.
(A) Uniform Manifold Approximation and Projection (UMAP) analysis of Control-STAMP filtered EPKM of merged cells from RPS2-STAMP conditions colored by mRNA expression Louvain clustering. Outline indicates the cluster of highest TurboRFP-expressing cells. Bar chart describes the fraction of each mRNA expression Louvain cluster in each EPKM Louvain cluster.
(B) UMAP analysis of Control-STAMP filtered EPKM of merged cells from RPS2-STAMP samples colored by EPKM Louvain clustering. Outline indicates the cluster of highest TurboRFP-expressing cells. Bar chart describes the fraction of each EPKM Louvain cluster contained in TurboRFP high- (n = 1,696) and low-expressing (n= 6,389) populations.
(C) Clustered heatmaps of enriched gene ontology (GO) terms extracted from differentially edited genes from each EPKM Louvain cluster (p < 0.05).
(D) Venn diagram overlaps of differentially edited genes between EPKM Louvain clusters A and C (top). Violin plots summarizing the normalized EPKM of differentially edited genes in EPKM Louvain cluster A (CST3), EPKM Louvain cluster C (CFL1) or both (RPL24). Cluster A, n = 3,656. Cluster C, n = 1,823.
(E) Heatmap of normalized EPKM signatures for merged RPS2-STAMP cells for the top seven differentially edited genes per EPKM Louvain cluster.
(F) Box plot comparing protein expression levels for CST3, RPL24 and CFL1 between TNBC and ER/PR/HER2 positive tumors downloaded from TCGA’s Cancer Proteome Study of Breast Tissue (Mertins et al., 2016). p* < 0.05, one-sided Student’s T-test.
See also Figure S7.
Discussion
Our RBP-focused CRISPR-Cas9 knockout library enabled the systematic discovery of factors involved in multiple stages of RNA processing, including RBPs that regulate transcription, mRNA stability, ribosome recruitment, and translation, as vulnerabilities in MYC-driven breast cancer, generalizing previous reports focusing on splicing (Hsu et al., 2015). Our observations support that deposition of m6A on RNA promotes cancer cell growth, survival and invasion (Lin et al., 2016) and we found that expression of m6A reader protein YTHDF2 is required to sustain MYC-driven cell growth and survival in both cells and tumors. Using eCLIP and m6A-seq, we identified specific, direct target mRNAs in TNBC cell lines belonging to MAPK/ERK signaling pathways, including several mRNAs encoding upstream activating growth factor and receptor families. Stabilization of these target mRNAs led to upregulation of cellular protein markers of EMT and considerably elevated translation rates causing ER stress from unfolded protein accumulation, culminating in the activation of programmed cell death. Since YTHDF2 maintains mRNA homeostasis by limiting the number of translating mRNAs in cancer cells, excessive translation can cause significant cellular stress, particularly in MYC-addicted cells.
YTHDF2 regulates mRNA localization (Ries et al., 2019) and translation (Zhou et al., 2015) in response to various cellular stressors and previous reports have attributed the function of YTHDF2 in various cancers to the regulation of singular target mRNAs (Chen et al., 2018; Dixit et al., 2020; Paris et al., 2019; Zhong et al., 2019). Our analysis revealed several candidate YTHDF2 targets, and we specifically found that co-depletion of YTHDF2 and its target mRNA, PRSS23 dampened cellular stress that led to the activation of programmed cell death. Nevertheless, our observations suggest that while single target mRNAs may be partially responsible for YTHDF2’s effect, a larger subset of its targets may be required for apoptosis in YTHDF2-depleted MYC-driven breast cancer cells.
Here, we applied scRibo-STAMP to measure mRNA translation in individual tumor cells, which to our knowledge, remains an outstanding challenge. scRibo-STAMP enabled the simultaneous quantification of transcriptomic and translational changes and provided an early snapshot of changes occurring within tumor cells following YTHDF2 depletion and prior to widespread apoptosis. Our dataset captured tumor cells in distinct states characterized by cell cycle phase, proliferation, and tumorigenesis. We observed increased editing on transcripts encoding secreted proteins regulating ECM composition in YTHDF2-depleted cells, which are known to be produced during EMT and metastasis and suggests mechanisms underlying crosstalk among cells with different levels of YTHDF2 expression. Our single cell studies demonstrate the utility of scribe-STAMP to reveal snapshots of translational landscapes and underlying gene expression profiles at critical periods in cancer progression.
Our results indicate that YTHDF2 is essential for the survival of TNBC cells to limit the availability of methylated transcripts during constitutively elevated levels of transcription and translation in cells with aberrantly high MYC activity, while it is dispensable for cells that are less reliant on elevated MYC expression. Our model is supported by gene expression analyses of TNBC tumors linking low YTHDF2 expression levels to longer patient survival rates and we demonstrate the efficacy and feasibility of depleting YTHDF2 as a potential therapeutic strategy by generating viable adult-life inducible systemic Ythdf2 knockout mice. Finally, recent evidence indicates that YTH-paralogs may play compensatory roles for one another, providing anticipation for minimal adverse side effects to YTHDF2 inhibition (Lasman et al., 2020; Zaccara and Jaffrey, 2020). Altogether, our studies reveal disease-promoting RBP-RNA interactions that are selectively essential for growth and survival of tumor cells but not somatic tissues, and that targeting RBPs holds great promise for minimally toxic and highly specific treatment modalities in specific cancer subtypes.
Limitations of study
Although we employed an isogenic cell line to interrogate RBP vulnerabilities in MYC-driven cancer, the cell line was derived from human mammary epithelial cells and all subsequent studies were performed using breast cancer cell lines of different subtypes. We did not elect to generalize our observations to cancers originating from tissues other than breast, however, several studies support our finding that YTHDF2 depletion triggers apoptosis in MYC-driven cancer cells. For example, an apoptotic phenotype was observed following YTHDF2 depletion in the AML cell line, THP-1 (Paris et al., 2019), prostate cancer cell lines, DU-145 and PC3 (Li et al., 2018a) and ovarian cancer cell line, SKOV3 (Li et al., 2020), all of which are highly sensitive to MYC depletion (Ai et al., 2013; Fan et al., 2016; Huang et al., 2014). Nevertheless, additional experimentation in MYC-amplified and MYC-normal cancer cell lines derived from different tissues of origin are necessary to generalize our findings across all MYC-driven cancers.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Gene W. Yeo, Ph.D. (geneyeo@ucsd.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
CRISPR screening, eCLIP, m6A-seq, and RNA-seq datasets generated during this study were deposited at GEO and are available under accession number GEO: GSE137258. Source data for dot plots generated from the in vivo shRNA screen are provided in Table S2.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines and Cell Culture
Immortalized human cell lines were used in this study. HEK293xT (Takara Bio), MDA-MB-231 (ATCC), MDA-MB-231-LM2 (gift from Thomas Westbrook, (Minn et al., 2005)), and MCF-7 (ATCC) cells were cultured in DMEM supplemented with 10% fetal bovine serum. SKBR3 (ATCC) cells were cultured in McCoy’s 5A Medium supplemented with 10% fetal bovine serum. Cells were passaged every 3 or 4 days with TrypLE EXPRESS (Life Technologies) using standard methods. MYC-ER HMECs (gift from Thomas Westbrook, (Kessler et al., 2012)) were cultured in Medium 171 supplemented with MEGS (Life Technologies). Cells were passaged every 3 or 4 days with TrypLE EXPRESS and Defined Trypsin inhibitor. Cells were maintained in a humidified incubator at 37°C with 5% CO2. Cells were tested weekly for mycoplasma. Experiments were performed within 5–10 passages after thaw.
Animal Studies
Animal protocols were approved by the Institutional Animal Care and Use Committee at Baylor College of Medicine and at University of California San Diego. Athymic Nude-Foxn1nu were purchased from Envigo International Holdings, Inc. CAG-CreERT mice (Jackson labs, stock number 004682) were mated with Ythdf2fl/fl mice (generous gift from Dr. Chuan He, University of Chicago) (Li et al., 2018b) to produce CAG-CreERT;Ythdf2fl/fl mice. To induce recombination at 8 weeks of age both CAG-CreERT;Ythdf2fl/fl and Ythdf2fl/fl littermates were injected with 75mg/kg body weight tamoxifen (Millipore Sigma; T5648) dissolved in corn oil daily for 5 days. Sex, genotype and age information is provided in figure legends and in the methods describing the experiment.
