Abstract
We apply feature-extraction and machine learning methods to multiple sources of contrast (acetic acid, Lugol’s iodine and green light) from the white Pocket Colposcope, a low-cost point of care colposcope for cervical cancer screening. We combine features from the sources of contrast and analyze diagnostic improvements with addition of each contrast. We find that overall AUC increases with additional contrast agents compared to using only one source.
I. INTRODUCTION
Each year over 500,000 women are diagnosed with cervical cancer and more than half of them die [1]. A disproportionate burden of incidence and mortality (>80%) occurs in low- and middle-income countries and this is projected to increase [1]. Reasons for disparities in mortality include barriers to early screening, detection and treatment which have been proven to significantly reduce incidence and loss of life in high-income countries. These barriers include cost of the screening tools, availability of expert interpretation of images, and distance to centralized facilities. In high-resource settings, cervical cancer management has multiple steps: (1) Screening with the Pap smear with or without HPV testing, (2) Diagnosis with Colposcopy (low-magnification imaging) guided biopsy which involves application of three main contrasts: acetic acid causes acetowhitening in abnormal regions, with CIN2+ showing textured regions in addition to whitening, whereas normal cervix regions remain a light pink color. Lugol’s iodine, turns normal regions dark brown, while abnormal areas appear pale/mustard yellow. Green filter/illumination highlights vasculature changes. To reduce loss to follow up, and due to barriers of limited resources, the WHO recommends visual inspection of the cervix with the naked eye after application of acetic acid (VIA) as a low-cost alternative to cervical cancer screening in low-resource settings [2]. However, it is subjective, dependent on user experience and has no image documentation for quality control. Additionally, it does not take advantage of other sources of contrast, visual inspection with Lugol’s iodine (VILI) and green illumination vascular imaging (GIVI) which are routinely used in high-income settings to supplement acetic acid contrast during colposcopy [3]. To address these barriers related to imaging limitations, we developed a low-cost, portable, tool, the Pocket colposcope for imaging lesions of the cervix at with magnification to reduce cost, improve sensitivity of diagnosis and enable screening at the point-of-care [4, 5]. The Pocket colposcope has been shown to enable cervical imaging quality on par with a high-end colposcope, while drastically reducing costs and increasing portability [6, 7]. The Pocket colposcope has onboard white LEDs at low and high brightness levels for capturing images from VIA and VILI respectively, and green LEDs for capturing GIVI images (Fig. 1). The Pocket Colposcope has been used in multiple clinical studies, yielding a database of images from VIA, VILI and GIVI. To address the limitations of subjectivity and user experience dependence, we propose algorithms for automated classification of images captured with the Pocket Colposcope. We have previously published on development of image processing algorithms to analyze VIA and VILI Pocket colposcope images of the cervix [8]. We described image processing methods to extract relevant color and texture features and train the features using a machine learning classifier. We also proposed methods to combine VIA and VILI images to enhance classification. In this work we expand our algorithms to include GIVI which reflects vascular features that are typically associated with high-grade dysplasias [3]. We look at individual performance of the algorithm on the three different sources of contrasts and on different combinations of contrast: VIA, GIVI, VILI, VIA+GIVI, VIA+VILI, VIA+VILI+GIVI. While this work builds on our previous work in terms of the automated algorithms we have developed, [8] it is distinct in that for the first time, it provides a comprehensive evaluation of combining the three different sources of contrast routinely used for cervical cancer detection. The manuscript is structured as follows: (1) description of the Pocket colposcope and images acquired, (2) feature extraction, and model training and validation, (3) results from the different combination of contrasts, and (4) discussion and conclusions.
Figure 1:
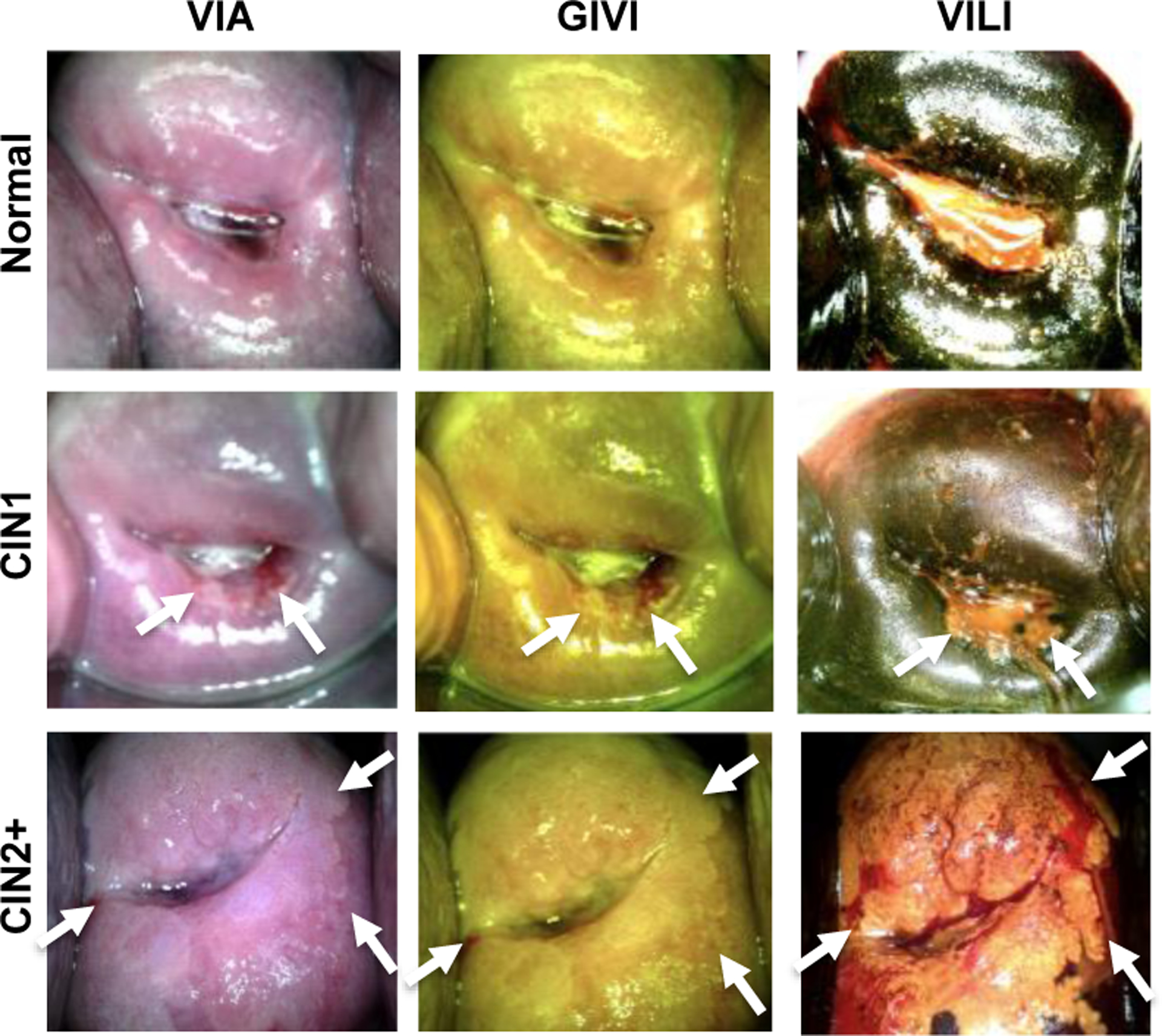
Representative normal, CIN1 and CIN2+ images of VIA, GIVI and VILI taken with the Pocket Colposcope. White arrows represent lesion locations.
II. Methods
A. Pocket Colposcope Image collection
Images, and pathology outcomes were retrospectively obtained from a database of Pocket colposcope images attained during various multi-site clinical studies over several years. This study was approved by Duke University Institutional Review Board (Pro00052865). The clinical sites included were Duke University Medical Center (DUMC), Durham, USA; La Liga Contra el Cancer (LLCC), Lima, Peru; Kilimanjaro Christian Medical Center (KCMC), Moshi, Tanzania; and University Teaching Hospital (UTH), Lusaka, Zambia. All sites collected VIA images as that is the primary source of contrast. LLCC collected VIA + VILI data and UTH only collected VIA+GIVI data. DUMC and KCMC collected images for all three sources of contrast. Only data from sites which had corresponding ground truth pathology for positives were included in the study. Per standard-of-care, no biopsies were taken for cervices which appeared normal during visual inspection as per standard-of-care. Table 1 summarizes the number of data points per contrast and per hospital sorted by diagnosis. For the purposes of the algorithm binary classification labels were used. Specifically, normal and CIN1s were grouped as “normal/benign”, as most CIN1 cases are non-malignant and regress on their own, whereas CIN2+ cases are is typically treated and thus grouped as abnormal.