METHOD DETAILS
Lentivirus production and purification
HEK293xT cells were seeded on twelve 15 cm plates at 40% confluency the day before transfection. One hour prior to transfection the media was removed and replaced with 8mL of pre-warmed OptiMEM. Transfections were performed using 62.5 μL Lipofectamine 2000, 125 μL Plus reagent, 12.5 μg lentiCRISPR plasmid library, 6.25 μg of pMD.2g, and 9.375 μg psPAX2. Media was changed 6 hours after transfection to DMEM + 10% FBS. After 48 hours, the supernatant was filtered through a 0.45 μm low protein binding membrane. The virus was then ultracentrifuged at 24,000 rpm for 2 hours at 4°C and resuspended overnight at 4°C in PBS. Virus aliquots were stored at −80°C.
Multiplicity of infection
For each new cell type, the volume of virus to achieve an MOI of 0.3 was determined by titrating virus in each well (between 5 and 50 μL). 1.5×106 cells per well of a 24-well plate were spinfected in medium supplemented with 8μg/mL polybrene at 2,000 rpm for 2 hours at 37°C. Media (without polybrene) was replaced after the spin and incubated overnight. Cells were split the next morning and half the cells from each condition were treated with puromycin (Thermofisher Scientific; A1113803). Cells were counted after 3–4 days and MOI was determined by the volume of virus allowing 30% cell survival.
MYC-ER HMEC RBP CRISPR screen
For each replicate, 3×106 cells were spinfected in 5 wells of a 12-well plate in medium supplemented with 8μg/mL polybrene (Millipore Sigma; TR-1003-G) and spun at 2,000 rpm for 2 hours at 37°C. 2X the amount of virus determined by MOI was added per well. After spinfection, media was replaced (without polybrene) and incubated overnight. The next morning, 5 wells from each replicate were pooled and split onto two 10 cm plates per replicate. Media was replaced containing 2μg/mL puromycin (Thermofisher Scientific; A1113803) after 6 hours. Media was changed every 2 days and puromycin was removed after 4 days. 4×106 cells were collected and snap frozen in an ethanol, dry-ice bath from each replicate as the day 0 timepoint. 4×106 cells were plated per 15 cm plate for a total of 2 plates per replicate. One plate from each replicate was treated with 15nM 4-Hydroxytamoxifen (4-OHT) (Millipore Sigma; H7904). Cells were cultured for an additional 16 days, changing media and 4-OHT every 2 days, and splitting cells every 4 days, always at a minimum of 4×106 cells per 15 cm plate. 4×106 cells were harvested on day 8 and day 16 per condition for each replicate and snap frozen.
Bulk sgRNA library preparation
DNA libraries were prepared using a targeted-enrichment approach as previously described (Wheeler et al., 2020). Briefly, gDNA was extracted from cell pellets using DNeasy Blood & Tissue kit (Qiagen; 69506). gDNA samples were sonicated to ~1000bp by Biorupter. sgRNA containing fragments were recovered with biotinylated RNA probes targeting the flanking region on the lenticrispr v2 backbone and pulled down with streptavidin beads. The gDNA fragments were purified and concentrated with DNA Clean and Concentrator-5 kit (Zymo Research; 11-303C). gDNA fragments were PCRed first with primers flanking the sgRNA (Forward (5’→3’): CCTACACGACGCTCTTCCGATCTTGTGGAAAGGACGAAACACCG; Reverse (5’→3’): GTTCAGACGTGTGCTCTTCCGATCTCCACTTTTTCAAGTTGATAACGGACTAGCC) and second with Illumina sequencing adapters. Libraries were analyzed for quality using an Agilent D1000 Screen Tape (Agilent Technologies) and then were sequenced to 6M reads per library on the Hi-Seq4000 in paired-end 55bp mode. Reads were aligned to the RBP library file and candidates were identified using the MaGeCK-v0.5.4 software package (Li et al., 2014).
Knockdown experiments
Cells were transduced with TRC lentiviral shRNA vector non-targeting control (NTC; Millipore Sigma; SHC002), and TRC lentiviral shRNA vector YTHDF2 (shYTHDF2–1; Millipore Sigma; TRCN0000168751), (shYTHDF2–2; Millipore Sigma; TRCN0000167813), TRC lentiviral shRNA vector METTL3 (shMETTL3; Millipore Sigma; TRCN0000034717), TRC lentiviral shRNA vector PRSS23 (shPRSS23; TRCN0000047042) or TRC lentiviral shRNA vector TNFRSF10B (shDR5; TRCN0000005929) for 24 hours before treatment with Puromycin (2 mg/mL). Cells were analyzed 6 days after the addition of lentivirus for all assays unless otherwise noted.
Annexin V/PI apoptosis assay
MYC-ER HMECs were transduced with NTC, shYTHDF2–1, shYTHDF2–2, or shMETTL3 virus at MOI > 1 and selected for 2–3 days with 2 μg/mL puromycin (Thermofisher Scientific; A1113803). MYC expression was induced with 15 nM 4-OHT for 24 hours. The Annexin V apoptosis assay was performed using the AnnexinV-FITC kit (BD Biosciences; BDB556547) according to manufacturer’s instructions. Cells were analyzed by flow cytometry using the BDSLRFortessa under the FITC (Annexin V) and PerCP-Cy5 (Propidium Iodide) channels with compensation. Analysis and gating were performed using FlowJo.
Cell Cycle Analysis
MYC-ER HMECs were transduced with NTC or shYTHDF2–2 virus at MOI > 1 and selected for 2–3 days with 2 μg/mL puromycin (Thermofisher Scientific; A1113803). MYC expression was induced with 15 nM 4-OHT for 72 hours prior to ethanol fixation and propidium iodide staining using the Propidium Iodide Flow Cytometry Kit (Abcam; ab139418) according to manufacturer’s instructions. Analysis and gating were performed using FlowJo.
Pooled screen in vivo tumor screen and analysis
MDA-MB-231-LM2 breast cancer cells were individually transduced at MOI of 1.2–1.5 with doxycycline-inducible shRNAs (shRNA targeting RNA metabolism genes were cloned from GIPZ plasmid to pINDUCER11 backbone (Meerbrey et al., 2011)):
GIPZ lentiviral shRNA vector YTHDF2 (Dharmacon; V2LHS_115143), (Dharmacon; V2LHS_115142), (Dharmacon; V3LHS_381614)
All cell lines were sequentially pooled in equal ratios. The obtained pool was subcutaneously transplanted (3 X 10^6 cells per mice) into athymic nude mice (female mice, 4–6 weeks old). Mice were randomized onto and maintained on 5% sucrose water (-Dox) or 5% sucrose water with 2 mg/mL dox (Sigma Aldrich; D9891) (+Dox) 3d post-transplantation. Tumors were measured using calipers and harvested when they reached 1000 mm3, approximately 2–3 weeks after engraftment and cells cultured in vitro were carried out for 12 population doublings. Genomic DNA from dissected tumors was isolated using the QIAamp DNA mini kit (Qiagen), and shRNA library was amplified using the following primers (5’–3’):
forward: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG TAGTGAAGCCACAGAGTA;
reverse: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGCGCGGAGGCCAGATCTT; The library was indexed using Nextera Index Kit (96 indices) (Illumina; KAPA#KK4824) and purified using PippinHT. The library was quantified using KAPA Library Quantification Kit (Illumina; FC-131–1096) and sequenced at Illumina HIseq platform (~10*10^6 reads per tumor with a read coverage of >10000 reads per shRNA per tumor).
Reads were processed to remove adapter sequences using Cutadapt (Martin, 2011) and then aligned to the reference library using Bowtie 2(Langmead and Salzberg, 2012) in end-to-end mode allowing up to a maximum of 3 mismatches/indels compared to the reference sequence. The raw number of reads mapping to each shRNA in each sample was then extracted from the SAM files and DESeq2-v.1.14.0 (Love et al., 2014) was used to determine the normalized abundance of each shRNA in the vehicle and DOX-treated tumors.