Table 1:
Summary of data collected from clinical sites
| VIA+VILI | VIA+GIVI | VIA+VILI+GIVI | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Normal | CIN1 | CIN2+ | Normal | CIN1 | CIN2+ | Normal | CIN1 | CIN2+ | |
| DUMC | 5 | 12 | 12 | 22 | 19 | 34 | 5 | 12 | 12 |
| LLCC | 51 | 41 | 23 | ||||||
| KCMC | 2 | 6 | 10 | 2 | 6 | 8 | 2 | 6 | 8 |
| UTH | 2 | 3 | 32 | ||||||
| TOTAL | 58 | 59 | 45 | 26 | 28 | 74 | 7 | 18 | 20 |
B. Image processing for feature extraction
We describe image processing methods used for feature extraction for VIA and VILI images in detail in our previous study (Asiedu et al, 2019). A similar methodology summarized below in Figure 1 was applied to combining VIA, VILI and GIVI images. Pre-processing was performed to crop the cervix from the image to reduce noise from vaginal walls or the speculum that may be present in the image. Images captured typically contain specular reflections, which are bright white spots from light saturation. We performed specular reflection attenuation to reduce effects from these areas, by separating each RGB image into individual channels, thresholding to identify pixels > 220 in each channel, and using a Laplacian infill method to replace bright white pixels with pixels similar in color to the surrounding cervix pixels. Once pre-processed, Gabor filters and Kmeans clustering were applied to automatically identify and extract out high texture regions of interest (ROIs) in the images. From these Gabor-segmented ROIs, summary statistics (mean, median, mode, variance and Otsu threshold level) were derived from individual channels of color transformations (greyscale, RGB, YCbCr, HSV and CIELab). Haralick’s texture features (contrast, correlation, energy and homogeneity) described in detail in Asiedu et al were also extracted. Feature extraction yielded 65 color-based features and 4 texture features, yielding a total of 69 features for each contrast.
C. Feature selection and classification
All features were passed through the forward sequential feature selection (FSFS) algorithm to select a subset of optimal features for each contrast. Feature subsets were used to train and cross-validate (5-fold cross validation) a Support Vector Machine, K-Nearest Neighbor and Logistic Regression Classifier to compare results. To prevent overfitting from combining features from two or three contrasts images, images were classified individually and assigned classification scores/probabilities on likelihood of being normal/benign or high-grade. The scores from the different contrasts were combined into a feature matrix of m × 2 features, in the case of the two contrast combinations and a matrix of m × 3 features for 3 contrast combinations where m is the number of examples. These features were used as inputs to train the combined contrast classifier. Receiver operating curves were constructed for the different combinations and the area under the curve compared to the individual contrasts.
III. RESULTS
A. Image processing and feature extraction
Fig. 2 shows representative images from the image processing steps for VIA, VILI and GIVI. Specular reflection attenuation results perform similarly qualitatively for all three sources of contrasts, preserving cervix tissue information while reducing the amount of bright white pixels. Gabor segments are obtained from applying Gabor filters to highlight regions of texture, and segmented using Kmeans clustering (n=2 clusters) and selecting the cluster with a high average pixel value post filtering. A bounding box around the Gabor segmented region is used for Haralick’s texture feature extraction, whereas the segment is used as is for color-based feature extraction. We observe qualitatively that the Gabor filter segments a region of high texture around the cervical os for a cervix with a lesion on it (Fig. 2). Since Haralick’s feature extraction requires a square/rectangular matrix of pixel values, a minimum bounding box around the Gabor segment is used for Haralick’s texture feature extraction. For color-based summary statistics, the segment is used as is. We make the assumption that a) segmenting a region of interest for feature extraction prevents dilution of features due to a larger area of non-lesion pixel values, and b) for color-based differences we can use the entire region of interest or a subset of the region of interest. For calculating summary statistics identifying pixels from the region of interest (ROI), specifically, pre-cancer is more relevant than including the entire ROI which might contain non-lesion pixels. For Haralicks texture features, the bounding box used may cause inclusion of a relatively smaller percentage non-lesion pixels. This would positively highlight textural differences between normal and abnormal regions.
Figure 2:
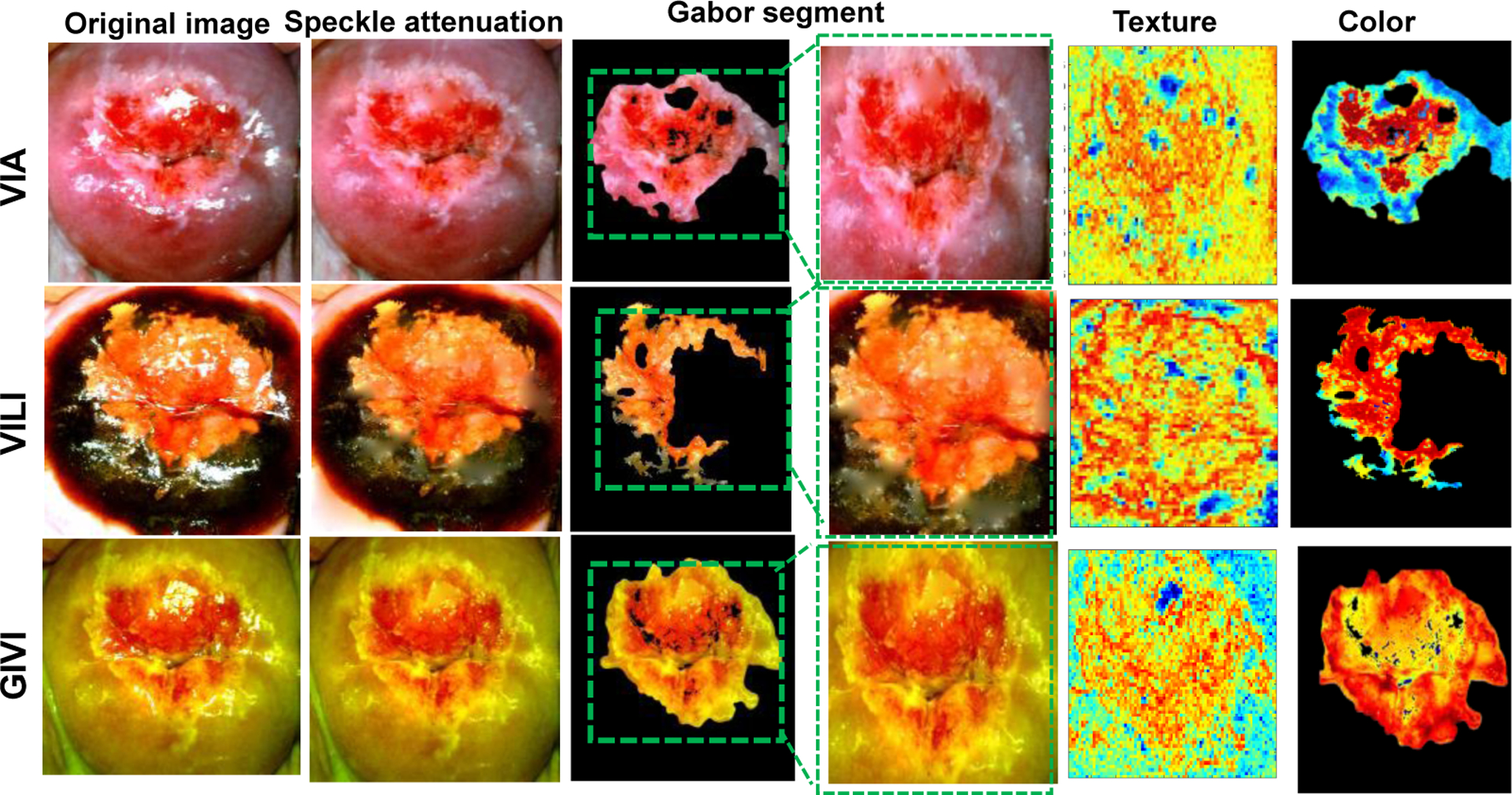
Representative images of a cervix with CIN2+ from image processing methods showing the original image, specular reflection attenuation, automatic Gabor segmentation showing original segment and segment with the bounding box for texture-based extraction. The figure also shows results from texture analysis and color transforms.