In vivo tumorgenicity assays
MDA-MB-231-LM2 breast cancer cells were transduced with validated YTHDF2 targeting pINDUCER11 shRNA 1 (Dharmacon; V2LHS_115143) and sorted for the top 10% of EGFP-expressing cells on a BD Influx Cell Sorter. Cells were expanded and then subcutaneously transplanted (3 X 106 cells per mouse) into athymic nude mice (female mice, 3–4 weeks old). Mice were randomized onto and maintained on 5% sucrose water (-DOX) or 5% sucrose water with 2 mg/mL dox (Sigma Aldrich; D9891) (+DOX) 14d post-transplantation. Tumors were measured twice, weekly using calipers and harvested when they reached 1000 mm3 on average and tumors were sectioned and either fixed in 4% paraformaldehyde for IHC analysis or snap frozen and cryopulvierized for RNA and protein extraction.
Immunohistochemistry
Tumor samples were fixed in 4% paraformaldehyde and then paraffin-embedded. Microtome sectioning and hematoxylin/eosin staining was performed by the Moores Cancer Center Histology Core. 5um thick sections were deparaffinized in Citrisol and rehydrated with graded alcohols. Epitope retrieval was performed by boiling slides for 10 min in sodium citrate buffer (10mM Sodium citrate, 0.05% Tween 20, pH 6.0). DAB staining was performed using Rabbit Specific HRP/DAB (ABC) Detection IHC Kit (Abcam; ab64261) according to manufacturer’s instructions. The following antibodies were incubated overnight in 5% goat serum in wash buffer containing 0.025% Triton X-100 in PBS: Rabbit pAb anti-Ki67 (Abcam; ab15580), Rabbit mAb anti-CD31 (Cell Signaling; 77699), Rabbit pAb YTHDF2 (Proteintech; 24744–1-AP), Rabbit pAb anti-RFP (Thermo Fisher; R10367). Hematoxylin (Vector Laboratories; H-3502) was used as a counter stain according to manufacturer’s instructions and slides were dehydrated before coverslipping.
Immunofluorescence
Cells were seeded on poly-D-lysine hydrobromide (PDL) (Millipore Sigma; P6407) coated 8-well chamber slides (Millipore Sigma). Cells were fixed in 4% paraformaldehyde in PBS, permeabilized and blocked 5% normal goat serum, 0.1% Triton-X in PBS for 1 hour at RT. Primary antibody: Rabbit mAb anti-Vimentin (Cell Signaling; 5741), was diluted in blocking buffer and incubated overnight at 4°C. Cells were washed 3 times in 0.1% Triton-X in PBS and incubated with secondary antibody: Goat anti-Rabbit IgG Alexa Fluor 488 (Invitrogen; A32731), for 1 hour at RT, followed by 3 washes and coverslip mounting with Prolong Diamond Antifade Mountant with DAPI (Thermofisher). Slides were imaged on a ZEISS Axio Vert.A1 inverted microscope.
eCLIP-seq library preparation and analysis
Experiments were performed as previously described in Van Nostrand et al., 2017 in biological duplicates. Briefly, 20M cells were UV-crosslinked at 400 mJ/cm2 constant energy, lysed in eCLIP lysis buffer on ice, and sonicated by BioRuptor. Lysate was treated with RNase I to fragment RNA, then protein-RNA complexes were immunoprecipitated (Sheep anti-rabbit Dynabeads) with a YTHDF2 antibody: Rabbit pAb anti-YTHDF2 (Aviva; ARP67917_P050). Inputs (2% of lysate) were saved and run alongside IP samples. IP samples were stringently washed, and for all samples the RNA was dephosphorylated with FastAP (NEB) and T4 PNK (NEB), followed by on-bead ligation of barcoded RNA adapters to the 3’ end (T4 RNA Ligase, NEB). RNA-protein complexes were run on standard protein gels and transferred to nitrocellulose membranes where the RNA in the region 65 kDa – 140kDa was excised off the membrane and proteinase K (NEB) treated. RNA was then reverse transcribed with Superscript III (Thermofisher) followed by treatment with ExoSAP-IT (Affymetrix) to remove excess oligonucleotides. Samples were cleaned up with Dynabeads MyOne Silane (Thermofisher) and subject to qPCR to determine the appropriate number of PCR cycles. Libraries were amplified with Q5 PCR mix (NEB), QCed using an Agilent D1000 Screen Tape (Agilent Technologies) and sequenced to 20M reads on the HiSeq4000 in single-end 75 bp mode.
Fastq files were run through eCLIP-v0.4.0 pipeline describe in Van Nostrand et al., 2017. Briefly, adapters and adapter-dimers were trimmed with cutadapt-v1.14.0, reads were mapped to repeat elements and filtered with STAR-v2.4.0, PCR duplicates were removed with umi_tools-v0.5.5, and enriched peak regions were called with CLIPPER-v1.2.2v. Peaks were input normalized, reproducible peaks were determined by irreproducible discovery rate (IDR) https://github.com/YeoLab/merge_peaks), and peaks were filtered using merge_peaks-v0.0.5. Peaks were annotated by gene and region. Motifs were analyzed using HOMER-v.4.9.1 (Heinz et al., 2010). Metagene plots were generated using MetaPlotR-v2.1.2 (Olarerin-George and Jaffrey, 2017).
m6A-seq library preparation and analysis
Experiments were performed as previously described in Dominissini et al., 2013 in biological duplicates. Briefly, 250 μg RNA was extracted using Trizol Reagent (Invitrogen) according to manufacturer’s instructions, rRNA depleted (RiboZero), and fragmented to ~100 nt. 1 μg fragmented RNA was saved as input and the rest was immunoprecipitated (Protein G sheep anti-mouse Dynabeads) with an m6A antibody: Mouse mAb anti-m6A (Synaptic Systems; 202 011). RNA was precipitated and libraries were prepared with TruSeq Stranded Total RNA Preparation Kit according to manufacturer’s instructions. Libraries were QCed using an Agilent D1000 Screen Tape (Agilent Technologies). Libraries were sequenced to ~20M reads on the HiSeq4000 on single-end 75 bp mode. Reads were subjected to cutadapt (Martin, 2011) to remove polyA tracts and adapter sequences, followed by removal of duplicates and alignment to the human genome build hg19 using the STAR-v2.4.0(Dobin et al., 2013). Uniquely mapped reads were subjected to peak-calling analysis using MACS2-v2.1.2 software (Zhang et al., 2008) with the following parameters: macs2 callpeak -t IP.sam -c Input.sam -f SAM --gsize=‘3137161264’ --tsize=50 --nomodel --extsize=50 -q 0.1 --down-sample Peaks were filtered based by -log10(q-value) > 3 and motifs were analyzed using HOMER-v.4.9.1 (Heinz et al., 2010). Metagene plots were generated using MetaPlotR-v2.1.2 (Olarerin-George and Jaffrey, 2017). Pairwise comparisons to YTHDF2 eCLIP was performed using HOMER-v.4.9.1 (Heinz et al., 2010).
RNA-seq library preparation and analysis
RNA was extracted with Direct-zol RNA Miniprep kit (Zymo Research; R2071) for two independent non-targeting control biological replicates and two independent shYTHDF2 biological replicates in MYC-ER HMECs induced with 15nM 4-OHT for 48 hours. 1μg total RNA was rRNA depleted (RiboZero) and processed using the TruSeq Stranded Total RNA Preparation Kit (Illumina; RS-122–2201) according to manufacturer’s instructions. Libraries were QCed using an Agilent D1000 Screen Tape (Agilent Technologies). Libraries sequenced to 20M reads on the HiSeq4000 in single-end 75 bp mode.
Adapters were trimmed and reads were mapped to the human genome build hg19 using STAR-v2.4.0. Differential expression was analyzed using DEseq2-v1.22.1 (with significance cutoffs of p < 0.001 and log2(fold change) > 1, with a minimum TPM of 1 in any sample).
Western Blot
Cells were lysed with cold RIPA buffer (Thermofisher) with 200X Protease inhibitor and 100X phosphatase inhibitor. Protein was quantified using Peirce BCA Protein Assay Kit. Total protein extracts were run on 4%–12% NuPAGE Bis-Tris gels in NuPAGE MOPS running buffer (Thermofisher) and transferred to PVDF membranes. Membranes were blocked in 5% nonfat milk in TBST for 1 hour, incubated overnight at 4°C with the following primary antibodies (5% BSA for phospho-antibodies): Rabbit pAb anti-YTHDF2 (Proteintech; 24744–1-AP), Rabbit mAb anti-YTHDF2 (for mouse tissues, Abcam; ab220163) Rabbit pAb anti-METTL3 (Proteintech; 15073–1-AP), Rabbit pAb anti-Phopho-p44/42 MAPK (Erk1/2) (Cell Signaling; 4370), Rabbit pAb anti-p44/42 MAPK (Erk1/2) (Cell Signaling; 4695), Rabbit Polyclonal anti-Phospho-eIF2α (Cell, Signaling; 9721). Rabbit mAb anti-eif4e (Cell Signaling; 9742), Rabbit pAb anti-eif2α (Cell Signaling; 9722), Rabbit mAb anti-c-Myc (Cell Signaling; 13987), Mouse mAb anti-GAPDH (Abcam; ab8245), Rabbit mAb anti-BiP (Cell Signaling; 3177), Rabbit mAb anti-Phospho-p38 MAPK (Cell Signaling; 4511), Rabbit mAb anti-p38 MAPK (Cell Signaling; 8690), Rabbit pAb anti-PARP (Cell Signaling; 9542), Rabbit pAb anti-Caspase-3 (Cell Signaling; 9662), Rabbit mAb anti-Cleaved Caspase-3 (Cell Signaling; 9664), Rabbit mAb anti-Phospho-SAPK/JNK (Cell Signaling; 4668), Rabbit pAb anti-SAPK/JNK (Cell Signaling; 9252), Rabbit pAb anti-IRE1α (Novus Biologicals; NB100–2324), Rabbit pAb anti-Phospho-IRE1α (Novus Biologicals; NB100–2323), Rabbit mAb anti-ZEB1 (Cell Signaling; 3396), Rabbit mAb anti-Snail (Cell Signaling; 3879), Rabbit pAb anti-PRSS23 (Abcam; ab201182), Rabbit pAb anti-DR5 (Cell Signaling; 3696), Rabbit mAb anti-Cleaved Caspase-8 (Cell Signaling; 9496), Rabbit mAb anti Cleaved Caspase-9 (Cell Signaling; 7237), Rabbit mAb anti-Bax (Cell Signaling; 5023), Rabbit pAb anti-RFP (Thermo Fisher; R10367), Rabbit pAb anti-GFP (Abcam; ab290), Rabbit mAb anti-HA-Tag (Cell Signaling; 3724), washed 3X for 5 minutes with TBST, incubated for 1 hour at RT in 5% nonfat milk in TBST with secondary HRP-conjugated antibody: Anti-mouse IgG, HRP-link Antibody (Cell Signaling; 7076), Anti-rabbit IgG, HRP-linked Antibody (Cell Signaling; 7074) at 1:5000 dilution, and washed 3X for 5 minutes with TBST. Membranes were developed using Thermo Pierce ECL detection reagents.
SUnSET Assay
MDA-MB-231-LM2 cells expression DOX-inducible YTHDF2-targeting harpin were treated with or without DOX for 3 days. Cells were treated with 10 μg/mL for 10 minutes and subsequently processed for Western blot analysis as above using the primary antibody, Mouse mAb anti-puromycin (clone 12D10) (Millipore Sigma; MABE343).
ER stress inhibition assay
MDA-MB-231-LM2 and MDA-MB-231 cells were transduced with NTC, shYTHDF2–1 and shYTHDF2–2, and selected for 2 days with 2 μg/mL puromycin (Thermofisher Scientific; A1113803). shYTHDF2 cells were treated with DMSO only, 1, 2, or 5 mM 4-phenylbutyric acid (4-PBA, Millipore Sigma; P21005) for 72 hours and subsequently processed for Western blot analysis as above.
Cellular ROS assay
MDA-MB-231-LM2 cells were transduced with NTC, shYTHDF2–2, or both shYTHDF2–2 and shPRSS23 virus at MOI > 1 and selected for 2 days with 2 μg/mL puromycin (Thermofisher Scientific; A1113803). The cellular ROS assay was performed (Abcam; ab186029) according to manufacturer’s instructions. Cells were analyzed by flow cytometry using the BDSLRFortessa under the APC-Cy7 (deep red) channel. Analysis and gating were performed in FlowJo.
RT-qPCR Analysis
RNA was extracted with Direct-zol RNA Miniprep kit (Zymo Research; R2071) for three biological replicates and cDNA synthesized from 1 μg total RNA using High-Capacity cDNA Reverse Transcription kit (Applied Biosystems; 4368814). Real-time PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems; 4367659). Values of gene expression were normalized to GAPDH expression using the ddCT method. Primer sequences can be found in Table S6.
Gene Ontology (GO) Analysis
GO analyses on single lists were conducted using the GOrilla tool (Eden et al., 2009) and multiple list analysis were conducted using the Metascape resource (Zhou et al., 2019). Expressed gene sets with TPM > 1 for each respective cell lines were used as background lists. GO terms were ranked by Bonferroni-corrected hypergeometric p-values.
Time Lapse Microscopy
Cells were seeded at 5K cells in Incucyte ImageLock plates (Essen BioSciences; 4379). The next day, plates were loaded into the IncucyteTM and imaged at 10X magnification for 84 hours every 12 hours. Phase images were analyzed using the Incucyte ZOOM Basic Analyzer to measure confluence.
TCGA data description
The publicly available dataset from The Cancer Genome Atlas Breast Invasive Carcinoma (TCGA-BRCA) was directly downloaded from the cBioPortal for Cancer Genomics at https://www.cbioportal.org/. For detailed information, refer to Liu et al., 2018. For gene expression data, we used mRNA expression z-scores calculated by RNA-Seq by Expectation Maximization (RSEM) and categorized the data by the tumor’s clinical subtype. For survival data, we determined the Kaplan-Meier probability using clinical overall survival status by first grouping by high/low MYC or RBP mRNA expression determined by median z-score (above/below).
We obtained proteomics and phosphoproteomics profiling data from the CPTAC at https://cptac-data-portal.georgetown.edu/cptac/s/S029. For detailed information, refer to Mertins et al., 2016. After removing the missing values, Pearson’s correlation tests were performed to analyze the correlation between each of the known players in the IRE1α branch of the UPR pathway and YTHDF2 expression for targets found in >50% of samples.
scRibo-STAMP assays and analysis
WT MDA-MB-231-LM2 cells or MDA-MB-231-LM2 cells transduced with YTHDF2 targeting pINDUCER11 shRNA 1, sorted for top EGFP expression (Dharmacon; V2LHS_115143) were each transduced with either RPS2-APOBEC1 or APOBEC1 control STAMP vectors at an MOI <0.3 to limit copy number variation. Lentivirus was made using pLIX403 Capture-1 control-STAMP-HA-P2A-mRuby or pLIX403 Capture-1 RPS2-STAMP-HA-P2A-mRuby plasmids (Brannan et al., 2021). Cells were expanded and then subcutaneously transplanted (3 X 106 cells per mouse) into athymic nude mice (female, 10 weeks old). Tumors were allowed to engraft and grow for 6 weeks. Tumors were measured twice, weekly using calipers and mice were given 5% sucrose water with 2 mg/mL dox when tumors were 200 mm3 on average for 3 days before harvesting. Tumors were washed with sterile PBS and minced with a sterilized blade into 2–3 mm cubes. Tumor pieces were dissociated in enzyme medium (~0.1 cm3 tumor/~2mL), shaking for 30 minutes at 37°C. The enzyme medium consisted of 10X enzyme mix (Collagenase IV, 1g/100mL HBSS; (Sigma, #C-5138), Hyaluronidase, 100mg/100mL HBSS; (Sigma, #H-6254), and Deoxyribonuclease, 20,000 U/100 mL HBSS; (Sigma, #D-5025)) in RPMI 1640 medium with 10% Pen/Strep. Cells were strained and spun down. Red blood cells were lysed in water for 20 s and immediately neutralized with PBS +Ca/Mg. Cells were resuspended in media with Pen/Strep and counted with approximately 90% cell viability.