B. Classification improves with combining contrasts
Logistic Regression Classifier consistently achieved higher performance in with 5-fold cross validation for the three contrast and was selected for classification analysis.
VIA and VILI:
VIA and VILI Cervigram pairs were obtained from patients from LLCC, UTH, and a subset of DUMC patients. This yielded a total of 162 image pairs, of which 58 were normal, 59 were CIN1 and 45 were CIN2+. Normal/CIN1 were binned in normal category for algorithm classification, while CIN2+ was binned in abnormal. ROC curves from 5-fold cross validation for VIA only, VILI only and VIA+VILI combinations specific to this subset of data are shown. ROC curves for VIA only, VILI only and VIA+ VILI are shown in Fig. 3A. Both VIA only and VILI only attain AUCs of 0.79, whereas combining the two result in an AUC of 0.86, a 7% increase, most notably improving the sensitivity at the specificity range 0.6–0.8 from ~0.7 to ~0.85.
Figure 3:
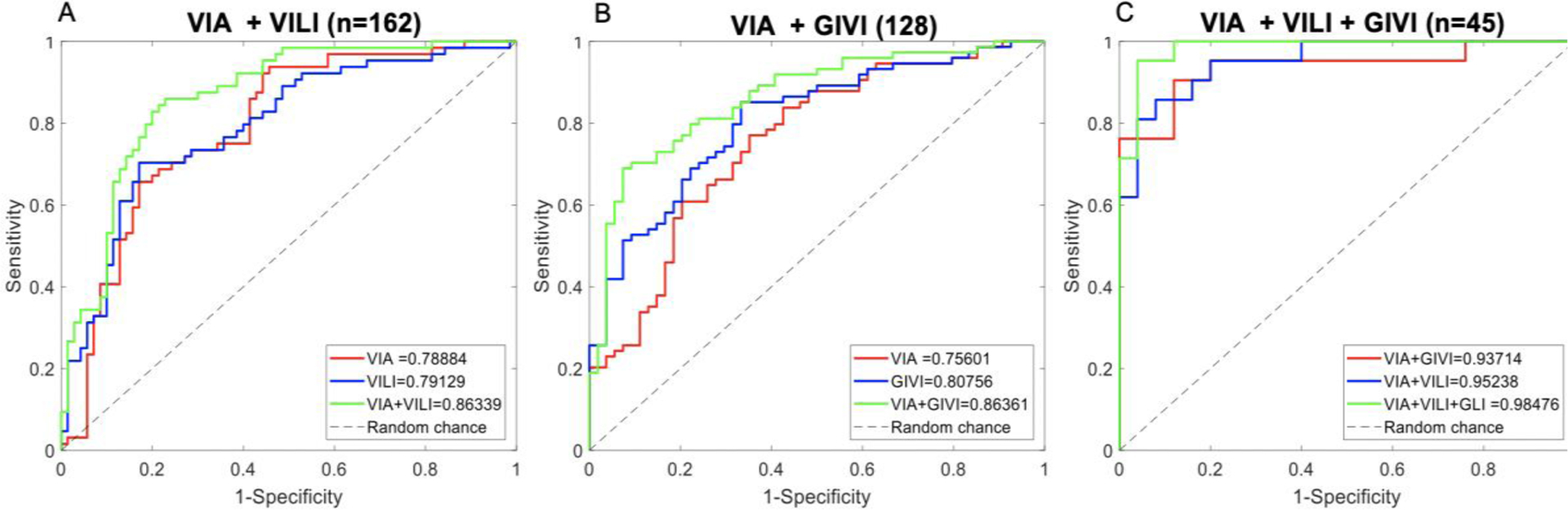
A) ROC curves for VIA and VILI datasets showing curves for VIA only, VILI only and VIA + VILI with corresponding AUCs. B) ROC curves for VIA and GIVI datasets showing curves for VIA only, GIVI only and VIA + GIVI with corresponding AUCs. C) ROC curves for subset of data with all three contrasts showing results from VIA + GIVI, VIA + VILI and VIA + VILI + GIVI.