Single cell library preparation and analysis was performed as previously described (Brannan et al., 2021). Cells were counted and resuspended at a density of 1,000 cell/μL in 0.04% BSA in PBS. Single cells were processed through the Chromium Single Cell Gene Expression Solution using the Chromium Single Cell 3’ Gel Bead, Chip, 3’ Library v3 (10X Genomics; #PN-1000079, #PN-1000075, #PN-120262) as per the manufacturer’s protocol. Sixteen thousand total cells were added to each channel for a target recovery of 10,000 cells. The cells were then partitioned into Gel Beads in Emulsion in the Chromium instrument, where cell lysis and barcoded reverse transcription of RNA occurred, followed by amplification, fragmentation, end-repair, A-tailing and 5’ adaptor and sample index attachment as indicated in the manufacturer’s protocol for 3’ expression capture. Agilent High Sensitivity D5000 ScreenTape Assay (Agilent Technologies) was performed for QC of the libraries and sequenced on an Illumina NovaSeq 6000. Reads were aligned to custom hg19 + lentiviral-genes transcriptomes + TurboRFP. Unique molecular identifier (UMI)-collapsing and sample aggregation were performed using the Cellranger-version 2.0.1 toolkit (Zheng et al., 2017) provided by 10X Genomics. Analysis of output digital gene expression matrices was performed using the Scanpy-v1.4.4 package (Wolf et al., 2018). Genes that were not detected in at least five single cells were discarded. Cells with fewer than 1,000 or more than 7,000 expressed genes as well as cells with more than 50,000 unique transcripts or 40% mitochondrial expressed genes were removed from the analysis. Transcripts per cell were normalized to 10,000, added a unit and logarithmized (“ln(TPM+1)”) and scaled to unit variance (z-scored). Top 2,000 variable genes were identified with the filter_genes_dispersion function, flavor=‘cell_ranger’. PCA was carried out, and the top 40 principal components were retained. With these principal components, neighborhood graphs were computed with 10 neighbors and standard parameters with the pp.neighbors function. Single-cell scores for comparisons with bulk DEG genes and cell cycle genes were computed with tl.score_genes and tl.score_genes_cell_cycle functions, respectively.
Single cell edits were called by first computing the MD tag from Cellranger outputs (possorted_genome_bam.bam) using Samtools-v1.3.1 calmd (Danecek et al., 2021) and splitting every read according to their cell barcode. Reads belonging to each cluster of barcodes were combined using a custom script and treated similarly. “Aggregated” edits (across reads that belong to a collection of cells instead of looking at edits per-cell) were used to assess the quality of scRibo-STAMP data, for example when looking at the correlation across regions by combining reads belonging to filtered barcodes into a single BAM file.
Analysis of output digital gene edit matrices was performed using the Scanpy-v1.4.4 package (Wolf et al., 2018). Matrices for all samples were concatenated and all genes that were not edited in at least 2 single cells were discarded. Cells with fewer than 10 edited genes were removed from the analysis. To calculate the Edits per kilobase of transcript per million mapped reads (EPKM) per gene metric, cumulative edit counts were used (overall ‘T’ coverage for each edit site called) as determined by SAILOR-v1.1.0 (Deffit et al., 2017). Region-specific (either CDS or CDS+3’UTR as defined by hg19 v19 Gencode annotations) edit counts per gene were summed and divided by “per million” mapped read counts that successfully aligned to either CDS or CDS+3UTR, respectively. For analyses, all genes with unique feature mapped counts greater than 0, as defined by Subread featureCounts-v1.5.3 (Liao et al., 2014), were included. This number was then normalized to the length of either the CDS or CDS+3’UTR of each gene in kilobases (kb) to determine per gene EPKM. EPKM for each cell were normalized to 10,000, added a unit and logarithmized (“ln(TPM+1)”) and scaled to unit variance (z-scored). PCA was carried out, and the top 40 principal components were retained. With these principal components, neighborhood graphs were computed with 10 neighbors and standard parameters with the pp.neighbors function. Louvain clusters were computed with the tl.louvain function and standard parameters. Following visual inspection, subsets of Louvain clusters were merged guided by their overlap (or lack thereof) with control-STAMP cells in order to define a Ribo-STAMP-specific clusters. Single cell and mean EPKM per sample heatmaps were generated with the pl.heatmap and pl.matrixplot functions, respectively. Differentially edited genes were determined for each set of Louvain (or modified) clusters with the tl.rank_gene_groups function (method=‘wilcoxon’) and up to 100 genes with p-value < 0.05 were identified per cluster. Gene ontology was performed using Metascape’s multiple gene list function (Zhou et al., 2019) with all edited genes as the background set.
QUANTIFICATION AND STATISTICAL ANALYSIS
Investigators responsible for monitoring and measuring the xenografts of individual tumors were not blinded. Simple randomization was used to allocate animals to experimental groups. All animal studies were performed per institutional and national animal regulations. Power analysis was used to determine the appropriate sample size to detect significant changes in animal survival, which were based on previous survival analyses in our laboratory. All healthy animals with successfully xenografted tumors were included in analyses. All statistical tests are described in figure legends including the test used, exact value of n, definition of center, and precision measures. P-values < 0.05 were considered significant and specific p-values corresponding to each figure are presented either on the figure itself or in the figure legend.
Supplementary Material
Table S1: Summary of MaGeCK results used to identify synthetic lethal RBP candidates, related to Figure 1.
Table S2: Normalized read counts of YTHDF2 hairpins from pooled shRNA screen in vitro and in vivo, Related to Figure 2.
Table S3: Quality control metrics for eCLIP and m6A-seq experiments, Related to Figure 3.
Table S4: Summary of RNA-seq results indicating high confidence YTHDF2 eCLIP and m6A target genes, Related to Figure 4.
Table S5: Differentially expressed and translated genes per cluster identified by scRibo-STAMP, Related to Figures 6 and 7.
Table S6: RT-qPCR primer sequences, Related to STAR Methods.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit pAb anti-YTHDF2 | Proteintech | Cat#24744-1-AP; RRID: AB_2687435 |
| Rabbit mAb anti-YTHDF2 | Abcam | Cat# ab220163; RRID: AB_2868573 |
| Rabbit pAb anti-METTL3 | Proteintech | Cat#15073-1-AP; RRID: AB_2142033 |
| Mouse mAb anti-m6A | Synaptic Systems | Cat#202 011; RRID: AB_2619890 |
| Rabbit pAb anti-YTHDF2 | Aviva Systems Biology | Cat#ARP67917_P050 |
| Rabbit pAb anti-Phopho-p44/42 MAPK (Erk1/2) | Cell Signaling | Cat#4370; RRID: AB_2315112 |
| Rabbit pAb anti-p44/42 MAPK (Erk1/2) | Cell Signaling | Cat#4695; RRID: AB_390779 |
| Rabbit mAb anti-eif4e | Cell Signaling | Cat#9742; RRID: AB_823488 |
| Rabbit pAb anti-Phospho-eIF2α (Ser51) Antibody | Cell Signaling | Cat#9721; RRID: AB_330951 |
| Rabbit pAb anti-eif2α | Cell Signaling | Cat#9722; RRID: AB_2230924 |
| Rabbit mAb anti-c-Myc | Cell Signaling | Cat#13987; RRID: AB_2631168 |
| Mouse mAb anti-GAPDH | Abcam | Cat#ab8245; RRID: AB_2107448 |
| Rabbit mAb anti-BiP | Cell Signaling | Cat#3177; RRID: AB_2119845 |
| Rabbit mAb anti-Phospho-p38 MAPK | Cell Signaling | Cat#4511; RRID: AB_2139682 |
| Rabbit mAb anti-p38 MAPK | Cell Signaling | Cat#8690; RRID: AB_10999090 |
| Rabbit pAb PARP | Cell Signaling | Cat#9542; RRID: AB_2160739 |
| Rabbit pAb anti-Caspase-3 | Cell Signaling | Cat#9662; RRID: AB_331439 |
| Rabbit mAb anti-Cleaved Caspase-3 | Cell Signaling | Cat#9664; RRID: AB_2070042 |
| Rabbit mAb anti-Phospho-SAPK/JNK | Cell Signaling | Cat#4668; RRID: AB_823588 |
| Rabbit pAb anti-SAPK/JNK | Cell Signaling | Cat#9252; RRID: AB_2250373 |
| Rabbit pAb anti-IRE1α | Novus Biologicals | Cat#NB100-2324; RRID: AB_10000972 |
| Rabbit pAb anti-Phospho-IRE1α | Novus Biologicals | Cat#NB100-2323; RRID: AB_10145203 |
| Rabbit mAb anti-Snail | Cell Signaling | Cat#3879; RRID: AB_2255011 |
| Rabbit mAb anti-ZEB1 | Cell Signaling | Cat#3396; RRID: AB_1904164 |
| Rabbit mAb anti-Vimentin | Cell Signaling | Cat#5741; RRID: AB_10695459 |
| Rabbit pAb anti-PRSS23 | Abcam | Cat#ab201182 |
| Rabbit pAb anti-DR5 | Cell Signaling | Cat#3696; RRID: AB_10692107 |
| Rabbit mAb anti-Cleaved Caspase-8 | Cell Signaling | Cat#9496; RRID: AB_561381 |
| Rabbit mAb anti-Cleaved Caspase-9 | Cell Signaling | Cat#7237; RRID: AB_10895832 |
| Rabbit mAb anti-BAX | Cell Signaling | Cat#5023; RRID: AB_10557411 |
| Rabbit pAb anti-RFP | Thermo Fisher | Cat# R10367; RRID: AB_2315269 |
| Rabbit pAb anti-GFP | Abcam | Cat# ab290; RRID: AB_303395 |
| Rabbit mAb anti-HA-Tag | Cell Signaling | Cat#3724; RRID: AB_1549585 |
| Rabbit pAb anti-Ki67 | Abcam | Cat#ab15580; RRID: AB_443209 |
| Rabbit mAb anti-CD31 | Cell Signaling | Cat#77699; RRID: AB_2722705 |
| Mouse mAb anti-puromycin (clone 12D10) | Millipore Sigma | Cat#MABE343; RRID: AB_2566826 |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling | Cat#7076; RRID: AB_330924 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling | Cat#7074; RRID: AB_2099233 |
| Goat anti-Rabbit IgG (H+L) Superclonal, Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat#A11034; RRID: AB_2536161 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Puromycin | Thermofisher Scientific | Cat#A1113803; CAS: 58-58-2 |
| Poly-D-lysine hydrobromide | Millipore Sigma | Cat#P6407; CAS: 27964-99-4 |
| (Z)-4-Hydroxytamoxifen | Millipore Sigma | Cat#H7904; CAS: 68047-06-3 |
| Polybrene | Millipore Sigma | Cat#TR-1003-G |
| Doxycycline hyclate | Millipore Sigma | Cat#D9891; CAS: 24390-14-5 |
| 4-Phenylbutyric Acid | Millipore Sigma | Cat#P21005; CAS: 1821-12-1 |
| Tamoxifen | Millipore Sigma | Cat#T5648; CAS: 10540-29-1 |
| Critical Commercial Assays | ||
| TruSeq Stranded Total RNA kit | Illumina | Cat#RS-122-2201 |
| Pierce BCA Protein Assay Kit | ThermoFisher Scientific | Cat#23225 |
| Annexin V: FITC Apoptosis Detection Kit I | BD | Cat# BDB556547 |
| PureLink™ HiPure Plasmid Filter Maxiprep Kit | Invitrogen | Cat#K2100-17 |
| DNeasy Blood & Tissue Kit | Qiagen | Cat#69506 |
| Direct-zol RNA Miniprep Kit | Zymo Research | Cat#R2071 |
| Nextera Index Kit | Illumina | Cat#KAPA#KK4824 |
| KAPA Library Quantification Kit | Illumina | Cat#FC-131-1096 |
| Chromium Single Cell B Chip Kit, 16 rxns | 10X Genomics | Cat#PN-1000075 |
| Chromium Single Cell 3’ Feature Barcode Library Kit | 10X Genomics | Cat# PN-1000079 |
| Chromium i7 Multiplex Kit, 96 rxns | 10X Genomics | Cat#PN-120262 |
| Rabbit Specific HRP/DAB (ABC) Detection IHC Kit | Abcam | Cat#ab64261 |
| Hematoxylin and Eosin Stain Kit | Vector Laboratories | Cat#H-3502 |
| Cellular ROS Assay Kit | Abcam | Cat#ab186029 |
| Propidium Iodide Flow Cytometry Kit | Abcam | Cat#ab139418 |
| Deposited Data | ||
| RBP CRISPR screen in MYC-ER HMECs | This paper | GEO: GSE137258 |
| YTHDF2 eCLIP-seq in human breast cancer cell lines | This paper | GEO: GSE137258 |
| m6A-seq in human breast cancer cell lines | This paper | GEO: GSE137258 |
| YTHDF2 knockdown RNA-seq in MYC-induced HMECs | This paper | GEO: GSE137258 |
| scRibo-STAMP in MDA-MB-231-LM2 cells with and without YTHDF2 depletion | This paper | GEO: GSE137258 |
| MYC MCF10A ChIP-seq data | ENCODE | ENCSR000DOS |
| TCF12 MCF-7 ChIP-seq data | ENCODE | ENCSR000BUN |
| Experimental Models: Cell Lines | ||
| Human: Lenti-X 293T | Takara | Cat#632180 |
| Human: Myc-ER HMECs | Kessler et al., 2012. | Gift from Thomas Westbrook |
| Human: MDA-MB-231-LM2 | Minn et al. 2005 | Gift from Thomas Westbrook |
| Human: MDA-MB-231 | ATCC | Cat#HTB-26 |
| Human: MCF-7 | ATCC | Cat#HTB-22 |
| Human: SKBR3 | ATCC | Cat#HTB-30 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Athymic Nude-Foxn1nu 3-4wk, female | Envigo International Holdings, Inc | Cat#6901 |
| Mouse: Ythdf2fl/fl | Li et al., 2018. | Gift from Chuan He |
| Mouse: CAG-CreERT mice | Jackson Labs | Cat#004682 |
| Mouse: CAG-CreERT;Ythdf2fl/fl | This paper | N/A |
| Oligonucleotides | ||
| shRNA Library Amplification Primer: Forward: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG TAGTGAAGCCACAGAGTA |
Integrated DNA Technologies | N/A |
| shRNA Library Amplification Primer: Reverse: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGCGCGGAGGCCAGATCTT |
Integrated DNA Technologies | N/A |
| qPCR Primers: See Table S6 for primers | Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| lentiCRISPR v2 | Addgene | Cat#52961 |
| pINDUCER11 (miR-RUG) | Meerbrey et al., 2011. | Gift from Trey Westbrook |
| TRC lentiviral shRNA vector non-targeting control | Millipore Sigma | SHC002V |
| TRC lentiviral shRNA vector YTHDF2 (shYTHDF2-1) | Millipore Sigma | TRCN0000168751 |
| TRC lentiviral shRNA vector YTHDF2 (shYTHDF2-2) | Millipore Sigma | TRCN0000167813 |
| TRC lentiviral shRNA vector PRSS23 | Millipore Sigma | TRCN0000047042 |
| GIPZ lentiviral shRNA vector YTHDF2 | Dharmacon | V2LHS_115143 |
| GIPZ lentiviral shRNA vector YTHDF2 | Dharmacon | V2LHS_115142 |
| GIPZ lentiviral shRNA vector YTHDF2 | Dharmacon | V3LHS_381614 |
| Software and Algorithms | ||
| MaGeCK-v0.5.4 | Li et al., 2015. | https://bitbucket.org/liulab/mageck-vispr |
| DESeq2-v1.14.0 | Love et al., 2014. | https://github.com/mikelove/DESeq2 |
| featureCounts-v1.5.3 | Liao et al., 2014 | http://subread.sourceforge.net/ |
| HOMER-v.4.9.1 | Heinz et al., 2010. | http://homer.ucsd.edu/homer/ |
| eCLIP-v0.4.0 | Van Nostrand et al., 2017. | https://github.com/YeoLab/eclip |
| Merge_peaks-v0.0.5 | Van Nostrand et al., 2017. | https://github.com/YeoLab/merge_peaks |
| MetaPlotR | Olarerin-George and Jaffrey, 2017. | https://github.com/olarerin/metaPlotR |
| MACS2-v2.1.2 | Zhang et al., 2008. | https://github.com/taoliu/MACS |
| SAILOR-v1.1.0 | Deffit et al. 2017 | https://github.com/YeoLab/sailor |
| Cellranger-v2.0.1 | Zheng et al. 2017 | https://github.com/10XGenomics/cellranger |
| Samtools-v1.3.1 | Danecek et al. 2021 | htslib.org |
| Scanpy-v1.4.4 | Wolf et al., 2018 | https://github.com/theislab/scanpy |
Highlights.