VIA and GIVI:
VIA and GIVI cervigram pairs were obtained from patients from KCMC, UTH and a subset of DUMC patients. This yielded a total of 128 image pairs with 26 normal, 28 low-grade and 74 high grades, with normal/CIN1 in normal category and CIN2+ in abnormal category. We show ROC curves from 5-fold cross validation for VIA only, GIVI only specific to this subset of data set, and VIA+GIVI combination of the individual classification scores (Fig 3B). We attain an AUC for VIA only of 0.76, GIVI only of 0.81 and VIA + GIVI of 0.86. GIVI outperforms VIA, however combing GIVI and VIA outperform the individual contrasts. Most notably improving the sensitivity at high specificity values. Results demonstrate that combining VIA and GIVI algorithms outperforms VIA only or GIVI only.
VIA, VILI and GIVI:
We show ROC curves for VIA+VILI, VIA+GIVI and VIA+VILI+GIVI for this specific data set (Fig 3C). For individual values, not shown in the graph, VIA only attained an AUC of 0.86, VILI only an AUC of 0.89 and GIVI only, an AUC of 0.93. VIA + GIVI attains an AUC of 0.94, and VIA + VILI and AUC of 0.95. We observe that combining all three contrasts provides the best results 0f 0.98. Only a small subset of DUMC images and a subset of KCMC images had all three sources of contrast. This yielded a total of 45 image trios with 7 normal, 18 CIN1 and 20 CIN2+. Since this is a small data set, results may be overly optimistic, seeing as the performance is higher than the algorithms performance on larger data sets. However, we focus on the trend in improvement with contrast addition rather than the absolute scores.
IV. Discussion
Several studies have looked at using machine learning and more recently deep learning for improving diagnostic performance of colposcopy [9–15]. However, these have mainly focused on VIA images without consideration for VILI and GIVI images. Even though VIA is the most widely used source of contrast, we demonstrate that combining additional contrasts, VILI, and GIVI, has the ability to improve accuracy, especially in situations where VIA is used for diagnostic and referral decisions. Most previous studies have also been done with high end colposcopes which have different image contents such as outer genitalia, and the speculum, compared to the Pocket colposcope which is closer to the cervix (~3cm) than standard colposcopes (~30cm). Deep learning methods used have been shown to have high performance on standard VIA colposcopy images [13]. However, these require large corpus of data sets on an order of thousands, which is currently not feasible with the Pocket colposcope, due to its novelty. Additionally, these algorithms have not been demonstrated to transfer to images captured with low-cost colposcopes.
V. Conclusions
We hereby demonstrate that combining multiple sources of contrast from colposcopy images for machine learning algorithm development, outperforms individual sources of contrast, and that using all three types of contrast has the potential to outperform pair-wise combinations. These algorithms though applied to the Pocket colposcope can easily be customized and transferred for other colposcopes. In addition, our group is developing the Callascope a device which will enable self-cervix imaging by women at home. Combining these algorithms with self-cervix imaging can potentially enable at home self-cervical cancer screening, however large-scale clinical validation is needed.
Clinical Relevance—
This establishes that development of algorithms for automated colposcopy may benefit from combining multiple sources of contrast. The Pocket colposcope is a low-cost tool that can capture all three standard-of-care sources of contrast. This technology has the potential to provide automated diagnosis of cervical pre-cancer in the hands of non-experts.
Acknowledgments
Research supported by the National Institutes of Health
Contributor Information
Mercy N. Asiedu, Department of Biomedical Engineering, and Department of Global Health, Duke University, Durham, NC, 27708, USA and is now at the Massachusetts Institute of Technology, Computer Science and Artificial Intelligence Lab, Cambridge, MA, 02139 USA..
Guillermo Sapiro, Department of Biomedical Engineering, and Department of Electrical Engineering and Computer Science, Duke University, Durham, NC, 27708, USA.
Nirmala Ramanujam, Department of Biomedical Engineering, and Department of Global Health, Duke University, Durham, NC, 27708, USA.