RBP CRISPR screening identifies 57 candidates that maintain MYC-driven cell survival
YTHDF2 depletion triggers apoptosis in triple negative breast cancer cells and tumors
YTHDF2 maintains mRNA homeostasis by limiting the number of translating mRNAs
scRibo-STAMP reveals proteomic heterogeneity in triple negative breast tumors in vivo
Acknowledgements
We thank Prof. Chuan He (U. of Chicago) for the generous gift of Ythdf2fl/fl mice. We are grateful to Dr. Kristopher Brannan and Ryan Marina in the Yeo Lab for providing reagents and guidance on implementing STAMP. We thank the Sanford Consortium Human Embryonic Stem Cell Core and UC San Diego Stem Cell Genomics Core for providing us use of their instruments and cell-sorting services (1S10OD025060). We acknowledge the Tissue Technology Shared Resource at Moores’ Cancer for histology processing. This publication includes data generated using an Illumina NovaSeq 6000 UC San Diego provided by the IGM Genomics Center (#S10OD026929). Data used in this publication were generated by the Clinical Proteomic Tumor Analysis Consortium (NCI/NIH) (Liu et al., 2018; Mertins et al., 2016). The results published here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. CHIP-seq datasets were downloaded from the ENCODE portal (Sloan et al., 2016) with the following identifiers: ENCSR000DOS and ENCSR000BUN. J.M.E is supported by the Ruth. L. Kirschstein F31 National Research Service Award (F31 CA217173) and Cancer Systems Biology Training Program (P50 GM085764 and U54 CA209891). M.P. is supported by the Ruth. L. Kirschstein F32 National Research Service Award (F32 HL143978). I.A.C. is a UC San Diego Chancellor’s Postdoctoral Fellow and a San Diego IRACDA Fellow supported by NIH/NIGMS (K12 GM068524). J.K.M. is supported by Susan G. Komen PDF Basic/Translational and Clinical Funding Program (PDF17487931). T.F.W. is supported by the NIH and NCI (U01CA214125, 1R01CA215226, and 1R01CA215452), the DOD (1W81XWH-18-1-0573), the McNair Medical Institute, and the CRUK Grand Challenge and the Mark Foundation for Cancer Research (C5470/A27144 to T.F.W. as a member of the SPECIFICANCER Team). This study was partially supported by NIH grants HG009889, HG004659, AI123202 and AI132122 to G.W.Y.
Disclosure of Financial Interests
G.W.Y. is a co-founder, a member of the Board of Directors, an equity holder, on the SAB, and a paid consultant for Locanabio and Eclipse BioInnovations. G.W.Y. is also a visiting faculty at the National University of Singapore, Singapore. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. J.M.E. and G.W.Y are inventors on a patent disclosure at UCSD related to this work.
Footnotes
The authors declare no other competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ai ZH, Wang J, Xu YL, Zhu XL, and Teng YC (2013). Suppression of RNA interference on expression of c-myc of SKOV3 ovarian carcinoma cell line. Eur Rev Med Pharmacol Sci 17, 3002–3006. [PubMed] [Google Scholar]
- Brannan KW, Chaim IA, Marina RJ, Yee BA, Kofman ER, Lorenz DA, Jagannatha P, Dong KD, Madrigal AA, Underwood JG, et al. (2021). Robust single-cell discovery of RNA targets of RNA-binding proteins and ribosomes. Nature Methods 18, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IH, Wang HH, Hsieh YS, Huang WC, Yeh HI, and Chuang YJ (2013). PRSS23 is essential for the Snail-dependent endothelial-to-mesenchymal transition during valvulogenesis in zebrafish. Cardiovasc Res 97, 443–453. [DOI] [PubMed] [Google Scholar]
- Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, et al. (2018). RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67, 2254–2270. [DOI] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. (2021). Twelve years of SAMtools and BCFtools. Gigascience 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deffit SN, Yee BA, Manning AC, Rajendren S, Vadlamani P, Wheeler EC, Domissy A, Washburn MC, Yeo GW, and Hundley HA (2017). The C. elegans neural editome reveals an ADAR target mRNA required for proper chemotaxis. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S, Tameire F, and Koumenis C (2013). PERK-ing up autophagy during MYC-induced tumorigenesis. Autophagy 9, 612–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, Qiu Z, Kidwell RL, Kim LJY, Xie Q, et al. (2020). The RNA m6A reader YTHDF2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Discov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, and Rechavi G (2013). Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc 8, 176–189. [DOI] [PubMed] [Google Scholar]
- Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, and Wu L (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun 7, 12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, and Yakhini Z (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Peng G, Sahgal N, Fazli L, Gleave M, Zhang Y, Hussain A, and Qi J (2016). Regulation of c-Myc expression by the histone demethylase JMJD1A is essential for prostate cancer cell growth and survival. Oncogene 35, 2441–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YX, Sokol ES, Del Vecchio CA, Sanduja S, Claessen JH, Proia TA, Jin DX, Reinhardt F, Ploegh HL, Wang Q, et al. (2014). Epithelial-to-mesenchymal transition activates PERK-eIF2alpha and sensitizes cells to endoplasmic reticulum stress. Cancer Discov 4, 702–715. [DOI] [PubMed] [Google Scholar]
- Green AR, Aleskandarany MA, Agarwal D, Elsheikh S, Nolan CC, Diez-Rodriguez M, Macmillan RD, Ball GR, Caldas C, Madhusudan S, et al. (2016). MYC functions are specific in biological subtypes of breast cancer and confers resistance to endocrine therapy in luminal tumours. Br J Cancer 114, 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Yang Y, Chen J, He X, Lv N, and Yan R (2019a). PRSS23 knockdown inhibits gastric tumorigenesis through EIF2 signaling. Pharmacol Res 142, 50–57. [DOI] [PubMed] [Google Scholar]
- Han H, Jain AD, Truica MI, Izquierdo-Ferrer J, Anker JF, Lysy B, Sagar V, Luan Y, Chalmers ZR, Unno K, et al. (2019b). Small-Molecule MYC Inhibitors Suppress Tumor Growth and Enhance Immunotherapy. Cancer Cell 36, 483–497 e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa T, Katayama A, Yokobori T, Yamane A, Fujii T, Obayashi S, Kurozumi S, Kawabata-Iwakawa R, Gombodorj N, Nishiyama M, et al. (2019). Carboxypeptidase A4 accumulation is associated with an aggressive phenotype and poor prognosis in triple-negative breast cancer. Int J Oncol 54, 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart LS, Cunningham JT, Datta T, Dey S, Tameire F, Lehman SL, Qiu B, Zhang H, Cerniglia G, Bi M, et al. (2012). ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J Clin Invest 122, 4621–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Arslan AD, Ho TT, Yuan C, Stampfer MR, and Beck WT (2014). Involvement of polypyrimidine tract-binding protein (PTBP1) in maintaining breast cancer cell growth and malignant properties. Oncogenesis 3, e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Castello A, Schwarzl T, and Preiss T (2018). A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol 19, 327–341. [DOI] [PubMed] [Google Scholar]
- Hsu TY, Simon LM, Neill NJ, Marcotte R, Sayad A, Bland CS, Echeverria GV, Sun T, Kurley SJ, Tyagi S, et al. (2015). The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 525, 384–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 20, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Spencer GJ, Lynch JT, Ciceri F, Somerville TD, and Somervaille TC (2014). Enhancers of Polycomb EPC1 and EPC2 sustain the oncogenic potential of MLL leukemia stem cells. Leukemia 28, 1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova I, Much C, Di Giacomo M, Azzi C, Morgan M, Moreira PN, Monahan J, Carrieri C, Enright AJ, and O’Carroll D (2017). The RNA m(6)A Reader YTHDF2 Is Essential for the Post-transcriptional Regulation of the Maternal Transcriptome and Oocyte Competence. Mol Cell 67, 1059–1067 e1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et al. (2012). A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science 335, 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon I, Carballes F, Brewer G, Poiret M, and Morello D (1998). Developmental expression of AUF1 and HuR, two c-myc mRNA binding proteins. Oncogene 16, 3413–3421. [DOI] [PubMed] [Google Scholar]
- Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, and Burge CB (2014). RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol Cell 54, 887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Q, Liu PY, Haase J, Bell JL, Huttelmaier S, and Liu T (2019). The Critical Role of RNA m(6)A Methylation in Cancer. Cancer Res 79, 1285–1292. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasman L, Krupalnik V, Viukov S, Mor N, Aguilera-Castrejon A, Schneir D, Bayerl J, Mizrahi O, Peles S, Tawil S, et al. (2020). Context-dependent functional compensation between Ythdf m(6)A reader proteins. Genes Dev 34, 1373–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Meng S, Xu M, Wang S, He L, Xu X, Wang X, and Xie L (2018a). Downregulation of N(6)-methyladenosine binding YTHDF2 protein mediated by miR-493–3p suppresses prostate cancer by elevating N(6)-methyladenosine levels. Oncotarget 9, 3752–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wu L, Pei M, and Zhang Y (2020). YTHDF2, a protein repressed by miR-145, regulates proliferation, apoptosis, and migration in ovarian cancer cells. J Ovarian Res 13, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, and Liu XS (2014). MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biology 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Qian P, Shao W, Shi H, He XC, Gogol M, Yu Z, Wang Y, Qi M, Zhu Y, et al. (2018b). Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res 28, 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lin S, Choe J, Du P, Triboulet R, and Gregory RI (2016). The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell 62, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, et al. (2018). An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 173, 400–416 e411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone MK, Smrekar K, Park S, Blakely B, Walter A, Nasta N, Park J, Considine M, Danilova LV, Pandey NB, et al. (2020). Cytokines secreted by stromal cells in TNBC microenvironment as potential targets for cancer therapy. Cancer Biol Ther 21, 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17. [Google Scholar]
- Mayr C, and Bartel DP (2009). Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, et al. (2011). The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci U S A 108, 3665–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, Wang X, Qiao JW, Cao S, Petralia F, et al. (2016). Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, and Massague J (2005). Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olarerin-George AO, and Jaffrey SR (2017). MetaPlotR: a Perl/R pipeline for plotting metagenes of nucleotide modifications and other transcriptomic sites. Bioinformatics 33, 1563–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, Mapperley C, Lawson H, Wotherspoon DA, Sepulveda C, et al. (2019). Targeting the RNA m(6)A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell 25, 137–148 e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Brugiolo M, Akerman M, Das S, Urbanski L, Geier A, Kesarwani AK, Fan M, Leclair N, Lin KT, et al. (2019). Differential Functions of Splicing Factors in Mammary Transformation and Breast Cancer Metastasis. Cell Rep 29, 2672–2688 e2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira B, Billaud M, and Almeida R (2017). RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer 3, 506–528. [DOI] [PubMed] [Google Scholar]
- Quintela-Fandino M, Arpaia E, Brenner D, Goh T, Yeung FA, Blaser H, Alexandrova R, Lind EF, Tusche MW, Wakeham A, et al. (2010). HUNK suppresses metastasis of basal type breast cancers by disrupting the interaction between PP2A and cofilin-1. Proc Natl Acad Sci U S A 107, 2622–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon YCS, Castellvi J, Hummer S, Peg V, Pelletier J, and Sonenberg N (2018). Beyond molecular tumor heterogeneity: protein synthesis takes control. Oncogene 37, 2490–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, Patil DP, Kwak H, Lee JH, and Jaffrey SR (2019). m6A enhances the phase separation potential of mRNA. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenow A, Noben JP, Bouwman FG, Mariman EC, and Renes J (2013). Hypoxia-mimetic effects in the secretome of human preadipocytes and adipocytes. Biochim Biophys Acta 1834, 2761–2771. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, Mori K, Sadighi Akha AA, Raden D, and Kaufman RJ (2006). Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol 4, e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan CA, Chan ET, Davidson JM, Malladi VS, Strattan JS, Hitz BC, Gabdank I, Narayanan AK, Ho M, Lee BT, et al. (2016). ENCODE data at the ENCODE portal. Nucleic Acids Res 44, D726–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. (2005). A human protein-protein interaction network: a resource for annotating the proteome. Cell 122, 957–968. [DOI] [PubMed] [Google Scholar]
- Struntz NB, Chen A, Deutzmann A, Wilson RM, Stefan E, Evans HL, Ramirez MA, Liang T, Caballero F, Wildschut MHE, et al. (2019). Stabilization of the Max Homodimer with a Small Molecule Attenuates Myc-Driven Transcription. Cell Chem Biol 26, 711–723 e714. [DOI] [PubMed] [Google Scholar]
- Tang X, Tu G, Yang G, Wang X, Kang L, Yang L, Zeng H, Wan X, Qiao Y, Cui X, et al. (2019). Autocrine TGF-beta1/miR-200s/miR-221/DNMT3B regulatory loop maintains CAF status to fuel breast cancer cell proliferation. Cancer Lett 452, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truitt ML, Conn CS, Shi Z, Pang X, Tokuyasu T, Coady AM, Seo Y, Barna M, and Ruggero D (2015). Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell 162, 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Nguyen TB, Gelboin-Burkhart C, Wang R, Blue SM, Pratt GA, Louie AL, and Yeo GW (2017). Robust, Cost-Effective Profiling of RNA Binding Protein Targets with Single-end Enhanced Crosslinking and Immunoprecipitation (seCLIP). In: Shi Y (eds) mRNA Processing. Methods in Molecular Biology, Vol 1648 (New York, NY: Humana Press; ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C, Gong C, Zhang H, Hua L, Li X, Chen X, Chen Y, Ding X, He S, Cao W, et al. (2016). beta2-adrenergic receptor signaling promotes pancreatic ductal adenocarcinoma (PDAC) progression through facilitating PCBP2-dependent c-myc expression. Cancer Lett 373, 67–76. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler EC, Vu AQ, Einstein JM, DiSalvo M, Ahmed N, Van Nostrand EL, Shishkin AA, Jin W, Allbritton NL, and Yeo GW (2020). Pooled CRISPR screens with imaging on microraft arrays reveals stress granule-regulatory factors. Nature Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Edell KA, Kehasse A, Scott GK, Yau C, Rothschild DE, Schilling B, Gabriel BS, Yevtushenko MA, Hanson IM, Held JM, et al. (2014). RPL24: a potential therapeutic target whose depletion or acetylation inhibits polysome assembly and cancer cell growth. Oncotarget 5, 5165–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Cheryan VT, Xu L, Rishi AK, and Reddy KB (2017). Myc mediates cancer stem-like cells and EMT changes in triple negative breast cancers cells. PLoS One 12, e0183578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S, and Jaffrey SR (2020). A Unified Model for the Function of YTHDF Proteins in Regulating m(6)A-Modified mRNA. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat Commun 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, Ma H, and Kang T (2019). YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett 442, 252–261. [DOI] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, and Qian SB (2015). Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Huang S, Zeng L, Ma J, Sun S, Zeng F, Kong F, and Cheng X (2017). HMOX-1 inhibits TGF-beta-induced epithelial-mesenchymal transition in the MCF-7 breast cancer cell line. Int J Mol Med 40, 411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Yan L, Zhu W, Song X, Yang G, and Wang S (2019). MMP2/3 promote the growth and migration of laryngeal squamous cell carcinoma via PI3K/Akt-NF-kappaB-mediated epithelial-mesenchymal transformation. J Cell Physiol. [DOI] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. (2011). RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Summary of MaGeCK results used to identify synthetic lethal RBP candidates, related to Figure 1.
Table S2: Normalized read counts of YTHDF2 hairpins from pooled shRNA screen in vitro and in vivo, Related to Figure 2.
Table S3: Quality control metrics for eCLIP and m6A-seq experiments, Related to Figure 3.
Table S4: Summary of RNA-seq results indicating high confidence YTHDF2 eCLIP and m6A target genes, Related to Figure 4.
Table S5: Differentially expressed and translated genes per cluster identified by scRibo-STAMP, Related to Figures 6 and 7.
Table S6: RT-qPCR primer sequences, Related to STAR Methods.
Data Availability Statement
CRISPR screening, eCLIP, m6A-seq, and RNA-seq datasets generated during this study were deposited at GEO and are available under accession number GEO: GSE137258. Source data for dot plots generated from the in vivo shRNA screen are provided in Table S2.