References
- [1].Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, and Jemal A, “Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries,” CA Cancer J Clin, vol. 68, no. 6, pp. 394–424, November 2018, doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- [2].WHO, in Comprehensive Cervical Cancer Control: A Guide to Essential Practice, nd Ed., (WHO Guidelines Approved by the Guidelines Review Committee. Geneva, 2014. [PubMed] [Google Scholar]
- [3].I. S. Group. “The modified Reid colposcopic index (RCI)*.” International Agency for Research on Cancer. http://screening.iarc.fr/colDoaDDendix5.DhD (accessed May 17, 2017). [Google Scholar]
- [4].Lam CT et al. , “Design of a Novel Low Cost Point of Care Tampon (POCkeT) Colposcope for Use in Resource Limited Settings,” PLoS One, vol. 10, no. 9, p. e0135869, 2015, doi: 10.1371/journal.pone.0135869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lam CT et al. , “An integrated strategy for improving contrast, durability, and portability of a Pocket Colposcope for cervical cancer screening and diagnosis,” PLoS One, vol. 13, no. 2, p. e0192530, 2018, doi: 10.1371/journal.pone.0192530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mueller JL et al. , “Portable Pocket colposcopy performs comparably to standard-of-care clinical colposcopy using acetic acid and Lugol’s iodine as contrast mediators: an investigational study in Peru,” BJOG, June 12 2018, doi: 10.1111/1471-0528.15326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mueller JL et al. , “International Image Concordance Study to Compare a Point-of-Care Tampon Colposcope With a Standard-of-Care Colposcope,” J Low Genit Tract Dis, vol. 21, no. 2, pp. 112–119, April 2017, doi: 10.1097/LGT.0000000000000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Asiedu MN et al. , “Development of algorithms for automated detection of cervical pre-cancers with a low-cost, point-of-care, Pocket Colposcope,” IEEE Trans BiomedEng, December 18 2018, doi: 10.1109/TBME.2018.2887208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cristoforoni PM, Gerbaldo D, Perino A, Piccoli R, Montz FJ, and Capitanio GL, “Computerized colposcopy: results of a pilot study and analysis of its clinical relevance,” Obstet Gynecol, vol. 85, no. 6, pp. 1011–6, June 1995, doi: 10.1016/0029-7844(95)00051-R. [DOI] [PubMed] [Google Scholar]
- [10].Garcia-Arteaga JD, Kybic J, and Li WJ, “Automatic colposcopy video tissue classification using higher order entropy-based image registration,” (in English), Comput Biol Med, vol. 41, no. 10, pp. 960–970, October 2011, doi: 10.1016/j.compbiomed.2011.07.010. [DOI] [PubMed] [Google Scholar]
- [11].Gordon S, Lotenberg S, Long R, Antani S, Jeronimo J, and Greenspan H, “Evaluation of uterine cervix segmentations using ground truth from multiple experts,” Comput Med Imaging Graph, vol. 33, no. 3, pp. 205–16, April 2009, doi: 10.1016/j.compmedimag.2008.12.002. [DOI] [PubMed] [Google Scholar]
- [12].Greenspan H et al. , “Automatic detection of anatomical landmarks in uterine cervix images,” IEEE Trans Med Imaging, vol. 28, no. 3, pp. 454–68, March 2009, doi: 10.1109/TMI.2008.2007823. [DOI] [PubMed] [Google Scholar]
- [13].Hu L et al. , “An Observational Study of Deep Learning and Automated Evaluation of Cervical Images for Cancer Screening,” J Natl Cancer Inst, vol. 111, no. 9, pp. 923–932, September 1 2019, doi: 10.1093/jnci/djy225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ji Q, Engel J, and Craine E, “Classifying cervix tissue patterns with texture analysis,” (in English), Pattern Recogn, vol. 33, no. 9, pp. 1561–1573, September 2000, doi: 10.1016/S0031-3203(99)00123-5. [DOI] [Google Scholar]
- [15].Pallavi V and Payal K, “Automated analysis of cervix images to grade the severity of cancer,” Conf Proc IEEE Eng Med Biol Soc, vol. 2011, pp. 3439–42, 2011, doi: 10.1109/IEMBS.2011.6090930. [DOI] [PubMed] [Google Scholar]


